Abstract
Interspecies genetic exchange is an important evolutionary mechanism in bacteria. It allows rapid acquisition of novel functions by transmission of adaptive genes between related species. However, the frequency of homologous recombination between bacterial species decreases sharply with the extent of DNA sequence divergence between the donor and the recipient. In Bacillus and Escherichia, this sexual isolation has been shown to be an exponential function of sequence divergence. Here we demonstrate that sexual isolation in transformation between Streptococcus pneumoniae recipient strains and donor DNA from related strains and species follows the described exponential relationship. We show that the Hex mismatch repair system poses a significant barrier to recombination over the entire range of sequence divergence (0.6 to 27%) investigated. Although mismatch repair becomes partially saturated, it is responsible for 34% of the observed sexual isolation. This is greater than the role of mismatch repair in Bacillus but less than that in Escherichia. The remaining non-Hex-mediated barrier to recombination can be provided by a variety of mechanisms. We discuss the possible additional mechanisms of sexual isolation, in view of earlier findings from Bacillus, Escherichia, and Streptococcus.
Bacteria from all major taxa are able to exchange genes across species by homologous recombination (26). While the various bacteria take up donor DNA by a diversity of mechanisms, all studied systems of homologous recombination share at least one homologous feature: recombination depends ultimately on the activity of the RecA protein and its homologues (26). Similarly, there is one system that hinders recombination across species in both the Proteobacteria and the gram-positive bacteria (3). This is the mismatch repair system encoded by mutS, mutL, and their homologues. Because some of the molecular basis for interspecies recombination is shared across disparate taxa, we might expect that recombination between species is constrained in similar ways throughout the bacterial world.
In addition, previous studies have shown that the frequency of homologous recombination decreases with the sequence divergence between donor and recipient, in a manner that is similar across a wide range of organisms. In Bacillus transformation as well as in Escherichia conjugation, the frequency of recombination decreases exponentially with the degree of DNA sequence divergence between donor and recipient (23, 28, 31). A similar exponential relationship has been observed for the frequency of intrachromosomal crossovers in Saccharomyces cerevisiae (5). Nevertheless, the major mechanisms producing recombinational barriers have been shown to differ in each of the above cases. In Escherichia coli, the predominant barrier to recombination is presented by the methylation-directed mismatch repair system (21). In Bacillus, mismatch repair is only marginally effective in preventing recombination between divergent sequences; the most significant barrier is that donor strand invasion and initiation of strand exchange require near identity between donor and recipient at both ends of the donor segment (15, 16). In yeast, the effects of mismatch repair and sequence divergence are both significant recombinational barriers (5).
Whereas an exponential relationship between interspecies recombination and sequence divergence appears to be universal, it is striking how much more mismatch repair contributes to recombination barriers in Escherichia than in Bacillus. It is not clear whether the role of mismatch repair in Escherichia or Bacillus is typical and under what circumstances mismatch repair is most likely to evolve to be a significant barrier to interspecies recombination. More comparative work is needed to understand the evolution of mismatch repair and its role as an interspecies recombinational barrier. In this paper, we approach this problem by investigating another system of microbial recombination: natural transformation in Streptococcus pneumoniae. We test whether the exponential relationship between sequence divergence and sexual isolation, observed in Bacillus and Escherichia, also holds for transformation in Streptococcus. We also investigate the mechanisms of sexual isolation between Streptococcus species.
S. pneumoniae is a gram-positive bacterium, naturally competent for transformation. The Streptococcus HexAB mismatch repair system is homologous to MutSL (3) and has been shown to correct single mismatches arising during recombination between otherwise identical substrates. The HexA protein recognizes mismatches in heteroduplex DNA, and, for transformation by mismatched DNA, it is able to remove the entire donor strand and thus prevent recombination (3). However, while the role of the Hex system in detecting single mismatches is well documented, its effectiveness in preventing recombination between divergent species is not yet fully known. There is evidence that the system is easily saturated in the presence of multiply mismatched DNA and may not be able to provide an efficient recombinational barrier (13).
In this study, we investigated the extent to which DNA sequence divergence reduces the rates of recombination between Streptococcus species, as well as the role that the HexAB mismatch repair system plays in producing sexual isolation (i.e., resistance to recombination across species). We transformed three recipient strains, two wild-type strains and one mismatch repair-deficient derivative, with chromosomal DNA isolated from related Streptococcus strains marked with rifampin resistance. Consequently, we determined the relationship between transformation frequencies and DNA sequence divergence, in the presence and absence of mismatch repair.
MATERIALS AND METHODS
Strains.
Strains used in this study are listed in Table 1. We used S. pneumoniae strains Pn16 (highly competent wild-type isolate) and R6 (unencapsulated laboratory strain) as recipients to determine the relationship between sequence divergence and sexual isolation in wild-type strains. We used strain R6Δ3, a mismatch repair mutant of strain R6 with a deletion in the hexA gene, as a recipient in investigating the role of mismatch repair in sexual isolation. The remaining strains, used as donors, were obtained from the National Collection of Type Cultures (NCTC; Colindale, United Kingdom) and the American Type Culture Collection (ATCC; Manassas, Va.).
TABLE 1.
List of strains
| Species | Strain | Source | Relevant genotype |
|---|---|---|---|
| S. pneumoniae | Pn16 | C. G. Dowson | wta |
| S. pneumoniae | R6 | C. G. Dowson | wt |
| S. pneumoniae | R6Δ3 | C. G. Dowson | ΔhexA |
| S. pneumoniae | 96129 | C. G. Dowson | wt |
| S. mitis | NCTC 10712 | NCTC | Type strain |
| S. oralis | NCTC 11427 | NCTC | Type strain |
| S. sanguis | NCTC 7863 | NCTC | Type strain |
| S. parasanguis | ATCC 15912 | ATCC | Type strain |
| S. crista | NCTC 12479 | NCTC | Type strain |
| S. anginosus | ATCC 9895 | ATCC | Type strain |
| S. constellatus | NCTC 11063 | NCTC | Type strain |
| S. intermedius | ATCC 27335 | ATCC | Type strain |
| S. adjacens | ATCC 49175 | ATCC | Type strain |
wt, wild type.
Isolation of rifampin-resistant mutants.
Strains used as DNA donors were streaked onto plates from glycerol stocks and incubated overnight. A single colony was picked and spread onto a 2- to 3-cm square on a fresh plate. Following a further 24 h of growth, the square was harvested onto a swab and spread over a fresh plate containing 10 μg of rifampin/ml. Resistant colonies appeared within 2 to 3 days.
Isolation of chromosomal DNA.
Bacteria were harvested from eight 90-mm-diameter brain heart infusion (BHI)-blood (5%)-agar plates using a plastic loop and resuspended in 1 ml of 50 mM Tris–10 mM EDTA, pH 8.0. After addition of 100 μl of 10-mg/ml lysozyme and 5 μl of RNase (Boehringer), the cells were incubated at 37°C for 30 to 60 min, followed by addition of 150 μl of 100-mg/ml proteinase K and a further 30 min of incubation. Cells were lysed by addition of 100 μl of 20% (wt/vol) sarcosyl and incubated at 37°C until lysis was complete, indicated by the clearing of the cell suspension, subject to a maximum time of 2 h. The resulting lysate was subjected to two extractions with phenol-chloroform and a further extraction with chloroform. The DNA was then precipitated by the addition of 1/10 volume of 3 M sodium acetate, pH 3.5, and 1 volume of isopropanol. The DNA was collected by centrifugation in a microcentrifuge at 13,000 × g for 15 min, and the resulting DNA pellet was washed with 70% ethanol, air dried briefly, and resuspended in 200 μl of deionized water. The quality and quantity of the DNA were determined by spectrophotometry and confirmed by visualization on an agarose gel. All samples had ratio of optical density at 260 nm to that at 280 nm of between 1.8 and 2.0.
Preparation of competent cells and transformation.
A single colony was inoculated into 3 to 4 ml of C medium (11a) and grown at 37°C until the solution became slightly cloudy (approximately 3 h). A 100-μl aliquot was then used to inoculate 7 ml of prewarmed C medium, and the mixture was incubated for 1 3/4 h. At 10-min intervals, 425 μl was removed and added to 75 μl of sterile glycerol; the solution was mixed, and a 20-μl sample was removed to a fresh tube. Both samples were frozen on dry ice. After 2 h, sampling was stopped.
Cells sampled at each time point were tested for competence as follows. Each 20-μl sample was thawed and added to 480 μl of C medium, and the mixture was split into two 250-μl aliquots. One microgram of chromosomal DNA from the rifampin-resistant isogenic strain was added to one sample. Cells were incubated for 2 h at 37°C, and 50 μl was plated onto BHI-agar supplemented with 10 μg of rifampin/ml. Competence was induced over a 40- to 60-min period, shown by an increasing number of transformants during the competent phase. The time of onset of competence depended upon the strain and the optical density of the original inoculum. In subsequent experiments, the competent cells were partially thawed on ice and 80 μl was added to 1,920 μl of C medium. The remaining cells were refrozen on dry ice and stored at −80°C. The diluted competent cells were split into 150-μl aliquots, and 0.6 μg of donor DNA was added (for the negative control, no DNA was added). Following incubation at 37°C for 2 h, cells were serially diluted in BHI broth and plated on selective medium. The sexual isolation between a recipient and a test donor was quantified as the frequency of homogamic transformation (the frequency at which the recipient was transformed by its own Rifr mutant) divided by the frequency of heterogamic transformation (the frequency at which the recipient was transformed by the test donor).
Estimate of sequence divergence at rpoB.
We used PCR to amplify two fragments of the rpoB genes from all the strains used in the experiments. The first fragment, extending from base 372 to 1160 (corresponding to base numbers of Streptococcus pyogenes ATCC 700294 rpoB [http://www.genome.ou.edu]), was amplified using degenerate primers 5′-GGG ACG TTC GTN ATH AAY GG-3′, for the leading strand, and 5′-CCA AGG TGG TCD ATR TCR TC-3′, for the lagging strand. The second fragment, base 1440 to 2380, was amplified using primers 5′-TTG TCA CAR TTY ATG GAY CA-3′, for the leading strand, and 5′-TCG CGA GTG ATY TCY TCM GG-3′, for the lagging strand. All primers were designed using the rpoB alignment of Bacillus subtilis (2) and S. pneumoniae (C. G. Dowson, unpublished data). PCRs were carried out using Taq DNA polymerase (Qiagen) and consisted of 95°C denaturation for 30 s and 57°C annealing for 30 s, followed by extension at 72°C for 1 min, for 25 cycles. The PCR products were purified using Qiaquick PCR purification columns (Qiagen) and adjusted to standard concentrations. They were sequenced using the external primers (described above) at the DNA Sequencing Facility at the University of Pennsylvania Medical Center, by the dRhodamine dye terminator method with ABI PRISM 373XL and 377 sequencers (Perkin-Elmer, Applied Biosystems Division). The result was a full double-stranded sequence of 510 bp for the first fragment, and 872 bp for the second fragment, for all tested strains. The combined sequence divergence of the two fragments was used as the estimate of DNA sequence divergence in the rpoB region. We have earlier found that, in Bacillus, the above fragments give a good estimate of interspecies divergence across the entire gene (15). In addition, rpoB is located within the highly conserved ribosomal gene cluster. In Streptococcus, rpoB is directly flanked by rpoC and is in close proximity to rpoA (the other subunits of RNA polymerase), along with several ribosomal protein genes (data from the ongoing S. pyogenes sequencing project is available at http://www.genome.ou.edu). Since all the above genes are highly conserved, the sequence divergence at rpoB is representative of divergence within the entire region throughout which the recombinant molecules are expected to extend.
Nucleotide sequence accession numbers.
The sequences obtained in this study have been assigned GenBank accession no. AF194507 to AF194528.
RESULTS
Sequence analysis of donor and recipient strains.
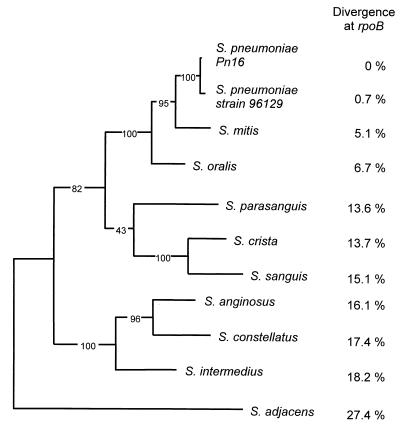
The sequence divergence values and phylogenetic relationships among the strains are shown in Table 2 and Fig. 1. We used maximum parsimony along with bootstrap analysis based on 1,000 replicates to determine the phylogenetic relationships among all strains, based on the partial rpoB sequence data. The phylogeny agrees with that determined earlier using 16S rRNA and sodA1 data (14, 20), with the exception of the clustering within the Streptococcus anginosus group. The analysis of rpoB shows Streptococcus constellatus to be most closely related to S. anginosus, whereas according to earlier data (16S rRNA and sodA1) S. anginosus forms a clade with Streptococcus intermedius. This discrepancy is supported by high bootstrap confidence values and might indicate a recombination event in the vicinity of rpoB.
TABLE 2.
Sequence identity matrix for Streptococcus strains based on a partial rpoB sequence
| Species or strain | % DNA sequence identity or divergencea compared to:
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| S. pneumoniae strain Pn16 | S. pneumoniae strain 91629 | S. mitis | S. oralis | S. parasanguis | S. crista | S. sanguis | S. anginosus | S. constellatus | S. intermedius | S. adjacens | |
| S. pneumoniae strain Pn16 | 99.3 | 94.9 | 93.3 | 86.4 | 86.3 | 85.0 | 83.9 | 83.4 | 81.8 | 72.6 | |
| S. pneumoniae strain 91629 | 0.7 | 94.6 | 93.3 | 86.1 | 86.1 | 84.7 | 83.6 | 83.0 | 81.7 | 72.4 | |
| S. mitis | 5.1 | 5.4 | 92.3 | 85.2 | 86.1 | 84.7 | 82.7 | 82.2 | 80.8 | 71.8 | |
| S. oralis | 6.7 | 6.7 | 7.7 | 85.2 | 85.6 | 85.3 | 83.3 | 81.9 | 81.5 | 72.4 | |
| S. parasanguis | 13.6 | 13.9 | 14.8 | 14.8 | 85.6 | 84.4 | 82.2 | 83.4 | 81.9 | 70.4 | |
| S. crista | 13.7 | 13.9 | 13.9 | 14.4 | 14.4 | 92.2 | 83.6 | 83.5 | 83.7 | 71.4 | |
| S. sanguis | 15.0 | 15.3 | 15.4 | 14.7 | 15.6 | 7.8 | 84.9 | 85.5 | 84.2 | 72.2 | |
| S. anginosus | 16.1 | 16.4 | 17.3 | 16.7 | 17.8 | 16.4 | 15.1 | 92.9 | 88.4 | 70.3 | |
| S. constellatus | 16.6 | 17.0 | 17.8 | 18.1 | 16.6 | 16.5 | 14.6 | 7.1 | 88.6 | 71.9 | |
| S. intermedius | 18.2 | 18.3 | 19.2 | 18.5 | 18.1 | 16.3 | 15.8 | 11.7 | 11.4 | 70.5 | |
| S. adjacens | 27.4 | 27.6 | 28.2 | 27.6 | 29.6 | 28.6 | 27.8 | 29.7 | 28.1 | 29.5 | |
Percentages above and to the right of the diagonal formed by the blank entries represent DNA sequence identity; those below and to the left represent sequence divergence.
FIG. 1.
Phylogeny of S. pneumoniae and related species, based on 1,380 bases of the rpoB sequence. The tree is the single most parsimonious phylogeny, created using the exhaustive search algorithm of PAUP, version 3.1.1 (D. L. Swofford, PAUP. Phylogenetic analysis using parsimony, version 4, Sinauer Associates, Sunderland, Mass., 1998). Branching confidence values are based on 1,000 bootstrap replicates. DNA sequence divergences between each donor strain and the S. pneumoniae recipients are shown on the right. The above phylogeny agrees with results obtained by analysis of 16S rRNA and sodA genes, with the notable exception that S. anginosus clusters here with S. constellatus, whereas earlier results show it to be more closely related to S. intermedius. The high bootstrap confidence value of this grouping suggests a possible recombination event at rpoB.
We were not able to detect any nucleotide differences at rpoB among the three recipient strains (Pn16, R6, and R6Δ3). Among the rifampin-resistant donors, we noted His526 substitutions, also known to confer resistance in Bacillus, Mycobacterium, and Escherichia (8, 30). We identified Asp526 (S. pneumoniae Pn16, S. intermedius) and Tyr526 (Streptococcus oralis, S. pneumoniae strain R6, S. pneumoniae strain 96129) substitutions.
Relationship between sexual isolation and DNA sequence divergence.
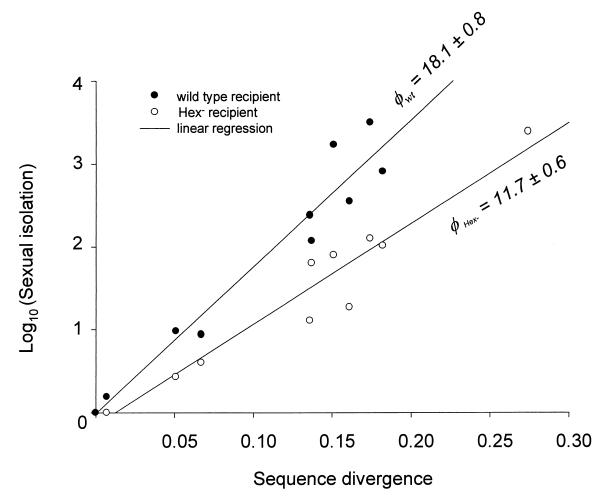
The transformation frequencies for each donor and recipient, along with the corresponding sexual isolation values, are given in Tables 3 and 4. The relationship between sexual isolation and sequence divergence between the donor and the recipient is shown in Fig. 2. We find that the fit of the log-transformed sexual isolation data (R2 = 0.94 for the wild type, and R2 = 0.93 for the Hex− recipient) is significantly better than a corresponding fit of raw, non-log-transformed data (R2 = 0.35 for the wild type; R2 = 0.31 for the Hex− recipient). That is, the relationship between sexual isolation (ρ) and sequence divergence (π) in Streptococcus is close to exponential and can be described by the formula
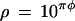 |
1 |
Values of the sensitivity parameter, φ, represented by the regression slopes in Fig. 1, are 17.82 and 18.39 for the Pn16 and R6 strains, respectively. Analysis of covariance shows that these two values are not significantly different (F = 0.6; P = 0.45). This suggests that the sensitivity of sexual isolation to sequence divergence is constant across wild-type S. pneumoniae strains, despite the fact that strain Pn16 exhibits a baseline (homogamic) transformation rate eightfold higher than that of strain R6 (Table 3).
TABLE 3.
Transformation frequencies, log10 transformed
| Source of donor DNA | Transformation frequencya for recipient strain:
|
||
|---|---|---|---|
| Pn16 | R6 | R6Δ3 | |
| S. pneumoniae Pn16 | −2.59 ± 0.32 | −3.26 ± 0.24 | −2.70 ± 0.34 |
| S. pneumoniae R6 | −2.79 ± 0.50 | −3.33 ± 0.27 | −2.81 ± 0.24 |
| S. pneumoniae 91629 | −2.64 ± 0.42 | −3.65 ± 0.22 | −2.59 ± 0.22 |
| S. mitis | −3.61 ± 0.23 | −4.59 ± 0.22 | −3.24 ± 0.11 |
| S. oralis | −3.37 ± 0.28 | −4.15 ± 0.07 | −3.30 ± 0.28 |
| S. parasanguis | −4.73 ± 0.50 | −5.97 ± 0.21 | −3.92 ± 0.31 |
| S. crista | −4.72 ± 0.62 | −5.89 ± 0.14 | −4.17 ± 0.50 |
| S. sanguis | −5.63 ± 0.54 | −6.80 ± 0.26 | −4.71 ± 0.52 |
| S. anginosus | −5.04 ± 0.22 | −6.00 ± 0.20 | −4.14 ± 0.20 |
| S. constellatus | −6.17 ± 0.29 | −6.74 ± 0.35 | −4.91 ± 0.34 |
| S. intermedius | −5.49 ± 0.35 | −6.26 ± 0.44 | −4.50 ± 0.12 |
| S. adjacens | No transformants | No transformants | −5.62 ± 0.12 |
Log10 (frequency of transformation) ± standard error, based on four or more experimental trials.
TABLE 4.
Sexual isolation (log10 transformed) between the S. pneumoniae recipients and the donor strains
| Source of donor DNA | Log10 (sexual isolation) ± SEb for recipient strain:
|
||
|---|---|---|---|
| Pn16 | R6 | R6Δ3 | |
| S. pneumoniae Pn16 | 0a | −0.08 ± 0.23 | −0.11 ± 0.16 |
| S. pneumoniae R6 | 0.21 ± 0.22 | 0 | 0 |
| S. pneumoniae 91629 | 0.06 ± 0.10 | 0.32 ± 0.11 | −0.04 ± 0.28 |
| S. mitis | 1.00 ± 0.19 | 0.95 ± 0.17 | 0.48 ± 0.26 |
| S. oralis | 1.16 ± 0.28 | 0.73 ± 0.23 | 0.88 ± 0.13 |
| S. parasanguis | 2.13 ± 0.23 | 2.64 ± 0.44 | 1.11 ± 0.23 |
| S. crista | 2.07 ± 0.27 | 2.56 ± 0.36 | 1.80 ± 0.08 |
| S. sanguis | 3.04 ± 0.42 | 3.47 ± 0.24 | 1.91 ± 0.28 |
| S. anginosus | 2.45 ± 0.16 | 2.67 ± 0.34 | 1.27 ± 0.36 |
| S. constellatus | 3.58 ± 0.22 | 3.41 ± 0.27 | 2.10 ± 0.42 |
| S. intermedius | 3.06 ± 0.33 | 2.78 ± 0.29 | 2.01 ± 0.40 |
| S. adjacens | No transformants | No transformants | 3.39 ± 0.15 |
The sexual isolation between a recipient and its own Rifr mutant is defined to be zero.
Sexual isolation values and standard errors are based on at least four experimental trials. Note that the values are obtained by dividing the homogamic transformation frequency by the heterogamic frequency for each trial for each donor-recipient pair and then averaged. Since not all donor-recipient comparisons were carried out in the same experimental trial, sexual isolation values will not in general be equal to the results of simply dividing the average transformation frequencies from Table 3.
FIG. 2.
Log10 (sexual isolation) versus sequence divergence between each donor and recipient. Sexual isolation values were calculated by dividing the frequency of homogamic recombination (using the donor's own Rifr DNA) by the frequency of heterogamic recombination (using a divergent donor's DNA). The sexual isolation shown for the wild-type recipients is the average value for the two wild-type recipient strains, Pn16 and R6. The sexual isolation in the absence of mismatch repair is the result of transformation of the R6Δ3 hexA mutant. φwt and φHex− are the slopes of the regression of log10 (sexual isolation) on sequence divergence for the wild type and the Hex− mutant, respectively.
The effect of mismatch repair on heterospecific transformation.
The mismatch repair-deficient mutant exhibits sexual isolation consistently lower than that of its isogenic wild-type strain: φ for the wild type (φwt) = 18.39; φ for the Hex− mutant (φHex−) = 11.70. Analysis of covariance shows these coefficients to be statistically significant (F = 53.8; P < 0.0001). Averaging over all donor strains, mismatch repair is responsible for 34% of the observed sexual isolation (i.e., [φwt − φHex−]/φwt · 100%). For every donor, with the exception of S. oralis, sexual isolation values are higher for the wild-type recipient than for the mismatch repair mutant. The influence of mismatch repair on integration frequencies varies across strains, increasing the sexual isolation by up to 36-fold, for Streptococcus sanguis. Transformation with Streptococcus adjacens (Abiotrophia adjacens) DNA, which is 27.4% divergent from that of the recipient at rpoB, produced no detectable transformants in the presence of mismatch repair, while the mismatch-deficient strain produced transformants at a sexual isolation of 3,800. Thus, mismatch repair plays a role in preventing recombination between divergent Streptococcus species, across the entire spectrum of sequence divergence tested (0.7 to 27.4%). However, the effectiveness of the system in detecting multiple mismatches is significantly reduced, compared to its effectiveness against single mismatches (see Discussion). Our results show this reduction to be most pronounced at intermediate sequence divergence levels (S. oralis). The lack of a detectable mismatch repair function in preventing integration of S. oralis DNA may represent the saturation effect experienced by the Hex system in the presence of partially divergent DNA, as described by Humbert et al. (13).
DISCUSSION
The relationship between sequence divergence and sexual isolation.
Our study shows that S. pneumoniae experiences sexual isolation from related strains and species. Sexual isolation in Streptococcus increases approximately exponentially with DNA sequence divergence, as has previously been observed within Bacillus and Escherichia. It is notable that Streptococcus and Bacillus share not only the exponential form of the relationship but also the same level of sensitivity of sexual isolation to sequence divergence. The sensitivities of sexual isolation to sequence divergence parameters (φ) for transformation of wild-type Streptococcus and Bacillus strains are 18.1 (average for Pn16 and R6 recipients) and 21.37 (15), respectively. These figures are remarkably close. To further illustrate this trend, the sexual isolation (ρ) between S. pneumoniae and S. sanguis is 1,920 at 15.1% sequence divergence. Within Bacillus, the corresponding sexual isolation between B. subtilis and Bacillus licheniformis is 2,344 at 14.5% divergence (15). It is possible that maintenance of comparable levels of sexual isolation arises from similar evolutionary pressures on recombination frequencies within each genus.
Possible barriers to interspecies recombination.
An interspecies recombination event may be broken down into the following steps: (i) donor DNA is taken up by the recipient cell, (ii) donor DNA escapes the recipient cell's restriction system, (iii) a donor-recipient DNA heteroduplex molecule is formed, (iv) the mismatched heteroduplex escapes the action of the mismatch repair system, and (v) the recombinant strand is replicated by the recipient's DNA replication machinery. We present a short discussion of the relevance of the above steps as mechanisms of sexual isolation in Streptococcus.
The initial DNA uptake step can be dismissed as a genetic barrier in our system. Although in some bacterial species, such as Haemophilus influenzae (26), DNA uptake has been shown to be sequence specific, in the known gram-positive species (Streptococcus and Bacillus) the rate of uptake is independent of the sequence and is hence not sensitive to the sequence divergence between the donor and the recipient (18, 26). Thus, in Streptococcus, DNA uptake is unlikely to be a source of sexual isolation.
The donor DNA must then escape the action of the recipient's restriction endonucleases. The recipient strains used in the experiments described in this paper do not harbor restriction/modification systems. Thus, in this particular case, neutralization of donor DNA by the recipient's restriction enzymes cannot control recombination rates. In general, the effect of restriction systems has been shown to introduce a constant reduction in recombination frequencies, independent of the divergence between the donor and the recipient (31). In Bacillus, this effect is small (possibly because the single-stranded donor DNA taken up on transformation is not subject to digestion), amounting to at most a sixfold reduction in recombination frequencies for restricting strains compared to those for nonrestricting strains (31). Similarly, in conjugation between Salmonella and Escherichia cells, sexual isolation is not significantly influenced by the presence of type II restriction (17). Therefore, in Streptococcus, restriction is not expected to be a source of the exponential relationship between sequence divergence and sexual isolation (18).
The two subsequent steps in the recombination process, namely, the formation of a heteroduplex molecule and the evasion of the recipient's mismatch repair system, present the most likely barrier to interspecies recombination. A wealth of information on the formation of recombinant DNA molecules is available from both in vivo and in vitro studies of E. coli and S. cerevisiae. In E. coli initial processing of the donor recombinant substrate is often necessary to produce free single-stranded ends. This is carried out by the RecBCD enzyme, which unwinds and digests double-stranded DNA until it encounters a characteristic χ sequence. The process results in the creation of free 3′ single-stranded DNA ends. All further recombinogenic processes depend on the action of the RecA protein in binding to the single-stranded DNA and initiating invasion into a homologous and highly conserved (24) region of the recipient's double-stranded DNA molecule. (Although 5′ donor ends can also be invasive, they cannot be extended and do not initiate recombination [10, 22].) The 3′ recombinant joints are then subject to extension and editing (see reference 27 for a recent discussion of the above processes). In E. coli, (27) all modes of DNA editing, while involving several different components, appear to be controlled by the MutS protein (homologue of the Streptococcus HexA protein). Hence, the HexA mutant used in our experiments is expected to lack all mismatch repair activity.
The importance of RecA and HexAB functions in promoting and preventing recombination between divergent substrates is well documented (18, 26), and we discuss those events in detail, as pertinent to Streptococcus transformation, in separate sections below. In fact, we argue that RecA-dependent and HexAB-dependent steps are likely to produce most of the sexual isolation in Streptococcus.
On the other hand, since during transformation the donor DNA enters the cell as a single-stranded molecule, no processing by RecBCD is necessary. In fact, no known homologues of the RecD protein exist in gram-positive bacteria, while the function of the AddA and AddB enzymes in Bacillus (RecB and RecC homologues) seems to be limited to UV damage repair and might not be required during transformation (31). Thus, it is unlikely that DNA processing prior to recombination is involved in preventing interspecies recombination in Streptococcus.
In later stages, postsynaptic events such as branch migration and DNA replication may influence the frequency of successful recombination. Branch migration, which extends the initial heteroduplex molecule, is promoted by the RuvA, RuvB, and RuvC proteins (26). RuvA mutants of E. coli exhibit a reduced frequency of conjugational recombination. However, the reduction is relatively small and appears not to be very sensitive to DNA sequence divergence. Matic et al. (17) demonstrated that, in intraspecific crosses, the recombination frequency is reduced by a factor of 4, whereas in interspecific conjugation (between E. coli and Salmonella enterica serovar Typhimurium) it is reduced by a factor of about 10 (27). This corresponds to a sexual isolation factor due to the RuvA pathway of at most 2.5. In addition, Majewski and Cohan (16) demonstrated that, in Bacillus, by providing regions of flanking identity to the recipient, they were able to force a divergent donor insert to recombine at a nearly homogamic frequency. Similarly, the RecG protein, which is involved in the resolution of Holiday junctions, has comparable effects in intraspecific and interspecific crosses in E. coli (17). Thus, branch migration is at most slightly impeded by mismatches and should not contribute considerably to sexual isolation between species.
Lastly, DNA synthesis may be initiated from the early heteroduplex intermediates, preventing dissociation of such potentially unstable joints. Stambuk and Radman (27) showed that mutations in E. coli priA and recF genes, which are known to be involved in recombination-associated DNA synthesis, have differential effects in interspecific and intraspecific crosses. It is possible that de novo DNA synthesis is necessary to circumvent the potential dissociation of early heteroduplex intermediates. It is possible that such DNA replication events may be impeded by mismatched heteroduplex joints. However, in E. coli, disrupting the RecF pathway reduces recombination frequencies by not more than sixfold in interspecies recombination and about twofold in intraspecies events. Hence the sexual isolation produced by mismatches impeding the RecF-mediated processes is at the most a factor of 3. In addition, Zawadzki et al. (31) found no measurable effect of RecF in preventing interspecific transformation in Bacillus. Thus, DNA synthesis induced by recombination is likely to be only a minor source of sexual isolation in Streptococcus.
Mismatch repair as recombination barrier.
Our results show that the Hex mismatch repair system plays a significant role in preventing recombination between divergent DNA sequences. The extent of its significance is intermediate between that observed in Escherichia, where mismatch repair is the major recombinational barrier, and Bacillus, where it plays only a minor part. It has been suggested (15) that the relative ineffectiveness of the Bacillus mismatch repair system may be caused by degradation of mismatch repair proteins, which occurs under starvation conditions (9) necessary for induction of competence. In Streptococcus, competence is not induced under starvation, and thus mismatch repair proteins are probably not degraded (13). This may explain the more significant role of mismatch repair in preventing interspecific recombination in Streptococcus than in Bacillus.
However, the Streptococcus mismatch repair is still a rather poor mechanism of sexual isolation. The Hex system has been shown to efficiently correct specific single-base pair mismatches, known as low efficiency (LE) markers, resulting from transformation, with an accuracy of over 90% (4). It is believed that once a mismatch is detected, the entire donor strand, which may be as large as several kilobases, is rejected (3). Let us consider a simple model, assuming that, in a donor strand containing several (m) mismatches, detection of every mismatch is an independent event. Hence the probability of the entire strand escaping detection is equal to the probability of m mismatches being undetected. We further assume that, on average, one in every four mismatches is an LE mismatch (4, 11), escaping detection with a probability of 0.1, while others are high-efficiency (HE) mismatches, always escaping detection. The probability of a foreign strand escaping the mismatch repair system can then be expressed as (0.1)m/4 or (0.1)Lπ/4, where L is the length of the donor strand and π is the sequence divergence. Taking into account that the length of donor DNA integrated on transformations is often greater than 8 kb (1, 3), we should expect that the reduction in transformation frequency between 15%-divergent DNA strands, caused by mismatch repair, should be of the order of 10300.
In our experiments, the Hex system reduces the recombination frequencies by not more than 36-fold (S. sanguis, 15% sequence divergence from recipient). It is obvious that the effectiveness of mismatch repair against multiply mismatched sequences is greatly reduced compared to its effectiveness against single mismatches. Hence, the simple model described above is incorrect, and detection events cannot be considered independent. We present three explanations for the decrease in efficiency of the Hex system in the presence of multiple mismatches.
First, it is possible, that a saturation of mismatch repair may be reducing the effectiveness of the Hex system. Humbert et al. (13) have previously shown that the concentrations of HexA and HexB proteins become limiting during transformation with divergent DNA. They found that mismatch repair is ineffective in preventing transformation by DNA with intermediate sequence divergence levels (1 to 10%) from S. pneumoniae and suggested that saturation of the system may be responsible. Independent recombination events at loci other than the selected locus may reduce the number of mismatch repair proteins available to prevent integration of the selectable marker. This would further suggest that organisms closely related to S. pneumoniae, such as S. oralis, that are quite likely to possess a wide range of genes with intermediate sequence divergence are potentially important donors in the evolution of the pneumococcal genome (6, 25; S. King, unpublished data). This finding suggests a potential problem for the development of novel pneumococcal targets for vaccination or chemotherapy that may become subject to a strong selective pressure (7).
Second, it has also been shown that the presence of a particular HE marker (namely, a C/C mismatch) in the immediate proximity of an LE marker reduces the probability of detection of the LE marker (11). Thus, some mismatches may have a “masking” effect on adjacent mismatches, possibly by destabilizing the DNA helix conformation. In multiply mismatched strands, extended destabilized regions may increase the overall probability of an entire strand escaping detection.
Finally, multiple mismatches may interfere with the search for nicks in the donor strand. Such nicks allow the Hex system to distinguish between donor and recipient and correct the appropriate strand. The nick-directed search needs to extend from the mismatch all the way to the end of the donor strand. Mismatches present along the way may impede the search. On the other hand, in Escherichia, mismatch repair uses methylation patterns to distinguish between the donor and the recipient strands. The search for methylation does not need to extend to the end of the strand and takes, on average, 256 bases. This may explain why, in E. coli, mismatch repair is less hampered by multiple mismatches (13).
It has been suggested that the mismatch repair system may be modulated so as to provide a strong barrier to recombination under normal growth conditions and a weak barrier under stressful conditions, when recombination may be particularly favorable (12, 28). Our results show that even under exponential growth, which is necessary for induction of competence in Streptococcus, mismatch repair is a poor barrier to interspecific recombination. Moreover, Humbert et al. (13) have shown that levels of mismatch repair proteins are not modulated during competence. In view of the above and the data from transformation in B. subtilis (15, 16), we believe that the role of mismatch repair as a barrier to interspecific recombination in gram-positive bacteria is minor and most probably incidental to that involved in the correction of errors introduced during DNA replication.
Recombinant joint formation as a genetic barrier.
According to data from recombination in Escherichia (27–29) and Bacillus, the reluctance to form mismatched DNA heteroduplex molecules is caused primarily by the scarcity of minimum efficiently processed segments (MEPS), short regions of near identity between the donor and the recipient (15, 16). Once such conserved blocks are located, the adjacent sequences seem to undergo recombination despite a high level of divergence between the donor and the recipient. In Bacillus, recombination requires mismatch-free segments of about 20 bp or more at both ends of the donor strand (16). Data from E. coli indicate that only one highly conserved, invasive end is needed in that system (22, 24). Such sequences are most probably necessary for initiation of RecA-mediated strand invasion, stabilization of the recombinant joint, and possibly the subsequent recombination-associated DNA synthesis.
In the preceding discussion of possible mechanisms of sexual isolation, we suggested that, other than mismatch repair, most intermediate processes are unlikely to have a major sequence divergence-specific effect on sexual isolation. We further propose that the remaining major barrier to recombination between diverged sequences is the availability of MEPS sequences. We can apply the model described by Majewski and Cohan (15) to determine the length of a MEPS in Streptococcus from the experimentally determined sensitivity of sexual isolation to sequence divergence, φHex−, in the absence of mismatch repair. The model assumes that a mismatch-free region of n nucleotides is necessary for a successful recombination. It can be shown from equation 1 that the length of such a region may be estimated as n = φHex− · ln (10). Thus, in Streptococcus, the minimum length of an invasive end would be 27 bp, if one such end was needed, or 14 bp, if two flanking ends were necessary (as for Bacillus). More work, for example, analysis of the recombinant junction produced on transformation with divergent DNA, needs to be done to determine which of the above models is correct. We also observe that the resistance to joint formation poses less of a barrier to recombination in Streptococcus (φHex− = 11.72) than in Bacillus (φMutSL− = 17.95) (15). This difference is significant by analysis of covariance (F = 33.4; P < 0.0001).
To conclude, we find that the frequency of transformation in Streptococcus is an approximately exponential function of the sequence divergence between the donor and the recipient. As in other recombinational systems, the two major factors producing sexual isolation are the mismatch repair system and the difficulty in heteroduplex formation.
ACKNOWLEDGMENTS
We thank J. P. Claverys for the supply of the hexA construct that was used to create the R6 hexA mutant strain.
This work was supported by U.S. Environmental Protection Agency grant R82-5348-01-2 and by the Wellcome Trust.
REFERENCES
- 1.Biswas G D, Ravin A W. Heterospecific transformation of Pneumococcus and Streptococcus. IV. Variations in hybrid DNA produced by recombination. Mol Gen Genet. 1971;110:1–22. doi: 10.1007/BF00276040. [DOI] [PubMed] [Google Scholar]
- 2.Boor K J, Duncan M L, Price C W. Genetic and transcriptional organization of the region encoding the beta subunit of Bacillus subtilis RNA polymerase. J Biol Chem. 1995;270:20329–20336. doi: 10.1074/jbc.270.35.20329. [DOI] [PubMed] [Google Scholar]
- 3.Claverys J P, Lacks S A. Heteroduplex deoxyribonucleic acid base mismatch repair in bacteria. Microbiol Rev. 1986;50:133–165. doi: 10.1128/mr.50.2.133-165.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Claverys J P, Mejean V, Gasc A M, Sicard A M. Mismatch repair in Streptococcus pneumoniae: relationship between base mismatches and transformation efficiencies. Proc Natl Acad Sci USA. 1983;80:5956–5960. doi: 10.1073/pnas.80.19.5956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Datta A, Hendrix M, Lipsitch M, Jinks-Robertson S. Dual roles for DNA sequence identity and the mismatch repair system in the regulation of mitotic crossing-over in yeast. Proc Natl Acad Sci USA. 1997;94:9757–9762. doi: 10.1073/pnas.94.18.9757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dowson C G, Coffey T J, Kell C, Whiley R A. Evolution of penicillin resistance in Streptococcus pneumoniae; the role of Streptococcus mitis in the formation of a low affinity PBP2B in S. pneumoniae. Mol Microbiol. 1993;9:635–643. doi: 10.1111/j.1365-2958.1993.tb01723.x. [DOI] [PubMed] [Google Scholar]
- 7.Dowson C G, Barcus V, King S, Pickerill P, Whatmore A, Yeo M. Horizontal gene transfer and the evolution of resistance and virulence determinants in Streptococcus. Soc Appl Bacteriol Symp Ser. 1997;26:42–51. [PubMed] [Google Scholar]
- 8.Enright M, Zawadzki P, Pickerill P, Dowson C G. Molecular evolution of rifampicin resistance in Streptococcus pneumoniae. Microb Drug Resist. 1998;4:65–70. doi: 10.1089/mdr.1998.4.65. [DOI] [PubMed] [Google Scholar]
- 9.Feng G, Tsui H C, Winkler M E. Depletion of the cellular amounts of the MutS and MutH methyl-directed mismatch repair proteins in stationary-phase Escherichia coli K-12 cells. J Bacteriol. 1996;178:2388–2396. doi: 10.1128/jb.178.8.2388-2396.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Friedman-Ohana R, Cohen A. Heteroduplex joint formation in Escherichia coli recombination is initiated by pairing of a 3′-ending strand. Proc Natl Acad Sci USA. 1998;95:6909–6914. doi: 10.1073/pnas.95.12.6909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gasc A M, Sicard A M, Claverys J P. Repair of single- and multiple-substitution mismatches during recombination in Streptococcus pneumoniae. Genetics. 1989;121:29–36. doi: 10.1093/genetics/121.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11a.Gurney T, Jr, Fox M S. Physical and genetic hybrids formed in bacterial transformation. J Mol Biol. 1968;32:83–100. doi: 10.1016/0022-2836(68)90147-2. [DOI] [PubMed] [Google Scholar]
- 12.Harris R S, Feng G, Ross K J, Sidhu R, Thulin C, Longerich S, Szigety S K, Winkler M E, Rosenberg S M. Mismatch repair protein MutL becomes limiting during stationary-phase mutation. Genes Dev. 1997;11:2426–2437. doi: 10.1101/gad.11.18.2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Humbert O, Prudhomme M, Hakenbeck R, Dowson C G, Claverys J P. Homeologous recombination and mismatch repair during transformation in Streptococcus pneumoniae: saturation of the Hex mismatch repair system. Proc Natl Acad Sci USA. 1995;92:9052–9056. doi: 10.1073/pnas.92.20.9052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kawamura Y, Hou X G, Sultana F, Miura H, Ezaki T. Determination of 16S rRNA sequences of Streptococcus mitis and Streptococcus gordonii and phylogenetic relationships among members of the genus Streptococcus. Int J Syst Bacteriol. 1995;45:406–408. doi: 10.1099/00207713-45-2-406. [DOI] [PubMed] [Google Scholar]
- 15.Majewski J, Cohan F M. The effect of mismatch repair and heteroduplex formation on sexual isolation in Bacillus. Genetics. 1998;148:13–18. doi: 10.1093/genetics/148.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Majewski J, Cohan F M. DNA sequence similarity requirements for interspecific recombination in Bacillus. Genetics. 1999;153:1525–1533. doi: 10.1093/genetics/153.4.1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matic I, Rayssiguier C, Radman M. Interspecies gene exchange in bacteria: the role of the SOS and mismatch repair systems in evolution of species. Cell. 1995;80:507–515. doi: 10.1016/0092-8674(95)90501-4. [DOI] [PubMed] [Google Scholar]
- 18.Mejean V, Claverys J P. DNA processing during entry in transformation of Streptococcus pneumoniae. J Biol Chem. 1993;268:5594–5599. [PubMed] [Google Scholar]
- 19.Mortier-Barrierre I, Humbert O, Martin B, Prudhomme M, Claverys J-P. Control of recombination rate during transformation of Streptococcus pneumoniae: an overview. Microb Drug Resist. 1997;3:233–242. doi: 10.1089/mdr.1997.3.233. [DOI] [PubMed] [Google Scholar]
- 20.Poyart C, Quesne G, Coulon S, Berche P, Trieu-Cuot P. Identification of streptococci to species level by sequencing the gene encoding the manganese-dependent superoxide dismutase. J Clin Microbiol. 1998;36:41–47. doi: 10.1128/jcm.36.1.41-47.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rayssiguier C, Thaler D S, Radman M. The barrier to recombination between Escherichia coli and Salmonella typhimurium is disrupted in mismatch-repair mutants. Nature. 1989;342:396–401. doi: 10.1038/342396a0. [DOI] [PubMed] [Google Scholar]
- 22.Reddy G, Burnett B, Radding C M. Uptake and processing of duplex DNA by RecA nucleoprotein filaments: insights provided by a mixed population of dynamic and static intermediates. Biochemistry. 1995;34:10194–10204. doi: 10.1021/bi00032a013. [DOI] [PubMed] [Google Scholar]
- 23.Roberts M S, Cohan F M. The effect of DNA sequence divergence on sexual isolation in Bacillus. Genetics. 1993;134:401–408. doi: 10.1093/genetics/134.2.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shen P, Huang H V. Homologous recombination in Escherichia coli: dependence on substrate length and homology. Genetics. 1986;112:441–457. doi: 10.1093/genetics/112.3.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sibold C, Henrichsen J, Konig A, Martin C, Chalkley L, Hakenbeck R. Mosaic pbpX genes of major clones of penicillin-resistant Streptococcus pneumoniae have evolved from pbpX genes of a penicillin-sensitive Streptococcus oralis. Mol Microbiol. 1994;12:1013–1023. doi: 10.1111/j.1365-2958.1994.tb01089.x. [DOI] [PubMed] [Google Scholar]
- 26.Smith G R. Homologous recombination in procaryotes. Microbiol Rev. 1988;52:1–28. doi: 10.1128/mr.52.1.1-28.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stambuk S, Radman M. Mechanism and control of interspecies recombination in Escherichia coli. I. Mismatch repair, methylation, recombination and replication functions. Genetics. 1998;150:533–542. doi: 10.1093/genetics/150.2.533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vulic M, Dionisio F, Taddei F, Radman M. Molecular keys to speciation: DNA polymorphism and the control of genetic exchange in enterobacteria. Proc Natl Acad Sci USA. 1997;94:9763–9767. doi: 10.1073/pnas.94.18.9763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Westmoreland J, Porter G, Radman M, Resnick M A. Highly mismatched molecules resembling recombination intermediates efficiently transform mismatch repair proficient Escherichia coli. Genetics. 1997;145:29–38. doi: 10.1093/genetics/145.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Williams D L, Spring L, Collins L, Miller L P, Heifets L B, Gangadharam P R, Gillis T P. Contribution of rpoB mutations to development of rifamycin cross-resistance in Mycobacterium tuberculosis. Antimicrob Agents Chemother. 1998;42:1853–1857. doi: 10.1128/aac.42.7.1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zawadzki P, Roberts M S, Cohan F M. The log-linear relationship between sexual isolation and sequence divergence in Bacillus transformation is robust. Genetics. 1995;140:917–932. doi: 10.1093/genetics/140.3.917. [DOI] [PMC free article] [PubMed] [Google Scholar]