Summary
Background
Inflammatory and immune responses are essential and dynamic biological processes that protect the body against acute and chronic adverse stimuli. While conventional protein markers have been used to evaluate systemic inflammatory response, the immunological response to stimulation is complex and involves modulation of a large set of genes and interacting signalling pathways of innate and adaptive immune systems. There is a need for a non-invasive tool that can comprehensively evaluate and monitor molecular dysregulations associated with inflammatory and immune responses in circulation and in inaccessible solid organs.
Methods
Here we utilized cell-free messenger RNA (cf-mRNA) RNA-Seq whole transcriptome profiling and computational biology to temporally assess lipopolysaccharide (LPS) induced and JAK inhibitor modulated inflammatory and immune responses in mouse plasma samples.
Findings
Cf-mRNA profiling displayed a pattern of systemic immune responses elicited by LPS and dysregulation of associated pathways. Moreover, attenuation of several inflammatory pathways, including STAT and interferon pathways, were observed following the treatment of JAK inhibitor. We further identified the dysregulation of liver-specific transcripts in cf-mRNA which reflected changes in the gene-expression pattern in this generally inaccessible biological compartment.
Interpretation
Using a preclinical mouse model, we demonstrated the potential of plasma cf-mRNA profiling for systemic and organ-specific characterization of drug-induced molecular alterations that are associated with inflammatory and immune responses.
Funding
Molecular Stethoscope.
Keywords: Cell free messenger RNA, Inflammation, Systemic inflammatory response, Liquid biopsy
Research in context.
Evidence before this study
Inflammatory and immune responses are essential and dynamic biological processes that protect the body against acute and chronic adverse stimuli. While there are several conventional blood-based protein markers that are used to evaluate systemic inflammatory response, the immunological response to stimulation is complex and involves modulation of a large set of genes and interacting signalling pathways of innate and adaptive immune systems. The understanding of inflammatory and immune responses is incomplete, we are unable to dissect systemic and solid organ contributors and we lack molecular tools to effectively monitor progressive disease and the efficacy of interventional approaches.
Added value of this study
While protein-based assays can evaluate a limited number of targets, we have developed a cell-free messenger RNA (cf-mRNA) RNA-Seq platform with computational biology interpretation and machine learning analysis which enables non-invasive, hypothesis-independent, whole transcriptome profiling. The approach permits comprehensive assessment of inflammatory and immune responses in both circulating and solid organ compartments. To the best of our knowledge, our study is the first to utilize cf-mRNA RNA-Seq to evaluate therapeutic responses in a well-characterized pre-clinical model. We sequenced a total of 174 plasma samples from mice that were treated with or without lipopolysaccharide (LPS) and/or a JAK inhibitor at various time points. We show that cf-mRNA profiling displayed a temporal pattern of systemic immune responses elicited by LPS and dysregulation of associated pathways. Furthermore, we show that the treatment of a JAK inhibitor suppressed LPS-induced inflammatory signals in plasma transcriptome. We also identified the dysregulation of liver-specific transcripts in cf-mRNA which reflected changes in the gene-expression pattern in the liver.
Implications of all the available evidence
Our data demonstrated the potential of plasma cf-mRNA profiling for systemic and organ-specific characterization of drug-induced molecular alterations that are associated with inflammatory and immune responses to potentially complement protein-based assays. Here we utilized cf-mRNA RNA-Seq, computational biology and machine learning to examine the dysregulation of cf-mRNA plasma transcriptome following LPS and JAK inhibitor treatments in mice. Our approach could be utilized in various stages of drug development including pre-clinical models and eventual clinical trials for the evaluation of drug efficacy, target engagement, as well as pharmacodynamic and pharmacokinetic assessment.
Alt-text: Unlabelled box
Introduction
Inflammation is an important self-defense mechanism that is typically triggered by invading pathogens, tissue injuries, tumor growth or the onset of pathological autoimmune activities. Although inflammation is crucial for protecting the body from harmful stimuli and initiating the healing process, chronic inflammation has been recognized to contribute to multiple non-infectious diseases including type 2 diabetes, non-alcoholic fatty liver disease (NAFLD), rheumatoid arthritis, inflammatory bowel disease, neurodegenerative diseases, cardiovascular diseases and certain types of cancer.1 Therefore, inflammation has become a key target for drug development in multiple diseases. For instance, several biologics such as infliximab, adalimumab, certolizuma pegol and golimumab are currently used to neutralize the inflammatory mediator TNF-α or to block its receptors.2 Moreover, currently several signalling pathways that regulate cytokine response have been targeted as a potential alternative approach to suppress inflammation.3 In particular, Janus kinase (JAK), a family of intracellular, non-receptor tyrosine kinases which transduce cytokine-mediated signals through the JAK-STAT pathway, has emerged as a potential therapeutic candidate for drug development.4, 5, 6 Accordingly, a number of JAK inhibitors have been approved for the treatment of rheumatoid arthritis (Baricitinib) and psoriasis (Tofacitinib), with several other drugs being tested in ongoing clinical trials.7,8 Considering the potential of immune modulators, such as JAK inhibitors, for the treatment of inflammation-related chronic diseases, there is a need for an approach that can comprehensively assess the efficacy of drugs through evaluation of target engagement and downstream target modulation.
Lipopolysaccharide (LPS) is an endotoxin derived from the outer membrane of Gram-negative bacteria which triggers a potent acute inflammatory reaction in vivo. LPS is known to stimulate multiple cell types including monocytes, dendritic cells, macrophages and B cells through binding to the Toll-like receptor 4 (TLR4) complex and activate downstream pathways including the IKK/NFκB pathway.9 The activated immune cells subsequently release cytokines and chemokines and results in further recruitment and reprograming of other immune cells, inducing systemic inflammation.9,10 Due to its well-characterized activation mechanisms, LPS has been widely used as a model to study acute inflammation in both in vivo and in vitro models.11, 12, 13, 14 Although there are several existing blood-based tests, such as high-sensitivity C-reactive protein (hsCRP) and fibrinogen, that are used clinically to evaluate systemic inflammation,15,16 there are currently no non-invasive approaches that could reliably and comprehensively assess the molecular dysregulation that are associated with inflammatory responses in the circulation or solid organs in a hypothesis-independent manner.
Accumulating evidence indicates that the molecular profile of circulating cell-free messenger RNA (cf-mRNA) reflects organ-specific molecular alterations.17, 18, 19, 20 Cf-mRNA is mRNA that is derived from plasma/sera portion of the blood compared to the blood portion that contains RNA from blood cells (i.e., whole blood, peripheral blood mononuclear cells (PBMC) and buffy coat). A previous study has shown that plasma contains substantially higher portion of mRNAs that are derived from organs compared to that of whole blood.17 We have developed a highly robust RNA-Seq based assay that can accurately quantify the cf-mRNA transcriptome. We also assessed transcriptional dysregulation of cf-mRNA in patients with Alzheimer's disease,20 liver disease19 and hematological cancers17 and demonstrated the prospect of clinical utility of the cf-mRNA platform. A previous study has shown that circulating mRNAs exist in extracellular vesicles (EVs) and particles (EPs), but are scarce as free form.21, 22, 23 EVs, including exosomes, microvesicles, exomeres, supermeres and apoptotic bodies, are expected to be primarily released through endosomal transport. The disparate heterogeneity and overlap of the physical properties as well as the lack of standardization of definitions precludes discrete purification of any one type of complex.24 While, different RNA biotypes are present within these various complexes, mRNAs have well characterized networks and biological processes facilitating interpretation compared to their noncoding RNA counterparts. In spite of the challenges noted, studies have reported the functional role of these complexes as intercellular communiques to adjacent and distal cells.25 Of particular note is communication of solid organ cells with tissue resident bespoke immune cells poised to mount immune and inflammatory responses.26 Sampling the cf-mRNA biological compartment therefore promises a new vista into not only the underlying pathological processes in inaccessible organs but also both passive and active signalling to the immediate cellular microenvironment and to other organs. Here, we examine the robustness of cf-mRNA profiling for the monitoring of LPS and immune modulator induced inflammatory responses using a preclinical mouse model. Our data demonstrate the potential utility of cf-mRNA profiling for the evaluation of drug-induced molecular dysregulations of systemic and organ-specific inflammatory responses and may be useful as a biomarker for drug development and clinical trials.
Methods
Study design and sample collection
For all studies six to ten-week-old male C57BL/6 mice were used. We conducted two independent studies. One of which is focused on the collection of blood sample following treatment of LPS and JAK inhibitor (Study 1) and the other focused on the collection of organ and blood samples following LPS insult (Study 2). For study 1, total of 38 animals were used. These animals were randomly assigned into treatment groups. The animals were treated with LPS (3 mg/kg body weight: VetOne) (time = 0). LPS was administered through intraperitoneal injection. Formulations were prepared by diluting LPS in sterile saline solution, and by combining AZD1480 (a JAK2 inhibitor) (30mg/kg body weight; Thermo Fisher) in 0.5% hydroxypropyl methylcellulose + 1% Tween-80, with the pH adjusted to 3.0 for AZD1480. Blood was collected from the mice via cardiac puncture at 2, 4, 8, 24, 48 and 72 hours post LPS administration, and control animals treated with vehicle. For the administration of AZD1480, two doses of AZD1480 were administered orally, 1 hour prior to and 3 hours post LPS administration. Animals used for the study 1 are summarized in Supplementary Table 1. Although there was no prior study examining the effects of cf-mRNA dysregulation in mice, we chose animals per time point to be approximately 5 animals, based on a previous mouse study where the protein levels of an inflammatory target was measured in both blood and liver following LPS stimulation.27 In addition, a resource equation approach of sample size calculation28 indicated that the minimum sample size for study 1 to be 3 per group.
For study 2, animals were treated with LPS (3 mg/kg body weight: VetOne and diluted in saline) (time = 0) via intraperitoneal injection and blood and selected organs were collected 4 24, 42 and 78 hours post LPS administration. Total of 18 animals were used for the study 2 and these animals were randomly assigned into treatment groups. Blood samples were collected by cardiac puncture and selected organs including lung, brain, kidney and liver were harvested from exsanguinated mice. The organs were dissected in half longitudinally and one section placed in trizol and the other section flash frozen on dry ice. A resource equation approach of sample size calculation estimated that the minimum sample size for the study 2 to be 3 per group. All samples were stored at -80 ⁰C. For each treatment group 2-4 biological replicates were evaluated (Supplementary Table 2). No exclusion criteria were set for the study and animals were randomly assigned to each group using number randomization sequence. All animals were kept under climate-controlled room with controlled conditions of light (12hr light and dark cycle). All animals were housed, and experiments were performed by BTS Research, San Diego, CA.
Ethics
All housing, handling and treatment protocols were reviewed and approved by BTS Research Institutional Animal Care and Use Committee. Study was conducted accordance to the National Institute of Health Guidance for the Care and Use of Laboratory Animals (8th Edition Institute for Laboratory Animal Research). Study reference number 17W-MCP-002 and 17W-MCP-004.
RNA extraction, library preparation and whole-transcriptome RNA-seq
RNA was extracted from 100 µl of plasma using QIA amp Circulating Nucleic Acid Kit (Qiagen) and eluted in 15 µl volume (RNA profile shown in Supplementary Figure 1). ERCC RNA Spike-In Mix (Thermo Fisher Scientific, Cat. # 4456740) was added to RNA as an exogenous spike-in control according to manufacturer's instructions (10−5 diluted ERCC was added to each sample). Agilent RNA 6000 Pico chip (Agilent Technologies, Cat. # 5067-1513) was used to assess the integrity of extracted RNA. RNA samples were converted into cDNA library using Swift 2S kit (Swift Biosciences, catalog no. 28096) according to manufacturer's instruction. We labelled the libraries with 2S Dual Index Kit (Swift Biosciences cat 28096). Subsequently, we performed whole-exome capture using SureSelect XT V6 whole exome+UTR capture probes (Agilent) according to the manufacturer's instruction. Qualitative and quantitative analysis of the NGS library preparation process was conducted using a chip-based electrophoresis and libraries were quantified using a qPCR-based quantification kit (Roche, Cat. # KK4824). Sequencing was performed using Illumina NextSeq 500 platform (Illumina Inc), using paired-end sequencing, 76-cycle sequencing. Base-calling was performed on an Illumina BaseSpace platform (Illumina Inc), using the FASTQ Generation Application. For sequencing data analysis, adaptor sequences were removed and low-quality bases were trimmed using cutadapt (v1.11). Reads shorter than 15 base-pairs were excluded from subsequent analysis. Read sequences greater than 15 base-pairs were aligned to the mouse reference genome GRCm38 using STAR (v2.5.2b) with GENCODE vM14 gene models. Duplicated reads were removed using the samtools (v1.3.1) rmdup command. Gene expression levels (in the unit of transcripts per million (TPM)) were calculated from de-duplicated BAM files using RSEM (v1.3.0). Key sequencing metrics for each sequencing files (total number of aligned reads, number of aligned reads and number of detected protein coding genes) are provided in Supplementary Table 3.
Identification of non-peripheral blood cell and tissue-specific cf-mRNA transcripts
In plasma, a large portion of cell-free transcripts are of peripheral blood cell origin.17 These transcripts that are derived from peripheral blood cells, in general, originate from the bone marrow, but not from the organs of interest. Therefore, in order to identify which transcripts are enriched in plasma, but not in peripheral blood cells, we compared the transcriptional profiles of plasma and compared to that of peripheral blood mononuclear cells (PBMC). To identify peripheral blood cell (PBC) transcripts, the following criterion was used. We define Rij as the ratio of the TPM in plasma over the TPM in PBMC fraction for gene i in mouse j. Similarly, we define rij as the ratio of the TPM in plasma over the TPM in RBC fraction for gene in mouse j. Let Li denote the number of plasma samples where gene i is detected (TPM > 3), Ni denotes the number of plasma samples where Rij > 3, and Mi denotes the number of plasma samples where rij > 3. Gene i is considered as “non-PBC” if Li >= 6 AND Mi > Li * 0.9 AND Ni > Li * 0.9. Tissue (cell-type) specific transcripts are defined as transcripts whose expression in a particular tissue (cell-type) is > 5-fold higher than all the other tissue types (cell-types). Tissue (cell-type) transcriptome expression levels were obtained from the following two datasets: BodyMap for gene expression across 17 human tissues and Immgen for gene expression in endothelial cell, epithelial cell and fibroblastic reticular cell. In this study we only considered the tissue/cell-type specificity for non-PBC transcripts. In addition, we used PanglaoDB cell type specific gene annotations that are derived from single cell sequencing data including Tabula Muris was used to assess cell type specific transcripts.29
Differential expression analysis and pathway enrichment analysis
Differential expression analysis was implemented with DESeq2 (v1.12.4)30 using read counts as input. Genes with fewer than 5 total reads across the entire cohort were excluded from subsequent analysis. Benjamin-Hochberg correction was used to correct for multiple testing and obtain adjusted p-values. Pathway enrichment analysis was conducted using Ingenuity Pathway Analysis (IPA) software version 47547484. The complete list of differentially expressed transcripts was uploaded to IPA and Expression Analysis was used to determine pathways that are highly enriched. IPA categories including Canonical pathways and “Top diseases and bio functions” were examined. In addition, the Metascape analysis tool was used to conduct Gene Ontology and Reactome pathway enrichment analysis with M. Musculus as the input species and H. Sapiens as the analysis species.31
Computational transcriptome deconvolution analysis using non-negative matrix factorization (NMF)
Normalization was first implemented whereby the expression level of each gene in each sample was divided by the gene's maximum value across the samples. This step is designed to rescale the expression levels among different genes to avoid a small number of highly expressed genes dominating the decomposition process. The normalized expression matrix was then subjected to NMF decomposition using sklearn.decomposition.NMF within the Python library Scikit-learn (Ver 0.22.2) (https://scikit-learn.org/stable/). NMF decomposition achieves a more parsimonious representation of the data by decomposing the expression matrix into the product of two matrices X = WH. X is the expression matrix with n rows (n samples) and m columns (m genes); W is the coefficient matrix with n rows (n samples) and p columns (p components); H is the loading matrix with p rows (p components) and m columns (m genes). W is in a sense a summarization of the original matrix H with reduced number of dimensions. H contains information about how much each gene contribute to the components. Biological interpretation of the derived components was achieved by performing pathway analysis on the top genes that contribute the most to each component. Subsequently, the list of genes in each component was used to conduct pathway enrichment analyses using IPA or Metascape for Gene Ontology and Reactome.
Statistical analysis
Pearson's correlation was used to examine the correlation between different bioanalytes. Within a condition/treatment group, a median of all the animals in the group was taken before correlation or fold change calculation. Student's t-test or Mann–Whitney test were used to evaluate the difference between the two groups according to the data distribution. For correlational analysis or Pearson or Spearman's correlations were used. The Benjamin-Hochberg method was used to correct for multiple testing. All statistical analyses were performed using R (3.4.4, R Development Core Team, https://cran.r-project.org/) unless otherwise stated.
Roles of funders
The study was funded by Molecular Stethoscope Inc. No external funding was used to fund the study.
Results
Identification of functionally relevant LPS associated cf-mRNA components
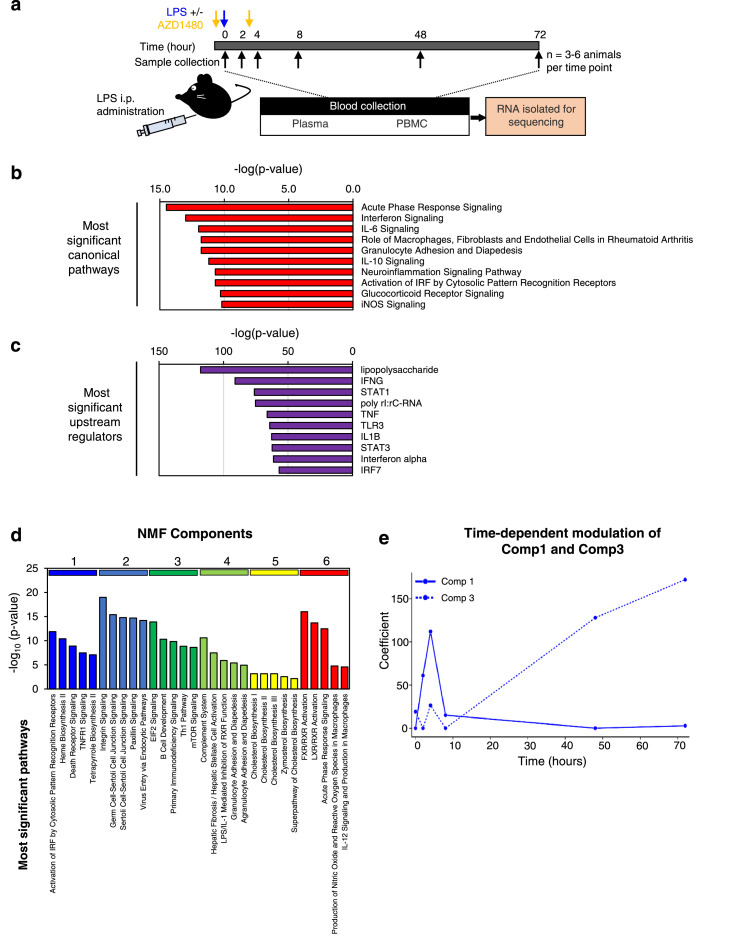
LPS stimulation is a widely utilized preclinical tool to induce an acute inflammatory response and the molecular signatures of the response have been well characterized in mice.11, 12, 13 Therefore, we first investigated whether the cf-mRNA profile is altered in response to acute LPS-induced stimulation. A single dose of LPS (3 mg/kg) was administrated to each C57bl6 mice intraperitoneally. Animals were sacrificed at 2, 4, 8, 48 and 72 hours following the LPS treatment and blood and organ samples were collected (Figure 1a and Supplementary Table 1). RNA was subsequently isolated from the plasma and sequenced. The evaluation of the gene-expression pattern in cf-mRNA showed induction of cytokine transcripts, as early as 2 hours after the LPS treatment, followed by a subsequent regression of cytokine transcripts 8 hours after the treatment (Supplementary Figure 2). We then compared the cf-mRNA transcriptome of untreated and LPS treated animals (4-hour post LSP treatment) and identified 750 differentially expressed cf-mRNA transcripts (476 upregulated and 274 downregulated, FDR < 0.05 was used as the cut-off criterion). We chose 4-hour post-treatment samples for the comparison as cytokine transcripts appear to be at the highest levels at this time point in the plasma compared to other time points. We performed canonical computational biology pathway analysis using IPA (Qiagen) using transcripts that were upregulated following LPS treatment. Pathway analysis identified several key inflammation-associated pathways including, acute phase response (p < 0.0001), interferon signalling (p < 0.0001), IL-6 signalling (p < 0.0001) and TLR signalling (p < 0.0001) as the most enriched pathways (Figure 1b and Supplementary Table 4). Furthermore, the most significant common upstream regulator identified by IPA was LPS (p < 0.0001), followed by major LPS response mediators such as Interferon-γ (p < 0.0001), Stat1 (p < 0.0001), TNF (p < 0.0001) and Toll-like receptor 3 (p < 0.0001) (Figure 1c and Supplementary Table 4). In addition, we performed alternative pathway analysis using Gene Ontology and Reactome (Supplementary Figure 3a and b and Supplementary Table 4). Consistent to the results of IPA analysis, we identified pathways such as “response to lipopolysaccharide” (p < 0.0001) and “inflammatory response” (p < 0.0001) for Gene Ontology and various cytokine associated pathways for Reactome, indicating the overall effect of LPS treatment.
Figure 1.
Identification of 6 cf-mRNA sub-clusters following LPS treatment. (a) A schematic overview of the experimental design. (b) Most significant IPA canonical pathways identified using 750 dysregulated cf-mRNA transcripts as inputs (Control vs 4 h. post LPS treatment). (c) Upstream regulators identified using 750 dysregulated cf-mRNA transcripts as inputs (Control vs 4 h. post LPS treatment). (d) Most significant canonical pathways for individual NMF components. (e) Temporal patterns of component 1 (solid line) and component 3 (dashed line) transcripts. For all pathway analyses, p-values were calculated using Fisher's exact test.
Collectively, these data indicate that prominent inflammatory signals that are typically dysregulated by LPS stimulation are recapitulated in the plasma cf-mRNA of mice treated with LPS.
Next, we performed unsupervised clustering of cf-mRNA transcripts using non-negative matrix factorization (NMF) to evaluate whether these transcripts share expression patterns across various time points and conditions and form gene clusters. NMF analysis resulted in identification of six clusters of gene components (Supplementary Figure 4 and Supplementary Table 5). IPA pathway analysis of the individual clusters showed that these gene components are enriched in distinct biological processes and pathways (Figure 1d and Supplementary Table 5). For example, Component 1 transcripts are enriched in biological processes such as “innate immune response” (p < 0.0001) and “response to cytokines” (p < 0.0001) (Figure 1d and Supplementary Table 5), while Component 3 transcripts are enriched in lymphocyte-specific markers and lymphocytic -related pathways (Figure 1d and Supplementary Table 5). Furthermore, we performed additional gene enrichment analyses by Gene Ontology and Reactome to confirm that component 1 is enriched immune pathway and Component 3 is associated with lymphocyte activation (Supplementary Figure 3c and d and Supplementary Table 5). We then evaluated the temporal patterns of expression level changes for each component following the LPS stimulation. The expression levels of Component 1 transcripts showed a substantial increase following LPS treatment with the highest expression level observed at 4-hours post LPS time point (Figure 1e). Subsequently, the Component 1 transcripts declined and returned to the basal levels by 48 hours. The response we observed for this cluster is consistent to that of cytokine response. In contrast, Component 3 transcripts were unaffected during the early stages of the LPS stimulation but increased substantially at 48-hour post LPS treatment time point, indicative of a delayed response of lymphocytic lineage cells (Figure 1e and Supplementary Figure 5). This observation is also consistent with previous reports that an LPS-insult leads to enhanced survival and hence the accumulation of lymphocytes.32, 33 Our results demonstrated that LPS-induced transcriptional changes in cf-mRNA are consistent with previously known cellular and molecular events triggered by LPS and highlights the ability of the assay to comprehensively monitor molecular alterations in cf-mRNA that are associated with inflammatory response.
Cf-mRNA profiling captures anti-inflammatory property of immunomodulators
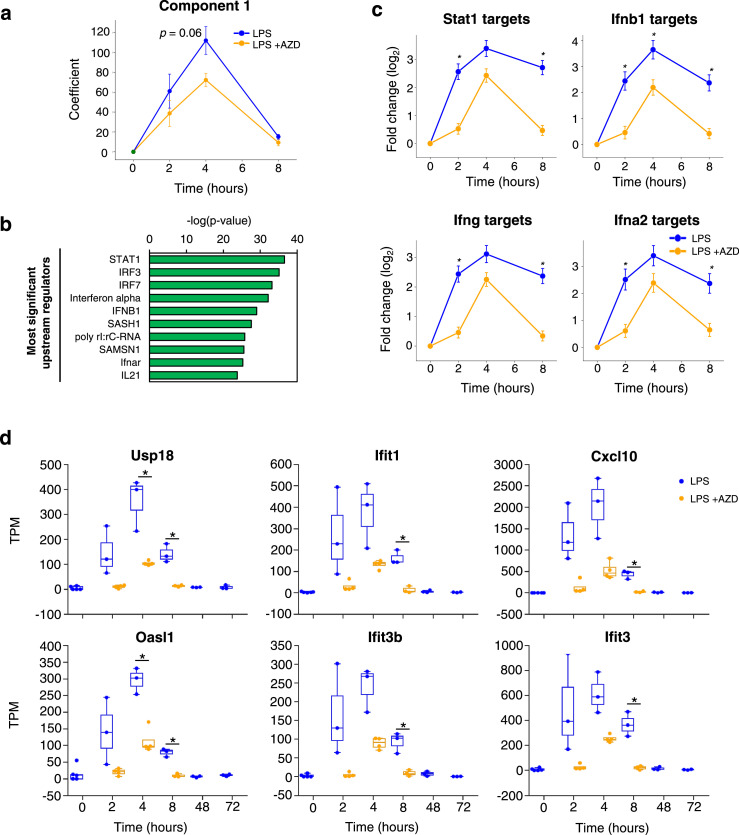
Given the increasing focus on immuno- and inflammatory modulations as a potential drug development strategy, we next evaluated whether cf-mRNA profiling can be used to monitor the effects of immune modulators. To assess the efficacy of immune modulators on LPS-induced transcriptional changes, we treated mice with AZD1480, orally twice, 1 hour prior and 3 hours after LPS treatment. AZD1480 is a JAK inhibitor which is known to preferentially inhibit JAK2. The AZD treatment showed an attenuating trend for the LPS-induced elevation of the Component 1 transcripts (p = 0.06) (Figure 2a), which was enriched for immune response and cytokine response pathways (Supplementary Table 5). The comparison between LPS treated samples with or without AZD resulted in identification of 87 transcripts that are significantly down-regulated in the cf-mRNA of AZD treated group at 2-hour post LPS time point. IPA upstream regulator analysis revealed that these dysregulated transcripts were enriched in downstream targets of interferons and cytokines (Figure 2b and Supplementary Table 6). Transcription factor STAT1, a key mediator of JAK signalling, ranked as the most enriched upstream regulator (p < 0.0001), indicating that AZD indeed inhibited the JAK/STAT pathway. When examining the median fold change of the downstream target genes of STAT1 and the interferons relative to the untreated control, AZD significantly attenuated the expression levels of these transcripts (p < 0.05 at time points 2h and 8 h for Stat1, Ifnb1, Ifng and Ifna2 targets) (Figure 2c). Furthermore, several notable interferon-γ signalling pathway-associated transcripts that were upregulated by LPS (USP18, IFIT1, CXCL10, OASL1, IFIT3B and IFIT3) were suppressed by AZD (p < 0.05 between LPS and LPS +AZD groups at various time points for USP18, IFIT1, CXCL10, OASL1, IFIT3B and IFIT3) (Figure 2d). Collectively, these data indicate that the transcriptional plasma cf-mRNA profile appears to reflect the anti-inflammatory effects of JAK inhibitor and suggest that the cell-free transcriptome can be used to monitor systemic molecular regulation of immunomodulators.
Figure 2.
Cf-mRNA profiling captures anti-inflammatory property of immunomodulators. (a) Time-dependent temporal patterns of component 1 transcripts following LPS treatment with (blue) or without (orange) AZD. (b) Upstream regulators identified using 87 dysregulated cf-mRNA transcripts as inputs (2 h. post LPS treatment with or without AZD). p-values were calculated using Fisher's exact test. (c) Average fold changes of target transcripts of JAK/STAT related upstream regulators relative to the untreated controls. (d) Temporal changes in expression levels of Interferon-γ related transcripts. Mann-Whitney test was used to compare LPS vs LPS+AZD groups (Figures, a, c and d) * represents p <0.05.
Identification of tissue-specific gene-expression signals for inflammatory responses
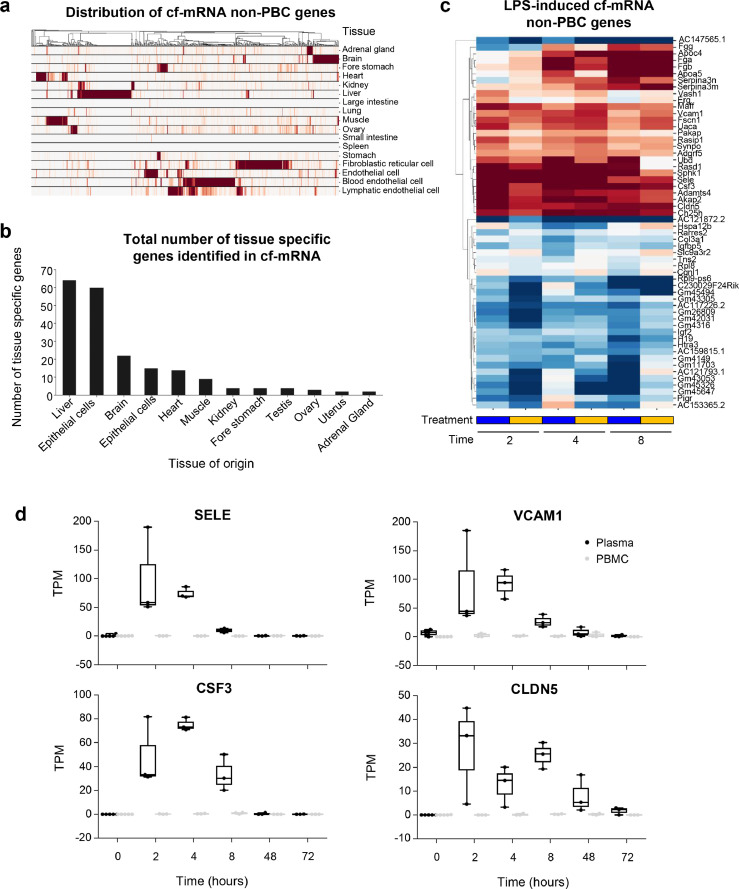
We previously reported that both circulating blood cells and resident tissue cells contribute to the composition of cf-mRNA transcriptome in humans.17 After demonstrating that cf-mRNA profiling can be used to monitor the systemic LPS-induced inflammatory and immune responses, we next investigated whether transcriptional alterations that are occurring in the solid tissues following LPS stimulation can be detected in cf-mRNA. First, we evaluated the transcriptome profiles of matched mouse plasma, PBMCs and red blood cells by separating different blood fractions using Ficoll–Paque and conducted RNA sequencing of each fraction. The comparison of gene-expression profiles between these fractions resulted in identification of 1,054 transcripts that are highly enriched in the plasma cf-mRNA fraction relative to both the PMBC and RBC fractions. We termed these transcripts “non-peripheral blood cell (PBC) transcripts” (non-PBC), since these gene transcripts are enriched in the cell-free fraction of the blood when compared to the peripheral blood cells. Next, we investigated the expression levels of these non-PBC transcripts across various organs and cell types that are published in well-established public datasets (BodyMap & Immgen). A portion of the non-PBC transcripts that we have identified in the cf-mRNA were indeed organ/tissue specific (Figure 3a). In particular, the quantification of organ-specific transcripts in cf-mRNA suggests that the majority of the organ-specific transcripts in the circulation appear to be derived from liver, brain, heart and muscle (Figure 3b). In addition, we used cell type annotation that is derived from single cell databases (Panglao DB)29 to estimate the number of cell type specific transcripts that are present in cf-mRNA (Supplementary Figure 6a). Following, fibroblast and endothelial cells, we identified hepatocytes to have the highest number of marker genes present in the plasma confirming our tissue type estimation. Summation of genes based on the tissue type showed that liver had the highest fraction and total number of tissue genes in plasma (Supplementary Figure 6b). Collectively, both quantification approaches indicate that there are high number of liver derived transcripts present in the plasma.
Figure 3.
Identification of tissue-specific gene-expression signals for inflammatory responses. (a) Expression patterns of non-peripheral blood cell (PBC) transcripts across tissues and cell types. Each row represents a tissue or cell type while each column represents a gene. Each column (gene) is normalized by its maximum value. A hierarchical clustering was performed on the columns. (b) Number of tissue specific transcripts. A transcript with expression level in a particular tissue >5 fold higher than any other tissue is considered specific to that particular tissue. (c) A heatmap depicting fold changes relative to baseline at various time points for non- PBC transcripts. Only transcripts that are significantly differentially expressed compared to baseline (FDR < 0.01) and with TPM > 30 in at least one condition are shown. Hierarchical clustering was performed on the transcripts (rows). Animals treated with LPS treatment with (blue) or without (orange) AZD. (d) Examples of LPS-induced expression changes for non-PBC transcripts. Expression levels in the plasma samples were denoted in black while those in the matching PBMC samples were denoted in grey.
Out of 1054 non-PBC transcripts that we identified, 167 transcripts were significantly dysregulated following LPS stimulation (FDR < 0.05). Most of the organ-specific transcripts dysregulated by LPS treatment were liver-specific (71%). These transcripts were significantly elevated in 4- and 8-hour post LPS treatment time points (Supplementary Figure 7). A collection of the most significantly differentially expressed transcripts (FDR < 0.01 with TPM > 30 in at least one condition) are shown in Figure 3c. In particular, vascular cell adhesion molecule (VCAM1) and E-selectin (SELE) showed a marked increase in plasma following LPS stimulation (Figure 3d). These two genes are known to encode cell surface adhesion molecules that play crucial roles in the adhering of circulating leukocytes to vascular endothelial cells and their subsequent extravasation to the site of inflammation.34 Both genes have been shown to be up-regulated following LPS treatment in human endothelial cells through an NF-κB dependent mechanism.34, 35 Furthermore, granulocyte colony-stimulating factor (CSF3) is known to encode a cytokine that stimulates hematopoiesis of the phagocytic neutrophils and its precursors and play an essential part in innate immune response.36 The levels of CSF3 increased significantly following LPS induction and returned to the basal level by 48-hour post-treatment time point (Figure 3d). Moreover, we showed that LPS stimulation increased the expression level of Claudin 5 (CLDN5) in the circulation, a gene which encodes tight junction protein that is highly specific to endothelial cells. In addition, CLDN4 has been shown to be a key regulator of endothelium permeability37; our data were consistent with a previous study38 (Figure 3d). Upregulation of these transcripts was only observed in plasma, not PBMC, suggesting that these tissue-specific transcripts are highly enriched in the plasma portion of blood. Collectively these data showed that cf-mRNA gene-expression profiles appear to reflect inflammatory processes in the tissues and could potentially be used for organ-specific assessment of inflammatory responses.
LPS induced transcriptional changes in the organs are reflected in the cf-mRNA fraction
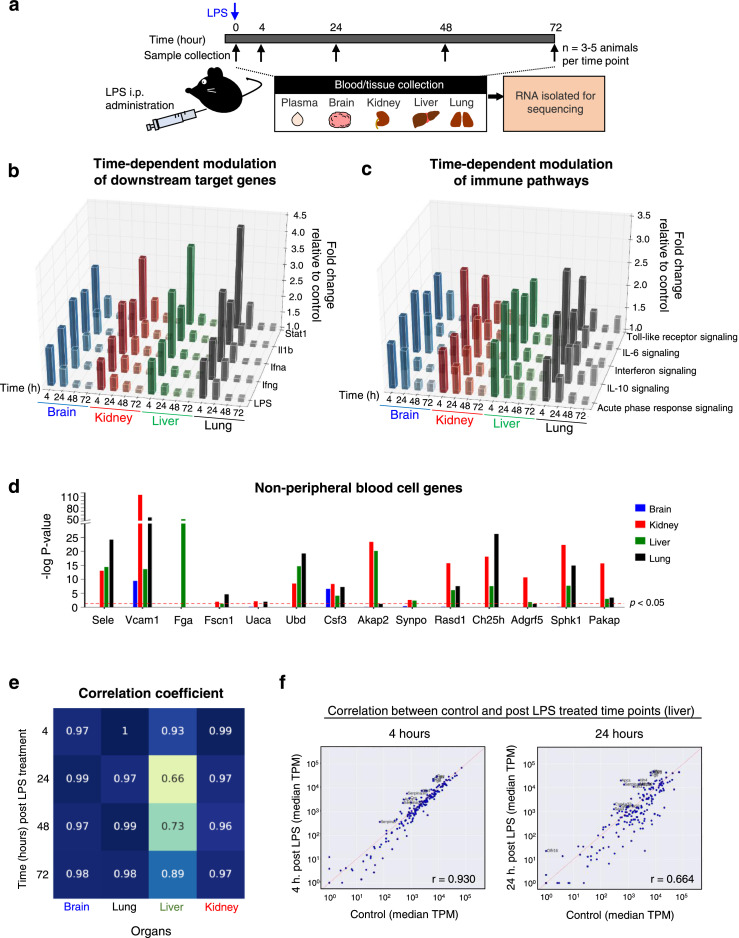
To investigate whether LPS-induced transcriptional alterations in specific organs are reflected in cf-mRNA, we examined gene-expression profiles of multiple organs following LPS stimulation. We conducted an additional experiment where we collected both plasma and corresponding tissue samples (liver, kidney, lung and brain tissues) from LPS treated mice 4, 24, 48 and 72 hours post-LPS treatment (Figure 4a). Differentially expressed transcripts were identified using the untreated animals as the reference and pathway enrichment analysis was performed on differentially expressed transcripts using IPA. In the organs we examined, transcripts up-regulated 4 hours after LPS stimulation were significantly enriched in “acute phase response”, “IL-6 signalling” and “Interferon signalling” pathways, suggesting that LPS-induced inflammatory effects were observed (Supplementary Table 7). Indeed, in all the organs analyzed, LPS was the most prominent upstream regulator identified by IPA, along with other well-known inflammation regulators such as Interferons and STAT1 (Supplementary Table 7). Furthermore, we assessed the time-dependent dynamics of the LPS-induced response by evaluating the average fold-changes of the downstream target genes of the inflammation regulators relative to untreated controls (Figure 4b). The analysis showed elevation of LPS-related immune pathways in the tested tissue types at 4-hour post treatment time point followed by steady attenuation (Figure 4b). Consistently, the time-dependent dynamics of genes involved in the major inflammatory pathways followed similar patterns (Figure 4c).
Figure 4.
Identification of organ-specific transcript dysregulation. (a) A schematic overview of the experimental design. (b) Bar plots showing the median fold changes of transcripts in inflammation related canonical pathways relative to untreated controls. (c) Bar plots showing the median fold changes of downstream target transcripts for immune related regulators Stat1, Il1b, Interferon-α, Interferon-β and LPS relative to untreated controls. (d) Bar plots showing the statistical significance (corrected p-value) of differential expression in the tissues 4 hours after LPS treatment. Those transcripts were chosen because they were significantly upregulated in the plasma and with TPM > 30. The red dashed line indicates the p = 0.05 significance level. (e) Heatmap showing the correlation of tissue specific transcript expression levels between LPS-treated samples and untreated control samples. The columns correspond to different tissues while rows correspond to different time points after treatment. The numbers in each grid represents the Pearson Correlation Coefficient for each comparison. (f) Scatter plots comparing expression levels of liver specific transcripts 4 hours (left) and 24 hours (right) after LPS treatment against untreated controls (x-axis).
In the previous section, we identified a number of cf-mRNA transcripts not expressed in peripheral blood cells that were dysregulated following LPS treatment. Considering that transcripts not expressed in peripheral blood cells likely to originate from solid tissues, we examined whether similar gene-expression dysregulation can be observed in the transcriptomes of solid tissues. We focused specifically on the transcripts not expressed in peripheral blood cells that displayed increased levels of cf-mRNA transcripts at 4-hour post LPS treatment time point, in order to conduct a direct comparison between cf-mRNA and tissue profiles at a matching time point. We showed that all of tissue-specific transcripts were significantly up-regulated in at least one of the studied organs, with most of transcripts being significant in multiple organs (Figure 4d).
Next, we investigated the LPS-induced molecular dysregulation in organs and examined transcriptional alterations of organ-specific transcripts. Transcriptional profiles of post-LPS stimulation time points were compared to that of untreated control. In the lung, brain and kidney, the expression levels of organ-specific transcripts at time points following LPS treatment correlated highly with its corresponding control time point, indicating that LPS treatment had minimal influence on transcriptional dysregulation on organ-specific genes in lung, brain and kidney (Figure 4e and Supplementary Figure 8). However, in the liver tissue we observed a substantial alteration of organ-specific transcripts, with the highest dysregulation observed at 24-hour post LPS. Of the dysregulated organ-specific transcripts, we identified several transcripts that were categorized in the “acute phase response” pathway (IPA) including complement system components (C3), haptoglobin (HP), fibrinogens (FGA, FGB and FGG) and serum amyloid A proteins (SAA1, SAA2, SAA3, SAA4) (Supplementary Figure 9). Dysregulation of these organ-specific genes in the liver tissues are consistent with previous reports.39,40 Collectively, these data indicate that a subset of organ-specific transcripts in the liver appear to be dysregulated following LPS-stimulation.
cf-mRNA profiling captures liver-specific LPS-induced inflammatory response
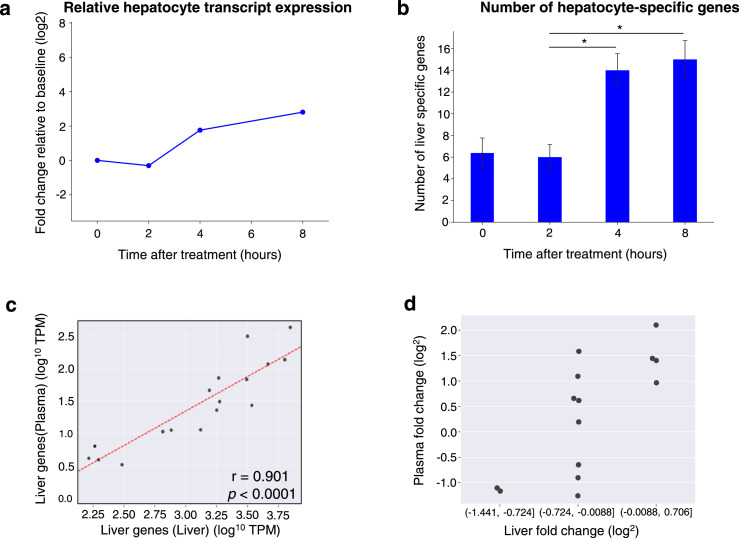
Considering that we identified dysregulation of several organ-specific transcripts in the liver following LPS stimulation, we further examined time-dependent gene-expression alterations of liver-specific transcripts in cf-mRNA following LPS stimulation. Since study 1 has more data within the first 8 hours for animals that were treated with LPS, we examined cf-mRNA profiles of animals that were treated with LPS in study 1. In general, the majority of liver-specific transcripts increased at 4-hour time point and were further elevated at 8-hour time point and this pattern was reflected in the median cf-mRNA liver-specific transcripts (Figure 5a). Concordantly, using a counting threshold of TPM > 5, the total number of detectable liver-specific transcripts significantly increased at 4- and 8-hour post-LPS treatment time points compared to the 2 hour-time point (Figure 5b). These data indicate that liver-specific genes are modulated by LPS-stimulation.
Figure 5.
LPS-induced transcriptional changes of liver-specific transcripts are reflected in cf-mRNA (a) Gene-expression changes of liver specific transcripts following LPS treatment (represented as fold change). Each light blue line represents one liver specific transcript, the blue curve represents the median of all the liver specific transcripts. The fold changes are relative to untreated controls. (b) Number of liver specific transcripts detected in plasma samples at specific time points (TPM > 5 was used as a detection cut off for individual genes). Mann-Whitney test was used to compared different time points. *represents p < 0.05. (c) Scatter plot directly comparing expression levels of liver specific transcripts in the liver tissue (x-axis) and the plasma sample (y-axis). The comparison shown was based on liver tissues and plasma samples harvested from mice 8 hours after LPS treatment. (d) Liver specific transcripts were grouped by their expression fold change in the liver tissue between 4-hour and 8-hour after LPS treatment. The group with higher fold change in the liver tissue also have higher fold change in the plasma and vice versa.
In order to compare the liver-specific gene expression profiles between liver tissue and plasma cf-mRNA, we conducted a correlation analysis of the TPM levels of liver-specific transcripts in the plasma and tissue at 8-hour post-LPS treatment. This time point was chosen for the analysis due to the highest number of liver-specific transcripts were detected in plasma cf-mRNA. We identified a significant linear correlation between the expression levels of liver-specific transcripts in cf-mRNA and that of liver tissue (Pearson's correlation: r = 0.78 and p < 0.0001) (Figure 5c), indicating that the transcriptional dysregulation of liver-specific transcripts in the liver appear to be reflected in the cf-mRNA profile. Furthermore, we examined relative fold-change of the individual liver-specific transcripts between the untreated time point and time points 4 and 8 hours post-LPS stimulation and evaluated the corresponding abundance of individual transcripts between liver tissue and plasma as a fold-change. As expected, the liver-specific transcripts with higher fold-change in the liver had correspondingly higher fold-change in the plasma (Figure 5d). Accordingly, there was a linear correlation between the fold-changes calculated from liver tissue and plasma (Pearson's correlation, r = 0.82, p < 0.001, Supplementary Figure 10). Taken together, these data suggest that liver-specific transcripts in the plasma compartment could be used to evaluate the molecular response in the liver to a therapeutic agent.
Discussion
Inflammatory and immune responses are complex biological responses involving dynamic modulation of a large set of genes and pathways of the innate and adaptive immune systems. In the present study, we performed RNA-Seq of cf-mRNA to characterize LPS-induced systemic and organ-specific inflammatory responses using a mouse model. We demonstrated that cf-mRNA RNA-sequencing is able to provide comprehensive temporal insights of LPS-induced systemic and organ-specific inflammation as well as the modulatory effects of a JAK inhibitor. Furthermore, the comparison between different blood fractions resulted in the identification of transcripts that are enriched in plasma, but not in peripheral blood cells and many of these transcripts appear to be dysregulated by LPS-stimulation. Finally, we discovered several unique liver-specific transcripts in cf-mRNA that reflected molecular alterations in the liver.
In the present study, LPS stimulation was used to evaluate the inflammatory response associated molecular dysregulation in cf-mRNA. We specifically chose LPS stimulation in a mouse model since molecular characteristics of LPS stimulation have been extensively studied and well-characterized in multiple organs.11, 12, 13 While there are existing blood-based tests to evaluate the inflammatory response such as C-Reactive Protein (CRP) and cytokine panels,41,42 a small number of proteins in the circulation is not sufficient to comprehensively assess a complex dynamic process which involves modulation of multiple signalling pathways in systemic and solid organ compartments. Although there are proteomics techniques including mass spectrometry and aptamer-based multiplexing platforms that allow simultaneous quantification of a limited number of proteins in the circulation,43,44 cf-mRNA profiling using RNA-sequencing technology offers an alternative and likely complementary approach to robustly quantify gene expression alterations of the entire transcriptome in a hypothesis-independent digital manner.17,20 The opportunity to discern allelic imbalance and splice variant isoforms because of the sequencing detection strategy casts an even broader net for discovery. Furthermore, we evaluated whether cf-mRNA sequencing can be used to assess the efficacy and target engagement of therapeutic agents using this in vivo model. Accordingly, we examined the effects of LPS stimulation as well as anti-inflammatory effects of a JAK inhibitor to suppress LPS-induced inflammation. Our data showed that a JAK inhibitor substantially suppressed inflammation associated cf-mRNA transcripts that were upregulated by LPS-stimulation. Our study is consistent with and extends the findings of a recent study where inflammatory response was suppressed by a JAK inhibitor in the hepatocyte following LPS stimulation.45 Our approach of simultaneous monitoring of a broad swath of transcripts in the circulation is ideally positioned to study the impact of drugs and drug combinations that target multiple genes or pathways for complex diseases. Collectively, our data demonstrated that the dysregulation of cf-mRNA in the plasma reflected the efficacy of the immune modulating drugs that are administered to the animals.
Many chronic complex diseases such as inflammatory bowel disease, NAFLD, diabetes, rheumatoid arthritis, Alzheimer's Disease and lupus are characterized by chronic inflammation of specific organs/tissues. Unfortunately, blood-based inflammatory biomarkers alone are generally unable to quantify organ-specific immune-related molecular alterations. We have shown previously that a portion of messenger RNAs are known to be specific or enriched in distinct organs.46, 47, 48 Therefore, we looked for tissue-specific transcripts that are dysregulated by LPS-stimulation. Our data from both cf-mRNA and tissue transcriptome profiling indicated that the liver is the organ most affected by the LPS treatment. A direct comparison between cf-mRNA and the matching liver tissue profiles indicated that a small portion of liver-specific transcripts in cf-mRNA appear to recapitulate LPS-induced molecular dysregulation in the liver. Although we demonstrated the significant overlap of dysregulated genes and pathways between plasma cf-mRNA and the corresponding organs in our previous clinical studies,19,20 the studies used molecular tissue profiles of independent studies and not matched tissues. In contrast, in this study we generated plasma cf-mRNA and tissue sequencing data from matched samples using a preclinical model increasing the robustness of our interpretation. Our study further supports that tissue-specific molecular dysregulations are reflected in the gene-expression profile of cf-mRNA. In addition, the comparison of transcriptomic profiles between cf-mRNA and PBMC showed that the tissue-specific transcripts are detectable in plasma, but not PBMC, confirming that the tissue-derived transcripts are present in cell-free portion of blood and not PBMC.
Some of the key questions for quantifying cf-mRNA are whether these mRNAs are actively and/or passively secreted into the circulation, how best to deconvolve the cf-mRNA bulk signal to cell specific signals and what are the turnover times in the plasma. Although several studies have suggested that cell free DNA is primarily released passively following cell death associated processes such as apoptosis and necrosis,49 we have observed in our previous study that cf-mRNA profiling did not change substantially following chemotherapy for both multiple myeloma and acute myeloid leukaemia subjects where the treatment typically results in significant amount of apoptosis and necrosis.17 However, we observed changes in cf-mRNA profile following bone marrow transplantation as well as erythropoietin stimulation. These outcomes suggest that cf-mRNA profile appears to be not influenced primarily by passive release of RNA following tissue/organ injury, but may be actively released during repopulation and stimulation of cells.17 Furthermore, the origin of cf-mRNA appears to be different than other biotypes of circulating RNAs and cfDNAs. Studies have begun to appear that attempt to delineate the major vesicle and particle contributors of extracellular RNA.21,23 Further, while non-invasive molecular biomarkers can be developed using both hypothesis dependent or hypothesis independent strategies, the present study focused on genes that are biologically relevant.
The key limitations of the study include insufficient tissue matched plasma samples for early time points and the limited organs that were assessed. Increased time points with matched tissue samples following LPS stimulation would have allowed us to better evaluate the time delays between the molecular changes occurring in the tissue and corresponding changes that are reflected in the plasma.
In summary, we used a mouse preclinical model to demonstrate the utility of cf-mRNA for comprehensive systemic and organ-specific transcriptomic profiling of LPS induced and JAK inhibitor modulated inflammatory and immune responses. Gruner and McManus systematic review of the evidence for extracellular RNA function in health and disease includes intercellular communication and sensors of homeostatic disruption but note the value of more definitive experimentation.50 The identification of new subclasses of extracellular complexes with extracelluar RNA cargo and heterogeneity of size and density of each subclass provide challenges that the NIH-funded Phase 2 of Extracellular RNA Communication Consortium hopes to address.51 Our data highlight the potential of cf-mRNA profiling for the evaluation of drug efficacy and safety and may inform various steps of drug development and clinical trials including target engagement, pharmacodynamic and pharmacokinetic assessment and toxicological examination.
Contributors
AI, AA, APK and ST performed the experiments. JZ performed bioinformatics analyses. JZ, AI and ST performed data analyses. JZ, AI, AA, APK, JA, MN, JJS, SRQ and ST interpreted the results of the experiments. JZ AI and ST prepared the figures. JZ AI, JJS and ST drafted the manuscript. JZ, AI, AA, APK, JA, MN, JJS, SRQ and ST edited and revised the manuscript. JA and MN were in charge of the project administration. AI, MN and ST supervised the study. JZ and ST have validated underlying data. All authors read and approved the final version of the manuscript.
Data sharing statement
All sequencing data generated in this study was deposited in Sequence Read Archive (SRA) under accession number PRJNA871361. Codes used for this study are available at Bitbucket: https://bitbucket.org/MS_JialiZhuang/mouselps_cfmrna_2022/src/master/.
Declaration of interests
JZ, AI, AA, APK, JA, MN JJS and ST are past/current employees at Molecular Stethoscope Inc. JJS, SRQ and ST have company stock options. SRQ is a founder of Molecular Stethoscope Inc. and a member of scientific advisory board.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.ebiom.2022.104242.
Appendix. Supplementary materials
References
- 1.Chen L, Deng H, Cui H, et al. Inflammatory responses and inflammation-associated diseases in organs. Oncotarget. 2018;9(6):7204–7218. doi: 10.18632/oncotarget.23208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lis K, Kuzawińska O, Bałkowiec-Iskra E. Tumor necrosis factor inhibitors - state of knowledge. Arch Med Sci. 2014;10(6):1175–1185. doi: 10.5114/aoms.2014.47827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rose-John S, Schooltink H. Cytokines are a therapeutic target for the prevention of inflammation-induced cancers. Recent Results Cancer Res. 2007;174:57–66. doi: 10.1007/978-3-540-37696-5_5. [DOI] [PubMed] [Google Scholar]
- 4.Rawlings JS, Rosler KM, Harrison DA. The JAK/STAT signaling pathway. J Cell Sci. 2004;117(Pt 8):1281–1283. doi: 10.1242/jcs.00963. [DOI] [PubMed] [Google Scholar]
- 5.Samadi A, Ahmad Nasrollahi S, Hashemi A, Nassiri Kashani M, Firooz A. Janus kinase (JAK) inhibitors for the treatment of skin and hair disorders: a review of literature. J Dermatolog Treat. 2017;28(6):476–483. doi: 10.1080/09546634.2016.1277179. [DOI] [PubMed] [Google Scholar]
- 6.Ferrajoli A, Faderl S, Ravandi F, Estrov Z. The JAK-STAT pathway: a therapeutic target in hematological malignancies. Curr Cancer Drug Targets. 2006;6(8):671–679. doi: 10.2174/156800906779010227. [DOI] [PubMed] [Google Scholar]
- 7.Fragoulis GE, McInnes IB, JAK-inhibitors Siebert S. New players in the field of immune-mediated diseases, beyond rheumatoid arthritis. Rheumatology (Oxford) 2019;58(Suppl 1):i43–i54. doi: 10.1093/rheumatology/key276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schwartz DM, Kanno Y, Villarino A, Ward M, Gadina M, O'Shea JJ. JAK inhibition as a therapeutic strategy for immune and inflammatory diseases. Nat Rev Drug Discov. 2017;17(1):78. doi: 10.1038/nrd.2017.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lu YC, Yeh WC, Ohashi PS. LPS/TLR4 signal transduction pathway. Cytokine. 2008;42(2):145–151. doi: 10.1016/j.cyto.2008.01.006. [DOI] [PubMed] [Google Scholar]
- 10.Pålsson-McDermott EM, O'Neill LA. Signal transduction by the lipopolysaccharide receptor, toll-like receptor-4. Immunology. 2004;113(2):153–162. doi: 10.1111/j.1365-2567.2004.01976.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Seeley JJ, Ghosh S. Molecular mechanisms of innate memory and tolerance to LPS. J Leukoc Biol. 2017;101(1):107–119. doi: 10.1189/jlb.3MR0316-118RR. [DOI] [PubMed] [Google Scholar]
- 12.Lee JC, Young PR. Role of CSB/p38/RK stress response kinase in LPS and cytokine signaling mechanisms. J Leukoc Biol. 1996;59(2):152–157. doi: 10.1002/jlb.59.2.152. [DOI] [PubMed] [Google Scholar]
- 13.Mukaida N, Ishikawa Y, Ikeda N, et al. Novel insight into molecular mechanism of endotoxin shock: biochemical analysis of LPS receptor signaling in a cell-free system targeting NF-kappaB and regulation of cytokine production/action through beta2 integrin in vivo. J Leukoc Biol. 1996;59(2):145–151. doi: 10.1002/jlb.59.2.145. [DOI] [PubMed] [Google Scholar]
- 14.Fadok VA, Bratton DL, Konowal A, Freed PW, Westcott JY, Henson PM. Macrophages that have ingested apoptotic cells in vitro inhibit proinflammatory cytokine production through autocrine/paracrine mechanisms involving TGF-beta, PGE2, and PAF. J Clin Invest. 1998;101(4):890–898. doi: 10.1172/JCI1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Davalos D, Akassoglou K. Fibrinogen as a key regulator of inflammation in disease. Semin Immunopathol. 2012;34(1):43–62. doi: 10.1007/s00281-011-0290-8. [DOI] [PubMed] [Google Scholar]
- 16.Luan YY, Yao YM. The clinical significance and potential role of C-reactive protein in chronic inflammatory and neurodegenerative diseases. Front Immunol. 2018;9:1302. doi: 10.3389/fimmu.2018.01302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ibarra A, Zhuang J, Zhao Y, et al. Non-invasive characterization of human bone marrow stimulation and reconstitution by cell-free messenger RNA sequencing. Nat Commun. 2020;11(1):400. doi: 10.1038/s41467-019-14253-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ngo TTM, Moufarrej MN, Rasmussen MH, et al. Noninvasive blood tests for fetal development predict gestational age and preterm delivery. Science. 2018;360(6393):1133–1136. doi: 10.1126/science.aar3819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chalasani N, Toden S, Sninsky JJ, et al. Noninvasive stratification of nonalcoholic fatty liver disease by whole transcriptome cell-free mRNA characterization. Am J Physiol Gastrointest Liver Physiol. 2021;320(4):G439–GG49. doi: 10.1152/ajpgi.00397.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Toden S, Zhuang J, Acosta AD, et al. Noninvasive characterization of Alzheimer's disease by circulating, cell-free messenger RNA next-generation sequencing. Sci Adv. 2020;6(50) doi: 10.1126/sciadv.abb1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Enderle D, Spiel A, Coticchia CM, et al. Characterization of RNA from exosomes and other extracellular vesicles isolated by a novel spin column-based method. PLoS One. 2015;10(8) doi: 10.1371/journal.pone.0136133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Winter J, Diederichs S. Argonaute proteins regulate microRNA stability: Increased microRNA abundance by Argonaute proteins is due to microRNA stabilization. RNA Biol. 2011;8(6):1149–1157. doi: 10.4161/rna.8.6.17665. [DOI] [PubMed] [Google Scholar]
- 23.Zhang Q, Jeppesen DK, Higginbotham JN, et al. Supermeres are functional extracellular nanoparticles replete with disease biomarkers and therapeutic targets. Nat Cell Biol. 2021;23(12):1240–1254. doi: 10.1038/s41556-021-00805-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gandham S, Su X, Wood J, et al. Technologies and standardization in research on extracellular vesicles. Trends Biotechnol. 2020;38(10):1066–1098. doi: 10.1016/j.tibtech.2020.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Raposo G, Stahl PD. Extracellular vesicles: a new communication paradigm? Nat Rev Mol Cell Biol. 2019;20(9):509–510. doi: 10.1038/s41580-019-0158-7. [DOI] [PubMed] [Google Scholar]
- 26.Sbierski-Kind J, Mroz N, Molofsky AB. Perivascular stromal cells: directors of tissue immune niches. Immunol Rev. 2021;302(1):10–31. doi: 10.1111/imr.12984. [DOI] [PubMed] [Google Scholar]
- 27.Zhong J, Deaciuc IV, Burikhanov R, de Villiers WJ. Lipopolysaccharide-induced liver apoptosis is increased in interleukin-10 knockout mice. Biochim Biophys Acta. 2006;1762(4):468–477. doi: 10.1016/j.bbadis.2005.12.012. [DOI] [PubMed] [Google Scholar]
- 28.Arifin WN, Zahiruddin WM. Sample size calculation in animal studies using resource equation approach. Malays J Med Sci. 2017;24(5):101–105. doi: 10.21315/mjms2017.24.5.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Franzén O, Gan LM, Björkegren JLM. PanglaoDB: a web server for exploration of mouse and human single-cell RNA sequencing data. Database (Oxford) 2019;2019 doi: 10.1093/database/baz046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou Y, Zhou B, Pache L, et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun. 2019;10(1):1523. doi: 10.1038/s41467-019-09234-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McAleer JP, Vella AT. Understanding how lipopolysaccharide impacts CD4 T-cell immunity. Crit Rev Immunol. 2008;28(4):281–299. doi: 10.1615/critrevimmunol.v28.i4.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schweighoffer E, Nys J, Vanes L, Smithers N, Tybulewicz VLJ. TLR4 signals in B lymphocytes are transduced via the B cell antigen receptor and SYK. J Exp Med. 2017;214(5):1269–1280. doi: 10.1084/jem.20161117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ulbrich H, Eriksson EE, Lindbom L. Leukocyte and endothelial cell adhesion molecules as targets for therapeutic interventions in inflammatory disease. Trends Pharmacol Sci. 2003;24(12):640–647. doi: 10.1016/j.tips.2003.10.004. [DOI] [PubMed] [Google Scholar]
- 35.Lush CW, Cepinskas G, Kvietys PR. LPS tolerance in human endothelial cells: reduced PMN adhesion, E-selectin expression, and NF-kappaB mobilization. Am J Physiol Heart Circ Physiol. 2000;278(3):H853–H861. doi: 10.1152/ajpheart.2000.278.3.H853. [DOI] [PubMed] [Google Scholar]
- 36.Martins A, Han J, Kim SO. The multifaceted effects of granulocyte colony-stimulating factor in immunomodulation and potential roles in intestinal immune homeostasis. IUBMB Life. 2010;62(8):611–617. doi: 10.1002/iub.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yuan L, Le Bras A, Sacharidou A, et al. ETS-related gene (ERG) controls endothelial cell permeability via transcriptional regulation of the claudin 5 (CLDN5) gene. J Biol Chem. 2012;287(9):6582–6591. doi: 10.1074/jbc.M111.300236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mayerhofer R, Fröhlich EE, Reichmann F, et al. Diverse action of lipoteichoic acid and lipopolysaccharide on neuroinflammation, blood-brain barrier disruption, and anxiety in mice. Brain Behav Immun. 2017;60:174–187. doi: 10.1016/j.bbi.2016.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pretorius E, Mbotwe S, Bester J, Robinson CJ, Kell DB. Acute induction of anomalous and amyloidogenic blood clotting by molecular amplification of highly substoichiometric levels of bacterial lipopolysaccharide. J R Soc Interface. 2016;13(122) doi: 10.1098/rsif.2016.0539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jirillo E, Caccavo D, Magrone T, et al. The role of the liver in the response to LPS: experimental and clinical findings. J Endotoxin Res. 2002;8(5):319–327. doi: 10.1179/096805102125000641. [DOI] [PubMed] [Google Scholar]
- 41.Young B, Gleeson M, Cripps AW. C-reactive protein: a critical review. Pathology. 1991;23(2):118–124. doi: 10.3109/00313029109060809. [DOI] [PubMed] [Google Scholar]
- 42.Hanada T, Yoshimura A. Regulation of cytokine signaling and inflammation. Cytokine Growth Factor Rev. 2002;13(4-5):413–421. doi: 10.1016/s1359-6101(02)00026-6. [DOI] [PubMed] [Google Scholar]
- 43.Williams SA, Kivimaki M, Langenberg C, et al. Plasma protein patterns as comprehensive indicators of health. Nat Med. 2019;25(12):1851–1857. doi: 10.1038/s41591-019-0665-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ignjatovic V, Geyer PE, Palaniappan KK, et al. Mass spectrometry-based plasma proteomics: considerations from sample collection to achieving translational data. J Proteome Res. 2019;18(12):4085–4097. doi: 10.1021/acs.jproteome.9b00503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Markotic A, Flegar D, Grcevic D, et al. LPS-induced inflammation desensitizes hepatocytes to Fas-induced apoptosis through Stat3 activation-the effect can be reversed by ruxolitinib. J Cell Mol Med. 2020;24(5):2981–2992. doi: 10.1111/jcmm.14930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Koh W, Pan W, Gawad C, et al. Noninvasive in vivo monitoring of tissue-specific global gene expression in humans. Proc Natl Acad Sci U S A. 2014;111(20):7361–7366. doi: 10.1073/pnas.1405528111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Consortium G. The genotype-tissue expression (GTEx) project. Nat Genet. 2013;45(6):580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Consortium G. Human genomics. The genotype-tissue expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015;348(6235):648–660. doi: 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Heitzer E, Auinger L, Speicher MR. Cell-Free DNA and apoptosis: how dead cells inform about the living. Trends Mol Med. 2020;26(5):519–528. doi: 10.1016/j.molmed.2020.01.012. [DOI] [PubMed] [Google Scholar]
- 50.Gruner HN, McManus MT. Examining the evidence for extracellular RNA function in mammals. Nat Rev Genet. 2021;22(7):448–458. doi: 10.1038/s41576-021-00346-8. [DOI] [PubMed] [Google Scholar]
- 51.Mateescu BJ, Jones JC, Alexander RP, et al. Phase 2 of extracellular RNA communication consortium charts next-generation approaches for extracellular RNA research. iScience. 2022;25 doi: 10.1016/j.isci.2022.104653. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.