Summary
Background
Glioblastoma (GBM) is the most aggressive type of primary brain tumor and is often resistant to current therapies. Tumor microenvironment-centered therapies may unleash new hope for GBM treatment. Therefore, an in-depth understanding of tumor-stroma communication is urgently needed to identify promising therapeutic targets.
Methods
We systematically analyzed GBM single-cell RNA sequencing (scRNA-seq), bulk RNA-seq and spatial scRNA-seq data from various human and mice studies to characterize the network within the microenvironment. Moreover, we applied ex vivo co-culture system, flow cytometry analysis and immunofluorescent staining to validate our findings.
Findings
Our integrative analyses revealed that highly heterogeneous GBM tumor cells can be classified into MES-like, AC-like, OPC-like and NPC-like subtypes based on molecular studying. Additionally, trajectory and regulatory network inference implied a PN to MES cell state transition regulated by specific transcriptional factor (TF) regulons. Importantly, we discovered that glycoprotein nonmetastatic B (GPNMB) derived from macrophages played a crucial role in this transition through immune cell-tumor interplay. Besides, through deep signal transduction analyses and cell co-culture studies, we further disclosed that these GPNMB-high macrophage subpopulations, originating from monocytes, could also ineffectively retain T cells from activating by dendritic cells (DCs).
Interpretation
Our study suggests that targeting this particular GPNMB-high macrophage subset may provide a new strategy to control GBM plasticity and facilitate T cell-based immunotherapy.
Funding
A full list of funding bodies that contributed to this study can be found in the Acknowledgements section.
Keywords: Glioblastoma, Macrophages, Glycoprotein NMB, Integrated analysis, Single cell
Research in context.
Evidence before this study
Glioblastoma (GBM) is one of the most lethal and treatment-refractory cancers. Intra-tumoral heterogeneity is one of the major reasons for therapy failure in GBM. The highly immunosuppressive microenvironment, including various types of immune and stromal cells, is another crucial factor that promotes tumor progression and orchestrates therapeutic resistance. Therefore, an in-depth understanding of the GBM ecosystem, particularly the interaction between different cell types using single-cell-based approaches will facilitate the development of new therapeutic opportunities.
Added value of this study
In this work, we reveal the heterogeneity and plasticity of GBM cells on the transcriptomic level and identify a dynamic transition from PN-to-MES subtype regulated by GPNMB derived from macrophages using scRNA-seq analysis. Besides, we constructed intercellular communication networks between myeloid cells and T cells, and further demonstrate spatial colocalization and ineffective retention of GPNMB+ macrophages with T cells.
Implications of all the available evidence
Our study systemically characterizes the intercellular interactions in the tumor microenvironment of GBM and provides a potential strategy targeting GPNMB-high macrophages for combination immunotherapy in the future.
Alt-text: Unlabelled box
Introduction
Glioblastoma (GBM) is the most aggressive primary brain tumor with a median survival less than 15 months despite surgery, chemotherapy, and radiotherapy.1,2 Immunotherapy has revolutionized cancer treatments in recent years, however, GBM, as an immunologically cold tumor, is generally resistant to this approach because of the severe immunosuppression.3,4 Molecular studies classified GBM into proneural (PN), classical (CL) and mesenchymal (MES) subtypes.5,6 MES subtype has the worst prognosis and can be transformed from PN subtype during tumor development.7,8 In fact, multiple subtypes exist within the same tumor, which makes GBM even more refractory to current therapies. To date, studies have examined intrinsic factors involved in tumor heterogeneity and cancer progression. However, how biological events extrinsically drive cancer development and shape immunosuppressive tumor immunity are still largely unknown. Thus, explicitly understanding the GBM microenvironment and the interplay between diverse cellular compositions will provide new therapeutic avenues for both targeted therapy and immunotherapy.
Macrophages can comprise 30–50% of tumor mass,9 constituting a distinct population with heterogeneous properties in GBM. Typically, tumor-associated macrophages have M2-like characteristics, which promote tumor development and immune suppression.10 In contrast, M1-like macrophages that produce inflammatory cytokines can support anti-tumor immune responses.11 Therefore, multiple strategies have been developed for targeting macrophages switching through particular signals (e.g., Pi3Kr, CSF1R, HDAC) in order to slow tumor growth and limit the immunosuppressive effects on T cells,12, 13, 14 but the benefits remain limited and controversial. Importantly, this binary subset cannot fully capture the complexity of macrophage-mediated immunity in the tumor microenvironment (TME), hence exploring the functional diversity of macrophage subsets remains imperative for promising targets development.
Recently, multi-omics approaches at single-cell resolution have been considered a promising method for tumor biology study. Increasing numbers of single-cell RNA sequencing data have revealed major characteristics of GBM and complex immune microenvironment.15, 16, 17, 18, 19, 20 Here, we analyzed various scRNA-seq data to discover a PN to MES cell state transition regulated by key transcriptional factor (TF)-regulons through trajectory and regulatory network inference. Moreover, using a computational method that predicts the ligands from sender-cell to interact with specific targets from receiver-cell based on gene expression and prior knowledge on regulatory signaling paths, we uncovered glycoprotein nonmetastatic B (GPNMB) derived from macrophages as the top ligand in regulating these TF-regulons implicated in the PN-MES transition.
GPNMB, a transmembrane glycoprotein, presents on the cell surface or resides in endosomes and lysosomes.21 The membrane-bound part can be cleaved by metalloproteinases to produce the soluble isoform,22 which binds multiple receptors to promote intercellular crosstalk, and its high expression level correlates with therapy resistance in cancer treatment.23 GPNMB protein was originally discovered in tumor cells and involved in tumor migration, invasion, metastasis,24,25 and also immune evasion through directly inhibiting T cell activation.26 However, in our study, we show that GPNMB is predominantly expressed on macrophages rather than dendritic or tumor cells in the GBM microenvironment. Through signal transduction networks investigation, ex vivo cell co-culture, flow cytometry and immunofluorescent staining, we demonstrate that GPNMB-high macrophages not only promote tumor progression by inducing PN-MES tumor cell state transition but also dampening T cell activation through non-effective retention. Thus, targeting GPNMB-high macrophages provides the susceptibility of GBM to molecular-targeted therapies and creates a favorable environment for T cell-based immune responses.
Methods
Ethics approval
Because our study used publicly available datasets, an ethical committee approval was not required.
Data accessibility
The scRNA-seq data of GBM samples (GSE103224, GSE138794, GSE139448 and GSE131928) were obtained from Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo/) database. The bulk RNA-seq and microarray data of GBM samples were obtained from TCGA and CGGA databases (http://gliovis.bioinfo.cnio.es/).
scRNA-seq data process and integration
The scRNA-seq data including GSE103224, GSE138794, GSE139448 and GSE131928 were explored using Seurat package in R 4.0.5.27 The first step was to filter out low-quality cells with a cutoff value of less than 200 total feature RNA and more than 5% mitochondrial RNA. After normalizing the gene expression in each cell, the data from each dataset were then integrated using SCTransform integration workflow (https://satijalab.org/seurat/articles/integration_introduction.html). A total of 2000 integration anchors representing nearest neighbors within datasets were identified, and the data were further scaled by regressing out cell cycle effect and total number of counts per cell, as well as percentages of mitochondria-expressed genes per cell. To reduce dimensions, principal component analysis (PCA) was performed (npcs=30) and followed by t-distributed stochastic neighbor embedding (tSNE) and uniform manifold approximation and projection (UMAP) algorithms (dims = 1:15). Afterward, 28 cell clusters were found in the first place after applying the FindNeighbors and FindClusters functions (resolution=0.8). Using the Wilcoxon rank-sum test, genes differentially expressed in each cluster were identified using FindAllMarkers function (cutoff:min.pct=0.25 and logfc.threshold=0.25). Then, small clusters were merged together by annotating with the singleR package28 and manual verification based on canonical markers. Doublets were removed using the doubletFinder R package.29 Last, integration yielded a total 54,534 cells across 40 samples with 11 major cell clusters named MES-like, AC-like, NPC-like, OPC-like tumor cells, unknown cells, oligodendrocytes (Oligo), Macrophages, Microglia cells, NK and T cells (Lymphocyte), endothelial cells (EC) and smooth muscle cells or GBM-associated endothelial cells (SMC/GBM-EC) for further analysis. Gene signatures for MES-like AC-like, NPC-like and OPC-like were listed in Supplemental Table 1, and signature scores of cells were performed with AddModuleScore function.
Inference of CNV
To distinguish malignant cells from normal cells, copy number variation was estimated using infercnv package.30 Low-expression genes that expressed less than 10 cells and a median expression below 0.1 were removed. Genes were then annotated according to chromosomal position, and the CNV score was estimated from the moving averages of 100 genes. Lastly, hierarchical clustering was used to distinguish non-malignant cells from malignant cells with clear chromosomal deletions or amplifications.
Trajectory analysis
Data collected from Seurat can be easily imported into Monocle 2 package31 using as.CellDataSet function for trajectory analysis. First, genes were excluded if the expression was less than 0.5 and expressed cells were fewer than 200. Based on different gene expression patterns, the dataset was re-ordered by highly different genes with a q value of 1e-40. Next, a DDRtree dimensional reduction algorithm was performed for data interpretation. Last, pesudotime analysis was undertaken to identify significant genes along with their trajectories, while cells in the same branch were considered to be at the same differentiation stage. Besides, the branch expression analysis modeling (BEAM) test was used to identify differential expression levels at a branch-dependent level. In addition, unsupervised inference of developmental directions of tumor cells was also confirmed using VECTOR algorithm.32 Briefly, all the UMAP dimensions regarding tumor cells were treated as an image, and then were split by pixels, and the largest connected pixel network by linking adjacent pixels in UMAP was generated to infer the developmental direction.
Gene set enrichment analysis GSEA and pathway analysis
Using the differentialGeneTest function in Monocle2, the top 1000 expressed genes between cell types were generated and further GSEA and pathway analysis was conducted using msigdbr33 and clusterProfiler packages.34 Gene signatures from Hallmark, GO-biological processes and REACTOME datasets were used as inputs to evaluate the pathway activity of different cell clusters.
SCENIC
Activated regulons in different tumor subsets were identified using SCENIC package.35 The raw count matrix used as input was calculated for co-expression activity using a Spearman correlation by GENIE3 (treeMethod= ”RF”, K=”sqrt”, nTrees = 1000). These filtered targets were then analyzed for motif enrichment using RcisTarget. Particularly, an AUCell with a Wilcoxon rank-sum test was used to estimate regulatory activity scores for gene motifs located 500 bp upstream of the TSS and 10 kb around the TSS. Last, important regulons modulated by key TFs were identified. For example, these targets, EPAS1-regulon (31g), FOSL2-regulon (30g), CEBPB-regulon (22g), SOX4-regulon (16g) and SOX11-regulon (19g) were then used as signature scores for visualization using AddModuleScore and FeaturePlot function in Seurat.
Survival analysis
Expression data of GBM were obtained from HG-UG133A, Agilent-4502A, and CGGA datasets in Gliovis. The expression of each gene is formatted as Log2 (TPM+1) scale with or without normalization. According to the subtype categories in the meta files, samples were divided into MES, PN, and CL groups. And a GSVA score for MES or PN high-or-low subtype with particular gene signatures was also analyzed in groups. Kaplan-Meier survival curves were fitted using the survival package36 in R with 50-50 percentiles, and survival plots were visualized using the ggsurvplot function.
Correlation analysis
In order to quantify the relative fractions of certain cell types such as macrophages, dendritic cells and T cells, deconvolution of bulk seq (TCGA and CGGA datasets) was used with xCell (http://xCell.ucsf.edu/). A Pearson's correlation test was used to determine the correlation between cells, target transcription factors, markers of cells and GPNMB-correlated genes. P-value lower than 0.05 and correlation coefficients more than 0.3 were considered significantly correlative.
Spatial transcriptomics data processing
The spatial transcriptomic data of GBM were downloaded from 10x Genomics (https://www.10xgenomics.com/cn/resources/datasets/human-glioblastoma-whole-transcriptome-analysis-1-standard-1-2-0), and analyzed in R using the Seurat 4.0 package according to the recommended data processing guidelines (https://satijalab.org/seurat/articles/spatial_vignette.html). In briefly, The SCTranform function was used for data normalization, followed by PCA and UMAP for dimension reduction, and clustering was conducted with the default resolution of the first 30 PCs. The gene expression features were visualized by SpatialFeaturePlot function, and signature scoring (Supplemental Table 1) for MES-like, AC-like, NPC-like and OPC-like malignant cells was performed with the AddModuleScore function.
Nichenet
Nichenet package collects the protein-protein interaction data from Omnipath, PathwayCommons, ConsensusPathDB, KEA, DEPOD and the regulator interaction data from TRRUST, HTRIDB, RegNetwork, Ontogenet, CHEA, ENCODE, JASPAR, MOTIFMAP, MSigDB to generate a network matrix.37 To estimate the interactive ligands from sender cells (such as microglia cells, macrophages, NK and T cells, ECs and GBM-ECs), all tumor cells were combined as receiver cell and gene signatures, such as EPAS1, FOSL2, and CEBPB as well as downstream signals targets (Supplemental Table 2) were used as targets. Only the top 15% expressed genes in sender cells were calculated by regulatory potentials and ligand activity was ranked with a cutoff of 0.5 using Pearson test. The predicted ligands from stromal cells as well as the potential targets and receptors of tumor cells are listed afterward.
CytoTalk
Signal networks between two specific cell types were analyzed using CytoTalk package.38 Cell-gene matrixes from total T cells, total dendritic cells, Gpnmb-high macrophages and monocytes along with a ligand-receptor paired list were used as input to infer the gene interactions. Genes expressed in less than 10% of each cell type were filtered out. This algorithm constructed a singling network using prize-collecting Steiner forest (PCSF) based on node prize (size of each node, representing the specificity in each cell type) and edge cost (lines connected to each node, corresponding to the potential interaction between two genes). In particular, PCSF yielded 175 signaling networks between macrophages and T cells, 250 signaling networks between monocytes and T cells, and 164 signaling networks between dendritic cells and T cells. The figures were also automatically exported and can be visualized using Cytoscape version 3.9.39 Cell color was used to distinguish cell type and color intensity indicated the expression levels.
GBM tumor induction
All studies were supervised and approved by the Shanghai University of Traditional Chinese Medicine Institutional Animal Care and Use Committee (IACUC). GL261 cell lines were kindly provided by Dr. Hao Duan (State Key Laboratory of Oncology in South China, Department of Neurosurgery/Neuro-oncology, Sun Yat-sen University Cancer Center, Collaborative Innovation Center for Cancer Medicine, Guangzhou, China. RRID: CVCL_Y003). This cell line has been performed short tandem repeat (STR) profiling analysis and tested negative for mycoplasma contamination using bioluminescent assays by Shanghai Fuheng Biology (Reagent Validation Files). GL261 cells were cultured in DMEM supplemented with 10% FBS and 2 mM L-glutamine (Life Technologies) and pretreated with mycoplasma removal agent (MP Biomedicals) before use. A syngeneic GBM mouse model was induced by orthotopically injecting 1 × 105 GL261 cells into wild-type mice on the C57BL/6J background (RRID: IMSR_JAX:000664) as previously described.40,41 After 3–4 weeks, mice with severe GBM symptoms including dome head and hemiparesis were euthanized and the single-cell suspensions of GBM were prepared with the gentleMACS Dissociator (Miltenyi Biotech, 130-093-235) and the collagenase II and dispase II enzymes (Millipore Sigma, 10269638001) for further analysis. All procedures were approved by the Animal Ethic Review Committee of Shanghai University of Traditional Chinese Medicine (#PZSHUTCM210604002).
Immune cell isolation
Mouse T cells were isolated from wild-type C57BL/6J mouse spleens using total an EasySep Mouse T Cell Isolation Kit (Stemcell Technologies; 19851) and cultured in RPMI 1640 medium supplemented with 10% FBS, 20 mM HEPES buffer and 50 U ml−1 interleukin-2 (Peprotech; 212-12). For monocytes isolation, the bone marrow cells were initially obtained from femurs and tibia by flushing RPMI-1640 medium (Life Technologies, 11875119), then passed through a 40 μm strainer, and subjected to an EasySep Mouse Monocytes Isolation Kit (Stemcell Technologies; 19861). Tumor-derived single-cells were incubated with FcBlock (Biolegend, 101319, RRID:AB_1574973), and followed by staining with a mixture of fluorochrome-conjugated antibodies including anti-CD45 (1:200, eBioscience, 48-0451-82, RRID:AB_1518806), anti-CD11b (1:200, eBioscience, 69-0112-80, RRID:AB_2637405), anti-CD11c (1:200, BioLegend, 117307, RRID:AB_313776), anti-F4/80 (1:200, BioLegend, 123107, RRID:AB_893500), anti-MHCII (1:200, eBioscience, 47-5321- 80, RRID:AB_1548792) and anti-GPNMB (1:100, Invitrogen, 50-5708-80, RRID:AB_2574238). All antibodies were validated by the commercial vendor. CD11b+F4/80+GPNMB+ macrophages and CD11c+MHCII+ dendritic cells were sorted on a FACSAria II instrument (BD Biosciences).
Realtime-PCR analysis
A PN-subtype of GBM cell line was authenticated and tested negative in mycoplasma contamination by supplying company (DSMZ, ACC 880, RRID: CVCL_A5ED). Cell was cultured in DMEM supplemented with 10% FBS and 2 mM L-glutamine (Life Technologies). Tumor-associated macrophages were freshly sorted from GL261-derived tumor cells using flow cytometer, and subsequently co-cultured with GBM spheroids using transwell inserts (upper chamber for macrophages and lower chamber for GBM cells) in presence of neutralizing antibody against GPNMB (0.5 µg/mL; Thermo Fisher; PA5-47301, RRID:AB_2605801). GBM spheroids were split every three days and then re-cultured with newly isolated macrophages again. After 15 days, the GBM cells at lower chamber were collected for RNA isolation using TRIzol (Invitrogen, 15596026), chloroform (Sigma–Aldrich, C2432) and 2-propanol (Fisher Scientific, A416-1). RNA was then transcribed into cDNA using SuperScript III First-Strand Synthesis SuperMix (Life Technologies, 18080400), and a Real-time PCR was performed using Fast SYBR Green Master Mix (Applied Biosystems, 4385612) with the following primer sequences: Gapdh (forward: 5’-aactttggcattgtggaagg-3’; reverse: 5’-acacattgggggtaggaaca-3’), Fosl2 (forward: 5’-ctttatcgcctcaagccaag-3’; reverse: 5’-atccatgctatgggcagaag-3’), Cebpb (forward: 5’-acttcagcccctacctggag-3’; reverse: 5’-gaggtcggagaggaagtcgt-3’) and Epas1 (forward: 5’-caacctgcagcctcagtgta-3’; reverse: 5’-gtgtggcttgaacagggatt-3’)
T cell activation and proliferation assay
GL261 cells were lysed by 3–5 cycles of freeze-thawing (−80 °C to 37 °C). The lysates were centrifuged at 2000 rpm for 10 min and the supernatants were subsequently incubated with different myeloid subsets overnight for further analysis. For T cell activation assay, monocytes, CD11c+MHCII+ dendritic cells or CD11b+F4/80+GPNMB+ cells were isolated using BD Influx (BD Biosciences) and subsequently pulsed with tumor lysates prior to co-culturing with naïve T cells (1:1) for 3 days (Supplementary Fig.3). After incubation, cell mixtures were stained with anti-IFNγ (1: 100, BioLegend, 505808, RRID:AB_315402), anti-GZMB (1: 100, BioLegend, 372211, RRID:AB_2728378), anti-CD69 (1: 100, BioLegend, 104511, RRID:AB_493565), anti-CD8a (1: 200, BioLegend, 100706, RRID:AB_312745) or control IgG antibodies for FACs analysis. All antibodies were validated by the commercial vendor. For T cell proliferation assay, freshly isolated T cells were labeled with CFSE dye (Thermo Fisher Scientific; C34554) for 20 min. After several wash steps, T cells were further co-cultured with pulsed cell populations for 3 days. The data were acquired using CantoII flow cytometer (BD Biosciences) and analyzed using FlowJo software.
siRNA treatment
Bone marrow-derived monocytes were obtained from femurs and tibia of wild-type C57BL/6J. 10 ng/ml CSF-1 was treated for 3 days to differentiate M0 macrophages. Macrophages were transfected with siRNA targeting Gpnmb (Thermo, 175440) using Amaxa 4D-Nucleofector (Lonza) with program EA-100. 100 ng/ml of IL-4 was treated for another 2 days to induce M2 macrophage polarization. After treatment, the expression of CD11b (1:100, BioLegend, 101211, RRID:AB_312794) and CD206 (1:100, BioLegend, 141710, RRID:AB_10900445) expression were determined by flow cytometry.
Immunofluorescence
Immunofluorescence was carried out as previously described.40 Briefly, after de-paraffinization and rehydration, section samples were then applied to antigen retrieval using Retrieve Solution (DAKO; S1699) at 95 °C for 25 min. Sections were further blocked with 5% horse serum and incubated with anti-GPNMB (1:50, R&D Systems, AF2330, RRID:AB_2112934), anti-Mac-3 (1:100, BD, 550292, RRID:AB_393587) and anti-CD3 (1:100, Abcam, ab11089/ab16669, RRID:AB_2889189/AB_443425) overnight at 4 °C. After washing in PBS, sections were stained with anti-488, anti-568, and anti-647 appropriate secondary IgGs (1:500, Life Technologies) for 1 h. All antibodies were validated by the commercial vendor. Images were acquired using an Axio Imager microscope (Zeiss).
Flow cytometry
Single-cell suspensions from mouse GBM were further stained with fluorochrome-conjugated antibodies for 30 min, including anti-CD45 (1:200, eBioscience, 48-0451-82, RRID:AB_1518806), anti-CD3 (1:100, BioLegend,100233, RRID:AB_2561387), anti-CD11b (1:200, BioLegend, 101228, RRID:AB_893232), anti-CD11c (1:200, BioLegend, 117309, RRID:AB_313778), anti-F4/80 (1:200, BioLegend, 123107, RRID:AB_893500), anti-MHCII (1:200, eBioscience, 47-5321-80, RRID:AB_1548792) or control IgG antibodies. All antibodies were validated by the commercial vendor. Data were collected using CantoII flow cytometer (BD Biosciences) and analyzed using FlowJo software.
Statistical analysis
All statistical analyses and plots were performed using R (4.0.5) and GraphPad Prism 8. Log-rank test was used for Kaplan-Meier survival analysis and for linear relationship evaluation, Pearson correlation coefficient was utilized. A one-way ANOVA test was used for multiple comparisons. P valued was marked within plots to indicate statistical significance.
Role of Funders
The Funders do not participate in study design, data collection, data analyses, interpretation or writing of report.
Results
Transcriptomic characterization of the TME and tumor heterogeneity in GBM by scRNA-seq analysis
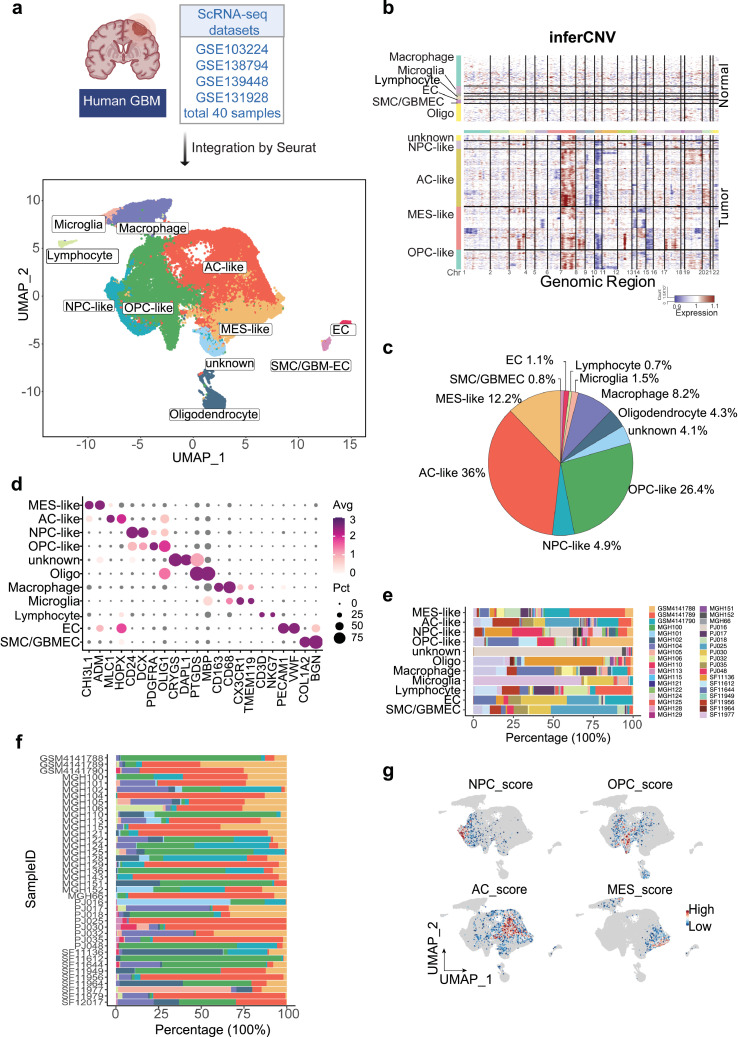
In order to study the heterogeneity of GBM, we integrated and analyzed scRNA-seq data from different independent datasets using Seurat. SCTransform algorithm was performed to remove the batch effect and the integrated data yields a total of 54,534 cells from 40 patients. After normalization of gene expression and principal component analysis (PCA), uniform manifold approximation and projection (UMAP) analyses revealed 28 unsupervised clusters across all cells (Supplementary Figure 1). By applying the annotation with singleR and cell-type-specific markers, we merged small cell clusters into big cell lineages based on the shared transcriptomes (Figure 1a, Supplementary Figure 2). Copy-number variation helps to distinguish the tumor cells from normal cells. Therefore, we observed the chromosome 7 gain along with chromosome 10 loss in tumor cells but not in normal cells by analyzing the average expression of 100 top genes in each cell type (Figure 1b), which is consistent with published WES data.42 Malignant cells including MES-like (12.2%, marked with CHI3L1 and ADM), AC-like (36%, marked with MLC1 and HOPX), NPC-like (4.9%, marked with CD24 and DCX) and OPC-like (26.4%, marked with PDGFRA and OLIG1) clustered together, whereas the non-tumor cells such as macrophages (8.2%, marked with CD163 and CD68), microglia cells (1.5%, marked with CX3CR1 and TMEM119), lymphocytes (0.7%, marked with CD3D and NKG7), endothelial cells (1.1%, marked with VWF and PECAM1), and tumor-associated endothelial cells (0.8%, marked with COL1A2 and BGN), oligodendrocytes (4.3%, marked with PTGDS and MBP) were scattered (Figure 1c-d). The percentage of cell types varied across patients, especially for different cellular states of tumor cells, indicating that heterogeneity is the major feature of GBM (Figure 1e-f). Especially for one cell cluster, here referred to as unknown cells, was highly dominated by one single patient (PJ016). Chromosomal 7 amplification indicated that this type of cell was more like tumor cells than matured oligodendrocytes cells, although both express PTGDS and MBP. Additionally, Gene set enrichment analysis (GSEA) analyses verified the different tumor subtypes of GBM by analyzing a panel of specific gene expressions as Z-score (Supplementary Table 1, Figure 1g). Collectively, molecular subtyping of single cells suggests that GBM patients show a high level of intratumor subtype heterogeneity.
Figure 1.
Transcriptomic characterization of the TME and tumor heterogeneity in GBM by scRNA-seq analysis.
(a) Integration of 40 human GBM scRNA-seq data collected from four individual datasets by Seurat. A total of 54,764 cells were analyzed using UMAP, and 10 significant cell clusters are color-coded and labeled as indicated. (b) Inference of copy number variation analysis based on average expression of 100 genes shows the chromosome 7 gain (red) and chromosome 10 (blue) loss in tumor cells compared with normal cells. Each row corresponds to a cell type. (c) A pie chart demonstrates the proportions of each cell type in GBM. (d) Dot plot displays the represented markers for each major cell cluster, including MES-like, AC-like, NPC-like, OPC-like, oligo, macrophages, microglia, lymphocyte, EC, and GBM-ECs. (e) Stacked bar plots show the percentage of each cell cluster by patient ID and (f) the distribution of cell population in each patient. Cell type colors match those represented in the UMAP plot, while each patient is represented by a distinct color as indicated. (g) Feature plot displays the heterogeneity of GBM subtypes by Z-score, as determined by calculating the geometric mean of gene signature expressions. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Trajectory interference indicates key regulons in proneural-mesenchymal subtype transition
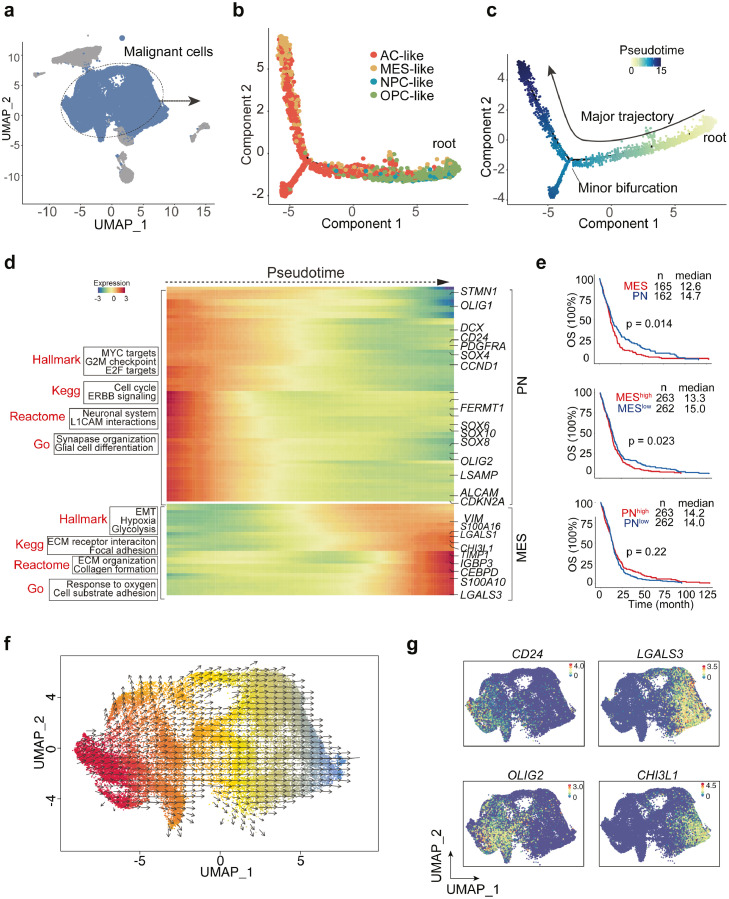
Next, to explore the transition between different subtypes of GBM (Figure 2a), we extracted all the tumor cells and applied unsupervised trajectory analysis using Monocle.31 Trajectory analysis revealed a major lineage starting from NPC- and OPC-like tumor cells, corresponding to the proneural (PN) type of GBM classified by TCGA,6 to AC- and MES-like tumor subtypes, while there was also a minor branch toward AC-like cell state (Figure 2b). Moreover, the pseudotime analysis data revealed the plasticity of GBM and a dynamic transition from proneural to mesenchymal subtype (Figure 2c). Also, the unsupervised developmental inference analysis confirmed this dynamic change (Figure 2f). Importantly, bioinformatic analysis showed the different gene expressions and pathway enrichments along the pseudotime. For instance, DCX, CD24, OLIG1/2, SOX family-related genes, corresponding to PN states, were gradually decreased, while VIM, CH3L1, and LGALS3, representing the mesenchymal state, increased dramatically (Figure 2d, 2g). Pseudotime-dependent pathway enrichment analyses indicated the presence of cell cycle, G2M checkpoints, and glial cell differentiation gene signatures within PN state cells (Figure 2d), suggesting proneural cells are highly proliferative and plastic. In contrast, the mesenchymal state was characterized by epithelial-mesenchymal transition (EMT), hypoxia, ECM organization and cell adhesion pathways (Figure 2d). Importantly, we analyzed the survival rates from TCGA dataset by dividing the patients into, MES-high or MES-low, PN-high or PN-low groups. Survival data showed that patients with MES-like transcriptome signatures predicted poor overall survival compared with PN subtype, while patients with higher MES-score (calculated by average expression of mesenchymal gene signatures) were always associated with poor prognosis compared to those with lower MES scores (Figure 2e). Besides, there was no survival rate difference between high or low PN scores (Figure 2e), all of which suggests that GBM patients with mesenchymal signatures have poor survival outcomes.
Figure 2.
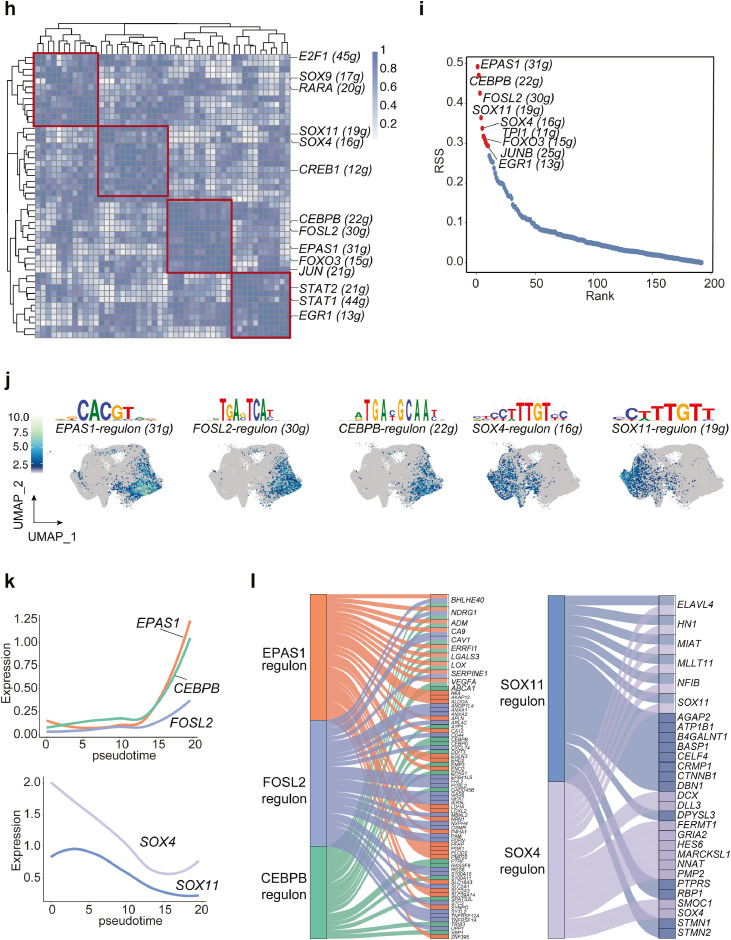
Trajectory interference reveals key TF-regulons in PN-MES transition.
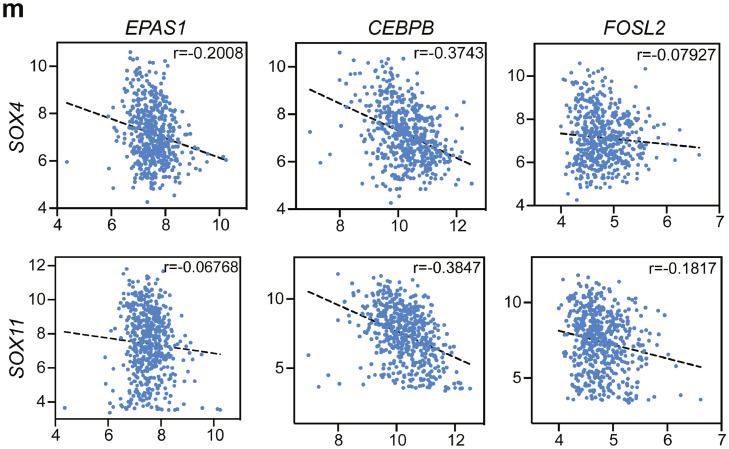
(a) Unsupervised trajectory analysis of tumor cells was conducted using monocle. (b-c) Pseudotime analysis demonstrates a major transition starting from OPC- and NPC-like to mesenchymal cells. Each dot represents a single cell, colored by cell type. (d) Heatmap plot of represented genes and pathways across trajectory states. Arrow corresponds to the direction of pseudotime. (e) Overall Kaplan-Meier survivals of different subtype patients based on Z-score of MES-or PN-like gene signatures. (f) Developmental inference analysis shows the dynamic shift in cell state. Arrow predicates the direction of cell state transition. (g) Feature plot of represented gene expressions (PN- or MES-like) in all tumor cells. (h) The SCENIC analysis identifies the master regulons in different cell states. (i) Top ranking regulons are listed. Z-score was determined by calculating the geometric mean of transcriptional factors and the corresponding cis-regulatory targets. (j) Feature plot showing the enriched expression of TF-regulons along with DNA binding motif sequences. (k) Pseudotime-ordered analysis of key regulators including EPAS1, FOSL2, CEBPB, SOX4 and SOX11. (l) Sankey plot showing the master regulatory modules. (m) The correlation of SOX4, SOX11 expression (PN-TFs) with EPAS1, CEBPB and FOSL2 (MES-TFs) was analyzed by linear regression using HU133-datasets from GlioVis. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Transcriptional factors are the key regulators in cell differentiation and transition, and TF-regulated networks are essential for tumor progression. Bioinformatic approaches are able to identify the cis-regulatory elements regulated by TFs among cell clusters, and these downstream targets are termed regulons. Here, we sought to discover the key regulatory network in PN or MES cell state by conducting a regulon activity analysis with single-cell regulatory network inference and clustering (SCENIC).35 SCENIC nominated a set of key regulons in tumor cell states driven by different TFs, for example, E2F1, SOX9, RARA, SOX11 and SOX4 were specially expressed in OPC and NPC cells, whereas EAPS1, CEBPB, FOSL2, STAT2 and EGR1 were predominant mediators in MES and AC cell states (Figure 2h). Additionally, scoring regulon activity for each cell type highlighted some master regulators, such as EAPS1 (31 regulated genes), CEBPB (22 regulated genes) and FOSL2 (30 regulated genes) in MES state, and SOX4 (16 regulated genes) and SOX11 (19 regulated genes) in PN state (Figure 2i). The module score activities verified the expression of dominant regulons in different cell states (Figure 2j). Among the downstream targets regulated by each important TF, we discovered shared targets mediated by mesenchymal TFs, such as VEGFA, LGALS3, and CAV1 (Figure 2l), which have been implicated in EMT.43, 44, 45 In addition, the pseudotime analysis confirmed the induction of EPAS1, CEBPB, and FOSL2 as well as the reduction of SOX4 and SOX11 along with the PN-MES transition (Figure 2k). Last, linear regression showed a negative correlation between MES-TFs and PN-TFs using TCGA dataset (Figure 2m). In sum, our data highlight a dynamic transition from PN to MES cell states mainly driven by key regulators.
Immune tumor microenvironment in PN- and MES-GBM tumors
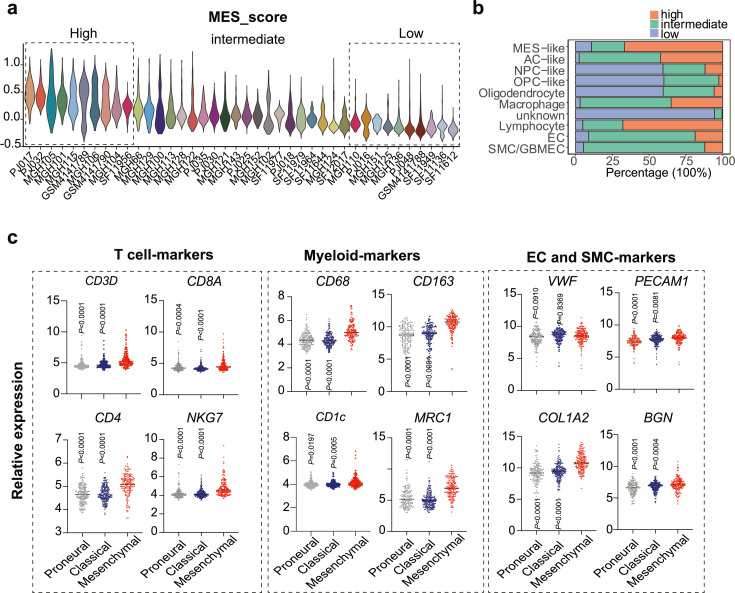
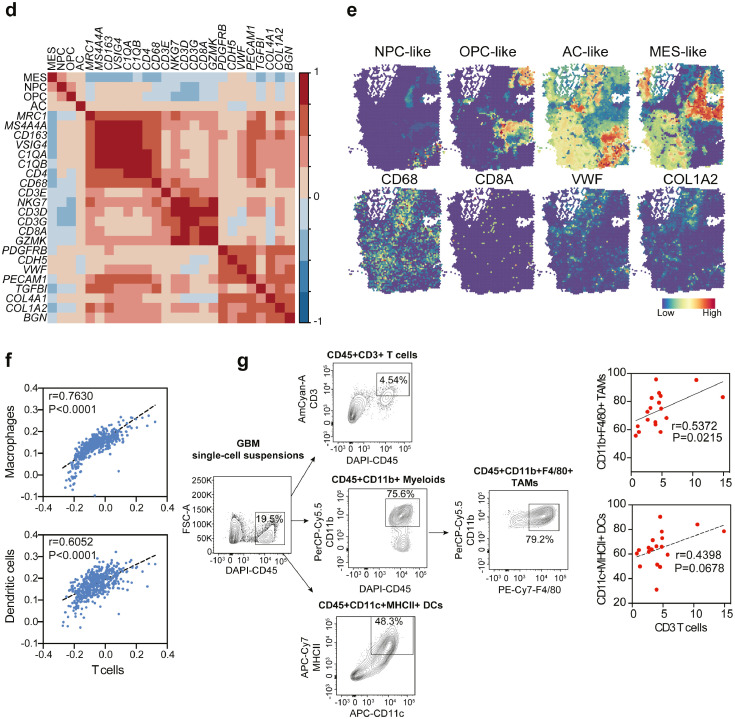
To assess the tumor microenvironment between PN- and MES-GBM, we assigned each patient to a high- or low-MES score group (Figure 3a). Importantly, patients with high MES scores were accompanied by enriched macrophages, T cells and endothelial cells infiltration in the tumor microenvironment (Figure 3b). In contrast to the well-established role of ECs and macrophages in mesenchymal transition, a higher infiltration of T cells is extremely unexpected due to the improved prognosis associated with multiple types of cancer.46, 47, 48 Bulk-seq analyses of TCGA datasets demonstrated that mesenchymal GBM subtypes expressed higher T cells and myeloid cell markers and revealed a strong correlation between T cells and macrophages (Figure 3c-d). Additionally, spatial scRNA-seq data confirmed the close contact between T cells and myeloid cells (Figure 3e), which may occur in vascular niches, as CD8+ T cells colocalized with CD68+ myeloid cells and endothelial cells. Deconvolution algorithms have been used to determine the relative expression of various cell types based on specific gene signatures in bulk RNA-seq data.49 Employing the deconvolution strategy with TCGA data, here we showed that T cells abundance was highly associated with macrophages as well as dendritic cells (DCs), which are two major myeloid cell lineages (Figure 3f). To further support this, we analyzed the immune cell profiles in mouse GBM using flow cytometric analysis and confirmed the positive correlation between CD11b+F480+ tumor-associated macrophages (TAMs) or CD11c+MHCII+ DCs with total T cells (Figure 3g). Thus, the higher proportion of T cells in mesenchymal subtypes is largely due to the infiltration of macrophages.
Figure 3.
Immune tumor microenvironment in PN- and MES-GBM tumors.
(a) MES gene signature was used to classify patients into the MES-high, MES-intermediate and MES-low groups. (b) The relative percentage of each cell type across different groups. (c) TCGA dataset reveals gene expression patterns associated with proneural, classical, and mesenchymal subtypes of GBM. Statistical analysis by one-way ANOVA test. (d) Pearson correlation between cell-type-specific genes in the GBM-TCGA dataset, with blue representing a negative correlation and red indicating a positive correlation. (e) Spatial distribution of cell-type-specific scores/genes defined by scRNA-seq. (f) Association of macrophage/dendritic cells with T cell fractions, which were estimated by a cellular deconvolution algorithm. The correlation was conducted using linear regression analysis in TCGA datasets. (g) Flow cytometry analysis of CD3+ T cells, CD11b+F4/80+ TAMs and CD11c+MHCII+ DCs in GBM mouse (n=18). Left, gating strategy. Right, a linear relationship was evaluated using Pearson correlation. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
GPNMB derived from macrophages contributes to PN-MES cell state transition
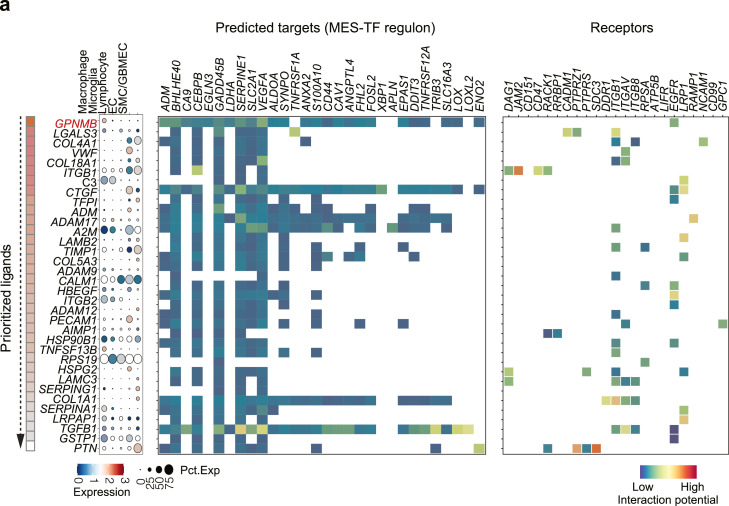
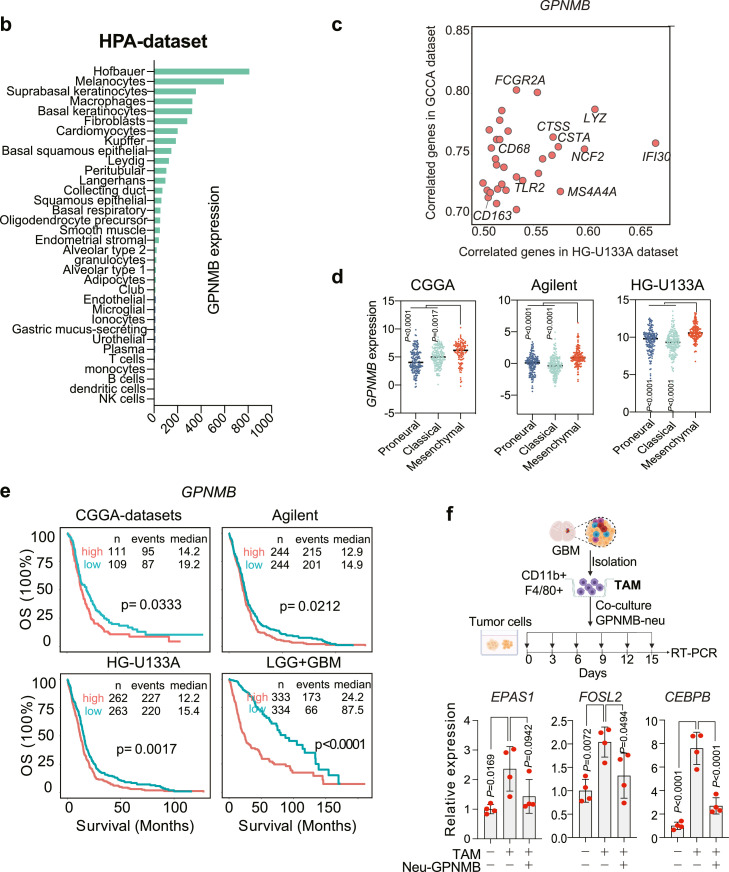
Cell fate is determined not only by intrinsic factors but also by extrinsic events. To explore the possible role of extrinsic regulations in the PN-MES transition, we analyzed the intercellular crosstalk within the tumor microenvironment using NicheNet.37 NicheNet is a computational approach that can predict intercellular ligand-target communications based on gene regulatory networks. First, we listed out the key regulons in mesenchymal transcriptome generated from our trajectory analysis above, including EPAS1, FOSL2, and CEBPB as well as downstream signals targets (Supplementary Table 2). We then calculated the potential ligands from the stromal cells (sending cells) that could interact with the targets in the tumor cells (receiving cells). Finally, we prioritized the top ligands and identified GPNMB derived from myeloid cells as the top 1 ligand that can interact with most of the mesenchymal targets (Figure 4a). HPA and TCGA datasets showed GPNMB to be mainly expressed in macrophages and highly correlated with macrophage markers, which indicates macrophages are the major source of GPNMB (Figure 4b-c). Our flow cytometry data confirmed the predominant expression of GPNMB on macrophages rather than dendritic or tumor cells (Supplementary Figure 3a). Moreover, GPNMB was also highly expressed in MES subtype patients (Figure 4d), and this higher expression predicted poor prognosis in both low- and high-grade gliomas (Figure 4e). These findings imply that macrophages with a high expression of GPNMB could induce a cell state transition toward MES phenotype, we therefore applied a co-culture system to investigate this hypothesis. This system includes the genetic resemblance to the proneural type of glioma cells50 and fresh isolated CD11b+ F4/80+ macrophages. In order to induce a dramatic transcriptome change, we freshly co-cultured macrophages every three days to eventually assess the expression of three master transcription factors in mesenchymal phenotypes (EPAS1, CEBPB, FOSL2) after continuous and long-term stimulation, as obtained by our analysis. Our results revealed that long-term macrophage contact induced mesenchymal regulator expressions in tumor cells, and importantly, treatment with a neutralizing antibody against GPNMB could partially abolish this effect, suggesting GPNMB as an essential mediator for PN-MES transition (Figure 4f).
Figure 4.
GPNMB derived from macrophages contribute to PN-MES cell state transition.
Intercellular communications between stromal and tumor cells were inferred using Nichenet. (a) Ligands ranking from the sender cells (Macrophages, Microglia, Lymphocyte, EC and SMC/GBMEC) was predicted to communicate with target cells (tumor cells) based on interacting with MES-regulons. Red heatmaps indicate the Pearson correlation of prioritized ligands from high to low. The ligand expressions across cell types are shown on the left. Color encodes the average expression level across all cells (blue is low, red is high), while the dot size corresponds to the percentage of cells expressing the feature gene. The right heatmap indicates the potential interaction between ligands and targets/receptors. Color indicates the high or low interaction, respectively. (b) The relative expression of GPNMB in each cell type by analyzing the HPA dataset. (c) The highly correlated genes with GPNMB from the TCGA and CGGA datasets. (d) Relative GPNMB expression in different GBM subgroups. (e) Kaplan-Meier analysis of survival in patients with high- and low-expressed GPNMB. (f) Tumor-associated macrophages were freshly sorted from GL261-derived tumor cells, and subsequently co-cultured with proneural type of GBM spheroids in presence of neutralizing antibody of GPNMB. After constant stimulation, the expressions of EPAS1, FOSL2, and CEBPB were determined using RT-PCR. Statistical analysis by one-way ANOVA (n=4 independent biological replicates, mean ± SD). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
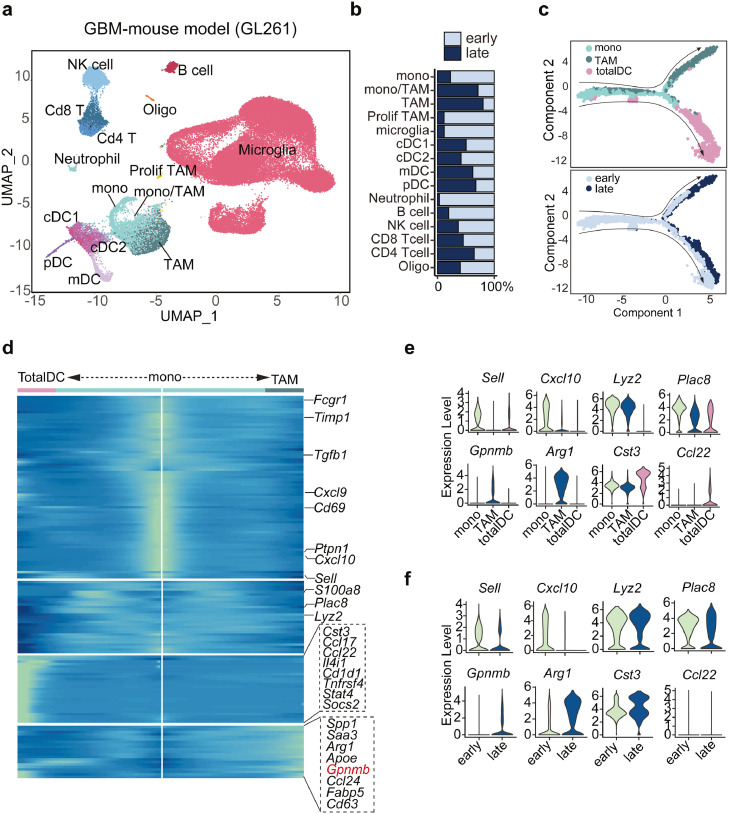
Monocytes differentiate into Gpnmb-high macrophages based on scRNA-seq analysis on GBM-mouse model
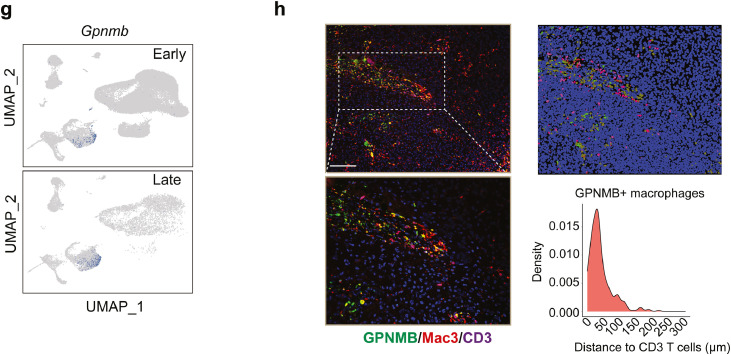
Due to the very few immune cells obtained in current human GBM scRNA-seq data for further analysis, we included another scRNA-seq dataset focusing on CD45+ immune cells collected from GBM mouse at early (day 7) and late (day 28) stages.51 After re-analyzing the data, we identified the main types of immune cells such as NK cells, Cd8+ T cells, Cd4+ T cells, B cells, neutrophils, cDC1, cDC2, pDC, mDC, monocytes, TAM, monocytes/TAM, and microglia cells based on cell marker expressions (Figure 5a). As expected, macrophages dramatically invaded tumor sites while microglia cells vanished during tumor development (Figure 5b), showing consistency with other findings.52 In addition, trajectory analysis inferred two major cell lineages, dendritic cells and macrophages, originated from monocytes, and progressed along with tumor development (Figure 5c). Interestingly, distinct pathways dominated the recruitment of tumor-infiltrating T cells: monocytes predominately expressed Cxcl10, Cxcl9 and DCs specifically expressed Ccl17, Ccl22, while macrophages expressed Ccl24 (Figure 5d). Besides, another set of important mediators including Saa3, Arg1, and Gpnmb presented exclusively in macrophages (Figure 5d). During tumorigenesis, expressions of Arg1 and Gpnmb were robustly induced in macrophages (Figure 5e-g), which was in accordance with the findings in human GBM datasets (Figure 4d). However, GPNMB is not a driving factor for M2 macrophage polarization, evidenced by unchanged CD206 expression (classic M2 macrophage marker) after Gpnmb knockdown (Supplementary Fig. 4). Particularly, immunofluorescence staining demonstrates the co-location of GPNMB+ macrophages with T cells (Figure 5h), suggesting a potential interaction between them.
Figure 5.
Monocytes differentiate into Gpnmb-high macrophages or dendritic cells in the tumor microenvironment based on scRNA-seq analysis on GBM-mouse model.
(a) Immune cells were re-clustered from GL261 mouse scRNA-seq and visualized in UMAP plot. Color encodes different cell clusters. (b) The percentage of each cell type in early and late stages of tumors. (c) Monocytes, dendritic cells, and macrophages were applied to trajectory analysis. Pseudo-temporal ordering of cells across cell types (top) and time points (bottom). (d) Heatmap plot showing the bifurcation of gene expression starting from monocytes. Arrows correspond to the direction of either dendritic cells or macrophages. Represented genes in this branched modeling are shown on the right. (e) The violin plot represents the gene expression levels in different transition states or (f) tumors collected at different time points. (g) Feature plot showing the expression of Gpnmb in early and late stages of tumors. (h) Left, immunofluorescence staining of GPNMB, Mac-3, and CD3 in GBM sections. Green color represents GPNMB, red color corresponds to Mac-3 and blue color indicates CD3 (Bar represents 200 μm). Right, quantitative analysis of the distance between GPNMB+ macrophages and CD3+ T cells (n=5 mice). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
GPNMB-high macrophages ineffectively retain T cells from activating by DCs
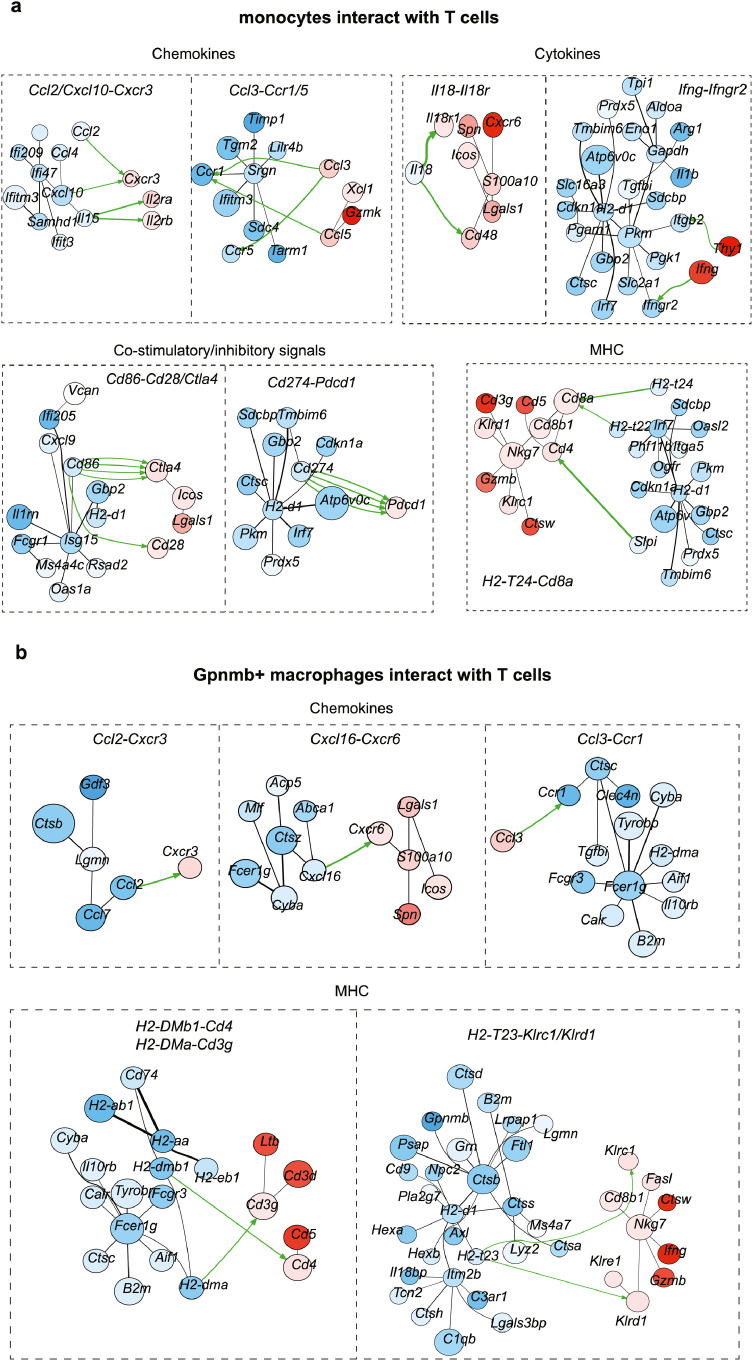
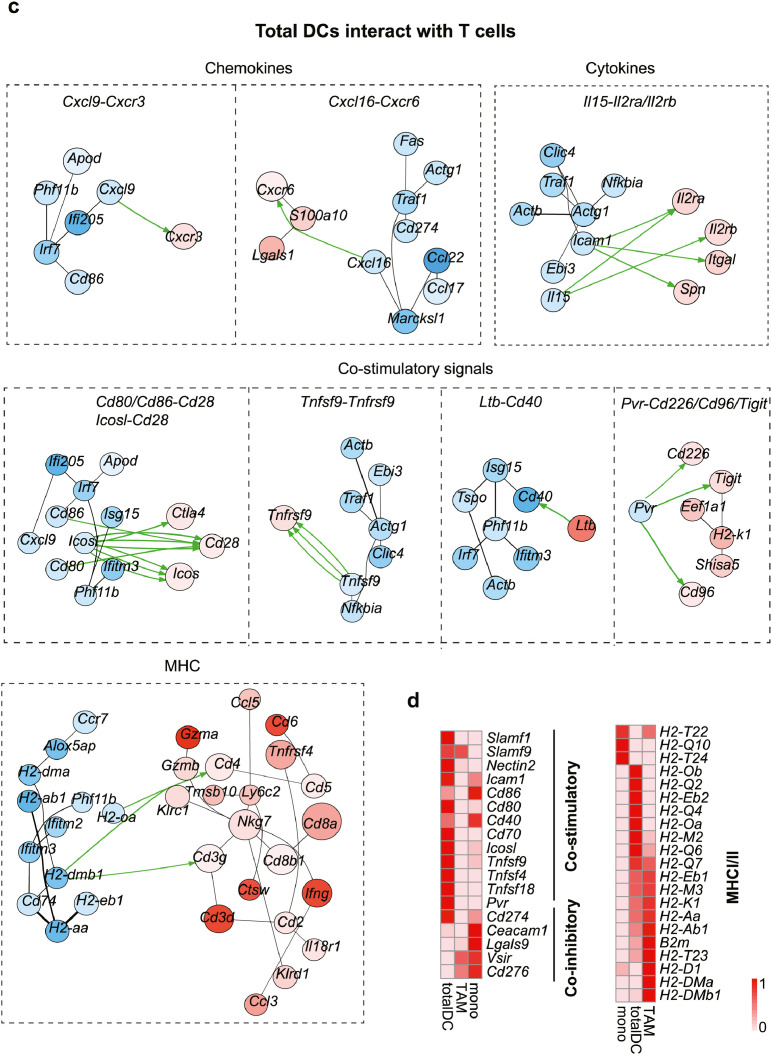
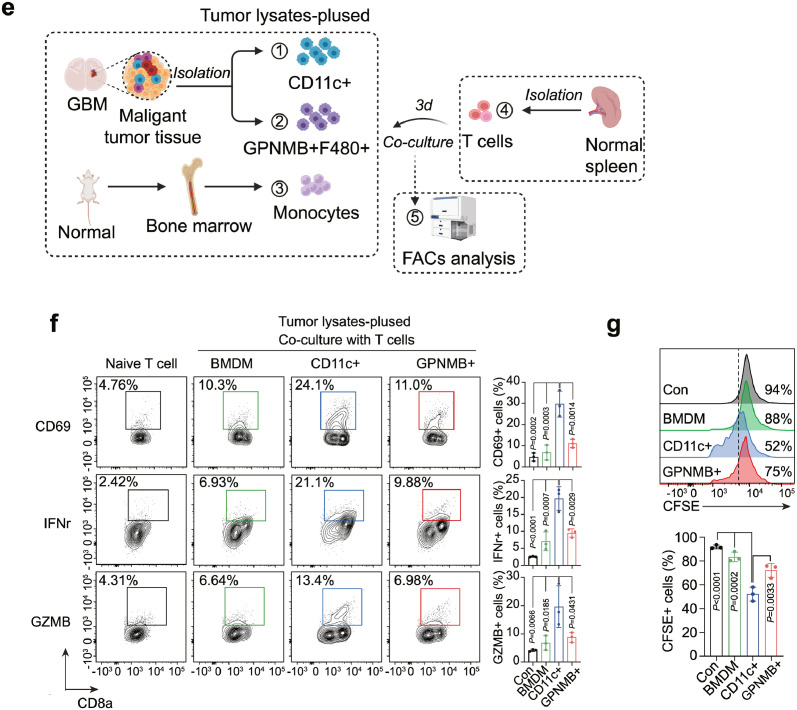
Current cell-cell interaction analyses examine the relative expression of ligands and receptors in different cell types without examining the overall signaling networks. To explicitly investigate the interaction of T cells with APC-like cells in GBM, we thus applied the CytoTalk algorithm to study the full signal transduction pathways.38 Chemokines, cytokines, co-stimulatory/inhibitory and antigen-presentation pathways provided insights into how T cells interact with monocytes, Gpnmb-high macrophages and dendritic cells (Figure 6a-c). For example, Gpnmb-high macrophages and DCs expressed high levels of but totally different MHCI/II molecules compared with monocytes, suggesting that in the GBM tumor microenvironment, macrophages and DCs are the dominant antigen-presenting cells. A number of chemokines and cytokines in monocytes, including Cxcl10, Il15, and Il18, could interact with Cxcr3, Il2ra/b, Il18r1/Cd48 on T cells to promote downstream signaling and infiltration. Well, Ccl2-Cxcr3, Cxcl16-Cxcr6 and Ccl3-Ccr1 were the predominate chemokines involved in the interaction of Gpnmb-high macrophages and T cells. Importantly, DCs shared the same Cxcl16-Cxcr6 chemokine signals for T cell recruitment, indicating a competent role within myeloid cells. Furthermore, a study of enriched co-stimulatory signals suggests that T cells can activate when encountering DCs rather than macrophages (Figure 6d). To confirm this potential non-effective retention by interacting with Gpnmb-high macrophages, we extracted the F4/80+GPNMB+ macrophages, CD11c+ DCs and bone marrow-derived monocytes from GBM tumor and normal mouse, and then pulsed them with tumor lysates prior to co-culturing with naïve T cells (Figure 6e). Our data showed that CD11c+ DCs induced robust T cell activation when compared with GPNMB+ macrophages or monocytes, evidenced by much higher levels of CD69, IFNγ and GZMB (Figure 6f). Similarly, according to the results of the T cell proliferation assay, CD11c+ DCs significantly enhance T cell proliferation, in contrast to monocytes or GPNMB+ macrophages (Figure 6g). Taken together, all of the data indicate that Gpnmb-high macrophages could not activate T cells due to the sustained co-stimulatory signals but instead exert non-effective retention.
Figure 6.
GPNMB-high macrophages ineffectively retain T cells from activating by DCs.
Gpnmb-high macrophages, monocytes, total dendritic cells and T cells were sub-clustered and applied to CytoTalk analysis. Major inter-and intracellular communications are shown between (a) monocytes and T cells, (b) Gpnmb-macrophages and T cells, (c) dendritic cells and T cells. Important categories including chemokines, cytokine, co-stimulatory signals, and antigen presentation machinery are displayed. Blue encodes monocytes/macrophages/dendritic cells, while red encodes T cells. Color shade of gene nodes represents the proportional to the cell-type specificity. The size of nodes indicates the average expression of genes in each cell type. (d) The relative expression of inhibitory and stimulatory molecules on monocytes, macrophages and dendritic cells. (e) F4/80+GPNMB+ macrophages, CD11c+ dendritic cells and bone marrow-derived monocytes were isolated and subsequently pulsed with tumor lysates prior to co-culturing with naïve T cells for 3 days. (f) The expressions of CD69, IFNγ and GZMB as well as the (g) proliferation of T cells were analyzed using flow cytometry. Statistical analysis by one-way ANOVA (n=3, independent biological replicates, mean ± SD). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
In sum, taking advantage of the analysis of scRNA-seq data, we identified a key GPNMB-high macrophage subset as a hub in cell-cell communication. Our ex vivo data verified that GPNMB+ macrophages not only induced the PN-MES tumor cell transition but also impaired T cell activation through competing with DCs. Therefore, targeting GPNMB+ macrophages could benefit in treating GBM combined with other therapies.
Discussion
GBM, a typically heterogeneous and immunologically cold tumor, is highly resistant to chemotherapy, radiotherapy and immunotherapy. Microenvironment-targeted immunotherapy holds new hope for GBM treatment.40,41 Therefore, explicitly dissecting the tumor microenvironment will help identify the determinants of GBM tumor evolution and promising targets for further combination therapy. Based on integrative analyses with multiple scRNA-seq data and ex vivo experiments, we identified a GPNMB-high subset of macrophages that could (1) induce key regulons-mediated PN to MES cell state transition, and (2) impede T cell activation by non-effectively retention.
GBM has been classified as proneural, classical and mesenchymal subtypes based on molecular classification.6 GBM can undergo a phenotypic shift from PN to MES upon stimulation, making them more aggressive and resistant.53 This transition involves a whole range of intrinsic molecular signals. Thus, taking advantage of the unbiased SCNEIC analysis, we identified the top three important transcriptional factors, FOSL2, CEBPB and EPAS1 dominant in mesenchymal cells. Similarly, previous studies have confirmed that CEBPB and STAT3, the master regulators, can cooperate with FOSL2 to mediate PN to MES transition through multiple downstream signals.54 Besides, FOSL2 independently participates in the PN-to-MES drift through epigenetic mechanism.7 Hypoxia is also associated with tumor progression and higher expression of HIF1a and HIF2a (EPAS1) were observed in MES patients as well.55,56 As such, targeting these regulators or the stimulatory signals from the tumor microenvironment may inhibit the transition and overcome the resistance to traditional therapies.
Besides exploring the intrinsic mechanism, we are more interested in how the extrinsic factors affected the transcriptomic plasticity of GBM. Tumor-associated macrophages have been associated with transcriptome shift toward mesenchymal signatures,5,57 although the direct evidence is still lacking. One particular study revealed NF-kB as a master signal transductor in mediating mesenchymal differentiation upon TNFα stimulation in GBM.58 Although they proposed that the source of TNFα could be macrophages or microglia, further investigation seems to be needed. Our ex vivo studies demonstrated that constant exposure of tumor-associated macrophages to proneural type of GBM triggered mesenchymal differentiation. The present study provides clear evidence that extrinsic events can affect the phenotypic plasticity of tumor cells. Furthermore, coupled with modeling intercellular crosstalk by NicheNet method, we investigated potential ligand-receptor-target interactions between tumor and non-tumor cells. Our unbiased analysis uncovered some interesting ligands, such as TGFB1, a master inducer in the mesenchymal transition,59,60 which can promote the majority of our curative targets. LGALS3, produced by a variety of different cells, including macrophages, and endothelial cells, acts as another major source of this transcriptome change. As shown in our results, mesenchymal regulon expression was induced via integrin B1, the well-known receptor that interacts with LGALS3 as well as CADM1 and PTPRZ1.45,61 In addition to macrophages, endothelial and transformed endothelial cells played an essential role in the crosstalk. For example, CTGF, highly expressed by these stromal cells, could induce this mesenchymal transcriptome. In different cancer types, CTGF has been reported to be involved in cancer cell migration, invasion, and epithelial-mesenchymal transition.62,63 And LRP receptor discovered here has been proved by other studies to be a coreceptor that binds with CTGF and activates WNT signaling.64 The TIMP-1 gene expressed by ECs can likewise be induced to upregulate mesenchymal markers as well through TWIST regulation, which is another intriguing target worth examining.65 Among our most interesting findings, we show that GPNMB is the top 1 ligand to interact with EGFR to induce most of the mesenchymal targets. Other studies also demonstrated that GPNMB engaged with EGFR as a heterodimer complex upon HB-EFG stimulation in the breast cancer model.66 It is intriguing to note that overexpressed GPNMB can cause cancer cells to migrate, invade, and undergo EMT, and EGFR inhibitor treatment can abolish these phenotype changes, which supports our findings.67 Also, anti-GPNMB antibody has been assessed in phase I/II trials in treating breast cancer and melanomas,24,25 making GPNMB an attractive target in cancer treatment. However, in our flow cytometry analysis study, GPNMB is primarily expressed by macrophages as opposed to tumor cells in other cancer types (Supplementary Figure 3a), and neutralizing antibody can attenuate the TAM-induced mesenchymal transition, further demonstrating the significance of both GPNMB protein and GPNMB-expressed macrophages to the tumor development.
According to our trajectory analysis, monocytes can differentiate into dendritic cells or macrophages, which express a high level of GPNMB, hence we conducted a transduction signal analysis to understand how these cell types interact with T cells. Our analysis revealed a large number of chemokines involved in this intercellular crosstalk, such as CXCR3, an important modulator of T cell trafficking and activation. Recently, T cell recruitment mediated by CXCR3 has been shown to be required for effective immune checkpoint blockade.68 CXCR3 is highly expressed on T cells and responds to a range of chemokines, such as CXCL9 and CXCL10, which are demonstrated to promote anti-tumor activities by attracting both CD4+ helper T cells and CD8+ effector T cells.69 CXCL9 and CXCL10 are produced in response to IFNγ stimulation,70 here, we clearly observe the signal transduction between monocytes and T cells induced by Infg-Infgr2 interaction. Myeloid cells, especially cDC1 cells, are the primary sources of CXCL9 and CXCL10 and are responsible for mediating antitumor immune responses in response to checkpoint inhibitors.71,72 Our analysis reveals the presence of Cxcl9 and Cxcl10 in monocytes or dendritic cells, but not in GPNMB-high macrophages, have a robust interaction with Cxcr3 in T cells. Another important chemokine receptor identified by our analysis is Cxcr6. CXCR6 is critical for CD8+ cytotoxic T cell-mediated tumor control as well as the ability to augment the anti-tumor efficacy of PD-1.73 Meanwhile, CXCR6 deficient CD8+ T cells exhibit markedly decreased retention in tumor tissues.74 As the ligand of CXCR6, CXCL16 is primarily expressed by myeloid cells and associated with antigen presentation genes. Most of the studies demonstrated the important role of CXCL16 on DCs in regulating T cell function and tumor growth,75,76 but how CXCL16+ macrophages impact T cell's recruitment and function is truly needed in further exploration. But one important finding supports our analysis: the interaction between CXCL16+ DCs and CXCR6+ T cells can promote the production of Il15,76 a potent immune-stimulating cytokine that enhances both reactive NK and T cell survival.77,78 Consist with our analysis, this Il15-Il2ra/rb signaling occurs in DC-T cell crosstalk, but not in GPNMB-macrophages. Moreover, GPNMB has been reported to directly inhibit T cell activation by competing with syndecan-4, thereby leading to immune evasion in melanoma models.26 And blockade of GPNMB is correlated with an increase in CD4 and CD8 T cell infiltration and the efficacy of immune checkpoint blockade.23 But we suggest here not only the GPNMB protein, but more importantly, the GPNMB-high macrophages can impede T cell activation. Another remarkable finding here reveals that GPNMB-macrophage interacts with T cells via H2-t23 and KLrc1/Klrd1. Several studies have suggested that HLA-E (homolog in mice is called Qa-1b, also known as H2-t23) negatively affects IL2 receptor-dependent proliferation and influences IFNγ-mediated antitumor responses through binding directly to the NKG2A (encoded by Klrc1)/CD94 (encoded by Klrd1) receptor on tumor-specific T cells.79 Additionally, current therapeutic vaccines that block NKG2A with Qa-1 can enhance CD8+ T cells immunity.80 NK- and T-cell mediated anti-tumor effects were also promoted by an anti-NKG2A monoclonal antibody.81 All of this suggests that this particular macrophage subset may attract T cells to tumor sites through competitive Cxcl16-Cxcr6 interaction with DCs; however, this interaction may not be able to induce co-stimulatory signals, but rather impair T cell function through CD94/NKG2A-MHC interactions. Our immunofluorescence staining verified the co-localization of GPNMB-macrophages and T cells, and ex-vivo isolation of GPNMB-high macrophages cannot activate T cells as dendritic cells or even GPNMB-low macrophages did (Supplementary Figure 3b), all of which fully support these findings conducted from the comprehensive analysis. Besides, our data also suggest that in the tumor microenvironment, tumor-associated dendritic cells still possess the ability but maybe not the chance to activate T cells, this in situ antigen presentation can be enhanced when T cells either gain more mobility or decrease the retention with non-effective interaction.
We thoroughly compared the MES- and PN-tumor microenvironment and showed the higher infiltration of T cells accompanied by macrophages and endothelial cells. The correlation was also supported by deconvolution of bulk-seq, spatial scRNA-seq as well as in flow cytometry analysis of the single-cell immune profiling on syngeneic GBM mouse model, while another group obtained the same results.82 Taken together, this raised an important question that why MES-subtype GBM has more T cell infiltration: (1) Can monocytes/macrophages release signals for T cells recruitment solely before entering tumor sites? (2) or T cells and macrophages are both attracted by endothelial cells at the vascular niche, where more vessels are present in MES subtype. As a result, simply targeting monocyte/macrophage depletion or anti-angiogenesis therapy may affect T cell recruitment to some extent. Interestingly, knockdown of GPNMB in macrophages did not reverse immunosuppressive phenotype, as shown by unchanged CD206 expression (Supplementary Figure 4), a classic M2 macrophage marker, indicating that GPNMB is not a driving factor in alternative macrophage polarization.
Thus, we proposed that in further study, depletion of this subset will be the best therapeutic option for sufficient release of T cells to exert its function when encountering dendritic cells as well as preventing the transcriptomic transition of GBM into a high-grade mesenchymal phenotype. In summary, our integrative analyses suggest that targeting GPNMB-high macrophages can reshape a favorable and sensitive tumor microenvironment for future combination therapy.
Contributors
A.X. and J.Z. performed and analyzed most of experiments. J.Z. and F.Y. produced figures. Y.C. and Y.Z. helped data analyses. F.Y. designed, supervised the project and wrote the manuscript. J.Z. and F.Y verified the underlying data used in this work. All authors have read and approved the final manuscript.
Data sharing statement
All data are available upon request to the corresponding authors.
Declaration of interests
The authors declare no conflict of interest relevant to the present manuscript.
Acknowledgments
This work was supported by National Natural Science Foundation of China (82004004) and Shanghai Sailing Program (18YF1421700).
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.ebiom.2022.104239.
Appendix. Supplementary materials
References
- 1.Stupp R, Mason WP, van den Bent MJ, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352(10):987–996. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 2.Huse JT, Holland EC. Targeting brain cancer: advances in the molecular pathology of malignant glioma and medulloblastoma. Nat Rev Cancer. 2010;10(5):319–331. doi: 10.1038/nrc2818. [DOI] [PubMed] [Google Scholar]
- 3.O'Rourke DM, Nasrallah MP, Desai A, et al. A single dose of peripherally infused EGFRvIII-directed CAR T cells mediates antigen loss and induces adaptive resistance in patients with recurrent glioblastoma. Sci Transl Med. 2017;9(399) doi: 10.1126/scitranslmed.aaa0984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Filley AC, Henriquez M, Dey M. Recurrent glioma clinical trial, CheckMate-143: the game is not over yet. Oncotarget. 2017;8(53):91779–91794. doi: 10.18632/oncotarget.21586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang Q, Hu B, Hu X, et al. Tumor evolution of glioma-intrinsic gene expression subtypes associates with immunological changes in the microenvironment. Cancer Cell. 2017;32(1):42–56.e6. doi: 10.1016/j.ccell.2017.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Verhaak RG, Hoadley KA, Purdom E, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17(1):98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Segerman A, Niklasson M, Haglund C, et al. Clonal variation in drug and radiation response among glioma-initiating cells is linked to proneural-mesenchymal transition. Cell Rep. 2016;17(11):2994–3009. doi: 10.1016/j.celrep.2016.11.056. [DOI] [PubMed] [Google Scholar]
- 8.Minata M, Audia A, Shi J, et al. Phenotypic plasticity of invasive edge glioma stem-like cells in response to ionizing radiation. Cell Rep. 2019;26(7):1893–1905.e7. doi: 10.1016/j.celrep.2019.01.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hambardzumyan D, Gutmann DH, Kettenmann H. The role of microglia and macrophages in glioma maintenance and progression. Nat Neurosci. 2016;19(1):20–27. doi: 10.1038/nn.4185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pollard JW. Tumour-educated macrophages promote tumour progression and metastasis. Nat Rev Cancer. 2004;4(1):71–78. doi: 10.1038/nrc1256. [DOI] [PubMed] [Google Scholar]
- 11.Sica A, Mantovani A. Macrophage plasticity and polarization: in vivo veritas. J Clin Invest. 2012;122(3):787–795. doi: 10.1172/JCI59643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kaneda MM, Messer KS, Ralainirina N, et al. PI3Kγ is a molecular switch that controls immune suppression. Nature. 2016;539(7629):437–442. doi: 10.1038/nature19834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pyonteck SM, Akkari L, Schuhmacher AJ, et al. CSF-1R inhibition alters macrophage polarization and blocks glioma progression. Nat Med. 2013;19(10):1264–1272. doi: 10.1038/nm.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guerriero JL, Sotayo A, Ponichtera HE, et al. Class IIa HDAC inhibition reduces breast tumours and metastases through anti-tumour macrophages. Nature. 2017;543(7645):428–432. doi: 10.1038/nature21409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Couturier CP, Ayyadhury S, Le PU, et al. Single-cell RNA-seq reveals that glioblastoma recapitulates a normal neurodevelopmental hierarchy. Nat Commun. 2020;11(1):3406. doi: 10.1038/s41467-020-17186-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Neftel C, Laffy J, Filbin MG, et al. An integrative model of cellular states, plasticity, and genetics for glioblastoma. Cell. 2019;178(4):835–849.e21. doi: 10.1016/j.cell.2019.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Patel AP, Tirosh I, Trombetta JJ, et al. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science. 2014;344(6190):1396–1401. doi: 10.1126/science.1254257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yuan J, Levitin HM, Frattini V, et al. Single-cell transcriptome analysis of lineage diversity in high-grade glioma. Genome Med. 2018;10(1):57. doi: 10.1186/s13073-018-0567-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang L, Babikir H, Müller S, et al. The phenotypes of proliferating glioblastoma cells reside on a single axis of variation. Cancer Discov. 2019;9(12):1708–1719. doi: 10.1158/2159-8290.CD-19-0329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang R, Sharma R, Shen X, et al. Adult human glioblastomas harbor radial glia-like cells. Stem Cell Rep. 2020;14(2):338–350. doi: 10.1016/j.stemcr.2020.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kobayashi M, Chung JS, Beg M, et al. Blocking monocytic myeloid-derived suppressor cell function via anti-DC-HIL/GPNMB antibody restores the in vitro integrity of T cells from cancer patients. Clin Cancer Res. 2019;25(2):828–838. doi: 10.1158/1078-0432.CCR-18-0330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rose AA, Annis MG, Dong Z, et al. ADAM10 releases a soluble form of the GPNMB/Osteoactivin extracellular domain with angiogenic properties. PLoS One. 2010;5(8):e12093. doi: 10.1371/journal.pone.0012093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chung JS, Ramani V, Kobayashi M, et al. DC-HIL/Gpnmb is a negative regulator of tumor response to immune checkpoint inhibitors. Clin Cancer Res. 2020;26(6):1449–1459. doi: 10.1158/1078-0432.CCR-19-2360. [DOI] [PubMed] [Google Scholar]
- 24.Bendell J, Saleh M, Rose AA, et al. Phase I/II study of the antibody-drug conjugate glembatumumab vedotin in patients with locally advanced or metastatic breast cancer. J Clin Oncol. 2014;32(32):3619–3625. doi: 10.1200/JCO.2013.52.5683. [DOI] [PubMed] [Google Scholar]
- 25.Ott PA, Hamid O, Pavlick AC, et al. Phase I/II study of the antibody-drug conjugate glembatumumab vedotin in patients with advanced melanoma. J Clin Oncol. 2014;32(32):3659–3666. doi: 10.1200/JCO.2013.54.8115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tomihari M, Chung JS, Akiyoshi H, Cruz PD Jr., Ariizumi K. DC-HIL/glycoprotein Nmb promotes growth of melanoma in mice by inhibiting the activation of tumor-reactive T cells. Cancer Res. 2010;70(14):5778–5787. doi: 10.1158/0008-5472.CAN-09-2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stuart T, Butler A, Hoffman P, et al. Comprehensive integration of single-cell data. Cell. 2019;177(7):1888–1902.e21. doi: 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aran D, Looney AP, Liu L, et al. Reference-based analysis of lung single-cell sequencing reveals a transitional profibrotic macrophage. Nat Immunol. 2019;20(2):163–172. doi: 10.1038/s41590-018-0276-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McGinnis CS, Murrow LM, Gartner ZJ. DoubletFinder: doublet detection in single-cell RNA sequencing data using artificial nearest neighbors. Cell Syst. 2019;8(4):329–337.e4. doi: 10.1016/j.cels.2019.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tickle TI, Georgescu C, Brown M, Haas B. inferCNV of the Trinity CTAT Project. 2019.
- 31.Trapnell C, Cacchiarelli D, Grimsby J, et al. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat Biotechnol. 2014;32(4):381–386. doi: 10.1038/nbt.2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang F, Li X, Tian W. Unsupervised inference of developmental directions for single cells using VECTOR. Cell Rep. 2020;32(8) doi: 10.1016/j.celrep.2020.108069. [DOI] [PubMed] [Google Scholar]
- 33.Bhuva D, Smyth G, Garnham A. msigdb: An ExperimentHub Package for the Molecular Signatures Database (MSigDB). R package version 1.4.0; 2022. https://bioconductor.org/packages/release/data/experiment/html/msigdb.html
- 34.Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. Omics. 2012;16(5):284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Aibar S, González-Blas CB, Moerman T, et al. SCENIC: single-cell regulatory network inference and clustering. Nat Methods. 2017;14(11):1083–1086. doi: 10.1038/nmeth.4463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Therneau TM, Grambsch PM. Modeling Survival Data: Extending the Cox Model. Springer; New York: 2000. [Google Scholar]
- 37.Browaeys R, Saelens W, Saeys Y. NicheNet: modeling intercellular communication by linking ligands to target genes. Nat Methods. 2020;17(2):159–162. doi: 10.1038/s41592-019-0667-5. [DOI] [PubMed] [Google Scholar]
- 38.Hu Y, Peng T, Gao L, CytoTalk Tan K. De novo construction of signal transduction networks using single-cell transcriptomic data. Sci Adv. 2021;7(16) doi: 10.1126/sciadv.abf1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ma W, Wang Y, Zhang R, et al. Targeting PAK4 to reprogram the vascular microenvironment and improve CAR-T immunotherapy for glioblastoma. Nat Cancer. 2021;2(1):83–97. doi: 10.1038/s43018-020-00147-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang F, He Z, Duan H, et al. Synergistic immunotherapy of glioblastoma by dual targeting of IL-6 and CD40. Nat Commun. 2021;12(1):3424. doi: 10.1038/s41467-021-23832-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brennan CW, Verhaak RG, McKenna A, et al. The somatic genomic landscape of glioblastoma. Cell. 2013;155(2):462–477. doi: 10.1016/j.cell.2013.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fantozzi A, Gruber DC, Pisarsky L, et al. VEGF-mediated angiogenesis links EMT-induced cancer stemness to tumor initiation. Cancer Res. 2014;74(5):1566–1575. doi: 10.1158/0008-5472.CAN-13-1641. [DOI] [PubMed] [Google Scholar]
- 44.Bailey KM, Liu J. Caveolin-1 up-regulation during epithelial to mesenchymal transition is mediated by focal adhesion kinase. J Biol Chem. 2008;283(20):13714–13724. doi: 10.1074/jbc.M709329200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Margadant C, van den Bout I, van Boxtel AL, Thijssen VL, Sonnenberg A. Epigenetic regulation of galectin-3 expression by β1 integrins promotes cell adhesion and migration. J Biol Chem. 2012;287(53):44684–44693. doi: 10.1074/jbc.M112.426445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Badoual C, Hans S, Rodriguez J, et al. Prognostic value of tumor-infiltrating CD4+ T-cell subpopulations in head and neck cancers. Clin Cancer Res. 2006;12(2):465–472. doi: 10.1158/1078-0432.CCR-05-1886. [DOI] [PubMed] [Google Scholar]
- 47.An N, Wang H, Jia W, et al. The prognostic role of circulating CD8(+) T cell proliferation in patients with untreated extensive stage small cell lung cancer. J Transl Med. 2019;17(1):402. doi: 10.1186/s12967-019-02160-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sato E, Olson SH, Ahn J, et al. Intraepithelial CD8+ tumor-infiltrating lymphocytes and a high CD8+/regulatory T cell ratio are associated with favorable prognosis in ovarian cancer. Proc Natl Acad Sci USA. 2005;102(51):18538–18543. doi: 10.1073/pnas.0509182102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Newman AM, Liu CL, Green MR, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 2015;12(5):453–457. doi: 10.1038/nmeth.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kosaka A, Ohkuri T, Okada H. Combination of an agonistic anti-CD40 monoclonal antibody and the COX-2 inhibitor celecoxib induces anti-glioma effects by promotion of type-1 immunity in myeloid cells and T-cells. Cancer Immunol Immunother. 2014;63(8):847–857. doi: 10.1007/s00262-014-1561-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Friedrich M, Sankowski R, Bunse L, et al. Tryptophan metabolism drives dynamic immunosuppressive myeloid states in IDH-mutant gliomas. [DOI] [PubMed]
- 52.Chen Z, Feng X, Herting CJ, et al. Cellular and Molecular Identity of Tumor-Associated Macrophages in Glioblastoma. Cancer Res. 2017;77(9):2266–2278. doi: 10.1158/0008-5472.CAN-16-2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fedele M, Cerchia L, Pegoraro S, Sgarra R, Manfioletti G. Proneural-mesenchymal transition: phenotypic plasticity to acquire multitherapy resistance in glioblastoma. Int J Mol Sci. 2019;20(11) doi: 10.3390/ijms20112746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Carro MS, Lim WK, Alvarez MJ, et al. The transcriptional network for mesenchymal transformation of brain tumours. Nature. 2010;463(7279):318–325. doi: 10.1038/nature08712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Majmundar AJ, Wong WJ, Simon MC. Hypoxia-inducible factors and the response to hypoxic stress. Mol Cell. 2010;40(2):294–309. doi: 10.1016/j.molcel.2010.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Keith B, Johnson RS, Simon MC. HIF1α and HIF2α: sibling rivalry in hypoxic tumour growth and progression. Nat Rev Cancer. 2011;12(1):9–22. doi: 10.1038/nrc3183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lu-Emerson C, Snuderl M, Kirkpatrick ND, et al. Increase in tumor-associated macrophages after antiangiogenic therapy is associated with poor survival among patients with recurrent glioblastoma. Neuro Oncol. 2013;15(8):1079–1087. doi: 10.1093/neuonc/not082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bhat KPL, Balasubramaniyan V, Vaillant B, et al. Mesenchymal differentiation mediated by NF-κB promotes radiation resistance in glioblastoma. Cancer Cell. 2013;24(3):331–346. doi: 10.1016/j.ccr.2013.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yan T, Tan Y, Deng G, et al. TGF-β induces GBM mesenchymal transition through upregulation of CLDN4 and nuclear translocation to activate TNF-α/NF-κB signal pathway. Cell Death Dis. 2022;13(4):339. doi: 10.1038/s41419-022-04788-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Joseph JV, Conroy S, Tomar T, et al. TGF-β is an inducer of ZEB1-dependent mesenchymal transdifferentiation in glioblastoma that is associated with tumor invasion. Cell Death Dis. 2014;5(10):e1443. doi: 10.1038/cddis.2014.395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nangia-Makker P, Hogan V, Raz A. Galectin-3 and cancer stemness. Glycobiology. 2018;28(4):172–181. doi: 10.1093/glycob/cwy001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jiang CG, Lv L, Liu FR, et al. Connective tissue growth factor is a positive regulator of epithelial-mesenchymal transition and promotes the adhesion with gastric cancer cells in human peritoneal mesothelial cells. Cytokine. 2013;61(1):173–180. doi: 10.1016/j.cyto.2012.09.013. [DOI] [PubMed] [Google Scholar]
- 63.Kim H, Son S, Ko Y, Shin I. CTGF regulates cell proliferation, migration, and glucose metabolism through activation of FAK signaling in triple-negative breast cancer. Oncogene. 2021;40(15):2667–2681. doi: 10.1038/s41388-021-01731-7. [DOI] [PubMed] [Google Scholar]
- 64.Mercurio S, Latinkic B, Itasaki N, Krumlauf R, Smith JC. Connective-tissue growth factor modulates WNT signalling and interacts with the WNT receptor complex. Development. 2004;131(9):2137–2147. doi: 10.1242/dev.01045. [DOI] [PubMed] [Google Scholar]
- 65.D'Angelo RC, Liu XW, Najy AJ, et al. TIMP-1 via TWIST1 induces EMT phenotypes in human breast epithelial cells. Mol Cancer Res. 2014;12(9):1324–1333. doi: 10.1158/1541-7786.MCR-14-0105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lin A, Li C, Xing Z, et al. The LINK-A lncRNA activates normoxic HIF1α signalling in triple-negative breast cancer. Nat Cell Biol. 2016;18(2):213–224. doi: 10.1038/ncb3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Okita Y, Kimura M, Xie R, et al. The transcription factor MAFK induces EMT and malignant progression of triple-negative breast cancer cells through its target GPNMB. Sci Signal. 2017;10(474) doi: 10.1126/scisignal.aak9397. [DOI] [PubMed] [Google Scholar]
- 68.Han X, Wang Y, Sun J, et al. Role of CXCR3 signaling in response to anti-PD-1 therapy. EBioMedicine. 2019;48:169–177. doi: 10.1016/j.ebiom.2019.08.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chow MT, Ozga AJ, Servis RL, et al. Intratumoral activity of the CXCR3 chemokine system is required for the efficacy of anti-PD-1 therapy. Immunity. 2019;50(6):1498–1512.e5. doi: 10.1016/j.immuni.2019.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.House IG, Savas P, Lai J, et al. Macrophage-derived CXCL9 and CXCL10 are required for antitumor immune responses following immune checkpoint blockade. Clin Cancer Res. 2020;26(2):487–504. doi: 10.1158/1078-0432.CCR-19-1868. [DOI] [PubMed] [Google Scholar]
- 71.Mikucki ME, Fisher DT, Matsuzaki J, et al. Non-redundant requirement for CXCR3 signalling during tumoricidal T-cell trafficking across tumour vascular checkpoints. Nat Commun. 2015;6:7458. doi: 10.1038/ncomms8458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Spranger S, Dai D, Horton B, Gajewski TF. Tumor-residing Batf3 dendritic cells are required for effector T Cell trafficking and adoptive T cell therapy. Cancer Cell. 2017;31(5):711–723.e4. doi: 10.1016/j.ccell.2017.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang B, Wang Y, Sun X, et al. CXCR6 is required for antitumor efficacy of intratumoral CD8(+) T cell. J Immunother Cancer. 2021;9(8):e003100. doi: 10.1136/jitc-2021-003100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Muthuswamy R, McGray AR, Battaglia S, et al. CXCR6 by increasing retention of memory CD8(+) T cells in the ovarian tumor microenvironment promotes immunosurveillance and control of ovarian cancer. J Immunother Cancer. 2021;9(10):e003329. doi: 10.1136/jitc-2021-003329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Veinotte L, Gebremeskel S, Johnston B. CXCL16-positive dendritic cells enhance invariant natural killer T cell-dependent IFNγ production and tumor control. Oncoimmunology. 2016;5(6) doi: 10.1080/2162402X.2016.1160979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Di Pilato M, Kfuri-Rubens R, Pruessmann JN, et al. CXCR6 positions cytotoxic T cells to receive critical survival signals in the tumor microenvironment. Cell. 2021;184(17):4512–4530.e22. doi: 10.1016/j.cell.2021.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pilipow K, Roberto A, Roederer M, Waldmann TA, Mavilio D, Lugli E. IL15 and T-cell stemness in T-cell-based cancer immunotherapy. Cancer Res. 2015;75(24):5187–5193. doi: 10.1158/0008-5472.CAN-15-1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Klebanoff CA, Finkelstein SE, Surman DR, et al. IL-15 enhances the in vivo antitumor activity of tumor-reactive CD8+ T cells. Proc Natl Acad Sci USA. 2004;101(7):1969–1974. doi: 10.1073/pnas.0307298101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Abd Hamid M, Wang RZ, Yao X, et al. Enriched HLA-E and CD94/NKG2A interaction limits antitumor CD8(+) tumor-infiltrating T lymphocyte responses. Cancer Immunol Res. 2019;7(8):1293–1306. doi: 10.1158/2326-6066.CIR-18-0885. [DOI] [PubMed] [Google Scholar]
- 80.van Montfoort N, Borst L, Korrer MJ, et al. NKG2A blockade potentiates CD8 T cell immunity induced by cancer vaccines. Cell. 2018;175(7):1744–1755.e15. doi: 10.1016/j.cell.2018.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.André P, Denis C, Soulas C, et al. Anti-NKG2A mAb is a checkpoint inhibitor that promotes anti-tumor immunity by unleashing both T and NK cells. Cell. 2018;175(7):1731–1743.e13. doi: 10.1016/j.cell.2018.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hara T, Chanoch-Myers R, Mathewson ND, et al. Interactions between cancer cells and immune cells drive transitions to mesenchymal-like states in glioblastoma. Cancer Cell. 2021;39(6):779–792.e11. doi: 10.1016/j.ccell.2021.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The scRNA-seq data of GBM samples (GSE103224, GSE138794, GSE139448 and GSE131928) were obtained from Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo/) database. The bulk RNA-seq and microarray data of GBM samples were obtained from TCGA and CGGA databases (http://gliovis.bioinfo.cnio.es/).