Abstract
Bacteriophage lambda integrase (Int) catalyzes at least four site-specific recombination pathways between pairs of attachment (att) sites. Protein-protein contacts between monomers of Int are presumed to be important for these site-specific recombination events for several reasons: Int binds to the att sites cooperatively, catalytic Int mutants can complement each other for strand cleavage, and crystal structures for two other recombinases in the Int family (Cre from phage P1 and Int from Haemophilus influenzae phage HP1) show extensive protein-protein contacts between monomers. We have begun to investigate interactions between Int monomers by three approaches. First, using a genetic assay, we show that regions of protein-protein interactions occur throughout Int, including in the amino-terminal domain. This domain was previously thought to be important only for high-affinity protein-DNA interactions. Second, we have found that an amino-terminal His tag reduces cooperative binding to DNA. This disruption in cooperativity decreases the stable interaction of Int with core sites, where catalysis occurs. Third, using protein-protein cross-linking to investigate the multimerization of Int during recombination, we show that Int predominantly forms dimers, trimers, and tetramers. Moreover, we show that the cysteine at position 25 is present at or near the interface between monomers that is involved in the formation of dimers and tetramers. Our evidence indicates that the amino-terminal domain of Int is involved in protein-protein interactions that are likely to be important for recombination.
Bacteriophage lambda integrase (Int) is a recombinase which inserts and excises the phage genome into and out of the Escherichia coli chromosome. It belongs to a large family of tyrosine recombinases whose members carry out site-specific recombination of phage, plasmid, and chromosomal sequences (6, 29). Int mediates recombination via four distinct pathways (summarized in Table 1), in which different protein-DNA complexes are assembled by Int and its accessory factors on four types of recombination target sites, known as attachment or att sites (the structure of attL is shown in Fig. 3). In addition to the catalytic domain, Int has two DNA binding domains: a relatively high-affinity binding domain contacts the “arm” binding sites, which are distal to the loci of strand exchange, while a low-affinity binding domain contacts the “core” binding sites, which directly flank the loci of strand exchange (32, 41).
TABLE 1.
Summary of the four pathways of bacteriophage λ site-specific recombination
| Component(s) | Pathwayc
|
|||
|---|---|---|---|---|
| Integration | Excision | Bent-L | Straight-L | |
| att substrates | attP, attB | attL, attR | attL (tenP′1)a | attL |
| Int requirement | Y | Y | Y | Y |
| Bending protein requirement | IHF | IHF or HU | IHF | Inhibitory |
| Xis requirement | Inhibitory | Y | N | N |
| Supercoiling requirement | Y | N | N | N |
| Efficiencyb | High | High | High | Low |
The bent-L pathway works equally well with wild-type attL and attL tenP′1 substrates in vivo but only with attL tenP′1 substrates in vitro.
High efficiency denotes >25% conversion of substrates to products. Low efficiency denotes <10% (and usually <5%) conversion of substrates to products. Note that integration and excision are unidirectional pathways and often convert 60 to 70% of substrates to products. In contrast, the bent-L pathway is bidirectional and should convert no more than 50% of substrates to products under normal circumstances.
Y, yes; N, no.
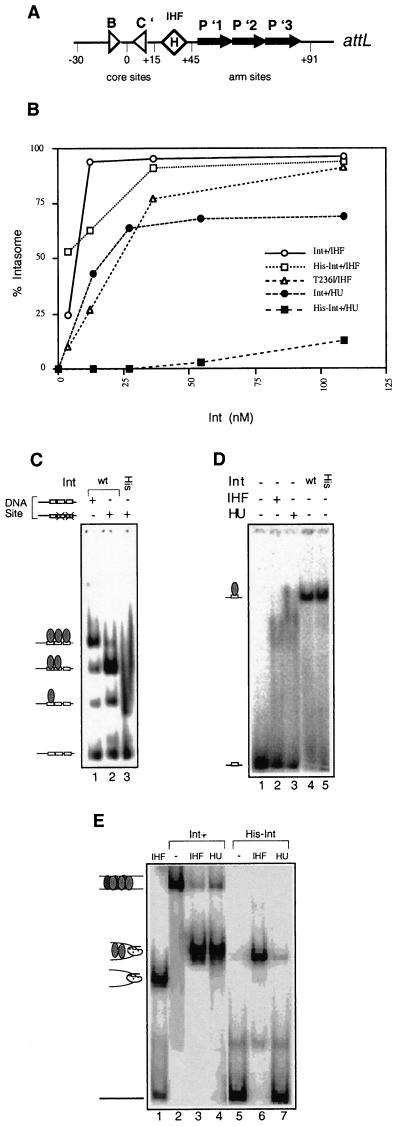
FIG. 3.
Comparison of cooperative interactions and intermediate assembly by His-tagged and non-His-tagged wild-type Int. (A) Structure of the attL site. Triangles represent the low-affinity core-type binding sites for Int which flank the points of strand exchange. The diamond labeled H denotes the IHF binding site. The black arrows denote the higher-affinity arm-type Int binding sites. (B) Percentages of total counts found as intasome complexes at different Int concentrations. These complexes contain one to several Int monomers as well as a bending protein (IHF or HU). (C) Ability of His-tagged and non-His-tagged Int proteins to bind to the arm sites of attL. The attL fragments comprised coordinates +12 to +110 on attL, with either wild-type (wt) arm binding sites or a wild-type P′1 arm site and mutated P′2 and P′3 sites. Reaction mixtures contained 3 nM labeled DNA fragment, 100 ng of salmon sperm DNA, and 120 nM Int. Bands representing arm sites occupied by one, two, or three Int monomers are depicted in the cartoons shown to the left (the cartoons refer strictly to the stoichiometry rather than the actual position occupied by Int monomers). (D) Ability of His-tagged and non-His-tagged Int proteins to bind to an individual arm site, the P′1 sequence (coordinates +54 to +63 of attL plus five flanking residues). All reaction mixtures contained 1 nM labeled DNA (nonspecific DNA was omitted). Lanes 2 and 3 show control reaction mixtures containing 37 nM IHF or HU, respectively. Lanes 4 and 5 show reaction mixtures containing a 7.5 nM concentration of the specified Int protein. (E) Formation of intasome and BMC complexes by wild-type or His-tagged Int. All reaction mixtures contained 1 nM labeled full-length attL (coordinates −69 to +110) and 1 μg of salmon sperm DNA. Lane 1 contains labeled attL and IHF only. Lanes 2 to 4 contain non-His-tagged wild-type Int. Lanes 5 to 7 contain His-tagged Int in the absence or presence of the specified bending protein. Reaction mixtures contained, when specified, IHF or HU at 37 nM and either of the Int proteins at 100 nM.
Int cleaves and rejoins four phosphodiester bonds in the host and phage DNAs via transient covalent intermediates between itself and DNA. Presumably because abortive reactions would be detrimental both to the phage and to the host, the recombinases have evolved to be very efficient, and very few abortive products are seen (30). At worst, aborted recombination events would generate breaks in both the bacterial and phage genomes; at best, integration or excision would be less efficient due to unproductive rounds of strand cleavage and joining and the lysogeny versus lysis decision of the phage would be impeded. Abortive recombination may be minimized by carrying out strand exchange in the context of specific synaptic complexes containing both of the DNA substrates and all of the protein subunits necessary for the reaction. This indeed appears to be the case (1, 31, 35, 36). Moreover, the DNA strand cleavage and ligation events in integrative and excisive recombination are highly concerted, for which interactions between Int monomers are presumed to be important. Protein-protein interactions appear to be a central feature of site-specific recombination reactions, including phase variation of flagellar antigens in Salmonella enterica (12, 22), resolution of γδ transposon-generated cointegrate structures (14, 25), and 2μ plasmid inversion mediated by Flp (reference 21 and references therein).
The DNA binding and catalytic properties of Int, as well as those of many protein-DNA intermediates assembled by Int in conjunction with its accessory factors, have been studied extensively (reviewed in references 20 and 27). However, no direct information is available on the interactions between Int monomers occurring during recombination. Two major observations suggest that protein-protein contacts between Int monomers are important. First, binding of Int to the att sites is highly cooperative (16, 36). Second, different catalytic mutants of the Int protein complement each other for strand cleavage activity (10), suggesting intimate interactions between at least two Int monomers. More recently, further information has become available from several crystal structures of Int family proteins. Four structures have been solved, namely, those of the catalytic domain of Int (19), the catalytic domain of the Haemophilus influenzae phage HP1 Int (13), the E. coli XerD protein (39), and the cocrystal of the bacteriophage P1 Cre protein bound to a Holliday junction (7, 9). The Int structure and the XerD structure were solved as monomers and thus provide information on protein-protein contacts only by homology alignments with the catalytic domains of HP1 Int, solved as a dimer, and with the Cre structure, which was solved as a pseudosymmetric tetramer (9). However, Int and the other bacteriophage integrases share little or no recognizable homology in the noncatalytic domains of their relatives (6). Therefore, while the Cre and HP1 Int structures give insight into protein-protein contacts lying within the catalytic domain, none of these structures is very informative about interactions occurring in the N-terminal half of the Int protein.
In order to obtain more information about protein-protein interactions necessary for Int's function, we have investigated interactions between Int molecules using several approaches. First, we have used a genetic assay to localize the regions where protein-protein contacts occur. Second, we have investigated cooperative interactions among Int monomers binding to DNA. Third, we have used protein-protein cross-linking as a physical assay to determine the predominant multimeric species assembled by Int. We find that residues which contribute to stable protein-protein contacts are spread throughout most of the Int primary sequence. The genetic and physical assays agree that Int's N-terminal domain, previously thought to comprise only the DNA binding domain necessary for contacting Int's high-affinity arm binding sites, also provides an important protein-protein interface between Int monomers. We show that cooperative interactions at the N terminus of Int affect the stable binding of Int to its core binding sites, the loci of strand exchange. Last, we show that Int forms several types of multimers and we test the effect on recombination of modifying cysteines and tethering Int protomers to each other.
MATERIALS AND METHODS
Strains.
The tester strain for examining the transcription-inducing activities of Int-AraC fusion proteins was M8834, F− Δ(araOC leu)1109 rpsL150 (Strr) (generously provided by Malcom Casadaban). It was constructed such that araC was deleted but a functional araBAD operon remained. Transcription of araBAD depends on appropriate dimerization of the AraC DNA binding domains (AraCD). An hsdR deletion was introduced by P1 transduction to facilitate direct cloning of PCR-generated ligation products into the M8834 strain. Mutations in lon and clpP were introduced by transducing M8834 with clpP::Tn5 linked to a Δlon allele (donor strain was kindly provided by S. Gottesman). Phage P1 transductions were performed according to standard protocols, and mucous Kanr transductants were used as recipients of the cloned fusion proteins (lon mutations confer a mucous phenotype). The clone expressing the His-tagged Int gene was the generous gift of J. Gardner, while the clone expressing the IntC25S gene was the generous gift of R. Tirumalai and A. Landy.
Generation of LacZ::Int::AraC chimeric genes.
Fusion proteins were constructed by introducing portions of the Int gene, generated by PCR, upstream of and in frame with the AraCD. Plasmid pKM19-C170 (from N. Lee via M. Casadaban) is a modified pUC19 plasmid containing the AraCD, consisting of amino acid residues 170 to 292 (23). Upstream of the truncated AraC protein are the translation start sites and the first five amino acids of LacZ, all of which are under the control of a ptac promoter. PCR primers were designed to introduce restriction enzyme sites upstream (HindIII) and downstream (BamHI) of appropriate Int sequences to allow cloning upstream of and in frame with the AraCD in pKM19-C170. The sequences of the primers used are summarized in Table 2. The PCR products, generated with VENT polymerase (New England Biolabs [NEB]) were purified with a Wizard PCR Preps kit (Promega), digested overnight with BamHI and HindIII (NEB), gel purified, and ligated to gel-purified, digested vector DNA. The ligated plasmids were recovered in DH5α cells and electroporated into M8834 or its relatives. Screens for araBAD activity were carried out on MacConkey agar (Difco) containing 1% l-arabinose (Sigma).
TABLE 2.
Oligomers used in this studya
| Primer or P′1 oligonucleotide | Sequence (5′ to 3′) |
|---|---|
| Primers | |
| 1–262 (HindIII) | CCGAAGCTTGATGGGAAGAAGGCGAAGT |
| 1–262 (BamHI) | GCGGATCCTCGCATTTATCAAGTGTTTC |
| 80–169 (HindIII) | CCAAGCTTGTCATGGCTTGATCGCTAC |
| 80–169 (BamHI) | GCGGGATCCGCGAGTGGCAGCGCAATG |
| 80–356 (BamHI) | GCCGGATCCTTTGATTTCAATTTTGTCCC |
| Oligonucleotides | |
| LP′1T | AGAACAGGTCACTATTCTT |
| LP′1B | CAAGAATAGTGACCTGTTC |
Primers were used in cloning Int::AraC fusions. Primers designated HindIII point towards the 3′ end of the Int sequence, while those designated BamHI point towards the 5′ end of the Int sequence. Combinations of these primers were used to generate the five fusions depicted in Fig. 2. The restriction enzyme sites are underlined. The oligonucleotides were used to assemble the P′1 arm substrate used in the experiment whose results are shown in Fig. 3D. The LP′1T and LP′1B oligonucleotides anneal over their whole length to form a duplex, with the exception of a single protruding nucleotide at the 5′ end, to increase labeling efficiency with polynucleotide kinase. The central nine bases comprise the P′1 arm binding site.
Cell extracts.
Extracts of cells expressing the fusion proteins were made by a freeze-thaw lysozyme procedure, as follows. Overnight cultures of the tester strain containing the fusion proteins were subcultured 1:10 into 40 ml of 1× NCE (4) supplemented with 1% casein digest (Sigma) and 1% arabinose with 100 μg of ampicillin (Sigma) per ml, grown at 37°C for 2.5 to 3 h to an optical density at 650 nm of 0.7 to 0.8, and pelleted. Cell pellets were resuspended in a solution containing 200 μl of 0.5 M Tris HCl (pH 7.4) and 10% sucrose (Sigma) and frozen at −80°C. Pellets were thawed on ice, and the following concentrations of protease inhibitors were added: 0.5 μl of leupeptin (5 mg/ml in H2O; Sigma), 0.25 μl of pepstatin (10 mg/ml in H2O; Sigma), 0.5 μl of soybean trypsin inhibitor (100 mg/ml in H2O; Sigma), and 5 μl of phenylmethylsulfonyl fluoride (100 mM in ethanol; Sigma). The cells were then lysed by incubating them on ice for 30 min with a 1/20 volume of 10-mg/ml lysozyme (in 10 mM Tris HCl [pH 7.4]). The cellular debris was removed by centrifugation at 15,000 rpm in a Sorvall RCSC centrifuge with an SA 600 rotor for 20 min. Total protein present in the extract was measured by a modified Bradford protein assay (Bio-Rad). The extracts were then frozen in a dry ice-ethyl alcohol bath and stored at −80°C.
Western blot analysis.
Approximately 120 ng of cell extracts was mixed 1:1 with Laemmli loading buffer (Bio-Rad), boiled for 10 min, and electrophoresed on 10 to 20% polyacrylamide gradient minigels (Novex). Electrophoresis was carried out for 2.5 h at 120 V. The gels were transferred to nitrocellulose (Bio-Rad) in a Novex Western Blot Module according to the manufacturer's instructions at 30 V for 2 h in transfer buffer. The nitrocellulose was probed with antibody, washed, and treated with 10 ml of SuperSignal peroxide substrate (Pierce) for 10 min before exposure to Kodak XAR film. For reprobing with a different primary antibody, each blot was gently shaken in stripping solution (2% sodium dodecyl sulfate [SDS; Sigma], 62.5 mM Tris-HCl [pH 6.8], 0.1 M β-mercaptoethanol [Sigma]) at 65°C for 30 min and was rinsed several times before the Western blot procedure described above was repeated.
Three primary antibodies were used. Int was detected with a polyclonal anti-Int antibody obtained from Susan Bear via Howard Nash or with polyclonal antibody obtained against gel-purified His-tagged Int whose tag had been cleaved off with thrombin (the latter antibody was raised by BABCo). The polyclonal antibody raised against the whole AraC protein does not detect the AraCD (23). Therefore, an AraC peptide representing amino acids 176 to 193 was synthesized, conjugated to keyhole limpet hemocyanin, and injected into rabbits. The peptide and the polyclonal antibody were made by Sigma Genosys.
Arabinose isomerase activity assay.
In order to quantitate the activity of the arabinose operon induced by the fusion proteins, arabinose isomerase (AraA) activity assays were performed using published methods (3, 33). The colorimetric assay measures the activity of the AraA gene at time zero and at several time points thereafter. The zero time point served as the reference, and activities were calculated from the linear part of the curve using the following equation: (2 × 1010) (optical density at 550 nm)/(minutes of incubation)(number of cells in assay).
Purification of His-tagged Int.
E. coli BL21 (λDE3 pLysS) cells (250 ml) carrying the Int gene cloned into the pET28a vector (Novagen) were grown in Luria broth supplemented with neomycin sulfate (100 μg/ml; Sigma) at 37°C to mid-log phase. Int expression was induced with 0.5 mM IPTG (isopropyl-β-d-thiogalactopyranoside; Bachem) for 3 to 4 h; during induction, cultures were shaken at room temperature. Cell pellets were resuspended in extraction buffer (50 mM TrisCl [pH 8], 0.8 M KCl, 10% sucrose) after one freeze-thaw cycle. Extracts were made by homogenization on ice in the presence of phenylmethylsulfonyl fluoride, pepstatin A, leupeptin, and soybean trypsin inhibitor (concentrations of protease inhibitors were as for the previous cell extracts). The supernatant was frozen at −80°C, and the pellet was reextracted in 400 μl of extraction buffer. The supernatants were combined and mixed with 200 to 300 μl of Talon metal-affinity resin (Clontech) which had been washed in binding buffer (50 mM Tris-Cl [pH 8], 300 mM KCl, 10% glycerol, 10 mM β-mercaptoethanol). The beads were rotated with the protein extracts for 2 h at 4°C and then washed five times (30 min each) with 500 μl of extraction buffer. Two sequential elution steps were performed, each with 150 μl of elution buffer (50 mM Tris-Cl [pH 8], 600 mM KCl, 10% glycerol, 0.6 M imidazole or histidine, 10 mM β-mercaptoethanol). The resin was stripped with elution buffer containing 1 M imidazole. Fractions showing the greatest activity were quantitated by Western blot analysis.
Purification of non-His-tagged Int.
E. coli BL21 (λDE3 pLysS) or HN1463 (IHF−HU−) cells (500 ml) carrying the Int gene cloned into a pT7 vector (41) or in a pLex 5B vector (5) were grown in Luria broth supplemented with ampicillin (100 μg/ml; Sigma) at 37°C to mid-log phase. Int expression was induced with 0.5 mM IPTG (Bachem), and cultures were shaken at room temperature for 4 h. Cell extracts were prepared as described above for His-tagged Int purification. Int was isolated from these extracts by mixing 200 μl of phosphocellulose equilibrated in buffer X (50 mM Tris-Cl [pH 8], 400 mM KCl, 1 mM EDTA, 10% glycerol, 10 mM β-mercaptoethanol) with 800 μl of cell extract. After being rotated for 2 h at 4°C, the resin was pelleted gently, the supernatant containing unbound proteins was removed, and the resin was washed with 400 μl of buffer X for 30 min. Two sequential elution steps were performed with 100 μl of buffer X. The first elution step was performed with buffer containing 600 mM KCl, while the second elution step was performed with buffer containing 1 M KCl.
Gel mobility shift and in vitro recombination assays.
Linear substrates containing the attL or attL tenP′1 (27) sequence were end labeled using T4 polynucleotide kinase (NEB) and [γ-32P]ATP (NEN). All recombination and binding reactions were performed in 20-μl total volumes. Recombination reaction mixtures contained 1 nM labeled att site (179 bp long) and 4 nM unlabeled att site (496 bp long), while binding reaction mixtures generally contained 1 to 4 nM labeled att site, as specified in figure legends. Each reaction mixture contained 0.1 to 1 μg of sonicated salmon sperm DNA, 44 mM Tris-Cl (pH 8), 60 mM KCl, 0.05 mg of bovine serum albumin per ml, 11 mM Tris-borate (pH 8.9), 1 mM EDTA, 13.6% (vol/vol) glycerol, and 5 mM spermidine. Int and integration host factor (IHF) were present at 50 and 35 nM, respectively. Recombination reactions were stopped by the addition of 5 μl of 2% SDS-containing bromophenol blue and heated at 65°C for 5 min. These samples were loaded onto 5% polyacrylamide Tris–SDS gels and electrophoresed in Tris-Tricine-SDS buffer at a 100-mA constant current. For the bent-L pathway, reaction mixtures were incubated at 30°C for 90 min. For the straight-L pathway, reaction mixtures were incubated at room temperature for 90 min. Gel mobility shift reactions were assembled as described above and were layered without loading dye directly onto 5% native polyacrylamide–0.5× Tris-borate-EDTA gels (29:1 acrylamide-bisacrylamide). Gels were run at 165 V at 4°C. Band shift assays using the oligomeric substrate coding for the individual P′1 arm site were loaded onto 10% native polyacrylamide gels, and reactions were carried out as described above but without nonspecific DNA competitor. All gels were dried and analyzed with a Molecular Dynamics PhosphorImager.
Reaction mixtures contained an excess of Int relative to specific att site DNA for several reasons. First, we do not know the fraction of active Int in the preparation. Second, the actual number of Int monomers required for proper assembly of intermediates and for catalysis for each recombination pathway is not known; although four Int monomers are expected to be required for catalysis (1, 7, 9), additional monomers may be recruited to serve an architectural role (36; L. Jessop and A. Segall, unpublished results). Third, since we used nonspecific DNA in our reaction mixtures to minimize nonspecific aggregation of Int, some Int molecules almost certainly bound to this DNA rather than to the att sites.
Protein-protein cross-linking assays.
Reaction mixtures for gel mobility shift assays were assembled as described above except that the concentration of the att site was 4 nM. After 0 to 30 min of incubation at room temperature, bismaleimidohexane (BMH; Pierce) was added to 0.3 mg/ml, disuccinimidyl glutarate (DSG; Pierce) was added to 0.25 mg/ml, or dithio-bis-maleimidoethane (DTME; Pierce) was added to 0.3 mg/ml. All cross-linkers were resuspended in dimethyl sulfoxide (Sigma). The reaction mixtures were incubated for an additional 10 to 30 min at room temperature (no difference in extents of cross-linking was seen over this time period). To quench the cross-linkers, dithiothreitol (DTT; Sigma) was added to a final concentration of 20 mM for BMH-containing reaction mixtures or Tris (pH 8) was added to a final concentration of 150 mM for DSG reactions. The DTME tether was cleaved by adding 20 mM DTT. Laemmli's sample buffer (Bio-Rad) was added to the samples, which were then boiled for 10 min (Int multimers are very stable and are dependent on cross-linker only when reaction mixtures are boiled for 10 min in the presence of a reducing agent [data not shown]). Cross-linking products were separated on a 4 to 12% Tris–glycine gel (Novex) at 125 V for approximately 2 h. Proteins were electroblotted to nitrocellulose (Bio-Rad) using a Novex blot module, and Int was detected with polyclonal Int antibody and horseradish peroxidase-conjugated goat anti-rabbit secondary antibody (Jackson Laboratories). SuperSignal or Super Signal West Dura substrates (Pierce) were used to develop the blot.
When we were looking at the effect of DSG on recombination, HEPES buffer was used. Although this buffer system is suboptimal for recombination and the formation of the synaptic intermediate, Tris buffer could not be used because it contains primary amines that interfere with efficient DSG cross-linking.
Assaying the effect of cross-linking on recombination and protein-DNA intermediates.
Reaction mixtures were assembled as for the in vitro recombination assays described above. Cross-linker was added either when the reaction mixtures were assembled, after 45 min of incubation at room temperature, or as specified in the figure legends. Following a total incubation time of 90 min, reaction mixtures were loaded onto a 5% polyacrylamide–Tris–Tricine–SDS gel to assay for recombination or a 5% native polyacrylamide gel to assay the formation of protein-DNA complexes. Samples assayed for recombination received 5 μl of 2% SDS plus dye and were heated at 65°C prior to electrophoresis.
RESULTS
Fusion assay for localizing intermonomer contacts among Int molecules.
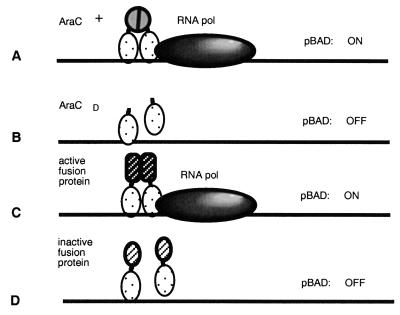
The fusion assay, developed by Malcolm Casadaban (unpublished results), takes advantage of properties of the transcriptional activator AraC (Fig. 1). This activator must bind two sites, araI1 and araI2, upstream of the paraBAD promoter in order to activate transcription from this promoter (reviewed in reference 34). The AraC activator consists of two independent domains, a dimerization domain and a DNA binding domain, connected by a hinge. The DNA binding domain by itself is sufficient to bind the araI sites and weakly activate paraBAD in the absence of arabinose (23). However, efficient activation of the natural promoter occurs only if a dimer of an AraCD contacts araI. This is accomplished in the wild-type AraC protein via protein-protein contacts mediated by the dimerization domain. The natural dimerization domain has been successfully replaced by heterologous multimerization domains such as leucine zippers (2) or by entire proteins which can dimerize (M. Casadaban, unpublished data).
FIG. 1.
Identification of Int regions involved in protein-protein contacts using the activation of paraBAD as an assay. The araC gene together with the upstream araC binding sites are deleted in the M8834 strain, and thus the repression loop cannot form. (A) Efficient transcription initiation of paraBAD requires the presence of an AraC dimer bound at the I1 and I2 sites. AraC protein consists of two domains linked by a flexible tether; one binds to DNA, and the other mediates protein-protein contacts between two AraC monomers. (B) The AraCD cannot bind to DNA stably in the absence of the multimerization domain, resulting in poor activation of the operon. (C) Fusion of heterologous multimerization domains to the AraCD results in efficient activation of the operon. (D) Fusion of proteins or protein fragments which mediate weak or inappropriate interactions between two monomers do not activate the operon.
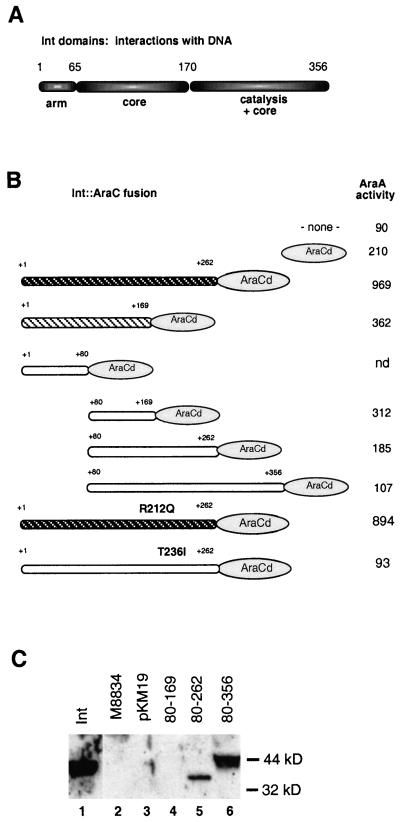
Our first goal was to identify the regions of Int involved in protein-protein contacts. Therefore, we constructed a series of fusion proteins between fragments of the Int protein and the AraCD. Those regions of Int which can fold appropriately and interact stably as a homodimer (or possibly higher species) should increase the transcriptional activity of the paraBAD promoter. This interaction will be productive only if the fusion proteins maintain the appropriate geometry of the AraCD with respect to the araI sites. Note that the endogenous araC gene and both araO sites are deleted and thus that the repression loop cannot be formed. Fusion proteins between Int fragments and the C-terminal 122 amino acids of AraC were expressed from the ptac promoter, using the translational start site and first five amino acids of LacZ. The constructs, diagrammed in Fig. 2B, were designed to cover overlapping portions of Int. The boundaries of the Int fragments were placed such that they did not interrupt highly conserved regions between the λ Int protein and its closest relative to date, the phage HK022 Int protein (26); we reasoned that these highly conserved regions would be part of a folding domain (the fusions predated the crystal structures). In addition, we took into account that amino acids in the central region of Int affected cooperative binding to att sites and were thus postulated to affect protein-protein contacts (11). The current model for the domain structure of Int, proposed by Landy and colleagues, is shown in Fig. 2A (40). The 169-amino-acid fragment of Int which we fused to the AraCD (Fig. 2B) coincides almost exactly with the boundary between the C-terminal catalytic domain of Int and the N-terminal half of the protein. Previous proteolysis studies showed that a small N-terminal domain corresponding to the first 65 amino acids of Int was generated (32). Since the number of identities between λ Int and HK022 Int decreased substantially between amino acids 60 and 80, we reasoned that a less conserved linker region might connect this domain to the rest of the Int protein and thus make the initial fusion breakpoint towards the end of the putative linker. Finally, the breakpoint at amino acid 262 is located near the end of alpha helix D of the Int catalytic domain (19, 29).
FIG. 2.
(A) Domain organization of the bacteriophage λ Int protein. The figure depicts the regions of the protein responsible for contacting the arm-type sites of the att recombination targets, for contacting the core-type sites which flank the points of strand exchange, or for catalytic activity (refer to Fig. 3A for the structure of the attL site). The numbers above the diagram refer to the amino acids in the protein. (B) Structures of eight fusion proteins between fragments of Int (rounded rectangles) and the AraCD (oval). The numbers above the proteins refer to the Int amino acids included in the fusion protein. The darker the Int portion, the darker the color on MacConkey agar plates of colonies carrying the fusion protein: the fusion represented by white hatching on a black background turned colonies bright red, the fusion represented by black hatching on a white background conferred a light pink color, while the fusions represented in white conferred no color. On the right is listed the activity of the AraA protein in units per cell (see Materials and Methods); the averages of triplicate measurements from one representative experiment are shown (all fusions were tested at least three independent times). In this assay, the activity of the wild-type AraC protein was 923 U. “none” refers to AraA activity in a strain which did not contain any part of the AraC protein. nd, not determined. (C) Western blot of selected strains with anti-Int polyclonal antibody. Lane 1 shows a purified Int protein control. Lane 2 shows an extract of the tester strain M8834, containing the araBAD operon and the araC deletion (see Materials and Methods), which was used to express all the fusion proteins. Lane 3 shows M8834 with the pKM19-C170 plasmid, which expresses only the AraCD on a multicopy plasmid. Lanes 4, 5, and 6 show extracts of M8834 expressing Int-AraCD fusion proteins containing the Int amino acids depicted above the figure.
The activities of the fusion proteins were screened on MacConkey agar plates containing 1% l-arabinose and were then quantitated using an assay for arabinose isomerase (AraA) activity (3, 33). These data are presented in Fig. 2B. The fusion protein containing Int amino acids 1 through 262 was very positive on MacConkey agar plates, and another, containing amino acids 1 to 169, was weakly positive (the phenotype on MacConkey agar is represented by different patterns for the fusion proteins in Fig. 2B). Among the fusion proteins that were negative on MacConkey agar plates, two had AraA activities that were reproducibly above the background level conferred by the vector itself (the vector contained the AraCD). Comparison of the AraA activities of Int1–169::AraCD and Int80–169::AraCD suggested that amino acids 80 to 169 are sufficient to confer significant dimerization. However, comparison of the activities of the fusion protein Int80–262::AraCD to Int1–262::AraCD showed that the presence of the N-terminal 80 amino acids confers significantly greater dimerization. Although it is formally possible that the breakpoint at amino acid 80 induces misfolding of the fusion protein Int80–262::AraCD, this is unlikely based on the activity shown by the Int80–169::AraCD fusion. Thus, the amino terminus of Int contributes to protein-protein contacts, although by itself it does not suffice to dimerize the AraCD fragment.
Extracts of strains expressing the fusion proteins were tested by Western blot analysis using two polyclonal antibodies, one made against an AraCD peptide (amino acids 176 to 193) (see Materials and Methods) and one made against the whole Int protein. Two of the fusion proteins, Int80–262::AraCD and Int80–356::AraCD, gave strong signals with both antibodies, despite being negative in the transcription activation assays (Fig. 2C). Neither of the two most positive fusion proteins were seen on Western blots (data not shown). This result is not entirely surprising since few molecules of wild-type AraC are present in the cell even under conditions of maximal expression of the araBAD operon (18). One of the transcriptionally negative fusion proteins, Int80–169::AraCD, was also invisible by Western blot analysis and thus may either be very unstable or, like the active fusion proteins, be made in amounts too small to detect. We expressed the fusion proteins in a strain deficient for both the Lon and ClpP proteins (encoding major proteases or protease subunits) but detected no additional paraBAD activity (data not shown). Nevertheless, other E. coli proteases may be responsible for degradation of some of the fusion proteins.
Fusion proteins that are expressed and stable but do not activate paraBAD may be negative for two reasons: they may not make sufficiently strong contacts between two monomers, or they may mediate contacts which confer the wrong geometry onto the AraCD, thus precluding activation of paraBAD. Given the crystallographic data obtained with the Haemophilus phage HP1 integrase and the E. coli phage P1 Cre protein, we expected the catalytic domain of Int, encoded in the carboxy-terminal half of the protein, to form fairly extensive protein-protein contacts (7, 9, 13). However, the Int80–356::AraCD fusion protein activated paraBAD less than the AraCD alone, although the fusion protein was expressed at the highest level with respect to those of the other fusion proteins. We speculated that this fusion protein may confer the wrong geometry for promoter activation but that it may dimerize stably and tested whether the fusion proteins convey contacts with wild-type Int protein. However, we saw no evidence of negative complementation of Int in vivo in a lysogeny assay and the Int80–356::AraCD fusion protein was not detected in a pull-down assay when it was coexpressed with a His-tagged Int protein (data not shown). We thus have no strong evidence either for or against stable protein-protein interactions in this fragment of Int.
We wanted to test the relationship between the transcriptional activation potential of the fusion proteins and the ability of the Int protein to carry out recombination. We introduced two point mutations which are known to confer a recombination defect into the most transcriptionally active fusion protein, Int1–262::AraCD. The R212Q mutation affects an active site residue and does not appear to affect protein folding or protein-protein interactions in several in vivo (11) and in vitro assays (J. Boldt and A. Segall, unpublished results). On the other hand, the T236I mutation is known to affect the formation of intermediates that depend on cooperative interactions between Int monomers in vitro (see below and Fig. 3B; T. Bankhead and A. Segall, unpublished results). Int1–262::AraCD fusion proteins carrying these two mutations were introduced into the tester strain and plated on MacConkey media. The R212Q mutation did not affect the phenotype on MacConkey agar, and it did not decrease the paraBAD promoter activity measured by AraA activity (Fig. 2B). In contrast, the T236I mutation rendered the fusion protein-carrying strain white on MacConkey agar and significantly decreased AraA activity. These data suggest that the property presented by the fusion proteins, namely, the ability of specific Int fragments to confer dimerization, is related to properties important during recombination. To summarize our fusion protein data, residues throughout the first 262 amino acids of Int contribute to multimerization contacts, although a smaller fragment comprising amino acids 80 to 169 can stimulate multimerization of the AraCD.
An N-terminal six-His tag confers a cooperativity defect.
Because we wanted to examine the properties of many Int mutant proteins and to perform pull-down assays, we investigated in detail the recombination and DNA binding activities of an N-terminal six-His-tagged Int protein. The N terminus of Int contacts the arm binding sites present outside the core region of three out of four att sites (32), and thus we were interested in how the His tag affects interactions of Int with DNA. Landy and colleagues have shown, using DNA footprinting, that the three arm binding sites on the attL site (Fig. 3A) are filled cooperatively and that Int forms a protein-DNA intermediate called an intasome consisting of IHF and several Int protomers (16). Comparison of the His-tagged wild-type Int protein with the non-His-tagged protein showed that the His tag itself decreased cooperative interactions of Int with the wild-type attL site in the presence of IHF (Fig. 3B). We tested the cooperativity of the two proteins in a more stringent assay. In bandshift assays with a DNA substrate comprising only the arm sites, wild-type Int formed three complexes, corresponding to the filling of each arm site (Fig. 3C, lane 1). This binding was cooperative. We compared the cooperativities of the two proteins using a mutant DNA substrate in which one arm site, P′1, was wild type and the two neighboring arm sites were defective for Int binding due to triple substitutions, known as ten mutations, within the arm site consensus sequence (the ten mutations were shown to prevent Int binding [28]). The wild-type Int protein made a very prominent complex corresponding to the filling of two arm sites rather than one, and even showed a small amount of a complex containing three filled arm sites (Fig. 3C, lane 2). In contrast, the His-tagged Int shifted at least 50% of the substrate, mostly to the position corresponding to a single Int protomer bound to the DNA substrate (Fig. 3C, lane 3). Thus, the His-tagged Int binds DNA but is very defective in cooperative interactions. In order to more definitively separate the ability of the His-tagged Int to bind DNA from its ability to mediate cooperative interactions, we compared the extents of DNA binding of the wild-type and His-tagged Int proteins to a short DNA duplex containing a single arm binding site, P′1, and found that the two proteins shifted this DNA substrate to comparable extents (Fig. 3D, lane 4 versus lane 5). This binding was very sensitive to nonspecific DNA competitor in the cases of both proteins, and the reactions in panel D were performed without any competitor. We thus tested the specificity of Int interactions with the short probe by comparing the levels of binding of IHF and HU (the latter is a well-known nonspecific DNA binding protein) to the same fragment. While both these proteins clearly bound to the fragment, they did so to a lesser extent and formed different complexes than either of the two Int proteins (compare Fig. 3D, lanes 2 and 3 with lanes 4 and 5). We conclude that the His tag has little or no effect on DNA interactions but that it predominantly affects cooperative binding to the arm sites. This conclusion supports our genetic evidence that the N-terminal domain of the Int protein is involved in protein-protein interactions in addition to protein-DNA interactions.
We further tested the effect of the His tag on two protein-DNA recombination intermediates which depend on cooperative interactions between Int monomers. Integrase mediates recombination through four distinct pathways, all of which have been detected in vivo as well as in vitro; the pathways are compared in Table 1 (38). The first of the intermediates we tested was the synaptic intermediate in the straight-L pathway of recombination, in which Int pairs attL sites in a stable, noncovalent complex known as the bimolecular complex (BMC). Assembly of this intermediate depends on cooperative binding of Int to the attL site (36). Figure 3E, lanes 2 and 5, shows that formation of the BMC was reduced drastically when His-tagged Int was used in place of wild-type Int. The second of the intermediates was the attL intasome, an intermediate in excisive recombination which can be assembled with IHF or, if interactions of Int with the core sites are stable, with the nonspecific DNA bending homolog of IHF, HU (37). Interactions of Int with the core were most stable when multiple Int molecules were bound to the arm sites. Both wild-type Int and His-tagged Int assembled similar levels of IHF-containing intasomes (Fig. 3B and 3E, lane 3 versus lane 6), although the His-tagged Int assembled a complex that corresponded to the faster of the two complexes assembled by wild-type Int (these two complexes probably differ by one Int monomer held in the complex via cooperative interactions [16]). The ability of His-tagged Int to assemble HU-containing intasomes is greatly reduced in comparison to that of the wild type (Fig. 3B and D, lane 4 versus lane 7). Thus, cooperativity defects of the His-tagged protein affect intermediate formation. These defects also correlate with defects in recombination: in our hands, the His-tagged Int protein was essentially unable to carry out either integration or the straight-L pathway, was down at least threefold in the bent-L pathway, and had only 50 to 70% of the activity of the wild-type protein in excision. Moreover, the effects of the His tag overshadowed the effects of several point mutations within the body of the protein (Table 3). For example, some mutations were more deleterious in the context of the His-tagged protein (e.g., K93E, S282F, and L331F) while another was less deleterious (T236I). We do not yet understand the basis for the differences in the effects of the His tag on these mutant proteins. The His tag has been shown to improve the nonspecific DNA binding properties of binding-defective human immunodeficiency virus type 1 integrase domains (13a), which may account for the effect on IntT236I.
TABLE 3.
Summary of the differences between His-tagged and non His-tagged proteinsa
| Protein | Test result in pathway:
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Excision
|
Bent-L
|
Straight-L
|
Arm binding
|
Core binding
|
||||||
| −HT | +HT | −HT | +HT | −HT | +HT | −HT | +HT | −HT | +HT | |
| Wild type | + | + | + | + | + | + | + | + | +++ | + |
| K93E | + | + | + | − | + | − | + | + | +/−− | − |
| R293Q | + | + | + | + | + | + | + | + | +/− | +/− |
| S282F | + | − | + | − | + | − | + | + | +/− | − |
| T236I | − | + | + | + | − | + | + | + | − | +/− |
| L331F | − | − | + | − | − | − | + | + | +/− | − |
Integration was not tested for the His-tagged mutant proteins. The His tag is denoted HT. Arm binding was tested using the same assay as that whose results are shown in Fig. 3C, while core binding was tested by comparing the abilities of the proteins to form BMCs or attL intasomes in the presence of HU rather than IHF, as shown in Fig. 3E. Core binding values were assigned plus or minus signs representing differences in binding. Boldface symbols highlight situations in which the His tag significantly affected the behavior of the protein. +, binding; −, no binding; +++, strong binding; +/−, intermediate binding; +/−−, very weak binding.
Cleavage of the His-tagged protein with thrombin to remove the tag does not restore wild-type recombination or binding activity (data not shown; G. Cassell and A. Segall, unpublished results). Polyacrylamide gel electrophoresis (PAGE) analysis showed that the tag was removed, although we cannot guarantee 100% cleavage. More to the point, perhaps, is the fact that the recombinant protein retained three new amino acids (Gly, Ser, and His) at the N terminus that are not present in wild-type Int; these amino acids may be sufficient to impair the activity of the protein.
Determination of Int's multimeric state during recombination.
We directly investigated the multimeric state of Int using several homobifunctional cross-linking reagents which react either with primary amines or with cysteines. The primary amine-reactive compound which gave the greatest extent of cross-linking was DSG, a nonreversible cross-linking agent with a 7.7-Å tether between reactive groups. The most effective sulfhydryl-reactive compound was BMH, also a nonreversible agent but with a 16.1-Å tether between reactive groups. We have obtained very similar results by oxidizing Int with glutathione (data not shown).
Recombination reaction mixtures were assembled and protein-DNA intermediates were allowed to form. Cross-linkers were then added, and the reactions were subjected to PAGE followed by Western blot analysis to determine the sizes of the cross-linked products. Based on the molecular weights of the multimeric species, Int appears to assemble predominantly dimers, trimers, and tetramers in the presence of either DSG or BMH (Fig. 4; however, see below). If the cross-linking reaction mixtures lacked DNA, the same multimers formed but to a far lesser extent (Fig. 4). The addition of nonspecific DNA increased the amounts of these multimers, but the presence of a specific att site generally led to the greatest extent of multimerization. The fact that Int multimerizes to a significant extent in solutions which do not contain specific DNA indicates that preformed Int multimers may serve as a scaffold for the assembly of the DNA-containing recombination complex.
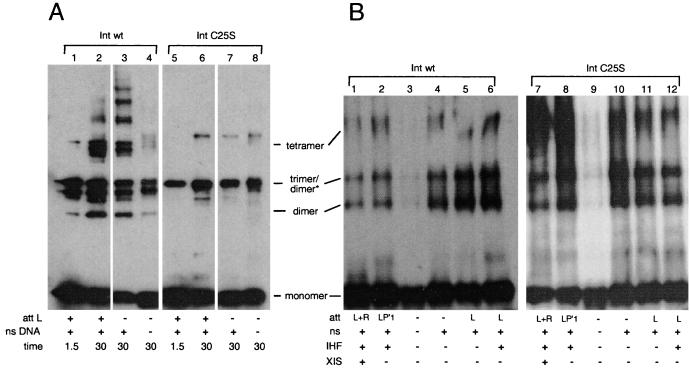
FIG. 4.
BMH and DSG cross-linking of wild-type Int and IntC25S. (A) Effect of specific attL DNA on multimerization as assayed by BMH cross-linking. Reaction mixtures were treated with 0.3 mg of BMH per ml for the number of minutes indicated and were then quenched with excess β-mercaptoethanol. Lanes 1 to 4 show reaction mixtures containing wild-type (wt) Int (see Materials and Methods). Lanes 5 to 8 show reaction mixtures containing IntC25S. At least one of the “trimer” species is in fact a dimer with slower mobility (refer to the text). ns DNA, nonspecific DNA (i.e., salmon sperm DNA). (B) Multimers formed in different recombination pathways as assayed by DSG cross-linking. Each reaction mixture was treated with 0.5 mg of DSG per ml for 10 min. Cross-linking was stopped by addition of excess Tris, pH 8. Lanes 1 to 6 show reaction mixtures containing wild-type Int, while lanes 7 to 12 show reaction mixtures containing IntC25S.
The extent of protein-protein cross-linking is far from complete. We estimate at best 50% cross-linking efficiency, and this changes relatively little when cross-linking is carried out for times between 15 min and 1 h. We assume that intramolecular cross-links compete significantly with intermolecular cross-links. In particular, the Int crystal structure shows that the cysteines at positions 197 and 262 are very close to each other (19) and that an intramolecular disulfide bridge or BMH-mediated bridge between these two residues could form easily. Alternatively, since we do not know the fraction of recombination-active Int in the reaction, some of the monomeric as well as some of the tethered Int may not have participated in recombination.
Int assembles different higher-order complexes depending on the att substrates and the accessory proteins necessary for each of the four different recombination pathways (Table 1). Therefore, we tested if the multimerization state of Int changes in the presence of different att substrates and accessory factors involved in three of the four recombination pathways (integrative recombination intermediates were not tested). We found that the tetramer forms regardless of the presence or absence of IHF or the identity of the att sites in the reaction mixture. Figure 4B, lane 1, represents conditions for excisive recombination, while lane 6 represents conditions for the assembly of the attL intasome, one of the two substrates in excision. Figure 4B, lane 2, represents recombination conditions for the bent-L pathway (Table 1) (38). Figure 4B, lane 5, represents recombination conditions optimal for the straight-L pathway, the simplest but least efficient of the reactions carried out by Int (36).
The N-terminal domain is involved in multimerization.
In order to test directly the involvement of the N-terminal domain in multimerization, we compared the cross-linking patterns of wild-type Int with those of a mutant protein, IntC25S. The IntC25S mutation, constructed in the Landy lab, was shown not to affect recombination via the integrative or excisive pathways (41). When the IntC25S protein was cross-linked with DSG, it yielded the same multimerization pattern as the wild-type protein (Fig. 4B). However, when the protein was cross-linked with BMH, the predominant cross-linked species migrated as a trimer (Fig. 4A). Although it migrates more slowly than expected, this trimer is in fact a dimer, since sulfhydryl-reactive cross-linkers tether this species in the cases of two Int proteins which contain a single cysteine each (IntC25S-C197A-C217A and IntC25S-C217A-C262A) (L. Jessop, J. Boldt, and A. Segall, unpublished data). Because IntC25S forms the same multimers as wild-type Int but BMH fails to tether some of them, the cysteine at position 25 must be at or near the interface involved in the formation of tetramers. These results provide further evidence that the N-terminal domain of Int is involved in protein-protein interactions.
Effect of cross-linking on Int activity.
How does tethering Int monomers to each other affect the assembly of recombination intermediates and recombination? Bent-L recombination reaction mixtures containing Int or IntC25S were assembled, and cross-linking reagents were added. The extent of recombination was determined by PAGE. Similar amounts of recombination products were obtained after 90 min from reaction mixtures with either wild-type Int or IntC25S (Fig. 5B, lanes 2 and 7; quantitation data are shown in Fig. 5C). When BMH was added to reaction mixtures containing either wild-type Int or IntC25S, recombination was inhibited significantly whether cross-linkers were quenched after 15 min (Fig. 5B, compare lane 2 with lane 3 and lane 7 with lane 8) or were active throughout the 90-min incubation (Fig. 5B, lanes 4 and 9). Experiments carried out with DSG gave similar results (data not shown).
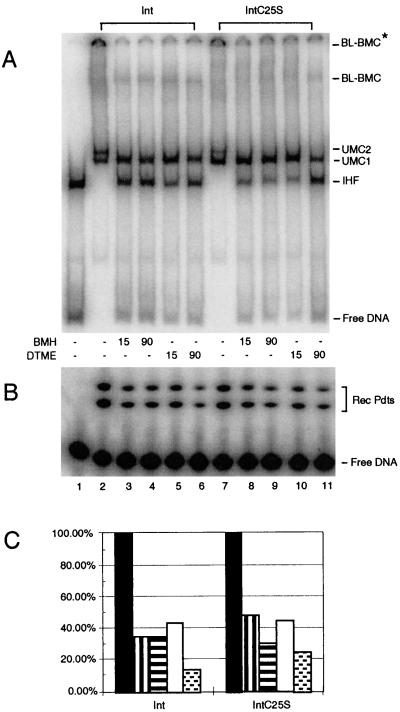
FIG. 5.
Effect of cross-linking on bent-L recombination and formation of pathway intermediates. Reaction mixtures in lanes 2 to 6 contain wild-type Int, while reaction mixtures in lanes 7 to 11 contain IntC25S. Cross-linkers in DMSO were added at time zero to the reaction mixtures in lanes 3 to 6 and 8 to 11. After the specified time, DTT was added to a final concentration of 20 mM to quench the cross-linkers. All reaction mixtures were incubated a total of 90 min at room temperature. (A) Separation of recombination intermediates on a native polyacrylamide gel in the presence or absence of cross-linking reagents. (B) Conversion of substrate DNA to recombinant products on a Tris-Tricine-SDS gel (see Materials and Methods). (C) Quantitation of the bands of the gel shown in panel B; percent recombination values were normalized such that the amount of recombination products (Rec Pdts) seen in the absence of cross-linker was 100%. Solid black bars represent reaction mixtures in lanes 2 and 7 (DMSO only); vertically striped bars represent reaction mixtures in lanes 3 and 8 (15-min BMH treatment); horizontally striped bars represent reaction mixtures in lanes 4 and 9 (90-min BMH treatment); open bars represent reaction mixtures in lanes 5 and 10 (15-min DTME treatment followed by breakage of the tether); and stippled bars represent reaction mixtures in lanes 6 and 11 (90-min DTME treatment followed by breakage of the tether). The actual values of recombination were as follows: lane 1, 0.4%; lane 2, 21.2%; lane 3, 7.3%; lane 4, 7.4%; lane 5, 9.2%; lane 6, 3.0%; lane 7, 15.7%; lane 8, 7.4%; lane 9, 4.8%; lane 10, 7.0%; and lane 11, 3.9%. This experiment is representative of 10 repetitions.
In order to analyze how cross-linking affected the formation of protein-DNA intermediates, parallel reactions with and without BMH treatment were compared by native PAGE. The bent-L pathway proceeds via the successive assembly of two unimolecular complexes known as UMC1 and UMC2 (35). Both of these complexes contain a single attL site with Int bound at the P′2 and P′3 arm sites and at the B core site (see Fig. 3A for the structure of attL). In addition, the C′ core site is unstably occupied in UMC2 (35). UMC2 is the direct precursor of the synaptic intermediate, the bent-L bimolecular complex (BL-BMC), which is a more stable intermediate than UMC2 (35). Because BL-BMC contains two att sites, two BL-BMCs are visible on native gels (Fig. 5A) (35, 36); the faster one contains two short labeled att sites, while the slower one contains one labeled att site and one longer, unlabeled att site (in Fig. 5A, this larger BMC did not enter the gel completely). When the att sites in the reaction are efficiently assembled into UMC and BMC species, no free att DNA and few if any att-IHF complexes are seen (Fig. 5A, lane 2). Int treated with BMH formed UMC1 but not UMC2 and less of BMC (Fig. 5A, lanes 3 to 7). This was the case whether reaction mixtures were assembled with wild-type Int, under which condition BMH captures dimers and tetramers, or with IntC25S, under which condition BMH captures predominantly the slower dimer (Fig. 4A). The decreased formation of UMC2 and BL-BMC is concomitant with an increase of the att-IHF complexes and free att sites, which fits with the proposed pathway of intermediate assembly (35). (We obtained similar results for excision reactions: treatment with BMH allowed formation of the lower UMC but blocked formation of the upper UMC [data not shown]. However, excision synaptic intermediates were not seen; thus, the effect of cross-linkers on their formation cannot be assessed under these conditions.) The results described show that cross-linking either wild-type Int or IntC25S through cysteines blocks formation of one intermediate (UMC2), decreases the level of another (BL-BMC), and decreases recombination levels by 70% or more.
Why does BMH inhibit recombination? One possibility we considered was that BMH treatment mimics the effect of N-ethyl maleimide (NEM) modification of Int. Treatment of wild-type Int with NEM, also a sulfhydryl-reactive compound, blocks Int-mediated recombination (15) by interfering with Int binding at the arm sites (24). Landy and colleagues showed that C25 was the only residue modified by NEM and that IntC25S-mediated recombination was immune to the effect of NEM (41). However, IntC25S was clearly not immune from inhibition by BMH (Fig. 5), despite the fact that treatment with sulfhydryl-reactive cross-linkers inhibited arm binding for wild-type Int but not for IntC25S (data not shown). Therefore, BMH must inhibit recombination mediated by IntC25S due to modification of one of the remaining cysteines. Unlike NEM, BMH (and its relative DTME; see below) must react with several cysteines in Int, as seen by the ability of these cross-linkers to capture multimers even when C25 is not present (Fig. 4). This difference in reactivity may be due to the hydrophobic tether present in BMH, which may allow these molecules to reach protein interfaces or pockets that are less accessible to NEM.
BMH cross-linking entails both addition of a chemical group to cysteine residues and tethering of Int protomers, and thus the block in recombination may be due to restricting movement within the Int multimer or to modification of cysteines within Int (or both). To separate these two factors, we compared BMH to a second sulfhydryl-reactive cross-linker, DTME; this cross-linker has a disulfide bridge in the middle of a hydrophobic 13.3-Å tether which is cleavable by addition of DTT or β-mercaptoethanol. When the tether is cleaved, the protein remains modified at the position of the original cross-link. The pattern of cross-linked proteins is the same with DTME treatment as with BMH treatment (data not shown). Like BMH, DTME blocked recombination and assembly of the UMC2 complex for both Int and IntC25S (Fig. 5, lanes 6 and 11). Cleaving the tether of DTME after 15 min did not increase the amount of recombination products recovered, and it did not permit assembly of UMC2 (Fig. 5, lanes 5 and 10). Modification of the remaining cysteines in the IntC25S protein must interfere either with protein-protein or with protein-DNA contacts in the C-terminal half of the protein. Because cleaving the DTME tether did not restore normal Int activity, we could not ascertain the necessity of movement during recombination.
DISCUSSION
We have described a series of experiments that begin to localize the regions of Int involved in protein-protein contacts and directly address the nature of the multimers assembled by Int during recombination. Taken together, our results from a genetic protein fusion assay, from biochemical assays of an Int protein modified at the N terminus, and from physical cross-linking assays show that the amino terminus of Int is involved in protein-protein interactions between Int monomers in addition to mediating protein-DNA interactions.
The genetic protein fusion assay identified amino acids 80 through 169 as the minimal region of Int (among the Int segments tested) that confers detectable protein-protein contacts. However, addition of the N-terminal 80 amino acids (Int fragment 1 to 169) improves the activity of the fusion protein, and comparison of the activities of the Int1–262::AraCD and Int80–262::AraCD fusion proteins confirms that the N terminus is involved in protein-protein contacts. Although we have not yet identified mutations in the N-terminal region of Int that affect multimerization, we have found that addition of a six-His tag (total of 20 amino acids) or even three new amino acids (Gly, Ser, and His) at the N terminus markedly impairs cooperative interactions between Int monomers without significantly affecting interactions between Int and DNA. Amino acid residues 169 to 262 also contribute to multimerization properties (compare the activities of fusions Int1–169::AraCD and Int1–262::AraCD). Within this segment of Int, we have identified one mutation, T236I, that decreases the activity of the Int1–262::AraCD protein to background level and decreases cooperative interactions between Int monomers in vitro. This mutation was isolated in a screen for recombination-defective Int proteins, although it remains catalytically active (11, 38). In contrast, a mutation that affects the active site of Int, R212Q, has no effect on the activity of the Int1–262::AraCD protein. These data suggest that protein-protein contacts are spread over at least two-thirds of the Int polypeptide.
Although the amino-terminal domain appears to mediate multimerization properties in vivo and in vitro, it is not essential for multimerization of Int in vitro. The His-tagged protein, although it has lost significant DNA binding cooperativity, can still multimerize as assayed by either DSG or BMH cross-linking (data not shown). It is not surprising that the N-terminal intermonomer contacts can be dispensable, since the fusion assay shows that amino acid residues throughout a large fraction of Int contribute to protein-protein contacts. Differences between the in vivo and in vitro effects of the N terminus on multimerization may be due to less stable folding on the part of the fusion proteins, lower concentration of the protein in vivo, or differences in the ionic environment in vivo versus in vitro. We are currently looking for mutations in the N terminus that affect multimerization. Crystallographic analysis of Int-related proteins shows that large protein-protein interfaces also exist in the catalytic domain (9, 13). Because the catalytic domains of the tyrosine recombinases are the most highly conserved (6, 29), these strong protein-protein interactions probably exist in Int as well (Fig. 2B and L. Jessop, J. Boldt, and A. Segall, unpublished data). (Fig. 2B).
Strong cooperativity at the arm sites is correlated with Int's ability to mediate strong contacts with the core sites and to assemble high levels of synaptic complexes (Fig. 3); both of these properties are necessary to carry out efficient recombination. Although the His-tagged protein can bind to arm sites, this binding is much less cooperative than that of the non-His-tagged protein and results in assembly of fewer Int-DNA complexes. In turn, cooperative interactions between the N termini of Int monomers bound at the arm sites of attL stabilize interactions of the catalytic domain with the core sites, near the loci of strand exchange. When these cooperative interactions are weakened, Int's recombination activity is compromised.
Cross-linking assays with the wild-type Int and IntC25S mutant proteins showed that Int assembles predominantly dimers, “trimers,” and tetramers during the recombination process (Fig. 4). At least one (and perhaps all) of the apparent trimers is in fact a dimer with slower mobility, based on the fact that proteins lacking all but Cys 197 (or all but Cys 262) can form this species (L. Jessop, J. Boldt, and A. Segall, unpublished data). Although multimerization is not triggered by DNA, att sites in particular and nonspecific DNA to a lesser extent do increase the extent of multimerization. The fact that the C25S mutation changes the observed cross-linking pattern—specifically, only one type of dimer and few if any tetramers are trapped with BMH—shows that this residue is found at an interface required to form a subset of Int dimers and that the amino terminus is involved in protein-protein interactions.
The phenotype of cross-linking the wild-type Int at C25 is in some ways similar to that caused by modification of the protein by NEM, which has been reported to occur exclusively at this N-terminal cysteine (41). Indeed, BMH and DTME treatments of wild-type Int strongly inhibit interactions between the arm binding domain of Int and the DNA, and the IntC25S protein becomes immune to the adverse effects of cross-linkers on arm binding (data not shown). Nevertheless, IntC25S is inactivated for recombination by either BMH or DTME; this loss of recombination activity must result from modification of one or more of the remaining three cysteines (C197, C217, and C262). We are currently investigating where this modification occurs and whether it depresses protein-protein and/or protein-DNA interactions.
Our results indicate that cross-linking IntC25S blocks recombination by preventing the assembly of UMC2 (Fig. 5). The UMC2 complex is formed by addition of an unstably bound Int monomer to the UMC1 intermediate (35), and we hypothesize that this Int is held in place by low-affinity protein-DNA interactions at the C′ core site and by protein-protein interactions. It is possible that modification of one or more of the three cysteines in the catalytic domain interferes with protein-protein interactions important for loading this incoming Int. While these interactions may be dispensable for cooperative binding to the arm sites, they might be essential for formation of UMC2 and for recombination. Alternatively, cross-linking may immobilize multimers, preventing rearrangements important for assembly of UMC2. A similar situation, in which tethering protein multimers resulted in destabilized DNA binding, was found in the case of the Hin recombinase (22). We disfavor this interpretation, however, because cleavage of the tether in DTME does not restore UMC2 formation.
Multimerization is an integral part of the mechanism of site-specific recombination (8, 12, 14, 21 [and references therein], 22, 25, 42). The coordination of catalytic events may involve communication between different protomers in the synaptic complex. This appears to be the case for the Cre protein, where the two monomers that have carried out cleavage occupy a similar position with respect to the complex but differ from the two monomers which have not carried out catalysis (9). Catalysis by individual Int protomers is less concerted in bent-L recombination than in excisive or integrative recombination (G. Cassell and A. Segall, unpublished data; 17). This may be due to subtle differences among the pathways in protein-DNA contacts, protein-protein contacts, or both. We are investigating these differences by fine-mapping the interfaces between Int protomers in different pathways of λ recombination. We hope that understanding differences in the conformations of intermediate complexes among the four pathways and the relationship between the geometry of intermediates and catalytic events will lead to an understanding of the molecular events that trigger catalysis. In turn, this will lead us to a more complete understanding of how the efficiency of site-specific recombination is achieved.
ACKNOWLEDGMENTS
Troy Bankhead and David Wong contributed equally to the work presented here.
We are indebted to Malcolm Casadaban for his advice, for providing strains for the AraC fusion assay, and for communicating results prior to publication. Robert Schleif helped us with his advice on AraA assays and on AraC. We thank Jeff Boldt for purification of the wild-type Int and IntC25S proteins and Geoffrey Cassell for his early experiments on the effect of thrombin cleavage on the activity of the His-tagged Int. Finally, we thank Forest Rohwer for comments on the manuscript.
This work was supported by NIH grant GM52847 to A.M.S.
REFERENCES
- 1.Burgin A B, Jr, Nash H A. Suicide substrates reveal properties of the homology-dependent steps during integrative recombination of bacteriophage λ. Curr Biol. 1995;5:1312–1321. doi: 10.1016/s0960-9822(95)00258-2. [DOI] [PubMed] [Google Scholar]
- 2.Bustos S A, Schleif R F. Functional domains of the AraC protein. Proc Natl Acad Sci USA. 1993;90:5638–5643. doi: 10.1073/pnas.90.12.5638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cribbs R, Englesberg E. l-Arabinose negative mutants of the l-ribulosekinase structural gene affecting the levels of l-arabinose isomerase in Escherichia coli. Genetics. 1964;49:95–108. doi: 10.1093/genetics/49.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Davis R W, Botstein D, Roth J R. Advanced bacterial genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1980. [Google Scholar]
- 5.Deiderich L, Roth A, Messer W. A versatile plasmid vector system for the regulated expression of genes in Escherichia coli. BioTechniques. 1994;16:916–923. [PubMed] [Google Scholar]
- 6.Esposito D, Scocca J J. The integrase family of tyrosine recombinases: evolution of a conserved active site domain. Nucleic Acids Res. 1997;25:3605–3614. doi: 10.1093/nar/25.18.3605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gopaul D N, Guo F, Van Duyne G D. Structure of the Holliday junction intermediate in Cre-loxP site-specific recombination. EMBO J. 1998;17:4175–4187. doi: 10.1093/emboj/17.14.4175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grindley N D F. Site-specific recombination: synapsis and strand exchange revealed. Curr Biol. 1998;7:608–612. doi: 10.1016/s0960-9822(06)00314-9. [DOI] [PubMed] [Google Scholar]
- 9.Guo F, Gopaul D N, Van Duyne G D. Structure of Cre recombinase complexed with DNA in a site-specific recombination synapse. Nature. 1997;389:40–46. doi: 10.1038/37925. [DOI] [PubMed] [Google Scholar]
- 10.Han Y W, Gumport R I, Gardner J F. Complementation of bacteriophage lambda integrase mutants: evidence for an intersubunit active site. EMBO J. 1993;12:4577–4584. doi: 10.1002/j.1460-2075.1993.tb06146.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Han Y W, Gumport R I, Gardner J F. Mapping the functional domains of bacteriophage lambda integrase protein. J Mol Biol. 1994;235:908–925. doi: 10.1006/jmbi.1994.1048. [DOI] [PubMed] [Google Scholar]
- 12.Haykinson M J, Johnson L M, Soong J, Johnson R C. The Hin dimer interface is critical for Fis-mediated activation of the catalytic steps of site-specific DNA inversion. Curr Biol. 1996;6:163–177. doi: 10.1016/s0960-9822(02)00449-9. [DOI] [PubMed] [Google Scholar]
- 13.Hickman A B, Waninger S, Scocca J J, Dyda F. Molecular organization in site-specific recombination: the catalytic domain of bacteriophage HP1 integrase at 2.7 Å resolution. Cell. 1997;89:227–237. doi: 10.1016/s0092-8674(00)80202-0. [DOI] [PubMed] [Google Scholar]
- 13a.Hickman A B, Palmer I, Engelman A, Craigie R, Wingfield P. Biophysical and enzymatic properties of the catalytic domain of HIV-1 integrase. J Biol Chem. 1994;269:29279–29287. [PubMed] [Google Scholar]
- 14.Hughes R E, Rice P A, Steitz T A, Grindley N D. Protein-protein interactions directing resolvase site-specific recombination: a structure-function analysis. EMBO J. 1993;12:1447–1458. doi: 10.1002/j.1460-2075.1993.tb05788.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kikuchi Y, Nash H. Nicking-closing activity associated with bacteriophage λ int gene product. Proc Natl Acad Sci USA. 1979;76:3760–3764. doi: 10.1073/pnas.76.8.3760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim S, Moitoso de Vargas L, Nunes-Duby S E, Landy A. Mapping of a higher order protein-DNA complex: two kinds of long-range interactions in λ attL. Cell. 1990;63:773–781. doi: 10.1016/0092-8674(90)90143-3. [DOI] [PubMed] [Google Scholar]
- 17.Kitts P A, Nash H A. An intermediate in the phage site-specific recombination reaction is revealed by phosphorothioate substitution in DNA. Nucleic Acids Res. 1988;16:6839–6856. doi: 10.1093/nar/16.14.6839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kolodrubetz D, Schleif R. Identification of the AraC protein on two-dimensional gels, its in vivo instability and normal level. J Mol Biol. 1981;156:53–66. doi: 10.1016/0022-2836(81)90265-5. [DOI] [PubMed] [Google Scholar]
- 19.Kwon H J, Tirumalai R, Landy A, Ellenberger T. Flexibility in DNA recombination: structure of the lambda integrase catalytic core. Science. 1997;276:126–131. doi: 10.1126/science.276.5309.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Landy A. Dynamic, structural, and regulatory aspects of lambda site-specific recombination. Annu Rev Biochem. 1989;58:913–949. doi: 10.1146/annurev.bi.58.070189.004405. [DOI] [PubMed] [Google Scholar]
- 21.Lee J, Whang I, Jayaram M. Assembly and orientation of Flp recombinase active sites on two-, three- and four-armed DNA substrates: implications for a recombination mechanism. J Mol Biol. 1996;245:219–227. doi: 10.1006/jmbi.1996.0183. [DOI] [PubMed] [Google Scholar]
- 22.Lim H M. Analysis of subunit interaction by introducing disulfide bonds at the dimerization domain of Hin recombinase. J Biol Chem. 1994;269:31134–31142. [PubMed] [Google Scholar]
- 23.Menon K P, Lee N C. Activation of ara operons by a truncated AraC protein does not require inducer. Proc Natl Acad Sci USA. 1990;87:3708–3712. doi: 10.1073/pnas.87.10.3708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moitoso de Vargas L, Pargellis C A, Hasan N M, Bushman E W, Landy A. Autonomous DNA binding domains of λ integrase recognize two different sequence families. Cell. 1988;54:923–929. doi: 10.1016/0092-8674(88)90107-9. [DOI] [PubMed] [Google Scholar]
- 25.Murley L L, Grindley N D. Architecture of the gamma delta resolvase synaptosome: oriented heterodimers identify interactions essential for synapsis and recombination. Cell. 1998;95:553–562. doi: 10.1016/s0092-8674(00)81622-0. [DOI] [PubMed] [Google Scholar]
- 26.Nagarajah R, Weisberg R A. Specificity determinants in the attachment sites of bacteriophages HK022 and λ. J Bacteriol. 1990;172:6540–6550. doi: 10.1128/jb.172.11.6540-6550.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nash H A. Site-specific recombination: integration, excision, resolution, and inversion of defined DNA segments. In: Neidhardt F C, et al., editors. (ed., Escherichia coli and Salmonella: cellular and molecular biology. Washington, D.C.: ASM Press; 1996. pp. 2363–2376. [Google Scholar]
- 28.Numrych T E, Gumport R I, Gardner J F. A comparison of the effects of single-base and triple-base changes in the integrase arm-type binding sites on the site-specific recombination of bacteriophage lambda. Nucleic Acids Res. 1990;18:3953–3959. doi: 10.1093/nar/18.13.3953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nunes-Duby S E, Kwon H J, Tirumalai R S, Ellenberger T, Landy A. Similarities and differences among 105 members of the Int family of site-specific recombinases. Nucleic Acids Res. 1998;26:391–406. doi: 10.1093/nar/26.2.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pargellis C A, Nunes-Duby S E, Moitoso de Vargas L, Landy A. Suicide recombination substrates yield covalent lambda integrase-DNA complexes and lead to identification of the active site tyrosine. J Biol Chem. 1988;263:7678–7685. [PubMed] [Google Scholar]
- 31.Richet E, Abcarian P, Nash H A. Synapsis of attachment sites during lambda integrative recombination involves capture of a naked DNA by a protein-DNA complex. Cell. 1988;52:9–17. doi: 10.1016/0092-8674(88)90526-0. [DOI] [PubMed] [Google Scholar]
- 32.Ross W, Landy A. Bacteriophage λ int protein recognizes two classes of sequence in the phage att site: characterization of arm-type sites. Proc Natl Acad Sci USA. 1982;79:7724–7728. doi: 10.1073/pnas.79.24.7724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schleif R F. Enzyme assays. In: Schleif R F, Wensink P, editors. Practical methods in molecular biology. New York, N.Y: Springer-Verlag; 1981. pp. 46–50. [Google Scholar]
- 34.Schleif R F. Two positively regulated systems, ara and mal. In: Neidhardt F C, et al., editors. (ed., Escherichia coli and Salmonella: cellular and molecular biology. Washington, D.C.: ASM Press; 1996. pp. 1300–1309. [Google Scholar]
- 35.Segall A M. Analysis of higher-order intermediates and synapsis in the bent-L pathway of bacteriophage λ site-specific recombination. J Biol Chem. 1998;273:24258–24265. doi: 10.1074/jbc.273.37.24258. [DOI] [PubMed] [Google Scholar]
- 36.Segall A M, Nash H A. Synaptic intermediates in bacteriophage lambda site-specific recombination: integrase can align pairs of attachment sites. EMBO J. 1993;12:4567–4576. doi: 10.1002/j.1460-2075.1993.tb06145.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Segall A M, Goodman S D, Nash H A. Architectural elements in nucleoprotein complexes: interchangeability of specific and nonspecific DNA binding proteins. EMBO J. 1994;13:4536–4548. doi: 10.1002/j.1460-2075.1994.tb06775.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Segall A M, Nash H A. Architectural flexibility in lambda site-specific recombination: three alternate conformations channel the attL site into three distinct pathways. Genes Cells. 1996;1:453–463. doi: 10.1046/j.1365-2443.1996.d01-254.x. [DOI] [PubMed] [Google Scholar]
- 39.Subramanya H S, Arciszewska L K, Baker R A, Bird L E, Sherratt D J, Wigley D B. Crystal structure of the site-specific recombinase, XerD. EMBO J. 1997;16:5178–5187. doi: 10.1093/emboj/16.17.5178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tirumalai R S, Healey E, Landy A. The catalytic domain of λ site-specific recombinase. Proc Natl Acad Sci USA. 1997;94:6104–6109. doi: 10.1073/pnas.94.12.6104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tirumalai R S, Pargellis C A, Landy A. Identification and characterization of the N-ethylmaleimide-sensitive site in λ integrase. J Biol Chem. 1996;271:29599–29604. doi: 10.1074/jbc.271.47.29599. [DOI] [PubMed] [Google Scholar]
- 42.Yang W, Mizuuchi K. Site-specific recombination in plane view. Structure. 1998;5:1401–1406. doi: 10.1016/s0969-2126(97)00290-6. [DOI] [PubMed] [Google Scholar]