Abstract
Staphylococcus epidermidis ATCC 14990 produces a wall-associated glycerol teichoic acid which is chemically identical to the major wall-associated teichoic acid of Bacillus subtilis 168. The S. epidermidis tagF gene was cloned from genomic DNA and sequenced. When introduced on a plasmid vector into B. subtilis 1A486 carrying the conditionally lethal temperature-sensitive mutation tagF1 (rodC1), it expressed an 85-kDa protein which allowed colonies to grow at the restrictive temperature. This showed that the cloned S. epidermidis gene encodes a functional CDP-glycerol:poly(glycerophosphate) glycerophosphotransferase. An amino acid substitution at residue 616 in the recombinant TagF protein eliminated complementation. Unlike B. subtilis, where the tagF gene is part of the tagDEF operon, the tagF gene of S. epidermidis is not linked to any other tag genes. We attempted to disrupt the chromosomal tagF gene in S. epidermidis TU3298 by directed integration of a temperature-sensitive plasmid but this failed, whereas a control plasmid containing the 5′ end of tagF on a similarly sized DNA fragment was able to integrate. This suggests that the tagF gene is essential and that the TagF and other enzymes involved in teichoic acid biosynthesis could be targets for new antistaphylococcal drugs.
Wall-associated teichoic acids are a heterogeneous class of phosphate-rich polymers that are covalently linked to the cell wall peptidoglycan of gram-positive bacteria (2). They consist of a main chain of phosphodiester-linked polyols and/or sugar moieties attached to peptidoglycan via a linkage unit (1). Glycerol and ribitol are the most commonly occurring polyols and are often substituted with d-alanine or various sugar residues. Glycosylated glycerol teichoic acids are present in coagulase-negative staphylococci (13, 14), while glycosylated ribitol teichoic acids have been found in Staphylococcus aureus and Staphylococcus saprophyticus (13). Teichoic acids containing glycosylpolyol phosphates or sugar phosphates alone as components of their main chains occur in Streptococcus pneumoniae (40) and some species of staphylococci (3), respectively. The physiological function of teichoic acids is still not clear. However, they have been implicated in the control of autolysin activity (21), cation assimilation (7, 12, 22), and the provision of a phosphate reserve (15).
Thus far, all of the studies concerning the genetics of teichoic acid biosynthesis have been performed in B. subtilis, particularly B. subtilis 168. The major wall teichoic acid of B. subtilis 168 is poly(glycerophosphate) [poly(groP)], and the genes involved in the biosynthesis and translocation (tag genes) are organized into three operons: tagAB, tagDEF, and tagGH (30).
There is substantial evidence to support the conclusion that the poly(groP) teichoic acid is essential for cell viability. First of all, there is the isolation of conditionally lethal temperature-sensitive mutants defective in the synthesis of poly(groP). Such tag mutants exhibit a reduction in growth rate and a pronounced disturbance in cell morphology at the nonpermissive temperature (8, 37, 38). Second, there is the failure to disrupt the tagAB, tagDEF, and tagGH operons by insertion mutagenesis (29, 31, 32). Finally, there is the controlled reduction in the expression of the tagGH operon, which resulted in a rod-to-sphere transition in cell morphology characteristic of the conditional lethal tag mutants grown under nonpermissive conditions (29). Several reports indicate that wall teichoic acid also plays an important role in cell wall integrity of gram-positive cocci (9, 23, 35), but there have been no genetic studies to determine whether these polymers are essential for survival.
Here we report the cloning and sequencing of the S. epidermidis tagF gene and show that it can complement the temperature-sensitive tagF1 (rodC1) mutation of B. subtilis 1A486. tagF encodes the CDP-glycerol:poly(groP) glycerophosphotransferase, which is responsible for the polymerization of the main chain of the teichoic acid by sequential transfer of glycerol-phosphate units from CDP-glycerol to the linkage unit lipid (38). We also present evidence which suggests that the tagF gene of S. epidermidis is essential.
MATERIALS AND METHODS
Bacterial strains and plasmids.
Bacterial strains and plasmids used in this study are listed in Tables 1 and 2, respectively.
TABLE 1.
Bacterial strains used in this study
| Strain | Relevant genotype | Properties | Source or reference |
|---|---|---|---|
| B. subtilis | |||
| 1A486 | leuB8 tagF1 (rodC1) | Mutant of 168 carrying a point mutation in the tagF gene, rendering the strain temperature sensitive | The Bacillus Genetic Stock Center, Ohio State University |
| BSS30 | leuB8 tagF1 CmramyEΩpDG268 | 1A486 with pDG268 integrated into the amyE locus, temperature sensitive | This study |
| BSS40 | leuB8 tagF1 CmramyEΩpSC2 | 1A486 with pSC2 integrated into the amyE locus, which allows growth at 43°C | This study |
| S. aureus RN4220 | tarF+ | Mutant of 8325-4 capable of stably maintaining recombinant plasmids | 27 |
| S. epidermidis | |||
| ATCC 14990 | tagF+ | Produces a glycerol teichoic acid chemically identical to that synthesized by B. subtilis 168 | SmithKline Beecham |
| TU3298 | tagF+ | Capable of being transformed with, and stably maintaining, recombinant plasmids | 4 |
TABLE 2.
Plasmids used in this studya
| Host strain and plasmid | Relevant markers and/or phenotype | Properties | Source or reference |
|---|---|---|---|
| E. coli | |||
| pGEM-7Z(f)+ | Apr | Cloning vector | Promega |
| pGN23 | Apr | 2.3-kb NsiI chromosomal fragment of the S. epidermidis tagF locus in pGEM-7Z(f)+ | This study |
| pGN333 | Apr | 333-bp NsiI chromosomal fragment of S. epidermidis tagF in pGEM-7Z(f)+ | This study |
| pGDH3 | Apr | 1.56-kb ClaI-NsiI internal fragment of the S. epidermidis tagF ORF in pGEM-7Z(f)+ | This study |
| pBluescript | Apr | Cloning vector | Stratagene |
| pBN23 | Apr | 2.3-kb insert from pGN23 in pBluescript | This study |
| pBH17 | Apr | 1.77-kb HindIII fragment from pBCP55 in pBluescript | This study |
| pBCP55 | Apr | 5.5-kb ClaI-PstI chromosomal fragment from S. epidermidis with the 3′ region of tagF and adjacent downstream sequence in pBluescript | This study |
| pBSKT13 | Apr | 2.35-kb HindIII Tcr fragment from pT181 in pBluescript | This study |
| pGEX-KG | Apr | GST fusion-protein expression vector | 16 |
| pGEX-2 | Apr | GST-′TagF fusion protein expression vector | This study |
| B. subtilis and/or E. coli | |||
| pHPS9 | Cmr | Shuttle vector for cloning in E. coli and B. subtilis | 17 |
| pFC10 | Cmr | 3.26-kb S. epidermidis tagF locus in pHPS9 | This study |
| pFCTH3 | Cmr | 3.26-kb tagF locus with a 2-bp mutation in the tagF ORF, in pHPS9 | This study |
| pDG268 | Cmr Apr | Integrational plasmid for complementation analysis in B. subtilis | P. Stragier, Institute de biologie physicochemique, Paris, France |
| pSC5 | Cmr Apr | 3.26-kb S. epidermidis tagF locus in pDG268 | This study |
| S. aureus, pT181 | Cmr Tcr | Carries a 2.35-kb HindIII Tcr fragment | 25 |
| S. aureus and/or S. epidermidis | |||
| pTS2 | Cmr, ts rep | Derived from pTV1ts | This laboratory, MSC from pBluescript |
| pTS2T | Cmr Tcr, ts rep | pTS2 containing the 2.35-kb HindIII Tcr fragment from pBSKT13 | This study |
| pTSTAG | Cmr Tcr, ts rep | 1.56-kb internal tagF fragment from pGDH3 cloned in the MCS of pTS2T | This study |
| pTSH17 | Cmr Tcr, ts rep | 1.77-kb fragment from pBH17, cloned in the MCS of pTS2T | This study |
MSC, multiple cloning site; ts rep, temperature-sensitive replication system.
Bacterial growth media and antibiotics.
The media used for culture of both Escherichia coli and B. subtilis strains was L broth (LB) or L agar (LA), supplemented, when required, with ampicillin (Ap) (100 μg ml−1) or chloramphenicol (Cm) (5 μg ml−1). S. aureus and S. epidermidis strains were grown in Trypticase soy broth (TSB) or on Trypticase soy agar (TSA) containing, when appropriate, Cm (5 μg ml−1) or tetracycline (Tc) (8 μg ml−1).
Manipulation of DNA.
Standard procedures were used for DNA manipulation (5, 42). DNA modifying enzymes were purchased from New England Biolabs and Promega.
Degenerate oligonucleotide PCR.
Degenerate oligonucleotide primers were designed by back-translation of the amino acid sequences ILYAPT (TP3 primer) and ITDYSSV (TP4 primer) shared between the aligned sequences of the TagB and TagF proteins of B. subtilis (31). The sequences of the TP3 and TP4 primers are 5′-AGCGAATTCATHYTNTAYGCNCCNAC-3′ and 5′-AGCGAATTCACNSWNSWRTARTCNGTDAT-3′, respectively (sequences incorporated into the primers for cloning of PCR fragments are underlined). The codes for degenerate positions are as follows: R, A+G; Y, C+T; S, G+C; W, A+T; H, A+T+C; D, G+A+T; and N, A+G+C+T. Reaction mixtures contained 100 ng of chromosomal DNA from S. epidermidis ATCC 14990, 2 μM concentrations of each oligonucleotide primer, 1.5 mM MgCl2 and 2.5 U of Taq polymerase) in a final volume of 100 μl. Thermal cycling parameters began with an initial denaturing step at 94°C for 4 min, followed by 30 cycles at 94°C for 1 min, 40°C for 2 min, and 72°C for 1 min.
Southern hybridization.
Transfer of DNA from agarose to Magna NT nytran membranes (Micron Separations, Inc.) and Southern hybridization with 32P-labeled probes were performed by standard procedures (5). Probes were prepared by random primer labeling of purified DNA fragments with [α-32P]dATP by using the Prime-A-Gene kit (Promega). Autoradiography was performed by using X-Omat S film (Eastman Kodak Co.).
Colony hybridization.
Colony hybridization was performed by the method of Hanahan and Meselson (18).
DNA sequencing and analysis.
Progressive unidirectional deletions of the tagF locus were constructed by using the Erase-a-Base Kit (Promega). Nested deletions were made in both directions in two overlapping clones, pBN23 and pBH17, which spanned the tagF locus. The sequencing reactions were carried out by the cycle sequencing method with the Flash Dye Primer Sequencing Kit (Genpak) and analyzed on a model 373A sequencer (Applied Biosystems). Homology searches were performed by using the various BLAST algorithms available at the National Center for Biotechnology Information (NCBI) site. Protein sequence alignments were performed by using the subprogram PALIGN of PC/GENE (Intelligenetics) and the CLUSTAL W algorithm accessible at the Baylor College of Medicine Human Genome Sequencing Center web site.
Transformation.
S. aureus and S. epidermidis cells were transformed by electroporation by the procedures of Oskouian and Stewart (33) and Augustin and Götz (4), respectively. Electroporation was performed with a Bio-Rad Gene Pulser equipped with a pulse controller (Bio-Rad Laboratories). Competent E. coli XL1-Blue (Stratagene) and B. subtilis cells were prepared and transformed by the procedures of Chung and Miller (10) and Karamata and Gross (24), respectively.
Construction of a site-directed mutation in the S. epidermidis tagF gene.
A 436-bp fragment was amplified by PCR from pFC10 template DNA by using the oligonucleotide primers 5′-ATAATGACGTCTCTGAATTATTTTTAATAttTGATTGTTTAATTAC-3′ (forward) and 5′-AATTTCAATTAAAATATTAAAAAGGGC-3′ (reverse), as well as VENT DNA polymerase. The forward primer contained altered bases (lowercase lettering) and an AatII restriction site close to the 5′ terminus (underlined).
The 436-bp PCR product was purified from an agarose gel by using the Wizard PCR Purification System (Promega) and blunt-end ligated into pBluescript, which had been digested with EcoRV, to form plasmid pBMA3. pBMA3 was subsequently digested with EcoRV and AatII to release a ca. 330-bp fragment that contained the altered bases and ligated with pFC10, which had also been digested with EcoRV and AatII to release the wild-type ca. 330-bp fragment. The 2-bp change in the mutant tagF gene created an SspI restriction site not present in the wild-type allele. Hence, the plasmid carrying the mutated gene (pFCTH3) could be distinguished from pFC10 by differences in the electrophoretic profile of restriction fragments produced by each construct following digestion with SspI. The mutated region of the tagF gene present in pFCTH3 was sequenced to ensure it was correct.
Overexpression and purification of the GST-TagF fusion protein.
In order to overexpress and purify a region of the S. epidermidis TagF protein, primers 5′-TACGGATCCAAAGTTAATCAATTTAG-3′ (GF1) and 5′-TACAAGCTTTTATCATTGTTCCTTG-3′ (GF2) were used to PCR amplify the region of the tagF gene encoding the 407 carboxyl-terminal amino acids of TagF and the two termination codons. Plasmid pFC10 was used as template DNA with VENT DNA polymerase and 30 temperature cycles of 94°C for 1 min, 55°C for 1 min, and 72°C for 2 min 30 s. Primers GF1 and GF2 have 9-bp extensions at their 5′ ends, which include restriction sites (underlined) to facilitate cloning. The PCR product was purified with the Wizard PCR Purification System (Promega), digested with HindIII and BamHI and ligated in frame with the glutathione S-transferase (GST) coding sequence of pGEX-KG to form pGEX-2 (GST-′tagF). pGEX-2 expressed a fusion protein in E. coli XL1-Blue of the predicted molecular mass following induction with isopropyl-β-d-thiogalactoside (IPTG).
The GST-′TagF fusion protein was expressed from a 200-ml culture of exponentially growing (A600 between 1 and 2) E. coli XL1-Blue by the addition of IPTG to a concentration of 100 μM and incubation at 37°C at 250 rpm for 4 h. Cells were harvested by centrifugation at 10,000 × g for 10 min and resuspended in 20 ml of ice-cold phosphate-buffered saline (PBS; Oxoid) containing DNase I (40 μg ml−1), RNase A (40 μg ml−1), and 2 mM phenylmethylsulfonyl fluoride. Cells were lysed by passage through a French press. Cell debris was removed by centrifugation at 30,000 × g for 10 min. The fusion protein was purified from the supernatant by batch affinity chromatography by using the Bulk GST Purification Module (Pharmacia) according to the manufacturer's instructions. Purified fusion protein was analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE).
Antibody generation and purification.
Next, 500-μl samples of the purified GST-′TagF fusion protein (60 μg of protein ml−1) were emulsified in equal volumes of Freund complete adjuvant and injected subcutaneously into two New Zealand White rabbits, from which preimmune serum had previously been taken. Following two booster injections of 20 μg of fusion protein emulsified in Freund incomplete adjuvant, at 14-day intervals, the rabbits were sacrificed and bled out. Antibodies were purified from the serum by the procedure described by Owen (34).
SDS-PAGE and Western immunoblotting.
SDS-PAGE was performed by standard procedures (28). The stacking and separating gels consisted of 4.5% (wt/vol) and 10% (wt/vol) 19:1 acrylamide-bisacrylamide, respectively. Following SDS-PAGE, gels were stained with Coomassie blue stain or transferred to nitrocellulose membranes (Millipore) by using a semidry blotter (Bio-Rad Transblot SD). Membranes were blocked overnight at 4°C in PBS containing 5% (wt/vol) skimmed milk (Marvel). After incubation with anti-GST-′TagF antibodies (1:2,000 in 5% skimmed milk) followed by incubation with horseradish peroxidase-conjugated anti-rabbit immunoglobulin G antibodies (Sigma) (1:1,000 in 5% skimmed milk), the TagF mutant and wild-type proteins were detected by using the enhanced chemiluminescence Western blotting reagent kit (Amersham) according to the manufacturer's instructions.
Detection of plasmid integration by PCR.
Oligonucleotide primers, TQ1 and TQ2, were designed to detect chromosomal integration by homologous recombination of plasmids pTSTAG and pTSH17. The nucleotide sequences of primers TQ1 and TQ2 are 5′-CGTTTAAGTGCTAAAGAAGTTGTAGG-3′ and 5′-GGAAATACAACGCATTTAC-3′, respectively. Primer HD1 hybridizes at a region 67 bp downstream from the last codon of tagF and was used in combination with primer TQ2 as a positive control. The nucleotide sequence of primer HD1 is 5′-AATTTCAATTAAAATATTAAAAAG-3′.
Efficiency of plating.
Strains of S. epidermidis TU3298 carrying the plasmids pTS2T, pTSH17, and pTSTAG were grown overnight at 30°C in TSB with Cm selection (5 μg ml−1) and then plated on TSA containing Tc (8 μg ml−1) at 45°C (restrictive temperature) and 30°C (permissive temperature). The efficiency of plating was determined as the proportion of colonies growing at 45°C compared to that growing at 30°C. At the restrictive temperature the plates needed to be incubated for 36 to 40 h before colonies were visible.
Nucleotide sequence accession numbers.
The GenBank accession numbers of the S. epidermidis tagF locus and the partial sequence of the lctP gene are AF162863 and AF162862, respectively.
RESULTS AND DISCUSSION
Identification of the S. epidermidis tagF gene.
S. epidermidis ATCC 14990 produces a wall-associated glycerol teichoic acid which is chemically identical to the major wall-associated teichoic acid produced by B. subtilis 168 (J. Lonsdale [SmithKline Beecham], personal communication). It is reasonable to assume that the biosynthetic pathway leading to the production of this teichoic acid is identical or very similar in both organisms. For this reason, strain ATCC 14990 was chosen for isolating genes involved in glycerol teichoic acid synthesis. Using degenerate primers corresponding to shared amino acid sequences in the B. subtilis TagB and TagF proteins, genomic DNA from S. epidermidis strain ATCC 14990 was amplified by PCR, and a 260-bp PCR product was obtained. Sequence analysis of translated open reading frames (ORFs) revealed significant similarity with parts of the ORFs of both the TagF protein (P = 6.4 × e−31) and to a lesser extent the TagB protein (P = 8.4 × e−8) of B. subtilis 168. It was therefore thought likely that this fragment represented part of the S. epidermidis tagF gene.
Cloning the S. epidermidis tagF gene.
The S. epidermidis tagF PCR product contains a single NsiI restriction site. When genomic DNA from S. epidermidis ATCC 14990 was cut with NsiI and hybridized with the 32P-labeled tagF PCR product, two reactive bands appeared. The larger was ca. 2.3 kb in length, and the smaller was ca. 330 bp in length. Both fragments were isolated from a plasmid gene bank which had been constructed in pBluescript from ATCC 14990 genomic DNA cut to completion with NsiI. Reactive clones were identified by colony hybridization, and plasmids pGN23 and pGN333 containing the 2.3-kb NsiI fragment and the 330-bp NsiI fragment, respectively, were isolated.
Mapping the tagF gene.
The orientation of the two NsiI fragments was determined by sequencing both ends of each fragment. The fragment cloned in pGN333 was 333 bp long and lay downstream from the 2.3-kb fragment, relative to the direction of tagF transcription. However, pGN333 did not contain the 3′ end of the gene. The fragment in pGN23 contained the 5′ part of tagF and was contiguous to the fragment in pGN333 (Fig. 1).
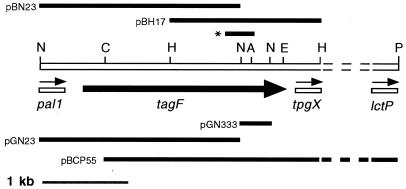
FIG. 1.
Physical map of the S. epidermidis chromosomal region containing the tagF locus. The tagF gene is represented by the thick black arrow. Sequence analysis of the 3′ end of the 5.5-kb fragment in pBCP55 revealed the presence of an ORF which shares significant homology (P = 4 × e−23) with the E. coli lctP gene encoding l-lactate permease (11). The partially sequenced genes pal1, tpgX, and lctP are represented by unshaded bars; the thin arrows above indicate the direction of their transcription. Regions of DNA cloned into pGN333, pGN23, pBCP55, pGDH3, pBH17, and pBN23 are represented by black bars. The position of the 260-bp tagF PCR product generated with the TP3 and TP4 primers is indicated with an asterisk. Reference restriction sites are indicated as follows: P, PstI; N, NsiI; H, HindIII; E, EcoRV; C, ClaI; A, AatII. Dotted lines mark the unsequenced region, which is not drawn to scale.
Sequence analysis of the fragment in pGN23 revealed the 3′ part of an ORF which had significant similarity (P = 5.2 × e−22) with the B. subtilis pai1 gene product (data not shown), a DNA-binding protein involved in transcription regulation (20). This S. epidermidis gene will be referred to as pal1 (the “pai one-like” gene). In B. subtilis 168 the tagE gene is located upstream of tagF within an operon comprised of the tagDEF genes (19, 31). Therefore, the organization of the tag genes is fundamentally different in these two organisms.
In order to obtain the entire tagF gene, a 5.5-kb fragment of chromosomal DNA which overlaps the two NsiI fragments and stretches past the 3′ end (of the sense strand) of the 333-bp fragment, was cloned in plasmid pBCP55. pBCP55 was identified by colony hybridization and isolated from a gene bank constructed in pBluescript from ATCC 14990 genomic DNA cut to completion with both ClaI and PstI. The 5.5-kb ClaI-PstI fragment contains the entire 333-bp NsiI fragment and about 1.5 kb of the 3′ end of the 2.3-kb NsiI fragment. Therefore, plasmids pGN23 and pBCP55 contain overlapping fragments of the entire tagF gene, the 3′ end of the pal1 gene and about 3.5 kb of DNA downstream of the 3′ end of tagF, respectively (Fig. 1).
Sequence analysis of the tagF locus.
Sequence analysis of the 3,263-bp region of DNA encompassed by the overlapping clones in pBH17 and pBN23 revealed the entire tagF gene, the 3′ end of the pal1 gene and the 5′ end of a third gene, which will be referred to as tpgX. The tagF ORF is 2,163 bp long and encodes a predicted protein of 721 amino acids. The distance between the 3′ end of the pal1 ORF and the 5′ end of the tagF ORF is 276 bp. The distance between the 3′ end of the tagF ORF and the first codon of the tpgX gene is 132 bp. The partial sequence of the tpgX ORF encodes the amino-terminal 140 amino acids of the predicted protein product and shares no significant similarity with any GenBank database sequences. The partial 5′ sequence of the pal1 gene encodes the carboxyl-terminal 91 amino acids of the predicted Pal1 protein. All three genes are transcribed in the same direction (Fig. 1). Twenty-four bases downstream from the UAA termination codon of tagF is a 37-base sequence capable of forming a potential hairpin loop structure in mRNA with a ΔG value of −21.4 kcal mol−1. Immediately following this hairpin loop is a 12-base sequence rich in rU residues (data not shown).
Analysis of the S. epidermidis tagF gene product.
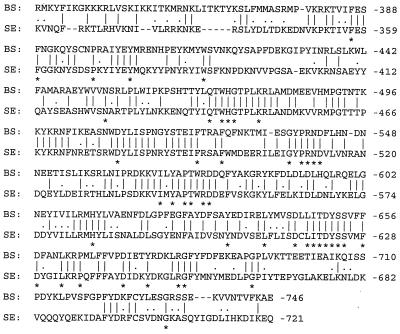
The protein encoded by the tagF gene from B. subtilis is 746 amino acids in length, which is 25 residues larger than the predicted product from the S. epidermidis TagF protein. Alignment of the two protein sequences shows 32.7% identity over their entire lengths. However, the carboxyl-terminal 410 amino acids of the proteins show higher levels of identity (45.9%; Fig. 2), while the remaining amino-terminal alignment possesses only 15.3% identity. The carboxyl-terminal region of the S. epidermidis TagF protein also shows sequence similarities with several other proteins (see legend to Fig. 2). It is reasonable to suggest that the more conserved C-terminal domain contains the catalytic activity and that conserved residues identified in Fig. 2 could be involved in catalyzing phosphodiester bonds during polymerization of polyol phosphate compounds. Honeyman and Stewart (19) suggested that the TagF protein in B. subtilis is cytoplasmic, and the same suggestion can be made for the TagF protein of S. epidermidis. However, this does not preclude association with the membrane by interaction with peripheral or integral membrane proteins involved with teichoic acid synthesis or translocation, for example, the TagG and TagH proteins (29).
FIG. 2.
Amino acid sequence alignment of the COOH-terminal regions of the TagF proteins of B. subtilis (BS) and S. epidermidis (SE). Identical amino acids are marked by bars, and conservative substitutions are indicated with dots. The BLASTP algorithm was used to conduct homology searches of the protein databases available at the NCBI site. Including the B. subtilis TagF, six gene products show sequence alignment scores of greater than or equal to 80 with the COOH-terminal region of the S. epidermidis TagF protein. In order of greatest similarity, the names, species of origin, accession numbers, and references for these gene products are as follows: TagF, B. subtilis, P13485 (19); Cps23fK, S. pneumoniae, AAC69534 (41); TasA, S. pneumoniae, CAA59773 (26); TagB, B. subtilis, P27621 (31); teichoic acid biosynthesis protein RodC related protein, M. thermoautotrophicum, AAB84867 (43); and hypothetical protein 3, H. influenzae, S49240 (44). Amino acid sequence alignment of these gene products with the S. epidermidis TagF protein was performed by using the CLUSTAL W algorithm. The asterisks correspond to amino acid residues common to the S. epidermidis and B. subtilis TagF proteins and at least three of the other five gene products examined.
Complementation of the tagF1 mutation in B. subtilis 1A486 with the S. epidermidis tagF gene.
B. subtilis 1A486 (tagF1) was transformed with the vector pHPS9 as a control and with the pHPS9-tagF+ plasmid pFC10. Thus, strain 1A486(pFC10) contained a chromosomally located mutant copy of the B. subtilis tagF gene and multiple copies of the pFC10-located S. epidermidis tagF gene. In contrast, strain 1A486(pHPS9) contained only the chromosomally located mutant B. subtilis tagF gene.
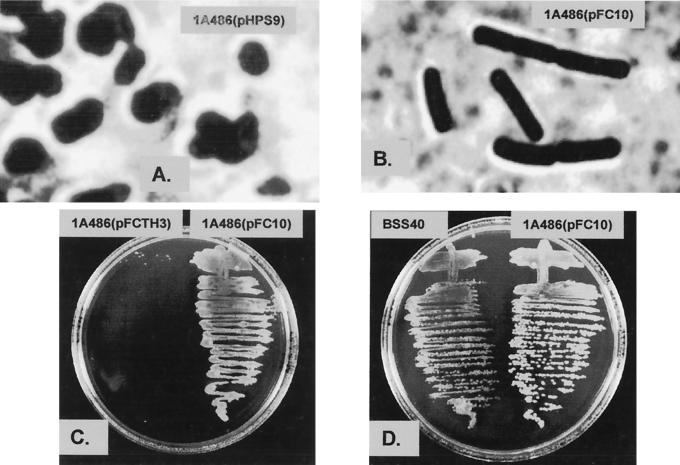
Strain 1A486(pHPS9) and strain 1A486(pFC10) grew normally at 30°C on agar, and examination of both strains under the light microscope revealed the normal rod-shaped morphology of wild-type B. subtilis cells. After incubation at 42°C for 16 h, strain 1A486(pHPS9) produced small areas of very limited growth (data not shown). Microscopic examination of material from these areas revealed the irregular coccoidal cell morphology characteristic of the TagF1 phenotype under restrictive growth conditions (Fig. 3A). Incubation of strain 1A486(pFC10) at 42°C for 16 h resulted in the growth of colonies, and microscopic examination revealed the rod-shaped morphology characteristic of wild-type B. subtilis cells (Fig. 3B). Therefore, the presence of the S. epidermidis tagF gene in pFC10 resulted in the complementation of the tagF1 mutation in strain 1A486(pFC10). This indicates that the gene we have cloned encodes a functional CGPTase. If TagF forms part of a multienzyme complex (6) the ability of the S. epidermidis protein to replace the defective TagF protein in B. subtilis 1A486, with which it has only 33% residue identity, suggests that the interactions are not too extensive.
FIG. 3.
Complementation analysis of B. subtilis 1A486 with the S. epidermidis tagF gene. Strains were grown for 16 h at 42°C on LA plates containing chloramphenicol (5 μg ml−1). (A) Photomicrograph of strain 1A486(pHPS9) exhibiting the coccoidal morphology characteristic of the TagF1 (RodC1) phenotype. (B) Photomicrograph of strain 1A486(pFC10) showing the restoration of the wild-type rod-shaped morphology by complementation with the S. epidermidis tagF gene. (C) Comparison of growth of strains 1A486(pFCTH3) and 1A486(pFC10). (D) Comparison of growth of strains BSS40 and 1A486(pFC10).
Transformation of B. subtilis 1A486 with a mutated S. epidermidis tagF gene.
A site-directed mutation in the S. epidermidis tagF gene was constructed. The nucleotide sequence of the tagF gene in pFCTH3 is identical to that of pFC10, except for a 2-bp mutation at positions 1846 and 1847 of the ORF. The mutation results in a single amino acid change, from serine to phenylalanine, in the TagF protein. This amino acid substitution is analogous to the tagF1 mutation in B. subtilis 1A486 which is responsible for the temperature-sensitive phenotype (19, 36, 38).
B. subtilis 1A486 was transformed with pFCTH3. Strain 1A486(pFCTH3) was incubated at 42°C for 16 h on agar. Growth was very poor and was limited to small patches, similar to that exhibited by strain 1A486(pHPS9) under the same conditions (Fig. 3C). Microscopic examination of material from one of these patches revealed the coccoidal cell morphology characteristic of the TagF1 phenotype at the restrictive temperature (data not shown). Thus, the mutation in the tagF gene present in pFCTH3 abolished complementation of the tagF1 mutation in strain 1A486(pFCTH3) and confirms the finding described above that the gene we describe is involved in S. epidermidis wall teichoic acid biosynthesis.
Integration of a single copy of the S. epidermidis tagF gene into the amyE locus of B. subtilis 1A486.
It was of interest to determine if a single copy of the S. epidermidis tagF gene, integrated into the chromosome of B. subtilis 1A486, would also complement the tagF1 mutation. For this purpose plasmid pSC5, which harbored S. epidermidis tagF on a 3.26-kb fragment, was linearized before being transformed into strain 1A486. A transformant possessing the desired Amy− Cmr phenotype was isolated and named BSS40. The vector pDG268 was also transformed into strain 1A486 as a control to form strain BSS30. Thus, strain BSS40 contained a single copy of the S. epidermidis tagF gene and a single copy of the cat gene integrated into the chromosomal amy locus, whereas strain BSS30 contained only a single copy of the cat gene integrated into this locus.
At the restrictive temperature of 42°C, no complementation of the tagF1 mutation was observed with strain BSS30. Strain BSS40 was able to grow and form small colonies, but it did not grow as well as strain 1A486(pFC10) at this temperature (Fig. 3D). Microscopic examination of strain BSS40 grown at 42°C revealed an oblate cell morphology intermediate between wild-type rods and the mutant coccoidal form (data not shown). Thus, the variation in the degree of complementation of the tagF1 mutation shown by strains 1A486(pFC10) and BSS40 is probably due to the difference in copy number, and hence the levels of the S. epidermidis TagF protein present.
Western immunoblotting analysis of B. subtilis 1A486 expressing the wild-type and mutant S. epidermidis TagF proteins.
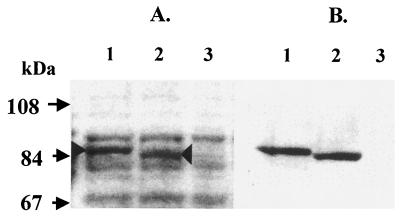
A protein band with an apparent molecular weight of 85 kDa was observed by SDS-PAGE analysis of a whole-cell lysate of B. subtilis 1A486(pFC10). The 85-kDa protein band corresponded to the size of the predicted TagF protein of S. epidermidis (85851 Da). This protein was present in relatively large amounts in the lysate of strain 1A486(pFC10) (Fig. 4A, lane 1). A whole-cell lysate of B. subtilis 1A486(pFCTH3) revealed the presence of a band that migrated slightly faster than that in 1A486(pFC10). This protein was present in relatively large amounts (Fig. 4A, lane 2). Both of these bands were absent from the lysate of B. subtilis 1A486(pHPS9) (Fig. 4A, lane 3). Western immunoblotting with anti-GST-′TagF antibody (Fig. 4B) indicates that the 85-kDa protein is S. epidermidis TagF.
FIG. 4.
SDS-PAGE and Western immunoblot analysis of whole-cell lysates of B. subtilis strains 1A486(pFC10), 1A486(pFCTH3), and 1A486(pHPS9). (A) Whole-cell lysates of B. subtilis strains 1A486(pFC10), 1A486(pFCTH3), and 1A486(pHPS9), separated by SDS-PAGE and stained with Coomassie blue stain, are shown in lanes 1, 2, and 3, respectively. A protein band of approximately 85 kDa (indicated by the arrowhead), present in lane 1 but absent from lanes 2 and 3, corresponds to the predicted molecular mass of the S. epidermidis TagF protein (85.9 kDa). A protein band with a molecular mass slightly less than 85 kDa is present in lane 2 (indicated by the arrowhead), corresponding to the mutated S. epidermidis TagF protein. (B) Western immunoblot analysis of whole-cell lysates of B. subtilis strains 1A486(pFC10), 1A486(pFCTH3), and 1A486(pHPS9). The polyclonal antiserum raised against the S. epidermidis GST-′TagF fusion protein is reactive against the wild-type and mutant S. epidermidis TagF proteins present in lanes 1 and 2, respectively. The lysate of strain 1A486(pHPS9) (lane 3) is nonreactive.
The relatively large amounts of the wild-type and mutant S. epidermidis TagF proteins present in these lysates can be accounted for by the fact that the tagF genes were cloned downstream from the strong P59 lactococcal promoter present in vector pHPS9 and are present on a multicopy plasmid (17).
Attempted disruption of the S. epidermidis tagF gene by plasmid integration.
The S. epidermidis tagF gene was cloned from strain ATCC 14990, but it proved impossible to introduce plasmid DNA into this strain by electroporation. Therefore, S. epidermidis TU3298, a strain capable of being transformed by electroporation (4), was used in the gene disruption experiments. Southern hybridization of the tagF locus from strain TU3298 indicated that it is very similar to that of strain ATCC 14990 (data not shown).
In order to determine if the S. epidermidis tagF gene is essential, an attempt was made to disrupt it by directed plasmid integration by using the temperature-sensitive (ts) plasmid pTSTAG, which carries a 1.56-kb internal fragment of tagF. As a control, plasmid pTSH17 carrying a similar-sized 1.77-kb fragment of DNA comprising the 3′ end of the S. epidermidis tagF gene and adjacent downstream sequences was constructed. Integration of pTSH17 by a single crossover will preserve a wild-type copy of tagF linked to its cognate promoter. This plasmid served as a positive control for plasmid integration in the tagF locus. Both pTSTAG and pTSH17 were derived from the temperature-sensitive vector pTS2T, which was also used as a control for reversion of the “ts rep” mutation (see Table 2) and nonspecific integration events. Colonies that could grow at 45°C on Tc agar occurred at frequencies of 1.2 × 10−6, 1.1 × 10−6, and 8.7 × 10−7 for strain TU3298 carrying the plasmids pTS2T, pTSH17, and pTSTAG, respectively.
PCR was used to determine if integration of pTSH17 and pTSTAG into the tagF locus had occurred. Thirty temperature-independent derivatives of both S. epidermidis TU3298(pTSTAG) and TU3298(pTSH17) were purified from five separate cultures of each strain. Oligonucleotide primers TQ1 and TQ2 were designed to detect chromosomal integration by homologous recombination of pTSTAG and pTSH17. Primer TQ1 hybridized downstream from the tet gene at the 5′ end of the 2.35-kb HindIII tet fragment (25). Primer TQ2 bound within pal1, 5′ to the tagF gene. Primers TQ1 and TQ2 will generate a PCR product in TU3298(pTSTAG) and TU3298(pTSH17) growing at 45°C only if the plasmids integrate into the tagF gene. Twenty-three out of thirty derivatives of TU3298(pTSH17) formed the expected PCR product, indicating that the plasmid was integrated in tagF as expected. In contrast, the PCR fragment was not detected in any of the 30 TU3298(pTSTAG) derivatives.
Genomic DNA was isolated from five TU3298(pTSTAG) derivatives and digested with EcoRV. Southern hybridization analysis with the 1.56-kb tagF fragment cloned in pGDH3 as a probe revealed that the ca. 8.5-kb chromosomal tagF fragment had not been disrupted and confirmed that pTSTAG had not integrated into the tagF gene (data not shown).
Genomic DNA from TU3298(pTSH17) derivatives was digested with ClaI and EcoRI. Southern hybridization analysis with the 1.77-kb tagF 3′ fragment cloned in pBH17 as a probe revealed that pTSH17 had undergone chromosomal integration at the tagF locus by homologous recombination to disrupt the ca. 5.5-kb tagF fragment of TU3298 in the two PCR-positive derivatives tested (data not shown), while the tagF gene was intact in the PCR-negative derivatives (data not shown). Thus, pTSH17 can integrate into the tagF locus at low frequency, indicating that the failure of pTSTAG to integrate is not due to polar effects on a cotranscribed 3′ gene. There is no evidence that cells could grow with the tagF gene disrupted by pTSTAG integration, a finding which is consistent with it being an essential gene. Thus, teichoic acids appear to be essential in the coccus as well as in the rod and are not just required for elongation of the cylindrical part of the bacillus wall as was suggested previously (39).
Conclusions.
The tagF gene of S. epidermidis has been identified by sequence similarities of the encoded TagF protein with that of B. subtilis 168 and because it can complement a temperature-sensitive B. subtilis tagF mutant.
The failure to inactivate the tagF gene of S. epidermidis by directed integration of a plasmid indicates that the gene, and hence teichoic acid biosynthesis, is essential in this organism.
ACKNOWLEDGMENTS
We thank SmithKline Beecham for financial support and John Hodgson, Elizabeth Lawlor, Alison Chalker, and Michael Young for helpful discussions.
REFERENCES
- 1.Araki Y, Ito E. Linkage units in cell walls of gram-positive bacteria. Crit Rev Microbiol. 1989;17:121–135. doi: 10.3109/10408418909105745. [DOI] [PubMed] [Google Scholar]
- 2.Archibald A R, Hancock I C, Harwood C R. Cell wall structure, synthesis, and turnover. In: Sonenshein A L, Hoch J A, Losick R, editors. Bacillus subtilis and other gram-positive bacteria: biochemistry, physiology, and molecular genetics. Washington, D.C.: American Society for Microbiology; 1993. pp. 381–410. [Google Scholar]
- 3.Archibald A R, Stafford G H. A polymer of N-acetylglucosamine 1-phosphate in the wall of Staphylococcus lactis 2102. Biochem J. 1972;130:681–690. doi: 10.1042/bj1300681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Augustin J, Götz F. Transformation of Staphylococcus epidermidis and other staphylococcal species with plasmid DNA by electroporation. FEMS Microbiol Lett. 1990;66:203–208. doi: 10.1016/0378-1097(90)90283-v. [DOI] [PubMed] [Google Scholar]
- 5.Ausubel F M, Brent R, Kingston R E, Moore D D, Smith J A, Seidman J G, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons; 1987. [Google Scholar]
- 6.Bertram K C, Hancock I C, Baddiley J. Synthesis of teichoic acid by Bacillus subtilis protoplasts. J Bacteriol. 1981;148:406–412. doi: 10.1128/jb.148.2.406-412.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beveridge T E, Murray R G E. Uptake and retention of metals by cell walls of Bacillus subtilis. J Bacteriol. 1976;127:1503–1518. doi: 10.1128/jb.127.3.1502-1518.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Briehl M, Pooley H M, Karamata D. Mutants of Bacillus subtilis 168 thermosensitive for growth and wall teichoic acid synthesis. J Gen Microbiol. 1989;135:1325–1334. [Google Scholar]
- 9.Chatterjee A N, Mirlman D, Singer H J, Park T. Properties of a novel pleiotropic bacteriophage-resistant mutant of Staphylococcus aureus H. J Bacteriol. 1969;100:846–853. doi: 10.1128/jb.100.2.846-853.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chung C T, Miller R H. A rapid and convenient method for the preparation and storage of competent bacterial cells. Nucleic Acids Res. 1988;16:3580. doi: 10.1093/nar/16.8.3580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dong J M, Taylor J S, Latour D J, Luchi S, Lin E C. Three overlapping lct genes involved in l-lactate utilization by Escherichia coli. J Bacteriol. 1993;175:6671–6678. doi: 10.1128/jb.175.20.6671-6678.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ellwood D C, Tempest D W. Effects of environment on bacterial wall content and composition. Adv Microb Physiol. 1972;7:83–117. [Google Scholar]
- 13.Endl J, Seidl H P, Fiedler F, Schleifer K H. Chemical composition and structure of cell wall teichoic acids of staphylococci. Arch Microbiol. 1983;135:215–223. doi: 10.1007/BF00414483. [DOI] [PubMed] [Google Scholar]
- 14.Fiedler F, Steber J. Structure and biosynthesis of teichoic acids in the cell walls of Staphylococcus xylosus DSM 20266. Arch Microbiol. 1984;138:321–328. doi: 10.1007/BF00410898. [DOI] [PubMed] [Google Scholar]
- 15.Grant W D. Cell wall teichoic acid as a reserve phosphate source in Bacillus subtilis. J Bacteriol. 1979;137:35–43. doi: 10.1128/jb.137.1.35-43.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guan K L, Dixon J E. Eucaryotic proteins expressed in Escherichia coli: an improved thrombin cleavage and purification procedure of fusion proteins with glutathione-S-transferase. Anal Biochem. 1991;192:262–267. doi: 10.1016/0003-2697(91)90534-z. [DOI] [PubMed] [Google Scholar]
- 17.Haima P, Van Sinderen D, Schotting H, Bron S, Venema G. Development of a β-galactosidase α-complementation system for molecular cloning in Bacillus subtilis. Gene. 1990;86:63–69. doi: 10.1016/0378-1119(90)90114-7. [DOI] [PubMed] [Google Scholar]
- 18.Hanahan D, Meselson M. Plasmid screening at high colony density. Gene. 1980;19:147–151. doi: 10.1016/0378-1119(80)90144-4. [DOI] [PubMed] [Google Scholar]
- 19.Honeyman A L, Stewart G C. The nucleotide sequence of the rodC operon of Bacillus subtilis. Mol Microbiol. 1989;3:1257–1268. doi: 10.1111/j.1365-2958.1989.tb00276.x. [DOI] [PubMed] [Google Scholar]
- 20.Honjo M, Nakayama A, Fukazawa K, Kawamura K, Ando K, Hori M, Furutani Y. A novel Bacillus subtilis gene involved in negative control of sporulation and degradative-enzyme production. J Bacteriol. 1990;172:1783–1790. doi: 10.1128/jb.172.4.1783-1790.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Höltje J V, Tomasz A. Specific recognition of choline residues in the cell wall teichoic acid by the N-acetylmuramyl-l-alanine amidase of pneumococcus. J Biol Chem. 1975;250:6072–6076. [PubMed] [Google Scholar]
- 22.Hoover D G, Gray R H. Function of cell wall teichoic acid in thermally injured Staphylococcus aureus. J Bacteriol. 1977;131:477–485. doi: 10.1128/jb.131.2.477-485.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Horne D S, Tomasz A. Possible role of a choline-containing teichoic acid in the maintainance of normal cell shape and physiology in Streptococcus oralis. J Bacteriol. 1993;175:1717–1722. doi: 10.1128/jb.175.6.1717-1722.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Karamata D, Gross J. Isolation and genetic analysis of temperature sensitive mutants of Bacillus subtilis defective in DNA synthesis. Mol Gen Genet. 1970;108:277–287. doi: 10.1007/BF00283358. [DOI] [PubMed] [Google Scholar]
- 25.Khan S A, Novick R P. Complete nucleotide sequence of pT181, a tetracycline resistance plasmid from Staphylococcus aureus. Plasmid. 1983;10:251–259. doi: 10.1016/0147-619x(83)90039-2. [DOI] [PubMed] [Google Scholar]
- 26.Kolkman M A B, Wakarchuk W, Nuijten P J M, van der Zeijst B A M. Capsular polysaccharide synthesis in Streptococcus pneumoniae serotype 14: molecular analysis of the complete cps locus and identification of genes encoding glycosyltransferases required for the biosynthesis of the tetrasaccharide subunit. Mol Microbiol. 1997;26:197–208. doi: 10.1046/j.1365-2958.1997.5791940.x. [DOI] [PubMed] [Google Scholar]
- 27.Kreiswirth B N, Löfdahl M S, Betley M J, OReilly M, Schlievert P M, Bergdoll M S, Novick R P. The toxic shock syndrome exotoxin structural gene is not detectably transmitted by a prophage. Nature. 1983;305:709–712. doi: 10.1038/305709a0. [DOI] [PubMed] [Google Scholar]
- 28.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 29.Lazarevic V, Karamata D. The tagGH operon of Bacillus subtilis 168 encodes a two-component ABC transporter involved in the metabolism of two wall teichoic acids. Mol Microbiol. 1995;16:345–355. doi: 10.1111/j.1365-2958.1995.tb02306.x. [DOI] [PubMed] [Google Scholar]
- 30.Lazarevic V, Mauël C, Soldo B, Freymond P P, Margot P, Karamata D. Sequence analysis of the 308° segment of the Bacillus subtilis 168 chromosome, a region devoted to cell wall metabolism, containing non-coding grey holes which reveal chromosomal rearrangements. Microbiology. 1995;141:329–335. doi: 10.1099/13500872-141-2-329. [DOI] [PubMed] [Google Scholar]
- 31.Mauël C, Young M, Karamata D. Genes concerned with the synthesis of poly(glycerol phosphate), the essential teichoic acid in Bacillus subtilis strain 168, are organised in two divergent transcription units. J Gen Microbiol. 1991;137:929–941. doi: 10.1099/00221287-137-4-929. [DOI] [PubMed] [Google Scholar]
- 32.Mauël C, Young M, Margot P, Karamata D. The essential nature of teichoic acids in Bacillus subtilis as revealed by insertional mutagenesis. Mol Gen Genet. 1989;215:388–394. doi: 10.1007/BF00427034. [DOI] [PubMed] [Google Scholar]
- 33.Oskouian B, Stewart G C. Repression and catabolite repression of the lactose operon of Staphylococcus aureus. J Bacteriol. 1990;172:3804–3812. doi: 10.1128/jb.172.7.3804-3812.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Owen P. Crossed immunoelectrophoresis in the study of outer membrane antigens. In: Korhenen T K, Dawes E A, Makela P H, editors. Enterobacterial surface antigens: methods for molecular characterisation. Amsterdam, The Netherlands: Elsevier; 1985. pp. 237–238. [Google Scholar]
- 35.Park J T, Shaw D R D, Chatterjee A N, Mirelman D, Wu T. Mutants of staphylococci with altered cell walls. Ann N Y Acad Sci. 1974;236:54–62. doi: 10.1111/j.1749-6632.1974.tb41481.x. [DOI] [PubMed] [Google Scholar]
- 36.Pollack J H, Neuhaus F C. Changes in wall teichoic acid during the rod-sphere transition of Bacillus subtilis 168. J Bacteriol. 1994;176:7252–7259. doi: 10.1128/jb.176.23.7252-7259.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pooley H M, Abellan F-X, Karamata D. A conditional-lethal mutant of Bacillus subtilis 168 with a thermosensitive glycerol-3-phosphate cytidylyltransferase, an enzyme specific for the synthesis of the major wall teichoic acid. J Gen Microbiol. 1991;137:921–928. doi: 10.1099/00221287-137-4-921. [DOI] [PubMed] [Google Scholar]
- 38.Pooley H M, Abellan F-X, Karamata D. CDP-glycerol:poly(glycerophosphate) glycerophosphotransferase, which is involved in the synthesis of the major wall teichoic acid in Bacillus subtilis 168, is encoded by tagF (rodC) J Bacteriol. 1992;174:646–649. doi: 10.1128/jb.174.2.646-649.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pooley H M, Abellan F-X, Karamata D. Wall teichoic acid, peptidoglycan synthesis and morphogenesis in Bacillus subtilis. In: de Pedro M A, Höltje J-V, Iöffelhardt W, editors. Bacterial growth and lysis: metabolism and structure of the bacterial sacculus. New York, N.Y: Plenum Press; 1993. pp. 385–392. [Google Scholar]
- 40.Poxton I R, Tarelli E, Baddiley J. The structure of C-polysaccharide from the walls of Streptococcus pneumoniae. Biochem J. 1978;175:1033–1042. doi: 10.1042/bj1751033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ramirez M, Tomasz A. Molecular characterization of the complete 23F capsular polysaccharide locus of Streptococcus pneumoniae. J Bacteriol. 1998;180:5273–5278. doi: 10.1128/jb.180.19.5273-5278.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 43.Smith D R, Doucette-Stamm L A, Deloughery C, Lee H-M, Dubois J, Aldredge T, Bashirzadeh R, Blakely D, Cook R, Gilbert K, Harrison D, Hoang L, Keagle P, Lumm W, Pothier B, Qiu D, Spadafora R, Vicaire R, Wang Y, Wierzbowski J, Gibson R, Jiwani N, Caruso A, Bush D, Safer H, Patwell D, Prabhakar S, McDougall S, Shimer G, Goyal A, Pietrokovski S, Church G M, Daniels C J, Mao J-I, Rice P, Nölling J, Reeve J N. Complete genome of Methanobacterium thermoautotrophicum delta H: functional analysis and comparative genomics. J Bacteriol. 1997;179:7135–7155. doi: 10.1128/jb.179.22.7135-7155.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Van Eldere J, Brophy L, Loynds B, Celis P, Hancock I, Carman S, Kroll J S, Moxon E R. Region II of the Haemophilus influenzae type b capsulation locus is involved in serotype-specific polysaccharide synthesis. Mol Microbiol. 1995;15:107–118. doi: 10.1111/j.1365-2958.1995.tb02225.x. [DOI] [PubMed] [Google Scholar]