Abstract
Purpose:
Oncogenic alterations of the phosphatidylinositol-3-kinase (PI3K)/AKT pathway occur in >40% of patients with metastatic castration-resistant prostate cancer (mCRPC), predominantly via PTEN loss. The significance of other PI3K pathway components in prostate cancer is largely unknown.
Experimental Design:
Patients in this study underwent tumor sequencing using the MSK-IMPACT clinical assay to capture single-nucleotide variants, insertions, and deletions, copy number alterations, and structural rearrangements, or were profiled through The Cancer Genome Atlas. The association between PIK3R1 alteration/expression and survival was evaluated using univariable and multivariable Cox proportional-hazards regression models. We used siRNA-based knockdown of PIK3R1 for functional studies. FDG-PET/CT examinations were performed with a hybrid PET/CT scanner for some prostate cancer patients in MSK-IMPACT cohort.
Results:
Analyzing 1417 human prostate cancers, we found a significant enrichment of PIK3R1 alterations in metastatic cancers compared to primary cancers. PIK3R1 alterations or reduced mRNA expression tended to be associated with worse clinical outcomes in prostate cancer, particularly in primary disease, as well as in breast, gastric, and several other cancers. In prostate cancer cell lines, PIK3R1 knockdown resulted in increased cell proliferation and AKT activity, including insulin-stimulated AKT activity. In cell lines and organoids, PIK3R1 loss/mutation was associated with increased sensitivity to AKT inhibitors. PIK3R1-altered patient prostate tumors had increased uptake of the glucose analogue 18F-fluorodeoxyglucose in PET imaging, suggesting increased glycolysis.
Conclusions:
Our findings describe a novel genomic feature in metastatic prostate cancer and suggest that PIK3R1 alteration may be a key event for insulin-PI3K-glycolytic pathway regulation in prostate cancer.
Keywords: Metastatic castration-resistant prostate cancer, phosphatidylinositol-3-kinase pathway, PIK3R1 alteration, Akt inhibitor, FDG-PET
INTRODUCTION
Prostate cancer is one of the leading causes of cancer deaths among men, with 34,500 deaths projected for the US in 2022 (1). Once prostate cancer becomes castration resistant and metastatic, it is considered to be incurable. Although systemic therapies for prostate cancer exist (eg, androgen deprivation therapy and androgen receptor signaling inhibitors), most prostate tumors develop resistance. No current treatment for metastatic castration-resistant prostate cancer is curative for even a subset of patients. The lack of curative treatment options points to a need for a better understanding of the genomic aberrations in prostate cancer that drive metastasis.
Alterations in the phosphoinositide 3-kinase (PI3K) pathway are common in advanced prostate cancer. PI3K proteins, when induced by activated receptor tyrosine kinases, phosphorylate phosphatidylinositol 4,5-bisphosphate to generate phosphatidylinositol 3,4,5-trisphosphate (PIP3), an activator of AKT. This phosphorylation can be reversed by the tumor suppressor PTEN; loss of PTEN is one of the most common alterations in primary prostate cancer and is a predictor of lethal prostate cancer (2). Thus, activation of PI3K and inactivation of PTEN both serve to activate AKT.
Clinical and laboratory studies have established the critical role of PTEN loss in activation of PI3K signaling and prostate cancer progression (2–4). Among the alterations that affect PI3K itself, the most attention has been devoted to those in PIK3CA, which encodes the catalytic subunit of PI3Kα. However, PIK3R1, which encodes the regulatory subunit of PI3K, has been reported to be altered in 1–6% of prostate cancers, and its expression is reduced in prostate cancer compared with normal prostate tissue (5,6). PIK3R1 represses PI3Kα activity in the absence of receptor tyrosine kinase stimulation, and it may also directly activate PTEN; therefore, alterations of PIK3R1 can increase activity of PI3Kα and AKT. The effects of PIK3R1 alterations are complex, since this protein also stabilizes the catalytic subunit, and because the three isoforms of the PIK3R1 protein (p85α, p55α, and p50α) have functional differences (7,8). Somatic mutation of PIK3R1 also increases activation of the MAPK pathway and confers sensitivity to MEK and JNK inhibitors (9).
The biological effects of loss of PIK3R1 activity can include reduced apoptosis, increased cell division, and increased insulin-stimulated uptake of glucose into cells. Indeed, elimination of Pik3r1 in mice results in increased insulin sensitivity as well as glucose uptake and lactate synthesis in tissues (7,10,11).
Because PI3K pathway alterations are found in a wide variety of cancers, several PI3K/AKT inhibitors have been developed (12). Some of these agents are currently in clinical trials. Results of the trials for prostate cancer have been mixed; however, in a phase III trial for patients with prostate cancer, the AKT inhibitor ipatasertib has been reported to prolong progression-free survival for a subset of patients (13).
Despite the potential importance of PI3Kα in prostate cancer, alterations in PIK3CA and PIK3R1 have not yet been linked to either metastasis or outcomes of patients with prostate cancer. In this study, we investigated whether genomic aberrations in PI3K pathway genes are associated with metastasis, using a large dataset of targeted sequencing results for primary and metastatic prostate cancer. Upon finding an association for PIK3R1 alterations, we investigated the clinical implications by examining the association of PIK3R1 alterations with outcomes among patients with prostate cancer and other cancers. We also examined the biological implications by testing the effects of reduced PIK3R1 activity using in vitro models of prostate and breast cancers.
METHODS
Study population
MSK-IMPACT, designed to test patients with advanced cancer, was ordered by treating physicians to identify genomic alterations that could potentially inform treatment decisions (14). The analysis of prostate cancer included men with prostate cancer who underwent paired tumor–normal sequencing as approved by the MSK institutional review board. We obtained written informed consent from all the patients in this study, which was conducted in accordance with the Declaration of Helsinki. All the patients in this study underwent tumor sequencing using the MSK-IMPACT clinical assay, which included between 340, 410, or 468 genes depending on its version, to capture single-nucleotide variants, insertions, and deletions, copy number alterations, and structural rearrangements. All panels included PIK3R1. Due to a high probability of passenger events in PIK3R1, samples were excluded if they had an MSI sensor score >10 and/or a tumor mutation burden (TMB) >20 mutations/Mb. For patients with multiple sequenced samples, only the first sample was analyzed. In addition, samples that did not pass FACETS quality control criteria were excluded as described in facets-preview (https://github.com/taylor-lab/facets-preview). Predefined QC metrics ensured reliable integer copy number calls, which is of particular interest when assessing the allelic composition at a specific loci (ie, PIK3R1).
Clinical annotations were extracted as described (15). For breast cancer samples, we analyzed the previously published MSK-IMPACT breast cancer cohort (16). Pan-cancer mRNA expression was analysed using GEPIA (17), and survival analysis was performed using a KM-plotter (18) dataset.
Genomics (MSK-IMPACT)
TMB was calculated as total number of non-synonymous mutations divided by number of bases sequenced. Somatic alterations were annotated using OncoKB to determine oncogenicity and clinical actionability (19) (Data version: v2.8, released on 2020–09-17). Canonical oncogenic pathway-level alterations were computed using curated pathway templates as previously reported (20,21). The fraction of copy number-altered genome (FGA) was defined as the fraction of the genome log2 copy gain and loss (>0.2 and < −0.2, respectively) relative to the size of the genome with copy number profiled. A GISTIC algorithm was used to detect recurrent somatic copy number variation by evaluating amplitude and frequency of events and to distinguish between broad and focal alterations in chromosome 5. FACETS algorithm was used to derive ploidy- and purity-corrected integer DNA copy number calls and cancer cell fraction from the data. Allelic integer copy number calls were used to determine chromosomal aberrations. A chromosomal loss was defined as any deviation from euploidy, where either the maternal and/or paternal allele was missing. Conversely, chromosomal gains were defined as one allele exceeding the number of its opposite allele. The clonality of each mutation (clonal or subclonal) was determined as described in facets-suite (https://github.com/mskcc/facets-suite).
Outcome analyses (MSK-IMPACT)
For patients with localized disease at the time of sequencing, metastasis-free survival was the outcome, with follow-up starting at sequencing and ending at metastasis or last clinic visit with a physician. For survival analyses in mCSPC and mCRPC, we followed patients for OS, with follow-up starting at sequencing and ending at death or last documented contact, using time since diagnosis of metastatic prostate cancer as the timescale. Baseline clinical characteristics and outcomes were available only for a subset of prostate cancer patients. For survival analyses in breast cancer, we followed patients for OS, with follow-up starting at sequencing and ending at death or last documented contact, using time since diagnosis of metastatic disease as the timescale, with stratification by breast cancer subtypes (HR+/HER2-, triple negative). For the survival analyses, follow-up time was censored to 72 months. To assess associations between genomic phenotypes and OS, we estimated each hazard ratio (HR) using Cox proportional hazards regression models.
Gene expression analysis
Publicly available RNA expression data from various prostate cancer clinical cohorts were extracted from cBioPortal (22), OncoMine (23), CANCERTOOL (24) and GEPIA (17),. All the gene expression comparative analyses between different groups of patients, as well as correlations among selected groups of genes, were done using R or CANCERTOOL. Kaplan-Meier survival curves (HR and 95% CI) were generated using R or KM-plotter (18), as mentioned accordingly.
Cell culture
Human prostate cancer cells LNCaP, 22RV1, DU145, PC3, VCaP, and triple-negative human breast cancer cell MDA MB-231 were obtained from ATCC (Manassas, VA). E006AA-T cells were provided by John T. Isaacs (The Johns Hopkins University School of Medicine, Baltimore, MD), PC3M cells were provided by Raymond C. Bergan (Knight Cancer Institute, Oregon Health & Science University, Portland, OR), and the LAPC4 cell line was provided by Charles Sawyers (MSK). Cells were cultured in RPMI1640 (LNCaP, LAPC4, 22RV1, and PC3M) or DMEM (DU145, E006AA-T, MDAMB-231) media supplemented with 10% FBS, 2 mM L-glutamine, and 1x antibiotic/antimycotic (Gemini Bio-Products, Sacramento, CA) at 37° C in 5% CO2. Human prostate epithelial cell RWPE1 was obtained from ATCC and cultured in keratinocyte serum-free medium (Thermo Fisher Scientific, Waltham, MA) at 37° C in 5% CO2. Cells were authenticated by human short tandem repeat profiling at the MSK Integrated Genomics Operation Core Facility.
Prostate organoids derived from patients with mCRPC were provided by Yu Chen (MSK) and cultured as described (25). Cells were acquired between 2017 and 2019; in general, passaged between two and six times between thawing (reviving), and experiments. Cells were authenticated by human short tandem repeat profiling at the MSK Integrated Genomics Operation Core Facility in December 2017 and June 2019. Mycoplasma testing was performed at the MSK Antibody & Bioresource Core Facility using the MycoAlert PLUS Assay.
Gene silencing
siRNAs (PIK3R1 and scrambled; listed in Supplementary Table S1) were transiently transfected to indicated cells using the TransIT-X2 system (Mirus, Madison, WI). Efficiency of knockdown was verified by qPCR and western blot.
Western blot
Cells were washed with Hank’s Balanced Salt Solution (HBSS) and lysed in RIPA (50 mM Tris-HCl pH 7.4, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 1% sodium deoxycholate, and 0.1% SDS) supplemented with protease and phosphatase inhibitors (Thermo Scientific). Protein concentrations were measured using the Bradford protein assay. Western blot was performed using specific antibodies (Supplementary Table S1). Western blots were performed in triplicate.
RNA extraction and qPCR
Total RNA was extracted using the Direct-zol RNA Kit (Zymo Research, Irvine, CA) and reverse transcribed with qScript cDNA SuperMix (Quantabio, Beverly, MA). cDNA corresponding to approximately 10 ng of starting RNA was used for one reaction. qPCR was performed with TaqMan Gene Expression Assay (Applied Biosystems, Waltham, MA). All quantifications were normalized to endogenous GAPDH. Each experiment was performed 3 times. Probes used for qPCR are listed in Supplementary Table S1.
Cell viability assay
Cell viability was measured by MTT (3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyl tetrazolium bromide; Invitrogen) assay. For the MTT assay, MTT crystals were dissolved in isopropanol. Cells (control and PIK3R1 knockdown) were plated at 2.5 × 103 per well in 96-well plates in complete media (with 10% FBS). Organoids (MSK-PCa1 and MSK-PCa3) were counted and plated to collagen-coated (type I collagen 50 μg/ml) 96-well plates (5× 103 cells/well in 100 μL media) in growth factor–enriched organoid-specific media as described previously (25). Cells were treated either with DMSO or with the indicated inhibitors. Drugs/inhibitors are listed in Supplementary Table S1. Each cell growth assay was peformed 4 times. After the indicated times, cells were incubated in 0.5 mg/mL MTT (Invitrogen) for 1 hour at 37° C, then photographed under a phase contrast microscope (Olympus). Absorbance was measured in a BioTek plate reader at 570 nm.
Suspension prostate cells sphere (prostatosphere) assay
Suspension tumorisphere/prostatosphere assays were performed as described (26). Briefly, 22RV1 cells (control or PIK3R1 knockdown) were detached using Accutase (Innovative Cell Technologies, San Diego, CA), collected using 70-μm cell strainers, counted, and resuspended in 1 mL of prostate organoid media (25). The cells (1×103 cell/well) were plated on ultra-low attachment plates (2 plates with 4 wells in each plate for each experimental condition) and allowed to grow for 7 days. Cell spheres were counted and photographed using GelCount colony counter (Oxford Optronix, Abingdon, England).
PET/CT imaging protocol and genomic analysis
FDG-PET/CT examinations were performed with a hybrid PET/CT scanner (Discovery; GE Healthcare, Milwaukee, Wis) about 60 minutes after intravenous injection of approximately 370 MBq of 18F-FDG (obtained from IBA Molecular North America Inc., Gilroy, CA, and calibrated by our in-house radiopharmacy) as described previously (27). Somatic alterations were annotated using OncoKB for oncogenicity and clinical actionability (19) (data version: v2.8, released on 09–17-2020). Recurrent oncogenic alterations were defined as oncogenic and present in at least 5% of samples. Canonical oncogenic pathway-level alterations were computed using curated pathway templates as previously reported (20,21). The FDG target peak distribution was normalized using a log10 transformation.
Statistical analyses
Comparisons between groups were done using a non-parametric Mann Whitney U test for continuous variables and the Fisher’s exact test for categorial variables. A binomial test was used to determine whether the ratio of truncating mutations present in the c-SH2 domain of PIK3R1 differed significantly. Time-to-event outcomes were analyzed using the Kaplan-Meier method and compared via the Mantel-Cox log rank test. The association between PIK3R1 alteration/expression and survival was evaluated using univariable and multivariable Cox proportional-hazards regression models. Student’s t-test or ANOVA were used to analyze normally distributed data. Pearson’s correlation coefficient was used to assess the association between two continuous variables. All statistical tests were defined as significant if the p value was < 0.05 or the false discovery rate (FDR) was < 0.05 for multiple comparisons. Analyses were performed using R v3.5.2 (www.R-project.org) and Prism 9.1.1 (Graphpad, USA) unless otherwise indicated.
Data availability
Genomic data from the MSK-IMPACT prostate cancer samples are available via cBioPortal for Cancer Genomics: https://www.cbioportal.org/study/summary?id=prad_pik3r1_msk_2021
RESULTS
PIK3R1 alterations are enriched in metastatic prostate tumors
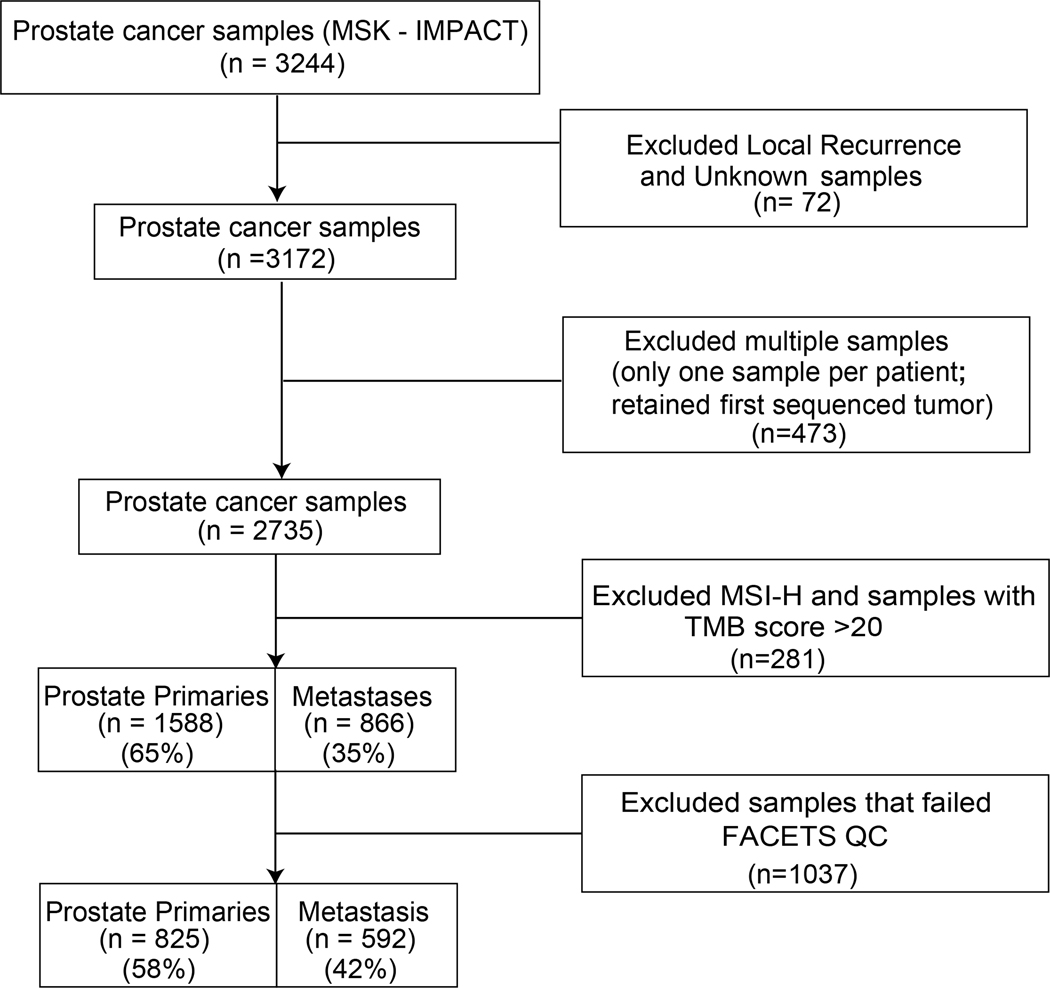
We analyzed panel sequencing data from 1417 samples obtained from patients with prostate cancer: 825 (58%) samples from primary tumors and 592 (42%) samples from metastases (Fig. 1). Each sample was tested for tumor and germline mutations using MSK-IMPACT, a capture-based next-generation sequencing assay that targets all coding exons and select introns of up to 505 key cancer genes. Patient characteristics are summarized in Supplementary Figs. S1 and S2.
Figure 1:
Flow chart of patients and samples in the MSK-IMPACT prostate cancer cohort analysis. Exclusion of 72 patients due to local recurrence and unknown sample types are on the basis of cBioportal annotation.
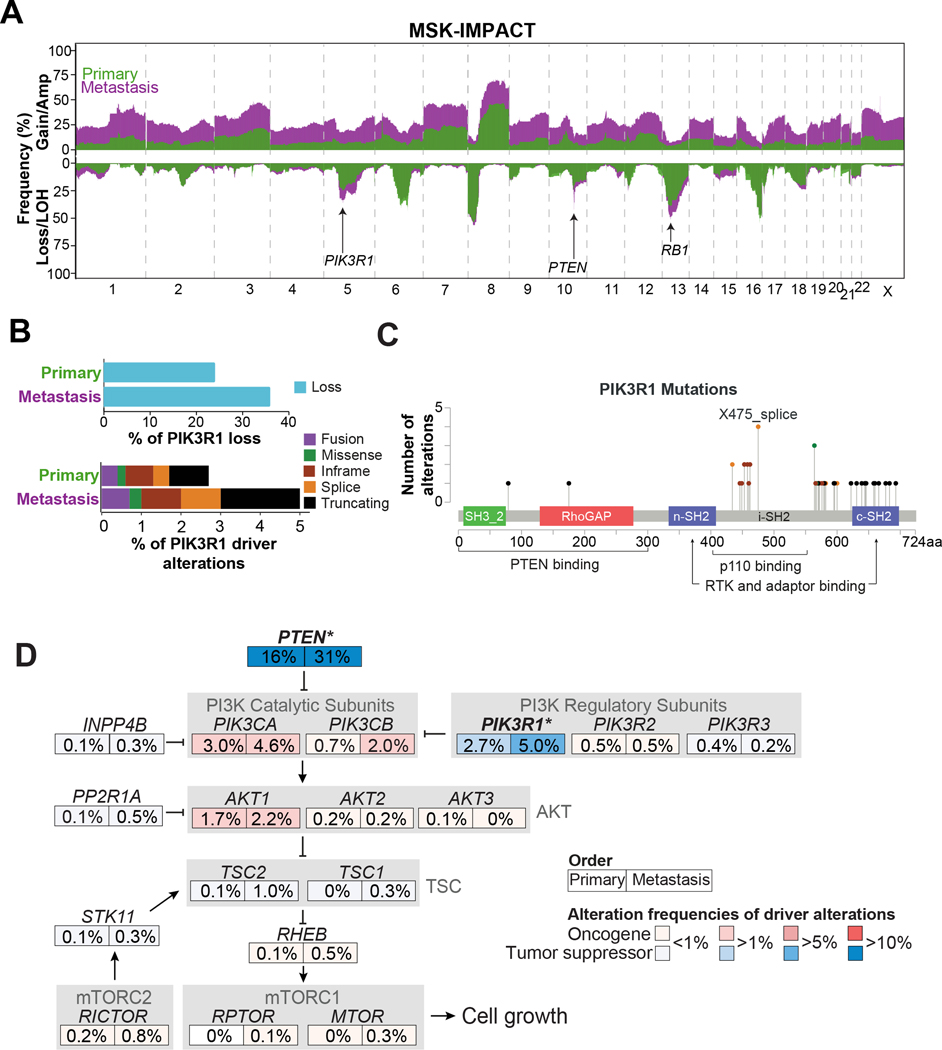
Regions of recurrent somatic copy number alterations were identified using the FACETS algorithm (28), revealing established focal deletions spanning PTEN and TP53, among others, and frequent genomic losses on chromosomes 5q, 8p, 13q, and 16q in the MSK-IMPACT prostate cancer dataset, of which many have been described previously (18,19) (Fig. 2A). We also observed global chromosomal amplification/gain in samples from prostate cancer metastases compared to primary prostate cancers (Fig 2A). Interestingly, we observed a higher degree of deletion in 13q (BRCA2 and RB1), focal deletion of chromosome 10q (PTEN), and deletion of chromosome 5q in prostate cancer metastases compared to primary prostate cancers.
Figure 2: Landscape of PIK3R1 and PI3K pathway in prostate cancer samples from MSK-IMPACT cohort.
(A) Overview of copy number prevalence identified in primary (green) and metastatic (purple) prostate cancer samples. Gains (top) and loss (bottom). (B) Bar charts representing frequency of PIK3R1 genomic losses in primary and metastatic samples (top). Stacked bar charts showing frequency of PIK3R1 driver alterations (mutations and fusions) in PIK3R1 mutated primary and metastatic samples. (C) Lollipop plot showing distribution of PIK3R1 driver mutations (truncating, splice, missense, and in-frame) found in both primary and metastatic samples. (D) Comparison of driver alteration frequency of patients with alterations in the corresponding canonical PI3K oncogenic signaling pathway members between primary and metastasis samples. Gradations of blue depicting tumor suppressor genes (TSG) and red depicting oncogenes. Statistically significant genes within the PI3K pathway enriched in PIK3R1 altered metastatic samples are indicated with an asterisk (* p < 0.05)
We observed that PIK3R1, a tumor suppressor gene located on the chromosome 5q13.1 locus, was more frequently lost in metastatic prostate cancer samples than in primary prostate cancers (36% vs 24%, p<0.001) (Fig. 2B, top). Further analysis using GISTIC also identified frequent genomic loss of 5q13.1/PIK3R1 (q < 0.001) in metastatic samples (Supplementary Fig. S3A). Additionally, we investigated whether PIK3R1 losses were specific or occurred in context on wider chromosome 5q deletions. We noticed that PIK3R1 losses varied in size (Supplementary Fig. S3B). Approximately 31% of samples that had losses were shorter than 25% of the entire chromosome 5q arm, and these could be considered as focal events. Genomic loss of PIK3R1 was found in 347 patients with complete clinical information (Supplementary Fig. S1 and S2). (Of a total of 407 patients, data on 26 with metastases and 34 with primary prostate cancer were not yet clinically annotated) We then sought to assess the prevalence of PIK3R1 driver alterations (predicted to be oncogenic driver alterations, as defined by OncoKB) (19), in prostate tumors. Driver alterations in PIK3R1 (mutations and fusions) were found in 51 of 1417 patients with prostate cancer (3.6%) (Fig. 2B, bottom). Complete clinical information was available for 40 patients with PIK3R1 driver alterations (11 out of a total of 51 patients (~22%); 4 patients with metastatic samples and 7 with a primary prostate tumor were missing complete clinical annotations) (Supplementary Fig. S1 and S2). Most of the PIK3R1 driver mutations were in the C-terminal Src homology 2 (cSH2) domain, which binds receptor tyrosine kinases (RTKs), or the inter-SH2 (iSH2) domain, which binds p110 (PIK3CA) (Fig. 2C) (29).
Because PIK3R1 mutations are most frequent in endometrial cancer (30), we compared the domain location of PIK3R1 mutations in prostate and endometrial cancers, analyzing 1769 microsatellite stable (MSS) endometrial cancers profiled with MSK-IMPACT. In addition, we also assessed the domain locations of PIK3R1 truncating mutations in 1756 breast cancer tumors profiled with MSK-IMPACT. We noticed that truncating mutations in prostate cancer samples clustered around the c-SH2 domain (11/19 truncating mutations), which was higher than expected by chance (p <0.001; Supplementary Fig. S3C); however, we did not see a similar association in endometrial or breast cancer (16/120 and 1/9 truncating mutational events clustering in the c-SH2 domain, p=0.22 and p>0.99 in endometrial and breast cancer, respectively). In endometrial cancer, the truncating mutational events clustered around the n-SH2 and i-SH2 domains, indicating that the mutational hotspot of PIK3R1 may vary between tumor types and may have differing functional significance. Therefore, domain hotspot analysis may provide leads linking PIK3R1 mutations to functional significance and cancer-specific phenotypes and could be an area for future investigation.
Recent comprehensive genomic characterization of prostate cancer has demonstrated recurrent alterations of genes involved in PI3K signaling, affecting approximately 40% of samples from metastatic castration-resistant prostate cancer (mCRPC) (31). We analyzed the alteration frequency of oncogenic driver alterations in major PI3K-AKT-mTOR pathway components (19 genes) in the MSK-IMPACT panel. As expected, loss of PTEN was one the most common alterations in prostate cancer and strongly enriched in metastatic samples compared to primary prostate cancers (p<0.001; Fig. 2D). Also, PIK3R1 driver alterations (5% of metastatic samples vs. 2.7% of primary samples, p=0.030) were significantly enriched in metastatic samples. Although 4.6% of samples from metastases harbored mutations in PIK3CA (encoding PI3K catalytic subunit p110a), PIK3CA alterations were not significantly enriched in metastatic samples compared to primary prostate cancers (3% driver alteration of PIK3CA in primary prostate cancers vs 4.6% in metastatic cancer; p=0.152; Fig. 2D). Alterations in other members of the PI3K-AKT-mTOR pathway were infrequent (<2% cases) and did not differ significantly between samples from primary tumor and metastases.
Next we explored the clonality of the mutations of PI3K-AKT-mTOR pathway components in the MSK-IMPACT prostate cancer cohort. Clonality was estimated for 21 of 44 mutations found in PIK3R1. We observed slightly more subclonal events in PIK3R1 and more clonal events in PTEN and PIK3CA (Supplementary Fig. S3D). Given the low number of samples with clonality calls, this observation has to be explored on a larger dataset.
Additionally, we assessed genes that frequently co-occured with PIK3R1 alterations (Supplementary Fig. S3E). PTEN, TP53, TMPRSS2, and APC had a tendency to co-occur; however, only co-occurrence of alterations in PTEN and PIK3R1 was statistically significant.
Collectively our data showed that PIK3R1-alteration frequencies (driver and loss) are significantly enriched in metastastatic prostate cancer samples compared to primary prostate cancer samples.
Clinical outcomes in PIK3R1-altered prostate cancer
We observed abundant PIK3R1 mRNA expression in various normal tissues, including the prostate (Supplementary Fig. S4A). Importantly, our proteomic analysis using the Human Proteome Map (32) showed that PIK3R1 and PIK3CA encode the only two class 1 PI3K subunits that are abundantly expressed in normal prostate tissue (Supplementary Fig. S4B).
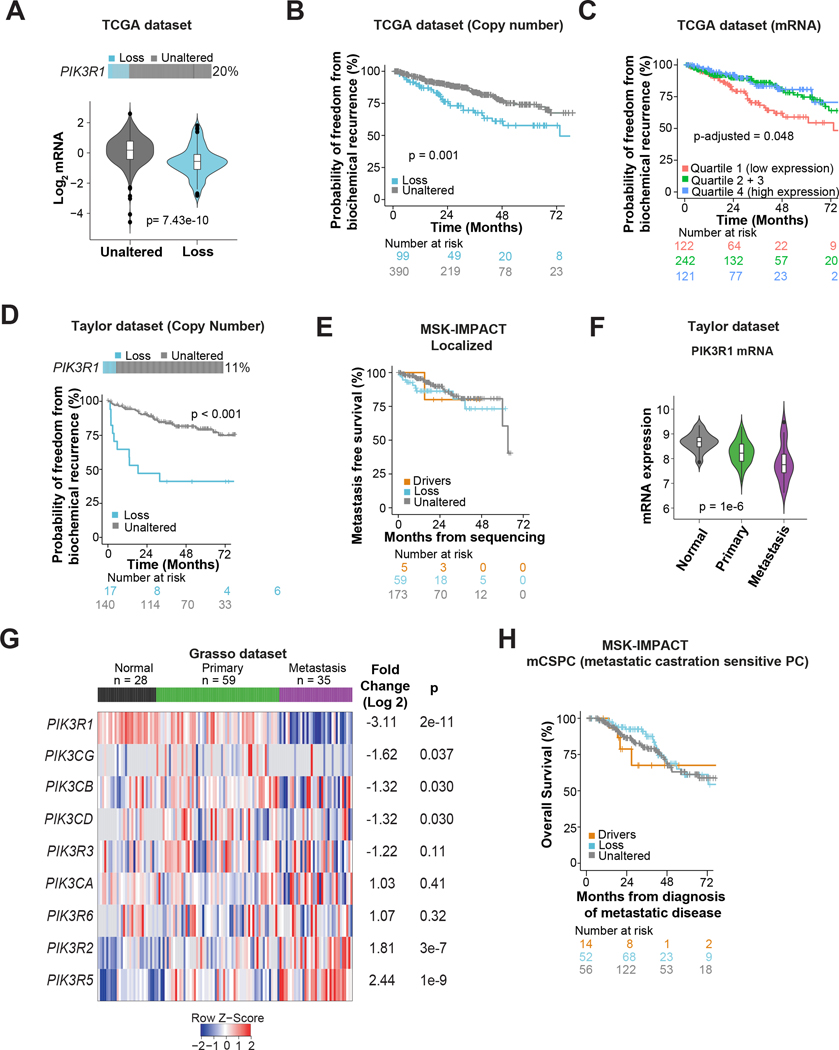
Next we assessed PIK3R1 RNA levels and their association with genomic copy number alterations and outcomes. To investigate the relationship between PIK3R1 genomic loss and mRNA expression, we used The Cancer Genome Atlas (TCGA; primary prostate cancer (33)), since MSK-IMPACT does not measure the transcriptome. Genomic PIK3R1 DNA copy number loss was found in ~20% of patients in the TCGA prostate cancer dataset (Fig. 3A, top), and was significantly associated with lower mRNA expression (p=7.43e–10; Fig. 3A, bottom). While the TCGA and Taylor (34) prostate cancer datasets were not designed to examine clinical outcomes such as overall survival (OS), in the available data, PIK3R1 loss was significantly associated with shorter disease/progression-free survival (TCGA: hazard ratio [HR] 2.04, 95% CI 1.32–3.15; p=0.001; Taylor: HR 4.13, 95% CI 2.01–8.50, p<0.001; Fig. 3B and 3D). In both datasets, decreased PIK3R1 mRNA expression was also associated with higher rates of tumor recurrence in models adjusted for collapsed Gleason Grade groups (6, 7, >=8), although estimates were attenuated (TCGA: HR 1.83, 95% CI 1.00–3.35, p=0.048, Fig. 3C; Taylor: HR 2.31, 95% CI 0.87–6.10, p=0.09; Supplementary Fig. S4E).
Figure 3: Loss of PIK3R1 is associated with disease progression in prostate cancer.
(A) Association between PIK3R1 mRNA expression and DNA copy number losses in the TCGA pan-cancer (prostate) dataset. p value was from Mann-Whitney U test. (B) Disease/progression-free survival (includes biochemical recurrence) according to PIK3R1 DNA copy number status among patients with primary prostate cancer in the TCGA pan cancer dataset. The log-rank p value is shown. (C) Freedom from biochemical recurrence according to PIK3R1 mRNA expression among patients with prostate cancer from the TCGA pan-cancer (prostate) dataset. Tumors were divided into 4 quartiles on the basis of PIK3R1 mRNA expression, and the middle two quartiles were grouped together. The p-value is from a Cox model adjusted for Gleason grade. (D) Disease/progression-free survival (includes biochemical recurrence) according to PIK3R1 DNA copy number status among patients in the Taylor et al prostate cancer dataset. The log-rank p value is shown. (E) Metastasis-free survival of patients with localized disease at the time of sequencing with PIK3R1 driver alteration (driver), genomic copy number loss (loss), and those with unaltered PIK3R1 tumor DNA in MSK-IMPACT cohort. (F) Violin plot showing the mRNA expression of PIK3R1 in different stages of prostate cancer (primary and metastasis) and in benign prostate tissue in Taylor et al prostate cancer dataset. The plot was generated using CANCERTOOL and p value was determined by ANOVA. (G) Heat map of mRNA expression of class-1 PI3K subunits in normal prostate and primary and metastatic CRPC samples in the Grasso prostate cancer dataset. The heat map was generated using Oncomine Suite. Genes are ranked on the basis of fold change (log 2; metastasis vs primary). (H) Overall survival of patients with metastatic castration-sensitive disease at the time of sequencing with PIK3R1 driver alteration, genomic copy number loss (loss), and unaltered PIK3R1 MSK-IMPACT cohort.
Among men profiled with MSK-IMPACT using tumors from primary prostate cancers (n=264), with complete follow-up for metastases available, results for associations between PIK3R1 alterations (drivers and loss) and metastasis-free survival were inconclusive (driver alterations: HR 1.40, 95% CI 0.18–10.46, p=0.74; loss: HR 1.53, 95% CI 0.69–3.38, p=0.28; Fig. 3E) due to the lower number of metastasis events (1 metastatic event among 5 men with PIK3R1 driver alterations; 9 metastatic events among 59 men with PIK3R1 copy number losses).
PIK3R1 expression was not associated with Gleason score in the TCGA dataset (Supplementary Fig. S4C). In an analysis of four publicly available datasets, we found that PIK3R1 mRNA expression was consistently significantly attenuated in metastatic prostate cancer compared to primary prostate cancer and normal prostate tissue (Fig. 3F and Supplementary Fig. S4D). Finally, to analyze the mRNA expression of all class 1 PI3K subunits in normal prostate tissue, primary prostate cancer, and metastatic prostate cancer, we analyzed a dataset reported by Grasso et al., because it has a sufficient number of samples of each type (35). Only PIK3R1 exhibited strongly attenuated expression in metastatic disease (9-fold decrease; p=2e–11; Fig. 3G). PIK3R1 mRNA expression is also positively correlated with PTEN mRNA in TCGA dataset (Supplementary Fig. S4F)
We also interrogated the association between PIK3R1 alteration status and OS in patients with metastatic prostate cancer in the MSK-IMPACT cohort. Results for associations between PIK3R1 driver alterations and OS were inconclusive (HR 1.67, 95% CI 0.62–4.42, p=0.30; 5 deaths among the 18 men with PIK3R1 driver alterations) in metastatic castration-sensitive disease (mCSPC; n=386) but would be compatible with worse prognosis. For copy number losses, results were not compatible with markedly worse prognosis (HR 0.88, 95% CI 0.53–1.46, p=0.63; Fig. 3H; 22 deaths among 115 men with copy number losses). Among men with mCRPC (n=376), results did not suggest strong associations (driver alteration: HR 0.77, 95% CI 0.38–1.41, p=0.36; loss: HR: 1.13, 95% CI 0.86–1.48; 10 deaths among 15 men with PIK3R1 driver alterations; 80 deaths among 116 men with PIK3R1 copy number losses). Therefore, in patients with metastatic disease, no statistically significant difference was found and therefore our data were inconclusive as to whether PIK3R1 genomic alterations would be associated with worse prognosis for metastatic prostate cancer.
Elimination of PIK3R1 induces activation of AKT signaling and augments cell growth
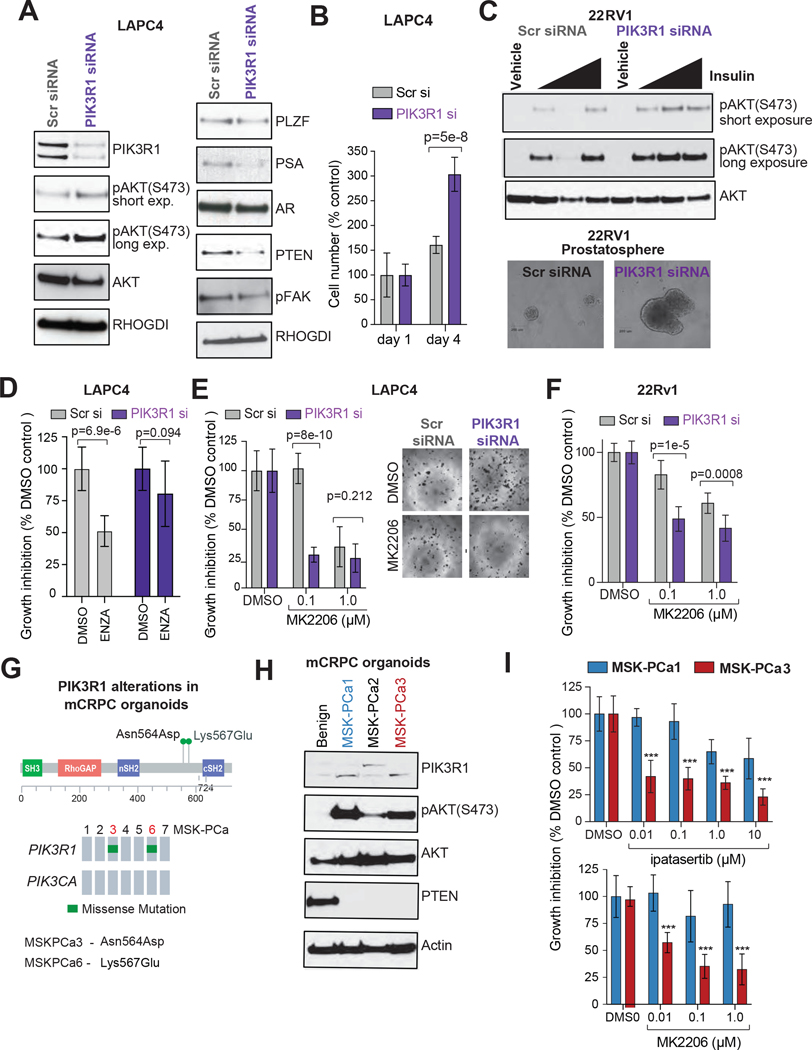
We directly examined the consequences of PIK3R1 loss via siRNA-mediated transient elimination of PIK3R1 in prostate cancer cells. Supplementary Fig. S5A shows the PI3K pathway gene mutations in various prostate cancer cell lines in data from the Cancer Cell Line Encyclopedia (36). In the LAPC4 cell line, a castration-sensitive prostate cancer cell line with wild-type PIK3R1 and PTEN (34) that also expresses high levels of PIK3R1 protein (Supplementary Fig. S5B), PIK3R1 siRNA reduced PIK3R1protein and mRNA levels (Fig 4A, Supplementary Fig. S5C) and induced AKT phosphorylation at serine-473 (S473) (Fig. 4A), which is required for maximal AKT kinase activation (37). We also observed that PIK3R1 knockdown results in lower levels of PTEN protein, which is consistent with a reported PTEN-stabilizing function of p85α (30). Importantly, even PIK3R1 transient knockdown reduced levels of androgen receptor (AR) downstream targets (PSA, PLZF, TMPRSS2, and NKX3.1) without affecting AR protein or mRNA expression (Fig. 4A; Supplementary Fig. S5C), which is also consistent with the previously reported AR transactivating function of p85α(38). In 22RV1, a castration-resistant prostate cancer cell line with wild-type PIK3R1 and PTEN but mutant PIK3CA (Supplementary Fig. S5A and S5B), PIK3R1 knockdown resulted in higher levels of AKT phosphorylation when serum-starved cells were treated with insulin (Fig. 4C; Supplementary Fig. S5D), consistent with the canonical role of PIK3R1 in downregulating insulin-mediated activation of the PI3K-AKT pathway, even in PTEN wild-type CRPC cells (39).
Figure 4. PIK3R1 loss augments AKT-pathway activation and prostate cancer progression.
(A) Western blot showing proteins in LAPC4 cells transfected with siRNA (SMARTPOOL; 72 hour post transfection) targeting PIK3R1. Cells transfected with scrambled (Scr) siRNA were used as control. RHoGDI served as loading control. (B) Effects of RIK3R1 knockdown on cell proliferation. PIK3R1- or Scr-siRNA-transfected LAPC4 cells (2.5× 103/well) were cultured in complete medium for 4 days after transfection. Cell growth was measured by MTT assay at the indicated times; P-values were determined by Student’s t-test. (C) Top, Overnight serum-starved 22RV1 PIK3R1 knockdown and Scr control cells (48-hour post siRNA transfection) were treated with insulin (0.01, 0.1 or 1 mM) for 2 hours. Cell lysates were analyzed for AKT activation using phospho AKT-S473 antibody by western blot. Total AKT was used as loading control. Bottom, Control and PIK3R1 knockdown 22RV1 cells (103 cells/well; 24-hour post transfection) were grown in low-adherence tissue culture plates for 7 days in growth factor–enriched media. The photographs show the 40X magnification images of representative cell spheres in suspension. (D) PIK3R1- or scrambled SMARTpool siRNA-transfected LAPC4 cells (2.5× 103/well) were cultured in complete medium supplemented with enzalutamide (ENZ; 10 μM) for 96 hours after transfection (bottom). Equivalent volume of DMSO was used as placebo treatment. Cell growth was measured by MTT assay; SD, P-values determined by Student’s t-test. (E) PIK3R1- or scrambled (Scr) SMARTpool siRNA-transfected LAPC4 cells (2.5× 103 /well) cells were cultured in complete medium supplemented with AKT-inhibitor (MK2206; 0.1 and 1 μM) or the equivalent volume of DMSO as control. After 72 hours, cells were treated with 0.5 mg/mL MTT and micrographed in 40x magnification (right). The bar graph shows the amount of cells as a percentage of the amount for Scr siRNA- and DMSO-treated controls (bottom). P values were calculated by Student t-test. (F) PIK3R1- or scrambled (Scr) SMARTpool siRNA-transfected 22RV1 cells (2.5× 103 /well) cells were cultured in complete medium supplemented with AKT-inhibitor (MK2206; 0.1 and 1 μM) for 72 hours after transfection. Cell growth was measured by the MTT assay. The bar graph shows the amount of cells as a percentage of the amount for Scr siRNA- and DMSO-treated samples (bottom). P values were calculated by Student t-test. (G) The lollipop plot (top) and Oncoprint (bottom) showing the PIK3R1 mutation in MSK mCRPC organoids (MSK-PCa1–7) (H) Status of PIK3R1, activated AKT and PTEN in mCRPC-derived organoids (MSKPCa1–3) were analyzed by western blot using the indicated antibodies. Lysate from parental benign organoid was used as control. b-Actin was used as loading control. (I) Organoids (MSK-PCa1 and MSK-PCa 3) were counted and plated (5000 cells/well in 100 μL media) to 96-well plates coated with collagen (Collagen I, 50 μg/ml), then treated with indicated concentrations of AKT inhibitors (ipatasertib and MK2206) or DMSO. After 7 days, cells were treated with 0.5 mg/mL MTT. The graphs represent the inhibition of cell viability by AKT-inhibitors treatment. P values were calculated by Student t-test.
In both the LAPC4 and 22RV1 cell lines, PIK3R1 knockdown increased cell proliferation, as evidenced by enhanced 2D cell growth compared to control cells (Fig. 4B; Supplementary Fig. S5C, S5E, and S5F). PIK3R1-knockdown 22RV1 cells also developed 3D spheres (prostatosphere) in anchorage-independent suspension culture (Fig. 4C, Supplementary Fig. S5G and S5H). Collectively, our results indicate that loss of PIK3R1 in prostate cancer cell lines promotes cell growth.
To investigate whether loss of PIK3R1 induces resistance to antiandrogen therapy, we performed a cell growth assay in media supplemented with an antiandrogen inhibitor (enzalutamide) using a control and PIKR31-knockdown LAPC4 cells (a castration-sensitive prostate cancer cell). Our results showed that RNAi-mediated transient silencing of PIK3R1 in LAPC4 cells exhibited relative resistance to antiandrogen therapy, as evidenced by growth in complete media supplemented with enzalutamide compared to control (scrambled siRNA transfected) LAPC4 cells (Fig. 4D).
PIK3R1 mutant cells are sensitive to AKT inhibitors
Alterations in the PI3K-signaling pathway in cancer have led to interest in the development of PI3K/AKT inhibitors, and many of these targeted therapies have shown promise for the treatment of PI3K-addicted tumors in clinical trials. Because PIK3R1 knockdown induces AKT activation, we next examined the effects of AKT-kinase inhibitor treatment on PIK3R1 knockdown prostate cancer cells. PIK3R1 knockdown LAPC4 and 22RV1 cells, compared with scrambled siRNA–treated control cells, exhibited greater sensitivity to the AKT kinase inhibitor MK2206, even at a very low concentration (100 nM) (Fig. 4E, 4F, and Supplementary Fig. S5I). These data indicate that inhibition of the AKT pathway reduces the viability of prostate cancer cells that express little PIK3R1.
We extended our study to include patient-derived organoids (40). Prostate organoids (MSK-PCa 1–7) were developed from specimens obtained from patients with mCRPC, and they retain the genetic characteristics of the original tumor (40). Most of the organoids are wild-type for PIK3R1, but 2 harbor missense mutations in PIK3R1 in the mutation hotspot of the iSH2 domain (Fig. 4G). We tested the PIK3R1, PTEN expression (protein and mRNA) and AKT-phosphorylation status in three mCRPC organoids (one PIK3R1 mutant and two wild type), all of which were derived from metastatic sites from castration-resistant tumors and harbor PTEN deletion (40). As a control, we analyzed a benign prostate organoid. All 3 prostate cancer organoids exhibited low PTEN expression and high AKT phosphorylation (Fig. 4H and Supplementary Fig. S5J). The PIK3R1-mutant organoid (MSK-PCa3; Asn564Asp mutation; likely pathogenic) exhibited strong growth reduction when treated with the AKT-kinase inhibitors MK2206 and ipatasertib (a small molecule AKT-kinase inhibitor currently in phase 2 and phase 3 clinical trials in multiple cancers incuding prostate), even at a very low concentration (10 nM: Fig. 4I). The PIK3R1–wild-type organoid MSK-PCa1, however, was only very modestly affected by the both AKT-kinase inhibitors. Interestingly, none of these PTEN-deleted organoids harbored any other known oncogenic mutations in PI3K-AKT pathway genes (source: cBioPortal for Cancer Genomics (22)), indicating that no other PI3K-Akt pathway mutation is required to confer sensitivity to Akt inhibitor treatment.
PIK3R1-mutant prostate tumors exhibit increased glucose uptake
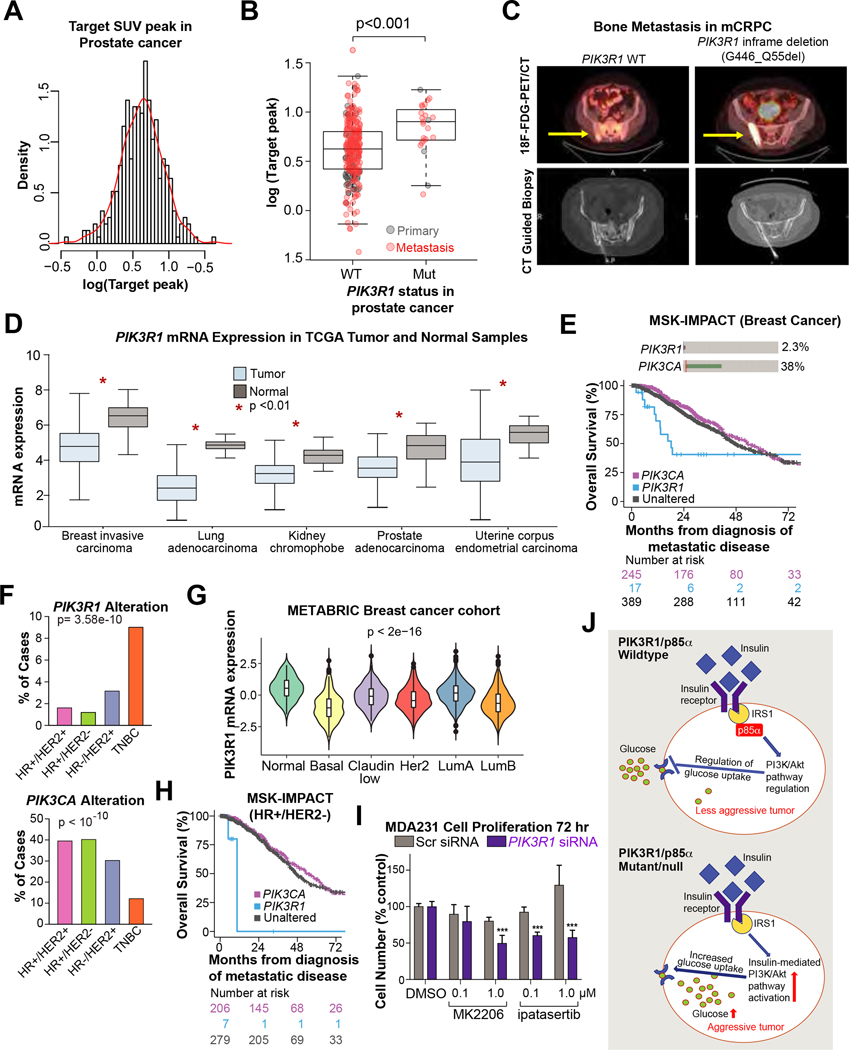
Fluorodeoxyglucose (18F) positron emission tomography (PET) combined with CT (FDG-PET/CT) is a well-established imaging tool for a variety of malignancies. FDG-PET signal is derived from increased uptake of FDG, an analog of glucose, reflecting tumor glycolytic activity (the Warburg effect) (41). For patients with prostate cancer, FDG-PET/CT is not widely used, given the low prevalence of FDG-avid tumors. We retrospectively analyzed data from prostate cancer patients in the MSK-IMPACT cohort who had undergone FDG-PET/CT. Patients’ demographics and inclusion criteria are summarized in Supplementary Fig. S6A. FDG-PET/CT data were available for 313 of the prostate cancer patients in our MSK-IMPACT cohort (corresponding to 71 primary prostate tumors and 242 metastasis samples). FDG uptake, measured by peak standardized uptake (Fig. 5A), was not significantly associated with known sequencing coverage and prognostic factors, including tumor purity, coverage, FGA, and TMB (Supplementary Fig. S6B and S6C). FDG uptake was higher in metastatic samples than in primary tumors (Supplementary Fig. S6D, p<0.001) and higher in mCRPC samples than in mCSPC samples (p<0.001).
Figure 5: PIK3R1 altered prostate cancer exhibit higher glycolytic activity and PIK3R1 associated with aggressive disease in multiple cancer types.
(A) Histogram of the normalized 18F-FDG peak density of prostate tumors (log 2 value). (B) Box-whisker plot of 18F-FDG uptake in PIK3R1-wildtype vs PIK3R1-altered (Mut) prostate cancer patients in the MSK-IMPACT cohort (primary and metastatic; n=313). Metastatic tumors are marked in red and primary prostate cancer are in black. (C) Representative fused 18F-FDG-PET/CT images (top) and CT-guided biopsy (bottom) of two patients with mCRPC and pelvic bone metastases (PIK3R1 wild type on the left, and mutant on the right). 18F-FDG uptake in was markedly higher in the PIK3R1-mutated tumor than in the PIK3R1-wildtype tumor (WT, SUVpeak 2.89 and PIK3R1 MUT, SUVpeak 13.23). (D) PIK3R1 mRNA expression (log2(TPM + 1)) in various tumor types compared to normal tissue in TCGA dataset. The graphs were generated using GEPIA tool. (E) Oncoprint showing PIK3R1 and PIK3CA oncogenic alteration in MSK-IMPACT breast cancer cohort (top). Kaplan-Meier curves displaying overall survival of metastatic breast cancer patients based on PIK3CA and PIK3R1 alterations (bottom). Tumors harboring oncogenic mutations in PIK3R1 are displayed in blue, those with known or putative functional alterations in PIK3CA are shown in pink, and those without either of these two alterations are shown in dark gray. (F) The alteration frequency of PIK3R1 (top) and PIK3CA (bottom) among various breast cancer subtypes in the MSK-IMPACT cohort. P value is from chi-squared test. (G) Violin plot showing mRNA expression of PIK3R1 in various breast cancer subtypes in the METABRIC breast cancer dataset. (H) Overall survival of HR+/ HER2- breast cancer patients in MSK-IMPACT cohort. Tumors harboring oncogenic mutations in PIK3R1 are displayed in blue, those with known or putative functional alterations in PIK3CA are shown in pink, and those without either of these two alterations are shown in dark grey. (I) PIK3R1 or scrambled (Scr) SMARTpool siRNA-transfected MDAMB-231 breast cancer cells (2.5× 103 /well) were cultured in complete medium supplemented with AKT-inhibitor (MK2206 and ipatasertib; 0.1 and 1 μM) or DMSO for 72 hours after transfection. Cells were then treated with 0.5 mg/mL MTT. The bar graph shows the quantity of cells as a percentage of the quantity for Scr siRNA- and DMSO-treated samples (bottom). P values were calculated by Student t-test. (J) Schematic representation showing the PIK3R1-mediated regulation of insulin-PI3K/AKT signaling pathway and glucose uptake in cells. IRS1; Insulin Receptor Substrate 1.
We next examined whether FDG uptake is associated with alteration of major oncogenic signaling pathways in mCRPC (n=146). Among the major known oncogenic signaling pathways in mCRPC (Supplementary Table S2), none were significantly associated with FDG uptake (Supplementary Fig. S6E). Similarly, most known driver mutations in mCRPC were not associated with FDG uptake (Supplementary Fig. S6E, Supplementary Table S2). Surprisingly, only PIK3R1 driver alterations were associated with significantly higher FDG values in mCRPC (n=10; p=0.002, FDR=0.039, Supplementary Fig. S6F, Supplementary Table S2 ). We extended our analysis to include primary tumors and mCSPC (n=313). FDG uptake was significantly higher in PIK3R1-altered prostate tumors (primary and metastatic) than in PIK3R1 wild-type tumors (p<0.001; Fig. 5B and 5C). Collectively, our data for the first time show that PIK3R1 mutation leads to increased glucose uptake in prostate tumor tissue, which can be detected noninvasively by FDG-PET/CT.
Pan-cancer analysis of PIK3R1 alteration
We assessed the prevalence of PIK3R1 driver alterations across 15 solid cancer types profiled within the MSK-IMPACT cohort. Multiple cancer types harbored PIK3R1 alterations (>2% of cases), including somatic mutations and rearrangements (Supplementary Fig. S6G). Driver alterations were most frequent in endometrial cancer (~25% of patients; Supplementary Fig. S6G), consistent with a previous report (30). Furthermore, using gene expression profiling and interactive analyses (GEPIA) (17), we found that PIK3R1 mRNA expression was markedly lower in tumor than in normal tissues across multiple cancer types (Fig. 5D). Moreover, in a large cohort of cancer patients (42), we found low PIK3R1 mRNA expression to be strongly associated with OS in multiple cancer types, including gastric, liver, and non–small cell lung cancer. Analysis was performed using KM-plotter in combined multiple publicly available datasets (Supplementary Fig. S6I) (18). Taken together, these results indicate that PIK3R1 loss may contribute to tumorigenesis and lethality in many cancers.
Because PI3K pathway alterations are prevalent in breast cancer, we analyzed the MSK-IMPACT breast cancer cohort (16) in greater depth. In total, 29 of the 1261 metastatic breast tumors (2.3%) had at least one PIK3R1 alteration (in patients with metastatic disease at diagnosis; Fig. 5E). Although the PIK3R1 interacting partner PIK3CA was more frequently altered among breast cancer patients (36%; n=452), patients with metastatic breast cancer in the PIK3R1-altered group had shorter OS than patients in the PIK3CA-altered group (HR 2.82, 95% CI 1.56–5.12, p<0.001; Fig. 5E, Supplementary Fig. S7A). PIK3R1 and PIK3CA mutations were mutually exclusive (p<0.001). The association was attenuated but still noticeable after adjustment for known prognostic covariates, age at diagnosis, and ER/PR/Her-2 status (HR 1.84, 95% CI 0.98–3.44, p=0.05; Supplementary Fig. S7A). We also observed that PIK3R1 alteration was significantly enriched in aggressive triple-negative breast cancer (TNBC; 16 of 176 patients [9.1%] with primary or metastatic; Fig. 5F) as compared to other subtypes (ER/PR/Her-2) of breast cancer. Moreover, PIK3R1 mRNA expression was attenuated in the basal/TNBC tumor subtype in the METABRIC cohort (2509 primary breast tumors (43); Fig. 5G). Interestingly, among patients with HR+/HER2- metastatic breast cancer, those with PIK3R1 alterations (n=11) had a shorter OS in an age-adjusted model than patients in the PIK3CA-altered group (HR 5.65, 95% CI 1.66–19.16, p=0.005; Fig. 5H and Supplementary Fig. S7A). Among patients with metastatic TNBC, PIK3R1 alteration was modestly associated with poor prognosis compared to PI3KCA alteration (H.R 1.86, 95% CI 0.58 – 6.04), even in an unadjusted analysis (Supplementary Fig. S7A and S7B).
In a large cohort of breast cancer patients (18,42), we observed that low PIK3R1 mRNA expression was significantly associated with worse relapse-free survival compared to patients with higher PIK3R1 mRNA expression (HR 1.43, 95% CI 1.30–1.60, p=6e-12; Supplementary Fig. S6H).
To test whether PIK3R1 loss induces sensitivity to AKT inhibitors in breast cancer, we used MDA-MB-231 cells, a TNBC cell line that expresses wild-type PIK3R1 and PTEN (36). After PIK3R1 knockdown, MDA-MB-231 cells exhibited enhanced sensitivity to the AKT inhibitors MK2206 and ipatasertib, compared to cells transfected with a scrambled siRNA (Fig. 5I). These data indicate that inhibition of the AKT pathway reduces the viability of at least some breast cancer cells with low PIK3R1 expression.
DISCUSSION
Our study demonstrates that metastatic prostate cancer has a high prevalence of alterations in PIK3R1/p85α, a regulatory subunit of phosphoinositide 3-kinase (driver alteration PI3KR1; 5% in metastatic disease compared to 2.7% in primary prostate cancer; genomic loss of 36% in metastatic disease compared to 24% in primary prostate cancer). To our knowledge, this association between PIK3R1 alterations and prostate cancer metastasis has not been previously reported. The PI3K family is essential to nearly all aspects of cell and molecular biology and central to human cancer, diabetes, and aging (44). Oncogenic alteration of the PI3K pathway is observed in approximately 40% of metastatic prostate cancers (31), and it is recognized as a hallmark survival and proliferative pathway in advanced prostate cancer. Several clinical and experimental studies have established the crucial role of PTEN loss in activation of PI3K signaling and aggressive prostate cancer progression (2–4). In our study of a large cohort of men with prostate cancer, we showed that driver alterations in PIK3R1 is second (after PTEN) among PI3K pathway genes in alteration frequency in metastatic prostate cancer. We were also able to estimate clonality for 21 of 44 PIK3R1 driver mutations in the MSK-IMPACT cohort and showed that a fraction of PIK3R1 mutations are subclonal (62%). Therefore, further analysis using larger cohorts will be necessary to fully understand the impact of the clonality of PI3K pathway driver mutations in mCRPC progression.
While previous publications have described the frequent PIK3R1 alterations in various cancers, including endometrial cancer (30), the clinical impact of these alterations has been poorly understood, particularly in prostate cancer. Our current study provides evidence suggesting that PIK3R1 alteration/loss may be particularly common as prostate cancer progresses. We observed that PIK3R1 alterations predominantly occur in metastatic prostate cancer. Findings from our prognostic analyses suggest that such events are harbingers of worse prognosis if they occur in primary disease. Patients with primary disease and low intratumoral PIK3R1 mRNA expression exhibited shorter biochemical recurrence-free survival, independent of Gleason grade, than those with higher PIK3R1. Similarly, in multiple independent prostate cancer datasets, we detected significantly lower PIK3R1 mRNA in metastatic samples compared to primary prostate cancer and benign prostate tissue. Our data were inconclusive as to whether PIK3R1 genomic alterations would be associated with worse prognosis in metastatic disease, but they were not strongly suggestive of such an association. Strikingly, PIK3R1 was the only class 1 subunit (either catalytic or regulatory)(44) of the PI3K pathway for which expression was significantly lower in metastatic prostate cancer. However, further experimental and clinical data are required to validate the hypothesis that PIK3R1 alterations contribute to cancer progression.
Previous studies in genetically engineered mouse models (GEMM) showed that even a single-copy loss of PIK3R1 could activate AKT and promote tumorigenesis (11,45,46). Similarly, data also showed that heterozygous splice mutation of PIK3R1 causes immunodeficiency due to hyperactivation of the PIK3 pathway (47). Therefore, in-depth somatic copy number alterations in the present study (MSK-IMPACT) were analyzed using FACETS. This approach enabled a detailed view of the allelic composition at the PIK3R1 locus (eg, determining loss of heterozygosity). PIK3R1 monoallelic losses were enriched in metastatic prostate cancer samples (36%) compared to primary tumors (26%, Fig. 1B). However, whether copy number alteration of PIK3R1 has any oncogenic potential is not well studied and is broadly unknown. Copy number loss of PIK3R1 was also frequent in other, independent primary prostate cancer cohorts (primarily heterozygous; 11% in Taylor and 20% in TCGA dataset), and heterozygous PIK3R1 loss was associated with significant downregulation of mRNA expression. Collectively, these data suggest that copy number alterations of PIK3R1 are a frequent event in both primary and metastatic samples. Follow-up studies are necessary to understand the clinical significance of these heterozygous losses in cancer progression and the effect of the disease state a patient is in at the time a sample is taken.
Although the MSK-IMPACT prostate cancer cohort consisted of 2,735 patients, we included samples from only 1,417 patients for downstream genomic and clinical analysis. This sample dropout rate is attributable to our use of the stringent FACETS quality control criteria (consisting of 11 quantitative and qualitative measures) for assessing inclusion of somatic copy number calls. The majority of samples that failed our quality assessment did so primarily because of ambiguous purity estimates and further incongruity in copy number states. We observed that many samples were “copy-number quiet” (ie, no detectable aberration from an euploid genomic configuration), a phenomenon that is well documented (20). Since sample purity estimation with FACETS predominately relies on the quantification of deviation from an euploid genome, we observed many ambiguous purity calls. We further note that purity estimates depict a crucial measure for genomic analysis (eg, clonality estimates); hence, we decided to concentrate on this reduced, quality-assessed cohort. Using less-stringent quality measures as well as choosing a different algorithm for purity estimation is possible, and this should be investigated further, but the potential implications of such an approach are difficult to predict.
In several other cancers (eg, breast, lung, kidney, and endometrial), PIK3R1 mRNA expression was reduced in tumor compared to normal tissue, and reduced expression was associated with poor OS, consistent with prior findings (48,49). Although only 2.3% of patients with breast cancer harbor alteration of PIK3R1, our data showed that PIK3R1 alteration was strongly enriched in the triple-negative subtype (9.1% of all TNBC cases). Similarly, our transcriptomic analysis revealed that the basal/TNBC subtype of breast cancer expresses significantly less PIK3R1 mRNA than other breast cancer subtypes, which to our knowledge has not been reported previously. A recent study also found that PIK3R1 was the most frequently mutated gene in Ewing sarcoma (at 59% of all cases) (50). Collectively, our data support previous observations (51) that loss/alteration of PIK3R1 may act as an oncogenic driver in multiple cancer types.
The major downstream effector of PI3K signaling is the protein serine/threonine kinase AKT, and phosphorylation of serine 473 (ser473) leads to its full activation (52). Activation of AKT plays a crucial role in prostate cancer progression, and AKT activation also confers resistance to therapy (53,54). PIK3R1 negatively regulates PI3K-AKT activation, and loss of PIK3R1 is known to result in increased AKT phosphorylation in various cell types and in GEMMs of cancer (11,30,39,45,46). The PI3K-AKT pathway is required for the insulin and insulin-like growth factor 1 (IGF1)-dependent cellular response (10,52,55). Canonically, insulin induces AKT activation (56). In a PIK3R1 wild-type cell, PIK3R1 protein/p85α limits insulin-induced AKT activation by binding and sequestering IGF1 (57). We showed that even transient knockdown of PIK3R1 (leading to a partial loss of PIK3R1 mRNA expression) alone is sufficient to induce AKT activation in a castration-sensitive and PIK3R1 and PTEN wild-type cell line (LAPC4). In a castration-resistant cell line, we observed that loss of PIK3R1 causes insulin-mediated AKT activation, indicating that the loss of PIK3R1 results in activation of the insulin-AKT pathway in prostate cancer cells. Even transient knockdown of PIK3R1 caused higher proliferation of cell lines, indicating these cells are more tumorigenic compared to wild-type cells. We also observed that knockdown of PIK3R1 reduces AR target gene expression (PSA and PLZF), indicating attenuation of AR function in response to PIK3R1 loss. Carver et al. reported a reciprocal feedback loop between the AR and PI3K pathways, in which AKT activation leads to inactivation of AR function and induces castration resistance (58). Previous data also showed that the PIK3R1 protein p85α directly interacts with the N-terminal region of AR (59). Our study also showed that treatment with the ARandrogen-receptor inhibitor enzalutamide treatment very modestly inhibits the growth of PIK3R1 knockdown LAPC4 cells (p=0.094) compared to vehicle (DMSO) tretated cells. However in the PIK3R1 proficient (control siRNA transfected) LAPC4 cells enzalutamide strongly reduces cell growth (p=6.9e-6) compared to vehicle treated cells. Together these data indicate that PIK3R1 loss may able to induce castration resistance by limiting AR function and activating Akt; however, in-depth studies using multiple experimental models are required to understand the molecular mechanisms associated with specific PIK3R1 driver mutation/copy number loss–induced castration resistance and to uncover the detailed molecular mechanisms underlying this process.
The critical role of PTEN in regulating PI3K pathway signaling raises the possibility that PI3K pathway inhibitors might be effective in PTEN-deficient prostate cancer (58). In fact, several PI3K/AKT inhibitors are currently in clinical trials(60). A recent phase 3 trial of ipatasertib (an AKT-inhibitor/AKTi) for patients with mCRPC (IPATential150, NCT03072238) showed that, in a subgroup of patients with mCRPC and PTEN loss, radiographic progression-free survival was slightly longer with ipatasertib plus the AR inhibitor abiraterone/prednisone compared with placebo plus abiraterone/prednisone (HR, 0.77; 95% CI, 0.61–0.98) (60). Moreover, ipatasertib monotherapy showed clinical benefit in heavily pretreated mCRPC patients and reduced growth of patient-derived xenografts (PDXs) that harbor AKT1 mutations (61). However, AKT inhibitors have never been explored in patients with known PIK3R1 alteration. Our results provide a rationale for such a study: we showed that PIK3R1 knockdown increases the sensitivity of prostate and breast cancer cells to AKT inhibitors. Moreover, an mCRPC organoid (MSK-PCa3) harboring a PIK3R1 oncogenic alteration showed greater sensitivity to AKT inhibitors (AKTi) than did a PIK3R1 wild-type organoid. Importantly, although both organoids harbor PTEN deletion (40), AKT selectively reduces growth only in PIK3R1-mutated organoids that also harbor PTEN deletion. Therefore, we hypothesize that AKTi-based therapy could have significant benefit for patients with mCRPC, and potentially also for patients with mCSPC or high-risk primary disease whose tumors show loss/alteration of PIK3R1. A previous study showed that a double-PIK3CA mutation predicted increased sensitivity to PI3kα inhibitors compared to a single hotspot mutation in patients with breast cancer (62). We observed strong co-occurence between PIK3R1 alteration/loss and PTEN mutation in both primary and metastatic prostate cancer. Therefore, it is plausible that prostate cancers that harbor dual alteration of PIK3R1 and PTEN may exhibit higher sensitivity towards AKT/PI3K inhibitors, a testable hypothesis for future studies.
FDG-PET/CT is a well-established, safe, and well-studied imaging tool that is FDA approved for use in staging various malignancies (41,63). The intensity of FDG uptake is a marker for the uptake of glucose by tumors (64), which in turn is closely correlated with certain types of tissue metabolism; and the intense FDG avidity of mCRPC associates with poor OS (27,65,66). Importantly, the correlation of FDG uptake with tumor genomic alteration/mutational status had not, to our knowledge, been previously addressed. In our analysis of patients in the MSK-IMPACT prostate cancer cohort, high FDG uptake was significantly associated only with alteration of PIK3R1. We found no statistically significant association of FDG uptake with any other mutations or signaling pathway. The canonical insulin-PI3K-AKT signaling acts as a primary driver of cellular glucose uptake via the transporter GLUT (67). Decades ago, a study in a GEMM showed that PIK3R1 deficiency increases insulin-mediated glucose uptake (7). Our finding of increased FDG uptake in PIK3R1-altered tumors gives clinical depth to this observation. Strikingly, we did not find any association between PTEN loss and glucose uptake, suggesting that PIK3R1 is the major player in the PI3K pathway in the regulation of insulin-mediated glucose uptake, which fuels cancer cell growth (68). The association of FDG uptake with a specific genomic alteration (ie, PIK3R1) may prove useful as a noninvasive screening test for potential eligibility in trials of therapy directed at the PI3K pathway.
Identifying vulnerabilities of PIK3R1-altered prostate cancer is particularly important given our finding of more aggressive clinicopathologic features associated with PIK3R1 alteration, including enrichment in metastatic prostate cancer and shorter biochemical recurrence-free survival. Our results also suggest that PIK3R1 alteration may define a distinct subtype in PI3K-dependent prostate cancer and may be associated with unique high glucose-uptake activity (summarized in Fig. 5J). In summary, our study for the first time identifies a direct association between PIK3R1 alterations and metastatic prostate cancer and demonstrates that men with mCRPC who harbor defective PIK3R1 have high FDG avidity and may benefit from PI3K/AKT inhibitors. Further in-depth studies are warranted to validate these findings and dissect therapeutic implications of PIK3R1-altered cancer.
Supplementary Material
STATEMENT OF TRANSLATIONAL RELEVANCE.
We found that PIK3R1 alterations in prostate cancer are associated with high PI3K-AKT-metabolic activity. Moreover, we identified frequent oncogenic alterations and reduced mRNA expression of PIK3R1 in advanced prostate cancer and several other tumor types including breast cancer and obtained evidence of its role in cancer progression and metastasis. Our study provides the first comprehensive evaluation of PIK3R1-alteration on activating PI3K-AKT signaling and resistance to anti-androgen therapy in prostate cancer. Thus, PIK3R1 alterations identify patients with possibly more aggressive disease, but more importantly, they identify patients for PI3K/AKT-inhibitor-based clinical trials.
Acknowledgements:
We thank Yu Chen (MSK) for the organoids, Cindy Lee of the MSK Human Oncology and Pathogenesis Program (HOPP) for organoid culture media, and Janet E. Novak (MSK) and Margaret McPartland (MSK) for editing.
Financial support:
This work was supported in part by a grant from the National Institutes of Health (NIH) to Memorial Sloan Kettering Cancer Center (P30 CA008748), a Department of Defense (DOD) Early Investigator Research Award (W81XWH-18-1-0330) to KHS, Prostate Cancer Foundation Young Investigator Awards to GC, KHS, WA, and LAM, and an NIH/NCI SPORE grant (P50 CA92629) to HIS. PWK was supported by a DOD Prostate Cancer Research Program (PCRP) award (W81XWH-19-1-0470) and NVKP was supported by a DOD PCRP award (W81XWH-19-1-0536). MGVH acknowledges support from the Emerald Foundation, the Lustgarten Foundation, a Faculty Scholar grant from the Howard Hughes Medical Institute, the MIT Center for Precision Cancer Medicine, the Ludwig Center at MIT, and the NCI (R35 CA242379, P30 CA14051). PWK and LAM were supported by a Prostate Cancer Foundation Challenge Award and by NCI grant P01 CA228696.
Conflict of interest disclosures:
H.I.S. reports the following support: compensated consultant/advisor to Ambry Genetics Corp, Konica Minolta Inc., Bayer, Pfizer Inc., Sun Pharmaceuticals, WCG Oncology; uncompensated consultant/advisor to Amgen, ESSA Pharma Inc., Janssen Research & Development, LLC, Janssen Biotech, Inc., Sanofi Aventis; has received research funding (to his institution) from Epic Sciences, Illumina, Inc., Janssen, Menarini Silicon Biosystems, Prostate Cancer Foundation, and ThermoFisher; intellectual property rights with BioNTech, Elucida Oncology, MaBVAX, and Y-mAbs Therapeutics, Inc., nonfinancial support from Amgen, Bayer, ESSA Pharma Inc., Menarini Silicon Biosystems, Phosplatin, Pfizer Inc., Prostate Cancer Foundation, and WCG Oncology.
M. J. M is an uncompensated consultant for Bayer, Advanced Accelerator Applications, Johnson and Johnson, Novartis, and Lantheus. He is a compensated consultant for Oric, Curium, Athenex, Exelexis, and Astra Zeneca. MSK receives funds for contracts for the conduct of clinical trials from Bayer, Advanced Accelerator Applications, Novartis, Corcept, Roche/Genentech, and Janssen.
T.T. is consultant/scientific advisory member for Genentech/Roche, Pfizer, AstraZenecam Merck, Puma Biotechnology, Athenex, Daiichi Sankyo, Ionis, Seattle Genetics, Eisai, Exact Science, Foundation Medicine, Ayala Pharmaceuticals, Gilead Sciences, Blueprint Medicines, Ellipses Pharma, Fuji Pharma, Iteos Therapeutics and Agendia; received grants/funding from Eisai, Pfizer, Novartis, Innocrin Pharma, AstraZeneca, Astellas Pharma, Immunomedics, Genentech/Roche, Daiichi Sankyo, Carrick Pharm, and Ayala Pharmaceuticals
P.R. received institutional grant/funding from Grail, Illumina, Novartis, Epic Sciences, and ArcherDx; and consultation/advisory board/honoraria from Novartis, Foundation Medicine, AstraZeneca, Epic Sciences, Inivata, Natera, and Tempus.
W.A. reports the following disclosures: he has received honoraria from Roche, Medscape, Aptitude Health, Clinical Education Alliance; he has a consulting/advisory role at Clovis Oncology, Janssen, MORE Health, ORIC Pharmaceuticals, and Daiichi Sankyo; he has obtained research funding (institutional) from AstraZeneca, Clovis Oncology, ORIC Pharmaceuticals, Epizyme, and Zenith Epigenetics.
J.C.D. is on the scientific advisory board and an investor in Adient Medical, Serpex Medical, and Verix Medical.
S.B.S. received grants/funding from Johnson & Johnson and GE Healthcare; and is a consultant for Johnson & Johnson, Advantagene, and Microbot; and is a shareholder of Johnson & Johnson, Lantheus, and Aperture Medical.
M.G.V.H. is a scientific advisory board member of Agios Pharmaceuticals, iTeos Therapeutics, Aeglea Biotherapeutics, and Faeth Therapeutics; is on the investment advisory board of DRIOA Ventures; and is a scientific founder and advisor for Auron Therapeutics.
L.A.M. receives grant/funding from Bayer and AstraZeneca, and has provided expert testimony for Bayer.
As of Feb 20, 2022 P.W.K. reports the following disclosures for the last 24-month period: he has had investment interest in Cogent Biosciences, Context Therapeutics LLC, Mirati, Placon, PrognomIQ, Seer Biosciences, SnyDevRx and XLink; he is a company board member for Context Therapeutics LLC; he is a company founder for XLink; he is co-founder and CEO of Convergent Therapeutics; he is/was a consultant/scientific advisory board member for Anji, Candel, Immunis, AI (previously OncoCellMDX), Janssen, Progenity, PrognomIQ, Seer Biosciences, SynDevRX, Tarveda Therapeutics, and Veru, and had served on data safety monitoring boards for Genentech/Roche and Merck. He reports spousal association with Bayer.
G.C., S.N., R.H., B.N., K.H.S., C.K., S.H.R., R.G., Y.Z.M., N.V.K.P., G.M.L., A.G.W. and N.S. have no conflicts of interest to declare.
REFERENCES
- 1.Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2022. CA Cancer J Clin 2022;72(1):7–33 doi 10.3322/caac.21708. [DOI] [PubMed] [Google Scholar]
- 2.Ahearn TU, Pettersson A, Ebot EM, Gerke T, Graff RE, Morais CL, et al. A Prospective Investigation of PTEN Loss and ERG Expression in Lethal Prostate Cancer. J Natl Cancer Inst 2016;108(2) doi 10.1093/jnci/djv346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mulholland DJ, Tran LM, Li Y, Cai H, Morim A, Wang S, et al. Cell autonomous role of PTEN in regulating castration-resistant prostate cancer growth. Cancer Cell 2011;19(6):792–804 doi 10.1016/j.ccr.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jamaspishvili T, Berman DM, Ross AE, Scher HI, De Marzo AM, Squire JA, et al. Clinical implications of PTEN loss in prostate cancer. Nat Rev Urol 2018;15(4):222–34 doi 10.1038/nrurol.2018.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.He Z, Tang F, Lu Z, Huang Y, Lei H, Li Z, et al. Analysis of differentially expressed genes, clinical value and biological pathways in prostate cancer. Am J Transl Res 2018;10(5):1444–56. [PMC free article] [PubMed] [Google Scholar]
- 6.Ye Y, Li SL, Wang SY. Construction and analysis of mRNA, miRNA, lncRNA, and TF regulatory networks reveal the key genes associated with prostate cancer. PLoS One 2018;13(8):e0198055 doi 10.1371/journal.pone.0198055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Terauchi Y, Tsuji Y, Satoh S, Minoura H, Murakami K, Okuno A, et al. Increased insulin sensitivity and hypoglycaemia in mice lacking the p85 alpha subunit of phosphoinositide 3-kinase. Nat Genet 1999;21(2):230–5 doi 10.1038/6023. [DOI] [PubMed] [Google Scholar]
- 8.Ueki K, Algenstaedt P, Mauvais-Jarvis F, Kahn CR. Positive and negative regulation of phosphoinositide 3-kinase-dependent signaling pathways by three different gene products of the p85alpha regulatory subunit. Mol Cell Biol 2000;20(21):8035–46 doi 10.1128/mcb.20.21.8035-8046.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheung LW, Yu S, Zhang D, Li J, Ng PK, Panupinthu N, et al. Naturally occurring neomorphic PIK3R1 mutations activate the MAPK pathway, dictating therapeutic response to MAPK pathway inhibitors. Cancer Cell 2014;26(4):479–94 doi 10.1016/j.ccell.2014.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen D, Mauvais-Jarvis F, Bluher M, Fisher SJ, Jozsi A, Goodyear LJ, et al. p50alpha/p55alpha phosphoinositide 3-kinase knockout mice exhibit enhanced insulin sensitivity. Mol Cell Biol 2004;24(1):320–9 doi 10.1128/mcb.24.1.320-329.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mauvais-Jarvis F, Ueki K, Fruman DA, Hirshman MF, Sakamoto K, Goodyear LJ, et al. Reduced expression of the murine p85alpha subunit of phosphoinositide 3-kinase improves insulin signaling and ameliorates diabetes. J Clin Invest 2002;109(1):141–9 doi 10.1172/jci13305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hanker AB, Kaklamani V, Arteaga CL. Challenges for the Clinical Development of PI3K Inhibitors: Strategies to Improve Their Impact in Solid Tumors. Cancer Discov 2019;9(4):482–91 doi 10.1158/2159-8290.CD-18-1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Bono J, Bracarda S, Sternberg C, Chi K, Olmos D, Sandhu S, et al. IPATential150: Phase III study of ipatasertib (ipat) plus abiraterone (abi) vs placebo (pbo) plus abi in metastatic castration-resistant prostate cancer (mCRPC). Annals of Oncology 2020;31(S4):S1153–S4. [Google Scholar]
- 14.Zehir A, Benayed R, Shah RH, Syed A, Middha S, Kim HR, et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat Med 2017;23(6):703–13 doi 10.1038/nm.4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Keegan NM, Vasselman SE, Barnett ES, Nweji B, Carbone EA, Blum A, et al. Clinical annotations for prostate cancer research: Defining data elements, creating a reproducible analytical pipeline, and assessing data quality. Prostate 2022. doi 10.1002/pros.24363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Razavi P, Chang MT, Xu G, Bandlamudi C, Ross DS, Vasan N, et al. The Genomic Landscape of Endocrine-Resistant Advanced Breast Cancers. Cancer Cell 2018;34(3):427–38 e6 doi 10.1016/j.ccell.2018.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res 2017;45(W1):W98–W102 doi 10.1093/nar/gkx247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nagy A, Munkacsy G, Gyorffy B. Pancancer survival analysis of cancer hallmark genes. Sci Rep 2021;11(1):6047 doi 10.1038/s41598-021-84787-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chakravarty D, Gao J, Phillips SM, Kundra R, Zhang H, Wang J, et al. OncoKB: A Precision Oncology Knowledge Base. JCO Precis Oncol 2017;2017 doi 10.1200/PO.17.00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ding L, Bailey MH, Porta-Pardo E, Thorsson V, Colaprico A, Bertrand D, et al. Perspective on Oncogenic Processes at the End of the Beginning of Cancer Genomics. Cell 2018;173(2):305–20 e10 doi 10.1016/j.cell.2018.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sanchez-Vega F, Mina M, Armenia J, Chatila WK, Luna A, La KC, et al. Oncogenic Signaling Pathways in The Cancer Genome Atlas. Cell 2018;173(2):321–37 e10 doi 10.1016/j.cell.2018.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 2013;6(269):pl1 doi 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, et al. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia 2004;6(1):1–6 doi 10.1016/s1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cortazar AR, Torrano V, Martin-Martin N, Caro-Maldonado A, Camacho L, Hermanova I, et al. CANCERTOOL: A Visualization and Representation Interface to Exploit Cancer Datasets. Cancer Res 2018;78(21):6320–8 doi 10.1158/0008-5472.CAN-18-1669. [DOI] [PubMed] [Google Scholar]
- 25.Gao D, Vela I, Sboner A, Iaquinta PJ, Karthaus WR, Gopalan A, et al. Organoid cultures derived from patients with advanced prostate cancer. Cell 2014;159(1):176–87 doi 10.1016/j.cell.2014.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gao H, Chakraborty G, Zhang Z, Akalay I, Gadiya M, Gao Y, et al. Multi-organ site metastatic reactivation mediated by non-canonical discoidin domain receptor 1 signaling. Cell 2016;166(1):47–62 doi 10.1016/j.cell.2016.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wibmer AG, Morris MJ, Gonen M, Zheng J, Hricak H, Larson SM, et al. Quantification of Metastatic Prostate Cancer Whole-Body Tumor Burden with Fdg Pet Parameters and Associations with Overall Survival after First Line Abiraterone or Enzalutamide: A Single- Center Retrospective Cohort Study. J Nucl Med 2021. doi 10.2967/jnumed.120.256602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shen R, Seshan VE. FACETS: allele-specific copy number and clonal heterogeneity analysis tool for high-throughput DNA sequencing. Nucleic Acids Res 2016;44(16):e131 doi 10.1093/nar/gkw520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marshall JDS, Whitecross DE, Mellor P, Anderson DH. Impact of p85alpha Alterations in Cancer. Biomolecules 2019;9(1) doi 10.3390/biom9010029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cheung LW, Hennessy BT, Li J, Yu S, Myers AP, Djordjevic B, et al. High frequency of PIK3R1 and PIK3R2 mutations in endometrial cancer elucidates a novel mechanism for regulation of PTEN protein stability. Cancer Discov 2011;1(2):170–85 doi 10.1158/2159-8290.CD-11-0039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Armenia J, Wankowicz SAM, Liu D, Gao J, Kundra R, Reznik E, et al. The long tail of oncogenic drivers in prostate cancer. Nat Genet 2018;50(5):645–51 doi 10.1038/s41588-018-0078-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim MS, Pinto SM, Getnet D, Nirujogi RS, Manda SS, Chaerkady R, et al. A draft map of the human proteome. Nature 2014;509(7502):575–81 doi 10.1038/nature13302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cancer Genome Atlas Research N. The Molecular Taxonomy of Primary Prostate Cancer. Cell 2015;163(4):1011–25 doi 10.1016/j.cell.2015.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Taylor BS, Schultz N, Hieronymus H, Gopalan A, Xiao Y, Carver BS, et al. Integrative genomic profiling of human prostate cancer. Cancer Cell 2010;18(1):11–22 doi 10.1016/j.ccr.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Grasso CS, Wu YM, Robinson DR, Cao X, Dhanasekaran SM, Khan AP, et al. The mutational landscape of lethal castration-resistant prostate cancer. Nature 2012;487(7406):239–43 doi 10.1038/nature11125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 2012;483(7391):603–7 doi 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Manning BD, Toker A. AKT/PKB Signaling: Navigating the Network. Cell 2017;169(3):381–405 doi 10.1016/j.cell.2017.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhu Q, Youn H, Tang J, Tawfik O, Dennis K, Terranova PF, et al. Phosphoinositide 3-OH kinase p85alpha and p110beta are essential for androgen receptor transactivation and tumor progression in prostate cancers. Oncogene 2008;27(33):4569–79 doi 10.1038/onc.2008.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Taniguchi CM, Winnay J, Kondo T, Bronson RT, Guimaraes AR, Alemán JO, et al. The phosphoinositide 3-kinase regulatory subunit p85alpha can exert tumor suppressor properties through negative regulation of growth factor signaling. Cancer Res 2010;70(13):5305–15 doi 10.1158/0008-5472.Can-09-3399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gao D, Vela I, Sboner A, Iaquinta PJ, Karthaus WR, Gopalan A, et al. Organoid cultures derived from patients with advanced prostate cancer. Cell 2014;159(1):176–87 doi 10.1016/j.cell.2014.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fox JJ, Schoder H, Larson SM. Molecular imaging of prostate cancer. Curr Opin Urol 2012;22(4):320–7 doi 10.1097/MOU.0b013e32835483d5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gyorffy B, Lanczky A, Eklund AC, Denkert C, Budczies J, Li Q, et al. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res Treat 2010;123(3):725–31 doi 10.1007/s10549-009-0674-9. [DOI] [PubMed] [Google Scholar]
- 43.Curtis C, Shah SP, Chin SF, Turashvili G, Rueda OM, Dunning MJ, et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature 2012;486(7403):346–52 doi 10.1038/nature10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jean S, Kiger AA. Classes of phosphoinositide 3-kinases at a glance. J Cell Sci 2014;127(Pt 5):923–8 doi 10.1242/jcs.093773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Luo J, Sobkiw CL, Logsdon NM, Watt JM, Signoretti S, O’Connell F, et al. Modulation of epithelial neoplasia and lymphoid hyperplasia in PTEN+/− mice by the p85 regulatory subunits of phosphoinositide 3-kinase. Proc Natl Acad Sci U S A 2005;102(29):10238–43 doi 10.1073/pnas.0504378102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thorpe LM, Spangle JM, Ohlson CE, Cheng H, Roberts TM, Cantley LC, et al. PI3K-p110alpha mediates the oncogenic activity induced by loss of the novel tumor suppressor PI3K-p85alpha. Proc Natl Acad Sci U S A 2017;114(27):7095–100 doi 10.1073/pnas.1704706114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lucas CL, Zhang Y, Venida A, Wang Y, Hughes J, McElwee J, et al. Heterozygous splice mutation in PIK3R1 causes human immunodeficiency with lymphoproliferation due to dominant activation of PI3K. J Exp Med 2014;211(13):2537–47 doi 10.1084/jem.20141759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cizkova M, Vacher S, Meseure D, Trassard M, Susini A, Mlcuchova D, et al. PIK3R1 underexpression is an independent prognostic marker in breast cancer. BMC Cancer 2013;13:545 doi 10.1186/1471-2407-13-545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Taniguchi CM, Winnay J, Kondo T, Bronson RT, Guimaraes AR, Aleman JO, et al. The phosphoinositide 3-kinase regulatory subunit p85alpha can exert tumor suppressor properties through negative regulation of growth factor signaling. Cancer Res 2010;70(13):5305–15 doi 10.1158/0008-5472.CAN-09-3399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jagodzinska-Mucha P, Sobczuk P, Mikula M, Raciborska A, Dawidowska A, Kulecka M, et al. Mutational landscape of primary and recurrent Ewing sarcoma. Contemp Oncol (Pozn) 2021;25(4):241–8 doi 10.5114/wo.2021.112234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jaiswal BS, Janakiraman V, Kljavin NM, Chaudhuri S, Stern HM, Wang W, et al. Somatic mutations in p85alpha promote tumorigenesis through class IA PI3K activation. Cancer Cell 2009;16(6):463–74 doi 10.1016/j.ccr.2009.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hemmings BA, Restuccia DF. PI3K-PKB/Akt pathway. Cold Spring Harb Perspect Biol 2012;4(9):a011189 doi 10.1101/cshperspect.a011189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Majumder PK, Febbo PG, Bikoff R, Berger R, Xue Q, McMahon LM, et al. mTOR inhibition reverses Akt-dependent prostate intraepithelial neoplasia through regulation of apoptotic and HIF-1-dependent pathways. Nat Med 2004;10(6):594–601 doi 10.1038/nm1052. [DOI] [PubMed] [Google Scholar]
- 54.Li B, Sun A, Youn H, Hong Y, Terranova PF, Thrasher JB, et al. Conditional Akt activation promotes androgen-independent progression of prostate cancer. Carcinogenesis 2007;28(3):572–83 doi 10.1093/carcin/bgl193. [DOI] [PubMed] [Google Scholar]
- 55.Fruman DA, Chiu H, Hopkins BD, Bagrodia S, Cantley LC, Abraham RT. The PI3K Pathway in Human Disease. Cell 2017;170(4):605–35 doi 10.1016/j.cell.2017.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mackenzie RW, Elliott BT. Akt/PKB activation and insulin signaling: a novel insulin signaling pathway in the treatment of type 2 diabetes. Diabetes Metab Syndr Obes 2014;7:55–64 doi 10.2147/DMSO.S48260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Luo J, Field SJ, Lee JY, Engelman JA, Cantley LC. The p85 regulatory subunit of phosphoinositide 3-kinase down-regulates IRS-1 signaling via the formation of a sequestration complex. J Cell Biol 2005;170(3):455–64 doi 10.1083/jcb.200503088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Carver BS, Chapinski C, Wongvipat J, Hieronymus H, Chen Y, Chandarlapaty S, et al. Reciprocal feedback regulation of PI3K and androgen receptor signaling in PTEN-deficient prostate cancer. Cancer Cell 2011;19(5):575–86 doi 10.1016/j.ccr.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Baron S, Manin M, Beaudoin C, Leotoing L, Communal Y, Veyssiere G, et al. Androgen receptor mediates non-genomic activation of phosphatidylinositol 3-OH kinase in androgen-sensitive epithelial cells. J Biol Chem 2004;279(15):14579–86 doi 10.1074/jbc.M306143200. [DOI] [PubMed] [Google Scholar]
- 60.Shi Z, Sweeney C, Bracarda S, Sternberg CN, Chi KN, Olmos D, et al. Biomarker analysis of the phase III IPATential150 trial of first-line ipatasertib (Ipat) plus abiraterone (Abi) in metastatic castration-resistant prostate cancer (mCRPC). Journal of Clinical Oncology 2020;38(6_suppl):182- doi 10.1200/JCO.2020.38.6_suppl.182. [DOI] [Google Scholar]
- 61.Herberts C, Murtha AJ, Fu S, Wang G, Schönlau E, Xue H, et al. Activating AKT1 and PIK3CA Mutations in Metastatic Castration-Resistant Prostate Cancer. Eur Urol 2020;78(6):834–44 doi 10.1016/j.eururo.2020.04.058. [DOI] [PubMed] [Google Scholar]
- 62.Vasan N, Razavi P, Johnson JL, Shao H, Shah H, Antoine A, et al. Double PIK3CA mutations in cis increase oncogenicity and sensitivity to PI3Kalpha inhibitors. Science 2019;366(6466):714–23 doi 10.1126/science.aaw9032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Larson SM, Schoder H. New PET tracers for evaluation of solid tumor response to therapy. Q J Nucl Med Mol Imaging 2009;53(2):158–66. [PubMed] [Google Scholar]
- 64.Sokoloff L, Reivich M, Kennedy C, Des Rosiers MH, Patlak CS, Pettigrew KD, et al. The [14C]deoxyglucose method for the measurement of local cerebral glucose utilization: theory, procedure, and normal values in the conscious and anesthetized albino rat. J Neurochem 1977;28(5):897–916 doi 10.1111/j.1471-4159.1977.tb10649.x. [DOI] [PubMed] [Google Scholar]
- 65.Fox JJ, Gavane SC, Blanc-Autran E, Nehmeh S, Gonen M, Beattie B, et al. Positron Emission Tomography/Computed Tomography-Based Assessments of Androgen Receptor Expression and Glycolytic Activity as a Prognostic Biomarker for Metastatic Castration-Resistant Prostate Cancer. JAMA Oncol 2018;4(2):217–24 doi 10.1001/jamaoncol.2017.3588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Vargas HA, Wassberg C, Fox JJ, Wibmer A, Goldman DA, Kuk D, et al. Bone metastases in castration-resistant prostate cancer: associations between morphologic CT patterns, glycolytic activity, and androgen receptor expression on PET and overall survival. Radiology 2014;271(1):220–9 doi 10.1148/radiol.13130625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Holman GD, Kasuga M. From receptor to transporter: insulin signalling to glucose transport. Diabetologia 1997;40(9):991–1003 doi 10.1007/s001250050780. [DOI] [PubMed] [Google Scholar]
- 68.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science 2009;324(5930):1029–33 doi 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Genomic data from the MSK-IMPACT prostate cancer samples are available via cBioPortal for Cancer Genomics: https://www.cbioportal.org/study/summary?id=prad_pik3r1_msk_2021