Abstract
pAD1 is a 60-kb hemolysin-bacteriocin plasmid in Enterococcus faecalis that encodes a conjugative mating response to a peptide sex pheromone, cAD1, secreted by plasmid-free bacteria. The pheromone response is regulated by two proteins: TraE1, which positively regulates all or most conjugative structural genes, and TraA, which negatively regulates traE1. TraA binds to pAD1 DNA at the iad (encoding the inhibitor peptide iAD1) promoter but is released upon binding to imported pheromone. This leads to enhanced transcription through two closely spaced downstream terminators (t1 and t2) into traE1. TraE1 is believed to then upregulate itself from a site located within t2; thus, a small amount of transcription through t1-t2 could lead to overall induction. It is important therefore that the t1-t2 terminators be tightly controlled to keep the response shut down in the absence of pheromone. A small (200-nucleotide) RNA molecule designated mD is encoded just upstream of t1 by a determinant (traD) oriented in the direction opposite to that of transcripts utilizing t1. mD is expressed at high levels in the uninduced state, but it decreases significantly upon induction. Here we present results of genetic studies relating to the activity of t1-t2 and show that mD strongly enhances transcriptional termination at t1. The mD activity is shown to influence transcription well downstream and can affect the determinant for aggregation substance asa1. The phenomenon is specific in that there is no effect of mD on the unrelated pheromone-responding plasmids pPD1 and pCF10.
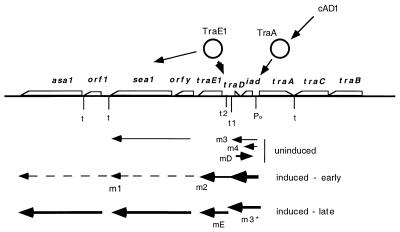
The conjugative plasmid pAD1 (60 kb), originally identified in the multiple-antibiotic-resistant Enterococcus faecalis DS16 (12, 40), encodes a hemolysin-bacteriocin (cytolysin) that contributes to virulence in animal models (6, 25, 27). Hemolytic clinical isolates from globally diverse geographical locations frequently harbor a highly similar, if not identical, plasmid (8, 9). pAD1 encodes a mating response to an octapeptide sex pheromone (cAD1) secreted by plasmid-free recipient cells (32), and investigations into the regulation of this response have been ongoing in our laboratory for a number of years. (For recent reviews of the enterococcal pheromone response phenomenon, see references 9, 15, and 29). The response leads to synthesis of a proteinaceous aggregation substance (21, 22) encoded by asa1; this protein binds to enterococcal binding substance which is present on the surface of both recipients and donors and may correspond in part to lipoteichoic acid (16). A second surface protein, Sea1, is expressed at a low level in the uninduced state but is upregulated upon exposure to pheromone and appears to affect surface exclusion (43). When donors are exposed to a culture filtrate of recipients (plasmid-free bacteria), they exhibit a clumping response which can even result in donor-donor mating, although at a reduced frequency compared to that involving transfer to plasmid-free cells (10). The clumping response provides the basis of a convenient assay for the quantitation of pheromone by serial dilution. The locations of asa1 and sea1, as well as other determinants relating to the regulation of the pheromone response, are shown in Fig. 1.
FIG. 1.
pAD1 map showing regulatory determinants and transcripts observed in the uninduced, early-induced (e.g., 5 min), and late-induced (e.g., 30 min) states. The arrows below the map represent the various transcripts and their orientations. Induction is viewed as the pheromone (cAD1) binding to TraA, causing a release of its binding to the iad (which encodes inhibitor iAD1) promoter, leading to enhanced transcription from the promoter as well as transcription through t1. The “induced-early” transcripts reflect various extensions resulting from read-through of the t1-t2 terminators giving rise to m1 and m2. The dashed line indicates that, while transcription may continue in the indicated direction, the relative degrees have not been determined. The thickness of the arrows corresponds qualitatively with the relative levels of expression. m3 terminates at t1, whereas m3* is an extension of m3 believed to terminate at t2. mD and m4 overlap at their 3′ ends by about 80 nt. mE is viewed as a transcript of traE1 which has been activated at its own promoter within t2 by TraE1 initially generated from the transient expression of m2. mD is generally expressed at a high level in the uninduced state but is downregulated by the pheromone-induced transcription from the opposing iad promoter.
When pAD1 is acquired by a recipient, it shuts down or masks the production of cAD1; the extent of the shutdown depends on the bacterial host (34). The transconjugants continue to secrete different pheromones which are specific for donors carrying different families of responsive plasmids (8, 13). pAD1 itself encodes an octapeptide (iAD1) which is secreted and acts as a competitive inhibitor (antipheromone) of cAD1 (24, 31); its purpose is believed to be the prevention of self-induction of donors that may still be releasing some endogenous cAD1. The iAD1 peptide corresponds to the last 8 amino acid residues of a 22-amino-acid amphipathic precursor resembling a lone signal sequence (11); its determinant is designated iad.
pAD1 encodes two primary regulatory proteins: TraE1, which positively regulates structural genes required for conjugation as well as itself, and TraA, which negatively regulates expression of TraE1 (Fig. 1). TraA acts to downregulate transcription from the iad promoter which governs expression of a transcript extending about 450 nucleotides (nt) to a termination region (t1-t2). t1 and t2 are separated by 125 nt, and based on energy considerations, the stem of t1 is stronger than that of the downstream t2 (37). The initiation of traE1 expression results from transcriptional readthrough of t1-t2 from the iad promoter (20, 37, 38). We have previously reported that TraA binds directly to imported cAD1, causing the protein to release its binding to DNA at the iad promoter, resulting in upregulation of transcription (18). This increase in transcription is assumed to contribute to readthrough into traE1.
Another factor has recently been identified which, like TraA, also negatively regulates traE1. This relates to a small open reading frame (traD), but there is evidence that it is the related transcript mD (200 nt) that is the active component (2, 3). mD is transcribed in the opposite direction from that of iad, from a promoter located downstream of iad. The mD transcript is present at a high level in uninduced cells; upon exposure of cells to cAD1, however, it becomes greatly reduced within 5 min and is almost undetectable after 10 min (3). It was suggested that mD acts negatively, along with TraA, to reduce transcription from the iad promoter and thus help prevent transcriptional readthrough into traE1. A temperature-sensitive mutant presumed to affect the conformation of mD was observed to cause induction at elevated temperature (32 to 42°C) as well as to enhance transcription from the iad promoter. Thermal induction of the conjugation response could be complemented in trans by plasmid clones carrying the mD (traD) determinant (2). It was hypothesized that reduction of mD synthesis upon pheromone exposure was related to the enhanced transcription from the iad promoter blocking transcriptional initiation from the downstream mD promoter on the opposite DNA strand. Based on structural considerations, both mD RNA and RNA transcribed from the iad promoter to t1 exhibit extensive secondary structure, and it is thus reasonable that point mutations might significantly affect folding.
In the present communication, we focus on the function of t1-t2 and show that, in addition to whatever influence mD may have at the iad promoter, it also has an effect independent of TraA on transcription through t1. We also show that, despite significant complementarity between the mD transcript and a potential transcript from a region just upstream of the asa1 determinant (3), mD does not appear to affect transcription here.
MATERIALS AND METHODS
Bacterial strains and plasmids.
Bacterial strains and plasmids used in this study are listed in Table 1. pAM714 (23) is a derivative of pAD1 carrying an insertion of the erythromycin resistance transposon Tn917 in a region (1) not involved in transfer. Its behavior with regard to transfer is therefore assumed to reflect the wild-type pAD1.
TABLE 1.
Strains and plasmids used in this studya
| Strain or plasmid | Relevant features | Source or reference(s) |
|---|---|---|
| Strains | ||
| E. faecalis | ||
| OG1X | str gel | 24 |
| FA2-2 | rif fus | 12 |
| JH2-2 | rif fus | 26 |
| E. coli | ||
| DH5α | F− φ80 lacZ ΔM15 endA1 recA1 hsdR17 (rK− mK+) supE44 thi-1 ΔgyrA96 Δ(lacZYA-argF)U169 | Bethesda Research Laboratories |
| Plasmids | ||
| pAD1 | 60-kb Hly-Bac | 12, 40 |
| pPD1 | 59-kb Bac | 19, 45 |
| pCF10 | 54-kb Tc(Tn925) | 14 |
| pAM714 | pAD1::Tn917; Hly-Bac erm; wild-type mating properties | 1, 23 |
| pAM714-d1HT | pAM714 derivative with point mutation (Met to Arg) in traD | 3 |
| pAM2703 | pAM714 with ts mutation in traD | 2 |
| pAM2120 | pAD1 with Tn917 insert in traA | 34 |
| pAM-HT | pAM401 with cloned 321-nt PCR fragment carrying traD | 3 |
| pAM-HTd1 | Same as pAM-HT except with the d1 (traD Met-to-Arg) mutation | 3 |
| pAM-HTts | Same as pAM-HT except with the ts lesion of pAM2703 | This study |
| pAM7304 | pAD1 with Tn917lac insertion between traD and iad | 36 |
| pAM7348 | pAD1 with Tn917lac insertion between traD and t1 | 36 |
| pAM2019 | pAD1 with Tn917lac insertion within t1 | 36 |
| pAM7343 | pAD1 with Tn917lac insertion between t1 and t2 | 36 |
| pAM2011 | pAD1 with Tn917lac insertion between t2 and traE1 | 36 |
| pAM7016 | pAD1 with Tn917lac insertion in sea1 | 35 |
| pAM7025 | pAD1 with Tn917lac insertion in asa1 | 35 |
| pAM2125 | pAD1 with Tn917lac insertion between traE1 and orfy and a Tn917lac insertion in traA | 41 |
| pBluescript KS(+) | E. coli plasmid vector; amp | Stratagene |
| pAM1.6SK | 1.6-kb fragment (iad through traE1) cloned in pBluescript | 11 |
| pSF141 | Cloning vector (Kmr and Cmr); suicidal in E. faecalis | 39 |
| pAM401 | E. coli-E. faecalis shuttle; cat tet | 44 |
| pALTER-1 | Mutagenesis vector; Ampr | Promega |
| pALTER::1.6SK | 1.6-kb KpnI/BamHI fragment from pAM1.6SK cloned in pALTER | This study |
| pSF141::1.6SK | 1.6 kb EcoRI fragment from pALTER::1.6SK cloned in pSF141 | This study |
| pAM1220 | pAM714 with point mutation, ATG to ATA, at orfTTS1 start site | This study |
| pAM1221 | pAM714 with AT insertion in t1 repeat | This study |
| pAM1222 | pAM714 with AGCT insert in XbaI site in mD (traD) promoter | This study |
| pAM1224 | pAM714 with HpaI/BstZ17I deletion destroying t1 | This study |
| pAM1231 | pAM714 with AccI/XbaI deletion destroying t1 | This study |
| pAM7016-dHB | pAM7016 also carrying the t1 deletion of pAM1224 | This study |
| pAM7025-dHB | pAM7025 also carrying the t1 deletion of pAM1224 | This study |
Hly, hemolysin; Bac, bacteriocin; ts, temperature sensitivity.
Media and reagents.
Media used were Todd-Hewitt broth (Difco Laboratories) or N2GT (nutrient broth no. 2 [Oxoid Ltd., London, United Kingdom] supplemented with 0.2% glucose and 0.1 M Tris-HCl [pH 7.5]) for E. faecalis and Luria-Bertani broth (28) for Escherichia coli. Solid media were prepared by including 1.5% agar. Cultures were grown at 37°C unless otherwise noted. Cell density was determined using a Spectronic colorimeter at a wavelength of 600 nm. General reagents were essentially as previously described (2, 3). The following antibiotics were used at the indicated concentrations; ampicillin, 100 μg/ml; erythromycin, 20 μg/ml; streptomycin, 500 μg/ml; kanamycin, 500 μg/ml; spectinomycin, 500 μg/ml; chloramphenicol, 50 μg/ml in the case of E. coli and 20 μg/ml in the case of E. faecalis; rifampin, 25 μg/ml; and fusidic acid, 25 μg/ml. X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) (Gibco BRL) was used at a concentration of 40 μg/ml for E. coli and 80 μg/ml for E. faecalis. Synthetic pheromone peptides were prepared at the University of Michigan Core Peptide Synthesis Facility.
Construction of plasmid chimeras for use in complementation studies.
pAM-HTts (pAM1240) was constructed using the same PCR method and primers for construction of pAM-HT and pAM-HTd1 as described in the work of Bastos et al. (3). BamHI sites were incorporated into the 5′ termini of the primers, allowing for cloning into the BamHI site of the shuttle vector pAM401. pAM-HTts has the same orientation in the vector as the corresponding segments in pAM-HT and pAM-HTd1.
Pheromone response and beta-galactosidase (LacZ) assays.
The LacZ activities were determined as previously described (41), with the following minor modifications. One hundred microliters of an overnight culture of the appropriate strain was subcultured in 2.5 ml of fresh N2GT medium and grown for 30 min at appropriate temperatures. Cultures were then exposed for 90 min with synthetic cAD1 at concentrations of 0, 20, 40, 100, and 200 ng/ml. Cells were harvested from 2.1-ml aliquots, suspended in 200 μl of 0.05 M sodium phosphate buffer (pH 7.5), and permeabilized with 100 μl of toluene for 15 min at 37°C. Samples were incubated in 0.1 ml of 3.2 mM reduced glutathione–0.25 ml of 0.01 M o-nitrophenyl-β-d-galactopyranoside–2.0 ml of 0.05 M sodium phosphate buffer (pH 7.5) for 30 min at 37°C. The reaction was stopped with 0.5 ml of 1 M Na2CO3, and cell debris was removed by centrifugation. Absorbancy was determined at 420 nm on a Spectronic colorimeter, and the results were expressed in Miller units (MU) (30).
Analysis of transcripts.
RNA was prepared as described previously (3). The probe representing the region between t1 and t2 corresponded to 124 nt and was generated as follows. A PCR-amplified segment between traD and just upstream of t2 was prepared using the upstream primer 5′-ATTTTGGATCCATTGATAACTTTATTTGTTGT-3′ and the downstream primer 5′-AAATTGGATCCACGGCTCTTACGAGTAGTTC-3′ (both contain BamHI sites near the 5′ ends) and was cloned into the BamHI site of pBluescript II KS(+). Then an HpaI/BamHI fragment of the plasmid containing the region between t1 and t2 was subcloned into the EcoRV/BamHI site of pBluescript II KS(+) (designated pAM1235). A HindIII/BamHI fragment of this construct was used as a probe in the Northern blot experiment. Northern blot analyses involved electrophoresis in a 1.2% agarose-MOPS (morpholinepropanesulfonic acid)-formaldehyde system. Hybridization was carried out at 42°C in the presence of 50% formamide. The probe was labeled by nick translation using a kit from Bethesda Research Laboratories.
Construction of the orfTTS1 mutation in pAM714.
The orfTTS1 mutation was constructed as follows using the Promega Altered Site II in vitro mutagenesis kit. pAM1.6SK is a pBluescript clone carrying a 1.6-kb pAD1 fragment extending from upstream of iad to downstream of traE1 (11). The 1.6-kb pAD1 DNA fragment was removed using BamHI and KpnI and cloned into the BamHI/KpnI site of the pALTER-1 E. coli vector; the derivative was designated pALTER::1.6SK. Site-directed mutagenesis of pALTER::1.6SK was done using the kit protocol with some minor modifications. A synthetic 21-mer primer (5′-AGCGGGGAATATATACAGTTC-3′) designed to make the desired orfTTS1 mutation (ATG to ATA; Met to Ile) was used. Mutated DNA was expected to be devoid of a BstZ17I cleavage site (GTATAC) that could be screened easily. A 1.6-kb EcoRI fragment from a mutated plasmid was subcloned into the EcoRI site of the pSF141 (encoding Kmr and Cmr determinants that are expressed in gram-positive bacteria [39]) vector that is suicidal in E. faecalis. The recombinant plasmid pSF141::1.6SK was introduced into E. faecalis OG1X/pAM714 by electroporation (17). Transformants (assumed to carry a cointegrate plasmid) were selected on plates containing erythromycin and kanamycin. A recombinant derivative was subcultured for 14 passages in Todd-Hewitt broth without drug and plated on medium containing erythromycin. Colonies were then replica plated on medium containing kanamycin, and candidates (Emr Kms) were screened using PCR amplification to obtain DNA to be examined for the absence of the BstZ17I cleavage site. The two primers used for PCR and cloning here were 5′-TTACGGAATTCAGATCATGCGTGTTGTGTACC-3′ (within traA) and 5′-ATTGCGAATTCACAGCAAACATCCCCTCAATT-3′ (within traE1). One candidate containing the expected mutation was designated pAM1220; the mutation was confirmed by sequencing.
Construction of mutations within the t1 region.
The 1.6-kb EcoRI fragment of pALTER::1.6SK was cloned into the EcoRI site of pSF141 (designated pSF141::1.6SK). Four kinds of mutations were constructed using pSF141::1.6SK by utilizing the XbaI, AccI, BstZ17I, and HpaI cleavage sites around the t1 sequence. pAM1213 has a deletion of the segment between the BstZ17I and HpaI cleavage sites. pAM1218 has a deletion of the segment between the XbaI and AccI cleavage sites. pAM1216 has an extra 4 bp generated by use of the Klenow enzyme to fill in the ends resulting from a cleaved XbaI site. pAM1217 has an additional 2 bp generated using the Klenow enzyme to fill in the ends resulting from a cleaved AccI site. The mutations in the four mutated plasmids were incorporated into pAM714 in the same manner as described above to generate pAM1220. The resulting plasmids were designated pAM1221, pAM1222, pAM1224, and pAM1231, respectively. In the cases where a t1-related mutation was incorporated into plasmid pAM7016 (Tn917lac inserted in sea1) and pAM7025 (Tn917lac inserted in asa1) to generate pAM7016dHB and pAM7025dHB, respectively, the pAM1213 plasmid (deletion between BstZ17I and HpaI) was utilized for the allelic exchange.
RESULTS
pAM-HT corresponds to the E. coli-E. faecalis shuttle vector pAM401 carrying a 321-nt PCR fragment containing traD; when pAM-HT is present in E. faecalis, a high level of expression of mD RNA has been observed (3). pAM-HT and a derivative, pAM-HTd1, unable to translate traD, were previously shown to be capable of complementing in trans a temperature-sensitive pAD1 point mutation (pAM2703) located in traD (3). mD RNA was therefore assumed to be the critical product in restoring the normal (temperature-stable) phenotype.
Table 2 shows that when E. faecalis cells carrying the wild-type pAD1 also carry pAM-HT, they become significantly less sensitive to cAD1; this was the case for two nonisogenic hosts, OG1X and FA2-2. To exhibit a clumping response, bacteria carrying pAM-HT required a pheromone concentration eight times higher than that when the cells were harboring the empty vector. The effect was specific for pAD1; no difference was observed when pAM-HT was present with plasmid pPD1 (which responds to cPD1), and only a twofold difference was noted for pCF10 (which responds to cCF10).
TABLE 2.
Effects of mD on the sensitivity of E. faecalis strains harboring pAD1, pPD1, or pCF10
| Strain/plasmid | Pheromone | Pheromone concn (ng/ml) required to generate clumping response after addition of plasmid:
|
Difference (fold) | |
|---|---|---|---|---|
| pAM401 (vector) | pAM-HT (cloned traD) | |||
| OG1X/pAD1 | cAD1 | 16 | 125 | 8 |
| FA2-2/pAD1 | cAD1 | 64 | 500 | 8 |
| JH2-2/pPD1 | cPD1 | 250 | 250 | 0 |
| FA2-2/pCF10 | cCF10 | 0.5 | 1.0 | 2 |
mD RNA influence at the transcription termination site t1.
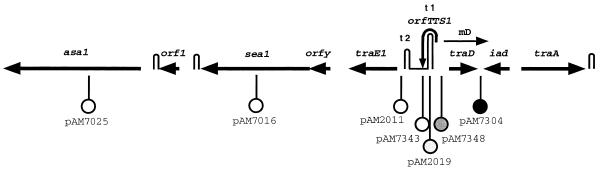
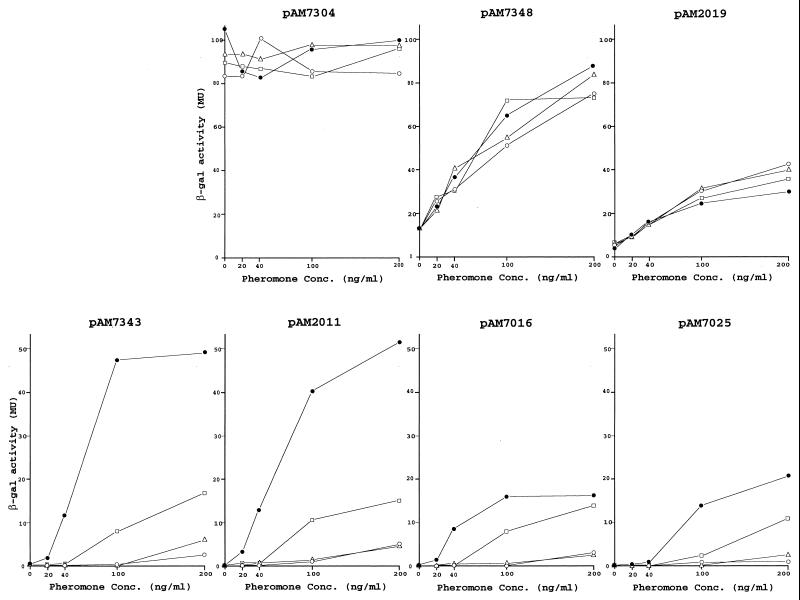
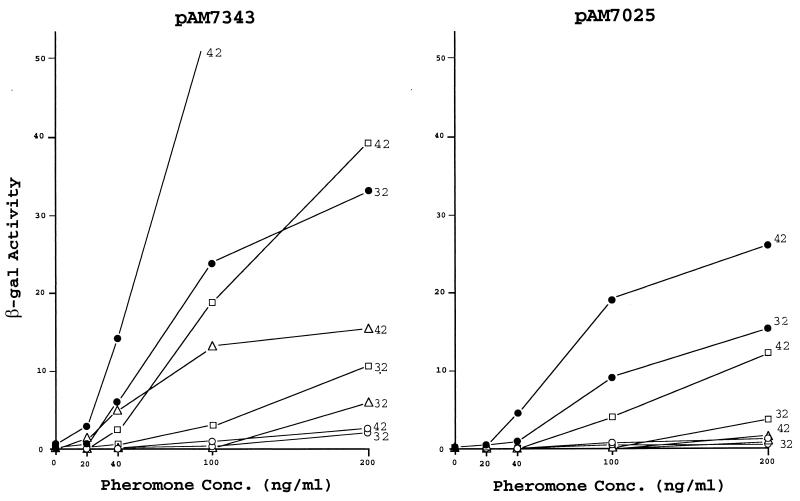
In an effort to determine how the mD product may affect transcription at different locations on pAD1, we examined a series of lacZ fusions corresponding to several pAD1::Tn917lac insertions. A map showing the location of these fusions, all of which are oriented to report transcription from right to left, is shown in Fig. 2. The data presented in Fig. 3 show that when excess mD is provided by pAM-HT or pAM-HTd1 there is a major effect on the expression of beta-galactosidase only on fusions that are located downstream of t1. For example, cells harboring pAM7343, containing the fusion just downstream of t1, were much less sensitive to cAD1, whereas there was no effect on pAM7348 with its insert just upstream of t1. With respect to pAM2019, where the insert is located within t1, presumably making the terminator nonfunctional, no effect was apparent. The effect of mD was observed well downstream as in the cases represented by the insertions in asa1 (pAM7025) and sea1 (pAM7016). Transcription in these determinants appeared lower than that for regions upstream. This is consistent with previous observations relating to Northern blot analyses (38) which showed that transcripts that extended to these genes were present at relatively low levels. The insertion between iad and traD (pAM7304) led to a full constitutive expression of LacZ, which is not complemented in trans—a result that we have noted previously (3).
FIG. 2.
Map showing the location of the Tn917lac insertions that are present in the indicated plasmid derivatives. All of the insertions are oriented such that the promoterless lacZ on the transposon will report transcription from right to left. The circles that are filled or contain shades of gray represent various levels of LacZ expression in the absence of pheromone. The unfilled circles mean that there is essentially no expression of LacZ in the uninduced state. The hairpin structures, in addition to t1 and t2, correspond to likely transcription terminators.
FIG. 3.
Induced expression of β-galactosidase by various pAD1::Tn917lac derivatives in response to increasing concentrations of cAD1 while in the presence of additionally provided mD in trans. The host strain was E. faecalis FA2-2 carrying the indicated plasmid. The plasmids provided in trans were pAM401 (●), pAM-HT (○), pAM-HTd1 (▵), and pAM-HTts1 (□). Exposure to cAD1 was for 1 h in each case.
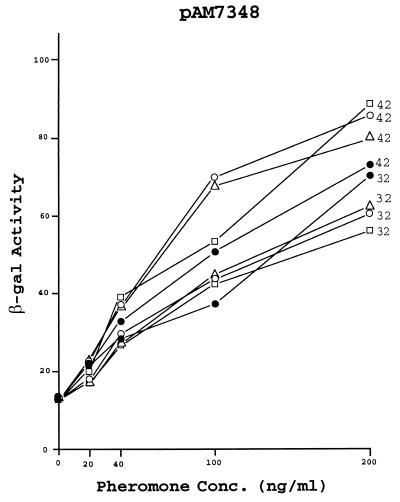
pAM-HTts1 differs from pAM-HT in that the 321-nt segment that includes traD contains the temperature-sensitive lesion of pAM2703. As shown in Fig. 3, cells carrying the mutated chimera are still reduced in their sensitivity to pheromone but to a much lesser extent than that for cells carrying the corresponding wild-type DNA. We note that these experiments along with those utilizing pAM-HT and pAM-HTd1 were conducted at 37°C, a temperature midway between 32 and 42°C, the differential that served to define temperature sensitivity. The data presented in Fig. 4 are related to similar experiments using three of the insertion derivatives, but in this case, cells grown at 32 or 42°C were compared. It is apparent that the inhibitory effect of the wild-type product (from pAM-HT) was not affected by temperature in the case of pAM7343 (insertion downstream of t1) and pAM7025 (insert in asa1), whereas when pAM-HTts1 was present, there was a much greater sensitivity to pheromone at the higher temperature. It is noteworthy that the cells in general appeared more sensitive to pheromone at the elevated temperature, even in the absence of additional traD product (i.e., see the case where the empty pAM401 was present). In the case of pAM7343, there was also an apparent temperature sensitivity related to the product of pAM-HTd1, which may be related to the nucleotide difference that resulted in altering the translational start codon, having an effect on RNA secondary structure. As anticipated, in the case of pAM7348 (insertion upstream of t1), none of the combinations significantly affected the sensitivity to pheromone at either temperature.
FIG. 4.
Effect of growth temperature on the cAD1 sensitivity of cells carrying various pAD1::Tn917lac derivatives in the presence of additionally provided mD in trans. The host strain was E. faecalis FA2-2 carrying the indicated plasmid. The plasmids provided in trans were pAM401, (●), pAM-HT (○), pAM-HTd1 (▵), and pAM-HTts1 (□). Cells were grown at 32 or 42°C as indicated. Exposure to cAD1 was for 1 h in each case. The enzyme activity was expressed in MU.
The effect of mD at t1 does not require functional TraA.
In a previous communication (3), we discussed the possibility that mD might be able to interact with TraA, although this was in the context of events believed to be occurring at the iad promoter. To examine the possibility that an interaction might somehow influence transcriptional readthrough of t1, we conducted the following experiments.
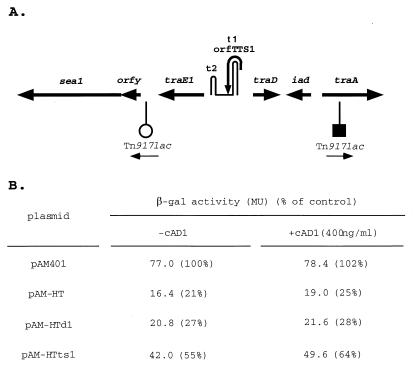
Plasmid pAM2125 contains two oppositely oriented transposon insertions, as illustrated in Fig. 5A. One corresponds to Tn917lac inserted into traA (within the N-terminal 25% of the determinant); the other is a Tn917lac transposon inserted just downstream of traE1. The two insertions are in opposite orientations. The Tn917lac insertion downstream of traE1 expresses beta-galactosidase at a high (constitutive) level because of the defective TraA, whereas the lacZ gene of the traA insertion is very low and not affected by pheromone (41). When pAM-HT or pAM-HTd1 was also present, there was a significant reduction (four- to fivefold) in enzyme expression (Fig. 5B), and this was not affected by exposure of the cells to cAD1. In the case where pAM-HTts1 was present with pAM2125, a less-than-twofold reduction was observed. These data imply that mD RNA effects a reduction of transcriptional readthrough at t1 without requiring an interaction with TraA. It is unlikely that the results could be related to an effect (upregulation) on expression from the traA promoter, since this would have been reflected in the data for pAM7348 in Fig. 3.
FIG. 5.
Inhibition of t1 transcriptional readthrough by mD in the absence of TraA. (A) The host strain was E. faecalis FA2-2 carrying the plasmid pAM2125 with two transposon insertions as indicated. (B) Level of expression of β-galactosidase when the cells, also carrying the indicated additional plasmid, were exposed to cAD1 for 1 h or were not exposed. The percent activities were based on comparison with pAM401, set at 100%.
Analyses of transcription through t1 and effects of structural alterations in the terminator.
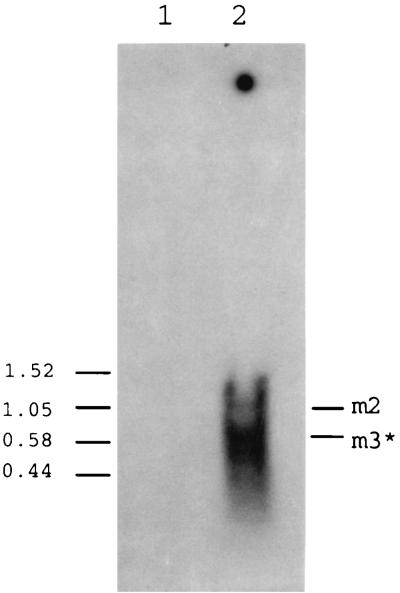
In an earlier report (38), it was shown that in the uninduced state there was significant transcription from the iad promoter to the t1-t2 region, giving rise to a transcript that was designated m3. Upon induction, there was a major increase in this RNA; however, at that time the extent to which m3 termination was occurring at t1 or t2 was not distinguished. Northern blot hybridization data shown in Fig. 6, for which a probe corresponding to the region between t1 and t2 was used, indicate the absence of detectable transcripts extending beyond t1 in the uninduced state (lane 1). This is supportive of the above data indicating that termination was occurring primarily at t1. Induction, however, resulted in detection of a transcript corresponding to a size in a range consistent with being about 150 nt longer than what would correspond to the ∼500-nt m3 (Fig. 6, lane 2). Such a transcript is consistent with termination at t2, and we now designate this m3*. The probe also revealed a longer (∼1,050-nt) transcript which we had earlier noted (38) and called m2; this RNA appears to terminate downstream of traE1. The regions representing these transcripts are illustrated in Fig. 1. Thus, while extensive transcriptional readthrough occurs during induction, significant, but not highly efficient, termination occurs at t2.
FIG. 6.
Northern blot hybridization of RNA from induced and uninduced OG1X cells carrying pAM714. The probe corresponded to a 124-nt segment of DNA representing the region between t1 and t2. Lane 1, uninduced cells. Lane 2, cells induced with synthetic cAD1 for 20 min.
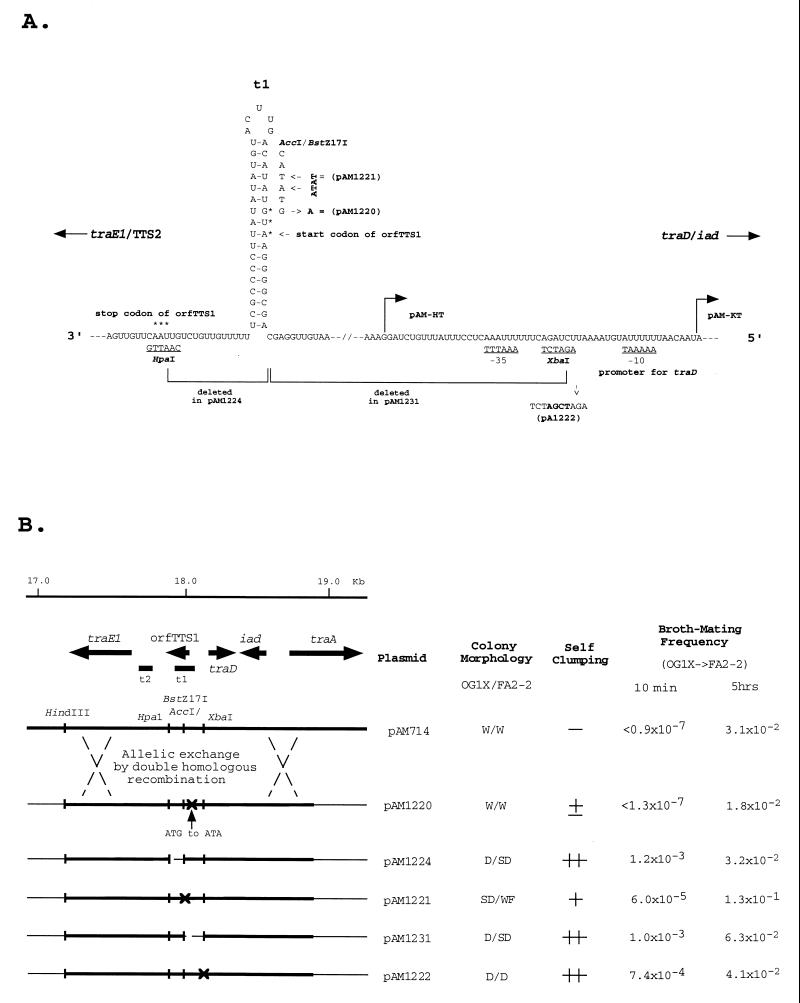
A number of variants altered in the structure of t1 were constructed and introduced into pAM714 (a pAD1::Tn917 derivative with essentially wild-type properties) by allelic exchange (see Materials and Methods). Figure 7 illustrates the nature of the modifications (Fig. 7A) and associated phenotypes (Fig. 7B). Note in Fig. 7B that pAM714 transfers at a very low frequency during the relatively short (10-min) time period. Since significant induction of the mating response generally requires at least 30 min, the 10-min data reflect only the extent to which the donors are expressing mating functions in the absence of pheromone. In addition, cells carrying pAM714 give rise to colonies with a watery morphology, in contrast to those expressing aggregation-mating functions which exhibit a dry colony morphology (42).
FIG. 7.
Analyses of mutations in and around t1. (A) Specific genetic modifications that were generated. (B) Various structural changes and the resulting phenotypes. W, watery (typical of uninduced cells); D, dry (typical of cells fully induced); SD, semidry; WF, watery-fracturable. +, clumping; ++, extensive clumping; ±, slight clumping; −, no clumping. These are all in the absence of added cAD1. The mating frequencies are indicated as number of transconjugants per donor.
We had previously noted a possible short open reading frame (orfTTS1) corresponding to 15 amino acids with a start site within the upstream repeat of t1 (37). pAM1220 is a derivative in which the start codon was eliminated by changing ATG to ATA (i.e., a methionine to an isoleucine codon); the G-to-A substitution at the third position should not significantly affect the stem-loop structure of the related RNA. Although these cells exhibited a slight clumping, their colony morphology and mating properties were very similar to those of the wild type (pAM714); thus, orfTTS1 would not appear to be of major significance, at least under the conditions examined, if indeed it is expressed as a peptide at all.
pAM1224 contained a deletion between the BstZ17I site and the downstream HpaI site, essentially eliminating the termination structure and resulting in (i) a dry or semidry colony morphology (depending on the host), (ii) clumping, and (iii) a high frequency of transfer during a 10-min mating. A similar result was found for pAM1231, which contained a deletion between the BstZ17I and upstream XbaI sites. pAM1221 contains a disruption in t1 as a result of a 2-nt insertion (AT) into the AccI site located within one of the repeated sequences; this significantly weakened the terminator, since clumping was evident and plasmid transfer occurred at a greatly increased frequency during a 10-min mating.
Finally, a derivative, pAM1222, not involving t1 but affecting the promoter site of mD via the insertion of 4 nt between the −10 and −35 promoter hexamers resulted in cells exhibiting full expression of mating functions. The promoter modification is presumed to eliminate the expression of mD, which in turn results in a reduction in t1 termination strength, allowing enough readthrough of t1 (and also t2) to upregulate the mating response.
Influence of mD on asa1 and sea1 transcription appears related only to t1 readthrough.
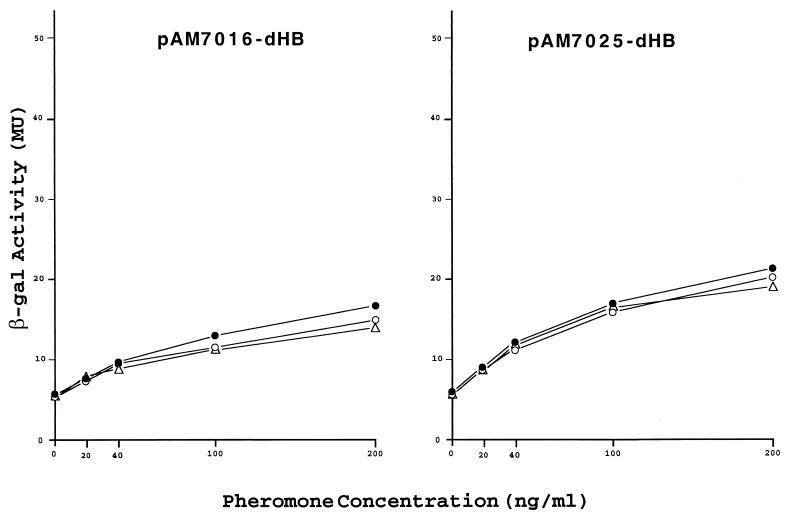
pAM7016-dHB and pAM7025-dHB correspond to pAD1::Tn917lac derivatives with insertions in asa1 and sea1, respectively, which also carry a deletion of a major portion of t1 (deletion between the HpaI and BstZ17I sites). As shown in Fig. 8, in the uninduced state expression of LacZ occurs at a level significantly higher than that for the corresponding derivatives pAM7016 and pAM7025, which have a normal t1 (Fig. 3). The elimination of t1 resulted in an expression of about 6 MU, compared to values of below 1 MU when t1 was present. This would be anticipated since, as we observed above, transcriptional readthrough in the case of t1 mutants leads to at least some expression of mating functions. Enzyme activity increased upon exposure to increasing concentrations of pheromone, reaching a maximum value similar to those observed for the wild-type t1 shown in Fig. 3 (i.e., pAM7016 and pAM7025); however, when mD was provided in trans (i.e., by pAM-HT or pAM-HTd1) there was no reduction in pheromone sensitivity. The data imply that, if mD RNA does target locations downstream of t1, it does not affect transcription. Like the case for the wild-type t1, the maximum levels of beta-galactosidase activities expressed are significantly less than those relating to Tn917lac insertions upstream of sea1 (Fig. 3). In the case of pAM7016-dHB and pAM7025-dHB, the transcription observed would appear to reflect initiation from the iad promoter, and inducibility is interpreted to relate only to cAD1-induced expression (TraA related) from this site.
FIG. 8.
Induced expression of β-galactosidase by pAD1::Tn917lac derivatives with a deleted t1 in response to increasing concentrations of cAD1 while in the presence of additionally provided mD in trans. The host strain was E. faecalis FA2-2 which carried the indicated plasmid with an insertion in sea1 (pAM7016) or asa1 (pAM7025). The plasmids provided in trans were pAM401 (●), pAM-HT (○), and pAM-HTd1 (▵). Exposure to cAD1 was for 1 h in each case.
DISCUSSION
We have shown here that the small pAD1-encoded RNA molecule mD plays a significant role in transcription termination at t1. Excess mD provided in trans by a multicopy plasmid resulted in strong inhibition of pheromone-induced beta-galactosidase expression by pAD1::Tn917lac fusions only if the transposon insertions were located downstream of t1. Since data also revealed that mD did not target regions near the 5′ end of sea1 or asa1, the observed influence on the expression of these genes likely reflects its activity at t1. The effect of mD on t1 was also present in the case of a traA mutant normally fully derepressed for conjugation functions, indicating that interaction between mD and TraA was not required to enhance termination. Consistent with these data was our observation that the inducible clumping response of cells carrying wild-type pAD1 was significantly less sensitive to pheromone when excess mD was provided in trans.
Analyses of the behavior of several mutants with altered t1 structures showed the importance of this terminator in preventing transcription into traE1. When t1 was made dysfunctional, t2 was still able to terminate to some extent, giving rise to m3*, but significant readthrough leading to expression of conjugation functions was clearly evident. At least one function of t2 is presumably to stop any transcription that has escaped normal t1 termination in the absence of induction. In this regard, it is important to recall that, since TraE1 upregulates itself at a site within t2 (3), it is critical to block even small amounts of transcription from upstream to avoid spontaneous induction.
In a previous report (3), we hypothesized that mD associated with an opposing transcript (m3 or m4) by its complementarity to form a complex that in turn acted at the iad promoter to help maintain the basal level of transcription that occurs in cells not exposed to pheromone. The notion of interaction by complementarity was based on the overlap predicted to occur which was partial in the case of m4 (an 80-nt overlap) and complete in the case of m3 (see Fig. 9). A transposon insertion between mD and m4 would be expected to prevent such an overlap, and the inability to form a complex would be consistent with the finding that transcription from the iad promoter became constitutive and could not be complemented by mD in trans. The phenomenon was reflected in the present study in the case of pAM7304 (Tn917::lac insert between traD and iad) and the data of Fig. 3, where mD provided in trans had no effect. It was hypothesized that the activity of the mD::m4(m3) RNA complex involved interaction with either TraA or RNA polymerase. Our present data would suggest that, if indeed mD acts at or near the site of the iad promoter, it is more likely to involve an association with RNA polymerase (see Fig. 9). A reasonable possibility is that mD, or mD complexed with m4 or m3, interacts with RNA polymerase in such a way as to enhance transcriptional termination upon its arrival at t1. A variation of this notion would be that mD anneals with nascent m3 in such a way as to cause premature release of RNA polymerase (close to t1) or to make the polymerase more sensitive to t1. In this regard, it is interesting that mD contains an 11-nt run, beginning at nt 50, that has complementarity to m3 in a region that would span the t1 loop region, a characteristic that brings to mind recent studies of Yarnell and Roberts (46) showing that oligonucleotides with complementarity to nascent RNA within its termination site can in some cases enhance termination.
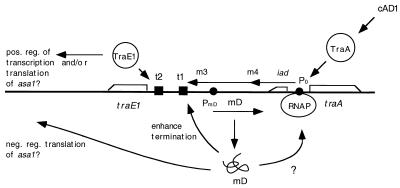
FIG. 9.
Current view of the effect of mD on regulation of the pAD1 pheromone response. A primary function of mD is to enhance termination of transcription at t1. This may relate to an interaction between mD and RNA polymerase (RNAP). Such an interaction could occur near the iad promoter (P0) or further downstream but prior to the RNA polymerase arrival at t1. PmD refers to the promoter of mD (traD). pos. reg., positive regulation; neg. reg., negatively regulated.
An important feature of the pAD1 control system is that in the uninduced state there is already enough expression from the iad promoter to synthesize the iAD1 inhibitor peptide. The detection of transcripts such as m3 and m4 in the absence of induction is presumed to reflect, at least in part, the need to produce and secrete this peptide (38). At the same time there must be a way to ensure that transcription does not go beyond t1-t2, since as noted above even a small amount of synthesis of TraE1 may significantly upregulate, via subsequent autoregulation, the pheromone response. mD would appear to be a key factor in maintaining tight control of this process. The overall circuitry must be designed to allow RNA polymerase to initiate transcription to a limited degree in the absence of pheromone. This could be manifest as TraA acting as a relatively weak repressor, or RNA polymerase may be affected by another factor (e.g., mD alone or mD complexed with m4 or m3) in such a way as to more efficiently compete with TraA for access to the promoter.
The initial evidence suggesting that mD plays some role in affecting transcriptional initiation from the iad promoter was the earlier observation that the temperature-sensitive mD variant resulting from a point mutation within traD gave rise to enhanced transcription from this site upon shifting cells from 32 to 42°C (3). Since we have shown in the present study that this mutation relates to control of transcriptional readthrough of t1 but not transcription up to t1, the question of how mD affects initiation at the iad promoter is puzzling. Although it is conceivable that the previously observed thermally enhanced transcription was due to an unrelated temperature effect on the transcription machinery, another possibility could be related to an effect on stabilization of m3 or possible extensions of mD into traA. (We note that there is no apparent intrinsic terminator relating to mD transcription.)
Finally, the observation that transcription of asa1 is regulated by mD activity at t1 and not at positions further downstream is interesting in connection with an earlier suggestion (3) that additional control may occur in closer proximity to asa1. The fact that the 5′ end of mD exhibits significant complementarity over about 70 nt of a possible untranslated transcript 37 nt upstream of the asa1 translational start site (3) is strongly suggestive of an interaction. If such an interaction indeed occurs, its main effect would now seem more likely to be on Asa1 translation rather than on transcription. And one might expect that the effect would be inhibitory, since mD is present at high levels in the uninduced state and at low levels after induction (Fig. 9). It is possible that mD prevents translation of small amounts of asa1 transcript that may be expressed from a promoter other than that of iad in the uninduced state—perhaps the one reported to occur just upstream of orf1 (33).
Muscholl et al. (33) reported that a segment of DNA containing only orf1 and asa1 cloned on the shuttle vector pWM401 was not able to express Asa1 in E. faecalis unless TraE1 was provided from a coresident plasmid clone in trans. The chimera that carried traE1 also carried iad and its promoter but not an intact traA. Thus, it also encoded mD (unknown at the time), which was probably not expressed since TraA was absent and expression from the iad promoter would therefore be expected to be constitutive. Good expression of TraE1 and greatly reduced expression of mD would seem to have been likely in the reported experiments (33). Although it was assumed that transcription was being regulated by TraE1, expression was being monitored by immunoblotting to detect Asa1; it is possible, therefore, that translation rather than transcription was being influenced. We have previously speculated on the possible interplay between TraE1 and mD (3); conceivably, both factors relate to translation.
The Dunny group, working with the pheromone-responding plasmid pCF10, has reported that induction results in transcription from a promoter analogous to that of iad through the determinant for aggregation substance (prgB) located at a distance similar to that of asa1 (7); indeed transcription appears to extend through several apparent terminators. (See reference 9 for a comparison of pAD1 and pCF10.) They also reported evidence that the large transcript is subsequently processed to give rise to smaller functional products (5), and there are data that support the view that PrgB is subsequently regulated at the translational level (4). A traE1 homologue is not present in the pCF10 system (in its place are prgR and prgS), and an mD equivalent has not been reported. It is very possible, however, that an equivalent mD determinant is present, since there is near identity between pAD1 and pCF10 with respect to a point midway within traD (of pAD1), though through t2. A pCF10 RNA equivalent to mD with an identical promoter and with identity for over ∼50% of the transcript can be easily envisioned. Such a transcript made by pCF10 would therefore be similar to the mD of pAD1 in only its 5′ half. A similar case can be made for pPD1, based on examination of its sequence in the corresponding region (19). Perhaps relating to such a difference was our finding that when mD (of pAD1) was provided in trans it did not significantly affect pCF10 or pPD1 sensitivity to cCF10 or cPD1, respectively. It is interesting that the pCF10 system exhibits constitutive expression from its icf (which encodes inhibitor peptide iCF10) promoter (equivalent to that of iad) (4), since it would suggest that an mD equivalent might never be expressed because of the converging transcription. If this is the case, it would be consistent with the report that transcription indeed occurs to a certain extent through the t1-t2 equivalent IRS1-IRS2 into prgR and prgS in the uninduced state (4). Induction of pCF10 is believed to involve a complex interaction of the internalized cCF10 with two regulatory factors (QL [an RNA equivalent to our m3*] and PrgS) and ribosomes. Such phenomena have not been identified in the pAD1 system, although they cannot be ruled out at this time.
Regulation of the pheromone response in E. faecalis is a complex process with both common and different features between various conjugative plasmids. The enhancement of transcription termination in trans by a small RNA molecule like mD is quite interesting and, to our knowledge, somewhat novel. Further characterization of this system and comparison to other systems such as pCF10 should provide new insights into the specific control mechanisms and their evolution.
ACKNOWLEDGMENTS
We thank Koichi Tanimoto, Maria Bastos, Shuhei Fujimoto, Keith Weaver, and Gary Dunny for helpful discussions.
This work was supported by National Institutes of Health grant GM33956 and the Clinical Research Center at the University of Michigan (MO1-RR00042).
REFERENCES
- 1.An F Y, Clewell D B. The origin of transfer (oriT) of the enterococcal, pheromone-responding, cytolysin plasmid pAD1 is located within the repA determinant. Plasmid. 1997;37:87–94. doi: 10.1006/plas.1996.1270. [DOI] [PubMed] [Google Scholar]
- 2.Bastos M C F, Tanimoto K, Clewell D B. Regulation of transfer of the Enterococcus faecalis pheromone-responding plasmid pAD1: temperature-sensitive transfer mutants and identification of a new regulatory determinant, traD. J Bacteriol. 1997;179:3250–3259. doi: 10.1128/jb.179.10.3250-3259.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bastos M C F, Tomita H, Tanimoto K, Clewell D B. Regulation of the Enterococcus faecalis pAD1-related sex pheromone response: analyses of traD expression and its role in controlling conjugation functions. Mol Microbiol. 1998;30:381–392. doi: 10.1046/j.1365-2958.1998.01074.x. [DOI] [PubMed] [Google Scholar]
- 4.Bensing B A, Manias D A, Dunny G M. Pheromone cCF10 and plasmid pCF10-encoded regulatory molecules act post-transcriptionally to activate expression of downstream conjugation functions. Mol Microbiol. 1997;24:285–294. doi: 10.1046/j.1365-2958.1997.3301710.x. [DOI] [PubMed] [Google Scholar]
- 5.Bensing B A, Meyer B J, Dunny G M. Sensitive detection of bacterial transcription initiation sites and differentiation from RNA processing sites in the pheromone-induced plasmid transfer system of Enterococcus faecalis. Proc Natl Acad Sci USA. 1996;93:7794–7799. doi: 10.1073/pnas.93.15.7794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chow J W, Thal L A, Perri M B, Vazquez J A, Donabedian S M, Clewell D B, Zervos M J. Plasmid-associated hemolysin and aggregation substance production contributes to virulence in experimental enterococcal endocarditis. Antimicrob Agents Chemother. 1993;37:2474–2477. doi: 10.1128/aac.37.11.2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chung J W, Dunny G M. Cis-acting, orientation-dependent, positive control system activates pheromone-inducible conjugation functions at distances greater than 10 kilobases upstream from its target in Enterococcus faecalis. Proc Natl Acad Sci USA. 1992;89:9020–9024. doi: 10.1073/pnas.89.19.9020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clewell D B. Sex pheromones and the plasmid-encoded mating response in Enterococcus faecalis. In: Clewell D B, editor. Bacterial conjugation. New York, N.Y: Plenum Press; 1993. pp. 349–367. [Google Scholar]
- 9.Clewell D B. Sex pheromone systems in enterococci. In: Dunny G M, Winans S C, editors. Cell-cell signaling in bacteria. Washington, D.C.: ASM Press; 1999. pp. 47–65. [Google Scholar]
- 10.Clewell D B, Brown B L. Sex pheromone cAD1 in Streptococcus faecalis: induction of a function related to plasmid transfer. J Bacteriol. 1980;143:1063–1065. doi: 10.1128/jb.143.2.1063-1065.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clewell D B, Pontius L T, An F Y, Ike Y, Suzuki A, Nakayama J. Nucleotide sequence of the sex pheromone inhibitor (iAD1) determinant of Enterococcus faecalis conjugative plasmid pAD1. Plasmid. 1990;24:156–161. doi: 10.1016/0147-619x(90)90019-9. [DOI] [PubMed] [Google Scholar]
- 12.Clewell D B, Tomich P K, Gawron-Burke M C, Franke A E, Yagi Y, An F Y. Mapping of Streptococcus faecalis plasmids pAD1 and pAD2 and studies relating to transposition of Tn917. J Bacteriol. 1982;152:1220–1230. doi: 10.1128/jb.152.3.1220-1230.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dunny G M, Craig R A, Carron R L, Clewell D B. Plasmid transfer in Streptococcus faecalis: production of multiple sex pheromones by recipients. Plasmid. 1979;2:454–465. doi: 10.1016/0147-619x(79)90029-5. [DOI] [PubMed] [Google Scholar]
- 14.Dunny G M, Funk C, Adsit J. Direct stimulation of the transfer of antibiotic resistance by sex pheromones in Streptococcus faecalis. Plasmid. 1981;6:270–278. doi: 10.1016/0147-619x(81)90035-4. [DOI] [PubMed] [Google Scholar]
- 15.Dunny G M, Leonard B A B. Cell-cell communication in gram-positive bacteria. Annu Rev Microbiol. 1997;51:527–564. doi: 10.1146/annurev.micro.51.1.527. [DOI] [PubMed] [Google Scholar]
- 16.Ehrenfeld E E, Kessler R E, Clewell D B. Identification of pheromone-induced surface proteins in Streptococcus faecalis and evidence of a role for lipoteichoic acid in formation of mating aggregates. J Bacteriol. 1986;168:6–12. doi: 10.1128/jb.168.1.6-12.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Flannagan S E, Clewell D B. Conjugative transfer of Tn916 in Enterococcus faecalis: trans activation of homologous transposons. J Bacteriol. 1991;173:7136–7141. doi: 10.1128/jb.173.22.7136-7141.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fujimoto S, Clewell D B. Regulation of the pAD1 sex pheromone response of Enterococcus faecalis by direct interaction between the cAD1 peptide mating signal and the negatively regulating, DNA-binding TraA protein. Proc Natl Acad Sci USA. 1998;95:6430–6435. doi: 10.1073/pnas.95.11.6430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fujimoto S, Tomita H, Wakamatsu E, Tanimoto K, Ike Y. Physical mapping of the conjugative bacteriocin plasmid pPD1 of Enterococcus faecalis and identification of the determinant related to the pheromone response. J Bacteriol. 1995;177:5574–5581. doi: 10.1128/jb.177.19.5574-5581.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Galli D, Friesenegger A, Wirth R. Transcriptional control of sex-pheromone-inducible genes on plasmid pAD1 of Enterococcus faecalis and sequence analysis of a third structural gene for (pPD1-encoded) aggregation substance. Mol Microbiol. 1992;6:1297–1308. doi: 10.1111/j.1365-2958.1992.tb00851.x. [DOI] [PubMed] [Google Scholar]
- 21.Galli D, Lottspeich F, Wirth R. Sequence analysis of Enterococcus faecalis aggregation substance encoded by the sex pheromone plasmid pAD1, Mol. Microbiol. 1990;4:895–904. doi: 10.1111/j.1365-2958.1990.tb00662.x. [DOI] [PubMed] [Google Scholar]
- 22.Galli D, Wirth R, Wanner G. Identification of aggregation substances of Enterococcus faecalis after induction by sex pheromones. Arch Microbiol. 1989;151:486–490. doi: 10.1007/BF00454863. [DOI] [PubMed] [Google Scholar]
- 23.Ike Y, Clewell D B. Genetic analysis of the pAD1 pheromone response in Streptococcus faecalis using transposon Tn917 as an insertional mutagen. J Bacteriol. 1984;158:777–783. doi: 10.1128/jb.158.3.777-783.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ike Y, Craig R C, White B A, Yagi Y, Clewell D B. Modification of Streptococcus faecalis sex pheromones after acquisition of plasmid DNA. Proc Natl Acad Sci USA. 1983;80:5369–5373. doi: 10.1073/pnas.80.17.5369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ike Y, Hashimoto H, Clewell D B. Hemolysin of Streptococcus faecalis subspecies zymogenes contributes to virulence in mice. Infect Immun. 1984;45:528–530. doi: 10.1128/iai.45.2.528-530.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jacob A E, Hobbs S J. Conjugal transfer of plasmid-borne multiple antibiotic resistance in Streptococcus faecalis var. zymogenes. J Bacteriol. 1974;117:360–372. doi: 10.1128/jb.117.2.360-372.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jett B D, Jensen H G, Nordquist R E, Gilmore M S. Contribution of the pAD1-encoded cytolysin to the severity of experimental Enterococcus faecalis endophthalmitis. Infect Immun. 1992;60:2445–2452. doi: 10.1128/iai.60.6.2445-2452.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1982. [Google Scholar]
- 29.Maqueda M, Quirants R, Martin I, Galvez A, Martinez-Bueno M, Valdivia E. Chemical signals in gram-positive bacteria: the sex-pheromone system in Enterococcus faecalis. Microbiologia. 1997;13:23–36. [PubMed] [Google Scholar]
- 30.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1972. [Google Scholar]
- 31.Mori M, Isogai A, Sakagami Y, Fujino M, Kitada C, Clewell D B, Suzuki A. Isolation and structure of Streptococcus faecalis sex pheromone inhibitor, iAD1, that is excreted by donor strains harboring plasmid pAD1. Agric Biol Chem. 1986;50:539–541. [Google Scholar]
- 32.Mori M, Sakagami Y, Narita M, Isogai A, Fujino M, Kitada C, Craig R, Clewell D, Suzuki A. Isolation and structure of the bacterial sex pheromone, cAD1, that induces plasmid transfer in Streptococcus faecalis. FEBS Lett. 1984;178:97–100. doi: 10.1016/0014-5793(84)81248-x. [DOI] [PubMed] [Google Scholar]
- 33.Muscholl A, Galli D, Wanner G, Wirth R. Sex pheromone plasmid pAD1-encoded aggregation substance of Enterococcus faecalis is positively regulated in trans by traE1. Eur J Biochem. 1993;214:333–338. doi: 10.1111/j.1432-1033.1993.tb17928.x. [DOI] [PubMed] [Google Scholar]
- 34.Nakayama J, Dunny G M, Clewell D B, Suzuki A. Quantitative analysis for pheromone inhibitor and pheromone shutdown in Enterococcus faecalis. Dev Biol Stand. 1995;85:35–38. [PubMed] [Google Scholar]
- 35.Pontius L T, Clewell D B. A phase variation event that activates conjugation functions encoded by the Enterococcus faecalis plasmid pAD1. Plasmid. 1991;26:172–185. doi: 10.1016/0147-619x(91)90041-t. [DOI] [PubMed] [Google Scholar]
- 36.Pontius L T, Clewell D B. Regulation of the pAD1-encoded pheromone response in Enterococcus faecalis: nucleotide sequence analysis of traA. J Bacteriol. 1992;174:1821–1827. doi: 10.1128/jb.174.6.1821-1827.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pontius L T, Clewell D B. Conjugative transfer of Enterococcus faecalis plasmid pAD1: nucleotide sequence and transcriptional fusion analysis of a region involved in positive regulation. J Bacteriol. 1992;174:3152–3160. doi: 10.1128/jb.174.10.3152-3160.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tanimoto K, Clewell D B. Regulation of the pAD1-encoded sex pheromone response in Enterococcus faecalis: expression of the positive regulator TraE1. J Bacteriol. 1993;175:1008–1018. doi: 10.1128/jb.175.4.1008-1018.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tao L, Ferretti J J. Cloning vectors with streptococcal resistance genes and an Escherichia coli origin of replication which can be used for cloning of streptococcal origins of replication or for gene inactivation in streptococci. In: Dunny G M, Cleary P P, McKay L L, editors. Genetics and molecular biology of streptococci, lactococci, and enterococci. Washington, D.C.: American Society for Microbiology; 1991. p. 305. [Google Scholar]
- 40.Tomich P K, An F Y, Damle S P, Clewell D B. Plasmid-related transmissibility and multiple drug resistance in Streptococcus faecalis subsp. zymogenes strain DS16. Antimicrob Agents Chemother. 1979;15:828–830. doi: 10.1128/aac.15.6.828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Weaver K E, Clewell D B. Regulation of the pAD1 sex pheromone response in Enterococcus faecalis: effects of host strain and traA, traB, and C region mutants on expression of an E region pheromone-inducible lacZ fusion. J Bacteriol. 1990;172:2633–2641. doi: 10.1128/jb.172.5.2633-2641.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weaver K E, Clewell D B. Control of Enterococcus faecalis sex pheromone cAD1 elaboration: effects of culture aeration and pAD1 plasmid-encoded determinants. Plasmid. 1991;25:177–189. doi: 10.1016/0147-619x(91)90011-k. [DOI] [PubMed] [Google Scholar]
- 43.Weidlich G, Wirth R, Galli D. Sex pheromone plasmid pAD1-encoded surface exclusion protein of Enterococcus faecalis. Mol Gen Genet. 1992;233:161–168. doi: 10.1007/BF00587575. [DOI] [PubMed] [Google Scholar]
- 44.Wirth R, An F Y, Clewell D B. Highly efficient protoplast transformation system for Streptococcus faecalis and an Escherichia coli-S. faecalis shuttle vector. J Bacteriol. 1986;165:831–836. doi: 10.1128/jb.165.3.831-836.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yagi Y, Kessler R, Shaw J, Lopatin D, An F, Clewell D. Plasmid content of Streptococcus faecalis strain 39-5 and identification of a pheromone (cPD1)-induced surface antigen. J Gen Microbiol. 1983;129:1207–1215. doi: 10.1099/00221287-129-4-1207. [DOI] [PubMed] [Google Scholar]
- 46.Yarnell W S, Roberts J W. Mechanism of intrinsic transcription termination and antitermination. Science. 1999;284:611–615. doi: 10.1126/science.284.5414.611. [DOI] [PubMed] [Google Scholar]