Abstract
Background
Analysis of circulating free DNA (cfDNA) is a promising tool for personalized management of colorectal cancer (CRC) patients. Untargeted cfDNA analysis using whole-genome sequencing (WGS) does not need a priori knowledge of the patient´s mutation profile.
Methods
Here we established LIquid biopsy Fragmentation, Epigenetic signature and Copy Number Alteration analysis (LIFE-CNA) using WGS with ~ 6× coverage for detection of circulating tumor DNA (ctDNA) in CRC patients as a marker for CRC detection and monitoring.
Results
We describe the analytical validity and a clinical proof-of-concept of LIFE-CNA using a total of 259 plasma samples collected from 50 patients with stage I-IV CRC and 61 healthy controls. To reliably distinguish CRC patients from healthy controls, we determined cutoffs for the detection of ctDNA based on global and regional cfDNA fragmentation patterns, transcriptionally active chromatin sites, and somatic copy number alterations. We further combined global and regional fragmentation pattern into a machine learning (ML) classifier to accurately predict ctDNA for cancer detection. By following individual patients throughout their course of disease, we show that LIFE-CNA enables the reliable prediction of response or resistance to treatment up to 3.5 months before commonly used CEA.
Conclusion
In summary, we developed and validated a sensitive and cost-effective method for untargeted ctDNA detection at diagnosis as well as for treatment monitoring of all CRC patients based on genetic as well as non-genetic tumor-specific cfDNA features. Thus, once sensitivity and specificity have been externally validated, LIFE-CNA has the potential to be implemented into clinical practice. To the best of our knowledge, this is the first study to consider multiple genetic and non-genetic cfDNA features in combination with ML classifiers and to evaluate their potential in both cancer detection and treatment monitoring.
Trial registration DRKS00012890.
Supplementary Information
The online version contains supplementary material available at 10.1186/s13045-022-01342-z.
Keywords: ctDNA, Colorectal cancer, Liquid biopsy, Whole-genome sequencing, Somatic copy number alterations, cfDNA fragmentation, Chromatin signatures
Background
Liquid biopsy (LB) is a highly promising tool for personalized patient management [1–5]. An important LB marker is circulating tumor DNA (ctDNA), which represents the fraction of circulating free DNA (cfDNA) released by tumor cells [6]. A major challenge in ctDNA analysis is the very low fractions of ctDNA in total cfDNA (commonly < 5%) [6, 7]. Therefore, methods with high analytical sensitivity and specificity are required [8–10], but to date, mainly methods targeting frequent hotspot variants have been validated [11–14]. However, this approach limits the application of LB to patients with known genetic tumor profiles. To extend the advantages of LB to all cancer patients, highly sensitive untargeted methods are required.
A commonly used approach for untargeted ctDNA detection is shallow whole-genome sequencing (WGS) (i.e., < 1× coverage) to identify genome-wide somatic copy number alterations (SCNAs) [15] However, this approach requires ctDNA fractions of at least 5% to 10% that may be present in a subset of CRC samples only [15–17]. Various studies suggest that enrichment of the ctDNA fraction in cfDNA by size selection, tumor-specific fragmentation patterns, and epigenetic signatures can enhance ctDNA detection [18–21].
In this study, we developed LIquid biopsy Fragmentation, Epigenetic signature and Copy Number Alteration analysis (LIFE-CNA) as an untargeted approach to detect ctDNA with high sensitivity in plasma samples of colorectal cancer (CRC) patients as a diagnostic, predictive and prognostic marker. To enable detection of ctDNA fractions < 5%, we increased coverage from shallow WGS to ~ 6× and combined the Illumina DRAGEN CNV (copy number variation) workflow with the Plasma-Seq pipeline for copy number profiling [15, 22], a fragmentation pipeline, and LIQUORICE, a tool for the identification of coverage in open-chromatin regions [20]. With this workflow, we integrated detection of multiple cfDNA features, including focal SCNAs, cfDNA fragmentation patterns and chromatin signatures, and established machine learning (ML) classifiers for the highly sensitive detection of ctDNA. Using LIFE-CNA, we aimed to establish cutoffs for ctDNA detection to facilitate translation of untargeted LB analysis into clinical practice. We further evaluated whether ctDNA analysis using LIFE-CNA is able to predict response or resistance to treatment. For analytical validation and a clinical proof-of-concept of LIFE-CNA, 259 cfDNA samples from 50 patients with stage I-IV CRC and 61 healthy controls were analyzed. To the best of our knowledge, this is the first study combining SCNA and fragmentation profiles for disease monitoring and providing a complete analytically validated workflow showing a clinical proof-of-concept that can be easily implemented into clinical practice to support CRC patient management.
Methods
Study design and participants
A total of 259 plasma samples were collected from 50 patients with UICC stage I-IV CRC (7 stage I, 14 stage II, 11 stage III, 18 stage IV) and 61 healthy individuals aged 20 to 88 years from March 2018 until April 2022 (Additional file 1: Table S1, Figure S1; Additional file 2: Table S2) [23]. 55 healthy controls were included in the reference set. Six healthy controls were used for external validation of LIFE-CNA. 198 plasma samples from 50 CRC patients were collected at diagnosis and during follow-up. These samples were categorized according to the time of sample collection during the course of disease (Additional file 1: Methods, Table S1). The course of disease was monitored by colonoscopies and imaging during routinely scheduled follow-up examinations. 134 of the 198 plasma samples from CRC patients collected during the course of disease with clinically diagnosed tumor burden served as positive controls. To identify molecular residual disease (MRD) following surgery, baseline blood samples were collected up to eight days pre-surgery and follow-up samples were collected one day up to six weeks post-surgery in 33 patients. For treatment monitoring, plasma samples from 15 patients were collected at several time points throughout the course of disease.
The study was approved by the ethics commission of the Bavarian Medical Association (No. 17059) and is registered with the German registry for clinical trials (trial registration ID: DRKS00012890). Neither clinicians nor patients were informed about the results. All participants provided informed written consent prior to blood and tissue specimen collection.
Clinical sample collection and categorization, DNA extraction, droplet digital PCR, CEA analysis, library preparation and in silico dilutions
Information on sample collection and categorization, DNA extraction, droplet digital PCR (ddPCR), carcinoembryonic antigen (CEA) analysis, library preparation, and in silico dilutions are provided in the Supplementary Methods (Additional file 1).
Whole-genome sequencing bioinformatics analysis
Following paired-end sequencing with 2 × 101 bp reads on the NovaSeq 6000 system (Illumina, San Diego, California, USA), demultiplexing of samples was performed using BCL Convert (Illumina), and raw sequencing data were processed using the DRAGEN DNA Pipeline on the Illumina DRAGEN Bio-IT Platform (Illumina) v3.9. After adapter trimming, sequencing reads were aligned to GRCh38/hg38. Duplicates and reads with a mapping quality < 30 were removed from analysis. A second bam file with 90–150 bp fragments only was generated for SCNA analysis. In all of the following analysis regions overlapping with ENCODE blacklist [24] and the UCSC gap track [25] were excluded.
Global and regional fragmentation analysis
Global and regional fragmentation of cfDNA was analyzed as described by Peneder et al. in 2021 [20]. Briefly, fragment length was determined using Picard CollectInsertSizeMetrics (version 2.26.6) and global fragmentation was derived as the fraction of fragments with distinct lengths. Regional fragmentation was established as the z-scored difference in the ratio of short (90–150 bp) to long (151–220 bp) fragments (S/L ratio) in 100 kb bins compared to the 55 healthy controls. Z-scores of the fragmentation of healthy controls were calculated by comparison to the other 54 healthy control samples. Data of genomic regions harboring SCNAs were excluded to avoid bias due to regionally enriched ctDNA. The computational analysis described by Peneder et al. in 2021 [20] was adapted that regions harboring SCNAs were identified based on the SCNA workflow, described below rather than ichorCNA. Furthermore, we used a significant enrichment of short fragments (90–150 bp) as indicator for ctDNA based on global, and significantly different z-scored S/L-ratios on at least one chromosome arm as indictor for ctDNA based on regional fragmentation.
Coverage in CRC-specific regions of interest
The LIQUORICE tool (v.0.5), developed by Peneder et al. in 2021 [20], was used to identify ctDNA based on significant coverage drops in CRC-specific transcriptionally active chromatin regions (epigenetic signatures). We analyzed the coverage in CRC-specific active chromatin regions, published by Chiara et al. in 2021 [26] and in universal DNase I hypersensitivity sites (DHS). The neighboring 20 kb of each region set were split into 500 bp bins to identify the mean coverage around the regions of interest. To correct for bias due to regionally enriched ctDNA, SCNAs, identified with the SCNA workflow, described below, were provided to LIQUORICE. Significant coverage drops compared to healthy controls in at least two of the analyzed region sets were considered as indicator for ctDNA.
Somatic copy number alterations
The CNV workflow provided with the Illumina DRAGEN Bio-IT Platform (Illumina) was performed based on 90–150 bp fragments, since higher sensitivity for SCNA calling was previously described for these short fragments [18, 20]. In detail, reads were counted in 50 kb bins, followed by GC bias correction and normalization based on a reference set containing data from 55 healthy control samples. Segmentation was performed by circular binary segmentation with disabled merging of two adjacent segments (merge-threshold = 0). Following the DRAGEN CNV workflow, SCNAs were identified according to the Plasma-Seq pipeline described by Heitzer et al. [15] applying chromosome specific thresholds (Additional file 1: Figure S3, Methods; Additional file 2: Tables S3 and S4).
Focal somatic copy number alterations
Focal SCNAs, identified within the Plasma-Seq pipeline were defined as described by Ulz and Belic et al. in 2016 [22]. SCNAs of < 20 Mb, overlapping with ≤ 100 genes of the COSMIC cancer gene census [27] with a higher or lower log2 ratio than the chromosome specific LOB compared to the neighboring 20 Mb were identified as focal SCNAs. In addition segments with a higher log2 ratio of 0.58 (~ three copies) compared to the neighboring 20 Mb were identified as focal amplifications, even if no gene of the COSMIC cancer gene census [27] overlapped.
Machine learning model for tumor detection
For ctDNA detection in samples collected from CRC patients, different machine learning (ML) classifiers were trained as described by Peneder et al. in 2021 [20]. Briefly, support vector machines, feed-forward neural networks, random forests and binomial generalized linear models with elastic-net regularization were trained and evaluated using 100 bootstrapping iterations with fivefold cross-validation in each training set.
We evaluated the performance of ML classifiers on the following feature sets: (i) Global fragmentation, (ii) regional fragmentation, and (iii) a meta-learner (Additional file 1: Methods; Additional file 2: Table S5) [18–20].
For each feature set the support vector machine was selected as best ML classifier to build a final ML model on the complete data.
Statistical analysis
Differences in global and regional fragmentation of healthy individuals and CRC patients were determined using a Mann–Whitney-U test. Bonferroni correction was used to adjust p-values for multiple testing. All statistical analyses were performed using statistical functions within the Python module SciPy v.1.8 (scipy.stats) with Python version 3.10.
Results
Tumor-specific global fragmentation pattern
To establish a comprehensive data set for LB analysis in all stages of CRC, we applied WGS with a median coverage of 6x (SD = 2.37) in 259 plasma samples of CRC patients (n = 50) and healthy controls (n = 61) (Additional file 2: Table S2). We first evaluated, whether the global fragmentation pattern of cfDNA may be a suitable marker for untargeted ctDNA detection. Fragmentation patterns are a result of various chromatin states that are associated with altered expression of tumor-associated genes [19, 21, 28, 29].
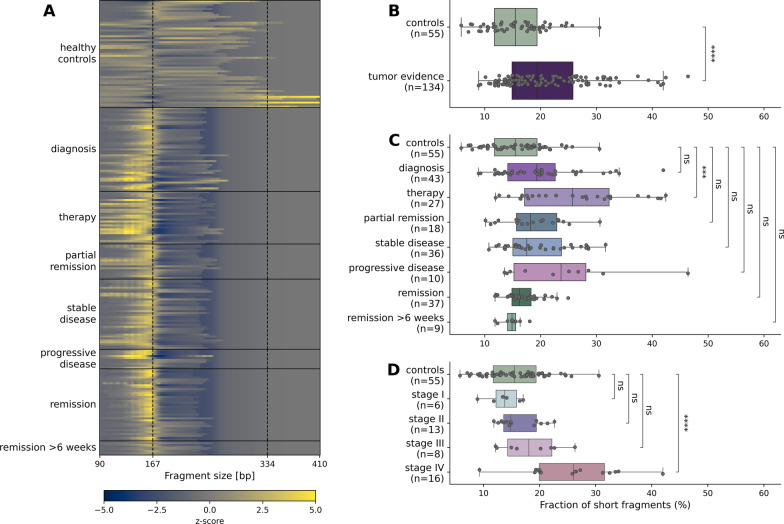
We compared the global fragmentation of cfDNA from CRC patients to cfDNA from healthy controls which typically present with a peak of ~ 167 bp corresponding to DNA bound by one nucleosome plus linker DNA [20] (Fig. 1A). We observed a significant enrichment of short fragments (90–150 bp) in CRC patient samples with clinically diagnosed tumor burden (n = 134) compared to healthy controls (n = 55) (Mann–Whitney-U test, p-value = 4.75*10–5) (Fig. 1B). When allocating CRC patient samples according to the course of disease, we observed a significant enrichment of short fragments during therapy (between surgery and adjuvant chemotherapy, or during chemotherapy before staging) (n = 27) (p-value = 1.48*10–4). A tendency (albeit not statistically significant) toward a higher proportion of short fragments could be identified in all other progression sample groups with clinically diagnosed tumor burden (Fig. 1C). When stratifying CRC patient samples collected at diagnosis according to their disease stage, we further observed a significant enrichment of short fragments in patients with stage IV CRC (n = 16) (p-value = 7.25*10–5) (Fig. 1D).
Fig. 1.
Differences in global fragmentation between cfDNA from CRC patients and healthy controls. A Heat map showing enrichment or decrease in cfDNA fragments from 90 to 410 bp according to their length as z-scores of each sample compared to healthy controls. B Short cfDNA fragments (90–150 bp) are significantly enriched in samples collected from CRC patients with clinically diagnosed tumor burden. C Only for samples collected in the beginning of therapy a significantly enriched fraction of short fragments can be observed. D At diagnosis a significant enrichment in short fragments was only observed in patients with stage IV CRC. (ns: p-value ≤ 1; *: p-value ≤ 5*10–2, **: p-value ≤ 1*10–2, ***: p-value ≤ 1*10–3, ****: p-value ≤ 1*10–4)
Interestingly albeit not statistically significant, we detected different fragmentation profiles due to enrichment of short fragments < 167 bp when analyzing samples from CRC patients in remission with no evidence of disease compared to healthy controls. When focusing on samples from CRC patients in remission more than six weeks post-treatment, we did no longer observe an enrichment of short fragments < 167 bp (Fig. 1A). The observed enrichment within the first weeks post-surgery is likely associated with the intake of low-molecular weight heparin, in accordance with previous findings [30, 31]. Taken together, our results indicate that cfDNA is more fragmented in CRC patients compared to healthy controls and can therefore support untargeted detection of ctDNA.
Tumor-specific regional fragmentation profiles
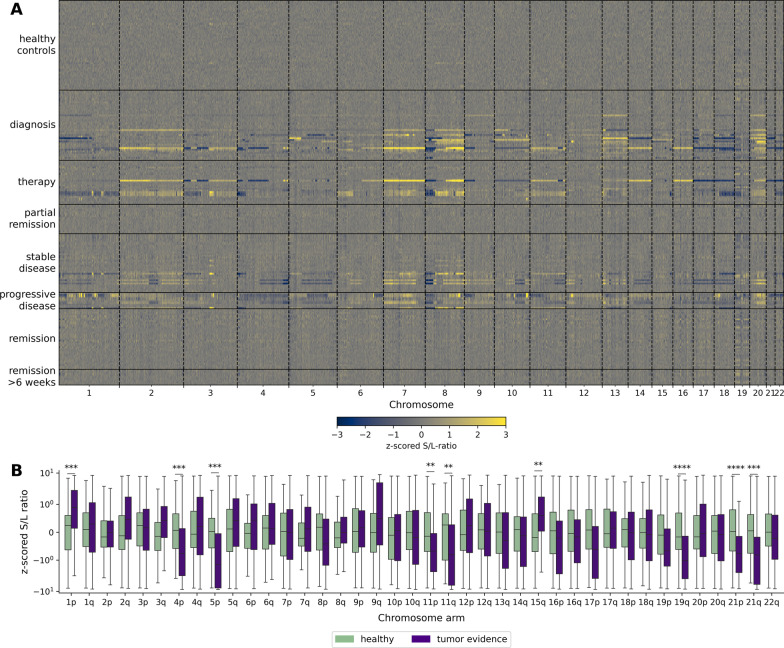
To assess whether regional fragmentation across the genome could serve as another non-genetic marker for ctDNA detection in CRC patients, we calculated the ratio of short (100–150 bp) to long (151–220 bp) fragments (S/L ratio) in 100 kb bins for each chromosome in CRC patients and healthy controls, as recently described [19, 20]. Notably, data of chromosome arms harboring SCNAs were excluded to avoid bias due to regionally enriched ctDNA. Compared to healthy controls, we observed distinct differences in the S/L ratio of CRC patients at diagnosis, during therapy, and with stable or progressive disease. In contrast, in CRC patients with partial remission or in remission, we did not observe such differences (Fig. 2A). Focusing on CRC patient samples with clinically diagnosed tumor burden, we observed a significant enrichment in short fragments on chromosome arms 1p and 15q, and significant enrichment of long fragments on chromosome arms 4p, 5p, 11p, 11q, 19q, 21p and 21q (Fig. 2B). Overall, we were able to detect ctDNA in 75% (100/134) of samples collected from CRC patients with clinically diagnosed tumor burden based on significantly different regional fragmentation on at least one chromosome arm.
Fig. 2.
Differences in regional fragmentation between cfDNA from CRC patients and healthy controls. A Heat map showing the z -scored of S/L-ratios in 100 kb bins of each sample compared to healthy controls. B Significant differences in z-scored S/L-ratios between samples collected from CRC patients with clinically diagnosed tumor burden and healthy controls were observed on multiple chromosome arms. (*: p-value ≤ 5*10–2, **: p-value ≤ 1*10–2, ***: p-value ≤ 1*10–3, ****: p-value ≤ 1*10–4)
The differences in regional fragmentation between CRC patients and healthy controls support recent findings identifying cfDNA fragmentation as independent biological feature representing chromatin profiles of the cells of origin [19, 20].
Combination of global and regional fragmentation analysis using machine learning
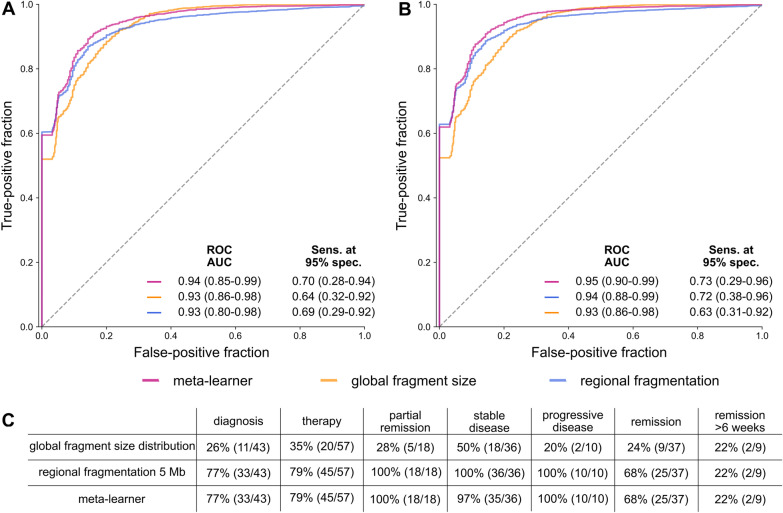
To test whether machine learning (ML) classifier based on global fragmentation and regional fragmentation in 5 Mb bins increase accurate detection of ctDNA, we trained four ML algorithms using 100 bootstrapping iterations with fivefold cross-validation (see Materials and Methods). For each iteration the prediction of the best model was stored and predictions for the two classifiers based on global and regional fragmentation were combined within a supervised meta-learner [20]. Samples collected from CRC patients with clinically diagnosed tumor burden (n = 134) served as positive cohort, and healthy individuals, including samples collected from patients in remission more than six weeks post-treatment without any known recurrence at a later time point (n = 63) served as control cohort for a better representation of biological variability (Additional file 1: Figure S1). All classifiers showed high prediction performance to distinguish cfDNA from CRC patients and healthy controls, with receiver operating characteristic (ROC) area under the curve (AUC) values of up to 94% and sensitivity at 95% specificity of up to 70% (Fig. 3A). Since our ultimate goal was to develop a workflow applicable in clinical practice, we trained a final model based on the best performing ML algorithm for each feature set. Evaluating the performance of ML classifiers using only the support vector machine, we observed ROC AUC values and sensitivity at 95% specificity of up to 95% and 75%, respectively (Fig. 3B). Eventually, we trained final ML models for both feature sets as well as the meta-learner including all data of CRC patients (n = 134) and controls (n = 63) without further subsetting. Applying these models with 95% specificity, ctDNA presence was correctly predicted in 36% (48/134) of samples based on global fragmentation (34/91 metastatic, 14/43 localized), and in 90% of samples based on regional fragmentation (121/134: 85/91 metastatic, 36/43 localized) and based on the meta-learner (120/134: 84/91 metastatic, 36/43 localized). However, also samples collected from patients in remission, especially within the first six weeks post-surgery were classified as ctDNA positive (Fig. 3C). These results in combination with the findings above indicate that the non-genetic cfDNA features analyzed within LIFE-CNA are not informative for the correct identification of ctDNA within the first six weeks post-surgery. However, the effects of surgery on cfDNA fragmentation seem to normalize after six weeks, indicating a potential use for recurrence monitoring starting at this time point.
Fig. 3.
Performance of ML classifiers based on global and regional fragmentation as well as a meta-learner. Performance was assessed over 100 bootstrapping iterations with fivefold cross validation A using the best performing model out of four classifiers for each iteration and B only a support vector machine over all iterations. C The three final classifiers detect ctDNA in CRC patients with high sensitivity
CRC-specific active chromatin for ctDNA detection
We evaluated whether CRC specific chromatin signatures can be detected based on coverage changes using the LIQUORICE tool [20] and whether these chromatin signatures represent an independent marker for ctDNA detection. Specifically, we analyzed five sets of enhancer regions identified to be active in CRC including (i) active distal ChromHMM-defined [32] enhancer regions, (ii) CRC-specific gained enhancers identified by Hi-C [33], (iii) gained enhancers occupied by the transcriptional coactivators YAP/TAZ, (iv) highly conserved enhancers occupied by YAP/TAZ, and (v) active transcriptional start sites (TSS) in CRC [26]. In addition, we analyzed the coverage in universal DHS. In total, we observed significantly stronger coverage drops in all region sets in samples collected from CRC patients compared to healthy controls. In 5% (3/55) of healthy controls significantly stronger coverage drops in one of the analyzed region sets were detected when comparing the coverage to all 54 other healthy control samples. Therefore, to ensure a specificity of ≥ 95% for ctDNA detection based on the coverage in CRC-specific active chromatin regions, significantly stronger coverage drops need to be identified in at least two of the analyzed region sets rather than one. Overall, we detected ctDNA based on differential coverage in 33% (44/134) of samples collected from CRC patients with clinically diagnosed tumor burden (Additional file 2: Table S6). However, we obtained similar values [32% (12/37)] for remission patients and [33% (3/9)] for remission patients more than six weeks post-treatment. Taken together, coverage-based chromatin site analysis for ctDNA detection is suitable at diagnosis, but not for recurrence (also not > 6 weeks).
Quantification of the ctDNA fraction in CRC patients
To quantify the ctDNA fraction as a complement to fragmentation and coverage-based chromatin site analysis, we used the ichorCNA tool [17], which led to correct prediction of ctDNA in only 35% (47/134) of samples with clinically diagnosed tumor burden, even when selectively enriching for ctDNA-associated 90–150 bp fragments (Additional file 1: Figure S3) [18, 20].
Detection of genome-wide and focal SCNAs in CRC patients
To identify genome-wide and focal SCNAs we applied a combination of the Illumina DRAGEN CNV workflow and Plasma-Seq [15, 22], considering ctDNA-associated 90–150 bp fragments (Additional file 1: Methods, Figures S4 and S5) [18, 20]. We analyzed paired tumor tissue and plasma samples collected at diagnosis to validate the SCNA pipeline. To correct for germline CNVs, constitutional DNA from saliva was additionally analyzed. In 44% (12/27) of patients with localized- and in 94% (15/16) of patients with metastatic CRC genome-wide SCNA profiles were highly concordant to the corresponding tissue. SCNAs unique to plasma were identified in 78% (21/27) of patients with localized- and 82% (13/16) of patients with metastatic CRC (Fig. 4A). In addition, we identified focal SCNAs in plasma matching tumor tissue in 4% (1/27) of patients with localized-, and in 63% (10/16) of patients with metastatic CRC, and focal SCNAs only in plasma in 15% (4/27) of patients with localized-, and in 63% (10/16) of patients with metastatic CRC (Fig. 4B). Certain genetic events found in plasma may not be present in tumor tissue because of the representation of only one site of the entire tumor mass rather than the complete tumor heterogeneity including metastatic sites. It is likely that low amplitude SCNAs may not be detected in plasma since ctDNA represents only a fraction of total cfDNA. Overall, although some SCNAs might be missed in plasma, with our approach we are able to detect genome-wide SCNAs in plasma from CRC patients over all stages, including subclonal events not identified in tumor tissue.
Fig. 4.
Matched plasma and tumor analysis. To validate the SCNA analysis integrated in LIFE-CNA we performed a matched analysis of plasma samples collected at diagnosis with tumor tissue. A Total SCNAs present in plasma (red) or tumor (blue) only or in both plasma and tumor (yellow) and B focal SCNAs present in plasma (pink) or tumor (violet) only or in both plasma and tumor (green) present on each chromosome for individual patients and summarized over all patients below. Since more than one SCNA can be present per chromosome, it is possible that on the same chromosome different SCNAs are detected in plasma only, tissue only or in both plasma and tumor tissue
Complementary ctDNA detection by combining cfDNA features
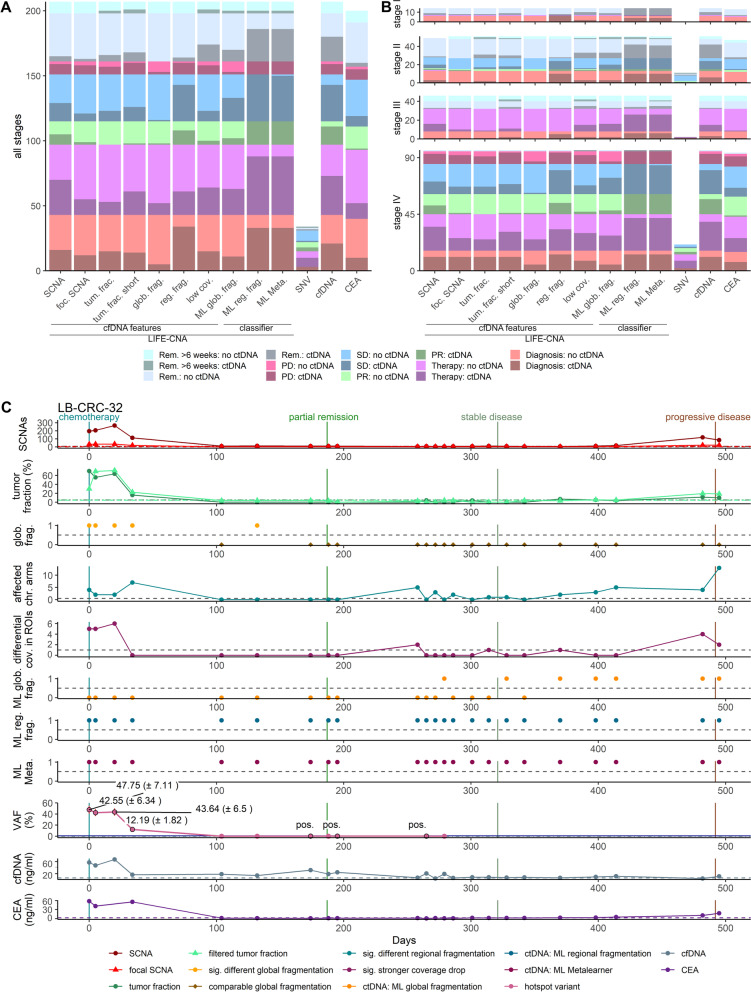
Based on our results showing that global and regional fragmentation as well as chromatin signatures, and SCNAs are capable to independently detect ctDNA, we compared the sensitivity of all features in CRC patients in general and across stages considering the time point of sample collection in the course of disease (Fig. 5A, B).
Fig. 5.
LIFE-CNA enables accurate disease monitoring in CRC patients. SCNAs, focal SCNAs (foc. SCNA), tumor fraction in all (tum. frac.) and filtered fragments (tum. frac. short), enrichment in fragments from 90 to 150 bp (glob. frag.), regional fragmentation (reg. frag.), and significantly stronger coverage drops (low cov.) were analyzed with LIFE-CNA. In addition ctDNA was predicted with machine learning classifiers based on global (ML glob. frag.) and regional fragmentation (ML reg. frag.), and a meta-learner (ML Meta.) integrated into LIFE-CNA. To assess performance of LIFE-CNA, hotspot variants (SNVs) cfDNA concentration (cfDNA) and CEA were analyzed A in samples from CRC patients collected at different time points during disease summarized over all samples and B stratified by disease stage. C LB-CRC-32 was used as one example to show response and resistance to treatment throughout the course of disease
Regional fragmentation and coverage in active chromatin enabled ctDNA detection in 77% (33/43) and 23% (10/43) of patients with localized- and in 74% (67/91) and 37% (34/91) of patients with metastatic CRC with clinically diagnosed tumor burden, respectively. As expected, increased numbers of called SCNAs as well as elevated tumor fractions (Additional file 1: Data) were mainly observed in patients with metastatic CRC (57%, 52/91 vs. 26%, 11/43 and 45%, 41/91 vs. 14%, 6/43, respectively). Enriched short cfDNA fragments enabled ctDNA detection only in a small number of patients with metastatic CRC (19%, 17/91). Considering the three ML classifiers integrated in our LIFE-CNA workflow, we observed that the classifiers based on regional fragmentation and the meta-learner have a higher sensitivity for ctDNA detection (90%, 121/134 and 120/134, respectively), compared to the classifier based on global fragmentation (36%, 48/134). However, when focusing on samples collected within the first six weeks post-surgery, we observed ctDNA predictions with all non-genetic cfDNA features besides the global fragmentation, with the highest numbers of 68% (25/37) being with the ML classifiers based on regional fragmentation and the meta-learner. When focusing on only those samples collected from patients in remission more than six weeks post-treatment ctDNA detection rates decreased.
LIFE-CNA for accurate treatment monitoring in CRC patients
The analysis of multiple ctDNA features improves the sensitivity of untargeted ctDNA detection. To assess the clinical validity of LIFE-CNA for disease monitoring, we assessed changes of our measures over a median follow-up time of 7.5 months (range 1–35.5 months) in 15 patients and correlated these changes with treatment outcome as a proof-of-concept (Additional file 2: Table S6). In addition to LIFE-CNA, we analyzed the commonly used serum protein marker CEA, plasma cfDNA concentration, and SNVs for patients with available hotspot variant data (n = 5). We were able to predict response to treatment in 77% (10/13) of patients (7/7 metastatic, 3/5 localized) by decreasing numbers of SCNAs, normalizing regional or global fragmentation, and/or normalizing coverage in regions of interest. CEA was informative in only 25% (3/12) of patients in two of those patients ~ 2 months later than LIFE-CNA, and decreasing plasma cfDNA concentrations could be correlated to treatment response in only 46% (6/13) of patients in one of those patients ~ 1 month later than LIFE-CNA. Further, LIFE-CNA correctly predicted progressive disease in 100% (5/5) of patients up to four months before clinical evidence with increasing differences to healthy controls of all analyzed cfDNA features. CEA was informative in only 80% (4/5) of patients in one of those patients ~ 3.5 months later than LIFE-CNA and cfDNA concentration was informative in only 20% (1/5) of patients ~ 9 months later than LIFE-CNA, respectively (Additional file 1: Figures S6–S20). For example, response and resistance to treatment could be detected with LIFE-CNA in patient LB-CRC-32 up to five and three months before clinical evidence, respectively (Fig. 5C). CEA identified response to treatment > 2 months later and resistance to treatment in parallel to LIFE-CNA. Although, decreasing cfDNA concentration was associated with response to treatment, at the time of progression no increase could be observed which is in line with previous reports showing low sensitivity and specificity of cfDNA concentration for treatment monitoring [34]. For SNVs, response to treatment could be identified in 3/4 samples, whereas no data were available to evaluate changing SNV levels for progression detection.
LIFE-CNA for cancer screening but not for MRD
To analyze whether LIFE-CNA could be applied for the detection of MRD post-surgery, plasma samples of 33 CRC patients collected up to 8 days pre-surgery and follow-up samples collected between 1 and 42 days post-surgery were analyzed (Additional file 1: Figure S21). Pre-surgery, we detected ctDNA in 92% (22/24) of patients with localized- and in 89% (8/9) of patients with metastatic CRC. Post-surgery, ctDNA was identified in 96% (23/24) of patients with localized- and in 100% (9/9) of patients with metastatic CRC, in particular due to the classifiers based on regional fragmentation and the meta-learner. Further, significant differences in coverage were observed in a large number of post-surgery samples (Additional file 1: Figures S21–S54). Decreasing ctDNA predictions more than six weeks post-treatment might enable the application of LIFE-CNA for recurrence monitoring (Fig. 5A&B, turquoise: remission more than six weeks post-treatment). In addition, the high sensitivity of ctDNA detection at diagnosis of patients with localized CRC (92%) suggests the great potential of LIFE-CNA for cancer screening.
Proof-of-principle of LIFE-CNA using six healthy controls and in silico dilutions
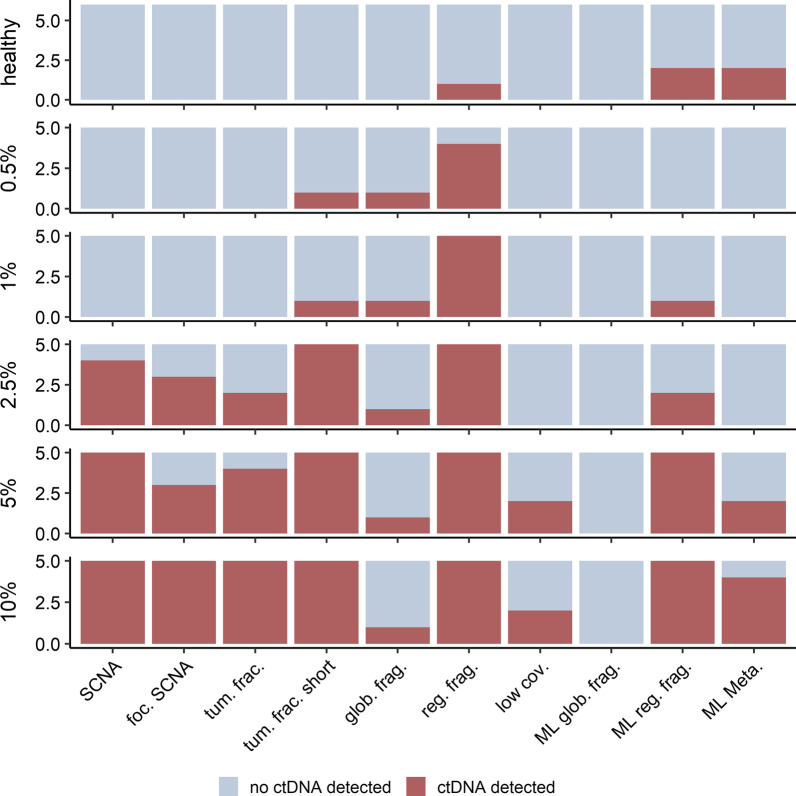
We evaluated the specificity of all cfDNA features by analyzing six additional healthy controls not included in the reference set. Of all analyzed cfDNA features only differential regional fragmentation was detected in 1/6 healthy controls while the remaining cfDNA features did not indicate ctDNA (Fig. 6). The ML classifiers based on regional fragmentation and the meta-learner, predicted ctDNA in 2/6 healthy controls. These results indicate low specificity of the regional-fragmentation and meta-learner based classifiers for ctDNA detection.
Fig. 6.
Proof-of principle showing the high sensitivity of LIFE-CNA. Focal SCNAs (foc. SCNA), tumor fraction (tum. frac.), tumor fraction in 90 to 150 bp fragments(tum. frac. short), enrichment in fragments from 90 to 150 bp (glob. frag.), differential regional fragmentation (reg. frag.), significantly stronger coverage drop in at least to region sets (low cov.), classifier based on global fragmentation (ML glob. frag.), classifier based on regional fragmentation (ML reg. frag.), and classifier based on meta-learner (ML Meta.) were analyzed in six additional healthy controls not included in the panel of normals and in in silico dilutions with 0.5%, 1%, 2.5%, 5% and 10% tumor fraction as a proof-of-principle for ctDNA detection using LIFE-CNA
In addition to specificity, we also assessed the sensitivity of LIFE-CNA for the detection of low ctDNA levels using in silico dilutions with tumor fractions of 0.5%, 1%, 2.5%, 5% and 10% (Additional file 2: Table S7). Analogous to disease monitoring, also for the in silico dilutions we observed the highest sensitivity for ctDNA detection based on regional fragmentation that correctly identified ctDNA in 4/5 samples with 0.5% tumor fraction and in all samples with 1% tumor fraction. At 0.5% tumor fraction, elevated tumor fractions based on ichorCNA and significant enrichment of short fragments could be predicted in one sample. Further, SCNAs could be detected in 4/5 samples with 2.5% tumor fraction. These results indicate that the sensitivity of our SCNA analysis could be increased compared to the previously described required tumor fractions above 5% to 10%. Focusing on the ML classifiers for ctDNA prediction, it was not possible to detect ctDNA based on global fragmentation in any of the in vitro dilutions. Using the classifier based on regional fragmentation, we detected ctDNA in 1/5 samples with 1% tumor fraction.
Discussion
Non-invasive and highly-sensitive ctDNA analyses allow real-time monitoring of patients throughout disease. The untargeted detection of ctDNA has the potential to extend the advantages of LB analysis to patients with cancer across all stages, and independently from knowledge about the presence of somatic hotspot variants. However, clinical validity of untargeted ctDNA analysis could so far mainly be shown for patients with metastatic cancer due to their high tumor fractions. Here, we developed LIFE-CNA for genome-wide ctDNA detection and disease monitoring based on multiple tumor-specific alterations across genetics, epigenetics and fragmentomics in patients with localized and metastasic CRC. We further provide analytical validation as well as a clinical proof-of-concept using a total of 259 plasma samples from 50 CRC patients and 61 healthy individuals. In contrast, a similar study conducted by Cristiano et al. [19] focused on one cfDNA feature (regional fragmentation analysis) for ctDNA detection only. Another study by Peneder et al. [20] also analyzed multiple cfDNA features in Ewing-sarcoma patients.
To facilitate clinical implementation of genome-wide ctDNA analysis suitable for all CRC patients, we defined distinct cutoffs or significance tests for each analyzed cfDNA feature. Establishing and validating definite criteria to report true ctDNA signals further are an important step towards the development of generic guidelines for the analytical validation of untargeted LB analyses, complementing the existing guidelines for targeted hotspot analyses [9, 35].
We evaluated performance of the various cfDNA features and of ML classifiers. CfDNA features achieved a higher sensitivity than ML classifiers for ctDNA detection at diagnosis of patients with localized and metastatic CRC, while false-positive predictions in external healthy controls were higher with the ML classifiers. Other applied ML classifiers reported in the literature achieved slightly better performance characteristics from training and testing procedures for early detection of ctDNA [20, 36]. One previous study performed external validation of a final ML classifier on a cohort of lung cancer patients and thereby achieved comparable sensitivity with slightly higher specificity compared to our ML classifier [37]. Although thorough external validation is required, considering an (albeit small) set of external samples indicates that our ML classifier might achieve a similar performance for CRC patients. Besides focusing solely on ML classifiers or the analysis of multiple cfDNA features, we also investigated whether a combination of ML classifiers with the analysis of multiple cfDNA features can improve the sensitivity of untargeted ctDNA detection. Concretely, combining the analysis of global and regional fragmentation, SCNAs and active chromatin coverage with the ML classifiers resulted in a slightly improved sensitivity for ctDNA detection at diagnosis of patients with localized and metastatic CRC and increased false-positive predictions in external healthy controls. We conclude that considering cfDNA features without ML classifiers may be favorable in cancer screening, as the number of false-positives is markedly reduced, with only a limited reduction in sensitivity, providing comparable performance to colonoscopies, the current gold standard in CRC screening [38]. However, before clinical implementation of LIFE-CNA, sensitivity and specificity needs to be externally validated in a larger cohort.
When evaluating the clinical sensitivity and specificity of LIFE-CNA for residual disease detection and treatment monitoring in a proof-of-concept study, we find a (too) high number of ctDNA positive predictions in R0-resected patients within the first six weeks post-treatment, showing that LIFE-CNA is probably not suited for residual disease detection. This may be explained by the fact that gene regulation and cfDNA fragment length, both factors being considered in the cfDNA features of LIFE-CNA, are perturbed after surgery. Multiple studies described altered gene regulation following surgery in response to cellular trauma [39–41] and the association of low-molecular weight heparin with increased levels of short cfDNA fragments [30, 31], which is given to patients directly after treatment.
There are some limitations that should be considered. Training the ML classifiers on a small a cohort of 134 CRC patient samples and 63 controls might cause overfitting. To overcome false-positive predictions caused by biological variability, larger control and positive cohorts to improve training and external validation for testing would be required before implementation of ML classifiers into clinical practice becomes feasible. Further, the median age of CRC patients (73) is much higher than the median age of healthy controls (32). With regard to the association between cfDNA fragmentation and nucleosome occupancy, which may change during life, future studies with age-matched healthy controls are highly important for validation of LIFE-CNA. Another limitation of this study is that a retrospective analysis of our rather small cohort enabled only the evaluation of a clinical proof-of-concept of LIFE-CNA but not the clinical validity. To establish the clinical utility a large prospective study would be required. If the clinical validity and utility of LIFE-CNA are demonstrated, simple blood sampling may allow rapid and non-invasive treatment monitoring, avoiding unnecessary colonoscopies and radiation introduced by imaging.
Conclusions
Taken together, we assume that considering multiple cfDNA features across different types of tumor-specific alterations in an untargeted genome-wide approach and evaluating them for various applications including screening and treatment monitoring, is an important step toward translating the high potential of liquid biopsy for future personalized medicine applications. Further, when analyzing active chromatin regions specific to other tumor entities we believe that LIFE-CNA can be easily transferred to all solid tumors.
Supplementary Information
Additional file 1: Methods and Data including Table S1 and Figures S1–S54. Methods contain more detailed information about the study cohort and methods required to reproduce the experiments. Data contain detailed information on results not shown in the main document and results obtained for each individual patient analyzed for residual disease detection or treatment monitoring.
Additional file 2: Data required to reproduce the results shown in Tables S2–S8.
Acknowledgements
We would like to thank all healthy individuals and colorectal cancer patients providing their written informed consent to participate in this study. We also thank all study secretaries, for their assistance in study data and patient sample collection.
Abbreviations
- CEA
Carcinoembryonic antigen
- cfDNA
Circulating free DNA
- CNV
Copy number variation
- CRC
Colorectal cancer
- ctDNA
Circulating tumor DNA
- ddPCR
Droplet digital PCR
- DHS
DNase I hypersensitivity sites
- LB
Liquid biopsy
- LIFE-CNA
LIquid biopsy Fragmentation, Epigenetic signature and Copy Number Alteration analysis
- ML
Machine learning
- MRD
Molecular residual disease
- S/L ratio
Short-to-long ratio
- SCNA
Somatic copy number alteration
- SNV
Single nucleotide variant
- TSS
Transcriptional start site
- WGS
Whole-genome sequencing
Author contributions
AH and JP researched the literature to identify the features that could improve sensitivity of data analysis. AH and AB-P developed the experimental procedures. AH performed the experiments. EH provided the Plasma-Seq pipeline. AH and FS performed initial tests to determine the required coverage. AH and TW developed the bioinformatics analysis. AH analyzed and interpreted the data, and performed statistical analysis. UM provided input to statistical analysis and machine learning. JP and VS-L designed the study. VS-L was responsible for obtaining the positive ethics vote for patient recruitment. MR, HV, MdW, and CH contributed to patient recruitment. JP supervised the work. EH-F provided financial and technical resources to enable conduction of the study. AH and JP wrote the manuscript. All authors read and approved the final manuscript.
Funding
The authors received no specific funding for this work.
Availability of data and materials
The sequence data have been deposited at the European Genome-phenome Archive (EGA) under accession number EGAS00001006490. These data are available under a controlled access regimen to ensure the protection of personally identifiable data; access can be obtained by contacting A.H. and J.P. Remaining data generated or analyzed during this study are included in this published article, and its supplementary information files or from the corresponding author on reasonable request. Code for sample analysis is modified from the publication by Heitzer et al. [15], Ulz et al. [22] and Peneder et al. [20] and is available from the corresponding author on reasonable request.
Declarations
Ethics approval and consent to participate
The study was approved by the ethics commission of the Bavarian Medical Association (No. 17059) and is registered with the German registry for clinical trials (trial registration ID: DRKS00012890). All participants provided informed written consent prior to blood and tissue specimen collection. The study was performed in accordance with the Declaration of Helsinki.
Consent for publication
All participants provided informed written consent to the publication of their data. This manuscript has been read and approved for publication by all named authors.
Competing interests
The authors declare no potential conflicts of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Ariane Hallermayr, Email: ariane.hallermayr@mgz-muenchen.de.
Tobias Wohlfrom, Email: tobias.wohlfrom@mgz-muenchen.de.
Verena Steinke-Lange, Email: verena.steinke-lange@mgz-muenchen.de.
Anna Benet-Pagès, Email: Anna.Benet-Pages@mgz-muenchen.de.
Florentine Scharf, Email: Florentine.Scharf@mgz-muenchen.de.
Ellen Heitzer, Email: ellen.heitzer@medunigraz.at.
Ulrich Mansmann, Email: mansmann@ibe.med.uni-muenchen.de.
Christopher Haberl, Email: christopher.haberl@klinikum-straubing.de.
Maike de Wit, Email: maike.dewit@vivantes.de.
Holger Vogelsang, Email: holger.vogelsang@klinikum-gap.de.
Markus Rentsch, Email: markus.rentsch@klinikum-ingolstadt.de.
Elke Holinski-Feder, Email: elke.holinski-feder@mgz-muenchen.de.
Julia M. A. Pickl, Email: julia.romic-pickl@mgz-muenchen.de
References
- 1.André F, Bachelot T, Commo F, Campone M, Arnedos M, Dieras V, et al. Comparative genomic hybridisation array and DNA sequencing to direct treatment of metastatic breast cancer: a multicentre, prospective trial (SAFIR01/UNICANCER) Lancet Oncol. 2014;15:267–274. doi: 10.1016/S1470-2045(13)70611-9. [DOI] [PubMed] [Google Scholar]
- 2.Stockley TL, Oza AM, Berman HK, Leighl NB, Knox JJ, Shepherd FA, et al. Molecular profiling of advanced solid tumors and patient outcomes with genotype-matched clinical trials: the Princess Margaret IMPACT/COMPACT trial. Genome Med. 2016;8:109. doi: 10.1186/s13073-016-0364-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moscow JA, Fojo T, Schilsky RL. The evidence framework for precision cancer medicine. Nat Rev Clin Oncol. 2018;15:183–192. doi: 10.1038/nrclinonc.2017.186. [DOI] [PubMed] [Google Scholar]
- 4.Heitzer E, Haque IS, Roberts CES, Speicher MR. Current and future perspectives of liquid biopsies in genomics-driven oncology. Nat Rev Genet. 2019;20:71–88. doi: 10.1038/s41576-018-0071-5. [DOI] [PubMed] [Google Scholar]
- 5.Pinzani P, D'Argenio V, Del Re M, Pellegrini C, Cucchiara F, Salvianti F, Galbiati S. Updates on liquid biopsy: current trends and future perspectives for clinical application in solid tumors. Clin Chem Lab Med. 2021;59:1181–1200. doi: 10.1515/cclm-2020-1685. [DOI] [PubMed] [Google Scholar]
- 6.Diehl F, Li M, Dressman D, He Y, Shen D, Szabo S, et al. Detection and quantification of mutations in the plasma of patients with colorectal tumors. PNAS. 2005;102:16368–16373. doi: 10.1073/pnas.0507904102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bettegowda C, Sausen M, Leary RJ, Kinde I, Wang Y, Agrawal N, et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med. 2014;6:224ra24. doi: 10.1126/scitranslmed.3007094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Merker JD, Oxnard GR, Compton C, Diehn M, Hurley P, Lazar AJ, et al. Circulating tumor DNA analysis in patients with cancer: American society of clinical oncology and College of American pathologists Joint review. J Clin Oncol. 2018;36:1631–1641. doi: 10.1200/JCO.2017.76.8671. [DOI] [PubMed] [Google Scholar]
- 9.Godsey JH, Silvestro A, Barrett JC, Bramlett K, Chudova D, Deras I, et al. Generic protocols for the analytical validation of next-generation sequencing-based ctDNA assays: a joint consensus recommendation of the BloodPAC's analytical variables working group. Clin Chem. 2020;66:1156–1166. doi: 10.1093/clinchem/hvaa164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Connors D, Allen J, Alvarez JD, Boyle J, Cristofanilli M, Hiller C, et al. International liquid biopsy standardization alliance white paper. Crit Rev Oncol Hematol. 2020;156:103112. doi: 10.1016/j.critrevonc.2020.103112. [DOI] [PubMed] [Google Scholar]
- 11.Diehl F, Schmidt K, Choti MA, Romans K, Goodman S, Li M, et al. Circulating mutant DNA to assess tumor dynamics. Nat Med. 2008;14:985–990. doi: 10.1038/nm.1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Taly V, Pekin D, Benhaim L, Kotsopoulos SK, Le Corre D, Li X, et al. Multiplex picodroplet digital PCR to detect KRAS mutations in circulating DNA from the plasma of colorectal cancer patients. Clin Chem. 2013;59:1722–1731. doi: 10.1373/clinchem.2013.206359. [DOI] [PubMed] [Google Scholar]
- 13.Zhang H, Liu R, Yan C, Liu L, Tong Z, Jiang W, et al. Advantage of next-generation sequencing in dynamic monitoring of circulating tumor DNA over droplet digital PCR in Cetuximab treated colorectal cancer patients. Transl Oncol. 2019;12:426–431. doi: 10.1016/j.tranon.2018.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smith T, Heger A, Sudbery I. UMI-tools: modeling sequencing errors in unique molecular identifiers to improve quantification accuracy. Genome Res. 2017;27:491–499. doi: 10.1101/gr.209601.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heitzer E, Ulz P, Belic J, Gutschi S, Quehenberger F, Fischereder K, et al. Tumor-associated copy number changes in the circulation of patients with prostate cancer identified through whole-genome sequencing. Genome Med. 2013;5:30. doi: 10.1186/gm434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Scheinin I, Sie D, Bengtsson H, van de Wiel MA, Olshen AB, van Thuijl HF, et al. DNA copy number analysis of fresh and formalin-fixed specimens by shallow whole-genome sequencing with identification and exclusion of problematic regions in the genome assembly. Genome Res. 2014;24:2022–2032. doi: 10.1101/gr.175141.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Adalsteinsson VA, Ha G, Freeman SS, Choudhury AD, Stover DG, Parsons HA, et al. Scalable whole-exome sequencing of cell-free DNA reveals high concordance with metastatic tumors. Nat Commun. 2017;8:1324. doi: 10.1038/s41467-017-00965-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mouliere F, Chandrananda D, Piskorz AM, Moore EK, Morris J, Ahlborn LB, Mair R, et al. Enhanced detection of circulating tumor DNA by fragment size analysis. Sci Transl Med. 2018 doi: 10.1126/scitranslmed.aat4921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cristiano S, Leal A, Phallen J, Fiksel J, Adleff V, Bruhm DC, et al. Genome-wide cell-free DNA fragmentation in patients with cancer. Nature. 2019;570:385–389. doi: 10.1038/s41586-019-1272-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peneder P, Stütz AM, Surdez D, Krumbholz M, Semper S, Chicard M, et al. Multimodal analysis of cell-free DNA whole-genome sequencing for pediatric cancers with low mutational burden. Nat Commun. 2021;12:3230. doi: 10.1038/s41467-021-23445-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ulz P, Perakis S, Zhou Q, Moser T, Belic J, Lazzeri I, et al. Inference of transcription factor binding from cell-free DNA enables tumor subtype prediction and early detection. Nat Commun. 2019;10:4666. doi: 10.1038/s41467-019-12714-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ulz P, Belic J, Graf R, Auer M, Lafer I, Fischereder K, et al. Whole-genome plasma sequencing reveals focal amplifications as a driving force in metastatic prostate cancer. Nat Commun. 2016;7:12008. doi: 10.1038/ncomms12008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wittekind C, Meyer H-J. TNM Klassifikation maligner Tumoren. 7. Weinheim, Germany: Wiley-VCH Verlag GmbH & Co. KGaA; 2010. [Google Scholar]
- 24.Amemiya HM, Kundaje A, Boyle AP. The ENCODE blacklist: identification of problematic regions of the genome. Sci Rep. 2019;9:9354. doi: 10.1038/s41598-019-45839-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.UCSC. Gap Locations. 2018. http://genome.ucsc.edu/cgi-bin/hgTrackUi?g=gap. Accessed 12 Mar 2022.
- 26.Della Chiara G, Gervasoni F, Fakiola M, Godano C, D'Oria C, Azzolin L, et al. Epigenomic landscape of human colorectal cancer unveils an aberrant core of pan-cancer enhancers orchestrated by YAP/TAZ. Nat Commun. 2021;12:2340. doi: 10.1038/s41467-021-22544-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.COSMIC - Catalogue of somatic mutations in cancer. Cancer Gene Census. https://cancer.sanger.ac.uk/census. Accessed 1 Oct 2021.
- 28.Snyder MW, Kircher M, Hill AJ, Daza RM, Shendure J. Cell-free DNA comprises an in vivo nucleosome footprint that informs its tissues-of-origin. Cell. 2016;164:57–68. doi: 10.1016/j.cell.2015.11.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sun K, Jiang P, Cheng SH, Cheng THT, Wong J, Wong VWS, et al. Orientation-aware plasma cell-free DNA fragmentation analysis in open chromatin regions informs tissue of origin. Genome Res. 2019;29:418–427. doi: 10.1101/gr.242719.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nakamura N, Sasaki A, Mikami M, Nishiyama M, Akaishi R, Wada S, et al. Nonreportable rates and cell-free DNA profiles in noninvasive prenatal testing among women with heparin treatment. Prenat Diagn. 2020;40:838–845. doi: 10.1002/pd.5695. [DOI] [PubMed] [Google Scholar]
- 31.Han DSC, Ni M, Chan RWY, Chan VWH, Lui KO, Chiu RWK, Lo YMD. The biology of cell-free DNA fragmentation and the roles of DNASE1, DNASE1L3, and DFFB. Am J Hum Genet. 2020;106:202–214. doi: 10.1016/j.ajhg.2020.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ernst J, Kellis M. ChromHMM: automating chromatin-state discovery and characterization. Nat Methods. 2012;9:215–216. doi: 10.1038/nmeth.1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van Berkum NL, Lieberman-Aiden E, Williams L, Imakaev M, Gnirke A, Mirny LA, et al. Hi-C: a method to study the three-dimensional architecture of genomes. Journal of Visualized Experiments : JoVE. 2010 doi: 10.3791/1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hallermayr A, Steinke-Lange V, Vogelsang H, Rentsch M, de Wit M, Haberl C, et al. Clinical validity of circulating tumor DNA as prognostic and predictive marker for personalized colorectal cancer patient management. Cancers. 2022 doi: 10.3390/cancers14030851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hallermayr A, Benet-Pagès A, Steinke-Lange V, Mansmann U, Rentsch M, Holinski-Feder E, Pickl JMA. Liquid Biopsy Hotspot Variant Assays: Analytical Validation for Application in Residual Disease Detection and Treatment Monitoring. Clin Chem. 2021 doi: 10.1093/clinchem/hvab124. [DOI] [PubMed] [Google Scholar]
- 36.Chen X, Gole J, Gore A, He Q, Lu M, Min J, et al. Non-invasive early detection of cancer four years before conventional diagnosis using a blood test. Nat Commun. 2020;11:3475. doi: 10.1038/s41467-020-17316-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mathios D, Johansen JS, Cristiano S, Medina JE, Phallen J, Larsen KR, et al. Detection and characterization of lung cancer using cell-free DNA fragmentomes. Nat Commun. 2021;12:5060. doi: 10.1038/s41467-021-24994-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lin JS, Piper MA, Perdue LA, Rutter CM, Webber EM, O'Connor E, et al. Screening for colorectal cancer: updated evidence report and systematic review for the US preventive services task force. JAMA. 2016;315:2576–2594. doi: 10.1001/jama.2016.3332. [DOI] [PubMed] [Google Scholar]
- 39.Sadahiro R, Knight B, James F, Hannon E, Charity J, Daniels IR, et al. Major surgery induces acute changes in measured DNA methylation associated with immune response pathways. Sci Rep. 2020;10:5743. doi: 10.1038/s41598-020-62262-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Caputi FF, Carboni L, Rullo L, Alessandrini I, Balzani E, Melotti RM, et al. An exploratory pilot study of changes in global DNA methylation in patients undergoing major breast surgery under opioid-based general anesthesia. Front Pharmacol. 2021;12:733577. doi: 10.3389/fphar.2021.733577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Henriksen TV, Reinert T, Christensen E, Sethi H, Birkenkamp-Demtröder K, Gögenur M, et al. The effect of surgical trauma on circulating free DNA levels in cancer patients-implications for studies of circulating tumor DNA. Mol Oncol. 2020;14:1670–1679. doi: 10.1002/1878-0261.12729. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Methods and Data including Table S1 and Figures S1–S54. Methods contain more detailed information about the study cohort and methods required to reproduce the experiments. Data contain detailed information on results not shown in the main document and results obtained for each individual patient analyzed for residual disease detection or treatment monitoring.
Additional file 2: Data required to reproduce the results shown in Tables S2–S8.
Data Availability Statement
The sequence data have been deposited at the European Genome-phenome Archive (EGA) under accession number EGAS00001006490. These data are available under a controlled access regimen to ensure the protection of personally identifiable data; access can be obtained by contacting A.H. and J.P. Remaining data generated or analyzed during this study are included in this published article, and its supplementary information files or from the corresponding author on reasonable request. Code for sample analysis is modified from the publication by Heitzer et al. [15], Ulz et al. [22] and Peneder et al. [20] and is available from the corresponding author on reasonable request.