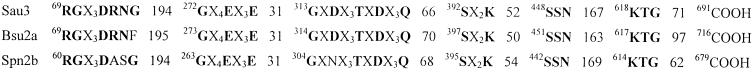Abstract
The gene pbpC from Staphylococcus aureus was sequenced: it encodes a 691-amino-acid protein with all of the conserved motifs of a class B high-molecular-weight penicillin-binding protein (PBP), including the transpeptidase conserved motifs SXXK, SXN, and KTG. Insertional inactivation of pbpC and introduction of the intact gene in a laboratory mutant missing PBP 3 showed that the pbpC gene encodes the staphylococcal PBP 3. Inactivation of pbpC caused no detectable change in the muropeptide composition of cell wall peptidoglycan and had only minimum, if any, effect on growth rates, but caused a small but significant decrease in rates of autolysis. Cells of abnormal size and shape and disoriented septa were produced when bacteria with inactivated pbpC were grown in the presence of a sub-MIC of methicillin.
Penicillin-binding proteins (PBPs) are enzymes involved in the final stages of peptidoglycan biosynthesis. They fall into two major groups: the low-molecular-weight (LMW) PBPs, monofunctional proteins that act mainly as dd-carboxypeptidases, and the high-molecular-weight (HMW) PBPs, which are multimodular enzymes anchored to the cytoplasmic membrane by a noncleavable pseudo signal peptide and which are normally composed of two modules localized on the outer side of the cytoplasmic membrane. The C-terminal module binds penicillin and catalyzes peptidoglycan cross-linking. The N-terminal domain can have transglycosylase activity (class A HMW PBPs), but in many cases, its function has not been clearly established (class B HMW PBPs) (12).
Staphylococcus aureus has four PBPs (PBPs 1 to 4), but the exact role of each one remains to be determined, with the exception of PBP 4, which has been shown to be responsible for the secondary cross-linking of peptidoglycan (33).
Besides their role in peptidoglycan synthesis, PBPs have attracted much attention because they are the targets of β-lactam antibiotics, which covalently bind to these proteins, inhibiting cell wall synthesis. Resistance to β-lactams can involve the acquisition of a new low-affinity PBP—PBP 2A—typical of methicillin-resistant S. aureus (MRSA) (16, 26), changes in the native PBPs (29), or the production of β-lactamase.
The gene sequences of PBPs 1, 2, and 4 of S. aureus are currently available (GenBank accession no. U94706, X62288, and U29454, respectively) and can be used for the study of the mechanism of resistance to β-lactams as well as to answer basic questions regarding the function and essentiality of each of the PBPs.
In the present study, we report the identification by sequencing and cloning of the gene pbpC. Evidence is described indicating that pbpC is the structural gene of S. aureus PBP 3.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth media.
The bacterial strains and plasmids used in this study are described in Table 1.
TABLE 1.
Strains and plasmids used in this study
| Strains | Relevant characteristics | Source or reference |
|---|---|---|
| Strain | ||
| S. aureus | ||
| RN4220 | Mcs restriction negative | R. Novick |
| M100 | Mcr laboratory step mutant | 30 |
| 27s | Mcs | 30 |
| COL | Homogeneous Mcr, MIC of 1,600 μg ml−1 | Rockefeller University collection |
| M100R130 | M100 (pbp2::Tn551) Emr | This study |
| RUSA 130 | COL Ω703 (pbp2::Tn551) Emr heterogenous, MIC of 12 μg ml−1 | 6, 23, 24 |
| RN4220pPBP3-B3 | RN4220/pPBP3B inserted into the chromosome Mcs Emr | This study |
| M100pPBP3-A | M100/pPBP3A inserted into the chromosome Mcr Emr | This study |
| COLpPBP3-B1 | COL/pPBP3B inserted into the chromosome Mcr Emr | This study |
| COLpPBP3-B4 | COL/pPBP3B inserted into the chromosome Mcr Emr | This study |
| E. coli DH5α | recA endA1 gyrA96 thi-1 hsdR17 supE44 relA1 φ80 ΔlacZΔM15 | Bethesda Research Laboratories |
| Plasmids | ||
| pSP64 | E. coli plasmid, Ampr | Promega |
| pSP64E | PSP64/1.2 kb with erm gene from Tn551 | This study |
| pPBP3A | pSP64E/2.0 kb encoding aa 61–691 of PBP 3 | This study |
| pPBP3B | pSP64E/1.4 kb encoding aa 61–544 of PBP 3 | This study |
S. aureus strains were grown on tryptic soy broth (TSB) (Difco Laboratories) with aeration as described previously (22). Escherichia coli strains were grown in Luria-Bertani broth (Difco) with aeration.
Antibiotics were purchased from Sigma and used at the following concentrations: erythromycin, 10 μg/ml; ampicillin, 100 μg/ml.
DNA methods.
Routine DNA manipulations were performed by standard methods (1, 27). All of the enzymes were purchased from either New England Biolabs or Boehringer Mannheim and used as recommended by the manufacturers. DNA sequencing was done at the Rockefeller University Protein/DNA Technology Center by the Taq fluorescent dye terminator sequencing method by using a PE/ABI model 377 automated sequencer.
Identification of pbpC.
The TIGR (The Institute for Genomic Research) S. aureus incomplete genome database (http://www.tigr.org) was searched for DNA fragments encoding peptides with homology to PBPs by using the known protein sequences of the PBPs from Bacillus subtilis. This search identified a 548-bp fragment that encoded a peptide 52% identical to an internal fragment of PBP 2A of B. subtilis. Chromosomal DNA of the MRSA strain COL was digested with several restriction enzymes, electrophoresed, and hybridized with the 548-bp fragment, a putative internal fragment of pbpC, obtained by PCR with the GeneAmp PCR reagent kit with AmpliTaq DNA polymerase (Perkin-Elmer) with primers PBP3P3 (5′-CTGCGAAGCTTCTTAATTTG-3′) and PBP3P4 (5′-CAGCAACTTTCCAAATTACC-3′) under the following conditions: 94°C for 4 min; 30 cycles of 94°C for 30 s, 53°C for 30 s, and 72°C for 1 min; and one final extension step of 72°C for 4 min. Restriction with PstI yielded a 6-kb fragment that hybridized with the pbpC probe and was cloned in pGEM3Z (Promega). A 3.8-kb portion of this fragment was sequenced by primer walking, and open reading frames (ORFs) were analyzed with DNASTAR software and compared to the known peptides from the EMBL and GenBank databases by using the BLAST algorithm.
Sequence analysis of pbpC from mutant M100.
A DNA fragment containing pbpC was amplified from the chromosomal DNA of mutant M100 (30) and its parental strain, 27s (30), with primers PBP3P11B (5′-TTGGAATGTAGTTAACTGGG-3′) and PBP3P12 (5′-CATGGTTATTCCTCCTTATC-3′) under the following conditions: 94°C for 4 min; 30 cycles of 94°C for 30 s, 53°C for 30 s, and 72°C for 3 min; and one final extension step of 72°C for 6 min. Sequences of pbpC from M100 and 27s were aligned and compared with DNASTAR software.
Isolation of RNA and Northern blot hybridization.
Overnight cultures of S. aureus strains 27s and COL were diluted 1:50 in TSB and grown to mid-log phase (optical density at 620 nm [OD620] of 0.8 to 1). The cells were pelleted and processed with the FastRNA Blue isolation kit (Bio 101, Inc.) in combination with FastPrep FP120 (Bio 101 Savant), according to the manufacturer's recommendations. RNA (5 μg) was electrophoresed through a 1.2% agarose–0.66 M formaldehyde gel in MOPS (morpholinepropanesulfonic acid) running buffer (Sigma). Blotting of RNA onto Hybond N+ membranes (Amersham) was performed with the Turboblotter alkaline transfer systems (Schleicher & Schuell). For the detection of specific transcripts, DNA probes were labeled with the Ready To Go labeling kit (Pharmacia Biotech) with [α-32P]dCTP (Amersham Life Sciences) and hybridized under high-stringency conditions. The blots were subsequently washed and autoradiographed.
Primer extension analysis.
Primer extension analysis was performed with primer PBP3P7 (5′-AAACGTAGTACTAGTACTGC-3′), which was end labeled with [γ-32P]ATP and purified by Sephadex G-25 spin columns (Boehringer Mannheim). RNA from COL (1.5 μg) or 27s (1.5 and 6 μg) was hybridized with the primer at 65°C for 90 min and slowly cooled to room temperature. Reverse transcription was carried out with SuperScript reverse transcriptase (Gibco BRL) at 42°C for 90 min, and the reaction mixture was heated at 65°C for 10 min to inactivate the enzyme. The reaction product was incubated with RNase H (3 U) at 37°C for 30 min, ethanol precipitated, resuspended in 1 μl of Sequenase stop solution, denatured, and applied to a 6% sequencing gel. Sequencing reaction mixtures prepared with the T7 Sequenase kit vs2.0 (Amersham Life Sciences), primed by an oligonucleotide identical to that used for primer extension, were also applied to the gel. The gel was dried and autoradiographed with an intensifying screen (Dupont) over the lanes corresponding to the primer extension bands.
pbpC cloning.
A suicide vector for S. aureus was constructed by cloning the erm gene from transposon Tn551 (accession no. Y13600) (32) into the E. coli plasmid pSP64 (Promega). A 1.1-kb DNA fragment containing the erm gene was amplified from the RUSA130 chromosomal DNA by high-fidelity PCR with the GeneAmp XL PCR kit (Perkin-Elmer), which includes rTth DNA polymerase XL, and with 20 pmol each of primers erm1 (5′-AATGGATCCAATCATGAGTATTGTCCGAG-3′) and erm2X (5′-GCTCTAGAACATTCCCTTTAGTAACGTG-3′). A hot start and the following conditions were used: 94°C for 2 min, 20 cycles of 94°C for 30 s and 55°C for 5 min, and one final extension step of 55°C for 10 min. The PCR fragment was purified with a QIAquick PCR purification kit (Qiagen), digested with BamHI and XbaI, ligated into plasmid pSP64, and transformed into E. coli DH5α. The resultant plasmid was called pSP64E.
To clone pbpC, without the promoter region and the first 60 codons, a 2.0-kb DNA fragment was amplified from the COL chromosomal DNA by high-fidelity PCR, as described above, with primers PBP3P19E (5′-GGCGAATTCCATTACAGTGAATGAGTCTG-3′) and PBP3P12A (5′-CGTGGATCCCATGGTTATTCCTCCTTATC-3′). This fragment was digested with EcoRI and BamHI and cloned into plasmid pSP64E, and the resultant plasmid was called pPBP3-A.
To clone an internal fragment of the pbpC gene, a 1.4-kb DNA fragment was amplified from the COL chromosomal DNA by high-fidelity PCR with primers PBP3P19E and PBP3P14 (5′-AGCGGATCCACCATCATTCGCTATAGTTG-3′). This fragment was digested with EcoRI and BamHI and cloned into plasmid pSP64E, and the resultant plasmid was called pPBP3-B.
Inactivation of pbpC in COL and RN4220 and complementation in M100.
For the complementation of M100 with a functional copy of pbpC, 18 μg of plasmid pPBP3-A was electroporated into RN4220 essentially as previously described (18). The correct insertion of the plasmid was confirmed by PCR, and it was subsequently introduced into strain M100 by transduction with phage 80α as described previously (22). The correct insertion of the plasmid in M100 was again confirmed by PCR. For the inactivation of pbpC, a similar procedure was done to insert plasmid pPBP3-B into the chromosome of RN4220 and afterwards transduce it to COL.
Inactivation of pbp2 in M100.
The Tn551 insertion in pbp2 described in RUSA130 (24) was transduced into the background of M100 with the phage 80α as previously described (22).
Membrane preparation and analysis of PBPs.
Membrane preparation and penicillin-binding assays were performed essentially as previously described (30). Membrane proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) in a 3.9% polyacrylamide stacking gel and an 8% separation gel.
Determination of growth and autolysis rates and population analysis profiles.
For growth rate determination, an overnight culture grown in TSB (control strains) or TSB with erythromycin (mutants) was diluted 1/1,500 into 50 ml of TSB and incubated at 37°C with aeration. The OD620 was observed over time. Triton X-100-induced autolysis (5) and population analysis (23) were performed as previously described. Values for growth and autolysis rates were determined based on three and four experiments, respectively.
Electron microscopy.
RN4220 and RN4220pPBP3-B3 were grown in TSB and/or in TSB with 0.3 μg of methicillin per ml (about 1/5 of the MIC) until the OD620 reached 0.7. Cells were harvested by low-speed centrifugation and fixed with 2.5% glutaraldehyde. Electron microscopy was done at the Electron Microscopy Service of The Rockefeller University.
Analysis of cell wall muropeptides.
The isolation of cell wall peptidoglycan and the analysis of the family of enzymatically released muropeptides, by reverse-phase high-pressure liquid chromatography (HPLC), were carried out essentially as previously described (4).
Nucleotide sequence accession number.
The complete nucleotide sequence determined in this study is available in the EMBL and GenBank databases under accession no. AJ243120.
RESULTS
Identification of the pbpC gene.
A 6-kb PstI fragment that hybridizes with an internal fragment of pbpC was cloned. Sequence analysis identified an ORF of 2,076 nucleotides (nt) that encodes a peptide of 691 amino acids (aa), 44% identical to the PBP 2A of B. subtilis (20) and 33% identical to the PBP 2B of Streptococcus pneumoniae (8), two HMW class B PBPs. This ORF, which was named pbpC, starts with a UUG codon instead of the more usual AUG codon. The hydrophilicity plot of Kyte-Doolittle shows a probable membrane anchor domain at the N terminus.
Alignment of PBP 3 encoded by pbpC from S. aureus, PBP 2A from B. subtilis, and PBP 2B from S. pneumoniae showed that not only are the transpeptidase conserved motifs present in PBP 3, but so are the conserved amino acids from the non-penicillin-binding module of similar class B PBPs (14), as well as the spacing between the various motifs (Fig. 1).
FIG. 1.
Amino acid sequence analysis of S. aureus PBP 3 (Sau3), S. pneumoniae PBP 2B (Spn2b), and B. subtilis PBP 2A (Bsu2a). The conserved motifs are common to HMW class B PBPs. The amino acid sequence signature of each module is in boldface.
Analysis of the transcription of pbpC.
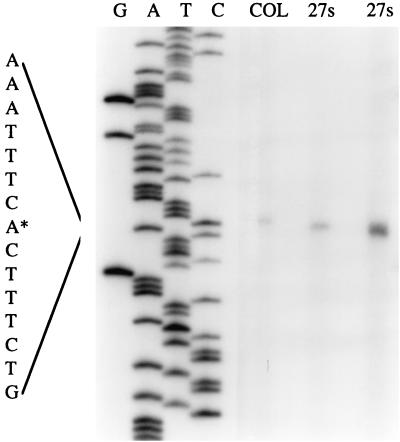
Primer extension analysis was performed to determine the transcription initiation site of pbpC (Fig. 2). The transcript initiates at a thymine residue located 29 nt upstream of the start codon (nt 148 of database sequence). The putative −10 and −35 regions of the promoter are located at nt 136 to 141 and 113 to 118, respectively. Northern blot analysis was also done (data not shown), indicating that the pbpC gene is transcribed alone from a single promoter and that the size of the transcript is 2.6 kb. This size of transcript indicates that pbpC mRNA may extend up to 0.5 kb downstream of the stop codon. Analysis of the 0.5-kb sequence downstream of pbpC showed that there is a small ORF encoding a protein homologue to the 50S ribosomal protein L33 from Bacillus stearothermophilus, which seems to have its own promoter region. Downstream of this ORF, there is a putative stem-loop structure followed by a stretch of five T's, features that are characteristic of rho-independent terminators. We were unable to find another terminator closer to the stop codon of pbpC, so it is possible that its transcription extends to the 50S ribosomal protein terminator, located 0.29 to 0.33 kb downstream of the end of pbpC.
FIG. 2.
Mapping of the 5′ end of the pbpC transcript by primer extension. The sequence encompassing the transcription start site (marked by an asterisk) is enlarged. Total RNA of S. aureus strains 27s (1.5 and 6 μg) and COL (1.5 μg) was used.
PBPs and sequence analysis of pbpC from mutant M100.
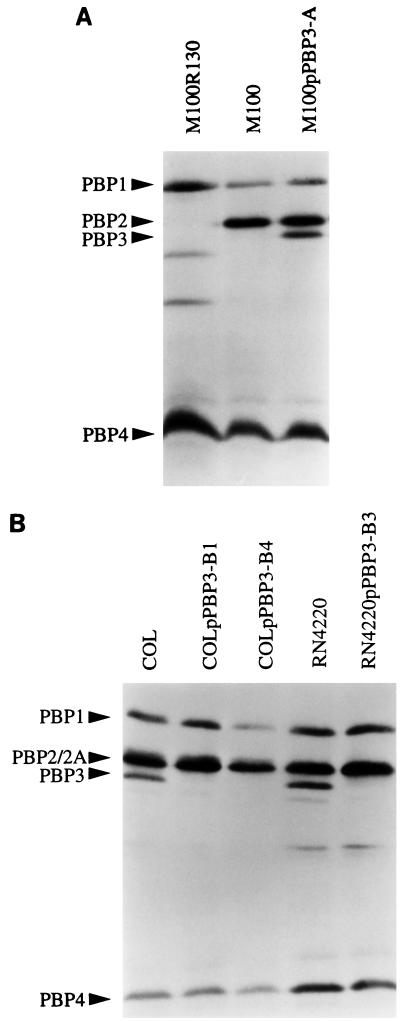
PBP fluorographic assays (Fig. 3A) showed that PBP 3 was not detectable in membrane preparations of M100, a laboratory step mutant selected for methicillin resistance from its parental strain, 27s, by growth in increasing concentrations of the antibiotic (30). The pbpC sequence from M100 was compared to the sequence of its parental strain, 27s. M100 has a single base change (C→T) at nt 471 from the sequence deposited in the databases, which causes the appearance of a premature stop codon. The pbpC of M100 encodes a peptide of only 98 aa.
FIG. 3.
(A) PBP fluorographic assay of M100, a laboratory mutant selected for methicillin resistance without PBP 3; M100pPBP3-A, in which a copy of pbpC was introduced; and M100R130, in which PBP 2 was selectively inactivated. (B) PBP fluorographic assay of MRSA strain COL; the two mutants in which pbpC was inactivated in COL background (COLpPBP3-B1 and -B4); methicillin-sensitive S. aureus strain RN4220; and the mutant RN4220pPBP3-B3, in which pbpC was inactivated in the RN4220 background.
pbpC complementation and inactivation.
A 2.0-kb fragment of pbpC that does not include the promoter region and the first 60 codons was cloned into the suicide vector pSP64E and inserted into the chromosomal DNA of RN4220. This results in a duplication of the pbpC gene in which a functional copy of the gene is followed by the plasmid pSP64E and by a second copy of pbpC encoding a protein truncated at its N terminus. This construct was transduced into M100, where it replaced the mutated copy of the pbpC gene. Figure 3A shows that the band corresponding to PBP 3 was now present in mutant M100pPBP3-A.
An internal fragment of pbpC was also cloned into the suicide vector pSP64E and used to disrupt the pbpC gene in the chromosome of strain RN4220 by Campbell-type integration. This construct was subsequently transduced to strain COL, where it replaced the functional copy of pbpC. The successful inactivation of PBP 3 penicillin-binding activity was confirmed by the fluorographic assay (Fig. 3B), showing that the band corresponding to PBP 3 has disappeared in the mutants in which pbpC was disrupted.
Characterization of pbpC mutants.
The inactivation of pbpC in the background of strain COL and RN4220 had only minimal if any effect on growth rates. However, it caused a reproducible decrease in rates of autolysis: 0.61 h−1 ± 0.06 for COL and 0.48 h−1 ± 0.02 for COLpPBP3-B1 and 1.88 h−1 ± 06 for RN4220 and 1.12 h−1 ± 0.05 for RN4220pPBP3-B3.
Cell wall muropeptide profiles were analyzed by HPLC (data not shown). The inactivation of pbpC did not cause any detectable change in the elution profile of muropeptides.
The effect on methicillin resistance was also tested. In the background of the MRSA strain COL, which produces the low-affinity PBP 2A, the methicillin resistance was not significantly altered by inactivation of pbpC. However when PBP 3 was reintroduced in the background of the laboratory mutant M100, it caused a twofold decrease of the methicillin MIC, from 50 μg/ml to 25 μg/ml.
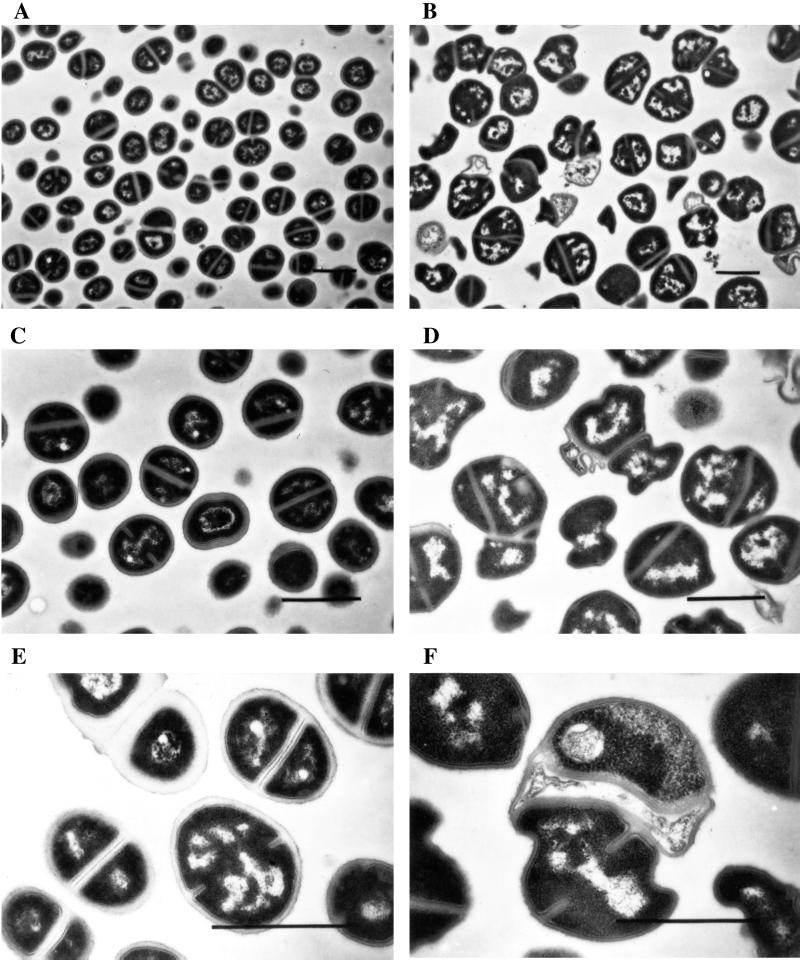
Strain RN4220 and its PBP 3 mutant, RN4220pPBP3-B3, were grown in TSB and observed by electron microscopy (data not shown). No striking changes in the morphology of the PBP 3 mutants could be observed, except that in some rare cases, we could observe cells with two septa formed in the same plane. However, when both the control strain and the PBP 3 mutant were grown in a sub-MIC concentration of methicillin (0.3 μg/ml), the control cells retained normal morphology, while the mutant had obvious changes (Fig. 4): the average size of the cells increased, and they had an abnormal shape; ruffling of the cell wall and membrane and irregularly placed septa were observable.
FIG. 4.
Electron microscopy of RN4220 (A, C, and E) and the PBP 3 mutant RN4220pPBP3-B3 (B, D, and F) grown in the presence of 0.3 μg of methicillin per ml. The bar corresponds to 1 μm.
DISCUSSION
Of the four normal PBPs of S. aureus, only the gene sequence of PBP 3 had not been described so far: the genetic determinants of PBP 1 (25), PBP 2 (15, 19, 23), and PBP 4 (7) had already been identified. Our observations indicate that pbpC described in this communication was the structural determinant of PBP 3, thus completing the genetic characterization of the entire set of major staphylococcal PBPs. The pbpC gene encodes a bimodular PBP with a C-terminal penicillin-binding domain, presumably responsible for peptidoglycan cross-linking, and an N-terminal non-penicillin-binding domain whose function remains unknown. Its highest-scoring homologues, PBP 2A of B. subtilis (44% identical to PBP 3 of S. aureus) (20) and PBP 2B of S. pneumoniae (33% identity) (8), belong to subclass B5 of the HMW PBPs (14). All of the conserved amino acids characteristic of both the non-penicillin-binding and penicillin-binding domains from class B PBPs, as well as the conserved spacing between them, are present in PBP 3. In E. coli, it has been shown that the non-penicillin-binding module of PBP 3, a key element in cell septation and a class B HMW PBP, is essential for the correct folding of the penicillin-binding domain, acting as a noncleaved intramolecular chaperone (13).
We inactivated the penicillin binding activity of PBP 3 in the background of both the MRSA strain COL (which produces PBP 2A) and the methicillin-sensitive strain RN4220 (which has the normal set of PBPs 1 to 4) by insertion duplication. The results of the inactivation, taken together with the fact that M100 has a premature stop codon that reduces the product of pbpC to a 98-aa peptide, indicate that PBP 3 is not an essential protein for the growth of S. aureus, contrary to what had been previously described based on the correlation between the antibacterial activities of different antibiotics and their binding affinities to each PBP (10). Mutants lacking PBP 4 were also described (3), suggesting that the remaining PBP 1, which seems to be essential for growth (31), and PBP 2 probably represent the lethal targets for β-lactam antibiotics in S. aureus.
Regarding the characteristics that we analyzed, namely growth and autolysis rates, cell wall muropeptide profile, and impact on methicillin resistance in an MRSA strain, there is no apparent phenotype associated with the inactivation of pbpC, except for a decrease in the autolysis rates. It has been reported that in E. coli, PBP 1A and 1B can take over the functions of each other (28). PBP 1 from S. aureus is also an HMW class B PBP, so it is conceivable that PBP 1 may substitute for PBP 3 when the latter is inactivated. It has been suggested on the basis of studies with selective inhibitors for each PBP (9, 21) that PBP 3 maybe involved with septum formation and cell separation. We did not observe any striking differences when comparing RN4220 and its PBP 3 mutant grown in TSB. However, when both control and mutant cells were grown in the presence of a sub-MIC concentration of methicillin, one could easily detect alterations in the mutant cells, such as an increase in cell size, abnormal shape, and, in some cases, a defect in cell separation. The fact that the control cells have a normal morphology in the presence of a 0.3-μg/ml concentration of methicillin, an antibiotic with selective high affinity for PBP 3, is in agreement with the observation that PBP 3 mutant cells seem normal when grown in TSB. However when the mutant with no PBP 3 is in the presence of the antibiotic and additional PBPs start being inhibited, the cells appear to be unable to maintain normal morphology. These data are consistent with the idea that the function of PBP 3 may be taken over by another PBP (presumably PBP 1) when PBP 3 is not available.
During the course of this study, a sequence identical to the one reported here was described as belonging to a gene encoding a new PBP of S. aureus (17). A recombinant form of that protein was reported to comigrate with PBP 2, and therefore it was tentatively named PBP 2B. Our observations do not support this conclusion: there was no PBP detectable in the position corresponding to that of PBP 2 in mutant M100R130, in which PBP 2 was selectively inactivated (Fig. 3A). The PBP that disappeared in cells with inactivated pbpC (COLpPBP3-B1 and -B4 and RN4220pPBP3-B3) (Fig. 3B) and reappeared in mutant M100 complemented by an intact copy of pbpC had the molecular size and position of migration in SDS-PAGE of the protein that has been assigned the name PBP 3 in the literature (11, 21).
Our observations indicate that inactivation of PBP 3 is part of the resistance mechanism of the step mutant M100. This observation is not limited to laboratory mutants. We have recently identified a clinical isolate with borderline resistance to methicillin and oxacillin, in which PBP 3 was undetectable (M. G. Pinho, S. R. Filipe, M. Struelens, C. Nonhoff, D. Baran, and H. de Lencastre, Abstr. 39th Intersci. Conf. Antimicrob. Agents Chemother., abstr. 1437, 1999). Why would it be an advantage for the bacteria not to have PBP 3 at all, instead of having it acylated by the β-lactam antibiotics? At least two possible explanations appear plausible. Acylation of PBP 3 may be involved with a pathway that eventually leads to the death of the bacteria, as suggested by the reduced rate of autolysis that accompanied inactivation of pbpC. Alternatively, replacement of the function of PBP 3 as part of a multienzyme cell wall synthetic complex (2) by another PBP may be possible only if the PBP 3 protein is physically absent. The fact that introduction of intact PBP 3 into strain M100 does not fully restore susceptibility to oxacillin suggests that the resistance mechanism in this mutant involves several additional changes as well.
ACKNOWLEDGMENTS
Partial support for this work was provided by contracts PRAXIS XXI 2/2.1/BIA/349/94 and PRAXIS XXI 2/2.1/BIO/1154/95 (Portugal) (H. de Lencastre) and by the Aaron Diamond Foundation and the Bodman Foundation (A. Tomasz). Mariana G. Pinho was supported by grant PRAXIS XXI/BD/9079/96.
Sequence data for the 548-bp internal fragment of pbpC were obtained from The Institute for Genomic Research website.
REFERENCES
- 1.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1996. [Google Scholar]
- 2.Ayala J A, Garrido T, de Pedro M A, Vicente M. Molecular biology of bacterial separation. In: Ghuysen J M, Hackenbeck R, editors. Bacterial cell wall. Amsterdam, The Netherlands: Elsevier Science; 1994. pp. 73–101. [Google Scholar]
- 3.Curtis N A C, Hayes M V, Wyke A W, Ward J B. A mutant of Staphylococcus aureus H lacking penicillin-binding protein 4 and transpeptidase activity in vitro. FEMS Microbiol Lett. 1980;9:263–266. [Google Scholar]
- 4.de Jonge B L, Chang Y S, Gage D, Tomasz A. Peptidoglycan composition of a highly methicillin-resistant Staphylococcus aureus strain. The role of penicillin binding protein 2A. J Biol Chem. 1992;267:11248–11254. [PubMed] [Google Scholar]
- 5.de Jonge B L M, de Lencastre H, Tomasz A. Suppression of autolysis and cell wall turnover in heterogeneous Tn551 mutants of a methicillin-resistant Staphylococcus aureus strain. J Bacteriol. 1991;173:1105–1110. doi: 10.1128/jb.173.3.1105-1110.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Lencastre H, Tomasz A. Reassessment of the number of auxiliary genes essential for expression of high-level methicillin resistance in Staphylococcus aureus. Antimicrob Agents Chemother. 1994;38:2590–2598. doi: 10.1128/aac.38.11.2590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Domanski T L, Bayles K W. Analysis of Staphylococcus aureus genes encoding penicillin-binding protein 4 and an ABC-type transporter. Gene. 1995;167:111–113. doi: 10.1016/0378-1119(96)82965-9. [DOI] [PubMed] [Google Scholar]
- 8.Dowson C G, Hutchison A, Spratt B G. Nucleotide sequence of the penicillin-binding protein 2B gene of Streptococcus pneumoniae strain R6. Nucleic Acids Res. 1989;17:7518. doi: 10.1093/nar/17.18.7518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Georgopapadakou N H, Dix B A, Mauriz Y R. Possible physiological functions of penicillin-binding proteins in Staphylococcus aureus. Antimicrob Agents Chemother. 1986;29:333–336. doi: 10.1128/aac.29.2.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Georgopapadakou N H, Liu F Y. Binding of β-lactam antibiotics to penicillin-binding proteins of Staphylococcus aureus and Streptococcus faecalis: relation to antibacterial activity. Antimicrob Agents Chemother. 1980;18:834–836. doi: 10.1128/aac.18.5.834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Georgopapadakou N H, Liu F Y. Penicillin-binding proteins in bacteria. Antimicrob Agents Chemother. 1980;18:148–157. doi: 10.1128/aac.18.1.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ghuysen J M. Serine beta-lactamases and penicillin-binding proteins. Annu Rev Microbiol. 1991;45:37–67. doi: 10.1146/annurev.mi.45.100191.000345. [DOI] [PubMed] [Google Scholar]
- 13.Goffin C, Fraipont C, Ayala J, Terrak M, Nguyen-Distèche M, Ghuysen J M. The non-penicillin-binding module of the tripartite penicillin-binding protein 3 of Escherichia coli is required for folding and/or stability of the penicillin-binding module and the membrane-anchoring module confers cell septation activity on the folded structure. J Bacteriol. 1996;178:5402–5409. doi: 10.1128/jb.178.18.5402-5409.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goffin C, Ghuysen J M. Multimodular penicillin-binding proteins: an enigmatic family of orthologs and paralogs. Microbiol Mol Biol Rev. 1998;62:1079–1093. doi: 10.1128/mmbr.62.4.1079-1093.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hackbarth C J, Kocagoz T, Kocagoz S, Chambers H F. Point mutations in Staphylococcus aureus PBP 2 gene affect penicillin-binding kinetics and are associated with resistance. Antimicrob Agents Chemother. 1995;39:103–106. doi: 10.1128/aac.39.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hartman B J, Tomasz A. Low-affinity penicillin-binding protein associated with β-lactam resistance in Staphylococcus aureus. J Bacteriol. 1984;158:513–516. doi: 10.1128/jb.158.2.513-516.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Komatsuzawa H, Choi G H, Ohta K, Sugai M, Tran M T, Suginaka H. Cloning and characterization of a gene, pbpF, encoding a new penicillin-binding protein, PBP2B, in Staphylococcus aureus. Antimicrob Agents Chemother. 1999;43:1578–1583. doi: 10.1128/aac.43.7.1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kraemer G R, Iandolo J J. High-frequency transformation of Staphylococcus aureus by electroporation. Curr Microbiol. 1990;21:373–376. [Google Scholar]
- 19.Murakami K, Fujimura T, Doi M. Nucleotide sequence of the structural gene for the penicillin-binding protein 2 of Staphylococcus aureus and the presence of a homologous gene in other staphylococci. FEMS Microbiol Lett. 1994;117:131–136. doi: 10.1111/j.1574-6968.1994.tb06754.x. [DOI] [PubMed] [Google Scholar]
- 20.Murray T, Popham D L, Setlow P. Identification and characterization of pbpA encoding Bacillus subtilis penicillin-binding protein 2A. J Bacteriol. 1997;179:3021–3029. doi: 10.1128/jb.179.9.3021-3029.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Okonogi K, Noji Y, Nakao M, Imada A. The possible physiological roles of penicillin-binding proteins of methicillin-susceptible and methicillin-resistant Staphylococcus aureus. J Infect Chemother. 1995;1:50–58. [Google Scholar]
- 22.Oshida T, Tomasz A. Isolation and characterization of a Tn551-autolysis mutant of Staphylococcus aureus. J Bacteriol. 1992;174:4952–4959. doi: 10.1128/jb.174.15.4952-4959.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pinho M G, de Lencastre H, Tomasz A. Transcriptional analysis of the Staphylococcus aureus penicillin binding protein 2 gene. J Bacteriol. 1998;180:6077–6081. doi: 10.1128/jb.180.23.6077-6081.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pinho M G, Ludovice A M, Wu S, De Lencastre H. Massive reduction in methicillin resistance by transposon inactivation of the normal PBP2 in a methicillin-resistant strain of Staphylococcus aureus. Microb Drug Resist. 1997;3:409–413. doi: 10.1089/mdr.1997.3.409. [DOI] [PubMed] [Google Scholar]
- 25.Pucci M J, Thanassi J A, Discotto L F, Kessler R E, Dougherty T J. Identification and characterization of cell wall-cell division gene clusters in pathogenic gram-positive cocci. J Bacteriol. 1997;179:5632–5635. doi: 10.1128/jb.179.17.5632-5635.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reynolds P E, Brown D F. Penicillin-binding proteins of beta-lactam-resistant strains of Staphylococcus aureus. Effect of growth conditions. FEBS Lett. 1985;192:28–32. doi: 10.1016/0014-5793(85)80036-3. [DOI] [PubMed] [Google Scholar]
- 27.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 28.Spratt B G, Jobanputra V. Mutants of Escherichia coli which lack a component of penicillin-binding protein 1 are viable. FEBS Lett. 1977;79:374–378. doi: 10.1016/0014-5793(77)80824-7. [DOI] [PubMed] [Google Scholar]
- 29.Tomasz A, Drugeon H B, de Lencastre H M, Jabes D, McDougall L, Bille J. New mechanism for methicillin resistance in Staphylococcus aureus: clinical isolates that lack the PBP 2a gene and contain normal penicillin-binding proteins with modified penicillin-binding capacity. Antimicrob Agents Chemother. 1989;33:1869–1874. doi: 10.1128/aac.33.11.1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tonin E, Tomasz A. β-Lactam-specific resistant mutants of Staphylococcus aureus. Antimicrob Agents Chemother. 1986;30:577–583. doi: 10.1128/aac.30.4.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wada A, Watanabe H. Penicillin-binding protein 1 of Staphylococcus aureus is essential for growth. J Bacteriol. 1998;180:2759–2765. doi: 10.1128/jb.180.10.2759-2765.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu S W, de Lencastre H, Tomasz A. The Staphylococcus aureus transposon Tn551: complete nucleotide sequence and transcriptional analysis of the expression of the erythromycin resistance gene. Microb Drug Resist. 1999;5:1–7. doi: 10.1089/mdr.1999.5.1. [DOI] [PubMed] [Google Scholar]
- 33.Wyke A W, Ward J B, Hayes M V, Curtis N A. A role in vivo for penicillin-binding protein-4 of Staphylococcus aureus. Eur J Biochem. 1981;119:389–393. doi: 10.1111/j.1432-1033.1981.tb05620.x. [DOI] [PubMed] [Google Scholar]