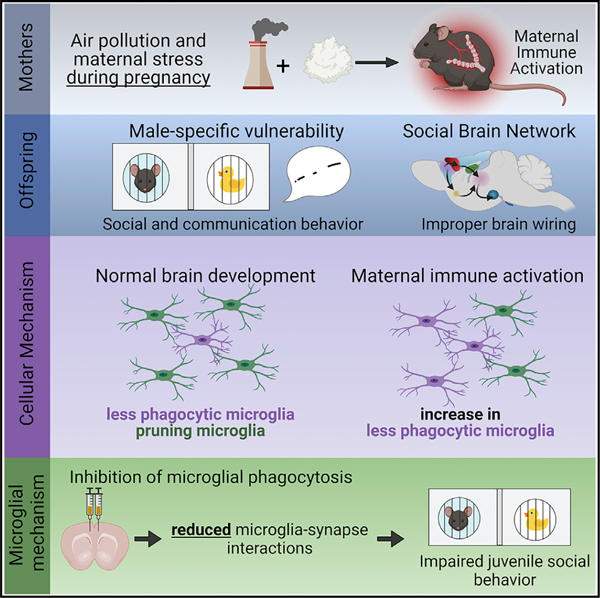
SUMMARY
Gestational exposure to environmental toxins and socioeconomic stressors is epidemiologically linked to neurodevelopmental disorders with strong male bias, such as autism. We model these prenatal risk factors in mice by co-exposing pregnant dams to an environmental pollutant and limited-resource stress, which robustly activates the maternal immune system. Only male offspring display long-lasting behavioral abnormalities and alterations in the activity of brain networks encoding social interactions. Cellularly, prenatal stressors diminish microglial function within the anterior cingulate cortex, a central node of the social coding network, in males during early postnatal development. Precise inhibition of microglial phagocytosis within the anterior cingulate cortex (ACC) of wild-type (WT) mice during the same critical period mimics the impact of prenatal stressors on a male-specific behavior, indicating that environmental stressors alter neural circuit formation in males via impairing microglia function during development.
Graphical abstract
In briefs
Block et al. show that combined exposure to air pollution and maternal stress during pregnancy activates the maternal immune system and induces male-specific impairments in social behavior and circuit connectivity in offspring. Cellularly, prenatal stressors diminish microglia phagocytic function, and inhibition of microglia phagocytosis phenocopies behavioral deficits from prenatal stressors.
INTRODUCTION
The incidences of neurodevelopmental disorders (NDDs) have been increasing in recent decades, suggesting a role for non-genetic environmental factors (Environmental Protection Agency, 2017; Boyle et al., 2011). Furthermore, sex is a significant risk factor for these disorders, with a strong male bias (Hanamsagar and Bilbo, 2016).
Air pollutant exposure during pregnancy or the first year of life is one of the most consistent environmental risk factors for NDDs (Oudin et al., 2019; Rafenberg and Annesi-Maesano, 2018; Raz et al., 2015; Roberts et al., 2013; Rossignol et al., 2014). However, the associations of single environmental agents with NDDs have been relatively weak, and thus causality has been difficult to determine. Non-chemical stressors such as limited resources or social support of the mother can increase the vulnerability of the fetus to toxic exposures, which could explain why certain populations are disproportionately affected (Larson, 2007; Rice et al., 2007). In fact, neighborhood quality is a significant modifier of air pollution risk (McGuinn et al., 2019), suggesting that environmental and social stressors synergize to increase vulnerability to pollutant exposure, but how these exposures alter fetal brain development and affect offspring behavior is largely unknown.
Inflammatory events during pregnancy, such as maternal infection with bacteria or viruses, lead to maternal immune activation (MIA), which is linked to NDDs in offspring (Lee et al., 2020; O’Callaghan et al., 1994; Rudolph et al., 2018). Recent transcriptome-wide studies in postmortem brains of individuals diagnosed with an NDD have identified expression modules with enrichment of genes involved in neuroinflammatory function, with a particular dysregulation of microglial genes (Gandal et al., 2018; Gilman et al., 2011; Quesnel-Vallieres et al., 2019; Velmeshev et al., 2019; Voineagu et al., 2011). Microglia are the primary immunocompetent cells of the brain and are exquisitely sensitive to perturbations of homeostasis and thus may be poised to act as immediate responders to environmental insults. Microglia are also essential regulators of activity-dependent synaptic remodeling during development (Kopec et al., 2018; Paolicelli et al., 2011; Schafer et al., 2012; Schafer and Stevens, 2013), in which they prune inappropriate/weak synapses while sparing appropriate/strong connections. Importantly, transcriptome studies have found that immune changes co-occur with gene enrichment modules affecting synaptic function, suggesting the possibility that neuroimmune changes during development could lead to aberrant synapse development by altering microglial function.
A recent analysis found that MIA was more common in male children with ASD than female children, suggesting that a sex difference in response to maternal inflammation may be one mechanism that underlies increased male vulnerability (Patel et al., 2020). Furthermore, we and others have found sex differences in microglial development, maturation, and function, including an increased relative expression of microglial genes in male brains, compared with females. Interestingly, the microglial genes enriched in male brains are also implicated in ASD (Hanamsagar et al., 2017; Werling et al., 2016). Together these data point to a mechanism by which sexually dimorphic microglial responses to prenatal stressors could lead to aberrant brain development, primarily in males.
Here, we demonstrate that a combination of air pollution and maternal stress exposures during pregnancy activate the maternal immune system of mouse dams leading to altered synaptic and microglial development, persistent changes in brain circuit function, and long-lasting alterations in social and communication behavior in male offspring.
RESULTS
Prenatal exposure to air pollution and maternal stress induces MIA
To model a combination of chemical and social stressor exposures in mice, we exposed pregnant dams to intermittent diesel exhaust particle (DEP) instillations to mimic chronic air pollution. DEP is a primary toxic component of air pollution and a potent neuroinflammatory stimulus (Jayaraj et al., 2017; Levesque et al., 2011, 2013). Then, we applied a maternal stressor of resource deprivation during the last trimester of pregnancy by limiting the bedding and nesting material (i.e., DEP + MS condition) (Bolton et al., 2013; Rice et al., 2008). Control dams received instillations of the vehicle solution (i.e., 0.05% Tween20 in PBS) and were housed in standard cages with full nesting material (CON). We measured segmented filamentous bacteria in fecal and cecal samples and found no evidence of it in our colony (Table S1).
The combined environmental stressors, hereafter called DEP + MS, did not alter weight gain during pregnancy or litter size in utero (Figures 1A–1C). To confirm the effects of maternal stress on dams, we measured the concentration of stress hormone, corticosterone (CORT), in serum from dams at embryonic day (E)17.5 and found an increase in DEP + MS dams compared with CON dams (Figure 1D, left). Importantly, CORT levels of CON dams were similar to baseline CORT levels of untreated pregnant dams (Figure 1D, right), indicating that our instillation method alone is not sufficient to induce significant stress in pregnant dams.
Figure 1. Combined prenatal stressors induce maternal immune activation.
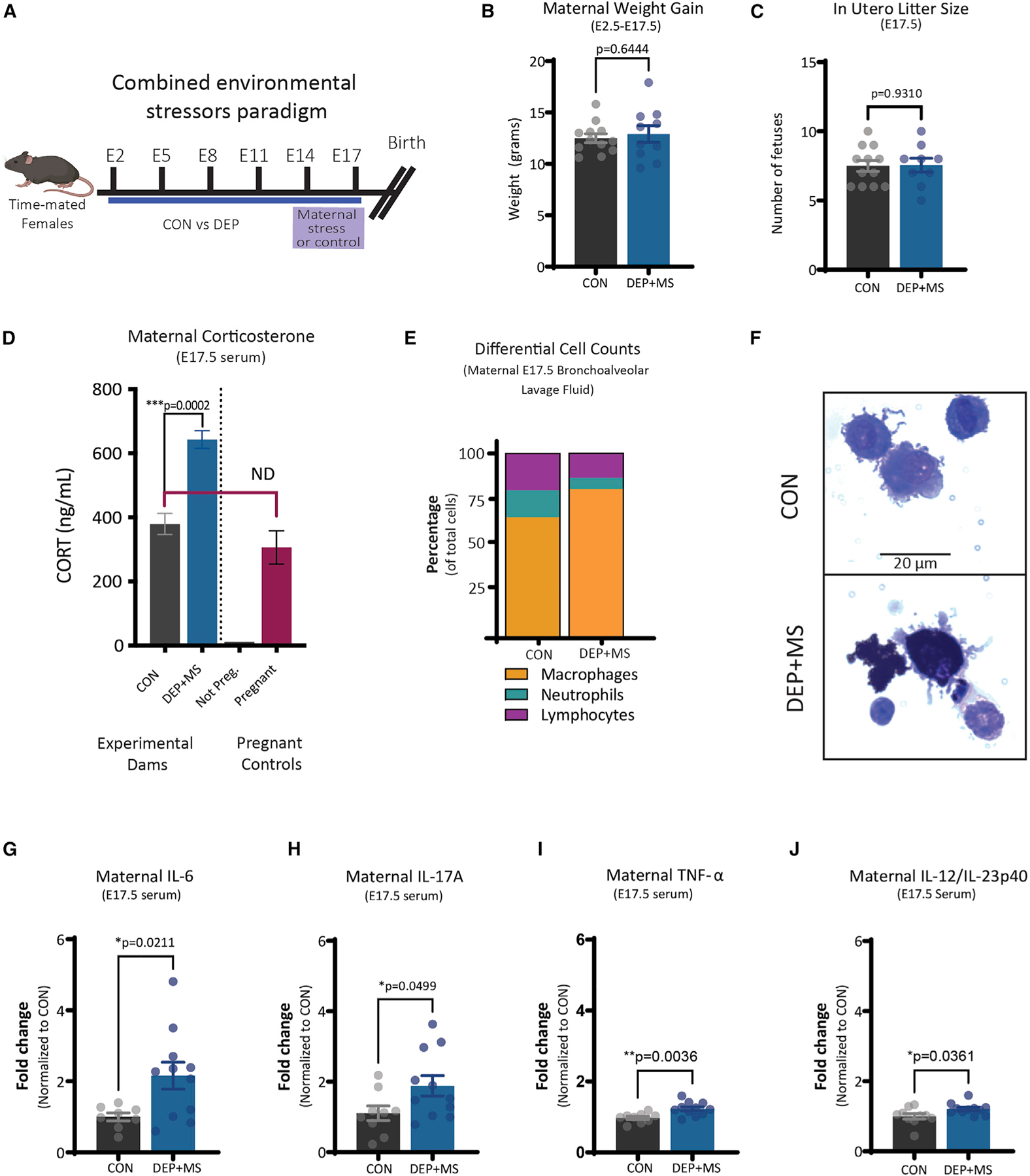
(A) Combined environmental stress paradigm.
(B) Pregnancy weight gain from E2.5 to E17.5 (n = 10–12 mice/condition, unpaired t test).
(C) In utero litter size at E17.5 (n = 10–12 mice/condition, unpaired t test).
(D) Serum concentrations of CORT at E17.5 (left, n = 5–6 mice/condition), and compared with non-pregnant and pregnant WT controls (right, n = 1–3 mice, unpaired t test; ND, not different).
(E) Differential cell count of BALF cells (n = 5 mice/condition).
(F) Representative image of CON versus DEP + MS alveolar macrophages.
(G–J) Serum concentrations of cytokines at E17.5 (n = 10–12 mice/condition, unpaired t test). Means ± SEM.
Chronic exposure to air pollution can impact lung function via inflammation or changes in cellular immunity. To examine inflammatory changes in the lung, we collected bronchoalveolar lavage fluid (BALF) from dams in each condition at E17.5 and performed histology on BALF cells. We found no significant group differences in the number of cells collected from BALF (Figure S1A) and no significant differences in the density of macrophages, neutrophils, and lymphocytes (Figures 1E, S1B, and S1C). When we examined macrophages from BALF in DEP + MS dams, we found that these cells are highly congested with particulates, in some cases filling more than 90% of the cell (Figure 1F).
Next, to examine the possibility that maternal exposures induce a systemic immune response in the dams, we analyzed a curated panel of pro- and anti-inflammatory cytokines linked to adverse outcomes in offspring. DEP + MS dams have significant increases in several proinflammatory cytokines in serum at E17.5, namely interleukin (IL)-6, IL-17A, tumor necrosis factor alpha (TNF-α), and IL-12/IL-23p40 (Figures 1G–1J). We found no significant differences in interferon gamma (IFN-ɣ), IL-5, monocyte chemoattractant protein-1 (MCP-1), or IL-10 (Figures S1D–S1G). One of the other widely-adopted models of MIA, the maternal viral infection model (poly I:C), also acutely increases IL-6, IL-17A, and TNF-α in dams (Choi et al., 2016; Hsiao and Patterson, 2011), demonstrating that diverse environmental factors converge onto similar pathways. Taken together, these results show that DEP + MS leads to robust systemic immune activation in pregnant dams.
Given the broad expression of cytokine changes we observe in our model, we tested the role of the pattern recognition receptor, toll-like receptor 4 (TLR4), which is upstream of many proinflammatory cytokines. Moreover, DEP binds to TLR4 in alveolar macrophages in the lung (Inoue et al., 2006), and we previously found the necessity of TLR4 in microglial changes in offspring in response to prenatal DEP exposure in the absence of MS (Bolton et al., 2017). Interestingly, loss of TLR4 did not prevent the increase in proinflammatory cytokines we see after combined DEP + MS exposure (Figure S1H). In contrast DEP + MS treatment in TLR4 KOs resulted in a greatly exaggerated IL-6 response in pregnant dams (Figure S1I). This result points to maternal stress as a critical component of our MIA model, which cannot be blocked by inhibiting a single innate immune activation pathway.
MIA produces lasting changes in communication and social behavior
To investigate the impact of combined prenatal stressors on the health and behavior of offspring, we measured litter outcomes and social and communication behavior across development (Figure 2A). DEP + MS did not alter litter size or sex composition postnatally (Figure S2A). Male and female DEP + MS offspring weigh significantly less beginning at P8 (Figure S2B). Importantly DEP + MS exposure does not alter maternal care (Bolton et al., 2013), suggesting that alterations in DEP + MS pups are driven by MIA and not by fractured maternal care.
Figure 2. Combined prenatal stressors induce lasting changes in offspring communication and social behavior.
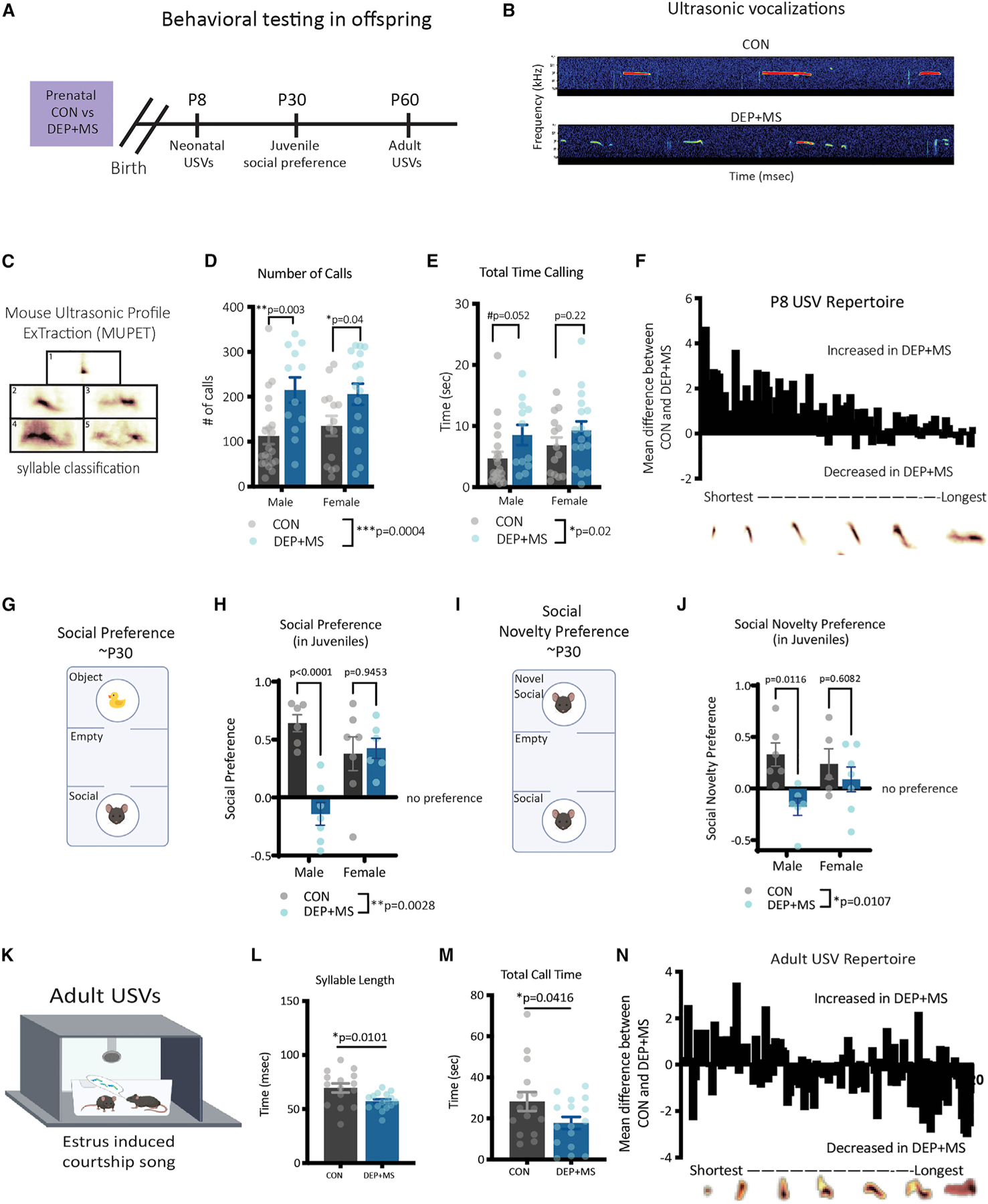
(A) Behavioral testing in offspring born to CON and DEP + MS dams.
(B) Representative spectrograms of USVs.
(C) MUPET syllable classification.
(D and E) USV number and total call time at P8 (n = 14–19 mice/condition/sex, two-way ANOVA with Holm-Sidak’s post hoc tests).
(F) Repertoire units were organized from shortest to longest and displayed as the absolute difference between group means.
(G and H) Schematic of three-chamber social preference test (n = 6–7 mice/condition/sex, two-way ANOVA with Sidak’s multiple comparisons post hoc tests).
(I and J) Schematic of three-chamber social novelty preference test. (n = 6–7 mice/condition/sex, two-way ANOVA, with Sidak’s multiple comparisons post hoc tests).
(K) Schematic of USV courtship assay.
(L and M) Adult USV syllable length and call time (n = 15–17 mice/condition, unpaired t-tests).
(N) Repertoire units organized from shortest to longest are displayed as the absolute difference between group means. Means ± SEM.
NDDs, mostly notably ASD, are characterized by deficits in communication and social behavior. In neonatal mice, communication behavior can be measured by briefly separating pups from the dam, which elicits ultrasonic vocalizations (USVs), an innate form of communication that promotes maternal care (D’Amato et al., 2005; Okabe et al., 2013). In wild-type (WT) C57BL/6J mice, peak USV production occurs at P8 (Figure S2C); thus, to probe for developmental changes, we recorded USVs from P7 to P9. We found that DEP + MS pups emit more calls and spend more time calling at P8 than CON pups (Figures 2B–2E), mimicking phenotypes reported in other MIA models and in a genetic mouse model of autism (Carlezon et al., 2019; Choi et al., 2016; Wang et al., 2011). This increased number of calls in DEP + MS offspring was also evident at P7 but was no longer significantly different by P9 (Figure S2D); furthermore, we found no differences in the frequency (kHz) of calls at any age (Figure S2E).
To probe whether the acoustic structure of USVs was altered we used a machine-learning tool, Mouse Ultrasonic Profile ExTraction (MUPET), to identify distinct repertoire/syllable units (Figure 2C) (Van Segbroeck et al., 2017). Using MUPET, we extracted 80 different repertoire units from P8 USVs and organized them from shortest to longest (Figure 2F). This analysis revealed that while DEP + MS offspring emit more USVs across the whole repertoire of syllables, there is a preferential increase in shorter, less complex calls (Figure 2F). Together, these results show that prenatal exposure to combined stressors alters pup-to-dam communication during a peak developmental window.
To probe whether more complex social behaviors are modified, we tested CON and DEP + MS mice in a three-chamber social preference task during the juvenile period (~P30). In this task, mice are placed into a three-chamber arena, where they are given a choice to interact with a novel object or a social stimulus (Figure 2G). As expected, we found that both CON male and female mice exhibit a normal preference for the social stimulus (Figure 2H). On the other hand, we found a significant reduction in the social preference score of DEP + MS offspring, with a significant interaction of sex by condition. Post hoc analyses revealed that while DEP + MS females exhibit a normal preference for the social stimulus, DEP + MS males show no preference for the social stimulus over the object and have significantly diminished sociability compared with CON males (Figures 2H and S2F).
Next, mice were tested in a social novelty preference task. In this task, juvenile mice are placed into the same three-chamber arena but are given a choice to interact with a sex-matched littermate or a novel mouse matched for age, sex, and condition (Figure 2I). Once again, male and female CON mice exhibit the expected preference for the novel stimulus, whereas DEP + MS male mice interact significantly less with the novel social stimulus (Figure 2J). Collectively these data reveal a male-specific effect of DEP + MS on social behavior.
We next tested whether changes in social and communication behavior persist into adulthood in DEP + MS offspring. Because social deficits were only apparent in males, we studied an adult male-specific communication behavior, ‘‘courtship song.’’ In this assay, males with sexual experience emit USVs in the presence of a sexually receptive females (i.e., females in estrus or proestrus) (Figure 2K), which drives female mate choices (Chabout et al., 2015; Hammerschmidt et al., 2009; White et al., 1998). We found no differences in the number or mean frequency of calls emitted (Figures S2G and S2H) in the courtship song of DEP + MS males compared with CON males. However, when we analyzed the acoustic properties of the calls, the individual calls were significantly shorter in DEP + MS mice, resulting in a significant reduction in the time spent vocalizing (Figures 2L and 2M).
As before, we performed MUPET analyses and identified 120 distinct repertoire units in both CON and DEP + MS males (Figure S2I). These analyses revealed that DEP + MS male offspring have a preferential increase in short and less complex calls and a reduction in longer and more complex call types (Figure 2N). Importantly, when presented with a choice, female mice prefer courtship songs that are longer and more complex (Chabout et al., 2015), suggesting DEP + MS males produce less competitive songs. This observed reduction in call complexity in adult DEP + MS males is similar to the changes in call complexity observed in neonatal DEP + MS offspring, which also preferentially emit calls that are shorter and less complex (Figure 2F). Collectively, these data reveal that changes in vocalization persist into adulthood in DEP + MS males.
MIA alters gene expression in the prefrontal cortices of neonatal offspring
To investigate the molecular changes underlying behavioral differences, we analyzed gene expression in the prefrontal cortices (PFCs) of P8 DEP + MS and CON male and female pups. We selected the PFC because it is a brain region that is dysfunctional in many NDDs and plays a critical role in regulating social and emotional behaviors (Schubert et al., 2015; Trakoshis et al., 2020). We identified 280 differentially expressed genes in DEP + MS males compared with CON males (Figures S3A and S3B). In littermate DEP + MS females, 145 genes were differentially expressed compared with CON females (Figures S3A and S3B). Interestingly, several microglial enriched genes were in the top 10 differentially expressed genes for both sexes (Figures S3C and S3D) and gene set enrichment analysis revealed a significant enrichment of hallmark immune pathways for both sexes (Figures S3E and S3F). On the other hand, gene ontology for the cellular compartment revealed a male-specific downregulation of genes involved in synaptic structure and function in DEP + MS offspring (Figure S3G). Interestingly, brain gene expression studies from subjects with NDDs show a similar downregulation in synaptic function genes and upregulation in immune response genes (Gandal et al., 2018; Velmeshev et al., 2019; Voineagu et al., 2011).
Combined prenatal stressors alter male behavior in an appetitive sociability task
Given the sex-specific changes in juvenile social behavior and adult communication we observed along with the striking downregulation of synaptic gene pathways in DEP + MS male offspring, we next asked whether offspring have lasting changes in brain functional connectivity. Since altered brain network activity can be a sensitive measure of social deficits in ASD and preclinical models (Dawson et al., 2005; Jokisch et al., 2005; Kroger et al., 2014; Pavlova et al., 2004; Ulloa and Pineda, 2007; Wang et al., 2016), we investigated whether DEP + MS alters the network activity that underlies appetitive social behavior. To do so, we implanted 54 total CON and DEP + MS mice of both sexes with electrodes targeting eight brain regions (Figure S4A). We then recorded electrical oscillations concurrently from cortical and subcortical regions as mice performed a social exploration task where they were given the choice to interact with a caged mouse or a caged inanimate object (Figure 3A). This assay was repeated for 10 sessions for each mouse, presenting novel stimuli each day. In total, we collected 100 min of concurrent behavioral and electrical recordings for each mouse, which enabled us to perform in-depth circuit activity analyses (Figures 3A and 3B). Overall, mice preferred the social stimulus (Figure 3C). Moreover, to our initial surprise, prenatal DEP + MS exposure resulted in a significant increase in the social preference scores of adult male mice compared with CON males (Figure 3C, left).
Figure 3. Functional activation of the social brain network is disrupted only in male offspring prenatally exposed to combined stressors.
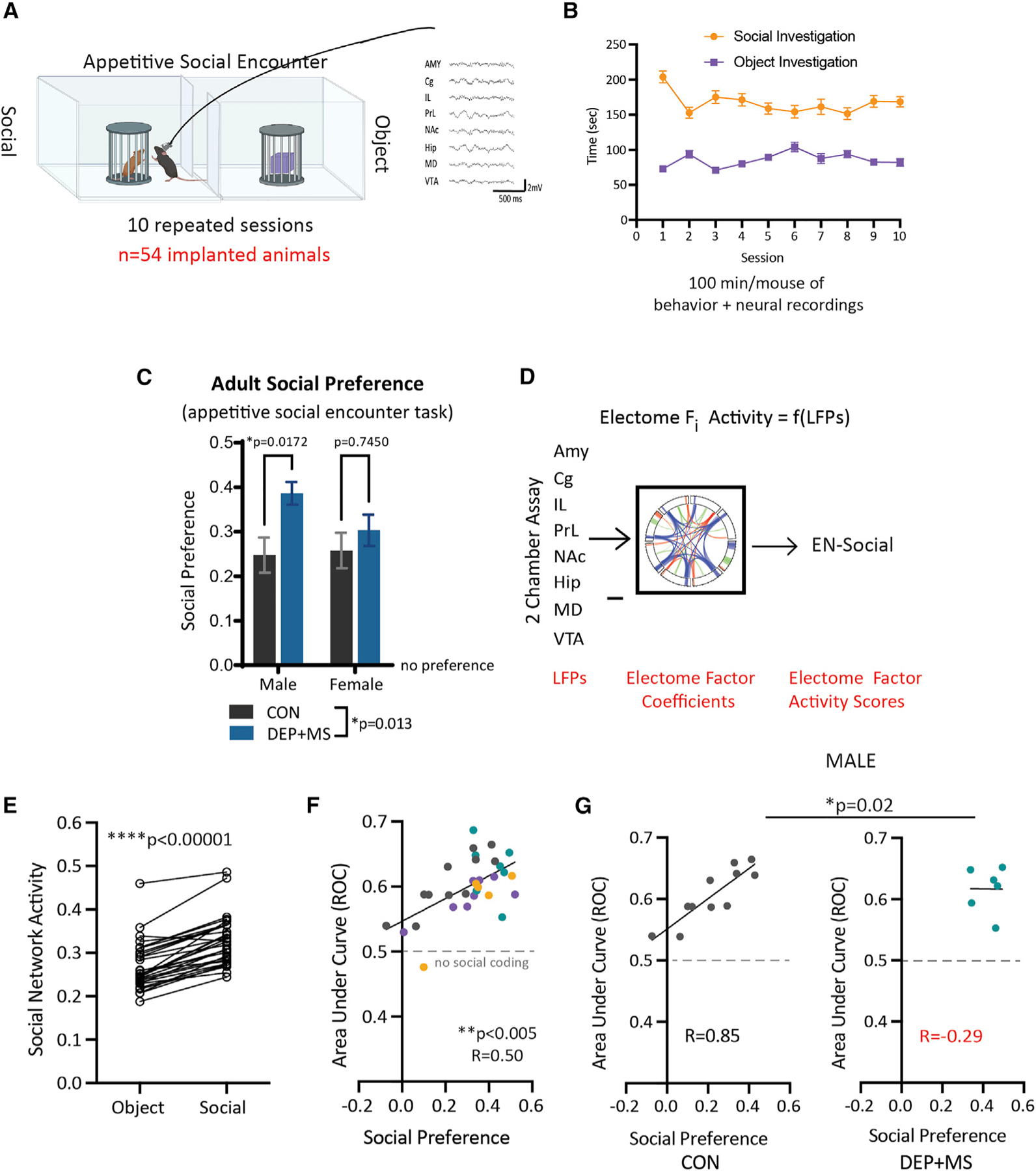
(A) CON and DEP + MS offspring were implanted with electrodes and underwent concurrent neural recording in an appetitive social encounter task.
(B) Mice were tested in 10 repeated sessions to collect 100 min of concurrent neural and behavioral data per mouse (n = 13–14 mice/condition/sex).
(C) Social preference in implanted adult mice (n = 13–14 mice/condition/sex, two-way ANOVA with Bonferroni’s multiple comparisons post hoc tests).
(D) LFP activity obtained during social behavior was projected into the learned electrical functional connectivity (electome) factor coefficients to generate the EN-social.
(E) Mice showed higher EN-social activity during social compared with object interactions (n = 32 animals, sign-rank test).
(F) The decoding accuracy of EN-social activity signaled social preference across the population of implanted mice (n = 32 animals, Spearman correlation).
(G) Prenatal DEP + MS exposure disrupted the relationship between EN-social activity and appetitive social behavior in male mice (n = 6–11 mice/condition, analysis of covariance with Box-Cox transformation with Spearman’s correlation). Means ± SEM.
Combined prenatal stressors induce male-specific deficits in adult socio-appetitive encoding
To address whether the increased preference ratio in adult DEP + MS males reflects altered social processing, we probed a brain network recently described to underlie normal social behavior in mice (Mague et al., 2022). Specifically, this network, hereafter referred to as EN-social, was characterized based on data collected from an untreated group of C57BL/6J WT mice performing an identical 10-day social appetitive task.
The EN-social network was discovered using a machine-learning approach that utilizes a discriminative cross-spectral factor analysis based on non-negative matrix factorization (Talbot et al., 2020). In brief, this method integrates local field potential (LFP) activity from multiple brain regions with concurrent behavior (Figure 3D) to generate electrical functional connectivity maps (or ‘‘electomes’’). The model features utilize LFP power, LFP synchrony, and spectral Granger causality (Geweke, 1982) resolved from 1 to 56 Hz. As reported by Mague et al. (2022), the network generalized on a mouse-by-mouse basis to encode individual socially appetitive behavioral outcomes. Critically, the activity of EN-social predicts an animal’s investigation of the social stimulus and correlates with an individual animal’s social preference, reflecting the rewarding nature of social encounters on a mouse-by-mouse basis (Mague et al., 2022). Notably, while the prominent signals, which compose EN-social, predict social behavior, the individual features (i.e., brain regions) do not strongly encode social versus object interactions on their own (Figure S4B). In other words, it is the circuit activity as a whole that meaningfully predicts behavior and not the action of any single brain region.
Using this network approach, we overlaid the brain activity of our 54 implanted mice onto this EN-social network. Overall, both DEP + MS and CON displayed higher EN-social network activity when interacting with the social stimulus versus the object (Figure 3E). This result is consistent with observations in a genetic mouse model of autism, for which the EN-social network continues to be activated when these mice are engaged with a social stimulus (Mague et al., 2022).
Moreover, across our entire group of mice, EN-social decoding strongly correlated with the social preference of mice (Figure 3F), demonstrating that the network was also encoding individual social and behavioral outcomes as predicted. However, when we performed within-sex comparisons between the treatment groups, we found that this brain activity-behavior relationship is disrupted in male DEP + MS mice (Figure 3G). Specifically, in CON males and females and DEP + MS females, higher social preference directly correlated with increased activation of the EN-social network in response to social encounters (Figures 3G and S4C); however, this correlation was abolished in DEP + MS male mice. Thus, EN-social failed to encode individual behavioral outcomes in DEP + MS males.
Notably, these findings are very similar to those found using a genetic mouse model of ASD in which the EN-social network activity-social preference relationship was also disrupted, despite a lack of change in social preference (Mague et al., 2022). Here, we detected atypical social preference in adult DEP + MS males after repeated presentations of a social stimulus that is not correlated with activity of brain circuits.
In summary, these findings reveal that prenatal DEP + MS exposure leads to long-lasting changes in socioemotional encoding and behavior in males. Notably, the incidence of NDDs is higher in males than females, and a recent study highlighted that a history of MIA is significantly higher in mothers of male children diagnosed with ASD than females (Patel et al., 2020). Our DEP + MS model captures this male-specific vulnerability to MIA, thus providing an important model for studying cellular and molecular mechanisms.
Combined prenatal stressors impair postnatal thalamocortical synapse development and microglial pruning in the anterior cingulate cortex
Our data thus far point to changes in synapse function, specifically in DEP + MS males, leading to circuit dysfunction. Our network approach indicates no single brain region is responsible for the observed behavioral changes (Figure S4B); however, the anterior cingulate cortex (ACC) is a critical node of EN-social (Mague et al., 2022) and is functionally linked to communication outcomes (Bennett et al., 2019; Guo et al., 2019; Heilbronner and Hayden, 2016). Therefore, since adult DEP + MS mice exhibited alterations in EN-social and behavioral changes, we hypothesized that prenatal DEP + MS exposure alters circuit formation within the ACC during a critical window of synaptic development.
The ACC receives excitatory synaptic inputs from several cortical and subcortical areas, including the thalamus. Thalamocortical synapses (TCs), which are formed from thalamic axonal inputs onto the cortical dendrites, can be identified by the juxtaposition of vesicular glutamate transporter-2 (VGlut2)-positive presynaptic terminals and PSD-95-positive postsynaptic densities (Figures 4A and 4B) (Nakamura et al., 2005). Thalamocortical pathways are critical for relaying subcortical sensory information to the cortex, and hypoconnectivity of these pathways is thought to underlie sensory processing issues in ASD patients (Iidaka et al., 2019; Nair et al., 2013; Ouhaz et al., 2018). We found that USVs are disrupted in DEP + MS pups at P8, and, in our transcriptome analyses, we found a DEP + MS male-specific downregulation of excitatory synapse genes at P8, a time point corresponding to heightened TC synaptogenesis. Therefore, we wondered whether TC synapse development is affected in these animals around this developmental period.
Figure 4. The second week of postnatal development is a critical window for ACC development.
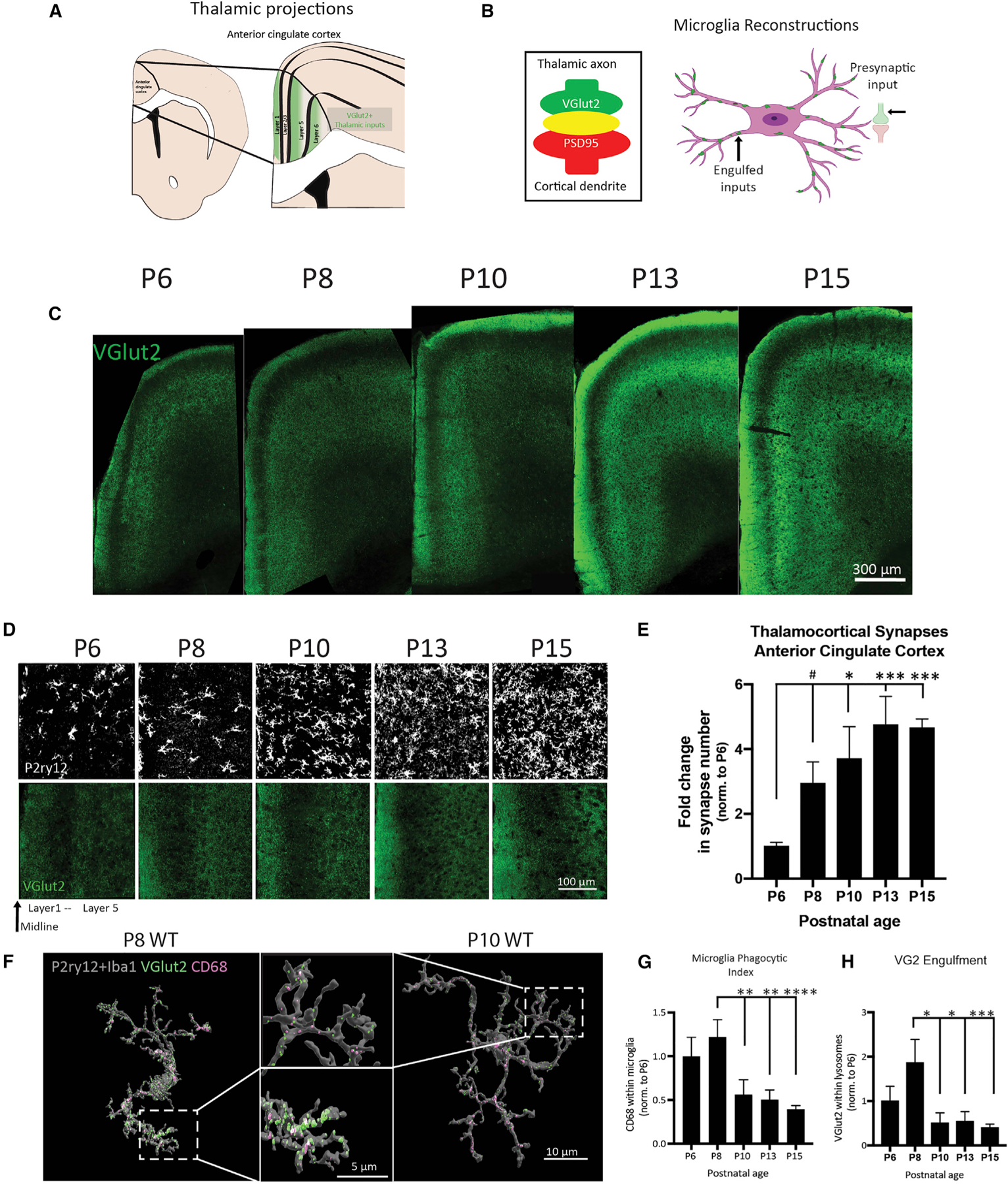
(A) Thalamic inputs onto the ACC are marked by VGlut2.
(B) Synapses can be quantified by the co-localization of VGlut2 and PSD95 (left). 3D cell reconstructions allow for visualizing internalized inputs inside microglia (right).
(C) Representative tile scan images of the ACC across development.
(D) Representative images of VGlut2 and P2ry12 in the ACC of WT mice from P6 to P15.
(E) Quantification of VGlut2 synapses in the ACC (n = 4 mice/age, 2M, 2F, three replicates/mouse, 300 images analyzed, data normalized to P6, one-way ANOVA with Holm-Sidak’s multiple comparisons post hoc tests).
(F) Representative Imaris 3D reconstructions of microglia from WT P8 and P10 male mice. Lysosomes (CD68) and TC inputs (VGlut2) can be visualized inside microglia and quantified.
(G and H) Quantification of CD68 and VGlut2 volume internalized in microglia (n = 6–7 cells/mice, two male mice/age, 75 microglia cells reconstructed, all values normalized to cell volume and engulfment normalized to P6, one-way ANOVA with Holm-Sidak’s multiple comparisons post hoc tests). ****p < 0.0001, ***p < 0.001, **p < 0.01, *p < 0.05; means ± SEM.
In early postnatal brain development, an exuberant period of synaptogenesis is closely followed by and overlaps with a period of synaptic pruning, where weak or unnecessary synapses are eliminated (Schafer et al., 2012). One mechanism of synaptic pruning and circuit refinement occurs via the activity-dependent engulfment of synaptic material by microglia (Paolicelli et al., 2011; Schafer et al., 2012). Microglia selectively phagocytose presynaptic structures, which are degraded through trafficking to lysosomal compartments (Weinhard et al., 2018). Increased microglial reactivity has been reported in several brain regions in ASD patients (Pardo et al., 2005; Vargas et al., 2005). Our transcriptome analyses identified enrichment of microglial genes and an upregulation of pathways involved in immune function, alongside a downregulation of synaptic genes in males (Figures S3C–S3G), suggesting a link between the two.
To first characterize the typical pattern of synaptic development in the ACC, which was unknown, we quantified synapse density and microglial engulfment during postnatal ages (P6–P15) in a naive group of WT mice (Figure 4). From P6 to P15, TC synapse density increases 5-fold (Figure 4E) and the ACC becomes increasingly organized and pseudo-laminated (Figure 4C). Moreover, we found that microglia undergo a period of rapid development, dramatically increasing in density and coverage between P8 and P10 (Figure 4D). To assess whether this period coincides with peak synapse elimination, we used Imaris to perform 3D reconstructions of microglia to visualize internalized VGlut2 within microglial lysosomal compartments (CD68) (Figures 4B and 4F). We found that lysosomal content/phagocytic activity was highest at P8 and was significantly diminished beginning at P10 (Figures 4F and 4G). Quantification of VGlut2+ within microglial lysosomes revealed that TC synapse engulfment peaks at P8 and is primarily completed by P10 (Figure 4H). These data show that the period between P8 and P10 represents a critical window of microglial engulfment of TC synapses.
Next, to determine if TC synaptic structures are altered in the ACC of DEP + MS mice, we quantified the number of TC synapses in male and female offspring at P8, at P15 when TC synapses reach their peak density, and in adulthood (>P60) when synapse density is relatively stable (Figures 5A–5D). At P8, we found a significant increase in the number of TC synapses in DEP + MS males, but not in females (Figure 5B). At P15, the peak of TC synapse abundance in WT mice, male DEP + MS offspring had a significant reduction in TC synapse number compared with CON males (Figure 5C), and this decrease persisted into adulthood in males (Figure 5D). Importantly, we found no significant differences in the density or distribution of neurons, astrocytes, and oligodendrocytes in P8 offspring (Figures S5C–S5E); thus, changes in synaptic connectivity cannot be attributed to increased cell number. Together these data show that DEP + MS males, but not females, have an overgrowth of TC synapses at P8; however, this initial overgrowth is rapidly lost by P15 and results in a reduction in TC connections in the ACC, a phenotype that persists into adulthood in males.
Figure 5. Prenatal DEP + MS induces enduring changes in thalamocortical synapse development and microglia elimination of synapses.
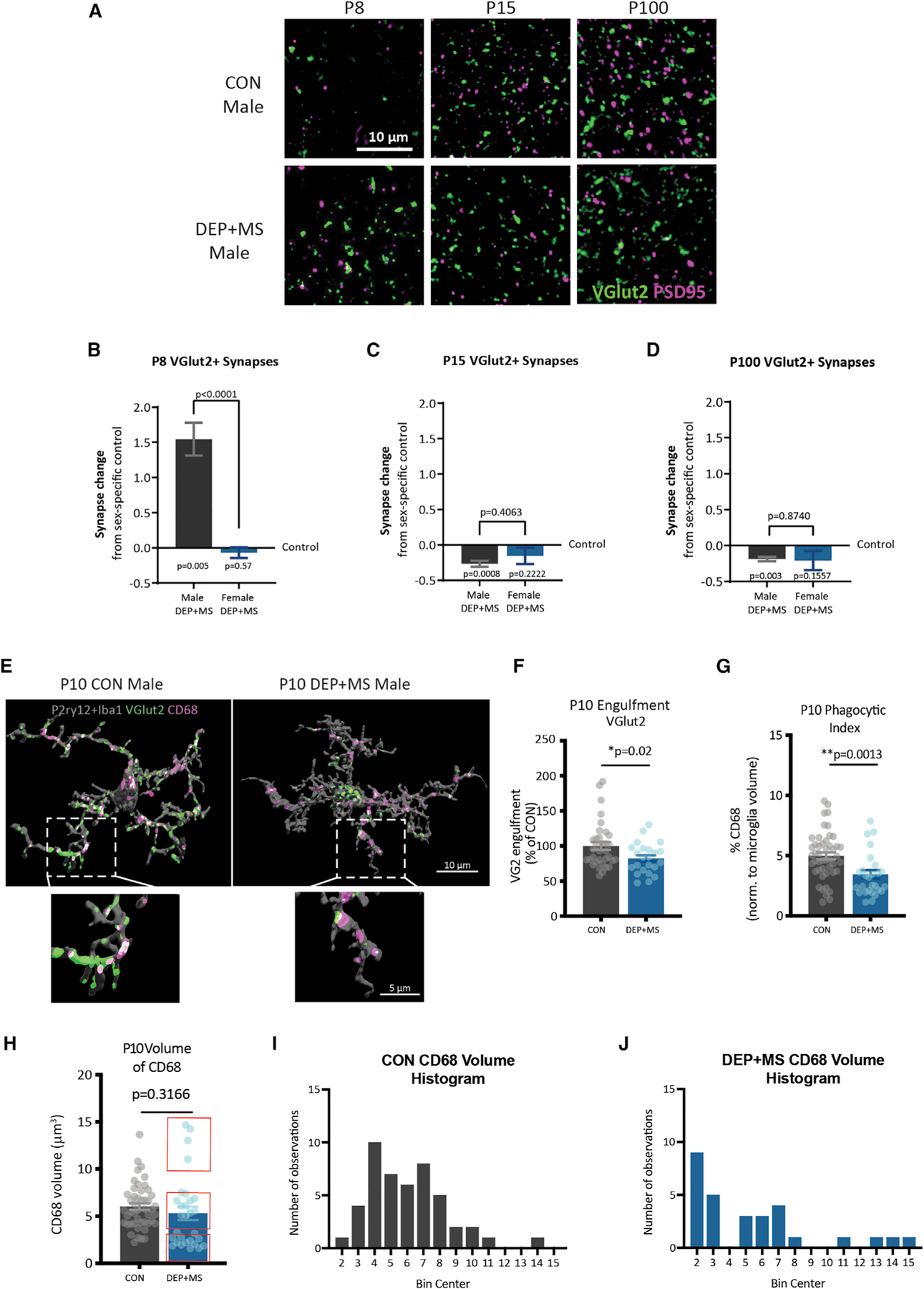
(A) Representative images of TC synapses in the ACC of P8, P15, and P100 CON and DEP + MS male mice.
(B–D) Quantification of TC synapses at P8, P15, and P100 in male and female offspring. Graphed as synapse change from sex-specific control (value of zero indicates no change from control). Three mice/condition/sex, n = 3 replicates/mouse, 540 images/age analyzed, one-sample t test comparison from control, unpaired t test between sexes.
(E) Representative surface rendered microglia from the ACC of P10 CON and DEP + MS male offspring labeled with P2ry12 + Iba1, VGlut2, and CD68.
(F and G) Microglial engulfment of VGlut2, and microglial CD68 content (n = 3–4 replicates/mouse, thee mice/condition, 110 total images analyzed, nested t test).
(H–J) Volume of CD68 in microglia. Histogram of CD68 distribution in CON (I, left) and DEP + MS (J, right) (Levene’s test for homogeneity of variance, p = 0.0117) (n = 3–4 replicates/mouse, three mice/condition, 110 total images analyzed, nested t test). Means ± SEM.
To determine whether rapid atrophy of TC synapses in males can be attributed to enhanced and prolonged microglial engulfment during this period, we next investigated whether DEP + MS male microglia had alterations in TC synapse engulfment at P10, when peak engulfment is completed. Surprisingly, we found that DEP + MS microglia engulfed significantly fewer synapses (Figures 5E and 5F) and phagocytic activity was also significantly diminished in microglia from DEP + MS males (Figure 5G). Intriguingly this change was not due to a consistent reduction in the volume of CD68 (Figure 5H); instead, we found that there was a significant difference in the distribution of CD68 volume, with subsets of high and low CD68-expressing cells (Figures 5I and 5J), suggesting heterogeneous functional changes in male DEP + MS microglia. In sum, microglia from male DEP + MS offspring engulf fewer TC synapses at P10, are less phagocytic, and have alterations in the distribution of CD68. Together our results indicate two unexpected phenomena. First, contrary to our initial hypothesis, microglia from DEP + MS males have diminished phagocytic function overall, and second, this diminished function only affects a subset of microglia.
Combined prenatal stressors lead to an increase in functional heterogeneity of male microglia
In DEP + MS males, we found diminished TC input engulfment by microglia at P10, suggesting that the reduction in TC synapse density by P15 cannot be attributed to enhanced microglial engulfment of synapses. Next, we wondered whether the atrophy of TC inputs could be attributed to an increase in microglia cell density. To investigate this possibility, we quantified microglia cell density in the ACC at P8, P15, and P25 in CON and DEP + MS offspring. To do so, we performed immunohistochemistry (IHC) using antibodies against P2ry12 and Iba1 and independently labeled these antigens by using separate fluorophores. Microglia were identified by P2ry12 and/or Iba1 signal, co-localizing with the nuclear marker DAPI (Figure 6A). There were no significant differences in the total density of microglial cells between CON and DEP + MS male offspring across all ages (Figure 6B), showing that changes in microglia numbers are not likely to underlie alterations in synaptic development. Intriguingly, while most microglia express high levels of both Iba1 and P2ry12, we identified a subset of cells that express high levels of one marker and not the other (Figure 6A, bottom).
Figure 6. Microglial development and synaptic engulfment are altered in male offspring prenatally exposed to combined stressors.
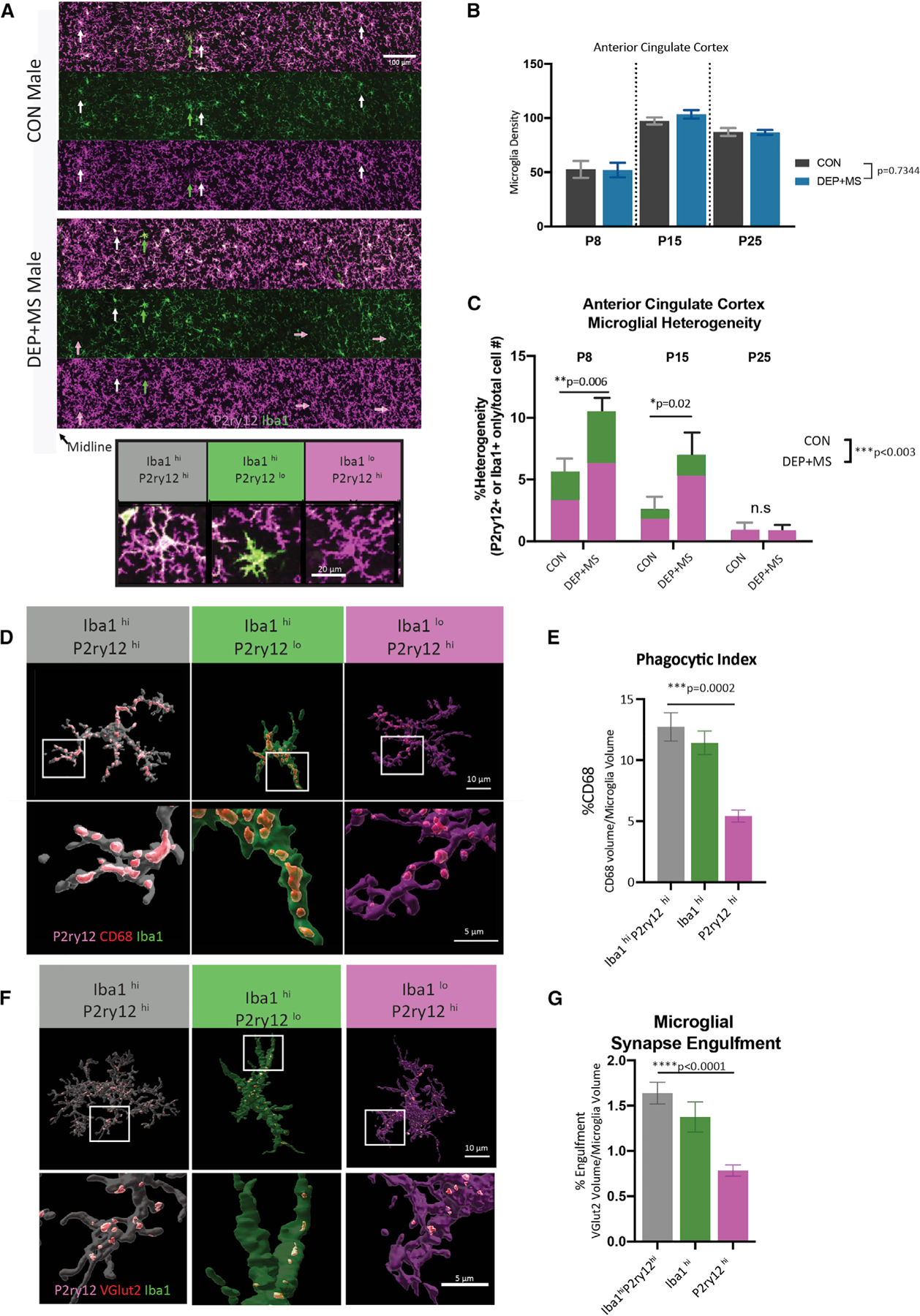
(A) Top: representative images of microglia labeled with P2ry12 and Iba1 in the ACC of P15 male offspring, arrows highlight microglia differentially marked by P2ry12 and Iba1 (white = both high; green = Iba1 high, magenta = P2ry12 high). Bottom: representative images of microglia with heterogeneous levels of expression of Iba1 and P2ry12 in the developing cortex.
(B) Quantification of microglial density across development in male offspring in the ACC (n = 3 replicates/mouse, three or four mice/condition/age, >6,000 cells counted, two-way ANOVA, condition × age).
(C) Quantification of microglial heterogeneity across development in male offspring in the ACC (n = 3 replicates/mouse, three or four mice/condition/age, >6,000 cells counted, two-way ANOVA condition × age, with Sidak’s post hoc test, n.s., not significantly different).
(D) Representative Imaris reconstructions of microglia at P8 with lysosomal content (CD68) in microglial subtypes.
(E) CD68 content in microglial subtypes (n = 5–10 cells/mouse/cell subtype, n = 3 animals/condition, a total of 120 cells analyzed, nested one-way ANOVA with Holm-Sidak’s post hoc test).
(F and G) Representative Imaris reconstructions and quantification of microglial engulfment of VGlut2 in different microglial subtypes (n = 5–12 cells/mouse/cell subtype, n = 3 mice/condition, a total of 168 reconstructed cells, nested one-way ANOVA with Holm-Sidak’s post hoc test). Means ± SEM.
P2ry12 and Iba1 each have essential roles in microglia function and are expressed at varying levels within microglia, reflecting different cellular states. For example, P2ry12, a G protein-coupled purinergic receptor, is necessary for ADP/ATP-mediated chemotaxis and microglial process extension to sites of brain injury (Haynes et al., 2006; Ohsawa et al., 2010). Furthermore, pharmacological block or deletion of P2ry12 during development leads to reduced critical period plasticity (Sipe et al., 2016). Moreover, immune activation severely diminishes P2ry12 expression in microglia (Eyo et al., 2014; Swiatkowski et al., 2016). On the other hand, Iba1, an ionized calcium-binding adaptor protein, is known to modulate actin reorganization, facilitates cell migration and phagocytosis, and has been implicated in synaptic development (Ito et al., 1998; Lituma et al., 2021).
In both CON and DEP + MS male offspring, we observed three types of microglia with respect to their differential expression of Iba1 and P2ry12. The majority of microglia highly expressed both Iba1 and P2ry12 (Figure 6A, Iba1hiP2ry12hi, bottom left) but some microglia expressed high levels of Iba1 but low levels of P2ry12 and had a strikingly different morphology (Figure 6A, Iba1hiP2ry12lo, bottom middle). We also found cells that expressed high levels of P2ry12 and low levels of Iba1, which were morphologically indistinguishable from Iba1hiP2ry12hi microglia (Figure 6A, Iba1loP2ry12hi, bottom right). Although these different types of microglia were more common in specific layers, they were often neighbored by the predominant microglia subtype, Iba1hiP2ry12hi.
To determine if prenatal DEP + MS exposure modifies the relative abundance of these microglial subtypes, we quantified the percentage of Iba1hiP2ry12lo or Iba1loP2ry12hi microglia, which we termed here as microglial heterogeneity. Early in development (P8–P15), microglial heterogeneity was higher in both CON and DEP + MS male offspring compared with a later developmental time point, P25 (Figure 6C). This observation suggests that these microglia subtypes do not reflect a pathological brain state but rather are part of a normal developmental process. Indeed, recent single-cell analyses of microglia across development have revealed that these cells are molecularly highly heterogeneous during very early postnatal ages (Hammond et al., 2019; Li et al., 2019; Masuda et al., 2019; Matcovitch-Natan et al., 2016). However, microglial heterogeneity was strikingly enhanced in DEP + MS male offspring ACC compared with CON, both at P8 and P15 (Figure 6C). Our data indicate that subtypes of microglia are present during early ACC development and that prenatal DEP + MS insult increases the relative abundance of heterogeneity.
At P8, when microglial heterogeneity is high, microglia are also actively pruning VGlut2 synapses (Figure 4H). Therefore, we next tested if the three subtypes of microglia differ in their phagocytic function and their ability to engulf VGlut2 synapses. To do so, we labeled the ACC microglia with Iba1, P2ry12, and CD68. Using Imaris, we reconstructed a total of 120 Iba1hiP2ry12hi, Iba1hiP2ry12lo, and Iba1loP2ry12hi microglia and quantified the lysosomal content, a proxy for phagocytic activity, within each microglial subtype (Figure 6D). There were no significant differences in the phagocytic activity of Iba1hiP2ry12lo cells compared with the Iba1hiP2ry12hi. However, Iba1loP2ry12hi cells had significantly lower CD68 content compared with the more prevalent Iba1hiP2ry12hi microglia type (Figures 6D and 6E). This pattern of reduced CD68 content was present in both CON and DEP + MS microglia (Figures S6A and S6C) and did not differ significantly between groups (Figures S6B and S6D). These results indicate that Iba1loP2ry12hi cells have lower phagocytic activity than the other two subtypes, suggesting this diminished phagocytic activity could alter their ability to eliminate synaptic inputs.
To investigate this possibility, we used Imaris to quantify the volume of VGlut2+ TC inputs within these distinct microglial subtypes (Figure 6F). Iba1loP2ry12hi microglia engulfed significantly fewer TC inputs compared with Iba1hiP2ry12hi cells (Figures 6F and 6G). Thus, Iba1loP2ry12hi cells have diminished lysosomal content and engulf fewer TC synapses. Importantly, these functional differences between the three microglial subtypes are present in both CON and DEP + MS male offspring brains (Figures S6E–S6H), showing that prenatal insults do not affect the per cell functional responses. Instead, specific subsets of cells (Iba1loP2ry12hi) are more abundant in DEP + MS offspring. In summary, we found evidence of a loss of normal microglial function in early postnatal DEP + MS male ACCs, reflected by increased heterogeneity and a net reduction in the ability of these cells to phagocytose synapses.
Loss of microglia synaptic pruning function during a critical postnatal period causes social deficits similar to those found in DEP + MS males
Does the transient loss of microglia function during early development (P8) underlie the enduring behavioral effects we observe in DEP + MS males? To test this possibility, we targeted microglia phagocytic function in the ACC by microinjecting neutrophil inhibitory factor (NIF) into the ACC of WT mice at P7, the period immediately before the peak in pruning. Microglial activation and phagocytosis of debris in the brain is in part mediated through the complement system via the activation of CR3 (CD11b/CD18) through opsonized proteins such as iC3b (Borucki et al., 2020). Several laboratories have demonstrated that developmental elimination of VGlut2+ synapses can be mediated by this same complement-dependent pathway (Salter et al., 2020; Schafer et al., 2012; Stevens et al., 2007). NIF, a glycoprotein produced by a canine hookworm, is a selective antagonist of the CR3-CD11b subunit, preventing the recognition of its endogenous ligands (Rieu et al., 1994). NIF effects are well characterized, including exploratory use in humans, and we and others previously found that NIF transiently (~48 h) inhibits rodent microglia phagocytosis both in vitro and in vivo without off-target effects (Kopec et al., 2018; Lees et al., 2003; Moyle et al., 1994).
In this experiment, WT neonatal male mice received bilateral microinjections of PBS or NIF (200 ng) into the ACC at P7, and brain tissue was collected 24 h later (Figure 7A). To confirm the effects of NIF on microglial phagocytic capacity, we quantified changes in the microglial lysosomal volume of CD68 (Figure 7B). As expected, microglia from animals microinjected with NIF had a significant reduction in the phagocytic index (~50%) and a significant decrease in the total lysosomal content within each microglia (Figures 7C and 7D). To determine whether this reduction in CD68 impaired microglial interactions with VGlut2 synapses, we once again performed Imaris reconstructions and quantified the volume of VGlut2 within microglia (Figure 7E). Microglia from NIF-treated animals are significantly smaller (~25%) than PBS control animals (Figure 7F); furthermore, this size reduction is accompanied by a significant decrease in the volume of internalized VGlut2 in microglia cells (Figure 7G). Last, we quantified the co-localization of VGlut2 and PSD95 and found that NIF-injected animals had about a 20% increase in VGlut2+ synapses (Figure 7H). Thus, NIF injections at P7 effectively reduce microglial phagocytic capacity and engulfment of VGlut2, which induces an abnormal increase in VGlut2 synapse density.
Figure 7. Early postnatal impairment of microglial phagocytic function is sufficient to induce social behavior impairments in male juvenile mice.
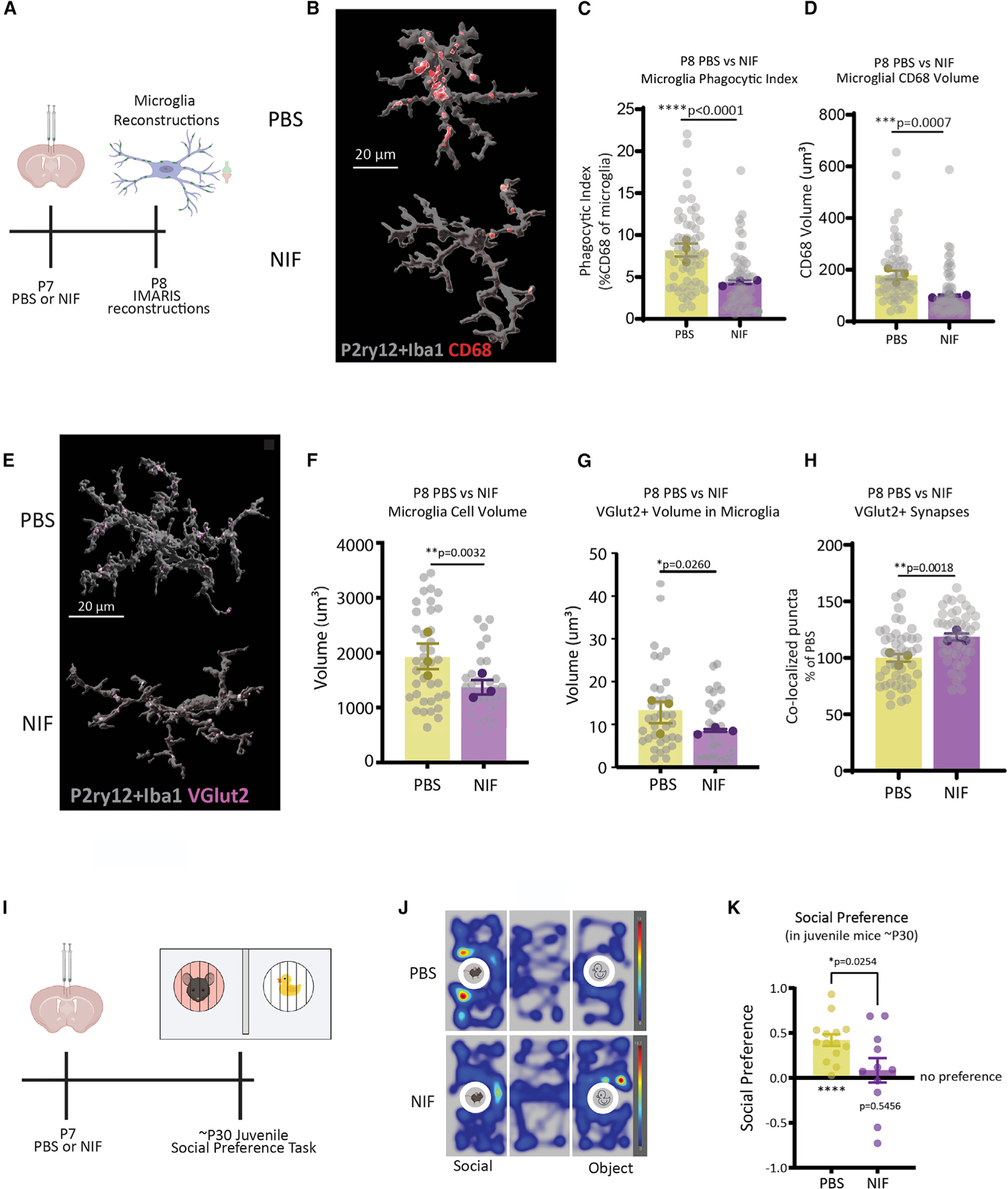
(A) PBS or NIF was microinjected into the ACC of P7 WT male mice. Microglial phagocytic content and engulfment were assessed at P8.
(B–D) Representative images and quantification of microglial phagocytic index and CD68 volume in PBS- versus NIF-injected mice (n = 15–20 cells/mouse/condition, n = 3 mice/condition, total of 114 cells analyzed, nested t test).
(E–G) Representative images and quantification of microglial volume and internalized VGlut2 in PBS- versus NIF-injected mice (n = 10–15 cells/mouse/condition, n = 3 mice/condition, total of 72 cells analyzed, nested t test).
(H) Quantification of VGlut2+ synapses in PBS versus NIF mice (n = 3 mice/condition, n = 3 replicates/mouse, 270 images analyzed, nested t test).
(I) PBS or NIF was microinjected into the ACC of P7 mice, then mice were tested in a social preference task at ~ P30.
(J) Representative heatmap of PBS versus NIF juvenile mouse exploration of social versus object.
(K) Quantification of social (n = 11–14 mice/condition, one-sample t test, unpaired t test). Means ± SEM.
Next, we tested whether reducing microglial synapse elimination during this early critical period could lead to a social deficit later in life. A separate cohort of WT male neonates was injected with PBS or NIF (pseudorandomized within litter) and raised into adolescence, where they were tested in a social preference task (Figure 7I). Juvenile males treated with PBS show a significant preference for the social stimulus compared with the novel object (Figures 7J and 7K, left bar), whereas littermates treated with NIF show no preference for the social stimulus and have a significant reduction in their social preference (Figure 7K, right bar). There were no significant differences in investigation time, distance traveled, or velocity (Figures S7A–S7D), demonstrating intact locomotor activity in NIF-injected animals. We also counted VGlut2 synapses in adolescent mice just after social preference testing (Figure S7D); interestingly, we did not find a significant decrease in the number of synapses, suggesting that the critical change in synapse number underlying the social deficit is the one occurring early in life, at P8. These data demonstrate that transient loss of function of microglia in the ACC is sufficient to induce changes in social behavior and demonstrate a cellular mechanism by which environmental stressors alter the development of neural circuits.
DISCUSSION
Immune dysfunction in pregnant mothers is increasingly implicated in the pathogenesis of NDDs and is strongly linked to male-offspring-specific behavioral outcomes (Patel et al., 2020). Here we show that prenatal co-exposure to two highly prevalent environmental factors, air pollution and maternal stress, is sufficient to induce MIA and significantly increase stress hormones in pregnant mice. Both male and female offspring born to these dams had altered USVs as neonates, whereas sociability and social novelty preference deficits during the juvenile period were observed only in DEP + MS male offspring. Notably, gene expression changes in the prefrontal cortices of neonatal mice differed by sex, and behavioral alterations only persisted into adulthood in male mice. These data indicate that prenatal environmental insults result in a distinct response in developing male brains compared with females.
Similarly, we found that the relationship between social investigation and the activation of EN-social was no longer behaviorally relevant in adult DEP + MS males. A possible interpretation of this difference is that, during development, synaptic circuits that encode social interactions form differently in male mice exposed to DEP + MS as fetuses. In agreement with this possibility, in the ACC, a critical brain region within this network, in DEP + MS male mice we found an early overabundance of TC synapses by the end of the first postnatal week. Changes in microglial function have been described in several neurological disorders. We found that microglia from DEP + MS males were less phagocytic and engulfed fewer synapses. Moreover, we discovered ACC microglia have a developmentally regulated antigenic and functional heterogeneity. This heterogeneity was strongly enhanced in males prenatally exposed to DEP + MS, leading to the overabundance of one specific subtype of microglia with a severely diminished phagocytic activity that eliminated fewer TC synapses. Taken together, our findings are in line with a loss-of-function phenotype within ACC microglia. Indeed, specific inhibition of phagocytic function within the ACC at P7 in WT mice decreased TC synapse elimination and induced a social deficit at P30, thus phenocopying the impact of the prenatal stressors. These data are consistent with previous findings on microglial heterogeneity within distinct brain regions; i.e., if microglial phagocytosis levels are not finely tuned to clearance requirements within a given brain region, this can result in aberrant brain development and altered behavior (Ayata et al., 2018).
In conclusion, our combined stressor model has allowed us to rigorously investigate the mechanisms underlying abnormal brain development in response to these pervasive environmental factors. Our findings elucidate a mechanism by which environmental pollutants can synergize with psychosocial stress in pregnant mothers and induce MIA, which has specific long-term effects on the development and function of male brains. This is particularly concerning, now more than ever, because ongoing climate change caused by increased economic activity and reduced environmental protection enforcements have led to a rapid worsening of air quality in recent years. Heightened air pollution is likely to synergize with social stressors in vulnerable populations, causing further disparities in the well-being of future generations. Therefore, our findings provide an important first step toward revealing the non-genetic causes for NDDs so that preventative and therapeutic approaches can be developed along with informed policy changes.
Limitations of the study
In disorders such as autism, early overgrowth of synaptic connectivity is often followed by atrophy, but the mechanism of the atrophy remains unknown. Here we see a similar phenotype, but we know that the loss of TC synapses cannot be explained by exuberant synapse elimination by microglia; thus, how these synapses are lost remains unclear. Synapses can be removed via multiple mechanisms, including astrocyte-mediated elimination (Chung et al., 2013). Astrocytes are macroglial cells that mediate synapse formation, functional maturation, and elimination. In particular, several studies revealed essential roles for astrocytes in controlling TC synapse formation and maturation (Risher et al., 2014; Singh et al., 2016; Stogsdill et al., 2017). Furthermore, neuroimmune insults are known to trigger different reactive profiles in astrocytes, which may also be happening in DEP + MS male brains, potentially underlying synapse loss and circuit dysfunction (Liddelow et al., 2017). Thus future investigations are needed to determine if astrocytes or other brain cell types are also involved in the dysfunctional synaptic development that we observe in DEP + MS male brains. Furthermore, we found a loss in the correlation between social investigation and the activation of EN-social. While we hypothesize this loss is due to a male-specific vulnerability in circuit development, an alternative possibility is that this correlation is lost due to ceiling effects, as low-preference animals are not present in the DEP + MS male group. Finally, future studies investigating the cellular and molecular mechanisms underlying male and female responses to MIA are needed to further understand why male brains are more vulnerable or female brains are protected (Patel et al., 2020).
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Staci Bilbo (Staci.bilbo@duke.edu).
Materials availability
This study did not generate a new unique reagent.
Data and code availability
RNA-sequencing data have been deposited at GEO and are publicly available as of the date of publication. Accession numbers are listed in the key resources table. Behavior and microscopy data reported in this paper will be shared by the lead contact upon request.
All original code has been deposited at GitHub and is publicly available as of the date of publication. DOIs are listed in the key resources table.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| NeuroTrace fluorescent Nissl Stain | Thermo Fisher Scientific | Cat#N21480 |
| Guinea pig anti-VGlut2 1:2000 | Synaptic Systems | Cat#135404; RRID: AB_887884 |
| Rabbit anti-PSD95 1:350 | Life Technologies | Cat#51–6900; RRID: AB_2533914 |
| Rat anti-CD68 1:500 | BioLegend | Cat#137002; RRID: AB_2044004 |
| Chicken anti-IBA1 1:1000 | Synaptic Systems | Cat#234006; RRID: AB_2619949 |
| Rabbit anti-P2y12 1:2000 | Anaspec | Cat#AS-55043A: RRID: AB_2298886 |
| Goat anti-Iba1 1:500 | Novus Biologicals | Cat#NB 100–1028; RRID: AB_521594 |
| Rabbit anti-Sox9 1:2000 | Millipore Sigma Aldrich | Cat#AB5535; RRID: AB_2239761 |
| Mouse anti-Olig2 1:2000 | Millipore Sigma Aldrich | Cat#MABN50; RRID: AB_10807410 |
| Mouse anti-NeuN 1:250 | Millipore Sigma Aldrich | Cat#MAB377; RRID: AB_2298772 |
| DAPI | Thermo Fisher Scientific | Cat#D1306 |
| Kwik-Diff™ | Thermo Fisher Scientific | Cat#99–907-00 |
|
Chemicals, peptides, and recombinant proteins | ||
| Diesel Exhaust Particles | EPA Gift from Ian Gilmour | N/A |
| TRIzol Reagent | Thermo Fisher Scientific | Cat#15596026 |
| Chloroform | Sigma-Aldrich | Cat#C2432 |
| Triton X-100 | Roche | Cat#11332481001 |
| 2-Propanol | Sigma-Aldrich | Cat#I9516–500ML |
| VECTASHIELD with DAPI | VWR | Cat#h-1200 |
| Normal Donkey Serum | Jackson ImmunoResearch | Cat#017–000-121 |
| Normal Goat Serum | Jackson ImmunoResearch | Cat#005–000-121 |
| Recombinant A. caninum NIF Protein, CF | R&D Systems | Cat#5845-NF-050 |
|
Critical commercial assays | ||
| U-Plex Proinflammatory Panel, Mouse | Meso Scale Discovery | Custom made |
| ELISA kit | Arbor Assays | Cat#K014-H1 |
| KAPA Stranded mRNA-Seq Kit | Roche | Cat#07962142001 |
|
Deposited data | ||
| Bulk RNA-sequencing of neonatal PFC in CON and DEP+MS male and females (Raw and analyzed data) | This paper | GEO: GSE160344 |
|
Experimental models: Organisms/strains | ||
| TLR4 KO Mice (C57BL/10ScNJ) | Jackson Labs | RRID:IMSR_JAX:003752 |
| WT Mice (C57BL/6J) | Jackson Labs | RRID:IMSR_JAX:000664 |
|
Software and algorithms | ||
| Avisoft-Ultrasound Gate recording software | Avisoft Bioacoustics | RRID:SCR_014436; http://www.avisoft.com/downloads/ |
| Avisoft SASLab Pro software | Avisoft Bioacoustics | RRID:SCR_014438 |
| MUPET v2.1 | Van Segbroeck et al., 2017 | https://github.com/mvansegbroeck/mupet |
| MATLAB Version 2017a | The MathWorks | RRID:SCR_001622; https://www.mathworks.com/ |
| R statistical programming environment | R Project for Statistical Computing | RRID:SCR_001905; https://www.r-project.org/ |
| PANTHER | Mi et al., 2019 | RRID:SCR_004869; http://www.pantherdb.org/ |
| FV31S-SW | Olympus Corporation | https://www.olympus-lifescience.com/en/support/downloads/ |
| ImageJ | NIH | RRID:SCR_003070; https://imagej.nih.gov/ij/ |
| Python 2019.3.3 | Python Programming Language | RRID:SCR_008394; http://www.python.org/ |
| Prism 8.0.0 | GraphPad Software, Inc. | RRID:SCR_002798; https://www.graphpad.com |
| Statistica Version 13.5.0.17 | TIBCO Statistica Software | RRID:SCR_014213; https://www.statisticssolutions.com/ |
| Imaris 9.5.1 | Oxford Instruments | RRID:SCR_007370; https://imaris.oxinst.com/ |
| Huygens Professional 19.10.0p3 64b | Scientific Volume Imaging | RRID:SCR_014237; https://svi.nl/Homepage |
| Cell count: U-net cell segmentation | Falk et al., 2019; Weigert et al., 2018; and this paper | Zenodo: https://doi.org/10.5281/zenodo.6800011 |
| Puncta Analyzer | Ippolito and Eroglu, 2010 | Zenodo: https://doi.org/10.5281/zenodo.6800214 |
| NMF Cross Frequency Spectral Analysis | Talbot et al., 2020 | Zenodo: https://doi.org/10.5281/zenodo.6078304 |
| Jwatcher | UCLA | RRID:SCR_017595; http://jwatcher.ucla.edu/ |
| Solomon Coder | Andras Peter | RRID:SCR_016041; https://solomon.andraspeter.com/ |
| Ethovision | Noldus Technology | RRID:SCR_000441; https://www.noldus.com/ethovision-xt |
|
Other | ||
| AlphaDri | Sheperd Specialty Papers | N/A |
| PicoLab Mouse Diet 5058 | Lab-Diet | N/A |
| Fine-gauge aluminum mesh platform | McNichols Co. | N/A |
EXPERIMENTAL MODEL AND SUBJECT DETAILS
All experiments were performed in accordance with the guidelines of the Division of Laboratory Animal Resources from Duke University School of Medicine and Institutional Animal Care and Use Committee. For experiments in wildtype animals, we obtained adult male and female C57BL/6J mice from Jackson Laboratories (Bar Harbor, ME) and maintained an internal colony of breeding animals for all experiments. For experiments in TLR4 KO dams, 6-week-old C57BL/10ScNJ female mice were acquired from Jackson Laboratories and were allowed to acclimate to the colony for at least two weeks prior to mating (Bar Harbor, ME).
Prenatal treatments were assigned randomly on the day of confirmed pregnancy. Male and female offspring born to mothers prenatally treated were used for behavioral, cellular, and molecular analyses. All behavioral analyses were conducted in both males and females, except for the adult USV courtship song, as female mice do not typically vocalize in this context. Behavior was collected as described below at P7, P8, P9, P30, and in adult animals (~P60-P100). Gene transcription analyses were performed at P8; 1 male and female littermate pair was randomly selected per litter (total of 4 litters/condition). For IHC, synapse analyses were performed in both males and females because the same phenotype was not observed in both sexes. Subsequent microglial analyses were performed only in male offspring.
METHOD DETAILS
Prenatal stressors
Diesel exhaust particle exposure
Performed as previously described (Bolton et al., 2013), briefly, adult females were time-mated and checked twice daily for the presence of a vaginal plug, considered to be E0. Females were paired in individually ventilated cages with specialized bedding (AlphaDri; Shepherd Specialty Papers, Milford, NJ) and ad libitum access to food (PicoLab Mouse Diet 5058; Lab-Diet, Philadelphia, PA) and filtered water. Females were treated with diesel exhaust particles (DEP) delivered via oropharyngeal aspiration. Beginning on E2, females were lightly anesthetized with 2% isoflurane and treated with either 50 μg of DEP suspended in 50 μL of PBS, 0.05% Tween-20, or vehicle alone (CON). Females received a total of six doses, with each dose administered every 3 days from E2-E17.
Maternal stress/nest restriction
To induce maternal stress, we utilized our adaptation of a previously described nest restriction model applied to the postnatal period (Bolton et al., 2013; Rice et al., 2008). Beginning at 5 pm on E13, control females receiving vehicle treatment were singly housed in a clean cage with a full-size nestlet (CON), and females exposed to DEP were housed in clean cages with a thin layer of bedding under an elevated fine-gauge aluminum mesh platform (mesh dimensions, 0.4 cm × 0.9 cm; McNichols Co., Tampa, FL) and provided with two-thirds of one square of felt-like nesting material (~1.9 g; MS group). This design results in two groups of dams: control dams (CON) and dams exposed to combined environmental stressors (DEP+MS). On E18.5 prior to the birth of pups, all dams were placed into a clean cage with a full-size nestlet and were treated identically. All pups were born into a standard caging environment and remained with the mother until tissue collection time point or weaning age ~P24, at which time mice were group-housed with same-sex littermates at a maximum of 5 animals per cage.
Maternal immune activation
Cytokines can vary as a function of ZT; thus, all experiments were tightly controlled for time-of-day effects. To determine the immune activation of pregnant dams at E17.5, prenatal instillations were performed 2 hours into the light phase (ZT2), and blood was collected 2–3 hours post-treatment (ZT4–5). A 5.5 mm lancet was used to pierce the submandibular vein, and cheek blood was collected into a sterile Eppendorf tube. To separate serum from red blood cells, blood was centrifuged twice at 16,000 xg at 4°C for 10 mins. Separated serum was collected into a clean Eppendorf tube and stored at −80°C until analysis.
Maternal cytokine analysis
For serum analysis, a multiplex electrochemiluminescence immunoassay kit (U-Plex Proinflammatory Panel, Mouse) was purchased from Meso Scale Discovery (Rockville, MD) and used according to manufacturer instructions to measure serum cytokine concentrations (pg/mL) of IFN-γ, IL-1β, IL-4, IL-5, IL-6, IL-10, IL-17A, IL-12/IL-23p40 MCP-1, and TNF-α. To prevent antibody cross-reactivity, IL-17A and IL-12/IL-23p40 were coated in 1 plate, and the remaining antibodies were coated onto a separate plate. Analysis of blinded samples was conducted by Duke Molecular Physiology Institute Metabolomics laboratory. Samples were run in duplicates, plates were read with a Sector Imager 2400 (Meso Scale Discovery), and data were analyzed using the Discovery Workbench 4.0 software (Meso Scale Discovery). Two separate cohorts of WT maternal serum were collected and analyzed. All data from each analysis were normalized to the CON values from the plate, and normalized values were combined for further analysis. Any values below the lower limit of detection (LLOD) were assigned 0 pg/mL. All samples were within the detection range for IFN-γ, IL-5, IL-6, IL-10, IL-17A, IL-12/IL-23p40, MCP-1, and TNF-α. For IL-1β and IL-4, more than half of the samples fell below the LLOD and were excluded from further analyses.
Maternal CORT measurement
Corticosterone serum levels were measured using a commercially available ELISA kit (K014-H; Arbor Assays, Ann Arbor, MI). To determine whether the method of instillation induced additional stress, control serum was rapidly collected from a non-pregnant female and three WT pregnant females without prenatal treatment. The optical density measurements (Bio-Tek Instruments) from the microplate reader were uploaded to https://www.myassays.com/arbor-assays-corticosterone-enzyme-immunoassay-kit.assay to calculate corticosterone concentration for each sample.
Lung analyses
Bronchoalveolar lavage fluid collection
After submandibular blood collection, dams were sacrificed using CO2. A small catheter was inserted into the trachea, and 3 mL of HBSS (without calcium and magnesium) was instilled and withdrawn from the lungs, and recovery volume was recorded for all samples.
Cell counts and BALF differentials
Collected fluid was processed for total and differential cell counts by the Duke Rodent Inhalation core, which was blinded to the condition. Collected fluid was centrifuged at 3000 rpm for 10 mins at 4C, cells were treated with 1XRBS lysis buffer, further centrifuged, and resuspended in PBS. Cells were counted with a hemocytometer (Hausser Scientific, Horsham, PA), and recovery volume was used to determine cell density. BALF cytology was performed by immobilizing 100uL of the cell suspension using a Cytospin 4 centrifuge (Thermo Fisher Scientific, Waltham, MA). Cells were stained with a Kwik-Diff™ kit (Thermo Fisher Scientific, Waltham, MA) according to manufacturer recommendations. Images were obtained using a 20x objective on an AxioImager M1 (Zeiss) microscope. Cell differential counts were determined by morphological analysis of acquired images and were used to identify and quantify macrophages, neutrophils, lymphocytes, and eosinophils. Out of the <2,000 cells counted, less than 5 were eosinophils. Thus eosinophils were excluded from further analyses.
Behavioral procedures
We assessed behavioral outcomes as a result of prenatal stressors in a cohort of neonatal (P7-P9) male and female offspring (n = 14–17 animals/sex from 4 litters per condition). A separate cohort of male offspring was utilized to assess outcomes in adulthood (n = 16–20 animals/condition from 4 litters per treatment group). A separate cohort of CON and DEP+MS mice were generated to assess neurophysiological ends points with concurrent behavior (n = 13–27 animals/sex per condition group). This cohort did not undergo behavioral testing during the neonatal or juvenile period and was undisturbed during the weanling stage.
Neonatal ultrasonic vocalizations
To determine the ultrasonic vocalization (USV) developmental timeline in C57BL/6J animals, USVs were collected from P4-P9. For experimental animals, USVs were collected on postnatal days 7–9. Pups were briefly separated from dams and placed into a sound-attenuating chamber for 3 minutes; USVs were recorded using an externally polarized condenser microphone with a frequency range of 10–200 kHz attached to the Avisoft-Ultrasound Gate recording software (Avisoft Bioacoustics, Berlin, Germany). Pup weight and toe clip identification were performed immediately after USV collection. WAV files for each pup were converted to spectrograms and analyzed with automated whistle tracking parameters by the Avisoft SASLab Pro software (Avisoft Bioacoustics) and manually validated for accuracy. For call complexity analysis, WAV files were analyzed using Mouse Ultrasonic Profile ExTraction Tool (MUPET) in MATLAB, which is an unsupervised machine-learning-based algorithm that analyzes vocalization parameters, classifies syllables into distinct repertoires, and compares vocalization patterns between test groups (Van Segbroeck et al., 2017). Repertoire units were sorted by length using a Python script.
Social preference and social novelty preference
Prior to behavioral testing in juvenile mice, all animals were gently handled five times. Social preference and social novelty preference tasks were performed in juvenile mice between postnatal days P27-P40. Males and females were tested using separate testing boxes on different days. Mice were habituated to the testing room (1 hour) and testing chambers (5 minutes) the day before testing. All behavioral testing was conducted during the second half of the light phase.
For both social preference and social novelty preference, a 3-chambered arena with doorways to allow for passage between the chambers was used, according to (Smith et al., 2015). In the social preference test, social vs. object stimuli (either novel sex-, treatment-, and age- matched conspecific or a novel rubber duck) were confined within smaller containers composed of Plexiglass rods in each of the opposite side chambers. Subject animals were placed into the middle chamber, and their movement and investigation of each of the stimuli were scored over the course of 5 min.
In the social novelty preference task, the same procedure was followed, except that stimuli consisted of either novel sex-, treatment-, and age-matched conspecific or a familiar cage mate (also matched for sex, treatment, and age), and their investigation behavior was quantified over the course of 10 min. The testing apparatus was cleaned with a disinfectant between each test.
All videos were scored using Jwatcher (JWatcher.ucla.edu) by a blinded observer. Scored elements included: time spent in each chamber, time spent investigating each stimulus (i.e., direct sniffing or nose-poking between the bars of the smaller stimulus containers), and time spent in the empty middle chamber.
The social preference for each session was defined as:
where InteractionTimeS is the total time spent proximal to the other mouse, and InteractionTimeO is the total time spent proximal to the object.
The social novelty preference for each session was defined as:
where InteractionTimeNovelStimulus is the total time spent proximal to the novel stimulus mouse, and InteractionTimecagemate is the total time spent proximal to the cage mate stimulus mouse.
Adult ultrasonic vocalizations
Prior to behavioral testing in adulthood, all animals were handled five times. Each day mice were habituated to the testing room (1 hour) and testing chambers (5 minutes) before introducing a stimulus animal. All behavioral testing was conducted during the second half of the light phase. Because males are the primary source of USVs in male-female encounters, adult USVs were only collected from male offspring using an estrous-induced courtship paradigm (Chabout et al., 2015, 2017). In this paradigm, male mice, after gaining sexual experience, are exposed to novel sexually receptive WT stimulus animals for 5 minutes for 3 days. To identify sexually receptive females, stimulus animals were vaginally swabbed, and cell morphology was assessed to identify females in estrous or proestrus (Byers et al., 2012). Females identified to be in estrous or proestrus were utilized as stimulus animals. USVs were recorded for 5 minutes using an externally polarized condenser microphone with a frequency range of 10–200 kHz attached to the Avisoft-Ultrasound Gate recording software (Avisoft Bioacoustics, Berlin, Germany). WAV files were analyzed using MUPET. To filter noise calls below 35 kHz were excluded, a noise-reduction value of 8.8 was utilized with a minimum syllable duration of 2.0 msec.
16S ribosomal RNA sequencing
Fecal boli samples were collected from CON and DEP+MS dams at E17.5 (n = 14 dams total). Bacterial taxa were identified using 16S rRNA sequencing of microbiome samples. Library preparation was conducted in accordance with standard protocols (earthmicrobiome.org). First, DNA was extracted from all samples using a DNeasy Powersoil Kit (Qiagen, Germantown, MD). Next, PCR with individually barcoded primers (515F-806R; (Apprill et al., 2015; Caporaso et al., 2012; Caporaso et al., 2011; Parada et al., 2016) was used to amplify the V4 hypervariable region of the 16S rRNA gene. PCR product was then purified (PCR Purification Kit, Qiagen, Germantown, MD), DNA concentration was measured using a Quant-iT Picogreen Assay (Thermofisher Scientific), and an equimolar pool of all samples was made and transferred to the core Duke Microbiome Core Facility for sequencing on an Illumina MiSeq Sequencer (Illumina, San Diego, CA, USA).
The Qiime2–2019.7 analysis platform was used to analyze 16S data. Briefly, forward and reverse reads were imported, demultiplexed, and quality filtered using DADA2. Amplicon sequence variants were then aligned with MAFFT, and a phylogenetic tree was generated. Taxonomy was assigned using a Naïve Bayes filtered classifier trained on the Greengenes database, version 13_8, at 99% sequence similarity.
Electrode implantation surgery
Mice were anesthetized with 1.5% isoflurane, placed in a stereotaxic device, and metal ground screws were secured above the cerebellum and anterior cranium. The recording bundles designed to target basolateral and central amygdala (AMY), medial dorsal thalamus (MD), nucleus accumbens core and shell (NAc), VTA, medial prefrontal cortex (mPFC), and VHip were centered based on stereotaxic coordinates measured from bregma (Amy: −1.4 mm AP, 2.9 mm ML, −3.85 mm DV from the dura; MD: −1.5 8 mm AP, 0.3 mm ML, −2.88 mm DV from the dura; VTA: −3.5 mm AP, ±0.25 mm ML, −4.25 mm DV from the dura; VHip: −3.3 mm AP, 3.0 mm ML, −3.75 mm DV from the dura; mPFC: 1.62 mm AP, ±0.25 mm ML, 2.25 mm DV from the dura; NAc: 1.3 mm AP, 2.25 mm ML, −4.1 mm DV from the dura, implanted at an angle of 22.1°). We targeted the prelimbic and infralimbic cortex using the PFC bundle by building a 0.5 mm and 1.1 mm DV stagger into our electrode bundle microwires, and animals were implanted bilaterally in mPFC and VTA; all other bundles were implanted on the left side. The NAc bundle included a 0.6 mm DV stagger such that wires were distributed across the NAc core and shell. We targeted BLA and CeA by building a 0.5mm ML stagger and 0.3 mm DV stagger into our AMY electrode bundle. In order to mitigate pain and inflammation related to the procedure, all animals received carprofen (5 mg/kg, s.c.) injections once prior to surgery and then once every 24 hours for three days following electrode implantation.
Histological confirmation
Histological analysis of implantation sites was performed at the conclusion of experiments to confirm recording sites used for neurophysiological analysis (Figure S4A). Animals were perfused with 4% paraformaldehyde, and brains were harvested and stored for 24 hours in PFA. Brains were cryoprotected with sucrose and frozen in OCT compound, and stored at −80C. Brains were sliced at 35 µm and stained using NeuroTrace fluorescent Nissl Stain (N21480, ThermoFisher Scientific, Waltham, MA). Floating sections were washed 3 times in PBST (0.1%). Sections were incubated in PBS with Nissl antibody (1:300) for 10 mins at room temperature and washed once in PBST (0.1%) and twice in PBS with azide (0.01% NaN3), after which the entire brain was mounted. Images were obtained using a Nikon Eclipse fluorescence microscope at 4× and 10× magnifications. Only animals in which all eight implantation sites were confirmed were included in the analysis. Multiple animals were removed due to tissue destruction during histological analysis, in which implantation could not be confirmed.
Neurophysiological data acquisition
Mice were connected to a headstage (Blackrock Microsystems, UT, USA) without anesthesia and placed in each behavioral arena. Neuronal activity was sampled at 30 kHz using the Cerebus acquisition system (Blackrock Microsystems Inc., UT). Local field potentials (LFPs) were bandpass filtered at 0.5–250 Hz and stored at 1000 Hz. Neurophysiological recordings were referenced to a ground wire connected to both ground screws.
Social interaction test with concurrent recording
Social preference was measured using a two-chamber assay in which animals explored a novel object or a novel mouse. The apparatus was a rectangular arena (61 cm’ 42.5cm’ 22 cm) constructed from clear plexiglass with a clear plexiglass wall dividing the arena into two equal chambers with an opening in the middle allowing free access between both chambers. The floor of the arena was constructed using a one-way mirror that allowed for video recording from beneath to avoid obstruction from the electrophysiological recording equipment. Plastic, circular holding cages (8.3 cm diameter and 12 cm tall) were centered in each of the two chambers and were used to house either a novel object or sex- and age-matched C3H target mouse. The arena was evenly lit with indirect white light (~125 lux). Test mice were handled and habituated to the social preference chambers and empty holding cages for a least three days prior to testing. Subsequently, mice underwent ten separate social preference test sessions, with at least one day off in between sessions, in which the test mice were allowed to freely explore the arena for ten minutes; the holding cages contained either a novel object or novel C3H target mouse. The side of the chamber holding the object/mouse was determined pseudorandomly, such that the object/mouse would not be placed in the same chamber on more than two consecutive sessions in order to prevent side biases and to distinguish target-specific effects from location-specific effects. Plastic toys and glass objects were used as novel objects, with the object being between 3–5 cm in all directions. Video data was tracked using Bonsai Visual Reactive Programming software, and the time spent in the proximity (4.98 cm) of either holding cage was used to determine social preference scores.
The social preference for each session was defined as:
where InteractionTimeS is the total time spent proximal to the other mouse, and InteractionTimeO is the total time spent proximal to the object.
LFP preprocessing to remove signal artifact
Rather than manually screening data, we used an automated heuristic strategy to remove recording segments with non-physiological signals. First, we estimated the envelope of the signal in each channel using the magnitude of the Hilbert transform. For any 1-second window where the envelope exceeds above a pre-selected low threshold, the entire segment is removed if the envelope exceeds a second, high threshold at any point within that window. The two thresholds were determined independently for each brain region. The high threshold was selected to be 5 times the median absolute deviation of the envelope value for that region. Five median absolute deviations were chosen as the high threshold because it is roughly equivalent to 3 standard deviations from the mean for normally distributed data but is robust to outliers in the data. The low threshold was empirically chosen to be 3.33% of the high threshold. If more than half the window was removed for a channel, we removed the rest of that window for that channel as well. In addition, any windows where the standard deviation of the channel is less than 0.01 were also removed. Using this approach, 13±3.5% of the data per mouse were excluded from this analysis. This conservative strategy optimized the potential of our learning model to discover a network that was uniquely related to appetitive social, emotional brain states.
Feature estimation
The LFPs were averaged across electrodes within each brain region to yield a more robust estimate of the LFP for each region. Each LFP recording was divided into 1-second windows with a univariate time series associated with each region. Feature extraction was performed with MATLAB (The MathWorks, Inc., Natick, MA). The three features of interest were frequency-based power within each region, frequency-based coherence between each pair of regions, and frequency-based Granger causality between each region.
For estimating power, we used the pwelch function in MATLAB, which averages multiple periodograms estimated using different segments of the window to obtain a denoised power spectrum. A sliding Fourier transform with a Hamming window was applied to the average LFPs (default pwelch settings), and the power was estimated at 1 Hz intervals. Estimating the frequency-based coherence was done using magnitude-squared coherence, defined as
which normalizes the cross-spectral estimates by the power spectra in each region, yielding a value between 0 and 1. This was done in MATLAB using the function mscohere with default settings, also at a resolution of 1 Hz.
The Granger causality is a measure of causal information flow between two signals (Geweke, 1982). While the original definition did not decompose this flow by frequency, work by (Barnett and Seth, 2014) developed the theory and toolbox to do this, known as the Multivariate Granger Causality (MVGC) MATLAB toolbox. We used the standard procedure as defined in the method; the non-stationary data went through a highpass Butterworth filter with a stopband at 1 Hz and a passband starting at 4 Hz. Granger causality values for each window were calculated using a 20-order AR model via the GCCA_tsdata_to_smvgc function of the MVGC toolbox. Once again, these causality values were estimated at the same frequency intervals as the power and coherence.
The Granger features themselves are not additive, a major drawback with most factor models. Rather than using the features directly, we used the exponential of all causality values, which can be interpreted as a ratio of total power to the unexplained power. That is,
where fY→X (λ)represents Granger causality at frequency λ from region Y to region X, SXX(λ) represents the spectral power in region X at frequency λ, and HXY (λ)ΣY|X HXY (λ)* represents the component of that power that is predicted by region Y. These values can be occasionally very large due to estimation error and were capped at 10 to prevent undue influence from single observations.
Discriminative cross-spectral factor analysis non-negative matrix factorization (dCSFA-NMF)
We used a non-negative matrix factorization to synthesize these estimated features into a network-based model of neural dynamics. This is termed Supervised Cross-Spectral Factor Analysis – Nonnegative Matrix Factorization (CSFA-NMF). This model is fully described elsewhere (Talbot et al., 2020), and the code to implement these models is publicly available at https://doi.org/10.5281/zenodo.6078304. To provide a succinct description of the methodology, CSFA-NMF assumes each window of data to be an independent stationary observation. Relevant dynamics and behavior occur at the timescale of windows rather than individual LFP measurements. In this work, we chose a 1-second window as a compromise between fast-changing dynamics in behavior and the extra stability in feature estimation provided by longer windows. Prior work has shown that shorter windows decrease predictive accuracy, and 5 second windows would not be fast enough to capture the rapidly changing behavioral dynamics needed for these experiments.
Each window of data consists of the estimated power, coherence, and exponential granger features totaling P distinct observations per window. These observations were vectorized. We use to denote a window within the N total windows. We describe the preprocessed data as (the P-dimensional non-negative domain) and the observed behavioral label as , where the binary indicates a social or non-social behavioral label. The objective function learned by this model is
where K is the number of different electomes. Each electome is described by a column in (e.g., W = [w1,…,wk]) that describes the multi-region spectral power and coherence relationships. The electome factor scores are given by the multi-output function and the relationship between the electome factor scores and the behavioral labels is given by balances the relative importance of prediction relative to reconstruction. d was defined to have a single non-zero element in order to limit the predictive capacity to a single latent network. This is a formulation of an NMF model that performs approximate inference using supervised autoencoders and requires the user to choose a parametrization for f(xn; φ). In our method, this is simply set to an affine function followed by a non-linearity, f(xn; φ) = softplus (Ax + b), where the parameters of the function are φ = {A; b} and the softplus means an element-wise operation of the operation softplus(a) = log(1 + exp(a)), which maps a real number to the non-negative space. While other rectifying functions are possible (such as the popular Rectified Linear Unit (ReLU)), we chose the softplus to prevent vanishing gradients in the parameter estimation.
This model is able to be learned through stochastic gradient descent and was implemented in TensorFlow 1.09 using the ADAM algorithm for learning. In addition to the benefits of increasing predictive ability, replacing explicit network score estimates with a predictive function allows for quicker inferences with stochastic rather than batch training. Furthermore, once a predictive function is learned, we can calculate the electome scores on new data simply by calling the function f(xn; φ). This contrasts with other methods, which typically require a potentially difficult optimization problem to estimate each new electome score. This allows for future applications requiring real-time estimation.
Hyper-parameter selection
This analysis requires us to choose several parameters, notably the number of electomes K, the supervision strength λ, the relative importance of the features, and the parameterization of the mapping function f(xn; φ). For the mapping function, we chose an affine transform with a softplus activation to avoid overfitting and to prevent vanishing gradients, respectively. Our analysis has two goals, to predict behavior in new animals well and to describe the brain dynamics accurately. These two goals are measured by the reconstruction error of the features and by the mean Area Under the Curve (mean AUC) on validation mice, respectively. Choosing the supervision strength was chosen to be a value found to work well in previous analyses. The number of networks K was chosen using an elbow analysis using an unsupervised NMF model, where we chose K to be the number of networks where minimal gains in the reconstructive loss were observed. This model’s parameters were learned elsewhere (Talbot et al., 2020). The electome scores on the mice in this paper are estimated by putting the extracted features through the previously trained function f(xn; φ). Thus, since the mice in this paper were not used for hyperparameter selection or training, they represent a true estimate of the accuracy and reconstructive ability of the model when applied to this novel population.
RNA-sequencing analysis of transcriptome
Tissue and sample preparation
Tissue samples were harvested from a cohort of behaviorally naïve P8 pups. Animals were anesthetized with avertin and perfused with saline (n = 4 animals/condition/sex). The brain was immediately extracted, and the prefrontal cortex was dissected before being flash-frozen in liquid nitrogen and stored at −80°C until RNA-extraction.
RNA-extraction
Frozen samples were homogenized in 1000 μL TRIzol Reagent (15596026, Thermo Fisher Scientific, Waltham, MA) and vortexed at 2000 rpm for 5 min. 200 μL of chloroform (Sigma-Aldrich, C2432, St. Louis, MO) was added to each tube and vortexed for an additional 2 min; samples were allowed to phase separate before being centrifuged at 11,900 rpm for 15 min at 4°C, after which the top clear aqueous phase was separated into a fresh tube. 500 μL of Isopropanol (Thermo Fisher Scientific, NY) was added, and samples were vortex at 2000 rpm for 1 min and incubated at room temperature for an additional 10 minutes and then centrifuged for 10 mins. The supernatant was discarded, and the RNA pellet was washed two times with 1 mL of ice-cold 75% ethanol, air-dried, and resuspended in 40 μL of RNase-free water.
Library prep and sequencing
All RNA samples were coded numerically. Sequencing was performed blind to sample identity by Sequencing and Genomic Technologies Shared Resource Duke Center for Genomic and Computational Biology. Extracted total RNA quality and concentration were assessed on Fragment Analyzer (Agilent Technologies) and Qubit 2.0 (ThermoFisher Scientific), respectively. RNA-seq libraries were prepared using the commercially available KAPA Stranded mRNA-Seq Kit (Roche). In brief, mRNA transcripts are first captured using magnetic oligo-dT beads, fragmented using heat and magnesium, and reverse transcribed using random priming. During the 2nd strand synthesis, the cDNA: RNA hybrid is converted into double-stranded cDNA (dscDNA) and dUTP incorporated into the 2nd cDNA strand, effectively marking the second strand. Illumina sequencing adapters are then ligated to the dscDNA fragments and amplified to produce the final RNA-seq library. The strand marked with dUTP is not amplified, allowing strand-specificity sequencing. Libraries were indexed using a dual indexing approach allowing for all the libraries to be pooled and sequenced on the same sequencing run. Before pooling and sequencing, fragment length distribution for each library was first assessed on a Fragment Analyzer (Agilent Technologies). Libraries were also quantified using Qubit. The molarity of each library was calculated based on qubit concentration and average library size. All libraries were then pooled in an equimolar ratio and sequenced. Sequencing was done on an Illumina NovaSeq 6000 sequencer. The pooled libraries were sequenced on an S-Prime flow cell at 50 bp paired-end. Once generated, sequence data were demultiplexed, and Fastq files were generated using bcl2fastq v2.20.0.422 file converter from Illumina.
Transcriptome data analysis methods
RNA-seq data was processed by the Genomic Analysis and Bioinformatics Shared Resource, Duke Center for Genomics and Computational Biology using the TrimGalore toolkit (http://www.bioinformatics.babraham.ac.uk/projects/trim_galore) which employs Cutadapt (Martin, 2011) to trim low-quality bases and Illumina sequencing adapters from the 3ʹ end of the reads. Only reads that were 20 nt or longer after trimming were kept for further analysis. Reads were mapped to the GRCm38v73 version of the mouse genome and transcriptome (Kersey et al., 2012) using the STAR RNA-seq alignment tool (Dobin et al., 2013). Reads were kept for subsequent analysis if they mapped to a single genomic location. Gene counts were compiled using the HTSeq tool (http://wwwhuber.embl.de/users/anders/HTSeq/). Only genes that had at least 10 reads in any given library were used in subsequent analysis. Normalization and differential expression were carried out using the DESeq2 (Love et al., 2014) and Bioconductor (Huber et al., 2015) package with the R statistical programming environment (www.r-project.org). We controlled for plates in each model that we ran. The false discovery rate was calculated to control for multiple hypothesis testing. Gene set enrichment analysis (Mootha et al., 2003) was performed to identify pathways associated with altered gene expression for each of the comparisons, PANTHER (http://www.pantherdb.org/) was used to perform a statistical overrepresentation test (Mi et al., 2019).
Immunohistochemistry
Mice used for IHC were anesthetized with 200 mg/kg tribromoethanol (avertin) and perfused with Tris-Buffered Saline (TBS, 25 mM Tris-base, 135 mM NaCl, 3 mM KCl, pH 7.6) supplemented with 7.5 mM heparin, followed by 4% PFA in TBS. Brains were extracted and post-fixed in 4% PFA in TBS overnight at 4C. After fixation, brains were washed 3 times with TBS and transferred to a 30% sucrose/TBS solution for cryoprotection. Brains were frozen and embedded into a solution containing 2 parts, 30% sucrose, and 1 part OCT (Tissue Tek, Sakura, Torrance, CA), and stored at −80°C. For synaptic staining, brains were sections at 20 µm thickness; for cell counting and microglia reconstructions, brains were sectioned at 40 µm, tissue sections were stored floating in a 1:1 mixture of TBS/glycerol at −20°C.
Synaptic staining
Free-floating sections were washed 3 times for 10 minutes with TBS with 0.2% Triton X-100 (Roche, Indianapolis, IN) and blocked in 5% Normal Goat Serum (NGS; Jackson ImmunoResearch, West Grove, PA) with 0.2% Triton X-100 in TBS for 1 hour at room temperature. Primary antibodies (see table below) were diluted in 5% NGS in TBS with 0.2% Triton X-100. Sections were incubated overnight at 4°C with primary antibodies and washed three times for 10 minutes with TBS the following morning. Secondary Alexa-fluorophore conjugated antibodies (Invitrogen, Carlsbad, CA) were diluted (1:200) in 5% NGS in TBS with 0.2% Triton X-100, and sections were incubated with secondary antibodies for 2 hours at room temperature, protected from light. After incubation, sections were washed three times for 15 minutes in TBS and mounted with VECTASHIELD with DAPI (Vector Laboratories, Burlingame, CA). Images were acquired on an Olympus FV3000 confocal laser-scanning microscope.
Acquisition and analysis of synaptic staining
Staining, image acquisition, and analysis were performed as in Ippolito and Eroglu (2010) (Ippolito and Eroglu, 2010) with adjustments. Synaptic staining was performed in two male/female littermate pairs at P6, P8, P10, P13, and P15 in WT C57BL6/J offspring to determine the normal developmental pattern. Synaptic staining was performed at P8, P15, and P100 in male and female offspring for CON and DEP+MS conditions. Image acquisition was performed in layer 1 (L1) of the ACC from P8, P15, and adult CON and DEP+MS animals. We chose to conduct our analyses in L1 because this layer contains sparse neuronal cell bodies and receives dense axonal inputs from both thalamic and neighboring regions. 5.1 mm-thick confocal images (optical section depth 0.33 µm, 15 sections/scan) were acquired at 60× magnification plus 1.4× optical zoom using an Olympus FV3000 confocal microscope or Zeiss 880.
Synapse quantification
Maximum projections of 3 consecutive optical sections were generated using ImageJ. The Puncta Analyzer Plugin (available at: https://doi.org/10.5281/zenodo.6800214) for ImageJ was used to count the number of colocalized synaptic puncta. The individual analyzing the images was always blinded to the experimental conditions. At least 5 maximum projections per brain, from 3 brain sections per animal, were analyzed using a nested t-test. For synapse quantification, sectioning, staining, imaging, and analysis were run side by side for each condition within specific sex and age. Because of the required pairing, data were graphed as the synapse change from sex-specific control ((DEP+MS- CON)/CON). To determine sex differences an unpaired t-test was performed between synapse change, to determine change from sex-specific control, a one-sample t-test was performed.
Cell staining
Free-floating sections were washed 3 times for 10 minutes with TBS with 0.5% Triton X-100 (Roche, Indianapolis, IN) and blocked in 5% Normal Goat Serum (NGS; Jackson ImmunoResearch, West Grove, PA) with 0.5% Triton X-100 in TBS for 1 hour at room temperature. Primary antibodies (see table) were diluted in 5% NGS in TBS with 0.5% Triton X-100. Sections were incubated overnight at 4°C with primary antibodies and washed three times for 10 minutes with TBS the following day. Secondary Alexa-fluorophore conjugated antibodies (Invitrogen, Carlsbad, CA) were diluted (1:500) in 5% NGS in TBS with 0.5% Triton X-100, and sections were incubated with secondary antibodies for 2 hours at room temperature, protected from light. During the last five minutes of secondary incubation, DAPI was added to achieve a dilution of 1:40,000 (ThermoFisher D1306). After incubation, sections were washed three times for 15 minutes in TBS and mounted with an in-house mounting media (20 mM Tris pH8.0, 90% Glycerol, 0.5% N-propyl gallate). Images were acquired on an Olympus FV3000 confocal laser-scanning microscope.
Microglia staining for cell counts
Free-floating sections were washed 3 times for 10 minutes with TBS with 0.5% Triton X-100 (Roche, Indianapolis, IN) and blocked in 5% Normal Donkey Serum (NDS; Jackson ImmunoResearch, West Grove, PA) with 0.5% Triton X-100 in TBS for 1 hour at room temperature. Primary antibodies (see table) were diluted in 5% NDS in TBS with 0.5% Triton X-100. Sections were incubated overnight at room temperature with primary antibodies and washed three times for 10 minutes with TBS the following day. Secondary Alexa-fluorophore conjugated antibodies (Invitrogen, Carlsbad, CA) were diluted (1:500) in 5% NDS in TBS with 0.5% Triton X-100, and sections were incubated with secondary antibodies for 2 hours at room temperature, protected from light. During the last five minutes of secondary incubation, DAPI was added to achieve a dilution of 1:40,000 (ThermoFisher D1306). After incubation, sections were washed three times for 15 minutes in TBS and mounted with an in-house mounting media (20 mM Tris pH8.0, 90% Glycerol, 0.5% N-propyl gallate). Images were acquired on an Olympus FV3000 confocal laser-scanning microscopes.
Acquisition and analysis for cell counts of neurons, astrocytes, and oligodendrocytes
Cell counts for NeuN, Olig2, and Sox9 were performed in male and female CON and DEP+MS offspring at P8. Coronal brain sections (40 mm) containing the ACC triple labeled with NeuN, Olig2, and Sox9. Confocal z-stacks of the ACC were acquired using the 30×objective on an Olympus FV3000 microscope. Tile scans of 10 µm z-stacks were acquired for the entire ACC using a 1.0 µm step size. To expedite imaging, a neural network was trained to denoise images from the resonant scanner (Weigert et al., 2018). Briefly, high-resolution images were acquired using the galvanometer scanner. Gaussian noise was added to reduce the signal-to-noise ratio to levels expected from the resonant scanner. These degraded images were used to train a neural network. This neural network model was applied to images acquired using a resonant scanner with the same objective and confocal. An example of pre-and post-processed images can be viewed in Figure S5A. Restored images were stitched using the grid/collection stitching feature in Fiji (1.52p). Images were max-projected, and an ROI of the ACC was applied. The ROI was pseudo-layered into bins of 160-micron lengths from the midline to layer 6 of the ACC. Incomplete layers were not counted. Automated cell counting was performed using the U-Net deep neural network; an example of automated segmentation for neuronal marker NeuN can be seen in Figure 5B (Falk et al., 2019). Separate models were trained for each individual marker (NeuN, Olig2, and Sox9) and were manually verified for accuracy. A lab-specific pipeline was generated using Python and is available at https://doi.org/10.5281/zenodo.6800011.
Acquisition and analysis for cell counts of microglia
Microglia cell counts of Iba1 and/or P2ry12 positive cells were performed in male and female CON and DEP+MS offspring at P8, P15, and P25. Coronal brain sections containing the ACC triple labeled with P2ry12, Iba1, and CD68. Confocal z-stacks of the ACC were acquired using the 30x silicone objective on an Olympus FV3000 microscope. Tile scans of 10-micron z-stacks were acquired for the entire ACC using a 1.0-micron step size. Images were stitched using Olympus software, images were maximum projected, and an ROI of the ACC was applied. The ROI was pseudo-layered into bins of 160-micron lengths from the midline to layer 6 of the ACC. Incomplete layers were not counted. Cell counts were manually performed in Fiji using the cell counter feature. Cells were counted when positive for DAPI and a microglial cell marker (Iba1 and/or P2ry12). A total number of microglia cells includes cells positive for DAPI and 1 or 2 microglial markers. Heterogenous microglia cells are considered cells that are expressing only high levels of one microglia marker (Iba1hiP2ry12lo, Iba1loP2ry12hi). Percent heterogeneity was quantified as the number of singly high labeled microglia over a total number of all microglia cells.
Acquisition and analysis for Cd68 quantification in heterogeneous microglia
CD68 content was quantified in heterogenous microglia at P8 in CON and DEP+MS male offspring. 40 µm coronal brain sections containing the ACC were triple labeled with Iba1, P2ry12, and CD68. Confocal z-stacks of the ACC were acquired using the 30× objective on an Olympus FV3000 microscope. Using the resonant scanner, tile scans of 20 µm z-stacks were acquired for the entire ACC using a 0.35 µm step size. Images were stitched using the grid/collection stitching feature of Fiji (1.52p). To enable the expedited acquisition of large tile scanned images, a deep neural network (U-Net) was utilized to denoise images (Weigert et al., 2018). Imaris software 9.5.1 was used to create surface renderings of individual microglia cells labeled with either Iba1 or P2y12, incomplete or poorly labeled cells were excluded from analyses, Cd68 content within microglia surface makers was quantified. After surface renderings, cells were identified as single or double-positive, and Cd68 content was normalized to cell volume.
Microglia engulfment staining
Free-floating sections were washed 3 times for 10 minutes with TBS with 0.5% Triton X-100 (Roche, Indianapolis, IN) and blocked in either 5% Normal Goat Serum or 5% Normal Donkey Serum (NGS; Jackson ImmunoResearch, West Grove, PA, NDS; Jackson ImmunoResearch, West Grove, PA) with 0.5% Triton X-100 in TBS for 1 hour at room temperature. Primary antibodies (see table below) were diluted in 5% NGS or NDS TBS with 0.5% Triton X-100. Sections were incubated overnight at room temperature with primary antibodies and washed three times for 10 minutes with TBS the following morning. Secondary Alexa-fluorophore conjugated antibodies (Invitrogen, Carlsbad, CA) were diluted (1:500) in 5% NGS or NDS in TBS with 0.5% Triton X-100, and sections were incubated with secondary antibodies for 2 hours at room temperature, protected from light. During the last five minutes of secondary incubation, DAPI was added to achieve a dilution of 1:40,000 (ThermoFisher D1306). After incubation, sections were washed three times for 15 minutes in TBS and mounted with an in-house mounting media (20 mM Tris pH8.0, 90% Glycerol, 0.5% N-propyl gallate). Images were acquired on an Olympus FV3000 confocal laser-scanning microscopes.
Acquisition and analysis of synaptic engulfment
Synaptic engulfment was performed in two pairs of WT C57BL6/J male offspring at P6, P8, P10, P13, and P15 to determine the normal developmental pattern of synapse elimination in the ACC. Synapse engulfment was quantified at P8 and P10 in CON and DEP+MS male offspring. For WT characterization and P10 analysis, 40 um coronal brain sections containing the ACC were stained for Iba1 and P2ry12 on the same fluorophore, Cd68, and VGlut2. For P8 engulfment analyses, coronal brain sections containing the ACC were stained for Iba1, P2ry12, and VGlut2, all on separate fluorophores. Due to the limit of fluorophores and inadequate stability of the 450 fluorophores, CD68 was excluded from these analyses. Confocal z-stacks of the ACC were acquired using the 60× oil objective on an Olympus FV3000 microscope (experimental groups) or Zeiss 880 (WT characterization), with 2.0× zoom. An entire microglia cell was captured with 0.35 um step size. Huygens Professional 19.10.0p3 64b was used to deconvolve images. Imaris 9.5.1 was used to create surface renderings of individual microglia cells, Cd68 within microglia surface and VGlut2 within microglia surface. The volume of phagocytes and engulfed synapses is normalized to cell volume.
Neonatal stereotaxic microinjection of NIF
Neutrophil inhibitory factor was prepared in sterile PBS according to the manufacturer’s recommendations at a concentration of 200 ng/uL (NIF; R&D Systems, 5845-NF-050, Minneapolis, MN).
Naïve WT (C57BL/6J) breeders were set up to generate WT pups. Stud males were removed prior to the birth of pups, and the day of birth was considered P0. Only male offspring were utilized for NIF experiments. Male pups from a single litter were pseudorandomized and divided into PBS or NIF conditions so that each litter would contain both PBS and NIF injected mice.
On postnatal day 7, mice were anesthetized and placed in a stereotaxic device specially adapted for neonatal mice. The ACC was targeted by experimentally determining bregma coordinates in neonatal offspring (P7 Anterior Cingulate Cortex: 0.0 mm AP, ±0.7mm ML, −1.5 mm DV). 1 uL of sterile PBS or NIF was injected into the ACC. Pups were recovered on a heating pad and, once alert and ambulating, were returned to their home cage and monitored until collection at P8 or until behavioral testing between P29-P35.
Immunohistochemistry in PBS vs. NIF animals
Mice used for IHC were collected at postnatal day 8, 24 hours after surgery or between P32-P40 after the completion of behavioral testing. Mice were anesthetized with 200 mg/kg tribromoethanol (avertin) and perfused with Tris-Buffered Saline (TBS, 25 mM Trisbase, 135 mM NaCl, 3 mM KCl, pH 7.6) supplemented with 7.5 mM heparin, followed by 4% PFA in TBS. Brains were extracted and post-fixed in 4% PFA in TBS overnight at 4C. After fixation, brains were washed 3 times with TBS and transferred to a 30% sucrose/TBS solution for cryoprotection. Brains were frozen and embedded into a solution containing 2 parts, 30% sucrose, and 1 part OCT (Tissue Tek, Sakura, Torrance, CA), and stored at −80°C. Brains were sectioned at 40 mm. Tissue sections were stored floating in a 1:1 mixture of TBS/glycerol at −20°C.
Microglial CD68 and VGlut2 staining
Free-floating sections were washed 3 times for 10 minutes with TBS with 0.3–0.5% (0.3 for VGlut2, 0.5% for CD68) Triton X-100 (Roche, Indianapolis, IN) and blocked in 5% Normal Donkey Serum (NDS; Jackson ImmunoResearch, West Grove, PA) with 0.3–0.5% Triton X-100 in TBS for 1 hour at room temperature. Primary antibodies (see table below) were diluted in 5% NDS TBS with 0.3–0.5% Triton X-100. Sections were incubated in primary antibody overnight at room temperature, followed by incubation at 4C for 24 hours. Sections were washed three times for 10 minutes with TBS the following morning. Secondary Alexa-fluorophore conjugated antibodies (Invitrogen, Carlsbad, CA) were diluted (1:500) in 5% NDS in TBS with 0.3–0.5% Triton X-100, and sections were incubated with secondary antibodies for 2 hours at room temperature, protected from light. During the last five minutes of secondary incubation, DAPI was added to achieve a dilution of 1:40,000 (ThermoFisher D1306). After incubation, sections were washed three times for 15 minutes in TBS and mounted with an in-house mounting media (20 mM Tris pH8.0, 90% Glycerol, 0.5% N-propyl gallate). Images were acquired on an Olympus FV3000 confocal laser-scanning microscopes.
Synaptic staining
Free-floating sections were washed 3 times for 10 minutes with TBS with 0.2% Triton X-100 (Roche, Indianapolis, IN) and blocked in 5% Normal Goat Serum (NGS; Jackson ImmunoResearch, West Grove, PA) with 0.2% Triton X-100 in TBS for 1 hour at room temperature. Primary antibodies (see table below) were diluted in 5% NGS in TBS with 0.2% Triton X-100. Sections were incubated overnight at 4°C with primary antibodies and washed three times for 10 minutes with TBS the following morning. Secondary Alexa-fluorophore conjugated antibodies (Invitrogen, Carlsbad, CA) were diluted (1:200) in 5% NGS in TBS with 0.2% Triton X-100, and sections were incubated with secondary antibodies for 2 hours at room temperature, protected from light. After incubation, sections were washed three times for 15 minutes in TBS and mounted with VECTASHIELD with DAPI (Vector Laboratories, Burlingame, CA). Images were acquired on an Olympus FV3000 confocal laser-scanning microscope.
Acquisition and analysis of synaptic staining
Staining, image acquisition, and analysis were performed as in Ippolito and Eroglu (2010) (Ippolito and Eroglu, 2010) with adjustments. Synaptic staining was performed in two male/female littermate pairs at P6, P8, P10, P13, and P15 in WT C57BL6/J offspring to determine the normal developmental pattern. Synaptic staining was performed at P8, P15, and P100 in male and female offspring for CON and DEP+MS conditions. Image acquisition was performed in layer 1 (L1) of the ACC from P8, P15, and adult CON and DEP+MS animals. We chose to conduct our analyses in L1 because this layer contains sparse neuronal cell bodies and receives dense axonal inputs from both thalamic and neighboring regions. 5.1 mm-thick confocal images (optical section depth 0.33 µm, 15 sections/scan) were acquired at 60× magnification plus 1.4× optical zoom using an Olympus FV3000 confocal microscope or Zeiss 880.
Synapse quantification
Maximum projections of 3 consecutive optical sections were generated using ImageJ. The Puncta Analyzer Plugin (available at: https://doi.org/10.5281/zenodo.6800214) for ImageJ was used to count the number of colocalized synaptic puncta. The individual analyzing the images was always blinded to the experimental conditions. At least 5 maximum projections per brain, from 3 brain sections per animal, were analyzed using a nested t-test.
Acquisition and analysis for Cd68 quantification in PBS and NIF microglia
CD68 content was quantified in microglia at P8 in PBS and NIF male offspring. 40 µm coronal brain sections containing the ACC were triple labeled with Iba1, P2ry12, and CD68. Confocal z-stacks of the ACC were acquired using the 30× objective on an Olympus FV3000 microscope. Using the Galvano scanner, tile scans of 30 µm z-stacks were acquired for the entire ACC using a 0.35 mm step size. Images were stitched using Olympus software. Imaris software 9.5.1 was used to create surface renderings of individual microglia cells labeled with both Iba1 or P2y12, incomplete or poorly labeled cells were excluded from analyses, Cd68 content within microglia surface makers was quantified. After surface renderings, cells were identified as single or double-positive, and Cd68 content was normalized to cell volume.
Acquisition and analysis of synaptic engulfment
VGlut2 engulfment was quantified in microglia at P8 in PBS and NIF male offspring. 40 µm coronal brain sections containing the ACC were triple labeled with Iba1, P2ry12, and VGlut2. Confocal z-stacks of the ACC were acquired using the 60× oil objective on an Olympus FV3000 microscope with 2.0× zoom. An entire microglia cell was captured with 0.35 um step size. Huygens Professional 19.10.0p3 64b was used to deconvolve images. Imaris 9.5.1 was used to create surface renderings of individual microglia cells, as well as VGlut2 within the microglia surface.
Juvenile social preference in PBS vs. NIF males
Mice microinjected with PBS or NIF at P7 were weaned with their sex-matched littermates and were tested in an identical social preference task as previously described. Briefly, mice were habituated to the testing room (1 hour) and testing chambers (5 minutes) the day before sociability testing. On the day of testing, mice were habituated to the testing room for 1 hour. Each test mouse was placed in the center of a 3-chambered test with an inanimate object (rubber duck) confined on one side of the test and a novel age- and sex-matched conspecific confined on the other side for 5 minutes. All behavior was recorded, and hand quantified using Solomon Coder by an observer blind to treatment.
The social preference for each session was defined as:
where InteractionTimeS is the total time spent proximal to the other mouse, and InteractionTimeO is the total time spent proximal to the object.
Ethovision was used to determine the total distance moved and average velocity for each test animal and to generate heatmaps.
QUANTIFICATION AND STATISTICAL ANALYSIS
We analyzed all data using GraphPad Prism version 8.0 (San Diego, CA), MATLAB Version 2017a (Natick, MA), or TIBCO Statistica Software version 13.5.0.17 (Palo Alto, CA). A one-way sample t-test was used to compare data sets against expected means, the student’s t-test was used to analyze data sets with two groups or a Rank-sum test for nonparametric data. One-way ANOVAs were used to analyze data sets with more than two groups. Two-way ANOVAs were used to analyze data sets with two independent variables. Nested analyses were performed for sets of data using biological replicates. Levene’s test for homogeneity of variance was used to determine differences in distribution. Spearman’s correlation was used to test the relationship between two variables, and analysis of covariance was used to test the regression pattern between two groups. All data are represented as mean ± SEM. See supplemental material for detailed statistical measures for all analyses.
Supplementary Material
Highlights.
Prenatal exposure to air pollution and stress (DEP + MS) induces MIA
DEP + MS induces social/communication and circuit deficits in male offspring
DEP + MS impairs microglial pruning of developing thalamocortical synapses
Inhibition of microglial phagocytosis phenocopies social deficits observed in DEP + MS
ACKNOWLEDGMENTS
We thank Dr. Ian Gilmour, Environmental Protection Agency, for the DEP. We thank Michael Muehlbauer from the Duke Molecular Physiology Institute Metabolomics Research Group for aiding in running the U-Plex MSD. We thank Duke Genomic Analysis and Bioinformatics Shared Resource for sequencing and data analysis assistance. We thank the Duke Light Microscopy Core Facility for access to microscopes and software for image analysis. We thank Michael D. Gunn for the BAC transgenic mice. BioRender was used to generate elements of main Figures 1, 3, 4, 7, and S7. This work was supported by a grant from the National Institute of Environmental Health Sciences (R01ES025549 awarded to S.D.B, C.E., and K.D.). C.L.B. was supported by the Dean’s Graduate Fellowship and an NSF GFRP DGE 1644868 fellowship. K.A.B. was funded by Trinity College of Arts & Sciences, Duke University. C.S. was supported by the Summer Undergraduate Research Fellowships in Cell Biology. K.D., D.E.C., and A.T. were supported by a WM Keck Foundation grant (awarded to K.D.). Animal housing and experimental supplies and effort for K.D. and S.D.M. were covered by an NIEHS grant R01ES025549 (awarded to S.D.B., C.E., and K.D.). K.D., C.B., and S.D.M. were covered by an NIH grant R01MH120158 (awarded to K.D.). K.D., D.E.C., and A.T. were covered by an NIH grant 1R01EB026937 (awarded to D.E.C. and K.D.). C.J.S. was covered by the National Institute of Environmental Health Sciences F32ES029912 (awarded to C.J.S.). A.M.C. was covered by the Eunice Kennedy Shriver National Institute of Child Health and Human Development F32HD104430 (awarded to A.M.C.). C.E. is an investigator of the Howard Hughes Medical Institute. This work was also supported by the Robert and Donna E. Landreth Family Fund.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2022.111161.
DECLARATION OF INTEREST
The authors declare no competing interests.
INCLUSION AND DIVERSITY
We worked to ensure sex balance in selecting non-human subjects. One or more of the authors of this paper identifies as an underrepresented ethnic minority in science. One or more of the authors of this paper received support from a program designed to increase minority representation in science.
REFERENCES
- Apprill A, McNally S, Parsons R, and Weber L (2015). Minor revision to V4 region SSU rRNA 806R gene primer greatly increases detection of SAR11 bacterioplankton. Aquat. Microb. Ecol 75, 129–137. [Google Scholar]
- Ayata P, Badimon A, Strasburger HJ, Duff MK, Montgomery SE, Loh YHE, Ebert A, Pimenova AA, Ramirez BR, Chan AT, et al. (2018). Epigenetic regulation of brain region-specific microglia clearance activity. Nat. Neurosci 21, 1049–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnett L, and Seth AK (2014). The MVGC multivariate Granger causality toolbox: a new approach to Granger-causal inference. J. Neurosci. Methods 223, 50–68. [DOI] [PubMed] [Google Scholar]
- Bennett PJG, Maier E, and Brecht M (2019). Involvement of rat posterior prelimbic and cingulate area 2 in vocalization control. Eur. J. Neurosci 50, 3164–3180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroger A, Bletsch A, Krick C, Siniatchkin M, Jarczok TA, Freitag CM, and Bender S (2014). Visual event-related potentials to biological motion stimuli in autism spectrum disorders. Soc. Cogn. Affect. Neurosci 9, 1214–1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolton JL, Huff NC, Smith SH, Mason SN, Foster WM, Auten RL, and Bilbo SD (2013). Maternal stress and effects of prenatal air pollution on offspring mental health outcomes in mice. Environ. Health Perspect 121, 1075–1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolton JL, Marinero S, Hassanzadeh T, Natesan D, Le D, Belliveau C, Mason SN, Auten RL, and Bilbo SD (2017). Gestational exposure to air pollution alters cortical volume, microglial morphology, and microglia-neuron interactions in a sex-specific manner. Front. Synaptic Neurosci 9, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borucki DM, Toutonji A, Couch C, Mallah K, Rohrer B, and Tomlinson S (2020). Complement-mediated microglial phagocytosis and pathological changes in the development and Degeneration of the visual system. Front. Immunol 11, 566892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle CA, Boulet S, Schieve LA, Cohen RA, Blumberg SJ, Yeargin-Allsopp M, Visser S, and Kogan MD (2011). Trends in the prevalence of developmental disabilities in US children. Pediatrics 127, 1034–1042. [DOI] [PubMed] [Google Scholar]
- Byers SL, Wiles MV, Dunn SL, and Taft RA (2012). Mouse estrous cycle identification tool and images. PLoS One 7, e35538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Lozupone CA, Turnbaugh PJ, Fierer N, and Knight R (2011). Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. USA 108, 4516–4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Huntley J, Fierer N, Owens SM, Betley J, Fraser L, Bauer M, et al. (2012). Ultrahigh-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J 6, 1621–1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlezon WA Jr., Kim W, Missig G, Finger BC, Landino SM, Alexander AJ, Mokler EL, Robbins JO, Li Y, Bolshakov VY, et al. (2019). Maternal and early postnatal immune activation produce sex-specific effects on autism-like behaviors and neuroimmune function in mice. Sci. Rep 9, 16928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chabout J, Sarkar A, Dunson DB, and Jarvis ED (2015). Male mice song syntax depends on social contexts and influences female preferences. Front. Behav. Neurosci 9, 76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chabout J, Jones-Macopson J, and Jarvis ED (2017). Eliciting and analyzing male mouse ultrasonic vocalization (USV) songs. J. Vis. Exp, 54137. [DOI] [PMC free article] [PubMed]
- Choi GB, Yim YS, Wong H, Kim S, Kim H, Kim SV, Hoeffer CA, Littman DR, and Huh JR (2016). The maternal interleukin-17a pathway in mice promotes autism-like phenotypes in offspring. Science 351, 933–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung WS, Clarke LE, Wang GX, Stafford BK, Sher A, Chakraborty C, Joung J, Foo LC, Thompson A, Chen C, et al. (2013). Astrocytes mediate synapse elimination through MEGF10 and MERTK pathways. Nature 504, 394–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Amato FR, Scalera E, Sarli C, and Moles A (2005). Pups call, mothers rush: does maternal responsiveness affect the amount of ultrasonic vocalizations in mouse pups? Behav. Genet 35, 103–112. [DOI] [PubMed] [Google Scholar]
- Dawson G, Webb SJ, and McPartland J (2005). Understanding the nature of face processing impairment in autism: insights from behavioral and electrophysiological studies. Dev. Neuropsychol 27, 403–424. [DOI] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, and Gingeras TR (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Environmental Protection Agency, U.S. (2017). Neurodevelopmental disorders. In America’s Children and the Environment, Third, EPA., ed. (Create-Space Independent Publishing Platform; ), pp. 233–251. [Google Scholar]
- Eyo UB, Peng J, Swiatkowski P, Mukherjee A, Bispo A, and Wu LJ (2014). Neuronal hyperactivity recruits microglial processes via neuronal NMDA receptors and microglial P2Y12 receptors after status epilepticus. J. Neurosci 34, 10528–10540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falk T, Mai D, Bensch R, Çiçek Ö, Abdulkadir A, Marrakchi Y, Böhm A, Deubner J, Jäckel Z, Seiwald K, et al. (2019). U-Net: deep learning for cell counting, detection, and morphometry. Nat. Methods 16, 67–70. [DOI] [PubMed] [Google Scholar]
- Gandal MJ, Zhang P, Hadjimichael E, Walker RL, Chen C, Liu S, Won H, van Bakel H, Varghese M, Wang Y, et al. (2018). Transcriptome-wide isoform-level dysregulation in ASD, schizophrenia, and bipolar disorder. Science 362, eaat8127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geweke J (1982). Measurement of Linear-Dependence and feedback between multiple time-series. J. Am. Stat. Assoc 77, 304–313. [Google Scholar]
- Gilman SR, Iossifov I, Levy D, Ronemus M, Wigler M, and Vitkup D (2011). Rare de novo variants associated with autism implicate a large functional network of genes involved in formation and function of synapses. Neuron 70, 898–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo B, Chen J, Chen Q, Ren K, Feng D, Mao H, Yao H, Yang J, Liu H, Liu Y, et al. (2019). Anterior cingulate cortex dysfunction underlies social deficits in Shank3 mutant mice. Nat. Neurosci 22, 1223–1234. [DOI] [PubMed] [Google Scholar]
- Hammerschmidt K, Radyushkin K, Ehrenreich H, and Fischer J (2009). Female mice respond to male ultrasonic ‘songs’ with approach behaviour. Biol. Lett 5, 589–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond TR, Dufort C, Dissing-Olesen L, Giera S, Young A, Wysoker A, Walker AJ, Gergits F, Segel M, Nemesh J, et al. (2019). Single-cell RNA sequencing of microglia throughout the mouse Lifespan and in the injured brain reveals complex cell-state changes. Immunity 50, 253–271.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanamsagar R, and Bilbo SD (2016). Sex differences in neurodevelopmental and neurodegenerative disorders: focus on microglial function and neuroinflammation during development. J. Steroid Biochem. Mol. Biol 160, 127–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanamsagar R, Alter MD, Block CS, Sullivan H, Bolton JL, and Bilbo SD (2017). Generation of a microglial developmental index in mice and in humans reveals a sex difference in maturation and immune reactivity. Glia 65, 1504–1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes SE, Hollopeter G, Yang G, Kurpius D, Dailey ME, Gan WB, and Julius D (2006). The P2Y12 receptor regulates microglial activation by extracellular nucleotides. Nat. Neurosci 9, 1512–1519. [DOI] [PubMed] [Google Scholar]
- Heilbronner SR, and Hayden BY (2016). Dorsal anterior cingulate cortex: a bottom-up view. Annu. Rev. Neurosci 39, 149–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiao EY, and Patterson PH (2011). Activation of the maternal immune system induces endocrine changes in the placenta via IL-6. Brain Behav. Immun 25, 604–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber W, Carey VJ, Gentleman R, Anders S, Carlson M, Carvalho BS, Bravo HC, Davis S, Gatto L, Girke T, et al. (2015). Orchestrating high-throughput genomic analysis with Bioconductor. Nat. Methods 12, 115–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iidaka T, Kogata T, Mano Y, and Komeda H (2019). Thalamocortical hyperconnectivity and amygdala-cortical hypoconnectivity in male patients with autism spectrum disorder. Front. Psychiatry 10, 252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ippolito DM, and Eroglu C (2010). Quantifying synapses: an immunocytochemistry-based assay to quantify synapse number. J. Vis. Exp, 2270. [DOI] [PMC free article] [PubMed]
- Ito D, Imai Y, Ohsawa K, Nakajima K, Fukuuchi Y, and Kohsaka S (1998). Microglia-specific localisation of a novel calcium binding protein. Brain Res. Mol. Brain Res 57, 1–9. [DOI] [PubMed] [Google Scholar]
- Jayaraj RL, Rodriguez EA, Wang Y, and Block ML (2017). Outdoor ambient air pollution and neurodegenerative diseases: the neuroinflammation hypothesis. Curr. Environ. Health Rep 4, 166–179. [DOI] [PubMed] [Google Scholar]
- Jokisch D, Daum I, Suchan B, and Troje NF (2005). Structural encoding and recognition of biological motion: evidence from event-related potentials and source analysis. Behav. Brain Res 157, 195–204. [DOI] [PubMed] [Google Scholar]
- Kersey PJ, Staines DM, Lawson D, Kulesha E, Derwent P, Humphrey JC, Hughes DST, Keenan S, Kerhornou A, Koscielny G, et al. (2012). Ensembl Genomes: an integrative resource for genome-scale data from non-vertebrate species. Nucleic Acids Res 40, D91–D97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopec AM, Smith CJ, Ayre NR, Sweat SC, and Bilbo SD (2018). Microglial dopamine receptor elimination defines sex-specific nucleus accumbens development and social behavior in adolescent rats. Nat. Commun 9, 3769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson CP (2007). Poverty during pregnancy: its effects on child health outcomes. Paediatr. Child Health 12, 673–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YH, Cherkerzian S, Seidman LJ, Papandonatos GD, Savitz DA, Tsuang MT, Goldstein JM, and Buka SL (2020). Maternal bacterial infection during pregnancy and offspring risk of psychotic disorders: variation by severity of infection and offspring sex. Am. J. Psychiatry 177, 66–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lees KR, Diener HC, Asplund K, and Krams M; UK-279276–301 Study Investigators (2003). UK-279, 276, a neutrophil inhibitory glycoprotein, in acute stroke: tolerability and pharmacokinetics. Stroke 34, 1704–1709. [DOI] [PubMed] [Google Scholar]
- Levesque S, Surace MJ, McDonald J, and Block ML (2011). Air pollution & the brain: subchronic diesel exhaust exposure causes neuroinflammation and elevates early markers of neurodegenerative disease. J. Neuroinflammation 8, 105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levesque S, Taetzsch T, Lull ME, Johnson JA, McGraw C, and Block ML (2013). The role of MAC1 in diesel exhaust particle-induced microglial activation and loss of dopaminergic neuron function. J. Neurochem 125, 756–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Cheng Z, Zhou L, Darmanis S, Neff NF, Okamoto J, Gulati G, Bennett ML, Sun LO, Clarke LE, et al. (2019). Developmental heterogeneity of microglia and brain myeloid cells revealed by deep single-cell RNA sequencing. Neuron 101, 207–223.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liddelow SA, Guttenplan KA, Clarke LE, Bennett FC, Bohlen CJ, Schirmer L, Bennett ML, Münch AE, Chung WS, Peterson TC, et al. (2017). Neurotoxic reactive astrocytes are induced by activated microglia. Nature 541, 481–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lituma PJ, Woo E, O’Hara BF, Castillo PE, Sibinga NES, and Nandi S (2021). Altered synaptic connectivity and brain function in mice lacking microglial adapter protein Iba1. Proc. Natl. Acad. Sci. USA 118. e2115539118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, and Anders S (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mague SD, Talbot A, Blount C, Walder-Christensen KK, Duffney LJ, Adamson E, Bey AL, Ndubuizu N, Thomas GE, Hughes DN, et al. (2022). Brain-wide electrical dynamics encode individual appetitive social behavior. Neuron 110, 1728–1741.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J 17, 10–12. [Google Scholar]
- Masuda T, Sankowski R, Staszewski O, Böttcher C, Amann L, Sagar, Scheiwe C, Nessler S, Kunz P, van Loo G, et al. (2019). Spatial and temporal heterogeneity of mouse and human microglia at single-cell resolution. Nature 566, 388–392. [DOI] [PubMed] [Google Scholar]
- Matcovitch-Natan O, Winter DR, Giladi A, Vargas Aguilar S, Spinrad A, Sarrazin S, Ben-Yehuda H, David E, Zelada González F, Perrin P, et al. (2016). Microglia development follows a stepwise program to regulate brain homeostasis. Science 353, aad8670. [DOI] [PubMed] [Google Scholar]
- McGuinn LA, Windham GC, Messer LC, Di Q, Schwartz J, Croen LA, Moody EJ, Rappold AG, Richardson DB, Neas LM, et al. (2019). Air pollution, neighborhood deprivation, and autism spectrum disorder in the Study to Explore Early Development. Environ. Epidemiol 3, e067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi H, Muruganujan A, Ebert D, Huang X, and Thomas PD (2019). PANTHER version 14: more genomes, a new PANTHER GO-slim and improvements in enrichment analysis tools. Nucleic Acids Res 47, D419–D426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J, Puigserver P, Carlsson E, Ridderstråle M, Laurila E, et al. (2003). PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet 34, 267–273. [DOI] [PubMed] [Google Scholar]
- Moyle M, Foster DL, McGrath DE, Brown SM, Laroche Y, De Meutter J, Stanssens P, Bogowitz CA, Fried VA, Ely JA, et al. (1994). A hookworm glycoprotein that inhibits neutrophil function is a ligand of the integrin CD11b/CD18. J. Biol. Chem 269, 10008–10015. [PubMed] [Google Scholar]
- Nair A, Treiber JM, Shukla DK, Shih P, and Müller RA (2013). Impaired thalamocortical connectivity in autism spectrum disorder: a study of functional and anatomical connectivity. Brain 136, 1942–1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura K, Hioki H, Fujiyama F, and Kaneko T (2005). Postnatal changes of vesicular glutamate transporter (VGluT)1 and VGluT2 immunoreactivities and their colocalization in the mouse forebrain. J. Comp. Neurol 492, 263–288. [DOI] [PubMed] [Google Scholar]
- O’Callaghan E, Sham PC, Takei N, Murray G, Glover G, Hare EH, and Murray RM (1994). The relationship of schizophrenic births to 16 infectious diseases. Br. J. Psychiatry 165, 353–356. [DOI] [PubMed] [Google Scholar]
- Ohsawa K, Irino Y, Sanagi T, Nakamura Y, Suzuki E, Inoue K, and Kohsaka S (2010). P2Y12 receptor-mediated integrin-beta1 activation regulates microglial process extension induced by ATP. Glia 58, 790–801. [DOI] [PubMed] [Google Scholar]
- Okabe S, Nagasawa M, Kihara T, Kato M, Harada T, Koshida N, Mogi K, and Kikusui T (2013). Pup odor and ultrasonic vocalizations synergistically stimulate maternal attention in mice. Behav. Neurosci 127, 432–438. [DOI] [PubMed] [Google Scholar]
- Oudin A, Frondelius K, Haglund N, Källén K, Forsberg B, Gustafsson P, and Malmqvist E (2019). Prenatal exposure to air pollution as a potential risk factor for autism and ADHD. Environ. Int 133, 105149. [DOI] [PubMed] [Google Scholar]
- Ouhaz Z, Fleming H, and Mitchell AS (2018). Cognitive functions and neurodevelopmental disorders involving the prefrontal cortex and mediodorsal thalamus. Front. Neurosci 12, 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paolicelli RC, Bolasco G, Pagani F, Maggi L, Scianni M, Panzanelli P, Giustetto M, Ferreira TA, Guiducci E, Dumas L, et al. (2011). Synaptic pruning by microglia is necessary for normal brain development. Science 333, 1456–1458. [DOI] [PubMed] [Google Scholar]
- Parada AE, Needham DM, and Fuhrman JA (2016). Every base matters: assessing small subunit rRNA primers for marine microbiomes with mock communities, time series and global field samples. Environ. Microbiol 18, 1403–1414. [DOI] [PubMed] [Google Scholar]
- Pardo CA, Vargas DL, and Zimmerman AW (2005). Immunity, neuroglia and neuroinflammation in autism. Int. Rev. Psychiatry 17, 485–495. [DOI] [PubMed] [Google Scholar]
- Patel S, Dale RC, Rose D, Heath B, Nordahl CW, Rogers S, Guastella AJ, and Ashwood P (2020). Maternal immune conditions are increased in males with autism spectrum disorders and are associated with behavioural and emotional but not cognitive co-morbidity. Transl. Psychiatry 10, 286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlova M, Lutzenberger W, Sokolov A, and Birbaumer N (2004). Dissociable cortical processing of recognizable and non-recognizable biological movement: analysing gamma MEG activity. Cereb. Cortex 14, 181–188. [DOI] [PubMed] [Google Scholar]
- Rafenberg C, and Annesi-Maesano I (2018). [Cost of hypertrophy due to intrauterine growth restriction attributable to air pollution in France]. Arch. Pediatr 25, 256–262. [DOI] [PubMed] [Google Scholar]
- Raz R, Roberts AL, Lyall K, Hart JE, Just AC, Laden F, and Weisskopf MG (2015). Autism spectrum disorder and particulate matter air pollution before, during, and after pregnancy: a nested case-control analysis within the Nurses’ Health Study II Cohort. Environ. Health Perspect 123, 264–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice F, Jones I, and Thapar A (2007). The impact of gestational stress and prenatal growth on emotional problems in offspring: a review. Acta Psychiatr. Scand 115, 171–183. [DOI] [PubMed] [Google Scholar]
- Rice CJ, Sandman CA, Lenjavi MR, and Baram TZ (2008). A novel mouse model for acute and long-lasting consequences of early life stress. Endocrinology 149, 4892–4900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieu P, Ueda T, Haruta I, Sharma CP, and Arnaout MA (1994). The A-domain of beta 2 integrin CR3 (CD11b/CD18) is a receptor for the hookworm-derived neutrophil adhesion inhibitor NIF. J. Cell Biol 127, 2081–2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risher WC, Patel S, Kim IH, Uezu A, Bhagat S, Wilton DK, Pilaz LJ, Singh Alvarado J, Calhan OY, Silver DL, et al. (2014). Astrocytes refine cortical connectivity at dendritic spines. Elife 3, e04047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts AL, Lyall K, Hart JE, Laden F, Just AC, Bobb JF, Koenen KC, Ascherio A, and Weisskopf MG (2013). Perinatal air pollutant exposures and autism spectrum disorder in the children of Nurses’ Health Study II participants. Environ. Health Perspect 121, 978–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossignol DA, Genuis SJ, and Frye RE (2014). Environmental toxicants and autism spectrum disorders: a systematic review. Transl. Psychiatry 4, e360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudolph MD, Graham AM, Feczko E, Miranda-Dominguez O, Rasmussen JM, Nardos R, Entringer S, Wadhwa PD, Buss C, and Fair DA (2018). Maternal IL-6 during pregnancy can be estimated from newborn brain connectivity and predicts future working memory in offspring. Nat. Neurosci 21, 765–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salter E, Lei G, Choi S, Ralph L, Zhang L, Jin F, Kadia A, Wang J, Georgiou J, and Collingridge G (2020). Complement C3-dependent glutamatergic synapse elimination in the developing hippocampus is region- and synapse-specific. Preprint at bioRxiv 10.1101/2020.05.20.106930. [DOI]
- Schafer DP, and Stevens B (2013). Phagocytic glial cells: sculpting synaptic circuits in the developing nervous system. Curr. Opin. Neurobiol 23, 1034–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer DP, Lehrman EK, Kautzman AG, Koyama R, Mardinly AR, Yamasaki R, Ransohoff RM, Greenberg ME, Barres BA, and Stevens B (2012). Microglia sculpt postnatal neural circuits in an activity and complement-dependent manner. Neuron 74, 691–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert D, Martens GJM, and Kolk SM (2015). Molecular underpinnings of prefrontal cortex development in rodents provide insights into the etiology of neurodevelopmental disorders. Mol. Psychiatry 20, 795–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh SK, Stogsdill JA, Pulimood NS, Dingsdale H, Kim YH, Pilaz LJ, Kim IH, Manhaes AC, Rodrigues WS Jr., Pamukcu A, et al. (2016). Astrocytes assemble thalamocortical synapses by bridging NRX1alpha and NL1 via hevin. Cell 164, 183–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sipe GO, Lowery RL, Tremblay MÈ, Kelly EA, Lamantia CE, and Majewska AK (2016). Microglial P2Y12 is necessary for synaptic plasticity in mouse visual cortex. Nat. Commun 7, 10905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens B, Allen NJ, Vazquez LE, Howell GR, Christopherson KS, Nouri N, Micheva KD, Mehalow AK, Huberman AD, Stafford B, et al. (2007). The classical complement cascade mediates CNS synapse elimination. Cell 131, 1164–1178. [DOI] [PubMed] [Google Scholar]
- Stogsdill JA, Ramirez J, Liu D, Kim YH, Baldwin KT, Enustun E, Ejikeme T, Ji RR, and Eroglu C (2017). Astrocytic neuroligins control astrocyte morphogenesis and synaptogenesis. Nature 551, 192–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swiatkowski P, Murugan M, Eyo UB, Wang Y, Rangaraju S, Oh SB, and Wu LJ (2016). Activation of microglial P2Y12 receptor is required for outward potassium currents in response to neuronal injury. Neuroscience 318, 22–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue KI, Takano H, Yanagisawa R, Hirano S, Ichinose T, Shimada A, and Yoshikawa T (2006). The role of toll-like receptor 4 in airway inflammation induced by diesel exhaust particles. Arch. Toxicol 80, 275–279. [DOI] [PubMed] [Google Scholar]
- Talbot A, Dunson D, Dzirasa K, and Carlson D (2020). Supervised autoencoders learn robust joint factor models of neural activity. Preprint at arXiv 10.48550/arXiv.2004.05209. [DOI]
- Trakoshis S, Martínez-Cañada P, Rocchi F, Canella C, You W, Chakrabarti B, Ruigrok AN, Bullmore ET, Suckling J, Markicevic M, et al. (2020). Intrinsic excitation-inhibition imbalance affects medial prefrontal cortex differently in autistic men versus women. Elife 9, e55684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulloa ER, and Pineda JA (2007). Recognition of point-light biological motion: mu rhythms and mirror neuron activity. Behav. Brain Res 183, 188–194. [DOI] [PubMed] [Google Scholar]
- Van Segbroeck M, Knoll AT, Levitt P, and Narayanan S (2017). MUPET-mouse ultrasonic profile ExTraction: a signal processing tool for rapid and unsupervised analysis of ultrasonic vocalizations. Neuron 94, 465–485.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vargas DL, Nascimbene C, Krishnan C, Zimmerman AW, and Pardo CA (2005). Neuroglial activation and neuroinflammation in the brain of patients with autism. Ann. Neurol 57, 67–81. [DOI] [PubMed] [Google Scholar]
- Velmeshev D, Schirmer L, Jung D, Haeussler M, Perez Y, Mayer S, Bhaduri A, Goyal N, Rowitch DH, and Kriegstein AR (2019). Single-cell genomics identifies cell type-specific molecular changes in autism. Science 364, 685–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voineagu I, Wang X, Johnston P, Lowe JK, Tian Y, Horvath S, Mill J, Cantor RM, Blencowe BJ, and Geschwind DH (2011). Transcriptomic analysis of autistic brain reveals convergent molecular pathology. Nature 474, 380–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, McCoy PA, Rodriguiz RM, Pan Y, Je HS, Roberts AC, Kim CJ, Berrios J, Colvin JS, Bousquet-Moore D, et al. (2011). Synaptic dysfunction and abnormal behaviors in mice lacking major isoforms of Shank3. Hum. Mol. Genet 20, 3093–3108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Bey AL, Katz BM, Badea A, Kim N, David LK, Duffney LJ, Kumar S, Mague SD, Hulbert SW, et al. (2016). Altered mGluR5-Homer scaffolds and corticostriatal connectivity in a Shank3 complete knockout model of autism. Nat. Commun 7, 11459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quesnel-Valliéres M, Weatheritt RJ, Cordes SP, and Blencowe BJ (2019). Autism spectrum disorder: insights into convergent mechanisms from transcriptomics. Nat. Rev. Genet 20, 51–63. [DOI] [PubMed] [Google Scholar]
- Weigert M, Schmidt U, Boothe T, Müller A, Dibrov A, Jain A, Wilhelm B, Schmidt D, Broaddus C, Culley S, et al. (2018). Content-aware image restoration: pushing the limits of fluorescence microscopy. Nat. Methods 15, 1090–1097. [DOI] [PubMed] [Google Scholar]
- Weinhard L, di Bartolomei G, Bolasco G, Machado P, Schieber NL, Neniskyte U, Exiga M, Vadisiute A, Raggioli A, Schertel A, et al. (2018). Microglia remodel synapses by presynaptic trogocytosis and spine head filopodia induction. Nat. Commun 9, 1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werling DM, Parikshak NN, and Geschwind DH (2016). Gene expression in human brain implicates sexually dimorphic pathways in autism spectrum disorders. Nat. Commun 7, 10717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White NR, Prasad M, Barfield RJ, and Nyby JG (1998). 40- and 70-kHz vocalizations of mice (Mus musculus) during copulation. Physiol. Behav 63, 467–473. [DOI] [PubMed] [Google Scholar]
- Smith CJW, Wilkins KB, Mogavero JN, and Veenema AH (2015). Social novelty investigation in the juvenile rat: modulation by the mu-opioid system. J. Neuroendocrinol 27, 752–764. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-sequencing data have been deposited at GEO and are publicly available as of the date of publication. Accession numbers are listed in the key resources table. Behavior and microscopy data reported in this paper will be shared by the lead contact upon request.
All original code has been deposited at GitHub and is publicly available as of the date of publication. DOIs are listed in the key resources table.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| NeuroTrace fluorescent Nissl Stain | Thermo Fisher Scientific | Cat#N21480 |
| Guinea pig anti-VGlut2 1:2000 | Synaptic Systems | Cat#135404; RRID: AB_887884 |
| Rabbit anti-PSD95 1:350 | Life Technologies | Cat#51–6900; RRID: AB_2533914 |
| Rat anti-CD68 1:500 | BioLegend | Cat#137002; RRID: AB_2044004 |
| Chicken anti-IBA1 1:1000 | Synaptic Systems | Cat#234006; RRID: AB_2619949 |
| Rabbit anti-P2y12 1:2000 | Anaspec | Cat#AS-55043A: RRID: AB_2298886 |
| Goat anti-Iba1 1:500 | Novus Biologicals | Cat#NB 100–1028; RRID: AB_521594 |
| Rabbit anti-Sox9 1:2000 | Millipore Sigma Aldrich | Cat#AB5535; RRID: AB_2239761 |
| Mouse anti-Olig2 1:2000 | Millipore Sigma Aldrich | Cat#MABN50; RRID: AB_10807410 |
| Mouse anti-NeuN 1:250 | Millipore Sigma Aldrich | Cat#MAB377; RRID: AB_2298772 |
| DAPI | Thermo Fisher Scientific | Cat#D1306 |
| Kwik-Diff™ | Thermo Fisher Scientific | Cat#99–907-00 |
|
Chemicals, peptides, and recombinant proteins | ||
| Diesel Exhaust Particles | EPA Gift from Ian Gilmour | N/A |
| TRIzol Reagent | Thermo Fisher Scientific | Cat#15596026 |
| Chloroform | Sigma-Aldrich | Cat#C2432 |
| Triton X-100 | Roche | Cat#11332481001 |
| 2-Propanol | Sigma-Aldrich | Cat#I9516–500ML |
| VECTASHIELD with DAPI | VWR | Cat#h-1200 |
| Normal Donkey Serum | Jackson ImmunoResearch | Cat#017–000-121 |
| Normal Goat Serum | Jackson ImmunoResearch | Cat#005–000-121 |
| Recombinant A. caninum NIF Protein, CF | R&D Systems | Cat#5845-NF-050 |
|
Critical commercial assays | ||
| U-Plex Proinflammatory Panel, Mouse | Meso Scale Discovery | Custom made |
| ELISA kit | Arbor Assays | Cat#K014-H1 |
| KAPA Stranded mRNA-Seq Kit | Roche | Cat#07962142001 |
|
Deposited data | ||
| Bulk RNA-sequencing of neonatal PFC in CON and DEP+MS male and females (Raw and analyzed data) | This paper | GEO: GSE160344 |
|
Experimental models: Organisms/strains | ||
| TLR4 KO Mice (C57BL/10ScNJ) | Jackson Labs | RRID:IMSR_JAX:003752 |
| WT Mice (C57BL/6J) | Jackson Labs | RRID:IMSR_JAX:000664 |
|
Software and algorithms | ||
| Avisoft-Ultrasound Gate recording software | Avisoft Bioacoustics | RRID:SCR_014436; http://www.avisoft.com/downloads/ |
| Avisoft SASLab Pro software | Avisoft Bioacoustics | RRID:SCR_014438 |
| MUPET v2.1 | Van Segbroeck et al., 2017 | https://github.com/mvansegbroeck/mupet |
| MATLAB Version 2017a | The MathWorks | RRID:SCR_001622; https://www.mathworks.com/ |
| R statistical programming environment | R Project for Statistical Computing | RRID:SCR_001905; https://www.r-project.org/ |
| PANTHER | Mi et al., 2019 | RRID:SCR_004869; http://www.pantherdb.org/ |
| FV31S-SW | Olympus Corporation | https://www.olympus-lifescience.com/en/support/downloads/ |
| ImageJ | NIH | RRID:SCR_003070; https://imagej.nih.gov/ij/ |
| Python 2019.3.3 | Python Programming Language | RRID:SCR_008394; http://www.python.org/ |
| Prism 8.0.0 | GraphPad Software, Inc. | RRID:SCR_002798; https://www.graphpad.com |
| Statistica Version 13.5.0.17 | TIBCO Statistica Software | RRID:SCR_014213; https://www.statisticssolutions.com/ |
| Imaris 9.5.1 | Oxford Instruments | RRID:SCR_007370; https://imaris.oxinst.com/ |
| Huygens Professional 19.10.0p3 64b | Scientific Volume Imaging | RRID:SCR_014237; https://svi.nl/Homepage |
| Cell count: U-net cell segmentation | Falk et al., 2019; Weigert et al., 2018; and this paper | Zenodo: https://doi.org/10.5281/zenodo.6800011 |
| Puncta Analyzer | Ippolito and Eroglu, 2010 | Zenodo: https://doi.org/10.5281/zenodo.6800214 |
| NMF Cross Frequency Spectral Analysis | Talbot et al., 2020 | Zenodo: https://doi.org/10.5281/zenodo.6078304 |
| Jwatcher | UCLA | RRID:SCR_017595; http://jwatcher.ucla.edu/ |
| Solomon Coder | Andras Peter | RRID:SCR_016041; https://solomon.andraspeter.com/ |
| Ethovision | Noldus Technology | RRID:SCR_000441; https://www.noldus.com/ethovision-xt |
|
Other | ||
| AlphaDri | Sheperd Specialty Papers | N/A |
| PicoLab Mouse Diet 5058 | Lab-Diet | N/A |
| Fine-gauge aluminum mesh platform | McNichols Co. | N/A |


