Abstract
Colony morphology has been used as an important identification and characterization criterion in bacteriology for many decades. However, the molecular mechanisms underlying the appearance of different colony types have been given little attention. The synthesis of O antigen is defunct in Escherichia coli K-12, and colonies should accordingly only appear to be rough. However, previous reports have noted the presence of different interchangeable colony morphology types. In this study we have addressed the influence of two phase-variable surface structures, antigen 43 and type 1 fimbriae, on colony morphology. Due to differential expression of these structures, four different colony phenotypes could be distinguished. By creating and studying defined mutants of the respective loci, i.e., flu and fim, we conclude that the presence or absence of the corresponding gene products on the cells correlates with the observed colony morphology forms. Interestingly, the habitat specificity of bacteria under static liquid conditions seems to correlate with the colony phenotypes.
In nature bacteria often grow as colonies on surfaces. Observation of colony development and morphology has for many years been an important tool for identification and characterization of bacteria because individual species often form colonies of characteristic size and appearance. However, even when a pure bacterial culture is plated out, different morphological colony types often arise. Traditionally, the colony morphology of Escherichia coli is identified as either a rough or a smooth form. The two forms are readily distinguished, as the colonies of the former are rough, flat, and irregular and colonies of the latter are smooth, high, and circular. E. coli K-12 strains are rough, their lipopolysaccharide having a complete core but no O antigen due to the insertion of an IS5 element in the rfb gene cluster controlling O antigen biosynthesis (23). Only extremely rarely are K-12 strains able to revert to the smooth form, as this requires the integrity of the rfb gene cluster to be restored. Nonetheless, K-12 strains have been observed to exhibit different interchangeable colony types.
Diderichsen (7) partially identified a mechanism controlling the colony morphology of K-12 strains. The observed phenotypes were somewhat similar to the ones seen for rough and smooth strains, but the strains frequently switched between the two. To distinguish these from the rough and smooth phenotypes related to lipopolysaccharide status, the rough, flat, and irregular variant was designated frizzy, or form 1, and the smooth, convex, circular variant was designated glossy, or form 2 (7, 14, 36). The locus responsible for this change in phenotype was mapped to 43 min on the E. coli K-12 chromosome and named flu (7).
In an independent study the product of the flu gene was investigated by virtue of its autoaggregative properties and association with the outer membrane and was termed antigen 43 (Ag43) (28). It was found to consist of two polypeptides termed α and β in a 1:1 ratio (29). Only recently was Ag43 unambiguously identified as the product of the flu gene (12, 15). Diderichsen (7) also in part identified the regulatory mechanism controlling the phase-variable expression of Ag43, since mutant strain BD1302 with a deletion in the 89-min region was locked in the frizzy (Ag43+) phenotype. This gene was later identified as the oxyR (mor) gene (13, 36). OxyR is an activator of a regulon of genes and additionally acts as an autorepressor (34, 35). OxyR is normally involved in protection against oxidative stress as it activates a regulon of peroxide-inducible genes (21, 22), but it also represses the flu gene (12, 15). Apart from OxyR, the Dam protein, responsible for methylating GATC sites of DNA, is also involved in the expression of Ag43. A dam mutant is, unlike an oxyR mutant, unable to express Ag43, whereas a double dam oxyR mutant is a constitutive Ag43 expressor (15). The combined effect of OxyR and Dam on flu results in phase-variable expression of Ag43.
Another surface feature of E. coli subject to phase variation is type 1 fimbriae encoded by the fim gene cluster, which confer mannose-sensitive adhesion to mannosylated receptor molecules. Type 1 fimbriae are found on the majority of E. coli strains and are widespread among members of the Enterobacteriaceae (reviewed in reference 19). The phase-variable expression of these rod-like appendages is quite different from that of Ag43. The fim gene cluster contains a promoter on a 314-bp invertible DNA fragment, viz., the fim switch (1, 8). Upstream of the switch are two recombinase-encoding genes, fimB and fimE (17). Under normal aerated growth conditions, FimB is able to catalyze inversion of the switch in both directions (10, 24), whereas FimE catalyzes only on-to-off inversion (2, 3, 10, 20), which leads to nonfimbriated cells.
A number of studies over the last decades have suggested a possible correlation between fimbriation and colony morphology in E. coli K-12 strains. Some strains gave rise to phase-variable colony forms: a small, convex, glossy form, which seemed to be concomitant with type 1 fimbriation, and a large, flat form, where the cells were nonfimbriated (3, 5, 7, 27). However, it was not clear whether type I fimbriation directly caused the observed colony morphology or whether expression of type 1 fimbriae was somehow concomitant with one of the forms. Furthermore, the Ag43 status of these colony types was not investigated. The data concerning the origin of phase variation of colony morphology in K-12 strains are therefore somewhat controversial: some reports relate the phenomenon to Ag43 expression and others to type 1 fimbriation. Against this background we decided to study the involvement of these two phase-variable systems with regard to colony morphology.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The strains and plasmids used in this study are described in Table 1. Cells were grown on solid medium or in liquid broth supplemented with the appropriate antibiotics unless otherwise stated.
TABLE 1.
Bacterial strains and plasmids
| Stain or plasmid | Relevant genotype and/or phenotype | Reference or construction |
|---|---|---|
| BD1428 | 7 | |
| CC118 | λpir on the chromosome | 16 |
| CSH50 | fimE::IS1 | 3 |
| HEHA4 | BD1428; Δfim::kan | This study |
| HEHA6 | BD1428; fimE+ | This study |
| HEHA9 | BD1428; ΔoxyR::Ω(Spr) | This study |
| HEHA11 | BD1428; flu::tet | This study |
| HEHA17 | HEHA11; Δfim::kan | This study |
| N9716 | GC4468; ΔoxyR::Ω(Spr) | G. Storz |
| PC31 | fim+ reference strain | 18 |
| pACYC184 | Camr Tetr | 6 |
| pBR322 | Ampr Tetr | 4 |
| pGP704 | Ampr; R6K-based origin (pir) | 16 |
| pHHA130 | Camr; oxyR+ | A 2,040-bp PCR fragment (primers 5 and 6) containing the oxyR gene from PC31 inserted into the HindIII site of pACYC184 |
| pHHA145 | Ampr Tetr; StyI site deleted | pBR322 cut with StyI, blunted with Klenow polymerase, and religated |
| pHHA146 | Ampr | A 3,522-bp PCR fragment (primers 1 and 2) containing the flu gene from PC31 inserted into EcoRI/BamHI site of pBR322 |
| pHHA154 | Ampr | A 3,550-bp PCR fragment containing the flu gene inserted into the EcoRV site of pHHA145 |
| pHHA159 | Ampr Tetr | A 1,724-bp BsaAI/SspI fragment containing the tet gene inserted into the (blunted) StyI site of pHHA154 |
| pHHA161 | Camr; fimE+-fimC+ | A 5,492-bp HindIII/EcoRI fragment containing the fimE to fimC genes inserted in the same sites of pMAK700oriT |
| pHHA165 | Ampr Tetr; flu::tet | A 5,450-bp EcoRI fragment containing the flu::tet construct inserted into the EcoRI site of pGP704 |
| pLBJ311 | Ampr | 32 |
| pMAK700oriT | Camr; Ts origin | T. Chakraborty |
| pMAS32 | Ampr; lacUV5::fimA-H | Part of the fim gene cluster (fimA to fimH) inserted behind the lacUV5 promoter |
DNA manipulations.
Isolation of plasmid DNA was carried out using the QIAprep Spin Miniprep kit (Qiagen). Restriction endonucleases were used according to the manufacturer's specifications (Biolabs). Chromosomal DNA purification was performed using the GenomicPrep cell and tissue DNA isolation kit (Amersham Pharmacia Biotech Inc.).
PCR methodology.
PCRs were performed as previously described (33). The primers used are listed in Table 2.
TABLE 2.
Primers used in this study
| Primer no. | Nucleotide sequence (5′–3′) |
|---|---|
| 1 | CCCGCGGCCGCGAATTCGTGACTGATGCCCTCCC |
| 2 | CCCGCGGCCGCGGATCCTGTGGCGTTGAAGATCCG |
| 3 | CGCTGAGCAATGACATCCG |
| 4 | AATGTCACCCTGAAGCAGG |
| 5 | GGGAAGCTTGCGGCCGCTTAGCAGGCTGGCTGGG |
| 6 | GGGAAGCTTGCGGCCGCAAAGGTGGCGGCAACAC |
| 7 | GGCGTCGACGACCGATTGAGGTTTCC |
| 8 | GCGCGGATCCGTTAAATCAAACCTCTTC |
Nucleotide sequencing.
The nucleotide sequences of PCR products and flanking regions in the genetic constructs were determined by using the ABI PRISMTM BigDye Terminator cycle sequencing ready reaction kit (PE Applied Biosystems). Samples were electrophoresed on a Perkin-Elmer ABI PRISM 310 genetic analyzer (PE Applied Biosystems) as described in the manufacturer's specifications.
Detection of type 1 fimbriae.
The capacity of bacteria to express a d-mannose-binding phenotype was assayed by determining their ability to agglutinate yeast cells on glass slides. Aliquots of liquid cultures grown to an optical density at 600 nm of 4.0 and a 5% (wt/vol) suspension of yeast cells were mixed, and the time until agglutination occurred was measured.
Construction of flu::tet mutants.
The flu gene was amplified by PCR from chromosomal DNA using primers 1 and 2. The resulting fragment was inserted directly into the EcoRI/BamHI site of pHHA145 to generate pHHA154. This plasmid was cut with StyI (1,138 bp inside the flu gene) and blunted, and an SspI/BsaAI fragment from pACYC184 containing the tetR gene and its promoter was inserted to generate plasmid pHHA159. Plasmid pHHA159 was subsequently cut with EcoRI, and the fragment containing the flu::tetR construct was inserted into plasmid pGP704 and amplified in the λpir-carrying strain CC118. After amplification, the plasmid was transformed into strain BD1428 and single-crossover mutants were selected on 8-μg/ml tetracycline plates. Double-crossover mutants were then screened by replica plating on 8-μg/ml tetracycline and 100-μg/ml ampicillin plates, and Tetr Amps colonies were picked for further work. Correct insertion of the flu::tetR construct on the chromosome was tested by PCR using primers 3 and 4 flanking the insertion point of the flu gene. Colonies in which PCR patterns had a shift in fragment size corresponding to the insertion were selected and tested for loss of autoaggregation ability. One representative strain with this genotype was designated HEHA11 and used in this study.
Construction of an oxyR::Ω(Spr) mutant.
A P1 phage lysate was made from oxyR::Ω(Spr) strain N9716 and used to transduce strain BD1428 as described by Miller (25). Double-crossover mutants were selected on Luria-Bertani (LB) plates containing spectinomycin (15 μg/ml), and correct inserts were tested by PCR with primers 5 and 6 (Table 2) annealing to regions flanking the oxyR gene. A clone with the same PCR fragment size as N9716 named HEHA9 was selected and used in this study.
Construction of Δfim mutants.
A Δfim variant of BD1428 was constructed using the λpir-dependent plasmid pLBJ311 containing a truncated fim gene cluster with a npt gene (Kanr) inserted between truncated versions of fimB and fimH, thus deleting all the fim genes. The insertion on the chromosome was done basically as previously described (33). Correct inserts were verified by PCR and Southern blotting as previously described (32). A clone carrying the correct insertion was designated HEHA4 and tested for loss of mannose-sensitive yeast agglutination. The same method was used on HEHA11 to construct the flu fim double mutant, designated HEHA17.
Allelic exchange of the fimE gene of BD1428.
In order to restore the fimE gene in strain BD1428, part of the fim gene cluster, i.e., the region downstream of fimB to the middle of fimD, was inserted into the integration vector pMAK700oriT by cutting plasmid pPKL4 with HindIII and EcoRI and inserting the relevant fragment into the same sites in pMAK700oriT. This plasmid carries a temperature-sensitive origin of replication, which permits it to replicate only at temperatures at or below 30°C. The resulting plasmid, pHHA161, was transformed into BD1428 and allowed to grow overnight at 30°C on 17-μg/ml chloramphenicol (CAM) plates. Single colonies, after being restreaked at 30°C for 2 days, were streaked out on 17-μg/ml CAM plates and placed at 42°C to select for recA-mediated single-crossover events. Camr colonies were then grown in liquid LB media containing 17 μg of CAM/ml at 42°C overnight. Overnight cultures were diluted 1:1,000 in liquid LB medium without antibiotics, grown to an optical density at 600 nm of 0.5, and then plated out on LB plates. Colonies from these plates were replica plated on 17-μg/ml CAM plates and LB plates, and chromosomal DNA from Cams colonies was tested by PCR employing primers 7 and 8 (flanking the fimE gene). A colony which had the normal fimE gene was selected and named HEHA6.
Colony morphology.
The colony morphology was assayed by employing a Carl Zeiss Axioplan epifluorescence microscope, and digital images were captured with a 12-bit cooled slow-scan charge-coupled device camera (KAF 1400 chip; Photometrics, Tucson, Ariz.) controlled by the PMIS software (Photometrics).
Immunofluorescence microscopy.
Surface presentation of type 1 fimbriae and Ag43 was assessed by immunofluorescence microscopy employing a monoclonal mouse antibody directed against FimA (33) or a polyclonal rabbit serum raised against the α-subunit of Ag43 (a kind gift from Peter Owen), respectively. As secondary antibodies a fluorescein isothiocyanate-labeled anti-rabbit serum and a tetramethyl rhodamine isocyanate-labeled anti-mouse serum from Sigma were used. Cell fixation, immunolabeling, and microscopy were carried out as previously described (30).
RESULTS
Colony types of E. coli K-12 strain BD1428.
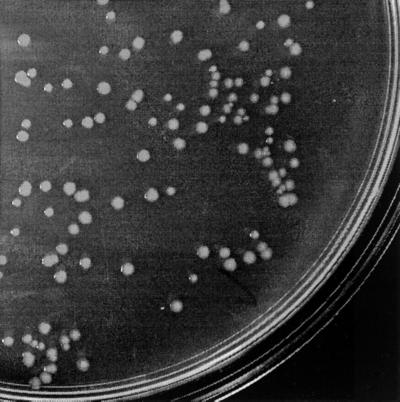
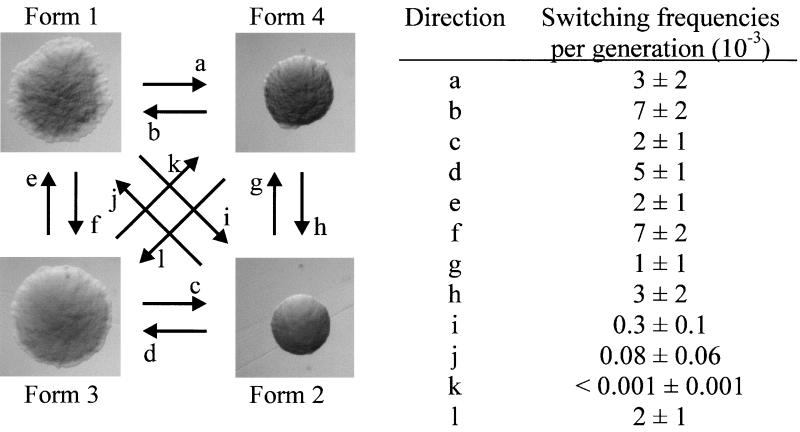
The E. coli reference strain BD1428, alias X474, undergoes colony morphology phase variation (Fig. 1). Two phase-variant forms of the strain were previously reported, with interconversion rates of about 10−3 (7). When we carefully investigated the morphology of colonies arising from BD1428, no less than four forms could be distinguished (Fig. 2). One was characterized by large, flat, frizzy colonies (form 1); another was characterized by small, convex, glossy colonies (form 2); a third was characterized by a flat irregular shape like form 1, but with a smooth surface (form 3); and finally there was a colony type with a small, convex appearance, similar to a form 2 colony, but with a frizzy surface (form 4). Interestingly, transition from a form 1 to a form 2 colony type and vice versa rarely occurred directly but rather via the form 3 or form 4 colony type. That is, restreaking of a form 1 colony resulted in form 1, form 3, and form 4 colonies but virtually never form 2 colonies. Conversely, restreaking of a form 2 colony type resulted in forms 2, 3, and 4 but only rarely form 1. Careful replating experiments with colonies of the four forms revealed the respective interconversion frequencies (Fig. 3).
FIG. 1.
Colony morphology variation of E. coli strain BD1428 from a representative section of an LB plate.
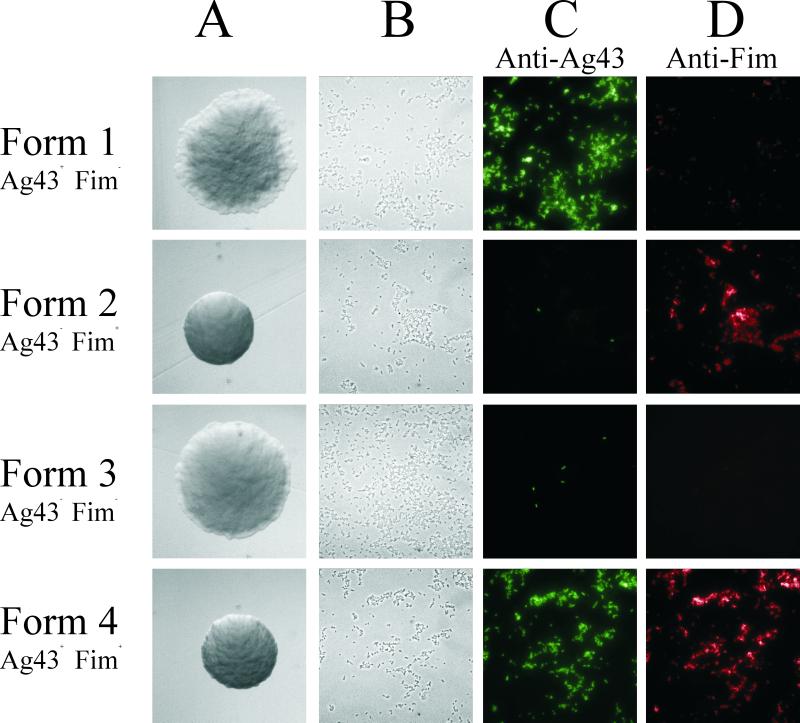
FIG. 2.
Phase-contrast microscopy of colonies (A) and cells (B) and immunofluorescence microscopy employing anti-Ag43 rabbit serum combined with fluorescein isothiocyanate-labeled anti-rabbit serum (C) and anti-Fim mouse serum combined with tetramethyl rhodamine isocyanate-labeled anti-mouse serum (D).
FIG. 3.
Switching frequencies between different colony forms of strain BD1428. Several individual colonies of each of the four forms were plated out twice. Subsequently, four colonies of each form were picked, diluted, and plated out. The switching frequencies were calculated by counting the fraction of each form.
Expression of type 1 fimbriae and Ag43 in colony types of strain BD1428.
Having established the presence of four colony morphology forms of strain BD1428, we proceeded to investigate a possible correlation between these and the expression of Ag43 and type 1 fimbriae. Strain BD1428 was reported to be capable of phase-variable expression of type 1 fimbriae and Ag43 (7). To expand on these preliminary findings, colonies of all four morphological types were picked directly from plates, prepared for immunofluorescence microscopy, and tested for expression of type 1 fimbriae and Ag43 (Fig. 2C and D). The data clearly indicated that there was a correlation between the expression of the two surface proteins and colony morphology. Most (>95%) of the cells from a form 1 colony expressed Ag43 but not type 1 fimbriae (Fig. 2). In contrast, cells from a form 2 colony showed the inverse relationship, i.e., virtually all cells (>99%) were fimbriated but only a few (<5%) expressed Ag43 (Fig. 2). Only a small fraction (<2%) of cells from a form 3 colony expressed Ag43 or fimbriae (Fig. 2), whereas virtually all cells from a form 4 colony expressed both (Fig. 2). These results indicate that cells from form 1 colonies are Fim− Ag43+, that cells from form 2 colonies are Fim+ Ag43−, that cells from form 3 colonies are Fim− Ag43−, and, finally, that form 4 cells are Fim+ Ag43+. The small convex appearance of form 2 and form 4 cells correlated with fimbriation, whereas the absence of fimbriae seemed to correlate with the characteristic flatness of form 1 and form 3 colonies. Expression of Ag43 seemed to give rise to the frizzy appearance of form 1 and form 4 colonies.
A flu null mutant is incapable of producing form 1 and form 4 colonies.
The apparent correlation between expression of Ag43 and the frizzy appearance of form 1 and form 4 colonies could be directly caused by either Ag43 or a secondary factor coregulated with flu. To investigate this issue further, a mutant of the BD1428 strain, HEHA11, in which the flu gene was knocked out by insertion of a tet cassette was made. Interestingly, HEHA11 exhibited both form 2 and form 3 colony morphology types; however, the ability to produce form 1 and form 4 colonies appeared to have been lost, even after consecutive restreaking of form 2 and form 3 colonies. This result was further corroborated by reintroducing the flu gene on a plasmid (pHHA146) in HEHA11, which reinstated the ability to form form 1 and form 4 colonies. Taken together the data indicate that the flu gene product is required in order to produce a frizzy phenotype but that, in line with our previous observations of the correlation of the Ag43 phenotype and colony morphology, the flu gene seems to play no role in the colony size phenotype.
Influence of fim status on colony morphology.
The correlation between fimbriation and form 2 and form 4 colony types suggests a direct role for type 1 fimbriae in colony size. To examine this further, fim deletion mutant HEHA4 of the BD1428 strain was made. Continuous restreaking of this strain revealed that it was still subject to phase variation between form 1 and form 3 colonies but not between form 2 and form 4 colonies. This was in agreement with our previous results and supported the notion that the small compact colony type exhibited by form 2 and form 4 colonies is uniquely due to the presence of type 1 fimbriae on the surfaces of the cells.
Strain BD1428 is a fimE null mutant.
Strain BD1302 has a chromosomal deletion encompassing the oxyR gene, which causes constitutive expression of Ag43. It is, however, capable of phase-variable expression of type 1 fimbriae (12). Accordingly, one would expect the strain to make form 1 and form 4 colonies. However, we observed colonies of BD1302 to be uniquely form 1. This inspired us to investigate the influence of OxyR on Ag43 expression and colony morphology in the BD1428 strain. Defined oxyR mutant HEHA9, in which the oxyR gene was replaced by an oxyR::Spec cassette was made (see Materials and Methods). When plated out, strain HEHA9 gave rise to form 1 colonies and, significantly, unlike BD1302, also form 4 colonies.
The apparent discrepancy between the colony types produced by BD1302 and HEHA9 prompted us to investigate the fim gene cluster of BD1428. It became clear that the BD1428 strain carried an insert of about 750 bp in the fimE gene. Subsequent sequencing of this region revealed the presence of an IS1 element after the 12th base in the fimE gene, rendering this gene nonfunctional. By allelic exchange, the truncated fimE gene in BD1428 was replaced with an intact fimE gene, resulting in strain HEHA6, thereby restoring the integrity of the fim gene cluster. Colonies of strain HEHA6 exhibited phase variation between form 1 and form 3, but notably form 2 and form 4 were not seen. This indicates that the presence of FimB and the absence of FimE account for the ability to vary between small and large colony sizes.
Population dynamics of cells originating from different colony forms under static liquid conditions.
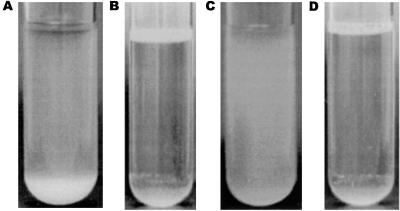
In order to probe a possible correlation between colony morphology and niche specificity under static liquid conditions, individual colonies of all four forms of BD1428 were incubated in static broth. The results indeed suggested a correlation. When a form 1 colony was picked and grown under static liquid conditions, most of the cells were observed to be present as a thick precipitate (Fig. 4A). In contrast, the progeny of a form 2 colony gave rise to a thick pellicle or surface film (Fig. 4B). Form 3 cells did not seem to have any preference for either surface or bottom life (Fig. 4C). Finally, cells from a form 4 colony essentially behaved like form 2 cells, i.e., most of the cells were located as a pellicle in the air-water interface (Fig. 4D).
FIG. 4.
Habitat specificity of E. coli BD1428 cells originating from a form 1 colony (A), a form 2 colony (B), a form 3 colony (C), and a form 4 colony (D) grown in static liquid medium.
DISCUSSION
Remarkably little attention has been given to colony morphology variation of E. coli (and other bacteria) and the underlying mechanisms. Two factors suspected to influence the colony morphology of certain E. coli K-12 strains are in fact both phase-variable surface components, viz., antigen 43 and type 1 fimbriae. Previous reports have suggested a correlation between these two surface structures and the ability of certain E. coli K-12 strains to produce two distinct colony morphologies (3, 5, 7, 27). However, it was not clear from these studies whether the expression of Ag43 and type 1 fimbriae directly caused the phenotypes or whether the apparent correlation was coincidental.
Because of the fact that strain BD1428 was reported to give rise to different colony types and had the ability to express Ag43 and type 1 fimbriae, it was selected for a detailed study on the potential correlation between colony morphology and the expression of Ag43 and type 1 fimbriae. Careful analysis of BD1428 colony types revealed the presence of four individual forms (Fig. 2). When cells originating from individual colony forms were investigated by immunofluorescence microscopy employing specific sera, it became clear that the four forms coincided with specific Ag43 and Fim phenotypes: form 1, Ag43+ Fim−; form 2, Ag43− Fim+; form 3, Ag43− Fim−; form 4, Ag43+ Fim+. The transition frequencies among the four morphological types were also investigated (Fig. 3). Shift rates associated with a change in either Ag43 or Fim status only were in the range of 10−3 to 7 × 10−3. Transition frequencies involving double phenotypic shifts were roughly in the expected range for transitions between form 1 and form 2. However, the rate of transition from form 4 to form 3 was several orders of magnitude higher than expected (2 × 10−3 compared to the expected 3 × 10−5), and the rate of transition from form 3 to form 4 was too small to be seen. It therefore seemed that the Ag43+ Fim+ phenotype of a form 4 type colony was selected against on solid medium.
Expression of type 1 fimbriae is phase variable, due to the inversion of the fimbrial phase switch. This inversion is catalyzed by the FimB and FimE recombinases. On solid medium the frequency of FimB-catalyzed inversion has been reported to be on the order of 10−3 per cell per generation, whereas FimE-catalyzed inversion (on to off) seems to be several orders of magnitude higher, i.e., 0.3 per cell per generation (9). From this point of view colonies involving a Fim+ phenotype, i.e., forms 2 and 4, were not to be expected. However, investigation of the fimE gene of BD1428 revealed that it was truncated by an IS1 element. Therefore, the observed transition rates involving changes in the Fim phenotype of BD1428 are uniquely FimB mediated and are in accordance with the reported switch inversion rates of this recombinase. Similar transition rates have been observed for E. coli K-12 strain CSH50, which also contains an IS1 element in the fimE gene, although in a different position (3). In line with this, replacement of the fimE::IS1 truncate in BD1428 with an intact fimE gene resulted in a strain, HEHA6, which only gave rise to form 1 and form 3 colonies, both of which are Fim−. The behavior of a Δfim version, HEHA4, of BD1428 was identical. That is, the strain was unable to express fimbriae and to make form 2 and form 4 colonies. Our results indicate that the small convex morphology of form 2 and form 4 colonies is caused by the physical presence of fimbriae on the bacteria. It should also be noted that plating of cells from form 2 and form 4 colonies revealed that such colonies contained only half as many cells as form 1 and form 3 colonies, indicative of the disadvantage of fimbriation during growth on solid medium.
In static liquid medium type 1 fimbriation mediates pellicle formation (26). Pellicle formation is abolished in the presence of the inhibitor α-methyl-d-mannoside, which is not readily metabolized by E. coli (26). Since Fim-specific adherence might be invoked to cause the compact appearance of form 2 and form 4 colonies, the effect of solid medium containing α-methyl-d-mannoside on colony morphology was tested. However, no significant effect was seen.
Several lines of evidence suggested that the frizzy phenotype of form 1 and form 4 colonies was directly caused by the physical presence of Ag43 on the surface of the cells. Cells of these colony types of BD1428 virtually all expressed Ag43, whereas cells from forms 2 and 3 did not (Fig. 2). Furthermore, a flu null mutant of BD1428 could only make form 2 and form 3 colonies. Finally, since OxyR is a repressor of the flu gene, an oxyR mutant is a constitutive Ag43 producer (12, 15, 36). In line with this finding, an oxyR version of BD1428 was observed to make only form 1 and form 4 colonies.
A recent study on the behavior of Pseudomonas fluorescens cells belonging to different morphological types identified a direct correlation between colony morphology and niche specificity in static liquid media (31). It is therefore interesting to correlate the colony phenotypes reported in this study with population dynamics in static liquid environments. Static liquid conditions are encountered by E. coli, for example, when mammals defecate in stagnant water bodies, i.e., pools, ponds, lakes, etc. From this perspective form 1 cells, characterized by an Ag43+ Fim− phenotype, would represent a bottom-dwelling community, since Ag43 confers autoaggregation and settling of the bacteria (12). Indeed, when a form 1 colony of BD1428 was picked and grown under static liquid conditions, most of the cells were observed to be present as a thick precipitate (Fig. 4). On the other hand, form 2 cells, characterized by an Ag43− Fim+ phenotype, would be surface dwellers due to the induction of surface pellicles by type 1 fimbriated cells (11, 26). In fact, when a form 2 colony was picked and grown under static liquid conditions, a thick pellicle formed (Fig. 4). Form 3 cells, Ag43− Fim−, did not seem to have any preference for either surface or bottom life and seem to represent a “pelagic” population (Fig. 4). We have recently shown that the presence of fimbriae abolishes Ag43-mediated autoaggregation (12). Therefore cells from a form 4 colony, Ag43+ Fim+, would be expected to behave like form 2 cells, which indeed turned out to be the case. The expression of Ag43 and type 1 fimbriae is phase variable, allowing members of a bacterial population to shift between different habitats in a static liquid environment as a function of their Ag43 and Fim phenotypes. This capacity would enable a portion of the population to be at the most favorable place at a given time and might well have far-ranging consequences for the ability of a bacterial strain to survive in nature. In conclusion, the Ag43 and Fim status of cells not only seems to determine colony morphology but also seems to correlate with habitat in static liquid environments.
The present report establishes the first conclusive correlation of colony morphology types of E. coli with well-defined surface proteins. Future research will reveal whether this approach can be used for other bacteria.
ACKNOWLEDGMENT
This work was supported by the Danish Natural Sciences Research Council (grant no. 9601682).
REFERENCES
- 1.Abraham J M, Freitag C S, Clements J R, Eisenstein B I. An invertible element of DNA controls phase variation of type 1 fimbriae of Escherichia coli. Proc Natl Acad Sci USA. 1985;82:5724–5727. doi: 10.1073/pnas.82.17.5724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blomfield I C, Calie P J, Eberhardt K J, McClain M S, Eisenstein B I. Lrp stimulates phase variation of type 1 fimbriation in Escherichia coli K-12. J Bacteriol. 1993;175:27–36. doi: 10.1128/jb.175.1.27-36.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blomfield I C, McClain M S, Princ J A, Calie P J, Eisenstein B I. Type 1 fimbriation and fimE mutants of Escherichia coli K-12. J Bacteriol. 1991;173:5298–5307. doi: 10.1128/jb.173.17.5298-5307.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bolivar F. Construction and characterization of new cloning vehicles. III. Derivatives of plasmid pBR322 carrying unique Eco RI sites for selection of Eco RI generated recombinant DNA molecules. Gene. 1978;4:121–136. doi: 10.1016/0378-1119(78)90025-2. [DOI] [PubMed] [Google Scholar]
- 5.Brinton C C J. The structure, function, synthesis and genetic control of bacterial pili and a molecular model for DNA and RNA transport in gram negative bacteria. Trans N Y Acad Sci. 1965;27:1003–1054. doi: 10.1111/j.2164-0947.1965.tb02342.x. [DOI] [PubMed] [Google Scholar]
- 6.Chang A C, Cohen S N. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J Bacteriol. 1978;134:1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Diderichsen B. flu, a metastable gene controlling surface properties of Escherichia coli. J Bacteriol. 1980;141:858–867. doi: 10.1128/jb.141.2.858-867.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Freitag C S, Abraham J M, Clements J R, Eisenstein B I. Genetic analysis of the phase variation control of expression of type 1 fimbriae in Escherichia coli. J Bacteriol. 1985;162:668–675. doi: 10.1128/jb.162.2.668-675.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gally D L, Bogan J A, Eisenstein B I, Blomfield I C. Environmental regulation of the fim switch controlling type 1 fimbrial phase variation in Escherichia coli K-12: effects of temperature and media. J Bacteriol. 1993;175:6186–6193. doi: 10.1128/jb.175.19.6186-6193.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gally D L, Leathart J, Blomfield I C. Interaction of FimB and FimE with the fim switch that controls the phase variation of type 1 fimbriae in Escherichia coli K-12. Mol Microbiol. 1996;21:725–738. doi: 10.1046/j.1365-2958.1996.311388.x. [DOI] [PubMed] [Google Scholar]
- 11.Harris S L, Elliott D A, Blake M C, Must L M, Messenger M, Orndorff P E. Isolation and characterization of mutants with lesions affecting pellicle formation and erythrocyte agglutination by type 1 piliated Escherichia coli. J Bacteriol. 1990;172:6411–6418. doi: 10.1128/jb.172.11.6411-6418.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hasman H, Chakraborty T, Klemm P. Antigen-43-mediated autoaggregation of Escherichia coli is blocked by fimbriation. J Bacteriol. 1999;181:4834–4841. doi: 10.1128/jb.181.16.4834-4841.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Henderson I, Owen P. The autoregulatory protein Mor and OxyR are identical. Microbiology. 1997;143:1482. doi: 10.1099/00221287-143-5-1482. [DOI] [PubMed] [Google Scholar]
- 14.Henderson I R, Meehan M, Owen P. Antigen 43, a phase-variable bipartite outer membrane protein, determines colony morphology and autoaggregation in Escherichia coli K-12. FEMS Microbiol Lett. 1997;149:115–120. doi: 10.1111/j.1574-6968.1997.tb10317.x. [DOI] [PubMed] [Google Scholar]
- 15.Henderson I R, Owen P. The major phase-variable outer membrane protein of Escherichia coli structurally resembles the immunoglobulin A1 protease class of exported protein and is regulated by a novel mechanism involving Dam and OxyR. J Bacteriol. 1999;181:2132–2141. doi: 10.1128/jb.181.7.2132-2141.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Herrero M, de Lorenzo V, Timmis K N. Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in gram-negative bacteria. J Bacteriol. 1990;172:6557–6567. doi: 10.1128/jb.172.11.6557-6567.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Klemm P. Two regulatory fim genes, fimB and fimE, control the phase variation of type 1 fimbriae in Escherichia coli. EMBO J. 1986;5:1389–1393. doi: 10.1002/j.1460-2075.1986.tb04372.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Klemm P, Jørgensen B J, van Die I, de Ree H, Bergmans H. The fim genes responsible for synthesis of type 1 fimbriae in Escherichia coli, cloning and genetic organization. Mol Gen Genet. 1985;199:410–414. doi: 10.1007/BF00330751. [DOI] [PubMed] [Google Scholar]
- 19.Klemm P, Krogfelt K A. Type 1 fimbriae of Escherichia coli. In: Klemm P, editor. Fimbriae adhesion, genetics, biogenesis, and vaccines. Boca Raton, Fla: CRC Press, Inc.; 1994. pp. 9–26. [Google Scholar]
- 20.Kulasekara H D, Blomfield I C. The molecular basis for the specificity of fimE in the phase variation of type 1 fimbriae of Escherichia coli K-12. Mol Microbiol. 1999;31:1171–1181. doi: 10.1046/j.1365-2958.1999.01257.x. [DOI] [PubMed] [Google Scholar]
- 21.Kullik I, Stevens J, Toledano M B, Storz G. Mutational analysis of the redox-sensitive transcriptional regulator OxyR: regions important for DNA binding and multimerization. J Bacteriol. 1995;177:1285–1291. doi: 10.1128/jb.177.5.1285-1291.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kullik I, Toledano M B, Tartaglia L A, Storz G. Mutational analysis of the redox-sensitive transcriptional regulator OxyR: regions important for oxidation and transcriptional activation. J Bacteriol. 1995;177:1275–1284. doi: 10.1128/jb.177.5.1275-1284.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu D, Reeves P R. Escherichia coli K12 regains its O antigen. Microbiology. 1994;140:49–57. doi: 10.1099/13500872-140-1-49. [DOI] [PubMed] [Google Scholar]
- 24.McClain M S, Blomfield I C, Eisenstein B I. Roles of fimB and fimE in site-specific DNA inversion associated with phase variation of type 1 fimbriae in Escherichia coli. J Bacteriol. 1991;173:5308–5314. doi: 10.1128/jb.173.17.5308-5314.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miller J H. A short course in bacterial genetics: a laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1992. [Google Scholar]
- 26.Old D C, Duguid J P. Selective outgrowth of fimbriate bacteria in static liquid medium. J Bacteriol. 1970;103:447–456. doi: 10.1128/jb.103.2.447-456.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Orndorff P E, Falkow S. Identification and characterization of a gene product that regulates type 1 piliation in Escherichia coli. J Bacteriol. 1984;160:61–66. doi: 10.1128/jb.160.1.61-66.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Owen P. Antigens of the Escherichia coli cell envelope. In: Bjerrum O J, editor. Electroimmunochemical analysis of membrane proteins. Amsterdam, The Netherlands: Elsevier Science Publishing, Inc.; 1983. pp. 347–373. [Google Scholar]
- 29.Owen P, Caffrey P, Josefsson L G. Identification and partial characterization of a novel bipartite protein antigen associated with the outer membrane of Escherichia coli. J Bacteriol. 1987;169:3770–3777. doi: 10.1128/jb.169.8.3770-3777.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pallesen L, Poulsen L K, Christiansen G, Klemm P. Chimeric FimH adhesin of type 1 fimbriae: a bacterial surface display system for heterologous sequences. Microbiology. 1995;141:2839–2848. doi: 10.1099/13500872-141-11-2839. [DOI] [PubMed] [Google Scholar]
- 31.Rainey P B, Travisano M. Adaptive radiation in a heterogeneous environment. Nature. 1998;394:69–72. doi: 10.1038/27900. [DOI] [PubMed] [Google Scholar]
- 32.Schembri M A, Pallesen L, Connell H, Hasty D L, Klemm P. Linker insertion analysis of the FimH adhesin of type 1 fimbriae in an Escherichia coli fimH-null background. FEMS Microbiol Lett. 1996;137:257–263. doi: 10.1111/j.1574-6968.1996.tb08115.x. [DOI] [PubMed] [Google Scholar]
- 33.Stentebjerg-Olesen B, Pallesen L, Jensen L B, Christiansen G, Klemm P. Authentic display of a cholera toxin epitope by chimeric type 1 fimbriae: effects of insert position and host background. Microbiology. 1997;143:2027–2038. doi: 10.1099/00221287-143-6-2027. [DOI] [PubMed] [Google Scholar]
- 34.Storz G, Altuvia S. OxyR regulon. Methods Enzymol. 1994;234:217–223. doi: 10.1016/0076-6879(94)34088-9. [DOI] [PubMed] [Google Scholar]
- 35.Storz G, Tartaglia L A, Ames B N. Transcriptional regulator of oxidative stress-inducible genes: direct activation by oxidation. Science. 1990;248:189–194. doi: 10.1126/science.2183352. [DOI] [PubMed] [Google Scholar]
- 36.Warne S R, Varley J M, Boulnois G J, Norton M G. Identification and characterization of a gene that controls colony morphology and auto-aggregation in Escherichia coli K12. J Gen Microbiol. 1990;136:455–462. doi: 10.1099/00221287-136-3-455. [DOI] [PubMed] [Google Scholar]