Abstract
The nuclear factor erythroid 2-related factor (Nrf2) and antioxidant response element (ARE) signaling pathway play an important role in the amelioration of cellular oxidative stress. Thus, assays that detect this pathway can be useful for identifying chemicals that induce or inhibit oxidative stress signaling. This chapter is to describe two cell-based Nrf2/ARE assays in a quantitative high throughput screening (HTS) format to test a large collection of chemicals for oxidative stress induction ability. The assay descriptions involve cell handling, assay preparation, instrument usage, and assay procedure.
Keywords: Antioxidant response element (ARE), KeratinoSens, β-Lactamase (bla), Luciferase (luc), Nuclear factor erythroid 2-related factor 2 (Nrf2), Reactive oxygen species (ROS)
1. Introduction
Oxidative stress, an imbalance of reactive oxygen species (ROS) and antioxidant defenses, plays a role in chemical-induced toxicity, cancer, and age-related diseases [1–3]. ROS can cause toxic effects through the production of peroxides and free radicals that damage all cellular components, including proteins, lipids, and DNA. ROS also acts as cellular messengers in redox signaling that can disrupt normal cellular signaling mechanisms [4]. The induction of many cytoprotective enzymes in response to ROS is mediated by antioxidant response elements (ARE), which activate the nuclear factor erythroid 2-related factor (Nrf2).
ARE is a cis-acting enhancer locating in the 5’ flanking region of many phase II detoxification genes. Nrf2 is a member of the Cap ‘n’ Collar family of transcription factors. Nrf2 is repressed by the cysteine-rich Keap1 protein, which targets Nrf2 for ubiquitination and subsequent degradation. When the cysteine residue of Keap1 protein is conjugated by chemicals, the Nrf2 protein is released and binds to ARE, activating downstream transcriptional pathway [5]. The Nrf2/ARE transcriptional pathway plays an important role in gene regulation and control of protein expression critical to the detoxification and elimination of ROS and electrophiles [6,7]. Several studies suggest the protective capabilities of Nrf2/ARE activation against environmentally induced oxidative stress and consider the Nrf2/ARE pathway as a potential therapeutic target [8,9]. Recently, the activation of Nrf2/ARE pathway in keratinocytes is considered the key event in skin sensitization adverse outcome pathway. The Nrf2/ARE-luciferase reporter gene assay is adapted to the skin sensitization test battery [10].
Here we describe two cell-based Nrf2/ARE reporter gene assays based on fluorescence and luminescence readouts. ARE β-lactamase (bla) reporter gene assay (ARE-bla assay) with Fluorescence Resonance Energy Transfer (FRET) technology can be used to identify the chemical compounds that modulate the ARE signaling pathway. The ARE-bla cell line contain an ARE-controlled β-lactamase reporter gene stably integrated into HepG2 cells. The assay basis for detection involves a β-lactamase substrate: CCF4-AM, a FRET-based fluorescent substrate. Once inside the cells, CCF4-AM is hydrolyzed by cytoplasmic esterases to form a polar molecule (CCF4). Upon excitation at 405 nm, the energy is transferred to the fluoresein moiety by FRET resulting in emission at 530 nm. In the presence of β-lactamase expression, the β-lactam ring in the CCF4 substrate is cleaved, which leads to excitation at 405 nm and emission at 460 nm [11] (Fig. 1). Thus, the fluorescence measured in these cells quantitatively corresponds to the activity change of ARE signaling. In contrast, the KeratinoSens™ assay uses immortalized human keratinocytes transfected with ARE-luc selectable plasmid [12]. ARE-luc reporter gene assay (Fig. 1) uses luciferin as the substrate to detect activation of ARE pathway which does not have the limitation of photo bleaching or autofluoresence associated with the ARE-bla assay. However, ARE-bla assay is more advantageous for visualizing live cells than bioluminescent proteins because fluorophores do not require cofactors or exogenous substrates for activity and are more stable than bioluminescent reporters. Therefore, the combination of ARE-luc and ARE-bla serve as a counter-screening system when detect hazards among different chemical classes. Quantitative data may be used for potency assessment within a specific structural class using a read-across approach with a comparison to related chemicals of known sensitization potential. The ARE-bla and ARE-luc assays are optimized in a quantitative high-throughput screening (qHTS) platform to detect the compounds that activate this pathway. This chapter describes the protocols for these assays in a 1536-well plate format.
Figure 1.
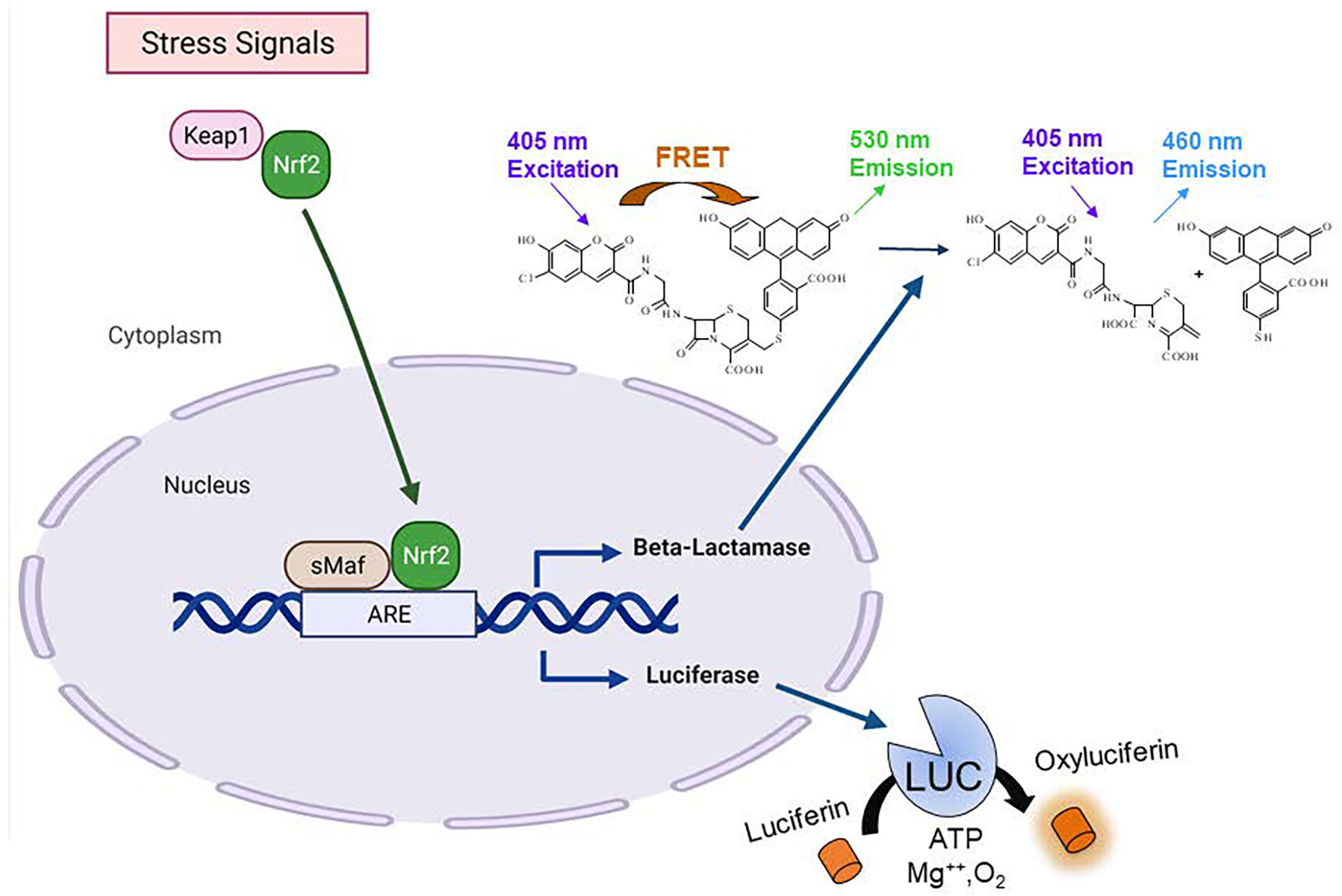
Schematics of ARE-bla and ARE-luc reporter gene assays (This figure was created with BioRender.com).
2. Materials
2.1. Cell line and cell culture condition
All the cell-culture-related medium and components (Table 1 and Table 2) are purchased from Life Technologies.
Cell line: A CellSensor® ARE-bla HepG2 cell line (Life Technologies) that contains three stably integrated copies of the ARE derived from the reduced form of human nicotinamide adenine dinucleotide phosphate (NADPH) quinone oxidoreductase 1 gene (NQO1) driving the expression of a downstream beta-lactamase reporter gene [9]. The cells were maintained in culture medium at 37°C under a humidified atmosphere and 5% CO2. KeratinoSens™ (Givaudan, Vernier, Switzerland) is an immortalized adherent cell line derived from HaCaT cells that contains luciferase gene under the transcriptional control of the AKR1C2 ARE sequence upstream of the SV40 promoter.
Culture medium: ARE-bla HepG2 cells were maintained in DMEM medium supplement with 10% dialyzed fetal bovine serum, 0.1 mM non-essential amino acids, 25 mM HEPES, 100U/mL penicillin, 100μg/mL streptomycin and 5μg/mL blasticidin (Table 1). KeratinoSens™ (ARE-luc HaCaT) cells were maintained in DMEM medium supplement with 10% fetal bovine serum, 100 U/mL penicillin, 100μg/mL streptomycin and 500 μg/ml G-418 (Table 2).
Thaw medium: same as culture medium but without antibiotics, blasticidin, or G-418.
Assay medium: ARE-bla HepG2 cells were suspended in DMEM medium supplement with 1% fetal bovine serum, 0.1mM non-essential amino acids, 25 mM HEPES, 100 U/mL penicillin and 100μg/mL streptomycin. KeratinoSens™ (ARE-luc HaCaT) cells were suspended in DMEM medium supplement with 1% fetal bovine serum, 100 U/mL penicillin, 100μg/mL streptomycin.
Freezing Medium: Recovery™ Cell Culture Freezing Medium (Life Technologies).
0.25% Trypsin/EDTA.
Dulbecco’s phosphate-buffered saline (DPBS) without calcium and magnesium.
Table 1:
Cell culture and assay medium for ARE-bla assay
| Components | Culture Medium | Assay Medium | Thaw Medium |
|---|---|---|---|
| DMEM | 90% | 99% | 90% |
| Dialyzed FBS | 10% | 1% | 10% |
| NEAA | 0.1 mM | 0.1 mM | 0.1 mM |
| HEPES | 25 mM | 25 mM | 25 mM |
| Penicillin | 100 U/mL | 100 U/mL | 100 U/mL |
| Streptomycin | 100 μg/mL | 100 μg/mL | 100 μg/mL |
| Blasticidin | 5 μg/mL | - | - |
Table 2:
Cell culture and assay medium for ARE-luc assay
| Components | Culture Medium | Assay Medium | Thaw Medium |
|---|---|---|---|
| DMEM | 90% | 99% | 90% |
| FBS | 10% | 1% | 10% |
| Penicillin | 100 U/mL | 100 U/mL | 100 U/mL |
| Streptomycin | 100 μg/mL | 100 μg/mL | 100 μg/mL |
| G-418 | 500μg/ml | - | - |
2.2. Assay Reagents and Chemicals
Reagents for fluorescence detection: LiveBLAzer™ FRET B/G Loading Kit (Life Technologies) including Solution A (CCF4-AM), Solution B and Solution C. Reagents for luminescence detection: One-Glo Luciferase assay system (Promega).
Solution D (Life Technologies) containing an anion transport inhibitor is used in conjunction with CCF4-AM-based assay to prevent active cellular export of the FRET-based substrates after loading. It is used in combination with Solution A, B, and C to create the CCF4-AM substrate mixture.
Dimethyl sulfoxide (DMSO) is used to resolve compounds and vehicle control for basal signal.
β-Naphthoflavone is used as the positive control.
2.3. Supplies and Equipment
T225 cell culture flasks.
Disposable, sterile centrifuge tubes.
1536-Well black clear bottom, cell culture treated plate for ARE-bla assay. 1536-Well white solid bottom, cell culture treated assay plate for ARE-luc assay.
Cell strainer: a receptacle with a 40 μm nylon filter that is used to remove clumped cells from cell suspensions.
Lids for assay and compound plate: these reusable lids are made from stainless steel and contain a rubber gasket that sits around the top outer edge. The cellular assay lid contains small evenly placed holes that allow air exchange necessary for cellular assays. The weight of the lid allows the gasket to form a strong barrier around the plate, virtually eliminating edge effects.
PinTool Workstation (Wako Automation): the PinTool performs transfer of 23 nL of compound from a 1536-well compound plate to a 1536-well assay plate (see Note 1).
Bioraptr FRD Workstation Beckman Coulter: a liquid handling system that can transfer of 0.2–10 μL of up to four different reagents or cells simultaneously into a 1536 well plate.
Multidrop Combi Dispenser: a high-speed dispenser capable of one reagent or cells using eight-channel detachable dispensing cassettes.
CyBi-well Vario Pipettor, a 96, 384 and 1536 channel simultaneous pipettor: it requires the use of disposable tips, which is more suitable for the preparation of positive control plate in 1536-well plate format.
Cellometer Auto T4 Cell Viability Counter (Nexcelom Bioscience) is used to count viable cells.
EnVision Multilabel Plate Reader (PerkinElmer) covers a wide range of fluorescence, absorbance and luminescence readouts commonly used in high throughput assays. The EnVision reader includes two detectors enabling simultaneous dual wavelength reading that is well suited for β-lactamase reporter assays. For an ARE-bla assay, reading requires dual bottom mirror and compatible filter sets (Table 3).
ViewLux Plate Reader (PerkinElmer, Waltham, MA)
Table 3:
Filter selection for ARE-bla assay
| Filters | Excitation | Emission |
|---|---|---|
| Channel 1 (Green) | 409/20 nm | 530/30 nm |
| Channel 2 (Blue) | 409/20 nm | 460/40 nm |
| Ratio (Ch2/Ch1) | 460 nm/530 nm | |
3. Methods
3.1. Cell Culture
Remove the cryovial containing the frozen cells from liquid nitrogen storage and immediately place it into a 37°C water bath with gentle agitation for 1–2 min.
Place the vial into a laminar flow hood. Before opening the vial, wipe the outside of the vial with 70% ethanol.
Transfer the thawed cells into a 50 mL conical sterile centrifuge tube with 30 mL prewarm thaw medium.
Centrifuge the cell supernatant for 4 min at 200 × g.
Carefully aspirate supernatant without disturbing the cell pellet
Gently re-suspend cell pellet in thaw medium.
Transfer the desired amount of the cells to a T225 tissue culture flask.
Place the flask in an incubator with a humidified atmosphere of 5% CO2 until passage. Maintain the cells at 30% to 90% confluence prior to passage. During the first passage, switch to culture medium.
After 48–72 h or cells with 80 – 90% confluence, aspirate medium and rinse once with 10 mL DPBS, followed by the addition of 10 mL of 0.25% Trypsin/EDTA and swirl to coat the cells evenly around flask.
Place the flask in incubator at 37°C for 2–3 min or until the cells detach.
Add 10 mL of culture medium to deactivate Trypsin.
Transfer the cells to a 50 mL conical tube and centrifuge at 200 × g for 4 min.
Carefully aspirate supernatant and re-suspend cell pellet in the culture medium.
Count cells using a Cellometer auto cell counter.
Transfer the cell suspension to a T225 tissue culture flask containing 4 × 10⁶ cells.
Incubate cell flask until next passage or assay. Cells are passaged twice a week to avoid over 90% cell confluence.
3.2. Preparation of Positive Control Plate
Assay-specific controls (positive and negative) are located on an additional 1536-well compound plate in columns 1–4. These controls will transfer simultaneously with the test compounds to the assay plate. The final concentration of DMSO in the assay is less than 0.5%.
To make a positive control plate, 5 μL of β-naphthoflavone stock solution in DMSO is dispensed into each well in columns 1, 2 and 3 (Fig. 2). CyBi-well Vario Pipettor can be used to prepare the control plate.
Figure 2.
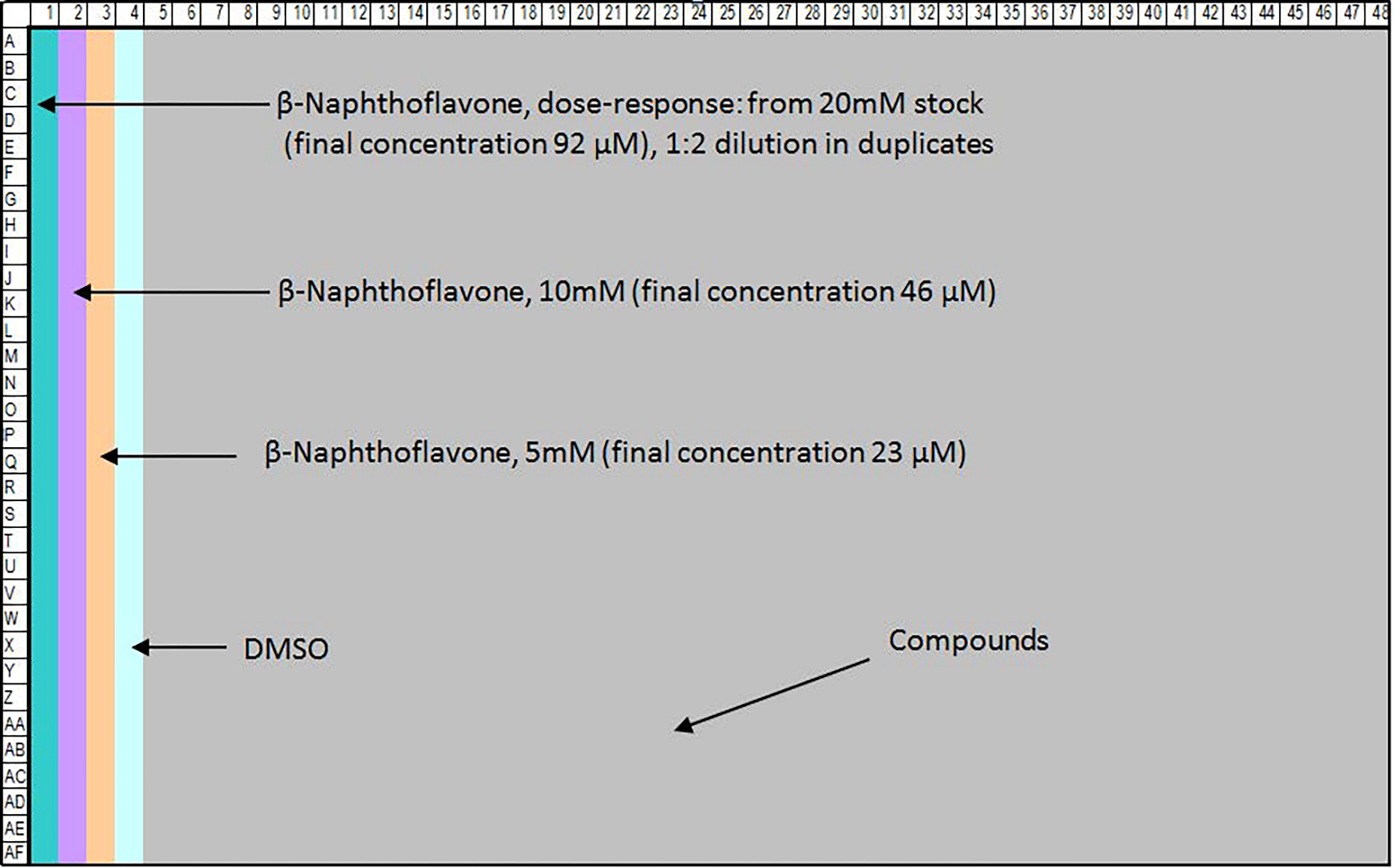
Control plate map in 1536-well format. Column 1 contains a sixteen-concentration titration ranging from 92 μM to 2.8 nM in duplicates. Columns 2 and 3 contain replicates for the β-naphthoflavone: 46 μM in column 2 and 23 μM in column 3 in 32 replicates for each concentration. DMSO, a negative control, is dispensed into column 4.
3.3. ARE-bla Assay
Harvest the ARE-bla HepG2 cells and resuspend the cell pellet in assay medium (see Note 2).
Place the cells on a cell strainer to remove clumped cells before counting cells.
Count cell number and determine cell viability. Cell viability of 95% or greater will have a better window of signal to basal level.
Prepare cell stock in assay medium at density of 0.4 × 10⁶ cells/mL.
Dispense 5 μL of cells prepared at step 4 into each well of a 1536-well black wall/clear-bottom, tissue culture-treated assay plates using a Multidrop Combi dispenser or Bioraptr FRD.
Place a pre-cleaned plate lid over the plate and incubate assay plates at 37°C under a humidified atmosphere and 5% CO2 for 5 hr to allow cells to attach.
Transfer 23 nL of compounds and controls to the assay plate by a PinTool.
Incubate the assay plates at 37°C, 5% CO2 for 16 h.
Freshly prepare 6X CCF4-AM Substrate Mixture prior to assay termination: add 6 μL 1mM of CCF4-AM substrate to 60 μL of Solution B and mix, and then add 874 μL of Solution C and 60 μL of Solution D to the combined solution and vortex.
Dispense 1 μL of 6X CCF4-AM Substrate Mixture to each well using a BioRAPTR dispenser after 16 hr compound treatment.
Incubate the plates for 2 h in the dark at room temperature for fluorescence development.
Measure fluorescence intensity at 460 and 530 nm emission and 405 nm excitation using an EnVision plate reader. Data is expressed as the ratio of 460nm/530nm emissions (see Note 3).
Percentage of activity of the compounds is calculated by normalizing the raw data to DMSO wells (0% activity) and maximal β-napthoflavone wells (100% activity).
The compound half maximum effective concentration (EC50) and maximum response (efficacy) values were calculated using a four-parameter Hill equation in GraphPad Prism software (Fig. 3).
Figure 3.
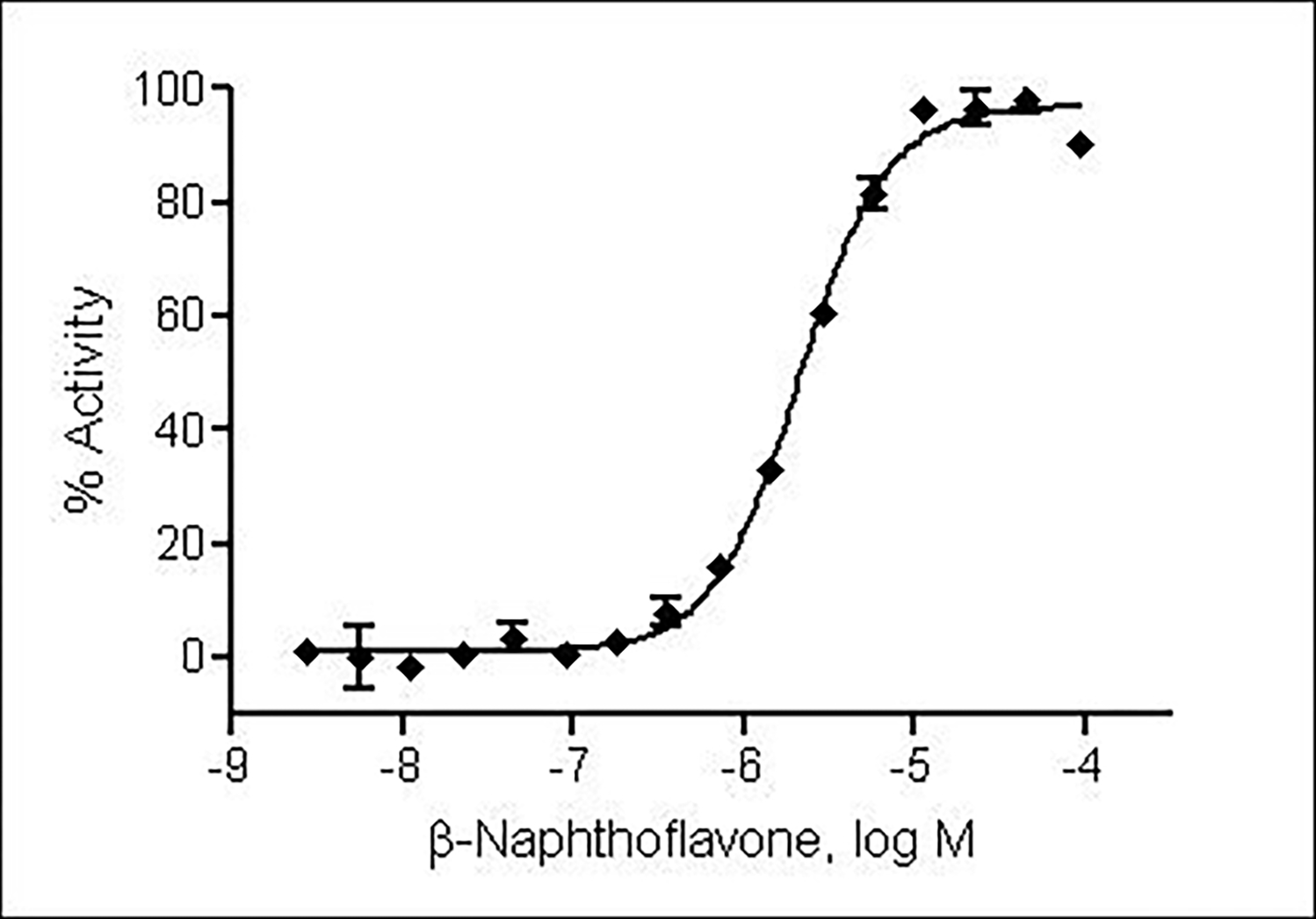
β-Naphthoflavone stimulated β-lactamase activity with EC50 of 2.1 μM in ARE-bla HepG2 cells in a 1536-well plate format.
3.4. ARE-luc Assay
Harvest the ARE-luc HaCaT cells and resuspend the cell pellet in assay medium (see Note 2).
Place the cells on a cell strainer to remove clumped cells before counting cells.
Count cell number and determine cell viability. Cell viability of 95% or greater will have a better window of signal to basal level.
Prepare cell stock in assay medium at density of 0.3 × 10⁶ cells/mL.
Dispense 5 μL of cells prepared at step 4 into each well of a 1536-well white solid bottom, tissue culture-treated assay plates using a Multidrop Combi dispenser.
Place a pre-cleaned plate lid over the plate and incubate assay plates at 37°C under a humidified atmosphere and 5% CO2 for 5 h to allow cells to attach.
Transfer 23 nL of compounds and controls to the assay plate by a PinTool station.
Incubate the assay plates at 37°C, 5% CO2 for 24 h.
Freshly prepare One-Glo solution and then add 4 μL into each well of the plate.
Incubate the plates in the dark at room temperature for 30 min.
Measure luminescence intensity using ViewLux reader. Data is expressed as intensity of luminescence units (see Note 4).
The compound half maximum effective concentration (EC50) and maximum response (efficacy) values were calculated using a four-parameter Hill equation in GraphPad Prism software (Fig. 4).
Figure 4.
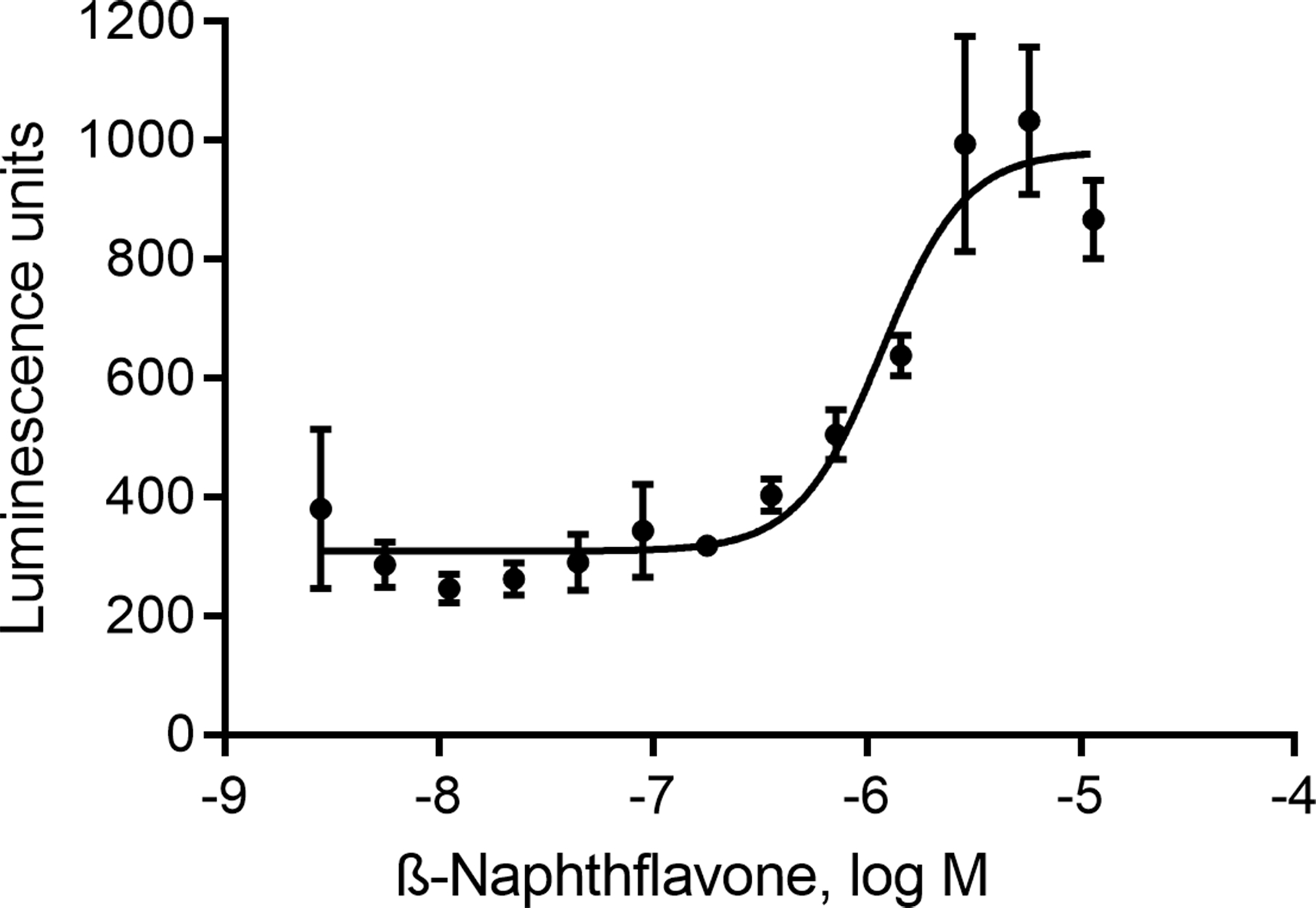
β-Naphthoflavone stimulated luciferase reporter activity with EC50 of 1.2 μM in ARE-luc HaCaT cells in a 1536-well plate format.
4. Notes
To avoid cross contamination, it is important to wash pins with appropriate reagents (e.g., DMSO and methanol) before and after use.
For screening, the cells that have passed three passages after thawing are recommended.
The ratiometric readouts from dual emissions (460 and 530 nm) can minimize well-to-well and plate-to-plate variation caused by differences in plating cell density.
Adjust exposure time and ensure luminescence intensity values are within the linear dynamic range of the plate reader used.
Acknowledgement
This work was supported in part by the Intramural research program of the NCATS, NIH.
References
- 1.Hu R, Saw CL, Yu R, Kong AN (2010) Regulation of NF-E2-related factor 2 signaling for cancer chemoprevention: antioxidant coupled with antiinflammatory. Antioxid Redox Signal 13 (11):1679–1698. doi: 10.1089/ars.2010.3276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Saw CL, Kong AN (2011) Nuclear factor-erythroid 2-related factor 2 as a chemopreventive target in colorectal cancer. Expert Opin Ther Targets 15 (3):281–295. doi: 10.1517/14728222.2011.553602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tufekci KU, Civi Bayin E, Genc S, Genc K (2011) The Nrf2/ARE Pathway: A Promising Target to Counteract Mitochondrial Dysfunction in Parkinson’s Disease. Parkinsons Dis 2011:314082. doi: 10.4061/2011/314082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Al-Dalaen SM (2014) Review Article: Oxidative Stress Versus Antioxidants. American Journal of Bioscience and Bioengineering 2 (5):60. doi: 10.11648/j.bio.20140205.11 [DOI] [Google Scholar]
- 5.Eggler AL, Small E, Hannink M, Mesecar AD (2009) Cul3-mediated Nrf2 ubiquitination and antioxidant response element (ARE) activation are dependent on the partial molar volume at position 151 of Keap1. Biochem J 422 (1):171–180. doi: 10.1042/BJ20090471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kensler TW, Wakabayashi N, Biswal S (2007) Cell survival responses to environmental stresses via the Keap1-Nrf2-ARE pathway. Annu Rev Pharmacol Toxicol 47:89–116. doi: 10.1146/annurev.pharmtox.46.120604.141046 [DOI] [PubMed] [Google Scholar]
- 7.Nguyen T, Nioi P, Pickett CB (2009) The Nrf2-antioxidant response element signaling pathway and its activation by oxidative stress. J Biol Chem 284 (20):13291–13295. doi: 10.1074/jbc.R900010200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shukla SJ, Huang R, Simmons SO, Tice RR, Witt KL, Vanleer D, Ramabhadran R, Austin CP, Xia M (2012) Profiling environmental chemicals for activity in the antioxidant response element signaling pathway using a high throughput screening approach. Environ Health Perspect 120 (8):1150–1156. doi: 10.1289/ehp.1104709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu RP, Hayashi T, Cottam HB, Jin G, Yao S, Wu CC, Rosenbach MD, Corr M, Schwab RB, Carson DA (2010) Nrf2 responses and the therapeutic selectivity of electrophilic compounds in chronic lymphocytic leukemia. Proc Natl Acad Sci U S A 107 (16):7479–7484. doi: 10.1073/pnas.1002890107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.OECD (2018) Test No. 442D: In Vitro Skin Sensitisation. doi:doi: 10.1787/9789264229822-en [DOI]
- 11.Zlokarnik G, Negulescu PA, Knapp TE, Mere L, Burres N, Feng L, Whitney M, Roemer K, Tsien RY (1998) Quantitation of transcription and clonal selection of single living cells with beta-lactamase as reporter. Science 279 (5347):84–88 [DOI] [PubMed] [Google Scholar]
- 12.Emter R, Ellis G, Natsch A (2010) Performance of a novel keratinocyte-based reporter cell line to screen skin sensitizers in vitro. Toxicol Appl Pharmacol 245 (3):281–290. doi: 10.1016/j.taap.2010.03.009 [DOI] [PubMed] [Google Scholar]


