Abstract
Intervening sequences (IVSs) were originally identified in the rrl genes for 23S rRNA (rrl genes, for large ribosomal subunit, part of rrn operon encoding rRNA) of Salmonella enterica serovars Typhimurium LT2 and Arizonae. These sequences are transcribed but later removed during RNase III processing of the rRNA, resulting in fragmentation of the 23S species; IVSs are uncommon, but have been reported in at least 10 bacterial genera. Through PCR amplification of IVS-containing regions of the rrl genes we showed that most Proteus and Providencia strains contain IVSs similar to those of serovar Typhimurium in distribution and location in rrl genes. By extraction and Northern blotting of rRNA, we also found that these IVSs result in rRNA fragmentation. We report the first finding of two very different sizes of IVS (113 bp and 183 to 187 bp) in different rrl genes in the same strain, in helix 25 of Proteus and Providencia spp.; IVSs from helix 45 are 113 to 123 bp in size. Analysis of IVS sequence and postulated secondary structure reveals striking similarities of Proteus and Providencia IVSs to those of serovar Typhimurium, with the stems of the smaller IVSs from helix 25 being similar to those of Salmonella helix 25 IVSs and with both the stem and the central loop domain of helix 45 IVSs being similar. Thus, IVSs of related sequences are widely distributed throughout the Enterobacteriaceae, in Salmonella, Yersinia, Proteus, and Providencia spp., but we did not find them in Escherichia coli, Citrobacter, Enterobacter, Klebsiella, or Morganella spp.; the sporadic distribution of IVSs of related sequence indicates that lateral genetic transfer has occurred.
In 1979 Winkler (30) discovered that the 23S rRNA of Salmonella enterica serovar Typhimurium is fragmented into several smaller fragments during 23S rRNA maturation. The basis for this fragmentation became clear in 1990, when Burgin et al. (6) reported novel intervening sequences (IVSs) in the 23S rRNA genes (rrl, for ribosomal subunit, large) of Salmonella serovars Typhimurium and Arizonae. These IVSs replace a short stem and loop in the putative secondary structure of the rRNA product (helix 25 and helix 45 in the case of Salmonella) with an extended stem-and-loop structure containing about 110 bp (Fig. 1A). This extended stem-and-loop structure becomes a substrate for RNase III which cleaves the IVS from the rRNA without religating it, resulting in fragmented rRNA. This fragmented rRNA appears to have no effect on the ability of the organism to grow and survive, since Escherichia coli cells (which do not normally contain IVSs) that contain a plasmid with a serovar Typhimurium rrl gene containing an IVS display fragmented 23S rRNA but maintain wild-type growth rates (7). Although RNase III is essential for removal of IVSs from the final product (6, 20), removal of IVSs is not essential for growth since RNase III-deficient strains of serovar Typhimurium retain the IVSs in all their ribosomes and yet are still viable (20).
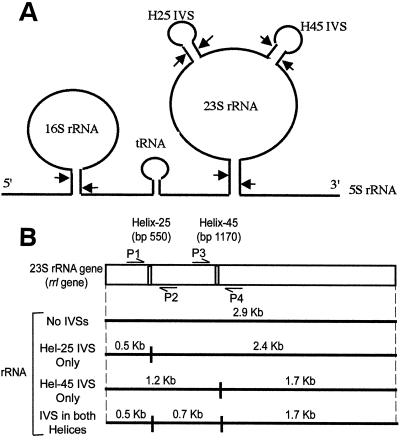
FIG. 1.
(A) Processing of the rRNA transcript, showing removal of IVSs. A typical 30S rRNA transcribed from an rrn operon is shown with 16S, spacer tRNA, 23S, and 5S rRNAs. The positions of helix 25 IVS (indicated as H25) at bp 550 and helix 45 IVS (indicated as H45) at bp 1170 are also shown as stem-loop structures on the 23S rRNA. The arrows indicate sites of cleavage by RNase III (an enzyme responsible for primary processing of the nascent 30S rRNA in E. coli and S. typhimurium [6]). (B) rrl gene for 23S rRNA, indicating the positions of helix 25 (ca. bp 550) and helix 45 (ca. bp 1170). Also indicated are primers used for PCR amplification; primers P1 and P2 amplify the helix 25 region, and primers P3 and P4 amplify the helix 45 region. The rRNA fragmentation patterns expected from excision of IVSs are also shown. The presence of 2.4- and 0.5-kb fragments indicates that an rrl gene contains an IVS in helix 25 only; 1.7- and 1.2-kb fragments indicate an IVS in helix 45 (ca. bp 1170) only, and 1.7-, 0.7-, and 0.5-kb fragments indicate IVSs in both helices. Mature 23S rRNA that lacks IVSs is 2.9 kb.
Perhaps the most intriguing aspect of IVSs is their sporadic distribution among bacteria. In the middle of the rRNA genes, which may be the most stable genetic element known, is an element that appears to have no function (other than to leave fragmented rRNA), is typically present in only a subset of genes for rRNA, and is unevenly distributed among bacterial genera. Since the early discoveries of Winkler (30) and of Burgin et al. (6), IVSs have been seen in at least nine other bacterial genera including: Agrobacterium (9), Brucella (9), and Rhizobium (25) (alpha group of Proteobacteria, as defined by 16S rRNA sequence) (31); Coxiella (1), Haemophilus (28), Salmonella (6, 18, 19), and Yersinia (26) (gamma group of Proteobacteria); Campylobacter (11, 13, 29) and Helicobacter (10) (epsilon group of Proteobacteria); and Leptospira (21) (spirochetes). Recently, Asai et al. (3) reported that rRNA genes cloned from a strain of Proteus vulgaris contained a large (200-bp) intervening sequence in helix 25, which resulted in fragmented 23S rRNA when the plasmid containing the IVS was cloned into E. coli K-12.
Another interesting observation is that, while most IVSs from different genera show little or no relatedness (based on nucleotide sequence comparisons), IVSs from Yersinia and Salmonella have been shown to be closely related (>80% nucleotide identity) (26) despite overall chromosomal unrelatedness (<20% overall chromosomal homology based on DNA-DNA reassociation) (4). This atypical relatedness combined with the sporadic distribution of IVSs among bacterial genera, suggest that IVSs are inherited by horizontal transfer rather than vertically from a common ancestor.
At this point, while very little is known about which bacteria possess IVSs, even less is known about which do not. It has been reported that E. coli, Shigella, and Citrobacter have normal, intact 23S rRNA (27). Using the enteric bacteria as a model system, we decided to take a further look at the distribution of IVSs. Within the Enterobacteriaceae, two distantly related genera—Salmonella and Yersinia—have been shown to contain IVSs that are related, while E. coli, which is more closely related to Salmonella (12), does not. During our study we have found that Proteus and Providencia spp. contain IVSs that are related to those of Salmonella in their nucleotide sequence. Other genera tested, including Citrobacter, Enterobacter, Klebsiella, and Morganella, do not have IVSs. Based on DNA-DNA reassociation, Proteus and Providencia are distinct genera, for species within each genus show more than 75% DNA relatedness, while species of Proteus show only about 40% DNA relatedness to species of Providencia (5). Furthermore, species of Proteus and of Providencia show less than 20% relatedness to other enteric bacteria (5). In this study we report the presence of IVSs in three species of Proteus and one species of Providencia. Unlike other IVSs previously reported, Proteus and Providencia spp. have two very different sizes of IVSs within helix 25 of the seven different rrl genes in the same strain.
MATERIALS AND METHODS
Bacterial strains and cultivation conditions.
All strains are maintained in 15% glycerol at −70°C in the collection of the Salmonella Genetic Stock Centre (University of Calgary, Calgary, Alberta, Canada [www.ucalgary.ca/∼kesander]). See Table 1 for a list of strains and their sources. The identity of the species of the strains was confirmed in selected cases by Helene Shaw and Chandar Anand, Southern Alberta Provincial Laboratory, Calgary. All strains were grown on Luria-Bertani (LB) medium at 37°C; LB agar also contains 1.5% agar. Prior to study, the bacteria were streaked on LB agar, and a single colony was isolated in all cases except for Proteus spp., which form swarms.
TABLE 1.
List of strains useda
| Species | Strain no. | Source |
|---|---|---|
| Proteus mirabilis | SA5448 | C. Anand, Southern Alberta Provincial Laboratory |
| Proteus mirabilis | SA5449 | C. Anand, Southern Alberta Provincial Laboratory |
| Proteus mirabilis | SA5450 | C. Anand, Southern Alberta Provincial Laboratory |
| Proteus mirabilis | SA5460 | J. Penner, University of Toronto |
| Proteus mirabilis | SA5461 | J. Penner, University of Toronto |
| Proteus mirabilis | SA5475 | Salmonella Genetic Stock Centre |
| Proteus mirabilis | SGSC3360 | A. Schryvers, University of Calgary |
| Proteus penneri | CDC2518-74 (SA5462) | J. Penner, University of Toronto |
| Proteus penneri (type strain) | CDC1808-73 (SA5463) | J. Penner, University of Toronto |
| Proteus vulgaris | SA5474 | Salmonella Genetic Stock Centre |
| Proteus vulgaris | ATCC 29905 (SA5464) | American Type Culture Collection |
| Proteus vulgaris | SGSC3359 | A. Schryvers, University of Calgary |
| Proteus vulgaris (type strain) | ATCC 13315 | American Type Culture Collection |
| Providencia rettgeri | ATCC 14505 | Salmonella Genetic Stock Centre |
| Providencia rettgeri | SA5473 | Salmonella Genetic Stock Centre |
| Citrobacter freundii | SGSC3347 | A. Schryvers, University of Calgary |
| Enterobacter aerogenes | SGSC3349 | A. Schryvers, University of Calgary |
| Enterobacter cloacae | SGSC3348 | A. Schryvers, University of Calgary |
| Escherichia coli K-12 | SAB1332 | Salmonella Genetic Stock Centre |
| Klebsiella oxytoca M5a1 | SA5451 | V. Stewart, Cornell University |
| Morganella morganii | SA5435 | C. Anand, Southern Alberta Provincial Laboratory |
| Morganella morganii | SA5436 | C. Anand, Southern Alberta Provincial Laboratory |
| Morganella morganii | SA5438 | C. Anand, Southern Alberta Provincial Laboratory |
| Salmonella serovar Typhimurium LT2 | SGSC1412 | L. Lilleengen via J. Lederberg |
The strains are maintained at the Salmonella Genetic Stock Centre, University of Calgary (www.ucalgary.ca/∼kesander).
Enzymes and chemicals.
RNasin was obtained from Promega; DNase and Taq polymerase were from Pharmacia Biotech. I-CeuI was obtained from New England Biolabs. Diethyl pyrocarbonate, dimethyl sulfoxide (DMSO), and glyoxal were from the Sigma Chemical Co., Phenol was from Fisher Scientific, and sodium iodoacetate was from Aldrich. All other chemicals, including LB media and agarose, were obtained from Gibco BRL.
Oligonucleotide primers.
PCRs were performed with the following primers, designed by Mattatall and Sanderson (19) and synthesized by Gibco BRL (P1 and P4) and University Core DNA Services (Health Sciences Centre, University of Calgary) (P2 and P3): P1 (5′-GCGTCGGAAGGTGATATG-3′), P2 (5′-GCTATCTCCCGGTTTGATTG-3′), P3 (5′-CCGATGCAAACTGCGAATAC- 3′), and P4 (5′-TTCTCTACCTGACCACCTG-3′). These primers were located at E. coli rrlB bases 74 to 92, 786 to 805, 901 to 920, and 1616 to 1634. The forward and reverse sequencing primers, 5′-TACTTCTGACTGACCGATAG-3′ and 5′-GGCTAGATCACCGGGTTTCG-3′ for helix 25 and 5′-CCTGCGCGGAAGATGTAACG-3′ and 5′-GCATTCGCACTTCTGATACC-3′ for helix 45 were also synthesized by University Core DNA Services. See Fig. 1B for the location of PCR primers.
RNA isolation.
A single colony isolate was transferred into a small test tube containing 3 ml of LB medium, which was shaken overnight at 37°C. The next day, a 250-ml flask containing 20 ml of LB medium was innoculated with 1 ml from the overnight culture, and this culture was shaken at 37°C for 4 h. A 10-ml aliquot of cell suspension was centrifuged for 15 min at 5,000 × g, and the pellet of cells was quickly resuspended in 3 ml of 10 mM sodium acetate–0.15 M sucrose extraction buffer (pH 4.8) and 1% sodium dodecyl sulfate. RNA was then extracted from the pellet of cells according to the procedure described by Mattatall and Sanderson (19). A sample of the extracted RNA was quantitated for amount of nucleic acid and amount of protein contamination by measuring the absorbance of the sample at 260 and 280 nm.
Denaturing electrophoresis and Northern blotting.
Approximately 10-μg samples of RNA were combined with 8.1 μl of 6 M glyoxal, 24 μl of DMSO, and 4.5 μl of 0.1 M sodium phosphate buffer (pH 7.0) (23). The mixture was incubated for 60 min at 50°C, after which 6 μl of 50% glycerol was added to each sample. The samples were loaded into a gel of 1.5% agarose in 10 mM sodium phosphate buffer (pH 7.0) and run for 4 to 4.5 h at 50 V. The gel and a Hybond-N+ membrane (Amersham) were equilibrated in 2× SSC (1× SSC is 0.15 M sodium chloride plus 0.015 M sodium citrate [23]), and Northern blotting was performed by using 10× SSC for the transfer (23). After blotting, the membrane was fixed in 5% acetic acid for 15 min (with shaking), followed by staining in 5% methylene blue for 10 min (19). The membrane was photographed by using Polaroid Polapan 57 black-and-white instant sheet film.
PCR amplification and gel electrophoresis.
PCR DNA templates were prepared by obtaining a small amount of cells from an isolated colony on a petri plate (using the tip of a toothpick) and boiling them in double-distilled H2O (ddH2O) for 5 min, followed by rapid cooling on ice. PCR reactions were carried out according to the instructions accompanying the Taq polymerase on a Techne Gene E Thermal Cycler. Thirty cycles of 1-min denaturation (94°C), 1-min annealing (56°C), and 1-min extension (72°C) were carried out, followed by a final extension at 72°C for 10 min.
The PCR products were run through a 1.5% agarose gel in 0.5× TBE buffer (1× TBE buffer contains 90 mM Tris, 90 mM boric acid, and 2 mM EDTA [pH 8.0]) with 0.5 μg of ethidium bromide per ml. The gel was illuminated under UV light and photographed by using Polaroid Polapan 57 black-and-white instant sheet film.
Preparation of genomic DNA, I-CeuI restriction digestion, pulsed-field gel electrophoresis (PFGE), and fragment isolation.
Bacteria were grown overnight at 37°C in LB broth. The cells were diluted 10-fold with fresh LB broth and incubated for another 3 h with vigorous shaking. Genomic DNA was prepared as described earlier (16) and digested with I-CeuI endonuclease as described previously (17). DNA digests were separated by a HULA GEL (Hoefer Scientific Instruments) or a CHEF MAPPER electrophoresis system according to published methods (16, 17). The DNA fragments were excised from the gel under long-wavelength UV light. During early experiments the DNA was purified by GlassMAX (Gibco BRL); during later experiments, gel slices containing the excised DNA were boiled to melt the agar and diluted 10× with ddH2O; the DNA was then used as a template for PCR.
DNA sequencing.
After PCR amplification, the amplicons were then purified by using the Wizard PCR Preps DNA Purification System. This DNA was then cycle sequenced by University Core DNA Services by using Automated Applied Biosystems (ABI) Sequencing and Taq Dye Deoxy Terminator Cycle Sequencing Kit (ABI).
Computer analysis of sequences.
Sequences were aligned using CLUSTAL X version 1.6 for the Macintosh. Predictions of secondary structure of rRNA were done using the “mfold server” available at Michael Zuker's home page, hosted by Washington University Institute for Biomedical Computing (32). Nucleotide identity comparisons were done by using DNASIS (Hitachi Software Engineering). BLASTN (Basic Local Alignment Search Tool), provided by the National Center for Biotechnology Information, was used to search GenBank for other sequences (2).
Nucleotide sequence accession numbers.
The nucleotide sequences reported here have been deposited in GenBank under the following accession numbers: AF176785 (SA5461 C&D, helix 25), AF176786 (CDC1808-73 [all], helix 25), AF176787 (SA5474 E, helix 45), AF176788 (SA5473 B&G, helix 45), AF176789 (SA5474 B, helix 25), AF176790 (SA5474 D&G, helix 25), AF176791 (SA5473 B, helix 25), AF176792 (SA5474 GB&C, helix 45), AF176793 (ATCC 29905 B, helix 25), AF176794 (ATCC 29905 GCD&F, helix 25), AF176795 (ATCC 29905 GC&E, helix 45), AF177051 (SA5461 B, helix 45), and AF177052 (SA5473 E, helix 25).
RESULTS
Survey of enteric bacteria.
We examined a number of enteric bacteria for the presence of IVSs using the methods described in Fig. 2. The following species, listed in Table 1, showed no evidence of IVSs using PCR, nor any evidence of fragmented RNA: Citrobacter freundii (one strain), Enterobacter aerogenes (one strain), Enterobacter cloacae (one strain), Klebsiella oxytoca (one strain), and Morganella morganii (three strains) (data not shown). IVSs and fragmented RNA were, however, detected in strains of Proteus and Providencia as described below.
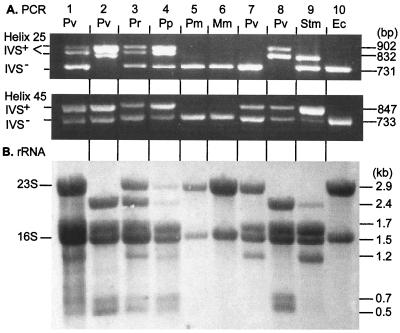
FIG. 2.
Agarose gel containing PCR products from amplification of regions of rrl genes containing IVSs and rRNA Northern blots from Proteus and Providencia strains. Lane 1, P. vulgaris (Pv, SGSC3360); lane 2, P. vulgaris (SA5474); lane 3, P. rettgeri (Pr, SA5473); lane 4, Proteus penneri (Pp, SA5462); lane 5, Proteus mirabilis (Pm, SA5450); lane 6, Morganella morganii (Mm, SA5438); lane 7, P. vulgaris (ATCC 13315, ATCC type strain); lane 8, P. vulgaris (ATCC 29905); lane 9, S. enterica serovar Typhimurium LT2 (Stm, SGSC1412); lane 10, E. coli K-12 (Ec, SA1332). (A) The regions of rrl genes containing IVSs were amplified by PCR, with primers P1 and P2 for helix 25 and primers P3 and P4 for helix 45 (as illustrated in Fig. 1). PCR products were separated by electrophoresis in agarose gels containing ethidium bromide and visualized under UV light. (B) rRNA was isolated by phenol extraction as described in Materials and Methods, separated by glyoxal–DMSO–1.5% agarose gel electrophoresis, blotted to a Hybond-N+ membrane, and stained with methylene blue (23).
Detection of IVSs using PCR.
Using primers developed by Mattatall and Sanderson (19), we used PCR to amplify the helix 25 and helix 45 regions of the rrl genes, which are known to contain IVSs in Salmonella species (Fig. 1B). Use of primers P1 and P2 in combination will amplify helix 25, while use of P3 and P4 will amplify helix 45. Whenever a gene contained an IVS in helix 25 or in helix 45, the PCR product containing that helix was larger, and a slower-running band was observed after electrophoresis.
E. coli K-12 (lane 10 of Fig. 2A) is known to contain no IVSs; PCR using the P1-P2 primer combination results in an amplicon that is predicted to be 731 bp in size based on the sequence of E. coli rrlB (from which the primers were designed) (19); the P3-P4 combination produces an amplicon that is predicted to be 733 bp in size.
Amplification products from helix 25 of serovar Typhimurium LT2 (lane 9) come in two sizes: a smaller product, corresponding to the non-IVS (IVS−) amplicon of E. coli, and a larger product that results from amplification of IVS-containing (IVS+) rrl genes (19). If all seven rrl gene copies are amplified equally, the relative intensities of the two bands should provide an estimate of how many genes contain IVSs and how many do not. Based on their relative intensity, one can estimate that two gene copies contain an IVS in helix 25 and five do not; this result is consistent with the conclusions of Mattatall and Sanderson (19).
The helix 45 amplicon for serovar Typhimurium also results in two product sizes: a smaller product corresponding to the 733-bp IVS− products of E. coli and a larger product (∼820 bp) resulting from amplification of genes that have an IVS in helix 45. The relative intensities of the two bands suggests that six genes have an IVS in helix 45 and one does not, which again confirms the conclusions of Mattatall and Sanderson (19).
The helix 45 amplicons from most strains of Proteus and both strains of Providencia that we tested came in two sizes corresponding to the IVS+ and IVS− products of Salmonella, indicating that these strains contain helix 45 IVSs that are similar in size. Analysis of the relative intensities of the helix 45 products of P. vulgaris ATCC 29905 (lane 8), for example, suggests it has four helix 45 IVS+ genes and three helix 45 IVS− genes. Table 2 contains the results of PCR analysis for 15 strains of Proteus and Providencia (including the 7 for which data are shown in Fig. 2).
TABLE 2.
Conclusions as to IVS content of Proteus and Providencia strains based on rRNA and PCR dataa
| Strain | rRNA datab
|
Total no. of IVSse based on rRNA and PCR data
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No. of fragments out of 7c
|
No. of rrl genes with IVSsd
|
|||||||||||
| 0.5 kb | 0.7 kb | 1.2 kb | 1.7 kb | 2.4 kb | 2.9 kb | Neither | H25 | H45 | Both | H25 | H45 | |
| P. vulgaris ATCC 13315 | 0 | 0 | 4 | 4 | 0 | 3 | 3 | 0 | 4 | 0 | 0 | 4 |
| P. rettgeri ATCC 14505 | 4 | 0 | 0 | 0 | 4 | 3 | 3 | 4 | 0 | 0 | 4 | 0 |
| P. vulgaris ATCC 29905 | 4 | 4 | 0 | 4 | 3 | 0 | 0 | 3 | 0 | 4 | 7 | 4 |
| P. penneri CDC1808-73 | 7 | 3 | 0 | 3 | 4 | 0 | 0 | 3 | 0 | 4 | 7 | 4 |
| P. penneri CDC2518-74 | 4 | 3 | 2 | 5 | 1 | 1 | 1 | 1 | 2 | 3 | 4 | 5 |
| P. rettgeri SA5473 | 4 | 2 | 1 | 3 | 2 | 2 | 2 | 2 | 1 | 2 | 4 | 3 |
| P. mirabilis SA5448 | 1 | 0 | 1 | 1 | 1 | 5 | 5 | 1 | 1 | 0 | 1 | 1 |
| P. mirabilis SA5449 | 0 | 0 | 2 | 2 | 0 | 5 | 5 | 0 | 2 | 0 | 0 | 2 |
| P. mirabilis SA5450 | 0 | 0 | 0 | 0 | 0 | 7 | 7 | 0 | 0 | 0 | 0 | 0 |
| P. mirabilis SA5460 | 4 | 0 | 0 | 0 | 4 | 3 | 3 | 4 | 0 | 0 | 4 | 0 |
| P. mirabilis SA5461 | 5 | 1 | 0 | 1 | 5 | 2 | 2 | 4 | 0 | 1 | 5 | 1 |
| P. vulgaris SA5474 | 7 | 4 | 0 | 4 | 3 | 0 | 0 | 3 | 0 | 4 | 7 | 4 |
| P. mirabilis SA5475 | 4 | 0 | 0 | 0 | 4 | 3 | 3 | 4 | 0 | 0 | 4 | 0 |
| P. vulgaris SGSC3359 | 1 | 0 | 1 | 1 | 1 | 5 | 5 | 1 | 1 | 0 | 1 | 1 |
| P. mirabilis SGSC3360 | 2 | 2 | 2 | 4 | 0 | 3 | 3 | 0 | 2 | 2 | 2 | 4 |
| Salmonella serovar Typhimurium LT2 SGSC1412 | 2 | 1 | 5 | 6 | 1 | 0 | 0 | 1 | 5 | 1 | 2 | 6 |
| E. coli SAB1332 | 0 | 0 | 0 | 0 | 0 | 7 | 7 | 0 | 0 | 0 | 0 | 0 |
Salmonella serovar Typhimurium LT2 and E. coli K-12, as previously reported (6, 19), have been included for comparison. H25, helix 25 in the proposed secondary structure of the rRNA produced by the rrl gene; H45, helix 45 in the proposed secondary structure of the rRNA produced by the rrl gene.
See Fig. 2 for pictures of sample rRNA and PCR gels (not all data has been shown). 23S rRNA quantitation was normalized to the intact 16S rRNA species.
The number of rrl genes coding for each fragment, measured as a fraction of the total amount of 23S rRNA expected, assuming all strains have seven rrl genes.
See Fig. 1 for a diagrammatic representation of an rrl gene; the data in Fig. 2 indicate the number of each type of rrl gene, based on the rRNA fragments observed.
Total number of helix 25 and helix 45 IVSs in the seven rrl genes as determined from the rRNA data and PCR data.
The helix 25 amplicons from most of these strains show two different sizes of IVS+ products in addition to the IVS− size. These results suggest that in Proteus and Providencia species, individual strains have IVSs of different sizes in their (predicted) seven rrl genes in helix 25. ATCC 29905 (lane 8) appears to have IVSs in all seven rrl genes: four of the helix 25 IVS+ genes that are similar in size to those of serovar Typhimurium and three helix 25 IVS+ genes that are ∼70 bp larger. ATCC 13315 (lane 7), in contrast, has no helix 25 IVS+ genes. Data for all 15 strains are summarized in Table 2.
Fragmentation of 23S rRNA due to IVS excision.
Cellular RNA was extracted with phenol, separated by denaturing agarose gel electrophoresis, and visualized by blotting to a Hybond-N+ membrane and methylene blue staining. E. coli (lane 10, panel B), which we know contains no IVSs, has intact 23S and 16S species. Serovar Typhimurium (lane 9), in contrast, lacks an intact 23S rRNA species, and several smaller fragments can be seen.
Figure 1B provides a diagrammatic representation of the rrl operon and the rRNA fragments that result from IVS cleavage. If an rrl gene contains an IVS in helix 25 and that IVS is removed during posttranscriptional processing (without religation of the rRNA), two smaller fragments of 2.4 and 0.5 kb result. If an rrl gene contains a helix 45 IVS, two fragments of 1.7 and 1.2 kb result. Finally, if an rrl gene contains IVSs at both helix 25 and helix 45, three fragments of 1.7, 0.7, and 0.5 kb result. We assume that the seven different operons are transcribed equally; the relative intensity of the various rRNA bands can be analyzed to obtain an estimate of IVS content, as we did with the PCR data.
Serovar Typhimurium (lane 9) is known to contain two helix 25 IVSs and six helix 45 IVSs, which are distributed as follows: one rrl gene contains a helix 25 IVS only, five contain a helix 45 IVS only, and one contains IVSs in both helices (19). From the rrl gene that contains only a helix 25 IVS, we expect a number of 2.4- and 0.5-kb rRNA molecules that is one-seventh the number of 1.5-kb rRNA molecules from the seven 16S rRNA genes. Similarly, from the five rrl genes containing helix 45 IVSs only, we expect a number of 1.7- and 1.2-kb rRNA molecules that is five-sevenths the number of 1.5-kb molecules. Finally, from the rrl gene that contains both helix 25 and helix 45 IVSs, we expect a number of 1.7-, 0.7-, and 0.5-kb rRNA molecules that is one-seventh the number of 1.5-kb molecules from the 16S rRNA genes. The ratios of the numbers of each molecule to the numbers of 1.5-kb molecules should be as follows: 0/7 for the 2.9-kb molecules (i.e., no intact 23S species), 1/7 for the 2.4- and 0.7-kb molecules, 2/7 for the 0.5-kb molecules, 5/7 for the 1.2-kb molecules, and 6/7 for the 1.7-kb molecules, as shown in Table 2. The intensity of the staining is also affected by the molecular weight of the fragment. With the exception of the bands corresponding to the 0.5- and 0.7-kb molecules, which were too faint to be easily distinguishable in this picture (but have been seen in other pictures), all bands can be seen, in the expected ratios, in lane 9 of Fig. 2B.
P. vulgaris ATCC 29905 (lane 8) contains no intact 23S species (no 2.9-kb molecules) and no 1.2-kb molecules, which indicates that all seven rrl genes contain an IVS in helix 25. The number of helix 45 IVSs can be estimated by comparing the intensity of the 2.4- and 1.7-kb bands. Since they are nearly equal in intensity, we conclude that there are three rrl genes that contain helix 25 IVSs only and four that contain IVSs in both helices (Table 2). The observed rRNA fragments and the estimated number of rrl genes with IVSs are summarized for all Proteus and Providencia strains in Table 2. The numbers of IVSs estimated for each strain, determined from both PCR and RNA analysis, are the same.
Sequence data from individual rrl genes.
In order to obtain sequence data from the IVSs in individual rrl genes, it is necessary to separate the seven gene copies; sequence data from genomic DNA containing a mixture of genes with or without IVSs would not yield meaningful data. I-CeuI is a restriction enzyme that recognizes a sequence that appears in all rRNA (rrn) operons near bp 2200 in the rrl genes (15). Digestion of genomic DNA of most enteric bacteria with this enzyme results in seven fragments and, if all genes are oriented in the same direction around the chromosome, each fragment will contain the sequence for one copy of helix 25 and one copy of helix 45. Genomic DNA from Providencia and Proteus strains was isolated in agarose blocks, and the DNA was digested with I-CeuI and separated by PFGE (data not shown); individual fragments were excised and used as DNA templates. PCR was used to amplify the helix 25 and helix 45 regions of the rrl genes, the products were purified by using the Wizard PCR Preps DNA Purification System, and the DNA was cycle sequenced by University Core DNA Service (University of Calgary).
The sequences were compared with each other for nucleotide identity with DNASIS (Tables 3 and 4) and aligned by using CLUSTAL X (Fig. 3), and secondary structure predictions were obtained from the mfold server (Fig. 4). The IVSs have been grouped into families where a family consists of all sequences that have 90% or greater nucleotide identity. IVSs from helix 25 fall into four families, with families E to G being quite similar in sequence (80 to 85% nucleotide identity) (Table 3), in length (183 to 187 bp) (Fig. 3A), and in putative secondary structure (Fig. 4, where only family G is shown). IVSs in family D have only 50% nucleotide identity to families E to G and are much shorter (only 113 bp), but they apparently have very similar secondary structure (Fig. 4).
TABLE 3.
Percent nucleotide identity between IVSs from helix 25 of the 23S rRNA of Proteus, Providencia, and Salmonella strainsa
| IVS family
|
% Nucleotide identity for (family, strain, gene):
|
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| E
|
F
|
G
|
D
|
A
|
B
|
||||||||||
| Strain | rrl gene | SA 5461
|
SA5473
|
SA5463, (All) | SA5474
|
SA5564, B | SA5564
|
LT2
|
|||||||
| C | D | B | B | D | G | C | D | F | G | rrlHb | rrlGb | ||||
| P. mirabilis SA5461 | C | 100 | 85 | 85 | 85 | 85 | 85 | 86 | 50 | 50 | 50 | 50 | 50 | 49 | |
| P. mirabilis SA5461 | D | 100 | 85 | 85 | 85 | 85 | 85 | 86 | 50 | 50 | 50 | 50 | 50 | 49 | |
| P. rettgeri SA5473 | B | 85 | 85 | 80 | 80 | 81 | 81 | 81 | 49 | 49 | 49 | 49 | 49 | 47 | |
| P. penneri SA5463 | (All) | 85 | 85 | 80 | 93 | 94 | 94 | 95 | 50 | 50 | 50 | 50 | 49 | 49 | |
| P. vulgaris SA5474 | B | 85 | 85 | 80 | 93 | 99 | 99 | 97 | 50 | 50 | 50 | 50 | 50 | 48 | |
| P. vulgaris SA5474 | D | 85 | 85 | 81 | 94 | 99 | 100 | 98 | 50 | 50 | 50 | 50 | 50 | 49 | |
| P. vulgaris SA5474 | G | 85 | 85 | 81 | 94 | 99 | 100 | 98 | 50 | 50 | 50 | 50 | 50 | 49 | |
| P. vulgaris SA5564 | B | 86 | 86 | 81 | 95 | 97 | 98 | 98 | 49 | 49 | 49 | 49 | 48 | 48 | |
| P. vulgaris SA5564 | C | 50 | 50 | 49 | 50 | 50 | 50 | 50 | 49 | 100 | 100 | 100 | 76 | 60 | |
| P. vulgaris SA5564 | D | 50 | 50 | 49 | 50 | 50 | 50 | 50 | 49 | 100 | 100 | 100 | 76 | 60 | |
| P. vulgaris SA5564 | F | 50 | 50 | 49 | 50 | 50 | 50 | 50 | 49 | 100 | 100 | 100 | 76 | 60 | |
| P. vulgaris SA5564 | G | 50 | 50 | 49 | 50 | 50 | 50 | 50 | 49 | 100 | 100 | 100 | 76 | 60 | |
| Salmonella serovar Typhimurium LT2 | rrlHb | 50 | 50 | 49 | 49 | 50 | 50 | 50 | 48 | 76 | 76 | 76 | 76 | 65 | |
| Salmonella serovar Typhimurium LT2 | rrlGb | 49 | 49 | 47 | 49 | 48 | 49 | 49 | 48 | 60 | 60 | 60 | 60 | 65 | |
Nucleotide identity comparisons were done by using DNASIS version 2.0 for the Macintosh. A family consists of IVS sequences with ≥90% nucleotide identity. rrl genes have been named for the I-CeuI fragments from which they were isolated, except in the case of serovar Typhimurium, where the gene designation is given. Strains shown within the boxes are in the same IVS family. “All” indicates that all seven rrl genes have identical intervening sequences.
TABLE 4.
Percent nucleotide identity between IVSs from helix 45 of the 23S rRNA of Proteus, Providencia, and Salmonella strainsa
| IVS family
|
% Nucleotide identity for (family, strain, gene):
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Q
|
R
|
M
|
|||||||||
| Strain | rrl gene | SA5473
|
SA5474
|
SA5564
|
LT2
|
||||||
| B | G | B | C | E | G | C | E | G | rrlHb | ||
| P. rettgeri SA5473 | B | 100 | 78 | 78 | 77 | 78 | 78 | 78 | 78 | 66 | |
| P. rettgeri SA5473 | G | 100 | 78 | 78 | 77 | 78 | 78 | 78 | 78 | 66 | |
| P. vulgaris SA5474 | B | 78 | 78 | 100 | 99 | 100 | 100 | 100 | 100 | 69 | |
| P. vulgaris SA5474 | C | 78 | 78 | 100 | 99 | 100 | 100 | 100 | 100 | 69 | |
| P. vulgaris SA5474 | E | 77 | 77 | 99 | 99 | 99 | 99 | 99 | 99 | 68 | |
| P. vulgaris SA5474 | G | 78 | 78 | 100 | 100 | 99 | 100 | 100 | 100 | 69 | |
| P. vulgaris SA5564 | C | 78 | 78 | 100 | 100 | 99 | 100 | 100 | 100 | 69 | |
| P. vulgaris SA5564 | E | 78 | 78 | 100 | 100 | 99 | 100 | 100 | 100 | 69 | |
| P. vulgaris SA5564 | G | 78 | 78 | 100 | 100 | 99 | 100 | 100 | 100 | 69 | |
| Salmonella serovar Typhimurium LT2 | rrlHb | 66 | 66 | 69 | 69 | 68 | 69 | 69 | 69 | 69 | |
Nucleotide identity comparisons were done by using DNASIS version 2.0 for the Macintosh. A family consists of IVS sequences with ≥90% nucleotide identity. rrl genes have been named for the I-CeuI fragments from which they were isolated, except in the case of Salmonella serovar Typhimurium, where the gene designation is given. Boxes are described in Table 3, footnote a.
The nucleotide sequences are all from this report except for Salmonella LT2 rrlH (GenBank accession no. U49926).
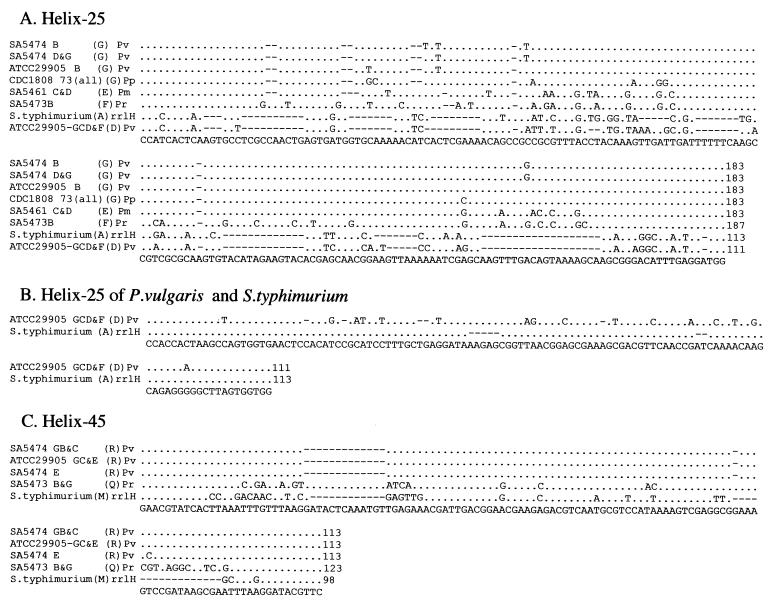
FIG. 3.
Intervening sequences from individual genes for rRNA of selected Proteus, Providencia, and Salmonella strains. Individual rrl genes were sequenced as described in Materials and Methods, and aligned using CLUSTAL X. The conserved residues are shown by dots, and deletions are shown by spaces. The consensus sequence is at the bottom. Each sequence is identified by the strain number and the I-CeuI fragment from which it was isolated, except in the case of serovar Typhimurium, where the individual rrl gene is indicated. Letters in parentheses indicate the family of IVSs to which each sequence belongs, where a family consists of sequences with 90% or greater nucleotide identity based on DNASIS (see Tables 3 and 4). We propose four families for helix 25 and two families for helix 45. The species are as follows: Pv, Proteus vulgaris; Pp, Proteus penneri; Pm, Proteus mirabilis; Pr, Providencia rettgeri, and S. typhimurium, Salmonella serovar Typhimurium. Numbers at the end of each sequence indicate the base pairs in the IVS.
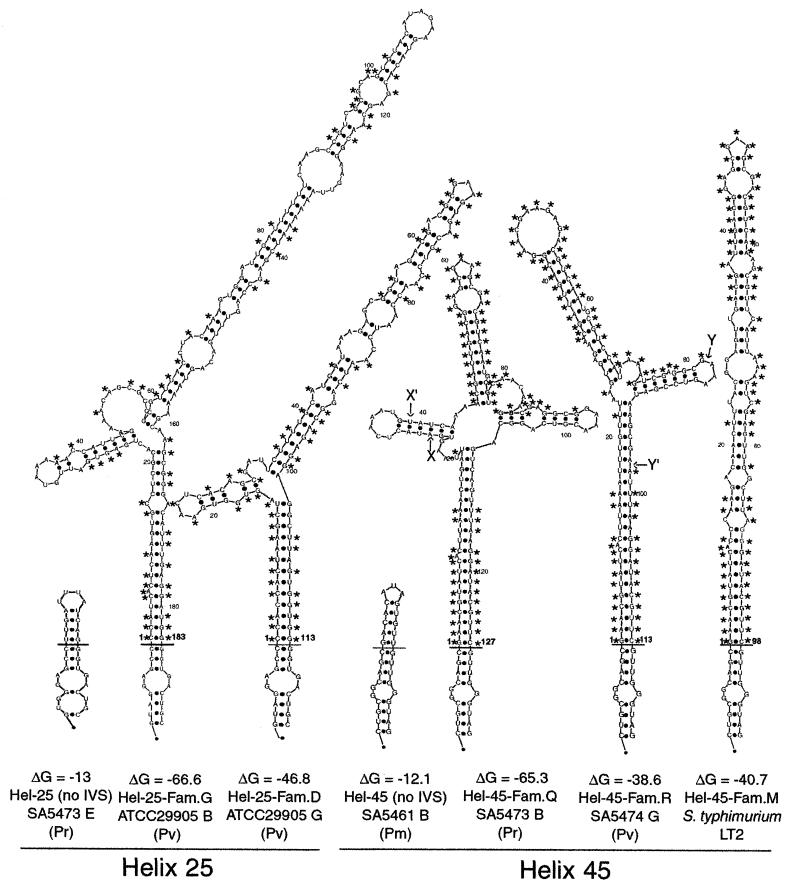
FIG. 4.
Proposed secondary structure of IVSs from Proteus, Providencia, and Salmonella rRNAs. Secondary structure predictions were obtained using the mfold server available at Michael Zuker's home page (www.ibc.wustl.edu/∼zuker/). One sequence from each of family G and family D of helix 25 IVSs and of the two families of helix 45, as well as an IVS− tetraloop of helix 25 and helix 45, are shown. The structures of families E and F of helix 25, which resemble that of family G, are not shown. The helix 45 IVS from serovar Typhimurium LT2 rrlH is included. The labels show the IVS helix and family, the strain number and I-CeuI fragment from which the gene was isolated, and the species (Pr, Providencia rettgeri; Pv, Proteus vulgaris; Pm, Proteus mirabilis; and S. typhimurium LT2, Salmonella serovar Typhimurium LT2). The positions of gaps indicated by the CLUSTAL X alignment of helix 45 IVSs are shown as X to X′ and Y to Y′. The short horizontal line indicates the position at which it is postulated that the rRNA at helix 25 or helix 45 was replaced by the IVS; the boldface number above this line indicates the number of base pairs in the IVS. The residues showing conservation for either helix 25 or helix 45 from the CLUSTAL alignment in Fig. 3 are marked by asterisks.
The IVSs from rrlH of serovar Typhimurium (Stm-rrlH) show 76% nucleotide identity to family D IVSs from Proteus, while those from rrlG of serovar Typhimurium (Stm-rrlG) show only 60% nucleotide identity (Table 3). The CLUSTAL X alignment of these sequences shows that the ends (corresponding to stems in secondary structure, Fig. 4) of Stm-rrlH and family D IVSs are nearly identical (Fig. 3B); these sequences are also highly related through the central loop region.
Based on nucleotide identity, IVSs from helix 45 of Providencia rettgeri were placed in family Q; members of family R from P. vulgaris are about 78% identical to family Q (Table 4). Substantial identity can be seen between families Q and R and IVSs of serovar Typhimurium LT2, shown as rrlH in the CLUSTAL X alignment (Fig. 3C), where 13 out of 15 bases at either end and 41 out of 55 bases in the middle are identical. In the secondary structure (Fig. 4), these regions correspond to the stems (near the site of IVS cleavage) and the ends of the loops.
DISCUSSION
In most of the Proteus strains and both Providencia strains examined here, the 23S rRNA contained as least some degree of fragmentation, a finding characteristic of rRNA transcribed from rrl genes that contain IVSs. PCR amplification of the helix 25 (∼bp 550) and helix 45 (∼bp 1170) encoding regions of rrl genes of these strains clearly shows the presence of IVSs. Based on nucleotide sequence data, the IVSs in helix 25 in Proteus and Providencia strains vary from 113 to 187 bp in length; although larger IVSs have been reported, this is the first time that two IVSs that are so different in size have been reported in helix 25 of different rrl genes in the same strain. IVSs in helix 45 varied from 113 to 123 bp.
The number of IVSs estimated from PCR (which corresponds to DNA sequences in the rrl genes) and the number of IVSs estimated from the rRNA fragmentation patterns (which indicates the sites at which rRNA is cut) are equal in all of the strains studied (Table 2). This indicates that there are no IVSs that are not excised (presumably by RNase III), and it also indicates that there is no fragmentation resulting from causes other than IVSs. Recently, Selenska-Pobell and Doring (24) discovered fragmentation of 23S rRNA in the family Rhizobiaceae that does not result from excision of IVSs; we find no evidence of this in the strains we examined.
In general, the Proteus and Providencia rrl genes are quite heterogeneous with respect to IVS content. In most cases, more than one rrl gene contains an IVS and some strains contain IVSs in all seven genes. Furthermore, most strains that possess helix 25 IVSs have two different sizes of IVSs. The possession of IVSs is not, however, a clear species characteristic; all four strains of P. vulgaris and two strains of Proteus penneri have IVSs, but only five out of six strains of Proteus mirabilis have IVSs. Based on nucleotide identity, the helix 25 IVSs represent four families, with families E, F, and G being larger (183 to 187 bp) than family D (113 bp), and they are quite similar to each other in sequence (>80% nucleotide identity) and in postulated secondary structure.
Although a BLASTN search of GenBank with sequences from families E to G revealed no significant hits with other IVSs, family D has nearly identical stems (and similar size) to helix 25 IVSs found in serovar Typhimurium (Stm-rrlH). Another group of helix 25 IVSs from serovar Typhimurium (Stm-rrlG) is similar in size to family D IVSs of Providencia rettgeri, but it shows lower nucleotide identity (60%). Furthermore, family D IVSs show greater similarity (76% nucleotide identity) to Stm-rrlH IVSs than the two IVSs from serovar Typhimurium show to each other (65%). These results lend evidence to the hypothesis that IVSs in bacteria are distributed by lateral transfer (19, 22, 26).
Further evidence of lateral transfer is provided by comparison of helix 45 IVSs from Proteus, Providencia, and Salmonella spp. Although the nucleotide identity between these IVSs is not high (66 to 78%), there is a striking similarity in the stems and terminal loop structures of the IVS secondary structure. As the sequence alignment in Fig. 3C shows, there is a high degree of conservation in the sequences at either end of the helix 45 sequences, as well as a high degree of conservation in the middle of these sequences. One block of 14 bases (X to X′ in Fig. 4) in helix 45 family Q (127 bases) has been deleted from family R (113 bases) and from Stm-rrlH (this deletion is shown as bases 28 to 39 in Fig. 3C). Another block of 17 bases (Y to Y′ in Fig. 4) is present in both family Q and R; this block, shown as bases 95 to 111 in Fig. 3C, has been deleted from Stm-rrlH. Thus, these sequences are strikingly similar, except for the insertions or deletions.
IVSs are sporadically distributed among bacterial genera, with some genera such as Salmonella, Yersinia, and Proteus having IVSs in most strains, while other genera such as Escherichia and Morganella appear to have none at all. Even within the genera that possess them, IVSs tend to be unevenly distributed, with some strains having IVSs in all seven rrl genes and others having only a few. Furthermore, the IVS sequence itself tends to vary within a genus as well as across genera. In some cases, such as in Yersinia and Salmonella, and in Proteus, Providencia, and Salmonella, there are sequences that are more closely related between two genera than with other IVSs within a genus.
In 1991, Skurnik and Toivanen (26) showed that the IVSs from helix 45 of Yersinia enterocolitica were remarkably similar to those in helix 45 of Salmonella serovars Typhimurium (89% nucleotide identity) and Arizonae (84% nucleotide identity), even though overall chromosomal DNA reassociation data show less than 20% similarity between Yersinia and Salmonella (4). In this report, we have also seen that helix 25 family D IVSs from Proteus spp. are more closely related to those from helix 25 of rrlH of serovar Typhimurium than they are to other families of Proteus and Providencia IVSs. This discrepancy suggests that IVSs are inherited by lateral transfer rather than vertically from a common ancestor. Under such a hypothesis, IVSs are transferred from one species to another (by phage-mediated transduction, transformation, or conjugation). These may then be copied from one rrl gene to another either by reciprocal recombination (recA dependent) or gene conversion (nonreciprocal) (19).
Despite the discovery of IVSs among several bacterial genera, there is no satisfactory explanation of their function. It was suggested that IVSs may protect the cell from unknown bacteriocins (26) or that having fragmented rRNA may provide the cell an advantage in stationary-phase growth (8). Whatever their origin, IVSs are being discovered in more and more bacterial genera. It will be interesting to see if a biological function is found for IVSs; they appear to have no effect on the ability of the bacterium to survive.
ACKNOWLEDGMENTS
We greatly appreciate the assistance of Ying-Mei Fu and Shu-Lin Liu in providing PFGE fragments of some of the bacteria.
The work was supported by an operating grant from the Natural Sciences and Engineering Research Council of Canada and by grants 34829-09A1 and 5R01-AI43283 from the National Instiute of Allergy and Infectious Diseases of the National Institutes of Health. W.L.M. is grateful for Summer Studentship grants from the Alberta Heritage Foundation for Medical Research.
REFERENCES
- 1.Afseth G, Mo Y-Y, Mallavia L P. Characterization of the 23S and 5S rRNA genes of Coxiella burnetii and identification of an intervening sequences within the 23S rRNA gene. J Bacteriol. 1995;177:2946–2949. doi: 10.1128/jb.177.10.2946-2949.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul S F, Madden T L, Schäffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Asai T, Zaporojets D, Squires C, Squires C L. An Escherichia coli strain with all chromosomal rRNA operons inactivated: complete exchange of rRNA genes between bacteria. Proc Natl Acad Sci USA. 1999;96:1971–1976. doi: 10.1073/pnas.96.5.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bercovier H, Brenner D J, Ursing J, Steigerwalt A G, Fanning G R, Alonso J M, Carter G P, Mollaret H H. Characterization of Yersinia enterocolitica sensu stricto. Curr Microbiol. 1980;4:201–206. [Google Scholar]
- 5.Brenner D J, Farmer III J J, Fanning G R, Stiegerwalt A G, Klykken P, Wathen H G, Hickman F W, Ewing W H. Deoxyribonucleic acid relatedness of Proteus and Providencia species. Int J Syst Bacteriol. 1978;28:269–282. [Google Scholar]
- 6.Burgin A B, Parodos K, Lane D J, Pace N R. The excision of intervening sequences from Salmonella 23S ribosomal RNA. Cell. 1990;60:405–414. doi: 10.1016/0092-8674(90)90592-3. [DOI] [PubMed] [Google Scholar]
- 7.Gregory S T, O'Connor M, Dahlberg A E. Functional Escherichia coli 23S rRNAs containing processed and unprocessed intervening sequences from Salmonella typhimurium. Nucleic Acids Res. 1996;24:4918–4923. doi: 10.1093/nar/24.24.4918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hsu D, Shih L-M, Zee Y C. Degradation of rRNA in Salmonella strains: a novel mechanism to regulate the concentrations of rRNA and ribosomes. J Bacteriol. 1994;176:4761–4765. doi: 10.1128/jb.176.15.4761-4765.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hsu D, Zee Y C, Ingraham J, Shih L M. Diversity of cleavage patterns of Salmonella 23S rRNA. J Gen Microbiol. 1992;138:199–203. doi: 10.1099/00221287-138-1-199. [DOI] [PubMed] [Google Scholar]
- 10.Hurtado A, Clewley J P, Linton D, Owen R J, Stanley J. Sequence similarities between large subunit ribosomal RNA gene intervening sequences from different Helicobacter species. Gene. 1997;194:69–75. doi: 10.1016/s0378-1119(97)00158-3. [DOI] [PubMed] [Google Scholar]
- 11.Konkel M E, Marconi R T, Mead D J, Cieplak W., Jr Identification and characterization of an intervening sequence within the 23S ribosomal RNA genes of Campylobacter jejuni. Mol Microbiol. 1994;14:235–241. doi: 10.1111/j.1365-2958.1994.tb01284.x. [DOI] [PubMed] [Google Scholar]
- 12.Krieg N R, Holt J G. Bergey's manual of systematic bacteriology. Baltimore, Md: The Williams and Wilkins Co.; 1984. [Google Scholar]
- 13.Linton D, Dewhirst F E, Clewley J P, Owen R J, Burnens A P, Stanley J. Two types of 16S rRNA gene are found in Campylobacter helveticus: analysis, applications and characterization of the intervening sequence found in some strains. Microbiology. 1994;140:847–855. doi: 10.1099/00221287-140-4-847. [DOI] [PubMed] [Google Scholar]
- 14.Liu S-L, Hessel A, Sanderson K E. Genomic mapping with I-CeuI, an intron-encoded endonuclease, specific for genes for ribosomal RNA, in Salmonella spp., Escherichia coli, and other bacteria. Proc Natl Acad Sci USA. 1993;90:6874–6878. doi: 10.1073/pnas.90.14.6874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu S-L, Hessel A, Sanderson K E. The XbaI-BlnI-CeuI genomic cleavage map of Salmonella typhimurium LT2 determined by double digestion, end labelling, and pulsed-field gel electrophoresis. J Bacteriol. 1993;175:4104–4120. doi: 10.1128/jb.175.13.4104-4120.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu S-L, Sanderson K E. A physical map of the Salmonella typhimurium LT2 genome made by using XbaI analysis. J Bacteriol. 1992;174:1662–1672. doi: 10.1128/jb.174.5.1662-1672.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu S-L, Sanderson K E. I-CeuI reveals conservation of the genome of independent strains of Salmonella typhimurium. J Bacteriol. 1995;177:3355–3357. doi: 10.1128/jb.177.11.3355-3357.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mattatall N R, Daines D A, Liu S-L, Sanderson K E. Salmonella typhi contains identical intervening sequences in all seven rrl genes. J Bacteriol. 1996;178:5323–5326. doi: 10.1128/jb.178.17.5323-5326.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mattatall N R, Sanderson K E. Salmonella typhimurium LT2 possesses three distinct 23S rRNA intervening sequences. J Bacteriol. 1996;178:2272–2278. doi: 10.1128/jb.178.8.2272-2278.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mattatall N R, Sanderson K E. RNase III-deficient Salmonella typhimurium LT2 contains intervening sequences (IVSs) in its 23S rRNA. FEMS Microbiol Lett. 1998;159:179–185. doi: 10.1111/j.1574-6968.1998.tb12858.x. [DOI] [PubMed] [Google Scholar]
- 21.Ralph D, McClelland M. Intervening sequence with conserved open reading frame in eubacterial 23S rRNA genes. Proc Natl Acad Sci USA. 1993;90:6864–6868. doi: 10.1073/pnas.90.14.6864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ralph D, McClelland M. Phylogenetic evidence for horizontal transfer of an intervening sequence between species in a spirochete genus. J Bacteriol. 1994;176:5982–5987. doi: 10.1128/jb.176.19.5982-5987.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 24.Selenska-Pobell S, Doring H. Sequences around the fragmentation sites of the large subunit ribosomal RNA in the family Rhizobiaceae. 23S-like rRNAs in Rhizobiaceae. Antonie Leeuwenhoek. 1998;73:55–67. doi: 10.1023/a:1000540023194. [DOI] [PubMed] [Google Scholar]
- 25.Selenska-Pobell S, Evguenieva-Hackenberg E. Fragmentations of the large-subunit rRNA in the family Rhizobiaceae. J Bacteriol. 1995;177:6993–6998. doi: 10.1128/jb.177.23.6993-6998.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Skurnik M, Toivanen P. Intervening sequences (IVSs) in the 23S ribosomal RNA genes of pathogenic Yersinia enterocolitica strains. The IVSs in Y. enterocolitica and Salmonella typhimurium have a common origin. Mol Microbiol. 1991;5:585–593. doi: 10.1111/j.1365-2958.1991.tb00729.x. [DOI] [PubMed] [Google Scholar]
- 27.Smith N H, Crichton P B, Old D C, Higgins C F. Ribosomal-RNA patterns of Escherichia coli, Salmonella typhimurium and related Enterobacteriaceae. J Med Microbiol. 1988;25:223–228. doi: 10.1099/00222615-26-3-223. [DOI] [PubMed] [Google Scholar]
- 28.Song X M, Forsgren A, Janson H. Fragmentation heterogeneity of 23S ribosomal RNA in Haemophilus species. Gene. 1999;230:287–293. doi: 10.1016/s0378-1119(99)00063-3. [DOI] [PubMed] [Google Scholar]
- 29.Trust T J, Logan S M, Gustafson C E, Romaniuk P J, Kim N W, Chan V L, Ragan M A, Guerry P, Gutell R R. Phylogenetic and molecular characterization of a 23S rRNA gene positions the genus Campylobacter in the epsilon subdivision of the Proteobacteria and shows that the presence of transcribed spacers is common in Campylobacter spp. J Bacteriol. 1994;176:4597–4609. doi: 10.1128/jb.176.15.4597-4609.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Winkler M E. Ribosomal ribonucleic acid isolated from Salmonella typhimurium: absence of the intact 23S species. J Bacteriol. 1979;139:842–849. doi: 10.1128/jb.139.3.842-849.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Woese C R. Bacterial evolution. Microbiol Rev. 1987;51:221–271. doi: 10.1128/mr.51.2.221-271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zuker M, Mathews D H, Turner D H. Algorithms and thermodynamics for RNA secondary structure prediction: a practical guide in RNA biochemistry and biotechnology. In: Barciszewski J, Clark B F C, editors. NATO ASI Series. Norwell, Mass: Kluwer Academic Publishers; 1999. pp. 11–43. [Google Scholar]