Abstract
The alkylbenzoate degradation genes of Pseudomonas putida TOL plasmid are positively regulated by XylS, an AraC family protein, in a benzoate-dependent manner. In this study, we used deletion mutants and hybrid proteins to identify which parts of XylS are responsible for the DNA binding, transcriptional activation, and benzoate inducibility. We found that a 112-residue C-terminal fragment of XylS binds specifically to the Pm operator in vitro, protects this sequence from DNase I digestion identically to the wild-type (wt) protein, and activates the Pm promoter in vivo. When overexpressed, that C-terminal fragment could activate transcription as efficiently as wt XylS. All the truncations, which incorporated these 112 C-terminal residues, were able to activate transcription at least to some extent when overproduced. Intactness of the 210-residue N-terminal portion was found to be necessary for benzoate responsiveness of XylS. Deletions in the N-terminal and central regions seriously reduced the activity of XylS and caused the loss of effector control, whereas insertions into the putative interdomain region did not change the basic features of the XylS protein. Our results confirm that XylS consists of two parts which probably interact with each other. The C-terminal domain carries DNA-binding and transcriptional activation abilities, while the N-terminal region carries effector-binding and regulatory functions.
The TOL plasmid of Pseudomonas putida encodes a pathway for the catabolism of toluene and xylenes (35, 36). The genes, which encode enzymes for catabolism of these hydrocarbons, are grouped into two operons. The upper pathway operon specifies oxidation of toluene to benzoate and xylenes to alkylbenzoates. The meta-pathway operon specifies further oxidation of these compounds, whereas the aromatic ring in catechols, the pathway intermediates, is cleaved in meta fission. Two regulatory proteins, XylR and XylS, positively regulate the catabolic operons (28). In the presence of upper pathway substrates, XylR activates the Pu promoter of the upper pathway operon and the Ps1 promoter of the xylS gene. Subsequently, overproduced XylS protein activates the Pm promoter of the meta-cleavage operon (10, 26) via binding to the operator-sequence Om (9, 12, 13). Furthermore, XylS protein is constitutively expressed at a low level from the weak Ps2 promoter (6) and in the presence of benzoates, i.e., the degradation products of the upper pathway and substrates for the meta pathway, it activates the Pm promoter at low protein concentrations. Therefore, transcriptional activation by XylS is stimulated by alkylbenzoates and modulated by the intracellular level of the protein. Expression of XylS from strong promoters has shown that overproduction of the protein, naturally mediated by XylR, is sufficient for activation of Pm in the absence of benzoate effector (11, 19, 34). On that basis, Mermod et al. (19) have suggested a hypothesis about a dynamic equilibrium between inactive and active, DNA-binding conformations of the protein in the cell. The putative role of effector would be to shift the equilibrium toward the active conformation. In vitro studies from our lab support the idea, as we have shown that alkylbenzoate slightly facilitates site-specific DNA binding but does not change the pattern of the DNA contacts made by XylS (12).
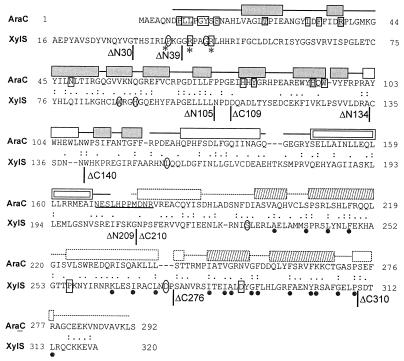
To identify which parts of the protein mediate the effect of benzoates, extensive mutagenesis of the xylS gene has been carried out. Several amino acid substitutions have been found that altered effector specificity of XylS (20, 25, 27) or produced a semiconstitutive phenotype. The latter was characterized by an increased basal level of transcriptional activity which was still inducible by benzoate (20, 37). Mutations of both types were scattered all over the xylS gene; however, many of them were clustered in a small glycine-rich N-terminal region P37-R45 (Fig. 1). It has been shown that some mutations in the C terminus are intra-allelically dominant over substitutions in the N terminus and, in contrast, a mutation in the N terminus (R45T) can restore the effector control that has been lost due to these C-terminal mutations. All these data suggest that the N and C termini of XylS may interact, and benzoate effectors may regulate the activity of the protein by modulation of that interaction (21).
FIG. 1.
Sequence alignment of E. coli AraC (accession no. P03021) and P. putida XylS (P07859) proteins. Identical residues are marked by colons and similar ones by periods. An 18.5% identity and a 51.2% similarity were found in a 248-residue overlap (amino acids 45 to 284 in AraC and 79 to 319 in XylS). A 21.2% identity and a 58.6% similarity were found in a 104-residue C-terminal region (amino acids 183 to 284 in AraC and 218 to 319 in XylS). Sequences were aligned by using the LALIGN program. Vertical lines indicate the endpoints of XylS truncations generated in this study. Mutations of boxed XylS residues affect effector specificity. Mutations of rounded XylS residues produce semiconstitutive transcriptional activation, and mutations of residues marked by an asterisk increase XylS stability. Mutations of R41 and R45 in XylS cause both semiconstitutive and altered effector specificity phenotypes (20, 27, 37). Dots under the sequence of XylS indicate conserved residues in the AraC-XylS family (7). The AraC residues, which interact directly with arabinose, are printed on a gray background, and those interacting indirectly are boxed. The elements of the AraC NTR secondary structure are shown above the sequence. Gray boxes indicate beta-sheets, and transparent boxes indicate helices. The dimerization helix, participating in a coiled coil, is shown by a double box (32). The linker region of AraC is underlined (4, 5). The secondary structure elements of homologous MarA protein are shown above the AraC CTD sequence with a dotted outline; the helix-turn-helix motifs are shaded (29).
XylS belongs to the AraC-XylS family of bacterial transcriptional activators (7). Proteins of this family are characterized by significant homology over a 100-residue stretch, a region that is proposed to be necessary for DNA binding and stimulation of transcription. Several small monomeric activators in the AraC-XylS family (e.g., MarA and SoxS from Escherichia coli) match the conserved sequence and do not contain additional domains (1, 3). A crystal structure for one of these proteins, MarA, in complex with its cognate binding site has been determined recently (29). AraC, the model protein of the family consists of two functional domains. The conserved C-terminal domain carries sequence-specific DNA-binding capability while the nonconserved N-terminal domain mediates effector responsiveness and carries dimerization capability (2, 16). The DNA-binding domain most probably contains all of the determinants necessary for transcriptional activation, since the separately expressed C-terminal domain of AraC has residual ability to activate transcription without the arabinose effector (18). In AraC, the N-terminal arm of the regulatory domain binds to the C-terminal domain and acts as an intramolecular modulator of transcriptional activation (31). Structural data show that binding of arabinose causes rearrangement of the arm, thereby releasing the protein to bind the target sites which are necessary for activation of transcription (32, 33).
Like the other regulators of carbon metabolism in the family, XylS contains the conserved region in its C terminus and could have the AraC-like modular organization. However, the modular structure of the XylS protein had not been demonstrated up to now, and the truncated variants of the protein have been reported to be completely inactive (14). XylS and AraC show some sequence homology not only in the C-terminal portion but also in the N-terminal region (Fig. 1). Therefore, we set a goal to specify which parts of XylS are responsible for the DNA-binding, transcriptional activation, and effector responsiveness of the protein and to clarify whether XylS and AraC proteins use similar mechanisms under which the effector controls the activity of the transcription factor. The results of our work show that XylS is a modular protein with a C-terminal DNA-binding–transcription activation domain and an N-terminal effector-binding–regulatory region.
MATERIALS AND METHODS
Construction of plasmids and strains.
For cloning and plasmid propagation, E. coli DH5α was grown in Luria-Bertani (LB) medium or on L agar at 37°C (30). Media were supplemented with 100 μg of ampicillin ml−1 and 7 μg tetracycline (Tc) ml−1 when required. DNA cloning and other common DNA manipulations were performed according to standard protocols (30).
Plasmid pBRSN217 for expression of XylS variants was constructed as follows. The lac operator sequence was generated by annealing of oligonucleotides 5′-AGCTTTAATGCGGTAATTGTGAGCGGATAACAATT-3′ and 5′-AGCTAATTGTTATCCGCTCACAATTACCGCATTAA-3′ (the underlined sequences denote the binding site of the lac repressor) and inserted into the single HindIII site in pBRSN117 (12). For expression of the N-terminal tag recognized by anti-BPV E2 monoclonal antibody 3F12 (15), oligonucleotides 5′-TATGGGTGTCTCATCCACCTCTTCTGATTTTAGAGATCGCT-3′ (coding strand) and 5′-CTAGAGCGATCTCTAAAATCAGAAGAGGTGATGAGACACCCA-3′ were annealed and inserted between the NdeI and XbaI sites to replace the sequence encoding the hemagglutinin epitope. The coding sequences of xylS variants were amplified by PCR with the primers listed in Table 1 and inserted into pBRSN217 between the XbaI and BamHI sites.
TABLE 1.
Nucleotide sequences and locations of PCR primers
| Primer | Sequence (5′→3′)a | Position in geneb | Added restriction site |
|---|---|---|---|
| Forward | |||
| xylS | CCCCTCTAGAGATTTTTGCTTATTGAACGAG | 4–24 (xylS) | XbaI |
| xylS-SpeI | GGACTAGTGATTTTTGCTTATTGAACGAG | 4–24 (xylS) | SpeI |
| xylS(L5R,L6K) | GCTCTAGAGATTTTTGCCGAAAGAACGAGAAAA | 4–28 (xylS) | XbaI |
| ΔN8 | GCTCTAGAAAAAGTCAGATCTTCGTCC | 25–43 (xylS) | XbaI |
| ΔN30 | CCCCTCTAGAACGCACTCTATTCGCCTGC | 90–118 (xylS) | XbaI |
| ΔN39 | CCCCTCTAGAGGGCGCCCGGCAGG | 117–130 (xylS) | XbaI |
| ΔN47 | GCCCACAGAATCTTCGGATGCCc | 141–159 (xylS) | |
| ΔN105 | CCCCTCTAGAAATCCGGATGACCAAGCC | 315–332 (xylS) | XbaI |
| ΔN134 | CCCCTCTAGATGCAGTGACAACAATTGG | 402–419 (xylS) | XbaI |
| ΔN209 | CCCCTCTAGAAACCCGTCTTTCGAGCGAG | 627–645 (xylS) | XbaI |
| λ cI CTD | GCTCTAGAACCTTTACCAAAGGTGATGCG | 360–369 (λ cI) | XbaI |
| 4GA | GCTAGCGTAAGAGATCAGGACCGATCC | 564–572 (BPV E2) | NheI |
| E2 hinge | GCTCTAGAGATCGCCCAGACGGAG | 618–636 (BPV E2) | XbaI |
| Reverse | |||
| xylS | GGGGATCCCTTCTTCGGCTACG | 988–973 (xylS) | BamHI |
| ΔC109 | GGGGGGATCCTCAATCCGGATTGAGCAG | 323–309 (xylS) | BamHI |
| ΔC140 | GGGGGGATCCTCAATTGTTGTCACTGCATGCCC | 416–397 (xylS) | BamHI |
| ΔC140-XbaI | GCTCTAGATCAATTGTTGTCACTGCATGC | 416–399 (xylS) | XbaI |
| ΔC174-XbaI | GCTCTAGACATACAAAGTCGATGCCT | 521–504 (xylS) | XbaI |
| ΔC210 | GGGGGATCCTCAACCTTTGCTGAAAATTTCACGG | 626–605 (xylS) | BamHI |
| ΔC209-XbaI | GCTCTAGAACCTTTGCTGAAAATTTCACGG-3′ | 623–605 (xylS) | XbaI |
| ΔC276 | GGGGGGATCCTCAGGGATCGTTCAAGCAGGC | 824–807 (xylS) | BamHI |
| ΔC310 | GGGGGGATCCTCAAGGCAACTCGCCGAACGC | 926–909 (xylS) | BamHI |
| λ cI CTD | GCTCTAGAGCCAAACGTCTCTTCAGGCCAC | 711–690 (λ cI) | XbaI |
| 4GA | ACTAGTATCATTGGTGGTGCGCCTG | 900–883 (BPV E2) | SpeI |
| E2 hinge | GCTCTAGACGGTACCGTGCCCTGCACG | 858–840 (BPV E2) | XbaI |
Restriction sites in noncomplementary overhangs are underlined.
Locations refer to positions within indicated open reading frames based on the first nucleotide of the initiation codon.
Bases of an added Ala codon are italicized.
The xylS variants were amplified from pUSR112 (12) DNA by standard PCR methods (30). For amplification of the xylS variants with N-terminal deletions and XylS(L5R,L6K), we used the xylS reverse primer and a suitable forward primer. For amplification of the xylS variants with C-terminal deletions, the xylS forward primer and a suitable reverse primer were used. Termination of polypeptides ΔC276 and ΔC310 appeared to be ineffective. Therefore, a BclI linker containing an additional stop codon was inserted into the BamHI site.
For construction of the xylS variants Δ140–209 and Δ174–209, we amplified fragments of xylS containing nucleotides (nt) 3 to 416 and 3 to 521, with the xylS forward primer and reverse primers ΔC140-XbaI and ΔC174-XbaI, respectively. The amplified fragments were cloned into the XbaI site of pBRSN217 with the coding sequence for ΔN209 between the XbaI and BamHI sites. For construction of Δ39–47, we amplified a xylS fragment containing nt 141 to 962 with ΔN47 forward primer and xylS reverse primer. pBRSN217 with the coding sequence for xylS was cut with StyI and BamHI, and the amplified fragment was inserted between these two sites.
xylS variants with insertions were constructed as following. The coding sequence for the λCI dimerization domain was amplified from λ phage DNA. The coding sequence for the bovine papillomavirus type 2 (BPV1) E2 protein hinge region was amplified from pET11 bearing the E2 gene. For construction of the GA/XylS expression plasmid, we amplified a sequence encoding a peptide, GS(GAGGGAGGAGAGARS)4, the tetrameric Gly-Ala (4GA) repeat, which was inserted into the BPV-1 E2 gene to replace the region encoding residues 192 to 311 (D. Örd, R. Kurg, and M. Ustav, unpublished data). Next, we amplified fragments of xylS containing nt 3 to 416 and 3 to 626, with forward primer xylS-SpeI and reverse primers ΔC140-XbaI and ΔC209-XbaI, respectively. The amplified xylS fragments were cloned into the XbaI site of pBRSN217 with the coding sequence for ΔN209 between the XbaI and BamHI sites. Thereafter, the heterologous sequences were inserted into the XbaI site of the resultant plasmids.
pBRSN317 is pBRSN217 with the Ps2 promoter inserted instead of modified Ptet. For construction of pBRSN317, first, a 568-bp Ps2-containing BglII fragment was inserted into the pUC18 polylinker, and the EcoRI-HindIII fragment of the resultant plasmid was cloned into pBRSN217 between the EcoRI and HindIII sites.
Characterization of XylS variants in vivo.
E. coli CC118Pm-lacZ (14) was transformed with plasmids producing XylS variants. The plasmids bearing bacteria were grown in LB medium overnight with appropriate antibiotics. Then, bacteria were diluted 1:100 in the same medium, in the presence or absence of 1 mM meta-toluate, and β-galactosidase levels were determined after 4 h according to the standard protocol (23). Cells were permeabilized with toluene.
DNA immunoprecipitation and DNase I footprinting.
Plasmid-bearing E. coli DH5α cells were grown at 20°C to an optical density at 600 nm (OD600) of ∼0.6, 1 mM IPTG was added, and the cells were incubated for an additional 2 h. Cells were washed with TBS and stored in high-salt lysis buffer (100 mM Tris-HCl [pH 7.5], 1.5 M NaCl, 5 mM EDTA, 20% [wt/vol] glycerol) at −70°C. For preparation of lysates, cells were thawed, and dithiothreitol (to 10 mM), phenylmethylsulfonyl fluoride (to 100 μg ml−1), aprotinin (to 1 μg ml−1), and CHAPS {3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate} (up to 0.2%) were added. Cells were incubated with lysozyme (0.5 mg ml−1) on ice for 20 min and then disrupted by sonication. Clarified lysate was applied for batchwise affinity enrichment with end-over-end agitation at 4°C for 1 h. Affinity beads were prepared by coupling 3F12 anti-BPV E2 monoclonal antibody to divinylsulfon-activated Toyopearl HW65 TSK-gel. The purified proteins were stored, the pUPM190 probes for DNA immunoprecipitation and footprinting were made, and the experiments were carried out as described earlier (12), except that polyethylene glycol 6000 was omitted from the binding buffer when complexes were formed for DNase I footprinting. For footprinting, the wild-type (wt) N-XylS-Om complexes were formed in the presence of 1 mM meta-toluate, and the ΔN209-Om complexes were formed without meta-toluate.
Immunoblots.
E. coli DH5α cells bearing the pBRSN217 series expression plasmids were grown in LB medium with ampicillin at 37°C to an OD600 of 0.6, 1 mM IPTG was added, and cells were incubated for additional 2 h. Equal amounts of cells, judged by the OD of the bacterial culture, were suspended in sodium dodecyl sulfate (SDS) sample buffer, supplemented with β-mercaptoethanol (30). Samples were boiled for 5 min, and proteins were separated by SDS–12% polyacrylamide gel electrophoresis (PAGE). The proteins were electroblotted onto nitrocellulose membrane filters. The filters were blocked for 1 h with 1% nonfat dry milk, probed with 3F12 anti-BPV E2 monoclonal antibody, and treated with goat anti-mouse alkaline phosphatase-conjugated secondary antibody.
RESULTS
Stimulation of Pm by truncated XylS proteins.
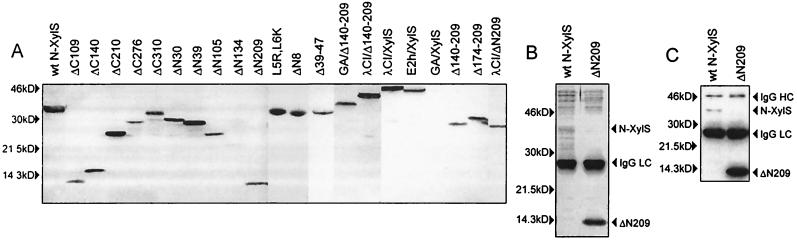
To examine whether the N and C termini of XylS constitute separable functional domains, we constructed two sets of progressing terminal deletions, from both ends of the coding sequence (Fig. 1). We truncated the protein in putative loop regions, indicated by a prediction of the secondary structure (not shown). The deletion mutants of xylS were generated by PCR and verified by sequencing. A peptide, GVSSTSSDFRDR, from BPV E2 protein was fused to the N terminus of wt XylS and the deletion mutants for monitoring and purification of the proteins. The tagged proteins were identified (Fig. 2A) by using the anti-BPV E2 monoclonal antibody 3F12 (15). The tag did not affect the meta-toluate responsiveness of wt XylS nor did it affect its ability to stimulate transcription from Pm. Therefore, we applied the tagged full-size XylS as a wt control (wt N-XylS). We have used previously the moderate tet promoter of pBR322 for production of the N-terminally tagged XylS protein (12). Since expression of several truncated XylS proteins (ΔN209 and ΔC310 in particular) from the tet promoter was apparently toxic to E. coli and caused plasmid instability, we inserted the lac operator sequence downstream of the promoter to reduce expression.
FIG. 2.
Expression and partial purification of tagged XylS variants. (A) Immunoblot analysis of expression of XylS proteins in E. coli DH5α. Equal amounts of bacteria bearing the pBRSN217 series expression plasmids were used for preparation of the total lysates in Laemmli sample buffer after 2 h of induction with IPTG (isopropyl-β-d-thiogalactopyranoside). Total protein extracts were separated by SDS-PAGE (12%) and analyzed by immunoblotting with the 3F12 monoclonal antibody. The proteins are schematically depicted on Fig. 3. (B) Silver-stained SDS-PAGE (12%) analysis of wt N-XylS and ΔN209 proteins retained on the 3F12 beads. (C) Immunoblot analysis of wt N-XylS and ΔN209 proteins retained on the 3F12 beads. Proteins were separated by SDS-PAGE (12%) and analyzed by immunoblotting by using the 3F12 monoclonal antibody. wt N-XylS and ΔN209 proteins, as well as the light chain (IgG LC) and heavy chain (IgG HC) of the 3F12 monoclonal antibody eluted from the TSK beads are indicated with arrows.
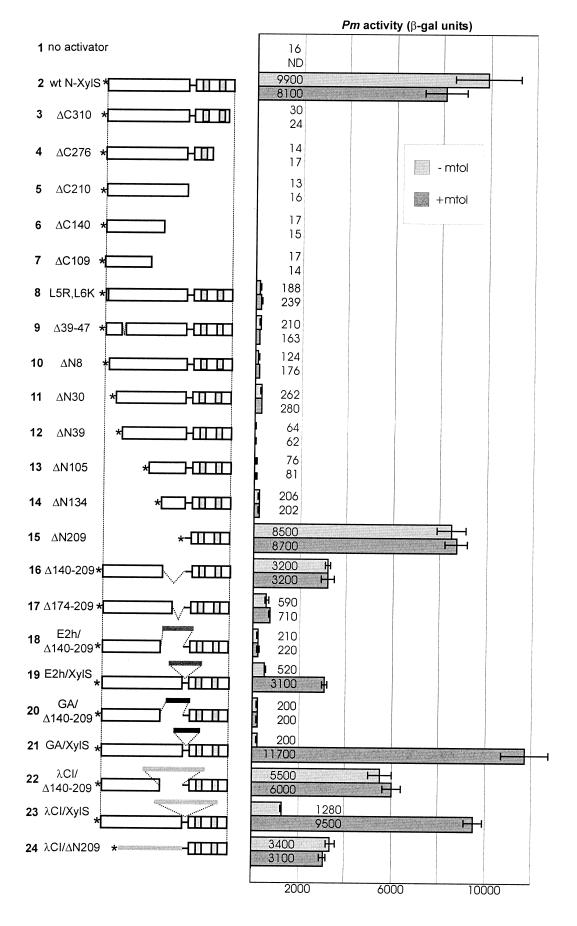
For assay of Pm activation, plasmids containing these xylS variants were transformed into E. coli strain CC118Pm-lacZ containing a chromosomal copy of the Pm promoter fused to the lacZ gene (14). Figure 3 shows β-galactosidase (β-Gal) levels, mediated by wt N-XylS and various deletion mutants, in the presence or absence of meta-toluate. Note that the level of expression of wt N-XylS from the modified tet promoter (labeled as Ptet* below) mimics the XylR mediated overexpression of XylS in P. putida—it produces full activation of Pm without the effector, and addition of the ligand has no further effect on the promoter activity (Fig. 3, line 2). We found that deletion mutant ΔN209, which corresponds to the putative DNA-binding domain, stimulates transcription from Pm as efficiently as wt N-XylS. When expressed from Ptet*, both mediated β-Gal levels close to 104 Miller units (Fig. 3, lines 2 and 15).
FIG. 3.
Activity of Pm when mediated by different XylS variants, overexpressed from the Ptet* promoter, in the presence or absence of meta-toluate. Transparent boxes on the diagram indicate the putative domains of XylS. Asterisks indicate the N-terminal tags, and gray boxes within the C-terminal domain indicate the putative helix-turn-helix regions. The hybrid XylS variants contain insertions of the hinge region of BPV E2 protein (lines 18 and 19), a synthetic Gly-Ala-rich region (lines 20 and 21), and the dimerization domain of the λCI protein (lines 22, 23, and 24) of E. coli CC118Pm-lacZ was transformed with plasmids for expression of XylS variants from the Ptet* promoter. Bacteria were grown in LB medium overnight, diluted 1:100 in the same medium in the absence (−mtol) or presence (+mtol) of 1 mM meta-toluate, and β-Gal levels were determined after 4 h. The values in Miller units are the averages of results from three to six assays. Error bars indicate the standard deviations.
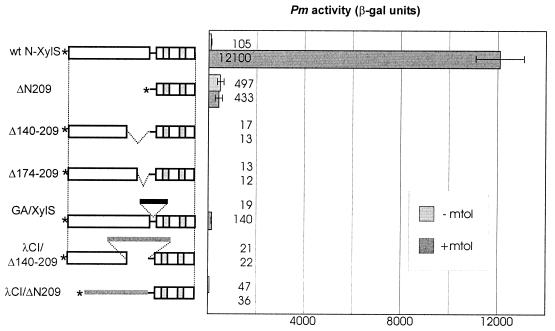
To probe whether ΔN209 is inducible by benzoates, ΔN209 and wt N-XylS were expressed from the weak Ps2 promoter (Fig. 4). The level of expression from Ps2 was so low that both proteins remained undetectable by the Western blotting of crude lysates even by enhanced chemiluminescence (ECL) detection. However, in the presence of meta-toluate, such a small amount of wt N-XylS was enough to produce the same β-Gal level as that produced by wt N-XylS overexpressed from Ptet*. We found that ΔN209 is not inducible by the effector and provides the phenotype of constitutive activator. When activator proteins were expressed from Ps2, the stimulation of Pm caused by ΔN209 was almost fivefold higher than that produced by wt N-XylS without effector. However, it reached only 4% of that produced by wt activator in the presence of meta-toluate (Fig. 4).
FIG. 4.
Activity of Pm when mediated by different XylS variants, expressed from the Ps2 promoter in the presence or absence of meta-toluate. E. coli CC118Pm-lacZ was transformed with plasmids for expression of XylS variants from the Ps2 promoter. Bacteria were grown in LB medium overnight, diluted 1:100 in the same medium in the absence (−mtol) or presence (+mtol) of 1 mM meta-toluate, and β-Gal levels were determined after 4 h. The values in Miller units are the averages of results from three to six assays. Error bars indicate the standard deviations.
Furthermore, we found that the other deletion mutants which retained the putative DNA-binding domain (ΔN8, ΔN30, ΔN39, ΔN105, and ΔN134) were able to stimulate transcription from Pm to some extent when overproduced from Ptet* (Fig. 3, lines 10 to 14). These N-terminally truncated proteins mediated 4- to 16-fold-higher β-Gal levels than the uninduced basal level of the strain. However, it makes only 0.6 to 2.6% of the β-Gal level produced by wt N-XylS. The XylS variants with longer N-terminal deletions than that of ΔN209 were unable to stimulate Pm and could not be detected by immunoblotting, obviously due to instability (data not shown). All deletions from the C terminus produced proteins which could not activate the Pm promoter (Fig. 3, lines 3 to 7). In conclusion, these results suggest that the C terminus of XylS indeed forms a DNA-binding domain and contains all the elements necessary for activation of transcription.
DNA binding by XylS CTD.
All deletion mutants which activate transcription from Pm in vivo should specifically bind to Om. In order to confirm the site-specific DNA binding of the C-terminal domain (CTD) of XylS in vitro, we purified both epitope-tagged ΔN209 and wt N-XylS proteins by single-step, batchwise immunoaffinity binding to the 3F12 monoclonal antibody, coupled to TSK beads. The DNA-binding activity was studied with the immobilized protein because purified XylS tends to aggregate in solution, making it impossible to use the regular gel-shift assays for this purpose. The cell extracts were prepared in a high-salt lysis buffer containing 1.5 M NaCl. High concentrations of sodium chloride do not hinder the specific interaction of 3F12 antibody with the epitope, while they reduce coimmunoprecipitation of the contaminating proteins. The protein preparations were examined by silver staining of SDS-PAGE and immunoblotting (Fig. 2B and C). We succeeded in isolating both wt N-XylS and ΔN209 that were functionally active in DNA binding; however, the yield of wt N-XylS was lower, and preparations contained some degradation products or contaminating proteins.
We used the matrix-bound N-XylS and ΔN209 in the specific DNA-binding and DNase I-footprinting assays. The mixture of end-labeled restriction fragments of the Om-containing plasmid pUPM190 (12) was incubated with the protein-loaded TSK beads in the presence or absence of meta-toluate. After the removal of free probe, only DNA that was bound to the immunopurified protein was retained on the beads. Since estimation of the amounts of proteins used in the assay was complicated, the experiment was carried out at oversaturating levels of the DNA probe so that less than 2% of the input labeled probe were retained on the beads. Figure 5A shows that both ΔN209 and wt N-XylS bind specifically the Om-containing 115-bp fragment of the pUPM190 HpaII-HinfI digest, whereas binding of any other fragment could not be observed. DNA binding by wt N-XylS was strongly (up to 100 times) induced by meta-toluate. Such a strong effect of meta-toluate could be observed only when wt N-XylS was purified from the high-salt lysate (1.5 M NaCl) and was not observed earlier when we used low-salt buffer conditions (220 mM KCl) (12). We did not detect any effect of meta-toluate on DNA binding by ΔN209. The 3F12 beads lacking XylS did not bind any DNA (data not shown).
FIG. 5.
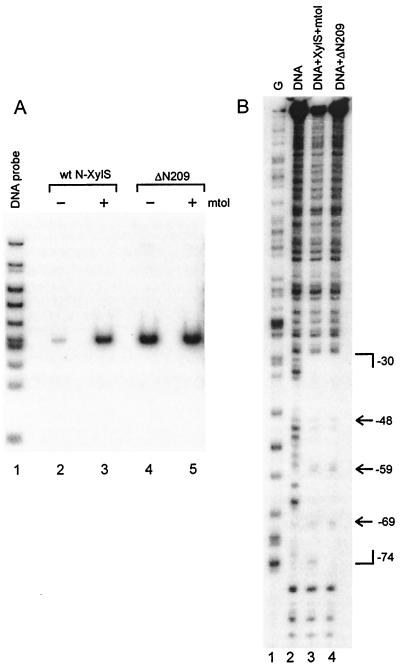
DNA immunoprecipitation (A) and DNase I footprinting (B) showing that wt N-XylS and XylS CTD bind specifically to Om. (A) A radiolabeled HpaII-HinfI digest of pUPM190 (lane 1) was incubated with 3F12 beads containing wt N-XylS (lanes 2 and 3) and ΔN209 (lanes 4 and 5) either in the absence (lanes 2 and 4) or in the presence (lanes 3 and 5) of 1 mM meta-toluate. Unbound DNA was removed by washing. Bound DNA was analyzed on nondenaturating TBE-PAGE (5%). (B) The XhoI (−117) to EcoRI (+242) Om-containing fragment from pUPM190 was end labeled in the lower strand at −117 and incubated with 3F12 beads containing wt N-XylS or ΔN209. Unbound DNA was removed by washing. Both free and protein-bound templates were subjected to DNase I cleavage. Lane 1, G-specific DNA sequence marker; lane 2, DNase I digest of the unbound DNA fragment; lane 3, DNase I digest of the fragment bound to wt N-XylS in the presence of meta-toluate; lane 4, DNase I digest of the ΔN209-bound DNA fragment. Brackets indicate a region protected by DNA-bound protein from DNase I cleavage. Arrows mark the hypersensitive sites within the protected region.
Further, we analyzed the interaction of both protein variants with Om by DNase I footprinting. The Om-containing DNA fragment was end labeled, and the DNA-protein complex was formed on the beads. With wt N-XylS the binding was done in the presence of meta-toluate. Again, the experiments were carried out at oversaturating levels of the DNA probe. After the removal of unbound DNA, the complex was subjected to DNase I cleavage. The treated DNA was extracted from the beads and analyzed on the sequencing gels. We found that both ΔN209 and wt N-XylS protect a 44-bp area, extending from positions −74 to −30 on the lower strand (Fig. 5B). The DNase I footprints of these two protein variants were almost identical, indicating that both have a similar mode of interaction with the binding site. The only difference is hypersensitive to DNase I at nucleotide −75, observed in the complex with wt N-XylS. Enhanced cleavage at that position was detected when the N-XylS-Om complex was formed in the presence of meta-toluate. That slight effect has been previously overlooked by us (12).
The results of DNA immunoprecipitation and DNase I footprinting confirm that the DNA-binding domain of XylS is located within the C-terminal 112 residues of the protein.
The complete N-terminal domain is required for effector responsiveness of XylS.
The N-terminal portion of XylS is believed to be necessary for effector binding and ligand-dependent regulation of the activity of the protein. We demonstrated that the truncated proteins ΔN8, ΔN30, ΔN39, ΔN105, and ΔN134, which contain a part of the putative regulatory domain in addition to the complete DNA-binding domain, are noninducible by effector and, when expressed from Ptet*, produce a much lower β-Gal activity than wt N-XylS or ΔN209 (Fig. 3, lanes 2 and 10 to 15). When placed under the control of Ps2 promoter, these deletion mutants were unable to stimulate the Pm promoter (data not shown). The deletion mutants ΔN8, ΔN30, ΔN39, ΔN105, and ΔN134 are expressed at the different intracellular concentrations, probably due to different stabilities of the truncated proteins (Fig. 2A). However, the activity of the N-terminal deletion mutants does not merely correlate with the protein levels seen in Fig. 2A. As the levels of these proteins are readily detectable, they must be far more abundant than wt N-XylS and ΔN209, expressed from the Ps2 promoter. As we have mentioned above, wt N-XylS and ΔN209 produce substantial Pm activation even at the levels of expression which are undetectable by Western blot analysis (Fig. 4). Thus, in vivo transcriptional activation data reflect, at least partially, the intrinsic properties of the truncated proteins and not only different levels of expression. Consequently, N-terminal deletions in the putative regulatory domain cause the loss of the regulatory function and reduce the activity of the C-terminal domain of XylS.
For further characterization of the regulatory portion of XylS and the requirements for its proper functioning, we constructed additional deletion and insertion mutants and tested their ability to stimulate Pm (Fig. 3, lines 16 to 23). Most of the mutants described below were expressed from Ptet* at a level comparable with that of wt N-XylS (Fig. 2A). Only GA/XylS and E2h/Δ140–209 were apparently very unstable. The latter could be detected on the Western blot only by ECL and is not shown on Fig. 2A.
We deleted two different portions of the central part of XylS from the other end of the putative regulatory domain. The deletion mutants Δ140–209 and Δ174–209 were more active than N-terminal truncations but also had lost their effector responsiveness and showed a reduced ability to stimulate Pm (Fig. 3, lines 16 and 17; Fig. 4). To ascertain whether that effect could be caused by the loss of hinge flexibility, which does not allow the domains to interact properly, we replaced the deleted residues 140 to 209 with long unstructured regions: the 80-residue hinge region of BPV1 E2 protein (8) and a 69-residue region, consisting mainly of glycine and alanine, which has been shown to replace effectively the BPV1 E2 hinge (Örd et al., unpublished). As we found that the region containing residues 140 to 209 is responsible for dimerization of XylS (N. Kaldalu and M. Ustav, unpublished data), it was also replaced with the dimerization domain of λCI repressor. The resultant proteins E2h/Δ140–209, GA/Δ140–209, and λCI/Δ140–209 remained unresponsive to meta-toluate. E2h/Δ140–209 and GA/Δ140–209 were much less active than wt N-XylS, whereas λCI/Δ140–209, when overexpressed, mediated about one-half of the β-Gal level produced by wt N-XylS (Fig. 3, lines 18, 20, and 22). Expression from the Ps2 promoter shows, however, that λCI/Δ140–209 is a much weaker transcription activator than wt N-XylS and ΔN209 (Fig. 4). When the same heterologous protein portions were inserted into the putative hinge region of XylS, retaining the N-terminal region intact, we found that these hybrid XylS activators were responsive to the effector. λCI/XylS, E2h/XylS, and GA/XylS, which carry the interdomain insertions, were weaker transcription activators than wt N-XylS but were clearly inducible by meta-toluate (Fig. 3, lines 19, 21, and 23; Fig. 4). Since XylS variants with the inserted λCI dimerization domain were relatively more active than the others, we substituted the entire N-terminal region with the λCI domain and expressed the resultant fusion protein λCI/ΔN209 from both Ptet* and Ps2 promoters. β-Gal levels mediated by λCI/ΔN209 remained several times lower than those produced by the C-terminal domain ΔN209 itself (Fig. 3, line 24; Fig. 4). These data suggest that the heterologous dimerization domain, as well as the incomplete N-terminal regions of XylS, interfered with transcriptional activation by the XylS CTD. Therefore, we do not have evidence of whether the λCI domain-containing proteins were more active due to their dimeric state or due to the lower level of interdomain interference.
In AraC protein, the N-terminal arm of the regulatory domain has been shown to interact with the C-terminal domain as an intramolecular repressor of binding to the adjacent binding sites, which is necessary for transcription activation. Leucines in the N-terminal arm were crucial for that activity (31). Using mutational analysis, we tried to specify a subdomain of a similar function within the N-terminal domain of XylS. Several mutations that caused the semiconstitutive behavior of XylS are clustered in a small glycine-rich N-terminal region, P37 to R45 (20, 37) (Fig. 1), which was expected to form a loop from a prediction of the secondary structure. To remove the putative loop, we produced a deletion mutant, Δ39–47. We also constructed a double mutant, XylS(L5R,L6K), to test whether the leucines in the extreme N terminus have a role in regulation of the XylS activity. Both Δ39–47 and XylS(L5R,L6K), as well as the N-terminal deletion mutant ΔN8, were unresponsive to the effector. However, these mutants were very weak and not constitutive activators (Fig. 3, lines 8 to 10). Therefore, we did not find any region in the XylS N-terminal region (NTR) which could be deleted to produce a constitutive phenotype, as has been shown in the case of AraC.
Thus, the data of the mutational analysis indicate that 210 N-terminal residues of XylS provide the ligand responsiveness to the protein, and the entire region is necessary for that activity. Since it has not been demonstrated that this part of XylS folds independently or functions (binds benzoate) independently of the CTD, it would be correct to use the term the NTR of XylS. All the examined deletions in the XylS NTR reduced the activity of the XylS CTD. Since the level of the Pm activation by the mutant proteins positively correlates with the strength of the promoter used for the expression of these proteins, the reduced activation must be an intrinsic characteristic of the mutants and not caused by aggregation due to overexpression.
DISCUSSION
We have shown here that XylS is comprised of a C-terminal DNA-binding–transcriptional-activation domain and an N-terminal regulatory region. Existence of separable domains has been confirmed earlier for AraC and MelR from the same protein family (2, 16, 18, 22) but not for XylS. Kessler et al. (14) have characterized several deletion mutants of XylS in vivo and found that all of them were unable to activate Pm or modulate the activity of wt XylS. These mutants were expressed as a result of the readthrough transcription at a very low level, one much lower than those produced from the Ptet* or Ps2 promoter, as deduced from the β-Gal activities mediated by wt XylS. Therefore, stimulation of Pm by the deletion mutants presumably remained undetectable by Kessler et al. (14) due to the low levels of expression, and their results principally match those obtained by us in the case of the XylS variants with deletions in the NTR.
In the present work, we demonstrated that the C-terminal DNA-binding domain lacks effector responsiveness and that to modulate the activity of the protein in ligand-dependent manner, the complete N-terminal region is necessary. It is possible that mutations in the N terminus which make XylS noninducible by effector and strongly reduce its ability to activate Pm disrupt the native structure of the N terminus of the protein. We suggest that the DNA-binding domain may contain a surface or surfaces that interact with the regulatory part of XylS. That idea is in agreement with the knowledge that mutations in both the N and C termini of XylS can yield the semiconstitutive phenotype (27) and that mutations in one domain can be suppressed by mutations in the other (21). It is possible that the misfolded N terminus provides a new site or sites for irreversible interaction with the CTD and, in that way, inhibits DNA binding and/or transcriptional activation.
Currently, we cannot make final conclusions about the mechanism by which ligand regulates the XylS activity. We have shown that meta-toluate strongly facilitates DNA binding by matrix-attached N-XylS but does not affect DNA binding by the XylS CTD in vitro (Fig. 4A). Therefore, stimulation of DNA binding must be at least one, if not the single, major effect of ligand. However, additional, cooperative effects should not be excluded. Stimulation of dimer formation by effector is possible, but this should not enhance DNA binding in our assay since we used immobilized wt N-XylS which was attached to the beads through the N terminus and presumably had not enough freedom to change its multimeric state. To account for our data, we propose that the regulatory region reversibly interacts with the DNA-binding–activator domain. The fact that ΔN209, presumably with a monomeric XylS CTD, mediated almost fivefold-higher Pm activation than wt N-XylS without effector, when these proteins were expressed from Ps2, suggests that the NTR works as an intramolecular repressor. Binding of the effector to the NTR causes a conformational change in it and releases the intramolecular repression. A similar mechanism has been validated for AraC (31). In the presence of effector, the XylS NTR may either passively release the inhibition or actively facilitate the function of CTD, e.g., by assistance in local folding of the DNA-binding regions.
In spite of the similarity of footprints produced by XylS in the presence or absence of ligand, we cannot conclude that liganded and unliganded XylS form identical complexes with Om or that these complexes behave identically in transcriptional activation. The data of in vivo DMS footprinting by Miura et al. (24) suggest that the complexes are not transcriptionally identical and that the benzoate effector modifies the interaction of XylS with RNA polymerase at the Pm promoter. RNA polymerase is retained on the Pm promoter by XylS in the absence of benzoate inducer and released by effector binding to XylS, with concomitant initiation of transcription (24). Recently, Marqués et al. (17) suggested that alkylbenzoates may have also an indirect role in Pm activation. They showed that transcription from the Pm promoter, which does not show similarity to the −10/−35 consensus sequence for binding of ς70 RNA polymerase, but matches in the −10 region with the consensus sequence of ς32 RNA polymerase, is dependent on ς32 in exponential-growth-phase E. coli. They also demonstrated activation of a ς32-dependent heat shock promoter as an indication of induction of heat shock response by meta-toluate. However, since meta-toluate did not affect the activation of Pm by the XylS CTD and other N-terminal deletion mutants of XylS, we did not observe any indirect effect of meta-toluate beyond its well-known role as an allosteric effector of XylS.
As we found, the XylS CTD, which is proposed to be a monomer, can recognize the Om binding site and activate transcription. Therefore, occupation of both half-sites, not necessarily the dimeric structure of the activator, appears to be essential for transcriptional activation. The DNase I footprinting results of the XylS CTD-Om complexes, as well as the earlier footprinting and methylation interference experiments with XylS (12), demonstrated equal occupation of both Om half-sites and suggest that the half-sites have similar affinities for XylS. The proposed dimerization of XylS certainly facilitates recognition of the operator by producing the dimeric protein with increased affinity. That may serve for an explanation of why wt N-XylS, in the presence of meta-toluate, stimulates the Pm promoter more effectively than ΔN209 when these proteins are expressed at low concentrations (Fig. 4).
ACKNOWLEDGMENTS
We are grateful to Jüri Parik for technical advice and assistance, to Daima Örd for the BPV E2 gene containing Gly-Ala repeats, and to Tanel Tenson, Reet Kurg, Maia Kivisaar, and Aare Abroi for critical reading of the manuscript.
This work was supported by grants 2496 and 2497 from the Estonian Science Foundation, grant HHMI 75195-541301 from the Howard Hughes Medical Institute, and EU grant CIPA-CT94-0154. N.K. was the recipient of an FEMS Young Scientist Award.
REFERENCES
- 1.Amábile-Cuevas C F, Demple B. Molecular characterization of the soxRS genes of Escherichia coli: two genes control a superoxide stress regulon. Nucleic Acids Res. 1991;19:4479–4484. doi: 10.1093/nar/19.16.4479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bustos S A, Schleif R F. Functional domains of the AraC protein. Proc Natl Acad Sci USA. 1993;90:5638–5642. doi: 10.1073/pnas.90.12.5638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cohen S P, Hachler H, Levy S B. Genetic and functional analysis of the multiple antibiotic resistance (mar) locus in Escherichia coli. J Bacteriol. 1993;175:1484–1492. doi: 10.1128/jb.175.5.1484-1492.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eustance R J, Bustos S A, Schleif R F. Reaching out: locating and lengthening the interdomain linker in AraC protein. J Mol Biol. 1994;242:330–338. doi: 10.1006/jmbi.1994.1584. [DOI] [PubMed] [Google Scholar]
- 5.Eustance R J, Schleif R F. The linker region of AraC protein. J Bacteriol. 1996;178:7025–7030. doi: 10.1128/jb.178.24.7025-7030.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gallegos M T, Marques S, Ramos J L. Expression of the TOL plasmid xylS gene in Pseudomonas putida occurs from a ς70-dependent promoter or from ς70- and ς54-dependent tandem promoters according to the compound used for growth. J Bacteriol. 1996;178:2356–2361. doi: 10.1128/jb.178.8.2356-2361.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gallegos M T, Schleif R, Bairoch A, Hofmann K, Ramos J L. AraC/XylS family of transcriptional regulators. Microbiol Mol Biol Rev. 1997;61:393–410. doi: 10.1128/mmbr.61.4.393-410.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Giri I, Yaniv M. Structural and mutational analysis of E2 trans-activating proteins of papillomaviruses reveals three distinct functional domains. EMBO J. 1988;7:2823–2829. doi: 10.1002/j.1460-2075.1988.tb03138.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.González-Pérez M M, Ramos J L, Gallegos M T, Marqués S. Critical nucleotides in the upstream region of the XylS-dependent TOL meta-cleavage pathway operon promoter as deduced from analysis of mutants. J Biol Chem. 1999;274:2286–2290. doi: 10.1074/jbc.274.4.2286. [DOI] [PubMed] [Google Scholar]
- 10.Inouye S, Nakazawa A, Nakazawa T. Expression of the regulatory gene xylS on the TOL plasmid is positively controlled by the xylR gene product. Proc Natl Acad Sci USA. 1987;84:5182–5186. doi: 10.1073/pnas.84.15.5182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Inouye S, Nakazawa A, Nakazawa T. Overproduction of the xylS gene product and activation of the xylDLEGF operon on the TOL plasmid. J Bacteriol. 1987;169:3587–3592. doi: 10.1128/jb.169.8.3587-3592.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kaldalu N, Mandel T, Ustav M. TOL plasmid transcription factor XylS binds specifically to the Pm operator sequence. Mol Microbiol. 1996;20:569–579. doi: 10.1046/j.1365-2958.1996.5381060.x. [DOI] [PubMed] [Google Scholar]
- 13.Kessler B, de Lorenzo V, Timmis K N. Identification of a cis-acting sequence within the Pm promoter of the TOL plasmid which confers XylS-mediated responsiveness to substituted benzoates. J Mol Biol. 1993;230:699–703. doi: 10.1006/jmbi.1993.1189. [DOI] [PubMed] [Google Scholar]
- 14.Kessler B, Timmis K N, de Lorenzo V. The organization of the Pm promoter of the TOL plasmid reflects the structure of its cognate activator protein XylS. Mol Gen Genet. 1994;244:596–605. doi: 10.1007/BF00282749. [DOI] [PubMed] [Google Scholar]
- 15.Kurg R, Parik J, Juronen E, Sedman T, Abroi A, Liiv I, Langel U, Ustav M. Effect of bovine papillomavirus E2 protein-specific monoclonal antibodies on papillomavirus DNA replication. J Virol. 1999;73:4670–4677. doi: 10.1128/jvi.73.6.4670-4677.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lauble H, Georgalis Y, Heinemann U. Studies on the domain structure of the Salmonella typhimurium AraC protein. Eur J Biochem. 1989;185:319–325. doi: 10.1111/j.1432-1033.1989.tb15118.x. [DOI] [PubMed] [Google Scholar]
- 17.Marqués S, Manzanera M, González-Pérez M M, Gallegos M T, Ramos J L. The XylS-dependent Pm promoter is transcribed in vivo by RNA polymerase with ς32 or ς38 depending on the growth phase. Mol Microbiol. 1999;31:1105–1113. doi: 10.1046/j.1365-2958.1999.01249.x. [DOI] [PubMed] [Google Scholar]
- 18.Menon K P, Lee N L. Activation of ara operons by a truncated AraC protein does not require inducer. Proc Natl Acad Sci USA. 1990;87:3708–3712. doi: 10.1073/pnas.87.10.3708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mermod N, Ramos J L, Bairoch A, Timmis K N. The xylS gene positive regulator of TOL plasmid pWWO: identification, sequence analysis and overproduction leading to constitutive expression of meta cleavage operon. Mol Gen Genet. 1987;207:349–354. doi: 10.1007/BF00331600. [DOI] [PubMed] [Google Scholar]
- 20.Michán C, Zhou L, Gallegos M T, Timmis K N, Ramos J L. Identification of critical amino-terminal regions of XylS. The positive regulator encoded by the TOL plasmid. J Biol Chem. 1992;267:22897–22901. [PubMed] [Google Scholar]
- 21.Michán C, Kessler B, de Lorenzo V, Timmis K N, Ramos J L. XylS domain interactions can be deduced from intraallelic dominance in double mutants of Pseudomonas putida. Mol Gen Genet. 1992;235:406–412. doi: 10.1007/BF00279387. [DOI] [PubMed] [Google Scholar]
- 22.Michán C M, Busby S J, Hyde E I. The Escherichia coli MelR transcription activator: production of a stable fragment containing the DNA-binding domain. Nucleic Acids Res. 1995;23:1518–1523. doi: 10.1093/nar/23.9.1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miller J H. A short course in bacterial genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1992. [Google Scholar]
- 24.Miura K, Inouye S, Nakazawa A. Protein binding in vivo to OP2 promoter of the Pseudomonas putida TOL plasmid. Biochem Mol Biol Int. 1998;46:933–941. doi: 10.1080/15216549800204482. [DOI] [PubMed] [Google Scholar]
- 25.Ramos J L, Stolz A, Reineke W, Timmis K N. Altered effector specificities in regulators of gene expression: TOL plasmid xylS mutants and their use to engineer expansion of the range of aromatics degraded by bacteria. Proc Natl Acad Sci USA. 1986;83:8467–8471. doi: 10.1073/pnas.83.22.8467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ramos J L, Mermod N, Timmis K N. Regulatory circuits controlling transcription of TOL plasmid operon encoding meta-cleavage pathway for degradation of alkylbenzoates by Pseudomonas. Mol Microbiol. 1987;1:293–300. doi: 10.1111/j.1365-2958.1987.tb01935.x. [DOI] [PubMed] [Google Scholar]
- 27.Ramos J L, Michán C, Rojo F, Dwyer D, Timmis K. Signal-regulator interactions. Genetic analysis of the effector binding site of xylS, the benzoate-activated positive regulator of Pseudomonas TOL plasmid meta-cleavage pathway operon. J Mol Biol. 1990;211:373–382. doi: 10.1016/0022-2836(90)90358-S. [DOI] [PubMed] [Google Scholar]
- 28.Ramos J L, Marqués S, Timmis K N. Transcriptional control of the Pseudomonas TOL plasmid catabolic operons is achieved through an interplay of host factors and plasmid-encoded regulators. Annu Rev Microbiol. 1997;51:341–373. doi: 10.1146/annurev.micro.51.1.341. [DOI] [PubMed] [Google Scholar]
- 29.Rhee S, Martin R G, Rosner J L, Davies D R. A novel DNA-binding motif in MarA: the first structure for an AraC family transcriptional activator. Proc Natl Acad Sci USA. 1998;95:10413–10418. doi: 10.1073/pnas.95.18.10413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 31.Saviola B, Seabold R, Schleif R F. Arm-domain interactions in AraC. J Mol Biol. 1998;278:539–548. doi: 10.1006/jmbi.1998.1712. [DOI] [PubMed] [Google Scholar]
- 32.Soisson S M, MacDougall-Shackleton B, Schleif R, Wolberger C. Structural basis for ligand-regulated oligomerization of AraC. Science. 1997;276:421–425. doi: 10.1126/science.276.5311.421. [DOI] [PubMed] [Google Scholar]
- 33.Soisson S M, MacDougall-Shackleton B, Schleif R, Wolberger C. The 1.6 Å crystal structure of the AraC sugar-binding and dimerization domain complexed with d-fucose. J Mol Biol. 1997;273:226–237. doi: 10.1006/jmbi.1997.1314. [DOI] [PubMed] [Google Scholar]
- 34.Spooner R A, Bagdasarian M, Franklin F C. Activation of the xylDLEGF promoter of the TOL toluene-xylene degradation pathway by overproduction of the xylS regulatory gene product. J Bacteriol. 1987;169:3581–3586. doi: 10.1128/jb.169.8.3581-3586.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Williams P A, Murray K. Metabolism of benzoate and the methylbenzoates by Pseudomonas putida (arvilla) mt-2: evidence for the existence of a TOL plasmid. J Bacteriol. 1974;120:416–423. doi: 10.1128/jb.120.1.416-423.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Worsey M J, Williams P A. Metabolism of toluene and xylenes by Pseudomonas putida (arvilla) mt-2: evidence for a new function of the TOL plasmid. J Bacteriol. 1975;124:7–13. doi: 10.1128/jb.124.1.7-13.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhou L M, Timmis K N, Ramos J L. Mutations leading to constitutive expression from the TOL plasmid meta-cleavage pathway operon are located at the C-terminal end of the positive regulator protein XylS. J Bacteriol. 1990;172:3707–3710. doi: 10.1128/jb.172.7.3707-3710.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]