Abstract
Motivation
Intercellular communication (i.e. cell–cell communication) plays an essential role in multicellular organisms coordinating various biological processes. Previous studies discovered that feedback loops between two cell types are a widespread and vital signaling motif regulating development, regeneration and cancer progression. While many computational methods have been developed to predict cell–cell communication based on gene expression datasets, these methods often predict one-directional ligand–receptor interactions from sender to receiver cells and are not suitable to identify feedback loops.
Results
Here, we describe ligand–receptor loop (LRLoop), a new method for analyzing cell–cell communication based on bi-directional ligand–receptor interactions, where two pairs of ligand–receptor interactions are identified that are responsive to each other and thereby form a closed feedback loop. We first assessed LRLoop using bulk datasets and found our method significantly reduces the false positive rate seen with existing methods. Furthermore, we developed a new strategy to assess the performance of these methods in single-cell datasets. We used the between-tissue interactions as an indicator of potential false-positive prediction and found that LRLoop produced a lower fraction of between-tissue interactions than traditional methods. Finally, we applied LRLoop to the single-cell datasets obtained from retinal development. We discovered many new bi-directional ligand–receptor interactions among individual cell types that potentially control proliferation, neurogenesis and/or cell fate specification.
Availability and implementation
An R package is available at https://github.com/Pinlyu3/LRLoop. The source code can be found at figshare (https://doi.org/10.6084/m9.figshare.20126138.v1). The datasets can be found at figshare (https://doi.org/10.6084/m9.figshare.20126021.v1).
Supplementary information
Supplementary data are available at Bioinformatics online.
1 Introduction
Multicellular organisms rely on cell–cell communication to coordinate various biological processes and respond to environmental stimuli (Bonnans et al., 2014; Zhou et al., 2018). Decades of research have accumulated a massive amount of information on signaling pathways and ligand–receptor interactions (Bich et al., 2019; Bonnans et al., 2014; Zhou et al., 2018). Recent sequencing technologies enable us to profile gene expression at the single-cell level (Montoro et al., 2018; Tammela and Sage, 2020). Many computational methods have been developed to mine single-cell RNA-seq (scRNA-seq) data for biologically important cell–cell signaling interactions. These methods have distinct features in terms of the usage of databases of ligands and receptors, the strategy for building lists of ligand–receptor interactions and the scoring systems to quantify the interactions based on gene expression (Almet et al., 2021; Armingol et al., 2021; Blencowe et al., 2019; Jin and Ramos, 2022; Shao et al., 2020).
If we focus on how these methods utilize the gene expression data, they can be classified into two major types. The first type predicts the cell–cell communication based on the expression of ligands and receptors. The methods include CellPhoneDB, SingleCellSignalR, Connectome, ICELLNET and NATMI (Cabello-Aguilar et al., 2020; Efremova et al., 2020; Hou et al., 2020; Noël et al., 2021; Raredon et al., 2022). If one cell type expresses a specific ligand and another cell type expresses its receptor(s), these two cell types are considered to be communicating with one another. The second class of methods (e.g. NicheNet and CCCExplorer) uses entire transcriptomes to predict cell–cell communication (Browaeys et al., 2020; Choi et al., 2015). These methods incorporate other genes beyond ligands and receptors, integrating the expression levels of downstream genes, transcription factors and signaling proteins. By constructing the signaling networks and transcriptional regulatory networks, the downstream genes can be identified for each ligand. Conversely, gene sets that are differentially expressed, including lowly expressed genes, in certain conditions can be collectively used to infer the upstream ligands. However, the two types of methods shared one feature that they predicted one-directional interactions at a time, from sender cells to receiver cells.
Previous studies discovered that intercellular feedback interactions were prevalent in many biological processes, including stem cell fate decision, development, regeneration and cancer biology. For example, some studies used mathematical modeling and cell population analysis to demonstrate the existence of feedback loops in stem cell signaling networks (Kirouac et al., 2009, 2010). Other studies identified specific feedback loops during developmental processes (Barone et al., 2017;Jing et al., 2021; Nilsson and Skinner, 2001). The feedback mechanism is necessary to ensure two-way communication between cell types. For instance, mesenchymal stem cells are found to confer a neuroprotective effect on injured retinal ganglion cells (RGCs) via platelet-derived growth factors (Johnson et al., 2014). This neuroprotective effect has to be triggered by a signal of ‘injury’ from RGCs to mesenchymal stem cells, demonstrating the importance of feedback communication between different cell types.
However, the currently existing methods were designed to predict individual one-directional ligand–receptor interactions. A computational method that is able to systematically predict the feedback loops is still lacking. In this work, we propose a new method, ligand–receptor loop (LRLoop), which is specifically designed to identify feedback loops from gene expression datasets. This method integrates the existing signaling and regulatory networks with the gene expression datasets. We assessed this in both bulk and scRNA-seq datasets and found that it outperformed other existing methods. Applying LRLoop to single-cell datasets obtained from developing mouse retina predicted many ligand–receptor interactions during retinal development.
2 Materials and methods
2.1 The overall design of LRLoop
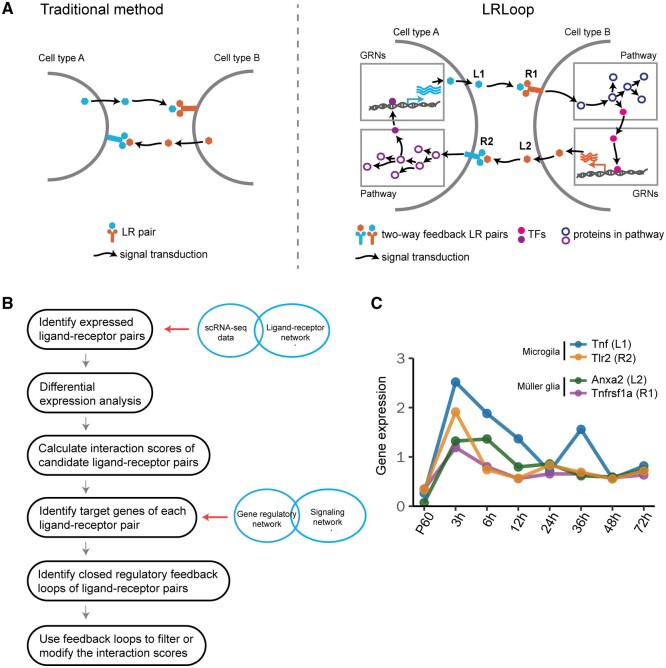
We integrate transcriptome, signaling pathways and regulatory networks to identify feedback for cell–cell communication. Specifically, for the two-way communication to occur, cell type A secretes ligand L1, which interacts with receptor R1 in cell type B. In turn, cell type B responds to the signal from cell type A by secreting ligand L2 that interacts with receptor R2 in cell type A. To make the L1–R1 and L2–R2 interactions responsive to each other, the ligands (L1 and L2) are required to be encoded by the genes that are regulated by receptors in the corresponding cells (Fig. 1A). Note that even though traditional methods can also predict bi-directional interactions, the interactions are not connected through signaling and regulatory networks. Therefore, these interactions are not responsive to each other (Fig. 1A).
Fig. 1.
LRLoop algorithm and its assessment. (A) Schematic of the LRLoop concept. Ligand 1 (L1) interacts with receptor 1 (R1), while ligand 2 (L2) interacts with receptor 2 (R2). Furthermore, L1 is the downstream target of R2 through signaling pathways and gene regulatory networks (GRNs). Similarly, L2 is the downstream target of R1. (B) The flowchart of LRLoop. (C) An example feedback loop between Muller glia and microglia
2.2 Sources of ligand–receptor pairs, signaling and gene regulatory networks
We combined literature-validated ligand–receptor pairs used in NicheNet (Browaeys et al., 2020) and connectomeDB2020 (Hou et al., 2020). Ligand–receptor pairs predicted based on protein–protein interactions are excluded. Only the ligand–receptor pairs that are literature supported are included. We further filtered these ligand–receptor pairs using ligand and receptor annotation from CellTalkDB and NATMI database (Hou et al., 2020; Shao et al., 2021). The ligand-receptor pairs were removed if the definition of the involved ligand or receptor was not supported by either of the two databases. After filtering, we retained 2512 ligand–receptor pairs, involving 859 ligands and 726 receptors, for subsequent downstream analysis.
To construct our default LRLoop network, we adopted the intracellular signaling network and gene regulatory network collected in NicheNet (Browaeys et al., 2020).
2.3 Construction of the LRLoop
The construction of LRLoop consists of three steps:
Construct the ligand/receptor-target regulatory potential matrices. With the collected ligand–receptor pairs, signaling and gene regulatory networks, we constructed the ligand/receptor-target regulatory potential matrices using NicheNet (Fig. 1B). For the calculation of these matrices, we adopted the functions construct_weighted_networks, apply_hub_corrections and construct_ligand_target_matrix in the R package nichenetr. We modified the function construct_ligand_target_matrix. Instead of calculating the regulatory potential score for each ligand, the modified version calculates the regulatory potential scores between each receptor and target gene. The algorithm was based on the idea of propagation of the signal from a ligand/receptor to the downstream proteins mediating receptor signaling, transcriptional regulators targeted by these factors and genes that are in turn regulated by these transcriptional regulators. Google’s Personalized PageRank algorithm was used to link the ligands/receptors to transcriptional regulators (Browaeys et al., 2020). The resulting ligand/receptor-transcriptional regulator matrices were then multiplied to the transcriptional regulator-target matrix of the gene regulatory network to obtain the ligand/receptor and target gene relationships.
Identifying the target genes of each ligand/receptor. With the ligand/receptor-target regulatory potential matrices, we identified the set of target genes of each ligand/receptor which were defined as the ones with relatively higher regulatory potential scores among all the potential target genes of the ligand/receptor (Fig. 1B). Specifically, for the construction of our default LRLoop network in the analysis, we used the function make_discrete_ligand_target_matrix of the nichenetr package with its default parameters (error_rate = 0.1, cutoff_method = ‘distribution’).
Finally, we identified all the [L1–R1]<->[L2–R2] pairs where L1 and L2 are among the target genes of each other for the feedback loops. Alternatively, we also identified the loops when L2 is a target gene of R1 and L1 is among the target genes of R2.
The interaction networks (ligand–receptor interactions, regulatory networks and signaling networks, ligand–target gene relationships) can be found in https://doi.org/10.6084/m9.figshare.20126021.v1.
2.4 Assessment of LRLoop using bulk datasets
Each of the 111 ligand treatment datasets collected by Browaeys et al. (2020) provides the treatment ligand, the expressed genes in the receiver cells and the differential expression information of these genes (including the logFC and q-value). For each of these datasets, based on our curated ligand–receptor network, we took the set of the ligand genes of all the expressed receptor genes in the receiver cells as the set of candidate ligands L1 and ranked these candidate ligands in the following ways:
max{|logFC of R1|}: We scored each candidate ligand gene L1 by the value of max{|logFC(R1)|: R1 the set of cognate receptor genes of L1}. The ligands L1 are then ranked by these scores in descending order.
CCCExplorer (L1 score): We replaced the ligand–receptor pairs and the gene regulatory network of CCCExplorer with the networks we used in the work so that the results obtained from different methods are comparable. As a required input of CCCExplorer, the built-in KEGG pathways remained the same. The candidate ligand genes L1 were then ranked based on a P-value score calculated by CCCExplorer (v1.0) in ascending order.
NicheNet (L1 score): We took all the genes expressed in the receiver cells as background and those with q-value < 0.1 and the absolute value of logFC greater than or equal to 1 as the set of differentially expressed target genes to calculate the ligand activity scores of the candidate ligand genes L1 using nichenetr package (v1.0.0). The ligand genes L1 were then ranked based on these scores in descending order.
For each method, we have also assessed the rank values for the treatment ligands using LRLoop filtering and random filtering. If a ligand did not form feedback, we assigned the rank of the ligand to the last among all the candidate ligands.
2.5 Application of LRLoop to scRNA-seq data
While the goal of LRLoop is to predict the feedback loop, we also developed an interaction score (SLR) to quantify the strength of individual ligand–receptor interactions in a particular biological condition. The score considered the contribution from both L1–R1 and looped L2–R2 interactions between two cell types. Suppose L1–R1 is a candidate ligand–receptor interaction expressed from cell type A to cell type B (by default setting, detected in at least 10% of the cells in each cell type, respectively). Let wL1R1 be the interaction strength (e.g. gene expression, NicheNet score or SingleCellSignalR score) of L1–R1 and wL2R2 the maximum of the interaction strength of its partners (L2–R2) expressed from cell type B to cell type A. If L1–R1 has no LRLoop partner, let wL2R2 = 0. The interaction score of the loop (L1–R1: L2–R2) was defined as
where 0 ≤ λ ≤ 1 can be understood as a weight for L2–R2 interaction, is a Hill function bounded between 0 and 1 that increases as the value of wL2R2 increases and the saturation curve becomes steeper as the Hill coefficient k > 0 gets larger. μ is the reflection point where the concavity of the function changes. When wL2R2 = μ, the Hill function is equal to 0.5. The detailed characterization and application of the Hill function can be found in Gesztelyi et al. (2012) and Weiss (1997).
We adopted four methods to calculate the L–R interaction strength (i.e. wL1R1 and wL2R2):
NicheNet (Pearson): In each cell type, we took the genes that are detected in at least 10% of the cells as ‘expressed’ genes and those whose average expression levels are above 75 percentile among all the expressed genes as ‘highly expressed’. We calculated the ligand activity score (the Pearson correlation coefficient) for each expressed ligand using nichenetr package (v1.0.0), taking all the expressed genes in the receiver cell type as background and the set of genes that are highly expressed in the receiver cell type as the target gene set of interest. An interaction L–R was counted if both L and R were expressed in the corresponding cell types and that the ligand activity score of L was above a threshold value.
SingleCellSignalR: The interaction strength of expressed (detected in at least 10% of the cells) ligand–receptor pairs was defined as in SingleCellSignalR (v1.6.0) (Cabello-Aguilar et al., 2020), where l and r are the average expression value of the ligand and receptor, respectively. is the average expression value of all genes in all cells.
CellPhoneDB: The interaction strength of expressed (detected in at least 10% of the cells) ligand–receptor pairs was defined as the mean of the average expression of the ligand and receptor genes in CellphoneDB (v2.1.7).
NATMI: The mean–expression weight of expressed (detected in at least 10% of the cells) ligand–receptor pairs was defined as the product of the mean expression of the ligand and the receptor genes in NATMI (v1.0).
We downloaded the high-quality batch-removed digital gene expression data (Mouse Cell Atlas) (Han et al., 2018). Tissues with only one cell type and cell types with less than 10 cells were removed for the analysis. For the remaining 36 tissues, we examined the proportions of between/within-tissue ligand–receptor interactions with or without consideration of L2–R2 contribution for each tissue pair at multiple cutoff score values. To find the optimal parameters, we calculated an overall reduction in the fraction of between-tissue interactions across different parameter settings (k = 0.2, 0.5, 1, 2, 4, 8, 16; λ = 0.1, 0.2, … , 0.8, 0.9, 1; μ = 0.1, 0.2, … , 0.8, 0.9). We found the parameter combination that yielded the largest reduction. For this dataset, the highest number of tissues pairs with an overall reduction was obtained at λ = 0.9, μ = 0.7 and k = 4 when using the SingleCellSignalR score for the values of wL1R1 and wL2R2 and λ = 0.9, μ = 0.08 and k = 4 using NicheNet ligand activity score. For the other two methods, the calculation was done with the same λ and k (0.9 and 4, respectively) and the μ value chosen around the minimum interaction score of expressed (detected in at least 10% of the cells) ligand–receptor pairs.
2.6 Prediction of L–R interactions between retinal cells
We separately analyzed the ligand–receptor interactions for the three retinal development stages. We identified 5, 6 and 6 retinal cell types in the early, intermediate and late stage, respectively. Based on the cell type annotations, we predicted the feedback LR loops and ranked them between each pair of these cell types using the LRLoop package.
Before running LRLoop, we first cleaned the ligand–receptor pairs according to their annotation in the Human protein atlas (Uhlén et al., 2015) and Uniprot (UniProt Consortium, 2021). We removed the ligand–receptor pairs if one of them has been annotated as ‘intracellular proteins’ in both datasets.
Next, we detected feedback loops between each pair of cell-types following the standard LRLoop analysis pipeline. Briefly, to calculate feedback loops between cell-type A and cell-type B, we first cleaned the LR networks based on the ligand expression (at least 2.5% cells expressed, the threshold can be adjusted based on the sequencing depth) in cell-type A and the receptors expression (at least 2.5% cells expressed) in cell-type B. We then cleaned the signaling networks based on the gene expression (at least 2.5% cells expressed) in cell-type B. With the cleaned LR network, cleaned signaling network and the gene regulatory network, we constructed two matrices: AtoB–ligand–target matrix and AtoB–receptor–target matrix. Conversely, with the same cleaning method, we constructed another two matrices: BtoA–ligand–target matrix and BtoA–receptor–target matrix. With the four matrices, we detected the feedback loops using the function: ‘PrepareBasics’ with min_pct = 0.025.
Finally, for each pair of cell-types, we calculated the S scores for all detected ligand–receptor pairs. To quantify the global interactions between each cell-type pair, we calculate the aggregated SC score, which was defined as
where C is the cutoff for SLR. To find the more specific LR pairs, we further filtered the ligand–receptor pairs with the following criteria: (1) we removed LR pairs if they are present at least 70% cell-type pairs. (2) We removed the LR pair if their maximum difference of SLR score is < 0.5 across all cell-type pairs.
3 Results
3.1 Closed feedback loop as a design principle for cell–cell communication
We propose a new method, LRLoop, which identifies feedback loop signaling interactions between individual cell types. For this purpose, we integrated the gene expression, signaling pathways and regulatory networks and extracted the feedback motifs in the composite networks. To illustrate the concept, we first show an example here (Fig. 1C). During injury-induced Muller glia reprogramming in mice (Hoang et al., 2020), microglial cells secrete ligand Tnf, which binds to receptor Tnfrsf1a, which in turn is expressed in Muller glial cells. At the same time, the ligand Anxa2 is secreted by Muller glial cells and its receptor Tlr2 is expressed in microglial cells. Furthermore, based on known signaling and regulatory networks, Tnf and Anxa2 are predicted to be downstream genes of Tlr2 in microglia and Tnfrsf1a in Muller glia, respectively. These four genes are found to be co-expressed in the time course; they are all upregulated at 3 h after retinal injury and gradually downregulated afterward (Fig. 1B; Hoang et al., 2020). The Tnf–Tnfrsf1a-mediated microglia–Muller glia communication was confirmed in mouse retina (Todd et al., 2020). Our analysis suggests that the feedback loops might be one type of signaling motif regulating cell–cell communication.
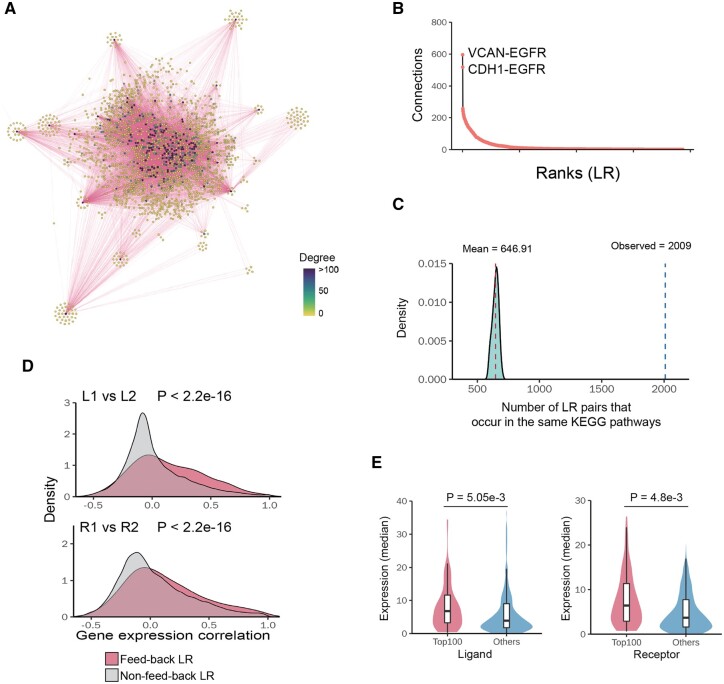
We next investigated the prevalence of the feedback loops based on known ligand–receptor pairs, signaling pathways and gene regulatory networks. In total, we identified 2512 experimentally supported ligand–receptor pairs based on literature. Among all 3 156 328 possible loops involving two known ligand–receptor pairs (L1–R1 and L2–R2, the loops could be from the same ligand–receptor pairs), we found that 0.55% (17 413) of them form feedback loops (see Section 2, Fig. 2A). These pairs involve 656 ligands and 647 receptors. Our identified feedback loops contained the majority of known ligand–receptor pairs (77%, 1935). Most ligand–receptor interactions form the feedback loops with a few other ligand–receptor interactions, while a few of them could partner with many interactions (Fig. 2B). The top ligand–receptor interaction identified is VCAN-EGFR, which pairs with 600 other ligand–receptor interactions to form feedback loops, probably in various biological contexts.
Fig. 2.
Properties of identified LRLoop. (A) Networks of feedback ligand–receptor pairs. Each node represents a ligand–receptor pair. Two nodes are connected if they form a feedback loop. (B) Ranked ligand–receptor pairs based on their connection degree in the feedback network (high to low). X-axis indicates their ranks. Y-axis indicates their number of connections. (C) Number of the feedback loops whose four members (L1, R1, L2 and R2) are in the same KEGG pathways. The distribution shows the same numbers if we randomly select two ligand–receptor pairs. (D) The Pearson correlation coefficients of connected L1–L2/R1–R2 are significantly higher than un-connected L1–L2/R1–R2. P-values were calculated using KS-test. (E) The median expression level of hub ligands/receptors is significantly higher than lowly connected ligands/receptors. P-values were calculated using t-test
Several lines of evidence suggest that the feedback loops identified using this approach are biologically relevant. First, we assessed whether the LR loops are more likely to be in the same biological pathways. In fact, members of 2009 feedback loops (L1, L2, R1 and R2) are found in the same KEGG pathways. For example, LAMA2–ITGB1 and PDGFB–PDGFRA interactions are found to form a feedback loop and both pairs of the proteins are part of the focal adhesion signaling pathway (Ogata et al., 1999). In contrast, randomly selected pairs of ligand–receptor interactions are much less likely to be in the same KEGG signaling pathways (Fig. 2C). Second, we examined whether the genes encoding the feedback loop members tend to be co-expressed. We analyzed the gene expression profiles across human tissues and calculated the correlation coefficients of gene expression (Carithers et al., 2015). Specifically, we calculated two correlation coefficients (L1–L2, R1–R2) and found that they were higher than those obtained from random pairs of ligand–receptor interactions (P < 2.2e−16) (Fig. 2D). Third, we examined whether ligand–receptor hubs have higher expression levels than non-hub genes, since these hubs formed feedback loops with many other ligand–receptor interactions and were more likely to be activated in multiple cell types or physiological conditions. We calculated the average gene expression of ligand–receptor pairs across human tissues and found that the genes in the hubs had higher expression levels than non-hub genes (P = 5.05e−3, P = 4.8e−3) (Fig. 2E). In summary, these results suggest that the feedback loops might be one widespread design principle regulating cell–cell communication.
3.2 Assessment of LRLoop using bulk datasets
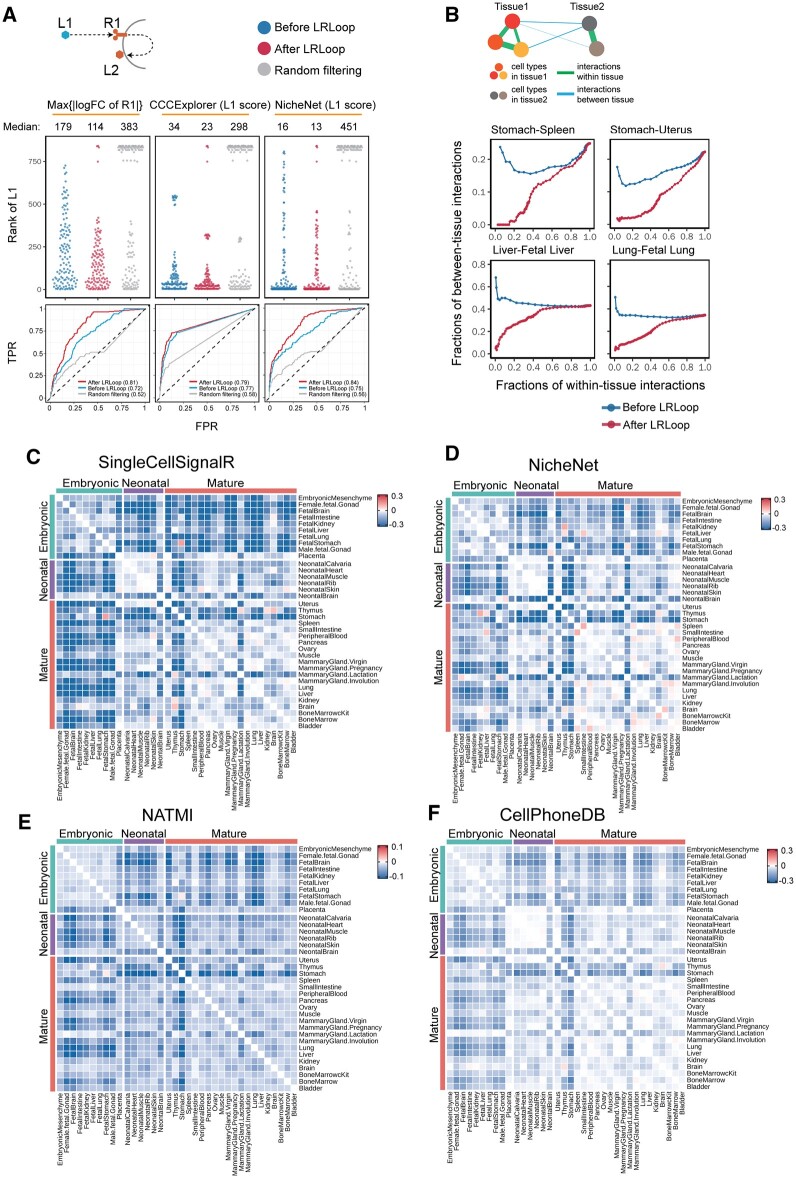
We next assessed whether the feedback loops could improve the prediction of cell–cell communication. We incorporated the identified feedback loops to different scoring methods (e.g. gene expression changes or ligand activity score from NicheNet (Browaeys et al., 2020)) and compared the performance with and without feedback loops. We applied our approach to a transcriptome dataset compiled by Browaeys et al. (2020) in which the gene expression from bulk samples was profiled before and after the cells were stimulated by one or two ligands. The dataset includes the transcriptomes from 111 treatments with a total 121 ligands. Note that the data only contain the expression profiles from the receiver cells, which include the differential expression data for R1 and L2 (Fig. 3A). We examined the rank of the treatment ligands (i.e. the expected ligands) among all ligands in the system using different scoring methods. When we ranked the ligands based on the gene expression changes of their interacting receptors (R1) before and after the treatment, the median of the rank for the expected ligands was 179, suggesting that on average 178 other ligands rank better than the expected ligand. If we required both R1 and L2 to be present and ranked the ligands based on the expression changes of R1, the rank of the expected ligands was improved, with a median value of 114. In contrast, if we used randomly created feedback loops as a filter, the median rank of expected ligands was 383 because many expected ligands (59 on average) did not form a feedback in the random dataset and were ranked at the bottom (Fig. 3A). Similar results were observed when we ranked the ligands based on the ligand activity scores using CCCExplorer (Choi et al., 2015) and NicheNet (Browaeys et al., 2020). When we used the LRLoop as a filter, our method improved the rank of the expected ligands. We also used a receiver operating characteristic (ROC) curve to evaluate the performance of LRLoop. It is clear that the area under the curve was larger than other three methods, suggesting that inclusion of looped ligand–receptor interactions could improve the prediction of targeted ligands (Fig. 3A). Unfortunately, we were not able to compare with other existing methods because these methods cannot be applied to this specific dataset due to the lack of ligand expression information for the sender cells. In summary, our results indicated that the feedback loops could substantially reduce the false positive rate.
Fig. 3.
Assessment of LRLoop using bulk and single-cell datasets. (A) Assessment using bulk expression datasets. Top panels: each dot represents an expected ligand. Bottom: the corresponding ROCs after applying LRLoop to each method. (B) Assessment using single-cell datasets. We used the between-tissue interactions as the indicator for false positive predictions. In the x-axis are the fractions of within-tissue interactions that remain in all expressed within-tissue interactions. The y-axis displays the fractions of between-tissue interactions in all interactions that remain at the cutoff score values. (C–F) Heatmap showing the reduction of between-tissue interactions, calculated as the ratio of the area under the ‘After LRLoop’ curve to the area under the ‘Before LRLoop’ curve as in B, and then minus one, among 630 tissue pairs. Four scoring methods were used
3.3 LRLoop enriched for within-tissue interactions
An ideal gold standard dataset to assess the performance should include cell type-specific, physiological condition-dependent ligand–receptor interactions. However, a challenge in this field is that there does not exist a comprehensive gold standard dataset to evaluate the performance of the predicted patterns of cell–cell communication. To benchmark the prediction performance, we propose an indirect evaluation based on the observation that gene expression tends to be coordinated in cells that are physically closer and thus most of the cell–cell interactions occur within a tissue where the cells are close to each other (Featherstone et al., 2016; Lander, 2013; Ren et al., 2020). Therefore, we reasoned that the cells that were either spatially or temporally separated are less likely to signal to one another than the cells within a tissue (Fig. 3B). For example, cells in the spleen and stomach are less likely to interact with each other because these cells are spatially separated. Likewise, cells in fetal lung and mature lung are also less likely to interact because they are temporally separated. For a pair of tissues, we calculated the numbers of predicted ligand–receptor interactions within tissues and between the two tissues, and used the fraction of between-tissue interactions among all interactions. We then compared the obtained fraction before and after applying LRLoop. The lower fraction indicated the reduced false positive rate.
To quantitatively include the contribution from both L1–R1 and L2–R2 interactions, we developed an empirical interaction score (SLR) to quantify the strength of ligand–receptor interactions (see Section 2). We then applied the method to scRNA-seq data from mouse tissues at embryonic, neonatal and mature stages (Han et al., 2018). We found that incorporating the feedback loops could reduce the rate of predicted between-tissue interactions. For instance, the stomach includes stomach epithelial cell, gastric mucosa cell and endocrine cell types, while the spleen includes neutrophil, monocyte, dendritic cell, macrophage and NK cell types. We then calculated the interaction score based on the SingleCellSignalR score (Cabello-Aguilar et al., 2020). When we used different SLR values as cutoffs, we found that the fraction of between-tissue interactions ranged from 0 to 0.25. If we do not consider the contribution from L2 to R2 interactions (i.e. wL2R2 = 0, see Section 2), the corresponding range was from 0.15 to 0.25 (Fig. 3B). The fraction of between-tissue interactions was substantially reduced if we considered the contribution from L2 to R2 interactions. Similarly, we also observed the reduction of between-tissue interactions for temporally separated tissues. For example, the interactions between fetal liver and adult liver as well as fetal lung and adult lung, the fraction of between-tissue interactions were significantly reduced after we included the looped L2–R2 (Fig. 3B). Overall, we observed 96.7% of 630 pairs of 36 tissues showed a reduction in the fraction of between-tissue interactions with an optimized parameter set (Fig. 3C). Similar results were observed when we applied the feedback loops to the NicheNet, NATMI and CellPhoneDB scoring methods (Fig. 3D–F). In summary, our results suggest that the LRLoop approach can successfully represent lower between-tissue interactions and higher within-tissue ones, perhaps because our method intrinsically considers a higher coordination of gene expression, which would be expected to occur in cells that are closer to each other.
3.4 Cell–cell communication during retinal development
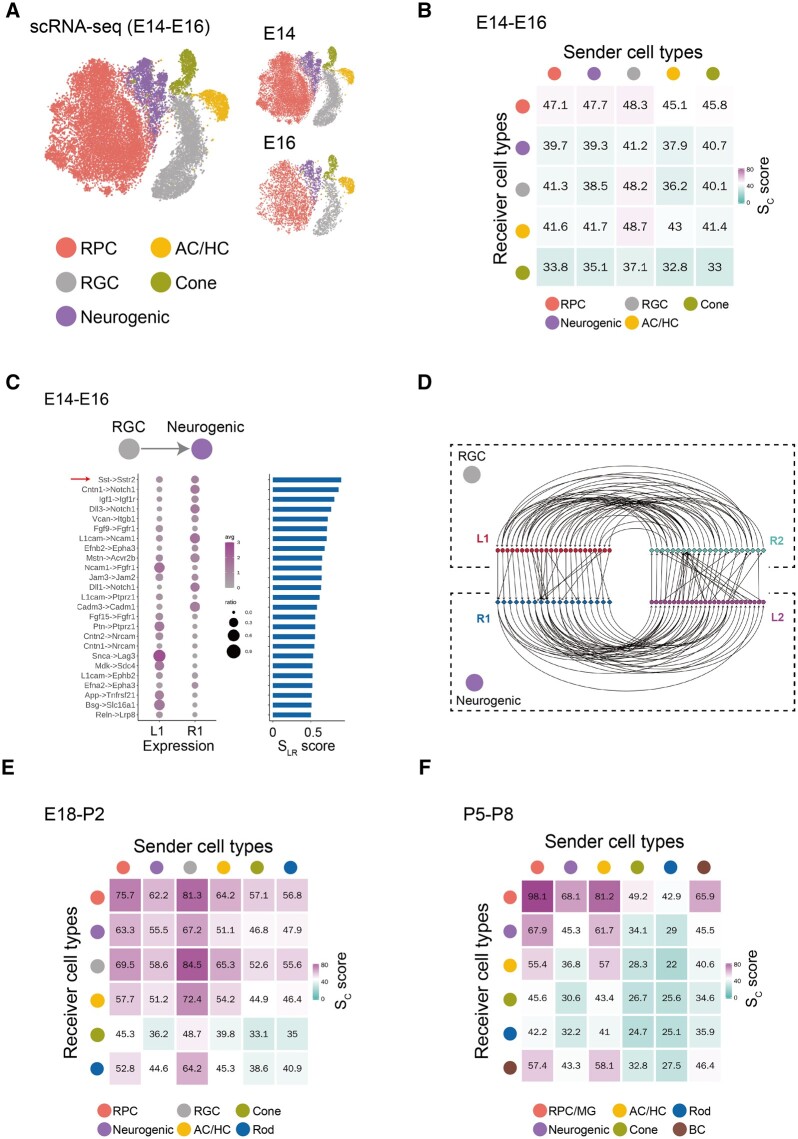
Previous studies have highlighted the importance of cell–cell signaling during retinal development, with extrinsic factors controlling progenitor proliferation, neurogenesis, neuronal differentiation and synapse formation (Sanes and Zipursky, 2020; Wallace, 2011). We applied our method to retinal scRNA-seq datasets and predicted cell–cell communication between individual retinal cell types during retinal development (Clark et al., 2019). The scRNA-seq datasets are grouped into three different stages of neurogenesis: early (E14–E16), intermediate (E18–P2) (Supplementary Fig. S1A) and late (P5–P8) (Supplementary Fig. S1B). During early stages of neurogenesis, five distinct cell types were identified, including retinal progenitor cells (RPCs), amacrine cells/horizontal cells (AC/HC), RGCs, cone photoreceptors and neurogenic cells (Fig. 4A). We identified the ligand–receptor feedback loops between each pair of cell types. A cell–cell communication map was obtained by summarizing all ligand–receptor interactions (Fig. 4B). Strong interactions were observed between RPCs and other cell types. Among these, 38 ligand–receptor interactions were identified for RPC–RPC signaling (Supplementary Fig. S1C), which included a number of previously identified signaling interactions. For example, fibroblast growth factor (FGF) signaling is known to play a key role in retinal progenitor proliferation and neurogenesis (da Silva and Cepko, 2017). Consistent with previous knowledge, Fgf9–Fgfr1, Fgf15–Fgfr1 and Fgf3–Fgfr1 interactions were found in RPCs. Similarly, 11 known ligand–receptor interactions were identified between RPC and neurogenic cells, including multiple Notch-Delta pathway members (Supplementary Fig. S1D; Mills and Goldman, 2017). Interactions identified between RGC and neurogenic progenitors included Sst-Sstr2, which is known to play a role in photoreceptor generation and regulation of retinal neurogenesis (Weir et al., 2021; Fig. 4C and D).
Fig. 4.
Application of the LRLoop to retinal development. (A) Five major cell types identified using scRNA-seq dataset during the early retinal developmental stages (E14 and E16). (B) Sc score between the five cell types calculated using LRLoop during early stages. (C) The unique ligand–receptor interactions predicted between RGCs and neurogenic cells during early stages. The red arrow indicates the known interactions. (D) Detailed feedback loops for interactions between RGCs and neurogenic cells during early stages of retinal neurogenesis. (E) Sc score between the six cell types calculated by LRLoop during intermediate stages. (F) Sc score between the six cell types calculated by LRLoop during late stages
The patterns of cell–cell communication changed substantially over the course of retinal development. While during early neurogenesis, signaling between RPCs and other cell types was most prominent, during intermediate stages of neurogenesis, the number of all interactions increased substantially. In particular, many interactions were observed between RGCs and AC/HC, as well as between RPCs and other cell types (Fig. 4E). During late neurogenesis, signaling between AC/HC and BC was prominent, as were reciprocal signaling between AC/HC and RPC/MG (Fig. 4F and Supplementary Table S1). Both the timing and pattern of cell–cell signaling at later developmental stages coincides with the establishment of synaptic connectivity (Sanes and Zipursky, 2020). As expected, signaling loops between GABAergic AC/HC and glutamatergic BC and RGCs include both multiple Neuroligin–Neurexin and Cadm gene family members, which mediate formation of GABAergic and glutamatergic synapses, respectively (Biederer et al., 2002; Huang and Scheiffele, 2008; Supplementary Table S1).
3.5 Evaluation of computational scalability
To evaluate the speed and memory usage of LRLoop, we tested one example dataset with Seurat objects of ∼1.6G and 6573 cells. It took ∼319 s on a computer running Microsoft Windows 11 Home OS, with Intel Core i7-8565U CPU with 1.80 GHz, 1992 MHz, 4 Core(s) and 16 GB installed physical memory. The total RAM used was 2.4 mebibytes and peak RAM used was 3803.5 mebibytes. This shows that LRLoop can be run on a personal computer with reasonable running speed and memory.
4 Discussion
Cell–cell communication plays an essential role in many biological processes in multicellular organisms. While many computational methods were developed for the prediction of ligand–receptor interactions, a recent survey of this field highlighted two major unsolved challenges (Dimitrov et al., 2022). First, a large discrepancy exists between the results obtained from different prediction methods. Second, we do not have a good gold standard dataset to assess the accuracy of these predictions. In this study, we developed a new computational method, LRLoop, to predict bi-directional ligand–receptor interactions. Compared with previous existing methods, the unique aspect of our method is that we require the presence of two ligand–receptor pairs that form a feedback signaling loop between two individual cell types. Furthermore, the expression regulation of the two ligands is dependent on the activity of the two receptors. This design principle of cell–cell communications ensures a robust bi-directional communication between cell types.
This feedback loop was identified in studies of cell–cell communication (Barone et al., 2017;Jing et al., 2021; Nilsson and Skinner, 2001). However, several key points are different in our analysis. First, in previous work, the two ligand–receptor interactions that form the feedback loop were considered to be independent of one another. The two ligand–receptor interactions were identified independently, using one-directional algorithms. In contrast, the two ligand–receptor interactions identified by LRLoop were responsive to one another and are interconnected through previously validated signaling networks and regulatory networks. Second, in previous work, the feedback loops were found only after the ligand–receptor interactions were identified, while the feedback loops were used as a prerequisite for the prediction of ligand–receptor interactions in this study.
Defining positive or negative feedback in gene regulation is relatively straightforward because the consequence of gene regulation is the gene expression, which is easy to define the up- or down-regulation and thus easy to define positive or negative feedback. In contrast, it is more complicated to determine positive or negative regulation in cell–cell communication. The consequence of cell–cell communication is the biological function (e.g. Tnf–Tnfrsf1a interaction promotes cell proliferation). One interaction might regulate multiple functions in a cell. These functions could have opposite directions (i.e. up- or down-regulation) with the same stimuli. Furthermore, two pairs of ligand–receptor interactions will act on two cell types in our feedback loop. Therefore, the consequence of the LRLoop is manifest in two cell types, which might correspond to two sets of biological functions. It is challenging to determine whether the looped ligand–receptor interactions will enhance or suppress the multiple biological functions in two cell types.
LRLoop and many existing methods (e.g. NicheNet) have distinct purposes. The goal of LRLoop is to identify the LR pairs that are connected through signaling and regulatory networks in two cell types. In contrast, the goal of the NicheNet is to predict ligands (or receptors) that are responsible for a set of differentially expressed genes in one cell type. LRLoop conveniently utilized the function of NicheNet to find the connection between a gene and its upstream receptors because NicheNet is a well-recognized program for this function. If we could improve the gene–ligand connection prediction in the future, we will replace the NicheNet with our own code.
We have three lines of evidence to demonstrate the quality of LRLoop predictions. (1) We used 111 experimentally validated datasets to evaluate the performance of LRLoop. One or two ligands were perturbated in each dataset and the corresponding altered gene expression profile was obtained. In these datasets, the targeted ligands are known. Therefore, the datasets could serve as the ground truth. (2) We used the fraction of between-tissue interactions as an indirect assessment for the false positive rate. Compared with existing methods, LRLoop produced lower between-tissue interactions when we applied the methods to 630 pairs of tissues. (3) We applied LRLoop to a retinal development dataset and predicted the potential ligand–receptor interactions in this specific system. We recovered many known interactions that supported our predictions.
We have several potential options to improve LRLoop. (1) We expect that the closed feedback loop is one design principle regulating cell–cell communication. Other design principles, potentially involving more than two cell types, are possible. (2) If a more detailed analysis of the spatial relationships between cell types is available, it should be possible to more accurately predict patterns of cell–cell communication. (3) In the current version of LRLoop, we did not include receptor complex information, we will integrate the complexes identified in CellPhoneDB to the next version of LRLoop.
Supplementary Material
Acknowledgements
We thank Drs. Mandeep Singh and Jeff Mumm for discussions.
Funding
The work was partially supported by National Institutes of Health grants (5R01EY029548 and 5P30EY001765 to J.Q.).
Conflict of Interest: none declared.
Contributor Information
Ying Xin, Department of Ophthalmology, Johns Hopkins University School of Medicine, Baltimore, MD 21287, USA.
Pin Lyu, Department of Ophthalmology, Johns Hopkins University School of Medicine, Baltimore, MD 21287, USA.
Junyao Jiang, CAS Key Laboratory of Regenerative Biology, Guangdong Provincial Key Laboratory of Stem Cell and Regenerative Medicine, Guangzhou Institutes of Biomedicine and Health, Chinese Academy of Sciences, Guangzhou 510530, China.
Fengquan Zhou, Department of Orthopedic Surgery, Johns Hopkins University School of Medicine, Baltimore, MD 21287, USA; Solomon H. Snyder Department of Neuroscience, Johns Hopkins University School of Medicine, Baltimore, MD 21287, USA.
Jie Wang, CAS Key Laboratory of Regenerative Biology, Guangdong Provincial Key Laboratory of Stem Cell and Regenerative Medicine, Guangzhou Institutes of Biomedicine and Health, Chinese Academy of Sciences, Guangzhou 510530, China.
Seth Blackshaw, Department of Ophthalmology, Johns Hopkins University School of Medicine, Baltimore, MD 21287, USA; Solomon H. Snyder Department of Neuroscience, Johns Hopkins University School of Medicine, Baltimore, MD 21287, USA.
Jiang Qian, Department of Ophthalmology, Johns Hopkins University School of Medicine, Baltimore, MD 21287, USA.
References
- Almet A.A. et al. (2021) The landscape of cell–cell communication through single-cell transcriptomics. Curr. Opin. Syst. Biol., 26, 12–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armingol E. et al. (2021) Deciphering cell–cell interactions and communication from gene expression. Nat. Rev. Genet., 22, 71–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barone V. et al. (2017) An effective feedback loop between cell–cell contact duration and morphogen signaling determines cell fate. Dev. Cell., 43, 198–211.e12. [DOI] [PubMed] [Google Scholar]
- Bich L. et al. (2019) Understanding multicellularity: the functional organization of the intercellular space. Front. Physiol., 10, 1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biederer T. et al. (2002) SynCAM, a synaptic adhesion molecule that drives synapse assembly. Science, 297, 1525–1531. [DOI] [PubMed] [Google Scholar]
- Blencowe M. et al. (2019) Network modeling of single-cell omics data: challenges, opportunities, and progresses. Emerg. Top. Life Sci., 3, 379–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnans C. et al. (2014) Remodelling the extracellular matrix in development and disease. Nat. Rev. Mol. Cell Biol., 15, 786–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browaeys R. et al. (2020) NicheNet: modeling intercellular communication by linking ligands to target genes. Nat. Methods, 17, 159–162. [DOI] [PubMed] [Google Scholar]
- Cabello-Aguilar S. et al. (2020) SingleCellSignalR: inference of intercellular networks from single-cell transcriptomics. Nucleic Acids Res., 48, e55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carithers L.J. et al. ; GTEx Consortium. (2015) A novel approach to high-quality postmortem tissue procurement: the GTEx project. Biopreserv. Biobank., 13, 311–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi H. et al. (2015) Transcriptome analysis of individual stromal cell populations identifies stroma–tumor crosstalk in mouse lung cancer model. Cell Rep., 10, 1187–1201. [DOI] [PubMed] [Google Scholar]
- Clark B.S. et al. (2019) Single-cell RNA-Seq analysis of retinal development identifies NFI factors as regulating mitotic exit and late-born cell specification. Neuron, 102, 1111–1126.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitrov D. et al. (2022) Comparison of methods and resources for cell–cell communication inference from single-cell RNA-Seq data. Nat. Commun., 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efremova M. et al. (2020) CellPhoneDB: inferring cell–cell communication from combined expression of multi-subunit ligand–receptor complexes. Nat. Protoc., 15, 1484–1506. [DOI] [PubMed] [Google Scholar]
- Featherstone K. et al. (2016) Spatially coordinated dynamic gene transcription in living pituitary tissue. eLife., 5, e08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gesztelyi R. et al. (2012) The Hill equation and the origin of quantitative pharmacology. Arch. Hist. Exact Sci., 66, 427–438. [Google Scholar]
- Han X. et al. (2018) Mapping the mouse cell atlas by Microwell-Seq. Cell, 172, 1091–1107.e17. [DOI] [PubMed] [Google Scholar]
- Hoang T. et al. (2020) Gene regulatory networks controlling vertebrate retinal regeneration. Science, 370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou R. et al. (2020) Predicting cell-to-cell communication networks using NATMI. Nat. Commun., 11, 5011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Z.J., Scheiffele P. (2008) GABA and neuroligin signaling: linking synaptic activity and adhesion in inhibitory synapse development. Curr. Opin. Neurobiol., 18, 77–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing J. et al. (2021) Reciprocal interaction between mesenchymal stem cells and transit amplifying cells regulates tissue homeostasis. Elife, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin S., Ramos R. (2022) Computational exploration of cellular communication in skin from emerging single-cell and spatial transcriptomic data. Biochem. Soc. Trans., 50, 297–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson T.V. et al. (2014) Identification of retinal ganglion cell neuroprotection conferred by platelet-derived growth factor through analysis of the mesenchymal stem cell secretome. Brain, 137, 503–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirouac D.C. et al. (2009) Cell–cell interaction networks regulate blood stem and progenitor cell fate. Mol. Syst. Biol., 5, 293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirouac D.C. et al. (2010) Dynamic interaction networks in a hierarchically organized tissue. Mol. Syst. Biol., 6, 417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander A.D. (2013) How cells know where they are. Science, 339, 923–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills E.A., Goldman D. (2017) The regulation of notch signaling in retinal development and regeneration. Curr. Pathobiol. Rep., 5, 323–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montoro D.T. et al. (2018) A revised airway epithelial hierarchy includes CFTR-expressing ionocytes. Nature, 560, 319–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson E., Skinner M.K. (2001) Cellular interactions that control primordial follicle development and folliculogenesis. J. Soc. Gynecol. Investig., 8, S17–S20. [DOI] [PubMed] [Google Scholar]
- Noël F. et al. (2021) Dissection of intercellular communication using the transcriptome-based framework ICELLNET. Nat. Commun., 12, 1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogata H. et al. (1999) KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res., 27, 29–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raredon M.S.B. et al. (2022) Computation and visualization of cell–cell signaling topologies in single-cell systems data using connectome. Sci. Rep., 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren X. et al. (2020) Reconstruction of cell spatial organization from single-cell RNA sequencing data based on ligand–receptor mediated self-assembly. Cell Res., 30, 763–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanes J.R., Zipursky S.L. (2020) Synaptic specificity, recognition molecules, and assembly of neural circuits. Cell, 181, 536–556. [DOI] [PubMed] [Google Scholar]
- Shao X. et al. (2021) CellTalkDB: a manually curated database of ligand–receptor interactions in humans and mice. Brief. Bioinformatics, 22. [DOI] [PubMed] [Google Scholar]
- Shao X. et al. (2020) New avenues for systematically inferring cell–cell communication: through single-cell transcriptomics data. Protein Cell., 11, 866–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Silva S., Cepko C.L. (2017) Fgf8 expression and degradation of retinoic acid are required for patterning a High-Acuity area in the retina. Dev. Cell., 42, 68–81.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tammela T., Sage J. (2020) Investigating tumor heterogeneity in mouse models. Annu. Rev. Cancer Biol., 4, 99–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todd L. et al. (2020) Microglia suppress Ascl1-Induced retinal regeneration in mice. Cell Rep., 33, 108507. [DOI] [PubMed] [Google Scholar]
- Uhlén M. et al. (2015) Proteomics. Tissue-based map of the human proteome. Science, 347, 1260419. [DOI] [PubMed] [Google Scholar]
- UniProt Consortium. (2021) UniProt: the universal protein knowledgebase in 2021. Nucleic Acids Res., 49, D480–D489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace V.A. (2011) Concise review: making a retina—from the building blocks to clinical applications. Stem Cells, 29, 412–417. [DOI] [PubMed] [Google Scholar]
- Weir K. et al. (2021) A potential role for somatostatin signaling in regulating retinal neurogenesis. Sci. Rep., 11, 10962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss J.N. (1997) The hill equation revisited: uses and misuses. FASEB J., 11, 835–841. [PubMed] [Google Scholar]
- Zhou X. et al. (2018) Circuit design features of a stable two-cell system. Cell, 172, 744–757.e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.