Abstract
Lactococcus lactis NCDO 2118 was grown in a simple synthetic medium containing only six essential amino acids and glucose as carbon substrates to determine qualitatively and quantitatively the carbon fluxes into the metabolic network. The specific rates of substrate consumption, product formation, and biomass synthesis, calculated during the exponential growth phase, represented the carbon fluxes within the catabolic and anabolic pathways. The macromolecular composition of the biomass was measured to distribute the global anabolic flux into the specific anabolic pathways. Finally, the distribution of radiolabeled substrates, both into the excreted fermentation end products and into the different macromolecular fractions of biomass, was monitored. The classical end products of lactic acid metabolism (lactate, formate, and acetate) were labeled with glucose, which did not label other excreted products, and to a lesser extent with serine, which was deaminated to pyruvate and represented approximately 10% of the pyruvate flux. Other minor products, keto and hydroxy acids, were produced from glutamate and branched-chain amino acids via deamination and subsequent decarboxylation and/or reduction. Glucose labeled all biomass fractions and accounted for 66% of the cellular carbon, although this represented only 5% of the consumed glucose.
Lactic acid bacteria (LAB) are of great importance in the food industry, mainly for lactic acid production from various substrates, but also for flavor compound or bacteriocin synthesis. The nutritional complexity of LAB is such that they are frequently cultivated in media containing complex nitrogen sources (MRS [9] or M17 [30]) or in natural media (milk or wine) for different applications. However, the complexity of these media is such that growth and metabolic behavior are difficult to characterize precisely. This is certainly the reason why it is generally considered that during LAB fermentation, sugar is only a catabolic substrate leading to metabolic end products and energy while biomass is formed from anabolic precursors, i.e., amino acids, nucleotides, etc., present in the culture broth. Whatever the medium used, more than 90% of the carbon in the sugar is normally converted into metabolic end products, generally, lactic acid. Moreover, the growth of LAB is characterized by poor growth yield, i.e., amount of biomass formed per amount sugar consumed, and hence, the quantity of the biomass formed is low compared to the quantity of lactic acid produced. However, this simplistic model is based on inaccurate carbon balances and is very controversial. A very small part of the sugar leading to anabolic reactions could represent a significant part of the biomass carbon, and this consideration has been totally neglected.
The study of radiolabeled substrate distribution into end products or biomass has been used to estimate the part of sugar leading to biomass. Sivakanesan and Dawes (29) reported that 0.5% of the glucose used as the substrate labeled the biomass of Staphylococcus epidermidis. Chauvet et al. (6) cultivated Leuconostoc oenos in wine supplemented with labeled substrates, e.g., glucose and malic and citric acids. They demonstrated that 2.1% of the glucose was recovered as biomass, while neither malic acid nor citric acid was a precursor of anabolic compounds. Later, Schmitt et al. (27) showed that during the cultivation of L. mesenteroides subsp. cremoris in complex MRS medium, [14C]glucose was not incorporated into biomass, while 1.1% of the citrate present was incorporated into biomass in the form of acetyl coenzyme A (acetyl-CoA). Benthin et al. (2) suggested that in steady-state and transient cultures of Lactococcus cremoris FD1 in a chemostat, a small part of the carbon in the sugar might contribute to biomass formation. Plihon et al. (22) performed batch experiments with L. mesenteroides in MRS medium, and they concluded that “the carbon balance confirmed that biomass was not created from sugar metabolism.” Clearly, this conclusion is subject to doubt since the experimental error observed in the substrate and product concentrations given by high-pressure liquid chromatography (HPLC) was 5%, illustrating that this kind of experiment is unable to give precise information about the precursors of biomass synthesis.
On the other hand, some evidence exists to support the idea that certain amino acids can be catabolized and metabolic products can be excreted into the culture medium. This is established for some amino acids converted to oxo or hydroxy acids involved in cheese flavor compounds (36). It has also been shown that serine can be deaminated to give pyruvate and then lactate (3, 18). In this case, the operation of this catabolic pathway modifies the calculated value of the lactate yield from the sugar and confirms that carbon balancing based solely upon sugar-to-product conversion cannot be used alone to draw conclusions about carbon flux distribution.
A precise study of carbon distribution into biomass requires that the macromolecular composition of the cells be known. The data previously published for the composition of L. lactis are difficult to generalize because they were obtained with a variety of strains and with various methods and, furthermore, only certain biomass components were assessed. Moreover, both growth conditions and growth rate can influence the percentage of each class of macromolecules or the chemical composition of a macromolecular fraction (5, 34). For example, the lipidic composition of Escherichia coli is affected by the pH, temperature, aeration, or growth rate (1). The macromolecular composition of L. lactis NCDO 712 is also dependent on the growth rate, particularly with regard to RNA/protein or RNA/DNA content ratios (4). Significant variation of protein content has been reported: Thomas and Batt (31) showed that L. lactis ML3 contains 48% protein, including the peptides of peptidoglycan, while Benthin et al. (2) reported a value of 45% for L. cremoris, not including the peptides of the cell wall. A very different value, between 23 and 30%, was obtained by Foucaud (10) for L. lactis subsp. lactis biovar diacetylactis CNRZ 125 or 141.
To date, no data have been reported on the detailed metabolic network of L. lactis, since these data necessitate quantitative determination of macromolecular biomass composition, precise determination of fermentation kinetics, and measurement of the distribution of 14C-labeled substrates into the cell, performed together with one strain and under one precise set of culture conditions. In this study, these various approaches have been combined to calculate the most complete carbon flux distribution pattern within the metabolic network of L. lactis NCDO 2118.
MATERIALS AND METHODS
Organism and culture media.
L. lactis subsp. lactis NCDO 2118, obtained from the collection held at the Institut National de la Recherche Agronomique (Jouy-en-Josas, France), was used throughout this study. The medium used for the growth of the inoculum was the synthetic MS10 medium described by Cocaign-Bousquet et al. (7), while the medium used for the experiments was the MCD medium described by Otto et al. (19) and modified by Poolman and Konings (23) or MS14 medium (18). These media were prepared from concentrated stock solutions stored at 4°C after filtration through cellulose nitrate membranes (0.22-μm pore size), except for the cysteine solution, which was freshly prepared. The media (pH 6.6) were sterilized by filtration through cellulose acetate membranes (0.22-μm pore size; Sartorius) directly into a sterilized (20 min at 121°C) culture vessel.
Culture conditions.
Fermentations were carried out under strictly anaerobic conditions in a 2-liter glass fermentor (Sétric Génie Industriel, Toulouse, France) or in anaerobic tubes at a temperature of 30°C and an agitation speed of 250 rpm. The bacteria were grown in a controlled gas environment created by flushing of both the vessel and the medium with nitrogen. The medium in the fermentor was aseptically gassed (30 min) immediately before inoculation and maintained under an N2 atmosphere at a positive pressure of 103 Pa. Fermentor cultures were maintained at pH 6.6 by automatic addition of 5 N KOH.
Inoculation was at 2% with a high-optical-density (optical density at 580 nm, 2.0) culture on MS10 medium with a high concentration of phosphates ([K2HPO4 = 15 g/liter and [KH2PO4] = 18 g/liter) to buffer acid production, and the culture was incubated overnight at 30°C in a butyl rubber-stoppered 250-ml shaken flask under N2. Precultures were washed twice with sterile phosphate buffer (100 mM, pH 6.6) to avoid carryover of essential nutrients and resuspended in the same buffer. All tube cultures were carried out in triplicate and repeated if experimental variation exceeded 5%.
Analytical methods.
Bacterial growth was monitored by spectrophotometric measurements at 580 nm and calibrated against cell dry weight measurements. Cells were harvested by filtration on 0.45-μm-pore-size nylon membranes, washed with 2 volumes of deionized water, and dried to a constant weight at 60°C under a partial vacuum (200 mm Hg [ca. 26.7 kPa]). A change of 1 U of optical density was shown to be equivalent to 0.31 g of dry matter · liter−1. The biomass formula was determined at ENSC (Toulouse, France) by elemental analysis of C, H, O, N, and ash. The ash fraction on MS14 medium corresponded to 9.2% (wt/wt) of the biomass. The biomass formula used to convert cell dry weights into molar cell carbon concentrations was C5.05H9.20O2.77N1.0, with a molar mass of 140 g · mol−1, including ash at 12 g · mol−1.
The carbon dioxide concentration in the gas phase was determined by gas chromatography using a Porapak Q column followed by a molecular sieve maintained at 40°C with helium as the carrier gas and catharometer detection.
Determination of sugars and organic acids from fermentation supernatants or from cell hydrolysates was performed by HPLC using an Aminex HPX-87H+ column (300 by 7.8 mm) and the following conditions: a temperature of 50°C, solvent H2SO4 (5 mM), a flow rate of 0.5 ml · min−1, and dual detection (refractometer and UV at 220 nm). Ethanol concentration measurements were also carried out with isocratic gas chromatography on a Porapak-Q packed column at 190°C with a flame ionization detector (Intersmat GC). Some compounds produced during the fermentation and visualized by peaks during HPLC analysis were identified by checking the retention time with standard solutions.
Ammonia concentrations were determined in filtered culture samples with an ammonia-selective electrode (Orion).
Concentrations of amino acids in the medium were determined with an AminoQuant 1090 HPLC apparatus (Hewlett-Packard) after derivatization by ortho-phtalaldehyde in the presence of 3-mercaptopropionic acid and by 9-fluorenylmethoxy carbonyl, separation with a C18 column, and spectrophotometric detection at 338 nm for ortho-phtalaldehyde derivatives or 266 nm for 9-fluorenylmethoxy carbonyl derivatives.
For the separation of radiolabeled amino acids, micellar HPLC on a Beckman Ultrasphere octyldecyl silane column (250 by 4.6 mm) was done by using a modification of the method of Saurina and Hernandez-Cassou (25). Eluent A contained H3PO4 (20 mM), Na2HPO4 (20 mM), and sodium dodecyl sulfate (10 mM) in deionized (MilliQ) water. In eluent B, water was replaced with a mixture of deionized water-acetonitrile (60/40). The flow rate was maintained constant at 0.15 ml/min throughout the entire chromatographic run, while the temperature was kept at 50°C. Separation of amino acids was performed by an initial isocratic elution (100% eluent A from 0 to 80 min), followed by a linear gradient from 80 min (0% B) to 560 min (100% B). The percentage of eluent B was linearly decreased afterward and then maintained at 0% until re-equilibration of the column. Detection of amino acids was performed by measurement of UV absorption at 200 nm, while in-flux radioactive countings were done as described below.
Determination of biomass macromolecular composition.
The cells were centrifuged (4°C, 10 min at 8,000 × g), washed three times with MgCl2 (1 to 5 mM), and then lyophilized. These cells were used for the quantification of each macromolecular biomass fraction.
Carbohydrates were measured by three methods, two colorimetric methods with anthrone or phenol (14), and one chromatographic method. For this last method, lyophilized cells were resuspended in distilled water, sonicated to disrupt the cell wall, and sealed in a tube under nitrogen with 25 to 50 mg of Dowex 50W×4 (200 to 400 mesh) resin. Hydrolysis was done at 115 to 125°C for 3 to 12 h. Sugars were separated by HPLC on an Aminex HPX-87H+ column (see the description of analytical methods).
After cell wall disruption by sonic treatment, lipidic compounds were extracted with n-hexane–isopropanol (3:2) as described by Hara and Radin (13) and weighed after drying.
Fatty acids were measured in cells incubated (15 min, 80°C) in concentrated HCl under argon. Methanol was added to this suspension and then esterified under argon (2 h, 80°C). Fatty acid esters were extracted with hexane, dried under argon flux, and recovered in a few milliliters of hexane. This extract was analyzed by gas chromatography (Hewlett-Packard 5890 Series II) by injection into a capillary column (Supelco SP2330, 30 m by 0.25 mm) at 200°C with nitrogen as the carrier gas.
Amino sugars constitutive of the peptidoglycan were measured after hydrolysis of the cell wall. Lyophilized cells were incubated (2 to 6 h, 105°C) in a sealed tube containing HCl (4 M) and under nitrogen. The hydrolysate was evaporated under vacuum, resuspended in distilled water, and passed through a Dowex 50W×4 column. The column was washed with distilled water and eluted with HCl (2 M), and the eluate was dried under vacuum. Amino sugars were measured spectrophotometrically after reaction with dimethylaminobenzaldehyde (11, 12) or by HPLC.
Nucleic acids were extracted from lyophilized cells by incubation with perchloric acid (0.5 M, 70 to 80°C, 15 to 20 min) and measured as described by Hanson and Phillips (12).
After solubilization of the cells with NaOH (1 M, 90°C, 10 min) proteins were determined by the biuret method, the Folin reagent, or the use of Coomassie blue (21). The amino acid content of the proteins was determined after hydrolysis (105°C, 24 to 36 h) under nitrogen by a modified form of the method of Ng et al. (17), the hydrolyzing solution containing 2-mercaptoethanol (0.2 or 1%), or a modified form of the method of Yano et al. (35) with a mixture of chlorhydric (7 M), trifluoroacetic (10%), and thioglycolic (20%) acids and phenol (1%). Lysozyme was used as a standard solution to quantify the loss of amino acids during hydrolysis.
Use of radiolabeled substrates.
All of the uniformly 14C-radiolabeled substrates were obtained from Amersham and used at 8 to 50 kBq/ml of medium.
Measurement of radioactivity in glucose and in the fermentation end products was done by passing the sample through an HPLC column (see the description of analytical methods), followed by detection with a continuous scintillation counter (Berthold 506A). The scintillation fluid was mixed with the HPLC eluant at a ratio of 2:1 and counted in the detector.
Radioactivity in the biomass macromolecules was determined by using cells washed three times with MgCl2 (5 mM) and counted in each fraction obtained after sequential fractionation of the biomass as described previously by Paalme et al. (20).
Balance and flux calculations.
The total carbon balance was calculated as the amount of the products and biomass formed during the entire duration of the culture period as a function of the concentration of glucose and amino acids consumed. The apparent catabolic balance was calculated as the ratio of the products formed, i.e., lactate, formate, acetate, and pyruvate, to the glucose consumed.
The specific rates of substrate consumption or product formation, expressed in millimoles per gram per hour, were calculated from the kinetic data of the substrate and product concentrations measured during the culture period. All of the specific rate values were maintained constant during the major part of the culture at their maximal value. These maximal values were used to calculate the fluxes through the metabolic pathways and correspond to the molecular fluxes at the input and output of the metabolic pathways. The conceptual basis and mathematical expression for such flux calculation have been described by Vallino and Stephanopoulos (32) and used previously for other bacteria (8). The metabolic fluxes were better expressed as carbon flux by taking into account the number of carbon atoms in each molecule. The biomass can be considered a macromolecule with molar mass M (in grams per mole) and macroelemental composition CaHbOcNd which is formed at specific growth rate μ (per hour). The flux of synthesis of a biomass macromolecule is the product of its concentration in the biomass (in millimoles per gram) and the growth rate, μ.
Calculation of carbon substrate incorporation into the carbon of a biomass macromolecular fraction was done with the equation I = Af · Mx · As−1 (ns/nx), where I is the micromoles of substrate carbon incorporated by the micromoles of biomass carbon, Af is the specific radioactivity of fraction f (in becquerels per milligram of biomass), As is the specific radioactivity of the substrate (in becquerels per micromole), Mx is the molar mass of the biomass (140 g · mol−1 in MS14 medium), ns is the number of carbon atoms in the substrate, and nx is the number of carbon atoms in a biomass molecule (5.05 in MS14 medium). These incorporation values were then converted to relative incorporation rates expressed as a function of the sum of the values obtained for the different fractions.
RESULTS AND DISCUSSION
Batch culture of L. lactis NCDO 2118 in MS14 medium.
The growth and metabolic behavior of L. lactis NCDO 2118 in MCD or MS14 medium were as previously described (15, 18). The metabolism was homolactic in both media and was characterized by a lactate yield of about 1.7 mol/mol of glucose. The identified minor fermentation products were formate, acetate, and ethanol in MCD medium, while in MS14 medium, pyruvate and ammonium were also excreted into the culture broth and no ethanol was produced (data not shown). Amino acid consumption was related to the amino acid composition of the medium. As previously described, large amounts of serine were consumed in the MS14 medium and associated with ammonia production (18).
Growth started immediately after inoculation and continued until glucose had been completely exhausted from the medium. Growth was more rapid in MCD medium, with a maximal specific rate of 0.8 h−1 instead of the 0.18 h−1 measured in MS14 medium. In this last medium, the maximal specific growth rate was the result of the absence of a number of amino acids from the medium (18).
The specific rates of growth, consumption of both sugar and amino acids, and product formation rapidly reached their maximal and were maintained constant for the major part of the culture period. These maximum specific rates, calculated during the exponential growth phase in MS14 medium, were as follows (expressed in millimoles per gram per hour): glucose consumption, 13.6; glutamate consumption, 0.3; serine consumption, 3.4; methionine consumption, 0.06; leucine consumption, 0.4; isoleucine consumption, 0.3; valine consumption, 0.2; lactate production, 24.2; formate production, 2.4; acetate production, 2.2; pyruvate production, 0.4, biomass production, 1.3 (corresponding to a growth rate of 0.18 g · g−1 · h−1).
The apparent catabolic balance, calculated by the ratio of lactate, formate, acetate, and pyruvate production to glucose consumption, showed 96% carbon recovery and might indicate that glucose was only catabolized for energy production, as currently assumed. However, since serine was massively deaminated to pyruvate and then reduced to lactate (18), this simple balance introduces flux errors. This consideration was confirmed by looking at the total carbon balance, i.e., products and biomass production from glucose and the consumed amino acids, which indicated that only 86% of the consumed carbon was recovered as biomass carbon or classical end products of lactic metabolism. It can therefore be assumed that other catabolic end products were formed from both glucose and amino acids.
Macromolecular composition of biomass.
The cells cultivated in MCD or MS14 medium were analyzed to determine the macromolecular composition of the biomass (Table 1). The polysaccharide fraction was greater in MS14 medium than in MCD medium, while the opposite was true of the RNA fraction. Other macromolecular fractions were identical in cells grown on either of the two media. The protein fraction, which represents the cellular proteins and the cell wall peptides, was by far the most important in the cell, with all other fractions representing between 3 and 15% of the biomass.
TABLE 1.
Macromolecular composition and amino acid composition of the protein fraction of L. lactis NCDO 2118 grown in MCD or MS14 mediuma
| Fraction or molecule | MCD | MS14 |
|---|---|---|
| Polysaccharides | 12 | 15 |
| Amino sugars | 5.5 | 5.5 |
| RNA | 8 | 6 |
| DNA | 3 | 3.3 |
| Lipids | 4.3 | 4 |
| Proteins | 45 | 45 |
| Teichoic acidb | 10 | 10 |
| Inorganic ions | 7 | ND |
| Alanine | 12.8 | 12.3 |
| Arginine | 3.8 | 3.4 |
| Asxc | 9.9 | 10.8 |
| Cysteine | 2.9 | 3.1 |
| Glxd | 10.5 | 12.0 |
| Glycine | 8.8 | 7.4 |
| Histidine | 1.4 | 1.3 |
| Isoleucine | 5.3 | 5.4 |
| Leucine | 7.3 | 7.9 |
| Lysine | 8.7 | 8.9 |
| Methionine | 2.2 | 2.1 |
| Phenylalanine | 3.2 | 3.4 |
| Proline | 2.9 | 3.3 |
| Serine | 4.4 | 4.6 |
| Threonine | 5.0 | 4.9 |
| Tryptophan | 2.2 | 0.7 |
| Tyrosine | 2.3 | 2.4 |
| Valine | 6.6 | 6.1 |
The values reported are averages of 5 to 10 measurements, and the accuracy varied from 5 to 10%, depending on the fraction or the molecule tested. Macromolecule composition is expressed as mass percentages, and amino acid composition is expressed as moles percent. ND, not determined.
Extrapolated from the percentage of amino sugars and data from the literature.
Sum of aspartic acid and asparagine.
Sum of glutamic acid and glutamine.
The values obtained for the polysaccharide fraction differed with the method used. In MCD medium, the anthrone method gave 7.1% while the phenol method led to 15.3%. A similar difference was obtained with MS14 medium, i.e., 9.4 and 17%, respectively. This difference was due to the insensitivity of anthrone to deoxyhexoses (rhamnose) and pentoses and to the reactivity of phenol with nucleotidic sugars. In view of this, the polysaccharide content of the cell was calculated by taking into account an estimation of the different sugars constituting this fraction. As a consequence, the values presented in Table 1 are not arithmetic averages of these two experimental values but corrected values. An apparent difference was observed between our polysaccharide content values and the values determined by Thomas and Batt (31) for L. lactis ML3 grown in a complex medium, but the 7.7% they found was obtained with anthrone and was similar to our value of 7.1% obtained with the same method during growth in MCD medium. Chromatographic analysis of this fraction has revealed that glucose and rhamnose are the predominant sugars while the cells have a lower galactose content, with a respective mass proportion of 5.5/5.1/1.0.
While the level of DNA measured was similar to that observed by Thomas and Batt (31), the RNA concentration was lower (6 to 8% instead of 21%). It is known that the RNA level of the cell is susceptible to variation with the metabolic activity and growth rate of the cell. This is probably the reason for the high RNA level observed by Thomas and Batt (31), since in their experiments, growth proceeded rapidly in a complex medium. Moreover, it has been previously observed for Saccharomyces cerevisiae (28) and Klebsiella aerogenes (16) that growth under nitrogen-limiting conditions, as would be the case in the media employed in this study, leads to a low RNA concentration.
The fatty acid composition of the cells determined by HPLC analysis showed that palmitic and oleic acids (C16:0 and C18:1, respectively) were present in similar amounts in the MCD medium used, while the oleic acid concentration was very low in the MS14 medium used. Hence, while the lipid concentrations were identical in the two media tested, and comparable to the data of Thomas and Batt (31), the composition differed with the medium. Moreover, lactobacillic acid (cyclopropane acid [C19]) was not identified in these cells, whatever the medium used, while it was detected in the type species NCDO 712 described by Schleifer et al. (26).
The protein fraction of our strain was in good agreement with the values generally reported. The amino acid content of proteins was determined after acidic hydrolysis of the total proteins of cells cultivated in MCD or MS14 medium. The different hydrolytic methods used gave very similar results for the majority of the amino acids. Only tryptophan and cysteine were not detected when hydrolysis was done with HCl and 3-mercaptopropionic acid (1%). The average percentage of each amino acid in cells cultivated in either MCD or MS14 medium is shown in Table 1. It appears that this amino acid composition was very conserved when the medium composition was varied, since the maximal difference between the two media used was only 1.5%, as observed for glutamate plus glutamine or for tryptophan. Alanine, aspartate-asparagine, glutamate-glutamine, lysine, leucine, and glycine were the predominant amino acids, each accounting for more than 7% of the protein fraction. The predominance of alanine, aspartate, glutamate, and lysine in the protein fraction was not surprising, since they are constituents of the peptidoglycan of L. lactis (26). On the other hand, tryptophan, histidine, and methionine are the amino acids least abundant in the cell.
Distribution of 14C-labeled substrates in fermentation end products.
Among the carbon substrates present in MS14 medium, glucose and five of the six amino acids (glutamate, serine, isoleucine, leucine, and valine) were used separately as uniformly 14C-labeled substrates during the cultures, and the radioactivity was measured in the fermentation end products. Only methionine was not tested because it was consumed in very low amounts and in proportion to the amount of methionine found in the cells.
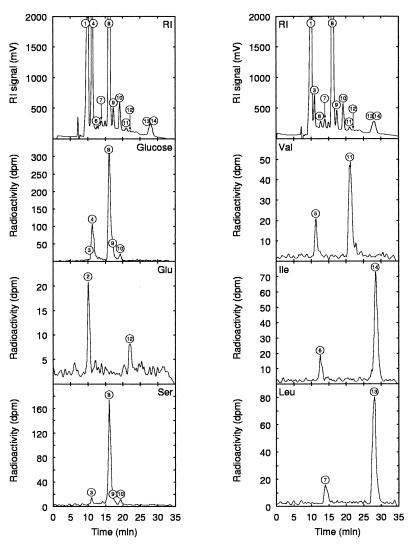
The HPLC profile obtained with a refractometer detector showed many peaks, some of them corresponding to glucose and the classical end products of lactic fermentation but others being unidentified (Fig. 1), as classically observed for LAB cultures. From the radioactivity measurements, it appeared that all of the substrates tested led to the production of excreted compounds. About 91% of the lactate, formate, acetate, and pyruvate found came from glucose, which did not give other products. The other 9% of these products derived from pyruvate, originating from serine. Hence, the classical products of lactic fermentation were derived specifically from glucose and serine.
FIG. 1.
Origin of fermentation end products of L. lactis NCDO 2118 grown in MS14 medium, determined by HPLC analysis, followed by radioactivity measurement. In the graphs at the top, the HPLC refractometric signal is plotted, showing all of the detected compounds, before (left) and after (right) glucose exhaustion. In the other graphs, the radioactivity signal after culture with various 14C-labeled substrates (written on the graph) is plotted, showing the compounds derived from the corresponding labeled substrates. Peaks: 1, phosphate; 2, 2-ketoglutarate; 3, pyruvate; 4, glucose; 5, 3-hydroxyisobutyrate; 6, 3-hydroxymethylbutyrate; 7, 3-hydroxyisovalerate; 8, lactate; 9, formate; 10, acetate; 11, 2-hydroxyisovalerate; 12, pyroglutamate; 13, 2-hydroxyisocaproate; 14, 2-hydroxymethylvalerate. RI, refractive index.
Glutamate gave two particular compounds produced in low concentrations, 2-ketoglutarate and pyroglutamate, a product of cyclization by dehydration between amine and δ-carboxylic groups of the molecule.
Each branched-chain amino acid produced two different compounds, some of them leading to important peaks observed by refractometry (peaks 13 and 14). None of these compounds were the keto acids produced by deamination (or transamination) of the amino acids (for example, 2-ketoisocaproate from leucine). However, the 2-hydroxy acids derived from each of these keto acids by a reduction step, e.g., 2-hydroxyisocaproate from leucine, 2-hydroxy-3-methylvalerate from isoleucine, and 2-hydroxyisovalerate from valine, were present in the medium. These hydroxy acids were identified by reducing in vitro the respective keto acids by sodium borohydride in phosphate buffer (pH 7.2) and checking the retention times of these compounds in HPLC (33). Moreover, this reduction was also achieved in vitro by lactate dehydrogenase in a phosphate buffer (pH 7.2) containing NADH. In this assay, not only did NADH absorption gradually decrease but hydroxy acids also appeared in the cuvette. The three other peaks observed from the three branched-chain amino acids (Fig. 1, peaks 5, 6, and 7) should correspond, from retention time data, to the 3-hydroxy acids obtained after successive deamination, decarboxylation, and oxidation of each amino acid. In conclusion, all amino acids not directly included in the biomass were deaminated into keto acids, a part of them being reduced with NADH, and this reduction could be catalyzed by lactate dehydrogenase.
Distribution of 14C-labeled substrates in biomass carbon.
Cells cultivated in MS14 medium with 14C-labeled substrates were harvested during the exponential growth phase and fractionated by a sequential procedure to obtain the various macromolecular fractions. The radioactivity present in each fraction was then counted and expressed as the percentage of carbon from the labeled substrate recovered in each fraction (Table 2). The macromolecular fractions obtained after sequential fractionation of the biomass differed from the fractions recovered by macromolecular analysis, since complete separation by the sequential method was impossible. This was particularly true for the last separation steps. Care should be taken in analyzing these results, since low radiolabeling values could be due to contamination of a fraction with other molecules. Nevertheless, this method offers the advantage of simplicity and gives results complementary to the analysis of biomass composition.
TABLE 2.
Distribution of 14C-labeled substrates (glucose and amino acids of MS14 medium) in biomass fractions obtained by sequential fractionationa
| Fraction | Glucose | Glu | Ser | Val | Ile | Leu | Total |
|---|---|---|---|---|---|---|---|
| Intracellular pools | 2.12 | 1.67 | 0.15 | 0.04 | 0.09 | 0.06 | 4.13 |
| Lipids | 2.96 | 0.19 | 0.22 | 0.03 | 0.05 | 0.05 | 3.50 |
| RNA, teichoic acid | 10.42 | 0.39 | 0.64 | 0.18 | 0.11 | 0.18 | 11.92 |
| DNA, sugars, peptidesb | 30.96 | 3.40 | 1.86 | 1.74 | 1.47 | 1.19 | 40.62 |
| Proteins, amino sugars | 19.50 | 7.83 | 2.34 | 3.01 | 2.97 | 4.15 | 39.80 |
| Total | 65.96 | 13.49 | 5.22 | 5.00 | 4.70 | 5.63 | 100.00 |
The contribution of these six substrates to biomass carbon is supposed to be 100%. For each substrate, the sum of the values reported in a column is the relative amount of that substrate in the total carbon biomass. For each fraction, the sum of the values reported on a horizontal line is the percentage of that fraction in the biomass.
Low-molecular-weight peptides soluble in hot perchloric acid.
The pool fraction represented 4.1% of the total biomass carbon. This fraction was issued for 92% of the molecules originating from two substrates, glucose (51.3%) and glutamate (40.4%). The other four substrates accounted for only 8.2% of the pool fraction. The very high concentration of glutamate or glutamate-derived products in the cytosol is in agreement with the previous observations of Thomas and Batt (31) for L. lactis ML3 or of Poolman et al. (24) for L. lactis subsp. cremoris Wg2. Glucose gave 84.6% of the lipidic fraction, glutamate gave 5.4%, and serine gave 6.3%, while the three branched-chain amino acids labeled no more than 4.0%. The fraction containing RNA and a part of teichoic acid was labeled predominantly by glucose (87.4%), and a small part was labeled by serine (5.4%), while the other four substrates labeled only 7.2%. The following fraction, containing DNA, polysaccharides, and also some peptides, was labeled as follows: glucose, 76.2%; glutamate, 8.4%; serine, 4.6%; valine, 4.3%; isoleucine, 3.6%; leucine, 2.9%. Finally, the protein fraction was labeled as follows: 49% by glucose, 19.7% by glutamate, 5.9% by serine, 7.6% by valine, 7.5% by isoleucine, and 10.4% by leucine. These results are coherent with the knowledge of biochemical pathways and the metabolic precursors of each class of macromolecules. Lipids originated predominantly from glucose via acetyl-CoA, RNA originated from glucose via the pentose pathway for the ribose moiety and via the tricarboxylic acid cycle and the serine family for the nucleotides, and teichoic acid originated from glucose via glycerol phosphate, galactose, and alanine (26). The large percentage of the DNA fraction and the presence of a significant amount of label from each amino acid indicate that important polysaccharide and soluble peptide cross-contamination might contribute to this fraction.
In MS14 medium, glucose was the predominant precursor of each macromolecular fraction and gave 66% of the total carbon biomass. The same experiment was realized in MCD medium with a complete amino acid composition and nucleotides with [14C]glucose as the substrate. In this case, glucose labeled 33% of the total carbon biomass (data not shown).
Since the protein fraction harvested from cells cultivated in MS14 medium was highly labeled by glucose, the distribution of radioactivity from each substrate to the amino acids of cellular proteins was determined (Table 3). Glucose gave the totality of the amino acids of the phosphoribosyl pyrophosphate pathway (His, Tyr, and Phe), about 90% of the alanine-and-aspartate family (Asp, Asn, Thr, and Lys), a small fraction of arginine, and, curiously, the majority of the glycine. Data for tryptophan were not available, since it was destroyed during protein hydrolysis, but it can be assumed that it was produced from glucose, as were Tyr and Phe, and also from serine. Glutamate labeled only the amino acids of its own family (Glu, Gln, Pro, and Arg). Serine labeled the amino acids coming from pyruvate (Ala) and the tricarboxylic acid cycle (Asp, Asn, Thr, and Lys) in a proportion of about 6 to 10%. Glycine, which belongs to the serine family, was, however, labeled at only 33% by serine. The branched-chain amino acids of the biomass originated directly from those present in the medium. Moreover, Ile and Leu labeled Arg and the amino acids of the aspartate family at a very low level.
TABLE 3.
Distribution of 14C-labeled substrates of MS14 medium in the amino acids of cellular proteins of L. lactis NCDO 2118
| Amino acid | Distribution (mol%) of:
|
|||||
|---|---|---|---|---|---|---|
| Glucose | Glu | Ser | Val | Ile | Leu | |
| His | 100 | |||||
| Tyr | 100 | |||||
| Phe | 100 | |||||
| Ser | 100 | |||||
| Gly | 67.7 | 32.3 | ||||
| Met | ||||||
| Ala | 90.3 | 9.7 | ||||
| Asxb | 88.5 | 7.7 | 0.8 | 3.0 | ||
| Thr | 93 | 5.5 | 1.5 | |||
| Lys | 87.4 | 6.6 | 3.9 | 2.1 | ||
| Glxc | 100 | |||||
| Arg | 13.3 | 83.8 | 1.0 | 2.0 | ||
| Pro | 100 | |||||
| Val | 100 | |||||
| Ile | 100 | |||||
| Leu | 100 | |||||
Asx = sum of aspartic acid and asparagine.
Glx = sum of glutamic acid and glutamine.
Metabolic network of L. lactis NCDO 2118 in MS14 medium.
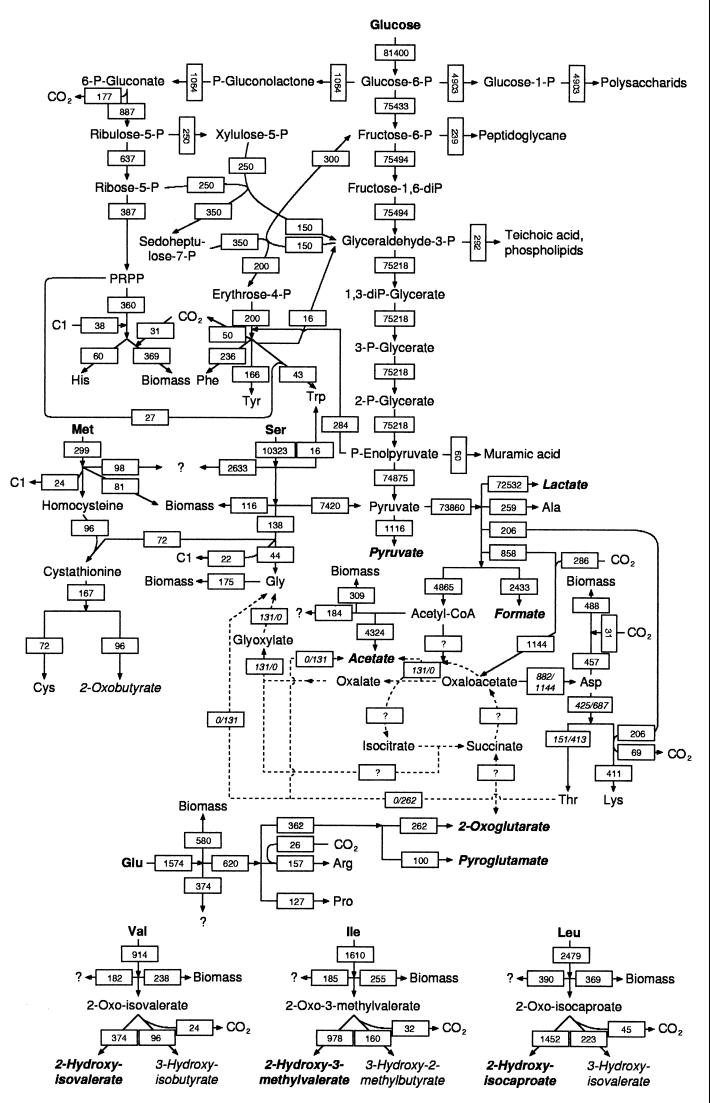
Data on 14C distribution into the macromolecular fractions enable the global metabolic scheme operating in this medium to be drawn. Taking into account kinetic values of flux of substrate consumption, biomass and product formation, and the relative quantity of each macromolecular fraction of the biomass has enabled the carbon flux through each metabolic pathway to be estimated (Fig. 2).
FIG. 2.
Carbon fluxes, expressed in micromoles of carbon per gram per hour, operating during growth of L. lactis NCDO 2118 in minimal synthetic medium MS14. Carbon substrates present in the culture medium are in boldface roman type, identified excreted products are in boldface italic type, supposed excreted products are in lightface italic type, and dashed arrows show possible pathways which could lead to glycine formation (maximum and minimum carbon flux values are given for the concerned products, depending on the operative pathway for glycine synthesis). P, phosphate; diP, diphosphate; PRPP, phosphoribosyl pyrophosphate.
The conclusions deduced from carbon balance estimations were confirmed by the study of distribution of 14C-labeled substrates in the metabolic network. The major catabolic process is the degradation of glucose by the glycolytic pathway. The specific rate of glucose consumption calculated during the exponential growth phase (13.6 mmol of glucose · g−1 · h−1) corresponded to a carbon flux input entering glycolysis of 81,400 μmol of C · g−1 · h−1 (Fig. 2). As shown in Tables 2 and 3, a part of the carbon derived from glucose and catabolized by glycolysis was used to give the totality of polysaccharides, amino sugars, phospholipids, glycerol, histidine, and aromatic amino acid fractions and a significant part of nucleotides, glycine, alanine, and the aspartate family. The sum of these carbon outputs toward biomass synthesis from glycolysis represented 8% of the initial glucose carbon input. Therefore, 92% of the initial carbon flux will pass through the entire glycolysis process to reach pyruvate. Another important carbon flux arises from serine at the level of pyruvate (72% of the initial serine flux) to give metabolites originating from pyruvate. The pyruvate flux coming from serine represents 10% of that originating from glucose, and the same proportion is observed at the level of lactate synthesis. Only 33% of the glycine came from serine, despite an apparently functional serine demethylation step via tetrahydrofolate. The pathway of glycine synthesis from glucose cannot be predicted from these data, since different possibilities could be operative. Glycine could be formed from threonine by threonine aldolase or 2-amino-3-oxobutyrate synthase or by transamination of glyoxylic acid, which could be produced by isocitrate lyase or from oxaloacetate via oxalate and oxalyl-CoA. Isocitrate lyase activity could not be detected in previous studies with this strain (15), but complementary studies are necessary to determine the nature of glycine synthesis in L. lactis.
Glutamic acid metabolism is distinct from the other metabolic pathways, because it gives only amino acids of its own family and two products, including ketoglutarate. Moreover, this last metabolite was not labeled by glucose, indicating that the tricarboxylic acid cycle was not operative in this medium, while it was shown to be functional, although at a low rate, in a similar synthetic medium lacking glutamate (15).
Each branched-chain amino acid contributed to biomass formation by itself in the protein fraction but also by participating in the carboxylation reactions operating in the biosynthesis of some metabolites (pyrimidines, Asp, Thr, Lys, and Arg), since CO2 was produced during decarboxylation of the corresponding keto acids. Pentose sugar formation from glucose and branched-chain amino acid metabolism under anaerobic conditions are the major CO2-generating pathways.
Despite the complex analysis of the metabolic network, some products originating from each consumed substrate remain unidentified. They represent 13.5% for isoleucine to 25.5% for serine, which can be considered low values relative to the total carbon fluxes but can be of industrial importance, since some pathways probably lead to flavoring compounds of interest in milk transformation.
Finally, 66% of the carbon biomass produced in this simple synthetic medium originating from glucose represented only 5% of the glucose carbon consumed, explaining the current assumption that glucose is only a catabolic substrate. The absence of glucose incorporation into biomass reported by Schmitt et al. (27) might be due to the consumption of other carbohydrates present in the complex medium they used. The part of glucose incorporated into biomass was reduced twofold in MCD medium containing all of the amino acids and nucleotides, in good agreement with the modified anabolic pathways operating in such a medium, for which synthesis of anabolic precursors present in the medium would no longer be necessary.
ACKNOWLEDGMENTS
We thank Muriel Cocaign-Bousquet, Nic Lindley, and Attila Szentirmai for useful discussions.
REFERENCES
- 1.Arneborg N, Salskov-Iversen A S, Mathiasen T E. The effect of growth rate and other growth conditions on the lipid composition of Escherichia coli. Appl Microbiol Biotechnol. 1993;39:353–357. [Google Scholar]
- 2.Benthin S, Schulze U, Nielsen J, Villadsen J. Growth energetics of Lactococcus cremoris FD1 during energy-, carbon-, and nitrogen-limitation in steady state and transient cultures. Chem Eng Sci. 1994;49:589–609. [Google Scholar]
- 3.Benthin S, Villadsen J. Amino acid utilization by Lactococcus lactis subsp. cremoris FD1 during growth on yeast or casein peptone. J Appl Microbiol. 1996;80:65–72. [Google Scholar]
- 4.Beresford T, Condon S. Physiological and genetic regulation of rRNA synthesis in Lactococcus. J Gen Microbiol. 1993;139:2009–2017. doi: 10.1099/00221287-139-9-2009. [DOI] [PubMed] [Google Scholar]
- 5.Bremer H, Dennis P P. Modulation of chemical composition and other parameters of the cell by growth rate. In: Neidhardt F C, Ingraham J L, Low K B, Magasanik B, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella typhimurium: cellular and molecular biology. Washington, D.C.: American Society for Microbiology; 1987. pp. 1527–1542. [Google Scholar]
- 6.Chauvet J, Brechot P, Dubois C, Dupuy P, Dorange J-L. Stimulation de la croissance dans le vin d'une flore malolactique par les acides malique et citrique. Sci Aliment. 1982;2:495–504. [Google Scholar]
- 7.Cocaign-Bousquet M, Garrigues C, Novák L, Lindley N D, Loubière P. Rational development of a simple synthetic medium for the sustained growth of Lactococcus lactis. J Appl Bacteriol. 1995;79:108–116. [Google Scholar]
- 8.Cocaign-Bousquet M, Lindley N D. Pyruvate overflow and carbon flux within the central metabolic pathways of Corynebacterium glutamicum during growth on lactate. Enzyme Microb Technol. 1995;17:260–267. [Google Scholar]
- 9.de Man J C, Rogosa M, Sharpe M E. A medium for the cultivation of lactobacilli. J Appl Bacteriol. 1960;23:130–135. [Google Scholar]
- 10.Foucaud C. Physiologie de Lactococcus lactis subsp. lactis biovar diacetylactis CNRZ 125 en conditions non optimales. Ph.D. thesis. Lyon, France: Université Claude Bernard Lyon I; 1990. [Google Scholar]
- 11.Ghuysen J M, Tipper D J, Strominger J L. Enzymes that degrade bacterial cell walls. Determination of total hexosamines. Methods Enzymol. 1966;8:692–693. [Google Scholar]
- 12.Hanson R S, Phillips J A. Chemical composition. In: Gerhardt P, Murray R G E, Costilow R N, Nester E W, Wood W A, Kroeg N R, Phillips G B, editors. Manual of methods for general bacteriology. Washington, D.C.: American Society for Microbiology; 1981. pp. 335–337. [Google Scholar]
- 13.Hara A, Radin N S. Lipid extraction of tissues with a low-toxicity solvent. Anal Biochem. 1978;90:420–426. doi: 10.1016/0003-2697(78)90046-5. [DOI] [PubMed] [Google Scholar]
- 14.Herbert D, Phipps P J, Strange R E. Chemical analysis of microbial cells. Methods Microbiol. 1971;5B:265–278. [Google Scholar]
- 15.Lapujade P, Cocaign-Bousquet M, Loubière P. Glutamate biosynthesis in Lactococcus lactis subsp. lactis NCDO 2118. Appl Environ Microbiol. 1998;64:2485–2489. doi: 10.1128/aem.64.7.2485-2489.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mulder M M, Van Der Gulden H M L, Postma P W, Van Dam K. Effect of macromolecular composition of micro-organisms on the thermodynamic description of their growth. Biochim Biophys Acta. 1988;936:406–412. doi: 10.1016/0005-2728(88)90017-5. [DOI] [PubMed] [Google Scholar]
- 17.Ng L T, Pascaud A, Pascaud M. Hydrochloric acid hydrolysis of proteins and determination of tryptophan by reversed-phase high-performance liquid chromatography. Anal Biochem. 1987;167:47–52. doi: 10.1016/0003-2697(87)90132-1. [DOI] [PubMed] [Google Scholar]
- 18.Novák L, Cocaign-Bousquet M, Lindley N D, Loubière P. Metabolism and energetics of Lactococcus lactis during growth in complex or synthetic media. Appl Environ Microbiol. 1997;63:2665–2670. doi: 10.1128/aem.63.7.2665-2670.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Otto R, Ten Brink B, Veldkamp H, Konings W N. The relation between growth rate and electrochemical proton gradient of Streptococcus cremoris. FEMS Microbiol Lett. 1983;16:69–74. [Google Scholar]
- 20.Paalme T, Olivson A, Vilu R. 13C-NMR of CO2-fixation during the heterotrophic growth in Chlorobium thiosulfatophilum. Biochim Biophys Acta. 1982;720:311–319. [Google Scholar]
- 21.Peterson G. Determination of total proteins. Methods Enzymol. 1983;91:95–119. doi: 10.1016/s0076-6879(83)91014-5. [DOI] [PubMed] [Google Scholar]
- 22.Plihon F, Taillandier P, Strehaiano P. Oxygen effect on lactose catabolism by a Leuconostoc mesenteroides strain: modeling of general O2-dependent stoichiometry. Biotech Bioeng. 1996;49:63–69. doi: 10.1002/(SICI)1097-0290(19960105)49:1<63::AID-BIT8>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 23.Poolman B, Konings W N. Relation of growth of Streptococcus lactis and Streptococcus cremoris to amino acid transport. J Bacteriol. 1988;170:700–707. doi: 10.1128/jb.170.2.700-707.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Poolman B, Smid E, Veldkamp H, Konings W N. Bioenergetic consequences of lactose starvation for continuously cultured Streptococcus cremoris. J Bacteriol. 1987;169:1460–1468. doi: 10.1128/jb.169.4.1460-1468.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Saurina J, Hernandez-Cassou S. Determination of amino acids by ion-pair liquid chromatography with post-column derivatization using 1,2-naphthoquinone-4-sulfonate. J Chromatogr. 1994;676:311–319. doi: 10.1016/0021-9673(94)80431-1. [DOI] [PubMed] [Google Scholar]
- 26.Schleifer K H, Kraus J, Dvorak C, Kilpper-Bälz R, Collins M D, Fischer W. Transfer of Streptococcus lactis and related streptococci to the genus Lactococcus gen. nov. Syst Appl Microbiol. 1985;6:183–195. [Google Scholar]
- 27.Schmitt P, Diviès C, Cardona R. Origin of end-products from the co-metabolism of glucose and citrate by Leuconostoc mesenteroides subsp. cremoris. Appl Microbiol Biotechnol. 1992;36:679–683. [Google Scholar]
- 28.Schulze U, Liden G, Nielsen J, Villadsen J. Physiological effects of nitrogen starvation in an anaerobic batch culture of Saccharomyces cerevisiae. Microbiology. 1996;142:2299–2310. doi: 10.1099/13500872-142-8-2299. [DOI] [PubMed] [Google Scholar]
- 29.Sivakanesan R, Dawes E A. Anaerobic glucose and serine metabolism in Staphylococcus epidermidis. J Gen Microbiol. 1980;118:143–157. doi: 10.1099/00221287-118-1-143. [DOI] [PubMed] [Google Scholar]
- 30.Terzaghi B, Sandine W E. Improved medium for lactic streptococci and their bacteriophages. Appl Microbiol. 1975;29:807–813. doi: 10.1128/am.29.6.807-813.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thomas T D, Batt R D. Degradation of cell constituents by starved Streptococcus lactis in relation to survival. J Gen Microbiol. 1969;58:347–362. doi: 10.1099/00221287-58-3-347. [DOI] [PubMed] [Google Scholar]
- 32.Vallino J, Stephanopoulos G. Metabolic flux distributions in Corynebacterium glutamicum during growth and lysine overproduction. Biotech Bioeng. 1993;41:633–646. doi: 10.1002/bit.260410606. [DOI] [PubMed] [Google Scholar]
- 33.Wann S R, Thorsen P T, Kreevoy M. Reduction of carboxylic acids derivatives by BH4− in acidic dimethyl sulfoxide. J Org Chem. 1981;46:2579–2581. [Google Scholar]
- 34.Wanner U, Egli T. Dynamics of microbial growth and cell composition in batch culture. FEMS Microbiol Rev. 1990;75:19–44. doi: 10.1111/j.1574-6968.1990.tb04084.x. [DOI] [PubMed] [Google Scholar]
- 35.Yano H, Aso K, Tsugita A. Further study on gas phase acid hydrolysis of protein: improvement of recoveries for tryptophan, tyrosine and methionine. J Biochem. 1990;108:579–582. doi: 10.1093/oxfordjournals.jbchem.a123245. [DOI] [PubMed] [Google Scholar]
- 36.Yvon M, Thirouin S, Rijnen L, Fromentier D, Gripon J C. An aminotransferase from Lactococcus lactis initiates conversion of amino acids to cheese flavor compounds. Appl Environ Microbiol. 1997;63:414–419. doi: 10.1128/aem.63.2.414-419.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]