Abstract
Crc (catabolite repression control) protein of Pseudomonas aeruginosa has shown to be involved in carbon regulation of several pathways. In this study, the role of Crc in catabolite repression control has been studied in Pseudomonas putida. The bkd operons of P. putida and P. aeruginosa encode the inducible multienzyme complex branched-chain keto acid dehydrogenase, which is regulated in both species by catabolite repression. We report here that this effect is mediated in both species by Crc. A 13-kb cloned DNA fragment containing the P. putida crc gene region was sequenced. Crc regulates the expression of branched-chain keto acid dehydrogenase, glucose-6-phosphate dehydrogenase, and amidase in both species but not urocanase, although the carbon sources responsible for catabolite repression in the two species differ. Transposon mutants affected in their expression of BkdR, the transcriptional activator of the bkd operon, were isolated and identified as crc and vacB (rnr) mutants. These mutants suggested that catabolite repression in pseudomonads might, in part, involve control of BkdR levels.
Pseudomonads play an important role in nature because of their ability to metabolize natural and manufactured organic chemicals. Many of these compounds are environmental pollutants, such as benzene, toluene, xylene, ethylbenzene, styrene, and chlorobenzoates (18), and their removal has been named bioremediation. Although the enzymic pathways responsible for degradation of these pollutants may be effective when the target compound is the sole growth-supporting substrate, in nature these compounds are present as mixtures, and some substrates may be degraded preferentially. Catabolite repression control refers to the ability of an organism to preferentially metabolize one carbon source over another when both are present in the growth medium. Because of the importance of pseudomonads to bioremediation efforts, understanding the control of catabolite repression is important so that more efficient, genetically modified organisms can be utilized in the removal of these environmental pollutants.
The molecular mechanisms of catabolite repression control have been extensively characterized in enteric bacteria, where glucose is the preferred carbon source. In these organisms, enzymes of the phosphoenolpyruvate-dependent phosphotransferase system mediate catabolite repression control by regulation of cyclic AMP (cAMP) concentration via adenylate cyclase activity (22). The strongest repressing substrates in Pseudomonas spp. are acetate, tricarboxylic acid cycle intermediates, and glucose (4, 10, 26). Unlike Escherichia coli, in Pseudomonas species adenylate cyclase activity, cAMP phosphodiesterase activity, and cAMP pools do not fluctuate with carbon source, nor does the addition of cAMP relieve repression of catabolite responsive pathways (21, 25). In addition, only one phosphotransferase system (fructose) has been identified in Pseudomonas (5), suggesting that PTS components are not involved in catabolite repression control in pseudomonads. The only protein thus far shown to be involved in catabolite repression in Pseudomonas is Crc of P. aeruginosa, but a function has not been identified for this protein (11). Crc has some sequence similarity (25 to 32% identity) to DNA repair enzymes of both prokaryotes and eukaryotes. However, Crc does not appear to have endonuclease activity or to bind DNA, suggesting that it has some other function.
Expression of branched-chain keto acid dehydrogenase (BCKAD) of P. putida is regulated by carbon and nitrogen sources (29). The bkd operon, which encodes BCKAD, and its regulation by BkdR, a positive transcriptional regulator, has been well characterized in P. putida (13–16, 29); therefore, P. putida BCKAD is a good model system for studying catabolite repression control of catabolic pathways in pseudomonads.
The objective of the present research was to determine if Crc played a role in catabolite repression control in P. putida, as well as in P. aeruginosa, by using the bkd operon as a model system. The phenotypes of the P. aeruginosa and P. putida crc mutants were compared, and although the carbon sources responsible for catabolite repression in the two species differ, the pathways regulated by Crc are identical in the two species.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. P. putida was grown with aeration at 30°C, while P. aeruginosa and E. coli were grown with aeration at 37°C. Starter cultures were grown in 2 ml of 2xYT medium (23). For BCKAD assays, P. putida and P. aeruginosa were grown in nitrogen-free valine-isoleucine medium (17) which contained 0.3% valine and 0.1% isoleucine (wt/vol) as the sole carbon and nitrogen sources (17).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristics | Source or reference |
|---|---|---|
| P. putida | ||
| JS386 | Φ(bkdR-lacZ)132(Hyb) | 14 |
| JS391 | Φ(bkdR-lacZ)132(Hyb) crc::Mini-Tn5 | This study |
| JS393 | Φ(bkdR-lacZ)132(Hyb) rnr::Mini-Tn5 | This study |
| JS394 | crc::Kmr | This study |
| PpG2 | Wild type | I. C. Gunsalus |
| P. aeruginosa | ||
| PAO1 | Wild type | P. V. Phibbs, Jr. |
| PAO8020 | Δcrc::Tcr | 11 |
| E. coli | ||
| DH5α | F− φ80d lacZΔM15 Δ(lacZYA-argF) endA1 recA13 supE44 hsdR17 | Gibco/BRL |
| HB101 | F−hsdS20 supE44 recA13 ara14 proA2ara14 proA2 rpsL20(Smr) xyl5 mlt-1λ− galK2 lacY1 | Gibco/BRL |
| JM109 | recA1 endA1 gyrA96 hsdR17 supE44 relA1 Δ(lac-proAB) (F′ traD36 proAB lacIqZΔM15) | |
| Plasmids | ||
| pJRS191 | 40-kb insert of P. putida DNA in pLAFR3; includes crc and rnr | This study |
| pJRS194 | crc+, 1.45-kb PCR insert in pUCPM19 | This study |
| pJRS196 | crc+, 2-kb insert in pUCPM19 | This study |
| pJRS197 | EcoRI/KpnI crc with Kmr at BsaAI in pUCPM19 minus stabilization fragment | This study |
| pLAFR1 | Broad-host-range cosmid cloning vector | 6 |
| pLAFR3 | Broad-host-range cosmid cloning vector | 28 |
| pPZ352 | PAO1 crc+ | 11 |
| pRK2013 | Helper plasmid; ColE1 mob+ tra+ (RK2) Kmr (8) | 8 |
| pUC19 | Cloning vector | 31 |
| pUCP19 | Escherichia-Pseudomonas shuttle vector derived from pUC19 | 24 |
| pUCPM18/19 | pUCP19 with mob fragment | This study |
| pUTKm | Tn5-based delivery plasmid in E. coli CC118 | 9 |
Both amino acids are supplied since growth on valine alone is toxic to the cell. For other assays, P. putida was grown in basal salts medium (BSM) (17) plus an inducing carbon source. For urocanase assays the carbon source was 10 mM histidine, for amidase assays the carbon source was 40 mM lactamide, and for glucose-6-phosphate dehydrogenase (G6PDH) the carbon source was 20 mM mannitol. The concentrations of catabolite repressors added to the medium were as follows: 20 mM glucose, 40 mM lactate, 40 mM succinate, 30 mM glutamate, and 20 mM gluconate. The concentrations of antibiotics used to inhibit growth of P. putida were as follows: carbenicillin, 2 mg/ml; kanamycin, 90 μg/ml; and tetracycline, 100 μg/ml. The concentrations of antibiotics used to inhibit growth of P. aeruginosa were as follows: carbenicillin, 1 mg/ml; kanamycin, 500 μg/ml; and tetracycline, 100 μg/ml. The concentrations of antibiotics used to inhibit growth of E. coli were as follows: ampicillin, 200 μg/ml; kanamycin, 90 μg/ml; and tetracycline, 50 μg/ml.
Enzyme assays.
Cultures for enzyme assays were grown in 100 ml of medium to an A660 of 0.6 to 0.8, harvested, and then treated as described previously for enzyme assays (17). The following enzyme assays were performed as described previously: BCKAD (27), amidase (26), urocanase (7), and G6PDH (10). One unit of BCKAD is 1 μmol of NADH formed/min/mg of protein, and one unit of G6PDH is 1 μmol of NADPH formed/min/mg of protein. Amidase assays were done on whole-cell suspensions, and the specific activity was measured as micromoles of lactamide per A660. The data in Fig. 5 are expressed as the percent activity relative to the lactamide plus lactate culture.
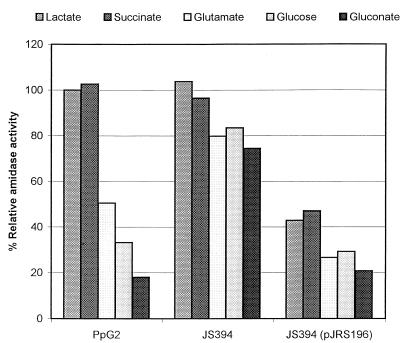
FIG. 5.
Effect of mutation in crc in catabolite repression of amidase in P. putida. P. putida strains PpG2, JS394, and JS394(pJRS196) were grown in BSM containing lactamide plus lactate, succinate, glutamate, glucose, or gluconate (columns in each group from left to right, respectively). Amidase activity is expressed as a percentage of the activity seen in P. putida PpG2 grown in lactamide plus lactate, which was included with each experiment.
Construction and screening of P. putida genomic cosmid library.
A complete BamHI digest of PpG2 chromosomal DNA was sized over a sucrose gradient, and fragments larger than 6 kb were ligated with BamHI-digested pLAFR3 (28) with T4 DNA ligase at a chromosome-to-vector ratio of 5:1. In vitro packaging and transformation of E. coli JM109 was performed according to the manufacturer's recommendations (Gigapack III Gold Packaging Extract; Stratagene). The library was screened with the 2.0 kb SstI fragment of pPZ352, which contains the PAO1 crc gene (11), and was labeled by using the RadPrime DNA labeling system (Life Technologies). The library was plated on 2xYT agar containing tetracycline, colonies were lifted by using Colony/Plaque Screen (Dupont/NEN Research Products), and hybridization and washing conditions were performed as suggested by the manufacturer.
Nucleic acid preparations and manipulations.
Chromosomal and cosmid DNAs used for library construction were purified by using cesium chloride gradients. Plasmid DNA was purified by using QIAprep Spin Miniprep kit (Qiagen). Chromosomal DNA was purified by using the Qiagen Midi kit or the Gentra Systems Puregene kit. Basic DNA manipulations were performed as described earlier (23). Transfer of plasmids from E. coli to P. putida was done by triparental mating with pRK2013 (8).
Oligonucleotide synthesis was performed by the University of Oklahoma Health Sciences Center Molecular Biology Resource Facility and Life Technologies.
Plasmid constructions.
pUCP18/19 are pUC-derived plasmids which contain a fragment from RP1 (19) which permits stable maintenance of these plasmids in Pseudomonas species (24). These plasmids were further modified in this study so that they could be used to transform Pseudomonas species by triparental mating. In order to accomplish this, the XmaIII fragment from pLAFRI, (6), which contains the mobilization (mob) site, was blunted and cloned into the SmaI site of pUC18. The 750-bp mob fragment was isolated after digestion with KpnI and BamHI and blunted. Next, pUCP18/19 was digested with SspI and blunted, and the 750-bp blunted mob fragment was cloned into the blunted vector, providing pUCPM18 and pUCPM19.
pJRS191 was obtained from the pLAFR3 cosmid library and contains a fragment of P. putida DNA, of approximately 40 kb, inserted into the BamHI. pJRS196 contains a 2.0-kb fragment from pJRS191 that includes the entire structural genes of crc and pyrE, cloned into the KpnI site of pUCPM19.
Mutant construction.
The coding region of crc was amplified from pJRS191 by using primers S163 (GAACAGGCCGGCATTGAAGAAATA) and S165 (GCGCTGGACATGAGCAAGCTGGGCG). The PCR product was digested with EcoRI and KpnI and ligated with pUCPM19 digested with the same restriction enzymes; this plasmid was designated pJRS194. The stabilization fragment was removed from plasmids which were to be used for homologous recombination. To remove the stabilization fragment of pUCPM19, pJRS194 was digested with NdeI and EcoRI, and the 4.6-kb fragment was religated. Transposon pUTKm (9) was digested with AlwNI, and the 1.25-kb fragment containing the Kmr gene was blunt ended and ligated with the construct described above digested with BsaAI. The resulting plasmid, pJRS197, contained the Kmr gene cloned in the opposite orientation to crc 447 bp downstream of the ATG of crc. Conjugal transfer of this suicide plasmid into PpG2 was accomplished by triparental mating with pRK2013 (8). Mutants were isolated by using Pseudomonas isolation agar containing kanamycin. Because all mutants isolated were also Cbr and therefore were single crossovers, restriction digests and Southern blotting were used to identify recombination events that occurred upstream of the Kmr gene insertion site. Double crossovers were detected by the loss of carbenicillin resistance, and this recombination event was confirmed by restriction digests and Southern blotting with the NdeI/AlwNI fragment of pUC19 containing bla as a probe. The mutant with crc inactivated by insertion of a Kmr cassette was named P. putida JS394.
Transposon mutagenesis and screening.
To isolate transposon mutants affected in bkdR expression, triparental matings of P. putida JS386, E. coli CC118(pUTKm), and E. coli HB101(pRK2013) were plated on Difco Pseudomonas Isolation Agar plus kanamycin plus 0.004% X-Gal (wt/vol; 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside). Approximately 10,000 colonies were screened on the basis of color, since JS386 is light blue on this medium. Both white and dark blue colonies were collected, and phenotypes were confirmed by β-galactosidase assays. Southern blots were used to identify mutants with insertions in the bkdR-bkdA1 intergenic region or in lacZ, and these mutants were discarded. Four white colonies and three dark blue colonies were selected for further study. Genomic DNA from each mutant was digested with several restriction enzymes and Southern blotted with the 3.0-kb EcoRI-Kmr fragment to identify fragments of a desired size for cloning. The transposon insertion sites for three of the white colonies and all of the dark blue colonies were identified by cloning into pUC19 and sequencing the DNA insert by the Oklahoma University Health Sciences Center DNA Sequencing Facility.
DNA sequencing and analysis of nucleotide sequences.
DNA Sequencing was performed by Bruce A. Roe's lab at the University of Oklahoma Advanced Center for Genomic Technology (Norman, Okla.). Cosmid DNA was purified by means of a cleared-lysate diatomaceous earth method (20). Sequencing was undertaken by using the double-stranded, shotgun-based approach (2). The resulting sequences were screened to eliminate vector, assembled into contiguous fragments, and proofread by using the Phred/Phrap/Consed system developed by P. Green (http://chimera.biotech.washington.edu/uwgc). Contigs larger than 2 kb were deposited before publication in the “unfinished” division of the high-throughput genome sequencing GenBank database and were given accession number AC004396.
RESULTS
Involvement of Crc in catabolite repression of the bkd operon of P. aeruginosa PAO1.
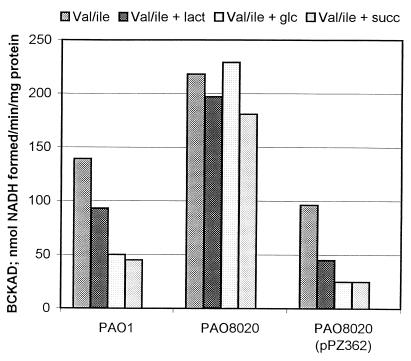
Each strain of P. aeruginosa was grown on valine-isoleucine medium alone or supplemented with the additional carbon sources lactate, glucose, or succinate. Addition of glucose or succinate to the inducing (valine-isoleucine) medium caused a 65 to 70% repression of BCKAD activity in the wild-type strain PAO1, and even lactate caused some repression (Fig. 1). Repression was completely relieved in the crc mutant PAO8020, where similar BCKAD activities were seen in inducing medium with or without glucose or succinate. Repression was restored when PAO8020 was complemented with crc+ from P. aeruginosa on a plasmid (pPZ352). These results indicate that crc is responsible for a significant fraction of repression of the P. aeruginosa bkd operon by glucose and succinate synthetic media. The levels of BCKAD activity in the crc mutant were elevated compared to the wild type under all growth conditions, suggesting that constitutive levels of Crc in P. aeruginosa PAO1 cause some repression of bkd operon expression in the absence of repressing carbon source.
FIG. 1.
Effect of mutation in crc on catabolite repression of BCKAD in P. aeruginosa. P. aeruginosa PAO1, PAO8020, and PAO8020(pPZ352) were grown in valine-isoleucine medium either alone or supplemented with lactate, glucose, or succinate (columns in each group from left to right, respectively) as described in Materials and Methods. Data are the averages of three separate experiments.
Isolation of P. putida crc.
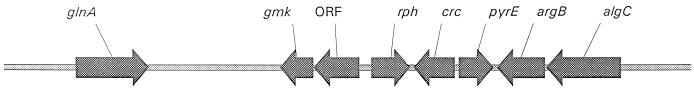
P. putida crc was isolated in order to determine if Crc was involved in catabolite repression of the bkd operon of P. putida. A PpG2 chromosomal DNA library was constructed in pLAFR3 and screened with a 2.0-kb SstI fragment of pPZ352 containing the P. aeruginosa crc gene. The positive clone with a ∼40-kb insert of P. putida, pJRS191, was partly sequenced, and the consensus sequence of a 13-kb contig containing the crc gene was obtained (Fig. 2).
FIG. 2.
Annotation of contig 270 from pJRS191. Open reading frames were identified by similarity searching by using the BLAST program (1). Similarities were found to the following proteins (identities at the protein level are given in parentheses): glnA, glutamine synthetase (61% to E. coli); gmk, GMP kinase (61% to E. coli); rph, RNase PH (84% to P. aeruginosa); crc, Crc (86% to P. aeruginosa); pyrE, orotate phosphoribosyltransferase (80% to P. aeruginosa); argB, acetylglutamate kinase (46% to Methanococcus jannaschii); and algC, phosphomannomutase (75% to P. aeruginosa). The only similarity found to the open reading frame (ORF) was to other reading frames of unknown function identified by genomic sequencing.
P. putida crc is 780 bp long, and the encoded protein is 86% identical and 93% similar to PAO1 Crc. The 3.1-kb PAO1 crc region includes three genes: pyrE upstream of, and rph downstream of, crc (11). This organization is conserved in the PpG2 crc region, with the exception of the crc-rph intergenic region, which is 479 bp in PAO1 but only 66 bp in PpG2 (Fig. 2).
A BLAST (1) search of GenBank was undertaken to identify open reading frames in the remaining 10 kb of the contig, and the annotation based on this similarity search is shown in Fig. 2. In addition to genes described in the legend to Fig. 2, a LysR family regulator appears to be encoded upstream of the open reading frame with similarity to glutamine synthetase, but the 3′ end of the open reading frame could not be identified. The 2-kb sequence between the open reading frames with similarity to glutamine synthetase and GMP kinase appears to contain two open reading frames based on sequence similarity to open reading frames of unknown function in the DNA databases. However, the 5′ and 3′ ends of each could not be defined due to low similarity and incomplete open reading frames obtained in the search.
Effect of Crc on catabolite repression of the bkd operon of P. putida.
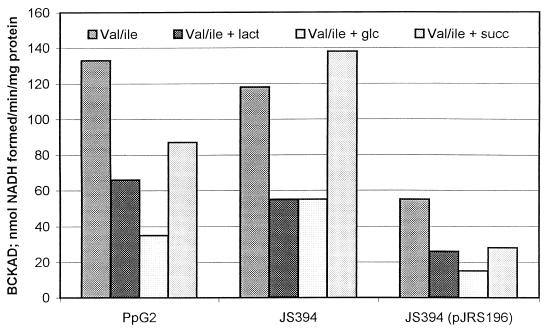
P. putida JS394, which possesses a chromosomal crc inactivated by insertion of a kanamycin cassette, was created by homologous recombination. For complementation studies, P. putida crc was cloned into pUCPM19. This construct was numbered pJRS196 and was used to transform P. putida JS394. P. putida PpG2, JS394, and JS394(pJRS196) were grown in valine-isoleucine medium alone and with lactate, glucose, or succinate (Fig. 3). BCKAD activity obtained in the presence of lactate and glucose was about one-half to one-fourth the activity obtained in cells grown in valine-isoleucine medium. BCKAD activity obtained in the presence of succinate was about three-fourths of the control value. There was some relief of catabolite repression in P. putida JS394 when succinate was the repressor (Fig. 3). However, when P. putida JS394 was complemented with crc in trans, there was distinct evidence of catabolite repression by lactate, glucose, and succinate, probably due the effect of multiple copies of crc (Fig. 3).
FIG. 3.
Effect of mutation in crc on catabolite repression of BCKAD in P. putida. P. putida strains PpG2, JS394, and JS394(pJRS196) were grown in valine-isoleucine medium either alone or supplemented with lactate, glucose, or succinate (columns in each group from left to right, respectively). Kanamycin was always added to medium in which JS394 was grown, and carbenicillin was added to medium in which strains containing pJRS196 were grown. Data are the averages of two separate experiments.
While it can be concluded that Crc plays a role in catabolite repression control of the bkd operon of both P. aeruginosa and P. putida, there are some differences. Crc does not have as much control over expression of the bkd operon of P. putida as it does in P. aeruginosa, and succinate is a much better repressor in P. aeruginosa. Another difference is that the elevated levels of BCKAD activity seen under all growth conditions in the P. aeruginosa crc mutant (Fig. 1) were not observed in the P. putida crc mutant.
Effect of P. putida Crc on catabolite repression of G6PDH and amidase.
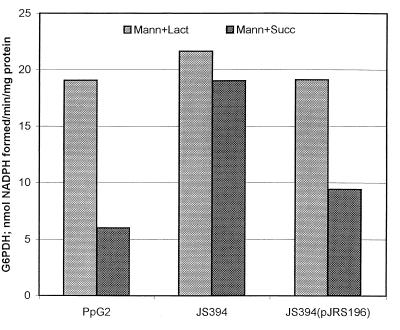
Crc also controls expression of G6PDH and amidase in P. aeruginosa (4), and the effect of Crc on expression of these enzymes was also studied in P. putida. G6PDH activities were studied in extracts of P. putida PpG2, JS394, and JS394(pJRS196) grown in BSM containing mannitol plus lactate or succinate. In this case, the control medium is mannitol plus lactate because growth in mannitol alone is very slow. As seen in Fig. 4, G6PDH activity in P. putida PpG2 is subject to strong catabolite repression by succinate; G6PDH activity in mannitol plus succinate was only about one-third that obtained in mannitol plus lactate medium. However, G6PDH activities in JS394 cultures grown in the presence of mannitol plus lactate and mannitol plus succinate were nearly identical, demonstrating relief of catabolite repression in the crc mutant. When JS394 was complemented with pJRS196, the repression by succinate was restored, demonstrating that P. putida Crc is involved in catabolite repression of G6PDH by succinate.
FIG. 4.
Effect of mutation in crc on catabolite repression of G6PDH of P. putida. P. putida PpG2, JS394, and JS394(pJRS196) were grown in BSM containing mannitol plus lactate or containing mannitol plus succinate (left and right columns in each group, respectively). Data are the averages of two separate experiments.
The effect of Crc on catabolite repression control of amidase in P. putida PpG2 is shown in Fig. 5. The control medium contained lactamide plus lactate, and the results are expressed as the percent activity obtained with P. putida grown in this medium. Succinate is a nonrepressing carbon source, which is in contrast to the situation in P. aeruginosa, where succinate is a strong repressor of amidase activity (12). Amidase activity obtained when gluconate was the repressor was less than 20% of the activity obtained in the control medium. Amidase activity obtained from cells grown in the presence of glutamate and glucose was 30 to 50% of the activity obtained in the control medium. Repression by glutamate, glucose, and gluconate was restored in the complemented mutant, P. putida JS394(pJRS196). Therefore, Crc plays a major role in catabolite repression control of amidase by glutamate, glucose, and gluconate in P. putida, as well as in P. aeruginosa, but again the carbon sources responsible for catabolite repression differ.
P. putida Crc is not involved in catabolite repression of urocanase activity.
Repression of urocanase and histidase activities by succinate was not relieved in P. aeruginosa crc mutants (P. Phibbs and C. MacGregor, unpublished data); therefore, Crc does not play a role in catabolite repression control of the hut operon. For comparison, P. putida PpG2 and JS394 were grown on BSM containing histidine plus lactate, succinate, or glucose and then assayed for urocanase activity. The specific activities of urocanase in PpG2 and JS394 grown on histidine plus glucose were the same for both strains and were about two-thirds that of the activity obtained when they were grown in histidine plus lactate. Thus, the results also suggest that Crc is not involved in catabolite repression control of the hut operon in P. putida, although repression was not very strong.
Crc and RNase R affect expression of bkdR.
Crc could act directly on transcription of the bkd operon or indirectly on expression of bkdR. To test the latter possibility, transposon mutagenesis of the bkdR-lacZ fusion of strain JS386 was undertaken to isolate and identify mutants affected in bkdR expression (see Materials and Methods). The strategy was to isolate dark blue colonies, which should be derepressed for expression of bkdR. Three dark blue colonies were isolated, two of which had insertions in crc. The other dark blue colony had an insertion in vacB, which encodes a protein involved in posttranscriptional regulation of virulence genes in Shigella flexneri (30). VacB has recently been identified as RNase R in E. coli (3), and this terminology will be used here. The structural gene for RNase R is rnr and was found on the same contig which contains crc (Table 1). The amino acid sequence of RNase R from P. putida is 50% identical to RNase R from E. coli and is 48% identical to RNase R from S. flexneri.
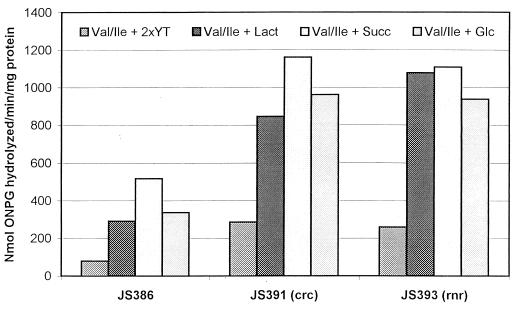
To examine the effects of Crc and RNase R on bkdR expression, β-galactosidase assays were performed on P. putida JS386, JS391 (crc), and JS393 (rnr) grown in valine-isoleucine plus lactate, succinate, or glucose (Fig. 6). There was no repression of β-galactosidase activity by glucose and succinate in P. putida JS386 which contains the bkdR-lacZ translational fusion. P. putida JS391 and P. putida JS393 had three- to fourfold-higher levels of β-galactosidase activity than JS386 under all growth conditions, indicating that Crc and RNase R had negative effects on the expression of bkdR. One interesting difference was that catabolite repression of amidase was not relieved in the rnr mutant.
FIG. 6.
Effect of mutations in crc and rnr on expression of bkdR in P. putida strains with a bkdR-lacZ translational fusion. P. putida strains JS386, JS391, and JS393 were grown in synthetic medium with valine-isoleucine plus 2xYT, lactate, succinate, or glucose (columns in each group from left to right, respectively). Data are the averages of at least three separate experiments.
DISCUSSION
Crc is involved in the control of catabolite repression of BCKAD (Fig. 1), G6PDH, and amidase in P. aeruginosa (4), but not of urocanase (Phibbs and MacGregor, unpublished). The same can be concluded for Crc of P. putida (Fig. 3 to 5), although the effect on catabolite repression of BCKAD is not as strong, and in P. aeruginosa it is weak (Fig. 1 and 3). While the same catabolic enzymes are affected in both species, the catabolite repressors have different degrees of effectiveness. Succinate is a good repressor in P. aeruginosa (Fig. 1) (4) but is a weak repressor in P. putida (Fig. 3 and 5). The high degree of sequence similarity between Crc from P. aeruginosa and P. putida and the results of the catabolite repression control studies (Fig. 3 to 5) support the conclusion that Crc carries out the same functions in both species. There are two differences which bear examination, namely, the small intergenic region between crc and rph in P. putida (69 bp) compared to that in P. aeruginosa (479 bp) and the increased expression of BCKAD relative to the wild type seen in P. aeruginosa (Fig. 1) but not P. putida (Fig. 3).
The fact that catabolite repression in pseudomonads does not involve cAMP makes the role of Crc in catabolite repression all the more interesting. Crc does not appear to be a DNA-binding protein and is therefore not likely to interfere with transcription. Other possibilities are that Crc acts posttranscriptionally by interfering with the expression of the mRNA or posttranslationally by modifying BkdR. However, the sequence similarity of Crc to endonucleases (11) makes the latter possibility remote. It has been pointed out that it was not possible to demonstrate the binding of Crc from P. aeruginosa DNA (11) and that has been our experience with P. putida DNA as well (K. L. Hester and J. R. Sokatch, unpublished data). The transposon mutants (Fig. 6) were isolated to test the possibility that Crc had an effect on expression of bkdR and, indeed, of the three colonies isolated with increased expression of LacZ from the crc-lacZ fusion, two were crc mutants. The other mutant with a similar phenotype was RNase R, which is interesting because Crc has some similarity to exonucleases (11). The accompanying study (9a) presents evidence that Crc acts posttranscriptionally.
ACKNOWLEDGMENTS
This research was supported by Public Health Service grant DK21737 and Presbyterian Health Foundation grant C5142801 to J.R.S., NSF EPSCoR program grant EPS-9550478 to B.A.R., and Environmental Protection Agency STAR fellowship grant U915028-01-0 to K.L.H.
REFERENCES
- 1.Altschul S F, Gish W. Local alignment statistics. Methods Enzymol. 1996;266:460–480. doi: 10.1016/s0076-6879(96)66029-7. [DOI] [PubMed] [Google Scholar]
- 2.Bodenteich A, Chissoe S L, Wang Y F, Roe B A. In: Automated DNA sequencing and analysis techniques. Adams M D, Fields C, Venter J C, editors. London, England: Academic Press; 1994. pp. 42–50. [Google Scholar]
- 3.Cheng Z F, Zuo Y, Li Z, Rudd K E, Deutscher M P. The vacB gene required for virulence in Shigella flexneri and Escherichia coli encodes the exoribonuclease RNase R. J Biol Chem. 1998;273:14077–14080. doi: 10.1074/jbc.273.23.14077. [DOI] [PubMed] [Google Scholar]
- 4.Collier D N, Hager P W, Phibbs P V., Jr Catabolite repression control in the pseudomonads. Res Microbiol. 1996;147:551–561. doi: 10.1016/0923-2508(96)84011-3. [DOI] [PubMed] [Google Scholar]
- 5.Durham D R, Phibbs P V., Jr Fractionation and characterization of the phosphoenolpyruvate:fructose 1-phosphotransferase system from Pseudomonas aeruginosa. J Bacteriol. 1982;149:534–541. doi: 10.1128/jb.149.2.534-541.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Friedman A M, Long S R, Brown S E, Buikema W J, Ausubel F M. Construction of a broad host range cosmid cloning vector and its use in genetic analysis of Rhizobium mutants. Gene. 1982;18:289–296. doi: 10.1016/0378-1119(82)90167-6. [DOI] [PubMed] [Google Scholar]
- 7.George D J, Phillips A T. Identification of α-ketobutyrate as the prosthetic group of urocanase from Pseudomonas putida. J Biol Chem. 1970;245:528–537. [PubMed] [Google Scholar]
- 8.Goldberg J B, Ohman D E. Cloning and expression in Pseudomonas aeruginosa of a gene involved in the production of alginate. J Bacteriol. 1984;158:1115–1121. doi: 10.1128/jb.158.3.1115-1121.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Herrero M, deLorenzo V, Timmis K N. Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in gram-negative bacteria. J Bacteriol. 1990;172:6557–6567. doi: 10.1128/jb.172.11.6557-6567.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9a.Hester K L, Madhusudan K T, Sokatch J R. Catabolite repression control by Crc in 2xYT medium is mediated by posttranscriptional regulation of bkdR expression in Pseudomonas putida. J Bacteriol. 2000;182:1150–1153. doi: 10.1128/jb.182.4.1150-1153.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hylemon P B, Phibbs P V., Jr Independent regulation of hexose catabolizing enzymes and glucose transport activity in Pseudomonas aeruginosa. Biochem Biophys Res Commun. 1972;48:1041–1048. doi: 10.1016/0006-291x(72)90813-3. [DOI] [PubMed] [Google Scholar]
- 11.MacGregor C H, Arora S K, Hager P W, Dail M B, Phibbs P V., Jr The nucleotide sequence of the Pseudomonas aeruginosa pyrE-crc-rph region and the purification of the crc gene product. J Bacteriol. 1996;178:5627–5635. doi: 10.1128/jb.178.19.5627-5635.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.MacGregor C H, Wolff J A, Arora S K, Phibbs P V., Jr Cloning of a catabolite repression control (crc) gene from Pseudomonas aeruginosa, expression of the gene in Escherichia coli, and identification of the gene product in Pseudomonas aeruginosa. J Bacteriol. 1991;173:7204–7212. doi: 10.1128/jb.173.22.7204-7212.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Madhusudhan K T, Hester K L, Friend V, Sokatch J R. Transcriptional activation of the bkd operon of Pseudomonas putida by BkdR. J Bacteriol. 1997;179:1992–1997. doi: 10.1128/jb.179.6.1992-1997.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Madhusudhan K T, Huang G, Sokatch J R. Characterization of BkdR-DNA binding in the expression of the bkd operon of Pseudomonas putida. J Bacteriol. 1995;177:636–641. doi: 10.1128/jb.177.3.636-641.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Madhusudhan K T, Huang N, Brasswell E H, Sokatch J R. Binding of L-branched chain amino acids causes a conformational change in BkdR. J Bacteriol. 1997;179:276–279. doi: 10.1128/jb.179.1.276-279.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Madhusudhan K T, Lorenz D, Sokatch J R. The bkdR gene of Pseudomonas putida is required for expression of the bkd operon and encodes a protein related to Lrp of Escherichia coli. J Bacteriol. 1993;175:3934–3940. doi: 10.1128/jb.175.13.3934-3940.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marshall V P, Sokatch J R. Regulation of valine catabolism in Pseudomonas putida. J Bacteriol. 1972;110:1073–1081. doi: 10.1128/jb.110.3.1073-1081.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nakazawa T, Furukawa K, Haas D, Silver S. Molecular biology of pseudomonads. Washington, D.C.: American Society for Microbiology; 1996. [Google Scholar]
- 19.Olsen R H, DeBusscher G, McCombie W R. Development of broad-host-range vectors and gene banks: self-cloning of the Pseudomonas aeruginosa PAO chromosome. J Bacteriol. 1982;150:60–69. doi: 10.1128/jb.150.1.60-69.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pan H Q, Wang Y P, Chissoe S L, Bodenteich A, Wang Z, Iyer K, Clifton S W, Crabtree J S, Roe B A. The complete nucleotide sequences of the SacBII Kan domain of the P1 pAD10-SacBII cloning vector and three cosmid cloning vectors: pTCF, svPHEP, and LAWRIST16. Genet Anal Tech Appl. 1994;11:181–186. doi: 10.1016/1050-3862(94)90039-6. [DOI] [PubMed] [Google Scholar]
- 21.Phillips A T, Mulfinger L M. Cyclic adenosine 3′,5′-monophosphate levels in Pseudomonas putida and Pseudomonas aeruginosa during induction and carbon catabolite repression of histidase synthesis. J Bacteriol. 1981;145:1286–1292. doi: 10.1128/jb.145.3.1286-1292.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Postma P W, Lengeler J W, Jacobson G R. Phosphoenolpyruvate: carbohydrate phosphotransferase systems of bacteria. Microbiol Rev. 1993;57:543–594. doi: 10.1128/mr.57.3.543-594.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1987. [Google Scholar]
- 24.Schweizer H P. Escherichia-Pseudomonas shuttle vectors derived from pUC18/19. Gene. 1991;97:109–112. doi: 10.1016/0378-1119(91)90016-5. [DOI] [PubMed] [Google Scholar]
- 25.Siegel L S, Hylemon P B, Phibbs P V., Jr Cyclic adenosine 3′,5′-monophosphate levels and activities of adenylate cyclase and cyclic adenosine 3′,4′-monophosphate phosphodiesterase in Pseudomonas and Bacteriodes. J Bacteriol. 1977;129:87–96. doi: 10.1128/jb.129.1.87-96.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smyth P F, Clarke P H. Catabolite repression of Pseudomonas aeruginosa amidase: the effect of carbon sources on amidase synthesis. J Gen Microbiol. 1975;90:81–90. doi: 10.1099/00221287-90-1-81. [DOI] [PubMed] [Google Scholar]
- 27.Sokatch J R, McCully V, Roberts C M. Purification of a branched-chain keto acid dehydrogenase from Pseudomonas putida. J Bacteriol. 1981;148:647–652. doi: 10.1128/jb.148.2.647-652.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Staskawicz B, Dahlbeck D, Keen N, Napoli C. Molecular characterization of cloned avirulence genes from race 0 and race 1 of Pseudomonas syringae pv. glycinea. J Bacteriol. 1987;169:5789–5794. doi: 10.1128/jb.169.12.5789-5794.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sykes P, Burns G, Menard J, Hatter K, Sokatch J R. Molecular cloning of genes encoding branched-chain keto acid dehydrogenase of Pseudomonas putida. J Bacteriol. 1987;169:1619–1625. doi: 10.1128/jb.169.4.1619-1625.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tobe T, Sasakawa C, Okada N, Honma Y, Yoshikawa M. vacB, a novel chromosomal gene required for expression of virulence genes on the large plasmid of Shigella flexneri. J Bacteriol. 1992;174:6359–6367. doi: 10.1128/jb.174.20.6359-6367.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]