Abstract
Aglaonema, commonly called Chinese evergreens, are widely used for ornamental purposes. However, attempts to identify Aglaonema species and cultivars based on leaf morphology have been challenging. In the present study, chloroplast sequences were used to elucidate the phylogenetic relationships of cultivated Aglaonema in South China. The chloroplast genomes of one green species and five variegated cultivars of Aglaonema, Aglaonema modestum, ‘Red Valentine’, ‘Lady Valentine’, ‘Hong Yan’, ‘Hong Jian’, and ‘Red Vein’, were sequenced for comparative and phylogenetic analyses. The six chloroplast genomes of Aglaonema had typical quadripartite structures, comprising a large single copy (LSC) region (91,092–91,769 bp), a small single copy (SSC) region (20,816–26,501 bp), and a pair of inverted repeat (IR) regions (21,703–26,732 bp). The genomes contained 112 different genes, including 79–80 protein coding genes, 28–29 tRNAs and 4 rRNAs. The molecular structure, gene order, content, codon usage, long repeats, and simple sequence repeats (SSRs) were generally conserved among the six sequenced genomes, but the IR-SSC boundary regions were significantly different, and ‘Red Vein’ had a distinct long repeat number and type frequency. For comparative and phylogenetic analyses, Aglaonema costatum was included; it was obtained from the GenBank database. Single-nucleotide polymorphisms (SNPs) and insertions/deletions (indels) were determined among the seven Aglaonema genomes studied. Nine divergent hotspots were identified: trnH-GUG-CDS1_psbA, trnS-GCU_trnS-CGA-CDS1, rps4-trnT-UGU, trnF-GAA-ndhJ, petD-CDS2-rpoA, ycf1-ndhF, rps15-ycf1-D2, ccsA-ndhD, and trnY-GUA-trnE-UUC. Additionally, positive selection was found for rpl2, rps2, rps3, ycf1 and ycf2 based on the analyses of Ka/Ks ratios among 16 Araceae chloroplast genomes. The phylogenetic tree based on whole chloroplast genomes strongly supported monophyletic Aglaonema and clear relationships among Aroideae, Lasioideae, Lemnoideae, Monsteroideae, Orontioideae, Pothoideae and Zamioculcadoideae in the family Araceae. By contrast, protein coding gene phylogenies were poorly to strongly supported and incongruent with the whole chloroplast genome phylogenetic tree. This study provided valuable genome resources and helped identify Aglaonema species and cultivars.
Introduction
Aglaonema, commonly called Chinese evergreens, belongs to the family Araceae and comprises 21 species [1–3]. They are native to southeast Asia, northeast India and southern China southward through Malaysia, New Guinea and the Philippines [1]. Aglaonema is a reliable ornamental crop for commercial growers, and plants readily adapt to low light and low relative humidity levels encountered under interior conditions [1,3–5]. Aglaonema cultivation in the United States started in the 1930s and soon became highly successful in Florida [2]. In China, Aglaonema modestum has been widely cultivated for ornamental and medical purposes in southern provinces. Historically, most new Aglaonema cultivars were introduced directly from the wild or were from established cultivars [3]. Over the past 20 years, both public and private breeders worldwide have generated many new Aglaonema cultivars by hybridization and tissue-cultured mutation selection [2–5].
Although Aglaonema has become a crucial ornamental foliage genus, identifying Aglaonema species and cultivars based on leaf morphology has been challenging; for example, Aglaonema ‘Red Valentine’ is closely analogous to Aglaonema ‘Lady Valentine’ in leaf morphology. Both are very popular in South China because of their beautiful foliage, good growth habit and excellent performance indoors. Commercially, tissue culture propagation is used to speed ‘Red Valentine’ plants. In our previous study, Aglaonema ‘Hong Yan’ and Aglaonema ‘Hong Jian’ are two mutations found among a population of tissue-cultured ‘Red Valentine’ plants. Although the genetic relationships of Aglaonema species and cultivars have been investigated using amplified fragment length polymorphism (AFLP) markers [2], the results have been limited in high resolution for Aglaonema species and cultivar identification. Recently, phylogenetic relationships among 13 Aroideae species have been evaluated using whole chloroplast genomes [6]. However, only one complete chloroplast genome of Aglaonema costatum has been reported in the genus Aglaonema [6], hindering molecular identification of Aglaonema species and cultivars, particularly failing to distinguish popular Aglaonema species and variegated cultivars in South China, namely, A. modestum, ‘Red Valentine’, ‘Lady Valentine’ and ‘Red Vein’. Therefore, we attempted to report six complete chloroplast genomes of these Aglaonema species and five cultivars, identify them, and explore their phylogenetic relationships.
Chloroplast genomes of angiosperms are highly conserved and have a typical quadripartite structure containing a large single copy (LSC) region, a small single copy (SSC) region, and two copies of inverted repeats (IRs) [7,8]. The size of chloroplast genomes ranges from 107 kb (Cathaya argyrophylla) to 280 kb (Pelargonium) and generally comprises 110–130 genes encoding ribosomal RNAs (rRNAs), transfer RNAs (tRNAs) and proteins [6–9]. The rapid development of high-throughput sequencing methods has made chloroplast genome sequencing faster and of higher quality. Complete chloroplast genomes have been widely used for phylogenetic analyses and molecular marker development in higher plants [6,9–11].
In this study, we sequenced and analyzed the chloroplast genomes of one species and five cultivars of Aglaonema: A. modestum, ‘Red Valentine’, ‘Hong Yan’, ‘Hong Jian’, ‘Red Vein’, and ‘Lady Valentine’. Our study aimed first to analyze the whole chloroplast genome structure characteristics among the tested Aglaonema species and cultivars. Second, we examined variations in long repeats and microsatellites among the six newly sequenced Aglaonema chloroplast genomes. Third, we screened divergent hotspot regions for use as potential DNA markers in genus Aglaonema. Fourth, we inferred the phylogenetic relationship of one species and five cultivars of Aglaonema along with one published species from the GenBank database (A. costatum, MN046881) and phylogenetic position in the family Araceae using the complete chloroplast genome and protein coding gene sequence alignments, respectively.
Materials and methods
Plant materials and DNA isolation
The fresh and healthy leaves of one green species and five variegated cultivars, namely, A. modestum, ‘Red Valentine’, ‘Hong Yan’, ‘Hong Jian’, ‘Lady Valentine’, and ‘Red Vein’ (Fig 1), were collected from the resource garden of the Environmental Horticulture Research Institute (23°23′N, 113°26′E) at the Guangdong Academy of Agricultural Sciences, Guangzhou, Guangdong Province, China. A. modestum is erect with a dark green stem and leaves, which is not variegated (Fig 1A). Both ‘Red Valentine’ and ‘Lady Valentine’ are variegated, with green and red along the midrib, along veins and throughout the leaf blade (Fig 1B and 1C). The leaves of ‘Hong Jian’ are narrow, oblong, dark pink to dark red, accounting for approximately 75% to 90% of the leaf area, and the shape of the apex of the leaves is strongly acute, with green and dark yellow green blotches at the margin and throughout the leaf blade (Fig 1D). The leaves of ‘Hong Yan’ are ovate-cordate, dark red, accounting for approximately 95% of the leaf area, and the shape of the apex of the leaves is obtuse to moderately acute, with green blotches at the leaf margin (Fig 1E). The leaves of ‘Red Vein’ are dark green with pink to red along the midrib and veins (Fig 1F). The collected leaf samples were quickly frozen in liquid nitrogen and then stored at -80°C until use. Genomic DNA was extracted using a modified sucrose gradient centrifugation method [12]. The DNA integrity of the extracted genomic DNA was examined by 1% agarose gel electrophoresis, and the concentration was checked using a NanoDrop 2000 microspectrometer (Wilmington, DE, USA).
Fig 1. Comparison of leaf morphologies among green species and variegated cultivars of genus Aglaonema.
(A) Aglaonema modestum, (B) Aglaonema ‘Red Valentine’, (C) Aglaonema ‘Lady Valentine’, (D) Aglaonema ‘Hong Yan’, (E) Aglaonema ‘Hong Jian’, and (F) Aglaonema ‘Red Vein’.
Chloroplast genome sequencing, assembly, annotation and structure analysis
For two variegated cultivars, ‘Lady Valentine’ and ‘Red Vein’, two libraries with insert sizes of 300 bp and 10 kb were constructed after purification and then sequenced on an Illumina HiSeq X Ten instrument (Biozeron, Shanghai, China) and a PacBio Sequel platform (Biozeron, Shanghai, China), respectively. Regarding the other four samples (A. modestum, ‘Red Valentine’, ‘Hong Yan’, and ‘Hong Jian’), a library with insert sizes of 300 bp was constructed for each sample after purification and then sequenced on an Illumina HiSeq X Ten instrument (Biozeron, Shanghai, China).
After sequencing, all raw reads were filtered using Trimmomatic v.0.3 with default parameters to obtain clean reads [13]. The assembly of the chloroplast genomes of ‘Lady Valentine’ and ‘Red Vein’ (Illumina data and PacBio data) was performed using SOAPdenovo v.2.04 software [14]. The clean Illumina sequences of the other four samples were assembled into complete chloroplast genomes using SOAPdenovo v.2.04 with default parameters [14] and the chloroplast genome of A. costatum (MN046881) as the reference. The GC content was calculated using Geneious version 11.0.4 software [15]. The genes of the chloroplast genomes were annotated using the online server Dual Organellar Genome Annotator (DOGMA) with default parameters [16]. BLAST searches for the complete chloroplast genome were performed against the following public databases: nonredundant protein databases, SwissProt, Gene Ontology, Clusters of Orthologous Groups, and Kyoto Encyclopedia of Genes and Genomes. Finally, verification of tRNAs and rRNAs was performed using tRNAscanSE with default parameters [17]. The map of the chloroplast genome was drawn using Organellar Genome DRAW v1.3.1 with default parameters [18]. The protein coding sequences of each sequenced chloroplast genome were extracted, and the relative synonymous codon usage (RSCU) values were calculated using MEGA7 software [19]. The clustered heatmap of RSCU values of six sequenced Aglaonema chloroplast genomes was drawn using R v.3.6.3 [20].
Analyses of long repeats and microsatellites
REPuter software (available online: http://bibiserv.techfak.uni-bielefeld.de/reputer/) [21] was used to identify and analyze the size and positions of long repeats, including the forward, palindrome, reverse and complement repeat units, within the six newly sequenced Aglaonema chloroplast genomes. The long repeat detection settings were a minimum repeat size of 30 bp and a Hamming distance of 3. MIcroSAtellite (MISA) was used to identify SSRs in the six newly sequenced Aglaonema chloroplast genomes (available online: http://pgrc.ipk-gatersleben.de/misa/) [22]. SSR detection parameters were set as follows: unit-size (nucleotide)_min-repeats: 1_8, 2_5, 3_4, 4_3, 5_3, 6_3.
Comparative genome and sequence divergence analyses
To calculate nucleotide variability (Pi) among seven chloroplast genomes within Aglaonema, six newly sequenced and one obtained from GenBank (A. costatum, MN046881), protein coding, intron and intergenic regions among these seven chloroplast genomes were extracted and then calculated using DnaSP version 6 software [23]. The sliding window length was set to 600 bp, and the step size was set to 200 bp. mVISTA software (http://genome.lbl.gov/vista/mvista/about.shtml) in the Shuffle-LAGAN mode [24] was used to compare the above seven complete chloroplast genomes of Aglaonema. The nonsynonymous (Ka) and synonymous (Ks) substitution rates of chloroplast genomes in seven members of Aglaonema and nine other members of Araceae (S1 Table) were estimated using KaKs_Calculator [25] with default parameters. The Pi values, mVISTA and Ka/Ks were analyzed using the complete chloroplast genome of A. modestum as the reference sequence. Based on genome annotations, the borders among the IR, LSC and SSC regions of the seven Aglaonema chloroplast genomes were also compared.
First, the seven Aglaonema complete genomes were aligned using MUMmer software [26] and adjusted manually where necessary using Se-Al 2.0 (available online: http://tree.bio.ed.ac.uk/software) [27], using the annotated A. modestum chloroplast genome as the reference. The single nucleotide polymorphisms (SNPs) and insertion/deletions (indels) were recorded separately, as well as their locations in the chloroplast genome. Second, the five variegated cultivars were analyzed to identify SNPs and indels using the annotated ‘Red Valentine’ chloroplast genome as the reference. Third, ‘Lady Valentine’ and ‘Red Vein’ were compared and analyzed to identify SNPs and indels.
Phylogenetic analyses
A total of 57 complete chloroplast genome sequences were used for phylogenetic analyses, including 6 newly sequenced genomes, 50 previously reported Araceae genomes, and Acorus americanus (EU273602), which was set as the outgroup taxa (S1 Table). The previously reported genome sequences were downloaded from GenBank database. The protein coding sequences of these 57 genomes were extracted using Geneious version 11.0.4 software [15]. Both the complete genome sequences and protein coding sequences were used for maximum likelihood (ML) tree construction. The nucleotide sequences were aligned using the MAFFT plugin [28] in Geneious version 11.0.4 with default settings. The complete alignment was used to construct an ML tree using PhyML version 3.0 [29]. The general-time-reversible, gramma distribution, and invariable sites (GTR+G+I) DNA substitution model was selected, and all the branch nodes were calculated with 1000 bootstrap replicates. Bootstrap values were classified as strong (> 85%), moderate (70–85%), weak (50–70%), or poor (< 50%) [30].
Results
General features of the six chloroplast genomes of Aglaonema
The total length of the six newly sequenced chloroplast genomes of Aglaonema ranged from 164,261 bp (‘red vein’) to 165,824 bp (‘Hong Yan’). The chloroplast genome size of the green species A. modestum was 165,626 bp, which was 198 bp shorter than that of ‘Hong Yan’ and 1365 bp longer than that of ‘Red Vein’. Interestingly, both ‘Red Valentine’ and ‘Hong Jian’ had the same genome size, which was 27 bp smaller than that of ‘Hong Yan’ (Tables 1 and S2). All six Aglaonema chloroplast genomes showed a typical quadripartite structure (Fig 2, Table 1 and S1 Fig), comprising a pair of inverted repeat (IR) regions (21,703–26,732 bp) separated by an LSC region (91,092–91,769 bp) and an SSC region (20,816–26,501 bp).
Table 1. Characteristics of six newly sequenced chloroplast genomes of genus Aglaonema.
| Genome characteristics | modestum | ‘Red Valentine’ | ‘Hong Yan’ | ‘Hong Jian’ | ‘Lady Valentine’ | ‘Red Vein’ |
|---|---|---|---|---|---|---|
| Genome size (bp) | 165,626 | 165,797 | 165,824 | 165,797 | 164,417 | 164,261 |
| LSC length (bp) | 91,269 | 91,092 | 91,092 | 91,092 | 91,135 | 91,769 |
| SSC length (bp) | 20,893 | 26,501 | 26,501 | 26,501 | 21,706 | 20,816 |
| IR length (bp) | 26,732 | 21,703 | 21,730 | 21,703 | 25,788 | 25,838 |
| Total genes (different) | 132 (112) | 132 (112) | 132 (112) | 132 (112) | 132 (112) | 132 (112) |
| Protein coding genes (different) | 87 (79) | 87 (79) | 87(79) | 87 (79) | 87 (80) | 87 (80) |
| tRNA genes (different) | 37 (29) | 37 (29) | 37 (29) | 37 (29) | 37 (28) | 37 (28) |
| rRNA genes (different) | 8 (4) | 8 (4) | 8 (4) | 8 (4) | 8 (4) | 8 (4) |
| Genes with introns | 18 | 18 | 18 | 18 | 19 | 19 |
| Genome GC (%) | 35.83 | 35.74 | 35.73 | 35.74 | 35.87 | 35.91 |
| Protein coding regions GC (%) | 37.69 | 37.69 | 37.71 | 37.69 | 37.72 | 37.90 |
| LSC GC (%) | 33.98 | 34.00 | 34.00 | 34.00 | 33.98 | 33.90 |
| SSC GC (%) | 29.12 | 28.53 | 28.49 | 28.53 | 29.25 | 30.17 |
| IR GC (%) | 41.62 | 41.67 | 41.67 | 41.67 | 41.98 | 41.78 |
| GenBank accession no. | OK094437 | OK094434 | OK094436 | OK094435 | MK262737 | MK262738 |
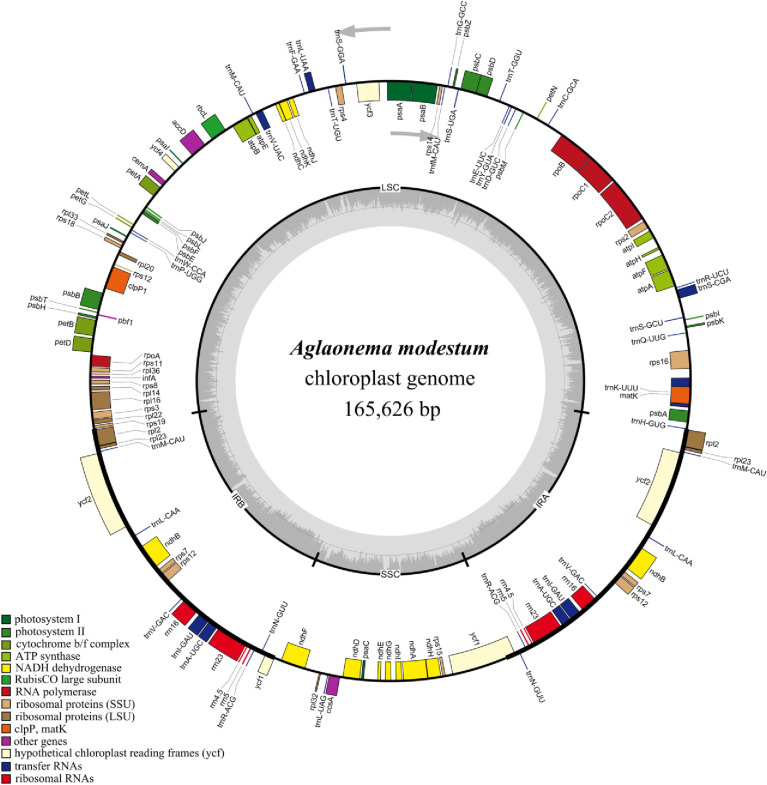
Fig 2. Chloroplast genome map of Aglaonema modestum.
The gray arrowheads indicate the direction of the genes. Genes shown inside the circle are transcribed clockwise and those outside are transcribed counterclockwise. Different genes are color coded. The innermost darker gray corresponds to GC content, whereas the lighter gray corresponds to AT content. IR, inverted repeat; LSC, large single copy region; SSC, small single copy region.
All six Aglaonema chloroplast genomes had GC contents varying from 35.73% to 35.91% (Tables 1 and S2). The IR region accounted for the highest GC content (41.62–41.98%), followed by the LSC region (33.90–34.00%), while the SSC region had the lowest GC content (28.49–30.17%) (Tables 1 and S2). The GC content of the protein coding regions varied slightly from 37.69% to 37.90%. The GC content at the third codon position (29.59–29.87%) was lower than that at the first (45.28–45.48%) and second (38.13–38.36%) positions in the protein coding genes of these six Aglaonema chloroplast genomes (S2 Table). The complete annotated chloroplast genomes were submitted to the GenBank database (accession numbers OK094437 for A. modestum, OK094434 for ‘Red Valentine’, OK094435 for ‘Hong Jian’, OK094436 for ‘Hong Yan’, MK262737 for ‘Lady Valentine’, and MK262738 for ‘Red Vein’) (Table 1).
The chloroplast genome annotation revealed 112 different genes in the six chloroplast genomes of Aglaonema (Table 1). Four different rRNA genes were identified in every Aglaonema chloroplast genome. However, the different protein coding genes and different tRNA genes in each Aglaonema chloroplast genome were unevenly distributed. The four Aglaonema chloroplast genomes (A. modestum, ‘Red Valentine’, ‘Hong Yan’, and ‘Hong Jian’) all comprised 79 different protein coding genes and 29 different tRNA genes, while the other two Aglaonema chloroplast genomes (‘Lady Valentine’ and ‘Red Vein’) both contained 80 different protein coding genes and 28 different tRNA genes (Tables 1, 2 and S3). Although most of the protein coding genes and tRNAs in the six newly sequenced Aglaonema chloroplast genomes were similar, slight differences were observed. For example, regarding protein coding genes, both chloroplast genomes of ‘Lady Valentine’ and ‘Red Vein’ had the ycf68 gene, while the other four sequenced genomes in this study lost this gene (S3 Table and S1 Fig). Furthermore, for tRNAs, the chloroplast genomes of ‘Lady Valentine’ and ‘Red Vein’ had one copy each of trnG-UCC and trnI-CAU, respectively, while trnS-CGA, trnS-GGA and trnT-GGU were missing; trnS-CGA, trnS-GGA and trnT-GGU showed one copy in the remaining four sequenced Aglaonema chloroplast genomes (Tables 2 and S3).
Table 2. Genes present in the six newly sequenced chloroplast genomes of genus Aglaonema.
| Category | Function | Genes |
|---|---|---|
| Photosynthesis | Photosystem Ⅰ | psaA, psaB, psaC, psaI, psaJ |
| Photosystem Ⅱ | psbA, psbB, psbC, psbD, psbE, psbF, psbH, psbI, psbJ, psbK, psbL, psbM, psbN, psbT, psbZ | |
| Cytochrome b/f | petA, petB*, petD*, petG, petL, petN | |
| ATP synthase | atpA, atpB, atpE, atpF*, atpH, atpI | |
| NADH dehydrogenase | ndhA*, ndhB (×2)*, ndhC, ndhD, ndhE, ndhF, ndhG, ndhH, ndhI, ndhJ, ndhK | |
| Rubisco | rbcL | |
| Self-replication | RNA polymerase | rpoA, rpoB, rpoC1*, rpoC2 |
| Large subunit ribosomal proteins | rpl2 (×2)*, rpl14, rpl16*, rpl20, rpl22, rpl23 (×2), rpl32, rpl33, rpl36 | |
| Small subunit ribosomal proteins | rps2, rps3, rps4, rps7 (×2), rps8, rps11, rps12 (×2)*, rps14, rps15, rps16*, rps18, rps19 (×2) | |
| Ribosomal RNAs | rrn4.5 (×2), rrn5 (×2), rrn16 (×2), rrn23 (×2) | |
| Transfer RNAs | trnA-UGC (×2)*, trnC-GCA, trnD-GUC, trnE-UUC, trnF-GAA, trnfM-CAU, trnG-GCC, trnG-UCC*&, trnH-GUG, trnI-GAU (×2)*, trnI-CAU&, trnK-UUU*, trnL-CAA (×2), trnL-UAA*, trnL-UAG, trnM-CAU (×3), trnN-GUU (×2), trnP-UGG, trnQ-UUG, trnR-ACG (×2), trnR-UCU, trnS-GCU, trnS-CGA*&&, trnS-GGA&&, trnS-UGA, trnT-GGU&&, trnT-UGU, trnV-GAC (×2), trnV-UAC*, trnW-CCA, trnY-GUA | |
| Others | Other proteins | accD, ccsA, cemA, clpP**, infA, matK |
| Proteins of unknown function | ycf1 (×2), ycf2 (×2), ycf3**, ycf4, ycf68 (×2)*& |
×2: Gene with two copies; ×3: Gene with three copies
*: Each gene containing only one intron
**: Each gene containing two introns
&: Only present in chloroplast genomes of ‘Lady Valentine’ and ‘Red Vein’
&&: Not present in chloroplast genomes of ‘Lady Valentine’ and ‘Red Vein’.
Regarding the genes duplicated in both IR regions in the chloroplast genomes of ‘Lady Valentine’ and ‘Red Vein’, 18 genes were identified, respectively, including seven protein coding genes (ndhB, rpl2, rpl23, rps7, rps12, ycf2, and ycf68), seven tRNA genes (trnI-CAU, trnL-CAA, trnV-GAC, trnI-GAU, trnA-UGC, trnR-ACG, and trnN-GUU), and all four rRNAs (rrn4.5, rrn5, rrn16 and rrn23) (S1 Fig and S3 Table). Regarding the genes in the IR regions in the remaining four chloroplast genomes, the total numbers of genes were the same as those in the above two genomes. However, ycf1 and trnM-CAU were present in the IR regions in the remaining four genomes, and ycf68 and trnI-CAU were absent in IR regions in the remaining four genomes.
In both chloroplast genomes of ‘Lady Valentine’ and ‘Red Vein’, 17 genes (trnA-UGC, trnI-GAU, trnG-UCC, trnK-UUU, trnL-UAA, trnV-UAC, atpF, ndhA, ndhB, rpoC1, petB, petD, rpl2, rpl16, rps12, rps16 and ycf68) contained one intron, while ycf3 and clpP each contained two introns (S4 Table). Among the 19 intron-containing genes, 5 genes (trnA-UAC, trnI-GAU, rpl2, ndhB and ycf68) occurred in both IRs, 12 genes (trnG-UCC, trnK-UUU, trnL-UAA, trnV-UAC, atpF, rpoC1, petB, petD, rpl16, rps16, ycf3 and clpP) were distributed in the LSC, one gene (ndhA) was in the SSC region, and one gene (rps12) was located in its first exon in the LSC region and the other two exons were located in both IR regions (S1 Fig and S4 Table). In comparison, in the remaining four sequenced genomes, trnS-CGA had one intron occurring in LSC regions, while no ycf68 genes and no introns were observed in trnG-UCC (S4 Table). Therefore, only 18 genes contained introns in the remaining four genomes, respectively.
Statistics of codon usage
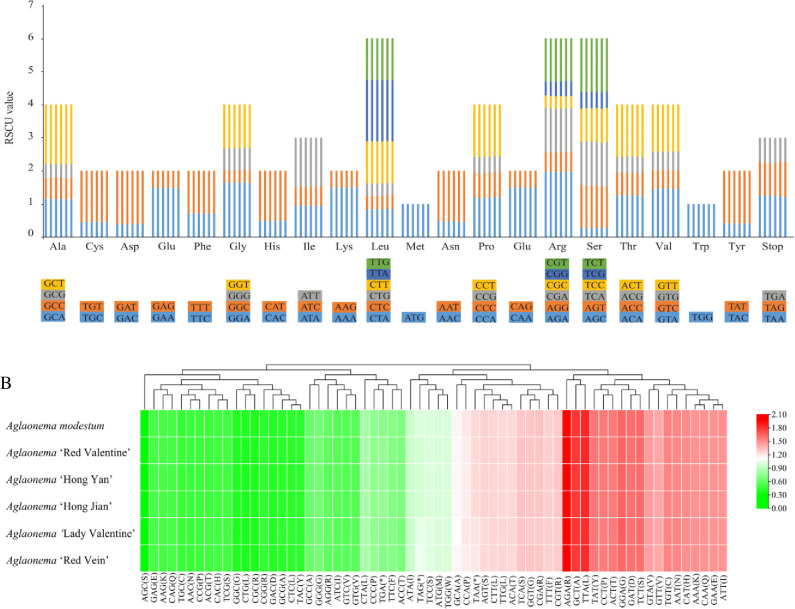
The six newly sequenced chloroplast genomes of Aglaonema were analyzed to investigate the codon usage, amino acid frequency, and relative synonymous codon usage (RSCU) (S5 Table). The total number of codons ranged from 25,800 (‘Lady Valentine’) to 26,670 (A. modestum) in six sequenced chloroplast genomes of Aglaonema. Although the total number of codons showed a tiny change, the types of codons and amino acids were the same. Methionine (Met) and tryptophan (Trp) were encoded by one codon usage, showing no codon bias both with RSCU values of 1.00, while others were encoded by multiple synonymous codons, ranging from two to six (Fig 3A). The codons with the four lowest RSCU values (AGC, GGC, CTG, and CGC) and three with the highest RSCU values (AGA, GCT, and TTA) were identified in the six newly sequenced genomes of Aglaonema (Fig 3B and S5 Table). Except for Met and Trp, thirty-two codons showed codon usage bias with RSCU > 1.00 in the chloroplast genes of all six newly sequenced genomes of Aglaonema (S5 Table). Interestingly, of the 32 codons, thirty-one were A/T-ending codons. Therefore, our RSCU results demonstrated that all six newly sequenced genomes had a higher usage frequency for A/T-ending than G/C-ending. Similar phenomena were found in Epipremnum aureum [31], Monstera adansonii [9], and Rhaphidophora amplissima [9].
Fig 3. Codon content of all protein coding genes of six newly sequenced Aglaonema chloroplast genomes.
(A) Codon content and codon usage of 20 amino acids and stop codons of all protein coding genes. Each histogram from left to right was A. modestum, ‘Red Valentine’, ‘Hong Yan’, ‘Hong Jian’, ‘Lady Valentine’, and ‘Red Vein’, respectively. (B) Heat map analysis for codon distribution of all protein coding genes in six newly sequenced chloroplast genomes. * indicates stop codons.
Long repeats and SSR analyses
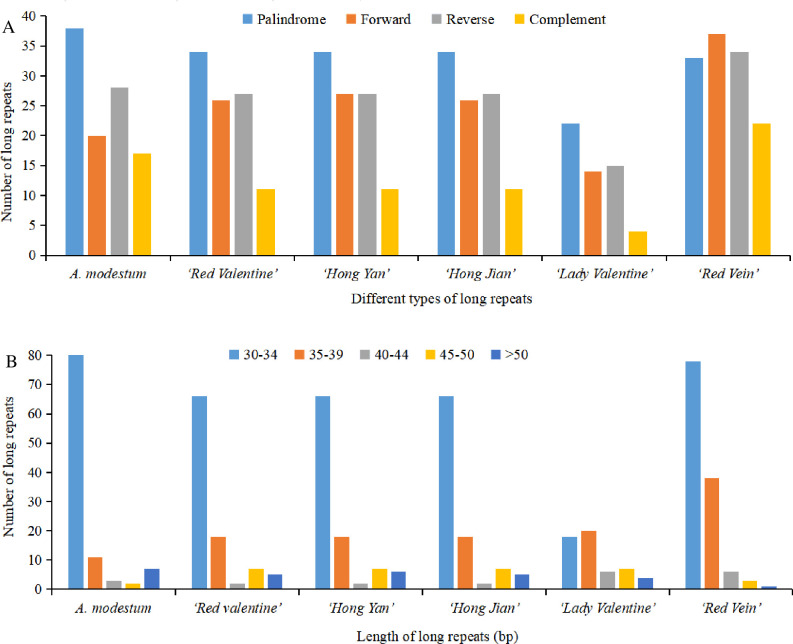
REPuter software was used to detect four different types of long repeats, including forward, complement, reverse, and palindromic repeats. The six sequenced chloroplast genomes had 579 long repeats, which contained 150 forward repeats, 76 complement repeats, 158 reverse repeats, and 195 palindromic repeats (Fig 4A and S6 Table). Among the six sequenced chloroplast genomes, ‘Red Vein’ had the largest number (126), and ‘Lady Valentine’ had the smallest number (55), of long repeats (Fig 4A and S6 Table). The number of forward repeats varied from 14 (‘Lady Valentine’) to 37 (‘Red Vein’), the number of complement repeats varied from 4 (‘Lady Valentine’) to 22 (‘Red Vein’), the number of reverse repeats varied from 15 (‘Lady Valentine’) to 34 (‘Red Vein’), and the number of palindromic repeats varied from 22 (‘Lady Valentine’) to 38 (A. modestum) (Fig 4A and S6 Table). The length of long repeats varied among the six sequenced chloroplast genomes, but most of the long repeats existed in the range of 30–34 bp (Fig 4B). The LSC region comprised more long repeats than the SSC and IR regions, and the details of the long repeats are also provided (S6 Table).
Fig 4. Analysis of long repeat sequences in six newly sequenced Aglaonema chloroplast genomes.
(A) Total number of four long repeat types. (B) Length distribution of long repeats in each sequenced chloroplast genome.
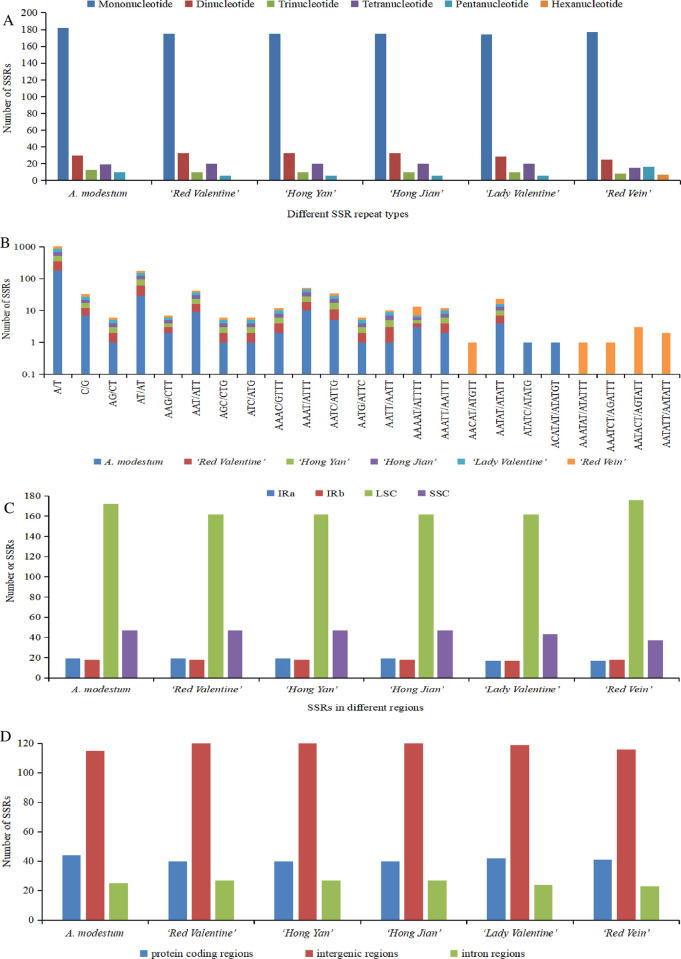
The SSRs in the six newly sequenced chloroplast genomes of Aglaonema were analyzed using MISA (Fig 5 and S7 Table). The number of detected SSRs ranged from 239 (‘Lady Valentine’) to 255 (A. modestum) (Fig 5A and S7 Table). Six types of SSRs were identified, inculding mononucleotide, dinucleotide, trinucleotide, tetranucleotide, pentanucleotide and hexanucleotide. Among these SSRs, only the chloroplast genomes of A. modestum and ‘Red Vein’ had hexanucleotide repeats (Fig 5A and S7 Table). Among the six sequenced chloroplast genomes, mononucleotide repeats were the most frequent, with numbers ranging from 174 to 182, followed by dinucleotides ranging from 25 to 33, tetranucleotides ranging from 15 to 20, trinucleotides ranging from 8 to 13, pentanucleotides ranging from 6 to 16, and hexanucleotides ranging from 0 to 7 (Fig 5A and S7 Table). Most of the mononucleotide SSRs were A/T repeats, which accounted for 68.63–70.71% of all SSRs among the six newly sequenced chloroplast genomes (Fig 5B and S7 Table). Regarding dinucleotide repeats, AT/AT repeats were observed most frequently, accounting for 9.68–13.11% of all SSRs (Fig 5B and S7 Table). Concerning the tetranucleotide category, the AAAT/ATTT repeats were the most abundant type, accounting for 2.42–3.92% of all SSRs (Fig 5B and S7 Table). Most SSRs were located in the LSC regions (162–176 loci; 66.39–70.97%) rather than in the SSC regions (37–47 loci; 14.92–19.26%) and IR regions (17–19 loci; 6.85–7.79%) of the six sequenced chloroplast genomes (Fig 5C and S7 Table). Additionally, SSRs were analyzed in the protein coding regions (exon, protein coding exon), intron regions and intergenic regions of the six sequenced chloroplast genomes, indicating that these six sequenced chloroplast genomes comprised 115 to 120 SSRs in intergenic regions, 40 to 44 SSRs in protein coding regions, and 23 to 27 SSRs in introns (Fig 5D and S7 Table). The gene ycf1 contained more SSR loci than the other genes in all six genomes (S7 Table).
Fig 5. Distribution of SSRs in six newly sequenced Aglaonema chloroplast genomes.
(A) Number of different SSR types detected in the six chloroplast genomes. (B) Frequency of identified SSR motifs in different repeat class types. (C) Frequency of SSRs in the LSC, IR and SSC regions. (D) Frequency of SSRs in the protein coding, intergenic and intron regions.
IR contraction and expansion analyses
Comparison details of the IR-LSC and IR-SSC boundaries were performed among the six sequenced chloroplast genomes of Aglaonema and one published chloroplast genome of A. costatum (Fig 6). Regarding IRa/LSC borders, the rpl2 and trnH-GUG genes were located at the boundaries of the IRa/LSC borders in all 7 Aglaonema chloroplast genomes. The distances between the ends of rpl2 and IRa/LSC borders ranged from 52 bp to 62 bp (Fig 6). trnH-GUG expanded into the IRa regions in all 7 Aglaonema chloroplast genomes, with distances ranging from 15 bp to 22 bp from the IRa/LSC borders (Fig 6). Among the 7 Aglaonema chloroplast genomes, the rps19 and rpl2 genes were located in the boundaries of the LSC/IRb regions, respectively (Fig 6). A total of 21–22 bp were found between the ends of rps19 and the LSC/IRb borders among the 7 Aglaonema chloroplast genomes, and the distances between the starts of rpl2 and the LSC/IRb boundaries ranged from 53 bp to 63 bp (Fig 6).
Fig 6. Comparison of the borders of the LSC, SSC, and IR regions among seven Aglaonema chloroplast genomes.
The six newly sequenced Aglaonema chloroplast genomes in this study are in bold.
The SSC/IRa border was located in the ycf1 coding region, which crossed into the IRa region in 5 Aglaonema chloroplast genomes, including A. modestum, ‘Red Valentine’, ‘Hong Yan’, ‘Hong Jian’, and A. costatum. However, the lengths of ycf1 in the IRa regions varied among the 5 Aglaonema chloroplast genomes, ranging from 399 bp to 868 bp (Fig 6). Two chloroplast genomes of ‘Lady Valentine’ and ‘Red Vein’, ycf1 and trnN-GUU, were found in the SSC/IRa borders; the distances between the ends of ycf1 and the SSC/IRa borders were 163 bp and 186 bp, respectively; and the distances between the starts of trnN-GUU and the SSC/IRa borders were 244 bp and 213 bp, respectively (Fig 6). The IRb/SSC borders of 4 chloroplast genomes (A. modestum, ‘Hong Yan’, ‘Hong Jian’, and A. costatum) were all located in ycf1, and ycf1 expanded into the SSC regions by 8 bp, 1 bp, 1 bp, and 40 bp, respectively (Fig 6). A total of 599 bp, 1124 bp, 1124 bp, and 671 bp were found between the starts of ndhF and the ends of ycf1 in A. modestum, ‘Hong Yan’, ‘Hong Jian’, and A. costatum, respectively (Fig 6). Regarding ‘Red Valentine’, a 230 bp distance was identified between the end of ycf1 and the IRb/SSC border and an 893 bp distance between the start of ndhF and the IRb/SSC border (Fig 6). Regarding ‘Lady Valentine’ and ‘Red Vein’, trnN-GUU and ndhF were located at the borders of IRb/SSC in both chloroplast genomes. A total of 243 bp and 212 bp distances were found between the ends of the trnN-GUU and IRb/SSC borders in ‘Lady Valentine’ and ‘Red Vein’, respectively. Furthermore, 440 bp and 190 bp were found between the starts of ndhF and the IRb/SSC borders in these two chloroplast genomes, respectively (Fig 6). trnN-GUU was duplicated in IR/SSC borders in the two genomes, ‘Lady Valentine’ and ‘Red Vein’, respectively, and ycf1 was duplicated in IR/SSC borders in the other five Aglaonema genomes. Overall, the IR/LSC boundary regions of the 7 chloroplast genomes of Aglaonema were highly conserved, but the IR/SSC boundary regions exhibited variations.
High variation regions and divergence analyses
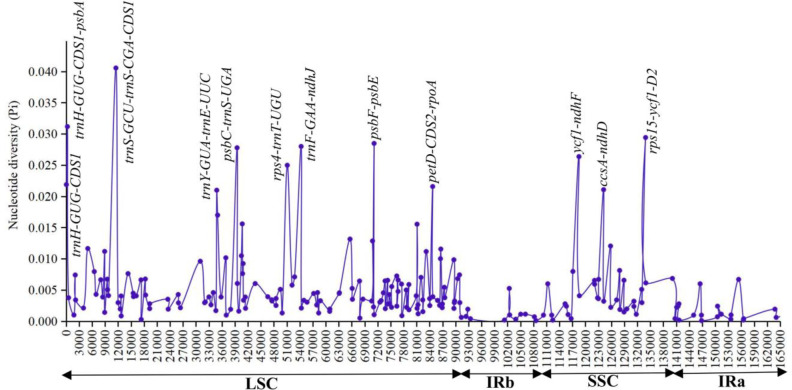
The highly divergent regions in the 7 chloroplast genomes of Aglaonema were analyzed using DnaSP by sliding window analysis (Fig 7 and S8 Table). The IR regions displayed lower variability than the LSC and SSC regions. Twelve regions that showed obviously higher Pi values (> 0.021) and were located in the LSC and SSC regions, which included trnH-GUG-CDS1, trnH-GUG-CDS1_psbA, trnS-GCU_trnS-CGA-CDS1, psbC-trnS-UGA, rps4-trnT-UGU, trnF-GAA-ndhJ, psbF-psbE, petD-CDS2-rpoA, ycf1-ndhF, rps15-ycf1-D2, ccsA-ndhD, and trnY-GUA-trnE-UUC (Fig 7 and S8 Table).
Fig 7. Plot of sliding window analysis of the whole chloroplast genomes for nucleotide diversity (Pi) compared among seven chloroplast genomes of Aglaonema.
Peak regions with a Pi value of > 0.021 were labeled with loci tags of names. x-axis shows the position of the midpoint of each window. y-axis shows Pi values of nucleotide diversity in a a sliding window analysis with window length 800 bp and step size 200 bp.
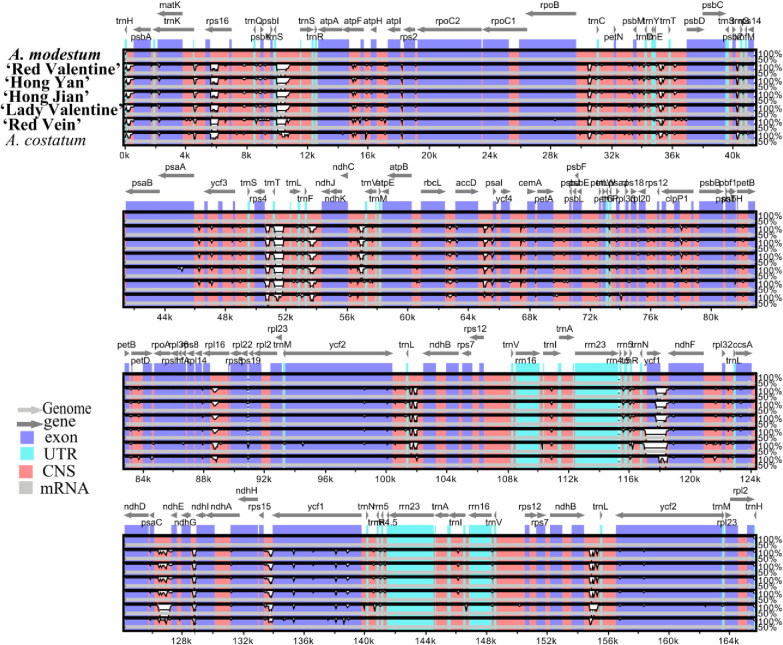
Furthermore, the 7 whole chloroplast genome alignments were examined using mVISTA (Fig 8). Most of the sequence variations were found in the LSC and SSC regions, while the two IR regions showed high sequence conservation of the 7 Aglaonema genomes. The main divergent regions were trnH-GUG-CDS1, trnH-GUG-CDS1_psbA, trnS-GCU_trnS-CGA-CDS1, rps4-trnT-UGU, trnF-GAA-ndhJ, petD-CDS2-rpoA, ycf1-ndhF, and rps15-ycf1-D2 (Fig 8). Combining the Pi values and mVISTA results, as well as considering the length of regions ≥ 200 bp for the selection of potential molecular markers of Aglaonema, 9 regions were found: trnH-GUG-CDS1_psbA, trnS-GCU_trnS-CGA-CDS1, rps4-trnT-UGU, trnF-GAA-ndhJ, petD-CDS2-rpoA, ycf1-ndhF, rps15-ycf1-D2, ccsA-ndhD, and trnY-GUA-trnE-UUC (S8 Table).
Fig 8. Complete chloroplast genome comparison of seven Aglaonema chloroplast genomes using A. modestum as a reference.
Gray arrows and thick black lines above the alignment indicate gene orientation. Purple bars represent exons, sky-blue bars represent untranslated regions (UTRs), red bars represent non-coding sequences (CNS), gray bars represent mRNA and white regions represent sequence differences among analyzed chloroplast genomes. The y-axis represents the identity percentage ranging from 50% to 100%. The six sequenced Aglaonema chloroplast genomes here are in bold.
Ka/Ks ratios among Araceae species
Using the 79 protein-coding gene sequences of A. modestum as references, Ka/Ks ratios were computed in the A. modestum chloroplast genome, six other chloroplast genomes of Aglaonema and nine other chloroplast genomes of Araceae (S9 Table). The Ka/Ks ratios were less than 1.00 and invalid for most comparison pairs (98.5%) (S9 Table). rpl2, rps2, rps3, ycf1 and ycf2 with Ka/Ks >1.00 and p < 0.05 were detected, indicating that these genes underwent positive selection (S9 Table). Furthermore, the Ka/Ks ratios of ycf2 in five pairwise comparisons, ycf1 in four pairwise comparisons, and rps2 in four pairwise comparisons were all > 1.00, indicating that the three genes ycf2, ycf1, and rps2 showed adaptation evolution under different environments.
SNPs and indels analyses of Aglaonema chloroplast genomes
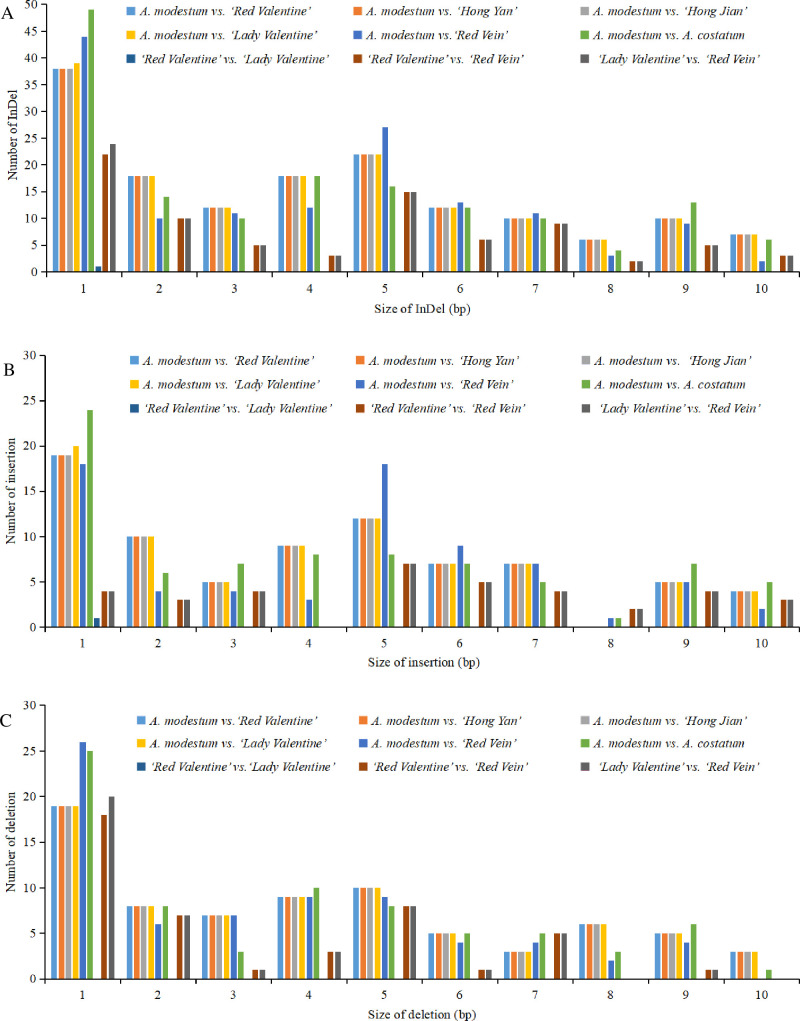
First, using the A. modestum chloroplast genome as the reference, SNP/indel loci of the chloroplast genomes of ‘Lady Valentine’, ‘Red Vein’, A. costatum, ‘Red Valentine’, ‘Hong Yan’, and ‘Hong Jian’ were detected. Three comparisons, A. modestum vs. ‘Red Valentine’, A. modestum vs. ‘Hong Yan’, and A. modestum vs. ‘Hong Jian’, revealed the same numbers of SNPs and indels, with 311 protein coding gene SNPs, 531 intergenic SNPs, and 153 indels, respectively (Fig 9, Tables 3, S10 and S11). Additionally, between A. modestum and ‘Lady Valentine’, 322 protein coding gene SNPs, 531 intergenic SNPs, and 154 indels were detected (Tables 3 and S11, and Fig 9), and the lengths of indels were mainly between 1 and 5 bp (Fig 9). Regarding A. modestum vs. ‘Red Vein’, 335 protein coding gene SNPs, 535 intergenic SNPs, and 142 indels were identified (Fig 9, Tables 3, S10 and S11). Concerning A. modestum vs. A. costatum, 333 protein coding gene SNPs, 452 intergenic SNPs, and 152 indels were found (Fig 9, Tables 3, S10 and S11).
Fig 9. Indels statistics of seven Aglaonema chloroplast genomes.
First, the A. modestum chloroplast genome was used as the reference sequence for indels analyses for the other six Aglaonema chloroplast genomes. Second, the five variegated cultivars were compared using chloroplast genome of ‘Red valentine’ as the reference. Third, ‘Lady valentine’ and ‘Red vein’ were compared. (A) Total indels statistics. (B) Insertion statistics. (C) Deletion statistics.
Table 3. Distribution of SNPs and indels among seven Aglaonema chloroplast genomes.
| Comparison pairs | Protein coding genes SNPs | Intergenic regions SNPs | Total SNPs | Insertions | Deletions | Indels |
|---|---|---|---|---|---|---|
| A. modestum vs. ‘Red Valentine’ | 311 | 531 | 842 | 78 | 75 | 153 |
| A. modestum vs. ‘Hong Yan’ | 311 | 531 | 842 | 78 | 75 | 153 |
| A. modestum vs. ‘Hong Jian’ | 311 | 531 | 842 | 78 | 75 | 153 |
| A. modestum vs. ‘Lady Valentine’ | 322 | 531 | 853 | 79 | 75 | 154 |
| A. modestum vs. ‘Red Vein’ | 335 | 535 | 870 | 71 | 71 | 142 |
| A. modestum vs. A. costatum | 333 | 452 | 785 | 78 | 74 | 152 |
| ‘Red Valentine’ vs. ‘Lady Valentine’ | 14 | 0 | 14 | 1 | 0 | 1 |
| ‘Red Valentine’ vs. ‘Red Vein’ | 164 | 422 | 586 | 36 | 44 | 80 |
| ‘Lady Valentine’ vs. ‘Red Vein’ | 173 | 426 | 599 | 36 | 46 | 82 |
Second, to detect SNPs/indels among five variegated cultivars of Aglaonema, using the ‘Red Valentine’ chloroplast genome as the reference, SNP/indel loci of the chloroplast genomes of ‘Lady Valentine’, ‘Hong Yan’, ‘Hong Jian’, and ‘Red Vein’ were analyzed. Two comparisons, ‘Red Valentine’ vs. ‘Hong Yan’ and ‘Red Valentine’ vs. ‘Hong Jian’, had no SNPs/indels. Fourteen SNPs and 1 indel were detected between ‘Red Valentine’ and ‘Lady Valentine’ (Fig 9, Tables 3 S10, and S11). Interestingly, these 14 SNPs were all in psaA (S10 Table), suggesting that psaA can be used to identify these two cultivars. One hundred sixty-four and 422 SNPs were detected between ‘Red Valentine’ and ‘Red Vein’ in protein coding genes and intergenic regions, respectively (Tables 3 and S10). Eighty indels were detected between ‘Red Valentine’ and ‘Red Vein’ (Fig 9, Tables 3 and S11). Third, SNPs/indels between ‘Lady Valentine’ and ‘Red Vein’ were also tested. A total of 173 and 426 SNPs in protein coding genes and intergenic regions were identified between ‘Lady Valentine’ and ‘Red Vein’, respectively (Tables 3 and S10). Eighty-two indels were also found between these two chloroplast genomes (Fig 9, Tables 3 and S11). The SNPs and indels analyzed in this study can be used for Aglaonema species and cultivar identification and phylogeny in the future.
Phylogenetic analyses
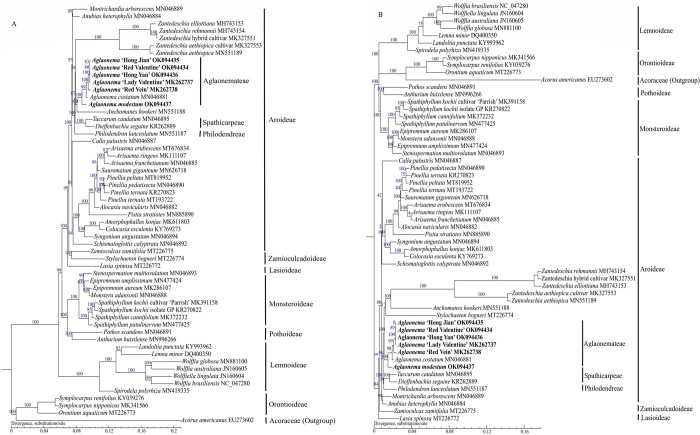
Two phylogenetic trees were constructed by ML method using whole chloroplast genomes and protein coding genes, respectively (Fig 10A and 10B). A. americanus was classified in the family Acoraceae [32], which was used as the outgroup. The 7 subfamilies within Araceae, including Aroideae, Lasioideae, Lemnoideae, Monsteroideae, Orontioideae, Pothoideae, and Zamioculcadoideae, were strongly identified using whole chloroplast genomes with bootstrap values of 89–100% (Fig 10A). By contrast, the phylogenetic tree constructed using protein coding genes showed moderate (bootstrap value = 84%) to strong identifications (bootstrap value = 100%) (Fig 10B). Within subfamily Aroideae, Aglaonema was strongly supported as monophyletic (bootstrap values = 100%) (Fig 10A and 10B). Aglaonema was seen as either sister to Anchomanes (bootstrap value = 100%), and Aglaonema + Anchomanes and Zantedeschia were strongly supported as sister genera (bootstrap value = 88%) (Fig 10A) or formed a poorly supported clade with Stylochaeton (bootstrap value = 17%) that was sister to the genera Anchomanes + Zantedeschia (bootstrap value = 100%) (Fig 10B). Within the genus Aglaonema, ‘Hong Jian’, ‘Red Valentine’, ‘Hong Yan’, and ‘Lady Valentine’ were clustered together with moderate to strong support (bootstrap values = 80–100%), and ‘Red Vein’ was sister to these four variegated cultivars with strong support (bootstrap value = 100%) (Fig 10A). A. costatum was sister to these five variegated cultivars with strong support (bootstrap value = 99%). A. modestum was sister to the clade of “A. costatum + five variegated cultivars” with strong support (bootstrap value = 100%) based on complete chloroplast genomes (Fig 10A). These seven Aglaonema species and cultivars showed similar topologies based on protein coding genes, but ‘Hong Jian’ and ‘Red Valentine’ were poorly supported in the ML analysis (bootstrap value = 0) (Fig 10B). The other five Aglaonema species and cultivars were strongly supported in ML analysis based on protein coding genes (bootstrap values = 89–100%) (Fig 10B).
Fig 10. Phylogenetic analyses of 56 Araceae chloroplast genomes with maximum likelihood, Acorus americanus was the outgroup taxa.
(A) The phylogenetic tree was constructed by the complete chloroplast genome sequences. (B) The phylogenetic tree was constructed using protein coding genes sequences. The six newly sequenced Aglaonema chloroplast genomes in this study were in bold.
Discussion
In the current study, the six newly assembled chloroplast genomes of Aglaonema species and variegated cultivars were highly conserved, possessing typical quadripartite structures and similar GC contents (35.73–35.91%) (Fig 2, Table 1 and S1 Fig). These quadripartite structures and similar GC contents were consistent with other reported Araceae plant chloroplast genomes, such as A. costatum [6], M. adansonii [9], E. aureum [31], Anthurium huixtlense [33], and Symplocarpus renifolius [34,35]. Interestingly, the variation in the chloroplast genome size of three variegated cultivars, ‘Hong Jian’, ‘Red Valentine’, and ‘Hong Yan’, was not significant. ‘Hong Jian’ and ‘Hong Yan’ were two mutations derived from tissue-cultured ‘Red Valentine’ plants (Fig 1). Both ‘Hong Jian’ and ‘Red Valentine’ showed the same genome size, while ‘Hong Yan’ was only 27 bp longer than these two cultivars (Table 1 and S1 Fig). Similar phenomena were also found in Hyacinthus and Utricularia cultivars [36,37]. In Hyacinthus cultivars, ‘Woodstock’, ‘Delft Blue’ and ‘Aiolos’ displayed the same chloroplast genome size of 154,640 bp [36]. In the case of two Utricularia amethystina cultivars, Purple and Yellow, only a 95 bp difference was found [37]. These chloroplast genome sizes were not changed substantially, possibly because the chloroplast genomes did not undergo recombination among the tested cultivars [36].
Furthermore, remarkable variations among the six Aglaonema species and variegated cultivars were gene loss and intron loss events. Both variegated cultivars, ‘Lady Valentine’ and ‘Red Vein’, had ycf68 and one intron in trnG-UCC, respectively, while the other four chloroplast genomes of A. modestum, ‘Hong Jian’, ‘Red Valentine’, and ‘Hong Yan’, lost ycf68 and introns in trnG-UCC (S3 Table and S1 Fig). Additionally, trnS-CGA, trnS-GGA and trnT-GGU were missing in the chloroplast genomes of ‘Lady Valentine’ and ‘Red Vein’, but these three tRNA genes showed one copy in the other four sequenced Aglaonema chloroplast genomes (Tables 2 and S3). In contrast to the present study, certain events of gene loss, intron loss, and gene duplication were also reported within Araceae chloroplast genomes; for example, gene loss events involved rpl23, rpl2, trnL-CCA, trnG-GCC, accD and psbE in the genus Amorphophallus [38].
Another variation among the tested Aglaonema species and variegated cultivars was the location of the boundaries among the four chloroplast genome regions. Location of the boundaries, particularly boundaries of the IR contraction and expansion, has been considered an important evolutionary event leading to gene duplication or reduction of duplicate genes to a single copy [6,9,39]. In contrast to previous studies, duplication of ycf1 and rps15 has been reported in the subfamily Lemnoideae [39], and conversion of duplicate genes to single-copy genes has occurred in rpl2 and rpl23 in Anchomanes hookeri of the family Araceae [6]. Our results revealed the duplication of rpl2 in IR/LSC boundary regions and integration of trnH-GUG into the IRa region (Fig 6), a finding that was also consistent with previous studies findings [6,9]. Additionally, our findings showed that trnN-GUU was duplicated in IR/SSC borders in two chloroplast genomes of ‘Lady Valentine’ and ‘Red Vein’, respectively, and ycf1 was duplicated in IR/SSC borders in the other five Aglaonema genomes (Fig 6). The same phenomenon exists in some species of the family Araceae [39–41].
Chloroplast genomes are rich in SSRs, long repeats, highly divergent regions, SNPs and indels [39–41]. They have been extensively used to identify species and cultivars [36] and for phylogenetic analyses [37,41]. In the present study, SSR detection results were the same as most other Araceae plants; for example, most SSRs were mononucleotide repeats and usually comprised short repeats of A/T [31,41,42]. Additionally, divergent analyses implemented by mVISTA and nucleotide diversity revealed 9 highly divergent regions among 7 Aglaonema chloroplast genomes, including rnH-GUG-CDS1_psbA, trnS-GCU_trnS-CGA-CDS1, rps4-trnT-UGU, trnF-GAA-ndhJ, petD-CDS2-rpoA, ycf1-ndhF, rps15-ycf1-D2, ccsA-ndhD, and trnY-GUA-trnE-UUC (Figs 7 and 8). In contrast to previous studies, six highly divergent regions (trnH-GUG_psbA, rps4-trnT-UGU, trnF-GAA-ndhJ, rps15-ycf1, ccsA-ndhD, and petD-rpoA) were used as DNA barcodes in Araceae plants or were in the marker development of DNA barcodes [9,41]. We suggest that the remaining three highly divergent regions of trnS-GCU_trnS-CGA-CDS1, ycf1-ndhF, and trnY-GUA-trnE-UUC can be used as special DNA barcodes for Aglaonema species and cultivar identification and phylogenetic analyses in the future.
Because ‘Red Valentine’ and ‘Lady Valentine’ were challenging to differentiate by their leaf morphological characteristics, developing molecular markers to identify these two Aglaonema cultivars is crucial. In a previous study, correlations among SNPs, indels, and repeats were investigated among 27 species from 7 subfamilies of Araceae, and strong/very strong correlations in most comparisons were observed [43]. Notably, in the present study, 14 SNPs and 1 insertion were found between these two cultivar genomes. These 14 SNPs were all located in psaA (S10 Table), and 1 insertion was found in rps12 (S11 Table). Hence, psaA and rps12 sequences can be used to differentiate these two cultivars. Additionally, the other two cultivar pairs, ‘Red Valentine’ vs. ‘Red Vein’ and ‘Lady Valentine’ vs. ‘Red Vein’, also contained many SNPs and indels (Tables 3, S10 and S11). These SNPs and indels could be used to identify these three variegated cultivars. However, both ‘Red Valentine’ vs. ‘Hong Jian’ and ‘Red Valentine’ vs. ‘Hong Yan’ had no SNPs/indels. These two comparisons indicated that the chloroplast genomes had not undergone recombination in two tissue culture-induced mutations of ‘Red Valentine’. The two mutations, ‘Hong Jian’ and ‘Hong Yan’, may be induced by nuclear gene variation and complex biological processes. The molecular regulation mechanisms of leaf shape and leaf color variations of ‘Hong Jian’ and ‘Hong Yan’ require further investigation.
In the present study, the Ka/Ks ratio showed low rates of evolution (Ka/Ks < 1) for most protein coding genes (74 of 79 genes) among the 16 Araceae chloroplast genomes tested. These findings agreed with those of other high plant chloroplast genomes [9,33,43–45]. However, our results contradicted a study in which majority (62 of 71 genes) had a Ka/Ks ratio of > 1, suggesting that these genes were positively selected within 14 Araceae species [35]. We demonstrated that rpl2, rps2, rps3, ycf1 and ycf2 were under positive selection among the 16 Araceae chloroplast genomes tested (S9 Table). By comparison, previous studies have revealed that rps3, ycf1 and ycf2 with positive selection in high plants may be very common [9,33,43,44]; ycf1 and ycf2 have been identified under positive selection in Eruca sativa [44]; ycf1 and ycf2 have also been reported under positive selection in Pothos scandens and Monsteroideae species of Araceae [9,33]; and rps3, ycf1 and ycf2 have been identified under positive selection in four Zingiber species [45].
Many studies have used complete chloroplast genomes or protein coding genes for phylogenetic studies in the family Araceae [6,9,31,33–35,40,41,46]. Specifically, chloroplast trnL-trnF has been used in Lemnaceae and Araceae phylogenetic relationship studies [47]. Additionally, previous molecular research based on complete chloroplast genomes identified eight subfamilies (excluding Gymnostachydoideae) within the family Araceae: Aroideae, Lasioideae, Lemnoideae, Monsteroideae, Orontioideae, Pothoideae, and Zamioculcadoideae [46]. Genetic relationships among the subfamilies of Orontioideae, Lemnoideae, Pothoideae and Monsteroideae were strongly supported [46]. Our phylogenetic tree obtained by whole chloroplast genomes was in broad agreement with previous studies [6,9,31,33,46]. However, the two phylogenetic trees, constructed using whole chloroplast genomes and protein coding genes, exhibited few inconsistencies in this study—for example, the shifting position of Stylochaeton in subfamily Aroideae in the phylogenetic tree based on protein coding genes (Fig 10A and 10B). The bootstrap value (17%) was too low in the latter tree to draw any meaningful conclusions about genetic relationships in subfamily Aroideae (Fig 10B). The reason for the shifting position of Stylochaeton may be (1) that the protein coding gene sequences were insufficient to resolve the relationships between Stylochaeton and the rest of subfamily Aroideae with strong support and (2) that more Stylochaeton and other Aroideae species chloroplast genomes must be sequenced to solve the uncertainties. Additionally, the position of Acorus contrasted that in previous studies [6,9,33]. Both of our trees indicated that Acorus was sister to the genera Orontium + Symplocarpus with strong support (bootstrap values = 100%) (Fig 10). Therefore, Acorus requires more chloroplast data or nuclear genomes to resolve its position.
Conclusions
In this study, the complete chloroplast genomes of A. modestum and five variegated cultivars of Aglaonema were sequenced and assembled. The six chloroplast genomes were highly conserved in terms of the gene content, codon usage, and genome structure but also distinguished the difference between IR-SSC boundary regions. ‘Red Vein’ differed significantly from the others in long repeat number and type. Comparative analyses of the chloroplast genomes identified 9 divergent hotspots (Pi > 0.021) with potential application as DNA barcodes. The Ka/Ks ratio analyses among 16 Araceae chloroplast genomes showed that rpl2, rps2, rps3, ycf1 and ycf2 were under positive selection. Phylogenetic trees based on whole chloroplast genomes and protein-coding genes strongly supported monophyletic Aglaonema. These assembled chloroplast genomes provided potential genomic resources and helped the identification and utilization of Aglaonema germplasm resources.
Supporting information
Genes shown inside the circle are transcribed clockwise, and those outside are transcribed counterclockwise. The gray arrowheads indicate the direction of the genes. Different genes are color coded. The innermost darker gray corresponds to the GC content, whereas the lighter gray corresponds to the AT content. The inner circle also indicates that the chloroplast genome contains a large single copy (LSC) region, a small single copy (SSC) region and two copies of the inverted repeat (IRA and IRB). (A) ‘Red Valentine’, (B) ‘Hong Yan’, (C) ‘Hong Jian’, (D) ‘Lady Valentine’, and (E) ‘Red Vein’. * in (D) and (E) indicates ycf68 and trnI-CAU only present in the IR regions of the chloroplast genomes of ‘Lady Valentine’ and ‘Red Vein’, respectively.
(DOC)
(XLSX)
(DOCX)
(DOCX)
(XLS)
(XLS)
(XLSX)
(XLSX)
(XLSX)
(XLS)
(XLS)
(XLS)
Data Availability
Data Availability Statement: The six newly assembled chloroplast genomes in this study have been deposited in GenBank with accession numbers MK262737, MK262738, OK094434, OK094435, OK094436, and OK094427.
Funding Statement
This research was financially supported by the Guangdong Basic and Applied Basic Foundation Project (No. 2021A1515010893), Special Financial Fund of Foshan-High-level Guangdong Agricultural Science and Technology Demonstration City Project (2022), and Guangdong Province Modern Agriculture Industry Technical System-Flower Innovation Team Construction Project (2021KJ121). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Nicolson DH. A revision of the genus Aglaonema (Araceae). Smithsonian Contributions to Botany, Smithsonian Institution Press, Washington, USA, 1969; pp 1–66. [Google Scholar]
- 2.Chen J, Devanand PS, Norman DJ, Henny RJ, Chao CCT. Genetic relationships of Aglaonema species and cultivars inferred from AFLP markers. Ann Bot. 2004; 93: 157–166. doi: 10.1093/aob/mch025 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Henny RJ, Chen J, Mellich TA, Brennan MS. ‘Moonlight Bay’ Aglaonema. HortScience. 2008; 43: 1598–1599. [Google Scholar]
- 4.Henny RJ, Chen J. ‘Key Lime’ Aglaonema. HortScience. 2009; 44: 1767–1768. [Google Scholar]
- 5.Henny RJ, Chen J. ‘Scenic Bay’ Aglaonema. HortScience. 2010; 45: 1281–1282. [Google Scholar]
- 6.Henriquez CL, Abdullah Ahmed I, Carlsen MM, Zuluaga A, Croat TB, McKain MR. Evolutionary dynamics of chloroplast genomes in subfamily Aroideae (Araceae). Genomics. 2020; 112: 2349–2360. doi: 10.1016/j.ygeno.2020.01.006 . [DOI] [PubMed] [Google Scholar]
- 7.Wicke S, Schneeweiss GM, DePamphilis CW, Muller KF, Quandt D. The evolution of the plastid chromosome in land plants: gene content, gene order, gene function. Plant Mol Biol. 2011; 76: 273–297. doi: 10.1007/s11103-011-9762-4 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Daniell H, Lin CS, Yu M, Chang WJ. Chloroplast genomes: diversity, evolution, and applications in genetic engineering. Genome Biol. 2016; 17: 134. doi: 10.1186/s13059-016-1004-2 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Henriquez CL, Abdullah Ahmed I, Carlsen MM, Zuluaga A, Croat TB, McKain MR. Molecular evolution of chloroplast genomes in Monsteroideae (Araceae). Planta. 2020; 251: 72. doi: 10.1007/s00425-020-03365-7 . [DOI] [PubMed] [Google Scholar]
- 10.Zheng G, Wei L, Ma L, Wu Z, Gu C, Chen K. Comparative analyses of chloroplast genomes from 13 Lagerstroemia (Lythraceae) species: identification of highly divergent regions and inference of phylogenetic relationships. Plant Mol Biol. 2020; 102: 659–676. doi: 10.1007/s11103-020-00972-6 . [DOI] [PubMed] [Google Scholar]
- 11.Abdullah Mehmood F, Shahzadi I, Waseem S, Mirza B, Ahmed I, Waheed MT. Chloroplast genome of Hibiscus rosa-sinensis (Malvaceae): comparative analyses and identification of mutational hotspots. Genomics. 2020; 112: 581–591. doi: 10.1016/j.ygeno.2019.04.010 . [DOI] [PubMed] [Google Scholar]
- 12.Li X, Hu Z, Lin X, Li Q, Gao H, Luo G, et al. High-throughput pyrosequencing of the complete chloroplast genome of Magnolia officinalis and its application in species identification. Acta Pharm Sin. 2012; 47: 124–130. . [PubMed] [Google Scholar]
- 13.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014; 30: 2114–2120. doi: 10.1093/bioinformatics/btu170 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Luo R, Liu B, Xie Y, Li Z, Huang W, Yuan J, et al. SOAPdenovo2: An empirically improved memory-efficient short-end de novo assembler. Gigascience. 2012; 1: 18. 10.1186/2047-217X-1-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kearse M, Moir R, Wilson A, Stoneshavas S, Cheung M, Sturrock S, et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012; 28: 1647–1649. doi: 10.1093/bioinformatics/bts199 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wyman SK, Jansen RK, Boore JL. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 2004; 20: 3252–3255. 10.1093/bioinformatics/bth352 . [DOI] [PubMed] [Google Scholar]
- 17.Lowe TM, Chan PP. tRNAscan-SE On-line: integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 2016; 44: W54–W57. doi: 10.1093/nar/gkw413 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Greiner S, Lehwark P, Bock R. Organellar Genome DRAW (OGDRAW) version 1.3.1: expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 2019; 47: W59–W64. doi: 10.1093/nar/gkz238 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kumar S, Stecher G, Tamura K. Mega7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016; 33: 1870–1874. doi: 10.1093/molbev/msw054 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.R Core Team. R: a language and environment for statistical computing. https://www.R-project.org. Accessed 20 May 2020.
- 21.Kurtz S, Choudhuri JV, Ohlebusch E, Schleiermacher C, Stoye J, Giegerich R. REPuter: The manifold applications of repeat analysis on a genomic scale. Nucleic Acids Res. 2001; 29: 4633–4642. doi: 10.1093/nar/29.22.4633 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Beier S, Thiel T, Münch T, Scholz U, Mascher M. MISA-web: a web server for microsatellite prediction. Bioinformatics. 2017; 33: 2583–2585. doi: 10.1093/bioinformatics/btx198 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rozas J, Ferrer-Mata A, Sánchez-DelBarrio JC, Guirao-Rico S, Librado P, Ramos-Onsins SE, et al. DnaSP 6: DNA sequence polymorphism analysis of large datasets. Mol Biol Evol. 2017; 34: 3299–3302. doi: 10.1093/molbev/msx248 . [DOI] [PubMed] [Google Scholar]
- 24.Frazer KA, Pachter L, Poliakov A, Rubin EM, Dubchak I. VISTA: computational tools for comparative genomics. Nucleic Acids Res. 2004; 32: W273–W279. doi: 10.1093/nar/gkh458 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang D, Zhang Y, Zhang Z, Zhu J, Yu J. KaKs Calculator 2.0: a toolkit incorporating gamma-series methods and sliding window strategies. Genom. Proteom. Bioinformatics. 2010; 8: 77–80. doi: 10.1016/S1672-0229(10)60008-3 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marcais G, Delcher AL, Phillippy AM, Coston R, Salzberg SL, Zimin A. MUMmer4: A fast and versatile genome alignment system. PLoS Comput Biol. 2018; 14: e1005944. doi: 10.1371/journal.pcbi.1005944 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rambaut A. Se-Al: Sequence Alignment Editor; Version 2.0. Available online: http://tree.bio.ed.ac.uk/software (accessed on 30 September 2017).
- 28.Rozewicki J, Li S, Amada KM, Standley DM, Katoh K. MAFFT-DASH: integrated protein sequence and structural alignment. Nucleic Acids Res. 2019; 47: W5–W10. doi: 10.1093/nar/gkz342 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010; 59: 307–321. doi: 10.1093/sysbio/syq010 . [DOI] [PubMed] [Google Scholar]
- 30.Kress WJ, Prince LM, Williams KJ. The phylogeny and a new classification of the gingers (Zingiberaceae) evidence from molecular data. Am J Bot. 2002;89:1682–1696. doi: 10.3732/ajb.89.10.1682 . [DOI] [PubMed] [Google Scholar]
- 31.Tian N, Han L, Chen C, Wang Z. The complete chloroplast genome sequence of Epipremnum aureum and its comparative analysis among eight Araceae species. PLoS ONE 2018; 13: e0192956. https://doi.org/0.1371/journal.pone.0192956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nauheimer L, Metzler D, Renner SS. Global history of the ancient monocot family Araceae inferred with models accounting for past continental positions and previous ranges based on fossils. New Phytol. 2012; 195: 938–950. doi: 10.1111/j.1469-8137.2012.04220.x . [DOI] [PubMed] [Google Scholar]
- 33.Abdullah Henriquez CL, Mehmood F Carlsen MM, Islam M Waheed MT, Poczai P, et al. Complete chloroplast genomes of Anthurium huixtlense and Pothos scandens (Pothoideae, Araceae): unique inverted repeat expansion and contraction affect rate of evolution. J Mol Evol. 2020; 88: 562–574. doi: 10.1007/s00239-020-09958-w . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Choi KS, Park KT, Park SJ. The chloroplast genome of Symplocarpus renifolius: a comparison of chloroplast genome structure in Araceae. Genes (Basel). 2017; 8: 324. doi: 10.3390/genes8110324 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim SH, Yang J, Park J, Yamada T, Maki M, Kim SC. Comparison of whole plastome sequences between thermogenic Skunk Cabbage Symplocarpus renifolius and Nonthermogenic S. nipponicus (Orontioideae; Araceae) in East Asia. Int J Mol Sci. 2019; 20: 4678. doi: 10.3390/ijms20194678 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wong KH, Wu HY, Kong BLH, But GWC, Siu TY, Hui JHL, et al. Characterisation of the complele chloroplast genomes of seven Hyacinthus orientalis L. cultivars: insights into cultivar phylogeny. Horticulturae 2022; 8: 453. 10.3390/horticulturae8050453. [DOI] [Google Scholar]
- 37.Silva SR, Pinheiro DG, Penha HA, Płachno BJ, Michael TP, Meer EJ, et al. Intraspecific Variation within the Utricularia amethystina Species Morphotypes Based on Chloroplast Genomes. Int J Mol Sci. 2019; 20: 6130. doi: 10.3390/ijms20246130 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu E, Yang C, Liu J, Jin S, Harijati N, Hu Z, et al. Comparative analysis of complete chloroplast genome sequences of four major Amorphophallus species. Sci Rep. 2019; 9: 809. doi: 10.1038/s41598-018-37456-z . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang W, Messing J. High-throughput sequencing of three Lemnoideae (duckweeds) chloroplast genomes from total DNA. PLoS ONE. 2011; 6: e24670. doi: 10.1371/journal.pone.0024670 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Abdullah Henriquez CL, Mehmood F, Shahzadi, Ali Z, Waheed MT, Croat TB, et al. Comparison of chloroplast genomes among species of unisexual and bisexual clades of the monocot family Araceae. Plants (Basel). 2020; 9: 737. doi: 10.3390/plants9060737 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li B, Liu T, Ali A, Xiao Y, Shan N, Sun J, et al. Complete chloroplast genome sequences of three aroideae species (Araceae): lights into selective pressure, marker development and phylogenetic relationships. BMC Genomics. 2022; 23: 218. doi: 10.1186/s12864-022-08400-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.He S, Yang Y, Li Z, Wang X, Guo Y, Wu H. Comparative analysis of four Zantedeschia chloroplast genomes: expansion and contraction of the IR region, phylogenetic analyses and SSR genetic diversity assessment. Peer J. 2020; 8: e9132. doi: 10.7717/peerj.9132 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Abdullah Henriquez CL, Croat TB, Poczai P, Ahmed I. Mutational dynamics of Aroid chloroplast genomes II. Front Genet. 2021; 11: 610838. doi: 10.3389/fgene.2020.610838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhu B, Qian F, Hou Y, Yang W, Cai M, Wu X. Complete chloroplast genome features and phylogenetic analysis of Eruca sativa (Brassicaceae). PLoS ONE. 2021; 16: e0248556. doi: 10.1371/journal.pone.0248556 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li DM, Ye YJ, Xu YC, Liu JM, Zhu GF. Complete chloroplast genomes of Zingiber montanum and Zingiber zerumbet: genome structure, comparative and phylogenetic analyses. PLoS ONE. 2020;15: e0236590. doi: 10.1371/journal.pone.0236590 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Henriquez CL, Arias T, Pires JC, Croat TB, Schaal BA. Phylogenomics of the plant family Araceae. Mol Phylogenet Evol. 2014; 75: 91–102. doi: 10.1016/j.ympev.2014.02.017 . [DOI] [PubMed] [Google Scholar]
- 47.Rothwell GW, Van Atta MR, Ballard HE Jr, Stockey RA. Molecular phylogenetic relationships among Lemnaceae and Araceae using the chloroplast trnL-trnF intergenic spacer. Mol Phylogenet Evol. 2004; 30: 378–385. doi: 10.1016/s1055-7903(03)00205-7 . [DOI] [PubMed] [Google Scholar]