Abstract
Spermidine and cadaverine were found to be constituents of the cell wall peptidoglycan of Anaerovibrio lipolytica, a strictly anaerobic bacterium. The peptidoglycan was degraded with the N-acetylmuramyl-l-alanine amidase and endopeptidase into two peptide fragments, peptide I and peptide II, at a molar ratio of 4:1. Peptides I and II were identified as l-alanine–d-glutamic acid(αcadaverine)γmeso-diaminopimelic acid (DAP)–d-alanine and l-alanine–d-glutamic acid(αspermidine)γmeso-DAP–d-alanine, respectively. The N1-amino group of spermidine was linked to the α-carboxyl group of the d-glutamic acid residue of peptide II.
We previously reported that the peptidoglycans of three bacterial species, Selenomonas ruminantium, Veillonella alcalescens, and Veillonella parvula, contain no lipoprotein but have cadaverine or putrescine (3–7) that and the diamines covalently linked to the peptidoglycan are essential for both cell surface integrity and normal cell growth (7, 8). We also reported that dl-α-difluoromethyllysine (DFML), which was shown to be a potent, irreversible, and specific inhibitor of lysine decarboxylase, markedly inhibited the growth of S. ruminantium and caused rapid cell lysis because of the synthesis of the abortive peptidoglycan without cadaverine (8). The inhibition of growth by DFML was completely reversed by adding either cadaverine or putrescine at 1 mM. However, spermidine, regardless of its concentration, did not result in recovery of cell growth (8). During studies of the effect of polyamine inhibitors on the cell growth of other bacteria, we noticed that the growth of Anaerovibrio lipolytica was completely inhibited by a 10 mM concentration of either DFML or dl-α-difluoromethylornithine (DFMO), which had been shown to be a specific and irreversible inhibitor of ornithine decarboxylase. The growth inhibition caused by either DFML or DFMO was completely reversed by adding 1 mM cadaverine, putrescine, or spermidine to the medium, and the diamines and spermidine were incorporated into the peptidoglycan of A. lipolytica (data not shown). These results suggest that spermidine and the diamines are covalently linked to the peptidoglycan in A. lipolytica and that spermidine plays a role in the maintenance of the integrity of the cell envelope of this bacterium. Here we show that spermidine and cadaverine are components of the peptidoglycan in A. lipolytica. We also demonstrate that the N1-amino group of spermidine is covalently linked to the α-carboxyl group of the d-glutamic acid residue of its peptidoglycan.
A. lipolytica (VPI 7553), which was a kind gift from M. P. Bryant of the University of Illinois, was grown in a yeast extract-glucose medium used for culture of S. ruminantium (8). The peptidoglycan preparation from this species was purified by a method described previously (5) and was hydrolyzed with 6 N HCl. The acid hydrolysate was analyzed for amino acids, amino sugars, and polyamines by high-performance liquid chromatography with an Amino Pack column (Tosoh, Tokyo, Japan). Glutamic acid, alanine, diaminopimelic acid (DAP), muramic acid, and glucosamine were evident, but no other amino acid was found, not even lysine, which is a representative amino acid of bacterial lipoproteins (data not shown). Two ninhydrin-positive compounds, A and B, were also detected, at positions which correspond to those of cadaverine and spermidine, respectively. Compound A was identified as cadaverine. Compound B was further analyzed by gas chromatography-mass spectrometry after acylation with heptafluorobutyric anhydride to give heptafluorobutyroyl derivatives. The mass spectrum showed that the heptafluorobutyroyl derivative of compound B was identical to that of authentic spermidine (data not shown). The molar ratios of alanine, meso-DAP, cadaverine, and spermidine to glutamic acid in the peptidoglycan preparation were 2.05, 1.00, 0.78, and 0.20, respectively.
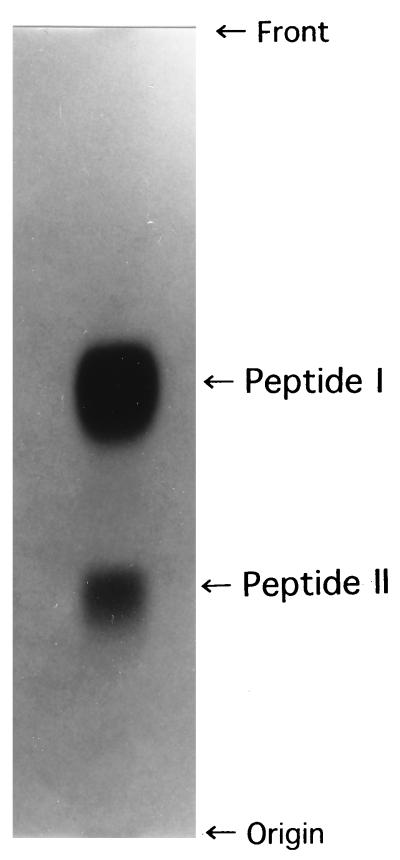
To elucidate the amino acid residue that is linked covalently to cadaverine and spermidine, and the determine which amino group of the spermidine residue which covalently links to the amino acid residue of the peptidoglycan, the purified peptidoglycan preparation (1 g) was incubated at 37°C with a Streptomyces albus G enzyme (500 μg of protein) which contains N-acetylmuramyl-l-alanine amidase and endopeptidase activities, the latter hydrolyzing the d-alanine–meso-DAP bond cross-linking the peptidoglycan. The degradation products were subjected to paper chromatography with an upper phase of n-butyl alcohol–acetic acid–water (4:1:5 [vol/vol/vol]). Two ninhydrin-positive peptides, I and II, were detected on the paper chromatogram, at a molar ratio of 4:1 (Fig. 1). Both peptides were eluted from the paper with distilled water, and they were hydrolyzed in 6 N HCl and subjected to amino acid and polyamine analyses by methods described previously (15). Both peptide I and peptide II were revealed to consist of glutamic acid, alanine, and meso-DAP at a molar ratio of 0.98:1.96:1.01. In addition, peptide I and peptide II contained cadaverine and spermidine at a molar ratio of 1.0:1.0 to the glutamic acid residue of each peptide, respectively (data not shown). The meso-DAP and cadaverine residues of peptide I and the meso-DAP and spermidine residues of peptide II were completely dinitrophenylated, whereas one-half of the alanine of both peptide preparations was dinitrophenylated. From the results, it was concluded that l- and d-alanine are the N- and C-terminal amino acids, respectively, in both peptides and that the l-alanine residue links covalently to the —COOH of the lactyl group of N-acetylmuramic acid. To elucidate which amino acid residue of the peptide moiety of the peptidoglycan is covalently linked to cadaverine and spermidine, the peptide I and II preparations were partially hydrolyzed with 2 N HCl and the degradation products were purified by ion-exchange chromatography followed by paper electrophoresis by methods described previously (7). From the peptide I preparation, two peptide fragments, consisting of meso-diaminopimelyl-d-alanine and l-alanyl-d-glutaminyl-cadaverine, were isolated (data not shown). Two peptide fragments were also obtained from the hydrolysate of the peptide II preparation. One was identified as meso-diaminopimelyl-d-alanine, and the other was determined to be l-alanyl-d-glutaminyl-spermidine. From these results, the primary structures of peptides I and II were identified as l-alanine–d-glutamic acid(αcadaverine)γmeso-DAP– d-alanine and l-alanine–d-glutamic acid(αspermidine)γmeso-DAP–d-alanine, respectively. Taken together with the fact, mentioned above, that the molar ratios of cadaverine and spermidine to glutamic acid in the peptidoglycan preparation were 0.78 and 0.20, respectively, it was concluded that the glutamic acid residue of the peptidoglycan of A. lipolytica was saturated with cadaverine and spermidine.
FIG. 1.
Isolation of the peptide I and II preparations by paper chromatography after treatment of the peptidoglycan preparation of A. lipolytica with N-acetylmuramyl-l-alanine amidase and the endopeptidase from S. albus G. Spots were detected by spraying 0.15% ninhydrin in acetone.
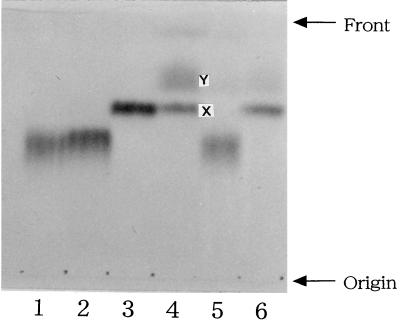
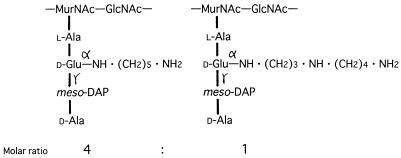
The chemical structure of spermidine is N1H2 · (CH2)3 · N4H · (CH2)4 · N8H2. To elucidate which amino group of the spermidine residue is covalently linked to the d-glutamic acid residue of the peptidoglycan, the peptide II preparation (210 nmol) was reacted with 2,4-dinitrofluorobenzene (DNFB; 160 nmol) in 500 mM sodium borate buffer (pH 10.0) at 25°C for 30 min and then 50°C for 10 min, by which time the free amino group(s) of the spermidine residue should be partially labeled with a dinitrophenyl group(s) to give monodinitrophenylated spermidine derivatives. The reaction mixture was hydrolyzed by addition of 1 ml of 6 N HCl and incubation at 110°C for 14 h. After the mixture was evaporated to dryness, the residue was suspended in 4 N NH4OH. Under alkaline conditions, the 2,4-dinitrophenylated spermidine derivatives were preferentially extracted with chloroform and analyzed on a Silica Gel 60 F254 thin-layer chromatography plate (E. Merck Ag, Darmstadt, Germany) together with authentic N1-, N4-, and N8-(2,4-dinitrophenyl)spermidine derivatives which were synthesized chemically (Fig. 2). When the solvent system for the separation of polyamine isomers, n-butyl alcohol–acetic acid–pyridine–36% formaldehyde (3:1:0.5:0.5 [vol/vol/vol]) (14), was used, N8-(2,4-dinitrophenyl)spermidine was successfully separated from N1- and N4-(2,4-dinitrophenyl)spermidine (Fig. 2, lanes 1 to 3). For the peptide II preparation, two dinitrophenylated spermidine derivatives were detected (Fig. 2, lane 4). One (compound X) had an Rf value identical to that of authentic N8-(2,4-dinitrophenyl)spermidine, and the other (compound Y) had a higher Rf value. The Rf values of these two spermidine derivatives were identical to those of the two derivatives of 2,4-dinitrophenylated N1-acetylspermidine (Fig. 2, lanes 1 to 3). The data indicate that compound Y is N4,N8-bis(2,4-dinitrophenyl)spermidine and that the amino group at position 8 of the spermidine residue of peptide II is free. From these findings, it was concluded that the amino group at the N1 position of the spermidine residue links covalently to the α-carboxyl group of the d-glutamic acid residue of the peptidoglycan from A. lipolytica. We propose that the primary structure of the peptidoglycan of A. lipolytica is as shown in Fig. 3, based on the following evidence: (i) the peptidoglycan of this strain consists of N-acetylglucosamine, N-acetylmuramic acid, l-alanine, d-glutamic acid, meso-DAP, cadaverine, spermidine, and d-alanine; (ii) two kinds of peptides, peptides I and II, at a molar ratio of 4 to 1, were obtained by digesting the peptidoglycan with N-acetylmuramyl-l-alanine amidase and endopeptidase; (iii) the primary structures of peptides I and II both contain l-alanine–d-glutamic acid–meso-DAP–d-alanine, and cadaverine and spermidine are covalently linked to the α-carboxyl group of the d-glutamic acid residues of peptides I and II, respectively; and (iv) one of two amino groups of cadaverine and the amino groups at positions 4 and 8 of spermidine are free.
FIG. 2.
Silica gel chromatogram of the dinitrophenylated spermidine preparations isolated from the 2,4-dinitrophenylated peptide II preparation. Lane 1, N1-(2,4-dinitrophenyl)spermidine; lane 2, N4-(2,4-dinitrophenyl)spermidine; lane 3, N8-(2,4-dinitrophenyl)spermidine; lane 4, 2,4-dinitrophenylated spermidine derivatives X and Y, isolated from the 2,4-dinitrophenylated peptide II preparation; lane 5, 2,4-dinitrophenylated spermidine derivative isolated from the 2,4-dinitrophenylated N8-acetylspermidine; lane 6, 2,4-dinitrophenylated spermidine derivative isolated from the 2,4-dinitrophenylated N1-acetylspermidine.
FIG. 3.
Proposed primary structure of peptidoglycan in A. lipolytica. MurNAc and GlucNAc, N-acetylmuramic acid and N-acetylglucosamine, respectively.
We mentioned above that 1 mM spermidine rescued the inhibition of cell growth of A. lipolytica by 10 mM DFML owing to the biosynthesis of abortive cadaverine-containing peptidoglycan. We also observed that spermidine added to the medium was incorporated into the peptidoglycan preparation. These results suggest that the spermidine incorporated into the peptidoglycan layer rescued the cell growth of A. lipolytica. To examine this possibility, the chemical compositions of the peptidoglycan of the cells which were grown in the medium containing 1 mM spermidine and 10 mM DFML were determined. The peptidoglycan was prepared from cells harvested at 16 h after the inhibitor and spermidine were added, and the amounts of amino acid, cadaverine, and spermidine in acid hydrolysates of the peptidoglycan preparations were measured. As shown in Table 1, the molar ratios of alanine, meso-DAP, cadaverine, and spermidine to glutamic acid were 1.9, 1.0, 0.03, and 0.28, respectively. The results indicate that the amount of cadaverine in the peptidoglycan preparation decreased drastically, but the spermidine content in the peptidoglycan preparation remained constant or increased, and that about a 30% saturation of the glutamic acid residues of the peptidoglycan with spermidine was sufficient for cell growth of this bacterium. We previously calculated the percent saturation of cadaverine in the peptidoglycan of cells of V. alcalescens (which was a stringent growth requirement for cadaverine) needed for normal cell growth and showed that at least a 40% saturation of cadaverine linked to the glutamic acid residue of the peptidoglycan was required (7). These results suggest that for the cell surface integrity of the bacteria, spermidine is more effective than diamines linked to the d-glutamic acid residue of the peptidoglycan.
TABLE 1.
Quantitative analysis of the peptidoglycan from A. lipolytica
| Component | nmol of component per mg of peptidoglycan prepn when grown:
|
|
|---|---|---|
| Without DFML | With DFML and spermidine | |
| Glutamic acid | 680 | 690 |
| Alanine | 1,390 | 1,340 |
| DAP | 685 | 704 |
| Cadaverine | 531 | 20 |
| Spermidine | 138 | 196 |
There are three characteristics that are shared by A. lipolytica and the bacteria which contain cadaverine or putrescine as a peptidoglycan constituent (4, 6, 7): (i) their outer and inner membranes all contain phosphatidylethanolamine and phosphatidylserine as predominant phospholipids (9); (ii) phosphatidylglycerol, which is known to be present widely in living cells and which has been shown to be the precursor of murein lipoprotein in gram-negative bacteria (1, 2), was not detected; and (iii) murein lipoprotein, which plays a role in the maintenance of the structural integrity of the outer membrane of the cell envelope in most gram-negative bacteria (16), was not detected. Therefore, the polyamines might connect the peptidoglycan to the outer membrane via interaction of their positively charged free amino groups with membrane phospholipids. Recently, Pfanzagl et al. reported the presence of N-acetylputrescine (NAP) covalently linked to the α-carboxyl group of the d-glutamic acid residue of the peptidoglycan of Cyanophora paradoxa (13) and showed that the existence of the NAP residue in the peptidoglycan layer is ubiquitous in cyanelle peptidoglycan of glaucocystophyte algae (10). The function of the NAP residue in cyanelle peptidoglycan remains unclear (13).
We previously reported the presence in the inner membrane fractions from S. ruminantium and the two Veillonella species of a lipid intermediate:diamine transferase, which catalyzes the addition of cadaverine or putrescine to a lipid intermediate during peptidoglycan synthesis. This enzyme can transfer diamines with carbon numbers of 3 to 7 to the α-carboxyl group of the d-glutamic acid residue of a lipid intermediate in vitro (7, 11). However, this enzyme is not capable of transferring spermidine to the lipid intermediate in vivo or in vitro (11). In this study, the presence of spermidine in peptidoglycan of A. lipolytica led us to postulate the existence of an enzyme which is involved in transferring spermidine preferentially to the peptidoglycan lipid intermediate in this organism.
REFERENCES
- 1.Braun V, Rehn K. Chemical characterization, spatial distribution and function of lipoprotein (murein lipoprotein) of the E. coli wall: the specific effects of trypsin on the membrane structure. Eur J Biochem. 1969;10:426–438. doi: 10.1111/j.1432-1033.1969.tb00707.x. [DOI] [PubMed] [Google Scholar]
- 2.Chattopadhyay P K, Lai J-S, Wu H C. Incorporation of phosphatidylglycerol into murein lipoprotein in intact cells of Salmonella typhimurium by phospholipid vesicle fusion. J Bacteriol. 1979;137:309–312. doi: 10.1128/jb.137.1.309-312.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kamio Y. Structural specificity of diamines covalently linked to peptidoglycan for cell growth of Veillonella alcalescens and Selenomonas ruminantium. J Bacteriol. 1987;169:4837–4840. doi: 10.1128/jb.169.10.4837-4840.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kamio Y, Itoh Y, Terawaki Y. Chemical structure of peptidoglycan in Selenomonas ruminantium: cadaverine links covalently to the d-glutamic acid residue of peptidoglycan. J Bacteriol. 1981;146:49–53. doi: 10.1128/jb.146.1.49-53.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kamio Y, Itoh Y, Terawaki Y, Kusano T. A new form of structural peptidoglycan in Selenomonas ruminantium: existence of polyamine in peptidoglycan. Agric Biol Chem. 1980;44:2523–2526. [Google Scholar]
- 6.Kamio Y, Itoh Y, Terawaki Y, Kusano T. Cadaverine is covalently linked to peptidoglycan in Selenomonas ruminantium. J Bacteriol. 1981;145:122–128. doi: 10.1128/jb.145.1.122-128.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kamio Y, Nakamura K. Putrescine and cadaverine are constituents of peptidoglycan in Veillonella alcalescens and Veillonella parvula. J Bacteriol. 1987;169:2881–2884. doi: 10.1128/jb.169.6.2881-2884.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kamio Y, Poso H, Terawaki Y, Paulin L. Cadaverine covalently linked to a peptidoglycan is an essential constituent of the peptidoglycan necessary for the normal growth in Selenomonas ruminantium. J Biol Chem. 1986;261:6585–6589. [PubMed] [Google Scholar]
- 9.Kamio Y, Takahashi H. Isolation and characterization of outer and inner membranes of Selenomonas ruminantium: lipid compositions. J Bacteriol. 1980;141:888–898. doi: 10.1128/jb.141.2.888-898.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kamio Y, Takahashi H. Outer membrane proteins and cell surface structure of Selenomonas ruminantium. J Bacteriol. 1980;141:899–907. doi: 10.1128/jb.141.2.899-907.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kamio Y, Terawaki Y, Izaki K. Biosynthesis of cadaverine-containing peptidoglycan in Selenomonas ruminantium. J Biol Chem. 1982;257:3326–3333. [PubMed] [Google Scholar]
- 12.Pfanzagl B, Allmaier G, Schmid E R, De Pedro M A, Löffelhardt W. N-Acetylputrescine as a characteristic constituent of cyanelle peptidoglycan in glaucocystophyte algae. J Bacteriol. 1996;178:6994–6997. doi: 10.1128/jb.178.23.6994-6997.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pfanzagl B, Zenker A, Pittenauer E, Allmaier G, Martinez-Torrecuadrada J, Schmid E R, De Pedro M A, Löffelhardt W. Primary structure of cyanelle peptidoglycan of Cyanophora paradoxa: a prokaryotic cell wall as part of an organelle envelope. J Bacteriol. 1996;178:332–339. doi: 10.1128/jb.178.2.332-339.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shirahata A, Takeda Y, Kawase M, Samejima K. Detection of spermine and thermospermine by thin-layer chromatography. J Chromatogr. 1983;262:452–454. [Google Scholar]
- 15.Takatsuka Y, Onoda M, Sugiyama T, Muramoto K, Tomita T, Kamio Y. Novel characteristics of Selenomonas ruminantium lysine decarboxylase capable of decarboxylating both l-lysine and l-ornithine. Biosci Biotechnol Biochem. 1999;63:1063–1069. doi: 10.1271/bbb.63.1063. [DOI] [PubMed] [Google Scholar]
- 16.Yem D W, Wu H C. Physiological characterization of an Escherichia coli mutant altered in the structure of murein lipoprotein. J Bacteriol. 1978;133:1419–1426. doi: 10.1128/jb.133.3.1419-1426.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]