Abstract
Nasopharyngeal carcinoma (NPC) is a common malignancy of the nasopharynx, and radioresistant represents the main obstacle in NPC treatment. Malignant transformation of normal cells is driven by genetic and epigenetic changes, which are primarily manifested as changes in miRNA levels and DNA methylation status. microRNA (miR)-613 plays an inhibitory role in several types of cancer. Herein, the current study sought to explore the roles of miR-613 in NPC cell radiosensitivity. miR-613 expression patterns in NPC tissues were detected, and its correlation with clinical indexes was analyzed. NP-69 and C666-1 cell lines were selected for cellular experimentation. Radioresistant cell line C666-1R was obtained by fractionated radiation. Cell viability, survival fraction, and apoptosis were detected by CCK-8, colony formation assay, and flow cytometry. The binding relation between miR-613 and DNMT3B was verified by dual-luciferase and RIP assays. miR-613 was lowly expressed in NPC tissues and cells, with lower expression levels in C666-1R than C666-1, and further correlated with lymph node metastasis, tumor size, and tumor metastasis. miR-613 overexpression reduced C666-1R cell viability and survival fraction and increased apoptosis, while C666-1 cells with silencing miR-613 presented the opposite trends. miR-613 targeted DNMT3B. miR-613 and DNMT3B overexpression led to enhanced C666-1R cell viability and survival fraction and decreased apoptosis. miR-613 reduced TIMP3 methylation and elevated TIMP3 protein level by inhibiting DNMT3B. miR-613 enhanced NPC radiosensitivity by inhibiting the DNMT3B/TIMP3/STAT1/FOXO1 pathway. Collectively, miR-613 inhibited DNMT3B, reduced TIMP3 methylation, and increased TIMP3 protein level, thus inhibiting the STAT1/FOXO1 pathway and enhancing the radiosensitivity of NPC cells.
1. Introduction
Nasopharyngeal carcinoma (NPC), defined as a head and neck cancer malignancy arising from the epithelium of the nasopharynx, is associated with bleak prognoses and increased lymphocyte infiltration [1, 2]. Interestingly, NPC has a dramatically skewed geographic distribution across the world, with significantly high incidence in East and Southeast Asia [3]. Epstein Barr virus represents the leading factor in the etiology of NPC; in addition, genetic factors and environmental and dietary causes can also precipitate NPC [4, 5]. Currently, radiotherapy is regarded as the gold-standard treatment for NPC [6]. However, the onset of radiation resistance poses a major hindrance in NPC treatment [7]. Accordingly, preventing radioresistance and increasing radiosensitivity are of the utmost importance for improving NPC patient prognosis and increasing 5-year survival rates [8]. In lieu of the same, it is imperative to expand on the potential mechanism of NPC radiosensitivity, provide novel effective strategies for clinical treatment, and prolong the survival time of patients with NPC.
microRNAs (miRNAs) are well-established as a group of small noncoding RNAs functioning in the posttranscriptional regulation of gene expression, also known for their crucial ability to regulate a variety of cellular activities, such as cell differentiation, growth, apoptosis, and development. Moreover, accumulating studies have indicated the association of miRNAs with various diseases, such that currently ongoing miRNA-mediated clinical trials have shown promising results in the treatment of viral infection and cancer [9]. As an important factor of posttranscriptional regulation, miRNAs are also capable of participating in ionizing radiation responses of cancer cells via the regulation of the cell cycle and apoptotic pathways [10–12]. What is more, hundreds of miRNAs have been discovered in diverse animals, and many of these are phylogenetically conserved [13]. miRNAs are 18-25 nucleotide long noncoding RNA molecules with the function of posttranscriptional gene regulation, which have attracted extensive attention as potential cancer regulators and biomarkers. Some miRNAs have tumor-suppressive or carcinogenic effects in oral carcinogenesis [14] and can be used as the biomarker for early oral cancer [15] and screening biomarkers for diagnosis and prognosis of colorectal carcinoma, sessile serrated lesions [16], and renal cell carcinoma patients [17]. Garnering our attention, miRNAs are associated with the malignant biological characteristics of cancer cells, including radiosensitivity [18, 19]. Furthermore, miRNA imbalances can contribute to tumor progression and metastasis. For instance, one such miRNA, namely, miR-613, confers an anticancer role in bladder cancer by targeting SphK1 [20]. In addition, miR-613 exhibits a tumor-suppressive role in several types of cancer [21]. One prior study uncovered that miR-613 plays an anticancer role in hepatocellular carcinoma by targeting tryptophan 5-monooxygenase activation protein [22]. Similarly, miR-613 was previously shown to inhibit proliferation, invasion, and tumor formation of retinoblastoma cells by targeting E2F transcription factor 5 [23]. Moreover, miR-613 can attenuate the Warburg effect in gastric cancer by targeting g6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 2 [24]. miR-613 targets sex-determining region Y-box 9 to play an antitumor role in glioma cells [25]. miR-613 inhibits the proliferation, migration, and invasion of thyroid papillary carcinoma cells by directly targeting transgelin 2 [26]. On the other hand, knockdown of miR-613 promotes the proliferation and metastasis of pancreatic cancer cells by facilitating Notch3 expression and regulating the Notch3 pathway [27]. Long noncoding RNA cytoskeleton regulator RNA has also been previously indicated to facilitate cell metastasis and invasion by titrating miR-613 in NPC [28]. Similarly, the family with sequence similarity 225 member B promotes the proliferation and metastasis of NPC via the miR-613/cyclin D2 axis [29]. Besides, miR-613 can further inactivate the AKT signal pathway by targeting fibronectin1, thus exerting antiangiogenic effects on NPC cells [30]. Together, these shreds of evidence highlight the potential involvement of miR-613 in the progression of NPC. However, whether miR-613 affects the radiosensitivity of NPC remains largely unknown.
The malignant transformation of normal cells is driven by genetic and epigenetic changes, and the dynamic regulation of DNA methylation serves as a key epigenetic mechanism for the occurrence, maintenance, and progression of cancer [31]. Moreover, epigenetic changes are primarily manifested as changes in miRNA levels and DNA methylation status [32]. DNA methyltransferase 3B (DNMT3B), as a major DNA methyltransferase, is known to function in de novo methylation of DNA, which exerts critical roles in various important biological functions, while also being involved in tumorigenesis, progression, and metastasis through DNA methylation [33–36]. What is noteworthy is that silencing DNMT3B restores and activates p53 and p21 through DNA demethylation, ensuing cell cycle arrest and apoptosis, thus improving the radiosensitivity of NPC [37]. Tissue inhibitor of matrix metalloproteinases-3 (TIMP3) is a special member of the TIMP family, which possesses the ability to regulate tumor growth, metastasis, angiogenesis, and other physiological progress through matrix metalloproteinases [38]. Furthermore, as an independent and promising biomarker, TIMP3 plays a regulatory role in a plethora of cancers [39–41] and is lowly expressed in NPC tissues [42]. In addition, TIMP3 methylation has been documented in several malignancies [34, 43, 44]. Strikingly, inhibition of DNMT3B reduces TIMP3 methylation levels and enhances TIMP3 protein levels, thus modulating the STAT1/FOXO1 pathway in breast cancer [34]. However, no studies have explored whether miR-613 regulates the DNMT3B/TIMP3/STAT1/FOXO1 pathway in the radiosensitivity of NPC cells. Accordingly, the current sets out to investigate the potential molecular mechanism of miR-613 in NPC cell radiosensitivity by targeting DNMT3B, affecting TIMP3 methylation level, and activating the STAT1/FOXO1 pathway.
2. Materials and Methods
2.1. Ethics Statement
The current study was approved by the ethics committee of the First People's Hospital of Chenzhou and performed in strict accordance with the code of ethics. Signed informed consents were obtained from all participants after a comprehensive understanding prior to specimen collection.
2.2. Sample Collection
A total of 121 pairs of adjacent normal tissues and tumor tissues from NPC patients (including 81 males and 40 females; aged 40-68 years with a mean calculated age of 51.3 ± 7.2 years) were collected at the First People's Hospital of Chenzhou from September 2016 to September 2020. The inclusion criteria [45] were as follows: (1) patients with NPC confirmed by biopsy and pathology; (2) patients who did not receive treatment prior to inclusion; (3) patients without distant metastasis; (4) patients receiving intensity-modulated radiotherapy; and (5) patient with complete clinical data. Patients not meeting any one of the above inclusion criteria were excluded and were not included for sample collection. In addition, none of the included patients received radiotherapy or chemotherapy before operation. According to the tumor, node, and metastasis (TNM) staging [46], there were 21 cases at stage I, 20 cases at stage II, and 80 cases at stages III and IV.
2.3. Cell Culture and Grouping
NP-69 and C666-1R cells were procured from ATCC (Manassas, VA, USA). C666-1R cells were prepared by in vitro fractionated radiation [47, 48]). All cell lines were cultured in RPMI-1640 medium (22400089, Gibco Company, Grand Island, NY, USA) supplemented with 10% fetal bovine serum and plated into 6-well plates (at a density of 1 × 105 cells/well), and subcultured in a humidified incubator with saturated humidity and 5% CO2 at 37°C.
Upon achieving 70-80% confluence, the C666-1R or C666-1 cells were plated into 6-well plates (at a density of 1 × 106 cells/well), cultured in a humidified incubator with saturated humidity and 5% CO2 at 37°C, and allocated and treated as according as follows: the NP-69 group, the C666-1 group, the C666-1R group, the C666-1R + miR mimi group (C666-1R transfected with 1 miR-613 mimics), the C666-1R + mimi NC group (C666-1R transfected with mimics NC), the C666-1 + miR inhi group (C666-1R transfected with miR-613 inhibitor), the C666-1 + inhi NC group (C666-1R transfected with inhibitor NC), the C666-1R + miR mimi + oe-DNMT3B group (C666-1R co-transfected with miR-613 mimics and oe-DNMT3B plasmid), the C666-1R + miR mimi + oe-NC group (C666-1R co-transfected with miR-613 mimics and oe-NC plasmid), the C666-1R + miR mimi+oe-DNMT3B + si-STAT1 group (C666-1R co-transfected with miR-613 mimics, oe-DNMT3B and si-STAT1 plasmid), and the C666-1R + miR mimi + oe-DNMT3B + si-NC group (C666-1R co-transfected with miR-613 mimics, oe-DNMT3B, and si-NC plasmid). Further experimentation was carried out after 12 h mimics NC, miR-613 mimics, oe-NC, oe-DNMT3B, si-NC, and si-STAT1 were transfected into cells using the Lipofectamine 2000 reagent (Invitrogen, Burlington, ON, Canada). The final transfection concentration was 100 nM. All the above-utilized plasmids were purchased from the Life Technologies Corporation.
2.4. Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
The total RNA content was extracted from 100 mg frozen tissues or cells using the TRIzol reagent (12183555, Thermo Fisher Scientific Inc., NY, USA) in a one-step manner. cDNA was synthesized by means of reverse transcription according to the instructions of PrimeScript RT-PCR kits (Takara, Tokyo, Japan). RT-qPCR was performed with SYBR FAST qRT-PCR Master Mix kits (Invitrogen). PCR reaction conditions were as follows: pre-denaturation for 15 min at 95°C, and 40 cycles of denaturation for 30 s at 94°C, annealing for 30 s at 55°C, and extension for 30 s at 72°C. U6 was adopted as the internal control for miR-613, and GAPDH was utilized as the internal control for DNMT3B, respectively. Following amplification, the dissolution curve was analyzed, and gene expression in each group was calculated using the 2-ΔΔCt method. Primer sequences are shown in Table 1.
Table 1.
PCR primer sequences.
| Gene | Primer sequences (5′-3′) |
|---|---|
| miR-613 | F 5′-AGGAATGTTCCTTCTTTGCC-3′ |
| R 5′-CAGTGCGTGTCGTGGAGT-3′ | |
| DNMT3B | F 5′-GTCAGGCGCTGACATAGTGA-3′ |
| R 5′-TCATGACTGGCAGAGGACAG-3′ | |
| GAPDH | F 5′-GGTGAAGGTCGGAGTCAACGG-3′ |
| R 5′-CCTGGAAGATGGTGATGGGATT-3′ | |
| U6 | F 5′-CTCGCTTCGGCAGCACA-3′ |
| R 5′-AACGCTTCACGAATTTGCGT-3′ |
2.5. Western Blot Analysis
C666-1 and C666-1R cell homogenates were added with protein lysate (R0278, Sigma-Aldrich, St. Louis, MO, USA) and centrifuged at a rate of 12000 r/min for 20 min at 4°C. Next, the supernatant was subpacked, and total protein concentration in homogenates was detected. Subsequently, 10% sodium dodecyl sulfate separating gel and stacking gel were prepared. Following mixing with the loading buffer, the protein samples were boiled for 5 min at 100°C. Afterwards, the proteins were placed in an ice bath, centrifuged, and equally added to each lane using a micropipette for electrophoresis. Thereafter, the proteins on the gel were transferred onto nitrocellulose membranes, which were blocked with 5% skim milk overnight at 4°C and probed with anti-DNMT3B (ab79822, dilution ratio of 1 : 1000, Abcam Inc., Cambridge, MA, USA), anti-FOXO1 (ab52857, dilution ratio of 1 : 1000, Abcam), anti-STAT1 (ab109461, dilution ratio of 1 : 1000, Abcam), and anti-GAPDH (ab9485, dilution ratio of 1 : 1000, Abcam) at room temperature for 1 h. Later, the membranes were rinsed 3 times with phosphate-buffered saline (PBS) at room temperature, 5 min each. After that, the secondary antibody goat antirabbit IgG H&L (HRP) (ab6721, dilution ratio of 1 : 2000, Abcam) was added for 1-h incubation at room temperature, followed by 3 PBS rinses at room temperature, 5 min each. Afterwards, the membranes were immersed in enhanced chemiluminescence solution (Pierce, Waltham, MA, USA) for 1 min at room temperature. The liquid was absorbed, and the membranes were covered with plastic film and exposed to X-ray film for observation. The gray value ratio of the target band to the internal reference band was regarded as the relative protein expression, with GAPDH serving as the internal reference.
2.6. Cell Counting Kit-8 (CCK-8) Assay
Cells (at a density of 5 × 103 cells/mL) were cultured in 96-well plates, with 6 duplicate wells set for each group, and each well was added with 100 μL cell culture medium. After 24 h, cells in the plates well adhered to the wall and then were irradiated with 6-MV X-ray using an accelerator (Elekta Instruments AB, Stockholm, Sweden) at a source-skin distance of 100 cm and a dose rate of 500 cGy/min. The irradiation doses of cells were set to 0, 2, 4, and 8 Gy, respectively. After irradiation, the cells were cultured in an incubator (51020241, Thermo Fisher) for 48 h. Afterwards, 100 μL medium and 10 μL CCK-8 reagent (C0037, Beyotime, Shanghai, China) were successively added to each well under the provided instructions with the CCK-8 kits. After 1–2-h incubation at 37°C, optical density (OD) values at 450 nm were detected using a microplate reader (Multiskan FC, Thermo Fisher). Three parallel wells were set for each group, with the mean value recorded. The cell proliferation curve was plotted with time as the abscissa and OD value as the ordinate.
2.7. Colony Formation Assay
Cells were cultured overnight on 6-well plates at a density of 1 × 106 and then treated with 4 Gy irradiation. After 10 days, the cells were fixed with 4% paraformaldehyde, stained with Giemsa stain kit (DM0002, LEAGE, Beijing Leagene Biotechnology, Beijing, China), and then counted under a microscope (CX23, Olympus, Tokyo, Japan).
2.8. Flow Cytometry
Cells were rinsed three times with PBS and suspended in the buffer and then subjected to staining with propidium iodide containing 50 μg/mL RNaseA (Sigma-Aldrich) and fluorescein isothiocyanate-AnnexinV (APOAF, Sigma-Aldrich). A FACScan flow cytometer (Beckman Coulter, Fullerton, CA, USA) was utilized for flow cytometric analysis. Data were analyzed using the FlowJo software (Tree Star, San Carlos, CA, USA).
2.9. Dual-Luciferase Reporter Gene Assay
The binding sites between miR-613 and DNMT3B were predicted and analyzed with the help of the StarBase database (http://starbase.sysu.edu.cn/index.php). Wild-type or mutant plasmids of DNMT3B (wt-DNMT3B/mut-DNMT3B) were constructed using pmirGLO vectors (GenePharma, Shanghai, China) and subsequently co-transfected with miR-613 mimic or miR-NC into C666-1 cells in strict accordance with the instructions of the Lipofectamine 2000 reagent (11668, Invitrogen). Luciferase activity was detected 48 h after transfection with the help of a dual-luciferase detection system (Promega, Madison, WI, USA).
2.10. Quantitative Real-Time Methylation-Specific PCR (qMSP)
DNA content was extracted from cells under provided instructions of the QIAamp DNA Mini Kit (51304, Qiagen, Hilden, Germany). Bisulfite modification was subsequently carried out in strict accordance with the manufacturer's instructions (Qiagen). DNA sequences were methylated (M) or unmethylated (U) with bisulfite-modified TIMP3, and the methylation level of TIMP3 in cells was detected by qMSP. TIMP3 methylated or unmethylated primer sequences are illustrated in Table 2 [34].
Table 2.
TIMP3 methylated (M) or unmethylated (U) primer sequences.
| Primer type | Primer sequences |
|---|---|
| M | Forward: 5′-TCGAGAGATAGAAATATTTTTACGA-3′ |
| Reverse: 5′-TAACATTAAAACAACAAAAACCGAA-3′ | |
| U | Forward: 5′-TTGAGAGATAGAAATATTTTTATGA-3′ |
| Reverse: 5′-TAACATTAAAACAACAAAAACCAAA-3′ |
2.11. Statistical Analysis
Data were processed and mapped using the GraphPad Prism 8.0.1 software (GraphPad Software Inc., San Diego, CA, USA). A minimum of three independent cell experiments were carried out per group. Measurement data were presented as mean ± standard deviation (SD). The t-test or repeated variance test was utilized for comparisons between two groups, and one-way analysis of variance (ANOVA) was adopted for comparisons between multiple groups, along with Tukey's multiple comparisons test. A value of p < 0.05 was regarded as statistically significant.
3. Results
3.1. miR-613 Was Poorly Expressed in NPC Tissues and Cells and Associated with NPC Radiosensitivity
Collection and subsequent RT-qPCR analyses illustrated that the clinical samples of 121 cases of NPC patients presented decreased expression levels of miR-613 relative to adjacent normal tissues (p < 0.01) (Figure 1(a)). To further elucidate the correlation between the expression of miR-613 and NPC patient clinicopathological characteristics and prognosis, the 121 NPC patients were assigned into the high expression group and low expression groups according to the median value of miR-613 expression in tumor tissues. There were no significant correlations between the expression of miR-613 and gender, age, smoking history, and clinical stages, whereas there were significant correlations between miR-613 expression and N classification, T classification, and distant metastasis (all p < 0.01) (Table 3), which underscored the involvement of miR-613 in the progression of NPC.
Figure 1.
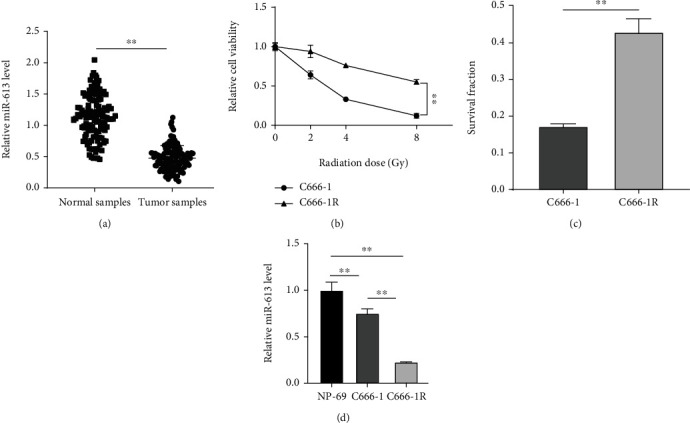
miR-613 was lowly expressed in NPC tissues and cell lines and was related to radiosensitivity. (a) miR-613 expression in adjacent normal tissues and tumor tissues of the 121 cases of NPC patients was detected using RT-qPCR; NP-69, NPC cell line C666-1, and the constructed radiation tolerant strain C666-1R were selected for the in vitro study. (b) Cell viability of NPC cells after radiation treatment was detected using the CCK-8 method. (c) Cell survival fraction was detected by colony formation assay. (d) miR-613 expression was detected using RT-qPCR. Three repeated tests were performed. Data were expressed as mean ± SD. T-test was used for comparisons between 2 groups in panels (a) and (c), repeated variance test was applied for comparisons between 2 groups in panel (b), and one-way-ANOVA was used for comparisons among multi-groups in panel (d), followed by Tukey's multiple comparisons test. ∗∗p < 0.01.
Table 3.
Correlation between NPC patient clinical features and miR-613 expression.
| Characteristics | Expression of miR-613 (%) | p | ||
|---|---|---|---|---|
| High expression | Low expression | |||
| (N = 60) | (N = 61) | |||
| Gender | Male | 35 (46.1%) | 41 (53.9%) | 0.3504 |
| Female | 25 (55.6%) | 20 (44.4%) | ||
| Age (y) | ≥ 50 | 36 (58.1%) | 26 (41.9%) | 0.0696 |
| < 50 | 24 (40.7%) | 35 (59.3%) | ||
| Smoking | Yes | 25 (61.0%) | 16 (39.0%) | 0.0858 |
| No | 35 (43.8%) | 45 (56.2%) | ||
| T classification | T1-T2 | 36 (39.1%) | 56 (60.9%) | < 0.0001 |
| T3-T4 | 24 (82.8%) | 5 (17.2%) | ||
| N classification | N0-N1 | 32 (62.7%) | 19 (37.3%) | 0.0169 |
| N2-N3 | 28 (40.0%) | 42 (60.0%) | ||
| Distant metastasis | Yes | 12 (29.3%) | 29 (70.7%) | 0.002 |
| No | 48 (60.0%) | 32 (40.0%) | ||
| Clinical stage | I-II | 26 (63.4%) | 20 (36.6%) | 0.2643 |
| III-IV | 34 (48.8%) | 41 (51.2%) | ||
Accumulating studies have further suggested the usage of miR-613 as an indicator of the radiosensitivity of cancer cells [49]. To investigate whether miR-613 could exert a similar role in the radiosensitivity of NPC, NPC cell line C666-1 was selected for in vitro experimentation, and the radioresistant NPC cell line C666-1R was obtained by in vitro fractionated radiation. CCK-8 was further carried out to detect the cell viability changes in C666-1 and C666-1R cells treated with radiation at the dose of 0, 2, 4, and 8 Gy. With the increase in radiation doses, the cell viability was significantly decreased in both C666-1 < C666-1R cells, and the change was the largest following 4 Gy irradiation (p < 0.01) (Figure 1(b)). In addition, C666-1R cells exhibited a higher survival fraction than C666-1 cells under 4 Gy radiation (p < 0.01) (Figure 1(c)). Furthermore, miR-613 level patterns in cells were detected using RT-qPCR, which revealed that compared with NP-69 cells, miR-613 was down-regulated in C666-1 and C666-1R cells; and the expression in tolerant C666-1R cells was relatively lower than that of the C666-1 cells (p < 0.01) (Figure 1(d)). Together, these findings highlighted that miR-613 might be involved in the regulation of NPC cell radiosensitivity.
3.2. Overexpression of miR-613 Improved NPC Cell Radiosensitivity
To further elaborate on the role of miR-613 in NPC cell radiosensitivity, we transfected C666-1R cells with miR-613 mimics to overexpress miR-613 expression and transfected C666-1 cells with miR-613 inhibitor to down-regulate miR-613 expression (p < 0.01) (Figure 2(a)). Subsequent experimentation demonstrated that C666-1R cell viability was decreased under 0, 2, 4, and 8 Gy radiation after miR-613 overexpression, the surviving fraction was decreased, and the apoptosis rate was increased under 4 Gy radiation (all p < 0.01); meanwhile, C666-1 cell viability was increased under 0, 2, 4, and 8 Gy radiation after miR-613 down-regulation, and the surviving fraction was increased, and the apoptosis rate was decreased under 4 Gy radiation (Figures 2(b)–2(d)). Overall, these findings indicated that overexpression of miR-613 exerted an enhancing effect on NPC cell radiosensitivity.
Figure 2.
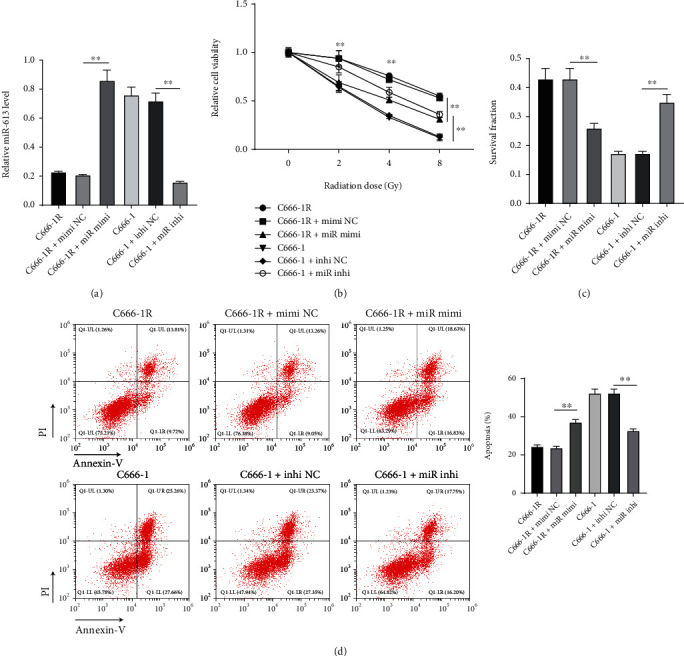
Overexpression of miR-613 increased NPC cell radiosensitivity. (a) miR-613 expression in cells was detected using RT-qPCR. (b) Cell viability was detected using the CCK-8 method. (c) Cell survival fraction was detected by colony formation assay. (d) Cell apoptosis was detected by flow cytometry. Three repeated tests were performed. Data were expressed as mean ± SD and analyzed using one-way ANOVA and Tukey's multiple comparisons test. ∗p < 0.05, ∗∗p < 0.01.
miR-613 increased NPC radiosensitivity by targeting DNMT3B.
DNMT3B was associated with NPC radiosensitivity [37]. Further detection illustrated that DNMT3B was highly expressed in NPC tissues (Figure 3(a), p < 0.01), and negatively correlated with miR-613 expression (Figure 3(b)). Subsequently, the targeted binding sites between miR-613 and DNMT3B were predicted using the StarBase database and verified with a dual-luciferase assay (Figure 3(c)). Moreover, the results of Western blot elicited higher protein levels of DNMT3B in C666-1R cell line relative to those in NP-69 and C666-1 cells (p < 0.01) (Figure 3(d)). After miR-613 overexpression, the protein levels of DNMT3B in C666-1R cells were diminished; whereas after miR-613 knockdown, the protein levels of DNMT3B in C666-1 cells were enhanced (Figure 3(e), p < 0.01). These findings suggested that miR-613 was capable of targeting DNMT3B. Subsequently, C666-1R cells were co-transfected with miR-613 mimics and oe-DNMT3B, followed by analyses of cell radiosensitivity. Subsequent results demonstrated augmented protein levels of DNMT3B in C666-1R cells (Figure 3(e), p < 0.05). Under 0, 2, 4, and 8 Gy radiation, cell viability was increased, cell surviving fraction under 4 Gy radiation was increased, and apoptosis was reduced (all p < 0.05) (Figures 3(f)–3(h)). Altogether, these findings indicated that miR-613 enhanced NPC radiosensitivity via inhibition of DNMT3B.
Figure 3.
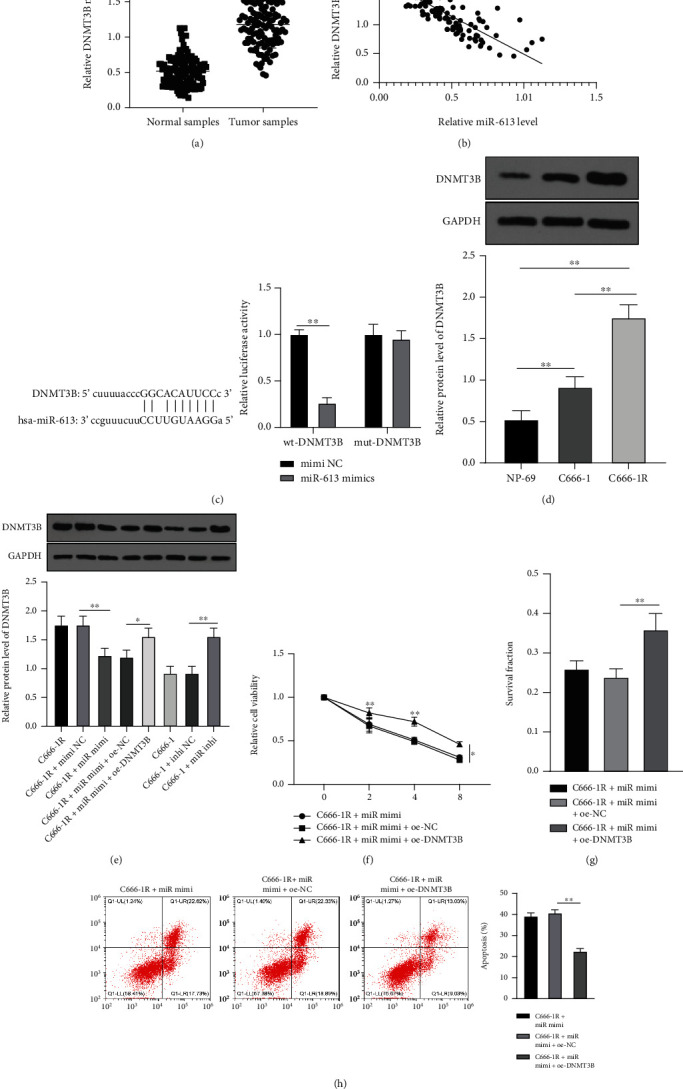
miR-613 increased NPC cell radiosensitivity by inhibiting DNMT3B. (a) mRNA levels of DNMT3B in cancer tissues and adjacent tissues of 121 NPC patients were detected by RT-qPCR. (b) Correlation between DNMT3B and miR-613. (c) Binding relationship between DNMT3B and miR-613 was verified by dual-luciferase assay. (d–e) Protein level of DNMT3B in cells was detected by Western blot. (f) Cell viability of C666-1R cells was detected using the CCK-8 method. (g) Cell clone formation and apoptosis in C666-1R cells were detected by colony formation and (h) flow cytometry. Three repeated tests were performed. Data were expressed as mean ± SD, and data in panels (a) and (c) were analyzed using independent t test; data in panels (d), (e), (g), and (h) were analyzed using one-way ANOVA, followed by Tukey's test; and data in panel (f) were analyzed using repeated variance test. ∗p < 0.05, ∗∗p < 0.01.
3.3. miR-613 Reduced TIMP3 Methylation by Inhibiting DNMT3B
TIMP3 was previously established to be lowly expressed in NPC tissues [42], and TIMP3 methylation is a common occurrence in many cancers [34, 43, 44]. DNMT3B further represents a major DNA methyltransferase regulated by miR-613. To explore whether miR-613 reduced the methylation level of TIMP3 in NPC cells by inhibiting DNMT3B expression, TIMP3 methylation level and its protein level patterns were detected in cells by qMSP and Western blot. Subsequent results illustrated that compared with NP-69 cells, TIMP3 methylation levels were elevated, and TIMP3 protein levels were suppressed in C666-1 and C666-1R cells; meanwhile, compared with C666-1 cells, TIMP3 methylation levels were enhanced, and TIMP3 protein levels were diminished in C666-1R cells, whereas after overexpression of miR-613, TIMP3 methylation levels were decreased, and TIMP3 protein levels were elevated in C666-1R cells. After further overexpression of miR-613 and overexpression of DNMT3B, TIMP3 methylation levels were enhanced, and TIMP3 protein levels were repressed in C666-1R cells (Figures 4(a) and 4(b), p < 0.01). Altogether, these findings highlighted that miR-613 reduced TIMP3 methylation levels by inhibiting the expression of DNMT3B.
Figure 4.
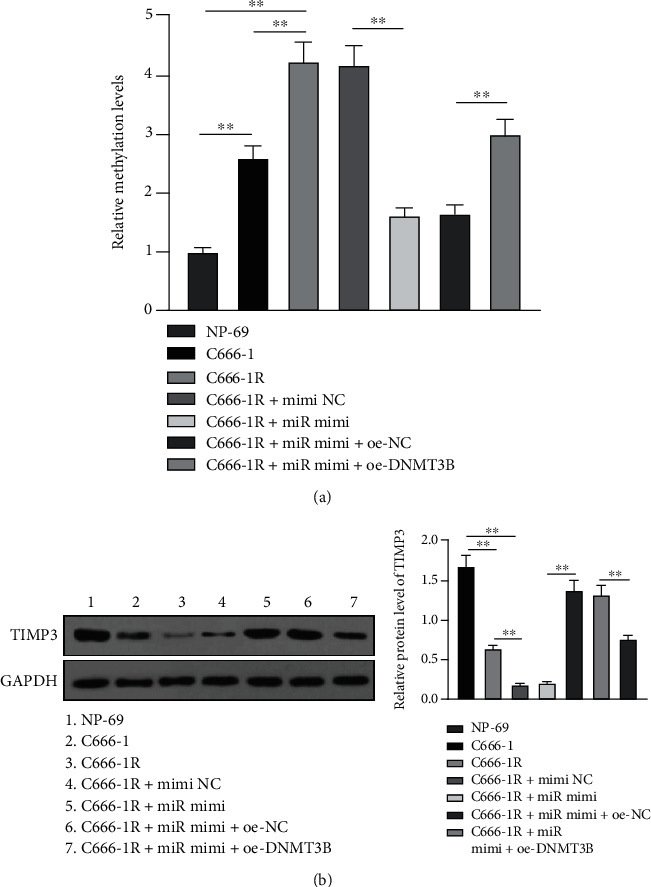
miR-613 reduced TIMP3 methylation by inhibiting DNMT3B. (a) The methylation level of TIMP3 in cells was detected by qMSP. (b) The protein level of TIMP3 was detected by Western blot. Cell experiment was repeated 3 times, and the data were expressed as mean ± SD and analyzed using one-way ANOVA, followed by Tukey's multiple comparisons test. ∗∗p < 0.01.
3.4. miR-613 Enhanced Radiosensitivity by Inhibiting the DNMT3B/TIMP3/STAT1/FOXO1 Pathway
DNMT3B is known to influence breast cancer development via regulation of the STAT1/FOXO1 pathway [34]. Moreover, STAT1 exerts a regulatory effect on the radiosensitivity of a variety of cancer cells [50–52]. To further explore whether miR-613 participated in the regulation of NPC radiosensitivity by inhibiting DNMT3B/TIMP3 and diminishing the activation of the STAT1/FOXO1 pathway, the protein level patterns of STAT1 and FOXO1 in NP-69, C666-1, and C666-1R were detected by Western blot. Subsequent results demonstrated that compared with NP-69 cells, the protein levels of STAT1 in C666-1 and C666-1R cells were increased, with C666-1R presenting with higher STAT1 levels than those in C666-1, and the protein levels of FOXO1 were decreased, with C666-1R presenting with lower STAT1 levels than those in C666-1 (Figure 5(a), p < 0.01). Moreover, C666-1R cells were co-transfected with miR-613 mimic, oe-DNMT3B, and si-STAT1. It was found that STAT1 levels were noticeably reduced, and FOXO1 levels were markedly elevated in C666-1R cells after miR-613 overexpression, while these trends were reversed in CNE-2R cells after miR-613 overexpression and DNMT3B overexpression. Lastly, STAT1 protein levels were significantly decreased, and FOXO1 protein levels were enhanced in C666-1R cells after transfection with miR-613 mimics, oe-DNMT3B, and si-STAT1, whereas C666-1R presented with decreased cell viability, reduced cell surviving fraction under 4 Gy radiation, and increased cell apoptosis under 2, 4, and 8 Gy radiation (Figures 5(a)–5(d), p < 0.05). Altogether, these findings highlighted that miR-613 increased the radiosensitivity of NPC cells by inhibiting the DNMT3B/TIMP3/STAT1/FOXO1 pathway.
Figure 5.
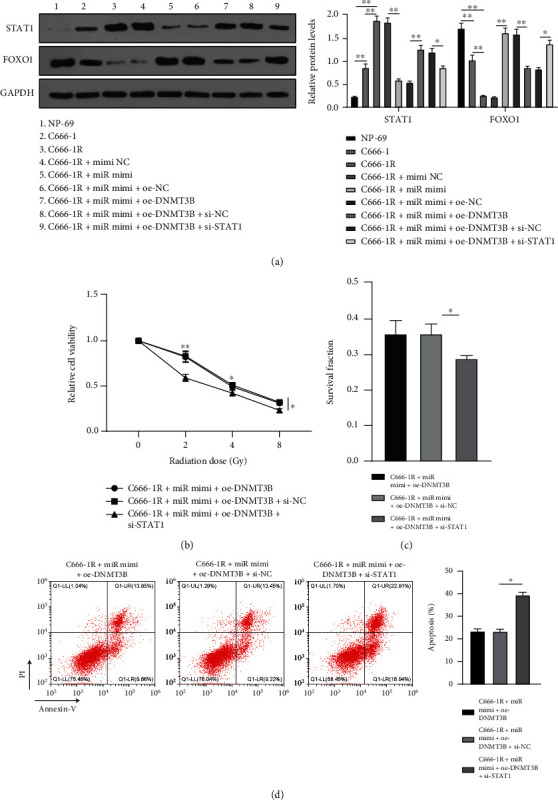
miR-613 enhanced NPC radiosensitivity by inhibiting the DNMT3B/TIMP3/STAT1/FOXO1 pathway. (a) Protein levels of STAT1 and FOXO1 in cells were detected by Western blot. (b) Cell viability changes of cells were detected using the CCK-8 method. (c) Cell survival fraction of cells was detected by cell colony formation assay. (d) Cell apoptosis was detected by flow cytometry. Three independent repeated tests were performed. Data were expressed as mean ± SD and analyzed using one-way ANOVA and Tukey's multiple comparisons test. ∗p < 0.05, ∗∗p < 0.01.
4. Discussion
NPC represents a common head and neck malignancy arising from the epithelium of the nasopharynx, associated with low survival rates in certain endemic areas [1]. Meanwhile, the last decade has witnessed the emergence of miRNAs as key regulators in diverse biological functions and disease progression [53]. Herein, the current study set out to elucidate the potential roles of miR-613 in NPC cell radiosensitivity, and our findings highlighted that miR-613 inhibited DNMT3B, reduced TIMP3 methylation, and increased TIMP3 protein level, thereby inhibiting the downstream STAT1/FOXO1 pathway and augmenting the radiosensitivity of NPC cells.
Our study is not the first of its kind to suggest the involvement of miRNA regulation of radiosensitivity in NPC cells [54]. Initial findings in our study demonstrated that miR-613 was poorly expressed in NPC tissues, and the aberrant expression of miR-613 was associated with N classification, T classification, and distant metastasis. Additional experimentation with increasing radiation doses revealed that there was a significant decrease in the cell viability of parental NPC cell line C666-1 and the radioresistant cell line C666-1R, and the change was the largest under 4 Gy irradiation, and C666-1 < C666-1R; meanwhile, C666-1R exhibited a higher survival fraction than C666-1 under 4 Gy radiation. Compared with NP-69 cells, miR-613 expression levels were down-regulated in C666-1 and C666-1R cells, while relative to the C666-1 cells, miR-613 expression levels were also diminished in the radiotolerant C666-1R cells. In accordance with our data, previous studies have documented that miR-613 expression is lowered in NPC cell lines and tissues [29]. Similarly, miR-613 was previously reported to exert effects on the cell radiosensitivity of cancers, such as nonsmall cell lung cancer, while also being implicated in cisplatin resistance of glioma [49, 55]. Altogether, these findings and evidence conclusively highlight the involvement of miR-613 in NPC cell radiosensitivity regulation.
Accumulating reports have further indicated the use of miR-613 as an indicator of the radiosensitivity of cancer cells [49]. To further elaborate on the role of miR-613 in NPC cell radiosensitivity, we overexpressed miR-613 expression in C666-1R cells and knocked down miR-613 expression in C666-1 cells. Subsequent experimentation demonstrated that following overexpression of miR-613, C666-1R cell viability was reduced under 0, 2, 4, and 8 Gy radiation, and the surviving fraction was repressed, and apoptosis rate was elevated under 4 Gy radiation, whereas C666-1 cells with silenced miR-613 presented with the opposite trends. Consistently, a prior report suggested that hepatoma cells with overexpressed miR-613 were more sensitive to sorafenib or cisplatin treatment [56]. Together, our findings in conjunction with existing evidence highlight that overexpression of miR-613 augments NPC cell radiosensitivity.
Another key component of our study, the DNMT3B gene is known to play roles in tumorigenesis, metastasis, progression, and NPC cell radiosensitivity [33, 35–37]. Further detection revealed that DNMT3B was highly expressed in NPC tissues and negatively correlated with miR-613. We further validated the targeting relationship between miR-613 and DNMT3B using a dual-luciferase assay and came across higher DNMT3B protein levels in C666-1R cell line relative to NP-69 and C666-1 cells. In addition, overexpression of miR-613 led to diminished DNMT3B protein levels in C666-1R cells, while knockdown of miR-613 brought about an increase in DNMT3B protein levels in C666-1 cells, which underscores that miR-613 targeted DNMT3B. In accordance with our data, elevated DNMT3B expression in NPC tissues has been reported before [37]. Furthermore, we verified the role of the miR-613/DNMT3B axis in NPC cell radiosensitivity that after co-transfection with miR-613 mimics and oe-DNMT3B, there was an increase in DNMT3B protein levels in C666-1R cells. Meanwhile, under 0, 2, 4, and 8 Gy radiation, cell viability was augmented, survival fraction was stimulated, and apoptosis was reduced under 4 Gy radiation. Consistently, DNMT3B induced by radiation was previously indicated to promote NPC radioresistance through methylation of p21 and p53 [37]. Likewise, another study indicated that overexpression of DNMT3B contributes to radiotherapy failure, such that its inhibition represents a promising radiosensitizing strategy for patients with metastatic or recurrent rhabdomyosarcoma tumors [57]. Exosomal miR-613 also exerts an inhibitory effect on lung cancer growth, while promoting the sensitivity to cisplatin [58]. Silencing has-circ0000199 enhanced breast cancer chemosensitivity by promoting miR-613 expression [59]. Overall, these findings and data indicate that miR-613 enhances NPC radiosensitivity by targeting DNMT3B.
Furthermore, various novel studies have come across TIMP3 methylation in a myriad of cancers [34, 43, 44]. Herein, our results illustrated that TIMP3 methylation levels were augmented and TIMP3 protein levels were suppressed in C666-1 and C666-1R cells, with C666-1R cells showing the more obvious alteration; meanwhile, after miR-613 overexpression, TIMP3 methylation levels were suppressed, and TIMP3 protein levels were facilitated in C666-1R cells, whereas further DNMT3B overexpression reversed the above trends in C666-1R cells. Consistent lowly expressed TIMP3 was previously reported in NPC tissues [42], while knockdown of TIMP3 methylation was associated with alleviation of breast cancer [34]. In short, these findings and data make it plausible to suggest that miR-613 reduces TIMP3 methylation levels by inhibiting DNMT3B.
The last focus of our study, STAT1, is widely established to regulate cell radiosensitivity in various cancers [50–52]. To investigate whether miR-613 regulated NPC radiotherapy by inhibiting DNMT3B/TIMP3 and the STAT1/FOXO1 pathway, we detected STAT1 and FOXO1 protein levels in NP-69, C666-1, and C666-1R cells, and discovered that STAT1 protein levels were promoted and FOXO1 protein levels were decreased in C666-1 and C666-1R cells, with C666-1R cells showing the more obvious alteration. Moreover, STAT1 levels were markedly diminished, and FOXO1 levels were augmented in C666-1R cells after miR-613 overexpression, which were reversed after further DNMT3B overexpression, and restored by further silencing STAT1, in addition to decreased cell viability and surviving fraction and enhanced cell apoptosis in C666-1R cells. Consistently, STAT1 silencing has previously been shown to sensitize NPC cell line CNE-2R radioresistant to radiotherapy [60]. Moreover, human glioma cell radiosensitivity can similarly be enhanced by LITAF via the FOXO1 pathway [61]. Unsurprisingly, there is prior evidence to suggest that DNMT3B affects the progression of breast cancer by targeting the STAT1/FOXO1 pathway [62]. Altogether, these findings illustrated that miR-613 diminishes the radiosensitivity of NPC cells by inhibiting the DNMT3B/TIMP3 via activation of the STAT1/FOXO1 pathway for the first time.
In summary, the current study highlighted that miR-613 inhibited DNMT3B, reduced TIMP3 methylation, and increased TIMP3 protein level, thus inhibiting the downstream STAT1/FOXO1 pathway and enhancing the radiosensitivity of NPC cells. However, we only carried out the relevant mechanism research in NPC cell line, and our findings warrant in vivo validation experiments in future studies.
Contributor Information
Liqiang Deng, Email: klsgn99xz50@163.com.
Debao Luo, Email: debaoluo1208@163.com.
Data Availability
The data used to support the findings of this study are available from the corresponding author upon request.
Conflicts of Interest
All authors declare that there is no conflict of interests in this study.
Authors' Contributions
Liqiang Deng and Qing Yin contributed equally to this work. All authors have contributed significantly, and all authors agree with the content of the manuscript.
References
- 1.Paul P., Deka H., Malakar A. K., Halder B., Chakraborty S. Nasopharyngeal carcinoma: understanding its molecular biology at a fine scale. European Journal of Cancer Prevention . 2018;27(1):33–41. doi: 10.1097/CEJ.0000000000000314. [DOI] [PubMed] [Google Scholar]
- 2.Zhang L., MacIsaac K. D., Zhou T., et al. Genomic analysis of nasopharyngeal carcinoma reveals TME-based subtypes. Molecular Cancer Research . 2017;15(12):1722–1732. doi: 10.1158/1541-7786.MCR-17-0134. [DOI] [PubMed] [Google Scholar]
- 3.Chen Y. P., Chan A. T. C., Le Q. T., Blanchard P., Sun Y., Ma J. Nasopharyngeal carcinoma. The Lancet . 2019;394(10192):64–80. doi: 10.1016/S0140-6736(19)30956-0. [DOI] [PubMed] [Google Scholar]
- 4.Petersson F. Nasopharyngeal carcinoma: a review. Seminars in Diagnostic Pathology . 2015;32(1):54–73. doi: 10.1053/j.semdp.2015.02.021. [DOI] [PubMed] [Google Scholar]
- 5.Roy Chattopadhyay N., Das P., Chatterjee K., Choudhuri T. Higher incidence of nasopharyngeal carcinoma in some regions in the world confers for interplay between genetic factors and external stimuli. Drug discoveries & therapeutics . 2017;11(4):170–180. doi: 10.5582/ddt.2017.01030. [DOI] [PubMed] [Google Scholar]
- 6.Sun X. S., Li X. Y., Chen Q. Y., Tang L. Q., Mai H. Q. Future of radiotherapy in nasopharyngeal carcinoma. The British Journal of Radiology . 2019;92(1102):p. 20190209. doi: 10.1259/bjr.20190209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tan Z., Xiao L., Tang M., et al. Targeting CPT1A-mediated fatty acid oxidation sensitizes nasopharyngeal carcinoma to radiation therapy. Theranostics . 2018;8(9):2329–2347. doi: 10.7150/thno.21451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gui S. J., Ding R. L., Wan Y. P., et al. Knockdown of annexin VII enhances nasopharyngeal carcinoma cell radiosensitivity in vivo and in vitro. Cancer Biomarkers . 2020;28(2):129–139. doi: 10.3233/CBM-190739. [DOI] [PubMed] [Google Scholar]
- 9.Saliminejad K., Khorram Khorshid H. R., Soleymani Fard S., Ghaffari S. H. An overview of microRNAs: biology, functions, therapeutics, and analysis methods. Journal of Cellular Physiology . 2019;234(5):5451–5465. doi: 10.1002/jcp.27486. [DOI] [PubMed] [Google Scholar]
- 10.Kang M., Xiao J., Wang J., et al. MiR-24 enhances radiosensitivity in nasopharyngeal carcinoma by targeting SP1. Cancer Medicine . 2016;5(6):1163–1173. doi: 10.1002/cam4.660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lin T., Zhou F., Zhou H., Pan X., Sun Z., Peng G. MicroRNA-378g enhanced radiosensitivity of NPC cells partially by targeting protein tyrosine phosphatase SHP-1. International Journal of Radiation Biology . 2015;91(11):859–866. doi: 10.3109/09553002.2015.1096028. [DOI] [PubMed] [Google Scholar]
- 12.Zhao L., Tang M., Hu Z., et al. miR-504 mediated down-regulation of nuclear respiratory factor 1 leads to radio-resistance in nasopharyngeal carcinoma. Oncotarget . 2015;6(18):15995–16018. doi: 10.18632/oncotarget.4138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ambros V. The functions of animal microRNAs. Nature . 2004;431(7006):350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 14.Min A., Zhu C., Peng S., Rajthala S., Costea D. E., Sapkota D. MicroRNAs as important players and biomarkers in oral carcinogenesis. BioMed Research International . 2015;2015:10. doi: 10.1155/2015/186904.186904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Crimi S., Falzone L., Gattuso G., et al. Droplet digital PCR analysis of liquid biopsy samples unveils the diagnostic role of hsa-miR-133a-3p and hsa-miR-375-3p in oral cancer. Biology . 2020;9(11):p. 379. doi: 10.3390/biology9110379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Peruhova M., Peshevska-Sekulovska M., Krastev B., et al. What could microRNA expression tell us more about colorectal serrated pathway carcinogenesis? World Journal of Gastroenterology . 2020;26(42):6556–6571. doi: 10.3748/wjg.v26.i42.6556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Spadaccino F., Gigante M., Netti G. S., et al. The ambivalent role of miRNAs in carcinogenesis: involvement in renal cell carcinoma and their clinical applications. Pharmaceuticals . 2021;14(4):p. 322. doi: 10.3390/ph14040322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Di Leva G., Garofalo M., Croce C. M. MicroRNAs in cancer. Annual Review of Pathology . 2014;9(1):287–314. doi: 10.1146/annurev-pathol-012513-104715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xie F., Xiao W., Tian Y., Lan Y., Zhang C., Bai L. MicroRNA-195-3p inhibits cyclin dependent kinase 1 to induce radiosensitivity in nasopharyngeal carcinoma. Bioengineered . 2021;12(1):7325–7334. doi: 10.1080/21655979.2021.1979356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu H., Duan P., Zhu H., Rao D. miR-613 inhibits bladder cancer proliferation and migration through targeting SphK1. American Journal of Translational Research . 2017;9(3):1213–1221. [PMC free article] [PubMed] [Google Scholar]
- 21.Ding D., Hou R., Gao Y., Feng Y. miR-613 inhibits gastric cancer progression through repressing brain derived neurotrophic factor. Experimental and Therapeutic Medicine . 2018;15(2):1735–1741. doi: 10.3892/etm.2017.5546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jiang X., Wu J., Zhang Y., et al. MiR-613 functions as tumor suppressor in hepatocellular carcinoma by targeting YWHAZ. Gene . 2018;659:168–174. doi: 10.1016/j.gene.2018.03.036. [DOI] [PubMed] [Google Scholar]
- 23.Zhang Y., Zhu X., Zhu X., et al. MiR-613 suppresses retinoblastoma cell proliferation, invasion, and tumor formation by targeting E2F5. Tumour Biology . 2017;39(3):p. 1010428317691674. doi: 10.1177/1010428317691674. [DOI] [PubMed] [Google Scholar]
- 24.Liu H., Chen K., Wang L., et al. miR-613 inhibits Warburg effect in gastric cancer by targeting PFKFB2. Biochemical and Biophysical Research Communications . 2019;515(1):37–43. doi: 10.1016/j.bbrc.2019.05.001. [DOI] [PubMed] [Google Scholar]
- 25.Sang Q., Liu X., Sun D. Role of miR-613 as a tumor suppressor in glioma cells by targeting SOX9. Oncotargets and Therapy . 2018;11:2429–2438. doi: 10.2147/OTT.S156608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang Y., Zhang H., Wang L., et al. MiR-613 inhibits the proliferation, migration, and invasion of papillary thyroid carcinoma cells by directly targeting TAGLN2. Cancer Cell International . 2021;21(1):p. 494. doi: 10.1186/s12935-021-02083-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cai H., Yao J., An Y., et al. LncRNA HOTAIR acts a competing endogenous RNA to control the expression of notch3 via sponging miR-613 in pancreatic cancer. Oncotarget . 2017;8(20):32905–32917. doi: 10.18632/oncotarget.16462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen W., Du M., Hu X., et al. Long noncoding RNA cytoskeleton regulator RNA promotes cell invasion and metastasis by titrating miR-613 to regulate ANXA2 in nasopharyngeal carcinoma. Cancer Medicine . 2020;9(3):1209–1219. doi: 10.1002/cam4.2778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dai W., Shi Y., Hu W., Xu C. Long noncoding RNA FAM225B facilitates proliferation and metastasis of nasopharyngeal carcinoma cells by regulating miR-613/CCND2 axis. Bosnian Journal of Basic Medical Sciences . 2022;22(1):77–86. doi: 10.17305/bjbms.2021.5691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gao R., Feng Q., Tan G. microRNA-613 exerts anti-angiogenic effect on nasopharyngeal carcinoma cells through inactivating the AKT signaling pathway by down-regulating FN1. Bioscience Reports . 2019;39(7) doi: 10.1042/BSR20182196. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 31.Lakshminarasimhan R., Liang G. The role of DNA methylation in cancer. Advances in Experimental Medicine and Biology . 2016;945:151–172. doi: 10.1007/978-3-319-43624-1_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Giambò F., Leone G. M., Gattuso G., et al. Genetic and epigenetic alterations induced by pesticide exposure: integrated analysis of gene expression, microRNA expression, and DNA methylation datasets. International journal of environmental research and public health . 2021;18(16):p. 8697. doi: 10.3390/ijerph18168697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen L. H., Hsu W. L., Tseng Y. J., Liu D. W., Weng C. F. Involvement of DNMT 3B promotes epithelial-mesenchymal transition and gene expression profile of invasive head and neck squamous cell carcinomas cell lines. BMC Cancer . 2016;16(1):p. 431. doi: 10.1186/s12885-016-2468-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li W., Yi J., Zheng X., et al. miR-29c plays a suppressive role in breast cancer by targeting the TIMP3/STAT1/FOXO1 pathway. Epigenetics . 2018;10(1):p. 64. doi: 10.1186/s13148-018-0495-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Linhart H. G., Lin H., Yamada Y., et al. Dnmt3b promotes tumorigenesis in vivo by gene-specific de novo methylation and transcriptional silencing. Genes & Development . 2007;21(23):3110–3122. doi: 10.1101/gad.1594007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang Y. C., Tang Y. A., Shieh J. M., Lin R. K., Hsu H. S., Wang Y. C. DNMT3B overexpression by deregulation of FOXO3a-mediated transcription repression and MDM2 overexpression in lung cancer. Journal of Thoracic Oncology . 2014;9(9):1305–1315. doi: 10.1097/JTO.0000000000000240. [DOI] [PubMed] [Google Scholar]
- 37.Wu C., Guo E., Ming J., et al. Radiation-induced DNMT3B promotes radioresistance in nasopharyngeal carcinoma through methylation of p53 and p21. Molecular Therapy-Oncolytics . 2020;17:306–319. doi: 10.1016/j.omto.2020.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang H. L., Liu Y. M., Sung T. Y., et al. TIMP3 expression associates with prognosis in colorectal cancer and its novel arylsulfonamide inducer, MPT0B390, inhibits tumor growth, metastasis and angiogenesis. Theranostics . 2019;9(22):6676–6689. doi: 10.7150/thno.34020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang Z., Wang J., Wang X., Song W., Shi Y., Zhang L. MicroRNA-21 promotes proliferation, migration, and invasion of cervical cancer through targeting TIMP3. Archives of Gynecology and Obstetrics . 2018;297(2):433–442. doi: 10.1007/s00404-017-4598-z. [DOI] [PubMed] [Google Scholar]
- 40.Qin S., Zhu Y., Ai F., et al. MicroRNA-191 correlates with poor prognosis of colorectal carcinoma and plays multiple roles by targeting tissue inhibitor of metalloprotease 3. Neoplasma . 2014;61(1):27–34. doi: 10.4149/neo_2014_005. [DOI] [PubMed] [Google Scholar]
- 41.Chen J., Gu Y., Shen W. MicroRNA-21 functions as an oncogene and promotes cell proliferation and invasion via TIMP3 in renal cancer. European Review for Medical and Pharmacological Sciences . 2017;21(20):4566–4576. [PubMed] [Google Scholar]
- 42.Zhao Y., Gu X., Wang Y. MicroRNA-103 promotes nasopharyngeal carcinoma through targeting TIMP-3 and the Wnt/β‐catenin pathway. The Laryngoscope . 2020;130(3):E75–E82. doi: 10.1002/lary.28045. [DOI] [PubMed] [Google Scholar]
- 43.Lim Y., Wan Y., Vagenas D., et al. Salivary DNA methylation panel to diagnose HPV-positive and HPV-negative head and neck cancers. BMC Cancer . 2016;16(1):p. 749. doi: 10.1186/s12885-016-2785-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Guilleret I., Losi L., Chelbi S. T., et al. DNA methylation profiling of esophageal adenocarcinoma using methylation ligation-dependent macroarray (MLM) Biochemical and Biophysical Research Communications . 2016;479(2):231–237. doi: 10.1016/j.bbrc.2016.09.049. [DOI] [PubMed] [Google Scholar]
- 45.Zhang Z., Huo H., Liao K., et al. RPA1 downregulation enhances nasopharyngeal cancer radiosensitivity _via_ blocking RAD51 to the DNA damage site. Experimental Cell Research . 2018;371(2):330–341. doi: 10.1016/j.yexcr.2018.08.025. [DOI] [PubMed] [Google Scholar]
- 46.Shepherd F. A., Crowley J., Van Houtte P., Postmus P. E., Carney D., Chansky K., et al. The International Association for the Study of Lung Cancer lung cancer staging project: proposals regarding the clinical staging of small cell lung cancer in the forthcoming (seventh) edition of the tumor, node, metastasis classification for lung cancer. Journal of Thoracic Oncology . 2007;2(12):1067–1077. doi: 10.1097/JTO.0b013e31815bdc0d. [DOI] [PubMed] [Google Scholar]
- 47.Wan F. Z., Chen K. H., Sun Y. C., et al. Exosomes overexpressing miR-34c inhibit malignant behavior and reverse the radioresistance of nasopharyngeal carcinoma. Journal of Translational Medicine . 2020;18(1):p. 12. doi: 10.1186/s12967-019-02203-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Du T., Jiang J., Chen Y., et al. MiR-138-1-3p alters the stemness and radiosensitivity of tumor cells by targeting CRIPTO and the JAK2/STAT3 pathway in nasopharyngeal carcinoma. Annals of Translational Medicine . 2021;9(6):p. 485. doi: 10.21037/atm-21-521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen X., Xu Y., Liao X., et al. Plasma miRNAs in predicting radiosensitivity in non-small cell lung cancer. Tumour Biology . 2016;37(9):11927–11936. doi: 10.1007/s13277-016-5052-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hui Z., Tretiakova M., Zhang Z., et al. Radiosensitization by inhibiting STAT1 in renal cell carcinoma. International Journal of Radiation Oncology • Biology • Physics . 2009;73(1):288–295. doi: 10.1016/j.ijrobp.2008.08.043. [DOI] [PubMed] [Google Scholar]
- 51.Khodarev N. N., Beckett M., Labay E., Darga T., Roizman B., Weichselbaum R. R. STAT1 is overexpressed in tumors selected for radioresistance and confers protection from radiation in transduced sensitive cells. Proceedings of the National Academy of Sciences of the United States of America . 2004;101(6):1714–1719. doi: 10.1073/pnas.0308102100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhu H., Wang Z., Xu Q., et al. Inhibition of STAT1 sensitizes renal cell carcinoma cells to radiotherapy and chemotherapy. Cancer Biology & Therapy . 2012;13(6):401–407. doi: 10.4161/cbt.19291. [DOI] [PubMed] [Google Scholar]
- 53.Tan L. P., Tan G. W., Sivanesan V. M., et al. Systematic comparison of plasma EBV DNA, anti-EBV antibodies and miRNA levels for early detection and prognosis of nasopharyngeal carcinoma. International Journal of Cancer . 2020;146(8):2336–2347. doi: 10.1002/ijc.32656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chang L. H., Yao Z. Z., Bao H. W., et al. miR-18a enhances the radiosensitivity of nasopharyngeal carcinoma cells through inducing autophagy. Zhonghua Er Bi Yan Hou Tou Jing Wai Ke Za Zhi . 2021;56(7):736–745. doi: 10.3760/cma.j.cn115330-20201027-00831. [DOI] [PubMed] [Google Scholar]
- 55.Ma Y., Zhou G., Li M., et al. Long noncoding RNA DANCR mediates cisplatin resistance in glioma cells via activating AXL/PI3K/Akt/NF-κB signaling pathway. Neurochemistry International . 2018;118:233–241. doi: 10.1016/j.neuint.2018.03.011. [DOI] [PubMed] [Google Scholar]
- 56.Li B., Liu D., Yang P., Li H. Y., Wang D. miR-613 inhibits liver cancer stem cell expansion by regulating SOX9 pathway. Gene . 2019;707:78–85. doi: 10.1016/j.gene.2019.05.015. [DOI] [PubMed] [Google Scholar]
- 57.Camero S., Vitali G., Pontecorvi P., et al. DNMT3A and DNMT3B targeting as an effective radiosensitizing strategy in embryonal rhabdomyosarcoma. Cells . 2021;10(11):p. 2956. doi: 10.3390/cells10112956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhai J. X., Zhang Z. X., Feng Y. J., et al. PDTC attenuate LPS-induced kidney injury in systemic lupus erythematosus-prone MRL/lpr mice. Molecular Biology Reports . 2012;39(6):6763–6771. doi: 10.1007/s11033-012-1501-7. [DOI] [PubMed] [Google Scholar]
- 59.Li H., Xu W., Xia Z., et al. Hsa_circ_0000199 facilitates chemo-tolerance of triple-negative breast cancer by interfering with miR-206/613-led PI3K/Akt/mTOR signaling. Aging . 2021;13(3):4522–4551. doi: 10.18632/aging.202415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Qu S., Guo Y., Huang S. T., Zhu X. D. Inhibition of STAT1 sensitizes radioresistant nasopharyngeal carcinoma cell line CNE-2R to radiotherapy. Oncotarget . 2018;9(9):8303–8310. doi: 10.18632/oncotarget.19690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Huang C., Chen D., Zhu H., Lv S., Li Q., Li G. LITAF enhances radiosensitivity of human glioma cells via the FoxO1 pathway. Cellular and Molecular Neurobiology . 2019;39(6):871–882. doi: 10.1007/s10571-019-00686-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang P., Xin X., Fang L., et al. HMGB1 mediates Aspergillus fumigatus-induced inflammatory response in alveolar macrophages of COPD mice via activating MyD88/NF-kappaB and syk/PI3K signalings. International Immunopharmacology . 2017;53:125–132. doi: 10.1016/j.intimp.2017.10.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data used to support the findings of this study are available from the corresponding author upon request.


