Abstract
To facilitate sequencing of the Sinorhizobium meliloti 1021 pSyma megaplasmid, a high-resolution map was constructed by ordering 113 overlapping bacterial artificial chromosome clones with 192 markers. The 157 anonymous sequence tagged site markers (81,072 bases) reveal hypothetical functions encoded by the replicon.
The symbiotic soil bacterium Sinorhizobium meliloti forms nitrogen-fixing nodules on the roots of leguminous host plants and displays a complex genome consisting of a 3.7-Mb chromosome and two megaplasmids, pSyma (1.4 Mb) and pSymb (1.7 Mb) (8, 27, 41). Genes required for symbiosis are located on all three replicons (18, 20), but are more frequently found on the megaplasmids. Genes involved in nodulation and nitrogen fixation are located on pSyma (21, 22, 39), whereas those essential for extracellular polysaccharide synthesis and other symbiotic functions are located on pSymb (16, 44). These two genetic elements have both chromosome-like and plasmid-like features: both are 3 orders of magnitude larger than many cloning vectors and carry some copies of housekeeping genes, such as groESL, and genes associated with other metabolic functions (33). On the other hand, the megaplasmids can be mostly or entirely cured without affecting growth and reproduction (at least in permissive conditions) (13, 24; M. Hynes, personal communication). Moreover, the megaplasmids can be transferred to and maintained in at least one heterologous genus, Agrobacterium (25, 43). Maps for the chromosome and pSymb of strain 1021 exist (9, 10, 12, 20, 23), but concerning pSyma, only three markers outside the 250-kb region containing symbiotic genes have been identified (3). As part of the international effort to sequence the entire S. meliloti genome, we constructed a high-resolution physical map of the pSyma megaplasmid, using PCR-based screening and assembly of recombinant bacterial artificial chromosome (BAC) clones. In addition to providing a valuable tool for the total genome sequencing project, the data reported here provide new insights into the genetic information contained on pSyma.
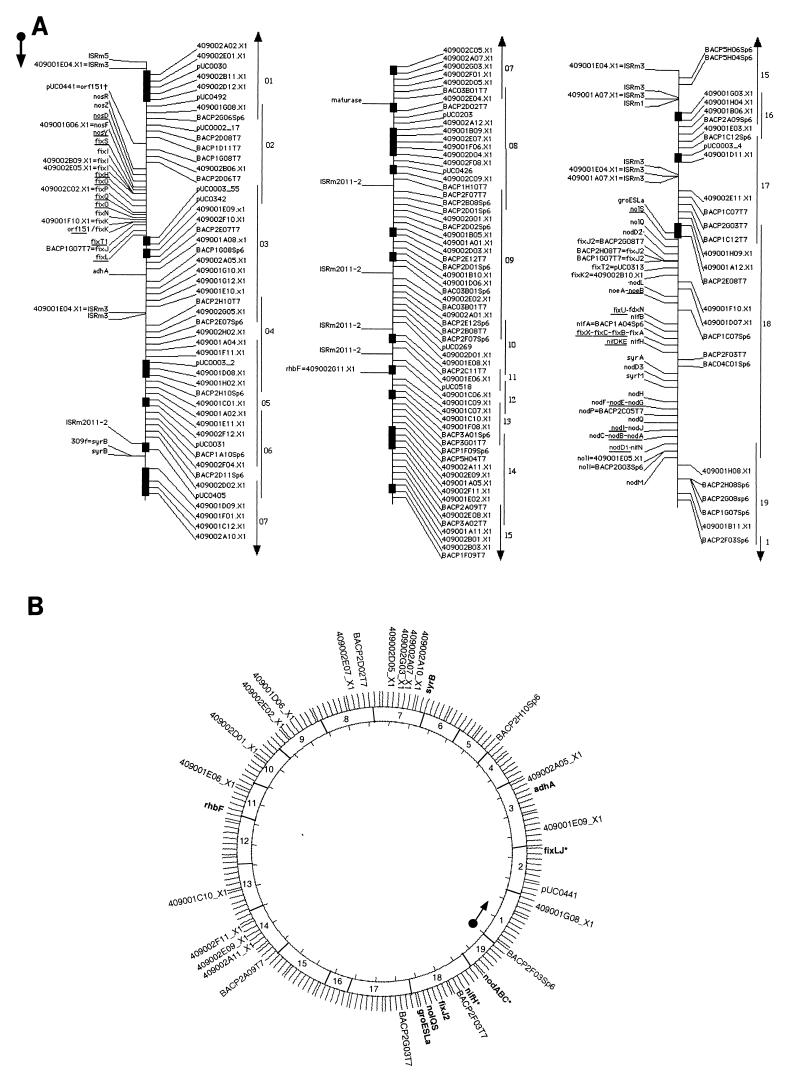
A high-resolution map of the S. meliloti pSyma megaplasmid was constructed by using the same materials and methods that were successfully used for chromosome mapping (9) except that additional random sequences from a pSyma-enriched library were incorporated into the BAC screening (methods are described at http://cmgm.stanford.edu/∼mbarnett/syma.htm). After screening 192 megaplasmid clones, we identified 88 pSyma clones. Additional clones from the total genomic BAC library were similarly screened to fill gaps in poorly represented regions of the pSyma contig. Thus, we assembled 113 BAC clones into a circular contig (Fig. 1A) encompassing the entire 1.4-Mb pSyma replicon, using a total of 192 markers, including 157 sequence tagged sites (STSs) (9) and 33 gene markers (Table 1) representing 14 individual genes, 15 operons (52 genes), and four insertion sequences available in the GenBank (7) and EMBL (42) databases. Assuming a random distribution of markers, average spacing was estimated at 7 kb, with a tiling path of 6.8 colinear BAC clones per marker. No region is represented by only one BAC, and we detected only five chimeric clones between pSyma and pSymb. All of the 108 other assembled BAC clones show an exact colinear distribution of markers, and pSyma is covered by a set of 19 BAC clones with minimal overlap. Assuming a map density of 7 kb, deletions or rearrangements on some clones should be smaller than 7 kb, if they do exist.
FIG. 1.
(A) High-resolution map of the pSyma megaplasmid of S. meliloti 1021. The map is presented in three linear and contiguous parts of approximately 500 kb for convenience. Identified S. meliloti genes (genetic database or BLASTX results) are indicated on the left side while anonymous STS markers are located on the right side. The positions of underlined genes were deduced from mapped genes in the operon; i.e., no PCR primers were designed. Some genes are listed more than once because several sets of primers were used. Black rectangles indicate pairwise invertable markers. ‡, partial similarity. The minimum set of BAC clones covering the replicon is also presented. (B) Simplified map oriented according to the Honeycutt et al. map (23) showing STS markers mentioned in the text, selected genes, and the minimum set of BAC clones. Lengths of BAC inserts are shown relative to the sizes determined by field inversion gel electrophoresis. Genes previously mapped by Honeycutt et al. are marked with asterisks. Corresponding BAC insert sizes are as follows: BAC01, 110 kb; BAC02, 75 kb; BAC03, 110 kb; BAC04, 75 kb; BAC05, 55 kb; BAC06, 85 kb; BAC07, 120 kb; BAC08, 65 kb; BAC09, 80 kb; BAC10, 75 kb; BAC11, 60 kb; BAC12, 110 kb; BAC13, 100 kb; BAC14, 140 kb; BAC15, 80 kb; BAC16, 25 kb; BAC17, 100 kb; BAC18, 80 kb; and BAC19, 60 kb.
TABLE 1.
Previously identified S. meliloti genes mapped on the pSyma megaplasmid
| Gene(s) | Encoded function or product |
|---|---|
| adhA | Alcohol dehydrogenase |
| fixABCX | Putative electron transport chain to nitrogenase |
| fixGHIS | Putative cation transport complex |
| fixJ2T2-fixK2 | Transcriptional activators |
| fixKorf151 | Transcriptional activator |
| fixLJT1 | Hemoprotein kinase; transcriptional activator |
| fixNOQP | Putative bacteroid oxidase |
| groESLa | Chaperonin |
| nifABfdxNfixU | Nitrogen fixation regulatory protein; ferredoxin-like protein |
| nifHDKE | Nitrogenase reductase |
| nifN | FeMo-cofactor biosynthesis |
| nodABCIJ | Acyltransferase; N acetylase; chitin synthase; transporter of nod factors |
| nodD1 | nod gene activator |
| nodD2 | nod gene activator |
| nodD3 | nod gene activator |
| nodFE | Acyl carrier protein; β-ketoacyl synthase |
| nodG | Putative dehydrogenase |
| nodH | Sulfotransferase |
| nodLnoeAB | O-Acetyltransferase |
| nodMnolFGHInodN | Glucosamine synthase; transport |
| nodPQ | ATP sulfurylase APS kinase |
| nolQS | Unknown function; similar to a thiamine biosynthetic enzyme |
| nosRZDFY | Nitrous oxide reduction proteins |
| maturase | Reverse transcriptase/maturase |
| rhbF | Siderophore biosynthesis in rhizobactin regulon |
| syrA | Increases exopolysaccharide abundance |
| syrB | Negatively affects syrM expression |
| syrM | nod gene activator |
The relative positions of the nodulation and nitrogen fixation genes are consistent with previous mapping data (22, 23, 39), in particular the (i) presence of two fixJ loci flanking the nod-nif region (5, 35), (ii) organization of nodulation genes (39), (iii) orientation between nos and fix gene clusters (11), (iv) location of syrA (4), and (v) location of groESLa (33). All of these genes are clustered in a well-known symbiotic region. Based on the insert size of the clones covering this region, the total length was estimated to be between 250 and 300 kb, which is also in agreement with Renalier et al. (35). The remaining 1.1 Mb of the replicon does not contain any known symbiotic genes, except syrB. We also positioned several previously unmapped genes: adhA, rhbF, and a gene encoding a maturase. Concerning insertion sequences, we detected one copy of ISRm1 (46), at least five copies of the widespread ISRm2011-2 (40), five copies of ISRm3 (45), and one copy of ISRm5 (28), compared with five copies on the chromosome (9). We did not obtain PCR products with primers designed from ISRm2, ISRm4, ISRm6, ISRm7, ISRm8, and ISRm9.
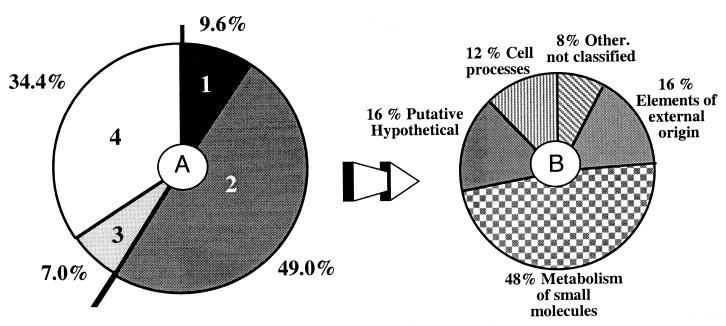
Each STS was analyzed by BLASTX (1) comparison with the nonredundant protein database from the National Center for Biotechnology Information, and results are available at the website http://www-recomgen.univ-rennes1.fr/meliloti. STS match results were divided into four categories (Fig. 2A), and the most significant homologies were divided into functional groups according to Riley's classification for orthologous Escherichia coli genes (36, 37) (Fig. 2B).
FIG. 2.
(A) Distribution of BLASTX results among four categories of significance. 1, strong similarity with S. meliloti proteins (E ≤ 1e−6; identity, ≥85%); 2, strong similarity with sequences available in the databases (E ≤ 1e−6); 3, local or weaker similarity with sequences available in the databases (1e−2 ≤ E ≤ 1e−6); 4, no similarity with sequences available in the databases. (B) Classification of the most significant matches (E ≤ 1e−6) using Riley's classification (34, 35).
This distribution shows many STS markers containing genes involved in the metabolism of small molecules, such as (i) 4-deoxy-l-threo-5-hexosulose-uronate-ketol-isomerase and succinate-semialdehyde dehydrogenase (encoded by gabD), which is involved in carbohydrate (C4-to-C6) degradation (32), and (ii) serine hydroxymethyltransferase, which is the key enzyme of C1 and C2 compound assimilation and is necessary for the formation of effective nodules in Bradyrhizobium japonicum (38). The next largest group of STSs is made up of those possibly involved in cellular processes (chemotaxis and transport); no matches with cell division proteins or general housekeeping genes were detected other than those for the previously reported groESLa (33). We also found matches to genes involved in nitrogen metabolism: the periplasmic nitrate reductase precursor of Paracoccus and Pseudomonas, the NifX-like protein of Rhizobium sp. strain NGR234, an arginine deiminase of Rhizobium etli (15), and the NifL nitrogen fixation regulator of Klebsiella and other bacteria (31), previously unknown in S. meliloti. Also, there are several less stringent intriguing matches: an STS identical to stage IV sporulation protein FB of Bacillus subtilis, required for spore formation (14), and a marker identical to protein AttB of the plant pathogen Agrobacterium tumefaciens, required for the attachment of bacteria to plant cells (30). One STS is similar to VirB4 from Agrobacterium and TraB from E. coli, both of which are required for DNA transfer and may represent part of a region involved in conjugative transfer of the pSyma replicon. We also detected one sequence similar to the adducin-like protein AddA of the obligate intracellular parasite Rickettsia prowazekii and to alpha-adducin, which promotes the assembly of the spectrin-actin network in eukaryotic cells (2, 26). In addition, some STSs have matches to transcriptional regulators from the LysR family of transcriptional regulators, the AraC family activators, the GntR family regulators, the trp repressor, and a repressor of the TetR-AcrR family. We did not find any STSs with matches to regulators of two-component systems.
The most relevant comparison for the pSyma sequence will be with that of the closely related Rhizobium sp. strain NGR234, which has a complex genome (17), including a symbiosis plasmid of 536 kb, for which the complete nucleotide sequence has been established (19). Given that the pSyma megaplasmid of S. meliloti is almost three times the size of the pSym megaplasmid of NGR234, it will be interesting to determine how related they are. In this regard, we noted that seven of the S. meliloti pSyma STS markers had a match with the pSym of NGR234, while 150 of the S. meliloti pSyma markers did not (for E ≤ 1e−4).
The elucidation of the S. meliloti total genome sequence will aid greatly our understanding of the ancestry and behavior of the pSyma replicon as well as provide insight into the genomic plasticity, the presence of multicopy genes, and the relative involvement of each replicon, both in symbiotic and free-living bacteria.
Acknowledgments
We thank Alain Billault and Catherine Soravito de Franceski (CEPH, Fondation Jean Dausset, Paris, France) for their involvement in the BAC library construction. We are also particularly grateful to Patricia Thebault and Jérôme Gouzy (UMR215 INRA-CNRS, Toulouse, France) for computer assistance.
This work has been supported by the CNRS through UPR41 and the CNRS Genome Project. Additional support came from U.S. Department of Energy grant DE-FG03-90ER20010 to S.R.L.
F.B.-H. and D.C. contributed equally to this work.
REFERENCES
- 1.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andersson S G, Zomorodipour A, Andersson J O, Sicheritz-Ponten T, Alsmark U C, Podowski R M, Naslund A K, Eriksson A S, Winkler H H, Kurland C G. The genome sequence of Rickettsia prowazekii and the origin of mitochondria. Nature. 1998;396:133–140. doi: 10.1038/24094. [DOI] [PubMed] [Google Scholar]
- 3.Barnett M J, Long S R. Identification and characterization of a gene on Rhizobium meliloti pSyma, syrB, that negatively affects syrM expression. Mol Plant-Microbe Interact. 1997;10:550–559. doi: 10.1094/MPMI.1997.10.5.550. [DOI] [PubMed] [Google Scholar]
- 4.Barnett M J, Swanson J A, Long S R. Multiple genetic controls on Rhizobium meliloti syrA, a regulator of exopolysaccharide abundance. Genetics. 1998;148:19–32. doi: 10.1093/genetics/148.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Batut J, Terzaghi B, Ghérardi M, Huguet M, Terzaghi E, Garnerone A M, Boistard P, Huguet T. Localization of a symbiotic fix region on Rhizobium meliloti pSym megaplasmid more than 200 kilobases from the nod-nif region. Mol Gen Genet. 1985;199:232–239. [Google Scholar]
- 6.Batut J, Daveran-Mingot M L, David M, Jacobs J, Garnerone A M, Kahn D. fixK, a gene homologous with fnr and crp from Escherichia coli, regulates nitrogen fixation genes both positively and negatively in Rhizobium meliloti. EMBO J. 1989;8:1279–1286. doi: 10.1002/j.1460-2075.1989.tb03502.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Benson D A, Boguski M S, Lipman D J, Ostell J, Ouellette B F F, Rapp B A, Wheeler D L. GenBank. Nucleic Acids Res. 1999;27:12–17. doi: 10.1093/nar/27.1.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burkardt B, Schillik D, Puhler A. Physical characterization of Rhizobium meliloti megaplasmids. Plasmid. 1987;17:13–25. doi: 10.1016/0147-619x(87)90004-7. [DOI] [PubMed] [Google Scholar]
- 9.Capela D, Barloy-Hubler F, Gatius M T, Gouzy J, Galibert F. A high-density physical map of Sinorhizobium meliloti 1021 chromosome derived from BAC library. Proc Natl Acad Sci USA. 1999;96:9357–9362. doi: 10.1073/pnas.96.16.9357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Casadesus J, Olivares J. Rough and fine linkage mapping of the Rhizobium meliloti chromosome. Mol Gen Genet. 1979;174:203–209. doi: 10.1007/BF00268356. [DOI] [PubMed] [Google Scholar]
- 11.Chan Y K, McCormick W A, Watson R J. A new nos gene downstream from nosDFY is essential for dissimilatory reduction of nitrous oxide by Rhizobium (Sinorhizobium) meliloti. Microbiology. 1997;143:2817–2824. doi: 10.1099/00221287-143-8-2817. [DOI] [PubMed] [Google Scholar]
- 12.Charles T C, Finan T M. Genetic map of Rhizobium meliloti megaplasmid pRmeSU47b. J Bacteriol. 1990;172:2469–2476. doi: 10.1128/jb.172.5.2469-2476.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Charles T C, Finan T M. Analysis of a 1600-kilobase Rhizobium meliloti megaplasmid using defined deletions generated in vivo. Genetics. 1991;127:5–20. doi: 10.1093/genetics/127.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cutting S, Roels S, Losick R. Sporulation operon spoIVF and the characterization of mutations that uncouple mother-cell from forespore gene expression in Bacillus subtilis. J Mol Biol. 1991;221:1237–1256. doi: 10.1016/0022-2836(91)90931-u. [DOI] [PubMed] [Google Scholar]
- 15.D'Hooghe I, Vander Wauven C, Michiels J, Tricot C, de Wilde P, Vanderleyden J, Stalon V. The arginine deiminase pathway in Rhizobium etli: DNA sequence analysis and functional study of the arcABC genes. J Bacteriol. 1997;179:7403–7409. doi: 10.1128/jb.179.23.7403-7409.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Finan T M, Kunkel B, De Vos G F, Signer E R. Second symbiotic megaplasmid in Rhizobium meliloti carrying exopolysaccharide and thiamine synthesis genes. J Bacteriol. 1986;167:66–72. doi: 10.1128/jb.167.1.66-72.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Flores M, Mavingui P, Girard L, Perret X, Broughton W J, Martinez-Romero E, Davila G, Palacios R. Three replicons of Rhizobium sp. strain NGR234 harbor symbiotic gene sequences. J Bacteriol. 1998;180:6052–6053. doi: 10.1128/jb.180.22.6052-6053.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Forrai T, Vincze E, Banfalvi Z, Kiss G B, Randhawa G S, Kondorosi A. Localization of symbiotic mutations in Rhizobium meliloti. J Bacteriol. 1983;153:635–643. doi: 10.1128/jb.153.2.635-643.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Freiberg C, Fellay R, Bairoch A, Broughton W J, Rosenthal A, Perret X. Molecular basis of symbiosis between Rhizobium and legumes. Nature. 1997;387:394–401. doi: 10.1038/387394a0. [DOI] [PubMed] [Google Scholar]
- 20.Glazebrook J, Meiri G, Walker G C. Genetic mapping of symbiotic loci on the Rhizobium meliloti chromosome. Mol Plant-Microbe Interact. 1992;5:223–227. doi: 10.1094/mpmi-5-223. [DOI] [PubMed] [Google Scholar]
- 21.Goldmann A, Boivin C, Fleury V, Message B, Lecoeur L, Maille M, Tepfer D. Betaine use by rhizosphere bacteria: genes essential for trigonelline, stachydrine, and carnitine catabolism in Rhizobium meliloti are located on pSym in the symbiotic region. Mol Plant-Microbe Interact. 1991;4:571–578. doi: 10.1094/mpmi-4-571. [DOI] [PubMed] [Google Scholar]
- 22.Holloway P, McCormick W, Watson R J, Chan Y K. Identification and analysis of the dissimilatory nitrous oxide reduction genes, nosRZDFY, of Rhizobium meliloti. J Bacteriol. 1996;178:1505–1514. doi: 10.1128/jb.178.6.1505-1514.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Honeycutt R J, McClelland M, Sobral B W. Physical map of the genome of Rhizobium meliloti 1021. J Bacteriol. 1993;175:6945–6952. doi: 10.1128/jb.175.21.6945-6952.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hynes M F, Quandt J, O'Connell M P, Puhler A. Direct selection for curing and deletion of Rhizobium plasmids using transposons carrying the Bacillus subtilis sacB gene. Gene. 1989;78:111–120. doi: 10.1016/0378-1119(89)90319-3. [DOI] [PubMed] [Google Scholar]
- 25.Hynes M F, Simon R, Puhler A. The development of plasmid-free strains of Agrobacterium tumefaciens by using incompatibility with a Rhizobium meliloti plasmid to eliminate pAtC58. Plasmid. 1985;13:99–105. doi: 10.1016/0147-619x(85)90062-9. [DOI] [PubMed] [Google Scholar]
- 26.Joshi R, Gilligan D M, Otto E, McLaughlin T, Bennett V. Primary structure and domain organization of human alpha and beta adducin. J Cell Biol. 1991;115:665–675. doi: 10.1083/jcb.115.3.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jumas-Bilak E, Michaux-Charachon S, Bourg G, Ramuz M, Allardet-Servent A. Unconventional genomic organization in the alpha subgroup of the Proteobacteria. J Bacteriol. 1998;180:2749–2755. doi: 10.1128/jb.180.10.2749-2755.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Laberge S, Middleton A T, Wheatcroft R. Characterization, nucleotide sequence, and conserved genomic locations of insertion sequence ISRm5 in Rhizobium meliloti. J Bacteriol. 1995;177:3133–3142. doi: 10.1128/jb.177.11.3133-3142.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Matthysse A G, Kijne J W. Attachment of Rhizobiaceae to plant cell. In: Spaink H P, Kondorosi A, Hooykaas P J J, editors. The Rhizobiaceae. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1998. pp. 235–249. [Google Scholar]
- 30.Matthysse A G, Yarnall H A, Young N. Requirement for genes with homology to ABC transport systems for attachment and virulence of Agrobacterium tumefaciens. J Bacteriol. 1996;178:5302–5308. doi: 10.1128/jb.178.17.5302-5308.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morett E, Kreutzer R, Cannon W, Buck M. The influence of the Klebsiella pneumoniae regulatory gene nifL upon the transcriptional activator protein NifA. Mol Microbiol. 1990;4:1253–1258. doi: 10.1111/j.1365-2958.1990.tb00704.x. [DOI] [PubMed] [Google Scholar]
- 32.Niegemann E, Schulz A, Bartsch K. Molecular organization of the Escherichia coli gab cluster: nucleotide sequence of the structural genes gabD and gabP and expression of the GABA permease gene. Arch Microbiol. 1993;160:454–460. doi: 10.1007/BF00245306. [DOI] [PubMed] [Google Scholar]
- 33.Ogawa J, Long S R. The Rhizobium meliloti groELc locus is required for regulation of early nod genes by the transcription activator NodD. Genes Dev. 1995;9:714–729. doi: 10.1101/gad.9.6.714. [DOI] [PubMed] [Google Scholar]
- 34.Pocard J A, Vincent N, Boncompagni E, Smith L T, Poggi M C, Le Rudulier D. Molecular characterization of the bet genes encoding glycine betaine synthesis in Sinorhizobium meliloti 102F34. Microbiology. 1997;143:1369–1379. doi: 10.1099/00221287-143-4-1369. [DOI] [PubMed] [Google Scholar]
- 35.Renalier M-H, Batut J, Ghai J, Terzaghi B, Gherardi M, David M, Garnerone A-M, Vasse J, Truchet G, Huguet T, Boistard P. A new symbiotic cluster on the pSym megaplasmid of Rhizobium meliloti 2011 carries a functional fix gene repeat and a nod locus. J Bacteriol. 1987;169:2231–2238. doi: 10.1128/jb.169.5.2231-2238.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Riley M. Functions of the gene products of Escherichia coli. Microbiol Rev. 1993;57:862–952. doi: 10.1128/mr.57.4.862-952.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Riley M. Systems for categorizing functions of gene products. Curr Opin Struct Biol. 1998;8:388–392. doi: 10.1016/s0959-440x(98)80074-2. [DOI] [PubMed] [Google Scholar]
- 38.Rossbach S, Hennecke H. Identification of glyA as a symbiotically essential gene in Bradyrhizobium japonicum. Mol Microbiol. 1991;5:39–47. doi: 10.1111/j.1365-2958.1991.tb01824.x. [DOI] [PubMed] [Google Scholar]
- 39.Schlaman H R M, Phillips D A, Kondorosi E. Genetic organization and transcriptional regulation of rhizobial nodulation genes. In: Spaink H P, Kondorosi A, Hooykaas P J J, editors. The Rhizobiaceae. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1998. pp. 361–386. [Google Scholar]
- 40.Selbitschka W, Arnold W, Jording D, Kosier B, Toro N, Puhler A. The insertion sequence element ISRm2011-2 belongs to the IS630-Tc1 family of transposable elements and is abundant in Rhizobium meliloti. Gene. 1995;163:59–64. doi: 10.1016/0378-1119(95)00371-c. [DOI] [PubMed] [Google Scholar]
- 41.Sobral B W, Honeycutt R J, Atherly A G, McClelland M. Electrophoretic separation of the three Rhizobium meliloti replicons. J Bacteriol. 1991;173:5173–5180. doi: 10.1128/jb.173.16.5173-5180.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stoesser G, Tuli M A, Lopez R, Sterk P. The EMBL nucleotide sequence database. Nucleic Acids Res. 1999;27:18–24. doi: 10.1093/nar/27.1.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Truchet G, Rosenberg C, Vasse J, Julliot J S, Camut S, Dénarié J. Transfer of Rhizobium meliloti pSym genes into Agrobacterium tumefaciens: host-specific nodulation by atypical infection. J Bacteriol. 1984;157:134–142. doi: 10.1128/jb.157.1.134-142.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Watson R J, Chan Y K, Wheatcroft R, Yang A F, Han S H. Rhizobium meliloti genes required for C4-dicarboxylate transport and symbiotic nitrogen fixation are located on a megaplasmid. J Bacteriol. 1988;170:927–934. doi: 10.1128/jb.170.2.927-934.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wheatcroft R, Laberge S. Identification and nucleotide sequence of Rhizobium meliloti insertion sequence ISRm3: similarity between the putative transposase encoded by ISRm3 and those encoded by Staphylococcus aureus IS256 and Thiobacillus ferrooxidans IST2. J Bacteriol. 1991;173:2530–2538. doi: 10.1128/jb.173.8.2530-2538.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wheatcroft R, Watson R J. Distribution of insertion sequence ISRm1 in Rhizobium meliloti and other gram-negative bacteria. J Gen Microbiol. 1988;134:113–121. doi: 10.1099/00221287-134-1-113. [DOI] [PubMed] [Google Scholar]