Graphical abstract
Keywords: Spatial transcriptomics, Deep learning, Tissue architecture visualization and identification
Abstract
Spatially resolved transcriptomics provides a new way to define spatial contexts and understand the pathogenesis of complex human diseases. Although some computational frameworks can characterize spatial context via various clustering methods, the detailed spatial architectures and functional zonation often cannot be revealed and localized due to the limited capacities of associating spatial information. We present RESEPT, a deep-learning framework for characterizing and visualizing tissue architecture from spatially resolved transcriptomics. Given inputs such as gene expression or RNA velocity, RESEPT learns a three-dimensional embedding with a spatial retained graph neural network from spatial transcriptomics. The embedding is then visualized by mapping into color channels in an RGB image and segmented with a supervised convolutional neural network model. Based on a benchmark of 10x Genomics Visium spatial transcriptomics datasets on the human and mouse cortex, RESEPT infers and visualizes the tissue architecture accurately. It is noteworthy that, for the in-house AD samples, RESEPT can localize cortex layers and cell types based on pre-defined region- or cell-type-enriched genes and furthermore provide critical insights into the identification of amyloid-beta plaques in Alzheimer's disease. Interestingly, in a glioblastoma sample analysis, RESEPT distinguishes tumor-enriched, non-tumor, and regions of neuropil with infiltrating tumor cells in support of clinical and prognostic cancer applications.
1. Introduction
Tissue architecture is the biological foundation of spatial heterogeneity within complex organs like the human brain [1] and is thereby essential in understanding the underlying pathogenesis of human diseases, including cancer [2] and Alzheimer's disease (AD) [3]. Unlike healthy and well-organized tissue architecture, tissues in a disease state such as cancer usually alternate the organization and lead to cytoarchitectural abnormalities with aberrant physiological processes [4], [5], [6]. Spatial transcriptomics is especially well-positioned to study such an abnormal organization and investigate its mechanism [7]. Recent advances in spatially resolved technologies such as 10x Genomics Visium provide spatial context together with high-throughput gene expression for exploring tissue domains, cell types, cell–cell communications, and their biological consequences [8].
Several computational methods have been developed for computational analyses of spatial transcriptomics [7], [9], [10]. Seurat [11] performs tissue architecture identification and interpretation based on variable gene selection, dimension reduction, and graph-based clustering (i.e., Louvain), followed by differentially expressed analysis. Giotto [12] is a comprehensive toolbox for spatial analysis and visualization, including spatially variable gene (SVG) pattern recognition, cell–cell communication inference, and tissue architecture identification, which uses a similar framework as Seurat. STUtility [13] uses non-negative matrix factorization to perform dimension reduction and then identifies tissue architecture based on Seurat and can integrate consecutive samples to obtain a more comprehensive three-dimensional view of tissue architectures. In addition, several deep learning methods were also introduced. SpaGCN [14] proposes a convolutional graph network to integrate gene expression, spatial location, and histology to define metagene (i.e., a group of genes sharing a common spatial pattern) and further characterize tissue architecture. stLearn [15] is also a comprehensive toolbox for spatial data analysis, including implementing the normalization method, performing tissue architecture identification, inferring pseudo-time analysis, and investigating cell–cell communication. Before clustering analysis for tissue architecture identification, stLearn firstly normalizes expression value based on the Spatial Morphological gene Expression normalization method (SME), which integrates gene expression, spatial location, and histology information via a transfer learning deep neural network model.
Moreover, statistical frameworks also play a pivotal role in spatial transcriptomics analysis. BayesSpace adopts a Bayesian statistical framework, uses the low-dimensional representation (e.g., PCA) of gene expression as input, employs the spatial smoothing (the Potts model) prior to model spatial correlation, and identifies tissue architecture using latent clusters based on the Metropolis-Hastings algorithm [10], [16]. BayesSpace can also be extended to computationally enhance resolution and bring insights at the sub-spot level. The hidden Markov random field (HMRF) is another approach to inform the organizational structure unbiasedly and has been mainly applied to image-based spatial transcriptomics. As the domain state of each cell spot (spot for short) was influenced by its gene expression pattern and the domain states of neighboring spots [17], HMRF considers gene expression information and spatial environment information simultaneously, which is essential to depict the heterogeneity and has been successfully integrated into Giotto.
Although these methods have been successfully implemented for tissue architecture identification, the prediction accuracy still has room to be improved, and the learned low-dimensional representations can seldom be visualized intuitively. The heterogeneity of tissue architecture cannot be fully viewed and characterized due to a lack of strong spatial representation for maximally retaining tissue heterogeneity. Therefore, it is still challenging to represent spatial heterogeneity, accurately characterize tissue architectures, and understand the underlying biological functions from spatial transcriptomics. We reasoned that three-dimensional embeddings from spatial transcriptomics could be transformed into RGB values for biologically-interpretable visualization and direct applications of state-of-the-art computer vision methods. RGB in computation graphics can resemble more than 16.7 million colors, while the human eyes can distinguish 2.3 million colors [18]. That means RGB values converted from three-dimensional embedding can intrinsically and intuitively reflect the human-eye distinguishable heterogeneity of tissue architecture. Moreover, we hypothesize that tissue architecture can be visualized and segmented from an RGB image converted by the low-dimensional representations embedding gene expression profiles and spatial topology of spots.
To this end, we formulate tissue architecture identification as an image segmentation problem in the computer vision field and introduce RESEPT (REconstructing and Segmenting Expression mapped RGB images based on sPatially resolved Transcriptomics), a framework for reconstructing, visualizing, and segmenting an RGB image from spatial transcriptomics to reveal tissue architecture and spatial heterogeneity. We highlight the unique features of RESEPT as follows: (i) to the best of our knowledge, RESEPT is a first-of-the-kind framework for identifying tissue architecture using the computer vision technique (i.e., segmentation). In detail, the image can also be sent to a pre-trained segmentation deep-learning model and an optional segmentation quality assessment protocol, which resists robustly to noises and artifacts. (ii) RESEPT enhances the interpretability for low-dimensional representation. Specifically, high-dimensional spatial transcriptomics data are converted as a human-eye distinguishable RGB image by mapping a low dimensional embedding to RGB color channels via a spatial retained graph neural network. It is noteworthy that the image intrinsically reflects tissue heterogeneity, and each RGB channel can associate with SVGs, which supports the basis of tissue architecture. (iii) With a defined panel of gene sets representing specific biological pathways or cell lineages, RESEPT can recognize the spatial pattern and detect the corresponding active functional regions. Specifically, the functional zonation boundaries of AD are determined effectively and flexibly by our segmentation model. (iv) RESEPT is capable of recognizing tumor, non-tumor, and tumor infiltration architectures in glioblastoma, and has demonstrated its applicative power in defining spatial information of human breast cancers and mouse brains.
2. Materials and methods
2.1. RESEPT pipeline
RESEPT is implemented in two major steps: (i) reconstruction of an RGB image of spots using gene expression or RNA velocity from spatial transcriptomics sequencing data; (ii) implementation of a pre-trained image segmentation deep-learning model to recognize the boundary of specific spatial domains and to perform functional zonation. Fig. 1, Fig. 2 demonstrate the pipeline with conceptual description and technical details, respectively.
Fig. 1.
The RESEPT schema. RESEPT takes gene expression or RNA velocity from spatial transcriptomics as the input. The input is embedded into a three-dimensional representation by a spatially constrained Graph Autoencoder, then linearly mapped to an RGB color spectrum to reconstruct an RGB image for visualization. A CNN image segmentation model is trained to obtain a spatially specific architecture (from whole-gene embedding) or spatial functional regions (from panel-gene embedding). Taking the human dorsolateral prefrontal cortex as an example (sample 151,510 in Supplementary Table 1), the adjusted rand index (ARI) is 0.839, which means the predictive result can faithfully reveal tissue architecture.
Fig. 2.
The RESEPT framework. (a) A spatial retained spot graph is established by spatial distances of spots and their expression or velocity matrix. The graph autoencoder takes the adjacent distance matrix of the spot graph as the input. Its encoder learns a 3-dimensional embedding of a spatial cell graph. The decoder reconstructs the adjacent correlations among all cells by dot products of the 3-dimensional embeddings followed by a sigmoid activation function. The graph autoencoder is trained by minimizing the cross-entropy loss between the input spatial and the reconstructed graphs. The learned 3-dimensional embeddings are mapped to a full-color spectrum to generate an RGB image revealing the spatial architecture. (b) The segmentation model takes the RGB image as the input, which may be processed with an imputation operation if missing spots exist. Its backbone network ResNet101 consists of one convolutional layer and a series of residual blocks, in which one type of residual block named convolutional block stacks three convolutional layers with a convolutional skip connection from the input signals to the output feature maps. The other type of residual block identity block stacks three convolutional layers with a direct skip connection from the input signals to the output feature maps. This extra deep network firstly extracts rich visual features of the input image. The encoder module further extracts multi-scale semantic features by applying four atrous convolutional with different rates and sizes of filters and one global pooling layer to the basic visual feature maps. And the decoder module up-samples the multi-scale features to the same size with basic visual feature maps and then concatenates them together. After a softmax activation function, the decoder module outputs a segmentation map classifying each spot into a specific spatial architecture.
2.2. Construct RGB image for spatial transcriptomics
An RGB image is constructed to reveal the spatial architecture of a tissue slice using three-dimensional embedding as the primary color channels. Besides gene expression, RESEPT can accept RNA velocity [19] as the input. RNA velocity unveils the dynamics of RNA expression at a given time by distinguishing the ratio of unspliced and spliced mRNAs, reflecting the kinetics and potential influences of transcriptional regulations in the present to the future cell state. The original BAM file of human studies is often unavailable to public users due to ethical reasons, and hence, in most cases, we only refer to expression-derived RGB images in our study. The scGNN [20] package is used to generate spatial embeddings for each spot based on the pre-processed expression matrix or RNA velocity matrix, along with the corresponding meta-data. In practice, RESEPT can adapt any type of low dimensional representations, such as embedding from UMAP, SEDR [21], and spaGCN [14]. On benchmarks, scGNN embedding obtained better results in most cases, so RESEPT uses scGNN in default.
2.2.1. Positional variational autoencoder
After log-transformation and library size normalization by count-per-million (CPM), the spatial transcriptome expression as the input is embedded into a low dimensional vector through an autoencoder. Both the encoder and the decoder consist of two symmetrically stacked layers of dense networks followed by the ReLU activation function. The encoder learns the embedding from the input gene expression matrix (selecting top 2000 highly variable genes by default), and the encoder reconstructs the matrix from the In addition, a positional encoding [22] as Eq. (1) is incorporated in the learning process to characterize the spatial coordinates and make the embedding space-aware.
| (1) |
where is set to the spot number along one dimension of the spatial slide [22]; is 2D Cartesian coordinates; and are coordinate indices, denotes the scale factor of positional encoding; denotes embedding matrix from autoencoder learned from expression matrix only. and , where is the number of input genes from the spatial transcriptome, is the dimension of the learned embedding (). is the number of spots on the spatial slide. The objective of the training is to achieve a maximum similarity between the original and reconstructed matrices measured by minimizing the mean squared error (MSE) as the loss function.
2.2.2. Generating Spatial retained Spot Graph
The cell graph is a powerful mathematical model to formulate cell–cell relationships based on similarities between cells. In single-cell RNA sequencing (scRNA-seq) data without spatial information, the classical K-Nearest-Neighbor (KNN) graph is widely applied to construct such a cell–cell similarity network in which nodes are individual cells, and the edges are relationships between cells in the gene expression space. With the availability of spatial information in spots as the unit of observation arranged on the tissue slice, our in-house tool scGNN adopts spatial relation in Euclidean distance as the intrinsic edge in a spot-spot graph. Each spot in the spatial transcriptomics data contains one or more cells, and the captured expression or the calculated RNA velocity is the summarization of these cells within the spot. Only directly adjacent spots in contact in the 2D spatial plane have edges between them, and hence, the lattice of the spatial spots comprises the spatial spot graph. For the generated spot graph , denoting the number of spots and representing the edges connecting with adjacent neighbors. is its adjacency matrix and is its degree matrix, i.e., the diagonal matrix of the number of edges attached to each node. The node feature matrix is the learned embedding from the dimensional reduction autoencoder. In the 10x Visium platform, each spot has six adjacent spots, so the spatial retained spot graph has a fixed node degree of six for all the nodes. Similar to the KNN graph derived from scRNA-seq, each node in the graph contains attributes.
2.2.3. Graph autoencoder
Given the generated spatial spot-spot graph, a graph autoencoder learns a node-wise three-dimensional representation to preserve topological relations in the graph. The encoder of the graph autoencoder composes two layers of graph convolution network (GCN) to learn the low dimensional graph embedding in Eq. (2).
| (2) |
where is the symmetrically normalized adjacency matrix and is a weight matrix learned from the training. The output dimensions of the first and second layers are set as 32 and 3, according to the three color channels as RGB, respectively. The learning rate is set at 0.001.
The decoder of the graph autoencoder is defined as an inner product between the graph embedding , followed by a sigmoid activation function:
| (3) |
where is the reconstructed adjacency matrix of .
The goal of graph autoencoder learning is to minimize the cross-entropy between the input adjacency matrix and the reconstructed matrix .
| (4) |
where and are the elements of adjacency matrix and , . As there are nodes as the number of spots in the slide, is the total number of elements in the adjacency matrix.
2.2.4. Reconstruct RGB Image
The learned embedding is capable of representing and preserving the underlying relationships in the modeled graph from spatial transcriptomics data. Meanwhile, the three-dimensional embedding can also be intuitively mapped to Red, Green, and Blue channels in the RGB space of the image. Normalized to an RGB color space accordingly to a full-color spectrum (pixel range from 0 to 255) as Eq. (5), the embedding of each spot is assigned a unique color for exhibiting the expression or velocity pattern in space.
| (5) |
where and is its transformed color of the -th spot in the -th channel, , . and represent the maximum and minimum of all embedding values in the RGB channels, respectively. With their coordinates and diameters at the full resolution provided from 10x Visium, we are able to plot all spots with their synthetic colors on a white drawing panel and reconstruct a full-size RGB image explicitly describing the spatial expression or velocity properties in the original spatial coordinate system. For the spatial transcriptomic data sequenced in lattice from other techniques, such as the ST platform, RESEPT allows users to specify a diameter to capture appropriate relations between spots in the RGB image accordingly.
2.3. RGB image segmentation model
The RGB image makes the single-cell spatial architecture perceptible in human vision. With the constructed image, we treat the potential functional zonation partition as a semantic segmentation problem, which automatically classifies each pixel of the image into a spatially specific segment. Such predictive segments reveal the functional zonation of spatial architecture.
2.3.1. Image segmentation model architecture
We trained an image-segmentation model based on a deep architecture DeepLabv3+[23], [24], which includes a backbone network, an encoder module, and a decoder module (Fig. 2).
Backbone network. The backbone network provides dense visual feature maps for the following semantic extraction by any deep convolutional network. Here, ResNet-101 [25] is selected as the underlying model for the backbone network, which consists of a convolutional layer with 64-channels in size of filters and 33 residual blocks, each of which stacks one convolutional layer with multi-channel (including 64, 128, 256, and 512) in size of filters and two convolutional layers with multi-channel (including 64, 128, 256, 512, 1024 and 2048) size of filters. The generated RGB image is mapped into a -channel feature map by the first convolutional layer and gradually fed into the following residual blocks to produce rich visual feature maps for describing the image from different perspectives. Here, equals 64. In each residual block, the feature map generated from the previous block is updated to in Eq. (5).
| (6) |
where
is the activation function, and we use ReLU [26] in this study.
represents the learning convolutional weights in the th block,.
represents the learning weights of the convolutional layer with 1 × 1 kernel size.
Element-wise addition operation in Eq. (6) enables a direct shortcut to avoid the vanishing gradient problem in this deep network. In the 1st, 4th, 8th, and 31st blocks of the 33 residual blocks, their input and output dimensions do not match up due to different filter settings from their previous layers. Accordingly, the projection shortcut with an additional 1 × 1 convolution in Eq. (6) is used to align dimensions in these blocks, which are also named identity blocks. The rest blocks stacked on the previous blocks with the same filter settings employ a direct shortcut. We leveraged ResNet-101 as a basic visual feature provider and sent the most informative feature maps from the last convolutional layer before logits to the following encoder module.
2.3.2. Encoder module
The aim of the encoder module is to capture multi-scale contextual information based on the dense visual feature maps from the backbone. To achieve the multi-scale analysis, atrous convolution [23] is adopted in the encoder to extend the size of the respective field. For the generated RGB image with width and length , the total number of spots . Given the input signal from Eq. (6) as with a -channel filter , the output feature signal is defined as follows:
| (7) |
where
represents the input signal at the location (i, j) with c-channel values. , . is the stride rate in atrous convolution.
represents the convolutional weights with c’-channel values, . K is the kernel size of the convolutional filter.
represents the output signal at the location (i, j) with c’-channel values.
Compared to the standard convolution, the atrous convolution samples the input signal with the stride r rather than using direct neighbors inside the convolutional kernel. Therefore, the standard convolution is a special case of atrous convolution with r = 1. By using multiple rate value settings (rate = 1, 6, 12, and 18), we separately apply-one standard convolutional layer with 256-channel size of filters (i.e., the atrous convolutional layer with rate = 1), three atrous convolutional layers with 256-channel size of filters and an additional average pooling layer to produce high-level multi-scale features. These semantic features are then merged into the decoder module.
2.3.3. Decoder module
In the decoder, the high-level input features are bilinearly up-sampled and concatenated with the basic visual features for recovering the segment boundaries and spatial dimension. A standard convolutional layer with 256-channel size filters is applied to outweigh the importance of the merged features and obtain sharper segmentation results. Eventually, an additional bilinear up-sampling operation forms the output of the decoder to a matrix, where and denote the width and height of the input image, respectively. The following convolution layer with predefined d-channel size of filters squeezes the feature matrix along the channel axis to shape, where each pixel is represented by a -dimensional features for the following inference. In the training stage, the softmax [27] function is then applied to generate a segment category of each pixel leading to a size segmentation map. The pixels falling into a certain category in the segmentation map point to a segmented spatial region. Our modeling objective is to minimize the cross-entropy [28] between the predictive segmentation map and labeled spatial functional regions :
| (8) |
where and are the segment categories of the pixel at the -th row and the -th column for the input images with pixels. , .
2.3.4. Training set data preparation
We performed scGNN using various autoencoder dimensions ( = 3, 10, 16, 32, 64, 128, and 254) and multiple positional encoding intensity parameters ( = 0.1, 0.2, 0.3, 0.5, 1.0, 1.2, 1.5, and 2.0), resulting in 56 embeddings used to generate diverse RGB images for each sample in the training set (see image results in “RGB image results” folder on https://github.com/OSU-BMBL/RESEPT/). In this study, we performed 16-fold Leave-One-Out Cross-Validation (LOOCV). In each fold, one sample was randomly extracted as the testing data, and the rest samples were treated as the training samples. For an unbiased evaluation, the mean of 16 ARIs from the 16-fold LOOCV was used as the comprehensive assessment metric, as shown in Fig. 3.
Fig. 3.
The RESEPT workflow and performance. (a) Mean and standard deviation of sequencing reads of 17 human brain datasets on 10x Visium platform. CT1 to 151,508 have manual annotations as the benchmark, CT2 & 151,674 for simulation for high mean and low standard deviations of read depth, G1, AD1, and AD2 for the case studies (more details in Supplementary Tables 1–2). (b) Performance of tissue architecture (with 7 clusters pre-defined) identification by seven existing tools and RESEPT on criteria ARI. (c) Stability of tissue architecture identification across sequencing depths on samples CT2 using different tools. The Y-axis shows ARI performance, and the X-axis represents the sequencing depth with subsampling. The lines are smoothed by the B-Spline smooth method. (d) Normalized performance vs sequencing depth on sample CT2. Performance of full sequencing depth is set as 1.0. RESEPT_E1 using the scGNN embedding, RESEPT_E2 using the spaGCN embedding. (e) and (f) The stability of ARI and normalized performance against the grid sequencing depth for samples CT2 and 151674. CT2 and AD2 results of HMRF were excluded due to failure to produce outcomes. (g) Ground truth of AD2. (h) Spatial domains on AD2 detected by RESEPT. (i) – (n) Tissue architecture results based on BayesSpace, Seurat, Giotto, stLearn, and SpaGCN, respectively.
2.3.5. Model training
We implemented the training procedure on the MMSegmentation platform [29], which is an open-source semantic segmentation toolbox based on PyTorch. The weights of DeepLabv3 + were initialized by the pre-trained weights from the Cityscapes dataset provided by MMSegmentation. To introduce diversity to the training data and improve the generalization of our model, we applied transforms defined in MMSegmentation, including the random cropping, rotation and photometric distortions, to augment the training RGB images. 400 × 400 sized patches are randomly cropped to provide different regions of interest from the whole RGB images. A random rotation (range from −180 degrees to 180 degrees) was further conducted to fit the potential irregular layout of spatial architectures. Some photometric distortions such as brightness, contrast, hue, and saturation changes were also utilized to augment training samples when loading to MMSegmentation. Stochastic gradient descent (SGD) [30] was chosen as the optimization algorithm, and its learning rate was set to 0.01. The training procedure iterated 30 epochs, and the checkpoint among all epochs with the best Moran’s I autocorrelation index [31] on the testing data was selected as the final model.
2.3.6. Image segmentation inference
Once a model completes training, it is capable of predicting the functional zonation on the tissue from its RGB images. On the inference, RESEPT performs scGNN with the same parameter combinations with the training settings resulting in 56 candidate RGB images for each input sample. The dimensional feature maps of each image before logits are extracted by DeepLabv3+ (see details in the encoder module). Then the k-means clustering algorithm [32] is applied to segment all pixels into clusters according to their dimensional features. RESEPT infers all the segmentation maps on these 56 images and scores them using the Moran’s I metric to assess the quality of segmentations. The segmentation maps of 5-top ranked images in terms of Moran’s I are returned for user selection. We found that such a quality assessment protocol results in segmentation results with higher accuracy than the default one and enhances the robustness of RESEPT. By setting the parameter , users can specify the number of segments to RESEPT. In the case of no user-specified k, RESEPT goes through a range of candidates and calculates their Moran’s I values for assessing the quality of segmentation result with each candidate . Eventually, the corresponding to the highest Moran’s I is selected as the default number of segments.
2.4. Experiment preparation, data generation, and processing
2.4.1. Experiment preparation and data generation
Four postmortem human brain samples of the middle temporal gyrus [33] were obtained from the Arizona Study of Aging and Neurodegenerative Disorders/Brain and Body Donation Program at Banner Sun Health Research Institute [34] and the New York Brain Bank at Columbia University Medical Center [35]. Two of them are from non-AD cases at Braak stage I-II, namely Samples CT1 and CT2 in the study, and the other two are from early-stage AD cases at Braak stage III-IV, namely Samples AD1 and AD2 in the study. The region of AD cases was chosen based on the presence of Aβ plaques and neurofibrillary tangles. Specifically, Visium is a spatial barcode-based technology based on a glass microscope slide with four capture areas (6.5 mm × 6.5 mm) [36]. Each capture area can profile up to 4992 spots, and the diameter of each spot is approximately 55 μm [9], [36], [37].
The 10x Genomics Visium Spatial Transcriptome experiment was performed according to the User Guide of 10x Genomics Visium Spatial Gene Expression Reagent Kits (CG00239 Rev D). All the sections were sectioned into 10 µm thick and mounted directly on the Visium Gene Expression (GE) slide for H&E staining and the following cDNA library construction for RNA-Sequencing. Besides the section mounted on the GE slide, one of the adjacent sections (20 µm away from GE section) from AD samples persevered for the Aβ immunofluorescence staining. The method of immunofluorescence staining of Aβ on persevered section was the same as previously described [38]. The image of Aβ staining was used as the ground truth and was aligned to H&E staining on GE slides using the “Transform/Landmark correspondences” plugin in ImageJ [39].
2.4.2. FASTQ generation, alignment, and count
BCL files were processed by sample with the SpaceRanger (v.1.2.2) to generate FASTQ files via spaceranger mkfastq. The FASTQ file was then aligned and quantified based on the reference GRCh38 Reference-2020-A via spaceranger count. The functions spaceranger mkfastq and spaceranger count were used for demultiplexing sample and transcriptome alignment via the default parameter settings.
2.5. Data preprocessing
To standardize the raw gene expression matrix and spot metadata, the different spatial transcriptomics data were preprocessed as follows.
For the 10x Visium data (Supplementary Table 1), the filtered feature-barcode matrix (HDF5 file) was reshaped into a two-dimensional dense matrix in which rows represent spots and columns represent genes. The dense matrix was further added with spots’ spatial coordinates by merging them with the ‘tissue_positions_list’ file, containing tissue capturing information, row, and column coordinates. The mean color values of the RGB channels for each spot’s circumscribed square and annotation label were also added to the dense matrix after processing the Hematoxylin-Eosin (H&E) image. The gene expression as part of the dense matrix was stored in a sparse matrix format. Other information describing the spots’ characteristics was stored as individual metadata.
For the HDST data, the expression matrix and spots’ coordinates were reshaped into the dense matrix, which was similar to 10x Visium preprocessing. The expression matrices from dense matrices were formed into the individual sparse matrices, and other information was stored as metadata.
For the ST data, the expression matrix was reshaped into the two-dimensional dense matrix, and spots’ spatial coordinates were added to the dense matrix by merging with the spot_data_selection file. The color values of each spot were added to the dense matrix after processing the H&E image (if available). The remaining steps were the same as for the 10x Visium data.
2.6. Data normalization and denoising
2.6.1. Data normalization
The raw read counts were used as formatted input to generate normalization matrices. Seven normalization methods were used in the study, including DEseq2 [40] (v.1.30.1), scran [41] (v.1.18.5), sctransform [42] (v.0.3.2), edgeR [43] (v.3.32.1), transcripts per million (TPM), reads per kilobase per million reads (RPKM), and log-transformed counts per million reads [44] (logCPM). We used Seurat (v.4.0.1) to generate the sctransform and the logCPM normalized matrices. edgeR was used to generate TMM [43] normalized matrices. The gene length was used for calculating TPM, and RPKM was obtained from biomaRt (v.2.46.3) by using useEnsemble function and parameters setting as dataset=“hsapiens_gene_ensembl” and GRCh = 38. All normalized matrices for whole transcriptomics were eventually calculated via the default settings and converted into sparse matrices. RNA velocity was calculated for the whole transcriptomics via velocyto [19] (v.0.17.17) and scVelo [45] (v.0.1), followed by their default settings. RNA velocity matrices were converted into sparse matrices.
2.6.2. Missing spots imputation
In practice, several spots may have a missing expression in some tissue slices due to imperfect technology, which leads to blank tiles at the locations of these spots on the RGB images. Such blank tiles as incompatible noises may skew the following boundary recognition of spatial architecture. We assume the near neighbors are more likely to have similar values to the missing spot and impute them by applying the weighted average to the pixels of their valid six neighboring spots. Since these missing spots are colored while in default as the same as the background out of tissue, we need to distinguish them from all-white pixels according to a topological structural analysis [46]. Firstly, all contours (including outer contours of tissue and inner contours caused by missing spots) of tissue are detected from the border following the procedure in [46]. The contour with the largest area is determined as the outer contour of tissue. Then, all pixels in white inside the tissue contour are replaced by imputations from their neighbors. Given missing spot coordinates, we search their nearest k valid spots (i = 1, 2,…, k) to calculate the imputation value of target missing spot s as:
| (9) |
where represents the Euclidean distance between target spot s and a certain neighbor in spatial space. The softmax function normalizes all distance reciprocals of s and its k (we set k = 6 by default) neighbors si to the weights ranging from 0 to 1. The imputation of s is the weighted average on all si. If a tissue slice is detected without missing spots, RESEPT skips this imputation process.
2.6.3. Parameter setting
Parameters in scGNN to generate embedding are referred to in the previous study [20]. In the case study of the AD sample, in analysis on cortical layers 2 & 3, the expressions of 8 well-defined marker genes were log-transformed and embedded by spaGCN with 0.65 resolution. In the analyses of cortical layer 2 to layer 6, PCA (n.PCs = 3) was firstly utilized to extract the principal components of their expressions of marker genes for highlighting the dominant signals, and then they were embedded by spaGCN with 0.65 resolution. In the exploration of tumor regions in glioblastoma samples, their marker gene expressions were preprocessed by logCPM normalization and PCA (n.PCs = 50). The processed data was embedded by spaGCN with 0.35 resolution. In the analyses of AD-associated critical cell types, marker gene expressions were preprocessed by log-transform and PCA (n.PCs = 3) as well and then embedded by spaGCN with 0.65 resolution. For investigating Aβ pathological regions, log-transform to the expressions of validated 20 upregulated genes was applied, and their embedding was generated by spaGCN with 0.65 resolution.
2.7. Benchmarking method
All the benchmarking tasks were run on a Red Hat Enterprise Linux 8 system with 13 T storage, 2x AMD EPYC 7H12 64-Core Processor, 1 TB RAM 1 TB DDR4 3200 MHz RAM, and 2x NVIDIA A100 GPU with 40 GB RAM. The usage of the existing tools and their parameter settings in our benchmarking evaluation are described below.
Seurat (v.4.0.1) identifies tissue architecture based on graph-based clustering algorithms (e.g., the Louvain algorithm). Creating a Seurat object, identification of highly variable features, and scaling of the data were performed using default parameters. The PCs were set to 128 to match our framework’s default setting. The FindNighbors and FindClusters functions with default parameters were used for tissue architecture identification. To further evaluate the robustness of the combination of the different parameters, we used 16 samples and selected three important parameters, including the number of PCs (dims = 10, 32, and 64), the value of for the FindNeighbor function (k.parm = 20, 50 and 100), and the resolution in the FindClusters function (res = 0.1 to 1, step as 0.1).
BayesSpace (v.1.0.0) identifies tissue architecture based on the Gaussian mixture model clustering and Markov Random Field at an enhanced resolution of spatial transcriptomics data. Creating the SingleCellExperiment object is implemented in the following analysis by loading normalized expression data and position information for barcodes. Then, we set 128 as the number of PCs in spatialPreprocess function and parameter log.normalize was set FALSE due to the normalized data input. Lastly, tissue architecture was identified by running qTune and spatialCluster functions. We followed the official tutorial and adopted k-means as the initial method, while other parameters were from the default based on prior information. In assessing the robustness of BayesSpace, we set the cluster number as seven, the parameter n.PCs in spatialPreprocess function (n.PCs = 10, 64, and 128), and the parameter nrep in spatialCluster function (nrep = 5000, 10000, and 150000) for 16 samples.
SpaGCN (v.0.0.5) can integrate gene expression, spatial location, and histology to identify spatial domains and spatially variable genes by graph convolutional network. SpaGCN was used to generate three-dimensional embedding and tissue architecture and includes three procedures, including loading data, calculating adjacent matrix, and running SpaGCN. In the first step, both expression data and spatial location information were imported. Second, adjacent matrices were calculated using default parameters. Lastly, we selected 128 PCs, the initial clustering algorithm as Louvain, and other parameters used default settings. To evaluate the robustness of the parameters and enable comparison with other tools, three parameters, the number of PCs (num_pcs = 20, 30, 32, 40, 50, 60, 64), the value of for the k-nearest neighbor algorithm (n_neighbors = 20, 30, and 40), and the resolution in the Louvain algorithm (res = 0.2, 0.3, and 0.4) for 16 samples were adjusted.
stLearn (v.0.3.2) is designed to comprehensively analyze ST data to investigate complex biological processes based on Deep Learning. stLearn highlights innovation to normalize data. Therefore, we input expression data, location information as well as images. stLearn consists of two steps, i.e., preparation and running stSME clustering. In preparation, loading data, filtering, normalization, log-transformation, preprocessing for spot image, and feature extraction were implemented. In the following module, PCA dimension reduction was set to 128 PCs, applying stSME to normalize log-transformed data and Louvain clustering on stSME normalized data using the default parameters. To evaluate the robustness of the parameters and enable comparison with other tools, three parameters were considered to be adjusted for 16 samples, the number of PCs (n_comps = 10, 20, 30, 32, 40, and 50), the value of for the kNN algorithm (n_neighbors = 10, 20, 30, 40, and 50), and the resolution in the Louvain algorithm (resolution = 0.7, 0.8, 0.9 and 1).
STUtility (v0.1.0) can be used to identify spatial expression patterns alignment of consecutive stacked tissue images and visualizations. We implemented STUtility as a tissue architecture tool based on the Seurat framework. RunNMF was carried out as the dimension reduction method. The number of factors was set to 128 to match our framework’s default setting. FindNeighbors and FindClusters were used to identify tissue architecture. To further evaluate the robustness of the combination of the different parameters, we used 16 samples and selected three important parameters for tuning, including the number of factors (nfactors = 10, 32, and 64), the value of for FindNeighbor function (k.parm = 20, 50, 100, 200, and 250), and the resolution in FindClusters function (res = 0.05, 0.1, 0.2, 0.3, 0.5, and 0.7, 0.9).
Giotto (v.1.0.3) is a comprehensive and multifunction computational tool for spatial data analysis and visualization. We implemented Giotto as the issue architecture identification tool in this study via using default settings. Giotto first identified highly variable genes via calculateHVG function, then performed PCA dimension reduction using 128 PCs, constructed the nearest neighbor network via createNearestNetwork, and eventually identified tissue architecture via doLeidenCluster. To further evaluate the robustness of the combination of the different parameters, we used 16 samples and selected three important parameters for tuning, including the number of PCs (npc = 10, 32, and 64), the value of for createNearestNetwork function (k = 20, 50 and 100), and the resolution in doLeidenCluster function (resolution = 0.1, 0.2, 0.3, 0.4, and 0.5).
smfishHmrf (v.1.3.3) can distinguish between intrinsic and extrinsic effects on global gene expression to dissect the cell-type- and spatial-domain-associated heterogeneity. smfishHmrf builts on the hypothesis that tissue is divided into domains with coherent gene expression patterns. To begin with the analysis, filtering genes, and selecting highly variable genes were performed using scanpy. Then, the gene expression matrix was used to compute the neighbor graph and calculate the silhouette score for each gene using the default parameters or recommended parameters, and the significant genes were preserved for the following analysis. After this preprocessing, HRMF is performed to assign a domain for each spot. To evaluate the robustness of the parameters and enable comparison with other tools, three parameters were considered to be adjusted according to silhouette score (n_genes = 40, 60, 80, 100, 120, and 140), the cutoff values in computing neighbor graph (cutoff = 0.3, 0.5, 0.7 and 1), and the beta values in the HRMF model (beta = 6, 9, 12 and 15).
Downsampling simulation for read depth. Comparing the mean and standard deviation of 16 10x Visium datasets, samples CT2 and 151,674 were selected to generate simulation data with decreasing sequencing depth. Let matrix be the expression count matrix, where is the number of spots and is the number of genes. Define the spot-specific sequencing depths , i.e., the column sums of . Thus, the average sequencing depth of the experiment is . Let be our target downsampled sequencing depth, and let be the downsampled matrix. We perform the downsampling as follows:
For each spot
Define the total counts to be sampled in the spot as .
Construct the character vector of genes to be sampled as.
Sample elements from without replacement and define as the number of times gene was sampled from for .
Let = .
Using this method, the average downsampled sequencing depth is:
as desired. Note also that this method preserves the relative total counts of each spot, i.e., spots with higher sequencing depths in the original matrix have proportionally higher depths in the downsampled matrix.
2.8. Benchmark performance evaluation criteria
Adjusted Rand Index (ARI), Rand index (RI), Fowlkes–Mallows index (FM), and Adjusted mutual information (AMI) are used to evaluate the performances between the ground truth and predicted results.
Adjusted Rand index (ARI) measures the agreement between two partitions. Given a set consisting of elements, and are two partitions of ; that is, and , so does . can be interpreted as a cluster generated by some clustering method. In this way, ARI can be described as follow:
| (10) |
where , denotes the number of objects in common between and ; and . Besides, , the higher reflects the higher consistency. The bs function of the splines package (v.4.0.3) was used for smoothing ARI generated from grid effective sequencing depth data via default settings.
Rand index (RI) is also a measure of the similarity between two data clustering results. If the ground truth is available, the can be used to evaluate the performance of one cluster method by calculating between the clustering produced by this method and the ground truth. Let be a set containing elements, which represents barcodes in this paper, and two partitions of , , ; that is, and ; so does . and are the subset of , representing one cluster produced by some clustering method and the ground truth, respectively. can be computed using the following formula:
| (11) |
where:
denote the number of pairs of elements in in the same subset in and in the same subset in , in different subsets in and in different subsets in , in the same subset in and in different subsets in , and in different subsets in and in the same subset in , respectively.
is the binomial coefficient. In addition, the range of is , and the higher , the higher similarity of the two partitions is.
The Fowlkes–Mallows index (FM) is an external evaluation method, which can measure the results’ consistency of two cluster algorithms. Not only can be implemented on two hierarchical clusterings, but also the clusters and the benchmark classifications. For the set of objects, and denote two clustering results (generated by two cluster algorithms, one for the clustering algorithm, one for the ground truth). In this paper, is produced by a clustering algorithm while the ground truth contributes . If the clustering algorithm performs well, then and should be as similar as possible. The calculation of can be described as:
| (12) |
where
is the number of true positives, representing the number of pair objects that are present in the same cluster in both and .
is the number of false positives, representing the number of pair objects that are present in the same cluster in but not in .
is the number of false negatives, representing the number of pair objects that are present in the same cluster in but not in .
is so-called while refers to In addition, . Therefore, in our cases, the closer it is to , the better the clustering algorithm will be.
Adjusted mutual information (AMI) is driven by probability theory and information theory and can be used for comparing clustering results. To introduce adjusted mutual information, the preliminary is necessary to present two conceptions of mutual information () and entropy. Given a set , and are two partitions of , that is, and , so does . between partition and is defined as:
| (13) |
where
measures the probability of one object belonging to and simultaneously.
The entropy associated with the partitioning is defined as:
| (14) |
where
refers to the probability that the object falls into the cluster .
and have analogous definitions.
The following formula shows the expected mutual information between two random clustering results:
| (15) |
where ; and , , represents the number of objects in common between and . Finally, can be obtained by
| (16) |
It should be pointed out that , the similarity between the two clusterings increases with the augment of .
2.9. Predicted segmentation map quality assessment
Differing from the Moran’s I auto-correlation index [31] used for revealing a single gene’s spatial auto-correlation, we modified Moran’s I in Geo-spatiality [47] to evaluate a predictive segmentation map without known ground truth. The metric analyzes the heterogeneity of predictive inter-segments by measuring the pixel contrast across any two predicted adjacent segments per channel. And then, the mode of Moran’s I from three RGB channels is computed:
| (17) |
where
is the binary spatial adjacency of the th segment and jth segment. ,
denotes the mean pixel values at channels in Red, Green, and Blue of the th segment, ,
denotes the mean pixel values at channels Red, Green, and Blue of the whole image.
2.10. RGB image and three-dimensional embedding evaluation
We reused the concept of Moran’s I to assess the color distribution of an RGB image and its annotated tissue architecture. In this case, defined in equation(17) is calculated according to a labeled segmentation map rather predicted one. Hence, such a Moran’s I score reflects the heterogeneity between any two adjacent regions on annotated tissue architectures. The larger Moran’s I from an RGB image illustrates that the better this RGB image can display biological tissue structures, and further implies the better quality its corresponding three-dimensional embedding can achieve.
2.11. Pixel correlation analysis between RGB channels and SVGs
SpatialDE [48] is used to detect the sample’s SVG, the SVGs with q-value < 0.0001 and Bayesian information criterion (BIC) greater than 0 are kept. Then k-means is conducted to cluster these SVGs into three groups, each of which is expected to contribute to a single R/G/B channel. Samples disentangling each group of genes are treated as the inputs and reconstructed as RGB images using RESEPT. These RGB images are further converted into gray-level images, which were treated as the encoding expression profile of the SVG groups in a single channel. Then, the pixel correlations (the Pearson correlation over the pixels from two images) cross each SVG expression profile image, and each of the three RGB channels was measured to observe their corresponding relationships.
2.12. Mouse cortex region annotation
Nine mouse brain cortex regions were cropped by Loupe Browser (V.5.0.1) manually. Following the SpaGCN’s annotation method [14], our neuroscience specialist referred to the Allen Brain Reference Atlases (https://atlas.brain-map.org/) [49], observed the cell density of the expected region (layers) based on the H&E image, and generated the mouse cortex architecture of nine samples for training the model and performance comparison.
2.13. Module score calculation and differential expression analysis
The module score for specific marker genes was calculated based on the Seurat function AddModuleScore. This function produces the gene module score to indicate whether the gene module has a higher mean expression in a group of spot subsets. The first step is to calculate the mean expression values of the input gene list as the targeted gene module for each spot. The second step is to generate a null distribution of gene module scores as the background. For this purpose, the average expression values across all spots for whole genes are calculated, sorted, and binned into 24 bins. Same as the targeted gene module size (i.e., number of genes in this targeted gene module), the null distribution of gene module scores will be generated by randomly selecting 100 times based on previously ranked and binned average expression values and then calculating the mean expression value as we did in the first step.
Finally, the gene module score is the targeted gene module mean expression value subtracted by the mean expression value of the null distribution. The DEG analysis was conducted by the Seurat function FindAllMarkers based on RESEPT predicted seven segments via default settings. Based on the identified DEGs, the enrichment analyses of GO terms (Biological Process) and KEGG were performed via the R package clusterProfile (v.3.18.0) using the functions of enrichGO and enrichKEGG. The enrichment analysis results were filtered out if the adjusted p-value was greater than 0.05. For KEGG analysis, gene database Org.Hs.eg.Db was used for transferring SYMBOL to ENREZID via function bitr. R package ggplot2 (v.3.3.2) was used for the visualizations.
3. Results
3.1. The architecture of RESEPT comprises representation learning and segmentation
We choose graph neural network (GNN)[50] to learn the low dimensional representation as a dimensional reduction step since GNN has demonstrated its power in modeling relations between cells in single-cell RNA-seq [20] and spots in spatial transcriptomics [14]. The learned low dimensional embedding in RESEPT enables reconstructing the graph's topology and inherently conserves the ambient gene expression relations in the 2D space of the sample slice, which empowers the reconstructed RGB image to faithfully depict the tissue heterogeneity. Compared to the traditional method of determining architectures of the human cerebral cortex by observing cell morphology and the density of high-resolution Hematoxylin-Eosin (H&E)-stained images, the RESEPT framework produces two major outputs describing tissue architectures from different angles. One is a reconstructed visualizable RGB image to display tissue heterogeneity using the low-dimensional representations of spatial transcriptomics. The other is a segmented image based on the reconstructed RGB, where the segmented regions reveal the tissue architecture of unknown samples with a similar structure (Fig. 1).
In RESEPT, spatial transcriptomics data are represented as a spatial spot-spot graph. Each observational unit within a tissue sample containing a small number of cells, i.e., “spot,” is modeled as a node. The measured gene expression values of the spot are treated as the node attributes, and the neighboring spots adjacent in the Euclidean space on the tissue slice are linked with an undirected edge. This lattice-like spot graph is modeled by a modified GNN framework, which learns a three-dimensional embedding to preserve the topological relationship between all spots in the spatial space of transcriptomics. The three-dimensional embedding of gene expression and cells’ spatial topology facilitates the visualization of tissue architecture by three RGB color channels Red, Green, and Blue in an RGB image, where spots in the same cell type tend to have similar colors. Then a semantic segmentation can be performed on the image to identify the spatial architecture by classifying each spot into a spatially specific segment with a supervised convolutional neural network (CNN) model.
In the 10x Visium Genome platform, each spot has six adjacent spots, so the spatial retained spot graph has a fixed node degree of six for all the nodes. On the generated spatial spot-spot graph, a graph autoencoder learns a node-wise three-dimensional representation to preserve topological relations in the graph. The encoder of the graph autoencoder composes two layers of graph convolution network (GCN) to learn the 3-dimensional graph embedding. The decoder of the graph autoencoder is defined as an inner product between the graph embedding, followed by a sigmoid activation function. The goal of graph autoencoder learning is to minimize the difference between the input and the reconstructed graph (Fig. 2a).
The segmentation architecture is comprised of a backbone network, an encoder module, and a decoder module. The backbone network employs an extra deep network ResNet101 [51]. ResNet101 consists of one convolutional layer and 33 residual blocks, each of which cascades three convolutional layers with a convolutional skip connection from the input signals to the output feature maps for extracting fine-grained features. The encoder module utilizes atrous convolutional layers with various rates and sizes of filters and one global pooling layer to detect multi-scale semantic features from ResNet101 feature maps. And the decoder module aligns the multi-scale features to the same size and outputs a segmentation map classifying each spot into a specific spatial architecture (Fig. 2b).
3.2. RESEPT accurately characterizes the spatial architecture of the human brain cortex region
Using manual annotations as the ground truth on 12 published samples [52] and four in-house samples [33] sequenced on the 10x Genomics Visium platform, RESEPT was benchmarked on both raw and normalized expression matrices of the 16 samples (not including the sample G1 in Supplementary Table 1 and Fig. 3a) following the leave-one-out cross-validation strategy. Our results demonstrate RESEPT outperforms six existing tools, namely Seurat [11], BayesSpace [16], SpaGCN [14], stLearn [15], STUtility [13], HMRF [17], and Giotto [12] on tissue architecture identification of which ARI is (Fig. 3b) based on tuned parameters (Supplementary Data 1). Additional benchmarking results in the default parameter settings with the other three evaluation matrices (i.e., Rand index (RI), Fowlkes–Mallows index (FM), and adjusted mutual information (AMI)), visualization of RESEPT outcome, running time, and memory usage can be referred to Supplementary Figs. 1, 2, and Supplementary Data 2, 3. The overall conclusion is that RESEPT outperforms the other seven tools regarding ARI (), RI (), FM (), and AMI () evaluation scores based on the LogCPM normalization and original data. To validate the stability of our model, we generated simulation data with gradient decreasing sequencing depth based on two selected datasets, CT2 and 151674. The RGB images at low read depth presented more intra-regional diversity in their color distributions (Supplementary Fig. 3 and Supplementary Data 4). To further demonstrate the RESEPT performance on different read depth data, we simulated two data with varying depths from samples CT2 and 151,674 by downsampling a gradient number of reads from the total transcripts across all spots. In the downsampling read depth gradients from low depth to full depth, RESEPT demonstrated its robustness by ARI on CT2, and ARI on 151,674 (Fig. 3c-3f). On the same sample, RESEPT reveals better tissue architecture than the other tools in ARI 0.409 (Fig. 3g-3n). More visualization results from different normalization methods can be referred to Supplementary Fig. 1 and Supplementary Data 5. All the data used in the study are summarized in Supplementary Tables 1–2, while datasets on 10x Genomics, Spatial Transcriptomics (ST), and High-Definition Spatial Transcriptomics (HDST)[53] platforms without manual annotations were analyzed by RESEPT detailed in Supplementary Fig. 4.
RESEPT also benefits from different embeddings using various dimension-reduction methods such as scGNN, SpaGCN, UMAP, and SEDR [21] (Supplementary Figs. 5 and 6 and Supplementary Table 3). Besides learned embeddings, the pre-trained segmentation model based on the sufficiently diverse training images with different parameters (Supplementary Fig. 7) and fine-gained visual features extracted from the extra deep CNN network also gives strong discerning power to our segmentation model. We then hypothesized and validated the performance improvement with an increasing number of annotated training data (Supplementary Fig. 8). This improvement implied that as more annotated spatial transcriptomic data comes out, RESEPT will enhance its robustness accordingly.
3.3. Reconstructed RGB image has biological interpretability and model generalizability
Both gene expression and RNA velocity [19], [45] are accepted by RESEPT to generate low dimensional embeddings as RGB images. These reconstructed images reveal spatial separation between segments from the identified architecture on AD2 (Moran’s I 0.920 for RNA velocity and 0.787 for gene expression), which is consistent with the cortical architecture of the human brain (Fig. 4a, b). More comparison results between gene expression and RNA velocity using various computational tools can be found in the Discussion section.
Fig. 4.
Model interpretation and generalizability for RESEPT. (a) RGB image generated from expression value (Moran’s I = 0.787). (b) RGB image generated from RNA velocity (Moran’s I = 0.920). (c) The figure Shows the ground truth of the 151,673 sample. (d) The RGB image was reconstructed from whole transcriptomics. (e) Visualization of Red channel from sample 151673′s reconstructed RGB image. (f) Visualization of Green channel from sample 151673′s reconstructed RGB image. (g) Visualization of Blue channel from sample 151673′s reconstructed RGB image. (h-j) Grayscale images reconstructed from genes in clusters 1–3 by k-means clustering, respectively. (k) Number of SVGs mapped on RGB channels, where the Red channel corresponds to 60 genes from cluster 3, the Green channel corresponds to 594 genes from cluster 1, and the Blue channel corresponds to 179 genes from cluster 2. (l) Mouse brain sagittal section from the Allen Brain Atlas [57]. (m) The mouse brain cortex region was cropped from the mouse brain sagittal posterior at the 10x official website (the region in black line). (n) The cropped mouse cortex was labeled based on annotation from the Allen Brain Atlas in figure panel l, where Blue represents layer 1, orange represents layers 2 and 3, Green represents layers 4 and 5, and Red represents layer 6 [14]. (o) The RGB image was reconstructed based on the mouse cortex transcriptome. (p) RESEPT’s results as a segmented image from the mouse brain cortex (ARI 0.336). (q) The figure shows ARI and spot-level RESEPT segmentation results. (r) The figure shows the impact of cluster numbers in human breast cancer (1160920F) results, where the x-axis is the number of predicted clusters and the y-axis indicates the ARI score. HMRF, stLearn, and STutility were excluded because of failing to find 3 to 8 clusters. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Herein, to explore how the RGB image can derive biological insight from spatial transcriptome, we explored the association between the reconstructed RGB images and the underlying SVG patterns in sample 151,673 (Fig. 4c). First, the RGB image constructed from the whole transcriptome (Fig. 4d) was split into the Red channel (Fig. 4e), the Green channel (Fig. 4f), and the Blue channel (Fig. 4g). Then, 836 significant SVGs were identified using spatialDE [48] from this dataset (see Supplementary Data 6). The k-means clustering confirmed three main SVG clusters based on expression patterns, where each cluster has 60, 594, and 179 SVGs. Each of the three SVG clusters was used for dimensional reduction and visualization, giving rise to three grayscale images (Fig. 4h-4j) with mono-color values. Finally, the pixel correlation, calculating the Pearson Correlation over the pixels from two images, analysis (Fig. 4k and Supplementary Fig. 9) indicates cluster 1 (60 SVGs) correlates with the Red channel (Pearson’s correlation (PCC) = 0.726), cluster 2 (594 SVGs) has a high correlation with the Green channel (PCC = 0.916), and cluster 3 (179 SVGs) also correlates with the Blue channel (PCC = 0.88).
Furthermore, the GO enrichment analysis results also supported that the channel-correlated SVGs are enriched with biological functions associated with specific human brain cortex architecture (Supplementary Data 6). For instance, the Red channel (Fig. 4e) visually corresponds to layers 2, 4, 5, and 6; and the Red-channel-correlated SVGs are enriched with ATP and ribonucleotide metabolic processes, which reflect the biological functions of layers 4 and 5 [33]. Similarly, the Green channel (Fig. 4f) can be mapped to layers 2, 3, and 4. And we observed that Green-channel-correlated SVGs are enriched in the synaptic vesicle cycle and modulation of chemical synaptic transmission, which matched the functions of the three layers (2–4)[54]. Finally, the Blue channel image was split into two regions, one corresponds to layers 1, 2, and 3, and the other region can be mapped to white matter (Fig. 4g). Interestingly, the Blue-channel-correlated SVGs are enriched with two kinds of pathways: (i) synaptic vesicle cycle and synaptic transmission representing the biological functions of layers 1, 2, and 3 [54]; and (ii) protein targeting to ER supporting the biological functions of white matter [33].
Next, we investigated the model's generalizability by collecting additional mouse data to test the RESEPT model. According to previous studies [14], nine mouse brain datasets were collected from the 10x official website [55]. Our neuroscience specialist manually annotated the mouse cortex region based on the Allen Brain Atlas and histological features (Fig. 4l-4n)[49]. With 12 healthy human brain cortex and nine healthy mouse brain cortex, the newly trained RESEPT model could identify both human and mouse cortex tissue architecture (Supplementary Fig. 10). The mouse cortex (Sagittal posterior 2), its RGB image, and the segmentation results are shown in Fig. 4o-q, respectively. Finally, one of the triple-negative human breast cancer samples (i.e., 1160920F)[56] was applied to test the generalizability of RESEPT on the non-brain sample. Due to the high heterogeneity of cancer tissue and for fairly comparing benchmarking tools, the number of the output clusters (or segments) is set from 3 to 8. RESEPT outperformed the other five tools on this well-annotated human breast cancer sample (Fig. 4r). Overall, the above results indicate RESEPT has good model generalizability in different tissue types and species, which showcases great potential in broad biological visualization and interpretability.
3.4. RESEPT interprets and discovers spatially related biological insights in AD
With two AD brain samples [33], human postmortem middle temporal gyrus (MTG) from AD cases (Sample AD1 and AD2) was spatially profiled on the 10x Visium platform, and RESEPT successfully identified the main architecture of the MTG compared with the manual annotation as the ground truth (AD1 ARI = 0.474; AD2 ARI = 0.409). With the RGB image generated from specific gene expression, we distinguished cortical layers 2 & 3 from other layers and identified regions enriched with excitatory neurons and amyloid-beta (Aβ) plaques. For the AD1 sample on cortical layers 2 & 3 (ground truth [33] as Fig. 5a), well-defined marker genes (C1QL2, RASGRF2, CARTPT, WFS1, HPCAL1 for layer 2, and CARTPT, MFGE8, PRSS12, SV2C, HPCAL1 for layer 3) from the previous study [52] were embedded and transformed to an RGB image instead of using whole transcriptomes (a full gene list in Supplementary Table 4). To validate the spatial specificity, module scores from Seurat [11] showed that these marker genes are statistically significantly enriched only on cortex layers 2 & 3 among all the layers (p < 0.0001 by Wilcoxon signed-rank test, Fig. 5b). Furthermore, RESEPT visually provided consistent colors for cortical layers 2 & 3 (Fig. 5c). These spatial patterns (Fig. 5d) were strengthened by selecting a specific segmentation number (set as 3). More RGB images from other layer-specific marker genes can be found in Supplementary Fig. 11.
Fig. 5.
RESEPT identifies spatial cellular patterns in the human postmortem middle temporal gyrus (MTG). (a) Layers 2 and 3 (cyan) of sample AD1. (b) The module score of the cortical layers 2 and 3 and other layers from sample AD1, where the x-axis shows layer categories and the y-axis represents scores. (c) RGB images where the yellow line points out the ground truth of layers 2 and 3 for samples AD1. (d) The segmented images were reconstructed by setting the number of segments as three for AD1. (e) Layers 2 to 6 architecture (yellow) of AD1, where excitatory neuron cells are distributed. (f) The module score of the cortical layers 2 to 6 and white matter for sample AD1, where the x-axis shows layer categories and the y-axis represents scores. (g) RGB images of AD1 were reconstructed from excitatory neuron cell markers. (h) The segmentation images via selecting the number of segments as two for samples AD1. (i)-(p) Similar analyses for AD2. (q) Aβ plaques are located by immunofluorescence assay. (r) The spots with the accumulation of Aβ plaques in red color. (s) Reconstructed RGB image from 20 genes relevant to the Aβ region. (t) Reconstructed RGB image cropped according to the Aβ region and marked by layers 2&3 (encircled by the red line). (u) The module score regarding the Aβ region and non-Aβ region for each layer, where red color represents the Aβ region, and gray color represents the non-Aβ region. (v) RGB channels show the color value dispersion, where the violin in the blue color represents RGB values in the Aβ region, and the violin in the orange color represents RGB values in the non-Aβ region. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
To reveal critical cell-type distribution (i.e., excitatory neuron) associated with selective neuronal vulnerability in AD [38], five well-defined excitatory neuron marker genes (SLC17A6, SLC17A7, NRGN, CAMK2A, and SATB2) in the cortex were obtained from our in-house database scREAD [58] (other cell-type marker genes in Supplementary Table 4). The excitatory neuron will majorly distribute in layer 2 to layer 6 (Fig. 5e). The module score and optimized RGB image (Fig. 5f) showed statistically significant enrichment of excitatory neuron marker genes in cortical layers 2–6 (p < 0.0001 by Wilcoxon signed-rank test), and the original and improved RGB image also localized the excitatory neurons (Fig. 5g, other cell types can be found in Supplementary Fig. 12). RESEPT model can also segment the excitatory neuron distribution pattern by selecting the segmentation number as 2 (Fig. 5h). We also performed similar analyses on the AD2 sample to visualize and segment layers 2 & 3 and the excitatory neuron region. The results can be reproduced as the same as AD1 (Fig. 5i-p).
Moreover, the RGB image can reflect an important AD pathology-associated region, i.e., Aβ plaques-accumulated region. We conducted an immunofluorescence staining of Aβ on the adjacent AD2 brain section and identified the brain region with Aβ plaques [33] (Fig. 5q–t). Among the gene module containing 57 Aβ plaque-induced genes discovered from the previous study [2], we validated those 20 upregulated genes showed specific enrichment in the Aβ region compared to the non-Aβ region in terms of layers 2 & 3 (p < 0.0001 by Wilcoxon signed-rank test, Fig. 5u). By comparing the color in Aβ region-associated spots with the RGB image (Fig. 5v and Supplementary Fig. 12), we observed Aβ region-associated spots behaved a consistent color in layers 2 & 3. These predicted results are consistent with our experimental observations, which showed Aβ region has a relatively higher proportion in layers 2 and 3 in the AD1 and AD2 samples (Supplementary Tables 5–6 and Supplementary Fig. 13).
To evaluate RGB value variation quantitatively, we investigated the value range of channels R, G, and B for the Aβ region and non-Aβ region (Fig. 5v). The result showed that the Aβ region had a relatively tighter dispersion than the non-Aβ region (p-value < 0.0001 by F-test), which proved the RGB image could indicate the pathological regions with Aβ plaques. Overall, with the evidence of images generated from hallmark panel genes, RESEPT can well reflect layer-specific, cell-type-enriched, and pathological region-specific architecture with marker genes and disease-associated genes. Overall, we concluded that, given a gene module with a known function, RESEPT could visually present the activity region of the gene module and potentially localize the important spatial architecture contributing to AD pathology.
3.5. The clinical and prognostic applications of RESEPT in cancer
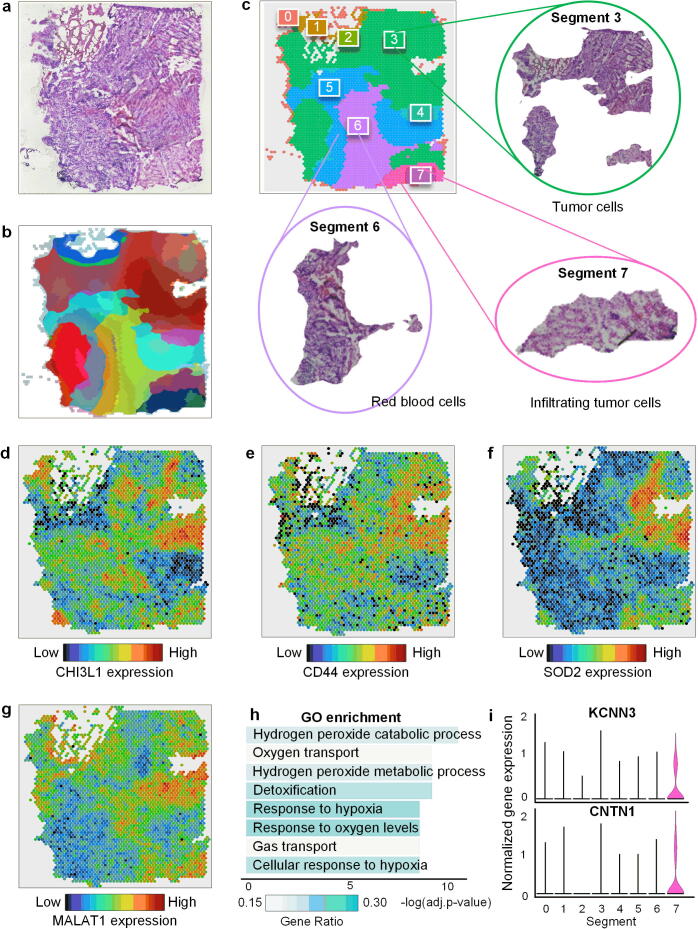
To demonstrate the clinical and prognostic applications of RESEPT in oncology, we analyzed a public glioblastoma dataset generated from the 10x Visium platform (Sample G1 in Fig. 6a) with 4,326 genes per spot, 43 million transcripts in total, and 33.7 Root Mean Square contrast (RMS contrast) over pixel intensities of H&E image. Glioblastoma, a grade IV astrocytic tumor with a median overall survival of 15 months [59], is characterized by heterogeneity in tissue morphologies which range from highly dense tumor cellularity with necrosis to other areas with single tumor cell permeation throughout the neuropil. Assessment of tissue architecture represents a key diagnostic tool for patient prognosis and diagnosis. RESEPT identified eight Segments (Fig. 6b-6c and Supplementary Fig. 14) and distinguished tumor-enriched, non-tumor, and regions of neuropil with infiltrating glioblastoma cells. These segmented areas show similarities to secondary structures of Scherer [60]. Based on the morphological features of Segment 3 in the H&E image (Fig. 6c), we observed cells with large cytoplasm and nuclei with prominent nucleoli, a morphology consistent with cortical pyramidal neurons, and many tumor cells located in this Segment showing neuronal satellitosis (Supplementary Fig. 15). Differentially expressed gene (DEG) analysis demonstrated that a pre-defined glioblastoma marker CHI3L1 [61], [62], which has been validated by the Allen brain atlas website(Supplementary Fig. 16), was highly expressed in most of the spots in Segment 3 (Fig. 6d, differentially expressed gene of each Segment can be found Supplementary Data 7).
Fig. 6.
RESEPT identifies tumor regions in glioblastoma samples (Sample G1). (a) Original H&E staining image from the 10x Genomics. (b) Tissue architecture was identified via the RESEPT pipeline. (c) Labeled segmentation by RESEPT and Segments 3, 6, and 7 are cropped according to the segmentation result. Based on morphological features, our physiologist found Segment 3 contains large tumors from morphological features; Segment 6 contains a large number of blood cells; Segment 7 contains infiltrating tumor cells. (d), (e), (f), and (g) show the expression of Glioblastoma markers CHI3L1, CD44, SOD2, and MALAT1 in all spots based on the logCPM normalization value. (h) The bar plot shows the results of GO enrichment analysis, indicating Segment 6 has a large proportion of blood cells with blood signature genes for gas transport. (i) Infiltrating glioblastoma signature marker genes KCNN3 and CNTN1 are highly expressed in Segment 7 based on the logCPM normalization.
Moreover, we observed that other tumor marker genes were also significantly enriched in Segment 3 based on DEGs results, including CD44 [62], SOD2 [63], and MALAT1 [64] (Fig. 6e-6 g). By exploring the H&E image of Segment 6, we found this prominent area of the Segment with erythrocytes, likely representing an area of acute hemorrhage during the surgical biopsy. This morphological observation was in line with the GO enrichment analysis, where DEGs were enriched in blood functionality pathways, such as oxygen transportation (Fig. 6h). Most interestingly, from the morphological features of Segment 7, we observed that this Segment belongs to infiltrating glioblastoma cells characterized by elongate nuclei admixed with non-neoplastic brain cells. Glioblastoma cells showing elongated nuclei are characteristic of invasion along white matter tracts [60]. Comparing DEGs with pre-defined infiltrating markers [65], we found that infiltrating tumor marker genes KCNN3 and CNTN1 were expressed specifically in Segment 7 (Fig. 6i). Furthermore, we found that the biological insights derived from this dataset (tumor, non-tumor, and infiltrating tumor regions) were robust and stable when changing the number of segments in the RESEPT framework (e.g., segmentation number equals 5 in Supplementary Fig. 17). Overall, RESEPT successfully recognized tumor architecture, non-tumor architecture, and infiltration tumor architecture. This tool augments the morphological evaluation of glioblastoma by enabling an improved understanding of glioblastoma heterogeneity. This objective characterization of the heterogeneity will ultimately improve oncological treatment planning for patients.
4. Discussion
Regarding tissue architecture identification tools for spatial transcriptomics, emerging computational tools have been developed based on either the statistical framework (BayesSpace [16]) or the deep learning framework (SpaGCN [14]). Unlike other spatial transcriptomics, the segmentation model of RESEPT is trained from the samples with known architectures in a supervised manner. The supervised image segmentation usually offers more accurate predictions with human guidance, while sufficiently diverse labeled data are required to increase its generalizability. In this study, we reduced the data-hunger of the supervised learning by applying the image augmentation strategy and a segmentation quality assessment protocol. Nevertheless, with the growth of available spatial transcriptomic data for training, the generalization of RESEPT is expected to be further enhanced. In practice, the pre-trained segmentation model of RESEPT as a base model paves the path for further model refinement with emerging annotated spatial data. When significant annotated spatial data are available, we will also explore classifying samples into different types and train a model for each type.
Regarding visualization, the core concept of converting three-dimensional representations to RGB images and being associated with SVGs may enable explainable AI. Such improvement may benefit from bench to bedside (e.g., clinical and prognostic purpose), visually and intuitively showing the natural tissue heterogeneity and architecture. In addition, RESEPT can be adjusted to most mixing color pallets in graphic design, such as CMYK (Cyan, Magenta, Yellow, and blacK), HSV (Hue, Saturation, and Value), and hexadecimal colors. These alternative color systems, as our future work, may provide a wide color spectrum and sufficient variation in hue and brightness to present more complex tissue and help color-blind users. With these styles of visualization layouts as options, tissue architectures might be more accessible and distinguishable in some cases.
As we observed in Fig. 4a and Fig. 4b, RNA velocity plays a complementary role with gene expression and sometimes brings more distinguishable features compared to gene expression in tissue architecture identification. With more in-depth analyses, we observed an enhanced performance from RNA velocity compared to gene expression on the 16 AD and control samples, if we assembled all the prediction results from the six different tools (Supplementary Fig. 18). However, when we targeted one specific tool (e.g., RESEPT, BayesSpace, or SpaGCN), the enhancement does not always apply. Although we do not have a large dataset to answer when and why velocity should be used instead of gene expression, we will carry out a full investigation of this interesting and challenging topic in the future.
In addition, RESEPT has a promising predictive power on lattice-based sequencing technologies (i.e., Visium) but may be limited by irregular distribution of fluorescence in situ hybridization (FISH) technology [66] and low-resolution spatial transcriptomics [67]. To overcome this limitation, we will investigate a granularity-based self-supervised graph framework, which diminishes the effects caused by the spot arrangement and resolution. With the availability of more tissue samples and spatial multi-omics, RESEPT can integrate more samples and other multi-modals of information as histology image pixels together with the spatial coordinates and gene expression to pursue a three-dimensional tissue architecture atlas. Meanwhile, RESEPT will be colorblind accessible with a ‘colorblind safe’ mode in visualization, in which all output images will be replaced with predefined color-blind palettes to avoid problematic color combinations. For different types of color blindness, RESEPT will offer corresponding narrow-down palettes accordingly. In addition, different patterns/labels instead of colors can be mapped in the image to distinguish among clusters.
RESEPT is also open to integrating other spatial transcriptomic features, especially cell morphological features from histology. Pathologists usually recognize functional zones by observing cell morphology on histology. In addition, recent research demonstrates that integrating morphological features and transcriptional features can identify novel cell types [68]. Hence, morphological features are expected to be complemented by gene expression to generate more informative RGB images reflecting tissue architectures. SpaGCN [14] has demonstrated the contributions of histological pixels to tissue identification and SVG detection. In the next version of RESEPT, we will define morphological descriptors of histology and integrate them into our graph encoder to upgrade the current RGB embedding.
5. Conclusions
Our results show that RESEPT is a robust and accurate tool for spatial transcriptomics data analysis, visualization, and interpretation. Empowered by GNN representation learning in a spatial spot-spot graph model, spatial transcriptomics is visualized as an RGB image. RESEPT formulates the problem as image segmentation and uses a deep-learning model to detect the tissue architecture. For best practice, RGB images can be used for visualizing tissue heterogeneity, especially for heterogeneous tissue. In another case, RESEPT also offers a pre-trained model for tissue segmentation and returns a clear boundary among different heterogeneous regions. As our trained model is based on the healthy human brain (cortical region), RESEPT has a promising performance on human brain architecture identification. Overall, RESEPT can provide specific spatial architectures in broad applications, including neuroscience, immuno-oncology, and developmental biology.
Data and Code Availability.
The 10x Visium datasets (10 from Spatial Gene Expression 1.0.0; 14 from Spatial Gene Expression 1.1.0, 13 from Spatial Gene Expression 1.2.0; including G1) can be accessed from https://www.10xgenomics.com/products/spatial-gene-expression. The datasets (12 samples) used for the training model and benchmarking can be accessed via endpoint “jhpce#HumanPilot10x” on the Globus data transfer platform at http://research.libd.org/globus/. The HDST datasets are available as accession number SCP420 in the Single Cell Portal via the link https://singlecell.broadinstitute.org/single_cell. The ST and 10x Visium data (squamous cell carcinoma) can be accessed from the GEO database with an accession number GSE144239. More details of datasets can be found in Supplementary Tables 1–2. The human breast cancer data (1160920F) can be downloaded at https://doi.org/10.5281/zenodo.4739739. RESEPT is freely available as an open-source Python package at https://github.com/OSU-BMBL/RESEPT.
CRediT authorship contribution statement
Yuzhou Chang: Methodology, Software, Validation, Visualization, Writing – original draft. Fei He: Methodology, Software, Validation, Visualization, Writing – original draft. Juexin Wang: Methodology, Writing - review & editing. Carter Allen: Validation. Bingqiang Liu: Writing – review & editing. Dongjun Chung: Writing – review & editing. Hongjun Fu: Writing – review & editing. Zihai Li: Writing – review & editing. Dong Xu: Conceptualization, Methodology, Writing – review & editing. Qin Ma: Conceptualization, Methodology, Writing – review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work was supported by awards R35-GM126985 and R01-GM131399 from the National Institute of General Medical Sciences and awards U54-AG075931, K01-AG056673, and R56-AG066782-01 from the National Institute on Aging of the National Institutes of Health. The work was also supported by the award NSF1945971 from the National Science Foundation and the award of AARF-17-505009 from the Alzheimer’s Association. We thank Hua Li and Shiyuan Chen from Stowers Institute, Liangping Li from the Ohio State University for helpful discussion, Kai Liu and Qiuyu Lv from Northeast Normal University for their technical support, and Paul Toth from Ohio State University for polishing the manuscript. Human de-identified brain tissues were kindly provided by the Banner Sun Health Research Institute Brain and Body Donation Program, supported by NIH grants U24-NS072026 and P30-AG19610 (TGB), the Arizona Department of Health Services (contract 211002, Arizona Alzheimer’s Research Center), the Arizona Biomedical Research Commission (contracts 4001, 0011, 05-901 and 1001 to the Arizona Parkinson's Disease Consortium) and the Michael J. Fox Foundation for Parkinson’s Research and the New York Brain Bank at Columbia University Irving Medical Center supported by the Taub Institute and NIH grants P50AG008702 and P30AG066462. This work was supported by the Pelotonia Institute of Immuno-Oncology (PIIO). The content is solely the responsibility of the authors and does not necessarily represent the official views of the PIIO.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2022.08.029.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Liao J., Lu X., Shao X., Zhu L., Fan X. Uncovering an organ’s molecular architecture at single-cell resolution by spatially resolved transcriptomics. Trends Biotechnol. 2020 doi: 10.1016/j.tibtech.2020.05.006. [DOI] [PubMed] [Google Scholar]
- 2.Chen W.T., Lu A., Craessaerts K., Pavie B., Sala Frigerio C., Corthout N., et al. Spatial transcriptomics and in situ sequencing to study Alzheimer's disease. Cell. 2020;182:976–991.e919. doi: 10.1016/j.cell.2020.06.038. [DOI] [PubMed] [Google Scholar]
- 3.Ji A.L., Rubin A.J., Thrane K., Jiang S., Reynolds D.L., Meyers R.M., et al. Multimodal analysis of composition and spatial architecture in human squamous cell carcinoma. Cell. 2020;182:497–514.e422. doi: 10.1016/j.cell.2020.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thrane K., Eriksson H., Maaskola J., Hansson J., Lundeberg J. Spatially resolved transcriptomics enables dissection of genetic heterogeneity in stage III cutaneous malignant melanoma. Cancer Res. 2018;78:5970–5979. doi: 10.1158/0008-5472.CAN-18-0747. [DOI] [PubMed] [Google Scholar]
- 5.Grauel A.L., Nguyen B., Ruddy D., Laszewski T., Schwartz S., Chang J., et al. TGFβ-blockade uncovers stromal plasticity in tumors by revealing the existence of a subset of interferon-licensed fibroblasts. Nat Commun. 2020;11:1–17. doi: 10.1038/s41467-020-19920-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berglund E., Maaskola J., Schultz N., Friedrich S., Marklund M., Bergenstråhle J., et al. Spatial maps of prostate cancer transcriptomes reveal an unexplored landscape of heterogeneity. Nat Commun. 2018;9:1–13. doi: 10.1038/s41467-018-04724-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rao A., Barkley D., França G.S., Yanai I. Exploring tissue architecture using spatial transcriptomics. Nature. 2021;596:211–220. doi: 10.1038/s41586-021-03634-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Method of the Year 2020: spatially resolved transcriptomics. Nature Methods 2021, 18:1-1. [DOI] [PubMed]
- 9.Lewis S.M., Asselin-Labat M.-L., Nguyen Q., Berthelet J., Tan X., Wimmer V.C., et al. Spatial omics and multiplexed imaging to explore cancer biology. Nat Methods. 2021;18:997–1012. doi: 10.1038/s41592-021-01203-6. [DOI] [PubMed] [Google Scholar]
- 10.Hu J., Schroeder A., Coleman K., Chen C., Auerbach B.J., Li M. Statistical and machine learning methods for spatially resolved transcriptomics with histology. Comput Struct Biotechnol J. 2021;19:3829–3841. doi: 10.1016/j.csbj.2021.06.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stuart T., Butler A., Hoffman P., Hafemeister C., Papalexi E., Mauck W.M., III, et al. Comprehensive integration of single-cell data. Cell. 2019;177:1888–1902.e1821. doi: 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dries R., Zhu Q., Dong R., Eng C.-H.-L., Li H., Liu K., et al. Giotto: a toolbox for integrative analysis and visualization of spatial expression data. Genome Biol. 2021;22(78) doi: 10.1186/s13059-021-02286-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bergenstråhle J., Larsson L., Lundeberg J. Seamless integration of image and molecular analysis for spatial transcriptomics workflows. BMC Genom. 2020;21:482. doi: 10.1186/s12864-020-06832-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hu J., Li X., Coleman K., Schroeder A., Ma N., Irwin D.J., et al. SpaGCN: Integrating gene expression, spatial location and histology to identify spatial domains and spatially variable genes by graph convolutional network. Nat Methods. 2021 doi: 10.1038/s41592-021-01255-8. [DOI] [PubMed] [Google Scholar]
- 15.Pham D, Tan X, Xu J, Grice LF, Lam PY, Raghubar A, Vukovic J, Ruitenberg MJ, Nguyen Q: stLearn: integrating spatial location, tissue morphology and gene expression to find cell types, cell-cell interactions and spatial trajectories within undissociated tissues. bioRxiv 2020:2020.2005.2031.125658.
- 16.Zhao E., Stone M.R., Ren X., Guenthoer J., Smythe K.S., Pulliam T., et al. Spatial transcriptomics at subspot resolution with BayesSpace. Nat Biotechnol. 2021 doi: 10.1038/s41587-021-00935-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhu Q., Shah S., Dries R., Cai L., Yuan G.C. Identification of spatially associated subpopulations by combining scRNAseq and sequential fluorescence in situ hybridization data. Nat Biotechnol. 2018 doi: 10.1038/nbt.4260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mollon J.D. Color vision: opsins and options. Proc Natl Acad Sci U S A. 1999;96:4743–4745. doi: 10.1073/pnas.96.9.4743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.La Manno G., Soldatov R., Zeisel A., Braun E., Hochgerner H., Petukhov V., et al. RNA velocity of single cells. Nature. 2018;560:494–498. doi: 10.1038/s41586-018-0414-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang J., Ma A., Chang Y., Gong J., Jiang Y., Qi R., et al. scGNN is a novel graph neural network framework for single-cell RNA-Seq analyses. Nat Commun. 1882;2021:12. doi: 10.1038/s41467-021-22197-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fu H, Xu H, Chong K, Li M, Ang KS, Lee HK, Ling J, Chen A, Shao L, Liu L, Chen J: Unsupervised Spatially Embedded Deep Representation of Spatial Transcriptomics. bioRxiv 2021:2021.2006.2015.448542. [DOI] [PMC free article] [PubMed]
- 22.Zhong E.D., Bepler T., Berger B., Davis J.H. CryoDRGN: reconstruction of heterogeneous cryo-EM structures using neural networks. Nat Methods. 2021;18:176–185. doi: 10.1038/s41592-020-01049-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen LC, Papandreou G, Schroff F, Adam H: Rethinking Atrous Convolution for Semantic Image Segmentation. 2017.
- 24.Chen LC, Zhu Y, Papandreou G, Schroff F, Adam HJS, Cham: Encoder-Decoder with Atrous Separable Convolution for Semantic Image Segmentation. 2018.
- 25.Deep Residual Learning for Image Recognition. In IEEE Conference on Computer Vision & Pattern Recognition. 2016.
- 26.Glorot X, Bordes A, Bengio Y: Deep Sparse Rectifier Neural Networks. In Proceedings of the Fourteenth International Conference on Artificial Intelligence and Statistics (Geoffrey G, David D, Miroslav D eds.), vol. 15. pp. 315--323. Proceedings of Machine Learning Research: PMLR; 2011:315--323.
- 27.Grave E, Joulin A, Cissé M, Grangier D, Jégou H: Efficient softmax approximation for GPUs. 2016.
- 28.de Boer P.-T., Kroese D.P., Mannor S., Rubinstein R.Y. A tutorial on the cross-entropy method. Ann Oper Res. 2005;134:19–67. [Google Scholar]
- 29.Xu JaC, Kai and Lin, Dahua: MMSegmenation. 2020, https://github.com/open-mmlab/mmsegmentation.
- 30.Zonca F, Chen L, Santoro RA: parallelized stochastic gradient descent. 1996.
- 31.Li H., Calder C.A., Cressie N. Beyond Moran's I: testing for spatial dependence based on the spatial autoregressive model. Geograph Anal. 2007;39:357–375. [Google Scholar]
- 32.Jin X, Han J: K-Means Clustering. In Encyclopedia of Machine Learning. Edited by Sammut C, Webb GI. Boston, MA: Springer US; 2010: 563-564.
- 33.Chen S, Chang Y, Li L, Acosta D, Morrison C, Wang C, Julian D, Hester ME, Serrano GE, Beach TG, et al: Spatially resolved transcriptomics reveals unique gene signatures associated with human temporal cortical architecture and Alzheimer’s pathology. bioRxiv 2021:2021.2007.2007.451554.
- 34.Beach T.G., Adler C.H., Sue L.I., Serrano G., Shill H.A., Walker D.G., et al. Arizona study of aging and neurodegenerative disorders and brain and body donation program. Neuropathology. 2015;35:354–389. doi: 10.1111/neup.12189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vonsattel J.P., Del Amaya M.P., Keller C.E. Twenty-first century brain banking. Processing brains for research: the Columbia University methods. Acta Neuropathol. 2008;115:509–532. doi: 10.1007/s00401-007-0311-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bassiouni R., Gibbs L.D., Craig D.W., Carpten J.D., McEachron T.A. Applicability of spatial transcriptional profiling to cancer research. Mol Cell. 2021;81:1631–1639. doi: 10.1016/j.molcel.2021.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nerurkar S.N., Goh D., Cheung C.C.L., Nga P.Q.Y., Lim J.C.T., Yeong J.P.S. Transcriptional spatial profiling of cancer tissues in the era of immunotherapy: the potential and promise. Cancers. 2020;12:2572. doi: 10.3390/cancers12092572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fu H.J., Possenti A., Freer R., Nakano Y., Villegas N.C.H., Tang M.P., et al. A tau homeostasis signature is linked with the cellular and regional vulnerability of excitatory neurons to tau pathology. Nat Neurosci. 2019;22:47-+ doi: 10.1038/s41593-018-0298-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schneider C.A., Rasband W.S., Eliceiri K.W. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lun ATL, McCarthy DJ, Marioni JC: A step-by-step workflow for low-level analysis of single-cell RNA-seq data with Bioconductor. F1000Research 2016, 5:2122-2122. [DOI] [PMC free article] [PubMed]
- 42.Hafemeister C., Satija R. Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression. Genome Biol. 2019;20:296. doi: 10.1186/s13059-019-1874-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Robinson M.D., McCarthy D.J., Smyth G.K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hao Y, Hao S, Andersen-Nissen E, Mauck WM, Zheng S, Butler A, Lee MJ, Wilk AJ, Darby C, Zagar M, et al: Integrated analysis of multimodal single-cell data. bioRxiv 2020:2020.2010.2012.335331. [DOI] [PMC free article] [PubMed]
- 45.Bergen V., Lange M., Peidli S., Wolf F.A., Theis F.J. Generalizing RNA velocity to transient cell states through dynamical modeling. Nat Biotechnol. 2020;38:1408–1414. doi: 10.1038/s41587-020-0591-3. [DOI] [PubMed] [Google Scholar]
- 46.Suzuki S. Topological structural analysis of digitized binary images by border following. Comput Vis Graph Image Process. 1985;30:32–46. [Google Scholar]
- 47.Fotheringham AS, Brunsdon CF, Charlton ME: Quantitative Geography: Perspectives on Modern Spatial Analysis. 2000.
- 48.Svensson V., Teichmann S.A., Stegle O. SpatialDE: identification of spatially variable genes. Nat Methods. 2018;15:343–346. doi: 10.1038/nmeth.4636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lein E.S., Hawrylycz M.J., Ao N., Ayres M., Bensinger A., Bernard A., et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature. 2007;445:168–176. doi: 10.1038/nature05453. [DOI] [PubMed] [Google Scholar]
- 50.Wu Z., Pan S., Chen F., Long G., Zhang C., Yu P.S. A comprehensive survey on graph neural networks. IEEE Trans Neural Networks Learn Syst. 2021;32:4–24. doi: 10.1109/TNNLS.2020.2978386. [DOI] [PubMed] [Google Scholar]
- 51.He K.M., Zhang X.Y., Ren S.Q., Sun J. Deep residual learning for image recognition. Ieee Conf Comput Vis Pattern Recognit (Cvpr) 2016;2016:770–778. [Google Scholar]
- 52.Maynard K.R., Collado-Torres L., Weber L.M., Uytingco C., Barry B.K., Williams S.R., et al. Transcriptome-scale spatial gene expression in the human dorsolateral prefrontal cortex. Nat Neurosci. 2021;24:425–436. doi: 10.1038/s41593-020-00787-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vickovic S., Eraslan G., Salmén F., Klughammer J., Stenbeck L., Schapiro D., et al. High-definition spatial transcriptomics for in situ tissue profiling. Nat Methods. 2019;16:987–990. doi: 10.1038/s41592-019-0548-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gómez-Isla T., Price J.L., McKeel D.W., Jr., Morris J.C., Growdon J.H., Hyman B.T. Profound loss of layer II entorhinal cortex neurons occurs in very mild Alzheimer's disease. J Neurosci. 1996;16:4491–4500. doi: 10.1523/JNEUROSCI.16-14-04491.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Genomics x: Mouse Brain Serial Section 1 (Sagittal-Anterior). (Genomics x ed. official website: V1; 2020.
- 56.Wu S.Z., Al-Eryani G., Roden D.L., Junankar S., Harvey K., Andersson A., et al. A single-cell and spatially resolved atlas of human breast cancers. Nat Genet. 2021;53:1334–1347. doi: 10.1038/s41588-021-00911-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sunkin S.M., Ng L., Lau C., Dolbeare T., Gilbert T.L., Thompson C.L., et al. Allen Brain Atlas: an integrated spatio-temporal portal for exploring the central nervous system. Nucleic Acids Res. 2013;41:D996–D1008. doi: 10.1093/nar/gks1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jiang J, Wang C, Qi R, Fu H, Ma Q: scREAD: A Single-Cell RNA-Seq Database for Alzheimer's Disease. iScience 2020, 23:101769. [DOI] [PMC free article] [PubMed]
- 59.Stupp R., Mason W.P., van den Bent M.J., Weller M., Fisher B., Taphoorn M.J., et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352:987–996. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 60.Zagzag D., Esencay M., Mendez O., Yee H., Smirnova I., Huang Y., et al. Hypoxia- and vascular endothelial growth factor-induced stromal cell-derived factor-1alpha/CXCR4 expression in glioblastomas: one plausible explanation of Scherer's structures. Am J Pathol. 2008;173:545–560. doi: 10.2353/ajpath.2008.071197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Steponaitis G, Skiriutė D, Kazlauskas A, Golubickaitė I, Stakaitis R, Tamašauskas A, Vaitkienė P: High CHI3L1 expression is associated with glioma patient survival. Diagnostic pathology 2016, 11:42-42. [DOI] [PMC free article] [PubMed]
- 62.Couturier C.P., Ayyadhury S., Le P.U., Nadaf J., Monlong J., Riva G., et al. Single-cell RNA-seq reveals that glioblastoma recapitulates a normal neurodevelopmental hierarchy. Nat Commun. 2020;11:3406. doi: 10.1038/s41467-020-17186-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chien C.H., Chuang J.Y., Yang S.T., Yang W.B., Chen P.Y., Hsu T.I., et al. Enrichment of superoxide dismutase 2 in glioblastoma confers to acquisition of temozolomide resistance that is associated with tumor-initiating cell subsets. J Biomed Sci. 2019;26:77. doi: 10.1186/s12929-019-0565-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chen W., Xu X.K., Li J.L., Kong K.K., Li H., Chen C., et al. MALAT1 is a prognostic factor in glioblastoma multiforme and induces chemoresistance to temozolomide through suppressing miR-203 and promoting thymidylate synthase expression. Oncotarget. 2017;8:22783–22799. doi: 10.18632/oncotarget.15199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Darmanis S., Sloan S.A., Croote D., Mignardi M., Chernikova S., Samghababi P., et al. Single-Cell RNA-Seq analysis of infiltrating neoplastic cells at the migrating front of human glioblastoma. Cell Rep. 2017;21:1399–1410. doi: 10.1016/j.celrep.2017.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Eng C.L., Lawson M., Zhu Q., Dries R., Koulena N., Takei Y., et al. Transcriptome-scale super-resolved imaging in tissues by RNA seqFISH+ Nature. 2019;568:235–239. doi: 10.1038/s41586-019-1049-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ståhl P.L., Salmén F., Vickovic S., Lundmark A., Navarro J.F., Magnusson J., et al. Visualization and analysis of gene expression in tissue sections by spatial transcriptomics. Science. 2016;353:78–82. doi: 10.1126/science.aaf2403. [DOI] [PubMed] [Google Scholar]
- 68.Bao F., Deng Y., Wan S., Shen S.Q., Wang B., Dai Q., et al. Integrative spatial analysis of cell morphologies and transcriptional states with MUSE. Nat Biotechnol. 2022 doi: 10.1038/s41587-022-01251-z. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.