Summary
Biomaterial-associated microbial contaminations in biologically conducive three-dimensional (3D) tissue-engineered constructs have significantly limited the clinical applications of scaffold systems. To prevent such infections, antimicrobial biomaterials are rapidly evolving. Yet, the use of such materials in bioprinting-based approaches of scaffold fabrication has not been examined. This study introduces a new generation of bacteriostatic gelatin methacryloyl (GelMA)-based bioinks, incorporated with varying doses of antibacterial superparamagnetic iron oxide nanoparticles (SPIONs). The SPION-laden GelMA scaffolds showed significant resistance against the Staphylococcus aureus growth, while providing a contrast in magnetic resonance imaging. We simulated the bacterial contamination of cellular 3D GelMA scaffolds in vitro and demonstrated the significant effect of functionalized scaffolds in inhibiting bacterial growth, while maintaining cell viability and growth. Together, these results present a new promising class of functionalized bioinks to 3D bioprint tissue-engineered scaffold with markedly enhanced properties for the use in a variety of in vitro and clinical applications.
Subject areas: Biomaterials, Nanoparticles, Tissue engineering
Graphical abstract
Highlights
-
•
Functionalized bioinks with bacteriostatic properties are developed and thoroughly characterized
-
•
The 200 μg/mL group yielded an optimal balance of printed scaffold properties
-
•
Incorporating nanoparticle also enabled noninvasive imaging of the bioprinted scaffold
Biomaterials; Nanoparticles; Tissue engineering
Introduction
Additive biomanufacturing techniques, such as three-dimensional (3D) bioprinting, have emerged in the tissue engineering field, offering many advantages in developing tissue models for use in regenerative medicine applications (Chan et al., 2020; Chung et al., 2020; Serpooshan and Guvendiren, 2020; Tomov et al., 2019a). By controlling the size, shape, and architecture of manufactured constructs, 3D bioprinting allows for the assembly of tissue analogs with highly biomimetic structural and functional properties. The bioink, which is typically a blend of biomaterials, biological and chemical components, and often times living cells, is a critical component of tissue bioprinting and structure fidelity (Hölzl et al., 2016; Ning et al., 2020; Tomov et al., 2020). Commonly, materials such as biodegradable hydrogels have been used as the base of bioinks as they inherently mimic the native tissue microenvironments and properties (Lee and Kim, 2018). Specifically, the use of gelatin methacryloyl (GelMA) as a bioink has recently gained increasing attention in the field due its facile tunability, adequate biocompatibility and bioactivity, and the ability to covalently crosslink via various light activated photoinitiators (Yue et al., 2015). GelMA hydrogel bioinks can be readily incorporated with factors, such as nanoparticles (NPs), to create composite functionalized bioinks for diverse tissue engineering applications (Tomov et al., 2020). This allows for the formation of a colloidal matrix that can further improve the structural stability and print fidelity along with increasing the desired biological properties of the hydrogel composite matrix (Bhattacharyya et al., 2021).
Clinical-scale tissue bioprinting and in vitro and in vivo applications of 3D printed scaffolds have been challenged by the difficulty to maintain these constructs infection free, particularly pre-operation (Busscher et al., 2012; Kuijer et al., 2007; Zimmerli and Trampuz, 2013). This is mainly due to their inherent nature as highly favorable culture systems. Biomaterial-related bacterial infection is a common cause of graft failure of implanted devices in vivo (Zimmerli and Trampuz, 2013). Staphylococcus aureus (S. aureus) is a member of the normal human skin microbiota and one of the most common causes of hospital-acquired infection and wound infection following surgery (Offerhaus et al., 2019). Increasing rates of antibiotic resistance make treatment of these infections difficult and have led to an emphasis on prophylaxis, infection control, and antisepsis (Johnson and Garcia, 2015). Commonly, organic antimicrobial compounds have been utilized to tackle this issue (Greenhalgh et al., 2019). Yet, such methods face major challenges due to their toxicity and/or antibiotic resistance rate, whereas the interest in inorganic disinfectants such as metal oxides is increasing (Abat et al., 2018; Hajipour et al., 2012). A variety of GelMA-based hydrogel systems with enhanced antibacterial properties have been created, including the polycaprolactone-GelMA-cephalexin electrospun nanofibrous scaffolds for wound healing applications (Bakhsheshi-Rad et al., 2019), a GelMA-based hydrogel drug delivery system used for local delivery of antibiotic cefazolin (Vigata et al., 2021), and a 3D printed GelMA scaffold laden with antibacterial tetrapodal zinc oxide microparticles (Siebert et al., 2021). While significant progress has been made, a GelMA-based bioink, with well-characterized antibacterial, printability, imaging, and cell support characteristics, has not been established for 3D bioprinting applications.
Recent advances in the use of nanobiomaterials for tissue engineering and cell therapy applications have shown great promise in eliciting desired cellular and tissue response (Kumar and Abrahamse, 2020; Serpooshan et al., 2018; Uskokovic, 2017). The versatile and unique properties of NPs have rapidly established their use in a wide range of clinical applications such as drug delivery, targeted therapy, thermal ablation, medical imaging, and more relevantly 3D printing (Hasan et al., 2018). More recent studies have uncovered key advantages of NPs over traditional antibiotics (e.g., penicillin, methicillin, erythromycin, and vancomycin) to combat microbial activity (Khezerlou et al., 2018; Wang et al., 2017). In particular, superparamagnetic iron oxide NPs (SPIONs) have attracted growing attention as contrast agents for MRI and as robust antibacterial agents (Ai et al., 2016; Hajipour et al., 2021; Nehra et al., 2018). The antibacterial properties of SPIONs are tunable by controlling their size, which is of high importance due to the differences in cell envelope between Gram-positive and Gram-negative bacterial species (Tran et al., 2010). In this study, we incorporated varying doses of SPIONs in 3D bioprinting of tissue engineering scaffolds and examined the NPs effect on biofilm formation and MRI visibility of constructs (Mahmoudi et al., 2016). Bioprinted GelMA constructs, functionalized with the antibacterial NPs, provided mechanical stability for the in vitro 3D culture, and supported endothelial cell viability and growth, while exhibiting significant bacteriostatic effect.
Results and discussion
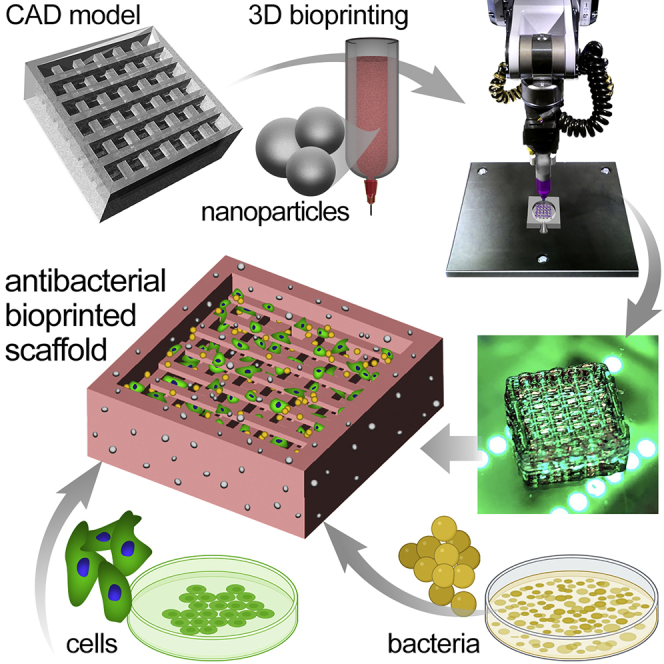
To date, the use of tissue-engineered scaffolds and medical devices is hindered by the biomaterial-associated infection as well as the impaired visibility of the 3D scaffolds in both in vitro and in vivo applications. We recently demonstrated that incorporation of SPIONs into a collagen type I-based cardiac patch could offer MRI-visibility, while impeding the growth of Salmonella bacteria in the patch (Mahmoudi et al., 2016). This work evaluated, for the first time, the use of these FDA-approved NPs in developing a novel generation of functionalized bioinks for 3D bioprinting of anti-infection and MRI-visible tissue constructs. As the fields of additive biomanufacturing and tissue bioprinting advance toward in situ fabrication of biological products into the patient body and other translational applications (Agarwal et al., 2020; Albanna et al., 2019; Singh et al., 2020), the need for anti-infection bioinks that also allow for noninvasive, longitudinal monitoring of engineered scaffolds is paramount. In this study, GelMA-based bioinks were supplemented with varying concentrations of SPIONs and their cytocompatibility, resistance to bacterial growth, and MR imaging properties were assessed in vitro (Figure 1). The specific range of SPION concentration (100–500 μg/mL) was selected based upon our prior experiences working with these engineered NP systems (Mahmoudi et al., 2011a, 2012a, 2012b; Sharifi et al., 2012). Doses lower than 100 μg/mL would not yield sufficient antibacterial or imaging effects (Mahmoudi et al., 2011a). Concentrations higher than 500 μg/mL often result in particle aggregation, which could be detrimental for the extrusion bioprinting processes; there could also be the risk of cytotoxicity at higher SPION doses (Mahmoudi et al., 2012a, 2012b; Sharifi et al., 2012).
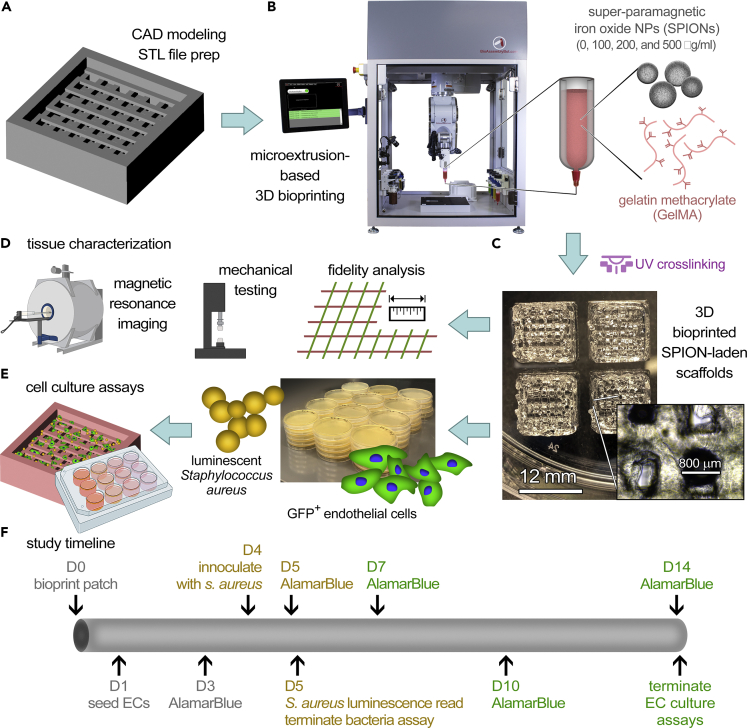
Figure 1.
Schematic overview of the experimental design for the fabrication and in vitro analysis of bacteriostatic 3D scaffold systems
(A–E) A CAD model of the 3D geometry of interest was designed (A) and bioprinted (B) using various GelMA bioinks containing 0 (control), 100, 200, and 500 μg/mL of superparamagnetic iron oxide nanoparticles (SPIONs) to create the 3D scaffolds (C). Bioprinted scaffolds were assessed for printing fidelity, mechanical, and MR imaging properties (D), and seeded with human cells (fluorescently tagged endothelial cells) and/or bacteria (luminescent Staphylococcus aureus) (E) for the in vitro 3D culture assays.
(F) The time line for the in vitro culture assays. Constructs were cultured either for 5 days (bacteria and human cell coculture) or 14 days (endothelial cell culture) and examined via AlamarBlue and imaging techniques. In panel (F), gray color represents the experimental steps for both bacteria and cell culture assays; brown: steps for the bacteria assays; and green: steps for the endothelial cell culture
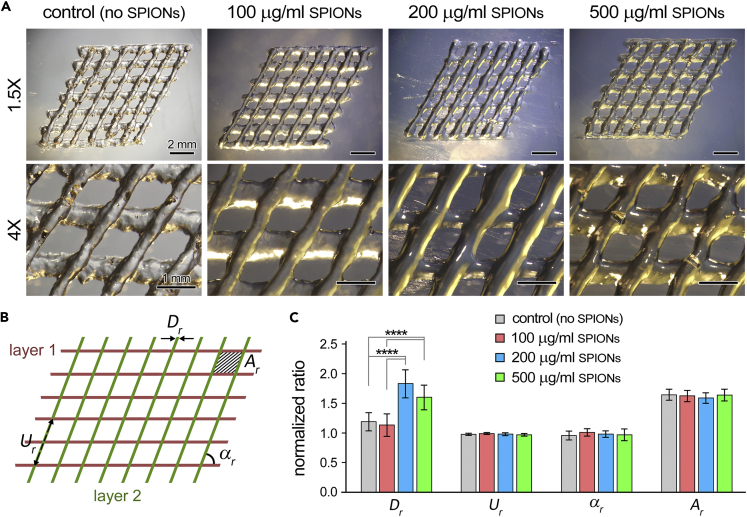
Printing fidelity measurements of a two-layer cross hatch design, at 1.5X and 4× magnifications, demonstrated a relative reproducibility for each bioink composition including the control GelMA group (10% w/v), and GelMA supplemented with 100, 200, and 500 μg/mL of SPIONs (Figures 2A and 2B, Table S1). The geometric parameters of each print were compared against those in the CAD model to obtain the strand diameter , strand uniformity , strand angle , and inter-strand area ratios (Equations (Equation 5), (Equation 6), (Equation 7), (Equation 8), Figure 2C). The strand diameter ratio () varied between 1.13 and 1.83 for the four study groups. The 200 and 500 μg/mL SPION groups showed significantly higher ratios (p < 0.0001), compared to the control and 100 μg/mL SPION groups, suggesting reduced printing fidelity once the NP concentration exceeds a certain (optimal) threshold. This is while strand uniformity () ranged from 0.97 to 0.99, and inter-strand angle () ratios ranged from 0.96 to 1.01 for all groups, with no significant differences (p > 0.05). The approximation of and ratios to 1 signifies the appropriate 2D and 3D printing accuracy in reference to the CAD model. The inter-strand area () ranged at higher values, however, from 1.59 to 1.64, while there were still no significant differences (p > 0.05) across the study groups. Overall, the no SPIONs and 100 μg/mL SPIONs bioinks exhibited the optimal printing fidelity characteristics. Qualitative evaluation of SPION-laden bioinks suggested no significantly adverse effect of the incorporated NPs in the photocrosslinking process of the GelMA hydrogel, as also reported by other groups (Kurian et al., 2022). Potential effect of SPIONs incorporation on the crosslinking of GelMA bioinks can be precisely examined, in the future works, using NMR analysis (Ning et al., 2020).
Figure 2.
Printing fidelity assessment of functionalized bioinks
(A) Representative optical (bright-field) images acquired for fidelity measurements for each GelMA bioink group: 0 (control), 100, 200, and 500 μg/mL SPIONs, at 1.5X (top) and 4× (bottom) magnifications. Scale bars in top and bottom rows represent 2 mm and 1 mm, respectively.
(B) Definition of the four geometric ratios quantified from microscopy images in (A) to assess fidelity of a 2-layer printed GelMA structure. These parameters include the strand diameter , strand uniformity , strand angle , and inter-strand area ratios (Equations (Equation 5), (Equation 6), (Equation 7), (Equation 8)).
(C) Comparison of quantified geometric ratios, defined in (B), for the four GelMA bioink compositions used at time zero of this study (immediately post printing). An n = 5 per experimental group was used for quantitative assays. ∗∗∗∗ p value < 0.0001.
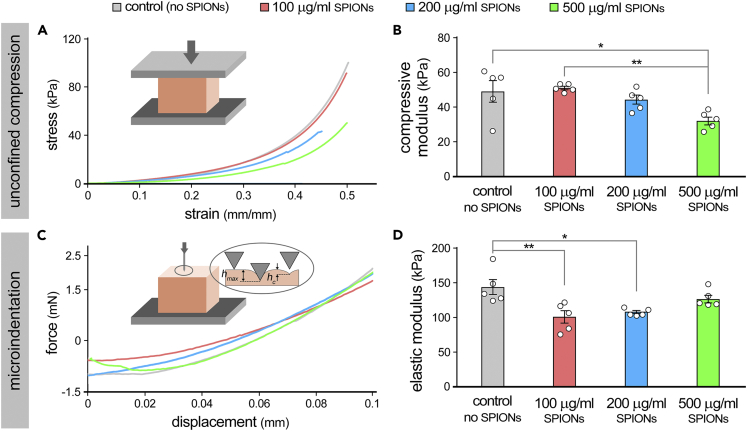
Incorporation of nanostructured materials into scaffolding biomaterials has been frequently used to tune or enhance the mechanical properties of engineered tissues (Corona-Gomez et al., 2016; Hasan et al., 2018; Shokouhimehr et al., 2021). To examine the effect of SPIONs on the mechanical properties of GelMA constructs, we conducted unconfined uniaxial compression and microindentation tests (Figure 3, Table S2). Unconfined compression demonstrated relatively high levels of elasticity for the control (no SPIONs) and SPION-loaded GelMA constructs (Figure 3A). Incorporation of SPIONs into the bioink resulted in a decrease in the compressive modulus of bioprinted constructs which became significantly lower in the 500 μg/mL SPIONs group (32.0 ± 5.0 kPa) in comparison to the control GelMA group (49.0 ± 14.0 kPa) Figure 3B). Microindentation tests showed a similar trend in the elasticity of SPION-free and SPION-loaded constructs (Figure 3C). Consistent with the compressive test, the elastic moduli measurements obtained from indentation showed a decreasing trend in the moduli by increasing the dose of SPIONs in the bioink, ranging from 143.8 ± 24.5 kPa (no SPIONs) to 100.8 ± 20.2 kPa (100 μg/mL SPIONs) (Figure 3D). Of note, however, the 500 μg/mL SPIONs group showed no significant difference in moduli (126.5 ± 12.5 kPa) in comparison to the GelMA control.
Figure 3.
Mechanical characterization of bioprinted constructs loaded with varying quantities of superparamagnetic iron oxide nanoparticles (SPIONs)
(A and B) Unconfined compression test conducted (at a 50% total strain at 20 μm/s) on bioprinted GelMA scaffolds containing no SPIONs (control), 100, 200, and 500 μg/mL SPIONs (n = 4 per group). Elastic moduli (B) were calculated from the slope of the stress-strain curves at the initial 0%–20% interval.
(C and D) Microindentation tests were conducted on GelMA constructs using a 500 μm probe, with a depth of 100 μm at 2 μm/s (n = 5 per group). Elastic moduli were calculated based on the force-displacement unloading curves as described in STAR Methods (the slope of the linear trend line at initial 5–20%). ∗ p value < 0.05, ∗∗ p value < 0.01.
Stiffness measurements from unconfined compression curves indicated no significant differences across four groups, and those obtained from microindentation showed significant difference in the stiffness only between control and the 100 μg/mL SPIONs groups (Figure S1). Consistent with our previous studies (Shokouhimehr et al., 2021), the overall reduction in elastic moduli (and partly stiffness) of SPION-laden scaffolds can be attributed to the potential role of NPs in blocking the bonds within the GelMA polymeric backbone, partially preventing the photocrosslinking of the LAP photoinitiator. Furthermore, the SPIONs may facilitate the formation of microcracks and also inhibit the fusion of micro-extruded GelMA layers, resulting in material instability (Shokouhimehr et al., 2021). The noticeable differences in the measured elastic moduli and stiffness of printed samples under compression vs. microindentation test (Figure 3 and S1) are expected due to the inherently distinct testing modalities, probing macro vs. micro-scale mechanical properties of hydrogels (Miller and Morgan, 2010).
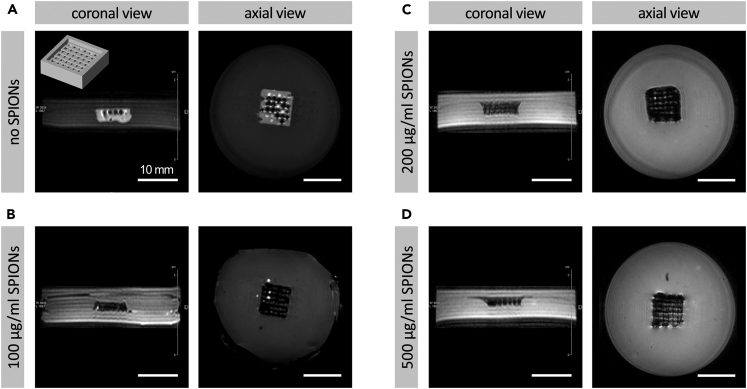
To date, SPIONs are extensively used as contrast agent for noninvasive labeling and tracking of a variety of biological reagents, including cells, scaffolds, and therapeutic macromolecules (Li et al., 2013; Mahmoudi et al., 2011b, 2016; Neuwelt et al., 2015). In this work, we evaluated the enhanced imaging properties of bioprinted scaffolds, laden with varying concentrations of SPIONs. Magnetic resonance (MR) images of printed structures embedded in 2% agarose (simulating soft tissue) demonstrated the remarkable effect of SPIONs incorporation on the in vitro MR visibility of bioprinted scaffolds (Figures 4A–4D). T2∗-weighted MR images showed a significant contrast induced by varying concentrations of magnetic NPs within the 3D constructs (Figures 4B–4D) in comparison to the control (no SPIONs) group (Figure 4A). No noticeable differences were observed in the resulting MR contrast across four study groups. The MR imaging results obtained here demonstrate a robust potential for conducting further quantitative analyses on bioprinted constructs in the future applications. The strong imaging contrast generated by the embedded SPIONs can enable precise fidelity measurements. Furthermore, comprehensive MRI imaging can be used to generate a standard curve for varying concentrations of SPIONs. Such data can then be used to predict/measure the SPION content within a given 3D construct in a rather noninvasive manner. We are currently working on utilizing a similar approach, based on computed tomography (CT) imaging and various CT contrast agents, to enable longitudinal and noninvasive analysis of 3D bioprinted GelMA constructs (Gil et al., 2021).
Figure 4.
Magnetic resonance imaging (MRI) of 3D bioprinted constructs loaded with varying quantities of superparamagnetic iron oxide nanoparticles (SPIONs)
(A–D) GelMA constructs laden with no SPIONs (control) (A), 100 (B), 200 (C), and 500 (D) μg/mL SPIONs were imaged in vitro via MRI. T1 weighted images from top (coronal, left) and T2∗ weighted images from side (axial, right) demonstrated the resulting significant contrast in the magnetic 3D scaffolds. Inset in panel (A, top left) shows the 3D CAD design used to bioprint the scaffolds. Scale bars represent 10 mm.
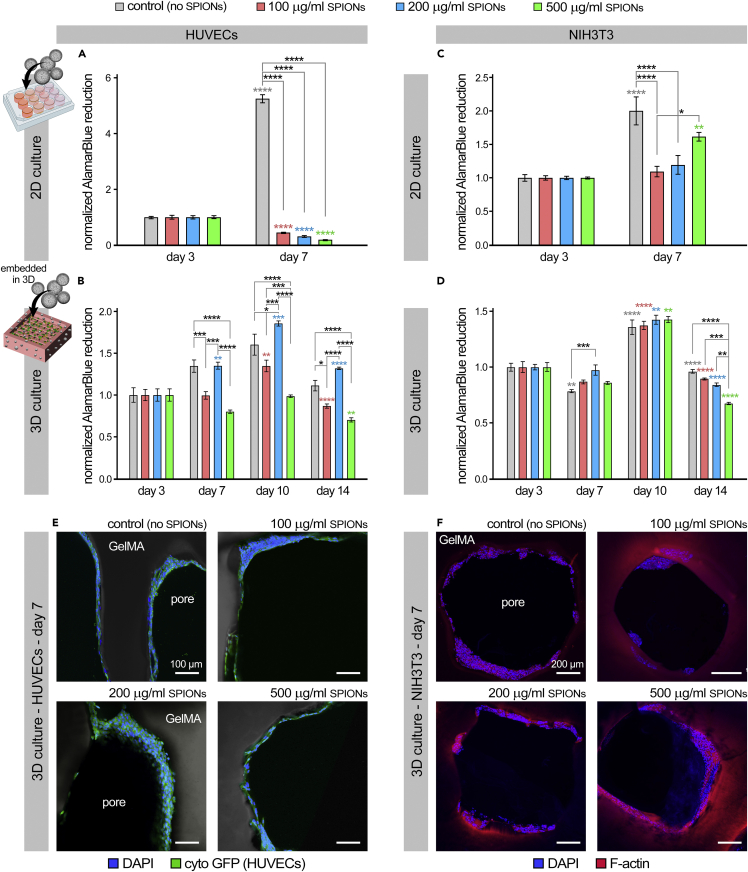
To evaluate the potential cytotoxic effect of SPIONs, a human umbilical vein cell (HUVEC) line and the NIH3T3 fibroblast cell line were seeded in a 2D environment (tissue culture plastic) with no SPIONs (control), or with 100, 200, or 500 μg/mL of SPIONs in the media. These two cell types were selected as some of the most commonly used cells in various tissue engineering and 3D bioprinting applications (Cetnar et al., 2019; Naderi et al., 2011; Scientific, 2015; Tomov et al., 2019b). All AlamarBlue readouts were normalized by the day 1 data for each group. In 2D culture, the SPION-free group showed a 5-fold significant increase in HUVEC viability and growth at day 7 (p < 0.0001), while the HUVECs cultured with varying concentrations of SPIONs all exhibited a significant decrease (p < 0.0001) (Figure 5A). The NIH3T3 cells seeded in 2D showed an increase in cell viability and growth over time in the control (no SPIONs) and the 500 μg/mL SPIONs groups Figure 5B). Interestingly, there were no significant decreases in cell viability at day 7 for the NIH3T3 cells in contrast to the HUVEC results, suggesting greater tolerance of the fibroblast line against the NP-associated cytotoxic effects (as reported before (Kong et al., 2011; Sahu et al., 2016)). Of note, in both EC and fibroblast cells, clathrin and caveolae-mediated endocytosis are the common receptors used for the uptake of NPs, including the SPIONs (Behzadi et al., 2017; Pottler et al., 2017; Rejman et al., 2004; Salingova et al., 2019). Thus, the SPION uptake in these cells can be regulated by manipulating these cell receptor pathways, as well as tuning the NP properties (e.g., size, protein corona, and surface charge) (Behzadi et al., 2017; Rejman et al., 2004).
Figure 5.
Evaluating cytotoxic effect of varying concentrations of superparamagnetic iron oxide nanoparticles (SPIONs) on cells in 2D and 3D conditions
(A and B) Evaluating the SPIONs cytotoxicity effect on HUVECs in 2D (A) and 3D bioprinted GelMA constructs (B). AlamarBlue reduction was quantified and normalized by the readout at day 1 for each group, as a measure of cell viability and growth (metabolic activity). Study groups included: no SPIONs (control), 100, 200, and 500 μg/mL SPIONs (n = 4 per group).
(C and D) AlamarBlue analysis of NIH3T3 fibroblasts cultured with varying concentrations of SPIONs in 2D (in culture media, C) and 3D bioprinted GelMA constructs (embedded within GelMA, D) (n = 4 per group).
(E and F) Immunohistochemical imaging of 3D bioprinted GelMA scaffolds following 7 days of 3D culture of HUVECs (E) and fibroblasts (F). In panel (E), HUVECs are cytoplasmic GFP positive (green) and DAPI stained nuclei (blue). In panel (F), F-actin (red) and DAPI (blue) staining were used. Scale bars in E and F represent 100 and 200 μm, respectively. ∗ p value < 0.05, ∗∗ p value < 0.01, ∗∗∗ p value < 0.001, and ∗∗∗∗ p value < 0.0001. Asterisks in color indicate statistical significance for each group in comparison to the prior time point.
Cytotoxicity effects were also studied in the 3D bioprinted GelMA constructs for up to 14 days (Figures 5C and 5D). Both HUVECs and fibroblast cells were seeded into the 3D constructs post bioprinting and crosslinking. HUVECs showed high viablility on the SPION-free, as well as the 100 and 200 μg/mL SPIONs constructs, with a maximum viability at day 10 (Figure 5C). Most favorable 3D substrates for HUVEC viability and proliferation were the control and 200 μg/mL SPION-laden GelMA constructs. NIH3T3 fibroblasts displayed a similar trend in 3D conditions with a maximum cell viability at day 10 (Figure 5D). At day 14, fibroblasts in the 500 μg/mL SPIONs group showed significantly lower viability than the other SPION-laden and SPION-free groups. Overall, the 200 μg/mL SPIONs group demonstrated the optimal support for cell viability and growth, while also providing adequate levels of printing fidelity (Figure 2), mechanical properties (Figure 3), and MRI contrast (Figure 4). The increased AlamarBlue readout in the 200 μg/mL groups for both cell types could be related to the increased cell metabolic activity to uptake the SPIONs (as reported before (Shokouhimehr et al., 2021)), while there is still no significant toxicity exerted by these particles. The noticeable difference observed in the 2D vs. 3D cell toxicity in response to SPION exposure (Figures 5A–5D) can be attributed to the significant effect of 3D bioprinted GelMA scaffold in sequestering the embedded SPIONs within the 3D bioink, hence, limiting/controlling the cytotoxic effect on cells.
Immunohistochemical staining and confocal imaging of bioprinted constructs following 7 days of 3D culture demonstrated significant cellularization of GelMA material with HUVECs (Figure 5E) and fibroblasts (Figure 5F). Both EC and fibroblast cells grew a multilayer coating onto the bioprinted GelMA scaffolds and demonstrated adequate attachment and morphology. In particular, ECs formed a rather continuous endothelium-like structure onto the porous structure Figure 5E).
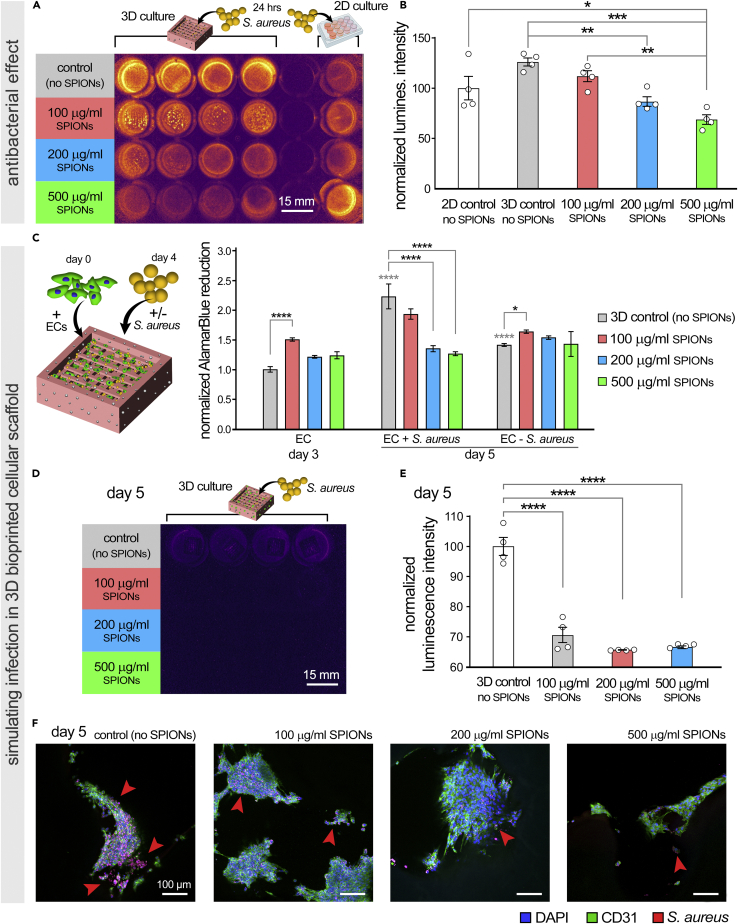
Scaffold biomaterial-associated infection, with pathogens such as S. aureus (Gristina, 1987), is a significant hurdle that has limited clinical applications of tissue-engineered implants (Darouiche, 2004; Johnson and Garcia, 2015; Qiu et al., 2007). While current strategies to prevent such infections, including antibiotic therapies, have been helpful, there still remain multiple challenges. Bacteria continuously develop resistance against common antibacterial agents (e.g., antibiotics) (Speert, 1996). Other drawbacks for conventional antimicrobial agents are the adverse side effects, primarily an intolerable toxicity (Damour et al., 1992; Mohamed et al., 2018). Thus, alternative strategies to treat biomaterial-associated bacterial infections have been developed. Among them, NPs have emerged as novel and highly effective antimicrobial agents, owing to their high surface area to volume ratio, which confers them distinct properties (Mahmoudi and Serpooshan, 2012; Mahmoudi et al., 2016; Wang et al., 2017). Multiple types of antimicrobial NPs, including SPIONs (Mahmoudi et al., 2016; Taylor et al., 2012), have shown great efficacy in treating infectious diseases, including antibiotic-resistant ones (Hajipour et al., 2012; Wang et al., 2017). In this study, for the first time, we examined the incorporation of SPIONs into GelMA bioinks as a novel approach to 3D bioprint antibacterial (i.e., bacteriostatic) scaffold systems. Bioprinted constructs were seeded with a luminescent S. aureus strain to assess the antibacterial effects (Figure 6). Embedded SPIONs within the constructs resulted in significant reduction of S. aureus bacteria, particularly in the 200 and 500 μg/mL SPION groups which showed 31% and 45% reductions in luminescence intensity, respectively, in comparison to the 3D control (Figures 6A and 6B). Of note, 3D printed GelMA constructs without SPIONs showed slightly (not significant) higher levels of bacterial activity and growth (∼26%) when compared to the 2D culture, which suggests the favorable environment of 3D scaffolds to grow bacteria as reported before (Mahmoudi et al., 2016). In comparison to the 3D constructs without SPIONs, the 500 μg/mL SPION group showed ∼57% reduction in bacteria growth (Figure 6B). The percentages of reduction in bacteria count reported here are consistent with previous reports (Mahmoudi et al., 2016; Shokouhimehr et al., 2021), indicating a ∼20%–40% reduction in bacterial growth, when comparable levels of SPIONs were incorporated into 3D scaffold systems. Thus, while such solutions do not fully eradicate bacteria in 3D culture, they are primarily proposed as a preventive measure to stop the initiation of bacterial films and control their growth in 3D scaffolds (Hajipour et al., 2012).
Figure 6.
Evaluating bacteriostatic effect of superparamagnetic iron oxide nanoparticles (SPIONs) in 2D cultures and 3D bioprinted scaffold systems
(A and B) Evaluating the antibacterial effect of varying concentrations of SPIONs in 2D culture (control) and 3D bioprinted GelMA scaffolds, seeded with luminescent S. aureus bacteria. Luminescence imaging of 2D controls and 3D printed constructs was performed after 24 h of incubation with bacteria (A) and the signal intensity was quantified and normalized based on the 2D control without SPIONs (B). Sample size (n) = 4 per group.
(C–E) We subsequently assessed the antibacterial function of nanoparticles in a cellular 3D culture of endothelial cells (ECs), simulating bacterial infection of in vitro culture specimens. AlamarBlue assay was performed on days 3 and 5, pre and post 24-h incubation of S. aureus to assess the effect of bacteria on EC viability and growth (C). At day 5, luminescence imaging was performed (D) and quantified (E) to examine the effect of incorporated SPIONs in inhibiting bacterial infection within the 3D bioprinted constructs. Sample size (n) = 4 per group.
(F) Confocal imaging of immunostained tissue slices at day 5, after 24 h of EC- S. aureus coculture, highlighting the colocalization of bacterial within 3D EC clusters in the GelMA matrix. Samples were sectioned and stained with anti-CD31 (green, ECs), anti-Staphylococcus aureus (red, bacteria), and DAPI (blue, nuclei). Red arrows point to localization of bacteria within the EC clusters in the 3D structures. Scale bars in (F) represent 100 μm. ∗ p value < 0.05, ∗∗ p value < 0.01, ∗∗∗ p value < 0.001, and ∗∗∗∗ p value < 0.0001. Asterisks in color indicate statistical significance for each group in comparison to the prior time point.
We subsequently simulated the bacterial infection of 3D HUVEC cultures in bioprinted GelMA constructs (Figures 6C–6E). On day 3, prior to addition of bacteria, AlamarBlue results demonstrated higher (significant for the 100 μg/mL group) EC viability and growth after 3 days of culture in the SPION-laden groups compared to the 3D control (empty GelMA) (Figure 6C). This confirmed the cytocompatibility of SPIONs at the selected concentrations while embedded within the 3D matrix (Saei et al., 2017; Wei et al., 2021). The increase in EC viability/growth observed here for the 100 μg/mL was not obtained in the prior cytotoxicity assay (Figure 5B). This rather small difference may be related to the inherent, slight variability of the AlamarBlue assay and the fluorescence readouts. On day 5, after 24 h of incubation with bacteria, there was a clear increase of AlamarBlue activity in the control GelMA constructs (without SPIONs) containing S. aureus bacteria, compared to those without the bacteria. This is attributed to the boosting effect of grown bacteria on reducing the AlamarBlue reagent (resazurin), as also evidenced in the contaminated cell cultures and other biological processes (e.g., milk contamination (O'Brien et al., 2000)). EC coculture with S. aureus in the constructs containing SPIONs showed a decrease in AlamarBlue readout at day 5. The 200 and 500 μg/mL SPION groups both demonstrated a significant (p < 0.0001) ∼41% decrease in AlamarBlue reduction which could be mainly attributed to the antibacterial function of SPIONs and the removal of bacteria. This is while the EC culture without S. aureus showed slightly higher or equal levels of cell viability and growth at day 5 (Figure 6C). To further investigate the bacteriostatic effect, we examined the luminescence signal of the S. aureus bacteria that were added to ongoing cultures on day 4. Significantly (p < 0.0001) distinct growth was observed after 24 h in the SPION-free control group versus the constructs loaded with varying levels of SPIONs (Figures 6D and 6E). By increasing the SPION concentration in the bioinks, the 100, 200, and 500 μg/mL SPION groups exhibited amplified bacteriostatic effects, resulting in 29%, 34%, and 33% decreases in the bacteria signal, respectively.
Immunohistochemical analysis of infected 3D constructs, after 24 h of incubation with S. aureus, revealed that the ECs remained viable and adherent to the 3D matrix, while the bacteria colonies colocalized with the EC clusters (Figure 6F). The formation of S. aureus biofilm was evident in the SPION-free control group, as well as some smaller areas in the SPION-laden constructs (red arrows, Figure 6F). Confocal imaging of the 500 μg/mL SPION group shows notably lower number of ECs, as well as bacteria, in the 3D constructs which is in agreement with the cytotoxicity results obtained via AlamarBlue assays (Figure 5). Overall, these results confirmed that the 200 μg/mL SPION-laden bioprints demonstrate the optimal functions (e.g., printability, cell viability and growth, mechanical properties, MRI contrast, and antibacterial effects) for in vitro modeling and screening applications, as well as use as implants for a variety of regenerative medicine applications.
Conclusions
This work demonstrated, for the first time, the use of engineered NP systems to develop a new class of hydrogel-based bioinks with enhanced antibacterial and imaging properties, while maintaining their printability and printing fidelity, mechanical properties, and bioactivity. The 3D bioprinted SPION-laden hydrogel constructs developed here can be used in a variety of soft and hard tissue engineering applications. Through preventing the biofilm formation, and simultaneously enabling longitudinal noninvasive tracking of the scaffolds, these engineered scaffolds would help alleviate several major complications related to biomaterial implants, particularly reducing the risk of implant infection and rejection. While our results suggest that a 200 μg/mL of SPIONs encapsulated in the GelMA bioink yields optimal in vitro functions of the scaffolds, the concentration and properties of NP systems will further require tuning for each specific NP type, hydrogel material, bacteria strain, and also for the specific biomedical application. Integrating these new functions of SPION-laden bioinks with the established advantages of bioprinting methods, such as patient/damage specificity and heterogeneous structures, would enable creation of robust multifunctional platforms for diverse clinical and translational applications.
Future studies could investigate the interplay between incorporated NPs with the immune system cells, such as macrophages and T cells, and its role in the elicited antibacterial effects as well as other functions of the bioprinted scaffold systems. Furthermore, incorporation of functional and tissue/patient-specific cells types, such as induced pluripotent stem cell-derived cardiomyocytes, within the NP-laden bioinks can usher in a new wave of personalized precision regenerative therapies.
Limitations of the study
Integration of functionalized antibacterial nanoparticles with microextrusion-based 3D bioprinting is one of the first attempts to generate antibacterial bioink formulations for antibacterial scaffold biofabrication. This research could still benefit from more in-depth characterization of the nanoparticle-bacteria interactions within the 3D structure. While this study showed relative success of the SPION-laden bioinks in inhibiting bacteria growth, a complete eradication of the bacteria was not achieved. Analysis and optimization of the SPIONs properties and encapsulation strategies (e.g., altering the particles composition, surface coating, and electrical charge to change affinity with the hydrogel bioink) could further improve the efficacy of the antibacterial properties. Furthermore, studying the potential impact of the released particles on cell/tissue response (e.g., cytotoxicity and clearance) in both in vitro and in vivo settings would be of great significance to assess the capacity of engineered bioinks for translation into clinical applications.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| anti-CD31 | Invitrogen™ | Cat: MA3100; RRID:AB_223516 |
| anti-connexin43 | Invitrogen™ | Cat: 71-070-0; RRID:AB_2533973 |
| polyclonal anti-Staphylococcus aureus | BioRad | Cat: 0300-0084; RRID:AB_619560 |
| Bacterial and virus strains | ||
| Staphylococcus aureus | ATCC | Cat:43300; RRID:WB-STRAIN:WBStrain00041949 |
| Chemicals, peptides, and recombinant proteins | ||
| Methacrylic anhydride | Millipore Sigma | Cas: 760-93-0 |
| lithium phenyl-2,4,6-trimethylbenzoylphosphinate (LAP) | Millipore Sigma | Cas: 85073-19-4 |
| Experimental models: Cell lines | ||
| human umbilical vein endothelial cell (HUVEC) | Lifeline Technology | Cat: FC-0044 |
| NIH-3T3 | ATCC | Cat: CRL-1658; RRID:CVCL_0594 |
| Software and algorithms | ||
| Autodesk Fusion 360 | Autodesk Inc., San Rafael, CA | |
| ImageJ | National Institutes of Health, USA | |
| JMP | JMP Statistical Discovery, SAS, USA | |
Resource availability
Lead contact
Further information and requests should be directed to and will be fulfilled by the Lead Contact, Vahid Serpooshan at vahid.serpooshan@bme.gatech.edu.
Materials availability
This study did not generate new unique reagents.
Experimental model and subject details
Details of the bioprinted in vitro model and cell culture model are completely described in method details.
Method details
3D bioprinting of constructs
3D constructs were prepared using GelMA synthesized from porcine gelatin type A and methacrylic anhydride (MA) as described before (88.9 ± 1.0% methacrylation degree) (Ning et al., 2020; Shirahama et al., 2016). SPIONs were purchased (Micromod Partikeltechnologie GmbH, Rostock, Germany) and incorporated into the GelMA bioink to obtain concentrations of 100 μg/mL, 200 μg/mL, and 500 μg/mL of NPs while maintaining the overall weight percentage of GelMA at 10% (g/mL). A 0.5% (w/v) concentration of lithium phenyl-2,4,6-trimethylbenzoylphosphinate (LAP) was used as a photoinitiator to crosslink the bioinks under a 10 mW/cm2 UV light source for 2 min. These crosslinking parameters were chosen based on our previous experiences (Li et al., 2022; ML Tomov et al., 2019; Ning et al., 2020; Ning et al., 2022; Ning et al., 2021) to generate GelMA constructs at biologically relevant stiffness (mimicking soft tissue stiffness, ∼20–140 kPa (Huang et al., 2012; Serpooshan et al., 2013; Wei et al., 2015)). 3D printed constructs were designed using Autodesk Fusion 360 CAD software (Autodesk Inc., San Rafael, CA) and converted to the standard STL file format for printing. Constructs were printed utilizing a BioAssemblyBot robotic arm 3D bioprinter (Advanced Solutions Life Sciences, Louisville, KY). One standard geometry was used for in vitro experiments. The scaffold geometry featured a 12 × 12 × 4 mm3 structure with an internal lattice structure with a pore size of 1 mm. This model was used for all in vitro cytotoxicity experiments, bacteria studies, and MR imaging. Bioprinting parameters used consistently to print all GelMA constructs for this study included: printing speed of 5 mm/s, pressure of 20 psi, temperature of 27°C, layer height of 0.2 mm, and a 27-gauge syringe needle.
Fidelity measurements
To assess fidelity of various bioinks developed in this study, we 3D bioprinted a simple two-layer cross hatch design and examined the accuracy of several geometric parameters. Bright-field microscopy images were acquired at 1.5X and 4× magnifications to assess the printing fidelity using ImageJ (National Institutes of Health, USA). Design factors such as the strand diameter ratio, strand uniformity ratio, strand angle ratio, and inter-strand area ratio were quantified as described before (Ning et al., 2020). In the CAD model, we used a strand diameter () of 0.3 mm, a length of strand () of 9 mm, a strand angle () of 60°, and an inter-strand area () of 1 mm2. These reference values were compared against the ones measured in the printed 2-layer structure to obtain the normalized ratios. These ratios are defined as the strand diameter (), strand uniformity (), strand angle (), and inter-strand area () ratios, obtained using Equations (5)–(8). ImageJ was used to perform and record all measurements.
| (Equation 5) |
| (Equation 6) |
| (Equation 7) |
| (Equation 8) |
Each fidelity test was repeated five times and three bright field microscopy images were acquired (randomly) from each construct and manually measured (n = 15).
Mechanical testing
Uniaxial unconfined compression and microindentation tests were performed on 3D printed cubic structures (7 × 7 × 4 mm3) composed of 10% (w/v) GelMA together with 100, 200, or 500 μg/mL SPIONs. A Mach-1 mechanical testing system (Biomomentum, QC, Canada) was utilized for the tests. Unconfined compression was conducted at a 50% total strain at 20 μm/s. The compressive modulus was derived from the slope of the linear trend line (initial 0–20%) of the stress-strain curve. Four replicates were prepared (n = 4) and the average value with standard deviation was recorded.
The localized mechanical behavior of printed constructs was further assessed using microindentation. A 500 μm probe was used to indent the surface of each construct (n = 5), with a depth of 100 μm at 2 μm/s. The force-displacement unloading curves were plotted and used to calculate the stiffness of the sample () from the linear trend line slope (initial 5–20%), and the reduced elastic modulus was next derived using the equation below (Oliver and Pharr, 2004):
| (Equation 1) |
where is a constant and equals 1 in this study and is projected contact area at the contact depth of , which can be obtained from the following equation:
| (Equation 2) |
where
| (Equation 3) |
and are the peak unloading displacement and unloading force, respectively, and is a constant with a value of 0.75 for a spherical probe (Oliver and Pharr, 1992).
The elastic modulus, , can be then calculated from using the Equation (4) (Oliver and Pharr, 2004):
| (Equation 4) |
where is the Poisson’s ratio of tested material with a value of 0.5 and is 0.5 for the indenter tip material. represents the elastic modulus of the probe, with a value of 2 GPa.
MR imaging of bioprinted scaffolds
The effect of SPION incorporation on the MR-visibility of bioprinted GelMA scaffolds was examined in vitro using T2∗ weighted images as described before (Mahmoudi et al., 2016). Briefly, bioprinted GelMA constructs containing varying concentrations of SPIONs were fixed, embedded in 2% agarose gel (Baek et al., 2019), and imaged using a 9.4T/20 cm Bruker animal MR imaging/spectroscopy system driven by LINUX workstation and Bruker ParaVision 5.1 imaging software (Magnuson et al., 2010).
Endothelial and NIH3T3cell culture
A well characterized human umbilical vein endothelial cell (HUVEC) line was purchased (Lifeline Cell Technology LLC, USA) and used for the in vitro cell culture experiments. The HUVECs were cultured with VascuLife VEGF Endothelial Medium Complete Kit (LL-0003) (Lifeline Cell Technology LLC, USA), with or without antibiotics and antimycotics, in T75 culture flasks in a humidified tissue culture incubator (37°C with a 5% CO2). 2D HUVEC cultures were seeded at a density of 10,000 cells/cm2 in a 6 well plate and cultured for up to 7 days. A density of 2 × 106 cells/construct were seeded onto 3D printed constructs and left to culture up to 5 or 14 days before fixation with 10% formalin. NIH 3T3 fibroblasts were cultured in 2D 10-cm Petri dishes using Eagle’s Minimum Essential Medium supplemented with 10% fetal bovine serum and 1% penicillin-streptomycin (all from ATCC, USA). Following confluency, fibroblasts were dissociated and seeded onto 3D bioprinted GelMA constructs at a density of 2 × 106 cells/construct.
Cell viability and growth measurements
Cellular viability was evaluated, longitudinally and in a noninvasive manner, using the AlamarBlue Cell Viability Reagent (BioRad, USA). The AlamarBlue reagent was added to fresh media in a 1:10 volumetric ratio and added to the culture wells. The samples were left to incubate in the solution for 4 h. Subsequently, 100-μL supernatant samples were collected from each well and transferred to a 96-well plate in triplicates. Absorbance was recorded at 550 and 600 nm utilizing a microplate reader (BioTek Instruments, USA). Readings were recorded at days 3 and 7, and days 3, 7, 10, and 14 for 2D and 3D SPION cytotoxicity studies, respectively.
To simulate bacterial infection of 3D cell culture, endothelial cells were seeded into the bioprinted GelMA constructs (2 × 106 cells/construct, seeded after printing and crosslinking) at varying SPION concentrations (0, 100, 200, and 500 μg/mL) and cultured for 3 days. On the day 3, S. aureus (S. aureus; ATCC 43300) bacteria were seeded into each construct (108 colony-forming-units/construct) as described below. Following 2 days of coculture, constructs were imaged via luminescence imaging and then fixed and analyzed via immunohistochemistry as explained below. AlamarBlue assay was conducted at days 3 and 5 as above.
Bacteria cultures and assays
S. aureus (S. aureus; ATCC 43300) was transduced with a modified luxABCDE operon from Photorhabdus luminescens optimized for Gram-positive expression (Xen29, Caliper) so that live bacteria emit bioluminescence and bacterial number can be quantified culture-independent and in real-time using a luminometer and an ICCD camera (Francis et al., 2001). S. aureus was cultured aerobically at 37° in Luria–Bertani (LB) medium (Difco) 18 h, then subcultured 1:40 to an optical density at 600 nm (OD600) of 0.4 via spectrophotometry. Subsequently, 108 colony-forming units of bacteria were added onto 3D printed constructs with and without SPIONs in a 24-well tissue culture plate (Costar) and co-incubated up to 24 h to determine remaining bacteria levels. Each plate was imaged on a ChemiDoc MP Imaging System (Bio-Rad Laboratories, USA) without external light source to specifically measure S. aureus bioluminescence. The luminescence signal was quantified using the analysis software ImageJ (NIH). The area of each SPION was selected and the total signal quantified from pixel intensity within this region, then averaged between technical replicates for each group.
Immunohistochemical analysis – Confocal microscopy
Harvested constructs were fixed in 10% neutral buffered formalin at room temperature for 30 min and washed immediately with PBS (5 min). Constructs were then sliced with a vibratome (Leica VT 1200S) to a thickness of 200 μm for further analysis. Sections were stained with a variety of primary antibodies including anti-CD31 (#ENMA3100, Invitrogen), anti-connexin43 (#71-070-0, Invitrogen), and polyclonal anti-S. aureus (#0300-0084, BioRad). Confocal imaging was performed using the Olympus FV1000 confocal laser microscope.
Quantification and statistical analysis
Statistical analysis was performed utilizing JMP (JMP Statistical Discovery from SAS, USA) software. Significant differences were determined with one-way ANOVA or two-way ANOVA if applicable. A post-hoc Tukey-Kramer test was performed for multiple comparisons and a p value of <0.05 was considered statistically significant (∗ p value < 0.05, ∗∗ p value < 0.01, ∗∗∗ p value < 0.001, and ∗∗∗∗ p value < 0.0001). Least square means connecting letters reports were also used to show significant differences between multiple comparisons. Levels not connected by the same letter are significantly different. Levels connected by the same letter are not significantly different.
Acknowledgment
The authors acknowledge the support from the National Institutes of Health (R00 HL127295 and R01 MH126195 to V.S., and NIAID R01 AI153071 to C.L.), National Science Foundation (NSF CAREER Award to V.S.), and Emory University Dean’s Imagine, Innovate and Impact (I3) Research Award (to V.S.). Research reported in this publication was supported in part by the Emory University Integrated Cellular Imaging (ICI) Core and Children’s Healthcare of Atlanta (CHOA). M.M. gratefully acknowledges financial support from the U.S. National Institute of Diabetes and Digestive and Kidney Diseases (grant DK131417).
Author contributions
Conceptualization, V.S.; Funding, V.S., C.L., M.M.; Original Draft, A.T., L.N., V.S.; Experiments: A.T., L.N., G.K.; Data Curation and Analysis, A.T., G.K., B.H., M.T., C.L.; Revision, V.S., H.B.H.; Resources, C.L., H.B.H., M.M., V.S.
Declaration of interests
Author V.S. is a Guest Editor on the Advanced Manufacturing of Tissue Constructs Special Issue published in iScience. Author M.M. discloses that (i) he is a co-founder and director of the Academic Parity Movement (www.paritymovement.org), a non-profit organization dedicated to addressing academic discrimination, violence and incivility; (ii) he is a Founding Partner at Partners in Global Wound Care (PGWC); and (iii) he receives royalties/honoraria for his published books, plenary lectures, and licensed patents.
Published: September 16, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2022.104947.
Supplemental information
Data and code availability
-
•
All data reported in this work will be shared by the lead contact upon reasonable request.
-
•
This work has no original code and custom computer code.
-
•
Any additional information required to reanalyze the data reported in this work is available from the Lead contact upon request.
References
- Abat C., Raoult D., Rolain J.M. Are we living in an antibiotic resistance nightmare? Clin. Microbiol. Infect. 2018;24:568–569. doi: 10.1016/j.cmi.2018.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agarwal S., Saha S., Balla V.K., Pal A., Barui A., Bodhak S. Current developments in 3D bioprinting for tissue and organ regeneration–A review. Front. Mech. Eng. 2020;6 doi: 10.3389/fmech.2020.589171. [DOI] [Google Scholar]
- Ai F., Ferreira C.A., Chen F., Cai W. Engineering of radiolabeled iron oxide nanoparticles for dual-modality imaging. Wiley Interdiscip Rev. Nanomed. Nanobiotechnol. 2016;8:619–630. doi: 10.1002/wnan.1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albanna M., Binder K.W., Murphy S.V., Kim J., Qasem S.A., Zhao W., Tan J., El-Amin I.B., Dice D.D., Marco J., et al. In situ bioprinting of autologous skin cells accelerates wound healing of extensive excisional full-thickness wounds. Sci. Rep. 2019;9:1856. doi: 10.1038/s41598-018-38366-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baek K., Jung S., Lee J., Min E., Jung W., Cho H. Quantitative assessment of regional variation in tissue clearing efficiency using optical coherence tomography (OCT) and magnetic resonance imaging (MRI): a feasibility study. Sci. Rep. 2019;9:2923. doi: 10.1038/s41598-019-39634-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakhsheshi-Rad H.R., Ismail A.F., Aziz M., Akbari M., Hadisi Z., Daroonparvar M., Chen X.B. Antibacterial activity and in vivo wound healing evaluation of polycaprolactone-gelatin methacryloyl-cephalexin electrospun nanofibrous. Mater. Lett. 2019;256:126618. doi: 10.1016/j.matlet.2019.126618. [DOI] [Google Scholar]
- Behzadi S., Serpooshan V., Tao W., Hamaly M.A., Alkawareek M.Y., Dreaden E.C., Brown D., Alkilany A.M., Farokhzad O.C., Mahmoudi M. Cellular uptake of nanoparticles: journey inside the cell. Chem. Soc. Rev. 2017;46:4218–4244. doi: 10.1039/c6cs00636a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya A., Janarthanan G., Noh I. Nano-biomaterials for designing functional bioinks towards complex tissue and organ regeneration in 3D bioprinting. Addit. Manuf. 2021;37:101639. doi: 10.1016/j.addma.2020.101639. [DOI] [Google Scholar]
- Busscher H.J., van der Mei H.C., Subbiahdoss G., Jutte P.C., van den Dungen J.J., Zaat S.A., Schultz M.J., Grainger D.W. Biomaterial-associated infection: locating the finish line in the race for the surface. Sci. Transl. Med. 2012;4:153rv110. doi: 10.1126/scitranslmed.3004528. [DOI] [PubMed] [Google Scholar]
- Cetnar A., Tomov M., Theus A., Lima B., Vaidya A., Serpooshan V. In: 3D Bioprinting in Medicine: Technologies, Bioinks, and Applications. Guvendiren M., editor. Springer International Publishing; 2019. 3D bioprinting in clinical cardiovascular medicine; pp. 149–162. [DOI] [Google Scholar]
- Chan W.W., Yeo D.C.L., Tan V., Singh S., Choudhury D., Naing M.W. Additive biomanufacturing with collagen inks. Bioengineering (Basel) 2020;7 doi: 10.3390/bioengineering7030066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung J.J., Im H., Kim S.H., Park J.W., Jung Y. Toward biomimetic scaffolds for tissue engineering: 3D printing techniques in regenerative medicine. Front. Bioeng. Biotechnol. 2020;8:586406. doi: 10.3389/fbioe.2020.586406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corona-Gomez J., Chen X., Yang Q. Effect of nanoparticle incorporation and surface coating on mechanical properties of bone scaffolds: a brief review. J. Funct. Biomater. 2016;7 doi: 10.3390/jfb7030018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damour O., Hua S.Z., Lasne F., Villain M., Rousselle P., Collombel C. Cytotoxicity evaluation of antiseptics and antibiotics on cultured human fibroblasts and keratinocytes. Burns. 1992;18:479–485. doi: 10.1016/0305-4179(92)90180-3. [DOI] [PubMed] [Google Scholar]
- Darouiche R.O. Treatment of infections associated with surgical implants. N. Engl. J. Med. 2004;350:1422–1429. doi: 10.1056/NEJMra035415. [DOI] [PubMed] [Google Scholar]
- Francis K.P., Yu J., Bellinger-Kawahara C., Joh D., Hawkinson M.J., Xiao G., Purchio T.F., Caparon M.G., Lipsitch M., Contag P.R. Visualizing pneumococcal infections in the lungs of live mice using bioluminescent Streptococcus pneumoniae transformed with a novel gram-positive lux transposon. Infect. Immun. 2001;69:3350–3358. doi: 10.1128/IAI.69.5.3350-3358.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil C., Evans C., Li L., Vargas M., Kabboul G., Fulton T., Veneziano R., Nick N., Bauser-Heaton H., Roeder R.K., Serpooshan V. Abstract MP207: a precision medicine approach for non-invasive, longitudinal, and quantitative monitoring of cardiac tissue-engineered scaffolds. Circ. Res. 2021;129:AMP207. doi: 10.1161/res.129.suppl_1.MP207. [DOI] [Google Scholar]
- Greenhalgh R., Dempsey-Hibbert N.C., Whitehead K.A. Antimicrobial strategies to reduce polymer biomaterial infections and their economic implications and considerations. Int. Biodeterior. Biodegrad. 2019;136:1–14. doi: 10.1016/j.ibiod.2018.10.005. [DOI] [Google Scholar]
- Gristina A.G. Biomaterial-centered infection: microbial adhesion versus tissue integration. Science. 1987;237:1588–1595. doi: 10.1126/science.3629258. [DOI] [PubMed] [Google Scholar]
- Hajipour M.J., Fromm K.M., Akbar Ashkarran A., Jimenez de Aberasturi D., Larramendi I.R.d., Rojo T., Serpooshan V., Parak W.J., Mahmoudi M. Antibacterial properties of nanoparticles. Trends Biotechnol. 2012;30:499–511. doi: 10.1016/j.tibtech.2012.06.004. [DOI] [PubMed] [Google Scholar]
- Hajipour M.J., Saei A.A., Walker E.D., Conley B., Omidi Y., Lee K.B., Mahmoudi M. Nanotechnology for targeted detection and removal of bacteria: opportunities and challenges. Adv. Sci. 2021;8:e2100556. doi: 10.1002/advs.202100556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasan A., Morshed M., Memic A., Hassan S., Webster T.J., Marei H.E. Nanoparticles in tissue engineering: applications, challenges and prospects. Int. J. Nanomed. 2018;13:5637–5655. doi: 10.2147/IJN.S153758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hölzl K., Lin S., Tytgat L., Van Vlierberghe S., Gu L., Ovsianikov A. Bioink properties before, during and after 3D bioprinting. Biofabrication. 2016;8:032002. doi: 10.1088/1758-5090/8/3/032002. [DOI] [PubMed] [Google Scholar]
- Huang G., Wang L., Wang S., Han Y., Wu J., Zhang Q., Xu F., Lu T.J. Engineering three-dimensional cell mechanical microenvironment with hydrogels. Biofabrication. 2012;4:042001. doi: 10.1088/1758-5082/4/4/042001. [DOI] [PubMed] [Google Scholar]
- Johnson C.T., Garcia A.J. Scaffold-based anti-infection strategies in bone repair. Ann. Biomed. Eng. 2015;43:515–528. doi: 10.1007/s10439-014-1205-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khezerlou A., Alizadeh-Sani M., Azizi-Lalabadi M., Ehsani A. Nanoparticles and their antimicrobial properties against pathogens including bacteria, fungi, parasites and viruses. Microb. Pathog. 2018;123:505–526. doi: 10.1016/j.micpath.2018.08.008. [DOI] [PubMed] [Google Scholar]
- Kong B., Seog J.H., Graham L.M., Lee S.B. Experimental considerations on the cytotoxicity of nanoparticles. Nanomedicine (London, England) 2011;6:929–941. doi: 10.2217/nnm.11.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuijer R., Jansen E.J., Emans P.J., Bulstra S.K., Riesle J., Pieper J., Grainger D.W., Busscher H.J. Assessing infection risk in implanted tissue-engineered devices. Biomaterials. 2007;28:5148–5154. doi: 10.1016/j.biomaterials.2007.06.003. [DOI] [PubMed] [Google Scholar]
- Kumar S.S.D., Abrahamse H. Advancement of nanobiomaterials to deliver natural compounds for tissue engineering applications. Int. J. Mol. Sci. 2020;21 doi: 10.3390/ijms21186752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurian A.G., Singh R.K., Patel K.D., Lee J.H., Kim H.W. Multifunctional GelMA platforms with nanomaterials for advanced tissue therapeutics. Bioact. Mater. 2022;8:267–295. doi: 10.1016/j.bioactmat.2021.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.-H., Kim H.-W. Emerging properties of hydrogels in tissue engineering. J. Tissue Eng. 2018;9 doi: 10.1177/2041731418768285. 2041731418768285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Gil C.J., Finamore T.A., Evans C.J., Tomov M.L., Ning L., Theus A., Kabboul G., Serpooshan V., Roeder R.K. Methacrylate-modified gold nanoparticles enable non-invasive monitoring of photocrosslinked hydrogel scaffolds. bioRxiv. 2022 doi: 10.1101/2022.01.26.477960. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Jiang W., Luo K., Song H., Lan F., Wu Y., Gu Z. Superparamagnetic iron oxide nanoparticles as MRI contrast agents for non-invasive stem cell labeling and tracking. Theranostics. 2013;3:595–615. doi: 10.7150/thno.5366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnuson M., Majeed W., Keilholz S.D. Functional connectivity in blood oxygenation level-dependent and cerebral blood volume-weighted resting state functional magnetic resonance imaging in the rat brain. J. Magn. Reson. Imag. 2010;32:584–592. doi: 10.1002/jmri.22295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahmoudi M., Hofmann H., Rothen-Rutishauser B., Petri-Fink A. Assessing the in vitro and in vivo toxicity of superparamagnetic iron oxide nanoparticles. Chem. Rev. 2012;112:2323–2338. doi: 10.1021/cr2002596. [DOI] [PubMed] [Google Scholar]
- Mahmoudi M., Saeedi-Eslami S.N., Shokrgozar M.A., Azadmanesh K., Hassanlou M., Kalhor H.R., Burtea C., Rothen-Rutishauser B., Laurent S., Sheibani S., Vali H. Cell "vision": complementary factor of protein corona in nanotoxicology. Nanoscale. 2012;4:5461–5468. doi: 10.1039/c2nr31185b. [DOI] [PubMed] [Google Scholar]
- Mahmoudi M., Sant S., Wang B., Laurent S., Sen T. Superparamagnetic iron oxide nanoparticles (SPIONs): development, surface modification and applications in chemotherapy. Adv. Drug Deliv. Rev. 2011;63:24–46. doi: 10.1016/j.addr.2010.05.006. [DOI] [PubMed] [Google Scholar]
- Mahmoudi M., Serpooshan V. Silver-coated engineered magnetic nanoparticles are promising for the success in the fight against antibacterial resistance threat. ACS Nano. 2012;6:2656–2664. doi: 10.1021/nn300042m. [DOI] [PubMed] [Google Scholar]
- Mahmoudi M., Serpooshan V., Laurent S. Engineered nanoparticles for biomolecular imaging. Nanoscale. 2011;3:3007–3026. doi: 10.1039/c1nr10326a. [DOI] [PubMed] [Google Scholar]
- Mahmoudi M., Zhao M., Matsuura Y., Laurent S., Yang P.C., Bernstein D., Ruiz-Lozano P., Serpooshan V. Infection-resistant MRI-visible scaffolds for tissue engineering applications. Bioimpacts. 2016;6:111–115. doi: 10.15171/bi.2016.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller G.J., Morgan E.F. Use of microindentation to characterize the mechanical properties of articular cartilage: comparison of biphasic material properties across length scales. Osteoarthritis Cartilage. 2010;18:1051–1057. doi: 10.1016/j.joca.2010.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ML Tomov A.T., Cetnar A., Bauser-Heaton H., Serpooshan V. 2019. Vascularized Multi-Tissue Platform – 3D Bioprinting an Organ Model in Vitro. [Google Scholar]
- Mohamed M.A., Nasr M., Elkhatib W.F., Eltayeb W.N. In vitro evaluation of antimicrobial activity and cytotoxicity of different nanobiotics targeting multidrug resistant and biofilm forming staphylococci. BioMed Res. Int. 2018:7658238. doi: 10.1155/2018/7658238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naderi H., Matin M.M., Bahrami A.R. Review paper: critical issues in tissue engineering: biomaterials, cell sources, angiogenesis, and drug delivery systems. J. Biomater. Appl. 2011;26:383–417. doi: 10.1177/0885328211408946. [DOI] [PubMed] [Google Scholar]
- Nehra P., Chauhan R.P., Garg N., Verma K. Antibacterial and antifungal activity of chitosan coated iron oxide nanoparticles. Br. J. Biomed. Sci. 2018;75:13–18. doi: 10.1080/09674845.2017.1347362. [DOI] [PubMed] [Google Scholar]
- Neuwelt A., Sidhu N., Hu C.A., Mlady G., Eberhardt S.C., Sillerud L.O. Iron-based superparamagnetic nanoparticle contrast agents for MRI of infection and inflammation. AJR Am. J. Roentgenol. 2015;204:W302–W313. doi: 10.2214/AJR.14.12733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ning L., Mehta R., Cao C., Theus A., Tomov M., Zhu N., Weeks E.R., Bauser-Heaton H., Serpooshan V. Embedded 3D bioprinting of gelatin methacryloyl-based constructs with highly tunable structural fidelity. ACS Appl. Mater. Interfaces. 2020;12:44563–44577. doi: 10.1021/acsami.0c15078. [DOI] [PubMed] [Google Scholar]
- Ning L., Shim J., Tomov M.L., Liu R., Mehta R., Mingee A., Hwang B., Jin L., Mantalaris A., Xu C., et al. A 3D bioprinted in vitro model of neuroblastoma recapitulates dynamic tumor-endothelial cell interactions contributing to solid tumor Aggressive behavior. Adv. Sci. 2022:e2200244. doi: 10.1002/advs.202200244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ning L., Zhu N., Smith A., Rajaram A., Hou H., Srinivasan S., Mohabatpour F., He L., McLnnes A., Serpooshan V., et al. Noninvasive three-dimensional in situ and in vivo characterization of bioprinted hydrogel scaffolds using the X-ray propagation-based imaging technique. ACS Appl. Mater. Interfaces. 2021;13:25611–25623. doi: 10.1021/acsami.1c02297. [DOI] [PubMed] [Google Scholar]
- O'Brien J., Wilson I., Orton T., Pognan F. Investigation of the Alamar Blue (resazurin) fluorescent dye for the assessment of mammalian cell cytotoxicity. Eur. J. Biochem. 2000;267:5421–5426. doi: 10.1046/j.1432-1327.2000.01606.x. [DOI] [PubMed] [Google Scholar]
- Offerhaus C., Balke M., Hente J., Gehling M., Blendl S., Höher J. Vancomycin pre-soaking of the graft reduces postoperative infection rate without increasing risk of graft failure and arthrofibrosis in ACL reconstruction. Knee Surg. Sports Traumatol. Arthrosc. 2019;27:3014–3021. doi: 10.1007/s00167-018-5323-6. [DOI] [PubMed] [Google Scholar]
- Oliver W.C., Pharr G.M. An improved technique for determining hardness and elastic modulus using load and displacement sensing indentation experiments. J. Mater. Res. 1992;7:1564–1583. [Google Scholar]
- Oliver W.C., Pharr G.M. Measurement of hardness and elastic modulus by instrumented indentation: advances in understanding and refinements to methodology. J. Mater. Res. 2004;19:3–20. [Google Scholar]
- Pottler M., Fliedner A., Schreiber E., Janko C., Friedrich R.P., Bohr C., Dollinger M., Alexiou C., Durr S. Impact of superparamagnetic iron oxide nanoparticles on vocal fold fibroblasts: cell behavior and cellular iron kinetics. Nanoscale Res. Lett. 2017;12:284. doi: 10.1186/s11671-017-2045-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu Y., Zhang N., An Y.H., Wen X. Biomaterial strategies to reduce implant-associated infections. Int. J. Artif. Organs. 2007;30:828–841. doi: 10.1177/039139880703000913. [DOI] [PubMed] [Google Scholar]
- Rejman J., Oberle V., Zuhorn I.S., Hoekstra D. Size-dependent internalization of particles via the pathways of clathrin- and caveolae-mediated endocytosis. Biochem. J. 2004;377:159–169. doi: 10.1042/BJ20031253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saei A.A., Yazdani M., Lohse S.E., Bakhtiary Z., Serpooshan V., Ghavami M., Asadian M., Mashaghi S., Dreaden E.C., Mashaghi A., Mahmoudi M. Nanoparticle surface functionality dictates cellular and systemic toxicity. Chem. Mater. 2017;29:6578–6595. doi: 10.1021/acs.chemmater.7b01979. [DOI] [Google Scholar]
- Sahu D., Kannan G.M., Tailang M., Vijayaraghavan R. In vitro cytotoxicity of nanoparticles: a comparison between particle size and cell type. J. Nanosci. 2016:4023852. doi: 10.1155/2016/4023852. [DOI] [Google Scholar]
- Salingova B., Simara P., Matula P., Zajickova L., Synek P., Jasek O., Veverkova L., Sedlackova M., Nichtova Z., Koutna I. The effect of uncoated SPIONs on hiPSC-differentiated endothelial cells. Int. J. Mol. Sci. 2019;20 doi: 10.3390/ijms20143536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scientific W. Bioprinting. 2015. Cell sources for bioprinting; pp. 165–177. [DOI] [Google Scholar]
- Serpooshan V., Guvendiren M. Editorial for the special issue on 3D printing for tissue engineering and regenerative medicine. Micromachines. 2020;11 doi: 10.3390/mi11040366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serpooshan V., Sheibani S., Pushparaj P., Wojcik M., Jang A.Y., Santoso M.R., Jang J.H., Huang H., Safavi-Sohi R., Haghjoo N., et al. Effect of cell sex on uptake of nanoparticles: the overlooked factor at the nanobio interface. ACS Nano. 2018;12:2253–2266. doi: 10.1021/acsnano.7b06212. [DOI] [PubMed] [Google Scholar]
- Serpooshan V., Zhao M., Metzler S.A., Wei K., Shah P.B., Wang A., Mahmoudi M., Malkovskiy A.V., Rajadas J., Butte M.J., et al. The effect of bioengineered acellular collagen patch on cardiac remodeling and ventricular function post myocardial infarction. Biomaterials. 2013;34:9048–9055. doi: 10.1016/j.biomaterials.2013.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharifi S., Behzadi S., Laurent S., Forrest M.L., Stroeve P., Mahmoudi M. Toxicity of nanomaterials. Chem. Soc. Rev. 2012;41:2323–2343. doi: 10.1039/c1cs15188f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirahama H., Lee B.H., Tan L.P., Cho N.-J. Precise tuning of facile one-pot gelatin methacryloyl (GelMA) synthesis. Sci. Rep. 2016;6:31036. doi: 10.1038/srep31036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shokouhimehr M., Theus A.S., Kamalakar A., Ning L., Cao C., Tomov M.L., Kaiser J.M., Goudy S., Willett N.J., Jang H.W., et al. 3D bioprinted bacteriostatic hyperelastic bone scaffold for damage-specific bone regeneration. Polymers. 2021;13 doi: 10.3390/polym13071099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siebert L., Luna-Cerón E., García-Rivera L.E., Oh J., Jang J., Rosas-Gómez D.A., Pérez-Gómez M.D., Maschkowitz G., Fickenscher H., Oceguera-Cuevas D., et al. Light-controlled growth factors release on tetrapodal ZnO-incorporated 3D-printed hydrogels for developing smart wound scaffold. Adv. Funct. Mater. 2021;31:2007555. doi: 10.1002/adfm.202007555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh S., Choudhury D., Yu F., Mironov V., Naing M.W. In situ bioprinting - bioprinting from benchside to bedside? Acta Biomater. 2020;101:14–25. doi: 10.1016/j.actbio.2019.08.045. [DOI] [PubMed] [Google Scholar]
- Speert D.P. Antimicrobial resistance: implications for therapy of infections with common childhood pathogens. Can. J. Infect Dis. 1996;7:169–173. doi: 10.1155/1996/431214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor E.N., Kummer K.M., Durmus N.G., Leuba K., Tarquinio K.M., Webster T.J. Superparamagnetic iron oxide nanoparticles (SPION) for the treatment of antibiotic-resistant biofilms. Small. 2012;8:3016–3027. doi: 10.1002/smll.201200575. [DOI] [PubMed] [Google Scholar]
- Tomov M.L., Gil C.J., Cetnar A., Theus A.S., Lima B.J., Nish J.E., Bauser-Heaton H.D., Serpooshan V. Engineering functional cardiac tissues for regenerative medicine applications. Curr. Cardiol. Rep. 2019;21:105. doi: 10.1007/s11886-019-1178-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomov M.L., Theus A., Sarasani R., Chen H., Serpooshan V. In: Cardiovascular Regenerative Medicine: Tissue Engineering and Clinical Applications. Serpooshan V., Wu S.M., editors. Springer International Publishing; 2019. 3D bioprinting of cardiovascular tissue constructs: cardiac bioinks; pp. 63–77. [DOI] [Google Scholar]
- Tomov M.L., Vargas M., Gil C.J., Theus A.S., Cetnar A.C., Pham Do K., Veneziano R., Serpooshan V. In: Nanomedicine for Ischemic Cardiomyopathy. Mahmoudi M., editor. Academic Press; 2020. Chapter 12 - nano-bioink solutions for cardiac tissue bioprinting; pp. 171–185. [DOI] [Google Scholar]
- Tran N., Mir A., Mallik D., Sinha A., Nayar S., Webster T.J. Bactericidal effect of iron oxide nanoparticles on Staphylococcus aureus. Int. J. Nanomed. 2010;5:277. doi: 10.2147/ijn.s9220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uskokovic V. Elsevier; 2017. Nanotechnologies in Preventive and Regenerative Medicine: An Emerging Big Picture. [Google Scholar]
- Vigata M., O'Connell C.D., Cometta S., Hutmacher D.W., Meinert C., Bock N. Gelatin methacryloyl hydrogels for the localized delivery of cefazolin. Polymers. 2021;13 doi: 10.3390/polym13223960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Hu C., Shao L. The antimicrobial activity of nanoparticles: present situation and prospects for the future. Int. J. Nanomed. 2017;12:1227–1249. doi: 10.2147/IJN.S121956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei H., Hu Y., Wang J., Gao X., Qian X., Tang M. Superparamagnetic iron oxide nanoparticles: cytotoxicity, metabolism, and cellular behavior in biomedicine applications. Int. J. Nanomed. 2021;16:6097–6113. doi: 10.2147/IJN.S321984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei K., Serpooshan V., Hurtado C., Diez-Cunado M., Zhao M., Maruyama S., Zhu W., Fajardo G., Noseda M., Nakamura K., et al. Epicardial FSTL1 reconstitution regenerates the adult mammalian heart. Nature. 2015;525:479–485. doi: 10.1038/nature15372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue K., Trujillo-de Santiago G., Alvarez M.M., Tamayol A., Annabi N., Khademhosseini A. Elsevier; 2015. Synthesis, Properties, and Biomedical Applications of Gelatin Methacryloyl (GelMA) Hydrogels. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerli W., Trampuz A. In: Biomaterials Associated Infection: Immunological Aspects and Antimicrobial Strategies. Moriarty T.F., Zaat S.A.J., Busscher H.J., editors. Springer New York; 2013. Biomaterial-associated infection: a perspective from the clinic; pp. 3–24. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
All data reported in this work will be shared by the lead contact upon reasonable request.
-
•
This work has no original code and custom computer code.
-
•
Any additional information required to reanalyze the data reported in this work is available from the Lead contact upon request.