Summary
Circulating tumor cells (CTCs) are tumor cells that shed from the primary tumor and intravasate into the peripheral blood circulation system responsible for metastasis. Sensitive detection of CTCs from clinical samples can serve as an effective tool in cancer diagnosis and prognosis through liquid biopsy. Current CTC detection technologies mainly reply on the biomarker-mediated platforms including magnetic beads, microfluidic chips or size-sensitive microfiltration which can compromise detection sensitivity due to tumor heterogeneity. A more sensitive, biomarker independent CTCs isolation technique has been recently developed with the surface-charged superparamagnetic nanoprobe capable of different EMT subpopulation CTC capture from 1 mL clinical blood. In this review, this new strategy is compared with the conventional techniques on biomarker specificity, impact of protein corona, effect of glycolysis on cell surface charge, and accurate CTC identification. Correlations between CTC enumeration and molecular profiling in clinical blood and cancer prognosis are provided for clinical cancer management.
Keywords: Liquid biopsy, Circulating tumor cells, Glycolysis, Surface charge, Nanoprobe, Biomarker
Introduction
Cancer is one of the terminal diseases that threaten human lives. According to the statistics from International Agency for Research on Cancer (IARC), there were 19.29 million new cancer cases and 9.96 million cancer deaths worldwide in 2020.1 Clinical reports have revealed that cancer metastasis is the primary cause of mortality and accounts for more than 90% cancer-related deaths.2,3 Since vast majority of cancer cases are found at an advanced stage, with metastasis at their first diagnosis, the current cancer managements face serious challenges despite of the advanced medical diagnosis and therapeutics, including surgery, chemotherapy, radiotherapy, immunotherapy, cellular therapy, proton-heavy ion therapy and other physical methods. If cancer can be detected at early stage, combined with appropriate therapeutics, the chance of cure will be significantly increased. In cancer management, the evaluation of treatment effect, post-treatment monitoring, and early warning of the risk of tumor recurrence and metastasis are critically important in cancer prevention, detection, and treatment. Therefore, different techniques on early detection of tumor biomarkers from various clinical samples have been developed and extensively researched. However, due to the complex nature of tumorigenesis and heterogeneity of the tumors, there have not been exclusive biomarkers with acceptable specificity for early cancer diagnosis. Although some serum markers, such as CEA, PSA, AFP, CA-199, etc. are currently applied as tumor screening markers in routine physical examination, none is guaranteed with absolute tumor specificity.4,5 To address this critical issue, the alternative methods for tumor liquid biopsy are emerging with great successes and promises.6
Circulating tumor cells (CTCs) are tumor cells that shed from the primary tumor or metastasis loci and intravasate into the peripheral blood circulation system spontaneously or even possibly resulting from surgical operations. Since its first discovery in 1869, critical and versatile roles of CTCs have been increasingly recognized in cancer biology, molecular profiling, and precision medicine via tumor liquid biopsy.7,8 In 1993, Professor Klaus Pantel from University of Hamburg, Germany, proposed a new concept of prototype which is equivalent to the current definition of “tumor liquid biopsy.”9 This concept was demonstrated with a toolbox refilled with circulating tumor cells, circulating tumor DNA, exosomes, and other markers in patients’ blood based on which the cancer patients were distinguished from healthy individuals, and classified in the different disease stages.10 However, due to significant variations among different tumor types, there has not been a universal tumor liquid biopsy technique that is valid for all cancer diagnosis and prognosis. Nonetheless, compared to ctDNA and exosomes, cell is a more complete biological entity that can provide dynamic information of proteins, DNA, RNA, etc. They can also be cultured in vitro to build CTC lines for further studies on metastasis mechanisms, prevention, and intervention.11 To this end, an ideal technology must be sufficiently versatile to target a heterogeneous subpopulation of CTCs and enable cell-friendly release for further in-vitro culture and analysis. More important, targeting CTCs via different interactions between nanoprobes and tumor cells must curb the influence of serum protein in the physiological environment. CTC detection and identification are also complicated by its rarity (one CTC out of a billion cells), cancer heterogeneity, and dynamic changes in cell biological properties during the metastasis process. Clinically, molecular profiling based on CTC studies provides critical information for developing treatment strategies and monitoring metastasis and recurrence.
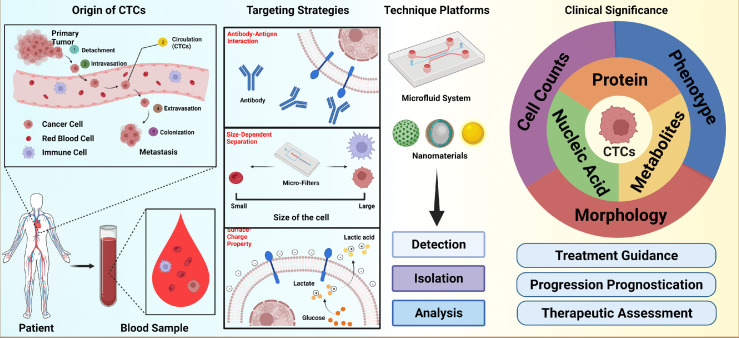
This review summarizes the most recent CTC research outcomes in both fundamental cancer biology and CTC detection technology. In particular, a new CTC isolation strategy based on the Warburg effect and cancer cell surface charge is introduced and compared with the conventional techniques that rely on immuno-affinity reaction and size dependent recognition. According to the Warburg effect, cancer cells do not produce energy via oxidative phosphorylation in the mitochondria, but through the process of aerobic glycolysis characterized by high level of glucose uptake and secretion of lactic acid. The cross-membrane movement of lactate results in removal of the labile inorganic cations, leaving a net of negative charges on cancer cell surfaces which distinguishes them from the normal blood cells except for red blood cells. The level of the negative change is dynamically regulated by glycolytic capacity. Taking advantage of this bio-electrical manifestation of the Warburg effect, various magnetic nanoprobes can be designed with surface positive charge enabling them to electrostatically bind onto the cancer cell surface for detection and magnetic separation. Compared to the conventional CTC isolation strategies, the charge-based approach does not require any biomarkers such as commonly used epithelial cell adhesion molecule (EpCAM), but much more sensitive due to strong electrostatic binding of the probes on cancer cells. Moreover, the glycolytic regulated surface charge is a hallmark characteristic of all cancer cells regardless of any phenotypic and genotypic differences. The biomarker independent cell targeting paves a new way to isolate a wide spectrum of CTC subpopulations for more accurate and sensitive cancer prognosis and feasible in vitro culture. This review places considerable emphasis on introduction to the charge-based CTC isolation strategy while giving historical assessment of previous achievements by technologies that rely on surface biomarkers and cellular morphologies (Figure 1).
Figure 1.
Schematic illustration of CTCs enrichment, analysis and clinical significance. Created with BioRender.com
Cancer biology fundamentals
Hanahan and Weinberg described the biological hallmarks of cancer that play the predominant roles in the processes of tumorigenesis, progression, and metastasis individually or synergistically.12,13 They showed that the biomarkers can be used to distinguish tumor cells from normal cells and attributed the origin of the biomarkers to carcinoma cells. Later, some of these biomarker characteristics were gradually recognized and utilized for identification of CTCs in blood samples. Numerous studies have shown that tumor cells are evolved from somatic cells, resulting from the synergistic effects of the accumulation of genetic damage, variations of genetic characteristics at the chromosomal level, specific environmental stimuli, and other factors. Among them, the classified proto-oncogenes and tumor suppressor genes play critical roles not only in regulating signal transduction pathways but also in reprogramming cellular metabolism.14 Moreover, human genes are complicated by variations of the physiological environments, thus typically characterized with heterogeneity, which is an intrinsic feature of tumors. As is well-known, differences at the genetic level can lead to differentiated expression of certain proteins in cells, and most solid tumors are of epithelial origin.15 During cancer development, cellular progression and metastasis are inherently linked to cellular communications and interactions. If the specific cancer cell surface molecules, such as protein and sugar molecules, are different from those on the normal cells, they can be used as biomarkers to identify CTCs.16
For detection of CTCs from the epithelial-derived tumors, cancer cell targeting through the differentiated EpCAM expression as the biomarkers has been extensively reported in the literature.16, 17 The CTC-based tumor metastasis is clinically defined with five important stages: 1) tumor cells detach from primary tumor tissue; 2) dissemination in extracellular microenvironment (ECM); 3) intravasation into the circulatory system, mostly referring to the blood circulation system; 4) extravasation from blood circulation system into the microenvironment of an organ, and 5) the cancer cells carry the genetic or molecular information of the primary tumor, regulate the environment by interacting with immune cells and extracellular matrix cells in the microenvironment to facilitate the colonization, invasion and proliferation for metastasis.18,19
Based on the breast cancer animal model, a proposed concept of “self-seeding tumor metastasis” that CTC metastasis in vivo is multidirectional and may move to the primary site.20 CTCs have been found to accelerate the growth, angiogenesis, and stromal recruitment of tumor of origin through tumor-derived cytokines such as IL6, IL8, and CXCL2. CTC enrichment is mainly executed in stages 3 of the metastasis. Studies have shown that Epithelial-Mesenchymal Transition (EMT) takes place during CTC detachment from primary or metastatic loci, which can promote tumor cell migration in the circulatory system, downregulate the expression of cell surface adhesive proteins, upregulate some mesenchymal markers, and provide CTCs with stem cell like biological features.21 Accordingly, a quantifiable, dual-colorimetric RNA–in situ hybridization (ISH) assay was established for a phenotype analysis of CTCs isolated from lung cancer patients’ clinical blood samples, in which three sub-phenotypes of CTCs were identified, exclusively epithelial, intermediate, and mesenchymal.22 However, EpCAM dependent CTC detection is usually found to have missed the partial CTC stem-cell like phenotypes, which compromises capture and recognition sensitivity.23 In CellSearch system, 7.5 mL blood is needed and the cut-off value for cancer prognosis is 2 CTCs per 7.5 mL sample. Besides the epithelial cell markers, there are some other cell membrane protein biomarkers for the specific tumors, but their clinical applicability still needs to be evaluated through clinical trials. Considering the limitations of protein biomarker-based strategy, researchers started looking for new CTCs detection strategy with higher sensitivity.24 Biophysical characteristics of tumor cell are independent of the surface protein markers during tumor development. The most direct biophysical feature is cell morphology, including cell size, nucleus-cytoplasmic ratio, cytoskeleton, and cell membrane plasticity.
Dr. Otto Heinrich Warburg first discovered that solid tumor tissue was a relatively hypoxic environment, in which tumor cells preferred to metabolize glucose through an anaerobic glycolysis consisting of high level of glucose uptake followed by secretion of lactic acid, even under the hypoxic circumstance.25 This tumor cell abnormal metabolic behavior is distinctively different from that of the normal cells which mainly undergo aerobic respiration. According to the “Warburg Effect,” tumor cells produce large amounts of lactate through aerobic glycolysis, which pass through the cellular membrane via the mono-carboxylate transporter protein family (MCT-1 to -4) on the membrane in a perpetual fashion.26,27 The continuous secretion of lactate through cellular membrane disrupts the equilibrium of membrane potential by taking away cations such as H+, Na+, K+ resulting in a net of negative surface charge. As the charge is a direct consequential effect of lactic secretion, its level is proportional to the rate of glycolysis. This relation was established in our previous work where the cancer cell surface charge was correlated to the glucose level and monitored by a positively charged magnetic nanoprobe.28 Due to electrostatic force, the positively-charged nanoprobes were found to strongly bind onto the negatively-charged cancer cells of all twenty-two different types.28 These nanoprobe-captured cancer cells were then separated magnetically and counted as cell capture efficiency.28 In addition, the cell capture efficiency was found to depend on the glucose level provided, reflected on the one to one relationship between glycolysis rate and cell surface charge. The lactate secreted from the cells was varied by reducing the supply of glucose in the culture media of the cancer cells for 48 h and well correlated to the negative surface charge via cell capture efficiency. For instance, significant drop (17%) in the K562 cancer cell capture efficiency was found when decreasing the glucose concentration from 10 to 0 mM. This correlation provides the experimental evidence that the cancer cell surface negative charge is a bio-electrical manifestation of the Warburg effect.
More profoundly, this glycolysis-driven negative change on cell surfaces was observed on twenty-two different cancer cell lines. Chen et al. reported the percentage of cancer cells captured by applying both positively and negatively charged nanoprobes (NPs) respectively.28 The cancer cells are mixed with the fluorescence labelled positively- or negatively- charged magnetic nanoprobes. In that case, HeLa cells were bound with numerous positive NPs due to the charge sign difference, while no significant binding was observed for the negative NPs, clearly indicating the cancer cell surfaces are negatively charged. It was found that although the binding efficiencies vary, all twenty-two different cancer cells were electrostatically bound to the positive NPs, and none is captured by the negative NPs, while 4 normal cells were left intact by either positive or negative NPs. Furthermore, positive NP cancer cell capture can also be reduced by inhibiting aerobic glycolysis and lactate production by 3-bromopyruvate (3BP) or Dichloroacetate acid (DCA), both are inhibitors of glycolysis. Those experimental results are consistent with the glycolytic-driven negative change on cancer cell surfaces.
Method of CTC recognition and impact of protein corona
There are several major approaches for CTC enrichment, which can be classified as, namely, magnetic beads platform, microfluidic chips systems, and hybrid systems which are designed and developed based on the different CTC capturing strategies. They can also be classified into positive, negative, and combined separation strategy according to the targeting cell types. Table 1 provides the main characteristics of different cell targeting strategies and representative CTC isolation methods from the spiked or clinical blood samples of cancer patients. Compared to the enumeration results of CTCs from the clinical blood samples, the isolation efficiency of the cancer cell line spiked into the donor's blood sample is more reasonably determined. However, reliability of the cell line as surrogate of the CTC still needs to be verified by further investigations. The advantages and shortages of various platforms are also listed in Table 1. Besides capture efficiency, purity, sample volume, and identification of the epithelial-to-mesenchymal transition (EMT) are key factors for evaluating cancer metastasis and recurrence.
Table 1.
Comparison of different CTC isolation strategies.
| Targeting Strategy | Isolation System Descriptions | Sample Type and Volume | CTCs capture number or Efficiency | Patients diagnosis accuracy | Capture Purity | EMT Identification | Ref. | |
|---|---|---|---|---|---|---|---|---|
| Immuno-affinity strategy | Positive selection | CTC-chip, a microfluidic platform with antibody (EpCAM)-coated microposts under precisely controlled laminar flow conditions | Whole blood from patients; Average 2.7 ml per sample (range, 0.9 to 5.1 ml) |
A range of 5–1281 CTCs per mL |
≥5 CTCs/mL were identified in 115 of 116 (>99%) patient samples | Approximately 50% | No | 29 |
| HBGO chip, a graphene oxide sheets functionalized with high-density anti-EpCAM antibodies and herringbone structures | Cancer cell spiked PBS buffer solution (50–200 cells/mL); 2 × 107 MCF-7 cells injected canine models; 5 mL |
≥ 90% at a flow rate of 1 mL/h in Cancer cell spiked PBS buffer solution; 212 MCF7 cells were isolated in ex vivo capture; 762 MCF7 cells were isolated in in vivo capture; |
- | - | No | 30 | ||
| Peptide H13 (HER2-positive CTCs targeting)-functionalized nanomaterials | Cancer cell spiked Whole blood Sample (50 cells/mL); 1 mL |
68.56% for SKBR3 cell; 20.04% for MCF7 cell; 79.26% for SKOV3 cell |
- | - | No | 31 | ||
| A controlled protein premodified immunomagnetic beads (IMBs) | MCF-7 cancer cell (2.5 × 104 cells) and J774A.1 mouse monocyte macrophage cells (2.5 × 106 cells) spiked 100% FBS; 0.5 mL |
94.00% in cell spiked FBS at 200 μg/mL; 84.0−90.0% MCF-7 cells from mimic blood samples contained 25−55 MCF-7 cells |
- | 85.4% in cell spiked FBS at 200 μg/mL; 99.996% leukocytes were depleted |
No | 32 | ||
| The rVAR2 protein conjugated streptavidin coated magnetic CELLection™, Biotin Binder Dynabeads® | Cancer cell spiked blood (Red blood cell lysed before isolation, 3-500 cells in 5 mL); Blood sample from patients (Red blood cell lysed before isolation); 5 mL |
Average recovery was 60.0% - 91.9% depends on the number of PDAC cells spiked in spiked samples; 12-23 cells were captured from 4 prostate patient samples |
CTCs detected from 100% (4/4) of patients | Contaminating PBMCs was 82% less compared to the EpCAM-based capture strategy | Yes | 33 | ||
| Negative selection | Human CD45 Depletion Kit (immunomagnetic nanobeads) |
SW620 cell spiked healthy blood samples (100 cells in 5 mL); Red blood cell lysis blood samples from patients 5mL for spiked blood samples; 20 mL for patients' blood samples |
Average 58% (range 50-66%) for spiked blood samples; | CTCs detected from 56% (47/84) of peripheral samples | Total number of leukocytes: Before enrichment: 3 × 107; After enrichment: 6.0 × 103. | Yes | 34 | |
| Biophysical feature strategy | Size dependent separation | Parsortix platform, utilizes a microfluidic cassette, which can separate cells based on size and deformability characteristics | HNSCC cells spiked whole blood samples (9–129 cells/mL); whole blood samples from HNSCC patients; 9 mL | Cell capture rate of 53.5% for unfixed cells;Cell capture rate of 65.3%, 73%, 73.1% at time points 0h, 24h, 72h using Transfix-collected samples; Cell capture rate of 51%, 32.3%, 6.0% at time points 0h, 24h, 72h using EDTA samples | CTCs detected from 50% (2/4) of patients | - | Yes | 35 |
| Optimized the Parsortix system | Cancer cell spiked whole blood samples (25, 50, 100 cells/ mL); whole blood from prostate patients 7.5 mL |
Capture rate of 25, 50, 100 PC3 cells spiked blood sample was 44.0% ± 4.0%, 48.0% ± 6.9%, 60.8% ± 5.2%; Capture rate of 25, 50 DU145 cells spiked blood sample was 57.3% ± 8.3%, 56.0% ± 8.7%; Capture rate of 25, 50 MCF-7 cells spiked blood sample was 54.7% ± 6.1%, 58.7% ± 13.3% |
More than five CTCs in 90% (9/10) | 3.1% ± 2.7% | Yes | 36 | ||
| Microfluidic devices with multiple arrays of crescent-shaped isolation wells in the microchannel | MCF-7, MDA-MB-231, HT-29 spiked PBS (1-3 cells/mL); 1 mL |
80% | - | - | Yes | 37 | ||
| Isolation by size of Tumor cells (ISET) filtration | Diagnostic leukapheresis (DLA) samples contain CTCs and mononuclear cell populations; 10 mL |
3.8 CTC/mL (IQR = 1.3–4.0) | CTCs detected from 88% (14/16) of patients | - | Yes | 38 | ||
| Biophysical feature strategy | Surface charge based targeting | Positively charged Fe3O4 Nanoparticle | Red blood cell lysis blood samples from patients; 1 mL |
2-8 CTCs cells/mL | CTCs detected from 100% (25/25) of patients | - | Yes | 39 |
| Others | Inertial microfluidic Labyrinth device | Cancer cell and WBC spiked PBS (each 1000 cells/mL); Whole blood samples from patients; 5 mL |
82% ± 5% of H1650 cell cancer cells from spiked PBS; Average of 417 CTC/mL (10.2–5068) from patient |
CTCs detected from 100% (25/25) of patients | 78% ± 18% of WBCs were removed from spiked PBS | Yes | 40 | |
| ApoStream, a dielectrophoretic device separate cells by dielectric property of the cells | SKOV3, MDA-MB-231 spiked peripheral blood mononuclear cells (PBMCs); 1 mL PBMCs (isolated from 7.5 mL whole blood samples) |
75.4 ± 63.1% SKOV3 cells (5000 cell spiked); 71.2 ± 61.6% MDA-MB-231 cells (500 cell spiked); Average 68.3% at low number of cancer cell spiked sample (4-23 cells spiked) |
- | 99.33% ± 0.56% of PBMCs were removed | Yes | [41 | ||
| Combined Strategy | 3D printed monolithic device with anti-CD45 anti- body functionalized surface to remove leukocytes followed by a commercial membrane filter | CD45+ Jurkat Clone E61 cell lines spiked PBS (7 × 106 cells/mL); Cancer cells spiked blood samples (3 × 103∼4 × 103/mL); whole blood samples from patients. 10 mL for patient blood samples |
∼90% tumor cell recovery rate for Cancer cells spiked blood samples | - | - | Yes | 42 | |
| Microfluidic chips with ISET filtration combined aptamer modified microbeads and Anti-CD45 modified nanoparticle | Cancer cell spiked human whole blood samples (10, 20, 50, 100 cells/mL); whole blood samples from patients. 1 mL for cancer cell spiked sample; 2 mL for whole blood sample from patients |
91% for cancer cells spiked samples; 0–35 CTCs can be detected from patient blood samples |
CTCs detected from 83% (5/6) of patients | Less than 0.01% of WBCs are detained | Yes | 43 | ||
| Combined Strategy | CTC-iChip, an integrated platform combined deterministic lateral displacement, inertial focusing and Magnetophoresis, for EpCAM positive cell line, anti-EpCAM antibody was functionalized on the magnetic beads, for negative depletion, anti-CD45 and anti-CD15 functionalized beads were used | Cancer cell spiked Whole blood and whole blood from patients Average 10 ml (range, 6 to 12 ml) |
98.6 ± 4.3% for SKBR3 human breast cancer cells; 89.7 ± 4.5% for PC3-9 human prostate cancer cells; 77.8 ± 7.8% for MDA-MB-231 “triple-negative” mesenchymal breast cancer cell line |
≥0.5 CTC/mL in 37 of 41 (90%) prostate cancer patients | Removed 96.7 ± 1.9% for MCF10A cell lines; 97.0 ± 1.7% for the MCF10A-LBX1 derivatives |
Yes | 44 | |
| A biomimetic microfluidic system combined with leukocyte membrane and SYL3C aptamer coated biomimetic nanoparticles | MCF-7 cells and J774A.1 cells spiked PBS (Ratio 1:107);cancer cell spiked blood sample from mice (104 cells); 20 μL |
Above 91% MCF-7 cells could be detected in PBS and blood sample; | - | Almost no leukocyte nonspecific adsorption | No for SYL3C aptamer | 45 | ||
| EpCAM-targeted nanoparticles combined with microfluidic devices contains X-shaped velocity valleys | Cancer cell spiked blood samples and blood samples from patients (Red blood cell lysed after isolation); 2 mL |
Near 100% when using 200 target VCap cells in spiked sample | CTCs detected from 100% (21/21) of patients | No more than 200 white blood cells were typically observed from patient samples |
Report on the status of EMT | 46 | ||
| Microchip-based immunomagnetic separation combined microchip and magnetic nanoparticles functionalized EpCAM antibody | Cancer cell spiked blood samples (5-2000 cells/mL, prepared in CellSave™ tubes); 2.5 mL |
Cancer cell capture rates are 90% for COLO205; 86% for SKBR3 | - | - | No | 47 | ||
| Combined Strategy | LPCTC-iChip combined inertial separation array devices (RBCs and platelets removal) and magnetic beads (WBCs depletion) | Cancer cell spiked leukapheresis products and leukapheresis-mimic samples (5000 and 1000 cancer cell spiked, respectively); 65 mL |
Recovered 89.2 ± 5.7% CTCs from spiked samples | - | Removed 99.96% WBCs, 99.998%RBCs, and 99.998% platelets from spiked samples | Yes | 48 | |
| SDI-Chip with immunocoated micropillar surfaces combined EpCAM antibody selection and size dependent selection | Cancer and control cell spiked PBS and blood samples (cancer cell from 10 cells/mL to 1 × 106 cells/mL, control cell, 1 × 106 cells/mL); Blood sample from Colorectal Cancer patient; 1 mL |
99.2 ± 3.5% in spiked PBS buffer 92.2 ± 6.4% in spiked blood CTCs detected in a range of 8–161 CTCs/mL |
CTCs detected from 100% (15/15) of patients | 92.4 ± 4.6% in spiked PBS buffer 82.3 ± 3.8% in spiked blood |
No | 49 | ||
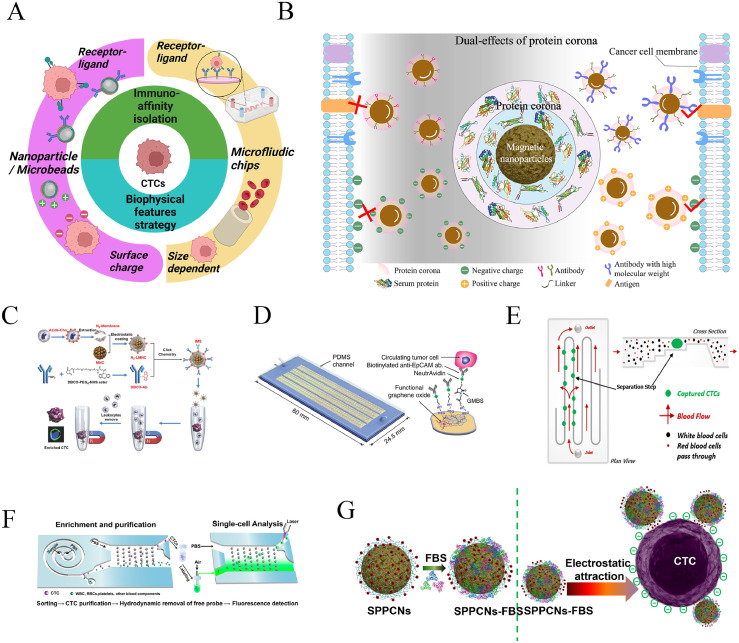
As shown in Figure 2A, CTC recognition can be divided into two categories of immuno-affinity strategy and biophysical factors which respective underlie the magnetic beads and microfluidic chip methods.50 When in contact with the clinical samples, the magnetic nanoparticles with either conjugated ligands or electrically-charged are inevitably modified to exhibit new surface morphologies and bio entities due to formation of protein corona. In this fashion, the newly grafted surface corona structures will significantly render the biophysiochemical properties of the original nanoparticles at the nano-bio interfaces and in turn affect cell targeting. It has been reported by our and other groups that the protein corona on the surface of functionalized nanoparticles plays a dual-effects on the targeting ability of the original nanoparticles, which is determined by the physiochemical features of the nanoparticles to large extent. The dual-effects of electrostatic interaction and immuno-affinity on the CTC targeting ability by the protein corona decorated nanoparticles are shown in Figure 2B. The characteristics of these targeting methodologies and the influence of protein corona on CTC targeting are described in the following sections.
Figure 2.
(A) Schematic diagram showing the main targeting strategies of CTCs isolation based on either nanoparticles or microfluidic chip platforms. Created with BioRender.com (B) Scheme showing the dual-effects of the formed protein corona on the binding ability of cancer cells by both immune-affinity and electrostatic interaction mediated magnetic nanoparticles, which can be tuned by the physiochemical feature of the functionalized nanoparticles. (C) Schematic illustration of construction of IMS and the procedure of CTC enrichment. Reprinted with permission from Ref.51 (D) Schematic of the HBGO chip and the conjugation chemistry between functional graphene oxide and anti-EpCAM antibody. Reprinted with permission from Ref.30 (E) Scheme of isolation principle inside the cassette, in which blood is forced along a series of channels and to flow through a 10 μm patented step which separates particles on the basis of size and compressibility. Reprinted with permission from Ref.36 (F) Schematic of CTC sorting microchip with rapid CTC separation relying on inherent centrifugal force. Reprinted with permission from Ref.52 (G) Schematic of electrostatic interaction mediated cancer cell binding by the FBS corona decorated positively-charged magnetic nanoparticles. Reprinted with permission from Ref.39
Receptor-ligand interactions
The CTCs antigen-antibody recognition analysis system employs EpCAM as the hallmark. To overcome the limited sensitivity for CTC capture due to EMT, several cell membrane protein markers have been developed that are independent of EMT. A new biomarker VAR2CSA malaria protein was found to be effective for capturing tumor cells which otherwise would have been missed by the EpCAM dependent methods.33 CD44 exon6 has also been used to recognize the phenotypes of cells that do not express EpCAM antigens.53 The other candidate, aptamer, with specific design produced with help of SELEX technology, can be used separately to target specific CTC phenotype or combined for targeting different phenotypes of CTCs with improved specificity.54,55 In some cases, peptide was developed to identify CTCs as well.31 Both the magnetic bead and microfluidic chip platforms can be effectively integrated into the aforementioned receptor-ligand recognition approach.56 In the magnetic bead system, the ligand can be directly conjugated onto the nanoparticle surface by chemical coupling, enabling cell binding for magnetic enrichment.
It should be noted that positive enrichment strategy aims at targeting tumor cells via surface-conjugated magnetic nanoparticles, while the negative enrichment removes other interference cells that are attached by the conjugated ligand.34 For example, anti-CD45 antibody-coupled magnetic beads are used to bind and remove leukocytes from the red blood cell in the lysed blood samples, recovering the tumor cells in the supernatant. The advantages of the magnetic bead systems include facile scaled-up synthesis, easy surface modification, available characterization, and acceptable cost. The main challenge is the difficulty in purifying the tumor cell from leukocytes, resulting in a lower purity of the isolated CTCs, except for the magnetic nanoprobes with pre-treatment of surface property by serum protein or cellular membrane.32,51 In that study, a biomimetic immuno-magnetosome (IMS) is prepared by coating a leukocyte membrane (decorated with anti-epithelial cell-adhesion molecule antibody) on a magnetic nanocluster, enable superior CTC recognition efficiency and undetectable leukocyte background (Figure 2C). However, multi-step preparation pathway may increase the processing complexity and compromise the advantage of the low cost of magnetic beads.
Besides magnetic beads, microfluidic chips have recently attracted extensive interests in association with the concept of “Lab on a chip”, which has been elucidated in some review articles.57,58 With this system, the whole blood sample flows through the channels of the microfluidic chip at a moderate speed controlled by microvalves and micropumps. In this process, only CTCs are captured by binding onto the ligands-grafted surface of the protruding side with other background cells rinsing away. One of the well-known examples of microfluidic chips 'for CTC isolation based on the EpCAM biomarker strategy is the CTC-chip developed by Haber's group at Harvard University in 2007. They reported a broad range of enumerated single CTCs isolated from 1 mL blood samples by the anti-EpCAM antibody-grafted micropillars of the chip from cancer patients’ peripheral blood sample, indicating good sensitivity of the method and reflection of heterogeneity of cancer progression.29 To overcome the shortcoming of limited accessible blood sample volume during the in-vitro CTC detection, a functional graphene oxide sheets decorated microfluidic system has been designed to continuously collect intravascular CTCs directly from a peripheral vein also based on the anti-EpCAM antibody targeting moiety (as shown in Figure 2D).30 Section shape and pattern of the micropillar of the chip influence the flow dynamics and biomolecules reaction dynamics. How to balance between the processing efficiency and ligand-receptor reacting efficiency is one of the key issues to be addressed. To this end, Chaoyong Yang's group at Xiamen University developed a tilted triangular micropillars array, which can change the flow pathway of both CTCs and normal blood cells to increase the collisions of the CTCs with ligands and reduce blocking of blood cells. As a result, capture efficiency, sensitivity, and selectivity of CTCs were considerably improved.49,59 Based on the foregoing, one can see various merits from the magnetic beads and microfluidic chips in the CTC isolation applications. The advantages of the microfluidic chip over the magnetic bead system lie on the enhanced purity of the captured CTCs, automated operation process, and the ability of dealing with whole blood sample directly.45 Despite of these advancements, overall efficiency of the microfluidic chip in processing liquid samples is still low, which remains to be improved for commercialized applications.60 Furthermore, although microfluidic chips can be designed with numerous of micro patterns for cell interaction, the interfacial efficacy is not as good as nanomaterials. Therefore, the combination of magnetic beads and microfluidic chips may provide a better performance by taking advantage of the higher surface-to-volume ratio of the nanoparticles and improving purity of the microfluidic system.47
Size dependent separation
Microfiltration membrane is a representative technology, such as the CelSee System, that sets a certain pore size to allow normal blood cells, which are generally smaller in size, to pass through the membrane, while tumor cells, larger in size, are trapped and separated.61 CTC enrichment using size and cell deformability as a marker has been extensively studied for the microfluidic chips. For example, the Parsortix system (as shown in Figure 2E) is a microfluidic chip with specific micron-sized microfluidic channels, which allows the cells with small size pass but block the larger-sized cells like CTCs.36 Another effective platform to separate CTCs is the inertial flow microfluidic devices with a spiral microchannel. The cancer cells are collected at the outlet of the channel according to different inertial forces on the cells due to size differences (Figure 2F).44,52, 62, 63, 64 In addition, Prof. Chwee Teck Lim's group at the National University of Singapore designed a microfluidic chip with well-arranged micropillars inside the channel, with three to four micropillars as a group, allowing normal cells to leak through the gaps between the pillars while efficiently trapping tumor cells of larger sizes by a group of adjacent micropillars.37 Except the microfluidic chips, it may be difficult for the filtering membrane to release the trapped CTCs due to cell deformation and entrapment in the membrane pore. Some commercial systems, such as the CanPatrol system, using immuno-negative selection and size principle for CTC isolation have been validated for several different cancerous species for metastasis recurrence prediction. These systems can separate leukocytes from blood samples by CD45 antibody followed by removal of smaller size cells via nano-filtration membrane, and further classification of the isolated CTCs with different EMT phenotypes using immune-fluorescence staining.65
CTC isolation via cancer cell surface negative charges – a non-biomarker approach
Due to tumor cell heterogeneity, there have been well-known limitations in the CTC capture strategies using either biomolecular markers or size-dependence. A cell surface electrical charge-based approach has been recently developed for highly sensitive and efficient broad-spectrum CTCs isolation from clinical blood (requires only 1 mL clinical blood sample). Cancer cells exhibit the so-called “Warburg Effect” characterized by high glycolysis capacity. High levels of glucose uptake and lactate secretion are the two most distinguishable metabolic behaviors commonly observed from all cancer cells. The continuous secretion of lactate through cellular membrane disrupts the equilibrium of membrane potential by removal of the cell surface cations such as H+, Na+, K+ resulting in a net of negative surface charge. Metabolically, the cancer cell surface negative charge is dynamically regulated by glycolysis capacity and can be well controlled by glucose uptake. Sufficient glucose facilitates high level of glycolysis that results in greater negative surface charge. Chen et al showed a close correlation between the cell surface charge and the lactate secreted from the cells into the culture media by varying the glucose concentration.28 Based on this unique metabolic behavior of cancer cells, unique magnetic nanoprobes (MNPs) were developed for cancer cell detection, capture, magnetic separation, and isolation. These MNPs were designed to be charge variable, label-free, fluorescent, and superparamagnetic for electrostatically binding onto the negative cancer cells.28,39,66, 67, 68 Recent works have shown that CTCs can be readily captured by positive NPs from clinical blood samples with high sensitivity without any protein biomarkers.39,66,67
Charge-mediated CTC isolation (CMCTCI) has been widely applied in cell specific targeting.69, 70, 71, 72 CMCTCI from the pretreated blood sample by red blood cell lysing has been proven to be an efficient strategy through EpCAM-independent tumor cells recognition.39,73 The charged NPs provide new bio-electricity-based cell targeting and capturing that is not relying on any protein-based biomarkers whose specificity has been a universal issue. Since the glycolytic-regulated surface negative charge is the hallmark characteristic of all cancer cells regardless of genotypic differences, the charge-based targeting will be unique, exclusive, and highly specific only to those that exhibit significant glycolysis. Therefore, this unique approach will address the universal non-specificity issue of cancer biomarkers and allow for development of new assay methods for early detection of cancer cells in clinical blood.
The captured CTCs can be magnetically separated and enumerated by immuno-fluorescence aided identification. These identified CTCs can be used for cancer diagnosis or screening, real-time long-term disease monitoring and even therapy guidance. Moreover, molecular analyses of CTCs can provide new insights into the biology of metastasis with important implications for the clinical management of cancers.
CMCTCI is known for several unique characteristics and advantages: 1) The cancer cell surface negative charge is directly regulated by high glycolytic metabolism which is the bio-electrical manifestation of the Warburg effect; 2) The cancer surface charge, as a bio-electrical analyte, can be quantitatively utilized for detection and capture of cancer cells without need of expensive monoclonal antibody which is required in the magnetic bead system and microfluidic chip platform; 3) CTC capture via glycolytic-regulated surface charge can address the problems of subpopulations with different EMT phenotypes, sizes, and chromosome ploidy with much greater sensitivity in a clinical setting, and 4) Only 0.5∼1 mL of blood sample is needed to complete the CTC capture and enumeration. Preliminary experimental results have shown the feasibility of the charge-based CTC detection in clinical blood of colorectal cancer (CRC) patients. Moreover, the sensitivity of the charge-based CTC capture has been compared with a commercialized inertial flow instrument using the same clinical blood samples from patients with lung, gastric, breast cancers, and melanoma.39
Impact of protein corona
As has been reported, the nanoparticles are inevitably decorated with biological macromolecules when they are dispersed in the physiological media (for instance, human serum) and subsequently developing the so-called “protein corona”.74 The biological decoration of the nanoparticles renders their bio-chemical-physical properties, ultimately altering the functionalities and behaviors in cell targeting, systemic circulation, fibrillation, cellular uptake, and biocompatibility. The protein coronas structurally and morphologically transform nanoparticles beyond recognition as compared to the original as-synthesized counterpart. Consequently, the effects of bio-decoration on a variety of nanoparticle properties have been extensively studied in many biomedical applications, such as bio-detection, nano-therapeutics, tissue engineering, and regenerative medicine.75, 76, 77, 78, 79 For cancer cell targeting, the protein corona on the surface of nanoprobes poses a dual-effects (Figure 2B). However, the level of corona influence during CTC isolation can be adjusted by modifying the physiochemical properties of the nanoprobes. Professor Caruso's group at the University of Melbourne demonstrated the impact of serum proteins on the targeting efficacy of the antibody-grafted inorganic microspheres.80 It was found that in the presence of human serum albumin (HSA) alone, protein coronas exhibited a favorable effect onligand-receptor interaction, while in the condition of many types of serum proteins, the targeting ability was eliminated due to alteration of the nanoprobe surface properties. In contrary, in the case of small molecular weight targeting ligands like transferrin or nanobody, the negative influence of the protein corona on the targeting ability is amplified to some degree (Figure 2B).81,82
Since most of the serum protein molecules display a negative potential, they tend to bind onto the positively-charged nanoparticles by forming a protein corona. This is a critical issue in charge-based CTC detection and isolation as the protein corona can significantly alter the overall nanoprobe charge conditions. We investigated the effects of hard and soft protein corona formed on the positively-charged (MNCs⊕) and the negatively-charged magnetic (MNCs⊖) nanoprobes in the PBS solution with different FBS concentrations and its influence on the capture efficiency of HeLa cells.68 At a moderate FBS concentration, Zeta potential of MNCs⊕ was rapidly reversed from +32 mV to -14 mV, and further changed to -15 mV after incubating in pure FBS solution. Interestingly, the FBS-MNCs⊕ complex with the reversed Zeta potential still kept nearly 78% HeLa cell capture efficacy, while both FBS-MNCs⊖ and MNCs⊖ showed negligible cancer cell binding efficiency (Figure 2B). With LC-MS analysis, different types of proteins were found to be absorbed on the positively- and negatively-charged magnetic nanoparticles with equal amounts of adsorptions.
However, when the Zeta potential of the FBS-MNCs⊕ complex was further dropped to -49.3 mV by introduction of polystyrene sulfonic (PSS), the HeLa cell capture efficiency dropped to the same level by the original negatively-charged counterpart.39 Those experimental results indicate the strong electrostatic interactions between the tumor cells and the positively charged particles (MNCs⊕) even with the particle surface-decorated corona. It is believed that the corona structure may not completely cover the surface of a positively-charged nanoparticle. Therefore, regions on the particle surface may locally appear positive resulting in considerable cell binding as shown in Figure 2G. Structurally, the conformation of the protein corona on the surface of MNCs⊕ may form islands rather than continuous layer.68 Commonly, in the aqueous dispersion of charged nanoparticles, there are the stern layer and the diffusion layer according the “Stern double layer” theory. The Zeta potential of nanoparticles measured by Dynamic Light Scattering (DLS) is calculated from the potential on the surface of shear. Therefore, the Zeta potential value is not equivalent to the available charge on the nanoparticle surface after protein adsorption, which is consistent with the conclusion in literature.83
Identification of the isolated CTCs
Analysis of CTCs in clinical blood samples includes two essential steps, isolation and identification. Most of the CTC nanotechnologies focused on developing methods for efficient isolation of single CTCs and CTC clusters (e.g. CTC-Neutrophil clusters). Immunofluorescence staining has been widely applied to determine the types of the isolated cells using multichannel fluorescence staining of cytokeratin CK, nuclear DAPI, and leukocyte CD45 markers. Usually, the captured cells with the CK and DAPI positive and CD45 negative fluorescence signals are recognized as CTCs.29 This method is quite repeatable, but the cells have to be fixed and mounted, a process that reduces the cell activity. Meanwhile, specificity of CK as a positive marker for CTC identification cannot be fully guaranteed.84 Therefore, staining for EMT-related protein markers has been added to improve the accuracy and reliability in CTCs identification, such as EpCAM, Vimentin, etc. Typically, normal somatic cells are diploid, and more triploid karyotypes are present in all malignant tumor types.85 In fluorescent photography, the cells with no less than three bright spots in the cell nucleus are considered as the cancer cells. This approach has been developed as an important tool known as RNA fluorescence in situ hybridization (FISH). Combined with immunofluorescence staining, iFISH is an effective method to increase the accuracy of CTC identification.86
For clinical applications, there are still some shortcomings with the traditional immunofluorescence staining and iFISH: 1) staining has certain requirements for the negative and positive control samples and staining techniques, 2) significant effort and time is required in searching for the positive cells via microscopy, but the results could be influenced by human errors, and 3) staining reagents for multiple protein or RNA marker are relatively expensive. It is, therefore, important to develop novel strategies to combine CTCs isolation and identification with high specificity and sensitivity. To this end, some techniques have been emerging87, 88, 89: 1) developing the integrated system for CTC capture, separation, in situ fluorescence detection, and ex-vivo expansion based on microfluidic chips, 2) using fluorescence loaded MOF materials for fluorescent molecule release detection and fluorescence imaging of captured CTCs, and 3) entrapping a single cell within micron-sized droplets and labelling the unique uptake or metabolic substance with fluorescent signals. In addition, based on the traditional immunofluorescence staining process, replacing manual reading with automated reading equipment has been shown to have practical advantages.
Several novel systems for CTC isolation, capture and identification have been developed and technically available.7,90,91 The ultimate goal is to develop a CTC-based liquid biopsy in clinical setting for prognosis. Despite the rapid progress in this field, however, there is still a gap between nanoscience and clinical practice due to the interdisciplinary boundaries.92 More collaborations are therefore needed between physical and medical sciences in jointly developing the tumor liquid biopsy. Precise information on enumeration and molecular profiling of the captured CTCs will be critical for determining a specific clinical stage of the cancer patient. The CTC numbers must be well correlated with the clinicopathological results for assessment of cancer recurrence and metastasis. The CTC data must also be acquired from a large number of clinical samples in predicting the overall survival and progression-free survival of patients. As approved by Food and Drug Administration (FDA), CellSearch has been the globally applied CTC detection system for prognostic monitoring of metastatic breast, colorectal and prostate cancers.93,94
On the other hand, molecular profiling of the CTCs can provide more significant insights into the tumor progression and metastasis.95 Correlation of CTCs biology with clinical implications is needed on the basis of whole subpopulation CTC isolation and appropriate analyzing packages. Although single cell RNA sequencing technique has been proved to be effective, its accessibility and accuracy will have to be resolved.44 Upon isolation, rapid CTC proliferation in vitro will enable more accurate sequencing, drug screening, and construction of CTC-based PDX model for drug resistance study.96,97 Currently, the CTC-friendly release is another important issue for the downstream molecular analysis and in-vitro proliferation.98
Cellular viability of isolated CTCs
Effect of geometry of nanoprobes on the cellular viability
In our previous work, we studied the electrostatic interactions between cells and the nanoprobes of different geometries. Cytotoxicity of the nanoprobes was investigated on cell viability, collapse of cell membrane, and destruction of the cell pseudopod. Considering the spherical geometry of the nanoparticles (∼100 nm) and the size of tumor cells (∼20-30 microns), hundreds of nanoparticles were found to bind onto the surfaces of the tumor cells either through ligand-receptor or electrostatic interaction strategy. Theoretically, a micron-scale magnetic graphene sheet may trap the cells with much less membrane surface area therefore reducing cellular internalization and cytotoxicity. The 2D graphene oxide nano-sheets were decorated with the charged nanoprobes for HeLa cell capture.98 It was found that the 2D graphene composites resulted in higher cells capture efficiency, but caused severe damages on cell membrane and pseudopods, leading to increased cytotoxicity compared with spherical nanoparticles. The cell damage was attributed to the sharp edges of the 2D thin films.99
Effect of serum protein on the cellular viability
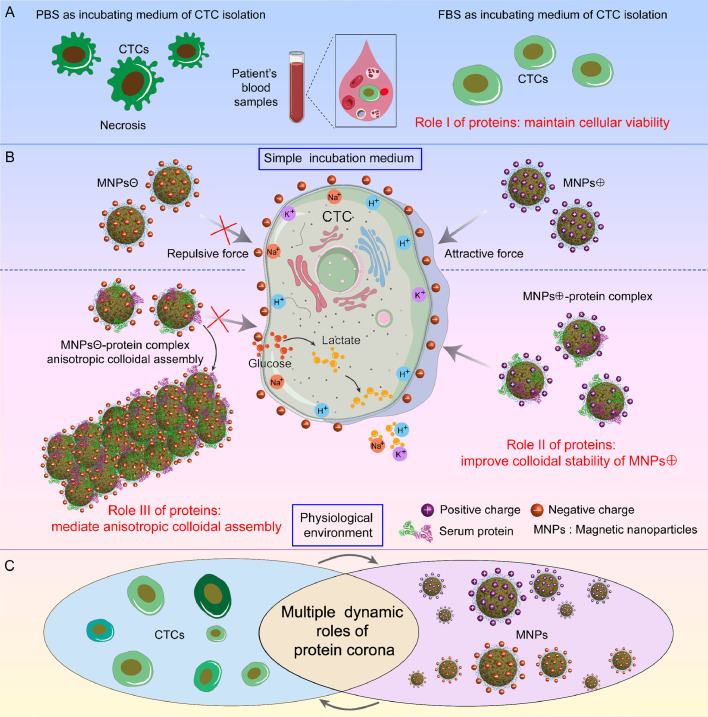
The roles of serum protein were investigated for the charge-mediated CTC isolation in our previous work.39 This study addressed the critical issue of protein decoration on the nanoparticles in the physiological environment for clinical application of CTC detection. In charge-mediated CTC detection, the nanoparticles are rendered positively charged via PEI surface functionalization. However, PEI (Mw=25 kd) as a representative polycationic polymer is well-known for its toxicity to tumor cells. It is widely-recognized that nanoparticles tend to aggregate at cost of its colloidal stability after administration into the physiological environment.100 Interestingly, the protein corona on the surface of MNCs⊕can reverse the nanoprobes’ Zeta potential from considerably positive to moderate negative, thus improving colloidal stability of the MNCs. In addition, FBS was found to provide certain protection for the rare tumor cells in the spiked blood samples (Figure 3A).39 Thus, the capture efficiencies of spiked cells at different concentrations in healthy donors’ blood samples when incubated in FBS and PBS after red blood cell lysing is apparently different. The well maintained cell viability contributes to the enhanced capture efficiency of HeLa cells in FBS.39
Figure 3.
Schematic diagrams showing multitude of dynamic roles of protein corona in the interaction of cancer cells with the surface-charged magnetic nanocomposites. (A) Role I: maintain cellular viability of rare cancer cells including CTCs. (B) Role II: improve colloidal stability of positively-charged magnetic nanocomposites, Role III: mediate anisotropic colloidal assembly based on negatively-charged magnetic nanocomposite with the help of temporary magnetic force. (C) The multiple “bridging roles” of the protein corona in cancer cells targeting and isolation.
Furthermore, it was found that the FBS-MNCs⊕ complexes efficiently captured 2-8 CTCs per mL clinical blood samples from 25 CRC patients, identified by immune-staining and iFISH staining. Using the same procedure, an average of 0.4 abnormal cells was captured and identified from 10 healthy donor's samples. More importantly, from 1 mL of clinical sample of CRC patient, the CTCs with different sizes, chromosome ploidy, and EMT phenotype were detected successfully. Comparison of CTC isolation sensitivity between the label-free capture method and a commercialized inertial microfluidic device indicated better performance of the former, both were evaluated by using CD45/iFISH staining.39
As for the negatively-charged magnetic nanocomposites, the capture efficiencies were much lower as compared to their positively-charged counterpart either in the medium of PBS or FBS. The bamboo-leaf like colloidal aggregates were formed after cell capturing due to protein corona decoration on the nanoparticles (Figure 3B). The formation mechanism of the colloidal nanoparticle assembly was identified involving multitude of parameters like protein types, protein concentration, magnetic force, and ions in medium. Using fibrinogen (Fg) and bovine serum albumin (BSA), two kinds of typical components of plasma, as models, the facile and reproducible colloidal assembly were investigated.101 It was found that the MNCs⊖-protein complex clusters acted as the seeds for the formation of the subsequent anisotropic self-assembly with the bamboo-leaf like morphology. This process was triggered by the temporary magnetic force and intermolecular electrostatic interaction of proteins on the nanocomposite surfaces. The one-dimensional fiber-shaped magnetic nanoparticle assembly was decorated by a variety of protein types. The formation mechanism provides the possibilities of constructing smart, dynamic, responsive, and biocompatible scaffolds for applications in tissue engineering and CTCs in-vitro culture.102
As shown in Figure 3, the surface protein corona plays majors roles in the dynamic interactions between the tumor cells and the surface-charged magnetic nanoprobes, which not only maintain the CTC viability in the blood sample but also enhance the colloidal stability of positively-charged nanoprobes (Figure 3B, 3C). These are critical functions to ensure highly efficient and whole subpopulation CTC isolation. On other hand, protein corona decorated nanoparticles may have important potential in biomedical applications that require both the protein structures and super-paramagnetic properties.
Cell release for molecular profiling and in-vitro proliferation
High cell viability can be well-achieved through efficient responsive release of the captured cells by the spherical magnetic nanoprobes. There have been extensive studies on the efficient release of the captured tumor cells using either the microfluidic chips or the nanoparticles platform through different release triggers, like light, sound, electricity, magnetism, heat, enzyme cleavage, pH value, hydrophobic force, polymer conformational changes, ligand substitution, etc.7,55,98,103, 104, 105 Among these approaches, three concerns remain to be addressed: 1) the influence of the trigger on the cellular viability, 2) the impact of the protein corona formed on the surface of nanoparticle, and 3) the substrate effect on releasing efficacy.
We have developed an inorganic core-shell calcium oxide magnetic nanocomposite with strong positive potential.66 The nanocomposite exhibits mild acidic circumstance responsive release property and high viable release efficiency without interference from the serum protein in the medium. It is worth noting that the pH value of the medium can be easily adjusted by changing the components’ ratio of PBS for the in vitro cell culture medium without introducing any new acidic substances. This system has been initially validated in breast cancer patients’ blood and ascites samples with peritoneal metastases.67
With the highly efficient CTCs release techniques, powerful single-cell-omics technologies can be developed. However, as a cell population, CTCs are inherently heterogeneous. Therefore, for drug screening and treatment assessment, the captured CTCs must typically represent the tumor characteristics of the cancer patients.106 Yu et al established brain metastasis mouse model based on ex-vivo cultured CTC lines isolated from blood samples of brain metastatic breast cancer patients. These data provided the direct experimental evidence on the promising role of CTCs as a prognostic factor for site-specific metastasis.107
Clinical implications and significance of the CTC analysis
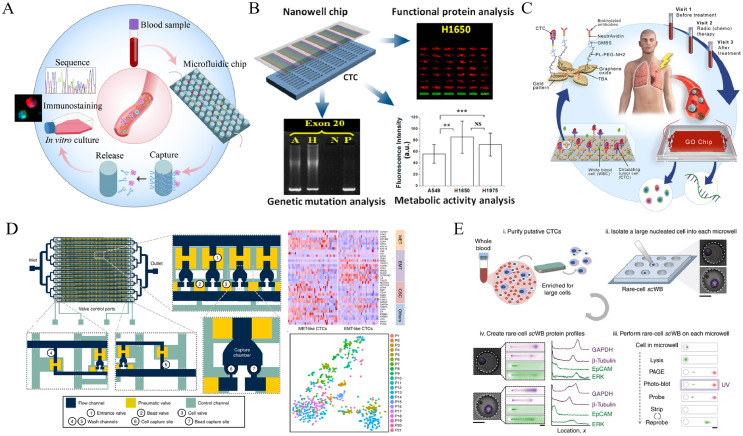
Since the early clinical applications of CTC enumeration from the patients’ peripheral blood samples, tremendous progresses have been made in three areas: 1)prognostic evaluation and risk estimation for metastatic relapse,108,109 2) therapeutic outcome assessment and medication guidance for oncology precision medicine,110,111 and 3) dynamic marker monitoring during the treatment.112 Cellsearch has been approved by FDA for clinical prognostic monitoring of patients with breast, colorectal and prostate cancers. As this is a mature and commercial analytical system that can provide stable results in different laboratories, it has been widely and clinically applied and validated consistently for many cancer types. With a large number of clinical studies based on this system, the value of CTC count in the prognostication of metastasis has been confirmed.113, 114, 115 In addition, a study based on 5 independent randomized clinical trials has proved that the changes in the number of CTCs in peripheral blood at a specific stage demonstrate better accuracy than the conventional method of using Prostate-Specific Antigen (PSA) protein for predicting drug efficacy in the metastatic castration-resistant prostate cancer (mCRPC).116 In addition to enumeration of CTC in the blood sample of cancer patients, correlation of cellular heterogeneity of CTC with clinical sequence via single CTCs molecular profiling and bioinformatics tools will be essential for translation of the fundamental understandings to clinical practice as illustrated by the scheme in Figure 4A.117, 118, 119
Figure 4.
(A) Schematic illustration of molecular profiling of the captured rare CTCs for some representative microfluidic platforms. (B) Multiplexed biological information provided by isolated CTCs. Reprinted with permission from Ref.120; (C) Real-time monitoring of immune activation of tumor microenvironment from isolated CTCs. Reprinted with permission from Ref.112; (D) Single-cell whole-transcriptome sequencing from isolated CTCs by a well-designed microfluidic chip. Reprinted with permission from Ref.121; (E) Multiplexed protein profiling of isolated CTCs through microfluidic western blot. Reprinted with permission from Ref.119
Studies have also shown that CTCs with different EMT phenotypes play different roles in the clinical cytopathology and prognostic monitoring of tumors. Based on animal models of breast cancer and clinical samples, Wang et al investigated the heterogeneity of four EMT-related CTC phenotypes and their relevance to migration, metastatic outgrowth and proliferation of CTCs or disseminated tumor cells (DTCs). These studies were carried out from the perspectives of morphological, molecular and phenotypic analyses, demonstrating the clinical value of EpCAM-positive CTC phenotypes for early detection and evaluation for the risk of metastasis in breast cancer.122 Another study employed the CanPatrol system for CTCs isolation and iFISH staining. Apart from CTC counts, the mesenchymal-CTC (M-CTC) percentage showed a clinical significance where the ovarian cancer patients with M-CTCs percentage ≥ 0.3 were associated with a 2.1-fold increase of recurrence rate when compared to those with M-CTC < 0.1.123 Besides, CTCs of ovarian cancer in liquid biopsy have been clinically applied for prognosis prediction and precision diagnosis.124
As one of the markers in tumor liquid biopsy, CTCs can provide a variety of biological information such as protein, RNA, DNA and metabolites as compared with ctDNA markers,125 which can be realized based on a microfluidic chip (Figure 4B).120 Therefore, in addition to CTC count for tumor development monitoring, clinicopathologists and oncologists are increasingly interested in obtaining more information on cell biology and molecular biology from CTCs for clinical efficacy assessment in recent years.126, 127, 128 For example, Nagrath and colleagues reported a new way to allow real-time monitoring of immune activation of tumor by measuring PD-L1 expression in isolated CTCs during the radiation therapy (Figure 4C). In this approach, gene expression analysis based on the CTCs revealed that higher levels of PD-L1 were associated with poor prognosis, which implied the critical role of monitoring the dynamic changes of PD-L1 as one of potentially prognostic markers for responsive treatment.112 There are two basic strategies to perform dynamic monitoring:1) single cell technique based on the isolated CTCs, including mass spectrometry (MS) analysis of protein, DNA, RNA and metabolites from a trace of CTCs,129 2) CTCs in vitro culture to obtain a large number of CTC clusters for analyzing their various biological characteristics.130 The former has the advantage of preserving the condition and biological properties of the original tumor cells. However, the rarity and weak signal intensity make the analysis difficult. The latter has a much higher number of cells with the increased signal intensity, but the changes at molecular level of the proliferated cells, as compared to the original CTCs, can create certain uncertainties. The microfluidic technology provides an excellent platform that integrates multiple functions including CTC isolation, enrichment, identification, and molecular analysis, for obtaining protein, gene, and metabolomics related information directly from blood samples. To improve the accuracy of CTC single cell sequencing by considering heterogeneity, it is necessary to effectively connect the different micro-regions with various functions in the microfluidic chip for suitable sequencing technology.
Yoon et al attempted a novel approach to address cellular heterogeneity in terms of gene expression and pathway regulation analysis using a massively parallel single-cell RNA sequencing (scRNA-seq) by using a microfluidic chip platform.121 They engineered the Hydro-Seq chips composed of 800 chambers per chip (can be expanded to an array of 12,800 chambers) with 16 branch channels. This Hydro-Seq chip utilizes size-based separation to isolate the CTCs from other blood cells. Using the Hydro-Seq chip, the captured CTCs can be lysed in the chambers with the mRNA released from CTCs that are uniquely labelled by a barcode bead and identified using single-cell whole-transcriptome sequencing (Figure 4D). With multiple processes and quake valves controlled by pneumatics, the design is capable of various functions, such as protein expression level analysis, which provides a wealth of information that can be used in breast cancer gene sequencing.
Meanwhile, microdroplet and microfluidic platforms for metabolism, fluorescence analysis and sequencing of single cells are the emerging methods in the CTC studies. Combined with hydrogel microdroplets, there is a potential to integrate molecular analysis of single cells with in-vitro cell cultures, enabling CTC-based precision diagnosis and drug evaluation.131 However, this system still needs to be validated in more clinical samples. Although these platforms are rapidly updated, it is critical to further explore the clinical potential of single-cell sequencing. Three major types of systems are currently available for single cell analysis: (1) microdroplet systems, (2) microfluidic chips, and (3) microchips. The single-cell analysis focuses on transcriptome and genome analysis, where the controlling of the side products from CTCs specific amplification will be the key in obtaining the accurate genetic information due to rarity of CTCs. Furthermore, the analysis of protein expression from a few CTCs is also critical for cancer diagnosis and prognostic monitoring. A microfluidic western blotting technique was developed capable of reporting protein expression on a per CTC basis and two statistically distinct GAPDH subpopulations within the patient-derived CTCs from estrogen receptor-positive (ER+) breast cancer patients (Figure 4E). This targeted single-CTC proteomics has shown a capacity for multiplexed protein analysis that offers a unique, complementary taxonomy for understanding CTC biology and ascertaining clinical impact.119 After microfluidic isolation, the CTC protein can also be recognized by using an aptamer probe based on inductively coupled plasma mass spectrometry (ICP-MS), which exhibits a 73% measurement efficiency for single CTCs.132
Conclusion and perspectives
As one of the important markers of tumor liquid biopsy, CTCs have been recognized to provide valuable information in both fundamental cancer biology studies and clinical diagnosis and prognosis. The single CTCs or CTC clusters isolated from peripheral blood can be used for prognostic monitoring and efficacy evaluation of many cancers. The molecular analysis of different CTC phenotypes associated with EMT can establish biological profiles of the metastatic tumors accurately. Longitudinal sampling and monitoring of tumor progression by CTC isolation before and after a specific treatment will provide dynamic molecular information for evaluating the driving cues for treatment resistance. Additionally, cancer heterogeneity, including different patients, stages, and cells, presents both challenges and opportunities for precision diagnosis and treatment. The current CTC studies call for development of stable, reproducible, and clinically viable CTC enrichment technologies for analyzing tumor cells at genomic, phenotypic, and cellular levels. Integrated with proteins, nucleic acids and metabolites analysis at the single cell level, CTC studies will deepen the understanding of the underlying mechanisms of tumor metastasis and recurrence. Nanoparticles are inevitably decorated with protein coronas when dispersed in the physiological solutions. The interfacial behaviors between the nanoparticles and the corona through the biological environmental dictate the outcomes of CTC detection, capture, and isolation. Ex-vivo culture of the captured CTCs can be used for establishing the mouse metastasis model and in-vitro drug screening. Therefore, there is an urgent need to achieve stable and efficient capture of CTCs for phenotypic analysis, single cell biological characterization, and nano-bio interface studies that are extremely valuable for both fundamental cancer biology research and downstream applications. To this end, some critical issues need to be addressed in future research for clinical practice: (1) develop clinically viable CTC isolation strategies against specific requirements of liquid biopsy. (2) Target those that have not been well reported such as head and neck cancer etc. (3) need more powerful and facile methods to identify CTCs without reducing its cellular viability. (4) With more advanced techniques for CTC ex vivo culture, calibrate different CTC isolation techniques by using cancer patients derived CTC-line as the standard base for evaluating the efficacy of CTC isolation.
Outstanding questions
Although CTC isolating technologies have been advanced in many areas, critical issues remain in detection sensitivity, technical and commercial viability, and CTC identification accuracy. The potential drawback of the CTC capture and detection system is the limitations in treating a small quantity of sample with multiple washes to increase sensitivity and the fractured identification of captured cells. The nano-probe based capturing may be tailored to simultaneous, small, multi-sample screening. For clinical settings, the key question is the possibility of developing a device that is capable of high throughput and large volume of blood sample filtering with synergy of CTC isolation, release, and molecular profiling. If CTCs can be efficiently captured from clinical blood, is it possible to develop a technique to remove CTCs from the blood in a continuous fashion, a process like Dialysis, for people with kidney failure.
Search strategy and selection criteria
The search strategy of this review replies on the most current research papers published in major medical and nanoscience journals focusing on circulating tumor cells (CTCs) and related cancer studies. We have applied explicit methods to perform a comprehensive literature search & critical appraisal of individual study searches of Web of Science, MEDLINE, Current Contents, PubMed, and references from relevant articles using the search terms such as “circulating tumor cells”, “Warburg Effect”, “protein corona” and “metastasis.” The selection criteria are based on research works published by well-known experts in the field of CTC studies, cancer biology, oncology, and pathology. Our own research on this topic is also included in this review.
Contributors
Writing - original draft preparation, Z.D., Y.W.; writing - review and editing, Y.W., Z.D., S.W., and D.S.; Figure - original draw preparation, Z.D., S.W.; Figure - review, Y.W. D.S.; All authors have read and agreed to the submission of the manuscript.
Declaration of interests
The authors declare no conflict of interest.
Acknowledgments
Y.W. and S.W. at Tongji University acknowledge the support from the National Natural Science Foundation of China (32071395). None of the funders had any role in paper design, data collection, data analysis, interpretation, and writing of the paper.
Contributor Information
Yilong Wang, Email: yilongwang@tongji.edu.cn.
Donglu Shi, Email: donglu.shi@uc.edu.
References
- 1.Latest global cancer data: Cancer burden rises to 19.3 million new cases and 10.0 million cancer deaths in 2020 – IARC n.d.https://www.iarc.who.int/news-events/latest-global-cancer-data-cancer-burden-rises-to-19-3-million-new-cases-and-10-0-million-cancer-deaths- in-2020/. Accessed 23 March 2022.
- 2.Guan X. Cancer metastases: challenges and opportunities. Acta Pharm Sin B. 2015;5:402–418. doi: 10.1016/j.apsb.2015.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lin Y, Xu J, Lan H. Tumor-associated macrophages in tumor metastasis: biological roles and clinical therapeutic applications. J Hematol Oncol. 2019;12:76. doi: 10.1186/s13045-019-0760-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Creaney J, Yeoman D, Demelker Y, et al. Comparison of osteopontin, megakaryocyte potentiating factor, and mesothelin proteins as markers in the serum of patients with malignant mesothelioma. J Thorac Oncol. 2008;3:851–857. doi: 10.1097/JTO.0b013e318180477b. [DOI] [PubMed] [Google Scholar]
- 5.Maestranzi S, Przemioslo R, Mitchell H, Sherwood RA. The effect of benign and malignant liver disease on the tumour markers CA19-9 and CEA. Ann Clin Biochem. 1998;35:99–103. doi: 10.1177/000456329803500113. [DOI] [PubMed] [Google Scholar]
- 6.Alix-Panabières C, Pantel K. Liquid biopsy: from discovery to clinical application. Cancer Discov. 2021;11:858–873. doi: 10.1158/2159-8290.CD-20-1311. [DOI] [PubMed] [Google Scholar]
- 7.Sharma S, Zhuang R, Long M, et al. Circulating tumor cell isolation, culture, and downstream molecular analysis. Biotechnol Adv. 2018;36:1063–1078. doi: 10.1016/j.biotechadv.2018.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu X, Jiang Z, Wang J, Ren Y, Wu A. Microfluidic applications on circulating tumor cell isolation and biomimicking of cancer metastasis. Electrophoresis. 2020;41:933–951. doi: 10.1002/elps.201900402. [DOI] [PubMed] [Google Scholar]
- 9.Pantel K, Schlimok G, Braun S, et al. Differential expression of proliferation-associated molecules in individual micrometastatic carcinoma cells. JNCI J Natl Cancer Inst. 1993;85:1419–1424. doi: 10.1093/jnci/85.17.1419. [DOI] [PubMed] [Google Scholar]
- 10.Pantel K, Alix-Panabières C. Real-time liquid biopsy in cancer patients: fact or fiction? Cancer Res. 2013;73:6384–6388. doi: 10.1158/0008-5472.CAN-13-2030. [DOI] [PubMed] [Google Scholar]
- 11.Liu J, Lian J, Chen Y, et al. Circulating tumor cells (CTCs): a unique model of cancer metastases and non-invasive biomarkers of therapeutic response. Front Genet. 2021;12 doi: 10.3389/fgene.2021.734595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/S0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 13.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 14.Ward PS, Thompson CB. Metabolic reprogramming: a cancer hallmark even warburg did not anticipate. Cancer Cell. 2012;21:297–308. doi: 10.1016/j.ccr.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mohtar MA, Syafruddin SE, Nasir SN, Low TY. Revisiting the roles of pro-metastatic EpCAM in cancer. Biomol. 2020;10 doi: 10.3390/biom10020255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martin TA, Ye L, Sanders AJ, Lane J, Jiang WG. Cancer invasion and metastasis: molecular and cellular perspective. In: Madame Curie Bioscience Database. Austin (TX): Landes Bioscience; 2000-2013. Available from: https://www.ncbi.nlm.nih.gov/books/NBK164700/
- 17.Patriarca C, Macchi RM, Marschner AK, Mellstedt H. Epithelial cell adhesion molecule expression (CD326) in cancer: a short review. Cancer Treat Rev. 2012;38:68–75. doi: 10.1016/j.ctrv.2011.04.002. [DOI] [PubMed] [Google Scholar]
- 18.Welch DR, Hurst DR. Defining the hallmarks of metastasis. Cancer Res. 2019;79:3011–3027. doi: 10.1158/0008-5472.CAN-19-0458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chemi F, Mohan S, Guevara T, Clipson A, Rothwell DG, Dive C. Early dissemination of circulating tumor cells: biological and clinical insights. Front Oncol. 2021;11:1–11. doi: 10.3389/fonc.2021.672195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim M-Y, Oskarsson T, Acharyya S, et al. Tumor self-seeding by circulating cancer cells. Cell. 2009;139:1315–1326. doi: 10.1016/j.cell.2009.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Willipinski-Stapelfeldt B, Riethdorf S, Assmann V, et al. Changes in cytoskeletal protein composition indicative of an epithelial-mesenchymal transition in human micrometastatic and primary breast carcinoma cells. Clin Cancer Res. 2005;11:8006–8014. doi: 10.1158/1078-0432.CCR-05-0632. [DOI] [PubMed] [Google Scholar]
- 22.Min Y, Aditya B, Ben S.W., et al. Circulating breast tumor cells exhibit dynamic changes in epithelial and mesenchymal composition. Science (80-) 2013;339:580–584. doi: 10.1126/science.1228522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grover PK, Cummins AG, Price TJ, Roberts-Thomson IC, Hardingham JE. Circulating tumour cells: the evolving concept and the inadequacy of their enrichment by EpCAM-based methodology for basic and clinical cancer research. Ann Oncol. 2014;25:1506–1516. doi: 10.1093/annonc/mdu018. [DOI] [PubMed] [Google Scholar]
- 24.Ignatiadis M, Sotiriou C, Pantel K. Springer Berlin Heidelberg; Berlin, Heidelberg: 2012. Minimal Residual Disease and Circulating Tumor Cells in Breast Cancer: Open Questions for Research. In: Ignatiadis M, Sotiriou C, Pantel K, eds; pp. 3–9. [DOI] [PubMed] [Google Scholar]
- 25.Warburg O. On the origin of cancer cells. Science (80-) 1956;123:309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- 26.Pinheiro C, Miranda-Gonçalves V, Longatto-Filho A, et al. The metabolic microenvironment of melanomas: prognostic value of MCT1 and MCT4. Cell Cycle. 2016;15:1462–1470. doi: 10.1080/15384101.2016.1175258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shi D. Cancer cell surface negative charges: a bio-physical manifestation of the warburg effect. Nano Life. 2017;07 doi: 10.1142/s1793984417710015. [DOI] [Google Scholar]
- 28.Chen B, Le W, Wang Y, et al. Targeting negative surface charges of cancer cells by multifunctional nanoprobes. Theranostics. 2016;6:1887–1898. doi: 10.7150/thno.16358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nagrath S, Sequist L V, Maheswaran S, et al. Isolation of rare circulating tumour cells in cancer patients by microchip technology. Nature. 2007;450:1235–1239. doi: 10.1038/nature06385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim TH, Wang Y, Oliver CR, et al. A temporary indwelling intravascular aphaeretic system for in vivo enrichment of circulating tumor cells. Nat Commun. 2019;10 doi: 10.1038/s41467-019-09439-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Peng J, Zhao Q, Zheng W, et al. Peptide-functionalized nanomaterials for the efficient isolation of HER2-positive circulating tumor cells. ACS Appl Mater Interfaces. 2017;9:18423–18428. doi: 10.1021/acsami.7b03905. [DOI] [PubMed] [Google Scholar]
- 32.Zhou X, Zhang Y, Kang K, et al. Controllable environment protein corona-disguised immunomagnetic beads for high-performance circulating tumor cell enrichment. Anal Chem. 2022;94:4650–4657. doi: 10.1021/acs.analchem.1c04587. [DOI] [PubMed] [Google Scholar]
- 33.Agerbæk MØ, Bang-Christensen SR, Yang M-H, et al. The VAR2CSA malaria protein efficiently retrieves circulating tumor cells in an EpCAM-independent manner. Nat Commun. 2018;9:3279. doi: 10.1038/s41467-018-05793-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu Z, Fusi A, Klopocki E, et al. Negative enrichment by immunomagnetic nanobeads for unbiased characterization of circulating tumor cells from peripheral blood of cancer patients. J Transl Med. 2011;9:70. doi: 10.1186/1479-5876-9-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Payne K, Brooks JM, Taylor GS, et al. Immediate sample fixation increases circulating tumour cell (CTC) capture and preserves phenotype in head and neck squamous cell carcinoma: towards a standardised approach to microfluidic CTC biomarker discovery. Cancers (Basel) 2021;13 doi: 10.3390/cancers13215519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu L, Mao X, Imrali A, et al. Optimization and evaluation of a novel size based circulating tumor cell isolation system. PLoS One. 2015;10 doi: 10.1371/journal.pone.0138032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tan SJ, Yobas L, Lee GYH, Ong CN, Lim CT. Microdevice for the isolation and enumeration of cancer cells from blood. Biomed Microdevices. 2009;11:883–892. doi: 10.1007/s10544-009-9305-9. [DOI] [PubMed] [Google Scholar]
- 38.Tamminga M, Andree KC, Hiltermann TJN, et al. Detection of circulating tumor cells in the diagnostic leukapheresis product of non-small-cell lung cancer patients comparing cellsearch® and ISET. Cancers (Basel) 2020;12 doi: 10.3390/cancers12040896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu S, Gu L, Qin J, et al. Rapid label-free isolation of circulating tumor cells from patients’ peripheral blood using electrically charged Fe 3 O 4 nanoparticles. ACS Appl Mater Interfaces. 2020;12:4193–4203. doi: 10.1021/acsami.9b16385. [DOI] [PubMed] [Google Scholar]
- 40.Zeinali M, Lee M, Nadhan A, et al. High-throughput label-free isolation of heterogeneous circulating tumor cells and CTC clusters from non-small-cell lung cancer patients. Cancers (Basel) 2020;12:1–17. doi: 10.3390/cancers12010127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gupta V, Jafferji I, Garza M, et al. ApoStreamTM, a new dielectrophoretic device for antibody independent isolation and recovery of viable cancer cells from blood. Biomicrofluidics. 2012;6 doi: 10.1063/1.4731647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chu CH, Liu R, Ozkaya-Ahmadov T, et al. Hybrid negative enrichment of circulating tumor cells from whole blood in a 3D-printed monolithic device. Lab Chip. 2019;19:3427–3437. doi: 10.1039/c9lc00575g. [DOI] [PubMed] [Google Scholar]
- 43.Sun N, Li X, Wang Z, Li Y, Pei R. High-purity capture of CTCs based on micro-beads enhanced isolation by size of epithelial tumor cells (ISET) method. Biosens Bioelectron. 2018;102:157–163. doi: 10.1016/j.bios.2017.11.026. [DOI] [PubMed] [Google Scholar]
- 44.Ozkumur E, Shah AM, Ciciliano JC, et al. Inertial focusing for tumor antigen-dependent and -independent sorting of rare circulating tumor cells. Sci Transl Med. 2013;5:179ra47. doi: 10.1126/scitranslmed.3005616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang F, Wu L, Nie W, et al. Biomimetic microfluidic system for fast and specific detection of circulating tumor cells. Anal Chem. 2019;91:15726–15731. doi: 10.1021/acs.analchem.9b03920. [DOI] [PubMed] [Google Scholar]
- 46.Mohamadi RM, Besant JD, Mepham A, et al. Nanoparticle-mediated binning and profiling of heterogeneous circulating tumor cell subpopulations. Angew Chemie Int Ed. 2015;54:139–143. doi: 10.1002/anie.201409376. [DOI] [PubMed] [Google Scholar]
- 47.Hoshino K, Huang YY, Lane N, et al. Microchip-based immunomagnetic detection of circulating tumor cells. Lab Chip. 2011;11:3449–3457. doi: 10.1039/c1lc20270g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mishra A, Dubash TD, Edd JF, et al. Ultrahigh-throughput magnetic sorting of large blood volumes for epitope-agnostic isolation of circulating tumor cells. Proc Natl Acad Sci. 2020;117:16839–16847. doi: 10.1073/pnas.2006388117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ahmed MG, Abate MF, Song Y, et al. Isolation, detection, and antigen-based profiling of circulating tumor cells using a size-dictated immunocapture chip. Angew Chemie Int Ed. 2017;56:10681–10685. doi: 10.1002/anie.201702675. [DOI] [PubMed] [Google Scholar]
- 50.Sawabata N. Circulating tumor cells: from the laboratory to the cancer clinic. Cancers (Basel) 2020;12:3065. doi: 10.3390/cancers12103065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xiong K, Wei W, Jin Y, et al. Biomimetic immuno-magnetosomes for high-performance enrichment of circulating tumor cells. Adv Mater. 2016;28:7929–7935. doi: 10.1002/adma.201601643. [DOI] [PubMed] [Google Scholar]
- 52.Pei H, Li L, Wang Y, et al. Single-cell phenotypic profiling of CTCs in whole blood using an integrated microfluidic device. Anal Chem. 2019;91:11078–11084. doi: 10.1021/acs.analchem.9b01647. [DOI] [PubMed] [Google Scholar]
- 53.Belthier G, Homayed Z, Grillet F, et al. CD44v6 defines a new population of circulating tumor cells not expressing EpCAM. Cancers (Basel) 2021;13:4966. doi: 10.3390/cancers13194966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dickey DD, Giangrande PH. Oligonucleotide aptamers: a next-generation technology for the capture and detection of circulating tumor cells. Methods. 2016;97:94–103. doi: 10.1016/j.ymeth.2015.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wu L, Wang Y, Xu X, et al. Aptamer-based detection of circulating targets for precision medicine. Chem Rev. 2021;121:12035–12105. doi: 10.1021/acs.chemrev.0c01140. [DOI] [PubMed] [Google Scholar]
- 56.Yoon HJ, Kozminsky M, Nagrath S. Emerging role of nanomaterials in circulating tumor cell isolation and analysis. ACS Nano. 2014;8:1995–2017. doi: 10.1021/nn5004277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yu L, Ng SR, Xu Y, Dong H, Wang YJ, Li CM. Advances of lab-on-a-chip in isolation, detection and post-processing of circulating tumour cells. Lab Chip. 2013;13:3163–3182. doi: 10.1039/C3LC00052D. [DOI] [PubMed] [Google Scholar]
- 58.Hejazian M, Li W, Nguyen N-T. Lab on a chip for continuous-flow magnetic cell separation. Lab Chip. 2015;15:959–970. doi: 10.1039/C4LC01422G. [DOI] [PubMed] [Google Scholar]
- 59.Song Y, Shi Y, Huang M, et al. Bioinspired engineering of a multivalent aptamer-functionalized nanointerface to enhance the capture and release of circulating tumor cells. Angew Chemie Int Ed. 2019;58:2236–2240. doi: 10.1002/anie.201809337. [DOI] [PubMed] [Google Scholar]
- 60.Lemaire CA, Liu SZ, Wilkerson CL, et al. Fast and label-free isolation of circulating tumor cells from blood: from a research microfluidic platform to an automated fluidic instrument, VTX-1 liquid biopsy system. SLAS Technol Transl Life Sci Innov. 2018;23:16–29. doi: 10.1177/2472630317738698. [DOI] [PubMed] [Google Scholar]
- 61.Gogoi P, Sepehri S, Zhou Y, et al. Development of an automated and sensitive microfluidic device for capturing and characterizing circulating tumor cells (CTCs) from clinical blood samples. PLoS One. 2016;11 doi: 10.1371/journal.pone.0147400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Smith KJ, Jana JA, Kaehr A, et al. Inertial focusing of circulating tumor cells in whole blood at high flow rates using the microfluidic CTCKeyTM device for CTC enrichment. Lab Chip. 2021;21:3559–3572. doi: 10.1039/d1lc00546d. [DOI] [PubMed] [Google Scholar]
- 63.Shiriny A, Bayareh M. Inertial focusing of CTCs in a novel spiral microchannel. Chem Eng Sci. 2021;229 doi: 10.1016/j.ces.2020.116102. [DOI] [Google Scholar]
- 64.Li Q, Cui S, Xu Y, et al. Consecutive sorting and phenotypic counting of CTCs by an optofluidic flow cytometer. Anal Chem. 2019;91:14133–14140. doi: 10.1021/acs.analchem.9b04035. [DOI] [PubMed] [Google Scholar]
- 65.Qi L-N, Xiang B-D, Wu F-X, et al. Circulating tumor cells undergoing EMT provide a metric for diagnosis and prognosis of patients with hepatocellular carcinoma. Cancer Res. 2018;78:4731–4744. doi: 10.1158/0008-5472.CAN-17-2459. [DOI] [PubMed] [Google Scholar]
- 66.Wu S, Wang Y, Shi D. Positively charged magnetic nanoparticles for capture of circulating tumor cells from clinical blood samples. Nano Life. 2020;10 doi: 10.1142/s1793984419710016. [DOI] [Google Scholar]
- 67.Wang P, Xue J, Wu S, Pei Y, Xu L, Wang Y. Cell-friendly isolation and pH-sensitive controllable release of circulating tumor cells by Fe 3 O 4 @CaCO 3 nanoplatform. Adv Mater Interfaces. 2021;8 doi: 10.1002/admi.202101191. [DOI] [Google Scholar]
- 68.Zhao J, Wu S, Qin J, Shi D, Wang Y. Electrical-charge-mediated cancer cell targeting via protein corona-decorated superparamagnetic nanoparticles in a simulated physiological environment. ACS Appl Mater Interfaces. 2018;10:41986–41998. doi: 10.1021/acsami.8b15098. [DOI] [PubMed] [Google Scholar]
- 69.Han X, Deng Z, Yang Z, et al. Biomarkerless targeting and photothermal cancer cell killing by surface-electrically-charged superparamagnetic Fe 3 O 4 composite nanoparticles. Nanoscale. 2017;9:1457–1465. doi: 10.1039/C6NR07161A. [DOI] [PubMed] [Google Scholar]
- 70.Deng Z, Kalin GT, Shi D, Kalinichenko VV. Nanoparticle delivery systems with cell-specific targeting for pulmonary diseases. Am J Respir Cell Mol Biol. 2021;64:292–307. doi: 10.1165/rcmb.2020-0306TR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Deng Z, Lin J, Bud'ko SL, et al. Dual targeting with cell surface electrical charge and folic acid via superparamagnetic Fe3O4@Cu2–xS for photothermal cancer cell killing. Cancers (Basel) 2021;13:5275. doi: 10.3390/cancers13215275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dunn AW, Kalinichenko VV., Shi D. Highly efficient in vivo targeting of the pulmonary endothelium using novel modifications of polyethylenimine: an importance of charge. Adv Healthc Mater. 2018;7 doi: 10.1002/adhm.201800876. [DOI] [PubMed] [Google Scholar]
- 73.Wang Y, Han X, Cui Z, Bioelectricity Shi D. Its fundamentals, characterization methodology, and applications in nano-bioprobing and cancer diagnosis. Adv Biosyst. 2019;3 doi: 10.1002/adbi.201900101. [DOI] [PubMed] [Google Scholar]
- 74.Lynch I, Dawson KA. Protein-nanoparticle interactions. Nano Today. 2008;3:40–47. doi: 10.1016/S1748-0132(08)70014-8. [DOI] [Google Scholar]
- 75.Fleischer CC, Payne CK. Nanoparticle–cell interactions: molecular structure of the protein corona and cellular outcomes. Acc Chem Res. 2014;47:2651–2659. doi: 10.1021/ar500190q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Dawson KA, Yan Y. Current understanding of biological identity at the nanoscale and future prospects. Nat Nanotechnol. 2021;16:229–242. doi: 10.1038/s41565-021-00860-0. [DOI] [PubMed] [Google Scholar]
- 77.Monopoli MP, Åberg C, Salvati A, Dawson KA. Biomolecular coronas provide the biological identity of nanosized materials. Nat Nanotechnol. 2012;7:779–786. doi: 10.1038/nnano.2012.207. [DOI] [PubMed] [Google Scholar]
- 78.Ren J, Cai R, Wang J, et al. Precision nanomedicine development based on specific opsonization of human cancer patient-personalized protein coronas. Nano Lett. 2019;19:4692–4701. doi: 10.1021/acs.nanolett.9b01774. [DOI] [PubMed] [Google Scholar]
- 79.Baimanov D, Cai R, Chen C. Understanding the chemical nature of nanoparticle–protein interactions. Bioconjug Chem. 2019;30:1923–1937. doi: 10.1021/acs.bioconjchem.9b00348. [DOI] [PubMed] [Google Scholar]
- 80.Dai Q, Yan Y, Ang CS, et al. Monoclonal antibody-functionalized multilayered particles: targeting cancer cells in the presence of protein coronas. ACS Nano. 2015;9:2876–2885. doi: 10.1021/nn506929e. [DOI] [PubMed] [Google Scholar]
- 81.Dai Q, Yan Y, Guo J, et al. Targeting ability of affibody-functionalized particles is enhanced by albumin but inhibited by serum coronas. ACS Macro Lett. 2015;4:1259–1263. doi: 10.1021/acsmacrolett.5b00627. [DOI] [PubMed] [Google Scholar]
- 82.Zuo L, Niu W, Li A. Isolation of circulating tumor cells of ovarian cancer by transferrin immunolipid magnetic spheres and its preliminary clinical application. Nano Life. 2019;09 doi: 10.1142/s1793984419400014. [DOI] [Google Scholar]
- 83.Wang X, Wang X, Wang M, et al. Probing adsorption behaviors of bsa onto chiral surfaces of nanoparticles. Small. 2018;14 doi: 10.1002/smll.201703982. [DOI] [PubMed] [Google Scholar]
- 84.Mikolajczyk SD, Millar LS, Tsinberg P, et al. Detection of EpCAM-negative and cytokeratin-negative circulating tumor cells in peripheral blood. J Oncol. 2011;2011 doi: 10.1155/2011/252361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Vainshelbaum NM, Zayakin P, Kleina R, Giuliani A, Erenpreisa J. Meta-analysis of cancer triploidy: rearrangements of genome complements in male human tumors are characterized by XXY Karyotypes. Genes. 2019;10 doi: 10.3390/genes10080613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ge F, Zhang H, Wang DD, Li L, Lin PP. Enhanced detection and comprehensive in situ phenotypic characterization of circulating and disseminated heteroploid epithelial and glioma tumor cells. Oncotarget. 2015;6:27049–27064. doi: 10.18632/oncotarget.4819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lin PP. Integrated EpCAM-independent subtraction enrichment and iFISH strategies to detect and classify disseminated and circulating tumors cells. Clin Transl Med. 2015;4:38. doi: 10.1186/s40169-015-0081-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Cheng S-BB, Chen M-MM, Wang Y-KK, Sun Z-HH, Xie M, Huang W-HH. Current techniques and future advance of microfluidic devices for circulating tumor cells. TrAC - Trends Anal Chem. 2019;117:116–127. doi: 10.1016/j.trac.2019.06.018. [DOI] [Google Scholar]
- 89.He G, Feng J, Zhang A, et al. Multifunctional branched nanostraw-electroporation platform for intracellular regulation and monitoring of circulating tumor cells. Nano Lett. 2019;19:7201–7209. doi: 10.1021/acs.nanolett.9b02790. [DOI] [PubMed] [Google Scholar]
- 90.Alix-Panabières C. Springer Berlin Heidelberg; Berlin, Heidelberg: 2012. EPISPOT Assay: Detection of Viable DTCs/CTCs in Solid Tumor Patients BT - Minimal Residual Disease and Circulating Tumor Cells in Breast Cancer. In: Ignatiadis M, Sotiriou C, Pantel K, eds; pp. 69–76. [DOI] [Google Scholar]
- 91.Cheng J, Liu Y, Zhao Y, et al. Nanotechnology-assisted isolation and analysis of circulating tumor cells on microfluidic devices. Micromachines. 2020;11 doi: 10.3390/mi11080774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Alix-Panabieres C. The future of liquid biopsy. Nature. 2020;579:S9. doi: 10.1038/d41586-020-00844-5. [DOI] [PubMed] [Google Scholar]
- 93.Heitzer E, Haque IS, Roberts CES, Speicher MR. Current and future perspectives of liquid biopsies in genomics-driven oncology. Nat Rev Genet. 2019;20:71–88. doi: 10.1038/s41576-018-0071-5. [DOI] [PubMed] [Google Scholar]
- 94.Lopes C, Piairo P, Chícharo A, et al. Her2 expression in circulating tumour cells isolated from metastatic breast cancer patients using a size-based microfluidic device. Cancers (Basel) 2021;13 doi: 10.3390/cancers13174446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Micalizzi DS, Maheswaran S, Haber DA. A conduit to metastasis: circulating tumor cell biology. Genes Dev. 2017;31:1827–1840. doi: 10.1101/gad.305805.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lim CT, Khoo BL. Liquid biopsy and expansion of patient derived circulating tumor cell spheroids for precision medicine. Proc Annu Meet Japanese Pharmacol Soc. 2018;WCP2018:SY80–SY83. doi: 10.1254/jpssuppl.WCP2018.0_SY80-3. [DOI] [Google Scholar]
- 97.Drapkin BJ, George J, Christensen CL, et al. Genomic and functional fidelity of small cell lung cancer patient-derived xenografts. Cancer Discov. 2018;8:600–615. doi: 10.1158/2159-8290.CD-17-0935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wu L, Xu X, Sharma B, et al. Beyond capture: circulating tumor cell release and single-cell analysis. Small Methods. 2019;3 doi: 10.1002/smtd.201800544. [DOI] [Google Scholar]
- 99.He Y, Qin J, Wu S, Yang H, Wen H, Wang Y. Cancer cell–nanomaterial interface: role of geometry and surface charge of nanocomposites in the capture efficiency and cell viability. Biomater Sci. 2019;7:2759–2768. doi: 10.1039/C9BM00037B. [DOI] [PubMed] [Google Scholar]
- 100.Cukalevski R, Ferreira SA, Dunning CJ, Berggård T, Cedervall T. IgG and fibrinogen driven nanoparticle aggregation. Nano Res. 2015;8:2733–2743. doi: 10.1007/s12274-015-0780-4. [DOI] [Google Scholar]
- 101.Pei Y, Wu S, Wang P, Qin J, Xu L, Wang Y. Path-dependent anisotropic colloidal assembly of magnetic nanocomposite–protein complexes. Langmuir. 2022;38:6265–6272. doi: 10.1021/acs.langmuir.1c02923. [DOI] [PubMed] [Google Scholar]
- 102.Narkar AR, Tong Z, Soman P, Henderson JH. Smart biomaterial platforms: controlling and being controlled by cells. Biomaterials. 2022;283 doi: 10.1016/j.biomaterials.2022.121450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Liu Y, Xu H, Li T, Wang W. Microtechnology-enabled filtration-based liquid biopsy: challenges and practical considerations. Lab Chip. 2021;21:994–1015. doi: 10.1039/D0LC01101K. [DOI] [PubMed] [Google Scholar]
- 104.Wu L, Zhu L, Huang M, et al. Aptamer-based microfluidics for isolation, release and analysis of circulating tumor cells. TrAC Trends Anal Chem. 2019;117:69–77. doi: 10.1016/j.trac.2019.05.003. [DOI] [Google Scholar]
- 105.Rushton AJ, Nteliopoulos G, Shaw JA, Coombes RC. A review of circulating tumour cell enrichment technologies. Cancers. 2021;13 doi: 10.3390/cancers13050970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Diamantopoulou Z, Castro-Giner F, Aceto N. Circulating tumor cells: ready for translation? J Exp Med. 2020;217:8–10. doi: 10.1084/JEM.20200356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Klotz R, Thomas A, Teng T, et al. Circulating tumor cells exhibit metastatic tropism and reveal brain metastasis drivers. Cancer Discov. 2020;10:86–103. doi: 10.1158/2159-8290.CD-19-0384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Heitzer E, Auer M, Gasch C, et al. Complex tumor genomes inferred from single circulating tumor cells by array-CGH and next-generation sequencing. Cancer Res. 2013;73:2965–2975. doi: 10.1158/0008-5472.CAN-12-4140. [DOI] [PubMed] [Google Scholar]
- 109.Alix-Panabières C, Pantel K. Clinical applications of circulating tumor cells and circulating tumor DNA as liquid biopsy. Cancer Discov. 2016;6:479–491. doi: 10.1158/2159-8290.CD-15-1483. [DOI] [PubMed] [Google Scholar]
- 110.Keller L, Pantel K. Unravelling tumour heterogeneity by single-cell profiling of circulating tumour cells. Nat Rev Cancer. 2019;19:553–567. doi: 10.1038/s41568-019-0180-2. [DOI] [PubMed] [Google Scholar]
- 111.Kujur PK, Flores BCT, Ramalingam N, Chinen LTD, Jeffrey SS. Springer International Publishing; Cham: 2020. Advances in the Characterization of Circulating Tumor Cells in Metastatic Breast Cancer: Single Cell Analyses and Interactions, and Patient-Derived Models for Drug Testing. In: Piñeiro R, (ed) pp. 61–80. [DOI] [PubMed] [Google Scholar]
- 112.Wang Y, Kim TH, Fouladdel S, Zhang Z, Soni P, Qin A, et al. PD-L1 expression in circulating tumor cells increases during radio(chemo)therapy and indicates poor prognosis in non-small. Cell Lung Cancer. Sci Rep. 2019;9:566. doi: 10.1038/s41598-018-36096-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Bidard F-C, Peeters DJ, Fehm T, et al. Clinical validity of circulating tumour cells in patients with metastatic breast cancer: a pooled analysis of individual patient data. Lancet Oncol. 2014;15:406–414. doi: 10.1016/S1470-2045(14)70069-5. [DOI] [PubMed] [Google Scholar]
- 114.Goldkorn A, Ely B, Quinn DI, et al. Circulating tumor cell counts are prognostic of overall survival in SWOG S0421: a phase III trial of docetaxel with or without atrasentan for metastatic castration-resistant prostate cancer. J Clin Oncol. 2014;32:1136–1142. doi: 10.1200/JCO.2013.51.7417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Scher HI, Heller G, Molina A, et al. Circulating tumor cell biomarker panel as an individual-level surrogate for survival in metastatic castration-resistant prostate cancer. J Clin Oncol. 2015;33:1348–1355. doi: 10.1200/JCO.2014.55.3487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Heller G, McCormack R, Kheoh T, et al. Circulating tumor cell number as a response measure of prolonged survival for metastatic castration-resistant prostate cancer: a comparison with prostate-specific antigen across five randomized phase III clinical trials. J Clin Oncol. 2018;36:572–580. doi: 10.1200/JCO.2017.75.2998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Lim SB, Lim CT, Lim W-T. Single-cell analysis of circulating tumor cells: why heterogeneity matters. Cancers. 2019;11 doi: 10.3390/cancers11101595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Ramalingam N, Jeffrey SS. Future of liquid biopsies with growing technological and bioinformatics studies: opportunities and challenges in discovering tumor heterogeneity with single-cell level analysis. Cancer J. 2018:24. doi: 10.1097/PPO.0000000000000308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Sinkala E, Sollier-Christen E, Renier C, et al. Profiling protein expression in circulating tumour cells using microfluidic western blotting. Nat Commun. 2017;8:14622. doi: 10.1038/ncomms14622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Zhang Y, Tang Y, Sun S, et al. Single-cell codetection of metabolic activity, intracellular functional proteins, and genetic mutations from rare circulating tumor cells. Anal Chem. 2015;87:9761–9768. doi: 10.1021/acs.analchem.5b01901. [DOI] [PubMed] [Google Scholar]
- 121.Cheng Y-H, Chen Y-C, Lin E, et al. Hydro-Seq enables contamination-free high-throughput single-cell RNA-sequencing for circulating tumor cells. Nat Commun. 2019;10:2163. doi: 10.1038/s41467-019-10122-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Liu X, Li J, Cadilha BL, et al. Epithelial-type systemic breast carcinoma cells with a restricted mesenchymal transition are a major source of metastasis. Sci Adv. 2019;5:eaav4275. doi: 10.1126/sciadv.aav4275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Yang J, Ma J, Jin Y, et al. Development and validation for prognostic nomogram of epithelial ovarian cancer recurrence based on circulating tumor cells and epithelial–mesenchymal transition. Sci Rep. 2021;11:6540. doi: 10.1038/s41598-021-86122-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Yang J, Cheng S, Zhang N, Jin Y, Wang Y. Liquid biopsy for ovarian cancer using circulating tumor cells: recent advances on the path to precision medicine. Biochim Biophys Acta - Rev Cancer. 2022;1877 doi: 10.1016/j.bbcan.2021.188660. [DOI] [PubMed] [Google Scholar]
- 125.Yu M, Stott S, Toner M, Maheswaran S, Haber DA. Circulating tumor cells: approaches to isolation and characterization. J Cell Biol. 2011;192:373–382. doi: 10.1083/jcb.201010021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Ahn JC, Teng P-C, Chen P-J, et al. Detection of circulating tumor cells and their implications as a biomarker for diagnosis, prognostication, and therapeutic monitoring in hepatocellular carcinoma. Hepatology. 2021;73:422–436. doi: 10.1002/hep.31165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Fabisiewicz A, Szostakowska-Rodzos M, Zaczek AJ, Grzybowska EA. Circulating tumor cells in early and advanced breast cancer; biology and prognostic value. Int J Mol Sci. 2020;21 doi: 10.3390/ijms21051671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Chen J, Ye C, Dong J, et al. Metabolic classification of circulating tumor cells as a biomarker for metastasis and prognosis in breast cancer. J Transl Med. 2020;18:59. doi: 10.1186/s12967-020-02237-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Zhang W, Li N, Lin L, Li H, Lin J-M. Metabolism-based capture and analysis of circulating tumor cells in an open space. Anal Chem. 2021;93:6955–6960. doi: 10.1021/acs.analchem.0c05155. [DOI] [PubMed] [Google Scholar]
- 130.Khoo BL, Grenci G, Jing T, et al. Liquid biopsy and therapeutic response: circulating tumor cell cultures for evaluation of anticancer treatment. Sci Adv. 2016;2 doi: 10.1126/sciadv.1600274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Zhu Z, Yang CJ. Hydrogel droplet microfluidics for high-throughput single molecule/cell analysis. Acc Chem Res. 2017;50:22–31. doi: 10.1021/acs.accounts.6b00370. [DOI] [PubMed] [Google Scholar]
- 132.Zhang X, Wei X, Men X, et al. Dual-multivalent-aptamer-conjugated nanoprobes for superefficient discerning of single circulating tumor cells in a microfluidic chip with inductively coupled plasma mass spectrometry detection. ACS Appl Mater Interfaces. 2021;13:43668–43675. doi: 10.1021/acsami.1c11953. [DOI] [PubMed] [Google Scholar]