Abstract
In the photosynthetic bacterium Rhodobacter capsulatus, a putative membrane-bound complex encoded by the rnfABCDGEH operon is thought to be dedicated to electron transport to nitrogenase. In this study, the whole rnf operon was cloned under the control of the nifH promoter in plasmid pNR117 and expressed in several rnf mutants. Complementation analysis demonstrated that transconjugants which integrated plasmid pNR117 directed effective biosynthesis of a functionally competent complex in R. capsulatus. Moreover, it was found that strains carrying pNR117 displayed nitrogenase activities 50 to 100% higher than the wild-type level. The results of radioactive labeling experiments indicated that the intracellular content of nitrogenase polypeptides was marginally altered in strains containing pNR117, whereas the levels of the RnfB and RnfC proteins present in the membrane were four- and twofold, respectively, higher than the wild-type level. Hence, the enhancement of in vivo nitrogenase activity was correlated with a commensurate overproduction of the Rnf polypeptides. In vitro nitrogenase assays performed in the presence of an artificial electron donor indicated that the catalytic activity of the enzyme was not increased in strains overproducing the Rnf polypeptides. It is proposed that the supply of reductants through the Rnf complex might be rate limiting for nitrogenase activity in vivo. Immunoprecipitation experiments performed on solubilized membrane proteins revealed that RnfB and RnfC are associated with each other and with additional polypeptides which may be components of the membrane-bound complex.
In Rhodobacter capsulatus, genes required for nitrogen fixation include the common nif genes found in other N2-fixing bacteria and an additional set of genes that are possibly involved in electron transport to nitrogenase (17). Seven of these genes are organized in one operon designated rnfABCDGEH (5). Genes homologous to the rnf genes have been identified in the genomes of nondiazotrophic bacteria, including Haemophilus influenzae and Escherichia coli (5, 12). Sequence comparisons revealed that three rnf gene products, RnfA, RnfD, and RnfE, had striking similarities with membrane components of an Na+-dependent NADH:ubiquinone oxidoreductase found in the bacterium Vibrio alginolyticus. On the other hand, the polypeptides encoded by rnfB and rnfC were predicted to be mainly hydrophilic and to bind iron-sulfur clusters. In addition, RnfC has potential binding sites for NADH and flavin mononucleotide and resembles in this respect the NADH-binding subunit of complex I of the respiratory chain (12). RnfB and RnfC were recently purified as Fe-S-containing proteins and were localized within the membrane of R. capsulatus (5). Taken together, these findings are consistent with the hypothesis that the rnf gene products form a membrane-bound complex which might function as a novel energy-coupling oxidoreductase (12).
The implication of the putative Rnf complex in electron transport to nitrogenase was deduced from biochemical analyses of mutants bearing insertions at various positions in the rnf operon (17). Mutants displayed less than 2% of the wild-type nitrogenase activity when assayed in vivo and around 20% when assayed in vitro. The lower than wild-type in vitro activities detected in rnf mutants was attributed to a secondary effect of the mutations on the stability of the nitrogenase components (17). Interestingly, a similar pattern of in vivo and in vitro nitrogenase activities was found in an fdxN mutant, bearing an insertion mutation in a ferredoxin-encoding gene. This ferredoxin, called FdI, has been shown to serve as electron donor to nitrogenase (7, 15, 16). In this study, strains of R. capsulatus overexpressing the rnf operon were constructed and used as a means to further investigate the role of the putative Rnf complex in nitrogen fixation and eventually facilitate its isolation. The results showed that the overproduction of the Rnf products promotes an enhancement of the in vivo nitrogenase activity. In addition, evidence is provided for the first time that RnfB and RnfC are physically associated with each other in a protein complex, which, as isolated from the membrane, may also contain three other polypeptides.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. E. coli strains were grown on Luria-Bertani medium supplemented with appropriate antibiotics. R. capsulatus strains were grown under anaerobic conditions in the light either in rich medium (YPS) or in mineral salts medium (RCV) as previously described (7).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Principal characteristics | Reference or source |
|---|---|---|
| R. capsulatus | ||
| B10S | Spontaneous Smr mutant of strain B10 | 11 |
| R363B | rnfA::Gmr Nif− | 17 |
| R386I | rnfB::lacZYA-Tcr Nif− | 17 |
| R368I | rnfC::Gmr Nif− | 17 |
| KS92I | rnfD::Gmr Nif− | 17 |
| R219BBII | rnfG::Gmr Nif− | 17 |
| Plasmids | ||
| pGEM-T | T-cloning vector | Promega |
| pR117 | pGEM-T derivative containing the rnfA-H operon | This study |
| pNF3 | Plasmid containing the nifH promoter of R. capsulatus | 14 |
| pNR117 | pNF3 derivative containing the rnfA-H operon | This study |
| pDPT51 | Helper plasmid for triparental conjugation | 14 |
For nitrogenase derepression experiments, resting cells from R. capsulatus were prepared and subjected to anaerobic derepression in N-free medium as follows. Bacteria were grown in RCV medium with 10 mM NH4Cl as a nitrogen source, harvested during the exponential growth phase (optical density at 660 of ∼1.0), and then washed three times in anaerobic N-free medium to remove fixed nitrogen. Bacteria were resuspended in N-free medium to give an optical density at 660 nm of ∼1.0. Resting cells were then incubated in the light at 30°C for the indicated time periods.
Cloning of the rnf operon and overexpression in R. capsulatus.
The rnfABCDGEH operon was amplified from R. capsulatus chromosomal DNA using a long-range PCR kit (Expand Long-Template PCR; Roche Diagnostics) and primers R501up (GGC cat atg CAA GAC TTC CTT CTC GTC) and R501dn (GGa agc ttC CAG CGC CGC CTG TGC CC) (lowercase letters denote NdeI and HindIII restriction sites introduced in R501up and R501dn, respectively). A PCR product with the expected size (5.7 kb) was cloned into pGEM-T (Promega) to give plasmid pR117. The insert was checked by restriction mapping. For expression of the cloned rnf genes in R. capsulatus, the 5.7-kb insert of pR117 was excised as an NdeI-HindIII fragment and cloned into the same sites of pNF3 (14). The resulting plasmid, called pNR117, as well as the control plasmid pNF3, were transferred into R. capsulatus via triparental conjugation using pDPT51 as the helper plasmid (7).
Southern blot analysis.
Isolation of R. capsulatus genomic DNA, electrophoretic analysis, and transfer onto a nylon Hybond-N+ membrane (Amersham) were carried out as described previously (16). Preparation of digoxigenin (DIG)-labeled rnf gene probes and detection of DNA fragments hybridizing to the probes were performed using a DIG labeling and detection kit (Roche Diagnostics).
Preparation of cell extracts for in vitro nitrogenase assays.
Cell extracts were prepared from derepressed resting cells inside an anaerobic glove box (Jacomex; O2 < 2 ppm). Bacteria were centrifuged and resuspended in 50 mM Tris-HCl buffer (pH 7.5) to a final protein concentration of approximately 3 mg/ml. Dithionite (2 mM) was added; cells were treated with lysozyme (0.2 mg/ml) and subjected to ultrasonication. Cell debris was removed by centrifugation at 14,000 × g for 30 min. Samples of the cell extract (100 μl) were transferred to assay vials, taken from the glove box, and subjected to acetylene reduction in a total volume of 0.5 ml containing 10 mM dithionite and 100 μl of an ATP-generating system (7).
Protein purification.
RnfB and RnfC were overproduced in E. coli and purified in recombinant form as described previously (5). Nitrogenase components were isolated from R. capsulatus according to published procedures (3, 8).
Nitrogenase and protein assays.
Nitrogenase activity was assayed by acetylene reduction as described previously (7), using 1 ml of resting cell suspension for in vivo assays or 100 μl of cell extract for in vitro assays. Protein concentration was determined by the bicinchoninic acid method (Pierce) with bovine serum albumin as a standard.
SDS-PAGE and Western blotting.
Proteins were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) on a 12.5% polyacrylamide gel in a Tris-glycine-SDS buffer system using a mini-slab gel apparatus (Mini-ProteanII; Bio-Rad) and revealed by Coomassie blue staining. Western blot analysis was carried out as described previously (5), using purified rabbit anti-RnfC or anti-RnfB antibodies at a 1:5,000 dilution. Blots were then incubated with peroxidase-coupled goat anti-rabbit immunoglobulin G as secondary antibody (1:10,000 dilution) and processed with an enhanced chemiluminescence kit (Amersham).
Preparation of chromatophores and solubilization of membrane-bound proteins.
Resting cells were harvested by centrifugation at 5,000 × g for 10 min at 4°C and resuspended in 10 ml of ice-cold TEN buffer (50 mM Tris-HCl [pH 7.5], 10 mM EDTA, 100 mM NaCl). Bacteria were subjected to lysozyme treatment (0.2 mg/ml, 15 min) and disrupted by ultrasonication. Cell debris were pelleted by centrifugation at 14,000 × g for 15 min, and the supernatant was centrifuged again at 200,000 × g for 30 min (TL100.2 rotor; Beckman) to separate the chromatophores from the cytosolic fraction. The chromatophores were resuspended in solubilizing buffer (50 mM potassium phosphate [pH 7.0], 100 mM NaCl, 10 mM EDTA, 10% glycerol) and either used immediately or kept frozen in liquid nitrogen.
Extraction of membrane-bound proteins was achieved by incubating chromatophores (1 mg of protein/ml) in solubilizing buffer containing 1% lauryl-maltoside (Sigma) and incubated at 4°C for 1 h with gentle agitation. After subsequent ultracentrifugation with an Airfuge centrifuge (Beckman) at 90,000 × g for 20 min, the solubilized proteins were recovered in the supernatant fraction.
In vivo radioactive labeling and immunoprecipitation.
Resting cells were prepared and incubated at 30°C in the light for nitrogenase derepression. After 1 h of derepression, 200 μCi of a mixture of [35S]methionine and [35S]cysteine (Easy-tag; NEN) was added and incubation was prolonged for 2 h. Cells were harvested by centrifugation and washed three times with 10 ml of TEN buffer. Chromatophores were prepared and treated with lauryl-maltoside as described above. The solubilized membrane proteins were incubated at 4°C for 1 h with either anti-RnfB or anti-RnfC antibodies coupled to protein A-agarose (see below) in solubilizing buffer containing 0.1% lauryl-maltoside. The agarose beads were washed three times with 0.5 ml of the same buffer. The adsorbed proteins were eluted by heating at 95°C for 5 min in 50 μl of buffered 2% SDS and then analyzed by SDS-PAGE. For detection of 35S-labeled proteins, gels were stained with Coomassie blue and dried under vacuum. Radioactive bands were visualized using an imaging system (PhosphorImager; Molecular Dynamics).
Preparation of immobilized purified antibodies.
Polyclonal anti-RnfB and anti-RnfC antibodies were purified from rabbit serum raised against each purified protein. The target antigen (purified RnfB or RnfC) was separated by SDS-PAGE and transferred onto a nitrocellulose membrane (Schleicher & Schuell). The membrane was stained with Ponceau S (Sigma), and the antigen band was cut off with a razor blade. The membrane pieces were incubated for 1 h at room temperature in blocking buffer, consisting of phosphate-buffered saline (PBS; 0.14 M NaCl, 2.7 mM KCl, 1.5 mM KH2PO4, 5.1 mM Na2HPO4 [pH 7.4]) containing 3% (vol/wt) bovine serum albumin. Serum (2 ml) was incubated with the corresponding antigen-coated membrane pieces for 16 h at 4°C with gentle shaking. The membrane pieces were washed extensively in PBS and then incubated for 20 min in a small volume of elution buffer (0.2 M glycine [pH 2.8], 1 mM EGTA). The protein eluate was neutralized by 0.1 volume of 1 M Tris base and subjected to immobilization on protein A-agarose (Sigma). For this purpose, 0.2 ml of purified antibodies was incubated with 0.1 ml of protein A-agarose for 1 h at room temperature, and the resin was washed three times with solubilization buffer containing 0.1% lauryl-maltoside. Agarose beads were then rinsed with borate buffer (0.2 M, pH 9), and the antibodies were covalently coupled to protein A-agarose with 20 mM dimethylpimelimidate (Sigma) for 30 min. The coupling reaction was stopped by 0.2 M ethanolamine (pH 8). The antibodies immobilized on agarose were washed three times in PBS and once in solubilizing buffer containing 0.1% lauryl-maltoside before use.
RESULTS
Cloning of the rnfABCDFEH operon and complementation of R. capsulatus rnf mutants.
The rnfABCDGEH operon comprises seven contiguous genes extending over approximately 5.7 kb of chromosomal DNA. A fragment of DNA beginning at the rnfA start codon and ending just after a putative transcriptional terminator located downstream of rnfH was amplified by PCR and cloned into pGEM-T. The cloned DNA was checked by restriction analysis and introduced as an NdeI-HindIII fragment into pNF3 to give plasmid pNR117. This plasmid was transferred by conjugation into five R. capsulatus rnf mutants, each bearing a unique insertion into the rnf operon (Table 1) (17). Transconjugants were selected on YPS plates containing spectinomycin and allowed to grow on RCV-N plates to test for nitrogen-fixing ability.
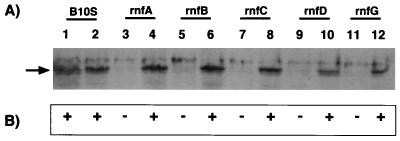
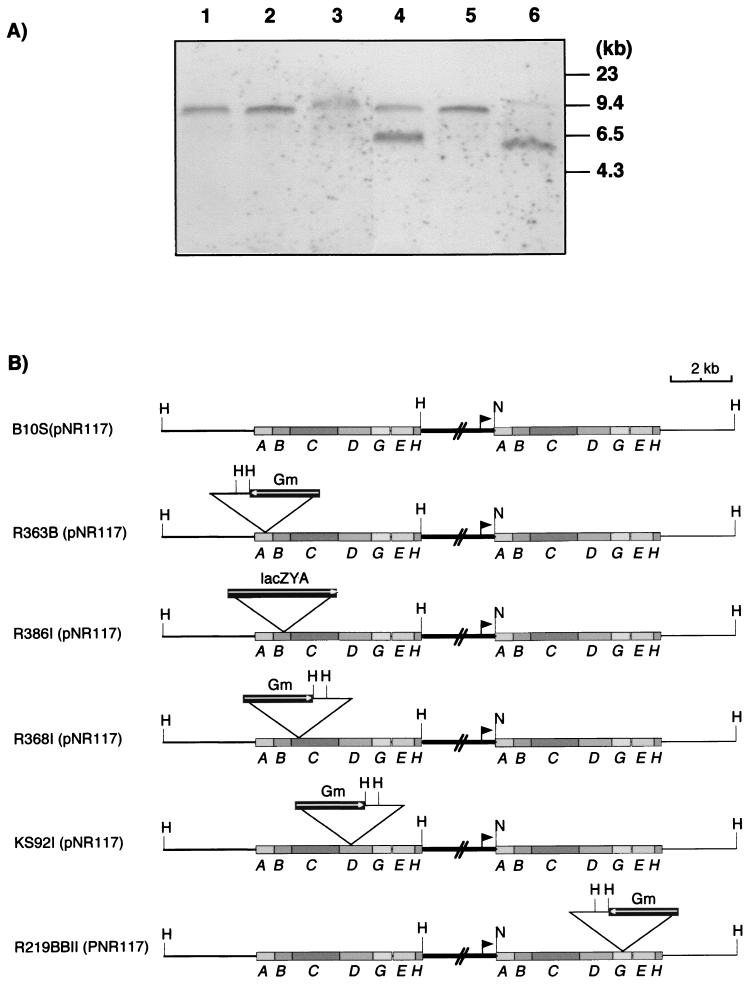
All selected transconjugants showed a Nif+ phenotype (Fig. 1), demonstrating that pNR117 complemented all five rnf insertion mutations and suggesting that at least five relevant genes (rnfA, rnfB, rnfC, rnfD, and rnfG) were successfully expressed in every mutant strain. Since pNR117 is derived from a plasmid (pNF3) which should not replicate in R. capsulatus, it was expected that transconjugants had integrated the plasmid into the chromosome by single crossover recombination. To test this hypothesis, genomic DNA from the recombinant strains was analyzed by Southern blotting with rnfB- and rnfG-specific probes. In a control experiment, we found that the digestion of strain B10S(pNF3) DNA by NdeI and HindIII yielded a single 11-kb band upon hybridization with either probe, consistent with the fact that the rnf operon is located on a HindIII chromosomal fragment of this size (data not shown). DNA analysis of pNR117 transconjugants from the rnfA, rnfB, rnfC, and rnfD mutants revealed an 8.0-kb fragment that hybridized to both probes (Fig. 2). No fragment matching the size of the pNR117 insert (5.7 kb) was detected in the NdeI/HindIII DNA digest of any strain. An additional 6.5-kb fragment was revealed by the rnfB probe in the DNA digest of strain R368I(pNR117). The 8.0-kb fragment could be explained only if plasmid pNR117 was integrated into the chromosome downstream of the site of insertion of the cassette (Fig. 2). In all transconjugants except that derived from the rnfG mutant, one copy of the entire rnfA-H operon must be under the control of the nifH promoter carried by the vector plasmid. In contrast, the hybridization pattern of strain R219BII(pNR117) showed two bands around 5.0 and 11 kb, consistent with integration of the plasmid occurring upstream of the intervening cassette. As a consequence, in this strain, only part of the rnf operon should be driven by the nifH promoter. Hybridization data obtained with the rnfG probe were consistent with the predicted maps shown in Fig. 2B (data not shown). Hence, overexpression of the rnf operon should be fully effective only in transconjugants derived from the rnfA, rnfB, rnfC, and rnfD mutants as well as in that derived from wild-type strain B10S.
FIG. 1.
pNR117-mediated complementation of rnf mutant strains. (A) Western blot analysis of whole-cell protein extracts using anti-RnfC antibodies. Odd numbers refer to strains B10S, R363B, R386I, R368I, KS92I, and R219BBII carrying pNF3 in this order; even numbers refer to the same strains carrying pNR117. (B) Nitrogen fixation abilities of the complemented strains. +, Nif+ phenotype; −, Nif− phenotype.
FIG. 2.
Southern blot analysis of genomic DNA from pNR117-containing transconjugants. Genomic DNA from each transconjugant was digested by HindIII and NdeI, electrophoresed on a 0.8% agarose gel, and transferred onto a Hybond-N+ membrane. (A) Southern blot with a DIG-labeled rnfB probe. Lane 1, B10S(pNR117); lane 2, R363B(pNR117); lane 3, R386I(pNR117); lane 4, R368I(pNR117); lane 5, KS92I(pNR117); lane 6, R219BBII(pNR117). Sizes of DNA markers are indicated at the right. (B) Predicted physical map of the region containing the rnf genes in each transconjugant, as deducted from Southern blot analysis. The orientation of the interposon cassette in each strain is shown as a boxed arrow. The nifH promoter from the pNR117 plasmid is indicated as a closed triangle. H, HindIII; N, NdeI; Gm, gentamicin.
Western blot analysis of whole-cell extracts showed that in all transconjugants, the RnfC polypeptide was synthesized at a level at least as high as in the parental strain B10S. In contrast, the RnfC polypeptide was undetectable in rnf mutant strains carrying the control plasmid pNF3 (Fig. 1). The absence of RnfC polypeptide was expected in strain R368I bearing an rnfC::Gm insertion but less obvious in mutants which carry a nonpolar insertion mutation in other rnf genes. In a previous study, we found that mutant strains bearing a cassette in rnfD and rnfG showed very low levels of RnfB and RnfC, whereas RnfC was undetectable in rnfA and rnfB mutants (5). Such results have led us and others (12) to suggest that RnfB, RnfC, and possibly other Rnf components mutually stabilize each other, so that when one of them is missing, no Rnf complex can be formed, and nonassociated Rnf polypeptides are degraded. On the other hand, the RnfC protein, and most likely the other rnf-encoded polypeptides, were successfully expressed in R. capsulatus mutants which integrated plasmid pNR117. In addition, complementation experiments further demonstrate that the Rnf polypeptides synthesized in this way were functionally competent.
Overexpression of the rnfABCDGEH operon results in higher levels of nitrogenase activity.
Previous work showed that the rnf mutants used in this study had residual in vivo nitrogenase activity not exceeding 2% of the wild-type level (17). It was therefore of interest to determine whether mutant strains harboring pNR117 had fully recovered nitrogenase activity. When assayed under identical derepression conditions, the nitrogenase activity measured in pNR117-containing strains was found to be 50 to 100% higher than in the control strain, B10S(pNF3) (Table 2). We also observed that the rnf mutants carrying pNR117 consistently displayed greater activities than strain B10S(pNR117). Strain R219BII(pNR117) was a notable exception, as it showed nitrogenase activities six- to sevenfold lower than levels for the wild-type strain. This observation led us to suppose that upon integration of pNR117 into the chromosome, DNA rearrangement might have altered the expression of some gene important for nitrogenase function. Recent DNA sequence analysis revealed that the region downstream of rnfH contains an open reading frame (ORF), 1,020 bp in length, potentially encoding a flavoprotein (preliminary sequence data on the Rhodobacter capsulatus genome project available from website http://rhodol.uchicago.edu/rhodo.html). Based on the DNA hybridization pattern shown in Fig. 2, the copy of the rnf operon of chromosomal origin is interrupted within rnfG in strain R219BII(pNR117); as a consequence, the additional putative gene is probably not expressed. The possible implication of this gene in nitrogen fixation is currently being investigated.
TABLE 2.
Nitrogenase activities in R. capsulatus transconjugants carrying pNR117a
| Strain | Nitrogenase activity (nmol of C2H4/min/mg [dry wt]) after derepression for:
|
|
|---|---|---|
| 2 h | 4 h | |
| B10S | 4.77 (±0.14) | 11.8 (±0.49) |
| B10S(pNF3) | 5.74 (±0.57) | 11.4 (±1.10) |
| B10S(pNR117) | 12.1 (±2.84) | 21.7 (±1.13) |
| R363B(pNR117) | 12.7 (±3.75) | 25.8 (±3.00) |
| R386I(pNR117) | 14.4 (±2.27) | 26.3 (±1.06) |
| R368I(pNR117) | 13.2 (±0.75) | 24.1 (±1.72) |
| KS92I(pNR117) | 12.4 (±2.84) | 23.5 (±0.81) |
| R219BII(pNR117) | 0.80 (±0.02) | 1.5 (±0.02) |
Resting cells were prepared from the indicated strains and incubated under identical derepression conditions (see Materials and Methods). Nitrogenase activity was assayed after 2 and 4 h of derepression. Values are means of three independent assays, each performed in duplicate. Numbers in parentheses are standard errors.
In other recombinant strains, the presence of plasmid pNR117 appeared to be responsible for the augmentation of in vivo nitrogenase activity. Assuming that dinitrogenase reductase was fully active under the derepression conditions used (see below), one is left with two possible explanations to account for such an augmentation of enzyme activity: either nitrogenase was synthesized at a higher level in pNR117-containing strains or the enzyme specific activity was enhanced.
To better understand the cause of the enhancement of nitrogenase activity in strains bearing pNR117, the relative nitrogenase content of the cells, as well as the RnfB and RnfC content, was analyzed by immunochemical detection using specific polyclonal antibodies. A comparison of the band intensities on Western blots indicated that the RnfB and RnfC polypeptides were more abundant in pNR117-containing strains than in control strains, whereas dinitrogenase (Rc1) and dinitrogenase reductase (Rc2) polypeptides seemed to reach similar levels (data not shown). To facilitate a quantitative comparison of the nitrogenase and Rnf polypeptides between the strains studied, in vivo radioactive labeling experiments were undertaken.
Overproduction of the rnf gene products in strains carrying pNR117.
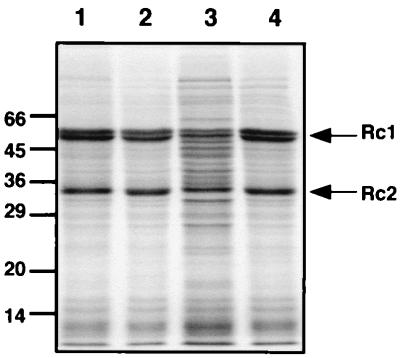
Two representative pNR117-containing transconjugants derived from strains B10S and R368I were subjected to in vivo radioactive labeling experiments. Strains carrying pNF3 were used as controls in these experiments. The labeling was performed by exposing resting cells to a mixture of [35S]Met and [35S]Cys 1 h after the beginning of derepression, at which time in vivo nitrogenase activity increases rapidly (7). Since the nitrogenase subunits are major components of the cell protein extract, the radioactivity incorporated into these polypeptides could be detected after a single analytical separation by SDS-PAGE (Fig. 3). On the other hand, the Rnf polypeptides, which appeared to be minor components of the membrane fraction, were not detectable in this way. They had to be first isolated from the membrane by a procedure involving solubilization of the proteins with a detergent, followed by immunocapture of Rnf polypeptides on immobilized anti-RnfB or anti-RnfC antibodies. Membrane proteins solubilized with lauryl-maltoside were incubated with purified anti-RnfB or anti-RnfC antibodies bound to protein A-agarose. The proteins recovered in this way were analyzed by SDS-PAGE and autoradiography (Fig. 4). From the comparison of the band patterns shown in Fig. 3, it appears that all strains except the rnfC mutant have similar nitrogenase contents. Quantification of the radioactivity incorporated into Rc1 (α and β subunits) and Rc2 indicated that the Rc1 content of strains carrying pNR117 differed by less than 17% from that of the control strain B10S(pNF3), while the Rc2 content differed by less than 10%. These minor variations likely reflect experimental errors and did not correlate with the differences in nitrogenase activity observed in vivo. On the other hand, the rnfC mutant showed a reduced level of both nitrogenase components (by 45% for Rc1 and 33% for Rc2), consistent with previous observations (17). It is concluded that the pNR117-dependent augmentation of nitrogenase activity is not due to an intracellular accumulation of the enzyme. In addition, it can be seen that the Rc2 component shows a single band pattern (33 kDa) indicating that the protein did not undergo significant inactivation by ADP-ribosylation (8, 9). Therefore, the enzyme was probably in the fully active form in all strains.
FIG. 3.
SDS-PAGE analysis of 35S-labeled cytoplasmic proteins from pNR117-carrying strains. Resting cells of strains containing or lacking pNR117 were subjected to 3 h of derepression in the presence of a mixture of 35S-labeled Met and Cys. Protein extracts were prepared and analyzed by SDS-PAGE as described in Materials and Methods. Lanes 1 to 4 compare the radioactive band patterns of the cytoplasmic proteins from strains B10S(pNF3), B10S(pNR117), R368I(pNF3), and R368I(pNR117), respectively. Arrows indicate the nitrogenase polypeptides, the α and β subunits of Rc1 (55 and 59.5 kDa), and the Rc2 subunit (33.5 kDa). Sizes of marker proteins are indicated in kilodaltons.
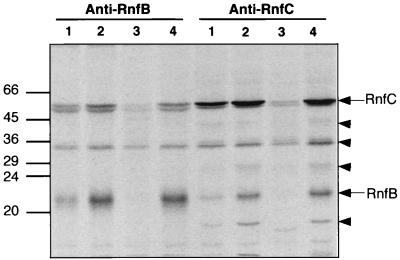
FIG. 4.
35S-labeled membrane polypeptides from pNR117-containing strains immunoprecipitated with anti-RnfB or anti-RnfC antibodies. Four strains were subjected to in vivo radioactive labeling as defined in the legend to Fig. 3. Chromatophores were isolated; proteins were solubilized and then incubated with immobilized anti-RnfB or anti-RnfC antibodies as described in Materials and Methods. Membrane proteins captured by immobilized anti-RnfB antibodies or anti-RnfC antibodies were analyzed by SDS-PAGE on a 12.5% polyacrylamide gel and visualized using a PhosphorImager system. Lanes 1 to 4 were loaded with proteins from strains B10S(pNF3), B10S(pNR117), R368I(pNF3), and R368I(pNR117), respectively. RnfB and RnfC polypeptides are marked with arrows. Triangles indicate coimmunoprecipitated polypeptides. Sizes of marker proteins are indicated in kilodaltons.
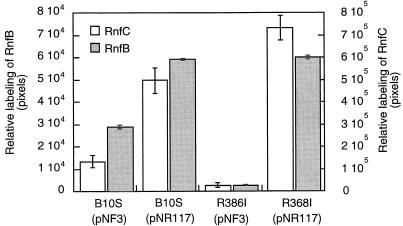
On the other hand, immunoprecipitation experiments using immobilized anti-RnfC antibodies revealed significant variations in the RnfC level in the same four strains (Fig. 4). Based on quantitative estimation of the radioactive bands, transconjugants carrying pNR117 contained twice as much of the RnfC polypeptide as the control strain did (Fig. 5). As expected, the rnfC mutant yielded just background radioactivity at a position corresponding to the migration distance of the RnfC polypeptide. Likewise, when anti-RnfB antibodies were used to probe the radioactivity incorporated in RnfB, the amount of radiolabeled polypeptide recovered from strains carrying pNR117 was fourfold greater than that found in the control strain (Fig. 4 and 5). The rnfC mutant did not contain detectable amount of RnfB, consistent with the immunoblot analysis shown in Fig. 1. These immunocapture experiments provide evidence that transconjugants carrying a copy of the rnfABCDGEH operon driven by the nifH promoter directed effective overproduction of the rnf gene products. The augmentation of the Rnf polypeptide content ranged from twofold for RnfC to fourfold for RnfB. A possible reason for this discrepancy might be that RnfC is only partially recovered upon solubilization from the membrane, in which case a twofold increase in the Rnf polypeptide content should be considered as a minimum.
FIG. 5.
Levels of 35S-labeled RnfB and RnfC polypeptides recovered from strains containing or lacking pNR117. In vivo-labeled membrane proteins from four strains were prepared and then subjected to immunoprecipitation with immobilized anti-RnfC and anti-RnfB antibodies as defined in the legend to Fig. 4. After separation by SDS-PAGE, the radioactivity incorporated in the RnfB and RnfC bands was quantified by integration of the PhosphorImager signals. Standard errors calculated from three independent experiments are shown.
Coimmunoprecipitation of putative Rnf polypeptides with RnfB and RnfC.
In addition to the target antigen, the anti-RnfC antibodies coimmunoprecipitated at least five other 35S-labeled polypeptides, which were absent or yielded much fainter bands in the control rnfC mutant (Fig. 4). One of these additional polypeptides with an apparent Mr of 21,000 was identified as RnfB by Western blot analysis (data not shown). The four other polypeptides had apparent Mrs of 40,000, 34,000, 27,000, and 18,000. These coimmunoprecipitated polypeptides yielded bands much fainter than the protein used as target antigen. This observation indicates that a large proportion of the RnfC protein was released alone from the membrane upon solubilization with detergent or during immunoprecipitation.
When solubilized membrane proteins were immunoprecipitated with anti-RnfB antibodies, only three labeled polypeptides were detectable, one of which was identified as RnfC. However, the amount of coimmunoprecipitated RnfC was low, suggesting that RnfB almost completely dissociated as a free component upon solubilization. This might also explain why the polypeptides coimmunoprecipitated with RnfC were not visible in the protein fraction captured by the anti-RnfB antibodies. The labeled polypeptide with an Mr of 34,000 might be unrelated to the Rnf proteins, as it was present in the control extract (rnfC mutant). These data provide preliminary evidence for the existence of a membrane-bound protein complex consisting of four (possibly five) polypeptides, including RnfB and RnfC.
In vitro nitrogenase activity is not enhanced in strains carrying pNR117.
From the data presented above, it appears that the pNR117-dependent increase of nitrogenase activity observed in vivo is not correlated with a commensurate augmentation of the cellular nitrogenase enzyme level. On the other hand, strains carrying pNR117 showed a higher level of at least two rnf gene products, RnfB and RnfC, suggesting that the greater abundance of the Rnf polypeptides is responsible for the enhancement of in vivo nitrogenase activity. Although this enhancement may be plausibly explained by a higher rate of electron transfer to nitrogenase mediated by the Rnf complex (see Discussion), the possibility remained that it resulted from an indirect effect of the Rnf proteins on the processing of nitrogenase. If this were the case, the level of Rnf proteins should have affected the specific activity of nitrogenase in vitro.
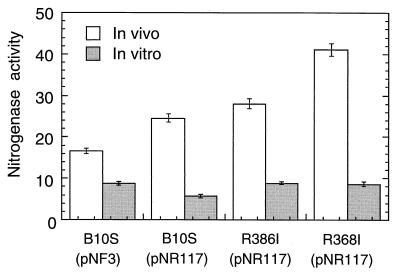
We therefore compared the in vivo and in vitro nitrogenase activities of cells containing or lacking plasmid pNR117 (Fig. 6). Transconjugants carrying pNR117 displayed in vivo nitrogenase activities 1.5- to 2.4-fold higher than that of the control strain B10S(pNF3). In contrast, the dithionite-supported nitrogenase activities measured in vitro were found to be fairly similar in the four strains tested, irrespective of the presence of pNR117. These results clearly show that the higher in vivo nitrogenase activity observed in pNR117-containing transconjugants could not be attributed to an equivalent augmentation of the enzyme specific activity as assayed in vitro.
FIG. 6.
In vivo and in vitro nitrogenase activity of pNR117-containing strains. Transconjugants of strains B10S, R386I (rnfB mutant), and R368I (rnfC mutant) carrying pNR117 were derepressed for 4 h, at which time nitrogenase was assayed both in vivo (white bars) and in vitro (gray bars). Strain B10S(pNF3) was used as a control. For in vitro assays, protein extracts were anaerobically prepared (see Materials and Methods), and sufficient purified Rc2 (1.5 nmol) was added to titrate the Rc1 component present in each extract. Activities are expressed as nanomoles of C2H2 reduced per minute per milligram of dry weight (in vivo assays) or per milligram of protein (in vitro assays). Standard errors calculated from at least five different experiments are shown.
DISCUSSION
In this study, we describe the successful overexpression of the whole rnfABCDGEH operon in R. capsulatus and observe that the augmentation of the level of the Rnf polypeptides resulted in a twofold enhancement of in vivo nitrogenase activity. Three possible explanations could account for this enhancement: (i) elevation of the nitrogenase protein level, (ii) reactivation of a partially inactive enzyme, or (iii) augmentation of the enzyme catalytic turnover.
Previous studies showed that nitrogenase derepression in R. capsulatus was influenced by physiological factors such as the balance of the nitrogen and carbon sources (N/C ratio) and light intensity (4, 6). Light was found to enhance in vivo nitrogenase essentially by stimulating the synthesis of the enzyme (10). A low N/C ratio during growth also promoted a high level of in vivo nitrogenase activity through overproduction of nitrogenase (1, 18). In comparison, results presented above showed that in cells overproducing the Rnf polypeptides, the higher level of nitrogenase was not correlated with an augmentation of the cellular content of the enzyme. Alternatively, an enhanced nitrogenase activity would be expected if the enzyme was partially inactive in control cells and assuming that this inactivation is relieved in rnf-overexpressing cells. It is well known that in R. capsulatus, nitrogenase is subjected to short-term regulation in response to various environmental stimuli. One regulatory mechanism involves ADP-ribosylation of dinitrogenase reductase (Rc2) which renders the enzyme inactive (8, 9). A source of fixed nitrogen (NH4+) or exposure to darkness has been shown to trigger nitrogenase inactivation by ADP-ribosylation (8, 13). Inactive nitrogenase can be distinguished from the active enzyme by SDS-PAGE, because ADP-ribosylated Rc2 gives a recognizable two band pattern (8). In addition, an independent NH4+-mediated inhibition of nitrogenase activity has been described for R. capsulatus (13, 20). Bearing this in mind, the question arose as to whether the observed enhancement of nitrogenase in rnf-overexpressing cells could result from an alteration of the regulation of nitrogenase activity. Although we cannot exclude this possibility, it appears to be unlikely because (i) nitrogenase assays were performed on cells derepressed in N-free medium in high light, conditions known to induce fully active enzyme, and (ii) molecular analysis of Rc2 indicated that it was not ADP-ribosylated. In addition, in vitro assays demonstrated that dinitrogenase had a fairly constant catalytic activity, irrespective of the cellular background. Finally, the enhancement of nitrogenase activity found in rnf-overexpressing cells can be plausibly explained by an increase in the supply of reductants to nitrogenase. This interpretation is consistent with the proposal that the Rnf products participate in electron transport to nitrogenase (5, 12, 17). In addition, our results suggest that because of the relatively low cellular level of the Rnf polypeptides, the Rnf-mediated electron transport is rate limiting for nitrogenase catalytic activity in wild-type R. capsulatus, at least under given experimental conditions.
Another operon located upstream of rnfA is also possibly involved in electron transport to nitrogenase (17). This operon includes two ferredoxin-encoding genes named fdxC and fdxN (2, 16), the rnfF gene, and two nonessential ORFs, ORF10 and ORF14. Only rnfF appeared to be absolutely required for nitrogen fixation (17). The products of the rnfF locus, which may in fact contain two genes (15), do not resemble a known electron transport protein. In addition, a mutation in rnfF does not affect the stability of RnfB and RnfC, suggesting that the product of this gene is not involved in the assembly of the Rnf complex considered in this study (5). Hence, the involvement of the rnfF gene product(s) in electron transport to nitrogenase is unclear. The fdxN gene encodes a ferredoxin (FdI) which was found to serve as electron donor to nitrogenase. Inactivation of fdxN resulted in a dramatic reduction of nitrogenase activity, although nitrogen fixation was not totally abolished (7, 17). FdI is synthesized at a high level in R. capsulatus, but strains producing 10-fold less of this ferredoxin were still capable of nitrogen fixation and displayed wild-type nitrogenase levels (7). Hence, the electron transfer reaction mediated by FdI is probably not rate limiting for nitrogenase activity in vivo. Finally, the product of ORF14 has recently been purified as a dimeric flavin mononucleotide-containing protein belonging to a novel family of bacterial flavoproteins, called FprA (19). FprA serves as electron acceptor for the fdxC gene product, a [2Fe-2S] ferredoxin, suggesting that the two proteins are physiological partners. Since both proteins have relatively high redox potentials (unpublished results), it is unlikely that they participate in electron transfer to nitrogenase.
Overproduction of the Rnf products should facilitate further functional and biochemical characterization of the membrane-bound complex that these proteins are thought to form. Preliminary experiments using membrane proteins solubilized in detergent suggested that RnfC was associated with five polypeptides. One of these proteins were identified as RnfB. It remains to be proven that the other coimmunoprecipitated polypeptides are also rnf gene products. It has so far been impossible to identify these polypeptides because they were recovered in very low amounts from the membrane. A major reason for this low recovery seems to be that RnfB and RnfC are rather loosely attached to the other components, once the membrane proteins have been solubilized with detergent. Since RnfB and RnfC had been found previously to be tightly bound to the chromatophore (5, 12), it seems likely that membrane protein solubilization destabilizes the Rnf complex.
ACKNOWLEDGMENTS
We thank John C. Willison for helpful discussions and critical reading of the manuscript.
This work was supported by grants from the Centre National de la Recherche Scientifique and the Commissariat à l'Energie Atomique and by a doctoral fellowship to H.-S.J. from the Korean government.
REFERENCES
- 1.Arp D J, Zumft W G. Overproduction of nitrogenase by nitrogen-limited cultures of Rhodopseudomonas palustris. J Bacteriol. 1983;153:1322–1330. doi: 10.1128/jb.153.3.1322-1330.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Grabau C, Schatt E, Jouanneau Y, Vignais P M. A new [2Fe-2S] ferredoxin from Rhodobacter capsulatus. Coexpression with a 2[4Fe-4S] ferredoxin in Escherichia coli. J Biol Chem. 1991;266:3294–3299. [PubMed] [Google Scholar]
- 3.Hallenbeck P C, Meyer C M, Vignais P M. Nitrogenase from the photosynthetic bacterium Rhodopseudomonas capsulata: purification and molecular properties. J Bacteriol. 1982;149:708–717. doi: 10.1128/jb.149.2.708-717.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hillmer P, Gest H. H2 metabolism in the photosynthetic bacterium Rhodopseudomonas capsulata: production and utilization of H2 by resting cells. J Bacteriol. 1977;129:732–739. doi: 10.1128/jb.129.2.732-739.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jouanneau Y, Jeong H S, Hugo N, Meyer C, Willison J C. Overexpression in Escherichia coli of the rnf genes from Rhodobacter capsulatus—characterization of two membrane-bound iron-sulfur proteins. Eur J Biochem. 1998;251:54–64. doi: 10.1046/j.1432-1327.1998.2510054.x. [DOI] [PubMed] [Google Scholar]
- 6.Jouanneau Y, Lebecque S, Vignais P M. Ammonia and light effect on nitrogenase activity in nitrogen-limited continuous cultures of Rhodopseudomonas capsulata. Role of glutamine synthetase. Arch Microbiol. 1984;139:326–331. [Google Scholar]
- 7.Jouanneau Y, Meyer C, Naud I, Klipp W. Characterization of an fdxN mutant of Rhodobacter capsulatus indicates that ferredoxin I serves as electron donor to nitrogenase. Biochim Biophys Acta. 1995;1232:33–42. doi: 10.1016/0005-2728(95)00106-x. [DOI] [PubMed] [Google Scholar]
- 8.Jouanneau Y, Meyer C, Vignais P M. Regulation of nitrogenase activity through Fe protein interconversion into an active and an inactive form in Rhodopseudomonas capsulata. Biochim Biophys Acta. 1983;749:318–328. [Google Scholar]
- 9.Jouanneau Y, Roby C, Meyer M, Vignais P M. ADP-ribosylation of dinitrogenase reductase in Rhodobacter capsulatus. Biochemistry. 1989;28:6524–6530. [Google Scholar]
- 10.Jouanneau Y, Wong B, Vignais P M. Stimulation of nitrogenase synthesis in cells of Rhodopseudomonas capsulata grown in N-limited continuous cultures. Biochim Biophys Acta. 1985;808:149–155. [Google Scholar]
- 11.Klipp W, Masepohl B, Puhler A. Identification and mapping of nitrogen fixation genes of Rhodobacter capsulatus: duplication of a nifA-nifB region. J Bacteriol. 1988;170:693–699. doi: 10.1128/jb.170.2.693-699.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kumagai H, Fujiwara T, Matsubara H, Saeki K. Membrane localization, topology, and mutual stabilization of the rnfABC gene products in Rhodobacter capsulatus and implications for a new family of energy-coupling NADH oxidoreductases. Biochemistry. 1997;36:5509–5521. doi: 10.1021/bi970014q. [DOI] [PubMed] [Google Scholar]
- 13.Pierrard J, Ludden P W, Roberts G P. Posttranslational regulation of nitrogenase in Rhodobacter capsulatus: existence of two independent regulatory effects of ammonium. J Bacteriol. 1993;175:1358–1366. doi: 10.1128/jb.175.5.1358-1366.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pollock D, Bauer C E, Scolnik P A. Transcription of the Rhodobacter capsulatus nifHDK operon is modulated by the nitrogen source. Construction of plasmid expression vectors based on the nifHDK promoter. Gene. 1988;65:269–275. doi: 10.1016/0378-1119(88)90463-5. [DOI] [PubMed] [Google Scholar]
- 15.Saeki K, Suetsugu Y, Tokuda K I, Miyatabe Y, Young D A, Marrs B L, Matsubara H. Genetic analysis of functional differences among distinct ferredoxins in Rhodobacter capsulatus. J Biol Chem. 1991;266:12889–12895. [PubMed] [Google Scholar]
- 16.Schatt E, Jouanneau Y, Vignais P M. Molecular cloning and sequence analysis of the structural gene of ferredoxin I from the photosynthetic bacterium Rhodobacter capsulatus. J Bacteriol. 1989;171:6218–6226. doi: 10.1128/jb.171.11.6218-6226.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schmehl M, Jahn A, Meyer zu Vilsendorf A, Hennecke S, Masepohl B, Schuppler M, Marxer M, Oelze J, Klipp W. Identification of a new class of nitrogen fixation genes in Rhodobacter capsulatus: a putative membrane complex involved in electron transport to nitrogenase. Mol Gen Genet. 1993;241:602–615. doi: 10.1007/BF00279903. [DOI] [PubMed] [Google Scholar]
- 18.Steinborn B, Jürgens U J, Oelze J. Control of nitrogenase in chemostat culture of Rhodobacter capsulatus grown on ammonium at different illuminations. Arch Microbiol. 1991;156:135–141. [Google Scholar]
- 19.Wasserfallen A, Ragettli S, Jouanneau Y, Leisinger T. A family of flavoproteins in the domains Archaea and Bacteria. Eur J Biochem. 1998;254:325–332. doi: 10.1046/j.1432-1327.1998.2540325.x. [DOI] [PubMed] [Google Scholar]
- 20.Yakunin A F, Hallenbeck P C. Short-term regulation of nitrogenase activity by NH4+ in Rhodobacter capsulatus: multiple in vivo nitrogenase responses to NH4+ addition. J Bacteriol. 1998;180:6392–6395. doi: 10.1128/jb.180.23.6392-6395.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]