Abstract
Mechanistic target of rapamycin complex 2 (mTORC2) is a multi-subunit kinase complex, central to multiple essential signaling pathways. Two core subunits, Rictor and mSin1, distinguish it from the related mTORC1 and support context-dependent phosphorylation of its substrates. mTORC2 structures have been determined previously; however, important questions remain, particularly regarding the structural determinants mediating substrate specificity and context-dependent activity. Here, we used cryo-EM to obtain high-resolution structures of the human mTORC2 apo-complex in the presence of substrates Akt and SGK1. Using functional assays, we then tested predictions suggested by substrate-induced structural changes in mTORC2. For the first time, we visualized in the apo-state the side chain interactions between Rictor and mTOR that sterically occlude recruitment of mTORC1 substrates and confer resistance to the mTORC1 inhibitor rapamycin. Also in the apo-state, we observed that mSin1 formed extensive contacts with Rictor via a pair of short α-helices nestled between two Rictor helical repeat clusters, as well as by an extended strand that makes multiple weak contacts with Rictor helical cluster 1. In co-complex structures, we found that SGK1, but not Akt, markedly altered the conformation of the mSin1 N-terminal extended strand, disrupting multiple weak interactions while inducing a large rotation of mSin1 residue Arg-83, which then interacts with a patch of negatively charged residues within Rictor. Finally, we demonstrate mutation of Arg-83 to Ala selectively disrupts mTORC2-dependent phosphorylation of SGK1, but not of Akt, supporting context-dependent substrate selection. These findings provide new structural and functional insights into mTORC2 specificity and context-dependent activity.
Keywords: mTORC2, structure, cryo-EM, Akt, SGK1, substrate specificity, conformation change
Abbreviations: CRIM, conserved region in the middle; FRB, FKBP12-rapamycin binding domain; HR, helical repeat; mTOR, mechanistic target of rapamycin; mTORC1, mTOR complex 1; mTORC2, mTOR complex 2; PH, pleckstrin homology; string, SGK-Targeting, Rictor-InteractING
Mechanistic target of rapamycin (mTOR) is a serine/threonine kinase, which belongs to the PI3K-related kinase family and is evolutionarily conserved from yeast to human (1, 2). mTOR signaling is central to a large array cellular processes, mediating responses to hormones and growth factors, nutrient availability, and extracellular ionic milieu (3). mTOR functions in cellular physiology through two structurally and functionally distinct multiprotein complexes, mTOR complex 1 (mTORC1) and mTOR complex 2 (mTORC2) (4, 5, 6). These two protein complexes share two conserved components, mTOR and mammalian lethal with SEC13 protein 8 (mLST8, also known as GβL) (7), as well as several complex-specific components. mTORC1 distinctively contains regulatory-associated protein of mTOR (Raptor) and proline-rich Akt substrate of 40 kDa (PRAS40) (8, 9), while mTORC2 specifically includes rapamycin-insensitive companion of mTOR (Rictor), mammalian stress-activated protein kinase-interacting protein 1 (mSin1). These compositional differences underlie the distinct substrate preferences of the two complexes, as well as their differential responses to the macrolide immunosuppressant, rapamycin; mTORC1 is acutely and potently inhibited by rapamycin, while mTORC2 responds only partially after long-term treatment (10, 11, 12).
The function of mTORC2 has received increasing attention during recent years. The representative downstream effectors of mTORC2 are mainly members of the AGC kinase family, including Akt (also known as protein kinase B), PKC, and serum- and glucocorticoid-induced kinases (SGK1 and SGK3) (13, 14, 15, 16, 17). Upon activation by mTORC2, these AGC kinases have been demonstrated to play fundamental roles in regulating cell metabolism, proliferation, survival, migration, and ion transport (18, 19, 20, 21, 22, 23). Dysregulation of mTORC2 signaling has been reported in some human diseases such as cancer, metabolic disorders, and neurodegenerative diseases (24, 25, 26, 27, 28).
In part, because it was discovered first and in part due to its inhibition by rapamycin, functional and structural research on mTORC1 has outpaced mTORC2. Cryo-EM and X-ray crystallographic structural studies of mTORC1 have substantially elucidated its structural features, in most domains at the atomic level. Important structural determinants of substrate specificity and rapamycin sensitivity have been elucidated. Notably, a domain within mTOR close to the catalytic cleft, termed the FKBP12-rapamycin binding (FRB) domain, binds both rapamycin (in complex with the endogenous FK-506 binding protein, FKBP12) and key mTORC1 substrates, 4EBP1 and S6-kinase (29).
mTORC2 structures have been determined by cryo-EM as well, but multiple key structural features, most notably those that confer context-dependent substrate phosphorylation, remain to be elucidated. Comparison of published structures indicates that the common subunits, mTOR and mLST8, assume very similar conformations in both mTORC1 and mTORC2, suggesting that specificity determinants lie elsewhere. Chen et al. used cross-linking mass spectrometry coupled with a 4.9 Å cryo-EM structure to identify interactions between the mTOR FRB domain and mTORC2 components, mSin1 and Rictor (30). Their data suggested that both of these mTORC2-specific subunits closely interacted with the FRB domain in a manner that could block binding of FKBP12-rapamycin, as well as that of the substrates, 4EBP1 and S6-kinase. However, structural detail was lacking. Scaiola et al. (31) published a higher resolution structure, which provided greater insight into the FRB domain, as well as clarifying the position and interactions of mSin1 with Rictor and mLST8. Furthermore, they showed that mSin1 did not interact directly with mTOR but rather provided an important link between Rictor and mLST8. However, significant detail of the structure of the mSin1 N-terminal region was not visualized. This is particularly important in the context of recruitment of specific substrates. Prior functional and biochemical studies had identified two distinct substrate interaction domains in mSin1: one in the N-terminal region of mSin1 was found to be specific for SGK1 and did not interact with Akt or PKC (32). Mutation of residues within the core of this domain disrupted mTORC2-dependent phosphorylation of SGK1 but not Akt or PKC. A second domain, the CRIM (conserved region in the middle) domain, was found to be required for binding and activation of all three of these substrates (14). The role of the CRIM domain was further elucidated in an NMR-based study (33). However, the N-terminal region has not been further characterized. Structural insights into the N-terminal region and CRIM domain have been elusive.
In this study, we present a representative cryo-EM structure of human mTORC2 apo-complex at overall resolution of 3.2 Å together with structures in the presence of Akt and SGK1. We describe the characteristic assembly that provides a basis for modeling local structural information including recognition of side chains in multiple regions. The structure reveals the basis for mTORC2 insensitivity to rapamycin, demonstrating specific side chain interactions through which Rictor occludes the mTOR FRB domain. Structural determinants of substrate preferences of mTORC1 and mTORC2 are also revealed. Of particular note, new insight into the structural basis for mTORC2 context-dependent regulation of SGK1 and Akt is revealed: in the presence of SGK1, but not Akt, the conformation of the mSin1 N-terminal region is altered, resulting in a large rotation of mSin1/Arg-83 to form a salt bridge with Rictor Asp-1679. The functional importance of this newly recognized structural feature is demonstrated in cell-based phosphorylation experiments.
Results
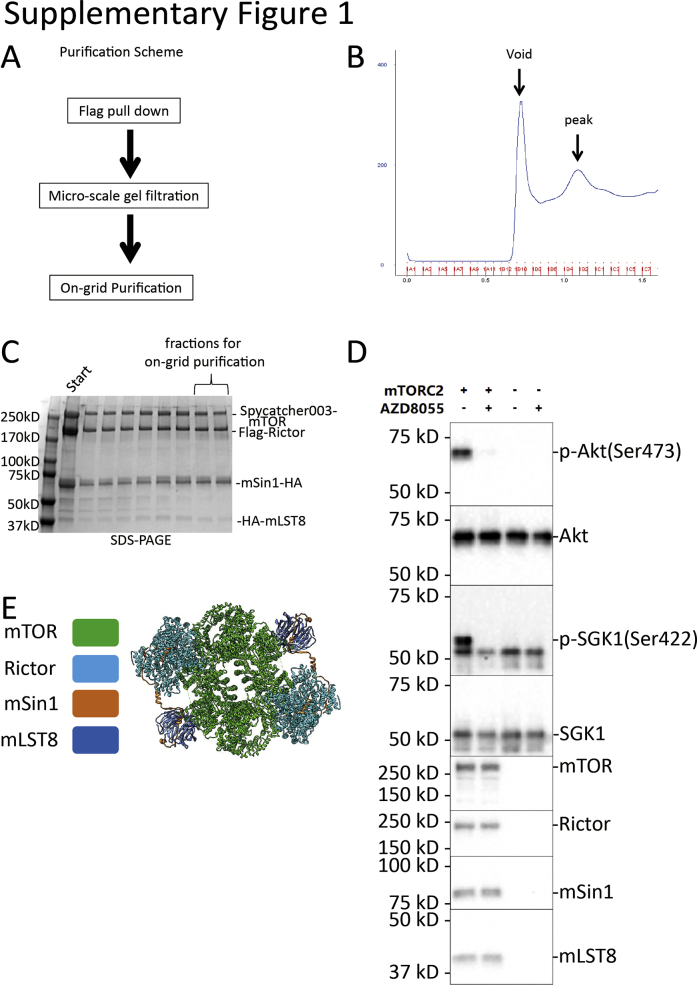
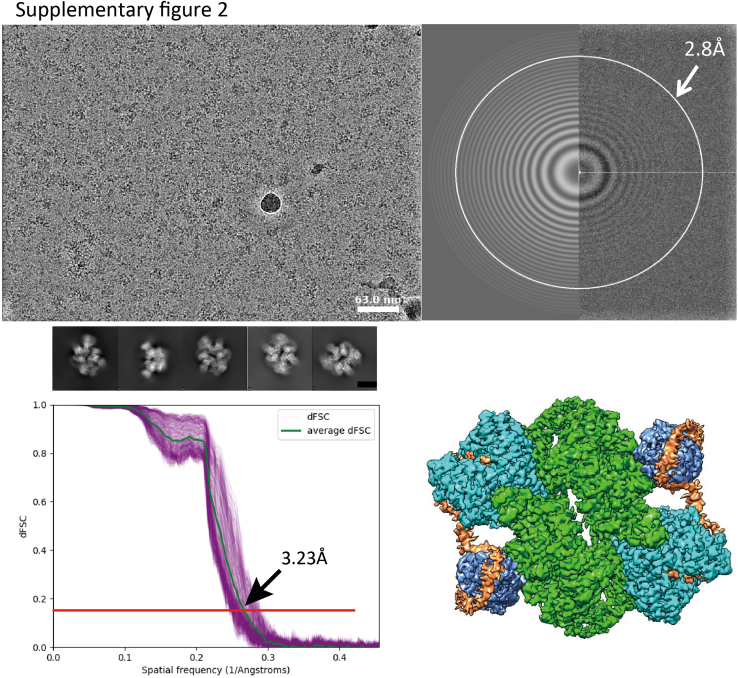
In order to obtain purified mTORC2 core complex for cryo-EM, Spycatcher003-mTOR, Flag-Rictor, HA-mLST8, and mSin1-HA (all of human origin) were coexpressed in Expi293F cells. The complex was purified using one-step flag pull-down, followed by size-exclusion chromatography (Fig. S1, A–C), and finally subjected to on-grid affinity binding using a new adaptation of the Spycatcher-Spytag system (34), before cryo-EM analysis. This combined approach allowed high quality cryo-EM samples to be obtained without needing large-scale expression and purification approaches. Our principal sample was prepared in the apo-state without small molecule additives or peptide substrates. The biochemical activities of purified mTORC2 were tested and verified (Fig. S1D). After 2D and 3D data analysis, approximately 200,000 particles were selected and used for the final refinement and reconstruction (Fig. S2). Based on the density map data, a model was built for the mTORC2 apo-state (Fig. S1E). Similar structures were solved for co-complexes of mTORC2 with two of its principal substrates, Akt and SGK1. We focused on these two substrates in light of their interconnected importance in regulating ion and solute transport, electrolyte and energy metabolism, and impact on renal disease and diabetes (3, 32, 35).
Global aspects of mTORC2 structure
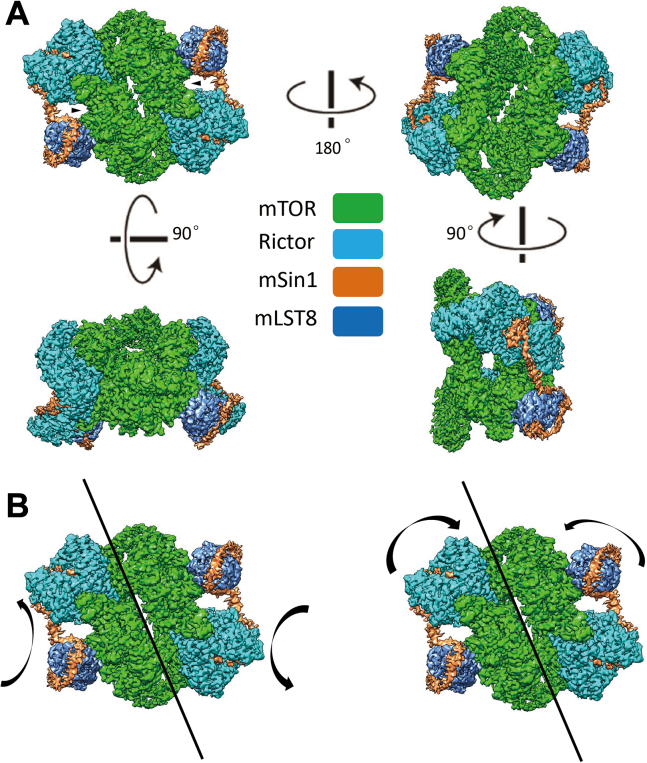
The mTORC2 structure comprises eight subunits consisting of two heterotetramers showing a 2-fold symmetry, similar to a prior report (30). The overall shape of the apo-complex is that of a rhomboid with a hollow center and a dimension of 220 × 160 × 140 (Å3) (Fig. 1, A and B). Each of the heterotetramers consists of mTOR, Rictor, mLST8, and mSin1 with a stoichiometry of 1:1:1:1. The two mTOR subunits form a dimer that serves as the foundation of the complex. mLST8 and Rictor sit in close proximity on its two distal ends. The structures of mLST8 and mTOR as well as their mutual interaction pattern are quite similar to that in mTORC1 (29, 36). The kinase loop of mTOR is located in the cleft between mLST8 and Rictor. Interestingly, as further addressed below, mSin1 forms a long bridge-like structure connecting mLST8 and Rictor, which has an extended α-helix that crosses over the deep cleft harboring the mTOR active site. 3D variability analysis showed that mTORC2 is not a rigid complex, and the two halves were found to “breathe” in two modes, mainly twisting or squeezing toward the geometric core of the complex (Fig. 1B). Both movements result in the altering of the interaction between the two symmetric mTOR subunits, as shown dynamically in supplemental movies (Movie S1 and S2).
Figure 1.
The overall structure of mTORC2. A, cryo-EM structure of mTORC2 at overall resolution 3.2 Å was reconstructed. The sample was prepared in the apo-state. Four views of the structure are presented, and the rotation axis and degree between views are indicated. The subunits are colored as indicated: mTOR (green), Rictor (cyan), mLST8 (blue), and mSin1 (orange). The kinase loop of mTOR is shown by arrows. B, movement of the two heterotetramers of mTORC2 along the central axis are in two directions, twisting (left panel) and squeezing (right panel) toward the geometric core of mTORC2. See also supplementary data for movies. mTORC2, mTOR complex 2; mTOR, mechanistic target of rapamycin.
Specific side chain interactions underlie Rictor contribution to mTORC2 substrate specificity and resistance to rapamycin
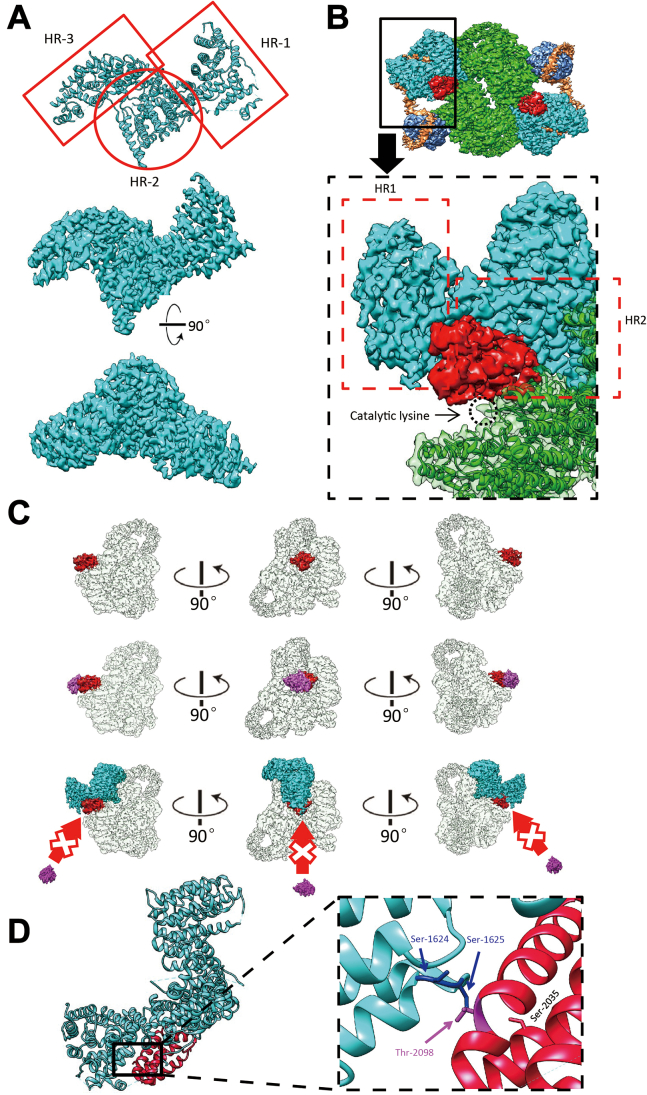
In the apo-complex, the two mTOR subunits are anchored to each other, forming the core of the complex and having a conformation quite similar to that in mTORC1 (Fig. 1A). The structure of Rictor includes three HEAT (Huntingtin, Elongation factor 3, protein phosphatase 2A, and the yeast kinase TOR) helical repeat (HR) clusters termed HR1, HR2, and HR3 (Fig. 2A), all of which make direct contacts with mTOR. HR1 and HR2 also interact with mSin1. Consistent with previous biochemical and lower resolution cryo-EM results, Rictor and mLST8 do not directly interact, but rather are held together by mSin1.
Figure 2.
Rictor interaction with mTOR blocks rapamycin accessibility and influences mTORC2 substrate specificity. A, the atomic model based on density map generated by cryo-EM (upper panel). The high-resolution density map ensures the accuracy of the model fitted from two representative views (mid and lower panel). B, the cryo-EM density of FRB domain is colored in red in the overall density map (upper panel). In the close-up view, HR1 and HR2 of Rictor are highlighted by red-dashed rectangle frames, and the mTOR catalytic lysine is highlighted by a black-dashed circle located in the kinase cleft as indicated. To focus on the interaction of Rictor with FRB domain, mSin1 is not shown here. C, upper panel: three views of the density map of mTOR (transparent except FRB domain shown in red). Middle panel: putative FKBP12-rapamycin binding with FRB domain regardless of Rictor. Lower panel: FRB domain becomes inaccessible to FKBP12-rapamycin in the presence of Rictor. D, the atomic model of Rictor (cyan) and FRB domain of mTOR (red), as shown in the left panel. The close-up view of the local area illustrates where Thr-2098 on FRB domain and its relevant Ser-1624 and Ser-1625 of Rictor are located (right panel). Ser-2035 is shown as well. mTORC2, mTOR complex 2; mTOR, mechanistic target of rapamycin; FRB, FKBP12-rapamycin binding domain; HR, helical repeat.
In Figure 2B, the FRB domain of mTOR is highlighted in red to call attention to its interaction with Rictor. In mTORC1, this domain is adjacent to both the kinase active site and to the Raptor N-terminal conserved) domain, which plays a central role in substrate recruitment (S6-Kinase and 4EBP (29)), and is involved in binding the FKBP12-rapamycin inhibitory complex (36). In mTORC2, the FRB domain is similarly located adjacent to the mTOR kinase active site, but also interacts with both HR1 and HR2 of Rictor. Importantly, HR1 of Rictor sterically prevents the FRB domain from interacting with the FKBP12-rapamycin complex (Fig. 2C), while in mTORC1, the FRB domain remains accessible (Fig. S6). This steric occlusion also prevents mTORC2 interaction with mTORC1-specific substrates and shielding a key interaction residue, Thr-2098 (37). Rictor/Ser-1624 and Ser-1625 were observed in close proximity to mTOR/Thr-2098, likely contributing to the interaction of Rictor HR1 and the FRB domain through hydrogen bond formation, and hence to the differential specificities of mTORC1 and mTORC2 (Fig. 2D). Furthermore, mTOR/Ser-2035, which was previously shown to play an important functional role in substrate recognition by mTORC1 (29, 38), is spatially shielded by Rictor, although there is no direct interaction between them (Fig. 2D). Finally, neither SGK1 nor Akt had any observable effect on the relationship of Rictor and mTOR.
mSin1 engages in key interactions with both Rictor and mLST8
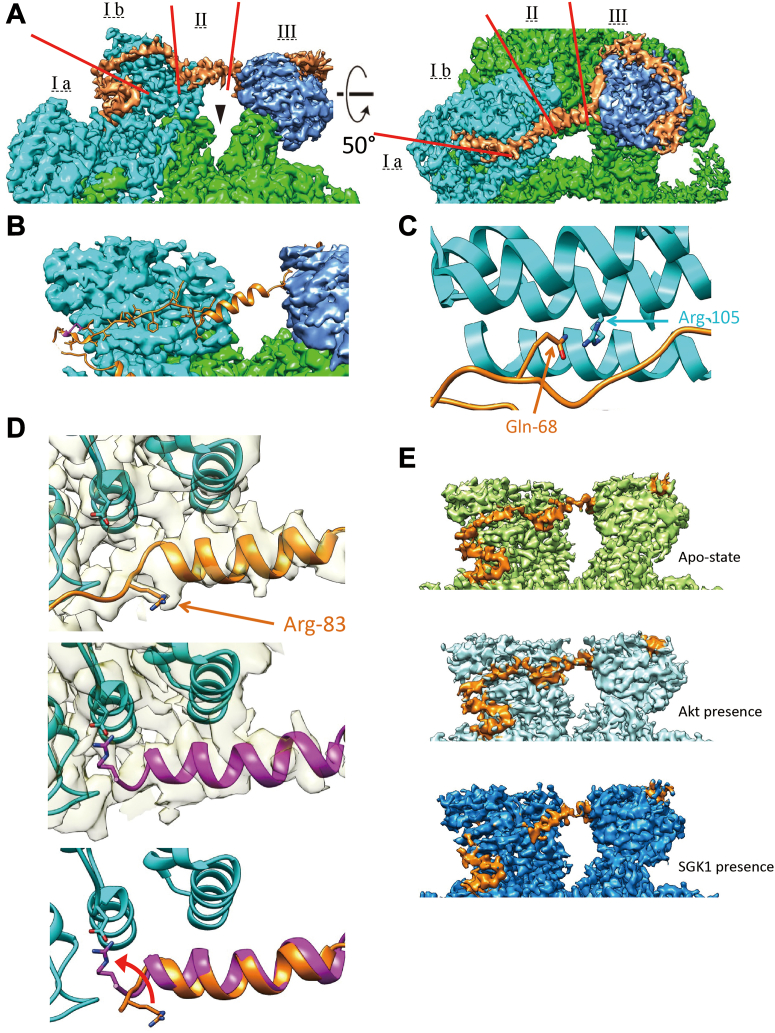
Based on functional and biochemical data, mSin1 contains four principal domains: an N-terminal domain, CRIM domain, Ras-binding domain, and PH (pleckstrin homology) domain (39). For the present apo-complex, we solved the major structure of the N-terminal domain, while the other three domains are invisible in the current density map likely due to their flexibility, as has been noted in prior publications (30, 31). Interestingly, one of the nonvisualized domains, the CRIM domain, has been implicated in binding of all three major mTORC2 substrates, Akt, SGK, and PKC. The N-terminal domain (aa 1–137) contains three distinct sections: a Rictor-interacting section, a bridge section connecting Rictor and mLST8, and an elongated section wrapping around mLST8 (Fig. 3A). The Rictor-interacting section includes a pair of short α-helices nestled between HR1 and HR2 of Rictor, followed by an extended strand, which runs along the periphery of HR1, making multiple weak contacts (Fig. 3, A and B). This region, which we have termed the string (SGK-Targeting, Rictor-InteractING) domain has not been visualized in prior density maps, which is notable in light of the role it plays in substrate binding. Gln-68, in particular, was previously shown to be necessary for SGK1—but not Akt or PKC—interaction with and phosphorylation by mTORC2 (32). In the apo-complex, the Gln-68 side chain is well resolved and seen to reach toward Rictor/Arg-105, consistent with a stabilizing interaction (Fig. 3C). At the C-terminal end of the string domain, we see Thr-86 embedded in a negatively charged pocket of the Rictor C-terminal region and specifically interacting with Rictor/Glu-1675. Interestingly, Thr-86 lies within the sequence RRRSNT, which conforms to the consensus substrate peptide for AGC kinase (sequence: RXRXXS/T), and has been shown previously to be phosphorylated by Akt and S6-kinase (40); however, in the present structures (including co-complexes), it is unphosphorylated. Downstream of the string domain, an α-helical “bridge” begins immediately C-terminal to Thr-86 and crosses the deep cleft harboring the mTOR catalytic site. After this traverse, mSin1 engages in extensive interactions with mLST8, beginning with a short complementary β-strand, followed by an extended section, which wraps approximately ¾ of the way around the peripheral surface of mLST8, and finally the whole visible part of mSin1 ends in a short α-helix before merging into the nonvisualized CRIM domain (Fig. 3A).
Figure 3.
Structural details of the N-terminal domain of mSin1 (orange).A, the N-terminus of mSin1 is shown in two views, comprising a Rictor-interacting section (I), a bridge section connecting Rictor and mLST8 (II) and an mLST8-interacting section (III). The Rictor-interacting section is divided into a pair of short α-helices (I a) and an extended “string” domain (I b, see text for details). The kinase cleft of mTOR is shown by arrowhead. B, string domain stays at the periphery of Rictor HR1, shown in the atomic model. C, mSin1/Gln-68, as a key amino acid to mediate SGK1 interaction with and phosphorylation by mTORC2, points toward Rictor/Arg-105 in the apo-state. D, the mSin1 Arg-83 side chain and nearby backbone with density map semi-transparent in the background are shown (upper and middle panels); nearby Rictor sequences are also seen (colored in cyan), including side chain of Asp-1679. Note marked difference in position of Arg-83 side chain in the absence (top panel) and presence (middle panel) of SGK1. mSin1 is modeled in orange in the apo-complex and purple in the SGK1 co-complex. Lower panel shows a superposition of the apo- and co-complexes (with density map hidden for clarity), emphasizing the SGK1-induced rotation in Arg-83 and formation of a salt bridge with Rictor/Asp-1679. E, mSin1 string domain is observed in the apo-complex (top panel) and co-complex with Akt (middle panel) of mTORC2 but becomes unobservable in the co-complex with SGK1 (bottom). mTORC2, mTOR complex 2; mTOR, mechanistic target of rapamycin; HR, helical repeat; string, SGK-Targeting, Rictor-InteractING.
The mSin1 N-terminal domain changes conformation in the presence of SGK1 but not Akt and a new side chain interaction with Rictor is observed
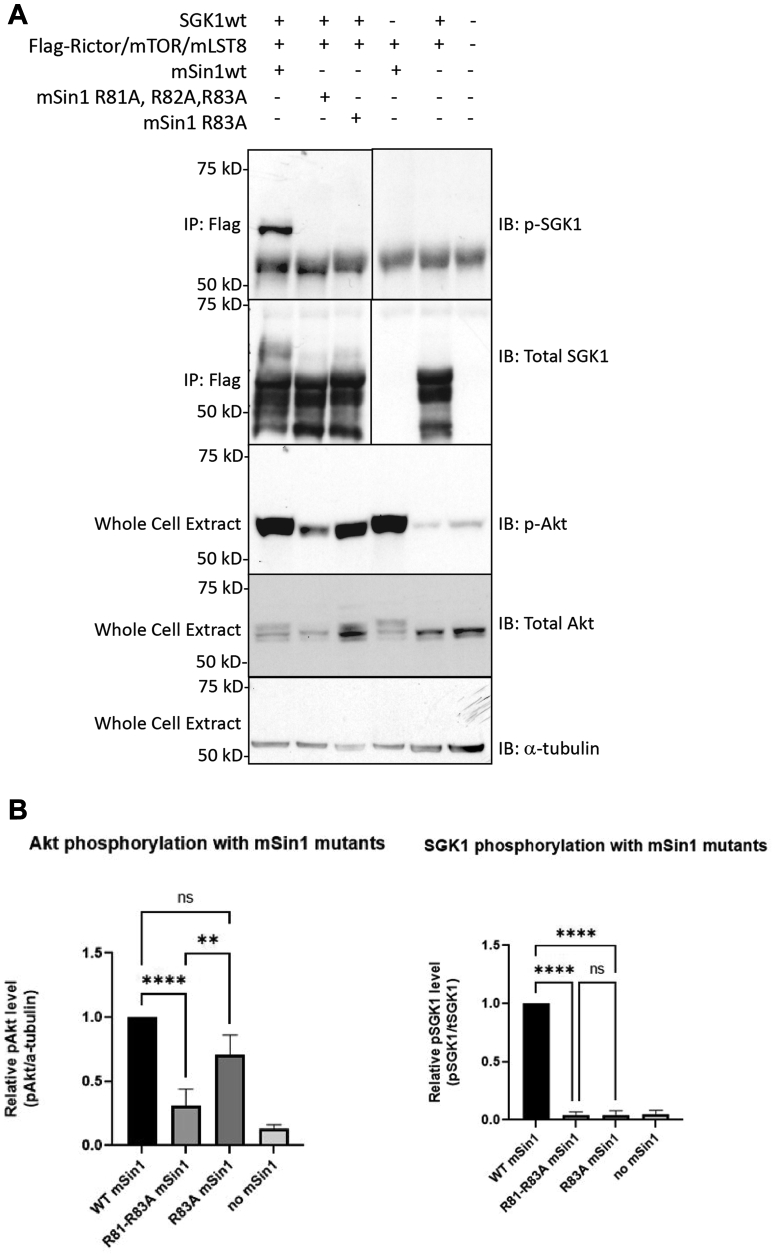
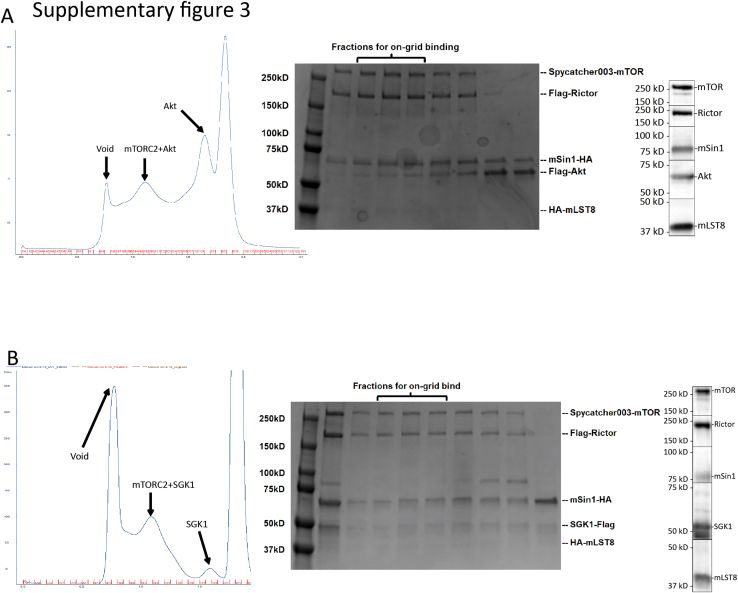
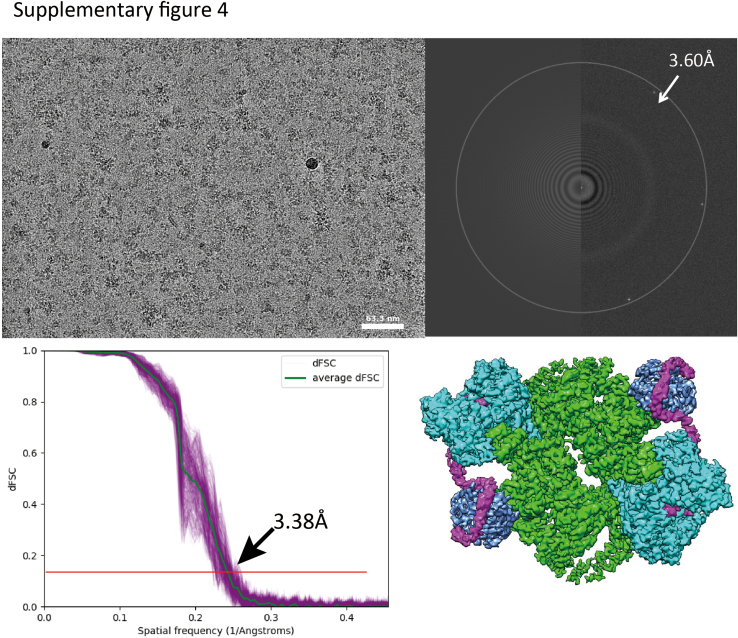
In the co-complexes with Akt and SGK1, the substrates themselves were not clearly resolved, likely due to movement and flexibility of the substrate-binding domains themselves or as a consequence of freezing. Although extra densities consistent with substrates were seen in the co-complexes after low pass filter at 8 Å in order to remove the noise from high frequency, these densities could not be clearly distinguished from densities seen in the apo-complex under similar conditions (not shown). Hence, it is also possible that the extra densities are part of downstream domains of mSin1, such as the CRIM or PH domains (which were not resolved in the high resolution structures). In contrast, biochemical data demonstrated the physical interaction of the substrates with mTORC2 (Fig. S3) and, although the overall volume and shape of the co-complexes were similar to the apo-complex (Figs. S4 and S5), the detailed relationship of the mSin1 N-terminal domain with Rictor was markedly altered in the SGK1 co-complex. First, the string domain upstream of Arg-83 including Gln-68 becomes unobservable, likely due to increased flexibility. Akt, in contrast, had no such effects: the string domain in its presence looks very similar to the apo-complex (Fig. 3E, second panel). Second, the Arg-83 side chain shows a large rotation toward a negatively charged patch of Rictor HR3, coming into close proximity to Asp-1679, with which it appears to form a salt bridge (Fig. 3D, middle and lower panel). In order to explore the functional role of Arg-83 in differential regulation of SGK1 and Akt, we tested the ability of mSin1/Arg-83-Ala to support mTORC2 kinase activity by using mSin1-deficient HEK-293T cells (41). These cells are defective in phosphorylation of both SGK1 and Akt, which is restored by WT mSin1 (41) (Fig. 4). Consistent with the specific role of mSin1/Arg-83 in SGK1 phosphorylation, Akt but not SGK1 phosphorylation was restored by mSin1/Arg-83-Ala. Together with prior literature (16, 32), these data support a model in which SGK1 binding to the string domain disrupts several weak interactions with Rictor (including mSin1/Gln-68 with Rictor/Arg-105) while concomitantly inducing a conformational change that results in a new stabilizing interaction.
Figure 4.
mSin1/Arg-83 is required for SGK1 but not Akt phosphorylation.A, Western blot analysis of FLAG-immunoprecipitates (IPs) and whole-cell extracts (WCEs) derived from mSin1 KO HEK293T cells cotransfected with Myc-tagged Rictor, SpyCatcher003-tagged mTOR, HA-tagged mLST8, HA-tagged mSin1 (WT, R81A/R82A/R83A triple mutant and R83A single mutant), and FLAG-tagged SGK1. Cells were transfected and then serum starved overnight and stimulated with 100 nM insulin for 60 min. Whole cell extracts were prepared and subjected to immunoprecipitation with anti-FLAG antibody followed by immunoblotting (IB) as shown and further described in “Experimental procedures”. Blots were probed with antibodies against phosphorylated SGK1 S422 (Fig. 4A-source data 1), total SGK1 (Fig. 4A-source data 2), phosphorylated Akt (Fig. 4A-source data 3), total Akt (Fig. 4A-source data 4), and α-tubulin (Fig. 4A-source data 5), respectively. B, quantification of band intensities from the Western Blot data in (A), presented as column graph of mean ± SD from three independent experiments. ∗∗p < 0.01; ∗∗∗∗p < 0.0001 by one-way ANOVA. NS, not significant. mTOR, mechanistic target of rapamycin.
Discussion
mTORC1 and 2 behave as distinct signaling kinases despite sharing a common core catalytic subunit, mTOR. Previous work on the mTORC1 structure has provided a detailed picture of key specificity determinants with high resolution, using a combination of X-ray crystallography and cryo-EM (29, 36). A previous structure of mTORC2 using cryo-EM reached a resolution of 4.9 Å (30), and a paper that was published while this article was in preparation reached a resolution of 3.0 Å (31). Our present reconstruction confirms key features identified in those structures and reveals structural features not previously visualized, providing new mechanistic insight. Instrumental in obtaining our more comprehensive density map was the use of a recently developed on-grid capture purification system utilizing the self-ligating “Spycatcher-Spytag system”. To our knowledge, this is the first adaptation of this technology to purifying a large protein complex (∼1.2 MDa) for cryo-EM study, as described in detail in Experimental procedures and Results. The approach significantly concentrates the sample on the grid, minimizes nonspecific binding, and protects the delicate mTORC2 particles from both the air–water interface and the graphene oxide surface. The net effect is to enhance the reconstruction resolution and to improve the density quality of peripheral parts of the complex.
In the overall structure, we show that mTORC2 is not a rigid structure, but rather using 3D variability analysis, we visualized the two halves moving in dynamic “breathing” modes. This movement likely represents a correlate of the dynamic flexibility of mTORC2 and may contribute to the differential behavior of mTORC1 and mTORC2. It seems likely to play a role in limiting the resolution of the overall structure of mTORC2 in the present and earlier structures (30, 31). Further studies are required to investigate the biological significance of these movements, which may answer some remaining questions such as whether the interaction patterns of the two mTOR monomers differ between mTORC1 and mTORC2 (30). Movement of the mSin1 N-terminal region is associated with its propensity to become increasingly flexible in response to one of its substrates, SGK1, as addressed further below.
Our structure provides novel insight into the atomic details of the interaction between Rictor and the mTOR FRB domain, which is critical both for the relative rapamycin insensitivity of mTORC2 and for its recruitment of S6-kinase and 4EBP1. In particular, specific contacts were observed between Ser-1624 and Ser-1625 of Rictor and mTOR/Thr-2098 (Fig. 2D), which preclude interaction with FKBP12-rapamycin or mTORC1 substrates (37). We also confirmed close apposition of Rictor/Val-1627 and mTOR/Ser-2035 that likely blocks access as well (Fig. 2D). Thus, Rictor plays a central role in mTORC2 specificity by limiting access to mTORC1 substrates and both endogenous and pharmaceutical small molecule ligands (10, 11).
While Rictor plays a central role in blocking access of mTORC1 substrates, mSin1 appears to be the principal mTORC2 component mediating substrate recruitment. The CRIM domain of mSin1 has been shown to interact with all three major mTORC2 substrates (14, 33), and we showed previously that the N-terminal region interacts selectively with SGK1 (32). Our structure now reveals for the first time the N-terminal extended strand or “string” domain of mSin1 (aa 66–86), which includes Gln-68 shown previously to be required specifically for activation of SGK1, but not Akt or PKC (32). Here, we show that in the apo-complex, the Gln-68 side chain reaches toward Rictor/Arg-105, consistent with a stabilizing interaction. Additionally, we observe multiple side chains between Gln-68 and Thr-86 forming weak interactions with Rictor HR1. Thr-86, the side chain of which interacts with Rictor/Glu-1675, constitutes the beginning of the α-helical bridge segment and provides an important anchor for the bridge, which traverses the deep catalytic cleft toward mLST8. It is notable in this context that phosphorylation of Thr-86 has been suggested to impair mTORC2 integrity (40). This issue is, however, controversial as others have found that phosphorylation of Thr-86 enhances mTORC2 activity (42). The structural correlates of this apparently context-dependent effect of Thr-86 phosphorylation are unclear at this time, as Thr-86 was unphosphorylated in our structures and those of others (31).
Comparison of apo-complex and co-complex structures reveals a marked effect of SGK1 on mTORC2 conformation in the string domain: Arg-83, which is quite far from any potential interacting residue in the apo-complex, undergoes a large rotation toward a negative patch in HR1, which includes Rictor/Asp-1679, with which it appears to form a salt bridge. Concomitant with this conformational change, the string domain upstream of Arg-83 becomes invisible, likely due to increased flexibility as it interacts with SGK1 and comes free from its moorings to Rictor HR1. We propose that the new Arg-83–Asp-1679 interaction replaces other (weaker) string domain interactions to maintain the stability of mTORC2 and further suggest that the substrate-induced increase in string domain flexibility allows it to coordinate with the CRIM domain in binding SGK1 and present the hydrophobic motif to the mTOR catalytic cleft. The functional importance of Arg-83 is further supported by the observation that an Ala mutant of that residue selectively disrupts SGK1 but not Akt phosphorylation. In the co-complexes with Akt and SGK1, the substrates themselves were not clearly resolved and hence this remains an important goal of future work. It is notable that a prior publication in which an Akt-mTORC2 co-complex was subjected to cryo-EM failed to visualize that substrate (31), consistent with high mobility of the mSin1 substrate-binding domain.
It is of interest to consider the implications of our structural observations for mTORC2 activation of its distinct substrates. Akt, SGK, and conventional PKCs are all mTORC2 targets; however, there are significant differences in the subcellular location of activation, relevant upstream effectors, and physiological activating conditions (3, 41). Indeed, recent data have identified hormonal and nonhormonal effectors that selectively stimulate mTORC2 phosphorylation of SGK1 but not Akt (22, 41). Akt regulation is substantially controlled by its N-terminal PH domain, which binds to phosphatidylinositol-3,4,5-triphosphate, mediates colocalization with mTORC2 at the plasma membrane, and together with CRIM domain interaction brings kinase and substrate together (14, 43). SGK1 has a distinct N-terminal domain, which binds monophosphorylated phosphoinositides but not phosphatidylinositol-3,4,5-triphosphate (44). Interaction with both the mSin1 string and CRIM domains is essential for its activation (14, 32). It seems likely that string domain binding and the ensuing conformation change in mSin1 play an important role in selective hormonal regulation of SGK1 (for example by angiotensin II) (41) and responses to internal physiological stressors such as severe cold (45) or rapid changes in electrolytes (22). Further studies will be needed to fully visualize the detailed interactions of SGK1 with mSin1 that support its selective regulation by mTORC2.
Experimental procedures
Reagents
Antibodies: anti-Rictor (2114, CST), anti-mTOR (2972, CST), anti-HA (3724, CST), anti-Akt (4691, CST), anti-phospho-Ser473 Akt (4060, CST), anti-phospho-Ser422 SGK1 (SC-16745-R, SCBT), and horseradish peroxidase–labeled anti-rabbit secondary antibodies (7074, CST).
Other reagents: anti-FLAG M2 agarose affinity resin (A2220, Sigma), 3×FLAG peptide (F4799, Sigma), Expi29 expression medium (A1435101, Thermo Fisher Scientific), ExpiFectamine 293 transfection kit (A14525, Thermo Fisher Scientific), Opti-MEM I reduced serum medium (31985062, Thermo Fisher Scientific), dulbecco’s modified eagle medium (DMEM) with high glucose (4.5 g/Liter) (CCFAA005, UCSF Cell Culture Facility), fetal bovine serum (11650, Atlanta Biologicals), penicillin-streptomycin (30-002-CI, Corning), L-glutamine (25-005-CI, Corning), trypsin (25-052-CI, Corning), PEI (24765, Polysciences), complete EDTA-free protease inhibitor cocktail (11697498001, Roche), PhosSTOP phosphatase inhibitor (04906837001, Roche), Bradford assay reagent (1856209, Thermo Fisher Scientific), ECL Western blotting detection reagent (RPN2106, GE Heathcare), Lambda protein phosphatase (P0753, NEB), AZD8055 (S1555, Selleckchem), Superose 6 increase 3.2/300 Gl (29091598, GE Healthcare), and Superdex 200 increase 3.2/300 Gl (28990946, GE Healthcare).
Protein expression and purification
To produce human mTORC2 protein, plasmids pcDNA3-FLAG-mTOR, pRK5-myc-Rictor, pcDNA3-mSin1(1.1)-HA, pRK5-HA-mLST8, and pDEST14-SpyCatcher003 were gifts from Jie Chen (Addgene plasmid #26603), David Sabatini (Addgene plasmid #11367), Jie Chen & Taekjip Ha (Addgene plasmid #73388), David Sabatini (Addgene plasmid #1865), and Mark Howarth (Addgene plasmid #133447), respectively. Site-directed mutagenesis was performed on pRK5-myc-Rictor to replace its original tag and generate pRK5-FLAG-Rictor. The ORF of SpyCatcher003 was subcloned from pDEST14-SpyCatcher003 into pcDNA3-FLAG-mTOR to replace its original tag and generate pcDNA3-SpyCatcher003-mTOR plasmid. All DNA constructs were verified by DNA sequencing.
Expi293F cells (A14527, Thermo Fisher Scientific) were grown in Expi29 expression medium in sterile polycarbonate Erlenmeyer flasks (89095, VWR) in a shaker (MaxQ 416 HP, Thermo Scientific Scientific) at 120 rpm at 37 °C and 8% CO2. The four plasmids, pcDNA3-SpyCatcheer003-mTOR, pRK5-FLAG-Rictor, pcDNA3-mSin1(1.1)-HA, and pRK5-HA-mLST8, were cotransfected in 100 μg per 100 ml Expi293F cells using ExpiFectamine 293 transfection kit and Opti-MEM I reduced serum medium, according to manufacturer's instructions. After cultured for 72 h, cells were harvested and lysed with lysis buffer (0.1% CHAPS, 40 mM Hepes pH 7.5, 1 mM EDTA, 120 mM NaCl, 50 mM NaF, 10 mM Na pyrophosphate, 10 mM glycerophosphate) containing complete protease inhibitor cocktail at 4 °C for 30 min. After centrifugation at 17,000g, 4 °C for 10 min, the supernatant was collected and added to anti-FLAG M2 agarose affinity resin. After incubation with rolling for 2 h at 4 °C, the resin was thoroughly washed with wash buffer (30 mM Hepes pH 7.4, 0.5 mM EDTA, 200 mM NaCl), and the protein complex was eluted from the resin with 0.25 mg/ml 3×FLAG peptide in elution buffer (30 mM Hepes pH 7.4, 0.5 mM EDTA, 200 mM NaCl, 1.0 mM TCEP) for 30 min at 4 °C. The protein was concentrated to 8.0 mg/ml using 100k molecular weight cutoff Amicon columns (UFC510024, Millipore) and further purified by gel filtration chromatography (Superose 6 increase 3.2/300 Gl) in the buffer containing 30 mM Hepes pH 7.4, 200 mM NaCl, and 0.5 mM EDTA. The peak fractions were concentrated to 1.1 mg/ml and ready for cryo-EM studies. For kinase assay, 10% (v/v) glycerol was added to purified protein aliquots for storage at −80 °C.
To produce inactive human kinase–dead Akt1 and WT SGK1 protein, site-directed mutagenesis was conducted on pcDNA3-HA-Akt1-K179M (Addgene plasmid #73409) and pcDNA3-SGK1-V5-6×His to change their original tags into FLAG tag. 293T cells were grown in DMEM supplemented with 10% fetal bovine serum, 1% L-glutamine, and 1% penicillin-streptomycin at 37 °C, 5% CO2 and transfected with the plasmid pcDNA3-FLAG-Akt1-K179M into 293T cells using PEI. Expi293F cells were cultured and transfected with the plasmid pcDNA3-SGK1-FLAG as described above. Both cells were harvested and lysed in lysis buffer (0.1% CHAPS, 40 mM Hepes pH 7.5, 1 mM EDTA, 120 mM NaCl, 50 mM NaF, 10 mM Na pyrophosphate, 10 mM glycerophosphate) containing complete protease inhibitor cocktail at 4 °C for 30 min. The clarified cell lysates were incubated with anti-FLAG M2 agarose affinity resin for 2 h at 4 °C and eluted with 0.25 mg/ml 3×FLAG peptide in elution buffer (30 mM Hepes pH 7.4, 0.5 mM EDTA, 200 mM NaCl, 1.0 mM TCEP) for 30 min at 4 °C. Akt1 and SGK1 were concentrated to 4.5 mg/ml and 3.5 mg/ml respectively, using 30k molecular weight cutoff Amicon columns (UFC503024, Millipore). To dephosphorylate them, proteins were incubated with lambda phosphatase for 1 h at room temperature and overnight at 4 °C. Dephosphorylated Akt1 and SGK1 were further purified by gel filtration chromatography (Superdex 200 increase 3.2/300 Gl) in the buffer containing 30 mM Hepes pH 7.4 and 200 mM NaCl. The peak fractions were concentrated to 1.7 mg/ml for Akt1 and 0.52 mg/ml for SGK1, directly forwarded to co-complex construction with mTORC2, or added with 10% (v/v) glycerol and stored at −80 °C for kinase assay.
To purify mTORC2+Akt co-complex and mTORC2+SGK1 co-complex, the raw mTORC2 protein after 3×FLAG peptide elution and concentration, ∼8.0 mg/ml, was incubated with Akt or SGK1 purified above at molar ratio of 1:2. After incubation for 2 h at 4 °C, the mixture was further purified by gel filtration chromatography (Superose 6 increase 3.2/300 Gl) in the buffer containing 30 mM Hepes pH 7.4, 200 mM NaCl, and 0.5 mM EDTA. The peak fractions of expected molecular weight were checked by SDS-PAGE and Coomassie blue staining, and fractions containing the correct co-complex were concentrated to 0.5 mg/ml and ready for cryo-EM studies.
In vitro phosphorylation assay
The in vitro kinase assays were performed in the reaction buffer containing 25 mM Hepes (pH 7.4), 100 mM potassium acetate, and 2 mM MgCl2. In a 45 μl reaction system, 0.55 μg–purified mTORC2 was mixed with 0.5 μg–purified Akt1 (K179M) or SGK1 and added with 10 μM AZD8055 where indicated. Reactions were initiated by adding 0.5 mM ATP and incubated for 30 min at 37 °C. Reactions were stopped by adding Laemmli sample buffer and followed by SDS-PAGE and immunoblotting.
Immunoprecipitation
Knock-out of mSin1 HEK293T cell was described before [ref Gleason 2019 JCS]. mSin1 KO HEK293T cells were transfected by the plasmids in this study by PEI following the provider’s instruction. Transfected cells were grown for protein expression, then lysed in binding buffer (50 mm Tris–HCl, pH 7.5, 10% glycerol, 1 mm EDTA, 2 mm DTT, 150 mm NaCl, and 0.1% CHAPS) for 30 min. After centrifugation, the supernatants were collected and incubated with the anti-FLAG M2 agarose affinity resin. The immunoprecipitates were collected by centrifugation, washed three times, and boiled for 5 min in 50 μl of cracking buffer (50 mm Tris–HCl, pH 7.0, 10% glycerol, 2% SDS, 2% β-mercaptoethanol). Immunoblotting was carried out by separating the immunoprecipitates on 10% polyacrylamide gels as described using a Bio-Rad Mini-Gel apparatus and transferring them electrophoretically to Hybond-C Extra membranes (GE Healthcare) using a Trans-Blot apparatus (Bio-Rad). The membranes were incubated to block nonspecific binding in 5% nonfat dry milk in T-PBS (1.5 mm KH2PO4, 8 mm Na2HPO4, 2.7 mm KCl, 130 mm NaCl, and 0.1% Tween 20) with gentle agitation for 1 h at room temperature and probed by Western blotting (for endogenous and transfected proteins, as described in the figure legends), using antibodies against SGK1 (Sigma), phospho-SGK1 (S422) (Santa Cruz Biotechnology), Akt, phospho-Akt, mSin1 (Bethyl Laboratories). After washing with T-PBS, the membranes were incubated with peroxidase-conjugated goat anti-rabbit IgG in T-PBS for 1 h, washed three times in T-PBS, and incubated with the ECL Western blotting detection reagents according to the manufacturer's instructions.
Immunoblotting
The protein samples were subjected to Laemmli sample buffer, run on SDS-PAGE, and transferred to a PVDF membrane pretreated by methanol for activation. After blocked with 5% nonfat milk in TBST for 30 min at room temperature, the membrane was probed with primary antibodies against HA tag, mTOR, Rictor, Akt, or phospho-Ser473 Akt (1:1000 dilution) overnight at 4 °C followed by horseradish peroxidase–conjugated anti-rabbit secondary antibodies (1:5000 dilution) for 1 h at room temperature. The protein bands were treated by ECL Western blotting detection reagents and visualized by ChemiDoc imaging system (Bio-Rad). Densitometry of the band intensity was further determined via Image J software.
Electron-microscopy data acquisition
For negative staining, 2.5 μl of the purified mTORC2 was applied to glow-discharged EM grids covered by a thin layer of continuous carbon film (TED Pella, Inc) and stained with 0.75% (w/v) uranyl formate solution as described. EM grids were imaged on a Tecnai T12 microscope (Thermo Fisher Scientific) operated at 120 kV with a camera UltraScan4000 (Gatan Inc). Images were recorded at a magnification of × 52,000, in a 2.23-Å pixel size on the specimen. Defocus was set to -1.5 μm. For cryo-EM, 3 μl of purified mTORC2 sample (∼0.8 mg/ml) was applied to Spytag003 immobilized affinity grids, which were fabricated as described previously (34). The gold Quantifoil grid is precoated with a layer of graphene oxide on its carbon side, further modified by PEG, maleimide, and a modified spytag that is immobilized on the graphene oxide surface. The spytag affinity grids are refrigerated at -20 degree until use. The sample was first applied on the surface of grid for 5-min incubation, then the grids were dried by paper and dip washed twice followed by adding 3 μl buffer onto grid to prevent drying. They were blotted by Whatman No. 1 filter paper and plunge-frozen in liquid ethane using a Mark IV Vitrobot (Thermo Fisher Scientific) with blotting times of 3 to 6 s at room temperature and over 90% humidity. Cryo-EM datasets were collected using SerialEM and EPU in different microscopes all equipped with Field Emission source and K3 camera (Gatan Inc) operated by SLAC national facility.
Image processing and model building
For all cryo-EM datasets, movies were motion-corrected by MotionCor237. Motion-corrected sums without dose weighting were used for defocus estimation by using CTFFIND4, incorporated in the cisTEM. Motion-corrected sums with dose weighting were used for all other image processing. All datasets were processed similarly. In summary, particles were picked automatically, refinement package created and 2D class averages, initial model generation and autorefinement by following the workflow in cisTEM (46, 47). Final maps were refined, reconstructed, and sharpened in cisTEM. Resolution was estimated by FSC = 0.143 criterion. Atomic model of mTORC2 (PDB: 5ZCS) was used as a starting model followed by manually adjusting in COOT to fit the density map, followed by iterative refinement with Phenix. real_space_refine and manually adjustment in COOT. This process was repeated until Ramachandran validation was satisfied. UCSF Chimera and Pymol were used to prepare images.
3D variability analysis
We carried out 3D variability analysis in cryoSPARC version 2 upon all particles used for the final 3D reconstruction from each dataset as input (48). Analytical computations were performed for the global structures. In each case, multiple modes of variability were solved and represented as conformational changes. To visualize the transformation of density, we calculated 20 reconstructions with a filter resolution of 6 Å. A movie that combines these reconstructions as frames was generated in Chimera for each dimension of motion.
Statistics
GraphPad Prism (GraphPad Software) and Microsoft Excel were used for data analysis. Comparisons of Western Blot densitometry values obtained via Image J software were analyzed by one-way ANOVA and a Tukey post-hoc test. Relative expression levels of phosphorylated proteins are presented as mean (SD) in bar charts. p < 0.05 was considered significant and symbols for p values are described in the figure legends.
Data availability
Further information and requests for resources and reagents should be directed to and will be fulfilled by the corresponding authors. There is no restriction on materials generated for this study and first reported here. The accession numbers for the data reported in this paper are EMDB: EMD-26213 (human mTORC2 complex) PDB: 7TZO, EMD-26212 (human mTORC2-Akt1) and EMD-26211 (human mTORC2-SGK1 (61–431 aa)). All other data are available from the corresponding authors upon request.
Conflict of interest
Y. C. is an Investigator of Howard Hughes Medical Institute. All other authors claim no conflict of interest.
Acknowledgments
Some of this work was performed at the Stanford-SLAC Cryo-EM Center (S2C2) supported by the NIH Common Fund Transformative High Resolution Cryo-Electron Microscopy program (U24 GM129541). We thank David P. Bulkley, Glenn Gilbert, and Matthew Harrington from UCSF Keck Foundation Advanced Microscopy Laboratory for their assistance. We also thank Louella Lee for administrative support. This work is supported by the UCSF cryoEM facility grants: 1S10OD026881, 1S10OD020054, 1S10OD021741.
Author contributions
Z. Y., J. C., and D. P. methodology; Z. Y., J. C., E. T., F. W., B. S., X. L., L.-M. J., C. E. G., M. J., and C. N. investigation; C. L. formal analysis; D. A., Y. C., and D. P. supervision.
Funding and additional information
This work is supported by the NIH grants R01-DK56695 (D. P.), R35GM118099 (D. A. A.), R35GM140847 (Y. C.) and Grant from the James Hilton Manning and Emma Austin Manning Foundation (D. P.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institues of Health.
Biographies

Zanlin Yu is a research specialist in the Department of Biochemistry and Biophysics at University of California San Francisco. His main interest is to understand the molecular mechanisms underlying the function of large protein complexes including the proteasome and mTORC2 using structural biology, particularly cryo-EM. Dr. Yu has made key contributions to developing new technologies for cryo-EM of multi-protein complexes, including using spytag-spycatcher affinity grids to stabilize complexes such as mTORC2 and acquire high-resolution structures. This approach was instrumental in the recognition of context-dependent phosphorylation of mTORC2 substrates in the present publication.

Junliang Chen is a Nephrologist who works now at SinoUnited Health in Shanghai, China. He was a postdoctoral researcher in the Departments of Medicine and Cellular and Molecular Pharmacology at University of California San Francisco with a longstanding interest in the epithelial sodium channel (ENaC). This drew him to pursue the molecular mechanisms of ENaC regulation by SGK1 and its upstream kinase, mTORC2. Toward this end, he initiated a collaboration with his colleague, cryo-EM expert Zanlin Yu, to study mTORC2 substrate specificity. He tried numerous approaches to tagging and purifying mTORC2 components, and finally sorted out the best combination to gain high purity and concentration and then optimized conditions for complex formation.
Edited by George DeMartino
Footnotes
Present address for Junliang Chen: Department of Internal Medicine, SinoUnited Health, Shanghai, China.
Present address for Catherine E. Gleason: Circle Pharma, South San Francisco, CA 94080.
Supporting information
Supplementary Figure 1.
Supplementary Figure 2.
Supplementary Figure 3.
Supplementary Figure 4.
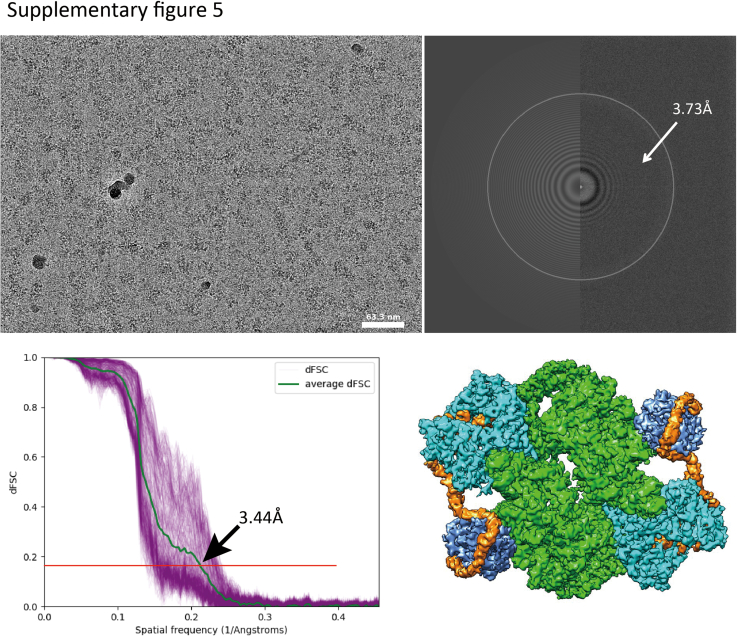
Supplementary Figure 5.
Supplementary Figure 6.
References
- 1.Heitman J., Movva N.R., Hall M.N. Targets for cell cycle arrest by the immunosuppressant rapamycin in yeast. Science. 1991;253:905–909. doi: 10.1126/science.1715094. [DOI] [PubMed] [Google Scholar]
- 2.Sabatini D.M., Erdjument-Bromage H., Lui M., Tempst P., Snyder S.H. RAFT1: a mammalian protein that binds to FKBP12 in a rapamycin-dependent fashion and is homologous to yeast TORs. Cell. 1994;78:35–43. doi: 10.1016/0092-8674(94)90570-3. [DOI] [PubMed] [Google Scholar]
- 3.Fu W., Hall M.N. Regulation of mTORC2 signaling. Genes (Basel) 2020;11:1045. doi: 10.3390/genes11091045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Saxton R.A., Sabatini D.M. mTOR signaling in growth, metabolism, and disease. Cell. 2017;169:361–371. doi: 10.1016/j.cell.2017.03.035. [DOI] [PubMed] [Google Scholar]
- 5.Luo Y., Xu W., Li G., Cui W. Weighing in on mTOR complex 2 signaling: the expanding role in cell metabolism. Oxid. Med. Cell Longev. 2018;2018 doi: 10.1155/2018/7838647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gaubitz C., Prouteau M., Kusmider B., Loewith R. TORC2 structure and function. Trends Biochem. Sci. 2016;41:532–545. doi: 10.1016/j.tibs.2016.04.001. [DOI] [PubMed] [Google Scholar]
- 7.Kim D.H., Sarbassov D.D., Ali S.M., Latek R.R., Guntur K.V., Erdjument-Bromage H., et al. GbetaL, a positive regulator of the rapamycin-sensitive pathway required for the nutrient-sensitive interaction between raptor and mTOR. Mol. Cell. 2003;11:895–904. doi: 10.1016/s1097-2765(03)00114-x. [DOI] [PubMed] [Google Scholar]
- 8.Kim D.H., Sarbassov D.D., Ali S.M., King J.E., Latek R.R., Erdjument-Bromage H., et al. mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell. 2002;110:163–175. doi: 10.1016/s0092-8674(02)00808-5. [DOI] [PubMed] [Google Scholar]
- 9.Wang L., Harris T.E., Roth R.A., Lawrence J.C., Jr. PRAS40 regulates mTORC1 kinase activity by functioning as a direct inhibitor of substrate binding. J. Biol. Chem. 2007;282:20036–20044. doi: 10.1074/jbc.M702376200. [DOI] [PubMed] [Google Scholar]
- 10.Loewith R., Jacinto E., Wullschleger S., Lorberg A., Crespo J.L., Bonenfant D., et al. Two TOR complexes, only one of which is rapamycin sensitive, have distinct roles in cell growth control. Mol. Cell. 2002;10:457–468. doi: 10.1016/s1097-2765(02)00636-6. [DOI] [PubMed] [Google Scholar]
- 11.Sarbassov D.D., Ali S.M., Kim D.H., Guertin D.A., Latek R.R., Erdjument-Bromage H., et al. Rictor, a novel binding partner of mTOR, defines a rapamycin-insensitive and raptor-independent pathway that regulates the cytoskeleton. Curr. Biol. 2004;14:1296–1302. doi: 10.1016/j.cub.2004.06.054. [DOI] [PubMed] [Google Scholar]
- 12.Sarbassov D.D., Ali S.M., Sengupta S., Sheen J.H., Hsu P.P., Bagley A.F., et al. Prolonged rapamycin treatment inhibits mTORC2 assembly and Akt/PKB. Mol. Cell. 2006;22:159–168. doi: 10.1016/j.molcel.2006.03.029. [DOI] [PubMed] [Google Scholar]
- 13.Wu Y.T., Ouyang W., Lazorchak A.S., Liu D., Shen H.M., Su B. mTOR complex 2 targets Akt for proteasomal degradation via phosphorylation at the hydrophobic motif. J. Biol. Chem. 2011;286:14190–14198. doi: 10.1074/jbc.M111.219923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cameron A.J., Linch M.D., Saurin A.T., Escribano C., Parker P.J. mTORC2 targets AGC kinases through Sin1-dependent recruitment. Biochem. J. 2011;439:287–297. doi: 10.1042/BJ20110678. [DOI] [PubMed] [Google Scholar]
- 15.Li X., Gao T. mTORC2 phosphorylates protein kinase Czeta to regulate its stability and activity. EMBO Rep. 2014;15:191–198. doi: 10.1002/embr.201338119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lu M., Wang J., Jones K.T., Ives H.E., Feldman M.E., Yao L.J., et al. mTOR complex-2 activates ENaC by phosphorylating SGK1. J. Am. Soc. Nephrol. 2010;21:811–818. doi: 10.1681/ASN.2009111168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Malik N., Macartney T., Hornberger A., Anderson K.E., Tovell H., Prescott A.R., et al. Mechanism of activation of SGK3 by growth factors via the Class 1 and Class 3 PI3Ks. Biochem. J. 2018;475:117–135. doi: 10.1042/BCJ20170650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chellappa K., Brinkman J.A., Mukherjee S., Morrison M., Alotaibi M.I., Carbajal K.A., et al. Hypothalamic mTORC2 is essential for metabolic health and longevity. Aging Cell. 2019;18 doi: 10.1111/acel.13014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Albert V., Svensson K., Shimobayashi M., Colombi M., Munoz S., Jimenez V., et al. mTORC2 sustains thermogenesis via Akt-induced glucose uptake and glycolysis in brown adipose tissue. EMBO Mol. Med. 2016;8:232–246. doi: 10.15252/emmm.201505610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kazyken D., Magnuson B., Bodur C., Acosta-Jaquez H.A., Zhang D., Tong X., et al. AMPK directly activates mTORC2 to promote cell survival during acute energetic stress. Sci. Signal. 2019;12 doi: 10.1126/scisignal.aav3249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gleason C.E., Frindt G., Cheng C.J., Ng M., Kidwai A., Rashmi P., et al. mTORC2 regulates renal tubule sodium uptake by promoting ENaC activity. J. Clin. Invest. 2015;125:117–128. doi: 10.1172/JCI73935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sorensen M.V., Saha B., Jensen I.S., Wu P., Ayasse N., Gleason C.E., et al. Potassium acts through mTOR to regulate its own secretion. JCI Insight. 2019;5 doi: 10.1172/jci.insight.126910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sato T., Ishii J., Ota Y., Sasaki E., Shibagaki Y., Hattori S. Mammalian target of rapamycin (mTOR) complex 2 regulates filamin A-dependent focal adhesion dynamics and cell migration. Genes Cells. 2016;21:579–593. doi: 10.1111/gtc.12366. [DOI] [PubMed] [Google Scholar]
- 24.Srivastava R.K., Li C., Khan J., Banerjee N.S., Chow L.T., Athar M. Combined mTORC1/mTORC2 inhibition blocks growth and induces catastrophic macropinocytosis in cancer cells. Proc. Natl. Acad. Sci. U. S. A. 2019;116:24583–24592. doi: 10.1073/pnas.1911393116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Venugopal S.V., Caggia S., Gambrell-Sanders D., Khan S.A. Differential roles and activation of mammalian target of rapamycin complexes 1 and 2 during cell migration in prostate cancer cells. Prostate. 2020;80:412–423. doi: 10.1002/pros.23956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yuan T., Rafizadeh S., Gorrepati K.D., Lupse B., Oberholzer J., Maedler K., et al. Reciprocal regulation of mTOR complexes in pancreatic islets from humans with type 2 diabetes. Diabetologia. 2017;60:668–678. doi: 10.1007/s00125-016-4188-9. [DOI] [PubMed] [Google Scholar]
- 27.Tang Y., Wallace M., Sanchez-Gurmaches J., Hsiao W.Y., Li H., Lee P.L., et al. Adipose tissue mTORC2 regulates ChREBP-driven de novo lipogenesis and hepatic glucose metabolism. Nat. Commun. 2016;7 doi: 10.1038/ncomms11365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Johnson J.L., Huang W., Roman G., Costa-Mattioli M. TORC2: A novel target for treating age-associated memory impairment. Sci. Rep. 2015;5 doi: 10.1038/srep15193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang H., Rudge D.G., Koos J.D., Vaidialingam B., Yang H.J., Pavletich N.P. mTOR kinase structure, mechanism and regulation. Nature. 2013;497:217–223. doi: 10.1038/nature12122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen X., Liu M., Tian Y., Li J., Qi Y., Zhao D., et al. Cryo-EM structure of human mTOR complex 2. Cell Res. 2018;28:518–528. doi: 10.1038/s41422-018-0029-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Scaiola A., Mangia F., Imseng S., Boehringer D., Berneiser K., Shimobayashi M., et al. The 3.2-A resolution structure of human mTORC2. Sci. Adv. 2020;6 doi: 10.1126/sciadv.abc1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lu M., Wang J., Ives H.E., Pearce D. mSIN1 protein mediates SGK1 protein interaction with mTORC2 protein complex and is required for selective activation of the epithelial sodium channel. J. Biol. Chem. 2011;286:30647–30654. doi: 10.1074/jbc.M111.257592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tatebe H., Murayama S., Yonekura T., Hatano T., Richter D., Furuya T., et al. Substrate specificity of TOR complex 2 is determined by a ubiquitin-fold domain of the Sin1 subunit. Elife. 2017;6 doi: 10.7554/eLife.19594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang F., Liu Y., Yu Z., Li S., Feng S., Cheng Y., et al. General and robust covalently linked graphene oxide affinity grids for high-resolution cryo-EM. Proc. Natl. Acad. Sci. U. S. A. 2020;117:24269–24273. doi: 10.1073/pnas.2009707117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rotin D., Staub O. Function and regulation of the epithelial Na(+) channel ENaC. Compr. Physiol. 2021;11:2017–2045. doi: 10.1002/cphy.c200012. [DOI] [PubMed] [Google Scholar]
- 36.Aylett C.H., Sauer E., Imseng S., Boehringer D., Hall M.N., Ban N., et al. Architecture of human mTOR complex 1. Science. 2016;351:48–52. doi: 10.1126/science.aaa3870. [DOI] [PubMed] [Google Scholar]
- 37.Yang H., Jiang X., Li B., Yang H.J., Miller M., Yang A., et al. Mechanisms of mTORC1 activation by RHEB and inhibition by PRAS40. Nature. 2017;552:368–373. doi: 10.1038/nature25023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McMahon L.P., Choi K.M., Lin T.A., Abraham R.T., Lawrence J.C., Jr. The rapamycin-binding domain governs substrate selectivity by the mammalian target of rapamycin. Mol. Cell Biol. 2002;22:7428–7438. doi: 10.1128/MCB.22.21.7428-7438.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Frias M.A., Thoreen C.C., Jaffe J.D., Schroder W., Sculley T., Carr S.A., et al. mSin1 is necessary for Akt/PKB phosphorylation, and its isoforms define three distinct mTORC2s. Curr. Biol. 2006;16:1865–1870. doi: 10.1016/j.cub.2006.08.001. [DOI] [PubMed] [Google Scholar]
- 40.Liu P., Gan W., Inuzuka H., Lazorchak A.S., Gao D., Arojo O., et al. Sin1 phosphorylation impairs mTORC2 complex integrity and inhibits downstream Akt signalling to suppress tumorigenesis. Nat. Cell Biol. 2013;15:1340–1350. doi: 10.1038/ncb2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gleason C.E., Oses-Prieto J.A., Li K.H., Saha B., Situ G., Burlingame A.L., et al. Phosphorylation at distinct subcellular locations underlies specificity in mTORC2-mediated activation of SGK1 and Akt. J. Cell Sci. 2019;132:jcs224931. doi: 10.1242/jcs.224931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang G., Murashige D.S., Humphrey S.J., James D.E. A positive feedback loop between Akt and mTORC2 via SIN1 phosphorylation. Cell Rep. 2015;12:937–943. doi: 10.1016/j.celrep.2015.07.016. [DOI] [PubMed] [Google Scholar]
- 43.Pearce L.R., Komander D., Alessi D.R. The nuts and bolts of AGC protein kinases. Nat. Rev. 2010;11:9–22. doi: 10.1038/nrm2822. [DOI] [PubMed] [Google Scholar]
- 44.Pao A.C., McCormick J.A., Li H., Siu J., Govaerts C., Bhalla V., et al. NH2 terminus of serum and glucocorticoid-regulated kinase 1 binds to phosphoinositides and is essential for isoform-specific physiological functions. Am. J. Physiol. Ren. Physiol. 2007;292:F1741–1750. doi: 10.1152/ajprenal.00027.2007. [DOI] [PubMed] [Google Scholar]
- 45.Allu P.K.R., Paulo E., Bertholet A.M., Situ G., Lee S.H., Wu Y., et al. Role of mTORC2 in biphasic regulation of brown fat metabolism in response to mild and severe cold. J. Biol. Chem. 2021;296 doi: 10.1016/j.jbc.2021.100632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Grant T., Rohou A., Grigorieff N. cis TEM, user-friendly software for single-particle image processing. Elife. 2018;7 doi: 10.7554/eLife.35383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zivanov J., Nakane T., Forsberg B.O., Kimanius D., Hagen W.J., Lindahl E., et al. New tools for automated high-resolution cryo-EM structure determination in RELION-3. Elife. 2018;7 doi: 10.7554/eLife.42166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Punjani A., Fleet D.J. 3D variability analysis: resolving continuous flexibility and discrete heterogeneity from single particle cryo-EM. J. Struct. Biol. 2021;213 doi: 10.1016/j.jsb.2021.107702. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Further information and requests for resources and reagents should be directed to and will be fulfilled by the corresponding authors. There is no restriction on materials generated for this study and first reported here. The accession numbers for the data reported in this paper are EMDB: EMD-26213 (human mTORC2 complex) PDB: 7TZO, EMD-26212 (human mTORC2-Akt1) and EMD-26211 (human mTORC2-SGK1 (61–431 aa)). All other data are available from the corresponding authors upon request.