Abstract
This study determined the frequency, clinicopathologic, and genetic features of colorectal carcinomas driven by oncogenic fusions of the anaplastic lymphoma kinase gene (ALK). Out of 8150 screened tumors, 12 (0.15%) were immunohistochemically ALK-positive with D5F3 antibody. These cancers harbored CAD-ALK (n=1), DIAPH2-ALK (n=2), EML4-ALK (n=2), LOC101929227-ALK (n=1), SLMAP-ALK (n=1), SPTBN1-ALK (n=4) and STRN-ALK (n=1) fusions as detected by an RNA-based NGS assay. ALK fusion carcinomas were diagnosed mostly in older patients with a 9:3 female predominance (median age, 72 years). All tumors, except a rectal one, occurred in the right colon. Most tumors were stage T3 (n=7) or T4 (n=3). Local lymph node and distant metastases were seen at presentation in 9 and 2 patients. These tumors showed moderate (n=6) or poor (n=3) glandular differentiation, solid medullary growth pattern (n=2), and pure mucinous morphology (n=1). DNA mismatch repair deficient phenotype was identified in 10 cases. Tumor-infiltrating lymphocytes were prominent in 9 carcinomas. In 4 carcinomas, tumor cells showed strong, focal (n=3) or diffuse PD-L1 immunoreactivity. CDX2 expression and loss of CK20 and MUC2 expression were frequent. CK7 was expressed in 5 tumors. Four patients died of disease within 3 years and 7 were alive with follow-up ranging from 1–8 years. No mutations in BRAF, RAS and genes encoding components of PI3K-AKT/MTOR pathway were identified. However, one tumor had a loss-of-function PTEN mutation. Aberration of p53 signaling, TP53 mutations and/or nuclear accumulation of p53 protein was seen in 9 cases. ALK fusion colorectal carcinomas are a distinct and rare subtype of colorectal cancers displaying some features of mismatch repair-deficient tumors.
Keywords: colorectal carcinoma, immunohistochemistry, ALK-expression, ALK fusion genes, next-generation sequencing
INTRODUCTION
Colorectal adenocarcinoma (CRC) is one of the major causes of cancer-related death in the United States and in the world. CRC is a highly heterogeneous disease distinguished by multiple genetic and epigenetic events critical for the tumor initiation and progression.1 Most CRCs are driven by oncogene or tumor suppressor gene mutations. However, a small subset is characterized by fusions involving receptor tyrosine kinase (RTKs) genes.2–4 Typically, the 3’ region of the RTK gene fuses to the 5’ region of a gene partner, forming a chimeric gene/oncogene that expresses a constitutively activated tyrosine kinase capable of inducing downstream signaling pathways that promote tumorigenesis.5
Anaplastic lymphoma kinase (ALK), originally identified as the kinase partner in the NPM1-ALK fusion in anaplastic large cell lymphoma, is encoded by a gene with high homology to the insulin receptor subfamily of transmembrane tyrosine kinases.6 ALK may also be involved in the development and function of the nervous system.7 Molecular ALK alterations – including amplification and activation of mutations and fusions with multiple 5’ partners – have been reported in different types of epithelial, hematopoietic, and mesenchymal malignancies.8
Recently, several distinct ALK inhibitors have been approved and show efficacy in the treatment of tumors harboring oncogenic ALK fusions.9–10 This opened an opportunity for targeted therapy of CRCs driven by oncogenic ALK fusions.11
Only a small number of CRCs harboring ALK fusions have been reported.3,11–23 Previously published ALK fusion-positive CRCs are listed in the supplemental data (Table S1). Nevertheless, the clinicopathologic profile of CRCs driven by oncogenic ALK fusions is not well established. This study presents a comprehensive clinicopathologic, immunohistochemical, and molecular genetic evaluation of 12 ALK fusion-driven tumors identified in a large cohort of CRCs screened with ALK immunohistochemistry.
MATERIALS and METHODS
Eight thousand one hundred fifty de-identified CRCs from Europe (Czech Republic, Finland, Germany, Italy and Poland), Japan, and the United States were studied. Tumor samples were assembled in tissue microarrays (TMAs) or multitumor blocks as previously reported.24
Tumor staging and histologic classification were based on The American Joint Committee on Cancer (AJCC) tumor/node/metastasis (TNM) classification and staging recommendation (http://cancerstaging.org) and WHO classification of tumors of the digestive system.25 The density of tumor infiltrating lymphocytes (TILs) was scored as previously described.26
Treatment and follow-up data were available in all cases. Each patient underwent surgery sometimes followed by adjuvant chemotherapy. However, no tyrosine kinase-inhibitor therapy was administrated.
Immunohistochemistry
Immunohistochemical ALK expression was evaluated using ALK antibody, clone D5F3 (Cell Signaling Technology, Inc., Danvers, MA), and Leica Bond-Max immunostainer (Leica Biosystems, Inc., Buffalo Grove, IL) with 25-minute heat induced epitope retrieval in ER2 buffer. All ALK-positive tumors were evaluated for the expression of several antigens including DNA-mismatch repair (MMR) proteins [MutL Homolog 1 (MLH1), PMS1 Homolog 2 (PMS2), MutS Homolog 2 (MSH2) and MutS Homolog 6 (MSH6)], Caudal Type Homeobox 2 (CDX2), Cytokeratin 7 and 20 (CK7, CK20), Mucin 2 (MUC2), Tumor Protein P53 (p53) and Programmed death-ligand 1 (PD-L1, CD274). Details of antibodies and immunohistochemical protocols are provided in the supplemental data (Table S2).
Fluorescence in situ hybridization
Interphase fluorescence in situ hybridization (FISH) was performed on 5μ-thick formalin-fixed paraffin-embedded (FFPE) tumor sections using Dual Color Break Apart SPEC ALK Probe (ZytoVision, Bremerhaven, Germany) and Histology FISH Acc Kit (Perlan Diagnostics, Gdynia, Poland) following the manufacturer’s protocol. Specimens and a probe were denatured for 5 minutes at 73°C and subsequently incubated at 37°C for approximately 12 hours in the hybridization chamber CytoHYB CT500 (CytoTest, Rockville, MD). Following standard washing procedures, sections were counterstained with 4′,6-diamidino-2-phenylindole (DAPI). Fluorescence microscope Axio Imager.Z2 (Carl Zeiss Microscopy, Jena, Germany) equipped with DAPI/Green/Red triple-band filter was used for visualization. One hundred cells were evaluated at 100x magnification in each case.
Molecular genetic studies
Detailed protocols of molecular procedures including extraction of nucleic acids, evaluation of MLH1 promotor hypermethylation staus and next-generation sequencing were previously reported.24
Briefly, nucleic acids were recovered from FFPE tumor samples using a Maxwell® RSC instrument (Promega, Madison, WI). Target-specific RNA libraries for next-generation sequencing (NGS) were constructed using Archer Universal® RNA Reagent Kit v2 (ArcherDx, Boulder, CO) and sequenced using a MiSeqDx instrument (Illumina, San Diego, CA). NGS data was analyzed using the Archer Analysis Pipeline Virtual Machine (https://archerdx.com). Genotyping was performed using the Ion Torrent™ (Life Technologies/Thermo Fisher Scientific, Waltham, MA) next-generation sequencing platform and either Ion AmpliSeq™ Comprehensive Cancer Panel (409 gene targets) or Ion AmpliSeq™ Cancer Hotspot Panel v2 Kit (50 gene targets).
Bioinformatics analysis of NGS-data was processed by Torrent Server Suite 5.12 and aligned to the human genome reference sequence, homo sapiens genome assembly GRCh37 (hg19) from Genome Reference Consortium (www.ncbi.nlm.nih.gov/grc). Variant calling was performed using Variant Caller v5.12, which is compatible with the Integrative Genomics Viewer (Broad Institute, Cambridge, MA), a high-performance visualization tool for interactive exploration of large, integrated data sets. Mutation nomenclature was based on recommendations from Human Genome Mutation Society (www.hgvs.org). The FATHMM (Functional Analysis Through Hidden Markov Models), SIFT (Sorting Intolerant from Tolerant) and PolyPhen (Polymorphism Phenotyping) scores predicting functional consequences of coding variants were either obtained from the COSMIC (Catalogue of Somatic Mutations in Cancer) at https://cancer.sanger.ac.uk or assessed during bioinformatic analysis.
RESULTS
ALK immunohistochemistry
ALK immunoreactivity showed three patterns of staining: diffuse, focal and luminal. The former appeared as either cytoplasmic or membrane staining. No nuclear staining was seen. All 12 CRCs characterized by diffuse ALK staining harbored ALK fusion genes. In contrast, no ALK fusions, ALK rearrangements and/or ALK mutations were identified in tumors showing focal or exclusively luminal ALK positivity.
The intensity of diffuse ALK staining varied. In general, tumors with moderate to strong cytoplasmic ALK immunoreactivity lacked membrane staining, while tumors with strong membrane immunoreactivity displayed weak cytoplasmic staining. Different patterns of diffuse ALK staining in colorectal carcinomas are listed in Table 1 and illustrated in Fig. 1A–D.
Table 1.
ALK immunoreactivity in 12 colorectal carcinomas harboring ALK fusion genes.
| ALK fusion gene transcript | No. of cases | ALK immunoreactivity | |
|---|---|---|---|
| Cytoplasmic | Membrane | ||
| CAD(e35)-ALK(e20) | 1 | Moderate to strong | Absent |
| DIAPH2(e17)-ALK(e20) | 1 | Weak | Strong |
| DIAPH2(e26)-ALK(e20) | 1 | Strong | Absent |
| EML4(e21)-ALK(e20) | 1 | Moderate to strong | Absent |
| EML4(e6)-ALK(e20) | 1 | Weak to moderate | Variable |
| LOC101929227(e2)-ALK(e10) | 1 | Weak | Absent |
| SLMAP(e14)-ALK(e20) | 1 | Moderate to strong* | Absent |
| SPTBN1(e7)-ALK(e20) | 4 | Weak | Strong |
| STRN(e3)-ALK(e20) | 1 | Moderate | Variable** |
Symbols:
= some cells with a dot-like perinuclear staining,
= stronger on luminal side.
Figure 1.
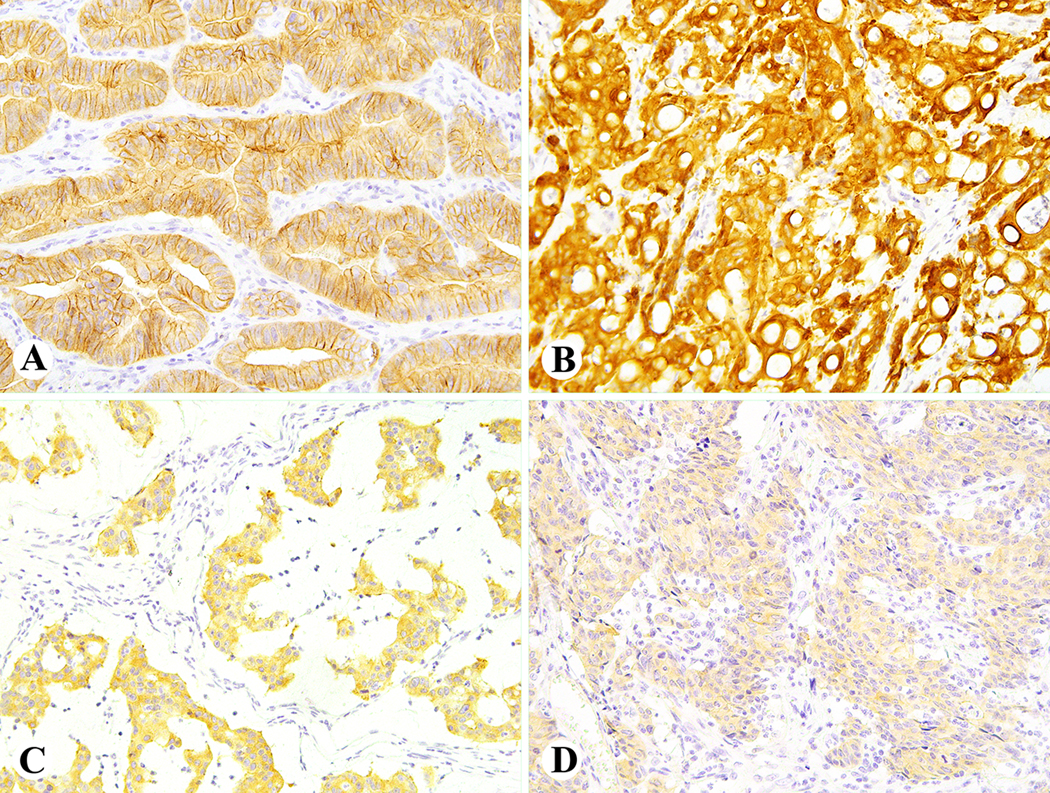
Example of diffuse ALK immunoreactivity in colorectal carcinomas. Strong membrane staining for ALK in Case 1 with a SPTBN1-ALK fusion (A); strong cytoplasmic ALK staining in Case 6 with a DIAPH2-ALK fusion (B); moderate cytoplasmic ALK staining in Case 8 with an EML4-ALK fusion (C); weak cytoplasmic ALK staining in Case 9 with a LOC101929227-ALK fusion (D).
The intensity of focal staining varied from strong to weak, while luminal staining was well demarcated in all positive cases. Examples of focal and luminal ALK staining are shown in Fig. 2A–B. There was no significant background staining in all but one case. The latter is illustrated and discussed in the supplemental data (Fig. S1A–D). Clinicopathologic and molecular profiles of selected colon carcinomas with focal and luminal ALK staining are presented in the supplemental data (Table S3).
Figure 2.
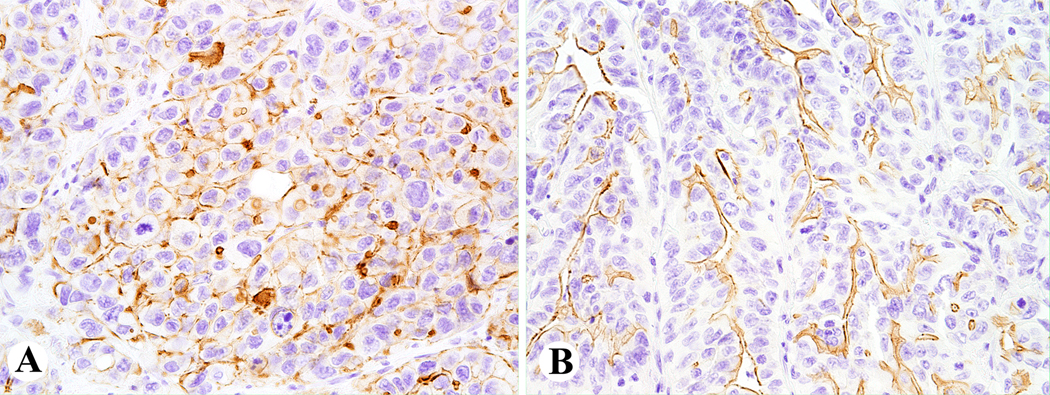
Example of focal (A) and luminal (B) ALK immunoreactivity in ALK fusion negative colon carcinomas.
ALK gene rearrangements and fusion gene transcripts
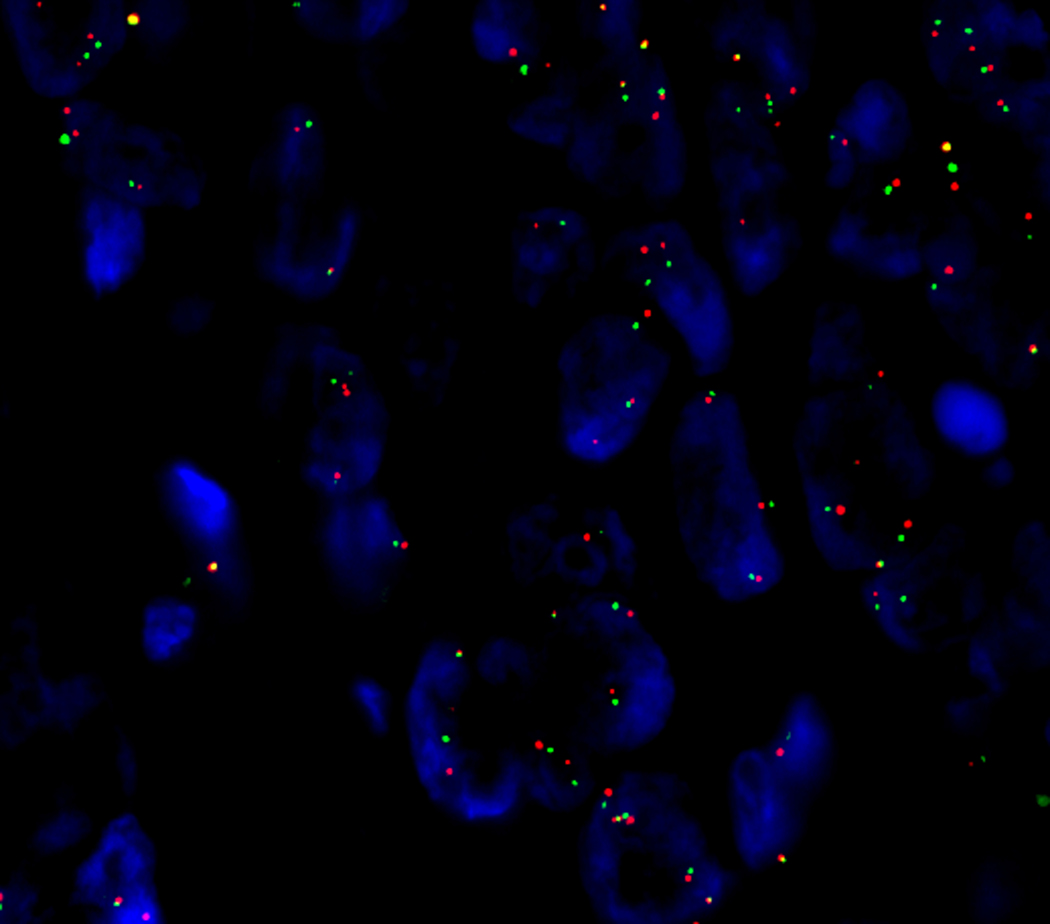
All tumors with diffuse immunopositivity for ALK showed ALK gene rearrangements and chimeric ALK fusion transcripts as evaluated by FISH with the break-apart probe and by ArcherDX assay, respectively. ALK fusion breaks occurred in exon (e) 20 in all but one case. Seven ALK fusion 5’partners were identified: CAD (Carbamoyl-Phosphate Synthetase 2, synonymous to Aspartate Transcarbamylase and Dihydroorotase, n = 1), DIAPH2 (Diaphanous Related Formin 2, n = 2), EML4 (Echinoderm Microtubule Associated Protein Like 4, n = 2), SLMAP (Sarcolemma Associated Protein, n = 1) and SPTBN1 (Spectrin Beta, Non-Erythrocytic 1, n = 4), STRN (Striatin, n=1) and uncharacterized gene locus LOC101929227 (n = 1). ALK-FISH of a colonic adenocarcinoma harboring a LOC101929227-ALK fusion is shown in Fig. 3. All ALK fusion gene transcripts are listed in Table 1.
Figure 3.
A dual-color FISH analysis performed on interphase nuclei using a break-apart ALK gene probe in CRC (Case 9) harboring a LOC101929227-ALK fusion. A split of orange and green signal indicates an ALK rearrangement.
Association between ALK staining patterns and ALK fusions
The association between the ALK staining pattern and the type of ALK fusion is highlighted in Table 1. Distinctive, strong membrane staining was seen in 4 SPTBN1(e7)-ALK(e20) and 1 DIAPH2(e17)-ALK(e20) fusion tumors. Variable membrane staining was seen in a rectal adenocarcinoma with an EML4(e6)-ALK(e20) fusion. Tumors harboring other ALK fusions involving CAD(e35), DIAPH2(e26), EML4(21), SLMAP(e14) and STRN(e3) showed moderate to strong cytoplasmic ALK immunoreactivity. Diffuse but weak cytoplasmic ALK staining was seen in LOC101929227(e2)-ALK(e10) fusion colon cancer. Also, intense perinuclear staining was noticeable in some tumor cells harboring SLMAP(e14)-ALK(e20) fusion. An example of perinuclear staining is shown in the supplemental data (Fig. S2).
Demographic and clinicopathologic features
The demographic and clinicopathologic features of ALK fusion CRCs are summarized in Table 2. Seventy-five % (9/12) of ALK fusion tumors were diagnosed in female patients with median age 72 years. The median age of the three male patients was 68 years.
Table 2.
Clinical characteristics of 12 colorectal carcinomas harboring ALK fusion genes.
| Case | Type of ALK fusion | Age | Sex | Tumor site | pTNM | Follow-up (in months) |
|---|---|---|---|---|---|---|
| 1 | SPTBN1(e7)-ALK(e20) | 53 | M | Cecum | pT3N1aM0 | ‡ANED (30m) |
| 2 | SPTBN1(e7)-ALK(e20) | 66 | F | Cecum | pT3N0M0 | ‡ANED (20m) |
| 3 | CAD(e35)-ALK(e20) | 87 | F | Cecum | pT4bN1bM1a | ‡DOD (1m) |
| 4 | SPTBN1(e7)-ALK(e20) | 64 | F | Ascending colon | pT3N2M0 | ⸸ANED (98m) |
| 5 | SLMAP(e14)-ALK(e20) | 69 | M | Ascending colon | pT3aN1cM1c | ⸸DOD (8m) |
| 6 | DIAPH2(e26)-ALK(e20) | 72 | F | Ascending colon | pT3N1bM0 | ‡DOD (34m) |
| 7 | SPTBN1(e7)-ALK(e20) | 75 | F | Ascending colon | pT4aN2bM0 | ‡ANED (78m) |
| 8 | EML4(e21)-ALK(e20) | 76 | M | Ascending colon | pT3N1M0 | ‡AWD (54m) |
| 9 | LOC101929227(e2)-ALK(e10) | 77 | F | Ascending colon | pT3pN1aM0 | ‡DOD (12m) |
| 10 | DIAPH2(e17)-ALK(e20) | 72 | F | Hepatic flexure | pT4aN2aM0 | §ANED (13m) |
| 11 | STRN(e3)-ALK(e20) | 65 | F | Splenic flexure | pT2N0M0 | ⸸ANED (15m) |
| 12 | EML4(e6)-ALK(e20) | 76 | F | Rectum | pT2N0M0 | §ANED (35m) |
Abbreviations and symbols: e = exon, ANED = alive, no avidance of disease, DOD = died of disease, NA = not available,
= adjuvant chemotherapy,
= no adjuvant chemotherapy,
= adjuvant chemotherapy status unknown.
All but one ALK fusion carcinoma developed in the right-sided colon including cecum (n=3), ascending colon (n=6), hepatic (n=1) and splenic (n=1) flexures. There was 1 rectal tumor. Based on the TNM classification, most (10/12) of the ALK fusion colon carcinomas were either T3 (n=7) or T4 (n = 3) with local lymph node (n=9) and distant (n=2) metastases diagnosed at presentation. In two cases, including a rectal tumor, the stage was T2N0M0.
Follow-up data were available in all cases. Four patients with local and distal metastases died of disease in a time span of 1–34 months. Seven patients were alive with (n=1) or without disease with follow-up ranging from 13 to 98 months (median follow-up 32.5 months).
Histologic features
The histologic features of ALK fusion CRCs analyzed in this study are summarized in Table 3 and illustrated in Fig. 4A–D. Most tumors showed moderate (n=6) or poor (n=3) glandular differentiation. Mucinous features were seen in three cases including 1 tumor with a pure mucinous morphology. Two carcinomas had solid growth patterns corresponding to medullary subtype. Lymphovascular invasion was present in 67% (8/12) tumors. Eight tumors had a high level (≥2/HPF) of intraepithelial lymphocytes.
Table 3.
Histopathological characteristics of 12 colorectal carcinoma harboring ALK fusion genes.
| Case | Type of ALK fusion | Degree of glandular differentiation | Mucinous component | Lymphovascular invasion | Tumor infiltrating lymphocytes |
|---|---|---|---|---|---|
| 1 | SPTBN1(e7)-ALK(e20) | Moderate | No | Yes | Low |
| 2 | SPTBN1(e7)-ALK(e20) | Poor | No | Yes | High |
| 3 | CAD(e35)-ALK(e20) | Moderate | No | Yes | High |
| 4 | SPTBN1(e7)-ALK(e20) | Moderate | Yes | No | High |
| 5 | SLMAP(e14)-ALK(e20) | Moderate | No | Yes | High |
| 6 | DIAPH2(e26)-ALK(e20) | Poor | No | Yes | High |
| 7 | SPTBN1(e7)-ALK(e20) | Poor | Yes | Yes | High |
| 8 | EML4(e21)-ALK(e20) | Mucinous | Yes | No | Low |
| 9 | LOC101929227(e2)-ALK(e10) | Solid (medullary) | No | Yes | High |
| 10 | DIAPH2(e17)-ALK(e20) | Moderate | No | Yes | Low |
| 11 | STRN(e3)-ALK(e20) | Moderate | No | No | High |
| 12 | EML4(e6)-ALK(e20) | Solid (medullary) | No | No | High |
Figure 4.
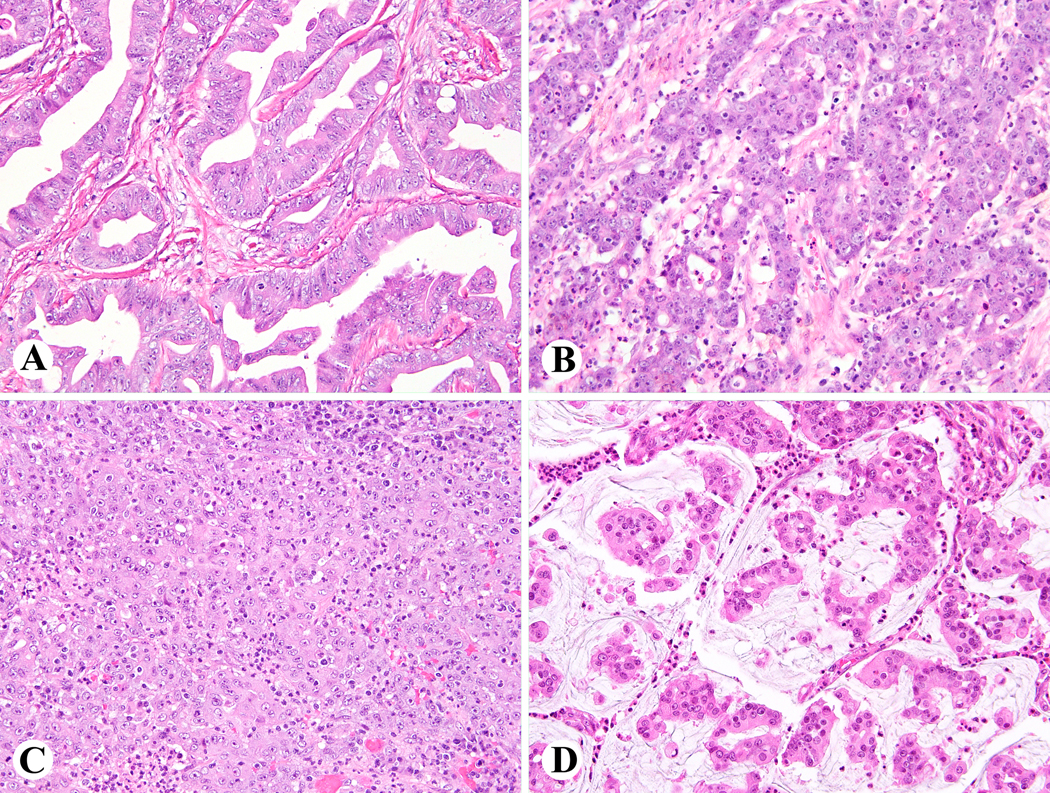
Histologic features of colorectal carcinomas with diffuse ALK immunoreactivity. Case 1, moderately differentiated CRC harboring SPTBN1-ALK fusion (A); Case 2, poorly differentiated CRC harboring SPTBN1-ALK fusion (B); Case 12, medullary CRC harboring EML4-ALK fusion (C); Case 8, mucinous tumor harboring EML4-ALK fusion (D)
Mismatch repair genes status
Mismatch repair (MMR) gene studies are summarized in Table 4. Nine colon carcinomas (75%) were MMR-deficient by immunohistochemistry. Eight tumors revealed loss of MLH1 and PMS2 expression (n=6) or heterogenous loss of MLH1 expression. (supplemental data Fig. S3) In one case, a loss of MSH2 expression was detected. All 12 CRCs expressed MSH6. Methylation studies performed on one of two cases displaying heterogenous loss of MLH1 expression revealed hypermethylation of the MLH1 promoter. No mutations in MLH1 and MSH2 were identified in MMR-deficient ALK fusion CRCs. However, 2 MMR-proficient tumors harbored PMS2 p.Lys647* and MSH6 p.Asn112Ser mutations, respectively.
Table 4.
Mismatch repair status, p53 alterations, CDX2, CK7, CK20, and MUC2 immunophenotypes, and PD-L1 expression in 12 colorectal carcinomas with ALK fusions.
| Case | ALK fusion partner gene | Mismatch repair status | p53 pathway alterations | Immunopositivity | PD-L1 expression | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MLH1 | PMS2 | MSH2 | MSH6 | p53 nuclear accumulation (TP53 mutation) |
CDX2 | CK7 | CK20 | MUC2 | Tumor cells | Tumor infiltrating immune cells | ||
| 1 | SPTBN1 | Retained | Retained | Loss | Retained | Focal (p.Arg273Cys) |
Diffuse | Diffuse | Negative | Focal | Negative | Focal |
| 2 | SPTBN1 | Loss | Loss | Retained | Retained | Focal (p.Gly199*) |
Diffuse | Negative | Negative | Negative | Focal | Diffuse |
| 3 | CAD | Retained | Retained | Retained | Retained | Diffuse (p.Met237Ile) |
Diffuse | Diffuse | Focal | Negative | Negative | Focal |
| 4 | SPTBN1 | Loss | Loss | Retained | Retained | Focal (WT) |
Diffuse | Diffuse | Diffuse | Focal | Negative | Focal |
| 5 | SLMAP | HLoss | Retained | Retained | Retained | Diffuse (p.Arg248Trp) |
Diffuse | Negative | Negative | Negative | Negative | Negative |
| 6 | DIAPH2 | Loss | Loss | Retained | Retained | Focal (p.Tyr126His) |
Diffuse | Negative | Focal | Focal | Focal | Focal |
| 7 | SPTBN1 | Loss | Loss | Retained | Retained | IPC (WT) |
Diffuse | Negative | Focal | Focal | Negative | Focal |
| 8 | EML4 | Retained | Retained | Retained | Retained | Focal (WT) |
Diffuse | Diffuse | Negative | Diffuse | Negative | Focal |
| 9 | LOC101929227 | Loss | Loss | Retained | Retained | Negative (WT) |
Focal | Diffuse | Focal | IPC | Focal | Focal |
| 10 | DIAPH2 | HLoss | Retained | Retained | Retained | Diffuse (p.Arg282Trp) |
Diffuse | Negative | Diffuse | Focal | Negative | Negative |
| 11 | STRN | Loss | Loss | Retained | Retained | IPC (WT) |
Diffuse† | Negative | Focal | Focal | Negative | Diffuse |
| 12 | EML4 | Loss | Loss | Retained | Retained | Focal (WT) |
Focal* | Negative | Negative | Negative | Diffuse | Diffuse |
Abbreviations and symbols: HLoss = heterogeneous loss, WT = wild type, IPC = isolated positive cells
= methylated promoter,
= weak staining. – = negative,
P53 pathway alterations
Immunohistochemical and molecular evaluation of p53 pathway is summarized in Table 4. Nuclear p53 immunoreactivity was detected in 11 cases. Six of nine tumors exhibiting focal (n=6) or diffuse (n=3) p53 staining harbored TP53 mutations. A variable number of scattered p53 positive nuclei were seen in 2 TP53-wild type carcinomas, while one cancer was negative for both p53 nuclear accumulation and TP53 mutation.
CDX2, CK7 and CK20, and MUC2 expression
Immunoprofiles of ALK fusion tumors are presented in Table 4. All tumors showed diffuse (n=10) or focal CDX2 expression. CK7 and CK20 were diffusely expressed in 5 and 2 cases, respectively. There was focal CK20 positivity in 5 tumors. Majority of tumors showed total (n=4) or focal loss (n=8) of MUC2 expression.
PD-L1 expression
The results of PD-L1 immunohistochemistry are summarized in Table 4 and illustrated in Fig. S4. Four CRCs showed strong, focal (n=3) or diffuse PD-L1 tumor cell positivity. Abundant PD-L1 positive tumor-infiltrating immune cells were seen in 3 cases. Seven cancers, including 2 with focal PD-L1 positivity of tumor cells, harbored scattered PD-L1 positive tumor-infiltrating immune cells.
Somatic mutations
All somatic mutations identified in this study are listed in the supplemental data Table S4. Tumors (Case 1–6, 8 and 11) with well-preserved DNA were evaluated with comprehensive cancer panel of 409 genes, while genotyping of three cases (Case 7, 9, 10 and 12) with insufficient DNA quality was limited to 50 genes.
None of the 12 ALK fusion tumors harbored mutations in the typical CRC drivers BRAF (B-raf proto-oncogene, serine/threonine kinase), K-RAS (Kirsten rat sarcoma viral oncogene homolog) and N-RAS (Neuroblastoma Ras viral oncogene homolog). Two tumors had subclones with additional pathogenic ALK mutations, p.Thr1151Met and p.Arg1275Gln.
Genes encoding components of the PI3K-AKT/MTOR (Phosphoinositide 3-kinases-serine/threonine protein kinase/mammalian target of rapamycin) signaling pathway, such as AKT, MTOR, PIK3CA (Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha), and TSC (Tuberous sclerosis complex), were not altered. However, a subclone with mutation affecting PIK3C2B (Phosphatidylinositol-4-phosphate 3-kinase catalytic subunit type 2 beta) encoding subunits of Class II PI3Ks was identified in 1 case. PTEN (Phosphatase and tensin homolog), a regulator of PI3K pathway, carried a loss-of-function mutation (p.Asn63fs) in 1 case.
TP53 was mutated in 50% (6/12) of analyzed CRCs. However, there were no mutations in genes encoding the Wnt/β-catenin signaling pathway components APC (Adenomatous polyposis coli protein) or CTNNB1(Catenin beta 1). FBXW7 (F-box and WD repeat domain containing 7), a tumor suppressor gene, was affected by the loss-of-function mutation (p.Arg367*) in one case.
DISCUSSION
This study analyzed clinicopathologic and genetic features of colorectal carcinomas (CRCs) characterized by immunohistochemical ALK expression. All eleven cases with diffuse cytoplasmic or membrane staining patterns harbored ALK fusions (0.15% of the screening cohort of 8150 cases). However, tumors with focal and luminal ALK expression revealed no ALK alterations. Thus, in CRC, diffuse cytoplasmic or membranous ALK expression predicts an ALK fusion.
Two previous immunohistochemical studies on CRC using anti-ALK 5A4 antibody (Novocastra) and a BenchMark Ultra autostainer (Ventana Medical Systems) and the clone D5F3 with a Leica Bond III autostainer reported a frequency of positive tumors between 0.05% (1 of 1889) and 0.1% (3 of 2980), respectively.14, 22 In this study, more sensitive detection of ALK positive tumors may have caused a slightly higher frequency of 0.15%.
Sensitivity of ALK immunohistochemistry in search of ALK fusions cannot be assessed in this investigation because ALK-negative CRCs were not genotyped. One hybrid capture-based next generation sequencing study found that 0.19% (6 of 3157) of analyzed tumors harbored ALK fusions.22 Another study reported CRC harboring a C2orf44-ALK fusion with 89.8-fold increase of ALK RNA and negative ALK immunohistochemistry.19 Although frequency of ALK fusion tumors among CRCs identified by immunohistochemical or molecular screening appears to be somewhat similar, it is possible that screening by ALK immunostain is not as accurate as sequencing.
In this study, ALK fusion partner genes included CAD, DIAPH2, EML4, LOC101929227, SLMAP, SPTBN1and STRN. However, no fusions with C2orf44 (Chromosome 2 Open Reading Frame 44), CENPF (Centromere Protein F), MAPRE3 (Microtubule Associated Protein RP/EB Family Member 3), PP4R3B (Protein Phosphatase 4 Regulatory Subunit 3B), PPP1R21 (Protein Phosphatase 1 Regulatory Subunit 21), PRKAR1A (Protein Kinase CAMP-Dependent Type I Regulatory Subunit Alpha) and PRKAR1B (Protein Kinase CAMP-Dependent Type I Regulatory Subunit Beta), previously identified ALK partners in CRC, were detected.20,22
EML4-ALK fusion, previously reported in CRC, was less frequent and accounted for 17% (2/12) of analyzed cases. Nevertheless, EML4 is the most common ALK fusion partner in the combined previously published and current fusions in colorectal cancer. The EML4-ALK fusion was first discovered in non-small cell lung carcinoma.27 Subsequently, it was found as an alternative molecular aberration in inflammatory myofibroblastic tumors and in a subset of breast, colorectal and non-small cell lung cancers.28–30 EML4 encodes for echinoderm microtubule-associated protein 4, which belongs to a family of proteins that stabilize microtubules.31 The involvement of EML4 protein in microtubule cytoskeleton might be responsible for the cytoplasmic and variable membrane ALK staining seen in EML4-ALK fusion tumors.
A SPTBN1(e7)-ALK(e20) gene was the most common (33%) fusion in our cohort (4/12 cases). This fusion was previously reported in CRC only once.23 However, an SPTBN1(e6)-ALK(e20) fusion was identified in a lung adenocarcinoma.32 In chronic myeloid leukemia and atypical myeloproliferative disorder, fusions with SPTBN1 are partnered with FLT3 and PDGFRB.33,34 SPTBN1, a member of the spectrin superfamily, encodes a 247-kDa cytoskeletal protein, which can bind to membrane phospholipids.35,36 This could explain a strong cell membrane ALK staining observed in SPTBN1-ALK fusion tumors.
One tumor harbored STRN(e3)-ALK(e20) fusion. Previously STRN has been shown to be an ALK fusion partner in carcinomas of colon, thyroid and kidney.15,21,22,37,38 STRN is a component of the striatin-interacting phosphatase and kinase (STRIPAK) complex. STRIPAK complexes are highly conserved in eukaryotic organisms and play a role in a variety of physiological cellular processes such as cell cycle control, signaling, migration, apoptosis and Golgi apparatus assembly.39 STRIPAK complex malfunction is associated with different pathological conditions including cancer.40,41 In this study, SLMAP, a member of tail-anchored proteins and another component of STRIPAK complex, has been shown to form with ALK chimeric protein.
SLMAP(e14)-ALK(e20) fusion protein has not been previously reported in CRC. However, two similar fusions SLMAP(e12)-ALK(e20) and SLMAP(e13)-ALK(e20) were identified in a lung adenocarcinoma.42 A subset of SLMAP-ALK fusion tumor cells revealed an intense perinuclear ALK staining spots probably indicating anchoring of the chimeric protein to Golgi apparatus.
The CAD(e35)-ALK(e20) fusion was identified in 1 case. An identical fusion was previously reported in 2 cases, ascending colon and rectal adenocarcinoma.11,16 CAD consists of 44 exons and encodes a multi-enzymatic protein associated with the enzymatic activity of the enzymes in the pathway of pyrimidine biosynthesis.43 A biallelic CAD loss-of-function has been associated with uridine-responsive epileptic encephalopathy.44 A patient with disseminated colon carcinoma harboring CAD-ALK fusion responded to ALK-inhibitor treatment.11
Fusions of ALK tyrosine kinase domain with new partners, DIAPH2 and LOC101929227, were identified in this study. A DIAPH2-ALK fusion occurred in two variants containing either 17 or 26 DIAPH2 exons. A protein encoded by DIAPH2 was indicated in the regulation of endosome dynamics and actin filament organization (https://www.uniprot.org). ALK immunohistochemistry revealed strong membrane expression in the tumor harboring DIAPH2(e17) variant and lack of membrane but strong cytoplasmic expression in the second case. Disruption of DIAPH2 due to a balanced translocation t(X;12)(q21;p13) has been linked to a family with a premature ovarian failure.45
LOC101929227, an uncharacterized RNA gene affiliated with the lncRNA class (www.genecards.org), partnered ALK in an unusual fusion involving the first two exons of LOC101929227 and twenty ALK exons (10–29) encoding part of the extracellular domain with second MAM (Meprin, A-5 protein, and receptor protein-tyrosine phosphatase Mu) domain, transmembrane (TM) and intracellular domains. In CRC, noncanonical ALK truncation has been identified once in the PPP4R3B(e11)-ALK(e2) fusion containing MAM and TM ALK domains (sequence available at The Cancer Genome Atlas under TCGA-F5–6864 code). The function of ALK extracellular domains, including MAM domains, is not well understood.46 This case was the only one that exhibited diffuse, weak cytoplasmic staining, but the fusion was confirmed by FISH.
Most ALK fusions reported in this study resulted from intrachromosomal rearrangements engaging ALK on chromosome 2p23.1 and fusion partner genes EML4 (ch2p21), CAD (ch2p23.3), SPTBN1 (ch2p16.2) and STRN (2p22.2) located in its vicinity. However, in three cases, DIAPH2 (Xq21.33)-ALK, LOC101929227 (11q23.3)-ALK and SLMAP (3p14.3)-ALK fusion genes were formed due to interchromosomal rearrangements. A few colorectal carcinomas harboring fusion genes, CENPF (1q41)-ALK, PRKAR1A (17q24.2)-ALK and PRKAR1B (7p22.3)-ALK, resulting from interchromosomal translocation have been described.13,22 Variants of ALK fusions reported in colorectal carcinomas in this study and previous studies are shown in Table 5.
Table 5.
Types of ALK fusions described in 38 colorectal carcinomas reported in this (n=12) and previously published studies.3,11––23
| Fusion gene partner | ALK | |||||
|---|---|---|---|---|---|---|
| Exon | 2 | 10 | 19 | 20 | NA | |
| C2orf44 | 4 | 1 | ||||
| CAD | 35 | 3 | ||||
| NA | 1 | |||||
| CENPF | 11 | 1 | ||||
| DIAPH2 | 17 | 1 | ||||
| 26 | 1 | |||||
|
EML4 |
2 | 1 | 1 | |||
| 6 | 2 | |||||
| 13 | 1 | |||||
| 20 | 2 | |||||
| 21 | 3 | |||||
| NA | 4 | |||||
| LOC101929227 | 2 | 1 | ||||
| MAPRE3 | 7 | 1 | ||||
| PPP4R3B | 11 | 1 | ||||
| PPP1R21 | 17 | 1 | ||||
| PRKAR1A | 2 | 1 | ||||
| PRKAR1B | 4 | 1 | ||||
| SLMAP | 14 | 1 | ||||
| SPTBN1 | 7 | 5 | ||||
| STRN | 3 | 4 | ||||
| Subtotal | 1 | 1 | 1 | 30 | 5 | |
| Total | 38 | |||||
ALK fusion tumors were diagnosed predominantly in women in their sixties or seventies (median age 72 years). In comparison, a median age of 50 years was previously reported for 11 patients with metastatic ALK fusion CRCs. 20 Because cases included in the current study were mostly from regional and university hospitals, this discrepancy might be related to the bias toward more advanced tumors in younger patients (in 4th or 5th decade) in cancer centers.
In this study, 11 ALK fusion tumors were classified as right sided based on whether they occurred before or after the splenic flexure, as this classification may better reflect tumor biology at different sites.47,48 Most of previously reported ALK fusion CRCs occurred in the right colon or rectum and only few were diagnosed in descending colon location. (Supplemental data Table S1).
Mucinous differentiation was seen in 25% (3/12) of tumors. Similarly, 31% (4/13) of previously reported ALK fusion tumors (Table S1) displayed mucinous component. This remains in the range of the previously published frequency of combined mucinous tumors and tumors with mucinous component in colorectal carcinomas.49 However mucinous histology was seen only in 1 of 8 metastatic colorectal carcinomas with ALK fusions. 20 ALK fusion colorectal carcinomas showed aberrant immunophenotypes with a frequent loss of CK7 and CK20 and MUC2 expression, occasionally accompanied by the partial loss of CDX2. Such antigenic patterns constituted a highly aggressive subgroup of poorly differentiated CRCs with early recurrences and shorter overall survivals.50–52
In colorectal cancer, a high level of tumor infiltrating lymphocytes has been considered a positive prognostic marker, particularly if paired with MMR-deficient status.53 In our cohort, 80% (8/10) of MMR-deficient tumors revealed high levels of tumor-infiltrating lymphocytes. Also, 4 MMR-deficient tumors showed diffuse or focal strong PD-L1 staining of tumor cells. A robust influx of PD-L1 positive tumor infiltrating immune cells were seen in 3 cases including one PD-L1 negative cancer. An association between PD-L1 expression and MMR-deficiency has been previously documented in CRCs.54 One study reported a durable response to anti PD-1 treatment in a case of MMR-deficient, EML4-ALK fusion colon cancer with partial PD-L1 positivity.20 Thus, screening of CRCs driven by ALK fusions for PD-L1 expression could be beneficial for the selection of checkpoint inhibitor therapy.
In this study, CRCs harboring ALK fusions revealed features characteristic of microsatellite-unstable colorectal tumors, such as female predominance (1:3 male to female ratio) and right colon location (>90%). Mismatch repair (MMR)-deficiency was identified by immunohistochemistry in 83% (10/12) of ALK fusion tumors analyzed in this study. A similar frequency of 73% (8/11) was previously recorded by a study on metastatic CRCs with ALK fusions.20 However, a review of previously reported ALK fusion CRCs with known MMR status revealed that 56% (9/16) were mismatch repair proficient tumors. 3,13–17,19,21,22 This questions the proposed rationale for ALK fusion screening for only MMR-deficient CRCs.21
BRAF mutations, typically seen in colon cancer with MMR-deficiency, were not detected in the ALK fusion tumors analyzed in this study. Additionally, other common colorectal carcinoma drivers such as K-RAS and, N-RAS were not involved. This highlights a primary oncogenic role of chimeric ALK fusion proteins. No BRAF p.V600E mutation has been identified in previously reported ALK fusion CRCs.14–16,20–22 However, 2 other BRAF mutations (one with biological significance) have been reported among 11 cases studied using next-generation sequencing. 20,22 Likewise, no oncogenic RAS mutations was identified in ALK fusion CRCs except KRAS p.G12A coexisting with EML4-ALK fusion in highly advanced, disseminated rectal tumor and KRAS p.G13D acquired during disease progression by STRN-ALK fusion cecal cancer.12,22 Thus, RAS involvement in that case appears secondary to an oncogenic ALK fusion.
In our cohort, genes encoding components of PI3K-AKT/MTOR signaling pathway, such as AKT, MTOR, PIK3CA, and TSC, were not affected by mutations. Previous studies reported similar results. 20,22
In most cases, TP53 mutations and nuclear accumulation of p53 protein (documented by immunohistochemistry) indicated pathologic modification of this pathway. This is in line with previously published studies reporting frequent TP53 missense mutations in ALK fusion colorectal carcinomas.20,22 One of TP53 mutants harbored a FBXW7p.Arg367* mutation. Mutational inactivation of FBXW7, a p53-dependent tumor suppressor gene, has been reported in approximately 10% of colorectal carcinomas and linked to tumor progression.55 Several ALK fusion CRCs have been reported to carry FBXW7 missense and truncating mutations.20,22
Molecular alterations of Wnt/β-catenin signaling pathway, common in non-hypermutated tumors, were not identified in this study. Also, previously published ALK fusion tumors infrequently carried Wnt/β-catenin signaling pathway mutations.20,21 However, relatively frequent mutations in the gene encoding RNF43, a protein considered to be negative regulator of the Wnt signaling, were reported.56 RNF43 was not included in the gene panel evaluated in this study.
Also, loss-of-function mutations affecting tumor suppressor genes ARID1A (AT-rich interactive domain-containing protein 1A) and ARID1B (AT-rich interactive domain-containing protein 1B), components of the human SWI/SNF chromatin remodeling complex, have been reported in ALK fusion tumors. 20 No ARID1A mutants were identified in this study, while ARID1B was not tested.
A recent study on ALK fusion colorectal carcinomas concluded extremely poor prognosis for these tumors. 20 In our cohort, 33% of patients died of the disease during a period of 2 and a half years. However, there were longer survivals (78 and 98 months alive without disease) including two patients with advanced (T3/T4) tumors and local metastases. Also, one recent study reported a patient with ALK fusion pT3N1M0 tumor as disease-free for 10 years following surgery and standard chemotherapy.17 Definitive conclusions of overall patient survival in ALK fusion CRC are currently limited by a small number of cases and relatively short follow-ups.
In summary, this study described clinicopathologic and molecular genetic features of rare colorectal cancers with oncogenic ALK fusions. Although these tumors had characteristics of MMR-deficient colon cancers, separation of ALK fusion tumors into a new molecular subtype is indicated due to the presence of additional therapeutic options.
Supplementary Material
REFERENCES
- 1.Menter DG, Davis JS, Broom BM, Overman MJ, Morris J, Kopetz S. Back to the Colorectal Cancer Consensus Molecular Subtype Future. Curr Gastroenterol Rep. 2019;21:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kumar-Sinha C, Kalyana-Sundaram S, Chinnaiyan AM. Landscape of gene fusions in epithelial cancers: seq and ye shall find. Genome Med. 2015;7:129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stransky N, Cerami E, Schalm S, et al. The landscape of kinase fusions in cancer. Nat Commun. 2014;5:4846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kloosterman WP, Coebergh van den Braak RRJ, Pieterse M, et al. A Systematic Analysis of Oncogenic Gene Fusions in Primary Colon Cancer. Cancer Res. 2017;77:3814–3822. [DOI] [PubMed] [Google Scholar]
- 5.Choi Y, Kwon CH, Lee SJ, et al. Integrative analysis of oncogenic fusion genes and their functional impact in colorectal cancer. Br J Cancer. 2018;119:230–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Morris SW, Kirstein MN, Valentine MB, Dittmer KG, Shapiro DN, Saltman DL, Look AT. Fusion of a kinase gene, ALK, to a nucleolar protein gene, NPM, in non-Hodgkin’s lymphoma. Science. 1994;263:1281–1284. [DOI] [PubMed] [Google Scholar]
- 7.Iwahara T, Fujimoto J, Wen D, et al. Molecular characterization of ALK, a receptor tyrosine kinase expressed specifically in the nervous system. Oncogene. 1997;14:439–449. [DOI] [PubMed] [Google Scholar]
- 8.Holla VR, Elamin YY, Bailey AM, et al. ALK: a tyrosine kinase target for cancer therapy. Cold Spring Harb Mol Case Stud. 2017;3:a001115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Menichincheri M, Ardini E, Magnaghi P, et al. Discovery of Entrectinib: A New 3-Aminoindazole As a Potent Anaplastic Lymphoma Kinase (ALK), c-ros Oncogene 1 Kinase (ROS1), and Pan-Tropomyosin Receptor Kinases (Pan-TRKs) inhibitor. J Med Chem. 2016;59:3392–3408. [DOI] [PubMed] [Google Scholar]
- 10.Rolfo C, Ruiz R, Giovannetti E, et al. Entrectinib: a potent new TRK, ROS1, and ALK inhibitor. Expert Opin Investig Drugs. 2015;24:1493–1500. [DOI] [PubMed] [Google Scholar]
- 11.Amatu A, Somaschini A, Cerea G, et al. Novel CAD-ALK gene rearrangement is drugable by entrectinib in colorectal cancer. Br J Cancer. 2015;113:1730–1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aisner DL, Nguyen TT, Paskulin DD, et al. ROS1 and ALK fusions in colorectal cancer, with evidence of intratumoral heterogeneity for molecular drivers. Mol Cancer Res. 2014;12:111–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eddy S, Toile M, Brick ME et al. Expanded clinical opportunities for crizotinib from an analysis of over 5,000 cancer patient exomes. AACR-NCI-EORTC International Conference on Molecular Targets and Cancer Therapeutics. 2013. abstract C256. [Google Scholar]
- 14.Houang M, Toon CW, Clarkson A, et al. ALK and ROS1 overexpression is very rare in colorectal adenocarcinoma. Appl Immunohistochem Mol Morphol. 2015;23:134–138. [DOI] [PubMed] [Google Scholar]
- 15.Lai AZ, Schrock AB, Erlich RL, et al. Detection of an ALK Fusion in Colorectal Carcinoma by Hybrid Capture-Based Assay of Circulating Tumor DNA. Oncologist. 2017;22:774–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee J, Kim HC, Hong JY, et al. Detection of novel and potentially actionable anaplastic lymphoma kinase (ALK) rearrangement in colorectal adenocarcinoma by immunohistochemistry screening. Oncotarget. 2015;6:24320–24332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee CH, Lee CT, Chen YL et al. Detection of anaplastic lymphoma kinase gene rearrangement in a patient with right colon cancer. J Cancer Res Pract. 2019;6:89–91 [Google Scholar]
- 18.Lin E, Li L, Guan Y, Rivers CS, Mohan S, Pandita A, et al. Exon array profiling detects EML4-ALK fusion in breast, colorectal, and non-small cell lung cancers. Mol CancerRes. 2009;7:1466–1476. [DOI] [PubMed] [Google Scholar]
- 19.Lipson D, Capelletti M, Yelensky R, et al. Identification of new ALK and RET gene fusions from colorectal and lung cancer biopsies. Nat Med. 2012;18:382–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pietrantonio F, Di Nicolantonio F, Schrock AB, et al. ALK, ROS1, and NTRK Rearrangements in Metastatic Colorectal Cancer. J Natl Cancer Inst. 2017;109(12). [DOI] [PubMed] [Google Scholar]
- 21.Wang J, Yi Y, Xiao Y, et al. Prevalence of recurrent oncogenic fusion in mismatch repair-deficient colorectal carcinoma with hypermethylated MLH1 and wild-type BRAF and KRAS. Mod Pathol. 2019;32:1053–1064. [DOI] [PubMed] [Google Scholar]
- 22.Yakirevich E, Resnick MB, Mangray S, et al. Oncogenic ALK Fusion in Rare and Aggressive Subtype of Colorectal Adenocarcinoma as a Potential Therapeutic Target. Clin Cancer Res. 2016;22:3831–3840. [DOI] [PubMed] [Google Scholar]
- 23.Ying J, Lin C, Wu J, et al. Anaplastic Lymphoma Kinase Rearrangement in Digestive Tract Cancer: Implication for Targeted Therapy in Chinese Population. PLoS One. 2015;10:e0144731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lasota J, Chłopek M, Lamoureux J, et al. Colonic Adenocarcinomas Harboring NTRK Fusion Genes: A Clinicopathologic and Molecular Genetic Study of 16 Cases and Review of the Literature. Am J Surg Pathol. 2020;44:162–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nagtegaal ID, Arends MJ, Salto-Tellez M. Colorectal adenocarcinoma. In: WHO Classification of Tumours: Digestive System Tumours, 5th ed., International Agency for Research on Cancer, Lyon, France, 2019, pp. 177–187. [Google Scholar]
- 26.Greenson JK, Huang SC, Herron C, et al. Pathologic predictors of microsatellite instability in colorectal cancer. Am J Surg Pathol. 2009;33:126–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Soda M, Choi YL, Enomoto M, et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature. 2007;448:561–566. [DOI] [PubMed] [Google Scholar]
- 28.Sokai A, Enaka M, Sokai R, et al. Pulmonary inflammatory myofibroblastic tumor harboring EML4-ALK fusion gene. Jpn J Clin Oncol. 2014;44:93–96. [DOI] [PubMed] [Google Scholar]
- 29.Lin E, Li L, Guan Y, et al. Exon array profiling detects EML4-ALK fusion in breast, colorectal, and non-small cell lung cancers. Mol CancerRes. 2009;7:1466–1476. [DOI] [PubMed] [Google Scholar]
- 30.Shen Q, Wang X, Yu B, et al. Comparing four different ALK antibodies with manual immunohistochemistry (IHC) to screen for ALK-rearranged non-small cell lung cancer (NSCLC). Lung Cancer. 2015;90:492–498. [DOI] [PubMed] [Google Scholar]
- 31.Houtman SH, Rutteman M, De Zeeuw CI, et al. Echinoderm microtubule-associated protein like protein 4, a member of the echinoderm microtubule-associated protein family, stabilizes microtubules. Neuroscience. 2007;144:1373–1382. [DOI] [PubMed] [Google Scholar]
- 32.Gu FF, Zhang Y, Liu YY, et al. Lung adenocarcinoma harboring concomitant SPTBN1-ALK fusion, c-Met overexpression, and HER-2 amplification with inherent resistance to crizotinib, chemotherapy, and radiotherapy. J Hematol Oncol. 2016;9:66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grand FH, Iqbal S, Zhang L, et al. A constitutively active SPTBN1-FLT3 fusion in atypical chronic myeloid leukemia is sensitive to tyrosine kinase inhibitors and immunotherapy. Exp Hematol. 2007;35:1723–1727. [DOI] [PubMed] [Google Scholar]
- 34.Gallagher G, Horsman DE, Tsang P, et al. Fusion of PRKG2 and SPTBN1 to the platelet-derived growth factor receptor beta gene (PDGFRB) in imatinib-responsive atypical myeloproliferative disorders. Cancer Genet Cytogenet. 2008;181:46–51. [DOI] [PubMed] [Google Scholar]
- 35.An X, Guo X, Gratzer W, et al. Phospholipid binding by proteins of the spectrin family: a comparative study. Biochem Biophys Res Commun. 2005;327:794–800. [DOI] [PubMed] [Google Scholar]
- 36.Liem RK. Cytoskeletal Integrators: The Spectrin Superfamily. Cold Spring Harb Perspect Biol. 2016. Oct 3;8(10). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kelly LM, Barila G, Liu P, et al. Identification of the transforming STRN-ALK fusion as a potential therapeutic target in the aggressive forms of thyroid cancer. Proc Natl Acad Sci U S A. 2014;111:4233–4238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kusano H, Togashi Y, Akiba J, et al. Two Cases of Renal Cell Carcinoma Harboring a Novel STRN-ALK Fusion Gene. Am J Surg Pathol. 2016;40:761–769. [DOI] [PubMed] [Google Scholar]
- 39.Tang Y, Chen M, Zhou L, et al. Architecture, substructures, and dynamic assembly of STRIPAK complexes in Hippo signaling. Cell Discov. 2019;5:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hwang J, Pallas DC. STRIPAK complexes: structure, biological function, and involvement in human diseases. Int J Biochem Cell Biol. 2014;47:118–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shi Z, Jiao S, Zhou Z. STRIPAK complexes in cell signaling and cancer. Oncogene. 2016;35:4549–4557. [DOI] [PubMed] [Google Scholar]
- 42.Pagan C, Barua S, Hsiao SJ, et al. Targeting SLMAP-ALK-a novel gene fusion in lung adenocarcinoma. Cold Spring Harb Mol Case Stud. 2019;5(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Simmer JP, Kelly RE, Rinker AG Jr, et al. Mammalian dihydroorotase: nucleotide sequence, peptide sequences, and evolution of the dihydroorotase domain of the multifunctional protein CAD. Proc Natl Acad Sci U S A. 1990;87:174–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Koch J, Mayr JA, Alhaddad B, et al. CAD mutations and uridine-responsive epileptic encephalopathy. Brain. 2017;140:279–286. [DOI] [PubMed] [Google Scholar]
- 45.Bione S, Sala C, Manzini C, et al. A human homologue of the Drosophila melanogaster diaphanous gene is disrupted in a patient with premature ovarian failure: evidence for conserved function in oogenesis and implications for human sterility. Am J Hum Genet. 1998;62:533–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Huang H.Anaplastic Lymphoma Kinase (ALK) Receptor Tyrosine Kinase: A Catalytic Receptor with Many Faces. Int J Mol Sci. 2018;19(11). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee GH, Malietzis G, Askari A, et al. Is right-sided colon cancer different to left-sided colorectal cancer? - a systematic review. Eur J Surg Oncol. 2015;41:300–308. [DOI] [PubMed] [Google Scholar]
- 48.Loree JM, Pereira AAL, Lam M, et al. Classifying Colorectal Cancer by Tumor Location Rather than Sidedness Highlights a Continuum in Mutation Profiles and Consensus Molecular Subtypes. Clin Cancer Res. 2018;24:1062–1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Langner C, Harbaum L, Pollheimer MJ, et al. Mucinous differentiation in colorectal cancer--indicator of poor prognosis? Histopathology. 2012;60:1060–1072. [DOI] [PubMed] [Google Scholar]
- 50.Kim JH, Rhee YY, Bae JM, et al. Loss of CDX2/CK20 expression is associated with poorly differentiated carcinoma, the CpG island methylator phenotype, and adverse prognosis in microsatellite-unstable colorectal cancer. Am J Surg Pathol. 2013;37:1532–1541. [DOI] [PubMed] [Google Scholar]
- 51.Yamagishi H, Imai Y, Okamura T, et al. Aberrant cytokeratin expression as a possible prognostic predictor in poorly differentiated colorectal carcinoma. J Gastroenterol Hepatol. 2013;28:1815–1822. [DOI] [PubMed] [Google Scholar]
- 52.Li C, Zuo D, Yin L, et al. Prognostic Value of MUC2 Expression in Colorectal Cancer: A Systematic Review and Meta-Analysis. Gastroenterol Res Pract. 2018;2018:6986870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Williams DS, Mouradov D, Jorissen RN, et al. Lymphocytic response to tumour and deficient DNA mismatch repair identify subtypes of stage II/III colorectal cancer associated with patient outcomes. Gut. 2019;68:465–474. [DOI] [PubMed] [Google Scholar]
- 54.Inaguma S, Lasota J, Wang Z, et al. Clinicopathologic profile, immunophenotype, and genotype of CD274 (PD-L1)-positive colorectal carcinomas. Mod Pathol. 2017;30:278–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yeh CH, Bellon M, Nicot C. FBXW7: a critical tumor suppressor of human cancers. Mol Cancer. 2018;17:115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Koo BK, Spit M, Jordens I, Low TY, Stange DE, van de Wetering M, van Es JH, Mohammed S, Heck AJ, Maurice MM, Clevers H. Tumour suppressor RNF43 is a stem-cell E3 ligase that induces endocytosis of Wnt receptors. Nature. 2012;488:665–669. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.