Abstract
The tbu regulon of Ralstonia pickettii PKO1 encodes enzymes involved in the catabolism of toluene, benzene, and related alkylaromatic hydrocarbons. The first operon in this regulon contains genes that encode the tbu pathway's initial catabolic enzyme, toluene-3-monooxygenase, as well as TbuT, the NtrC-like transcriptional activator for the entire regulon. It has been previously shown that the organization of tbuT, which is located immediately downstream of tbuA1UBVA2C, and the associated promoter (PtbuA1) is unique in that it results in a cascade type of up-regulation of tbuT in response to a variety of effector compounds. In our efforts to further characterize this unusual mode of gene regulation, we discovered another open reading frame, encoded on the strand opposite that of tbuT, 63 bp downstream of the tbuT stop codon. The 1,374-bp open reading frame, encoding a 458-amino-acid peptide, was designated tbuX. The predicted amino acid sequence of TbuX exhibited significant similarity to several putative outer membrane proteins from aromatic hydrocarbon-degrading bacteria, as well as to FadL, an outer membrane protein needed for uptake of long-chain fatty acids in Escherichia coli. Based on sequence analysis, transcriptional and expression studies, and deletion analysis, TbuX seems to play an important role in the catabolism of toluene in R. pickettii PKO1. In addition, the expression of tbuX appears to be regulated in a manner such that low levels of TbuX are always present within the cell, whereas upon toluene exposure these levels dramatically increase, even more than those of toluene-3-monooxygenase. This expression pattern may relate to the possible role of TbuX as a facilitator of toluene entry into the cell.
Ralstonia pickettii PKO1 has been investigated by our laboratory for several years as a model microorganism representative of those bacteria capable of metabolizing alkylaromatic hydrocarbons in oxygen-limited (hypoxic) aquifer environments (23, 34, 42, 43). The tbu pathway of R. pickettii PKO1, which encodes enzymes for utilization of benzene, toluene, and related alkylaromatic hydrocarbons as well as enabling this strain to transform trichloroethylene (TCE), has been cloned as a 26.5-kbp DNA fragment designated pRO1957 (41). The genes encoding enzymes for this catabolic pathway have been shown previously to be organized into three operons: the tbuA1UBVA2C and tbuT operon encoding the initial toluene-3-monooxygenase and the transcriptional activator TbuT (6), the tbuD operon encoding phenol/cresol hydroxylase (24, 26), and the tbuWEFGKIHJ operon encoding enzymes of the meta-cleavage pathway for conversion of catechol and methylcatechols to tricarboxylic acid cycle intermediates (25). We have previously shown through physiological analysis as well as through transcriptional fusion analysis of promoter regions that TbuT controls transcription of each of these operons in response to aromatic effector compounds. Moreover, it has been shown that the unique organization of tbuT, which is located immediately downstream of tbuA1UBVA2C, and the associated promoter (PtbuA1), results in a cascade type of up-regulation of tbuT in response to a variety of effector compounds, including toluene and TCE (6). In our efforts to further characterize this unusual mode of gene regulation, we sequenced the DNA region immediately downstream of tbuT and identified an open reading frame whose deduced amino acid sequence shared significant homology with a number of putative outer membrane proteins from aromatic hydrocarbon-degrading bacteria as well as homology to FadL (4), an outer membrane protein needed for uptake of long-chain fatty acids in Escherichia coli. We report here an analysis of the function of this gene, designated tbuX, in toluene utilization.
MATERIALS AND METHODS
Bacterial strains, plasmids and culture conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. E. coli JM109 and DH5α were used for routine maintenance and construction of plasmids and for construction of fragments used for DNA sequence analysis. The mobilization plasmid pRK2013 (12) carried in E. coli MM294 was used to mobilize plasmid pKRZ1 and its derivatives from E. coli DH5α into Pseudomonas aeruginosa PAO1c via triparental matings. P. aeruginosa transconjugants resulting from triparental matings were selected on the minimal medium of Vogel and Bonner (59) supplemented with 0.5% glucose. E. coli cells containing recombinant plasmids were always maintained on Luria-Bertani (LB) medium (49) supplemented with the antibiotics ampicillin (50 μg/ml), tetracycline (25 μg/ml), or kanamycin (75 μg/ml) as needed. P. aeruginosa PAO1c cells containing recombinant plasmid constructs were maintained on plate count medium (TNA [39]) containing carbenicillin (500 μg/ml), kanamycin (600 μg/ml), or tetracycline (50 μg/ml) as needed. When cells were grown for enzyme assays or for high-pressure liquid chromatography (HPLC) analyses of metabolites, all strains were routinely grown in a basal salts medium (BM [34]) containing (per liter) 2.49 g of Na2HPO4, 3.05 g of KH2PO4, 0.1995 g of MgSO4, 0.995 g of CaCl2, 0.00005 g of FeSO4, 0.00025 g of NaMoO4, 1.0 ml of Hunter's trace metal solution (7), 1.0 g of (NH4)2SO4, 1.0 g of KNO3, 0.1% Casamino Acids (Difco Laboratories, Detroit, Mich.), and 0.25% glucose. When used for enzyme induction experiments, liquid toluene was added directly to BM to a final concentration of 2.8 mM. Growth of liquid cultures and incubation of agar plates was carried out at 37°C except when cultures contained aromatic hydrocarbons, in which case incubations were done at 30°C.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristicsa | Reference or source |
|---|---|---|
| Strains | ||
| E. coli | ||
| DH5α | F− φ80dlacZΔM15 Δ(lacZYA-argF)U169 deoR recA1 endA1 phoA hsdR17 (rK− mK+) supE44 λ− thi-1 gyrA96 relA1 | 15 |
| JM109 | recA1 relA1 thi Δ(lac-proAB) gyrA96 hsdR17 endA1 supE44 | Promega |
| MM294 | supE44 hsdR endA1 pro thi | D. Clewell |
| R. pickettii PKO1 | Tol+ Phl+ | 41 |
| P. aeruginosa PAO1c | Prototroph | 18 |
| Plasmids | ||
| pBluescript II KS+(SK+) | lacZ Apr, 2.96-kb cloning vector | Stratagene |
| pKRZ1 | AprKmr; broad-host-range promoter probe vector containing lacZ as a reporter gene | A. Chakrabarty |
| pRO1727 | Tcr Cbr; 3.77-kb Pseudomonas cloning vector | 8 |
| pRO1614 | Tcr Cbr; 6-kb E. coli-Pseudomonas cloning vector | 38 |
| p352X/S | Apr Kmr; pKRZ1:: 352-bp XhoI-SmaI fragment; PtbuA1-lacZ fusion | 6 |
| p3.4kbX/B | Apr Kmr; pKRZ1:: 3.4-kbp XhoI-BglII fragment; PtbuD-lacZ fusion | 40 |
| p454X/S | Apr Kmr; pKRZ1:: 454-bp XhoI-SmaI fragment; PtbuX-lacZ fusion | This study |
| pRO1959 | Tcs Cbr; pRO1727::HindIII-BamHI (15.1-kbp) fragment of R. pickettii PKO1 containing tbuD, tbuA1UBVA2C, tbuT, and tbuX | 24 |
| pRO1966 | Tcs Cbr; pRO1727::ClaI-BamHI (10-kbp) fragment of pRO1959 containing tbuA1UBVA2C, tbuT, and tbuX | 41 |
| pHYK1000 | Tcs Cbr; pRO1727::ClaI-PvuII (1.96-kbp) fragment of pRO1966 containing tbuA1UBVA2C and tbuT | This study |
| pHYK1001 | Tcr Cbs pRO1614::EcoRI-PvuII (3.1-kbp) fragment of pRO1966 containing tbuT | 6 |
Abbreviations: Tol, toluene; Phl, phenol; Ap, ampicillin; Cb, carbenicillin; Tc, tetracycline; Km, kanamycin.
Toluene utilization assays.
Cultures were grown in 100 ml of BM with 0.1% Casamino Acids, 0.25% glucose, and 2.8 mM toluene in tightly stoppered 500-ml bottles at 30°C with shaking until they reached the late log phase. Cells were harvested by centrifugation at 10,000 × g at 4°C and were washed twice with 40 mM potassium-sodium phosphate buffer (pH 6.8). Washed cells were transferred to 10 ml of the same buffer containing toluene (2.8 mM) to produce an A425 of 1.0. These washed cell suspensions were incubated with shaking at 30°C for 24 h, and 500-μl samples were taken every 4 h, mixed with 500 μl of methanol in 1.5-ml microcentrifuge tubes, and then centrifuged at 4°C for 10 min to remove cells. The resulting supernatants were carefully transferred to autosampler vials and were analyzed by reverse-phase HPLC. Uninoculated bottles, which served as controls, were incubated under the same conditions, and results were corrected for toluene losses from the controls (which were always <5% of the initial toluene concentration). Reverse-phase chromatography was performed with a PhaseSep H4726 column (4.6 by 250 mm) filled with Spherisorb ODS2 (particle diameter, 5 μm) preceded by a Whatman CSKI guard column (6.5 by 65 mm) coupled to a Shimadzu SCL-6B solvent delivery system and a CR501 Chromatopac computing integrator. A methanol-water (90:10) solvent was used at a flow rate of 1 ml/min. Toluene was detected by monitoring at A254, and concentration was calculated by comparison with a standard curve as described previously (41).
Deletion mutagenesis and general molecular techniques.
For construction of a DNA fragment lacking tbuX, plasmid pRO1959 (Table 1) was digested with restriction endonucleases ClaI and BamHI. The resultant 10-kb ClaI-BamHI DNA fragment (which when cloned into vector pRO1727 was designated pRO1966 [Table 1]) was extracted following electrophoretic separation in a 1.2% agarose gel, partially digested with PvuII (Fig. 1), and then ligated with ClaI- and EcoRV-digested vector plasmid pRO1727. The ligation mixture was introduced into P. aeruginosa PAO1c by electroporation using the method of Smith and Iglewski (56), and electrotransformants were selected on TNA medium containing carbenicillin (500 μg/ml). Plasmids with the expected deletion were confirmed by ClaI and BamHI digestion patterns of purified DNAs obtained from selected clones. The desired tbuX deletion construct was designated pHYK1000 (Table 1). Other DNA restriction digests, ligations, and transformation procedures were performed according to previously published methods (38, 40, 49).
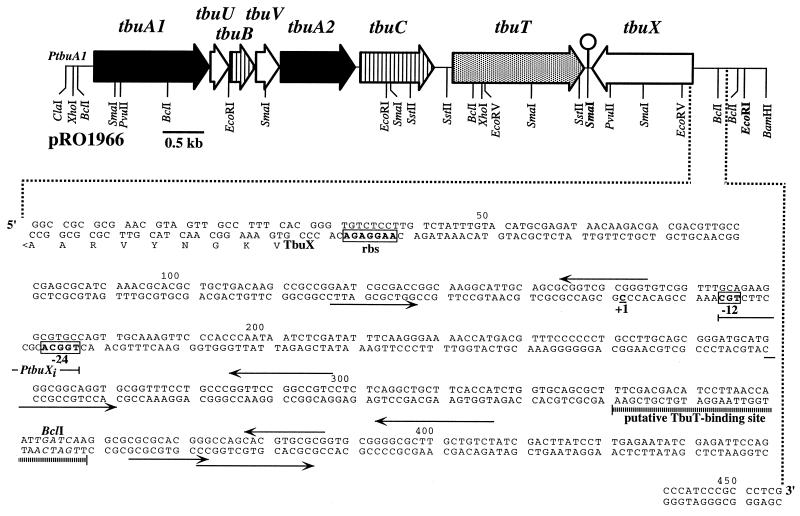
FIG. 1.
Physical and genetic maps of the 9.4-kb ClaI-BamHI fragment of pRO1966. The scale bar represents 0.5 kbp. The large arrows represent the locations and orientations of the genes (identified at the top) which comprise the toluene-3-monooxygenase operon (tbuA1UBVA2C and the positive regulator tbuT) and the tbuX gene. The lollipop symbol between the tbuT and tbuX genes indicates the location of a putative transcriptional terminator. The location of the toluene-3-monooxygenase promoter, PtbuA1, is also shown. Below the restriction map, the putative tbuX promoter region with relevant flanking DNA is expanded. The putative TbuT-binding site is demarcated by the horizontal hatched bar. The −12 and −24 sequences of the tbuX promoter, PtbuX, are boxed and labeled accordingly. The putative ribosome-binding site is boxed and labeled rbs. The first nine amino acids from the deduced amino acid sequence of the tbuX gene (labeled TbuX) are shown in one-letter code beneath the corresponding DNA coding region. The underlined C residue with a +1 designation identifies the tbuX transcriptional start site as determined by primer extension analyses. The BclI restriction endonuclease recognition sequence is shown in italics. The location and lengths of various sets of inverted repeats are shown by pairs of arrows.
DNA sequence analysis.
Nucleotide sequencing was initially carried out manually by the dideoxy-chain termination method of Sanger et al. (50), with a Sequenase version 1.0 kit (U.S. Biochemical, Cleveland, Ohio), [α-32P]dATP, and T7, T3, or specific synthetic oligonucleotide primers, as previously described (6). Later sequencing efforts were carried out using an ABI 373A automated sequencer. For PCR amplification of fragments to be sequenced, a total 10-μl reaction mixture containing 0.2 μg of template DNA, 1.6 pmol of primer, and 1 U of Amplitaq FS (Gibco BRL, Gaithersburg, Md.) was used for 25 cycles of 10 s at 96°C, 5 s at 50°C, and 4 min at 60°C. Sequence analysis was done with Lasergene software (DNA STAR, Inc., Madison, Wis.), MacVector version 4.5.3 (Oxford Molecular, Campbell, Calif.), and the Genetics Computer Group (GCG; University of Wisconsin, Madison) software package, version 8.1. Searches of the GenBank database and pairwise sequence comparisons were carried out with GCG programs TFASTA and BESTFIT, respectively. Similarity searches were also performed with the BLAST program and the National Center for Biotechnology Information databases. Nucleotide sequence alignments were done by using the GCG multiple sequence alignment program PILEUP or the Lasergene program MEGALIGN.
PCR.
PCRs for amplification of a DNA fragment containing the putative tbuX promoter region were carried out in 100-μl volumes using a GeneAmp kit (Perkin-Elmer, Branchburg, N.J.) and thermal cycler 480 (Perkin-Elmer Cetus, Norwalk, Conn.) essentially as previously described (6. A. M. Byrne and R. H. Olsen, Abstr. 96th Gen. Meet. Am. Soc. Microbiol. 1996, abstr. Q-356, p. 447, 1996). The following oligonucleotide primers, synthesized by the DNA Core facility at the University of Michigan, were used: 5′-GGCGCTCGAGCGAGGGCGGGATGGGCTGG (nucleotides 455 to 437 [Fig. 1]) and 5′-AGTTCCCGGGGGCCGCGCGAACGTAGTTGC (nucleotides 1 to 20 [Fig. 1]). The incorporation of an XhoI or SmaI restriction endonuclease site at the 5′ ends (underlined) of the primers allowed for directional cloning of the PCR product into SalI-SmaI cleaved pKRZ1 vector, resulting in plasmid p454X/S (Table 1).
Protein analysis.
Cells of P. aeruginosa PAO1c carrying pRO1966 or pHYK1000 (Table 1) were grown overnight at 37°C with shaking in 50 ml of BM with 0.1% Casamino Acids, 0.25% glucose, 500 μg of carbenicillin per ml, and 1 mM toluene. Cells harvested by centrifugation at 4°C were washed twice in 10 ml of 40 mM sodium-potassium phosphate buffer (pH 6.8) and then transferred in 10 ml of this buffer containing 2.8 mM toluene. The cells were incubated with shaking at 30°C for 12 h to ensure tbu pathway gene expression. Following this induction period, cells were harvested by centrifugation at 4°C. The harvested cells were lysed directly in a lysis buffer consisting of 25% glycerol, 14.4 mM β-mercaptoethanol, 2% sodium dodecyl sulfate (SDS), 60 mM Tris-HCl (pH 6.8), and 0.1% bromophenol blue. Lysis was achieved by incubation at 95°C for 5 min. The cell debris was removed by centrifugation at 14,000 × g for 20 min, and the supernatant was used for SDS-polyacrylamide gel electrophoresis (SDS-PAGE) using a Mini-Protean II system (Bio-Rad, Hercules, Calif.) according to the method of Laemmli (30). Gels were run for 3 h at 120 V through a 5% acrylamide stacking gel and 15% acrylamide separating gel. Prestained protein molecular weight standards (Gibco BRL) were used for molecular mass estimation. Gels were stained with Coomassie brilliant blue, destained in methanol-glacial acetic acid-water (1:1:8, vol:vol:vol), blotted onto Whatman 3-MM paper, and dried under vacuum at 70°C.
In vivo analysis of promoter activity.
Promoter activity, assessed by in vivo transcomplementation, was determined by assaying β-galactosidase activity in cells of P. aeruginosa PAO1c essentially as described previously (6, 40). P. aeruginosa PAO1c cells harboring pKRZ1 and its derivatives together with pRO1614 and its derivatives were grown overnight in LB broth containing kanamycin (600 μg/ml) and tetracycline (50μg/ml) to maintain plasmids. Cells were subsequently diluted in the same medium and grown for 8 h in the absence or presence of toluene. β-Galactosidase activity was assayed as described by Miller (35) except that cells were permeabilized by addition of chloroform and SDS. Reported β-galactosidase activity values, which represent the average of at least three independent experiments, are expressed in units as specified by Miller (35).
RNA preparation and primer extension analysis.
Total RNA was isolated from toluene-induced and uninduced cells of P. aeruginosa PAO1c carrying p454X/S in trans with pHYK1001, which has a 3.1-kb EcoRI-StuI DNA fragment expressing tbuT carried on plasmid pRO1614 (Table 1). Cells were grown under the conditions used for β-galactosidase assays, described above. Total RNA was extracted from 2 ml of the cultures using Trizol reagent (Gibco BRL) according to the manufacturer's recommended protocol, except that after the cells were mixed with the Trizol reagent, the suspension was incubated at 68°C for 10 min and then allowed to cool to room temperature. The remainder of the RNA extraction protocol was carried out as previously described (6, 27, 40). The concentration and purity of the RNA were estimated from the A260/A280 ratio and from visual inspection of RNA bands following electrophoretic analysis.
The 5′ ends of transcripts were determined by primer extension analysis with oligonucleotide primer 5′-GGCCGCGCGAACGTAGTTGC, which was complementary to nucleotides 8 to 27 of the deduced coding sequence of tbuX (Fig. 1). The primer was labeled at its 5′ end with [γ-32P]ATP (ICN Biomedicals, Costa Mesa, Calif.), using T4 polynucleotide kinase as described previously (49). The oligonucleotides (2 × 105 cpm) were annealed with 50 μg of RNA in 10 μl of hybridization buffer (50 mM Tris-HCl [pH 7.7], 100 mM KCl) at 95°C for 3 min, transferred to 65°C for 5 min, and then slowly cooled to 42°C over a 30-min interval. The samples were chilled on ice, and 2 μl of 5× Superscript RT buffer (250 mM Tris-HCl [pH 8.3], 375 mM KCl, 15 mM MgCl2), 25 mM dithiothreitol, 2.5 mM each deoxynucleoside triphosphate, and 6 U of RNasin RNase inhibitor (Promega Corp., Madison, Wis.) were added. The samples were then warmed to 42°C, and 1 μl (25 U) of Superscript RT RNase H− reverse transcriptase (Gibco BRL) was added. Extension reaction mixtures were incubated at 42°C for 1 h, after which 5 μl of stop solution containing 95% formamide, 20 mM EDTA, 0.05% bromophenol blue, and 0.05% xylene cyanol FF was added. The products of the extension reactions were resolved on 8% polyacrylamide gels containing 8 M urea.
Chemicals and reagents.
All chemicals used in this study were of the highest purity commercially available. Aromatic hydrocarbons were purchased from Sigma Chemical Co. (St. Louis, Mo.). Components for cell growth were purchased from Difco, Aldrich Chemical Co. (St. Louis, Mo.), and Gibco BRL. Enzymes and reagents used for nucleic acid manipulations were purchased from Promega, Gibco BRL, and Stratagene (La Jolla, Calif.).
Nucleotide sequence accession number.
The sequence data in this report have been submitted to the GenBank data library under accession no. AF100782.
RESULTS
Identification and nucleotide sequence analysis of the tbuX gene of R. pickettii PKO1.
Our previous studies on regulation of the tbu pathway genes from R. pickettii PKO1 reported an unusual organization of tbuT, the gene encoding the transcriptional activator, TbuT. The unusual feature involves the organization of the toluene-3-monooxygenase genes (tbuA1UBVA2) and the regulator tbuT with respect to PtbuA1. This organization allows for a cascade type of regulation where the presence of an effector molecule (an inducer) and TbuT results in induced expression of tbuA1UBVA2C and simultaneously increases expression of tbuT via readthrough transcription through tbuA1UBVA2C. This further elevates expression of tbuA1UBVA2C if additional effector molecules are present (6). In our efforts to further characterize the overall regulation of the tbu pathway of strain PKO1, we sequenced the region 3′ of the translational stop of tbuT and in the process identified an incomplete open reading frame whose deduced amino acid sequence shared significant homology with several putative membrane proteins from other aromatic hydrocarbon-degrading bacteria. To further explore this DNA region, we determined the complete nucleotide sequence of a 2,093-bp SmaI-EcoRI fragment of pRO1966 (Fig. 1), which is a 10-kb ClaI-BamHI subclone of pRO1957 that contains the genes encoding toluene-3-monooxygenase and the transcriptional activator, TbuT (5). Analysis of the nucleotide sequence revealed a 1,374-bp open reading frame that we have designated tbuX. The tbuX gene is encoded on the opposite DNA strand from tbuT and is located 63 bp downstream of the tbuT stop codon (Fig. 1). This open reading frame encodes a putative peptide of 458 amino acids with an estimated molecular mass of 47.8 kDa and estimated pI of 8.7. Determination of the probable 5′ end of tbuX was based on the identification of a putative ribosome-binding site (5′-AAGGAGA-3′) and the extensive homology with genes from other organisms (as discussed below). As found for the other genes of the tbu pathway, the tbuX coding region is G+C rich (63.8%) and accordingly displays a preferential use of codons with either a G or a C residue in the third position.
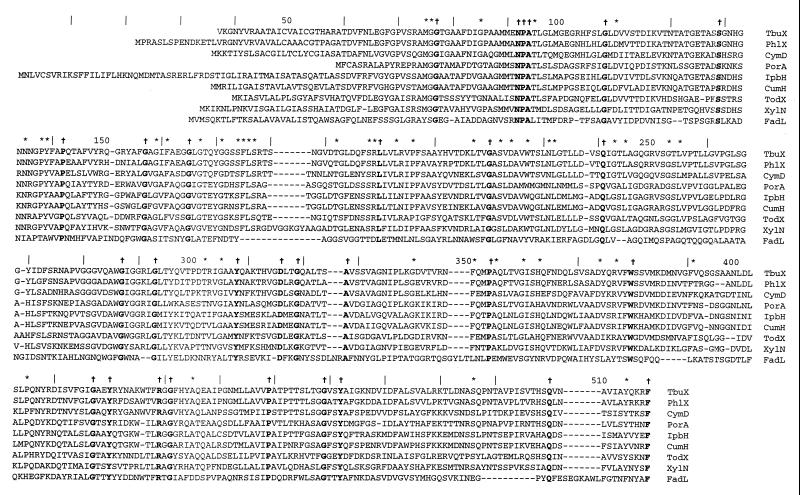
Comparisons of the deduced amino acid sequence of TbuX with translated sequences from the GenBank database revealed that TbuX shares homology with a group of putative membrane-associated proteins from several hydrocarbon-degrading bacteria, including PhlX from Ralstonia eutropha JMP134 (GenBank accession no. AF065891; unpublished), CymD from Pseudomonas putida F1 (10), PorA from Pseudomonas sp. strain Y2 (58), IpbH from P. putida RE204 (GenBank accession no. AF006691; unpublished), CumH from P. fluorescens IP01 (14), TodX from P. putida F1 (60), and XylN from P. putida PaW1 (GenBank accession no. D63341; unpublished). TbuX and these related proteins all shared a low degree of overall homology with FadL from E. coli (4), which is an outer membrane protein required for binding and transport of long-chain fatty acids. Pairwise comparisons of the deduced amino acid sequence of TbuX with those of PhlX, CymD, PorA, IpbH, CumH, TodX, XylN, and FadL revealed 60.3, 43.2, 37.4, 36.9, 36.5, 33.5, 31.4, and 12.8% identities, respectively, and 77.1, 55.2, 47.8, 47.8, 46.6, 44.4, 40.2, and 14.3% similarities, respectively (Fig. 2).
FIG. 2.
Multiple sequence alignment of TbuX from R. pickettii PKO1, PhlX from R. eutropha JMP134 (GenBank accession no. AF065891 [unpublished]), CymD from P. putida F1 (10), PorA from Pseudomonas sp. strain Y2 (58), IpbH from P. putida RE204 (GenBank accession no. AF006691 [unpublished]), CumH from P. fluorescens IP01 (14), TodX from P. putida F1 (60), XylN from P. putida PaW1 (GenBank accession no. D63341 [unpublished]), and FadL from E. coli (4). The amino acid residues conserved in all nine sequences are indicated in boldface and marked with daggers. Residues conserved in TbuX, PhlX, CymD, PorA, IpbH, CumH, TodX, and XylN are indicated with asterisks. Gaps are represented by dashes and were introduced to maximize the alignment. The multiple sequence alignment analysis was carried out using the Clustal method within the MEGALIGN program of Lasergene.
Analysis of the tbuX promoter and regulatory region.
Examination of the nucleotide sequence upstream of the tbuX gene revealed a promoter containing the invariant −24GG/−12GC sequence unique to ς54 (rpoN)-dependent promoters (Fig. 3A). A putative TbuT-binding site was also identified approximately 150 bp upstream of the putative tbuX promoter (Fig. 3B) based on its similarity to the promoter for tbuA1. Comparisons of the nucleotide sequences of both the putative tbuX promoter and the putative TbuT-binding site upstream of tbuX with similar sequences found within the Pu promoter of the upper TOL operon from P. putida mt-2 (47), the Po promoter of the dmp operon from Pseudomonas sp. strain CF600 (57), the Ptbm promoter upstream of the toluene and benzene monooxygenase operon from Burkholderia cepacia JS150 (20), the PtbuD promoter upstream of the phenol/cresol hydroxylase structural gene (40), and the PtbuA1 promoter upstream of the toluene-3-monooxygenase structural genes (6) are shown in Fig. 3B. Interestingly, the spacing between the islands of homology observed in the putative TbuT-binding site upstream of tbuX is similar to that observed in the putative DmpR-binding site upstream of its cognate promoter Po.
FIG. 3.
DNA sequence similarities among regions within and upstream of the PtbuX promoter sequence; promoters and UASs with homology to PtbuX. Nucleotide sequence identities among the compared sequences are indicated in boldface. The consensus sequences established from comparison of the region within and upstream of the PtbuX, PtbuD, Po, Ptbm, Pu, and PtbuA1 sequences are displayed below each alignment. (A) DNA sequence alignment of the ς54-dependent promoter sequences of PtbuX, PtbuA1, PtbuD, Pu, and Po. The −24 and −12 sequences within the consensus sequence are labeled accordingly. (B) DNA sequence alignment of the region upstream of tbuX with palindromic regions identified as the XylR-binding site upstream of Pu and those proposed for DmpR binding upstream of Po, TbuT binding upstream of tbuD and tbuA1, and TbmR binding upstream of tbmA. Gaps, indicated by dashes, were introduced to maximize homology.
To ascertain whether the region upstream of tbuX contained TbuT- and toluene-dependent promoter activity, a tbuX-lacZ fusion, designated p454X/S, was constructed using the promoter probe plasmid pKRZ1. To provide the trans-activating function of TbuT, the compatible plasmid pHYK1001 carrying the constitutively expressed tbuT gene was used. Plasmid p454X/S was mobilized into P. aeruginosa PAO1c strains carrying pRO1614 or pHYK1001. Expression from the tbuX-lacZ fusion was monitored by measuring β-galactosidase levels in these P. aeruginosa PAO1c strains grown in the presence or absence of the effector toluene. The P. aeruginosa PAO1c strains carrying the PtbuA1-lacZ (p352X/S) and PtbuD-lacZ (p3.4kbX/B) fusions in trans with pRO1614 or pHYK1001 were also used for purposes of comparison. The levels of β-galactosidase obtained are listed in Table 2. As expected, TbuT- and toluene-dependent promoter activity was observed from p454X/S. Surprisingly, the level of TbuT- and toluene-dependent promoter activity obtained from cells carrying p352X/S and p3.4kbX/B was two- to threefold lower than that observed from p454X/S. In addition, a significant level of promoter activity was observed when cells carrying p454X/S were grown in the absence of toluene or when the cells did not carry TbuT. These results suggest that tbuX might be expressed from either a leaky yet strong TbuT-dependent toluene-inducible promoter or perhaps from two promoters, one strong, TbuT dependent, and toluene inducible and the other weak and constitutively expressed.
TABLE 2.
Expression of putative tbuX promoter regiona
| Plasmid | Promoter | TbuTb | β-Galactosidase (U)c
|
Induction ratiod (toluene/none) | |
|---|---|---|---|---|---|
| None | Toluene | ||||
| pKRZ1 | None | − | 2 | 1 | 1 |
| + | 2 | 2 | 1 | ||
| p454X/S | PtbuX | − | 29 | 23 | 1 |
| + | 23 | 321 | 14 | ||
| p352X/S | PtbuA1 | − | 3 | 3 | 1 |
| + | 3 | 111 | 37 | ||
| p3.4kbX/B | PtbuD | − | 3 | 2 | 1 |
| + | 3 | 144 | 48 | ||
P. aeruginosa PAO1c cells carrying plasmids were grown overnight in LB medium containing kanamycin (600 μg/ml) and tetracycline (50 μg/ml) to maintain plasmids. Cells were subsequently diluted (1:50) in the same medium and grown for 8 h in the absence or presence of toluene.
+, presence of pHYK1001 in trans with the listed plasmid; −, pRO1614 is in trans.
Determined as described by Miller (35). Data are averages of three independent determinations, each conducted in duplicate samples. The variability between values did not exceed 10%.
Ratio of β-galactosidase activities under uninduced (none) versus induced (toluene) conditions.
Transcriptional analysis of tbuX.
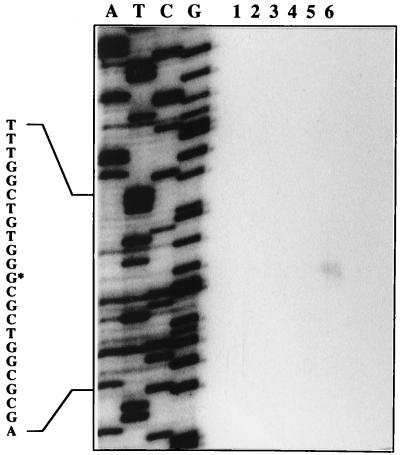
A transcriptional start site for tbuX was mapped by primer extension analysis just downstream of the ς54-dependent promoter sequence (Fig. 1). This single transcript was observed only from the primer extension reaction using total RNA extracted from toluene-induced cells carrying p454X/S and TbuT (Fig. 4, lane 6). These results provide evidence that the −24/−12 promoter sequence, designated PtbuX, is the TbuT-dependent toluene-inducible promoter identified in the tbuX-lacZ fusion studies, described above. The absence of a similar transcript from RNA extracted from uninduced cells carrying p454X/S and TbuT (Fig. 4, lane 3) suggests that the constitutive expression observed from the tbuX-lacZ fusion studies may not be from a leaky PtbuX promoter but rather from a second promoter elsewhere upstream of tbuX.
FIG. 4.
Determination of the 5′ end of the tbuX transcript by primer extension analysis. RNA was isolated from P. aeruginosa PAO1c(pKRZ, pHYK1001), P. aeruginosa PAO1c(p454X/S, pRO1614), and P. aeruginosa PAO1c(p454X/S, pHYK1001) grown in the absence (lanes 1 to 3, respectively) and presence (lanes 4 to 6, respectively) of toluene. A sequence ladder using the same primer and p454X/S is also shown (A, T, C, and G). To the left, an expanded view of the nucleotide sequence surrounding the transcriptional start site (marked with an asterisk) is shown.
Analysis of PtbuX promoter activity.
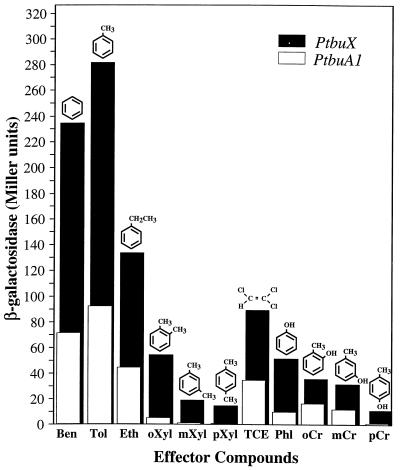
Given the differences observed in β-galactosidase levels for the tbuX-lacZ fusion in contrast to the tbuA1- and tbuD-lacZ fusions with toluene as an inducer (Table 2), we set out to investigate the possibility of differential promoter response from PtbuX and PtbuA1 for a range of effector compounds. These experiments were carried out using P. aeruginosa PAO1c carrying either p454X/S (the tbuX-lacZ fusion) or p352X/S (the PtbuA1-lacZ fusion) in trans with TbuT expressed from pHYK1001, as described above. Comparisons of promoter activity obtained from PtbuX and PtbuA1 in response to the presence of various effector compounds revealed that PtbuX is a stronger promoter (Fig. 5). Although the differences in promoter activity (PtbuX versus PtbuA1) in response to different effectors varied between 2- and 12-fold, in general the relative activation profiles from the various effectors were maintained. The rank order for effector activation of PtbuX was toluene > benzene > ethylbenzene > TCE > o-xylene > phenol > o-cresol > m-cresol > m-xylene > p-xylene > p-cresol. The rank order for PtbuA1 activation was toluene > benzene > ethylbenzene > TCE > o-cresol > m-cresol > phenol > o-xylene > m-xylene > p-cresol > p-xylene. Overall, the effector activation profile was similar for the two promoters with the exceptions of phenol and o-xylene. For PtbuX, o-xylene and phenol were better effectors than o- or m-cresol, whereas for PtbuA1 o- and m-cresol were better effectors.
FIG. 5.
Activation of the PtbuX and PtbuA1 promoters by TbuT in response to the presence of effectors. P. aeruginosa PAO1c cells containing pHYK1001 and either the PtbuX-lacZ transcriptional fusion plasmid p454X/S or the PtbuA1-lacZ transcriptional fusion plasmid p352X/S were grown and treated as described in Materials and Methods. The histogram represents the accumulation of β-galactosidase after 8 h of exposure of the cultures to the different effectors. Values are averages of duplicate determinations from three independent experiments. The variability between triplicate values did not exceed 10%. Abbreviations for effector compounds: Ben, benzene; Tol, toluene; Eth, ethylbenzene; oXyl, o-xylene; mXyl, m-xylene; pXyl, p-xylene; TCE, trichloroethylene; Phl, phenol; oCr, o-cresol; mCr, m-cresol; pCr, p-cresol.
Effect of tbuX deletion on expression of the tbu pathway genes.
To elucidate a possible functional role of tbuX in utilization of toluene by the toluene-3-monooxygenase encoded by tbuA1UBVA2C, a tbuX-deletion derivative of pRO1966 (Table 1) was constructed as described in Materials and Methods. The deletion derivative, designated pHYK1000, was introduced into P. aeruginosa PAO1c, and the cells carrying either pHYK1000 or pRO1966 were then analyzed for toluene utilization and protein synthesis.
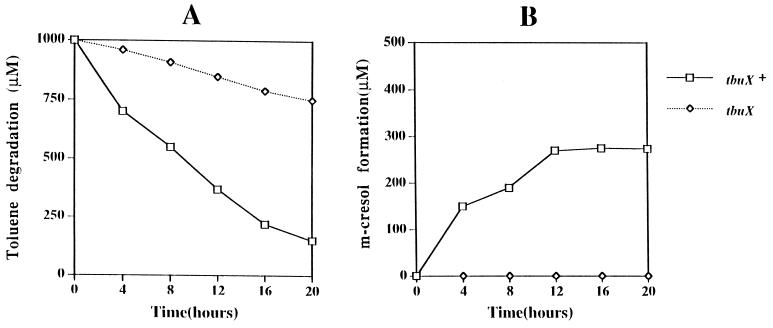
Analysis of toluene utilization and product formation by reverse-phase HPLC showed that cells carrying pHYK1000, the construct containing tbuA1UBVA2C and tbuT but lacking tbuX, were significantly impaired in the ability to utilize toluene (Fig. 6A). In addition, m-cresol, the first intermediate in the toluene degradation pathway of R. pickettii PKO1, was also not detected for this construct, whereas cells of P. aeruginosa PAO1c carrying pRO1966, which contains tbuA1UBVA2C, tbuT, and tbuX, were able to produce significant amounts of m-cresol from toluene (Fig. 6B).
FIG. 6.
Toluene utilization (A) and m-cresol production (B) for cells of P. aeruginosa PAO1c carrying pRO1966 (tbuX+) or pHYK1000 (tbuX).
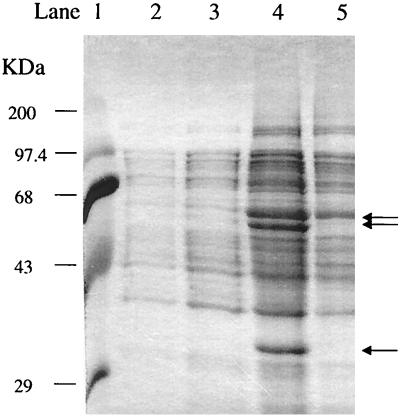
From one-dimensional SDS-PAGE analysis of the soluble peptides produced from P. aeruginosa PAO1c cells containing either pHYK1000 or pRO1966, it was clearly evident that toluene-grown cells carrying pRO1966 synthesized novel peptides of the sizes expected for components of the tbuA1UBVA2C and tbuT operon, whereas cells carrying pHYK1000 produced only barely detectable amounts of these peptides (Fig. 7).
FIG. 7.
SDS-PAGE profile of soluble cellular proteins of P. aeruginosa PAO1c carrying pRO1966 (lanes 2 and 4) or pHYK1000 (lanes 3 and 5), grown on 0.1% Casamino Acids and 0.25% glucose (lanes 2 and 3) or 0.1% Casamino Acids, 0.25% glucose, and 1 mM toluene (lanes 4 and 5). Sizes of molecular weight markers are shown to the left; arrows indicate the positions of TbuA1UBVA2C and TbuT components, as described in the text.
DISCUSSION
In this study, we have identified and characterized a new gene, tbuX, associated with the tbu pathway for toluene and benzene utilization in R. pickettii PKO1. The tbuX gene was identified downstream of and on the DNA strand opposite that of tbuT, the NtrC-like positive regulator that controls transcription of the tbu regulon (6). Analysis of the deduced amino acid sequence of TbuX demonstrated that this protein shared significant identity with a group of proteins associated with pathways for hydrocarbon degradation in a variety of gram-negative bacteria. These pathways included styrene utilization in Pseudomonas sp. strain Y2; isopropylbenzene utilization in P. putida RE204 and P. fluorescens IP01; toluene utilization in P. putida PaW1 which carries the TOL plasmid pWW0; two separate pathways in P. putida F1, one for toluene and one for p-cymene utilization; and phenol utilization in R. eutropha JMP134. The group of homologous proteins found in these organisms all shared a low but significant level of similarity to FadL, an outer membrane protein required for binding and transport of long-chain fatty acids in E. coli. Except for FadL, none of the members of this group has been analyzed in detail with regard to subcellular location. However, the provisional identification of TbuX as an outer membrane protein in R. pickettii PKO1 appears to be reasonable given the following observations derived from comparative sequence analysis: (i) the presence of a putative signal sequence (37) (residues 1 to 23 [VKGNYVRAATAICVAICGTHARA]) containing a charged amino acid (K, position 2) followed by a mostly hydrophobic segment and a peptidase cleavage site adjacent to three conserved residues (ARA, positions 21 to 23); (ii) the absence of cysteinyl residues in the putative mature peptide; and (iii) an abundance of hydrophobic amino acids (37.4% by frequency) in the putative mature peptide (16). In addition, comparative hydropathy analyses of TbuX, PhlX, CymD, PorA, IpbH, CumH, TodX, and XylN using the algorithms of Hopp and Woods (data not shown) revealed the presence of similar regions of hydrophobicity in each of these proteins, as previously demonstrated for FadL (4, 28). It is noteworthy that the putative outer membrane protein with which TbuX shares the greatest overall similarity is PhlX, which is associated with a catabolic pathway for phenol degradation in R. eutropha JMP134 (22). The tbu pathway of R. pickettii PKO1 can accommodate both hydroxylated aromatic compounds such as phenol and cresols as well as unactivated aromatic hydrocarbons such as benzene and toluene as both effectors and substrates (5, 24, 25, 41). It is possible that the similarities between PhlX and TbuX relate, in part, to their potential roles in phenol utilization in these two strains, in contrast to the other members of this protein group (CymD, PorA, IpbH, CumH, TodX, and XylN) which are all associated with pathways for utilization of unactivated aromatic hydrocarbons. In this regard, it would also be of interest to determine whether PhlX and the associated phl pathway of strain JMP134 can accommodate toluene and benzene as effectors and as substrates.
Deletion of TbuX severely affected toluene utilization in cells of P. aeruginosa PAO1c carrying tbu pathway genes (Fig. 6). This result can be explained by a model in which TbuX plays a role in the entry of toluene into the cell. A possible scenario might involve the facilitated passage of toluene through the outer membrane as a consequence of interaction with the TbuX protein. Such facilitated entry could provide toluene to TbuT, which in turn would result in activation of the various tbu pathway promoters. Wang et al. (60) have suggested that TodX, a homolog of TbuX that occurs in P. putida F1, may function as a facilitator of toluene entry into this strain. A necessary feature of this model is the constant presence of the TbuX protein in the cell's outer membrane as a consequence of a low basal level of expression. This prediction is consistent with our observations on the behavior of the PtbuX promoter, namely that a significant level of PtbuX promoter activity was observed when cells were grown in the absence of the effector, toluene, or when the cells did not carry the transcriptional activator, TbuT (Table 2), suggesting that there may be partially constitutive expression of tbuX. Our transcriptional analysis using primer extension did not reveal any message initiating from the major TbuT-dependent and toluene-responsive tbuX promoter, PtbuX, when analyzed in the absence of TbuT or toluene (Fig. 4, lanes 2 and 3); therefore, it is possible that the low basal level of tbuX expression is dependent on a second weakly constitutive promoter located further upstream and that was not contained in p454X/S. Inspection of the DNA sequence 5′ of the putative translational start of TbuX to the BamHI site at the right-hand terminus of the pRO1966 clone (Fig. 1) did not reveal any obvious ς70-like promoter (17), which might be expected to allow for partially constitutive expression; therefore, additional research will be required to further elucidate this.
In addition to affecting toluene utilization, deletion of tbuX also affected expression of peptides necessary for toluene utilization (Fig. 7). Analysis of the expression pattern for soluble proteins obtained from toluene-grown cells of P. aeruginosa PAO1c carrying pHYK1000, which has been deleted for tbuX, shows (Fig. 7, lane 5) the absence of bands at estimated molecular masses of 62, 57, and 37 kDa. In contrast, protein pattern analysis for toluene-grown P. aeruginosa PAO1c carrying pRO1966, which contains tbuA1UBVA2C, tbuT, and tbuX (Fig. 1), clearly shows (Fig. 7, lane 4) the presence of novel peptide bands with these estimated molecular masses. Peptides of these sizes would correspond to the predicted molecular masses for TbuT (65.2 kDa) and TbuA1 (57.6 kDa) and for TbuA2 (37.5 kDa) and TbuC (36.1 kDa), which are similar to one another in size. The other peptides that would be expressed from pRO1966, TbuB, TbuU, and TbuV, would not be expected to be detectable in this analysis since the molecular masses of these components are 12.3, 9.6, and 11.7 kDa, respectively. These results are consistent with the toluene-3-monooxygenase peptide expression patterns that we have previously determined from related work on regulation of the tbu pathway (5, 24, 40, 41), and they support the model that deletion of tbuX impaired the ability of toluene to enter the cell, which in turn negatively affected the expression of the toluene-3-monooxygenase structural genes as a result of the lack of effector binding to the transcriptional activator, TbuT. Therefore, Pseudomonas cells harboring pHYK1000 could not utilize toluene even though the toluene-3-monooxygenase structural genes and the transcriptional regulatory gene were present.
PtbuX is a stronger TbuT-dependent and toluene-responsive promoter than is PtbuA1. This finding is also consistent with our overall model of TbuX as a participant in facilitated entry of toluene into the cell, inasmuch as rapid and high-level synthesis of TbuX from a strong promoter would allow for more toluene entry and hence would provide more substrate for toluene-3-monooxygenase which is transcribed from the comparatively weaker PtbuA1 promoter. Such a regulatory scheme might be part of a general mechanism by which carbon flow could be controlled during toluene utilization by strain PKO1. In this regard, the tbu regulon appears to be unique among bacterial regulatory systems in that the binding of a single transcriptional activator, TbuT, to multiple promoter-upstream regions of different architecture (Fig. 3B) appears to allow for differential promoter response. The catabolic pathways that have been studied in the greatest detail and with which the tbu pathway shares the greatest homology with regard to transcriptional activators are the xyl pathway of P. putida PaW1 carrying the TOL plasmid, pWW0 (1, 2, 9, 19), and dmp pathway of Pseudomonas sp. strain CF600 (55). These pathways, although they show some similarity in overall mode of regulation to the tbu pathway, are substantially different in the detailed mechanisms by which the transcriptional units are controlled. The xyl regulatory system is controlled by two separate transcriptional activators, XylR (an NtrC-like activator) and XylS (a member of the AraC family of activators), that respond to separate effectors and that bind to separate and distinct promoter-upstream regions for each operon (48). In contrast, the dmp pathway is controlled by a single NtrC-like activator, DmpR, that binds to a single promoter-upstream region that controls the single dmp operon (55).
There seems to be some similarity in regulatory organization between the tbu pathway of strain PKO1 and the tbm pathway of B. cepacia JS150, inasmuch as the tbm system appears to controlled by one transcriptional activator, TbmR, that regulates expression of two different monooxygenases expressed from divergent promoters (20, 21), but this regulatory system has not yet been investigated in sufficient detail to determine the extent of its similarity to the tbu model system. Other catabolic pathways that have significant homology to the tbu pathway with respect to the structural genes, such as the pathways for toluene utilization in Pseudomonas mendocina KR1 (61) or in B. cepacia G4 (51–53), have not yet been investigated with regard to operonic organization or gene regulation. Therefore, it is not clear what regulatory paradigm would be expected to occur in these organisms, although data from an analysis of the kinetics of toluene degradation indicate that there are similarities between strains G4 and PKO1 with respect to the effect of oxygen availability on the rate of toluene degradation for the two strains (32). This suggests that these strains might share similar features for regulation of their toluene biodegradative pathways. The overall picture that is emerging from comparative analysis of these regulatory systems suggests that regulatory units have evolved independently of catabolic genes (11) and that regulatory units can be recruited in a modular fashion. Further evidence for the latter observation comes from our recent work on comparative organization of the regulatory systems controlling the phenol hydroxylase in R. pickettii PKO1 and the toluene/benzene-2-monooxygenase in Burkholderia sp. strain JS150, in which it appears that a regulatory module and associated promoter structure has been recruited, in the case of strain PKO1, to be associated with a unit peptide phenol hydroxylase, whereas in strain JS150 a similar regulatory module is associated with a multicomponent toluene/benzene-2-monooxygenase (40).
A central part of our studies on tbuX have focused on regulation of this operon as part of the overall tbu regulon in strain PKO1. Consistent with previous observations, we have shown that expression from PtbuX is dependent on the presence of the regulatory protein, TbuT, and appropriate effector compounds. Sequence analyses have suggested that the PtbuX promoter is dependent on the alternative sigma factor, ς54, and that activation of PtbuX is dependent on a DNA region 178 bp upstream of the transcription start site. Taken together, these findings indicate that the organization and regulation of the tbuX operon are similar to what has been found previously for the other operons in the tbu regulon (5, 6, 25–27, 40). The tbu pathway promoters are regulated by TbuT, which is an NtrC-like transcriptional activator that belongs to the large family of ς54-dependent activators (29, 33, 36, 54). These transcriptional activators function by binding to DNA upstream activation sequences (UAS) located 100 to 200 bp upstream of the promoters that they regulate. For the tbu pathway UAS shown in Fig. 3B, these sequences are found approximately 150 to 200 bp upstream of the transcriptional start site for each promoter. The tbu UAS regions have significant sequence homology to the palindromic regions upstream of Pu, the upper pathway promoter from the TOL plasmid, to which the transcriptional activator XylR has been shown to bind (9, 19). The sequences recognized by XylR in the Pu UAS region include the motif 5′-TTGANCAAATC-3′ (9). A sequence highly similar to this is present within each arm (Fig. 3B) of the inverted repeat upstream of PtbuX; thus, it is likely that this is the target site for TbuT binding for the PtbuX promoter. It is of interest that the spacing between the conserved inverted repeats in the PtbuX UAS region is similar to that found for the UAS region of the PtbuD promoter and not the PtbuA1 promoter (Fig. 3B). This difference in UAS palindrome architecture is correlated with promoter activity; i.e., greater promoter activity is observed when tbuT is in trans with promoter/UAS ′lacZ fusions of PtbuX or PtbuD than when it is in trans with fusions of PtbuA1. This suggests that promoter function can be affected by the structure of the site to which the transcriptional activator binds. Such an effect was seen previously by Leahy and coworkers (31) for cross-regulation of the PtbuA1 promoter of R. pickettii PKO1 and the PtbmA promoter of B. cepacia JS150 by either TbuT or TbmR. These researchers observed that the cognate and noncognate activators were capable of transcriptionally activating each promoter in response to the same set of effectors, but that the magnitude of the transcriptional response was greater for PtbmA than for PtbuA1 for each of the effectors tested. As shown in Fig. 3B, PtbmA is similar in architecture to PtbuX and not to PtbuA1.
As indicated above, members of the ς54-dependent family of promoters require an interaction between the transcriptional activator bound at the UAS and RNA polymerase bound at the −24/−12 region in order for an open transcriptional complex to form. This process necessarily requires DNA bending in order to achieve close physical contact between the UAS-bound activator and the promoter-bound ς54-RNA polymerase (45). In some ς54-dependent promoters, the required DNA bending is mediated by specific DNA-bending proteins such as integration host factor (IHF), which recognizes and binds to specific DNA sequences and induces sharp bends in the DNA. IHF-dependent DNA bending has been shown to be essential for promoter function for the XylR-dependent Pu promoter (1, 9, 46) as well as the DmpR-dependent Po promoter (55, 57). Moreover, in the case of the Pu promoter, IHF binding has been shown to specifically stimulate recruitment of RNA polymerase to the −24/−12 promoter region as a consequence of interaction of the carboxy-terminal domain of the α subunit of RNA polymerase with specific sequences upstream of the IHF-binding site (3). Inspection of the DNA sequence between the putative TbuT-binding site and the −24/−12 promoter region of the PtbuX promoter did not reveal a consensus IHF-binding site; therefore, it seems unlikely that IHF could contribute to the necessary promoter architecture for PtbuX. However, protein-assisted DNA bending necessary for ς54 promoter function can be achieved by mechanisms that are independent of IHF, as has been shown for the XylR-dependent Ps promoter of the TOL plasmid of P. putida (13, 44). With this promoter, the combined effects of intrinsically curved DNA and HU protein-mediated DNA bending have been shown to be necessary to achieve full transcriptional activity. Since the DNA region between the putative TbuT-binding site and the −24/−12 promoter region of the PtbuX promoter contains a greater number of dT stretches (Fig. 1) than found in the rest of the tbuX operon, it is possible that PtbuX is similar to Ps, relying on both the intrinsic curvature of promoter DNA and a nonspecific DNA-bending protein, such as HU, to achieve the promoter architecture required for transcription.
ACKNOWLEDGMENTS
This research was supported by the National Institute of Environmental Health Sciences through Superfund Basic Research Program grant P42-ES-04911. Some DNA sequence analyses were supported in part by the General Clinical Research Center at the University of Michigan, funded by grant M01RR00042 from the National Center for Research Resources, National Institutes of Health.
We thank Don Clewell for providing E. coli MM294(pRK2013) and Ananda Chakrabarty for providing plasmid pKRZ1. Technical assistance provided by Lisa Ramey and Andrew Berger is also gratefully acknowledged.
REFERENCES
- 1.Abril M A, Buck M, Ramos J L. Activation of the Pseudomonas TOL plasmid upper pathway operon: identification of binding sites for the positive regulator XylR and for integration host factor protein. J Biol Chem. 1991;266:15832–15838. [PubMed] [Google Scholar]
- 2.Abril M A, Ramos J L. Physical organization of the Pseudomonas TOL plasmid upper pathway operon promoter. Sequence and positional requirements for XylR-dependent activation of transcription. Mol Gen Genet. 1993;239:281–288. doi: 10.1007/BF00281629. [DOI] [PubMed] [Google Scholar]
- 3.Bertoni G, Fujita N, Ishihama A, de Lorenzo V. Active recruitment of ς54-RNA polymerase to the Pu promoter of Pseudomonas putida: role of IHF and αCTD. EMBO J. 1998;17:5120–5128. doi: 10.1093/emboj/17.17.5120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Black P N. Primary sequence of the Escherichia coli fadL gene encoding an outer membrane protein required for long-chain fatty acid transport. J Bacteriol. 1991;173:435–442. doi: 10.1128/jb.173.2.435-442.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Byrne A M, Kukor J J, Olsen R H. Sequence analysis of the gene cluster encoding toluene-3-monooxygenase from Pseudomonas pickettii PKO1. Gene. 1995;154:65–70. doi: 10.1016/0378-1119(94)00844-i. [DOI] [PubMed] [Google Scholar]
- 6.Byrne A M, Olsen R H. Cascade regulation of the toluene-3-monooxygenase operon (tbuA1UBVA2C) of Burkholderia pickettii PKO1: role of the tbuA1 promoter (PtbuA1) in the expression of its cognate activator, TbuT. J Bacteriol. 1996;178:6327–6337. doi: 10.1128/jb.178.21.6327-6337.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cohen-Bazaire G, Sistrom W R, Stanier R Y. Kinetic studies of pigment synthesis by non-sulphur purple bacteria. J Cell Comp Physiol. 1957;49:25–68. doi: 10.1002/jcp.1030490104. [DOI] [PubMed] [Google Scholar]
- 8.Cuskey S M, Pecoraro V, Olsen R H. Initial catabolism of aromatic biogenic amines by Pseudomonas aeruginosa PAO: pathway description, mapping of mutations, and cloning of essential genes. J Bacteriol. 1987;169:2398–2404. doi: 10.1128/jb.169.6.2398-2404.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Lorenzo V, Herrero M, Metzke M, Timmis K N. An upstream XylR- and IHF-induced nucleoprotein complex regulates the ς54-dependent Pu promoter of TOL plasmid. EMBO J. 1991;10:1159–1167. doi: 10.1002/j.1460-2075.1991.tb08056.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eaton R W. p-Cymene catabolic pathway in Pseudomonas putida F1: cloning and characterization of DNA encoding conversion of p-cymene to p-cumate. J Bacteriol. 1997;179:3171–3180. doi: 10.1128/jb.179.10.3171-3180.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fernández S, Pérez-Martín J, de Lorenzo V. Specificity and promiscuity of catabolic promoters of Pseudomonas spp. In: Nakazawa T, Furukawa K, Haas D, Silver S, editors. Molecular biology of pseudomonads. Washington, D. C.: ASM Press; 1996. pp. 165–175. [Google Scholar]
- 12.Figurski D H, Helinski D R. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc Natl Acad Sci USA. 1979;76:1648–1652. doi: 10.1073/pnas.76.4.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gomada M, Imaishi H, Miura M, Inouye S, Nakazawa T, Nakazawa A. Analysis of DNA bend structure of promoter regulatory regions of xylene-metabolizing genes on the Pseudomonas TOL plasmid. J Biochem. 1994;116:1096–1104. doi: 10.1093/oxfordjournals.jbchem.a124633. [DOI] [PubMed] [Google Scholar]
- 14.Habe H, Kasuga K, Nojiri H, Yamane H, Omori T. Analysis of cumene (isopropylbenzene) degradation genes from Pseudomonas fluorescens IP01. Appl Environ Microbiol. 1996;62:4471–4477. doi: 10.1128/aem.62.12.4471-4477.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hanahan D. Techniques for transformation of E. coli. In: Glover D M, editor. DNA cloning, a practical approach. Vol. 1. Oxford, England: IRL Press, Ltd.; 1985. pp. 109–136. [Google Scholar]
- 16.Hancock R E W. Model membrane studies of porin function. In: Inouye M, editor. Bacterial outer membranes as model systems. New York, N.Y: John Wiley & Sons, Inc.; 1987. pp. 187–225. [Google Scholar]
- 17.Harley C B, Reynolds R P. Analysis of E. coli promoter sequences. Nucleic Acids Res. 1987;15:2347–2351. doi: 10.1093/nar/15.5.2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Holloway B W, Krishnapillai V, Morgan A F. Chromosomal genetics of Pseudomonas. Microbiol Rev. 1979;43:73–102. doi: 10.1128/mr.43.1.73-102.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Inouye S, Gomada M, Sangodkar U M X, Nakazawa A, Nakazawa T. Upstream regulatory sequences for transcriptional activator XylR in the first operon of xylene metabolism of the TOL plasmid. J Mol Biol. 1990;216:251–260. doi: 10.1016/S0022-2836(05)80317-1. [DOI] [PubMed] [Google Scholar]
- 20.Johnson G R, Olsen R H. Nucleotide sequence analysis of genes encoding a toluene/benzene-2-monooxygenase from Pseudomonas sp. strain JS150. Appl Environ Microbiol. 1995;61:3336–3346. doi: 10.1128/aem.61.9.3336-3346.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johnson G R, Olsen R H. Multiple pathways for toluene degradation in Burkholderia sp. strain JS150. Appl Environ Microbiol. 1997;63:4047–4052. doi: 10.1128/aem.63.10.4047-4052.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim Y, Ayoubi P, Harker A R. Constitutive expression of the cloned phenol hydroxylase gene(s) from Alcaligenes eutrophus JMP134 and concomitant trichloroethylene oxidation. Appl Environ Microbiol. 1996;62:3227–3233. doi: 10.1128/aem.62.9.3227-3233.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kukor J J, Olsen R H. Diversity of toluene degradation following long term exposure to BTEX in situ. In: Kamely D, Chakrabarty A, Omenn G S, editors. Biotechnology and biodegradation. Houston, Tex: Gulf Publishing Co.; 1990. pp. 405–421. [Google Scholar]
- 24.Kukor J J, Olsen R H. Molecular cloning, characterization, and regulation of a Pseudomonas pickettii PKO1 gene encoding phenol hydroxylase and expression of the gene in Pseudomonas aeruginosa PAO1c. J Bacteriol. 1990;172:4624–4630. doi: 10.1128/jb.172.8.4624-4630.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kukor J J, Olsen R H. Genetic organization and regulation of a meta cleavage pathway for catechols produced from catabolism of toluene, benzene, phenol, and cresols by Pseudomonas pickettii PKO1. J Bacteriol. 1991;173:4587–4594. doi: 10.1128/jb.173.15.4587-4594.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kukor J J, Olsen R H. Complete nucleotide sequence of tbuD, the gene encoding phenol/cresol hydroxylase from Pseudomonas pickettii PKO1, and functional analysis of the encoded enzyme. J Bacteriol. 1992;174:6518–6526. doi: 10.1128/jb.174.20.6518-6526.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kukor J J, Olsen R H. Catechol 2,3-dioxygenases functional in oxygen-limited (hypoxic) environments. Appl Environ Microbiol. 1996;62:1728–1740. doi: 10.1128/aem.62.5.1728-1740.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kumar G B, Black P N. Bacterial long-chain fatty acid transport. J Biol Chem. 1993;268:15469–15476. [PubMed] [Google Scholar]
- 29.Kustu S, North A K, Weiss D. Prokaryotic transcriptional enhancers and enhancer-binding proteins. Trends Biochem Sci. 1991;16:397–402. doi: 10.1016/0968-0004(91)90163-p. [DOI] [PubMed] [Google Scholar]
- 30.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 31.Leahy J G, Johnson G R, Olsen R H. Cross-regulation of toluene monooxygenases by the transcriptional activators TbmR and TbuT. Appl Environ Microbiol. 1997;63:3736–3739. doi: 10.1128/aem.63.9.3736-3739.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Leahy J G, Olsen R H. Kinetics of toluene degradation by toluene-oxidizing bacteria as a function of oxygen concentration, and the effect of nitrate. FEMS Microbiol Ecol. 1997;23:23–30. [Google Scholar]
- 33.Merrick M J. In a class of its own—the RNA polymerase sigma factor ς54 (ςN) Mol Microbiol. 1993;10:903–909. doi: 10.1111/j.1365-2958.1993.tb00961.x. [DOI] [PubMed] [Google Scholar]
- 34.Mikesell M D, Kukor J J, Olsen R H. Metabolic diversity of aromatic hydrocarbon-degrading bacteria from a petroleum-contaminated aquifer. Biodegradation. 1993;4:249–259. doi: 10.1007/BF00695973. [DOI] [PubMed] [Google Scholar]
- 35.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 36.North A K, Klose K E, Stedman K M, Kustu S. Prokaryotic enhancer-binding proteins reflect eukaryote-like modularity: the puzzle of nitrogen regulatory protein C. J Bacteriol. 1993;175:4267–4273. doi: 10.1128/jb.175.14.4267-4273.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Oliver D. Protein secretion in Escherichia coli. Annu Rev Microbiol. 1985;39:615–648. doi: 10.1146/annurev.mi.39.100185.003151. [DOI] [PubMed] [Google Scholar]
- 38.Olsen R H, DeBusscher G, McCombie W R. Development of broad-host-range vectors and gene banks: self-cloning of the Pseudomonas aeruginosa PAO chromosome. J Bacteriol. 1982;150:60–69. doi: 10.1128/jb.150.1.60-69.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Olsen R H, Hansen J. Evolution and utility of a Pseudomonas aeruginosa drug resistance factor. J Bacteriol. 1976;125:837–844. doi: 10.1128/jb.125.3.837-844.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Olsen R H, Kukor J J, Byrne A M, Johnson G R. Evidence for the evolution of a single component phenol/cresol hydroxylase from a multicomponent toluene monooxygenase. J Indust Microbiol Biotechnol. 1997;19:360–368. doi: 10.1038/sj.jim.2900453. [DOI] [PubMed] [Google Scholar]
- 41.Olsen R H, Kukor J J, Kaphammer B. A novel toluene-3-monooxygenase pathway cloned from Pseudomonas pickettii PKO1. J Bacteriol. 1994;176:3749–3756. doi: 10.1128/jb.176.12.3749-3756.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Olsen R H, Mikesell M D, Kukor J J. Enumeration and characterization of BTEX-degrading bacteria from hypoxic environments functional with mixed electron acceptors. Res Microbiol. 1994;145:47–49. doi: 10.1016/0923-2508(94)90068-x. [DOI] [PubMed] [Google Scholar]
- 43.Olsen R H, Mikesell M D, Kukor J J, Byrne A M. Physiological attributes of microbial BTEX degradation in oxygen-limited environments. Environ Health Perspect. 1995;103:49–51. doi: 10.1289/ehp.95103s449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pérez-Martin J, de Lorenzo V. The ς54-dependent promoter Ps of the TOL plasmid of Pseudomonas putida requires HU for transcriptional activation in vivo by XylR. J Bacteriol. 1995;177:3758–3763. doi: 10.1128/jb.177.13.3758-3763.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pérez-Martin J, de Lorenzo V. Clues and consequences of DNA bending in transcription. Annu Rev Microbiol. 1997;51:593–628. doi: 10.1146/annurev.micro.51.1.593. [DOI] [PubMed] [Google Scholar]
- 46.Pérez-Martin J, Timmis K N, de Lorenzo V. Co-regulation by bent DNA. J Biol Chem. 1994;269:22657–22662. [PubMed] [Google Scholar]
- 47.Ramos J L, Marques S, Timmis K N. Transcriptional control of the Pseudomonas TOL plasmid catabolic operons is achieved through an interplay of host factors and plasmid-encoded regulators. Annu Rev Microbiol. 1997;51:341–373. doi: 10.1146/annurev.micro.51.1.341. [DOI] [PubMed] [Google Scholar]
- 48.Salto R, Delgado A, Gallegos M-T, Manzanera M, Marqués S, Ramos J L. Fine control of expression of the catabolic pathways of TOL plasmid of Pseudomonas putida for mineralization of aromatic hydrocarbons. In: Nakazawa T, Furukawa K, Haas D, Silver S, editors. Molecular biology of pseudomonads. Washington, D.C.: ASM Press; 1996. pp. 207–216. [Google Scholar]
- 49.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 50.Sanger F, Nicklen S, Coulsen A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shields M S, Montgomery S O, Chapman P J, Cuskey S M, Pritchard P H. Novel pathway of toluene catabolism in the trichloroethylene-degrading bacterium G4. Appl Environ Microbiol. 1989;55:1624–1629. doi: 10.1128/aem.55.6.1624-1629.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shields M S, Reagin M J. Selection of a Pseudomonas cepacia strain constitutive for the degradation of trichloroethylene. Appl Environ Microbiol. 1992;58:3977–3983. doi: 10.1128/aem.58.12.3977-3983.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shields M S, Reagin M J, Gerger R R, Campbell R, Somerville C. TOM, a new aromatic degradative plasmid from Buirkholderia (Pseudomonas) cepacia G4. Appl Environ Microbiol. 1995;61:1352–1356. doi: 10.1128/aem.61.4.1352-1356.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shingler V. Signal sensing by ς54-dependent regulators: derepression as a control mechanism. Mol Microbiol. 1996;19:409–416. doi: 10.1046/j.1365-2958.1996.388920.x. [DOI] [PubMed] [Google Scholar]
- 55.Shingler V, Bartilson M, Moore T. Cloning and nucleotide sequence of the gene encoding the positive regulator (DmpR) of the phenol catabolic pathway encoded by pVI150 and identification of DmpR as a member of the NtrC family of transcriptional activators. J Bacteriol. 1993;175:1596–1604. doi: 10.1128/jb.175.6.1596-1604.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Smith A W, Iglewski B H. Transformation of Pseudomonas aeruginosa by electroporation. Nucleic Acids Res. 1989;17:10509. doi: 10.1093/nar/17.24.10509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sze C C, Moore T, Shingler V. Growth phase-dependent transcription of the ς54-dependent Po promoter controlling the Pseudomonas-derived (methyl)phenol dmp operon of pVI150. J Bacteriol. 1996;178:3727–3735. doi: 10.1128/jb.178.13.3727-3735.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Velasco A, Alonso S, García J L, Perera J, Díaz E. Genetic and functional analysis of the styrene catabolic cluster of Pseudomonas sp. strain Y2. J Bacteriol. 1998;180:1063–1071. doi: 10.1128/jb.180.5.1063-1071.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vogel H J, Bonner D M. Acetylornithinase of Escherichia coli: partial purification and some properties. J Biol Chem. 1956;218:97–106. [PubMed] [Google Scholar]
- 60.Wang Y, Rawlings M, Gibson D T, Labbé D, Bergeron H, Brousseau R, Lau P C K. Identification of a membrane protein and a truncated LysR-type regulator associated with the toluene degradation pathway in Pseudomonas putida F1. Mol Gen Genet. 1995;246:570–579. doi: 10.1007/BF00298963. [DOI] [PubMed] [Google Scholar]
- 61.Yen K-M, Karl M R, Blatt L M, Simon M J, Winter R B, Fausset P R, Lu H S, Harcourt A A, Chen K K. Cloning and characterization of a Pseudomonas mendocina KR1 gene cluster encoding toluene-4-monooxygenase. J Bacteriol. 1991;173:5315–5327. doi: 10.1128/jb.173.17.5315-5327.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]