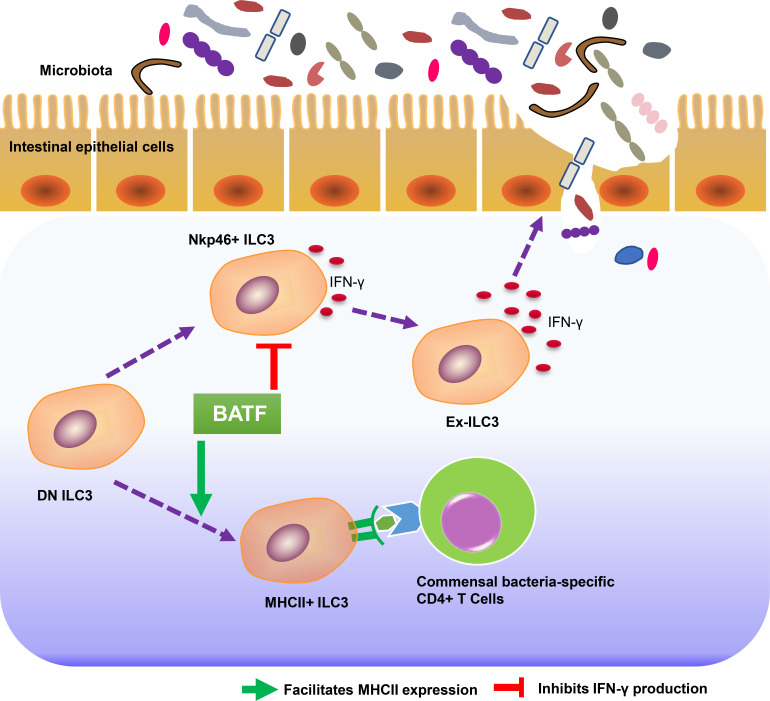
ILC3s are crucial for the maintenance of host–microbiota homeostasis in gastrointestinal mucosal tissues. This study shows that BATF globally shapes the chromatin landscape of intestinal ILC3s, which in turn orchestrates microbiota and mucosal CD4+ T cell immunity.
Abstract
Group 3 innate lymphoid cells (ILC3s) are crucial for the maintenance of host–microbiota homeostasis in gastrointestinal mucosal tissues. The mechanisms that maintain lineage identity of intestinal ILC3s and ILC3-mediated orchestration of microbiota and mucosal T cell immunity are elusive. Here, we identified BATF as a gatekeeper of ILC3 homeostasis in the gut. Depletion of BATF in ILC3s resulted in excessive interferon-γ production, dysbiosis, aberrant T cell immune responses, and spontaneous inflammatory bowel disease (IBD), which was considerably ameliorated by the removal of adaptive immunity, interferon-γ blockade, or antibiotic treatment. Mechanistically, BATF directly binds to the cis-regulatory elements of type 1 effector genes, restrains their chromatin accessibility, and inhibits their expression. Conversely, BATF promotes chromatin accessibility of genes involved in MHCII antigen processing and presentation pathways, which in turn directly promotes the transition of precursor ILC3s to MHCII+ ILC3s. Collectively, our findings reveal that BATF is a key transcription factor for maintaining ILC3 stability and coordinating ILC3-mediated control of intestinal homeostasis.
Graphical Abstract
Introduction
Inflammatory bowel diseases (IBD) display chronic relapsing inflammation of the intestine and are mainly represented by Crohn’s disease (CD) and ulcerative colitis (UC; Kaplan, 2015; Molodecky et al., 2012). Although the precise etiology remains unclear, mounting evidence suggests that inappropriate host immune responses to commensal microbiota are a primary trigger in IBD (Sartor, 2011; Round and Mazmanian, 2009; Comito and Romano, 2012; Mirsepasi-Lauridsen et al., 2018). Disruption of the intestinal epithelial barrier results in dysbiosis, which is considered to be a cause of such aberrant immune responses (Martini et al., 2017; Rescigno, 2011). Therefore, safeguarding epithelial barrier integrity to establish an equilibrium between the microbiota and immune system is fundamental to intestinal homeostasis (Khor et al., 2011; Vereecke et al., 2011). However, the relative contribution of innate immune cells to barrier impairments and dysbiosis, as well as the mechanisms by which microbiota and T cell–mediated mucosal immunity in the gut are orchestrated remain poorly understood.
Innate lymphoid cells (ILCs), particularly group 3 ILCs (ILC3s), are enriched in the gut and have emerged as a central hub to orchestrate lymphoid organogenesis, promote tissue repair, facilitate host defense, directly regulate adaptive immunity, and maintain gut commensal bacteria (Guo et al., 2014; Hepworth et al., 2013; Mortha et al., 2014; Lee et al., 2011; Kiss et al., 2011; Qiu et al., 2012; Castellanos et al., 2018). Intestinal ILC3s are highly heterogeneous (Robinette et al., 2015; Cella et al., 2019; Gury-Benari et al., 2016) and can be divided into two major subclasses based on the expression of the natural cytotoxicity receptor NKp46 (Nkp44 in human, also known as Ncr1; Vonarbourg et al., 2010). In response to type 1 immunity stimuli, Ncr1+ ILC3s can further increase expression of the transcription factor (TF) T-bet, decrease expression of the TF RORγt, and fully differentiate into ILC1-like or natural killer (NK)–like ILC3s, known as ex-ILC3s (Rankin et al., 2013; Artis and Spits, 2015; Nagasawa et al., 2019). Excessive and/or persistent production of IFN-γ, IL-22, and cytotoxic molecules by ex-ILC3s leads to the enhancement of epithelial permeability and dysbiosis and is thus implicated in IBD (Takayama et al., 2010; Zeng et al., 2019). Studies in humans and mice have revealed that the fetal and newborn gut is dominated by ILCs (Gomez de Aguero et al., 2016; Stras et al., 2019; Miller et al., 2018; Bando et al., 2015). More importantly, ILC3s can supervise the colonization of commensal microbiota and gut immunity during early life (van De Pavert et al., 2014; Mao et al., 2018). Whether IFN-γ–producing ILC3s-mediated barrier integrity attributes to the homeostasis of the gut microbiome remains unclear. In contrast to IFN-γ–producing ILC3s, MHCII-expressing ILC3s are CCR6+Ncr1− and lack T-bet expression (Eberl et al., 2004). Selective deletion of MHCII expression on ILC3s resulted in enhanced intestinal commensal specific CD4+ T cell responses and promoted spontaneous intestinal inflammation (Hepworth et al., 2013; Hepworth et al., 2015), indicating that MHCII-expressing ILC3 negatively regulated CD4+ T cell proliferation and activation. Unlike Ncr1+ ILC3 and CCR6+ ILC3, double negative ILC3s (DN ILC3s) harbor plasticity and precursor properties (Vonarbourg et al., 2010; Klose et al., 2013; Rankin et al., 2013). However, the transcription programs that control the heterogeneity and lineage stability of gut ILC3s remain largely unexplored.
BATF belongs to the activator protein-1 (AP-1) TF family, contains a basic region-leucine zipper domain, and has been recognized as a critical regulator of lineage signature genes expression in T follicular helper (Tfh; Ise et al., 2011) and Th2 cells (Ubel et al., 2014). BATF is also a pioneer TF controlling epigenetic landscape in CD8+ T cells, Th17 cells, regulatory T cells, and effector CD4+ T cells (Kuroda et al., 2011; Schraml et al., 2009; Delacher et al., 2020; Pham et al., 2019; Chen et al., 2021). Recently, BATF was found to regulate homeostasis of ILCs (Liu et al., 2020b) and also found to be required for the ILC2 response to IL-25 (Miller et al., 2020) or IL-33 (Wu et al., 2022) and IL-17A production by ILC3s (Liu et al., 2020a). Notably, through single-cell RNA sequencing (scRNA-seq), BATF was found to be enriched in specific subsets of tonsil ILC3s in humans and intestinal ILCs in mice (Cella et al., 2019; Gury-Benari et al., 2016), suggesting a potential role of BATF in ILC3 development and function. Nevertheless, the precise role of BATF in regulating the plasticity of intestinal ILC3s, as well as ILC3-orchestrated microbiota homeostasis and T cell immunity, has remained largely unknown.
Results
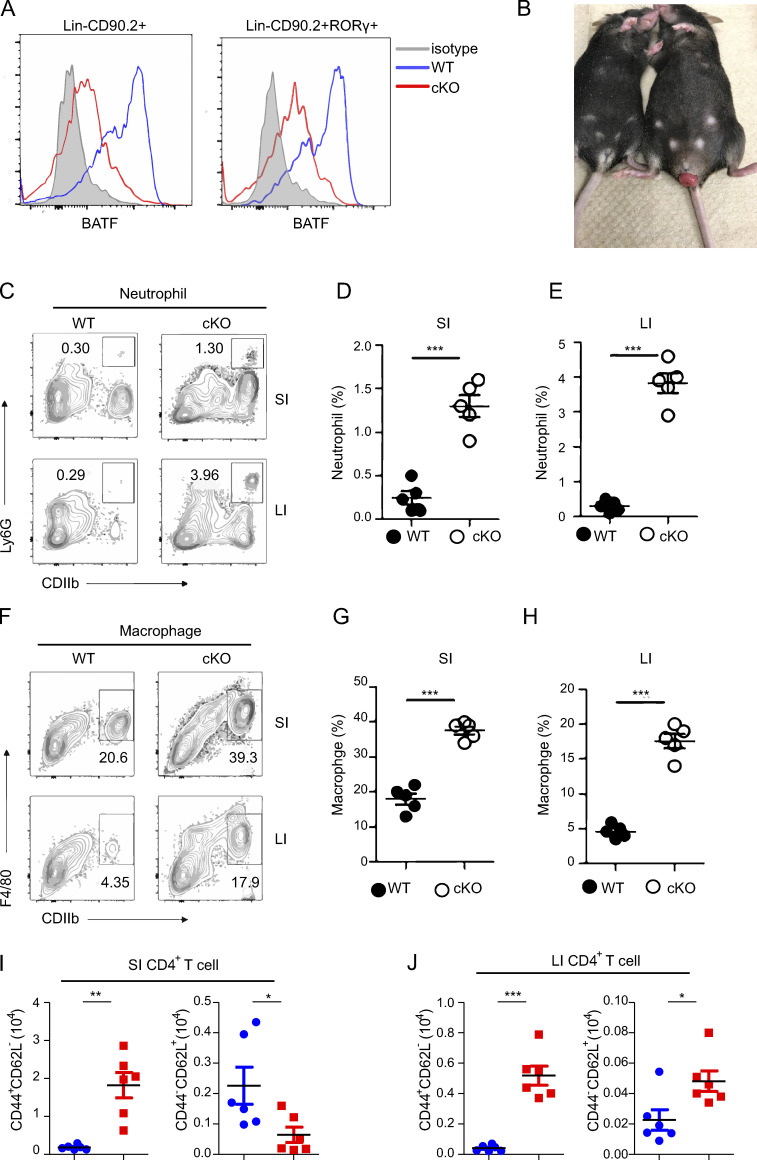
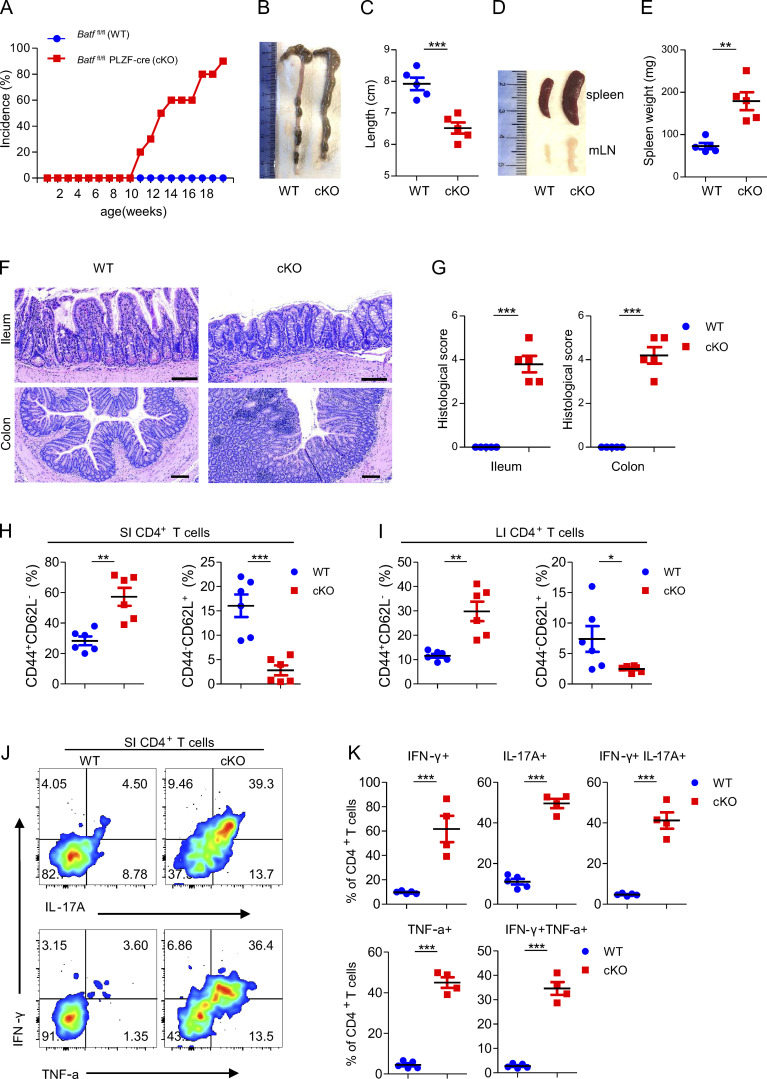
BATF deficiency in ILCs causes spontaneous colitis
To investigate the physiological role of BATF in ILCs, we crossed mice with loxP-flanked BATF alleles (Batffl/fl) mice with mice expressing Cre recombinase under the common helper innate lymphoid progenitor (CHILP)-specific Zbtb16 promoter (PLZF-Cre) to generate Batf ILC-conditional knockout Batffl/fl Plzf-Cre+ mice (referred to as cKO) and Batff/f Plzf-Cre− WT mice. We confirmed BATF protein deficiency in ILCs (Lin−CD90.2+) and ILC3s (Lin−CD90.2+RORrt+) in the small intestine (SI) by flow cytometry (Fig. S1 A). Notably, the cKO mice spontaneously developed rectal prolapse beginning at the age of 11–12 wk under specific pathogen–free conditions (Fig. 1 A and Fig. S1 B). Chronic colitis became more severe in aged cKO mice, as evidenced by splenomegaly, lymphadenopathy, and shortened colon length (Fig. 1, B–E). Histological analysis of SI and colon (large intestine, LI) sections stained with H&E showed crypt abscesses, cellular infiltrates, epithelial destruction, and higher histological scores in the ileum and colon of cKO mice compared with littermate WT controls (Fig. 1, F and G). Notably, among infiltrating cells, a portion of myeloid cells, including neutrophils and macrophages, were significantly increased in the SI and LI of cKO mice (Fig. S1, C–H). However, such immune infiltration has only been observed in the gut, but not in other peripheral tissues, such as liver and lung (data not shown). IBD is, in part, characterized by increased T cell activation (Funderburg et al., 2013). Consistent with this phenotype, cKO mice harbored fewer naive (CD62L+CD44−) but greater activated (CD62L−CD44+) CD4+ T cells restricted in the gut, except for the small increase in the number of colonic naïve CD4+ T cells, which was brought by increased total cellular number in the colon of cKO mice (Fig. 1, H and I; and Fig. S1, I and J). Concordantly, in comparison to WT mice, a greater frequency of CD4+ T cells in the gut of cKO mice produced IFN-γ, IL-17A, and TNF-α after stimulation with PMA and ionomycin (Fig. 1, J and K), indicating more CD4+ T cells with pathogenic capacity existed in the gut of cKO mice. Collectively, these results demonstrate that BATF serves a critical role in ILCs for maintaining intestinal immunological homeostasis.
Figure S1.
BATF deficiency in ILCs causes spontaneous colitis. Related to Fig. 1. (A) Flow cytometry histograms analyzing the expression of BATF in the SI ILCs (Lin−CD90.2+) and ILC3s (Lin−CD90.2+RORγt+) of WT and cKO mice. (B) Representative image of rectal prolapse of cKO (right) and littermate control WT (left) mice aged 12 wk. (C) Flow cytometry analysis of the frequency of total neutrophils (CD11b+Ly6G+) in the SI and LI of cKO and littermate control WT mice aged 12 wk. (D and E) Quantified frequencies of neutrophils in the SI (D) and LI (E) as assessed in C. (F) Flow cytometry analysis of frequency of total macrophages (CD11b+F4/80+) in the SI and LI of cKO and littermate control WT mice aged 12 wk. (G and H) Quantified frequencies of neutrophils in the SI (G) and LI (H) as assessed in C. (I and J) Quantified cell number of naive and effector CD4+ T cells in the SI (I) and LI (J) as assessed in C. Data shown as the mean ± SEM. ***P < 0.001 (two-tailed unpaired t test). Each dot represents one mouse, n = 4–6 mice per group. Data are pooled from two independent experiments.
Figure 1.
BATF deficiency in ILCs causes spontaneous colitis. (A) Batffl/fl (WT, n = 10) and littermate control Batffl/flPLZF-cre (cKO, n = 18) mice were monitored for incidence of rectal prolapse. (B and C) Representative images of the colon (B) from WT and littermate control cKO mice aged 12 wk old and quantification of clone length (C). (D and E) Representative images of the spleen and mLN (D) from WT and littermate control cKO mice aged 12 wk as well as quantification of spleen weight (E). (F and G) H&E staining of ileum and colon sections (F) from WT and littermate control cKO mice aged 12 wk and quantification of histological scores (G). Scale bar, 100 µm. (H and I) Frequency of total activated (CD44+CD62L−) and naive (CD44−CD62L+) CD4+ T cells from the SI (H) and LI (I) of WT and littermate control cKO mice aged 12 wk. (J) Representative flow plots of intracellular staining of IL-17A, IFN-γ, and TNF-α in SI CD4+ T cells after stimulation with PMA plus ionomycin ex vivo from WT and littermate control cKO mice aged 12 wk. (K) Quantification of plots in J. Data are shown as mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001 (two-tailed unpaired t test). Each dot represents one mouse, n = 4–6 mice per group. Data are pooled from two (C, E, and G) or three (H and I) experiments or are representative of at least two independent experiments.
BATF regulates the transcriptional program of ILC3s in the SI
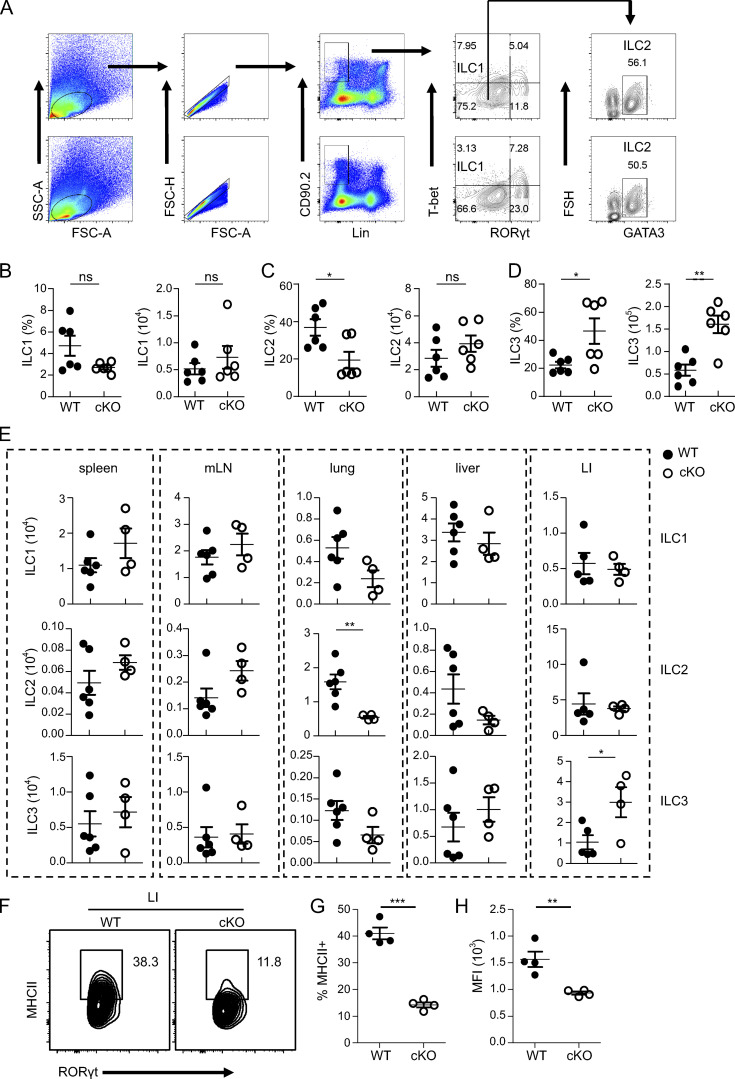
To investigate whether the spontaneous colitis that emerged in BATF-deficient cKO mice was associated with an altered composition of enteric ILCs, first, we compared the proportion and cell numbers of ILCs in SI of cKO mice with WT mice (Fig. S2 A). Notably, the proportion of ILC1s did not significantly change in cKO mice, while the proportion of ILC2s decreased, but the proportion of ILC3s increased in cKO mice (Fig. S2, B–D). Furthermore, the absolute number of ILC3s, but not ILC1s or ILC2s, was significantly increased in cKO mice compared with WT controls (Fig. S2, B–D). Additionally, the cell number of ILC subsets has been checked in different tissues of WT versus cKO mice to understand the regulatory role of BATF. The number of ILC1, ILC2, and ILC3 was comparable in different tissues between cKO mice and WT, except for two different subsets in cKO, pulmonary ILC2, and colonic ILC3, which showed a decrease and an increase, respectively, compared to WT (Fig. S2 E). This suggests that BATF may regulate the subsets of ILCs differently due to their tissue specificity.
Figure S2.
BATF deficiency in ILCs alters the heterogenicity of SI ILCs. (A) Flow cytometry gating strategy and analysis of different ILC subsets in the SI of WT and cKO mice. (B–D) Cell frequency and numbers of ILC1s (Lin−CD90.2+T-bet+Rorγt−; B), ILC2s (Lin−CD90.2+T-bet−Rorγt−GATA3+; C) and ILC3s (Lin−CD90.2+Rorγt+; D) in the SI of WT and cKO mice. (E) Cell numbers of ILCs in various tissues of WT and cKO mice. (F) Flow cytometry analysis of the frequency of MHCII+ ILC3 in the colon of cKO and littermate control WT mice aged 13 wk. (G) Quantified frequencies of MHCII+ ILC3 in the colon as assessed in (F). (H) Quantification of MFI of MHCII on ILC3 in the colon as assessed in (F). Data are shown as mean ± SEM. Statistical differences were tested using a paired t test (two-tailed; B and C) or two-way ANOVA for D. **P < 0.01. Each dot represents one mouse, n = 6 mice per group. Data are pooled from two independent experiments.
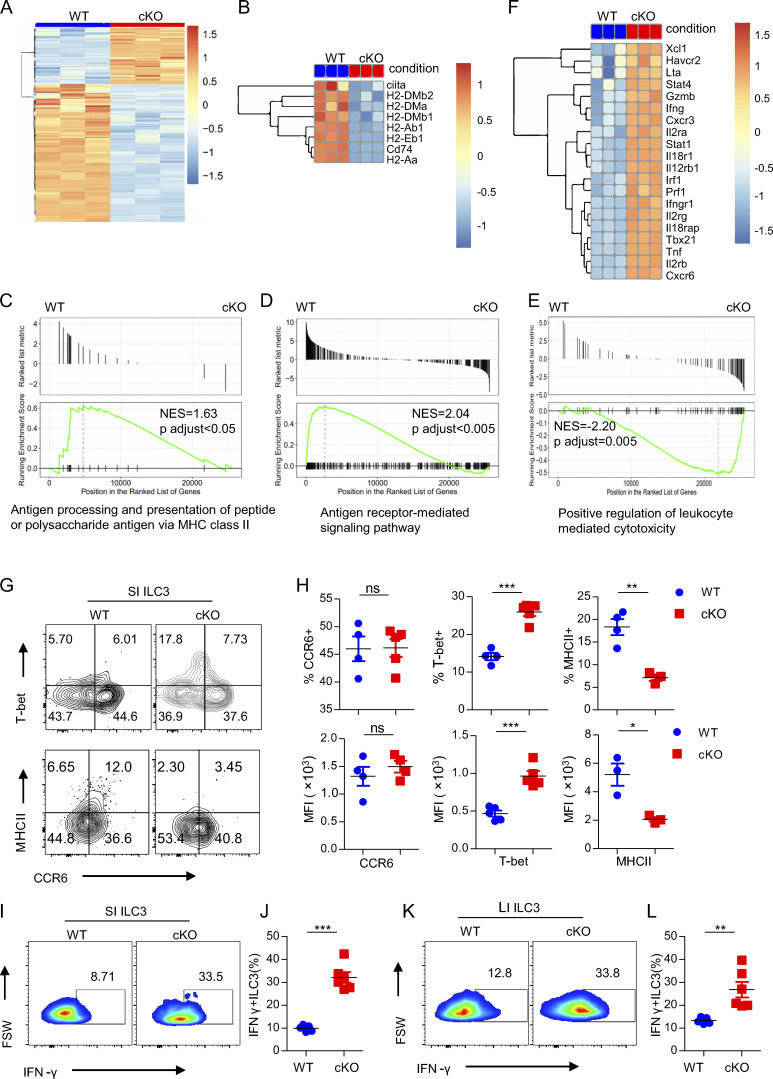
Given the specificity and majority of changes in SI ILC populations in cKO mice that occurred in ILC3s, we were, therefore, intrigued the role of BATF in SI ILC3s. To this end, we performed bulk RNA-seq analysis of sorted SI ILC3s from three independent WT or cKO mice. More than 4,748 genes were significantly differentially expressed in Batf-deficient ILC3s (false discovery rate <0.05, fold change >2.0; Fig. 2 A). The large proportion of downregulated DEGs (3,368 out of 4,748) in cKO mice suggests that BATF functions predominantly as a transcriptional activator in ILC3s, which is consistent with its function in lymphocytes (Li et al., 2012). Compared with WT ILC3s, Batf-deficient ILC3s exhibited downregulated genes related to the MHCII pathway (H2-Aa, H2-Ab, H2-Dma, H2-DMb1, H2-DMb2, Ciita, and Cd74) and wound healing (Tbxa2r, Insl3, Mylk, and Plpp3; Fig. 2 B and Fig. S3 A). Moreover, Gene Ontology (GO) annotation of these genes and gene set enrichment analysis (GSEA) corroborated these findings by revealing that antigen processing and presentation of peptide or polysaccharide antigen via MHC class II, antigen receptor–mediated signaling pathway, regulation of epithelial cell proliferation, and wound healing are significantly enriched in WT ILC3s (Fig. 2, C and D; and Fig. S3, B and C). Deletion of MHCII in ILC3s is known to lead to upregulated effector T cell responses, which drives spontaneous colitis (Hepworth et al., 2013). Consistently, we found that positive regulation of T cell–mediated cytotoxicity was significantly enriched in Batf-deficient ILC3s (Fig. 2 E). Additionally, genes associated with Th1 and cytotoxic immune responses (Tbx21, Ifng, Cxcr6, Il12rb1, Il18rap, Tnf, Gzmb, and Prf1) were significantly upregulated in Batf-deficient ILC3s (Fig. 2 F). Kyoto Encyclopedia of Genes and Genomes analysis also revealed that IBD was a pathway significantly enriched in Batf-deficient ILC3s (Fig. S3 D). Next, we validated T-bet and MHCII protein level changes in WT and Batf-deficient SI ILC3s by flow cytometry. As expected, loss of BATF led to significantly increased T-bet but reduced MHCII, both in their frequency and level of expression (Fig. 2, G and H; and Fig. S2, F and H). However, both frequency and expression of CCR6 were not affected (Fig. 2, G and H), most likely because our PLZF-cre system did not affect the formation of the CCR6+ ILC3 subset in the SI, as CCR6+ ILC3 has previously been thought to not originate from PLZFhi precursors (Constantinides et al., 2014). Intriguingly, the expression of BATF was deficient in all three subsets of intestinal ILC3s of cKO mice (Fig. S3, E–G), suggesting that CCR6+ ILC3s may in fact somehow be derived from PLZFhi precursors. Moreover, the expression of IFN-γ in SI and LI Batf-deficient ILC3s was significantly upregulated in response to PMA and ionomycin stimulation (Fig. 2, I–L). Taken together, these results validated that Batf-deficient ILC3s have a distinct gene expression program compared to WT ILC3s, one which was characterized by profound induction of Th1 transcriptional programs and decreased programs involving wound healing and antigen presentation via MHCII.
Figure 2.
BATF regulates the SI ILC3 transcriptional program. (A) Heatmap of bulk RNA-seq data showing significantly upregulated (1,380) and downregulated (3,368) genes of SI ILC3s from cKO and littermate control WT mice. (B) Heatmaps of expression of genes involved in MHCII antigen presentation from cKO versus WT SI ILC3s. (C–E) GSEA of selected pathways enriched in WT (C and D) and cKO (E) SI ILC3s. Normalized enrichment scores (NES) and adjusted P values are indicated for each gene set. (F) Heatmaps of expression of genes involved in type 1 effector genes from cKO versus WT SI ILC3s. (G) Representative flow plots of SI ILC3s (Lin−CD90.2+RORγt+) of WT and littermate control cKO mice. (H) Representative percentage (upper panel) and quantification of mean fluorescence intensity (MFI; bottom panel) of indicated markers on or in SI ILC3 of WT and littermate control cKO mice. (I and K) Representative flow plots of intracellular staining of IFN-γ–expressing ILC3s isolated from SI (I) and LI (K) of WT and littermate control cKO mice after stimulation with PMA plus ionomycin ex vivo. FSW, forward side pulse width. (J and L) Quantification of the frequency of IFN-γ+ ILC3s from SI (J) and LI (L). Data are shown as mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001 (two-tailed unpaired t test). Each dot represents one mouse, n = 4–6 mice per group. Data are pooled from two independent experiments. See also Fig. S5.
Figure S3.
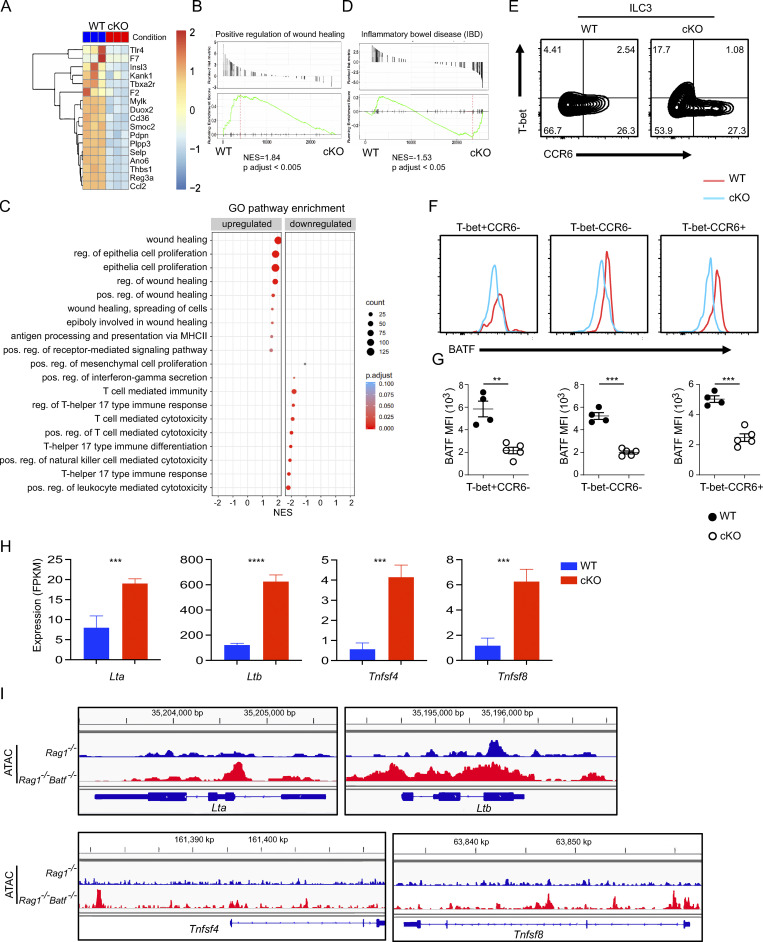
BATF regulates SI ILC3s transcriptional program. Related to Fig. 2. (A) Heatmap of wound healing signature genes from WT versus cKO SI ILC3s. (B–D) GSEA analysis of pathways differentially enriched in WT or cKO SI ILC3s. Normalized enrichment score (NES) and adjusted P value are indicated for each gene set. (B) GSEA plot of the positive regulation of wound-healing pathway. (C) GSEA dot plot of select GO pathways enriched in WT and cKO SI ILC3s. The size and color of each dot indicate the number of genes in a pathway and the adjusted P value, respectively. (D) GSEA plot of the IBD pathway. (E) Flow cytometry analysis subsets of intestinal ILC3 (Lin−CD90.2+Rorγt+) of cKO and littermate control WT mice aged 13 wk. (F) Flow cytometry histograms analyzing the expression of BATF in three subsets of ILC3 as assessed in E. (G) Quantification of MFI of BATF on ILC3s subsets as assessed in E. (H) The expression of indicated genes from cKO versus WT SI ILC3s via RNA-seq analysis. (I) Genomic snapshots showing accessibility (ATAC-seq) of genes as indicated in H in Rag1−/− and Rag1−/−Batf−/− ILC3s. Data are shown as mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001 (two-tailed unpaired t test). Each dot represents one mouse, n = 4–5 mice per group. Data are representative of at least two independent experiments.
BATF deficiency reshapes heterogeneity of enteric ILC3s
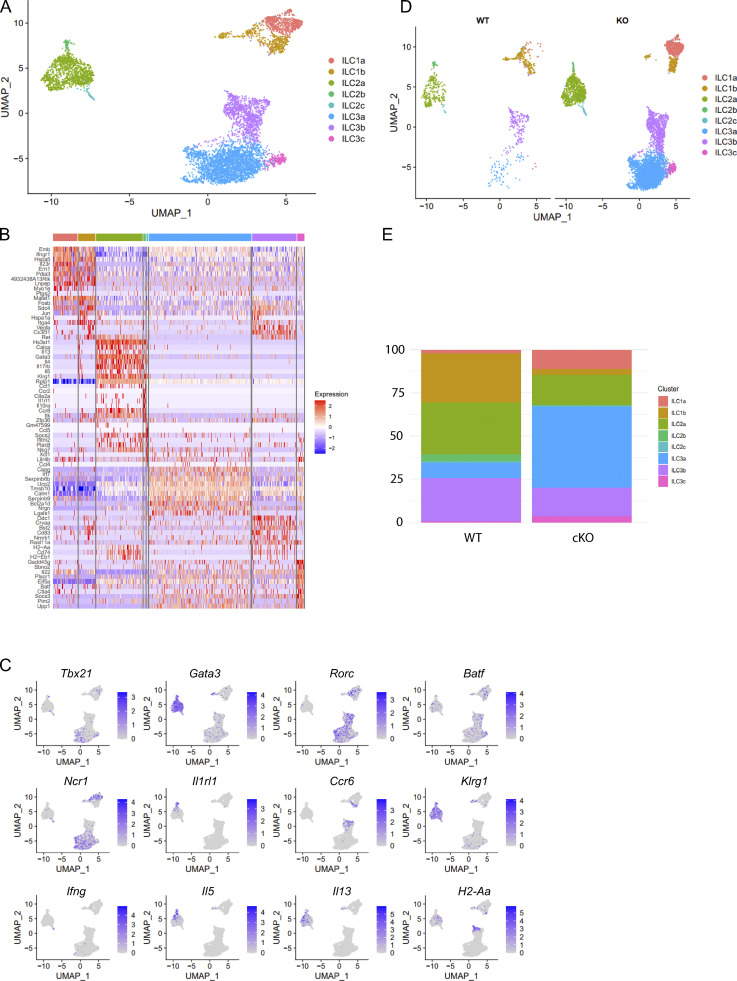
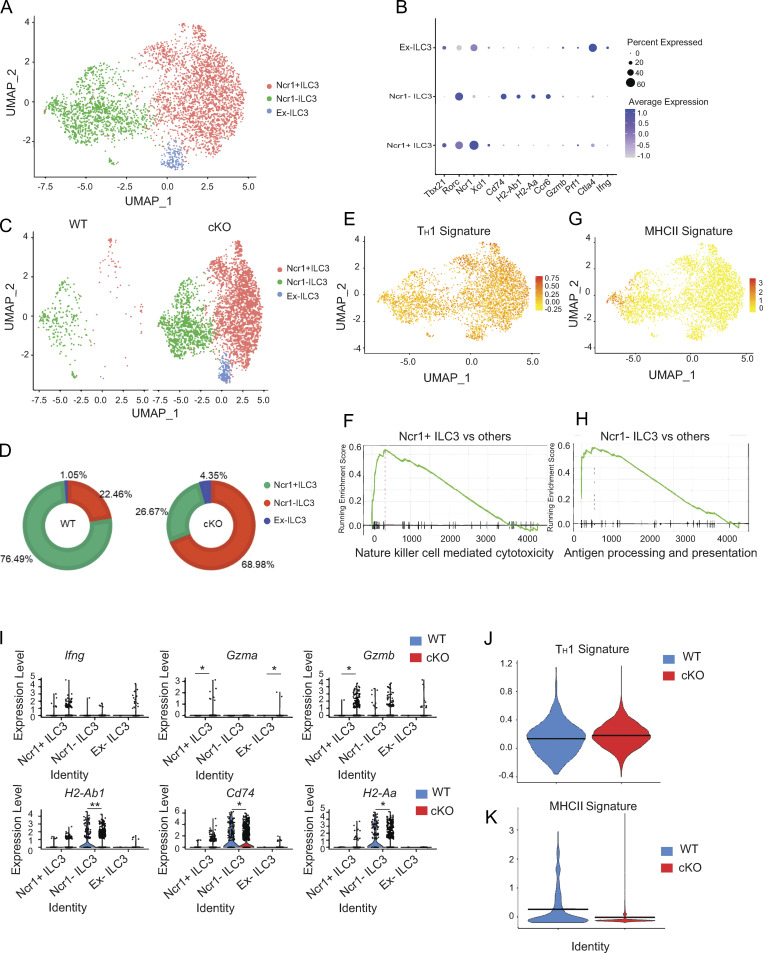
BATF deficiency increased Th1 transcriptional programs but decreased MHCII-associated programs in SI ILC3s, and this prompted us to investigate whether BATF maintains the heterogeneity of enteric ILC3s. We performed scRNA-seq on Lin−CD90.2+ ILCs sorted from the SI of WT and littermate cKO mice. Eight clusters were identified following visualization on a uniform manifold approximation and projection (UMAP) plot (Fig. S4, A and B). Clusters C1 and C2 were characterized as ILC1s (expressing Tbx21, Ncr1, and Ifng), C3–C5 were characterized as ILC2s (expressing Gata3, Il1rl1, klrg1, Il5, and Il13), and C6–C8 were characterized as ILC3s (expressing Rorc, H2-Aa, Ncr1, and Ccr6; Fig. S4 C). Notably, compared with ILCs in WT mice, the proportion of ILC1s and ILC2s decreased, but the proportion of ILC3s increased in cKO mice (Fig. S4, D and E). Furthermore, we computationally isolated and reclustered ILC3s by UMAP analysis, which formed three clusters (Fig. 3 A). Cluster 1 represented Ncr1+ ILC3s as they expressed Ncr1, Tbx21, and Xcl1. Cluster 2 represented Ncr1– ILC3s due to the lack of Ncr1 expression, but a positive expression of Ccr6 and antigen processing and presentation genes (H2-Aa, Ha-Ab1, and Cd74). Cluster 3 was defined to be ex-ILC3s as this cluster resembled cluster 1 but had lower levels of Rorc (encodes Rorγt) and high expression of cytotoxicity-related genes (Ifng, Gzma and Gzmb; Fig. 3 B). Notably, ILC3 heterogeneity, as indicated by the proportion of subsets within the ILC3 compartment, was significantly altered by BATF deficiency (Fig. 3, C and D). The proportion of Ncr1+ ILC3s and ex-ILC3s, both of which exhibited strong Th1 signature gene expression scores, increased by 4.1- and 3.1-fold, respectively, in the absence of BATF (Fig. 3, D and E). Moreover, GSEA revealed that genes correlated with NK cell–mediated cytotoxicity were significantly enriched in Ncr1+ ILC3s (Fig. 3 F), suggesting that BATF is required to prevent ILC3s from acquiring type 1 ILC features and converting to a cytotoxic ILC1- or NK-like fate. By contrast, the proportion of Ncr1− ILC3s, which exhibited strong MHCII signature gene expression scores and were highly enriched by GSEA for genes involved in antigen processing and presentation, decreased by a fold of 2.8 in the absence of BATF (Fig. 3, D, G, and H). The altered enrichment of ILC3 subtypes would likely be reflected in changes in gene expression. Consistent with this, BATF-deficient ILC3s increased Th1-associated effector genes (e.g., Ifng, Gzma, and Gzmb) and decreased MHCII signature genes (e.g., H2-Ab1, CD74, and H2-Aa) in their distinct clusters (Fig. 3 I). Collectively, BATF-deficient ILC3s have an increased Th1 signature but a decreased MHCII signature (Fig. 3, J and K). Taken together, our scRNA-seq analysis revealed significant alterations in enteric ILCs due to BATF deficiency, with the most noticeable changes occurring in ILC3s.
Figure S4.
BATF deficiency alters heterogeneity of SI ILCs. Related to Fig. 3. (A) UMAP plot of scRNA-seq data of SI-derived ILCs from cKO and littermate control WT mice. Cells are colored by cluster. (B) Heatmap reporting scaled expression of genes differentially upregulated by each cluster. (C) Expression of select marker genes as visualized by UMAP plots. (D) UMAP plot split by sample of origin. Cells are colored by cluster. (E) Stacked bar plots displaying the proportion of clusters in WT and cKO mice as shown in D.
Figure 3.
BATF deficiency in ILCs alters heterogeneity of SI ILC3s. (A) UMAP plot of combined scRNA-seq data of SI-derived ILC3s from WT and littermate control cKO mice. Cells are colored by cluster. (B) Representative differentially expressed genes between clusters. Dot size represents the percentage of cells in a cluster expressing the gene and color shows the average relative expression of the gene. (C) UMAP representation of SI ILC3s isolated from WT and littermate control cKO mice. Cells are colored by cluster. (D) Pie charts displaying the proportion of subsets of ILC3s as described in C. (E and G) UMAP visualization showing a Th1 (E) and antigen presentation via MHCII signature (G) score on SI ILC3s as described in A. (F and H) GSEA for NK cell–mediated cytotoxicity (F) and antigen processing and presentation (H) signature genes in a given cluster compared to the rest of the cells. (I) Violin plots showing the expression of indicated genes among the three ILC3 clusters. (J and K) Violin plot showing Th1 (J) and antigen presentation via MHCII (K) signature score comparison between WT and cKO SI ILC3s. Two-sided Wilcoxon rank-sum tests were used (*P < 0.05, **P < 0.01, ***P < 0.001).
BATF deficiency in ILCs alters gut microbial composition
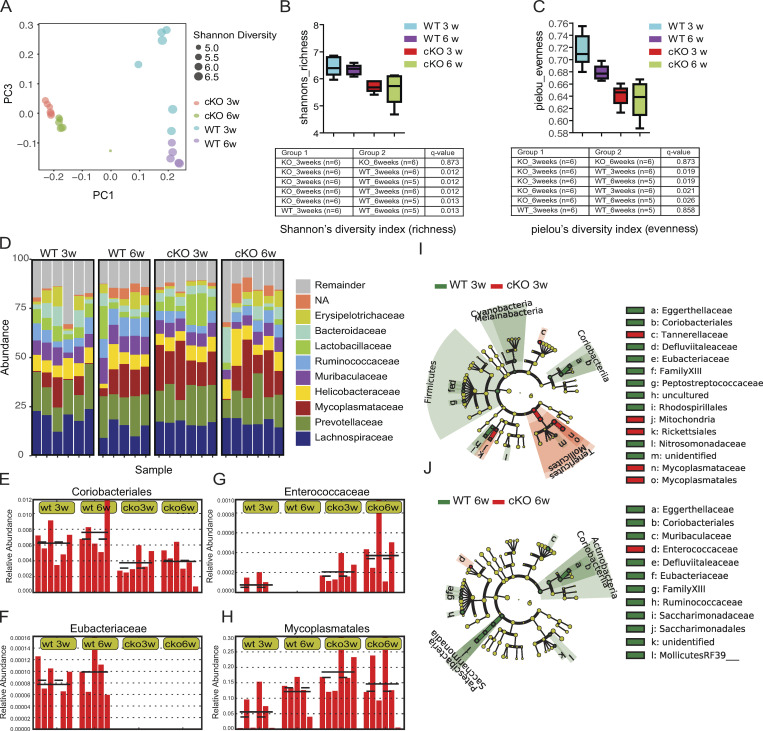
ILCs have been found to play an essential role in shaping the gut microbiota (Mao et al., 2018). To determine whether BATF ablation in ILCs altered the composition of commensal microbiota, we employed 16S ribosomal RNA (rRNA) sequencing to characterize fecal bacteria isolated from Batf-deficient cKO mice and co-housed littermate WT controls. To investigate the dynamics of the microbial communities, we collected samples at two-time points, one at 3 wk of age (following weaning) and the other at 6 wk of age. Jaccard principal coordinate analyses (PCoA) revealed that samples were separated largely by genotype, implying that Batf-deficient cKO mice harbored a distinct microbiome composition (Fig. 4 A). However, samples from cKO mice at different time points clustered more tightly than those from WT mice, indicating BATF deficiency in ILCs affected the variability of the microbiome (Fig. 4 A). We, therefore, compared bacterial diversity within each group by α diversity analysis. Shannon’s index and Pielou’s evenness index revealed a significant decrease in bacterial richness and diversity in Batf-deficient cKO mice at both 3 and 6 wk of age (Fig. 4, B and C). Further analysis of the relative abundance of predominant bacteria at the family level showed that Mycoplasmataceae family members, which can cause pelvic inflammatory disease and are associated with IBD (Chen et al., 2001), were more abundant in 3-wk-old cKO mice than age-matched WT mice (Fig. 4 D). By contrast, the probiotic bacterial family Lactobacillaceae, which was reported to have reduced abundance in IBD patients (Salem et al., 2019) and mice with ILC-intrinsic deletion of MHCII (Hepworth et al., 2013), was under-represented in cKO mice at 6 wk of age (Fig. 4 D). We further investigated how the abundance of different bacterial families changed between WT and cKO mice. Coriobacteriales and Eubacteriaceae, highly prevalent in WT mice, were significantly less abundant in Batf-deficient cKO mice at both time points (Fig. 4, E and F). Conversely, Enterococcaceae and Mycoplasmatales, which are known to have an increased abundance in pediatric IBD patients (Bhatt et al., 2018), were more abundant in samples from cKO mice compared with littermate controls (Fig. 4, G and H). The dysbiosis in the 3-wk-old and 6-wk-old cKO mice further recapitulated dysbiosis in IBD patients as it caused a reduction in the abundance of the Firmicutes phylum, as well as that of its constituent family Enterococcaceae (Zhou et al., 2018; Fig. 4, I and J). Collectively, these data suggested that Batf deficiency in ILCs led to the development of dysbiosis, which may initiate and/or exacerbate the pathogenesis of intestinal inflammation.
Figure 4.
BATF deficiency in ILCs alters gut microbial composition. (A) Unweighted UniFrac PCoA of fecal microbiota composition of co-housed WT and littermate control cKO mice. PC1 and PC3, principal components 1 and 3, respectively. Each symbol represents an individual mouse, with the size indicating Shannon diversity. (B and C) Boxplot representation of richness (Shannon diversity index; B) and evenness (Pielou’s index; C) for microbiome composition. Boxes represent the interquartile range and the horizontal line inside the box is the median. Whiskers span the most extreme data points within 1.5× the interquartile range. Statistical significance was determined using the Kruskal–Wallis test or one-way ANOVA with post-hoc Tukey’s correction. q value < 0.05 was considered statistically significant. (D) Representation of relative abundances of specific bacteria at the family level in individual mice. (E–H) Plots showing relative abundances of selected bacterial families in individual mice. Data are shown as mean (solid black line) and standard deviation (dotted black line), each bar represents an individual mouse. (I and J) Cladograms displaying differences in taxa composition determined by microbiome profiling of mice as described in A. Samples were collected at the age of 3 wk (I) and 6 wk (J). Each dot represents one mouse, n = 5–6 mice per group. Data are pooled from two independent experiments.
Dysregulation of the microbiome promotes intestinal inflammation
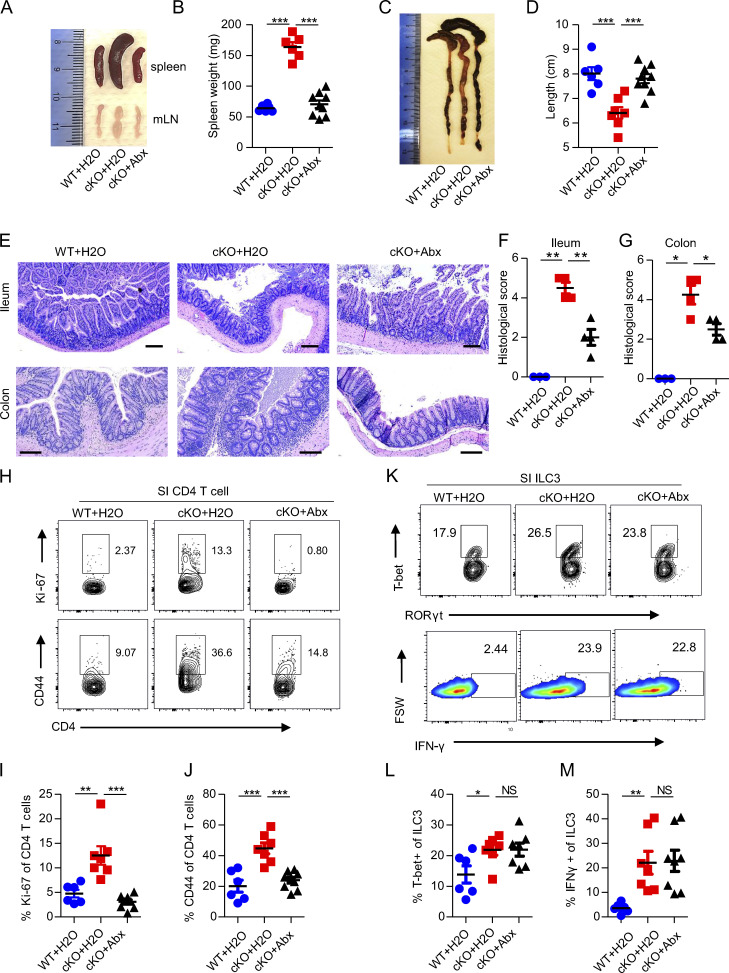
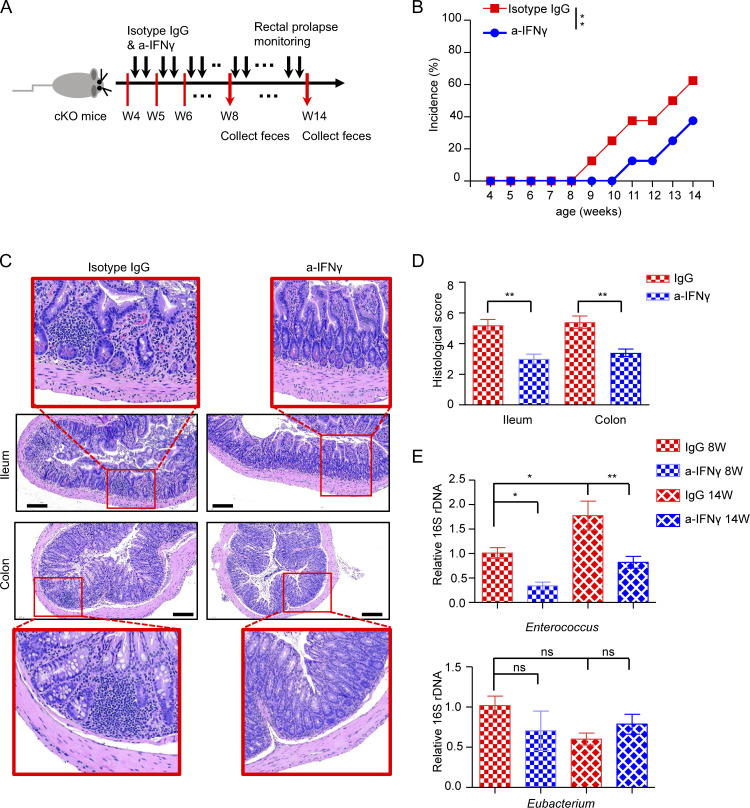
Alterations in microbiota contribute to the pathogenesis of colitis (Nishida et al., 2018). To functionally address whether the progression of spontaneous colitis seen in Batf cKO mice is indeed driven by dysbiosis, we treated cKO mice with a mix of broad-spectrum antibiotics in drinking water for 5–6 wk after weaning. As compared to cKO mice given untreated water, both WT mice given untreated water and cKO mice given antibiotics had markedly decreased spleen and mesenteric lymph node (mLN) sizes and spleen weight, as well as a significant increase in colon length (Fig. 5, A–D). Furthermore, histological examination suggested that antibiotic treatment significantly improved intestinal inflammation in cKO mice, as characterized by lower epithelial irregularity, reduced leukocyte infiltration, and overall decreased histological score (Fig. 5, E–G). Given that cKO mice have more activated, and thus potentially pathogenic CD4+ T cells in the gut than WT mice, we next evaluated whether depleting intestinal microbes can impact intestinal CD4+ T cell activation and/or proliferation. As expected, antibiotic treatment of cKO mice markedly reduced the frequencies of proliferative (Ki-67+) and activated (CD44+) CD4+ T cells in the SI (Fig. 5, H–J). Although GI tract inflammation in cKO mice was improved by antibiotic treatment, histological analysis of SI and LI tissues still showed elevated leukocyte infiltration as compared with untreated WT mice (Fig. 5, E–G). We therefore hypothesized that ablation of bacterial influx primarily limited pathogenic T cell–mediated chronic colitis, but was unable to treat the underlying onset of colitis, which was at least partially caused by persistent IFN-γ secretion by Batf-deficient ILC3s. In accordance with our hypothesis, we found that antibiotic treatment did not reduce the elevated expression of T-bet and IFN-γ in Batf-deficient ILC3s (Fig. 5, K–M), suggesting a dispensable role for the microbiome in the Th1 program inhibited by BATF in gut ILC3s. Of note, the persistent production of IFN-γ leads to epithelial permeability, as has been noted in patients with IBD (Takayama et al., 2010), which suggests that blockade of IFN-γ function may play a role in the development or progression of IBD. To further determine the relevance of IFN-γ in initiation and/or driving gut inflammation, we next analyzed the function of this cytokine by the administration of antibody blockade. For this, we treated co-housed cKO mice with anti–IFN-γ or an isotype-IgG control antibody two times per week starting after weaning (Fig. 6 A). Mice treated with anti–IFN-γ had significantly delayed development of rectal prolapse than did mice treated with isotype-IgG control antibody (Fig. 6 B). Histologic analyses of intestinal tissue showed better barrier integrity and less colitis in mice treated with anti–IFN-γ than in mice treated with isotype-IgG control antibody (Fig. 6, C and D). Furthermore, to determine whether IFN-γ blockade might affect the composition of intestinal microbiota in cKO mice, we collected feces from both groups after antibody blockade for 4 wk (before rectal prolapse onset) and 10 wk (after rectal prolapse onset). Two bacteria genera, Enterococcus and Eubacterium, which showed overgrowth and underrepresentation in cKO mice, respectively, were examined through quantitative PCR (qPCR). Intriguingly, IFN-γ blockade significantly inhibited the growth of pathogenic opportunistic bacteria Enterococcus in cKO mice but had little effect on the composition of Eubacterium (Fig. 6 E). Collectively, these data demonstrate that an altered microbiome exacerbates the severity of colitis, but does not fully account for intestinal inflammation, in Batf-deficient cKO mice.
Figure 5.
Antibiotics ameliorate spontaneous colitis in cKO mice. (A and B) Representative images of spleens and mLNs (A) and spleen weight (B) of WT and littermate control cKO mice treated or not treated with antibiotics. (C and D) Representative images of colons (C) and colon length (D) of WT and littermate control cKO mice treated or not treated with antibiotics. (E) H&E staining of ileum and colon of WT and littermate control cKO mice treated or not treated with antibiotics. Scale bar, 100 µm. (F and G) Quantification of a histological score of the ileums (F) and colons (G) of mice as in E. (H) Representative flow plots of proliferating Ki-67+ and activated CD44+ CD4+ T cells from SI of WT and littermate control cKO mice treated or not treated with antibiotics. (I and J) Quantification of the frequency of proliferating Ki-67+ (I) and activated CD44+ (J) CD4+ T cells as in (H). (K) Representative flow plot of T-bet+ ILC3s (upper panel) and IFN-γ production by ILC3s after stimulation with PMA plus ionomycin ex vivo. ILC3s were isolated from SI of WT and littermate control cKO mice treated or not treated with antibiotics. FSW, forward side pulse width. (L and M) Quantification of the frequency of T-bet+ (L) and IFN-γ+ (M) SI ILC3s as in K. Antibiotics (Abx) were continuously administered in the drinking water of WT and littermate control cKO mice at weaning until 11–13 wk of age. Data shown as mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001 (two-tailed unpaired t test). Each dot represents one mouse, n = 3–8 mice per group. Data are pooled from two independent experiments.
Figure 6.
IFN-γ blockade delays spontaneous intestinal inflammation in BATF-deficient cKO mice. (A) Experiment design showing the timeline for co-housed cKO mice intraperitoneally injected with anti–IFN-γ (a-IFNγ) or isotype-IgG control antibody, started at week 4. (B) Mice injection with anti–IFN-γ or isotype-IgG control antibody were monitored for incidence of rectal prolapse. (C) H&E staining of ileum and colon sections from mice injection with anti–IFN-γ or isotype-IgG control antibody at week 14. Scale bar, 100 µm. (D) Quantification of the histological score of ileum and colon as assessed in C. (E) qRT-PCR analysis of relative abundance of fecal microbiome from mice injection with anti–IFN-γ or isotype-IgG control antibody at week 8 and 14. Statistical differences were tested using two-way ANOVA for B, unpaired t test for D, and one-way ANOVA for E, n = 5–8 mice per group. Data are pooled from two independent experiments. *P < 0.05, **P < 0.01.
The effect of BATF on ILC3s is cell intrinsic
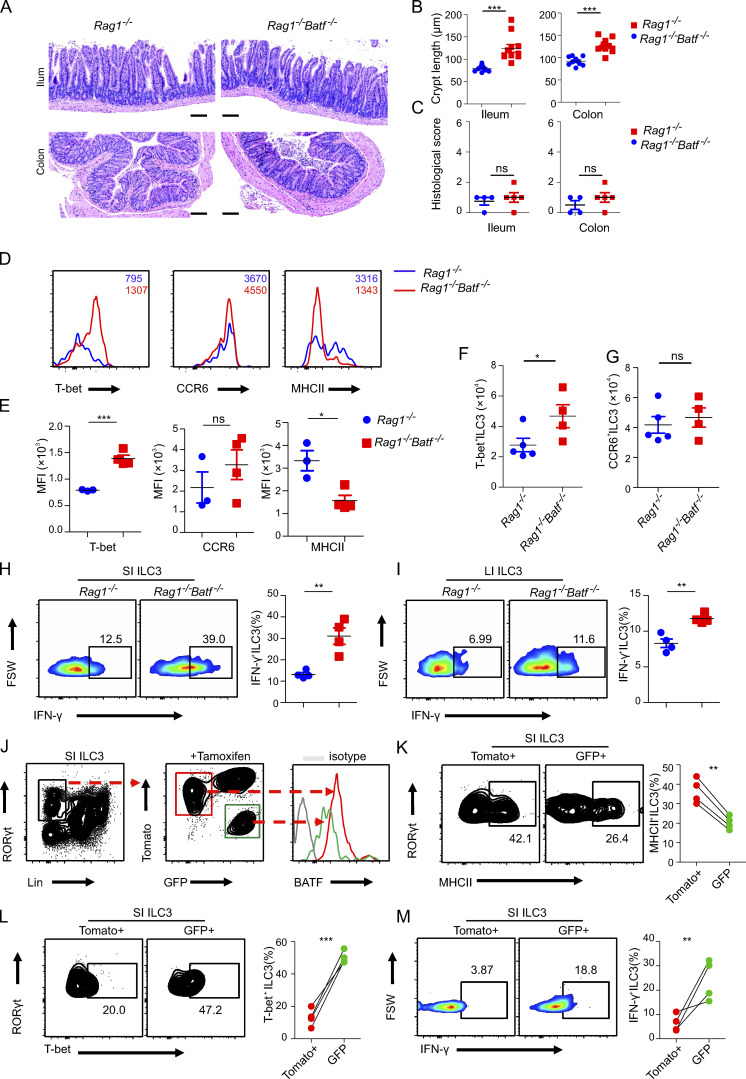
Having demonstrated that certain effects of BATF on ILC3s were independent of the microbiome, we sought to determine whether BATF functions intrinsically in ILC3s. First, to eliminate the potential impact of adaptive immunity, we crossed Batf−/− mice with Rag1−/− mice to obtain Batf−/−Rag1−/− double-deficient mice. Consistent with a previous report, Batf−/−Rag1−/− mice did not develop rectal prolapse (Liu et al., 2020a), even up to old age (data not shown). However, histological analyses of the intestinal tissue of aged Batf−/−Rag1−/− mice revealed atrophy of the villi in the ileum and crypt deformation (Fig. 7 A). Furthermore, the length of crypts of the ileum and colon was significantly higher in Batf−/−Rag1−/− mice than in Rag1−/− littermate control mice (Fig. 7 B), both Rag1−/− and Batf−/−Rag1−/− mice harbor low and comparable histological score in the gut (Fig. 7 C). Moreover, similar to altered expression between WT and cKO ILC3s, Batf−/−Rag1−/− ILC3s also exhibited higher expression of T-bet, but lower expression of MHCII as compared with Batf-sufficient Rag1−/− ILC3s (Fig. 7, D and E). Moreover, Batf−/−Rag1−/− mice harbored greater cellular numbers of T-bet+ ILC3s but comparable numbers of CCR6+ ILC3s in the gut (Fig. 7, F and G). Significantly more IFN-γ secretion was observed in both SI and LI Batf−/−Rag1−/− ILC3s under PMA and ionomycin stimulation (Fig. 7, H and I). All these data suggested an intrinsic and inhibitory role of BATF in regulating gene expression in ILC3s, independent of the effects of BATF on adaptive immunity. To further validate the intrinsic and dual role of BATF on ILC3s, we crossed Batffl/fl and ERT2CreRosamTmG color tracing mice to generate Batffl/fl ERT2CreRosamTmG mice. In this model, administration of tamoxifen (TAM) causes nuclear accumulation of Cre recombinase and concomitant excision of the floxed Batf locus as well as the floxed reporter locus. This causes membrane-bound tdTomato in Batf-sufficient cells to be replaced with a GFP once Cre recombinase is activated and the cells become Batf deficient. TAM was administered for three consecutive days by intraperitoneal injection, and the samples were analyzed by flow cytometry 10 d after the last injection. We detected three distinct populations in SI ILC3s and found that BATF was dramatically decreased in GFP+ ILC3s when compared to intact Tomato+ ILC3s from the same mice, which confirmed the efficiency of TAM-mediated BATF deletion (Fig. 7 J). Further analysis revealed reduced levels of MHCII but increased levels of T-bet and IFN-γ in Batf-deficient ILC3s (Fig. 7, K–M). These data in combination with scRNA-seq strongly support the notion that BATF works as a regulatory switch in ILC3s. While it promotes the stability of protective MHCII+ ILC3 subsets, it also suppresses the formation of pathogenic T-bet+ ILC3 in the gut. Thus, the regulation of BATF in ILC3s is crucial to control ILC3-mediated homeostasis in the gut.
Figure 7.
BATF acts on ILC3s in a cell-intrinsic manner. (A) Representative H&E staining of ileum and colon of Rag1−/− and littermate control Rag1−/−Batf−/− mice. Scale bar, 100 µm. (B) Quantification of crypt length of ileum and colon as in A. (C) Quantification of the histological score of ileum and colon as assessed in A. (D) Representative histogram flow plot of SI ILC3s (Lin−CD90.2+RORγt+) of Rag1−/− and littermate control Rag1−/−Batf−/− mice. (E) Quantification of MFI of indicated markers on or in SI ILC3s from Rag1−/− and littermate control Rag1−/−Batf−/− mice. (F and G) Quantification of cell number of T-bet+ ILC3 (F) and CCR6+ ILC3 (G) in SI from Rag1−/− and littermate control Rag1−/−Batf−/− mice. (H and I) Representative flow plots and quantification of intracellular staining of ILC3s isolated from SI (H) and LI (I) after stimulation with PMA plus ionomycin ex vivo. ILC3s were isolated from Rag1−/− and littermate control Rag1−/−Batf−/− mice. (J) Gating strategy of flow cytometry analysis of SI ILC3s from Batffl/fl ERT2CreRosamTmG mice treated with TAM (left) and intrinsic differences of BATF expression between Tomato+ WT (red) and GFP+ BATF-deficient (green) ILC3s. (K) Representative flow plot (left) and quantified frequency (right) of MHCII+ ILC3s gated on Lin−RORγt+Tomato+ and Lin−RORγt+GFP+ cells. (L) Representative flow plot (left) and quantified frequency (right) of T-bet+ ILC3s gated on Lin−RORγt+Tomato+ and Lin−RORγt+GFP+ cells. (M) Representative flow plot (left) and quantified frequency (right) of IFN-γ+ ILC3s gated on Lin−RORγt+Tomato+ and Lin−RORγt+GFP+ cells. ILC3s were isolated from SI and stimulated with PMA plus ionomycin ex vivo. For B–C and E–G, data are shown as mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001 (two-tailed unpaired t test). Each dot represents one mouse, n = 3−4 mice per group. Data are representative of at least two independent experiments. For K–M, connected dots indicate one individual mouse (n = 4). Statistical differences were tested using a paired t test (two-tailed). *P < 0.05, **P < 0.01, ***P < 0.001. Data are representative of two independent experiments.
IL-6 and TGF-β induce BATF expression in ILC3s
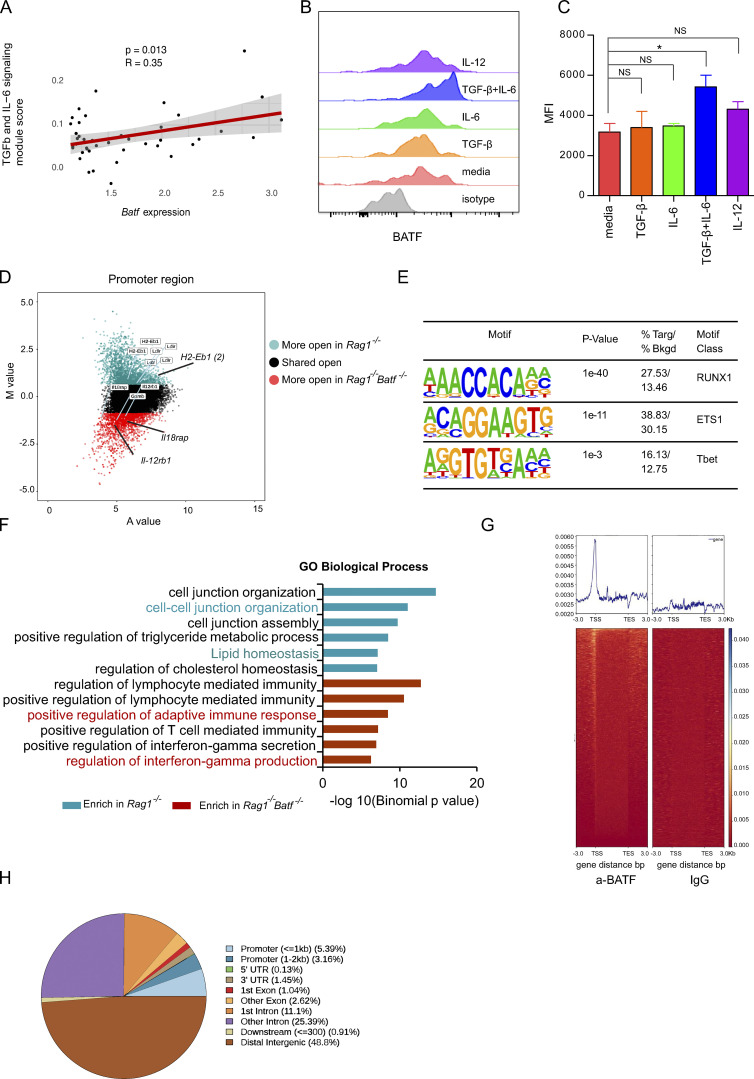
BATF expression is regulated by cytokine and antigen receptor sigaling. In particular, IL-6 has been shown to induce BATF expression in Th17 cells (Ciofani et al., 2012). This prompted us to test whether the same mechanism can be utilized by ILC3s. First, we performed pathway analysis on our scRNA-seq dataset and found that IL-6/TGF-β signaling module score positively correlated with BATF expression in ILC3s (Fig. S5 A). Next, we tested if TGF-β/IL-6 would induce BATF expression in ILC3s. To this end, we cultured intestinal ILC3s from Rag1−/− in the presence of IL-6 and TGF-β either alone or in combination. In addition, IL-12, another known cytokine that induces BATF expression, was also included. Strikingly, the addition of IL-6 with TGF-β significantly induced BATF expression, whereas the addition of IL-6, TGF-β, or IL-12 alone had little or no effect (Fig. S5, B and C). This indicates that ILC3s respond to similar stimuli, as adopted by their adaptive ortholog Th17 cells, to modulate BATF expression. Collectively, these data demonstrate that IL-6 in synergy with TGF-β induces the expression of BATF, which in turn intrinsically regulates ILC3 function by suppressing pro-inflammatory gene expression and promoting antigen presentation via MHCII.
Figure S5.
The expression of BATF is regulated by cytokines which maintains the chromatin landscape in ILC3s. (A) Linear correlation between Batf expression in ILC3s of scRNA-seq and a Seurat module score calculated using genes pooled from a gene list for TGF-β and IL-6 signaling. (B) Flow cytometry plot showing expression of BATF in ILC3s under different cytokines stimulation in vitro. Media include IL-2, IL-7, and SCF only. Isotype, isotype control. (C) Quantification of MFI of BATF in ILC3s as cultured in A. *P < 0.05, **P < 0.01 (one-way ANOVA analysis). (D) Scatter plot (MA plot) showing M value (log2[read density in Rag1−/− ÷ read density in Rag1−/−Batf−/−]) vs. A value (0.5 × log2[read density in Rag1−/− × read density in Rag1−/−Batf−/−]) of the merged set of Rag1−/− and Rag1−/−Batf−/− promoter ATAC-seq peaks after normalization. The top 3,000 peaks are highlighted for Rag1−/− (dark green) and Rag1−/−Batf−/− (dark red) ILC3s. Selected genes are labeled. (E) Selected Th1-associated TF binding motifs significantly enriched in the top 3,000 specific promoters of Rag1−/−Batf−/− ILC3s. (F) GO analysis of gene sets with differential chromatin accessibility between WT (dark green) and cKO (dark red) for the top six Molecular Signature Database pathways enriched in WT or cKO-specific promoters was performed using GREAT. Pathways of interest are highlighted in color. (G) Heatmap for genome-wide distribution of anti-BATF and control rabbit IgG binding signals at peak centers in ILC3s sorted from the SI of Rag1−/− mice using CUT&Tag sequencing. (H) Distribution of BATF binding peaks across the genome of ILC3s as assessed in G.
BATF maintains the chromatin landscape of ILC3s
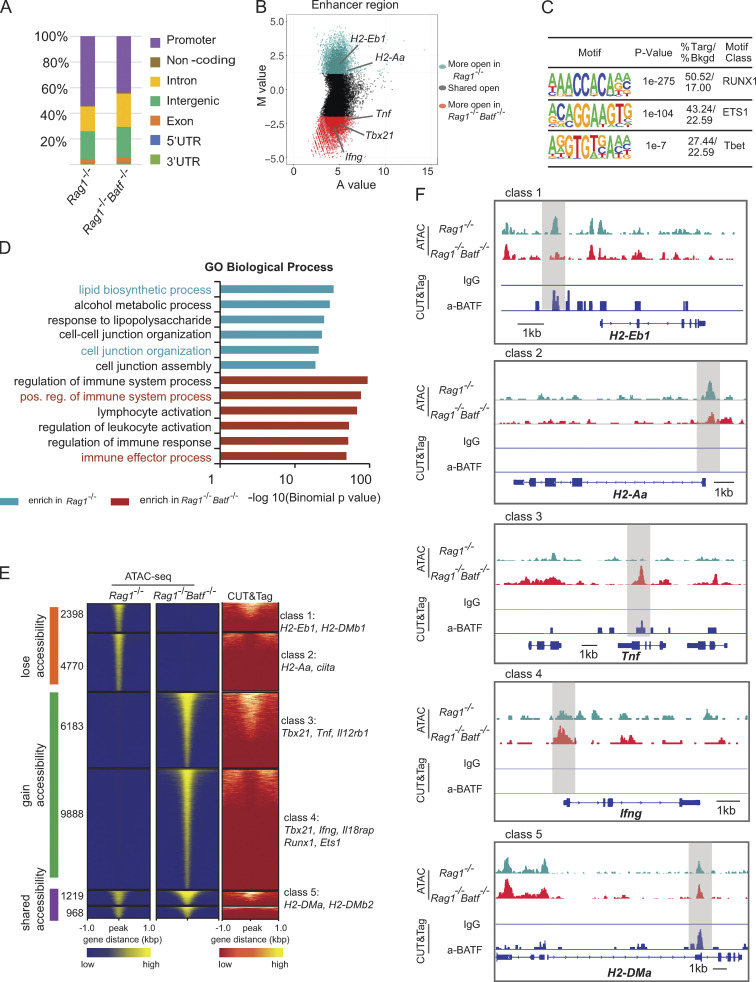
The molecular mechanism that controls ILC3s plasticity, in particular for conversion of ILC3 to ILC1, is accompanied by dynamic changes in chromatin landscape (Parker et al., 2020). BATF functions as a pioneer TF by regulating chromatin accessibility in various lymphocytes. To gain further insight into whether Batf deletion could affect chromatin accessibility in ILC3s, we performed an assay for transposase-accessible chromatin with sequencing (ATAC-seq) analysis of SI ILC3s sorted from Batf+/+Rag1−/− and Batf−/−Rag1−/− littermate mice. Globally, the analysis of ATAC-seq peak distribution across the genome revealed no significant changes between BATF-sufficient and BATF-deficient ILC3s (Fig. 8 A), with most peaks mapped to promoters (40–50%), introns (21–24%), and intergenic regions (19–26%; Fig. 8 A). Next, we compared changes in open chromatin status located in enhancer and promoter regions. Of note, genes encoding proteins involved in antigen presentation via MHCII, including H2-Aa, H2-Ab1, and Cd74, contained at least one locus that was selectively more accessible in Batf +/+ ILC3s than their counterpart Batf−/− ILC3s (Fig. 8 B; and Fig. S5 D). By contrast, the expression of Th1-related genes (such as Tbx21, Ifng, Tnf, Lta, Ltb, Tnfsf4, and Tnfsf8) was increased, and their chromatin accessibility was more accessible in BATF-deficient ILC3s (Fig. 8 B; and Fig. S3, H and I; and Fig. S5 D). In agreement with our RNA-seq data, these genes exhibited simultaneous differential chromatin accessibility and gene expression, indicating BATF may serve as a chromatin remodeler to regulate gene expression in ILC3s. To further identify potential TFs that mediate BATF-dependent alterations in chromatin accessibility, we analyzed consensus TF motif enrichment and found that Runx, ETS, and T-bet family TFs were significantly enriched in chromatin regions selectively opened in BATF-deficient ILC3s (Fig. 8 C and Fig. S5 E). This finding is in line with the described roles for Runx and ETS as T-bet coregulators in facilitating Th1 or Th17 gene regulation (Wang et al., 2014) and is indicative of a shift toward a pro-inflammatory chromatin landscape in BATF-deficient ILC3s. In support of this, Genomic Regions Enrichment of Annotation Tools (GREAT) analysis showed that Batf-deficient ILC3s were selectively enriched for GO pathways involving positive regulation of immune system process, lymphocyte activation, positive regulation of lymphocyte-mediated immunity, and positive regulation of IFN-γ secretion in enhancer and promoter regions (Fig. 8 D and Fig. S5 F). Chromatin accessibility is associated with transcriptional alteration and TF occupancy (Zaret and Carroll, 2011). To further investigate the involvement of BATF binding linked to chromatin accessibility, we performed genome-wide chromatin immunoprecipitation with sequencing (ChIP-seq) via cleavage under targets and tagmentation (CUT&Tag). Anti-BATF antibody, but not control IgG, binding peaks were strongly enriched within 1 kb of promoter regions across the whole genome of ILC3s from Rag1−/− mice (Fig. S5 G). Distribution of BATF binding peaks across the genome of ILC3s was mainly located in intergenic and intron regions (Fig. S5 H). Next, we sought to evaluate the relationship between BATF binding and altered chromatin accessibility in the absence of BATF. By integrating ATAC-seq and BATF CUT&Tag-seq datasets, we defined six distinct classes of enhancer regulatory elements that represent dynamic chromatin landscapes in ILC3s (Fig. 8 E). We identified more than 33% (2,398 of 7,168) of peaks that lost accessibility in Rag1−/− Batf−/− ILC3s (e.g., H2-Eb1 and H2-DMb1, class 1) and showed BATF binding in the CUT&Tag-seq dataset (Fig. 8, E and F), whereas another 66% (4,770 of 7,168) of peaks lost accessibility in Rag1−/− Batf−/− ILC3s (e.g., H2-Aa and ciita, class 2) but were not bound by BATF (Fig. 8, E and F). Furthermore, we found that 38% (6,183 of 16,071) of regulatory elements (e.g., Tbx21, Tnf, and Il12rb1, class 3) gained accessibility in Rag1−/−Batf−/− ILC3s and were accompanied by BATF binding (Fig. 8, E and F), whereas 61% (9,888 of 16,071) of regulatory elements (e.g., Ifng, Il18rap, Runx1 and Ets1, class 4) gained accessibility but without BATF binding (Fig. 8, E and F). In addition, we observed genomic regions that remained accessible and had BATF binding (e.g., H2-Dma and H2-DMb2, class 5; Fig. 8, E and F). Collectively, these findings underscore the fact that BATF epigenetically controls distinct sets of genes that contribute to maintaining ILC3s functional identity and plasticity.
Figure 8.
BATF maintains the chromatin landscape of ILC3s. (A) Distribution of ATAC-seq peaks across the genome in Rag1−/− and Rag1−/−Batf−/− SI ILC3s. (B) Scatter plot (MA plot) showing M value (log2[read density in Rag1−/− ÷ read density in Rag1−/−Batf−/−]) vs. A value (0.5 × log2[read density in Rag1−/− × read density in Rag1−/−Batf−/−]) of the merged set of Rag1−/− and Rag1−/−Batf−/− enhancer ATAC-seq peaks after normalization. The top 5,000 peaks are highlighted for Rag1−/− (dark green) and Rag1−/−Batf−/− (dark red) ILC3s. Selected genes are labeled. (C) Selected Th1-associated TF binding motifs significantly enriched in the top 5,000 specific enhancers of Rag1−/−Batf−/− ILC3s. (D) GO pathway analysis of genes with differential chromatin accessibility between Rag1−/− (dark green) and Rag1−/−Batf−/− (dark red) was performed using GREAT. The −log10(binomial P value) is shown for the top six Molecular Signature Database pathways enriched in Rag1−/− and Rag1−/−Batf−/−-specific enhancers, with pathways of interest highlighted based on sample enrichment. (E) Heatmap of all atlas peaks of ATAC-seq of enhancers in Rag1−/− and Rag1−/−Batf−/− ILC3s, including peaks that gain and lose accessibility between Rag1−/− and Rag1−/−Batf−/− ILC3s and those in common between the two (left and middle panels). The right panel shows a heatmap of BATF binding peaks corresponding to atlas peaks in the left and middle panels. Combinatorial peak distribution patterns generated distinct six major classes of locus accessibility and binding. Representative annotation of nearby genes for class 1–5 is shown. (F) Genomic snapshots showing accessibility (ATAC-seq) in Rag1−/− and Rag1−/−Batf−/− ILC3s as well as anti-BATF and control IgG (CUT&Tag) binding in Rag1−/− ILC3s. Representative loci of class 1–5 are shown.
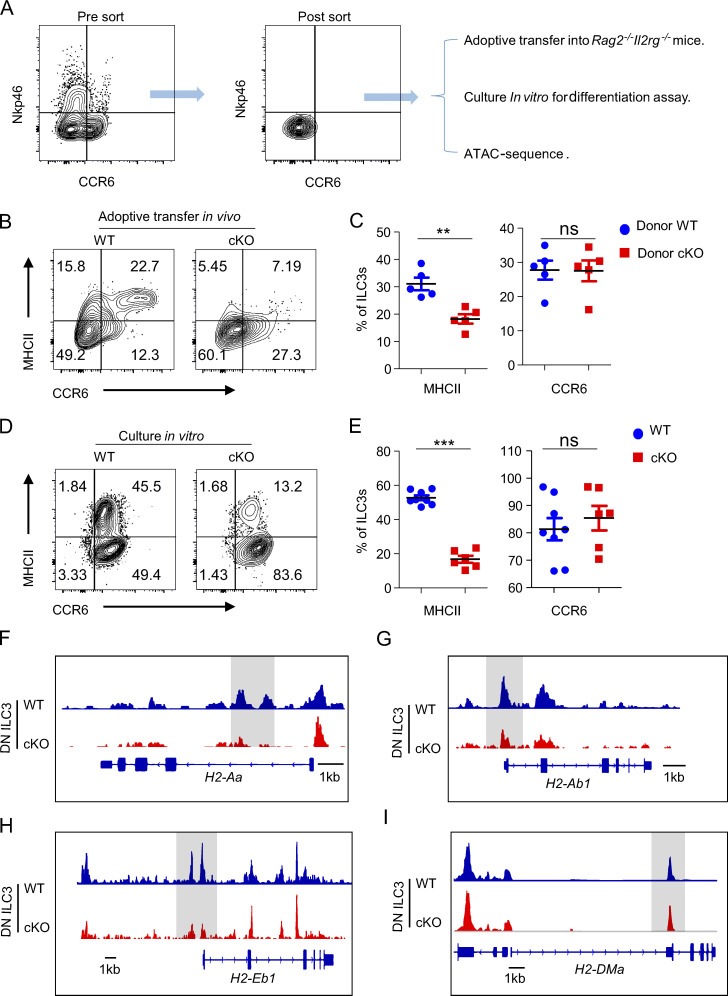
BATF directly promotes the development of DN ILC3 precursors into MHCII+ ILC3s
Several studies have demonstrated that DN ILC3s represent precursors amongst intestinal ILC3s (Rankin et al., 2013; Klose et al., 2013; Vonarbourg et al., 2010). To further elucidate whether this process requires BATF, we FACS-sorted DN ILC3s from WT and cKO mice and transferred them into immune-deficient Rag2−/−Il2rg−/− mice (Fig. 9 A). At 5 wk after transfer, we assessed their fate in the intestine. Intriguingly, we observed that both WT and BATF-deficient donor DN ILC3s had a comparable capacity of differentiating into CCR6+ ILC3s. However, the formation of donor-derived MHCII+ ILC3s in the host that received BATF-deficient DN ILC3s was significantly reduced, indicating that BATF selectively promotes the expression of MHCII, but not CCR6, during the differentiation of DN ILC3 precursors in the intestine (Fig. 9, B and C). As the combination of IL-6 and TGF-β can induce BATF expression in ILC3s, we next assessed whether this cytokine milieu can facilitate the differentiation of DN ILC3s. To this end, we cultured WT and BATF-deficient DN ILC3 in the presence of IL-6 and TGF-β for 4–5 d in vitro. Strikingly, both WT and BATF-deficient DN ILC3 showed strong and comparable abilities to develop into CCR6+ ILC3. However, similar to our observations in vivo, the formation of MHCII+ ILC3s was significantly impaired in the absence of BATF (Fig. 9, D and E). We further assayed whether these phenotypic changes could be explained by changes in chromatin accessibility caused by ablation of BATF in ILC3s. Thus, we FACS-sorted intestinal DN ILC3 from WT and cKO mice and performed ATAC-seq. Indeed, ATAC-seq analysis revealed that BATF-deficient DN ILC3s had a pronounced loss of open chromatin in close proximity to the H2-Aa, H2-Ab1, and H2-Eb1 loci (Fig. 9, F and H), while the gene loci of H2-DMa showed no apparent changes (Fig. 9 I). A similar trend was observed in ATAC-seq analysis of total Rag1−/−Batf−/− ILC3s versus Rag1−/− ILC3s (Fig. 8 F). These data further demonstrate that BATF directly controls the chromatin accessibility of major MHCII machinery genes, which in turn facilitates the transition of DN ILC3s into MHCII+ ILC3s.
Figure 9.
BATF promotes the conversation of the precursors DN ILC3 to MHCII+ ILC3. (A) Sorting strategy for DN ILC3s and downstream of the experimental setup. (B) Representative flow plots of intestinal ILC3s (Lin−CD90.2+RORγt+) of host Rag2−/−Il2rg−/− mice received donor WT or BATF-deficient DN ILC3 for 5 wk. (C) Quantification of frequency of MHCII + ILC3s and CCR6+ of plot in B. (D) Representative flow plots of ILC3s (Lin−CD90.2+RORγt+), which were differentiated from WT or BATF-deficient DN ILC3 cultured in presence of IL-6 plus with TGF-β for 4–5 d. (E) Quantification of the frequency of MHCII + ILC3s and CCR6+ of plot in D. (F–I) Genomic snapshots showing chromatin accessibility of the gene loci of H2-Aa (F), H2-Ab1 (G), H2-Eb1 (H), and H2-DMa (I) in WT and BATF-deficient DN ILC3. Data are shown as mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001 (two-tailed unpaired t test). Each dot represents one mouse in C, n = 5 mice per group, or one biological replicate in E. Data are pooled from two independent experiments.
Discussion
In the present study, we demonstrate the pivotal role of BATF in regulating SI ILC3 homeostasis and plasticity. Through scRNA-seq, we unveiled significant and distinct heterogeneity in ILC3 subsets in WT and BATF-deficient cKO mice. Our data show that BATF ablation causes proportionally higher levels of ILC1-like ILC3s (Ncr1+ ILC3s and ex-ILC3s), but lower proportions of MHCII+ ILC3s in the gut. Mechanistically, BATF directly binds to the loci of key genes in ILC3s to maintain or inhibit chromatin accessibility. By orchestrating the epigenomic landscape and ILC3 lineage stability, BATF acts as a rheostat in ILC3s to promote microbial homeostasis and limit T cell–mediated mucosal inflammation in the gut.
ILC3s have been suggested to drive the pathogenesis of colitis through the production of the inflammatory cytokines, IFN-γ and TNF-α (Vonarbourg et al., 2010; Verrier et al., 2016). ILC3 production of IFN-γ and TNF-α is mainly driven by the TF T-bet (Klose et al., 2013). Moreover, IFN-γ production by T-bet+ ILC3s has been reported to be positively correlated with inflammatory progress in the ileum and colon of patients with CD and in mouse models of bacteria-driven colitis (Buonocore et al., 2010; Geremia et al., 2011). In the context of intestinal inflammation, the potential of ILC3s to acquire an inflammatory phenotype is associated with increased T-bet and decreased RORγt expression (Klose et al., 2013). These inflammatory ILC3s have been called “ex-ILC3s”; they are characterized by high IFN-γ expression and drive colitis pathology in CD patients and mice (Bernink et al., 2013). Similarly, we identified increased ex-ILC3 levels in the SI of BATF-deficient cKO mice compared to their counterpart WT mice. However, lack of BATF led to a reduction of T-bet+ ILC3s in recipient mice that are reconstituted with competitive bone marrow cells from WT and BATF−/− mice, as well as a reduction in T-bet+ ILC3s in some certain tissues of paired WT and BATF−/− mice in parabiosis experiments (Liu et al., 2020b). However, when a Rag1−/−BATF−/− model was used, no significant reduction of T-bet+ ILC3s was found in various tissues of Rag1−/−BATF−/− mice as compared with Rag1−/− mice (Liu et al., 2020b). Differences between our data and data previously described in the literature are therefore likely due to different mouse models or experimental approaches used. Moreover, we found that this ex-ILC3 subset highly expresses Ifng, Tnf, Csf2, and Il12a as well as the cytotoxic markers Gzmb and Prf1. GM-CSF (encoded by Csf2) is secreted by both human and murine ILC3s, and it can recruit myeloid cells and further promote intestinal inflammation (Pearson et al., 2016); our data corroborate this, as we found that cKO mice had increased myeloid cell infiltration in the gut compared with WT controls. Furthermore, IFN-γ has been reported to reinforce the pro-inflammatory milieu by orchestrating the replacement of resident mononuclear phagocytes (MNPs) by circulating monocytes (Mortha and Burrows, 2018). Moreover, IFN-γ signaling in Paneth cells in the intestinal crypts has been coupled with cell death (Eriguchi et al., 2018), which probably promotes the permeability of mucosal barriers. Indeed, prolonged IFN-γ production can harm the host, resulting in severe gut inflammation (Lazarevic and Glimcher, 2011; Buonocore et al., 2010). In line with this, the IFN-γ blockade can significantly delay and ameliorate the symptom of colitis spontaneously developed in BATF deficient cKO mice. Given that the elevated expression of IFN-γ could also be derived from pathogenic T cells, we cannot fully attribute the promotion of colitis to IFN-γ from BATF-deficient ILC3s sources, even though we proposed the ILC3-derived IFN-γ in cKO mice could mainly act at an early stage of the disease. In another aspect, the pro-inflammatory cytokines IL-1β, IL-12, and IL-15 are implicated in driving the conversion of ILC3s to ex-ILC3s or even cytotoxic ILC3s (NK-like ILC3s; Colonna, 2018). Our findings suggest that the elevated expression of cytokine receptors in BATF-deficient ILC3s (e.g., Il1r, Il12rb, and Il18rap) may facilitate this conversion, as targeting cytokine receptors in the inflamed intestine of IBD patients appears to have therapeutic promise in clinical trials (Penny et al., 2018). Therefore, we propose a model in which BATF restricts the type 1 program of intestinal ILC3s and thereby serves as a critical gatekeeper for mucosal barrier integrity.
Intestinal ILC3-derived IL-22 also plays a critical role in maintaining mucosal barrier integrity (Longman et al., 2014). Since one of the limitations of the genetic model used in this study is that it could not determine the exact role of IL-22 during colitis development in cKO mice, we restrained our study on the role of excessive IFN-γ–mediated barrier integrity loss. Such loss of barrier integrity can account for the location-specific bacterial influx, which correlates with the production of pro-inflammatory cytokines and overactivated MNPs (Zareie et al., 2001). MNPs integrate microbial signals to activate ILC3s via the production of IL-23 and IL-1β and also present antigen and induce adaptive effector immunity (Longman et al., 2014; Panea et al., 2015). A range of inflammatory circuits further exacerbates intestinal inflammation and dysbiosis. The dysbiosis we observed in BATF-deficient cKO mice was characterized by the reduced abundance of beneficial commensal bacteria, such as Coriobacteriaceae, and the increased abundance of potential pathogens including Mycoplasmatales and Enterococcaceae. Significantly, the analysis of fecal samples collected from co-housed BATF-deficient cKO and their littermate WT mice revealed that distinct microbiota formed even at early weaning timepoints and were maintained even following co-housing for several weeks. Given that ILC3s can pre-empt colonization of the intestine by symbiotic microbiota (van De Pavert et al., 2014), it is reasonable to propose that BATF plays a critical function in ILC3s for shaping the landscape of the gut microbiome.
A previous study has reported that only a small proportion of the intestinal LTi cells fate map for PLZF and their development were independent of the bone marrow PLZFhi ILCPs (Constantinides et al., 2014). Therefore, it is plausible to speculate that the LTi cells might not be affected by using PLZF-cre mediated deletion. Alternatively, we and the others found that the highly efficient deletion indeed occurs in LTi cells by using PLZF-cre system (Zindl et al., 2022). To avoid any confusion that may have been raised by these observations, we further identified that the MHCII+CCR6+ ILC3 can be directly differentiated from intestinal precursors DN ILC3s, but not limited to LTi progenitors in bone marrow as proposed previously by Constantinides et al. (2014). Moreover, we found that Batf deficiency specifically impacts the expression of MHCII, but not CCR6 on ILC3s. All these could properly explain the seemingly contradictory findings between us and others. ILC3s have been also reported to play a critical role in shaping adaptive immunity via MHCII expression (Hepworth et al., 2013). However, the regulation of CD4+ T cells responses by MHCII-expressing ILC3s is context dependent and varies based on factors ranging from the microenvironment to cell activation status (Lehmann et al., 2020; Von Burg et al., 2014). Here, we show that Batf-deficient ILC3s significantly downregulated genes involved in MHCII antigen processing and presentation, such as H2-Aa, H2-Ab1, H2-DMa, H2-DMb2, H2-Eb1, Cd74, and Ciita. Importantly, Batf-deficient cKO mice spontaneously developed rectal prolapse similar to aged mice with ILC-intrinsic deletion of MHCII, as reported previously (Hepworth et al., 2013). Moreover, both MHCII- and BATF-deficient cKO mice exhibit significantly increased frequencies of activated CD44hiCD62Llow and proliferating Ki-67+ CD4+ T cells, as well as significantly increased numbers of pathogenic IFN-γ+ IL-17A+ CD4+ T cells in the gut. This indicates that BATF-deficient ILC3s enhances the activation of intestinal CD4+ T cells, possibly due to the lower expression of MHCII. Strikingly, aberrant CD4+ T cell activation in BATF-deficient cKO mice was abrogated upon oral administration of antibiotics. Additionally, the lack of conventional co-stimulatory molecule expression (e.g., Cd40, Cd80, and Cd86) on BATF-deficient ILC3s (data not shown) recapitulated the phenotype observed in MHCII-deficient cKO mice and further supported the notion that restriction of commensal bacteria-responsive CD4+ T cells by MHCII-expressing ILC3s at least partially requires BATF. In addition to MHCII, ILC3s can regulate CD4+ T cell activation via OX40L (Castellanos et al., 2018). Moreover, ILC3s broadly cross-talk with various lymphocytes in the context-dependent manner. Human splenic ILC3s can regulate marginal zone B cell response (Magri et al., 2014). In draining lymph nodes, ILC3s limit Tfh and B cells’ interaction through antigen presentation (Melo-Gonzalez et al., 2019). Of note, ILC3s are mainly tissue-resident cells and are more prone to imprinting by local cues such as cytokines, growth factors, and metabolites (Vivier et al., 2018). Our studies found that BATF regulates MHCII expression on SI ILC3s, but whether this is sufficient to promote MHCII expression and/or independent of tissue-specific niches remains to be investigated. It is well known that Ciita is a major transcriptional coactivator controlling MHCII expression (Masternak et al., 2000); we found that Ciita expression was downregulated in BATF-deficient ILC3s, suggesting that the regulation of MHCII expression by BATF may be indirect since BATF was not bound to the loci of certain MHCII genes such as H2-Aa and H2-Ab in our CUT&Tag assays.
Based on comprehensive transcriptomic and epigenomic data, we conclude that BATF serves two distinct and pivotal roles in SI ILC3s. BATF not only upregulates core genes involved in antigen processing and presentation via MHCII pathways but also inhibits the expression of Th1-associated genes. In agreement with our data, a dual function of BATF has also been implicated in CD8+ T cells and Th17 cells (Ciofani et al., 2012; Kurachi et al., 2014). Previous studies have shown that dynamic changes in the chromatin landscape were involved in the conversion of ILC3s into ILC1s (ex-ILC3s; Parker et al., 2020). Our data underscore an epigenetic role of BATF in this process. First, BATF directly binds and inhibits the Tbx21 locus, which encodes the Th1 lineage-defining TF T-bet, to prevent ILC3s from acquiring type 1 features. Notably, a similar mechanism is known to be employed by another AP-1 family TF, c-Maf, in ILC3s (Parker et al., 2020; Tizian et al., 2020). Therefore, it will be intriguing to further characterize how BATF interacts with c-Maf to regulate ILC3-ILC1 conversion, as BATF is known to cooperate with c-Maf in controlling lineage programs in Tfh cells (Sahoo et al., 2015). Conversely, in effector CD8+ T cells, BATF not only binds to the Tbx21 locus to upregulate T-bet expression but also suppresses IFN-γ expression (Kurachi et al., 2014), implying that the role of BATF in epigenetic programming has distinct, cell type-restricted functions. Additionally, similar to motif enrichment in c-Maf-deficient ILC3s (Parker et al., 2020), Runx and T-bet motifs were enriched within regions that gain accessibility in the absence of BATF, further suggesting that BATF restrains Runx and T-bet from driving Th1-like programs in ILC3s. In support of this, Runx3 collaborates with T-bet to regulate Th1 genes and promote Th17 cell conversion to an IFN-γ+ pathogenic fate (Wang et al., 2014). Finally, BATF is required to globally restrain the chromatin accessibility of Il1r, Il18rap, Il12rb1, and Il12rb2, all of which encode inflammatory cytokine receptors that are critical for the full transition of ILC3s to ILC1s (Bernink et al., 2015; Cella et al., 2010; Vonarbourg et al., 2010). Thus, it is tempting to speculate that BATF globally constrains lineage-defined TF (such as Tbet) and effectors (such as IFN-γ) to prevent ILC3 commitment to a pro-inflammatory fate.
In summary, we show that BATF acts as a rheostat in regulating the functional plasticity of SI ILC3s. BATF controls ILC3 identity by inhibiting cytotoxic type 1 immunity and maintaining an antigen-presentation program, thus governing gut homeostasis by shaping the microbiome and T cell-mediated mucosal immunity. In this regard, our study provides new insight into BATF-dependent programs in ILC3s and has profound therapeutic implications for targeting the ILC3–microbiome–T cell axis in IBD immunotherapy.
Materials and methods
Mice
Batffl/fl mice were kindly provided by E.J. Taparowsky (Betz et al., 2010). PLZF-Cre recombinase mice (stock #:024529) were purchased from the Jackson Laboratory and crossed to one another to generate age-matched Batffl/flPLZF-Cre− (WT) and Batffl/flPLZF-Cre+ (Batf innate lymphoid cell-conditional knockout, cKO) mice. Batf knockout mice (stock #: 013758) and Rag1 knockout mice (stock #:129S7) were purchased from Jackson Laboratory. Rag1−/−Batf−/− mice were generated by crossing Rag1−/− with Batf−/− mice. R26-CreERT2 mice (stock #: 008463) and RosamT/mG reporter mice (stock #: 007576) were purchased from the Jackson laboratory. Batffl/fl was crossed with ERT2CreRosamTmG to generate Batffl/fl ERT2CreRosamTmG mice. Mice were bred and maintained in a closed breeding facility, and mouse handling conformed to the requirements of the Institutional Animal Care and Use Guidelines of the Medical College of Wisconsin.
Tissue process and cells isolation
Myeloid cells and lymphocytes (include T cells and innate lymphoid cells) were isolated from tissues including mLN, SI, and LI. For the mLN, the tissues were grounded and homogenized in a 70-μm cell strainer. After spinning down, red blood cells were removed by the addition of ammonium-chloride-potassium buffer (Lonza) for 10 min on ice. Subsequently, the single-cell suspension was washed with Dulbecco's PBS containing 0.5% bovine serum albumin (Thermo Fisher Scientific) and 2 mM EDTA (Lonza). For the gut, intestines were cut open longitudinally, the tissues were fit, and Peyer’s patches were removed. Intestines were then washed with cold PBS and cut into pieces. Epithelial layers were removed by shaking incubation in 5 mM EDTA (Lonza) Ca2+ and Mg2+ free 1640 medium (Life Technologies) for 30 min each at 37°C. Then, the intestines were cut into fine pieces and digested twice for 40 min each at 37°C with Collagenase II and III (1 mg/ml; Worthington), and DNase I (200 μg/ml; Roche). Cells were isolated with 30–60% Percoll gradient and washed twice with cold Dulbecco's PBS.
ILC3s cells sorting and in vitro culture
For innate lymphoid cells sorting, the ILCs were isolated from the gut of indicted mice, then stained with lineage markers including biotin-labeled antibodies, including CD3, CD8a, TCRβ, TCRγ/δ, CD11b, CD11c, B220, Gr1, NK1.1, and TER119. After incubation for 15 min on ice, the cells were washed with cold PBS two times. Lineage-negative cells were then purified through Streptavidin-RapidSpheres 50001 (STEMCELL) through immunomagnetic separation. The purified ILCs were stained for 15 min on ice with antibodies against CD90.2 and KLRG1, live/dead cell staining, and streptavidin. Stained cells were washed and sorted using a FACSAria III cytometer (BD Bioscienes). ILC3s were sorted from the SI and cultured in vitro with RPMI-1640 medium (Invitrogen) with 10% heat-inactivated FBS (HyClone Fisher), 100 U/ml penicillin, and 0.1 mg/ml streptomycin (Thermo Fisher Scientific), 50 μM 2-mercaptoethanol (Sigma-Aldrich), 10 ng/ml of IL-2 (Peprotech), 10 ng/ml of IL-7 (Peprotech), and 10 ng/ml SCF (stem cell factor; Peprotech) to expand the cells for 15–20 d. For the short-term intracellular staining experiment, cells were cultured in IL-7, SCF, and IL-2 (10 ng/ml, Peprotech) for 2–3 d of expansion before stimulation with PMA plus ionomycin and intracellular staining. For cytokines stimulation, intestinal Rag1−/− ILC3s were sorted and cultured in the presence of 10 ng/ml of IL-2 (Peprotech), 10 ng/ml of IL-7 (Peprotech), and 10 ng/ml SCF with or without additional cytokines as indicated for 48 h. The following concentrations of TGF-β (5 ng/ml), IL-6 (20 ng/ml) and IL-12 (10 ng/ml) were used.
Flow cytometry analysis
Filtered single-cell suspensions were isolated from the above-mentioned tissues or ILC3s from in vitro cultured system and then stained with fluorophore-conjugated antibodies against cell surface antigens for 30 min at 4°C. For intracellular staining, cells were incubated with 50 ng/ml phorbol 12-myristate 13-acetate (Sigma-Aldrich) and 1 μM ionomycin (Sigma-Aldrich) for 30 min at 37°C. Then brefeldin A (2 mg/ml, Biolegend) was added and cultured for 4.5 h. The BD Fixation/Permeabilization Solution Kit (BD Biosciences) was used for fixation and staining, and the Foxp3 staining buffer kit (eBioscience) was used for nuclear TF staining. The dead cells were stained by Live and Dead violet viability kit (Invitrogen). The cells were stained with the following antibodies: BATF (D7C5), T-bet (4B10), GATA3 (16E10A23), RORγt (B2D), CD90.2 (30-H12), CD11b (ICRF44), CD4 (GK1.5), CD8α (53-6.7), IFN-γ (XMG1.2), IL-17A (BL168), TNFα (MAb11), CD45.2 (104), CD127 (A7R34), Ly6C (HK1.4), CD11c (N418), Siglec-F (S17007L), F4/80 (BM8), Ly6G (1A8), Ki-67 (16A8), KLRG1 (2F1/KLRG1), NK1.1 (PK136), B220 (RA3-6B2), TER-119 (TER-119), TCRβ (H57-597), TCRγδ (GL3), streptavidin (Z2), CCR6 (QA17A37), Nkp46 (29A1.4), MHCII (M5/114.15.2), CD44 (C44Mab-5), CD62L (MEL-14), and isotype control antibody (RTK2758). All flow cytometry data were acquired on an LSRII (BD Biosciences) and analyzed by FlowJo (Treestar).
scRNA-seq and analysis
ILCs separately FACS-sorted from the SI of WT and BATF cKO mice (four to five mice per genotype) were loaded on the Chromium Controller (10x Genomics). scRNA-seq libraries were prepared using the Chromium Single Cell 3′ v2 Reagent Kit (10x Genomics) according to the manufacturer’s protocol. Libraries were loaded onto an Illumina NextSeq with the NextSeq 500/550 High Output Kit v2 (150 cycles; FC-404-2002; Illumina) with the following conditions: 26 cycles for read 1, 98 cycles for read 2, and 8 cycles for i7 index. Basemount (Illumina) was used to download raw sequencing data. Cell Ranger (10x Genomics) functions “mkfastq” and “count” were used to demultiplex the sequencing data and generate gene-barcode matrices, respectively. Data were imported into R (version 3.6.1) using the Seurat package (version 3.1.1; Satija et al., 2015), and integrated Seurat analysis was used to combine the samples. UMAP was used to visualize the cells, which formed nine separate clusters based on the Louvain algorithm. We removed contaminating cells based on the expression of Cd3e and Cd74 and kept ILCs for further analysis. Reclustering the cells provided seven separate clusters consisting of ILC1s, ILC2s, and ILC3s. ILC3s were isolated for further analysis based on the expression of Rorc (encodes RORγt) and formed three separate clusters after reclustering. GSEA for scRNA-seq data was performed using the clusterProfiler package in R.
IFN-γ blockade
For in vivo blockade of IFN-γ function, weaned and co-housed cKO mice were injected with anti–IFN-γ or isotype-IgG control antibody at 250 μg/mouse intraperitoneally two times per week, followed by monitoring of rectal prolapse development.
qPCR
Fecal samples of mice were collected after antibody blockade at indicated times, and bacterial DNA was extracted using ZymoBIOMICS DNA Miniprep Kit (ZYMO research) as per the kit instructions. A qPCR analysis of equal amounts of DNA samples using SYBR Green was run on an ABI 7500 Fast Real-Time PCR System (Applied Biosystems). Samples were normalized to isotype-IgG control at 8 wk of age. The primer set for detection of bacterial 16S rRNA was used as previously reported (Liang et al., 2020).
DN ILC3 differentiation assay
For adoptive transfer of DN ILC3s, 200,000 intestinal DN ILC3 were sorted from WT and cKO mice and adoptively transferred into Rag2−/−Il2rg−/− mice. Host mice were sacrificed for analysis 5 wk after transfer. For in vitro culture assays, sorted DN ILC3 were cultured with 10 ng/ml IL-2, 10 ng/ml IL-7, 10 ng/ml SCF, 5 ng/ml TGF-β, and 20 ng/ml IL-6 for 4–5 d before flow cytometry analysis.
Bulk RNA-seq and analysis
A total of 2000 ILC3s (Lin−CD90.2+KLRG1−) from the SI of WT and littermate cKO mice directly sorted into lysis buffer containing Protector RNase Inhibitor (Roche). Three replicates from each group were subsequently sequenced. About 2 ng of purified RNA was used as the input for a modified SMART-Seq2 protocol entailing RNA secondary structure denaturation at 72°C for 3 min, reverse transcription with Maxima Reverse Transcriptase (Life Technologies), and KAPA HiFi HotStart ReadyMix 2X (Kapa Biosystems) was used for whole transcriptome amplification (WTA) for 12 cycles. After purification with Ampure XP beads (Beckman Coulter), the WTA products were quantified with a High Sensitivity DNA Chip run on a Bioanalyzer 2100 system (Agilent). A total of 0.2 ng of purified WTA product was used as input for the Nextera XT DNA Library Preparation Kit (Illumina). Uniquely barcoded libraries were pooled and sequenced with a NextSeq 500 sequencer using a high output V2 75 cycle kit (Illumina) and 2x38 paired-end reads. The Nextflow RNA-seq pipeline (nf-core/rnaseq 1.4.2) was used to conduct quality trimming of bulk RNA-seq reads, align to the Mus musculus mm10 genome, and calculate quality control statistics (Ewels et al., 2020). Using gene expression values quantified by Salmon (Patro et al., 2017), RNA-seq libraries were normalized and Wald tests were used within DESeq2 v1.24.0 (Love et al., 2014) to test for differential expression between BATF Ilc3 knock-out and WT samples. Shrunken fold change values were used for pre-ranked gene set enrichment analyses as facilitated by clusterProfiler v3.12.0 (Yu et al., 2012). Reactome (Fabregat et al., 2018), KEGG (Kanehisa and Goto, 2000), and GO (Ashburner et al., 2000) databases were used to provide input gene sets for the gene set enrichment analyses. All P values were adjusted for multiple testing using the Benjamini-Hochberg method. Expression values were transformed using the regularized logarithm transformation for display within figures (Love et al., 2014).
ATAC-seq and analysis
ATAC-seq was performed according to a published protocol with minor modification (Buenrostro et al., 2015). 50–80,000 ILC3 cells were sorted from the SI of Rag1−/− and their littermate Rag1−/−Batf−/− mice. The sorted ILC3s were spin down and washed with 50 μl cold PBS, followed by treatment with 50 μl lysis buffer (10 mM Tris-HCl [pH 7.4], 10 mM NaCl, 3 mM MgCl2, 0.1% IGEPAL CA-630). After centrifuging at 500 g for 10 min, the pellets were resuspended in a 40 μl transposition reaction with 2 μl Tn5 transposase (FC-121-1,030; Illumina) to tag and fragmentize accessible chromatin. Then they were incubated at 37°C for 30 min. The tagmented DNAs were then purified with a Qiagen MinElute kit and amplified with 12 cycles of PCR based on the amplification curve. Once the libraries were purified and quantified, the quantified libraries were sequenced using Nextseq 500/550 kit v2 with 75 cycles and 400 million reads with paired-end sequencing. Reads were aligned to M. musculus mm10 genome using Bowtie2 (Langmead and Salzberg, 2012). The reads were then filtered using SAMtools (-F 1796; Li et al., 2009). Picard was used to mark duplicates and thus, removed. Additionally, reads those were aligned to the mitochondrial genome were removed. Peaks were then called using MACS2 (--no model --shift -100 --ext size 200 -q 0.01; version 2.1.1). Normalization was performed on data from different subsets, followed by the identification of subset-specific peaks using MAnorm (version 0.1.0). HOMER (-h to calculate enrichment using cumulative hypergeometric distribution; version 4.10.4) was used to perform the motif analysis. To perform gene ontology analysis of subset-specific enhancers, GREAT (version 4.0.4) was used.
CUT&Tag ChIP-seq and analysis
ILC3s were sorted from the SI of Rag1−/− mice and cultured for 2 wk to expand the cells. 100,000 ILC3s were processed and library construction according to CUT&Tag ChIP-seq approach mentioned (version 2; Kaya-Okur et al., 2019; step-wise protocol can be found at https://www.protocols.io/view/bench-top-cut-amp-tag-z6hf9b6). The rabbit IgG1 from normal serum was used as a control. The sequence was performed on a Nextseq 550 with 37 cycles of paired-end sequencing, and about 5–10 million reads were generated for each sample. For data analysis, each dataset was first downsampled to the same read depth. For standardization between experiments, Escherichia coli DNA derived from transposase protein production was used to normalize sample read counts based on the recommendation of the CUT&Tag protocol. Reads were aligned to mouse reference genome mm10 and E. coli (strain K12) using Bowtie2 (version 2.2.5) with options: --local --very- sensitive-local --no-unal --no-mixed --no-discordant --phred33 -I 10 -X 700. Peaks were called and annotated by using SEACR (version1.3) with options: 0.01 non-stringent and HOMER (version 4.9.1), respectively. Visualizations were created using deep Tools (version 3.3.0) and IGV (version 2.8.2). k-means clustering of BATF ChIP-seq data was performed using seqMINER (version 1.3.3).
16S rRNA-sequence and analysis
Feces samples of mice were collected and stored at −20°C immediately. Bacterial DNA was extracted using ZymoBIOMICS DNA Miniprep Kit (ZYMO research). Briefly, samples were suspended with lysis solution and secured in a bead beater fitted with a 2 ml tube holder assembly. Samples were lysed and homogenized at maximum speed for more than 5 min. After centrifugation and washing, the filtered DNA was resolute with DNase/Rnase-free water and frozen at −20°C or directly processed to the next step. 16S rRNA-sequence library construction and preparation using Quikc-16S NGS library prep. kit (catalog no. D6410; ZYMO research). In short, The V3/V4 regions of the 16S ribosomal RNA gene were amplified and proceeded to reaction clean-up. After barcode addition, the library was quantified using fluorescence real-time PCR system. The pooled library was suspended with select-a-size magbead buffer and cleaned up with a magnetic rack. Samples were sequenced on an Illumina MiSeq platform using version 3 of MiSeq Reagent Kit (600-cycle, MS-102-3003) with 300 cycles from read1 and read 2. Fastq files were generated by bcl2fastq2 (version 2.20) and imported into QIIME 2 (version 2020.6) for further analysis. First of all, 16S primers and low-quality sequencing bases (QC score below 25) were removed from both forward and reverse reads. Then, DADA2 was used for quality filtering of the reads. To generate a phylogenetic tree, we have used “align-to-tree-mafft-fasttree” pipeline from the “q2-phylogeny” plugin. For α and β diversity analysis, a metrics was generated using the sampling depth of 162,618. PCoA plots were generated using Jaccard distance. Shannon’s diversity index was calculated to test the community richness. Pielou’s evenness was calculated to test the community evenness. Taxonomic analysis was performed by the “q2-feature-classifier” plugin and used a pre-trained Naive Bayes classifier on the SILVA-132-99 16S rRNA database (Quast et al., 2013) to obtain taxonomical placement of the operational taxonomic units (99% similarity). Family level taxonomy bar plot was generated using qiime2R (version 0.99.35) in R. To determine differences in microbial communities between microbiota settings, the LEfSe method was used as previously described (Segata et al., 2011). The α value for the factorial Kruskal–Wallis test among samples is 0.05, and the threshold on the logarithmic LDA score for discriminative features is 1.
Histology
Colons and ileum were excised and washed with PBS before fixation in 4% buffered formaldehyde and then embedded in paraffin. Tissue sections were stained with H&E. Histological scores were determined by a grading system that evaluates inflammatory cell infiltration and tissue damage as described previously (Onizawa et al., 2009). Briefly, the inflammatory cell infiltration scored as follows, occasional inflammatory cells present in the lamina propria was scored as 0, increased numbers of inflammatory cells in the lamina propria was scored as 1, and confluence of inflammatory cells extending into the submucosa was scored as 2, and transmural extension of the infiltrate was scored as 3. Tissue damage was scored as follows, no mucosal damage was scored as 0, discrete lymphoepithelial lesions were scored as 1, surface mucosal erosion or focal ulceration was scored as 2, and extensive mucosal damage and extension into deeper structures of the bowel wall were scored as 3. The combined histological score of inflammatory cell infiltration (score 0–3) and tissue damage (score 0–3) ranged from 0 to 6.
Antibiotic treatment
Co-housed littermates of WT and cKO mice (3–4 wk old) were treated with autoclaved water supplemented with or without antibiotics (1 g/liter ampicillin, 1 g/liter gentamicin, 1 g/liter neomycin, 1 g/liter metronidazole, and 0.5 g/liter vancomycin). After 5–6 wk, mice were sacrificed and tissues were collected for flow cytometry analysis or histological examination.
TAM treatment
For depletion of recombinations in creER transgenic mice, TAM (Sigma-Aldrich) was dissolved at 10 mg/ml in warm corn oil (Sigma-Aldrich) and administered to mice intraperitoneally. Mice were injected via intraperitoneal (i.p.) every other day three times. 10 d after the last injection, mice were sacrificed and the tissues were processed for flow cytometry detection.
Statistical analyses
Statistical tests were performed using Graphpad Prism software. To compare two groups of samples, P values were calculated using either two-tailed paired or unpaired Student’s t-tests and P < 0.05 were considered statistically significant. To compare multiple groups of samples, two-way multiple-range ANOVA test was used and P < 0.05 (confidence interval of 95%) was considered statistically significant. Figure legends specified the tests used, statistical significance, and the numbers of experimental and biological replicates.
Online supplemental material
The supplemental figures mainly include the spontaneous colitis phenotype of BATF-deficient cKO mice, the flow cytometry, and scRNA-seq analysis of the heterogeneity of intestinal ILC3s in WT and cKO mice. Moreover, The ATAC-seq and CUT&Tag ChIP-seq analyze the function of BATF in ILC3s and the cytokines that regulate the expression of BATF in ILC3s. Fig. S1 shows how BATF deficiency in ILCs causes spontaneous colitis. Fig. S2 shows how BATF deficiency in ILCs alters the heterogenicity of SI ILCs. Fig. S3 shows how BATF regulates SI ILC3s transcriptional program. Fig. S4 shows how BATF deficiency alters the heterogeneity of SI ILCs. Fig. S5 shows that an expression of BATF is regulated by cytokines that maintain the chromatin landscape in ILC3s.
Acknowledgments
The authors thank Dr. S. Henikoff (Fred Hutchinson Cancer Center, Seattle, WA) for providing 3XFlag-pA-Tn5-Fl plasmid and Dr. Nan Zhu (Versiti Blood Research Institute, Milwaukee, WI) for providing Protein A–Tn5 fusion protein. This research was completed in part with computational resources and technical support provided by the Research Computing Center at the Medical College of Wisconsin.
This work is supported by National Institutes of Health grants AI125741 (W. Cui), AI148403 (W. Cui), and DK127526 (M.Y. Kasmani); by an American Cancer Society research scholar grant (W. Cui); and by an Advancing a Healthier Wisconsin Endowment grant (W. Cui). M.Y. Kasmani is a member of the Medical Scientist Training Program at the Medical College of Wisconsin, which is partially supported by a training grant from the National Institute of General Medical Sciences (T32-GM080202).
Author contributions: X. Wu and W. Cui conceptualized and designed the study. X. Wu performed most of the experiments, analyzed the data, and wrote the manuscript A. Khatun, M.Y. Kasmani, Y. Chen, S. Zheng, S. Atkinson, and R. Burns performed and interpreted bioinformatics analyses. A. Khatun, Y. Chen, and C. Nguyen provided assistance for experiments. E.J. Taparowsky provided BATFflox/flox mice. X. Wu, A. Khatun, M.Y. Kasmani, Y. Chen, N.H. Salzman, T.W. Hand, and W. Cui revised and edited the manuscript. W. Cui supervised the study.
Data availability
The scRNA-seq, bulk RNA-seq, ATAC-seq, and CUT&Tag ChIP-seq data are deposited in the GEO database with the accession code GSE168080. All other raw data are available from the corresponding author upon reasonable request.
References
- Artis, D., and Spits H.. 2015. The biology of innate lymphoid cells. Nature. 517:293–301. 10.1038/nature14189 [DOI] [PubMed] [Google Scholar]
- Ashburner, M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., et al. 2000. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 25:25–29. 10.1038/75556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bando, J.K., Liang H.E., and Locksley R.M.. 2015. Identification and distribution of developing innate lymphoid cells in the fetal mouse intestine. Nat. Immunol. 16:153–160. 10.1038/ni.3057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernink, J.H., Krabbendam L., Germar K., De Jong E., Gronke K., Kofoed-Nielsen M., Munneke J.M., Hazenberg M.D., Villaudy J., Buskens C.J., et al. 2015. Interleukin-12 and -23 control plasticity of CD127(+) group 1 and group 3 innate lymphoid cells in the intestinal lamina propria. Immunity. 43:146–160. 10.1016/j.immuni.2015.06.019 [DOI] [PubMed] [Google Scholar]
- Bernink, J.H., Peters C.P., Munneke M., Te Velde A.A., Meijer S.L., Weijer K., Hreggvidsdottir H.S., Heinsbroek S.E., Legrand N., Buskens C.J., et al. 2013. Human type 1 innate lymphoid cells accumulate in inflamed mucosal tissues. Nat. Immunol. 14:221–229. 10.1038/ni.2534 [DOI] [PubMed] [Google Scholar]
- Betz, B.C., Jordan-Williams K.L., Wang C., Kang S.G., Liao J., Logan M.R., Kim C.H., and Taparowsky E.J.. 2010. Batf coordinates multiple aspects of B and T cell function required for normal antibody responses. J. Exp. Med. 207:933–942. 10.1084/jem.20091548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatt, B., Zeng P., Zhu H., Sivaprakasam S., Li S., Xiao H., Dong L., Shiao P., Kolhe R., Patel N., et al. 2018. Gpr109a limits microbiota-induced IL-23 production to constrain ILC3-mediated colonic inflammation. J. Immunol. 200:2905–2914. 10.4049/jimmunol.1701625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buenrostro, J.D., Wu B., Chang H.Y., and Greenleaf W.J.. 2015. ATAC-seq: A method for assaying chromatin accessibility genome-wide. Curr. Protoc. Mol. Biol. 109:21.29.1–21.29.9. 10.1002/0471142727.mb2129s109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buonocore, S., Ahern P.P., Uhlig H.H., Powrie F., Littman D.R., Littman D.R., and Maloy K.J.. 2010. Ivanov, IiInnate lymphoid cells drive interleukin-23-dependent innate intestinal pathology. Nature. 464:1371–1375. 10.1038/nature08949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellanos, J.G., Woo V., Viladomiu M., Putzel G., Lima S., Diehl G.E., Marderstein A.R., Gandara J., Perez A.R., Withers D.R., et al. 2018. Microbiota-induced TNF-like ligand 1A drives group 3 innate lymphoid cell-mediated barrier protection and intestinal T cell activation during colitis. Immunity. 49:1077–1089.e5. 10.1016/j.immuni.2018.10.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cella, M., Gamini R., Secca C., Collins P.L., Zhao S., Peng V., Robinette M.L., Schettini J., Zaitsev K., Gordon W., et al. 2019. Subsets of ILC3-ILC1-like cells generate a diversity spectrum of innate lymphoid cells in human mucosal tissues. Nat. Immunol. 20:980–991. 10.1038/s41590-019-0425-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cella, M., Otero K., and Colonna M.. 2010. Expansion of human NK-22 cells with IL-7, IL-2, and IL-1beta reveals intrinsic functional plasticity. Proc. Natl. Acad. Sci. USA. 107:10961–10966. 10.1073/pnas.1005641107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, W., Li D., Paulus B., Wilson I., and Chadwick V.S.. 2001. High prevalence of Mycoplasma pneumoniae in intestinal mucosal biopsies from patients with inflammatory bowel disease and controls. Dig. Dis. Sci. 46:2529–2535. 10.1023/a:1012352626117 [DOI] [PubMed] [Google Scholar]
- Chen, Y., Zander R.A., Wu X., Schauder D.M., Kasmani M.Y., Shen J., Zheng S., Burns R., Taparowsky E.J., and Cui W.. 2021. BATF regulates progenitor to cytolytic effector CD8(+) T cell transition during chronic viral infection. Nat. Immunol. 22:996–1007. 10.1038/s41590-021-00965-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciofani, M., Madar A., Galan C., Sellars M., Mace K., Pauli F., Agarwal A., Huang W., Parkhurst C.N., Muratet M., et al. 2012. A validated regulatory network for Th17 cell specification. Cell. 151:289–303. 10.1016/j.cell.2012.09.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colonna, M. 2018. Innate lymphoid cells: Diversity, plasticity, and unique functions in immunity. Immunity. 48:1104–1117. 10.1016/j.immuni.2018.05.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comito, D., and Romano C.. 2012. Dysbiosis in the pathogenesis of pediatric inflammatory bowel diseases. Int. J. Inflamm. 2012:687143. 10.1155/2012/687143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constantinides, M.G., Mcdonald B.D., Verhoef P.A., and Bendelac A.. 2014. A committed precursor to innate lymphoid cells. Nature. 508:397–401. 10.1038/nature13047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delacher, M., Imbusch C.D., Hotz-Wagenblatt A., Mallm J.P., Bauer K., Simon M., Riegel D., Rendeiro A.F., Bittner S., Sanderink L., et al. 2020. Precursors for nonlymphoid-tissue treg cells reside in secondary lymphoid organs and are programmed by the transcription factor BATF. Immunity. 52:295–312.e11. 10.1016/j.immuni.2019.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberl, G., Marmon S., Sunshine M.J., Rennert P.D., Choi Y., and Littman D.R.. 2004. An essential function for the nuclear receptor RORgamma(t) in the generation of fetal lymphoid tissue inducer cells. Nat. Immunol. 5:64–73. 10.1038/ni1022 [DOI] [PubMed] [Google Scholar]
- Eriguchi, Y., Nakamura K., Yokoi Y., Sugimoto R., Takahashi S., Hashimoto D., Teshima T., Ayabe T., Selsted M.E., and Ouellette A.J.. 2018. Essential role of IFN-gamma in T cell-associated intestinal inflammation. JCI Insight. 3:121886. 10.1172/jci.insight.121886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewels, P.A., Peltzer A., Fillinger S., Patel H., Alneberg J., Wilm A., Garcia M.U., Di Tommaso Pa., and Nahnsen S.. 2020. The nf-core framework for community-curated bioinformatics pipelines. Nat. Biotechnol. 38:276–278. 10.1038/s41587-020-0439-x [DOI] [PubMed] [Google Scholar]
- Fabregat, A., Jupe S., Matthews L., Sidiropoulos K., Gillespie M., Garapati P., Haw R., Jassal B., Korninger F., May B., et al. 2018. The Reactome Pathway Knowledgebase. Nucleic Acids Res. 46:D649–D655. 10.1093/nar/gkx1132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funderburg, N.T., Stubblefield Park S.R., Sung H.C., Hardy G., Clagett B., Ignatz-Hoover J., Harding C.V., Fu P., Katz J.A., Lederman M.M., and Levine A.D.. 2013. Circulating CD4(+) and CD8(+) T cells are activated in inflammatory bowel disease and are associated with plasma markers of inflammation. Immunology. 140:87–97. 10.1111/imm.12114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geremia, A., Arancibia-Carcamo C.V., Fleming M.P.P., Rust N., Singh B., Mortensen N.J., Travis S.P.L., and Powrie F.. 2011. IL-23-responsive innate lymphoid cells are increased in inflammatory bowel disease. J. Exp. Med. 208:1127–1133. 10.1084/jem.20101712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez De Aguero, M., Ganal-Vonarburg S.C., Fuhrer T., Rupp S., Uchimura Y., Li H., Steinert A., Heikenwalder M., Hapfelmeier S., Sauer U., et al. 2016. The maternal microbiota drives early postnatal innate immune development. Science. 351:1296–1302. 10.1126/science.aad2571 [DOI] [PubMed] [Google Scholar]
- Guo, X., Qiu J., Tu T., Yang X., Deng L., Anders R.A., Zhou L., and Fu Y.X.. 2014. Induction of innate lymphoid cell-derived interleukin-22 by the transcription factor STAT3 mediates protection against intestinal infection. Immunity. 40:25–39. 10.1016/j.immuni.2013.10.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gury-Benari, M., Thaiss C.A., Serafini N., Winter D.R., Giladi A., Lara-Astiaso D., Levy M., Salame T.M., Weiner A., David E., et al. 2016. The spectrum and regulatory landscape of intestinal innate lymphoid cells are shaped by the microbiome. Cell. 166:1231–1246 e13. 10.1016/j.cell.2016.07.043 [DOI] [PubMed] [Google Scholar]
- Hepworth, M.R., Fung T.C., Masur S.H., Kelsen J.R., Mcconnell F.M., Dubrot J., Withers D.R., Hugues S., Farrar M.A., Reith W., et al. 2015. Immune tolerance. Group 3 innate lymphoid cells mediate intestinal selection of commensal bacteria-specific CD4+ T cells. Science. 348:1031–1035. 10.1126/science.aaa4812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hepworth, M.R., Monticelli L.A., Fung T.C., Ziegler C.G.K., Grunberg S., Sinha R., Mantegazza A.R., Ma H.L., Crawford A., Angelosanto J.M., et al. 2013. Innate lymphoid cells regulate CD4+ T-cell responses to intestinal commensal bacteria. Nature. 498:113–117. 10.1038/nature12240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ise, W., Kohyama M., Schraml B.U., Zhang T., Schwer B., Basu U., Alt F.W., Tang J., Oltz E.M., Murphy T.L., and Murphy K.M.. 2011. The transcription factor BATF controls the global regulators of class-switch recombination in both B cells and T cells. Nat. Immunol. 12:536–543. 10.1038/ni.2037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa, M., and Goto S.. 2000. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28:27–30. 10.1093/nar/28.1.27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan, G.G. 2015. The global burden of IBD: From 2015 to 2025. Nat. Rev. Gastroenterol. Hepatol. 12:720–727. 10.1038/nrgastro.2015.150 [DOI] [PubMed] [Google Scholar]
- Kaya-Okur, H.S., Wu S.J., Codomo C.A., Pledger E.S., Bryson T.D., Henikoff J.G., Ahmad K., and Henikoff S.. 2019. CUT&Tag for efficient epigenomic profiling of small samples and single cells. Nat. Commun. 10:1930. 10.1038/s41467-019-09982-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khor, B., Gardet A., and Xavier R.J.. 2011. Genetics and pathogenesis of inflammatory bowel disease. Nature. 474:307–317. 10.1038/nature10209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss, E.A., Vonarbourg C., Kopfmann S., Hobeika E., Finke D., Esser C., and Diefenbach A.. 2011. Natural aryl hydrocarbon receptor ligands control organogenesis of intestinal lymphoid follicles. Science. 334:1561–1565. 10.1126/science.1214914 [DOI] [PubMed] [Google Scholar]
- Klose, C.S.N., Kiss E.A., Schwierzeck V., Ebert K., Hoyler T., D’hargues Y., Goppert N., Croxford A.L., Waisman A., Tanriver Y., and Diefenbach A.. 2013. A T-bet gradient controls the fate and function of CCR6-RORγt+ innate lymphoid cells. Nature. 494:261–265. 10.1038/nature11813 [DOI] [PubMed] [Google Scholar]
- Kurachi, M., Barnitz R.A., Yosef N., Odorizzi P.M., Diiorio M.A., Lemieux M.E., Yates K., Godec J., Klatt M.G., Regev A., et al. 2014. The transcription factor BATF operates as an essential differentiation checkpoint in early effector CD8+ T cells. Nat. Immunol. 15:373–383. 10.1038/ni.2834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroda, S., Yamazaki M., Abe M., Sakimura K., Takayanagi H., and Iwai Y.. 2011. Basic leucine zipper transcription factor, ATF-like (BATF) regulates epigenetically and energetically effector CD8 T-cell differentiation via Sirt1 expression. Proc. Natl. Acad. Sci. USA. 108:14885–14889. 10.1073/pnas.1105133108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead, B., and Salzberg S.L.. 2012. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 9:357–359. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarevic, V., and Glimcher L.H.. 2011. T-bet in disease. Nat. Immunol. 12:597–606. 10.1038/ni.2059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, J.S., Cella M., Mcdonald K.G., Garlanda C., Kennedy G.D., Nukaya M., Mantovani A., Kopan R., Bradfield C.A., Newberry R.D., and Colonna M.. 2011. AHR drives the development of gut ILC22 cells and postnatal lymphoid tissues via pathways dependent on and independent of Notch. Nat. Immunol. 13:144–151. 10.1038/ni.2187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann, F.M., Von Burg N., Ivanek R., Teufel C., Horvath E., Peter A., Turchinovich G., Staehli D., Eichlisberger T., Gomez De Aguero M., et al. 2020. Microbiota-induced tissue signals regulate ILC3-mediated antigen presentation. Nat. Commun. 11:1794. 10.1038/s41467-020-15612-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., and 1000 Genome Project Data Processing Subgroup . 2009. The sequence alignment/map format and SAMtools. Bioinformatics. 25:2078–2079. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, P., Spolski R., Liao W., Wang L., Murphy T.L., Murphy K.M., and Leonard W.J.. 2012. BATF-JUN is critical for IRF4-mediated transcription in T cells. Nature. 490:543–546. 10.1038/nature11530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang, W., Peng X., Li Q., Wang P., Lv P., Song Q., She S., Huang S., Chen K., Gong W., et al. 2020. FAM3D is essential for colon homeostasis and host defense against inflammation associated carcinogenesis. Nat. Commun. 11:5912. 10.1038/s41467-020-19691-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, B., Ye B., Zhu X., Yang L., Li H., Liu N., Zhu P., Lu T., He L., Tian Y., and Fan Z.. 2020a. An inducible circular RNA circKcnt2 inhibits ILC3 activation to facilitate colitis resolution. Nat. Commun. 11:4076. 10.1038/s41467-020-17944-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Q., Kim M.H., Friesen L., and Kim C.H.. 2020b. BATF regulates innate lymphoid cell hematopoiesis and homeostasis. Sci. Immunol. 5:eaaz8154. 10.1126/sciimmunol.aaz8154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longman, R.S., Diehl G.E., Victorio D.A., Huh J.R., Galan C., Miraldi E.R., Swaminath A., Bonneau R., Scherl E.J., and Littman D.R.. 2014. CX₃CR1⁺ mononuclear phagocytes support colitis-associated innate lymphoid cell production of IL-22. J. Exp. Med. 211:1571–1583. 10.1084/jem.20140678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love, M.I., Huber W., and Anders S.. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550. 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magri, G., Miyajima M., Bascones S., Mortha A., Puga I., Cassis L., Barra C.M., Comerma L., Chudnovskiy A., Gentile M., et al. 2014. Innate lymphoid cells integrate stromal and immunological signals to enhance antibody production by splenic marginal zone B cells. Nat. Immunol. 15:354–364. 10.1038/ni.2830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao, K., Baptista A.P., Tamoutounour S., Zhuang L., Bouladoux N., Martins A.J., Huang Y., Gerner M.Y., Belkaid Y., and Germain R.N.. 2018. Innate and adaptive lymphocytes sequentially shape the gut microbiota and lipid metabolism. Nature. 554:255–259. 10.1038/nature25437 [DOI] [PubMed] [Google Scholar]
- Martini, E., Krug S.M., Siegmund B., Neurath M.F., and Becker C.. 2017. Mend your fences: The epithelial barrier and its relationship with mucosal immunity in inflammatory bowel disease. Cell. Mol. Gastroenterol. Hepatol. 4:33–46. 10.1016/j.jcmgh.2017.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masternak, K., Muhlethaler-Mottet A., Villard J., Zufferey M., Steimle V., and Reith W.. 2000. CIITA is a transcriptional coactivator that is recruited to MHC class II promoters by multiple synergistic interactions with an enhanceosome complex. Genes Dev. 14:1156–1166. 10.1101/gad.14.9.1156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melo-Gonzalez, F., Kammoun H., Evren E., Dutton E.E., Papadopoulou M., Bradford B.M., Tanes C., Fardus-Reid F., Swann J.R., Bittinger K., et al. 2019. Antigen-presenting ILC3 regulate T cell-dependent IgA responses to colonic mucosal bacteria. J. Exp. Med. 216:728–742. 10.1084/jem.20180871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, D., Motomura K., Garcia-Flores V., Romero R., and Gomez-Lopez N.. 2018. Innate lymphoid cells in the maternal and fetal compartments. Front. Immunol. 9:2396. 10.3389/fimmu.2018.02396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, M.M., Patel P.S., Bao K., Danhorn T., O’connor B.P., and Reinhardt R.L.. 2020. BATF acts as an essential regulator of IL-25-responsive migratory ILC2 cell fate and function. Sci. Immunol. 5:eaay3994. 10.1126/sciimmunol.aay3994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirsepasi-Lauridsen, H.C., Vrankx K., Engberg J., Friis-Moller A., Brynskov J., Nordgaard-Lassen I., Petersen A.M., and Krogfelt K.A.. 2018. Disease-specific enteric microbiome dysbiosis in inflammatory bowel disease. Front. Med. 5:304. 10.3389/fmed.2018.00304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molodecky, N.A., Soon I.S., Rabi D.M., Ghali W.A., Ferris M., Chernoff G., Benchimol E.I., Panaccione R., Ghosh S., Barkema H.W., and Kaplan G.G.. 2012. Increasing incidence and prevalence of the inflammatory bowel diseases with time, based on systematic review. Gastroenterology. 142:46–54.e42; quiz e30. 10.1053/j.gastro.2011.10.001 [DOI] [PubMed] [Google Scholar]
- Mortha, A., and Burrows K.. 2018. Cytokine networks between innate lymphoid cells and myeloid cells. Front. Immunol. 9:191. 10.3389/fimmu.2018.00191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortha, A., Chudnovskiy A., Hashimoto D., Bogunovic M., Spencer S.P., Belkaid Y., and Merad M.. 2014. Microbiota-dependent crosstalk between macrophages and ILC3 promotes intestinal homeostasis. Science. 343:1249288. 10.1126/science.1249288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagasawa, M., Heesters B.A., Kradolfer C.M.A., Krabbendam L., Martinez-Gonzalez I., De Bruijn M.J.W., Golebski K., Hendriks R.W., Stadhouders R., Spits H., and Bal S.M.. 2019. KLRG1 and NKp46 discriminate subpopulations of human CD117(+)CRTH2(−) ILCs biased toward ILC2 or ILC3. J. Exp. Med. 216:1762–1776. 10.1084/jem.20190490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida, A., Inoue R., Inatomi O., Bamba S., Naito Y., and Andoh A.. 2018. Gut microbiota in the pathogenesis of inflammatory bowel disease. Clin. J. Gastroenterol. 11:1–10. 10.1007/s12328-017-0813-5 [DOI] [PubMed] [Google Scholar]
- Onizawa, M., Nagaishi T., Kanai T., Nagano K.i., Oshima S., Nemoto Y., Yoshioka A., Totsuka T., Okamoto R., Nakamura T., et al. 2009. Signaling pathway via TNF-alpha/NF-kappaB in intestinal epithelial cells may be directly involved in colitis-associated carcinogenesis. Am. J. Physiol. Gastrointest. Liver Physiol. 296:G850–G859. 10.1152/ajpgi.00071.2008 [DOI] [PubMed] [Google Scholar]
- Panea, C., Farkas A.M., Goto Y., Abdollahi-Roodsaz S., Lee C., Koscso B., Gowda K., Hohl T.M., Bogunovic M., and Ivanov I.I.. 2015. Intestinal monocyte-derived macrophages control commensal-specific Th17 responses. Cell Rep. 12:1314–1324. 10.1016/j.celrep.2015.07.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker, M.E., Barrera A., Wheaton J.D., Zuberbuehler M.K., Allan D.S.J., Carlyle J.R., Reddy T.E., and Ciofani M.. 2020. c-Maf regulates the plasticity of group 3 innate lymphoid cells by restraining the type 1 program. J. Exp. Med. 217:e20191030. 10.1084/jem.20191030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patro, R., Duggal G., Love M.I., Irizarry R.A., and Kingsford C.. 2017. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods. 14:417–419. 10.1038/nmeth.4197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson, C., Thornton E.E., Mckenzie B., Schaupp A.L., Huskens N., Griseri T., West N., Tung S., Seddon B.P., Uhlig H.H., and Powrie F.. 2016. ILC3 GM-CSF production and mobilisation orchestrate acute intestinal inflammation. Elife. 5:e10066. 10.7554/eLife.10066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penny, H.A., Hodge S.H., and Hepworth M.R.. 2018. Orchestration of intestinal homeostasis and tolerance by group 3 innate lymphoid cells. Semin. Immunopathol. 40:357–370. 10.1007/s00281-018-0687-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham, D., Moseley C.E., Gao M., Savic D., Winstead C.J., Sun M., Kee B.L., Myers R.M., Weaver C.T., and Hatton R.D.. 2019. Batf pioneers the reorganization of chromatin in developing effector T cells via Ets1-dependent recruitment of Ctcf. Cell Rep. 29:1203–1220 e7. 10.1016/j.celrep.2019.09.064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu, J., Heller J.J., Guo X., Chen Z.m. E., Fish K., Fu Y.X., and Zhou L.. 2012. The aryl hydrocarbon receptor regulates gut immunity through modulation of innate lymphoid cells. Immunity. 36:92–104. 10.1016/j.immuni.2011.11.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quast, C., Pruesse E., Yilmaz P., Gerken J., Schweer T., Yarza P., Peplies J., and Glockner F.O.. 2013. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 41:D590–D596. 10.1093/nar/gks1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rankin, L.C., Groom J.R., Chopin M., Herold M.J., Walker J.A., Mielke L.A., McKenzie A.N.J., Carotta S., Nutt S.L., and Belz G.T.. 2013. The transcription factor T-bet is essential for the development of NKp46+ innate lymphocytes via the Notch pathway. Nat. Immunol. 14:389–395. 10.1038/ni.2545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rescigno, M. 2011. The intestinal epithelial barrier in the control of homeostasis and immunity. Trends Immunol. 32:256–264. 10.1016/j.it.2011.04.003 [DOI] [PubMed] [Google Scholar]
- Robinette, M.L., Fuchs A., Cortez V.S., Lee J.S., Wang Y., Durum S.K., Gilfillan S., Colonna M., and Immunological Genome Consortium . 2015. Transcriptional programs define molecular characteristics of innate lymphoid cell classes and subsets. Nat. Immunol. 16:306–317. 10.1038/ni.3094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Round, J.L., and Mazmanian S.K.. 2009. The gut microbiota shapes intestinal immune responses during health and disease. Nat. Reviews. Immunol. 9:313–323. 10.1038/nri2515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahoo, A., Alekseev A., Tanaka K., Obertas L., Lerman B., Haymaker C., Clise-Dwyer K., Mcmurray J.S., and Nurieva R.. 2015. Batf is important for IL-4 expression in T follicular helper cells. Nat. Commun. 6:7997. 10.1038/ncomms8997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salem, F., Kindt N., Marchesi J.R., Netter P., Lopez A., Kokten T., Danese S., Jouzeau J.Y., Peyrin-Biroulet L., and Moulin D.. 2019. Gut microbiome in chronic rheumatic and inflammatory bowel diseases: Similarities and differences. United Eur. Gastroenterol. J. 7:1008–1032. 10.1177/2050640619867555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sartor, R.B. 2011. Key questions to guide a better understanding of host-commensal microbiota interactions in intestinal inflammation. Mucosal Immunol. 4:127–132. 10.1038/mi.2010.87 [DOI] [PubMed] [Google Scholar]
- Satija, R., Farrell J.A., Gennert D., Schier A.F., and Regev A.. 2015. Spatial reconstruction of single-cell gene expression data. Nat. Biotechnol. 33:495–502. 10.1038/nbt.3192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schraml, B.U., Hildner K., Ise W., Lee W.L., Smith W.A.E., Solomon B., Sahota G., Sim J., Mukasa R., Cemerski S., et al. 2009. The AP-1 transcription factor Batf controls T(H)17 differentiation. Nature. 460:405–409. 10.1038/nature08114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segata, N., Izard J., Waldron L., Gevers D., Miropolsky L., Garrett W.S., and Huttenhower C.. 2011. Metagenomic biomarker discovery and explanation. Genome Biol. 12:R60. 10.1186/gb-2011-12-6-r60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stras, S.F., Werner L., Toothaker J.M., Olaloye O.O., Oldham A.L., Mccourt C.C., Lee Y.N., Rechavi E., Shouval D.S., and Konnikova L.. 2019. Maturation of the human intestinal immune system occurs early in fetal development. Dev. Cell. 51:357–373 e5. 10.1016/j.devcel.2019.09.008 [DOI] [PubMed] [Google Scholar]
- Takayama, T., Kamada N., Chinen H., Okamoto S., Kitazume M.T., Chang J., Matuzaki Y., Suzuki S., Sugita A., Koganei K., et al. 2010. Imbalance of NKp44(+)NKp46(−) and NKp44(−)NKp46(+) natural killer cells in the intestinal mucosa of patients with Crohn’s disease. Gastroenterology. 139:882–892, 892.e1-3. 10.1053/j.gastro.2010.05.040 [DOI] [PubMed] [Google Scholar]
- Tizian, C., Lahmann A., Holsken O., Cosovanu C., Kofoed-Branzk M., Heinrich F., Mashreghi M.F., Kruglov A., Diefenbach A., and Neumann C.. 2020. c-Maf restrains T-bet-driven programming of CCR6-negative group 3 innate lymphoid cells. Elife. 9:e52549. 10.7554/eLife.52549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ubel, C., Sopel N., Graser A., Hildner K., Reinhardt C., Zimmermann T., Rieker R.J., Maier A., Neurath M.F., Murphy K.M., and Finotto S.. 2014. The activating protein 1 transcription factor basic leucine zipper transcription factor, ATF-like (BATF), regulates lymphocyte- and mast cell-driven immune responses in the setting of allergic asthma. J. Allergy Clin. Immunol. 133:198–199. 10.1016/j.jaci.2013.09.049 [DOI] [PubMed] [Google Scholar]
- Van De Pavert, S.A., Ferreira M., Domingues R.G., Ribeiro H., Molenaar R., Moreira-Santos L., Almeida F.F., Ibiza S., Barbosa I., Goverse G., et al. 2014. Maternal retinoids control type 3 innate lymphoid cells and set the offspring immunity. Nature. 508:123–127. 10.1038/nature13158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vereecke, L., Beyaert R., and Van Loo G.. 2011. Enterocyte death and intestinal barrier maintenance in homeostasis and disease. Trends Mol. Med. 17:584–593. 10.1016/j.molmed.2011.05.011 [DOI] [PubMed] [Google Scholar]
- Verrier, T., Satoh-Takayama N., Serafini N., Marie S., Di Santo J.P., and Vosshenrich C.A.J.. 2016. Phenotypic and functional plasticity of murine intestinal NKp46+ group 3 innate lymphoid cells. J. Immunol. 196:4731–4738. 10.4049/jimmunol.1502673 [DOI] [PubMed] [Google Scholar]
- Vivier, E., Artis D., Colonna M., Diefenbach A., Di Santo J.P., Eberl G., Koyasu S., Locksley R.M., Mckenzie A.N.J., Mebius R.E., et al. 2018. Innate lymphoid cells: 10 Years on. Cell. 174:1054–1066. 10.1016/j.cell.2018.07.017 [DOI] [PubMed] [Google Scholar]
- Von Burg, N., Chappaz S., Baerenwaldt A., Horvath E., Bose Dasgupta S., Ashok D., Pieters J., Tacchini-Cottier F., Rolink A., Acha-Orbea H., and Finke D.. 2014. Activated group 3 innate lymphoid cells promote T-cell-mediated immune responses. Proc. Natl. Acad. Sci. USA. 111:12835–12840. 10.1073/pnas.1406908111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vonarbourg, C., Mortha A., Bui V.L., Hernandez P.P., Kiss E.A., Hoyler T., Flach M., Bengsch B., Thimme R., Holscher C., et al. 2010. Regulated expression of nuclear receptor RORγt confers distinct functional fates to NK cell receptor-expressing RORγt(+) innate lymphocytes. Immunity. 33:736–751. 10.1016/j.immuni.2010.10.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y., Godec J., Ben-Aissa K., Cui K., Zhao K., Pucsek A.B., Lee Y.K., Weaver C.T., Yagi R., and Lazarevic V.. 2014. The transcription factors T-bet and Runx are required for the ontogeny of pathogenic interferon-gamma-producing T helper 17 cells. Immunity. 40:355–366. 10.1016/j.immuni.2014.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, X., Kasmani M.Y., Zheng S., Khatun A., Chen Y., Winkler W., Zander R., Burns R., Taparowsky E.J., Sun J., and Cui W.. 2022. BATF promotes group 2 innate lymphoid cell-mediated lung tissue protection during acute respiratory virus infection. Sci. Immunol. 7:eabc9934. 10.1126/sciimmunol.abc9934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, G., Wang L.G., Han Y., and He Q.Y.. 2012. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 16:284–287. 10.1089/omi.2011.0118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zareie, M., Singh P.K., Irvine E.J., Sherman P.M., Mckay D.M., and Perdue M.H.. 2001. Monocyte/macrophage activation by normal bacteria and bacterial products: Implications for altered epithelial function in Crohn’s disease. Am. J. Pathol. 158:1101–1109. 10.1016/S0002-9440(10)64057-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaret, K.S., and Carroll J.S.. 2011. Pioneer transcription factors: Establishing competence for gene expression. Genes Dev. 25:2227–2241. 10.1101/gad.176826.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng, B., Shi S., Ashworth G., Dong C., Liu J., and Xing F.. 2019. ILC3 function as a double-edged sword in inflammatory bowel diseases. Cell Death Dis. 10:315. 10.1038/s41419-019-1540-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, Y., Xu Z.Z., He Y., Yang Y., Liu L., Lin Q., Nie Y., Li M., Zhi F., Liu S., et al. 2018. Gut microbiota offers universal biomarkers across ethnicity in inflammatory bowel disease diagnosis and infliximab response prediction. mSystems. 3:e00188-17. 10.1128/mSystems.00188-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zindl, C.L., Witte S.J., Laufer V.A., Gao M., Yue Z., Janowski K.M., Cai B., Frey B.F., Silberger D.J., Harbour S.N., et al. 2022. A nonredundant role for T cell-derived interleukin 22 in antibacterial defense of colonic crypts. Immunity. 55:494–511.e11. 10.1016/j.immuni.2022.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The scRNA-seq, bulk RNA-seq, ATAC-seq, and CUT&Tag ChIP-seq data are deposited in the GEO database with the accession code GSE168080. All other raw data are available from the corresponding author upon reasonable request.