Abstract
Expression of the Anabaena sp. strain PCC 7120 RNA helicase gene crhC is induced by cold shock. crhC transcripts are not detectable at 30°C but accumulate at 20°C, and levels remain elevated for the duration of the cold stress. Light-derived metabolic capability, and not light per se, is required for crhC transcript accumulation. Enhanced crhC mRNA stability contributes significantly to the accumulation of crhC transcripts, with the crhC half-life increasing sixfold at 20°C. The accumulation is reversible, with the cells responding more rapidly to temperature downshifts than to upshifts, as a result of the lack of active mRNA destabilization and the continuation of crhC transcription, at least transiently, after a temperature upshift. Translational inhibitors do not induce crhC expression to cold shock levels, indicating that inhibition of translation is only one of the signals required to activate the cold shock response in Anabaena. Limited amounts of protein synthesis are required for the cold shock-induced accumulation of crhC transcripts, as normal levels of accumulation occur in the presence of tetracycline but are abolished by chloramphenicol. Regulation of crhC expression may also extend to the translational level, as CrhC protein levels do not correlate completely with the pattern of mRNA transcript accumulation. Our experiments indicate that the regulation of crhC transcript accumulation is tightly controlled by both temperature and metabolic activity at the levels of transcription, mRNA stabilization, and translation.
Microorganisms respond to decreases in growth temperature via a process termed the cold shock response (7, 10, 24). In Escherichia coli, where cold shock has been most extensively studied, the cold shock response is initiated when the organism experiences a decrease in growth temperature of greater than 13°C (10). Physiologically, the response is divided into two phases, an initial lag in the growth or acclimation period followed by a resumption in growth. Sixteen gene products, termed cold shock proteins (CSPs), are specifically and transiently expressed during the acclimation phase in E. coli (24). CSPs include a diverse group of proteins involved in the transcription, translation, and function of mRNA, including CspA, polynucleotide phosphorylase (PNPase), RecA, initiation factor 2, CsdA, RbfA, NusA, histone-like nucleoid-structuring protein (H-NS), trigger factor, and GyrA (7, 10, 24).
Regulation of CSP expression in E. coli occurs at numerous levels, including transcription, mRNA stabilization, and translation, and has been best characterized for the most abundant CSP gene, cspA. At the transcriptional level, cspA is regulated positively by an AT-rich upstream element and negatively by a cold box (8, 15). cspA expression is regulated differentially at the posttranscriptional level by an unusually long 5′ untranslated region (5′ UTR) and a downstream box required for translation initiation (15). mRNA stabilization also plays a significant role in cspA transcript accumulation (2, 6, 10). Cold shock induction of cspA transcript accumulation does not require protein synthesis (4), as is indicated by the induction of CSP expression in E. coli cells exposed to translational inhibitors which interfere with ribosome function at 37°C (9, 25).
Although the mechanism regulating CSP expression is not known, the rate-limiting step under cold shock conditions is the initiation of translation (10, 13). A cold shock ribosome adaptation model which allows this rate-limiting step to be overcome by the association of three CSPs, translation initiation factor 2, CsdA (RNA helix destabilization), and RbfA (ribosome binding factor A), with the ribosome, converting it into a cold-resistant translatable state, has been proposed (11, 24). In this scenario, csdA, which possesses RNA helix-destabilizing activity, is proposed to remove secondary structures in the highly structured 5′ UTRs of E. coli CSP mRNAs, thereby facilitating translation initiation (12).
In cyanobacteria, a group of gram-negative photoautotrophic prokaryotes, only fatty acid desaturases (14, 18), ribosomal protein S21 (22), heat shock protein ClpB (17), and a family of RNA binding proteins (20, 21) are known to be expressed in response to cold shock. Investigation of the desaturase (14, 18) and RNA binding protein (20, 21) gene family members which respond to cold shock indicates that temperature-induced changes in mRNA stability and rates of transcription play major roles in the regulation of their expression.
We have recently reported that a cyanobacterial RNA helicase gene, crhC, is specifically induced by cold shock (3). Here we describe the regulation of crhC expression by temperature at the transcriptional, posttranscriptional, and translational levels and discuss the possible role(s) which CrhC may perform during cold shock adaptation in cyanobacteria.
MATERIALS AND METHODS
Growth and maintenance of organisms.
Anabaena sp. strain PCC 7120 (referred to hereafter as Anabaena) was grown axenically in BG-11 medium at 30°C with a constant illumination of 150 microeinsteins/m2/s (3). Liquid cultures (300 ml) were aerated by a combination of shaking at 150 rpm on a rotary shaker and bubbling with air. Aliquots (50 ml) were aerated by shaking at 150 rpm on a rotary shaker and were treated as described in the figure legends. Cultures were cold induced for 1 h at 20°C unless otherwise stated. Antibiotics were obtained from Sigma, except for tetracycline (Boehringer Mannheim).
RNA manipulations.
Total RNA was isolated from Anabaena using glass bead lysis (20). Northern blots containing 15 μg of total RNA per lane were generated and probed as previously described (3). Radioactive probes for detection of the crhC and the constitutively expressed Anabaena RNase P (as a control for RNA loading) gene transcripts by Northern blot analysis were produced as previously described (3). Autoradiograms were produced on X-ray film, while for quantification, crhC signals were corrected for RNA loading by comparison with those obtained with the constitutive RNase P probe using a PhosphorImager (Molecular Dynamics) and analyzed using ImageQuant 4.0 software.
Protein manipulations.
Total protein was isolated from Anabaena by vortexing it in the presence of glass beads, in a buffer containing 50 mM Tris (pH 8), 100 mM EDTA, 0.5% Triton X-100, 0.5% Sarkosyl, and 0.4% sodium dodecyl sulfate, and quantified by the Bradford assay (Bio-Rad) using bovine serum albumin as the standard. Polypeptides (50 μg) were separated on sodium dodecyl sulfate–10% polyacrylamide gels and transferred to nitrocellulose membranes (Bio-Rad) using a semidry apparatus (Tyler), and Western blot analysis was performed as described previously (1) with rabbit anti-CrhC antiserum (E. Yu and G. W. Owttrim, unpublished data) and goat anti-rabbit immunoglobulin G horseradish peroxidase conjugate (Cappel). Polypeptide size was determined by comparison with Kaleidoscope prestained molecular weight markers (Bio-Rad). The degree of inhibition of protein synthesis by antibiotics was determined by in vivo pulse-labeling of cells with a [35S]methionine-cysteine mixture (Amersham ProMix) as described previously (1). Briefly, mid-log-phase Anabaena cells, grown at 30°C, were harvested and resuspended in an equal volume of sulfate-free BG-11. Cultures were incubated at the either 20 or 30°C for 5 min before the addition of antibiotics at the concentrations indicated in the figure legends, and incubation continued for 5 min. [35S]Met-Cys (10 μCi/ml) was added, and incubation continued for 25 min (30 min total in the presence of antibiotics). Duplicate aliquots were treated, and 35S incorporation into acid-insoluble material was quantitated as described previously (1). Control cultures, grown at either 20 or 30°C, were treated as described above except that the antibiotics were omitted.
RESULTS
crhC transcript accumulation is temperature and metabolism dependent.
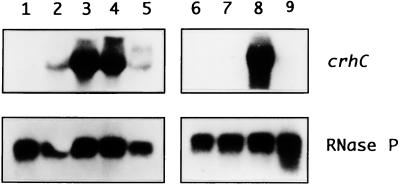
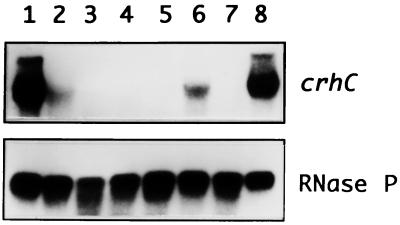
We have previously shown that crhC transcripts are specifically detected after exposure of Anabaena cells to cold shock conditions (20°C) (3). In order to determine how growth temperature and light-dark cycles influence crhC transcript accumulation, Northern blot analysis was performed. The results indicate that crhC transcripts are not detectable at 30°C but that their abundance is differentially induced at temperatures below this level, with maximum abundance observed between 20 and 15°C (Fig. 1). crhC transcripts do not accumulate at temperatures higher than 25°C, including 30°C (Fig. 1, lane 1), 37°C (data not shown), and 43°C (3). Thus, crhC transcript abundance is finely regulated by minor changes in growth temperature.
FIG. 1.
Cold shock-induced increases in crhC transcript abundance is regulated by temperature and metabolic activity. Northern blots of total RNA (15 μg) extracted from Anabaena exposed to different temperatures and/or light-dark conditions were hybridized with either the crhC or the RNase P gene. The autoradiograms are shown. Lane 1, 1 h at 30°C; lane 2, 1 h at 25°C; lane 3, 1 h at 20°C; lane 4, 1 h at 15°C; lane 5, 1 h at 10°C; lane 6, 3 h at 30°C in the dark, followed by 1 h at 20°C in the dark; lane 7, 4 h at 30°C in the dark; lane 8, 3 h at 30°C in the light, followed by 1 h at 20°C in the dark; lane 9, 4 h at 30°C in the light. The blots were probed with crhC, stripped, and reprobed with RNase P, as indicated.
Since Anabaena is an obligate photoautotroph, relying on photosynthetic light harvesting for growth and metabolism, we asked if light was also required for the cold shock-induced accumulation of crhC transcripts. This appears to be the case, as crhC transcripts were not detected in cells which were incubated in the dark for 3 h before transfer to the cold (Fig. 1, lane 6), results identical to those obtained from cells incubated continuously at 30°C in the dark or light (Fig. 1, lane 7 or 9, respectively). Light is not required, however, for the accumulation of crhC transcripts if the Anabaena culture is transferred to the cold and dark simultaneously (Fig. 1, lane 8). Thus, it is not light per se but light-derived metabolic capability which is required for crhC transcript accumulation in response to cold shock.
crhC transcript and protein accumulation patterns differ.
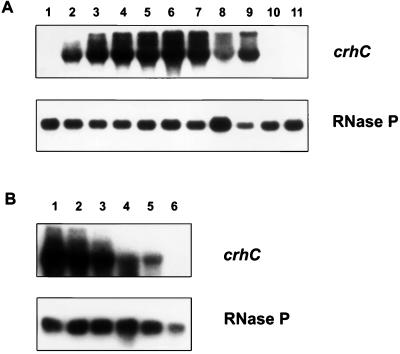
In order to determine the pattern of cold-induced crhC transcript accumulation, a time course of crhC induction was determined. Northern blot analysis indicated that crhC transcripts are not detectable in cells grown at 30°C (Fig. 2A, lane 1) but that they accumulate within 15 min after transfer to 20°C (Fig. 2A, lane 2). crhC transcripts were expressed constitutively during growth at 20°C, reaching half-maximal levels 30 min after a temperature downshift. We have consistently observed that crhC transcript levels decrease fourfold 24 h after cold shock initiation (Fig. 2A, lane 8) but recover after 48 h (Fig. 2A, lane 9). crhC transcripts were not detectable in cells grown for 24 or 48 h at 30°C (Fig. 2A, lanes 10 and 11). Three RNA transcripts were detected by the crhC probe, a major transcript whose size corresponds to that expected for a full-length crhC transcript and two longer ones. Although all three transcripts were observed at all times during cold stress, the relative abundance of each transcript varied depending on the length of exposure to cold shock. Early in cold shock the abundance of the longest transcript was low relative to that of the middle transcript, while after prolonged exposure the relative abundances of these transcripts reversed (Fig. 2A, compare lanes 3 and 9).
FIG. 2.
Time course of cold-induced accumulation and warmth-induced decay of crhC transcripts. (A) Fifteen micrograms of total RNA extracted from each sample was subjected to Northern blot analysis using crhC and RNase P probes, as indicated. RNA was obtained from Anabaena cells grown at 30°C and then cold shocked at 20°C for the following lengths of time: 0 h (lane 1), 0.25 h (lane 2), 0.5 h (lane 3), 1 h (lane 4), 2 h (lane 5), 3 h (lane 6), 6 h (lane 7), 24 h (lane 8), and 48 h (lane 9). Lanes 10 and 11 contain RNAs from control cultures grown at 30°C for 24 and 48 h, respectively. (B) Northern blot analysis of total RNA (15 μg) extracted from Anabaena exposed to 20°C for 1 h and then to 30°C for 0 h (lane 1), 0.25 h (lane 2), 0.5 h (lane 3), 1 h (lane 4), 2 h (lane 5), and 3 h (lane 6). The blot was probed with crhC, stripped, and reprobed with RNase P, as indicated.
To determine if the cold-induced accumulation of crhC transcripts is reversible, a time course of crhC mRNA transcript decay after cold-induced cultures were transferred to 30°C was determined (Fig. 2B). crhC transcript levels declined after a temperature upshift and were no longer detectable within 3 h (Fig. 2B, lanes 2 to 6). crhC transcript abundance declined by 50% within 90 min of a temperature upshift. The results indicate that the cold-induced accumulation of crhC transcripts is fully reversible. Furthermore, crhC transcripts accumulate more quickly after a temperature downshift than they decay after a temperature upshift.
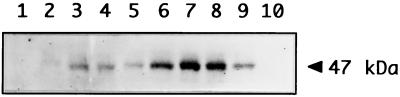
To determine if translational control plays a role in crhC expression, the relationship between the abundance of crhC mRNA transcripts and CrhC protein levels was determined. Western blot analysis of a time course of cold-induced CrhC protein production (Fig. 3) shows that the induction of CrhC polypeptide corresponds to transcript accumulation early in cold shock but not during prolonged exposure to low temperature. The crhC gene product was not detected in cells grown continuously at 30°C (Fig. 3, lanes 1 and 10) but was detectable within 15 min after transfer to 20°C (Fig. 3, lane 2) and remained elevated for the duration of the cold treatment (Fig. 3, lanes 2 to 9). While transcript levels decreased after 24 h of cold shock and recovered by 48 h, protein levels were reversed, being maximal after 24 h, and decreased at 48 h. It should be noted that growth at 20°C for longer than 48 h resulted in the cells becoming achlorotic, as was also seen for Anabaena variabilis strain M3 (21).
FIG. 3.
CrhC protein expression mimics transcript accumulation. Total protein was extracted from Anabaena exposed to 20°C for various lengths of time. The Western blot, containing 50 μg of protein per lane and immunodecorated with rabbit anti-CrhC antiserum, is shown. Lane 1, 0 min; lane 2, 15 min; lane 3, 30 min; lane 4, 1 h; lane 5, 2 h; lane 6, 3 h; lane 7, 6 h; lane 8, 24 h; lane 9, 48 h; lane 10, 48 h at 30°C. The position of the 47-kDa CrhC protein is indicated by an arrow.
crhC transcripts are stabilized in the cold.
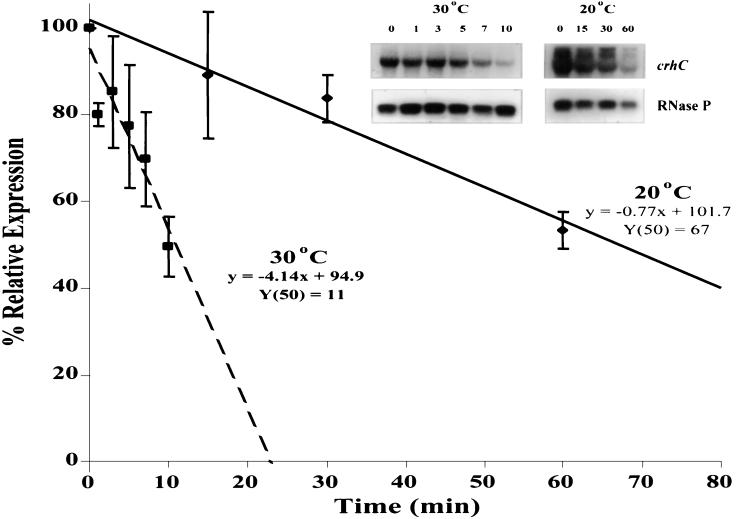
The crhC transcript half-life was determined to ascertain the contribution that temperature-induced changes in mRNA stability make to crhC transcript accumulation. The transcriptional inhibitor rifampin was added to cold-induced cultures before continued incubation at 20°C or transfer to 30°C, and crhC transcript levels were determined at the indicated times thereafter (Fig. 4). Linear regression analysis of changes in crhC transcript levels over time indicated that the crhC transcript has a sixfold longer half-life at 20°C (67 min) than at 30°C (11 min). mRNA stability, therefore, plays a significant role in the regulation of crhC transcript accumulation.
FIG. 4.
crhC transcripts are stabilized at 20°C. Total RNA (15 μg) was extracted from Anabaena exposed to 20°C for 1 h, followed by the addition of rifampin (400 μg/ml) and either continued exposure at 20°C or transferred to 30°C for the indicated times. crhC transcript levels, determined by Northern blot analysis, were quantitated to determine their half-lives at the respective temperatures. Average values from triplicate, independent repetitions of the experiments were subjected to linear regression analysis and are plotted (squares and hatched line, 30°C; diamonds and solid line, 20°C). Shown in the inset are representative autoradiograms of Northern blots used to generate the data points.
Temperature-induced alterations in the rate of crhC transcription may also play a role in the observed effects on crhC transcript accumulation. To test this possibility, we used PhosphorImager quantitation to compare the rates of crhC transcript degradation at 30°C, after a temperature upshift, in the presence and absence of rifampin. crhC transcript levels decreased to half-maximal levels within 90 min in the absence of rifampin, while similar levels were reached within 11 min in the presence of rifampin. These results suggest that crhC transcription continues after a temperature upshift. Continued transcription of crhC after a temperature upshift may occur only temporarily, as transcript accumulation was no longer detectable within 3 h after a temperature upshift (Fig. 2B, lane 6).
Protein synthesis is required for crhC transcript accumulation.
Temperature-induced changes in crhC mRNA stability and transcription may require protein synthesis. Since inhibitors of protein synthesis mimic the cold shock response in E. coli, we first determined crhC transcript levels in Anabaena cells grown at 30°C after exposure to five translational inhibitors. All of the inhibitors induced crhC transcript levels marginally at 30°C, although some were at a level which was too low to be reproduced photographically (Fig. 5, lanes 2 to 6). Ethanol, which was used to dissolve the inhibitors, did not induce expression (Fig. 5, lane 7). Although chloramphenicol and tetracycline, at 10 μg/ml, induced crhC transcript accumulation most efficiently of the five translational inhibitors tested, the level of accumulation was only 15 to 20% of that observed under cold shock conditions (Fig. 5, compare lanes 1 and 8 with 2 and 6). At 30°C, the effects of the inhibitors appeared to be concentration dependent, as comparable results were obtained using tetracycline at 10 μg/ml (Fig. 5, lane 6) and 40 μg/ml (data not shown), while chloramphenicol did not induce expression at 40 μg/ml (data not shown). Thus, inhibition of translation only partially mimics the cold shock response and protein synthesis may be required for the partial induction of crhC transcript accumulation by translational inhibitors at 30°C.
FIG. 5.
Cold shock-mimicking translational inhibitors partially induce crhC transcript accumulation at 30°C. The autoradiogram of a Northern blot of total RNA (15 μg) extracted from Anabaena exposed to various cold shock-mimicking translational inhibitors for 30 min at 30°C is shown. Lanes 1 and 8, 30 min at 20°C, no inhibitor; lane 2, chloramphenicol (10 μg/ml); lane 3, erythromycin (500 μg/ml); lane 4, fusidic acid (0.5 μg/ml); lane 5, spiramycin (800 μg/ml); lane 6, tetracycline (10 μg/ml); lane 7, ethanol (0.8%) as a control without antibiotics. The blots were probed with crhC, stripped, and reprobed with RNase P, as indicated.
To ensure that these observations were a result of translational inhibition, we determined the level of protein synthesis in Anabaena cells grown at 20 and 30°C in the presence of chloramphenicol and tetracycline at 10 and 40 μg/ml. At 20°C, chloramphenicol inhibited protein synthesis by 69% at 10 μg/ml while it inhibited synthesis by 91% at a concentration of 40 μg/ml. Tetracycline inhibited protein synthesis by 94 and 97% at final concentrations of 10 and 40 μg/ml, respectively. Temperature did not affect the level of inhibition, as comparable results were observed at 30°C.
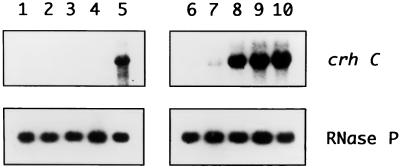
We then asked whether protein synthesis is required for crhC transcript accumulation under cold shock conditions. Northern blot analysis was performed on RNAs isolated from cultures exposed to either chloramphenicol or tetracycline at 20°C (Fig. 6). Inhibition of protein synthesis by chloramphenicol at 40 μg/ml suppressed the cold shock-induced accumulation of crhC transcripts (Fig. 6, lanes 1 to 4). As a control, the partial inhibition of protein synthesis by chloramphenicol at 10 μg/ml did not suppress crhC transcript accumulation (data not shown). Conversely, cold-induced crhC transcript accumulation was not affected by tetracycline at 10 μg/ml (Fig. 6, lanes 6 to 9) and was only marginally reduced by 40 μg/ml (data not shown), inhibitor concentrations which essentially abolished protein synthesis. Furthermore, both the time dependency and the degrees of cold-induced crhC transcript accumulation were found to be comparable in the presence and absence of tetracycline. Thus, a limited amount of protein synthesis is required for cold shock induction of crhC transcript accumulation in Anabaena.
FIG. 6.
Translational inhibitors differentially affect crhC induction at 20°C. The autoradiograms of Northern blots of total RNA (15 μg) extracted from Anabaena exposed to either chloramphenicol (40 μg/ml, lanes 1 to 4) or tetracycline (10 μg/ml, lanes 6 to 9) at 20°C for the indicated lengths of time are shown. Lanes 1 and 6, 0 min; lanes 2 and 7, 10 min; lanes 3 and 8, 20 min; lanes 4 and 9, 30 min; lanes 5 and 10, 30 min at 20°C without inhibitor. The blots were probed with crhC, stripped, and reprobed with RNase P, as indicated.
DISCUSSION
Free-living prokaryotic organisms must have the capacity to respond rapidly to fluctuations in growth temperature. These responses are regulated at the molecular level and have been characterized for heat shock but not nearly as well for cold shock (24). The majority of cold shock-induced gene products affect the translational machinery or membrane fluidity (24). Recently, we have shown that crhC, which encodes a cyanobacterial RNA helicase, is expressed under cold shock conditions (3). In this report we investigate the regulation of cold-induced crhC expression at the transcriptional, posttranscriptional, and translational levels.
At the metabolic level, the cold-induced expression of crhC does not require light directly, as transcript accumulation occurs if the cells are transferred to cold and dark conditions simultaneously. Similar results have been reported for the cold-induced expression of the A. variabilis M3 RNA binding protein gene rbpA1 (21). crhC expression, however, does require light-derived metabolic capability, since crhC transcript accumulation does not occur if the temperature downshift occurs 3 h after initiation of the dark treatment. During this dark period, cyanobacterial metabolism is downregulated to a significant degree, corresponding to the time when photosynthetically derived glucose levels are depleted (16).
In the presence of adequate metabolic activity, multiple crhC transcripts accumulate after a temperature downshift and remain elevated for the duration of the cold stress. The relative abundance of the individual crhC transcripts is altered by the length of exposure to cold stress. This observation suggests that there may be two phases of crhC cold shock regulation in Anabaena, a rapid initial response within 3 h and a long-term response after 24 h. The presence of three hybridizing transcripts may also indicate that crhC is expressed as part of a cold shock operon. Multiple transcripts, whose origins have not been determined, have also been observed for both the Synechocystis sp. strain PCC 6803 (14) and the Synechococcus sp. strain PCC 7002 (18) desC desaturase genes, whose constitutive expression is enhanced by cold shock. Although crhC is constitutively expressed for the duration of the cold stress, transcript levels decrease fourfold 24 h after initiation of the cold treatment but recover after 48 h. This observation may indicate the additional involvement of a secondary stress or circadian rhythm in the regulation of crhC transcript accumulation during prolonged exposure to low temperature. The pattern of crhC transcript induction after a temperature downshift is similar to that observed for the A. variabilis M3 cold-regulated RNA binding proteins rbpA1 and rbpA2 over a 25-h time course (20).
The half-rise time of crhC transcript accumulation is similar to that observed for the cold-induced accumulation of desA transcripts in Synechocystis sp. strain PCC 6803 (14). Furthermore, the cold-induced accumulation of crhC transcripts is completely reversible, with a time frame of decay which is slower than that required for crhC transcript accumulation to reach half-maximal levels after a temperature downshift. This reduction in the rate of crhC transcript decay after a temperature upshift may result from the continuation of transcription and/or lack of active crhC mRNA destabilization at 30°C (see below).
Changes in mRNA stability are important for the temperature regulation of crhC expression, as the crhC transcript half-life is enhanced sixfold by cold stress. This degree of crhC mRNA stabilization is similar to that observed for other cold-induced cyanobacterial genes, whose stability increases between 3.5- and 15-fold upon a temperature downshift (14, 18, 21). Temperature-induced changes in mRNA stability cannot be accounted for solely by thermodynamic considerations, as the mRNA half-lives of other cold-enhanced gene family members are differentially affected by temperature, for example desC, whose half-life is not affected by growth temperature (14, 18). In addition, cyanobacterial RNase activity does not appear to be affected by alterations in environmental conditions, as it has recently been shown that varied light regimens do not affect RNase activity in Synechococcus sp. strain PCC 7002 (19).
The regulation of temperature-induced changes in cold shock gene mRNA stability may involve either stabilization at 20°C or destabilization at 30°C. RNA stabilization is known to play a major role in the regulation of mRNA levels for a number of CSP genes, although the proposed mechanisms responsible for this control vary (2, 6, 7, 14, 18, 21, 24). The decrease in crhC mRNA stability when transcription is inhibited by rifampin is consistent with crhC mRNA stabilization at 20°C and rules out the involvement of an mRNA-destabilizing factor, as has been proposed to regulate desB levels in Synechococcus sp. strain PCC 7002 at 38°C (18).
Temperature-induced changes in crhC transcription may also be involved in crhC transcript accumulation. crhC transcription occurs at least transiently after a temperature upshift, as crhC transcripts are more stable in the absence of rifampin than in its presence. Regulation of cold-induced transcript accumulation by changes in transcriptional activity has been proposed for other prokaryotic genes, including cspA from E. coli (6), rbpA1 from A. variabilis M3 (21), and desA and desB from Synechococcus sp. strain PCC 7002 (18) and Synechocystis sp. strain PCC 6803 (14).
Translational inhibitors only marginally mimicked the cold-induced accumulation of crhC transcripts at 30°C. The limited induction may be concentration dependent, as chloramphenicol at concentrations which abolished translation did not elicit crhC transcript accumulation. Protein synthesis, therefore, may be required for a limited amount of crhC transcript accumulation at 30°C. These results suggest that the inhibition of protein synthesis is only one component of the signal required for the induction of crhC transcript accumulation by cold stress in Anabaena. In contrast, translational inhibitors and cold shock induce CSP expression by similar mechanisms in E. coli, as a low concentration of inhibitor mimicked CSP expression to cold shock levels at 37°C (24) while high concentrations of chloramphenicol only marginally induced expression (8). Thus, while similar mechanisms exist for CSP expression induced by cold shock and antibiotic inhibition of translation in E. coli (8), the two mechanisms appear to operate via different mechanisms in Anabaena.
A limited amount of protein synthesis is required for maximal crhC transcript accumulation at 20°C, as increased crhC transcript abundance is observed in the presence of tetracycline but not chloramphenicol at levels which reduce translation by greater than 90%. Cold shock-induced accumulation of crhC transcripts in the presence of chloramphenicol at levels which only partially inhibit translation is also consistent with the requirement for protein synthesis. The requirements for protein synthesis for the cold induction of transcript accumulation differ between the CSP genes. Protein synthesis is required for the accumulation of desA and desB transcripts in Synechococcus sp. strain PCC 7002 (18) but not for desC in Synechococcus sp. strain PCC 7002 (18), rbpA1 in A. variabilis strain M3 (21), or cspA in E. coli (4).
The antibiotic effects are not a result of the level of protein inhibition, as both antibiotics inhibit translation to comparable levels. They may, however, be a reflection of the underlying mechanism regulating crhC transcript accumulation. Tetracycline and chloramphenicol differentially affect translation; tetracycline inhibits initiation and not elongation, while chloramphenicol blocks only elongation (23). Cold-induced crhC transcript accumulation in the presence of tetracycline and not chloramphenicol therefore suggests that a factor whose expression is required for crhC transcript accumulation is transcribed and translated at 30°C but that its gene products are unstable at this temperature. Potential candidates for this factor include a derepressor of transcription, a possibility that is consistent with our preliminary observation of a protein associated with the crhC promoter at 30°C but not 20°C (R. Blush and G. W. Owttrim, unpublished data). Furthermore, the crhC promoter contains sequences (3) known to regulate transcription of cold shock genes in E. coli (5, 15). A model involving derepression of transcription by protein modification of a repressor has been proposed to regulate the cold-induced expression of rbpA1 in A. variabilis strain M3 (21).
Posttranscriptional regulation of crhC expression may also occur at the level of translation, as CrhC protein accumulation does not mimic crhC transcript abundance during prolonged exposure to low temperature. It is also possible that CrhC RNA helicase activity is required for its own translation in that it may unwind the secondary structure in its highly structured 5′ UTR (3), as has been proposed for CsdA in the E. coli cold shock ribosome adaptation model (11, 24).
The results presented here indicate that the cold-induced accumulation of crhC transcripts is tightly regulated by fine changes in growth temperature at the levels of metabolic activity, transcription, mRNA stabilization, and translation. Although the pattern of crhC transcript accumulation is similar to temperature-induced alterations in the levels of abundance of transcripts of other cold shock genes, the mechanisms by which this is accomplished appear to differ. We are currently investigating the factors regulating crhC expression during acclimation to cold stress in Anabaena and the role performed by CrhC in this process.
ACKNOWLEDGMENTS
We are grateful to A. Vioque for providing the Anabaena RNase P gene, E. Yu for generating the anti-CrhC antiserum, and S. Kujat for critical review of the manuscript.
This work was supported by a Natural Sciences and Engineering Research Council of Canada (NSERC) operating grant to G.W.O.
REFERENCES
- 1.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1994. [Google Scholar]
- 2.Brandi A, Pietroni P, Gualerzi C O, Pon C L. Post-transcriptional regulation of CspA expression in Escherichia coli. Mol Microbiol. 1996;19:231–240. doi: 10.1046/j.1365-2958.1996.362897.x. [DOI] [PubMed] [Google Scholar]
- 3.Chamot D, Magee W C, Yu E, Owttrim G W. A cold shock-induced cyanobacterial RNA helicase. J Bacteriol. 1999;181:1728–1732. doi: 10.1128/jb.181.6.1728-1732.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fang L, Jiang W, Bae W, Inouye M. Promoter-independent cold-shock induction of cspA and its derepression at 37°C by mRNA stabilization. Mol Microbiol. 1997;23:355–364. doi: 10.1046/j.1365-2958.1997.2351592.x. [DOI] [PubMed] [Google Scholar]
- 5.Fang L, Hou Y, Inouye M. Role of the cold-box region in the 5′ untranslated region of the cspA mRNA in its transient expression at low temperature in Escherichia coli. J Bacteriol. 1998;180:90–95. doi: 10.1128/jb.180.1.90-95.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goldenberg D, Azar I, Oppenheim A B. Differential mRNA stability of the cspA gene in the cold-shock response of Escherichia coli. Mol Microbiol. 1996;19:241–248. doi: 10.1046/j.1365-2958.1996.363898.x. [DOI] [PubMed] [Google Scholar]
- 7.Graumann P, Marahiel M A. Some like it cold: response of microorganisms to cold shock. Arch Microbiol. 1996;166:293–300. doi: 10.1007/s002030050386. [DOI] [PubMed] [Google Scholar]
- 8.Jiang W, Fang L, Inouye M. The role of the 5′-end untranslated region of the mRNA for CspA, the major cold-shock protein of Escherichia coli, in cold-shock adaptation. J Bacteriol. 1996;178:4919–4925. doi: 10.1128/jb.178.16.4919-4925.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jiang W, Jones P, Inouye M. Chloramphenicol induces the transcription of the major cold shock gene of Escherichia coli, cspA. J Bacteriol. 1993;175:5824–5828. doi: 10.1128/jb.175.18.5824-5828.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jones P G, Inouye M. The cold-shock response—a hot topic. Mol Microbiol. 1994;11:811–818. doi: 10.1111/j.1365-2958.1994.tb00359.x. [DOI] [PubMed] [Google Scholar]
- 11.Jones P G, Inouye M. RbfA, a 30S ribosomal binding factor, is a cold-shock protein whose absence triggers the cold-shock response. Mol Microbiol. 1996;21:1207–1218. doi: 10.1111/j.1365-2958.1996.tb02582.x. [DOI] [PubMed] [Google Scholar]
- 12.Jones P G, Mitta M, Kim Y, Jiang W, Inouye M. Cold shock induces a major ribosomal-associated protein that unwinds double-stranded RNA in Escherichia coli. Proc Natl Acad Sci USA. 1996;93:76–80. doi: 10.1073/pnas.93.1.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jones P G, VanBogelen R A, Neidhardt F C. Induction of proteins in response to low temperature in Escherichia coli. J Bacteriol. 1987;169:2092–2095. doi: 10.1128/jb.169.5.2092-2095.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Los D A, Ray M K, Murata N. Differences in the control of the temperature-dependent expression of four genes for desaturases in Synechocystis sp. PCC 6803. Mol Microbiol. 1997;25:1167–1175. doi: 10.1046/j.1365-2958.1997.5641912.x. [DOI] [PubMed] [Google Scholar]
- 15.Mitta M, Fang L, Inouye M. Deletion analysis of cspA of Escherichia coli: requirement of the AT-rich UP element for cspA transcription and the downstream box in the coding region for its cold shock induction. Mol Microbiol. 1997;26:321–335. doi: 10.1046/j.1365-2958.1997.5771943.x. [DOI] [PubMed] [Google Scholar]
- 16.Myers J. Photosynthetic and respiratory electron transport in a cyanobacterium. Photosynth Res. 1986;9:135–147. doi: 10.1007/BF00029739. [DOI] [PubMed] [Google Scholar]
- 17.Porankiewicz J, Clarke A K. Induction of the heat shock protein ClpB affects cold acclimation in the cyanobacterium Synechococcus sp. strain PCC 7942. J Bacteriol. 1997;179:5111–5117. doi: 10.1128/jb.179.16.5111-5117.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sakamoto T, Bryant D A. Temperature-regulated mRNA accumulation and stabilization for fatty acid desaturase genes in the cyanobacterium Synechococcus sp. strain PCC 7002. Mol Microbiol. 1997;23:1281–1292. doi: 10.1046/j.1365-2958.1997.3071676.x. [DOI] [PubMed] [Google Scholar]
- 19.Samartzidou H, Widger W R. Transcriptional and posttranscriptional control of mRNA from lrtA, a light-repressed transcript in Synechococcus sp. PCC 7002. Plant Physiol. 1998;117:225–234. doi: 10.1104/pp.117.1.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sato N. A family of cold-regulated RNA-binding protein genes in the cyanobacterium Anabaena variabilis M3. Nucleic Acids Res. 1995;23:2161–2167. doi: 10.1093/nar/23.12.2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sato N, Nakamura A. Involvement of the 5′-untranslated region in cold-regulated expression of the rbpA1 gene in the cyanobacterium Anabaena variabilis M3. Nucleic Acids Res. 1998;26:2192–2199. doi: 10.1093/nar/26.9.2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sato N, Tachikawa T, Wada A, Tanaka A. Temperature-dependent regulation of the ribosomal small-subunit protein S21 in the cyanobacterium Anabaena variabilis M3. J Bacteriol. 1997;179:7063–7071. doi: 10.1128/jb.179.22.7063-7071.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Spahn C M T, Prescott C D. Throwing a spanner in the works: antibiotics and the translation apparatus. J Mol Med. 1996;74:423–439. doi: 10.1007/BF00217518. [DOI] [PubMed] [Google Scholar]
- 24.Thieringer H A, Jones P G, Inouye M. Cold shock adaptation. Bioessays. 1998;20:49–57. doi: 10.1002/(SICI)1521-1878(199801)20:1<49::AID-BIES8>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 25.VanBogelen R A, Neidhardt F C. Ribosomes as sensors of heat and cold shock in Escherichia coli. Proc Natl Acad Sci USA. 1990;87:5589–5593. doi: 10.1073/pnas.87.15.5589. [DOI] [PMC free article] [PubMed] [Google Scholar]