Abstract
One of the major proteins secreted by Pseudomonas aeruginosa is a 43-kDa protein, which is cleaved by elastase into smaller fragments, including a 30-kDa and a 23-kDa fragment. The N-terminal 23-kDa fragment was previously suggested as corresponding to a staphylolytic protease and was designated LasD (S. Park and D. R. Galloway, Mol. Microbiol. 16:263–270, 1995). However, the sequence of the gene encoding this 43-kDa protein revealed that the N-terminal half of the protein is homologous to the chitin-binding proteins CHB1 of Streptomyces olivaceoviridis and CBP21 of Serratia marcescens and to the cellulose-binding protein p40 of Streptomyces halstedii. Furthermore, a short C-terminal fragment shows homology to a part of chitinase A of Vibrio harveyi. The full-length 43-kDa protein could bind chitin and was thereby protected against the proteolytic activity of elastase, whereas the degradation products did not bind chitin. The purified 43-kDa chitin-binding protein had no staphylolytic activity, and comparison of the enzymatic activities in the extracellular medium of a wild-type strain and a chitin-binding protein-deficient mutant indicated that the 43-kDa protein supports neither chitinolytic nor staphylolytic activity. We conclude that the 43-kDa protein, which was found to be produced by many clinical isolates of P. aeruginosa, is a chitin-binding protein, and we propose to name it CbpD (chitin-binding protein D).
The opportunistic pathogen Pseudomonas aeruginosa is able to secrete many proteins, including the exoenzymes S, T, and U, exotoxin A, lipase, phospholipase C, the proteases alkaline protease and elastase (LasB), and the staphylolytic proteases LasA and LasD, into the extracellular medium. Most of these proteins contribute to the virulence of the bacteria, as they are associated with epithelial cell and tissue damage or disfunctioning of infected host cells. These proteins are secreted across the bacterial cell envelope by three entirely different mechanisms. Exoenzymes S, T, and U are secreted by a type III secretion system and are actually injected directly into eukaryotic target cells (12, 51). Alkaline protease is secreted by a type I secretion machinery (8). The other proteolytic enzymes mentioned above and exotoxin A are secreted via the type II secretion pathway, encoded by the xcp genes (for a review, see reference 11). The major proteolytic enzyme, elastase, is synthesized as a preproenzyme in the cytosol. During or directly after translocation across the cytoplasmic membrane, the signal sequence is removed, and the proenzyme is folded in a process that requires the propeptide as an intramolecular chaperone (5, 35). After autoproteolytic processing, the propeptide remains noncovalently associated with the mature enzyme and inhibits enzymatic activity in the periplasm (27, 34). The propeptide dissociates from the enzyme only after translocation across the outer membrane (6). LasA, a staphylolytic protease, is synthesized as a preproenzyme of 42 kDa and is processed into a 21-kDa mature protein by either elastase, alkaline protease, or a lysine-specific endopeptidase in the extracellular medium (6, 28).
Elastase is also involved in the extracellular processing of the 43-kDa proform of the LasD protein into a 23-kDa form. However, in this case, the 23-kDa form corresponds to the N-terminal domain and, hence, the putative propeptide is located at the C terminus (6). It has been suggested that the 23-kDa LasD protein is functioning as a staphylolytic protease (38) and that it plays a role in the processing of LasA (39). However, we demonstrate here that the 43-kDa protein is a chitin-binding protein.
MATERIALS AND METHODS
Bacterial strains, growth conditions, plasmids, and DNA manipulations.
Bacterial strains and plasmids used in this study are listed in Table 1. Plasmid isolation and DNA manipulations were performed according to standard procedures (33). Genomic DNA of P. aeruginosa was isolated as described (7). The lasD gene—now called cbpD—was amplified by PCR with SacII-digested genomic DNA of strain PAO25 as a template. The PCR was carried out in the presence of 7% dimethyl sulfoxide with Goldstar polymerase (Eurogentec, Seraing, Belgium) according to the manufacturer's protocol. The primers (Gibco BRL) used were D1 (5′-CGCCGCCTGGAAGAGTTC-3′), which contains an internal EarI site (underlined) and D2 (5′-CAGGCTCTGGTGGACGATG-3′), which hybridize 374 bp upstream of the ATG start codon and 213 bp downstream of the TAA stop codon, respectively. Thirty amplification cycles of 1 min at 95°C for denaturation, 1 min at 51°C for primer annealing, and 2.5 min at 72°C for DNA synthesis were used. After the last cycle, DNA synthesis was prolonged for 10 min at 72°C. To remove 3′ overhanging ends generated by Goldstar polymerase, the PCR product was restricted with EarI and SphI (within the amplified DNA), was made blunt with Klenow polymerase, and was ligated into HincII-linearized pUC19, resulting in plasmid pJF29. To inactivate cbpD, pJF29 was digested with SalI, resulting in the excision of an internal fragment of 57 bp, which was substituted by the kanamycin resistance cassette of pBSL99, creating pJF30. A chromosomal cbpD mutant was constructed via insertional inactivation by using the special cloning vector pKNG101 (25), which requires the pir gene product for replication. For this purpose, an EcoRI/SphI fragment of pJF30, carrying the cbpD gene with the inserted kanamycin resistance cassette was made blunt with T4 DNA polymerase and was cloned into the SmaI site of pKNG101. The resulting plasmid, pJF31, was maintained in strain CC118(λpir). Plasmid pJF31 was introduced into PAO25 by triparental mating by using the conjugative helper plasmid pRK2013 as described (25). Single crossover transconjugants were selected on King's B medium (KB) (29) containing streptomycin, kanamycin, and nalidixic acid. These transconjugants were restreaked on KB plates containing nalidixic acid, kanamycin, and 5% (wt/vol) sucrose and were incubated overnight once at 37°C and once at room temperature, in order to select double crossover mutants. The cbpD mutation of the isolated mutant PAN17 was confirmed by PCR with primers D1 and a primer internal in the kanamycin resistance cassette (5′-GCCCTGAGTGCTTGCGGCA-3′), which yielded the expected 1,310-bp fragment in the case of the cbpD mutant and no fragment in the case of the wild-type strain (data not shown).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristicsa | Source or reference |
|---|---|---|
| E. coli strains | ||
| DH5α | F− Δ(lacZYA-algF)U169 thi-1 hsdR17 gyrA96 recA1 endA1 supE44 relA1 phoA φ80 dlacZΔM15 | 20 |
| CC118(λpir) | Δ(ara-leu) araD ΔlacZ74 galE galK phoA20 thi-1 rpsE rpoB argE(Am) recA1, λpir | 21 |
| P. aeruginosa strains | ||
| 7NSK2 | Wild type | 22 |
| PAK | Wild type | 4 |
| PAO1 | Wild type | 23 |
| PAO25 | PAO1 leu arg | 18 |
| PAN1 | PAO25 xcpQ::Gmr | 3 |
| PAN8 | PAO25 lasB::KmraprE | 6 |
| PAN9 | PAO25 xcpQ::GmrlasB::KmraprE::Hgr | 6 |
| PAN10 | PAO25 lasB::Kmr | 6 |
| PAN17 | PAO25 cbpD::Kmr | This study |
| PAN101-124 | Clinical isolates of P. aeruginosa | This study |
| P. fluorescens strains | ||
| WCS374 | Wild type | 16 |
| WCS417 | Wild type | 31 |
| En401 | Wild type | 48 |
| RS111 | Wild type | 48 |
| P. putida WCS358 | Wild type | 16 |
| S. aureus | Wild type | Laboratory strain |
| Plasmids | ||
| pBSL99 | AmpR, KmR, kanamycin-resistance cassette | 1 |
| pKNG101 | SmR, oriR6K, mobRK2, sacBR | 25 |
| pMMB67EH | AmpR, RSF replicon (IncQ), tac promoter | 14 |
| pRK2013 | KmR, Tra+, Mob+ | 10 |
| pUC18 | AmpR, ColE1, φ80dlacZ, lac promoter | 35 |
| pUC19 | AmpR, ColE1, φ80dlacZ, lac promoter | 52 |
| pJF29 | AmpR, cbpD gene in pUC19 opposite orientation relative to Plac | This study |
| pJF30 | AmpR, KmR kanamycin-resistance cassette insertion in cbpD of pJF29 | This study |
| pJF31 | KmR, SmR, cbpD::Kmr in pJF30 in pKNG101 | This study |
| pJF38 | AmpR, cbpD gene in pMMB67EH | This study |
Abbreviations: Amp, ampicillin; Gm, gentamycin; Hg, mercury; Km, kanamycin; Sm, streptomycin.
To be able to express cbpD from a plasmid in P. aeruginosa, the cbpD gene was excised from pJF29 with EcoRI and HindIII and was recloned in the broad-host-range vector pMMB67EH under control of the tac promoter, resulting in pJF38.
Unless stated otherwise, all strains were grown in Luria-Bertani (LB) broth at 37°C. The antibiotic concentrations used for selection and for maintenance of plasmids were 100 μg of ampicillin per ml, 25 μg of kanamycin per ml, and 100 μg of streptomycin per ml for Escherichia coli and 20 μg of kanamycin per ml and 20 μg of nalidixic acid per ml for P. aeruginosa.
Computer programs.
Sequence analysis was performed with DNasis V2.1 (Hitachi Software Engineering Co., Ltd.). Database searches were performed with the BLAST 2.0 service from the National Centre for Biotechnology Information World Wide Web server. Amino acid sequence alignments were carried out with AlignPlus version 3.0 (Scientific & Educational Software) and were optimized by hand.
Polysaccharide-binding assay.
Colloidal chitin was prepared from crab shell chitin (Sigma) as described (43). The chitin was freeze-dried and suspended in water to a concentration of 60 mg/ml. The suspension was homogenized by ultrasonication and was sterilized for 15 min at 120°C. Colloidal chitin was used at a final concentration of 0.15% (wt/vol) in binding assays. Cell-free supernatants were prepared by centrifugation of 1.5 ml of culture for 3 min at 4,000 × g followed by 3 in at 20,000 × g. Chitin was added to the supernatant, followed by incubation at room temperature for 30 min. Chitin was bound proteins was pelleted by centrifugation at 20,000 × g for 2 min and was washed with a physiological salt solution. Unbound proteins in the cell-free supernatant were precipitated with 5% (wt/vol) trichloroacetic acid. Binding of proteins in the cell-free culture supernatant to Avicel (Fluka), lichenan (Sigma), and xylan (Sigma) was examined similarly.
Alternatively, the binding of proteins to chitin was studied during growth. After overnight growth in 0.15% (wt/vol) chitin-containing medium, the cultures were transferred to 15-ml tubes. The chitin was precipitated by gravity, and the cell suspension was decanted. The chitin with bound proteins was washed several times with physiological salt solution, until it was cell free. The chitin-bound proteins were released by boiling for 10 min in sample buffer (30), which contains 10% (wt/vol) β-mercaptoethanol and 2% (wt/vol) sodium dodecyl sulfate (SDS), prior to electrophoresis on an SDS–11% polyacrylamide gel (32).
Purification of CbpD.
Fifteen milligrams of colloidal chitin were added to 250 ml of cell-free culture supernatant of an overnight-grown culture of PAN10 (lasB) or PAN8 (lasB aprE). The suspension was incubated with agitation for 30 to 60 min at room temperature. The colloidal chitin was collected by centrifugation (10 min, 6,000 × g), was washed twice with 0.9% NaCl solution, and was incubated for 5 min in 250 μl of 0.05 M HCl. After pelleting the chitin by centrifugation, the clear supernatant was transferred directly into a tube containing 500 μl of 0.1 M Tris-HCl, pH 8.0. The CbpD protein released from chitin (about 50%) was in the native conformation, since it could rebind chitin (data not shown). The purified protein was used to raise a polyclonal rabbit antiserum and was used in the staphylolytic assay.
Chitinolytic and staphylolytic assays.
Chitinase activity was detected by streaking colonies on an LB plate containing 0.05% (wt/vol) colloidal chitin. The plates were incubated at 37°C until halos were visible around the colonies. A colorimetric chitinase assay was adapted from that described by Wirth and Wolf (50). Carboxymethyl-chitin-Remazol brilliant violet aqueous solution (Blue Substrates, Göttingen, Germany) (250 μl, 2 mg/ml) was added to a mixture of 250 μl of cell-free supernatant and 250 μl of 0.1 M sodium acetate, pH 5.2. After 1 h of incubation at 37°C, the reaction was terminated by the addition of 250 μl of 1 M HCl. The reaction tubes were cooled on ice for 10 min and were centrifuged for 10 min at 20,000 × g. The absorbance of the supernatant was measured at 550 nm.
Staphylolytic activity was determined as previously described (38) with a few adaptations. Staphylococcus aureus cells were resuspended in 25 mM diethanolamine buffer, pH 9.3, and were heat killed by incubation for 10 min at 100°C. The cells were diluted with the same buffer to a turbidity of 3.0 to 4.0 at 595 nm. Cell-free culture supernatants of P. aeruginosa strains were incubated with the S. aureus cell suspension at a 1:1 ratio, and the turbidity was determined at 595 nm over a period of 3 h at 30-min intervals. Purified CbpD was added to fresh LB and was incubated with the S. aureus cell suspension.
Fungal-growth-inhibition assay.
Spots of 5 μl from overnight cultures of P. aeruginosa were inoculated on a KB plate supplemented with 200 μM FeCl3. After 2 days of incubation at 30°C, a plug of either Rhizoctonia solani or Fusarium oxisporum was inoculated at the center of the plate. The plates were incubated at room temperature, and inhibition of fungal growth around the bacteria was monitored for 1 week.
Nucleotide sequence accession number.
The nucleotide sequence of cbpD has been submitted to GenBank under accession no. AF196565.
RESULTS
Homology of the putative staphylolytic enzyme LasD to chitin- and cellulose-binding proteins.
The N-terminal 10 amino acid residues of the 23-kDa mature form of LasD and the 43-kDa proform of the protein have been shown to be identical (6). This N-terminal sequence, HGSMETPPSR, was used as a probe to search for the complete amino acid sequence of LasD in the almost-completed genome bank of P. aeruginosa. A single protein with an exact match, consisting of 389 amino acid residues, was found. This protein has a putative signal peptide of 25 amino acid residues followed by a 364-residue large domain, of which the first 10 amino acids exactly matched the probe sequence (Fig. 1A). The N-terminal 15-amino-acid sequence of LasD determined by Park and Galloway (38) was found to be identical to the first 15 residues of the 364-residue large domain.
FIG. 1.
Alignment of the amino acid sequence of the putative LasD with (A) CHB1 of S. olivaceoviridis (accession no. X78535), CBP21 of S. marcescens (accession no. AB015998), and p40 of S. halstedii (accession no. U51222) and (B) a C-terminal part of ChiA of V. harveyi (U81496), which is a chitin-binding domain. Amino acids identical to LasD are boxed, the arrowhead indicates amino acid residue +1 of LasD. The amino acid sequence of the putative LasD is deduced from the nucleotide sequence released by the Pseudomonas Genome Project. After sequencing the putative lasD gene, we found a 1-bp difference, resulting in the replacement of amino acid residue Gln 305 by Arg.
The full-length amino acid sequence of this open reading frame was used to screen databases for related proteins, resulting in the identification of three proteins with significant homology to the N-terminal part of LasD, i.e., p40 of Streptomyces halstedii (15), CHB1 of Streptomyces olivaceoviridis (45), and CBP21 of Serratia marcescens (46) (Fig. 1A). p40 is a cellulose-binding protein of 40 kDa, whereas CHB1 and CBP21 are both chitin-binding proteins of approximately 20 kDa with high affinities for α- and β-chitin, respectively. Neither of these proteins has a clear hydrolytic activity (45, 46). The extreme C terminus shows homology to the chitin-binding domain of chitinase A (ChiA) of Vibrio harveyi (47) (Fig. 1B), especially with respect to the position of aromatic residues, which are considered to be important for polysaccharide binding (45, 47).
Putative LasD is a chitin-binding protein.
Since the putative LasD is homologous to polysaccharide-binding proteins rather than to proteases, we decided to test whether the protein can actually bind polysaccharides. Cell-free culture supernatants of the wild-type strain PAO25 and of the lasB mutant strain PAN10, which has been shown to be defective in the putative processing of the 43-kDa form of supposed LasD (6), were incubated with crystalline cellulose (Avicel), colloidal chitin, xylan, or lichenan. The 43-kDa form in the supernatant of PAN10 was found to bind specifically to colloidal chitin and not to any of the other substrates tested (Fig. 2). In the supernatant of strain PAO25, small amounts of the chitin-binding 43-kDa form were occasionally detected, depending on the growth incubation period (results not shown), but no chitin-binding 23-kDa form was observed. Since the putative LasD protein is homologous to polysaccharide-binding proteins and could indeed bind the chitin polysaccharide, we propose to rename the putative LasD chitin-binding protein D (CbpD).
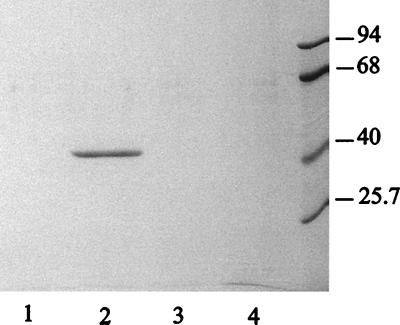
FIG. 2.
Characterization of polysaccharide-binding proteins in P. aeruginosa culture supernatants. Extracellular proteins of lasB mutant PAN10 were incubated with various polysaccharides, and proteins bound to Avicel (lane 1), colloidal chitin (lane 2), lichenan (lane 3), or xylan (lane 4) were analyzed by SDS-PAGE. Molecular mass marker proteins (in kDa) are shown at the right.
To study the fate of the 43-kDa form in wild-type culture supernatants, antibodies were raised against the chitin-binding protein. An immunoblot of PAO25 culture supernatant revealed a variety of degradation products of CbpD (Fig. 3, lane 2). The major degradation product had an apparent molecular mass of 30 kDa, and one of the minor degradation products migrated at the position of 23 kDa. Apparently, neither of these degradation products bound chitin (Fig. 3, lane 4). After prolonged growth (>24 h) of PAO25, no full-length CbpD or degradation products could be detected on immunoblots (data not shown), indicating that CbpD is not only processed, but is eventually totally degraded by elastase.
FIG. 3.
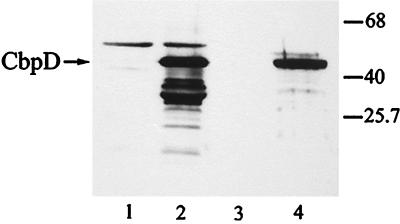
Immunoblot containing culture supernatant proteins of the cbpD mutant strain PAN17 (lanes 1 and 3) and PAO25 (lanes 2 and 4), incubated with antibodies directed against the 43-kDa chitin-binding protein. Strains were grown overnight in LB. Equal amounts of cell-free supernatant proteins (lanes 1 and 2) and chitin-bound proteins (lanes 3 and 4) were loaded. In this experiment, a background band was observed in the supernatant of both strains with a mobility slightly lower than that of CbpD on SDS-PAGE. Molecular mass markers (in kDa) are indicated at the right.
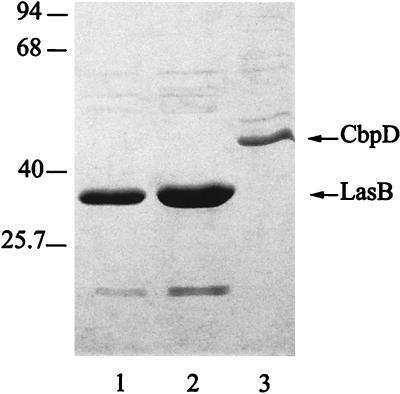
Since the 43-kDa form of CbpD binds chitin, we considered the possibility that this form could be protected from the proteolytic activity of elastase by chitin. To test this possibility, strain PAO25 was grown for 20 h in the presence or absence of colloidal chitin. Subsequently, the proteins in the supernatant of both cultures and the chitin-bound proteins were analyzed by SDS-polyacrylamide gel electrophoresis (PAGE) (Fig. 4). The supernatant of the strain grown in the absence of chitin did not contain the 43-kDa chitin-binding protein (lane 1). In contrast, in the colloidal chitin-supplemented culture, the 43-kDa form of CbpD was present in large amounts as a chitin-bound protein (lane 3). Apparently, by binding chitin, the 43-kDa form of the CbpD protein is protected from proteolytic activity of elastase.
FIG. 4.
Cultures of PAO25 were grown for 20 h in the absence (lane 1) or presence (lanes 2 and 3) of colloidal chitin. Cell-free supernatants were directly analyzed by SDS-PAGE (lane 1) or after separation of the proteins that had not (lane 2) or had (lane 3) bound to chitin. Molecular mass markers (in kDa) are shown on the left.
Possible functions of CbpD.
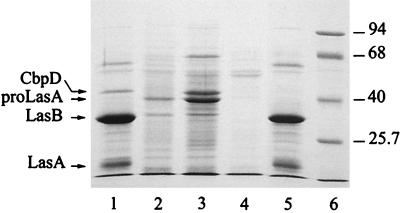
To gain further insight into the possible function of CbpD, the cbpD mutant strain PAN17 was constructed. This mutant did not produce the 43-kDa chitin-binding protein (Fig. 5, lane 5), a defect that was complemented by introduction of the cbpD-containing plasmid pJF38 (results not shown). This result demonstrates that the disrupted gene indeed encodes the 43-kDa chitin-binding protein. To test whether the chitin-binding protein plays a role in the hydrolysis of colloidal chitin, the wild-type strain PAO25 and PAN17 were streaked on an LB plate containing colloidal chitin. After 5 days, a halo was formed by the wild-type strain, indicating the presence of a chitinolytic enzyme (results not shown). However, the cbpD mutant formed a halo of similar size, indicating that CbpD is not responsible for the observed chitinolytic activity. The supernatants of both strains were also subjected to a colorimetric assay by using the soluble substrate CM-chitin-RBV, but no difference in chitinolytic activity was observed (data not shown). The results of these assays are in agreement with the fact that CbpD shows homology only with polysaccharide-binding proteins and with the chitin-binding domain of a chitinase, but not with the catalytic domain of chitin-hydrolyzing enzymes.
FIG. 5.
Culture supernatant protein profiles of wild-type P. aeruginosa PAO25 (lane 1), xcpQ mutant PAN1 (lane 2), aprE lasB mutant PAN8 (lane 3), lasB aprE xcpQ mutant PAN9 (lane 4), and cbpD mutant PAN17 (lane 5) analyzed by SDS-PAGE. The positions of CbpD, the unprocessed LasA (proLasA), elastase (LasB), and LasA are indicated at the left, and molecular mass standard proteins (in kDa; lane 6) are indicated at the right.
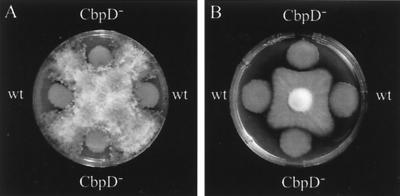
Proteins consisting of only chitin-binding domains but without chitinolytic activity can still inhibit fungal growth (2, 35, 40). Strain PAO25 was able to inhibit the growth of R. solani and F. oxysporum in a plate assay (Fig. 6). However, the cbpD mutant PAN17 inhibited the growth of these fungi to a similar extent.
FIG. 6.
Plugs of R. solani (A) or F. oxysporum (B) were placed on a KB plate on which cultures of strain PAO25 (wt) and the chitin-binding-protein-deficient strain PAN17 (CbpD−) were spotted. Inhibition of fungal growth around the bacterial spots was recorded after 1 week.
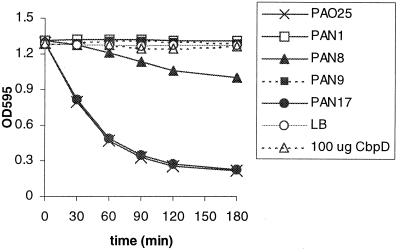
Park and Galloway (38) have purified a 23-kDa protein with the same N-terminal sequence as the chitin-binding protein, but this enzyme showed staphylolytic activity. The pH optimum of this enzyme was reported to be 9.3 (38). To test whether the chitin-binding 43-kDa protein has staphylolytic activity, 100 μg of purified CbpD was incubated with S. aureus cells. After 3 h of incubation, no significant decrease in optical density was observed (Fig. 7). Even incubation times up to 22 h did not reveal any significant staphylolytic activity for the purified CbpD.
FIG. 7.
Staphylolytic activity of purified CbpD and culture supernatants of various P. aeruginosa strains (indicated in the inset), measured by the decline in optical density of S. aureus cells. LB medium is used as negative control.
To test whether the CbpD protein or the breakdown products in the culture supernatant contribute to the total staphylolytic activity of the cells, supernatant fractions of several P. aeruginosa strains were tested for their staphylolytic activities. The staphylolytic activity in the supernatant fraction of the cbpD mutant was similar to that of the wild-type strain (Fig. 7), demonstrating that the chitin-binding protein does not contribute to the staphylolytic activity. Significant activity was detected in the supernatant of the lasB aprE mutant strain PAN8, albeit much less than in the wild-type strain (Fig. 7). Since in this mutant strain the inactive 41-kDa proform of LasA accumulates (6) (Fig. 5, lane 3), the observed activity might be derived from another staphylolytic protease. The presence of another staphylolytic enzyme is consistent with the report of Park and Galloway (38), but this putative enzyme is not the chitin-binding protein. This second staphylolytic enzyme is secreted via the type II secretion pathway, since no staphylolytic activity at all was observed in the supernatant of lasB aprE xcpQ triple-mutant strain PAN9 or xcpQ mutant strain PAN1 (Fig. 7), both of which are deficient in the secretion of proteins via the type II pathway (Fig. 5, lanes 2 and 4). Alternatively, the staphylolytic activity observed in the supernatant of strain PAN8 might be due to limited processing of proLasA.
On the basis of experiments with purified protein, Park and Galloway (39) also suggested that the 23-kDa form of LasD is involved in the processing of proLasA. However, we did not observe the accumulation of the 41-kDa proform of LasA protein in the supernatant of the cbpD mutant strain PAN17 (Fig. 5, lane 5), consistent with our recent observation that elastase and alkaline protease are responsible for the processing of proLasA into the 21-kDa form (6) (Fig. 5, lane 3).
Distribution of chitin-binding proteins among pseudomonads.
Elastase, which is secreted via the type II secretion pathway, is a virulence factor widely distributed among clinical isolates of P. aeruginosa (19). To gain insight into the distribution of CbpD among pseudomonads, several Pseudomonas strains were grown in the presence of chitin, and the chitin-bound proteins were analyzed by SDS-PAGE. At least 17 out of 23 clinical isolates of P. aeruginosa tested, as well as the laboratory strains PAK and PAO1, produced a 43-kDa protein that bound chitin (results not shown). With one exception, the secretion of the chitin-binding protein was accompanied by the secretion of elastase. No chitin-binding protein was detected in the cases of the plant-growth-promoting P. aeruginosa 7NSK2, four strains of Pseudomonas fluorescens, and a Pseudomonas putida strain (results not shown).
DISCUSSION
One of the major proteins secreted by P. aeruginosa is a 43-kDa protein, which is secreted via the type II secretion pathway (6). The N-terminal 10 amino acids of this protein were reported to be identical to those of the 23-kDa staphylolytic protease LasD (6, 38). In this paper, we show that the secreted 43-kDa protein consists of two domains, which both show homology to polysaccharide-binding proteins, and that the full-length mature protein is a chitin-binding protein. This form can be detected in large amounts in the cell-free supernatant of a P. aeruginosa lasB mutant strain. Elastase digests this protein in the supernatant of the wild-type strain, resulting in the appearance of several fragments, including a 30-kDa and a 23-kDa form. Since these fragments do not bind colloidal chitin, the intact 43-kDa protein is probably the functional form, which is degraded rather than processed by elastase. This degradation process probably does not take place under natural conditions, since the binding of the 43-kDa protein to its substrate, chitin, protects it from proteolysis.
The reported N-terminal sequence identity between the 23-kDa staphylolytic enzyme LasD and the 43-kDa chitin-binding protein could be explained by assuming (i) that there are two proteins secreted by P. aeruginosa with exactly the same N-terminal 15 amino acid residues, (ii) a degradation product of the chitin-binding protein shows staphylolytic activity, or (iii) the isolated staphylolytic enzyme was contaminated with the 23-kDa degradation product of the chitin-binding protein. The first option is probably not correct, since the virtually completed P. aeruginosa gene bank revealed only one perfect match with the N-terminal sequence of the chitin-binding protein. The second option is unlikely as well, since the N-terminal domain shows the highest sequence similarity to polysaccharide-binding proteins and is therefore probably involved in this function. Furthermore, this N-terminal domain shows no homology to proteases, and the staphylolytic activity in the supernatant of the chitin-binding-protein-deficient strain was identical to that of the wild-type strain. Therefore, we conclude that the third option is the only reasonable one, and that the N terminus of the isolated LasD protein (38) was probably blocked for protein sequencing, thus yielding the sequence only of the contaminant present in the protein preparation. We showed that the purified 43-kDa protein bound chitin and did not have staphylolytic activity. Hence we propose to name the new protein CbpD (chitin-binding protein D) and preserve LasD for the 23-kDa staphylolytic protease with a pH optimum of 9.3, as described by Park and Galloway (38). The existence of a second staphylolytic enzyme was suggested by the presence of staphylolytic activity in the culture supernatant of a lasB aprE double-mutant strain, which is defective in the processing of proLasA in the staphylolytic enzyme LasA. In a homology search in the P. aeruginosa genome bank, we found two new genes coding for proteins with homology to the staphylolytic proteins LasA and lysostaphin of Staphylococcus simulans (42) (data not shown). In mutual comparisons, the similarity between the various proteins was rather low (15 to 22% identical residues), but it was very high in the region in which the active site of LasA has been localized (17). One of these open reading frames could encode the second staphylolytic enzyme. However, it should be noted that no residual staphylolytic activity has been reported to be present in the culture supernatant of a lasA mutant (26). Therefore, the residual activity detected in the supernatant of the lasB aprE mutant could be the result of elastase- and alkaline protease-independent processing of proLasA, rather than the result of a second staphylolytic enzyme.
As described for the chitin-binding proteins CHB1 and CBP21 (45, 46), no chitinolytic activity could be detected for CbpD. Therefore, the physiological function of this major secreted protein of P. aeruginosa is not clear. The production of the type II secretion system of P. aeruginosa, by which CbpD is secreted, is under control of a quorum-sensing system, which indicates that CbpD is only needed under high-density conditions, such as in biofilms and during pathogenesis.
A search for CbpD in different Pseudomonas spp. demonstrated that it is present in many clinical isolates of P. aeruginosa and not in the soil pseudomonads tested. These data suggest a role for CbpD in pathogenicity, e.g., as an adhesin, mediating colonization of eukaryotic cells. The CbpD protein is secreted by the Xcp machinery into the extracellular medium, a property not directly expected of an adhesin. However, a proportion of the molecules might stick to the outer membrane of the bacteria and mediate attachment to chitin-containing substrates. Bordetella pertussis and Vibrio cholerae have been shown to secrete hemagglutinins, which are involved in the adherence of bacteria to epithelial cells. The filamentous hemagglutinin of B. pertussis and pertussis toxin contain carbohydrate recognition domains critical for binding to glycoconjugates on cell membranes (41, 49). One of the hemagglutinins of V. cholerae has been reported to be specific for N-acetyl-d-glucosamine, the monomeric subunit of chitin, and mediates binding of the bacterial cells to rabbit intestinal epithelial cells (44). Furthermore, it has been shown that Klebsiella pneumoniae is internalized efficiently by cultured human epithelial cells, presumably via an N-glycosylated receptor containing N-acetyl-glucosamine residues (13). Speculatively, a glycosylated epithelial cell surface molecule might exist as receptor for CbpD.
ACKNOWLEDGMENTS
We thank André Fleer (Wilhelmina Child Hospital, Utrecht, The Netherlands) for providing the clinical strains and the Pseudomonas Genome Project (http://www.pseudomonas.com) for releasing sequence information important for this study.
This study was supported by the Netherlands Technology Foundation (STW).
REFERENCES
- 1.Alexeyev M F, Shokolenko J N, Croughan T P. Improved antibiotic-resistance gene cassette and omega elements for Escherichia coli vector construction and in vitro deletin/insertion mutagenesis. Gene. 1995;160:63–67. doi: 10.1016/0378-1119(95)00108-i. [DOI] [PubMed] [Google Scholar]
- 2.Beyer M, Diekmann H. The chitinase system of Streptomyces sp. ATCC 11238 and its significance for fungal cell wall degradation. Appl Microbiol Biotechnol. 1985;23:140–146. [Google Scholar]
- 3.Bitter W, Koster M, Latijnhouwers M, de Cock H, Tommassen J. Formation of oligomeric rings by XcpQ and PilQ, which are involved in protein transport across the outer membrane of Pseudomonas aeruginosa. Mol Microbiol. 1998;27:209–219. doi: 10.1046/j.1365-2958.1998.00677.x. [DOI] [PubMed] [Google Scholar]
- 4.Bradley D E. The adsorption of Pseudomonas aeruginosa pilus-dependent bacteriophages to a host mutant with non-retractile pili. Virology. 1974;58:149–163. doi: 10.1016/0042-6822(74)90150-0. [DOI] [PubMed] [Google Scholar]
- 5.Braun P, Tommassen J, Filloux A. Role of the propeptide in folding and secretion of elastase of Pseudomonas aeruginosa. Mol Microbiol. 1996;19:297–306. doi: 10.1046/j.1365-2958.1996.381908.x. [DOI] [PubMed] [Google Scholar]
- 6.Braun P, de Groot A, Bitter W, Tommassen J. Secretion of elastinolytic enzymes and their propeptides by Pseudomonas aeruginosa. J Bacteriol. 1998;180:3467–3469. doi: 10.1128/jb.180.13.3467-3469.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen W, Kuo T-T. A simple and rapid method for the preparation of gram-negative bacterial genomic DNA. Nucleic Acids Res. 1993;21:2260. doi: 10.1093/nar/21.9.2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Duong F, Lazdunski A, Cami B, Murgier M. Seqeunce of a cluster of genes controlling synthesis and secretion of alkaline protease in Pseudomonas aeruginosa: relationships to other secretion pathways. Gene. 1992;121:47–54. doi: 10.1016/0378-1119(92)90160-q. [DOI] [PubMed] [Google Scholar]
- 9.Elliot B W, Jr, Cohen C. Isolation and characterization of a lysine-specific protease of Pseudomonas aeruginosa. J Biol Chem. 1986;261:11259–11265. [PubMed] [Google Scholar]
- 10.Figurski D H, Helinski D R. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc Natl Acad Sci USA. 1979;76:1648–1652. doi: 10.1073/pnas.76.4.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Filloux A, Michel G, Bally M. GSP-dependent protein secretion in gram-negative bacteria: the Xcp system of Pseudomonas aeruginosa. FEMS Microbiol Rev. 1998;22:177–198. doi: 10.1111/j.1574-6976.1998.tb00366.x. [DOI] [PubMed] [Google Scholar]
- 12.Finck-Barbançon V, Goranson J, Zhu L, Sawa T, Wiener-Kronisch J P, Fleiszig S M J, Wu C, Mende-Mueller L, Frank D W. ExoU expression by Pseudomonas aeruginosa correlates with acute cytotoxicity and epithelial injury. Mol Microbiol. 1997;25:547–557. doi: 10.1046/j.1365-2958.1997.4891851.x. [DOI] [PubMed] [Google Scholar]
- 13.Fumangalli O, Tall B D, Schipper C, Oelschlaeger T A. N-glycosylated proteins are involved in efficient internalization of Klebsiella pneumoniae by cultured human epithelial cells. Infect Immun. 1997;65:4445–4451. doi: 10.1128/iai.65.11.4445-4451.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fürste J P, Pansegrau W, Frank R, Blöcker H, Scholz P, Bagdasarian M, Lanka E. Molecular cloning of the plasmid RP4 primase region in a multi-host-range tacP expression vector. Gene. 1986;48:119–131. doi: 10.1016/0378-1119(86)90358-6. [DOI] [PubMed] [Google Scholar]
- 15.Garda A L, Fernandez-Abalos J M, Sanchez P, Ruiz-Arribas A, Santamaria R I. Two genes encoding an endoglucanase and a cellulose-binding protein are clustered and co-regulated by a TTA codon in Streptomyces halstedii JM8. Biochem J. 1997;324:403–411. doi: 10.1042/bj3240403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Geels F P, Schippers B. Reduction of yield depressions in high frequency potato cropping soil after seed tuber treatments with antagonistic fluorescent Pseudomonas spp. Phytopathol Z. 1983;108:207–214. [Google Scholar]
- 17.Gustin J K, Kessler E, Ohman D E. A substitution at His-120 in the LasA protease of Pseudomonas aeruginosa blocks enzymatic activity without affecting propeptide processing or extracellular secretion. J Bacteriol. 1996;178:6608–6617. doi: 10.1128/jb.178.22.6608-6617.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haas D, Holloway B W. R factor variants with enhanced sex factor activity in Pseudomonas aeruginosa. Mol Gen Genet. 1976;144:243–251. doi: 10.1007/BF00341722. [DOI] [PubMed] [Google Scholar]
- 19.Hamood A N, Griswold J. DNA hybridization analysis of the Pseudomonas aeruginosa elastase gene (lasB) from different clinical isolates. Can J Microbiol. 1995;41:910–917. doi: 10.1139/m95-125. [DOI] [PubMed] [Google Scholar]
- 20.Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 21.Herrero M, de Lorenzo V, Timmis K N. Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in gram-negative bacteria. J Bacteriol. 1990;172:6557–6567. doi: 10.1128/jb.172.11.6557-6567.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Höfte M, Mergeay M, Verstraete W. Marking rhizopseudomonas strain 7NSK2 with Mu d(lac) element for ecological studies. Appl Environ Microbiol. 1990;56:1046–1052. doi: 10.1128/aem.56.4.1046-1052.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Holloway B W. Genetics of Pseudomonads. Bacteriol Rev. 1969;33:419–443. doi: 10.1128/br.33.3.419-443.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jones J D G, Grady K L, Suslow T V, Bedbrook J R. Isolation and characterization of genes encoding two chitinase enzymes from Serratia marcescens. EMBO J. 1986;5:467–473. doi: 10.1002/j.1460-2075.1986.tb04235.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kaniga K, Delor I, Cornelis G R. A wide-host-range suicide vector for improving reverse genetics in gram-negative bacteria: inactivation of the blaA gene of Yersinia enterocolitica. Gene. 1991;109:137–141. doi: 10.1016/0378-1119(91)90599-7. [DOI] [PubMed] [Google Scholar]
- 26.Kessler E, Safrin M, Olson J C, Ohman D E. Secreted LasA of Pseudomonas aeruginosa is a staphylolytic protease. J Biol Chem. 1993;268:7503–7508. [PubMed] [Google Scholar]
- 27.Kessler E, Safrin M. The propeptide of Pseudomonas aeruginosa elastase acts as an elastase inhibitor. J Biol Chem. 1994;269:22726–22731. [PubMed] [Google Scholar]
- 28.Kessler E, Safrin M, Gustin J K, Ohman D E. Elastase and the LasA protease of Pseudomonas aeruginosa are secreted with their propeptides. J Biol Chem. 1998;273:30225–30231. doi: 10.1074/jbc.273.46.30225. [DOI] [PubMed] [Google Scholar]
- 29.King E O, Ward M K, Raney D E. Two simple media for the demonstration of pyocyanin and fluorescein. J Lab Clin Med. 1954;44:301–307. [PubMed] [Google Scholar]
- 30.Laemmli U K. Cleavage of structural proteins during the assembly of the head of the bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 31.Lamers J G, Schippers B, Geels F P. Soil-borne disease of wheat in the Netherlands and seed bacterization with pseudomonads against Gaeumannomyces graminis var. tritici. In: Jorna M L, Slootmaker L A J, editors. Cereal breeding related to integrated cereal production. Wageningen, The Netherlands: Pudoc; 1988. p. 244. [Google Scholar]
- 32.Lugtenberg B, Meijers J, Peters R, van der Hoek P, van Alphen L. Electrophoretic resolution of the “major outer membrane protein” of Escherichia coli K12 into four bands. FEBS Lett. 1975;58:254–258. doi: 10.1016/0014-5793(75)80272-9. [DOI] [PubMed] [Google Scholar]
- 33.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1982. [Google Scholar]
- 34.McIver K, Kessler E, Ohman D E. Substitution of active-site His-223 in Pseudomonas aeruginosa elastase and expression of mutated lasB alleles in Escherichia coli show evidence for autoproteolytic processing of proelastase. J Bacteriol. 1991;173:7781–7789. doi: 10.1128/jb.173.24.7781-7789.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McIver K S, Kessler E, Olsen J C, Ohman D E. The elastase propeptide functions as an intramolecular chaperone required for the elastase activity and secretion in Pseudomonas aeruginosa. Mol Microbiol. 1995;18:877–889. doi: 10.1111/j.1365-2958.1995.18050877.x. [DOI] [PubMed] [Google Scholar]
- 36.Nielsen K K, Nielsen J E, Madrid S M, Mikkelsen J D. Characterization of a new antifungal chitin-binding peptide from sugar beet leaves. Plant Physiol. 1997;113:83–91. doi: 10.1104/pp.113.1.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Norrander J, Kempe T, Messing J. Construction of improved M13 vectors using oligodeoxynucleotide-directed mutagenesis. Gene. 1983;26:101–106. doi: 10.1016/0378-1119(83)90040-9. [DOI] [PubMed] [Google Scholar]
- 38.Park S, Galloway D R. Purification and characterization of LasD: a second staphylolytic proteinase produced by Pseudomonas aeruginosa. Mol Microbiol. 1995;16:263–270. doi: 10.1111/j.1365-2958.1995.tb02298.x. [DOI] [PubMed] [Google Scholar]
- 39.Park S, Galloway D R. Pseudomonas aeruginosa LasD processes the inactive LasA precursor to the active protease form. Arch Biochem Biophys. 1998;357:8–12. doi: 10.1006/abbi.1998.0787. [DOI] [PubMed] [Google Scholar]
- 40.Ponstein A S, Bres-Vloemans S A, Sela-Buurlage M B, van den Elzen P J, Melchers L S, Cornelissen B J. A novel pathogen- and wound-inducible tobacco (Nicotiana tabacum) protein with antifungal activity. Plant Physiol. 1994;104:109–118. doi: 10.1104/pp.104.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Prasad S M, Yin Y, Rodzinski E, Tuomanen E I, Masure H R. Identification of a carbohydrate recognition domain in filamentous hemagglutinin from Bordetella pertussis. Infect Immun. 1993;61:2780–2785. doi: 10.1128/iai.61.7.2780-2785.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Recsei P A, Gruss A D, Novick R P. Cloning, sequence, and expression of the lysostaphin gene from Staphylococcus simulans. Proc Natl Acad Sci USA. 1987;84:1127–1131. doi: 10.1073/pnas.84.5.1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Roberts W K, Selitrennikoff C P. Plant and bacterial chitinases differ in antifungal activity. J Gen Microbiol. 1988;134:169–176. [Google Scholar]
- 44.Sasmal D, Guhathakurta B, Ghosh A N, Pal C R, Datta A. N-acetyl-d-glucosamine-specific lectin purified from Vibrio cholerae 01. FEMS Microbiol Lett. 1992;98:217–224. doi: 10.1016/0378-1097(92)90159-l. [DOI] [PubMed] [Google Scholar]
- 45.Schnellmann J, Zeltins A, Blaak H, Schrempf H. The novel lectin-like protein CHB1 is encoded by a chitin-inducible Streptomyces olivaceoviridis gene and binds specifically to crystalline α-chitin of fungi and other organisms. Mol Microbiol. 1994;13:807–819. doi: 10.1111/j.1365-2958.1994.tb00473.x. [DOI] [PubMed] [Google Scholar]
- 46.Susuki K, Suzuki M, Taiyoji M, Nikaidou N, Watanabe T. Chitin-binding protein (CBP21) in the culture supernatant of Serratia marcescens 2170. Biosci Biotechnol Biochem. 1998;62:128–135. doi: 10.1271/bbb.62.128. [DOI] [PubMed] [Google Scholar]
- 47.Svitil A L, Kirchman D L. A chitin-binding domain in a marine bacterial chitinase and other microbial chitinases: implications for ecology and evolution of 1,4-β-glycanases. Microbiology. 1998;144:1299–1308. doi: 10.1099/00221287-144-5-1299. [DOI] [PubMed] [Google Scholar]
- 48.Van Peer R, Punte W L M, de Weger L A, Schippers B. Characterization of the root surface endorhizosphere pseudomonads in relation to their colonization of roots. Appl Environ Microbiol. 1990;56:2462–2470. doi: 10.1128/aem.56.8.2462-2470.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Van't Wout J, Burntette W N, Mar V L, Rodzinski E, Wright S D, Tuomanen E I. Role of carbohydrate recognition domains of pertussis toxin in adherence of Bordetella pertussis to human macrophages. Infect Immun. 1992;60:3303–3308. doi: 10.1128/iai.60.8.3303-3308.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wirth S J, Wolf G A. Dye-labelled substrates for the assay and detection of chitinase and lysozyme activity. J Microbiol Methods. 1990;12:197–205. [Google Scholar]
- 51.Yahr T L, Goranson J, Frank D W. Exoenzyme S of Pseudomonas aeruginosa is secreted by a type III pathway. Mol Microbiol. 1996;22:991–1003. doi: 10.1046/j.1365-2958.1996.01554.x. [DOI] [PubMed] [Google Scholar]
- 52.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]