Abstract
Background
Nonalcoholic steatohepatitis (NASH) occupies a substantial proportion of chronic liver disease worldwide, of which pathogenesis needs further research. Recent studies have demonstrated the significant roles of circular RNAs (circRNAs) in NASH, while the function of a novel type of circRNAs, namely mitochondria-encoded circRNAs (mecciRNAs), remains elusive. Therefore, we aimed to investigate their potential to regulate the progression of NASH in this study.
Methods
GSE134146 was used to screen for differentially expressed mecciRNAs in NASH, while GSE46300 was used to identify NASH-related genes. To establish the mecciRNA-miRNA-mRNA networks, circMINE and miRNet databases were used for predicting downstream targets. Then, consensus clustering analysis was used to determine immune subtypes of NASH. Finally, we successfully validated our findings in vitro (LPS-treated hepatic stellate cells [HSCs]) and in vivo (MCD-diet mice) NASH models.
Results
We confirmed that circRNomics balance is disrupted in HSCs of NASH, while two mecciRNAs (hsa_circ_0089761 and hsa_circ_0089763) could function as competing for endogenous RNAs (ceRNAs) to regulate fibrosis-related signals. Furthermore, we constructed two ceRNA networks based on mecciRNAs for the first time. Cell and animal NASH models validated our findings that c-MYC and SMAD2/3 were upregulated in HSCs, while THBS1 and p-STAT3 were upregulated in hepatocytes. Moreover, we identified 21 core genes by overlapping the differentially expressed genes (NASH vs. Normal) with mecciRNA-targeted genes. According to their expression profiles, NASH patients could be divided in 2 different clusters, in which proinflammatory signals (TNF and IL-17 pathways) are significantly activated in Cluster 1.
Conclusion
We successfully established two novel mecciRNA-miRNA-mRNA networks in HSCs and hepatocytes, which were further confirmed by in vitro and in vivo models. Meanwhile, the novel immunotyping model revealed the heterogeneity of NASH, thereby might guiding treatment options. Altogether, our study brought a distinct perspective on the relationship between mecciRNAs and NASH.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12967-022-03595-1.
Keywords: NASH, HSC, circRNA, mecciRNA, miRNA, Immunotyping
Background
Nonalcoholic steatohepatitis (NASH) is the inflammatory subtype of nonalcoholic fatty liver disease (NAFLD), mainly caused by excess lipid accumulation in the liver [1]. According to statistical data, about 30% of the patients with fatty liver would progress to NASH [2]. Histologically, NASH is the manifestation of a wound-healing response to hepatocyte lipotoxicity [3]. Hence, NASH patients might benefit from treatment with preventing lipotoxicity or attenuating repair response effects. Currently, there are several innovative drugs for NASH. For instance, OCA, elafibranor, selonsertib, and CVC have entered phase III trials, despite the controversy about their long-term safety and effects [4–7].
The past two decades have witnessed dramatic advances in understanding the pathogenesis of NASH, in which hepatic stellate cells (HSCs) were identified as the major fibrogenic cells [8]. In the inactive state, HSCs maintain a non-proliferative, quiescent phenotype. However, HSCs become activated upon liver injury, transdifferentiating from vitamin-A-storing cells to myofibroblasts, which are proliferative, contractile, inflammatory, and chemotactic, while also characterized by enhanced ECM production [9]. In addition to this, the role of crosstalk between HSCs and hepatocytes also cannot be ignored, such as the activation of HSCs in response to apoptotic hepatocytes [10]. In summary, further studies exploring the cellular and molecular mechanisms of NASH will help to develop new treatment strategies.
Circular RNAs (circRNAs) are a class of non-coding RNAs that play important roles in several liver diseases, including NASH [11–14]. Among them, mitochondria-encoded circRNAs (mecciRNAs) are a novel type of circRNAs identified recently [15]. Our group demonstrated that mecciRNAs are distributed both inside and outside the mitochondria, despite the mechanism that how they shuttle in and out of mitochondria remains unclear [15, 16]. A striking mecciRNA, namely circSCAR, has been reported that could alleviate NASH via reducing mROS output [17]. This meaningful work strongly suggested the functional roles of mecciRNAs in NASH.
Here, combining bioinformatics analysis with experimental validation, we provided several lines of evidence that revealed the potential of mecciRNAs in HSCs to regulate NASH progression. Hopefully, this study could broaden our knowledge of circRNAs’ function, especially for mecciRNAs, and contribute to the development of treatment strategies for NASH.
Materials and methods
Materials
MCD (methionine/choline deficient) feed and standard feed were purchased from Nantong Trophy Feed Technology Company, MCD feed contains (per 1000 g): amino acid premix (methionine free) 175.7 g, methionine 0 g, choline chloride 0 g, sucrose 431.9 g, dextrin 50 g, corn starch 150.0 g, corn oil 100.0 g, cellulose 30.0 g, mineral mix 52.4 g. All the primers used in this study were synthesized by Tsingke Company and CWBIOTECH, their sequences were shown in Supplementary Materials. All of antibodies used in western blot, including c-MYC (ab32072), SMAD2 (ab40855), SMAD3 (ab40854), THBS1 (ab267388), STAT3 (ab68153), p-STAT3 (Y705, ab267373) and GAPDH (ab8245), were purchased from Abcam.
Animal samples
8 weeks male C57BL/6 mice were kept in a controlled environment (24 ± 2 °C, 12/12 h day/dark cycle). And mice were randomly divided into 2 subgroups (n = 6): Group 1 was fed with standard diet for 6 weeks (control group); Group 2 was fed with MCD diet for 6 weeks (MCD group). After the treatment mentioned above, mice were fasted for 12 h before being sacrificed. The liver was excised and perfused with saline. One portion of the liver from each mouse was fixed in 4% paraformaldehyde solution for histological analysis. Another portion of the liver was used to prepare liver homogenate for the biochemical analysis.
Cell lines and culture
Human hepatic stellate cells cell lines LX-2 were gifted by Dr. Xinping Huang, Guangzhou Institute of Biomedicine and Health, Chinese Academy of Sciences (purchased from the Advanced Research Center of Central South University). LX-2 was cultured in Dulbecco's Modified Eagle’s Media (Thermo Fisher Scientific, USA) supplemented with 10% fetal bovine serum (FBS, Thermo Fisher Scientific, USA) in a 5% CO2 humidified incubator at 37 °C. All the experiments were performed within 1 months of resuscitation and the cell passage was less than 3 generations from initial resuscitation (avoid activation of LX-2 cells).
RNA extraction and quantitative real-time polymerase chain reaction (RT-qPCR)
Total RNAs were isolated using Trizol reagent (Invitrogen, USA), either from cultured cells or liver tissue. 2 ug of total RNA was subjected to reverse transcription using Superscript III transcriptase (Invitrogen, USA). RT-qPCR was conducted using a Bio-Rad CFX96 system (Bio-Rad, USA) with SYBR green to determine the expression level of targets of interest. In addition, miRNA cDNA Synthesis Kit (CWBIOTECH, CN) and miRNA qPCR Assay Kit (CWBIOTECH, CN) were used for miRNA detection. Expression levels of circRNAs were normalized to the expression levels of GAPDH, while small RNA RNU6 (U6) was used for miRNA. And GAPDH and MTCO2 served as the cytosolic and mitochondrial control, respectively.
Mitochondria isolation
We conducted this experiment according to the kit protocol (Cat. no. 37612, QIAGEN, GER). First, LX-2 cells were suspended in Lysis Buffer (selectively disrupts the plasma membrane without solubilizing it, resulting in the isolation of cytosolic proteins), and incubated for 10 min. After that, plasma membranes and compartmentalized organelles, such as nuclei, mitochondria, and endoplasmic reticulum, remained intact and were pelleted by centrifugation (1000×g, 10 min). Then, the pellet was resuspended in Disruption Buffer, repeatedly passed through a narrow-gauge needle, and recentrifuged (1000×g, 10 min) to pellet nuclei, cell debris, and unbroken cells. The supernatant (contains mitochondria) was recentrifuged (6000×g, 10 min) to pellet mitochondria. After removal of the supernatant, mitochondria are washed and resuspended in Mitochondria Storage Buffer.
Screening of differentially expressed circRNAs and genes
We downloaded high-throughput sequencing and noncoding RNA expression profiling data (GSE134146 and GSE46300) from the GEO database. Among them, GSE134146 was used to screen for differentially expressed mecciRNAs in NASH, while GSE46300 was used to identify NASH-related genes. Differential expressions of all genes were calculated using the R package “limma”, and significance was evaluated by one-way analysis of variance (ANOVA).
Prediction of circRNAs and miRNAs downstream targets
We used the circMINE database (http://hpcc.siat.ac.cn/circmine/), which is based on 3 well-annotated databases, including miRanda [18], miRBase [19], and circBase resources [20], to predict the potential miRNAs targeted by mecciRNAs.
In addition, miRNet database (https://www.mirnet.ca/) was based on the 14 open-source databases, which could analyze and generate miRNA-mRNA network online. Specifically, the miRNA target gene data were collected from three well-annotated database miRTarBase [21], TarBase [22], and miRecords [23]. The miRNA to molecule interaction data were collected from SM2miR [24] and Pharmaco-miR [25]. The miRNA to disease interaction data were collected from miR2Disease [26] and PhenomiR [27]. The miRNA to epigenetic modifier interaction data were collected from EpimiR [28]. And, the exosomal miRNA annotation data were collected from ExoCarta [29]. It should be emphasized that Protein–Protein Interaction (PPI) database was included in the analysis during the construction of miRNA-mRNA network.
GO enrichment and KEGG pathway enrichment analysis
GO terms in three categories (GO: biological process, GO: cellular component and GO: molecular function) were used for pathway-enrichment analysis and biological interpretation. We used the GO enrichment analysis and visualization tool (GOrilla) to identify GO terms that were significantly enriched in the target gene list. KEGG pathway enrichment analysis was performed with clusterProfiler with a background set of all entrez IDs mapped to a KEGG pathway.
Construction of protein–protein interaction network
Given that searching for protein interactions and interaction networks is integral to further exploration of cellular states, biological processes and functions, relevant targets were entered into STRING (version 11.5, https://string-db.org/) [30]. Searching by gene name, selecting Homo sapiens as the species and selecting suitable confidence, the purpose of in-depth study of protein–protein interactions can be achieved. Network nodes and edges represent proteins and protein–protein associations, respectively.
Classification of NASH immune subtypes
On the basis of the expression profile, 21 mecciRNA-related genes were clustered by the R packages. Consensus Cluster Plus with reps = 100, p Item = 0.8, and p Feature = 1 [31]. By comprehensively analyzing the consistency matrix and the consistency cumulative distribution function, the optimal partition is defined.
Calculation of immune infiltration score
To differentiate immune levels between 2 clusters, we used single-sample gene set enrichment analysis (ssGSEA) to characterize 23 types of immune cells in the liver tissues, based on the specific gene signatures of immune cells [32].
Statistical analysis
The applicable statistical methods were used depending on the type of data. The student’s t-test was used for comparisons between groups. ANOVA for multiple comparisons was used to detect differences amongst the various treatments. All data from three separate experiments at least are presented as mean ± SD. Differences were considered significant for P-values less than 0.05. *P < 0.05, **P < 0.01, and ***P < 0.001.
Results
circRNomics balance is disrupted in HSCs of NASH
circRNAs have become a novel research hotspot in RNA biology. They localize in specific subcellular compartments, and play different biological roles [33]. However, the functions of circRNAs in NASH is still elusive. As activation of HSCs is well regarded as the central driver of fibrosis in NASH, we decided to explore the differentially expressed circRNAs in HSCs of NASH patients [8].
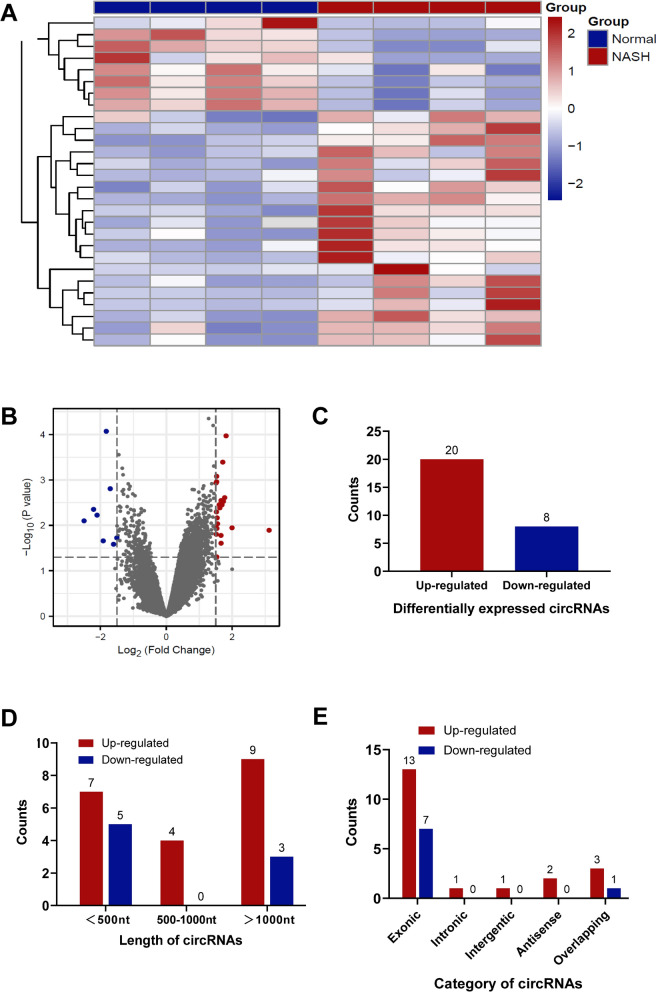
Thus, we downloaded the microarray gene profiling dataset (GSE134146, including 4 patients with NASH cirrhosis and 4 patients without NASH) from Gene Expression Omnibus (GEO) for bioinformatics analysis [17]. The circRNAs with P-value < 0.05 and |Log2FC|≥ 1.5 from t-test were identified as differentially expressed circRNAs (Fig. 1A–B). In total, 20 circRNAs were upregulated, while 8 circRNAs were downregulated in NASH (Fig. 1C). Among them, 12 circRNAs are less than 500nt in length (inclined to function as molecular scaffolds), 4 are between 500 and 1000 nt, and 12 are longer than 1000nt (inclined to function as miRNA sponges) (Fig. 1D). Consistent with previous reports, most of them (20 out of 28) are encoded by exons (Fig. 1E). These results indicated that circRNomics homeostasis of HSCs is disrupted in NASH patients. Consequently, we speculated that there might be an association between circRNA imbalance and NASH progression.
Fig. 1.
circRNomics Balance is Disrupted in HSCs of NASH. A, B The heatmap (A) and the volcano plot (B) show differentially expressed circRNAs in HSCs of NASH patients (GSE134146). P-value < 0.05 and |Log2FC|≥ 1.5. C The number of upregulated and downregulated circRNAs, respectively. D The length of upregulated and downregulated circRNAs, respectively. E The number of upregulated and downregulated circRNAs according to their parental genes
mecciRNAs account for half of downregulated circRNAs in HSCs of NASH
During the development of NASH, there is a constant dysfunction of mitochondria, including alterations in enzyme activities, protein expression, and signaling networks [34]. Thus, we wondered whether the mecciRNAs expression profile was altered in NASH progression.
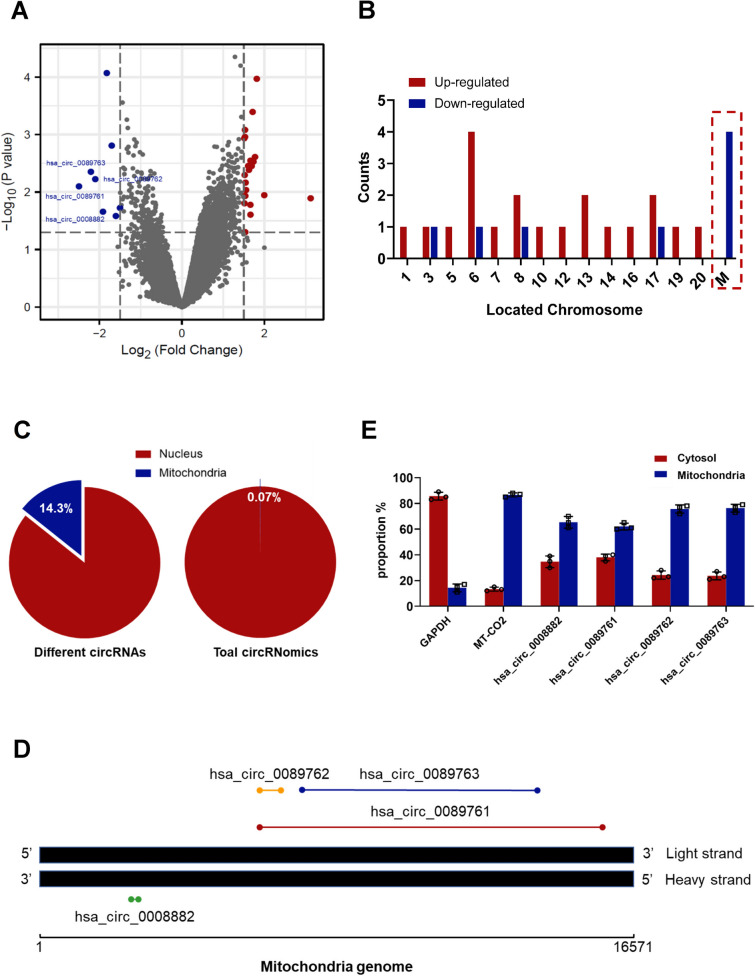
Surprisingly, we found that half (4 out of 8) of downregulated circRNAs were encoded by the mitochondrial genome, namely hsa_circ_0089761, hsa_circ_0089762 (also known as circSCAR [17]), hsa_circ_0089763 and hsa_circ_0008882 (Fig. 2A, B). Given the majority of circRNomics are occupied by nuclear-encoded circRNAs (99.93%), it is noteworthy that mecciRNAs accounted for 14.3% of the differentially expressed circRNAs in NASH (Fig. 2C) [35]. Among them, hsa_circ_0008882 are encoded by the heavy strand of mitochondrial genome, while hsa_circ_0089761, hsa_circ_0089762 and hsa_circ_0089763 are generated from the light strand (Fig. 2D). Additionally, their sizes span a wide range, in which lengths of hsa_circ_0089761 and hsa_circ_0089763 are over 5000 nt, while hsa_circ_0089762 and hsa_circ_0008882 are less than 400 nt (Fig. 2D). Then, we used mitochondrial-cytoplasmic separation assays to determine their cellular localization. The results demonstrated that these mecciRNAs were mainly located in mitochondria, while a part of them were also presented in cytoplasm (Fig. 2E).
Fig. 2.
mecciRNAs Account for Half of Downregulated circRNAs in HSCs of NASH. A The volcano plot shows upregulated (red) and downregulated (blue) circRNAs in HSCs of NASH patients (GSE134146), in which 4 mecciRNAs were marked. P-value < 0.05 and |Log2FC|≥ 1.5. B The chromosome locations of differentially expressed circRNAs, in which mecciRNAs were highlighted. C The numbers of differentially expressed nucleus-encoded and mitochondria-encoded circRNAs were 24 and 4, respectively (left panel). And the proportion of mecciRNAs in the total circRNomics is shown (right panel). D The name, length, location of differentially expressed mecciRNAs in the mitochondrial genome. E The expression levels of 4 mecciRNAs in cytosolic and mitochondrial fractions of LX-2 cells were determined by RT-qPCR. GAPDH and MTCO2 mRNA served as the cytosolic and mitochondrial control, respectively
mecciRNAs regulate fibrosis-related signaling pathways in HSCs
circRNAs could function as miRNA decoys, which are defined as miRNA sponges that bind miRNAs and prevent them from suppressing their target mRNAs [36]. Noteworthy, circRNAs are generally with relatively few binding sites for miRNAs, the idea of controlling the stability and quantity of miRNAs by circRNAs and achieving measurable effects should be considered with caution [37, 38]. However, hsa_circ_0089761 and hsa_circ_0089763 are highly possible to possess powerful miRNA-sponge capacity, resulted from their long sequence lengths (over 5000 nt). The copy number ratio between specific circRNA and many different targeted miRNAs is relatively small [39]. Hence, considering the great abundance of substrates (miRNAs), hsa_circ_0089761 and hsa_circ_0089763 are not likely to have competitive binding. On the contrary, it is reasonable to speculate that these two circRNAs might have double sponge capacity for specific miRNAs, due to the fact that hsa_circ_0089761's sequence completely contains hsa_circ_0089763's sequence (Fig. 2D).
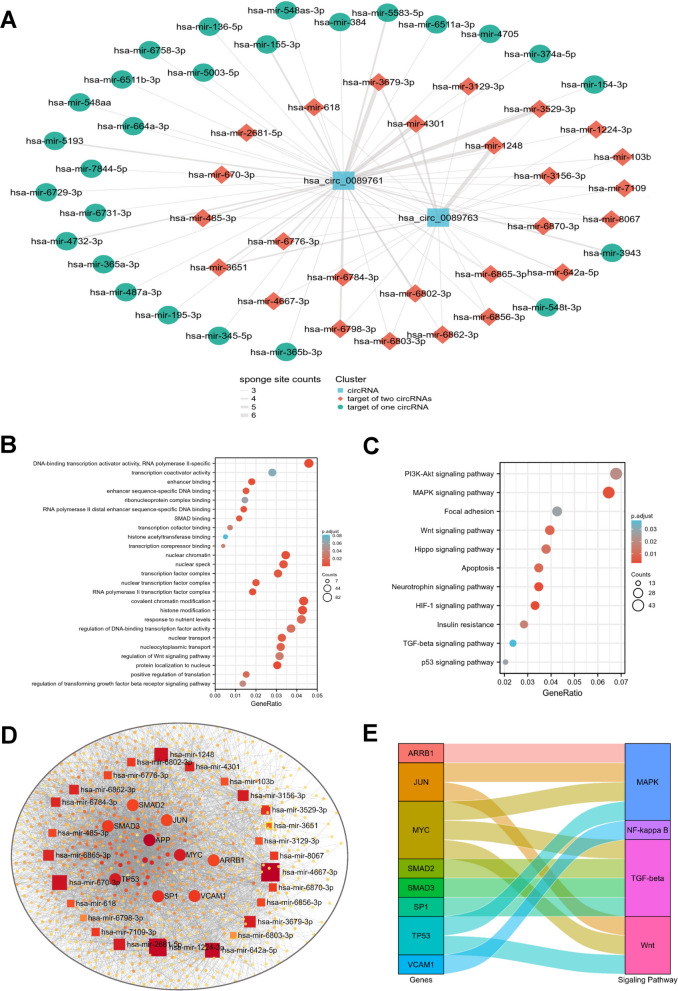
First, utilizing the circMINE database, we predicted the miRNAs targeted by hsa_circ_0089761 and hsa_circ_0089763 [40]. According to the filter criteria (a. Score cutoff > 150; b. Energy cutoff < –7; c. Amount of specific miRNA-binding sites ≥ 3), we identified 52 and 26 mecciRNA-miRNA pairs respectively (Fig. 3A). As expected, hsa_circ_0089761 and hsa_circ_0089763 shared 26 identical miRNA targets (hsa-mir-136-5p, hsa-mir-154-3p, hsa-mir-155-3p, hsa-mir-195-3p, hsa-mir-345-5p, hsa-mir-365a-3p, hsa-mir-365b-3p, hsa-mir-374a-5p, hsa-mir-384, hsa-mir-3943, hsa-mir-4705, hsa-mir-4732-3p, hsa-mir-5003-5p, hsa-mir-5193, hsa-mir-548aa, hsa-mir-548as-3p,hsa-mir-548t-3p, hsa-mir-5583-5p, hsa-mir-6511a-3p, hsa-mir-6511b-3p, hsa-mir-664a-3p, hsa-mir-6729-3p, hsa-mir-6731-3p, hsa-mir-6758-3p, hsa-mir-7844-5p, hsa-mir-487a-3p). Then, we identified 2000 mRNAs targeted by the 26 miRNAs in miRNet database (Additional file 4: Table S1) [41]. It is worth mentioning that we also took the PPI database into account when identifying targeted mRNAs. Therefore, among these targets, there may not only be changes in mRNA or protein expression levels, but also changes in protein modification status. Next, To further enhance the reliability, we only selected the part with degree frequency > 1 for subsequent analysis (Additional file 5: Table S2, Additional file 1: Figure S1A). Finally, we successfully established a novel mecciRNA-miRNA-mRNA network in HSCs, including 2 mecciRNA nodes, 26 miRNA nodes and 1343 mRNA nodes (Additional file 2: Figure S1B).
Fig. 3.
mecciRNAs Regulate Fibrosis-related Signaling Pathways in HSCs. A mecciRNA-centered ceRNA network is shown. The thickness of the line represents the number of sponge sites. Color represents the different clusters of ncRNAs. B, C The results of GO (B) and KEGG (C) enrichment analysis for mecciRNAs-related genes were shown, respectively. D ceRNA network shows that a set of fibrotic genes (such as SMAD2, SMAD3, MYC, etc.) received the highest enrichment scores. E The Sankey diagram shows the relationships between core genes of ceRNA network and NASH-related pathways
To achieve a better understanding of the functions associated with these mecciRNAs-related genes, we performed GO Term and KEGG pathway enrichment analysis. GO analysis showed an enrichment of GO terms indicative of transcription regulation, such as transcription coactivator activity, enhancer binding, and histone modification (Fig. 3B). Meanwhile, KEGG analysis results revealed the enrichment of several pro-fibrotic signaling pathways, such as Hippo signaling pathway and TGF-β signaling pathway (Fig. 3C) [10, 42]. Strikingly, the top 20 most enriched genes contained many famous NASH-related genes, such as c-MYC, SMAD2 and SMAD3 (Fig. 3D–E) [43–46]. Taken together, these results indicated that mecciRNAs might have the potential to regulate fibrosis-related signaling pathways in HSCs.
Exosomes-mediated crosstalk between HSCs and hepatocytes in NASH
Death of hepatocytes could induce liver inflammation and then directly or indirectly promote the activation of HSCs [47]. Conversely, activated HSCs also further aggravated the damage and death of hepatocytes [48]. Hence, the crosstalk between HSCs and hepatocytes performs important functions in NASH progression, in which exosome-mediated intercellular communication cannot be ignored [49]. The exosomal cargo consists of proteins, lipids and ncRNAs, while many of the biologic effects of exosomes have been attributed to miRNAs [50]. Therefore, we were particularly interested in whether the changes in mecciRNA-miRNA profile of HSCs could influence the signaling pathways of hepatocytes.
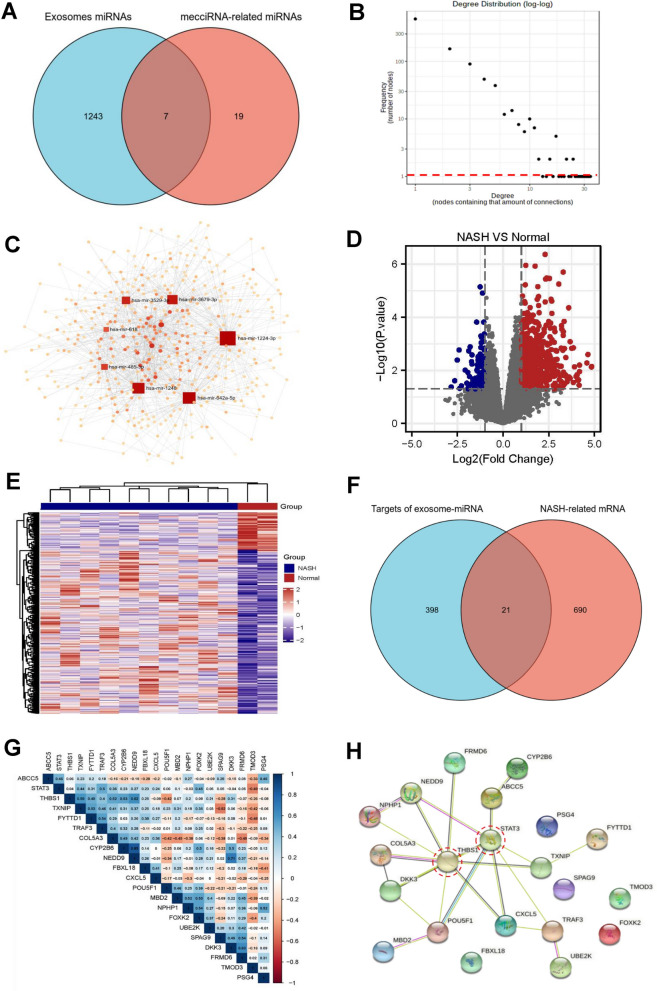
First, using the exosomal miRNAs dataset (ExoCarta, a well-known database of exosomal proteins, RNA and lipids) in miRNet, we screened out 7 candidates (hsa-mir-485-3p, hsa-mir-618, hsa-mir-642a-5p, hsa-mir-1224-3p, hsa-mir-1248, hsa-mir-3529-3p and hsa-mir-3679-3p) among the 26 mecciRNA-targeted miRNAs, which had been reported that can be secreted to extracellular microenvironment through exosomes to function (Additional file 6: Table S3, Fig. 4A) [41]. Then, utilizing the same analytical methods and filter criteria, we finally identified 419 mRNAs in hepatocytes, which might be regulated by the mecciRNA-miRNA network (Additional file 7: Table S4, Fig. 4B, C).
Fig. 4.
Exosomes-mediated Crosstalk Between HSCs and Hepatic Cells in NASH. A The Venn diagram shows that only 7 specific miRNAs possess the potential to regulate NASH-related pathways in hepatocytes. B, C State of degree distribution in the ceRNA network (B, frequency [number of nodes] = 1 was highlighted with red dotted line), consisting of 7 miRNAs and 419 mRNAs (C). D, E The volcano plot (D) and heatmap (E) shows the differentially expressed genes in NASH liver tissues vs. normal liver tissues (GSE46300, red: upregulated genes, blue: downregulated genes). P-value < 0.05 and |Log2FC|≥ 1.0. F The Venn diagram shows that there are 21 shared genes between the NASH-related gene set and exosomal-miRNA targeted gene set. G Protein interaction network diagram shows the interaction between 21 overlapped proteins. H PPI network of 21 overlapped proteins, in which STAT3 and THBS1 (labeled with red circles) are localized at the core of network
Meanwhile, we downloaded the raw sequencing data of GSE46300 from GEO, which includes liver tissues with or without NASH, and obtained a set of differentially expressed genes between NASH and normal liver (|Log2FC|≥ 1, P < 0.05, Fig. 4D, E) [51]. In total, 21 overlapped genes were identified (STAT3, POU5F1, TRAF3, UBE2K, CXCL5, SPAG9, MBD2, THBS1, TMOD3, NPHP1, NEDD9, COL5A3, ABCC5, FRMD6, FBXL18, TXNIP, FYTTD1, PSG4, DKK3, CYP2B6, FOXK2), in which STAT3 and THBS1 were localized at the core of protein interaction network (Fig. 4F–H). Despite several reports about their functions in liver steatosis, in-depth molecular mechanisms about STAT3 and THBS1 regulating NASH progression still deserve further study [52–54].
A novel immunotyping of NASH based on mecciRNA-related network
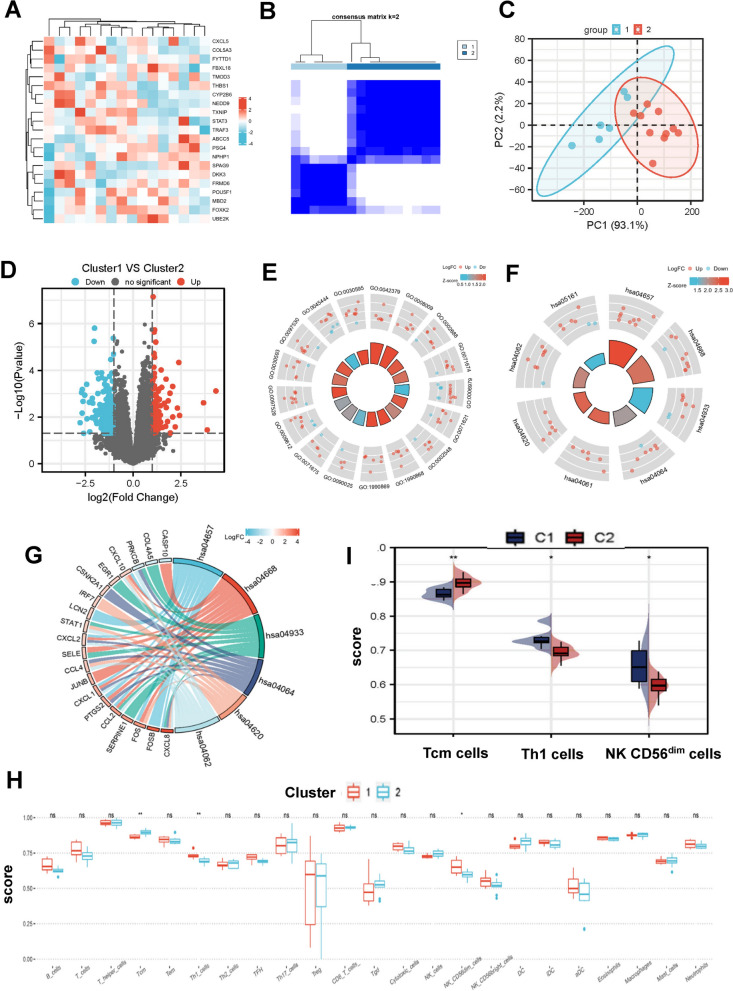
Classification of specific disease based on different cellular and molecular features could deepen our understanding of this disease and even provide valuable information for treatment. Hence, in order to evaluate the effects of these 21 genes on NASH, we extracted the expression profiles of these targets from GEO46300 to construct a consensus cluster (Fig. 5A). Two subtypes (Cluster 1 and Cluster 2) were obtained with the minimum variance within the group and the maximum variance across the groups (Fig. 5B, Additional file 2: Fig. 2A–D). Meanwhile, the results of principal component analysis (PCA) confirmed that there are significant differences between the two clusters (Fig. 5C).
Fig. 5.
A Novel Immunotyping of NASH Based on mecciRNA-related Network. A The heatmap shows the expression profiles of 21 specific targets from 16 steatotic liver tissues. B NASH patients clustering heatmap, consensus matrix k = 2. C PCA diagram shows the significant differences between 2 clusters of NASH patients (Cluster 1 labeled with blue, Cluster 2 labeled with red). D The volcano plot shows the upregulated (red) and downregulated (blue) genes in Cluster 1 vs. Cluster 2. P-value < 0.05 and |Log2FC|≥ 1.0. E The results of GO enrichment analysis for differentially expressed genes between Cluster 1 and Cluster 2. The names of GO terms are shown in Table.1. F, G The results of KEGG enrichment analysis for differentially expressed genes between Cluster 1 and Cluster 2. The names of KEGG terms are shown in Table.2. H, I Differential enrichment scores of 23 immune cell signatures in Cluster 1 and Cluster 2. *P < 0.05, **P < 0.01, n.s. means none was statistically significant
We then performed differential gene expression analysis across the transcriptomes of these two subtypes. In total, we identified 273 differentially expressed genes (Additional file 8: Table S5, Fig. 5D). GO analysis revealed that these genes were highly enriched for functions related to chemokine activity and immune cell chemotaxis (Table. 1, Fig. 5E). And KEGG analysis demonstrated that, in addition to the upregulation of immune cell chemotaxis-related pathways, IL-17 signaling pathway and TNF signaling pathway were also significantly activated in Cluster 1, which could facilitate the development of NAHS by exerting proinflammatory effects (Table. 2, Fig. 5F–G) [55, 56].
Table 1.
Results of GO enrichment analysis between Cluster 1 and Cluster 2
| Ontology | ID | Description | GeneRatio | BgRatio | P value | p.adjust | Q value |
|---|---|---|---|---|---|---|---|
| BP | GO:0002688 | Regulation of leukocyte chemotaxis | 8/224 | 114/18670 | 7.01e−05 | 0.066 | 0.064 |
| BP | GO:0071674 | Mononuclear cell migration | 7/224 | 90/18670 | 1.05e−04 | 0.066 | 0.064 |
| BP | GO:0006979 | Response to oxidative stress | 16/224 | 451/18670 | 1.16e−04 | 0.066 | 0.064 |
| BP | GO:0071621 | Granulocyte chemotaxis | 8/224 | 123/18670 | 1.20e−04 | 0.066 | 0.064 |
| BP | GO:0002548 | Monocyte chemotaxis | 6/224 | 65/18670 | 1.28e−04 | 0.066 | 0.064 |
| BP | GO:1990868 | Response to chemokine | 7/224 | 97/18670 | 1.68e−04 | 0.066 | 0.064 |
| BP | GO:1990869 | Cellular response to chemokine | 7/224 | 97/18670 | 1.68e−04 | 0.066 | 0.064 |
| BP | GO:0090025 | Regulation of monocyte chemotaxis | 4/224 | 25/18670 | 2.09e−04 | 0.066 | 0.064 |
| BP | GO:0071675 | Regulation of mononuclear cell migration | 5/224 | 46/18670 | 2.18e−04 | 0.066 | 0.064 |
| BP | GO:0009612 | Response to mechanical stimulus | 10/224 | 210/18670 | 2.32e−04 | 0.066 | 0.064 |
| BP | GO:0097529 | Myeloid leukocyte migration | 10/224 | 210/18670 | 2.32e−04 | 0.066 | 0.064 |
| BP | GO:0030593 | Neutrophil chemotaxis | 7/224 | 104/18670 | 2.58e−04 | 0.068 | 0.065 |
| BP | GO:0097530 | Granulocyte migration | 8/224 | 141/18670 | 3.06e−04 | 0.074 | 0.071 |
| BP | GO:0045444 | Fat cell differentiation | 10/224 | 223/18670 | 3.75e−04 | 0.081 | 0.078 |
| BP | GO:0030595 | Leukocyte chemotaxis | 10/224 | 224/18670 | 3.88e−04 | 0.081 | 0.078 |
| MF | GO:0042379 | Chemokine receptor binding | 7/218 | 66/17697 | 1.64e−05 | 0.008 | 0.007 |
| MF | GO:0008009 | Chemokine activity | 6/218 | 49/17697 | 2.93e−05 | 0.008 | 0.007 |
Table 2.
Results of KEGG enrichment analysis between Cluster 1 and Cluster 2
| ID | Description | GeneRatio | BgRatio | P value | p.adjust | Q value |
|---|---|---|---|---|---|---|
| hsa04657 | IL-17 signaling pathway | 9/97 | 94/8076 | 1.64e−06 | 3.41e−04 | 3.07e−04 |
| hsa04668 | TNF signaling pathway | 9/97 | 112/8076 | 7.09e−06 | 7.37e−04 | 6.64e−04 |
| hsa04933 | AGE-RAGE signaling pathway in diabetic complications | 8/97 | 100/8076 | 2.43e−05 | 0.002 | 0.002 |
| hsa04064 | NF-kappa B signaling pathway | 7/97 | 104/8076 | 2.38e−04 | 0.012 | 0.011 |
| hsa04620 | Toll-like receptor signaling pathway | 6/97 | 104/8076 | 0.002 | 0.039 | 0.035 |
| hsa04062 | Chemokine signaling pathway | 8/97 | 192/8076 | 0.002 | 0.043 | 0.038 |
Considering the differences between 2 subtypes were mainly focused on immune-related pathways, we further utilized ssGSEA scores to characterize the immune cell components in the 2 subtypes [32]. Despite most of the differences did not reach statistical significance, immune cell infiltration was higher in Cluster 1 than in Cluster 2 generally, which was consistent with the GO and KEGG analysis (Fig. 5H). Noteworthy, infiltration of T helper 1 (Th1) cells and NK CD56dim cells was significantly higher in Cluster 1, while central memory T (Tcm) cells infiltration was reduced (Fig. 5I). It is universally agreed that Th1 cells are proinflammatory cells and aggravate liver fibrosis development [57–59]. However, the roles of NK CD56dim cells and Tcm cells in NASH remain elusive and require further study.
In general, we established a novel immunotyping of NASH based on mecciRNA-related network, in which Cluster 1 presents increased immune cell infiltration and activation of proinflammatory signaling pathways.
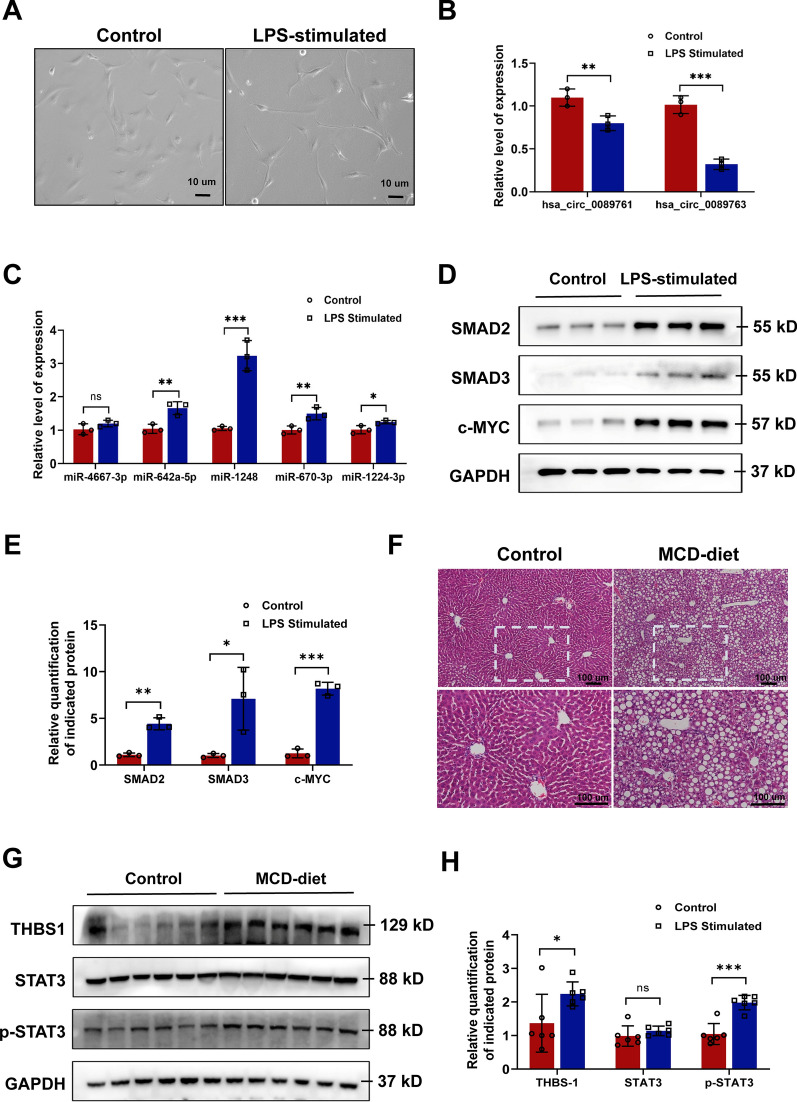
Validation of mecciRNAs networks in vitro and in vivo
First, to validate the bioinformatic conclusions, we successfully established HSC-activation model by exposing LX-2 cells (human hepatic stellate cell line) to lipopolysaccharide (LPS) at a concentration of 100 ng/ml for 48 h (Fig. 6A). We confirmed that both hsa_circ_0089761 and hsa_circ_0089763 was detected in a lower amount in activated HSCs compared to wild-type HSCs (Fig. 6B). To further validate our computational predictions, we detected the expression levels of 5 core miRNAs (as test cases) in the mecciRNA-miRNA network. The results largely conformed to our predictions that miR-642a-5p, miR-1248, miR-670-3p and miR-1224-3p was upregulated in activated HSCs, while only miR-4667-3p level was comparable between the two groups (Fig. 6C). As shown in Fig. 3D, SMAD2, SMAD3 and c-MYC are core targets in HSCs mecciRNA-miRNA network, which have been reported to play vital roles in NASH progression [43–46]. The western blot results demonstrated the significant upregulation in expression levels of these proteins in activated HSCs as expected (Fig. 6D).
Fig. 6.
Validation of mecciRNAs Networks in vitro and in vivo. A Under the stimulation of LPS (100 ng/ml, 48 h), LX-2 cells were significantly activated, and their cell morphology changed. Scale bar = 10 μm. B RT-qPCR result shows that the expression levels of hsa_circ_0087761 and hsa_circ_0087763 were decreased in activated LX-2 cells. The expression of GAPDH was used as the reference in the RT-qPCR analyses. **P < 0.01, ***P < 0.001. C RT-qPCR result shows that miR-642a-5p, miR-1248, miR-670-3p, and miR-1224-3p were upregulated significantly in activated LX-2 cells, while the expression level of miR-4667-3p was not changed. The expression of U6 was used as the reference in the RT-qPCR analyses. *P < 0.05, **P < 0.01, ***P < 0.001, n.s. means none was statistically significant. D, E Endogenous levels of SMAD2, SMAD3, and c-MYC from control or LPS-stimulated LX-2 cells (n = 3) were determined by Western blot (D). GAPDH was used as a loading control. The relative ratios of indicated proteins over GAPDH and statistical analysis results are shown (E). *P < 0.05, **P < 0.01, ***P < 0.001. F HE stained images shows that the mice in MCD-diet group had significant hepatic steatosis compared to the mice in standard diet group. Scale bar = 100 μm. G, H Endogenous levels of THBS1, STAT3, and p-STAT3 from control or MCD-diet mice (n = 6) were determined by Western blot (G). GAPDH was used as a loading control. The relative ratios of indicated proteins over GAPDH and statistical analysis results are shown (H). *P < 0.05, **P < 0.01, ***P < 0.001, n.s. means none was statistically significant
Meanwhile, a set of candidates were considered as the downstream effectors of mecciRNAs-miRNAs network in hepatocytes, in which THBS1 and STAT3 occupied key positions. As feeding mice an MCD diet is a widely accepted model to study relevant mechanisms in NASH, we established this animal model, and detected the expression levels of THBS1 and STAT3 in control group and MCD-diet group (Fig. 6E) [60, 61]. The results revealed that the expression of THBS1 was increased in MCD-diet group, despite the similar expression levels of STAT3 between the two groups (Fig. 6F). Considering PPI database was also included in mecciRNA network analysis, we further detected the expression level of phosphorylated STAT3. As shown in Fig. 6F, the results demonstrated the elevated level of phosphorylated STAT3 in MCD-diet group.
Based on these experimental results, we confirmed the reliability of mecciRNA-miRNA networks preliminarily. Additionally, our study also brought a distinct perspective on the understanding of relationship between mecciRNAs and NASH.
Discussion
With the development of society and economy, the incidence of NASH has gradually increased, becoming one of the main chronic diseases globally [62]. Therefore, clarification the cellular and molecular mechanism of NASH has become a research hotspot. Over the course of research, it was found that circRNAs might function significantly in progression of NASH, in which a novel type of circRNAs, namely mecciRNAs, have attracted much interest recently [14, 17, 63]. Through bioinformatic analysis and experimental verification, this study has proposed the possibility that mecciRNAs might function as ceRNAs to regulate NASH progression.
Although our group demonstrated the existence of mecciRNAs and provided solid evidence to confirm that mecciRNAs are also localized outside of mitochondria [15], additional studies are needed to further illuminate several issues.
First, the mechanism by which mecciRNAs shuttle in and out of mitochondria is not well understood. Our previous study revealed that mecciRNAs could interact with PNPase, an enzyme that has been shown to be critical for the mitochondrial import of several noncoding RNAs [15, 64–66]. Hence, we speculated that the mitochondrial export of mecciRNAs might require the participation of PNPase. In addition, members of the mitochondrial carrier family (SLC25) provide the transport steps for nucleotides across the mitochondrial inner membrane, approximately one-third of which are currently orphan transporters, with no known substrate [67]. Hence, it is reasonable to speculate that SLC25 family might mediate mecciRNAs transport.
Second, the biosynthesis and metabolism of mecciRNAs require more in-depth research. mecciRNAs and nuclear-encoded circRNAs possess similar junction motifs, suggesting a mechanism of back splicing might exist in mecciRNA biogenesis. Although previous studies believed that introns and linear splicing events cannot occur in the mitochondria of multicellular animals, recent study has found that splicing factors may exist in mammalian mitochondria [68]. Additionally, our data showed that nuclear-transfected plasmids harboring the corresponding mitochondrial DNA fragments (with the flanking sequences) can successfully overexpress mecciRNA [15]. Then it may be speculated that there is a back splicing form of mitochondrial splicing in multicellular animals to generate mecciRNAs. Meanwhile, the discovery of nuclear-encoded circRNAs generated from single-exon genes by back splicing, and no linear splicing is involved in the biogenesis of mRNA from single-exon genes also make us reasonably suspect that mecciRNAs may be generated through a unique splicing-independent mechanism [20].
Both of hsa_circ_0089761 and hsa_circ_0089763 possess strong ability of miRNA sponges, due to their surprising sequence lengths and partial cytoplasmic localization. And incorporating PPI database into analysis made the bioinformatic prediction much closer to reality, which set this work apart from the other ceRNA network studies. It also meant that the targets of mecciRNA-miRNA networks may not only experience changes in expression levels, but also may undergo changes in protein modification due to the effect of other proteins (such as phosphorylation, etc.). Meanwhile, as the tools matures for ceRNA network analysis, utilizing bioinformatic methods to predict the potential functions of circRNAs is becoming more and more reliable [69]. Building on this theoretical foundation, we carried out this study.
Completely different from the two mecciRNAs mentioned above, hsa_circ_0089762 and hsa_circ_0008882 is really tiny, so that might act as molecular scaffolds to regulate specific complex functions or serve as molecular chaperones in the folding of mitochondria-imported proteins [15, 36]. Further study is certainly required to confirm this speculation. As of now, there is only one mecciRNA has been reported that could perform significant effect on NASH [17]. Considering the major contribution of mitochondria dysfunction in NASH, it is reasonable to speculate that a substantial proportion of functional mecciRNAs remain unidentified [70].
In this study, we have divided NASH patients into 2 subtypes according to 21 mecciRNA-related genes. Unexpectedly, we found large differences in immune-related signaling between the two subtypes. As we all know, NASH is essentially a chronic disease of immunometabolism, whose progression is associated with the liver immune microenvironment [71]. From this perspective, it is of significant interest that a set of mecciRNA-related genes could guide immunophenotyping in NASH, of which molecular mechanism deserves further research. Unfortunately, GSE46300 lacks clinical information of patients, such as liver enzymes. Hence, it is not clear whether the clinical manifestations of cluster 1 patients are more severe than those of cluster 2, in addition to the up-regulation of proinflammatory signals.
However, our study still has some limitations. On the one hand, due to the significant differences existing between human and mice, especially in transcript level, mice model of NASH could not fully simulate the complexity of human illnesses [72, 73]. Interestingly, there are currently models using humanized liver in mice that allow to study NASH development, which might make study more closely with the reality [74]. On the other hand, compared with the next generation sequencing used in this study, single-cell RNA sequencing, as a novel method to comprehensively characterize the cells, could better reflect the changes of hepatocytes and hepatic stellate cells during the progression of NASH, which have a broad application prospect in NASH-related research.
Conclusion
We successfully established two mecciRNA-miRNA-mRNA networks based on bioinformatic analysis. Moreover, LPS and MCD-diet induced NASH model supported our prediction to some extent. Meanwhile, utilizing 21 NASH-related genes targeted by mecciRNAs, a novel immunotyping model for NASH was built for the first time, directly reflecting the state of liver immune microenvironment, which might guide treatment option in future. In summary, our study brought a distinct perspective on the relationship between mecciRNAs and NASH.
Supplementary Information
Additional file 1: Supplementary Figure. 1 mecciRNAs Regulate Fibrosis-related Signaling Pathways in HSCs
Additional file 2: Supplementary Figure. 2 A Novel Immunotyping of NASH Based on mecciRNA-related Network
Additional file 3: Supplementary Figure Legends and Primer Sequence.
Additional file 4: Supplementary Table. 1 The 2000 mRNAs Targeted by 26 miRNAs in miRNet Database.
Additional file 5: Supplementary Table. 2 The 1343 mRNAs Targeted by 7 exosomal miRNAs in miRNet Database.
Additional file 6: Supplementary Table. 3 . The Evidence Indicates The Presence of These Following 7 miRNAs in Exosomes.
Additional file 7: Supplementary Table. 4 The 419 mRNAs Regulated by mecciRNA-miRNA Network in Hepatocytes.
Additional file 8: Supplementary Table. 5 The 273 Differentially Expressed Genes Between Cluster 1 and Cluster 2.
Acknowledgements
We wish to express gratitude for the contributions of all authors of the research articles corresponding to GSE134146 and GSE46300.
Author contributions
XC and GS conceived and designed the research. BL and YT performed most of the experiments. BL and JH performed all of bioinformatics analysis. QG and BJ completed the construction of animal models. BL wrote the manuscript with the assistance from HS and WL, LS and HY provided administrative support and revision suggestion. All authors read and approved the final manucsript.
Funding
Professor Xiujun Cai receives funding support from National Natural Science Foundation of China (No. 81827804), Zhejiang Clinical Research Center of Minimally Invasive Diagnosis and Treatment of Abdominal Diseases (No. 2018E50003) and Key Research and Development Project of Zhejiang Province (No. 2018C03083).
Professor Ge Shan receives funding support from National Key R&D Program of China (2019YFA0802600), National Natural Science Foundation of China (No. 31725016, No. 31930019, and No. 91940303).
Professor Hong Yu receives funding support from "Ten Thousand Plan" Innovation Leader of Zhejiang Province (No. 2020R52007), Major Science and Technology Project of Zhejiang Province of China (No. 2021C306) and Key Project of Province and Ministry (No. WKJ-ZJ-2124).
Doctor Liang Shi receives funding support from Clinical Top-notch Personnel Support of Zhejiang University.
Declarations
Ethics approval and consent to participate
All methods were carried out in accordance with relevant guidelines and regulations. All animal experimental procedures were approved by the Ethics Committee of Sir Run Run Shaw Hospital of Zhejiang University School of Medicine.
Consent for publication
Not applicable.
Availability of data and materials
The data sets analysed during this study are available in public, open access repositories listed in this article.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Boqiang Liu, Yuanshi Tian and Jing He contributed equally to this work
Contributor Information
Ge Shan, Email: shange@srrsh.com.
Xiujun Cai, Email: srrsh_cxj@zju.edu.cn.
References
- 1.Angulo P. Nonalcoholic fatty liver disease. N Engl J Med. 2002;346:1221–1231. doi: 10.1056/NEJMra011775. [DOI] [PubMed] [Google Scholar]
- 2.Williams CD, Stengel J, Asike MI, Torres DM, Shaw J, Contreras M, Landt CL, Harrison SA. Prevalence of nonalcoholic fatty liver disease and nonalcoholic steatohepatitis among a largely middle-aged population utilizing ultrasound and liver biopsy: a prospective study. Gastroenterology. 2011;140:124–131. doi: 10.1053/j.gastro.2010.09.038. [DOI] [PubMed] [Google Scholar]
- 3.Machado MV, Diehl AM. Pathogenesis of nonalcoholic steatohepatitis. Gastroenterology. 2016;150:1769–1777. doi: 10.1053/j.gastro.2016.02.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Neuschwander-Tetri BA, Loomba R, Sanyal AJ, Lavine JE, Van Natta ML, Abdelmalek MF, Chalasani N, Dasarathy S, Diehl AM, Hameed B, et al. Farnesoid X nuclear receptor ligand obeticholic acid for non-cirrhotic, non-alcoholic steatohepatitis (FLINT): a multicentre, randomised, placebo-controlled trial. Lancet. 2015;385:956–965. doi: 10.1016/S0140-6736(14)61933-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ratziu V, Harrison SA, Francque S, Bedossa P, Lehert P, Serfaty L, Romero-Gomez M, Boursier J, Abdelmalek M, Caldwell S, et al. Elafibranor, an agonist of the peroxisome proliferator-activated receptor-alpha and -delta, induces resolution of nonalcoholic steatohepatitis without fibrosis worsening. Gastroenterology. 2016;150(1147–1159):e1145. doi: 10.1053/j.gastro.2016.01.038. [DOI] [PubMed] [Google Scholar]
- 6.Loomba R, Lawitz E, Mantry PS, Jayakumar S, Caldwell SH, Arnold H, Diehl AM, Djedjos CS, Han L, Myers RP, et al. The ASK1 inhibitor selonsertib in patients with nonalcoholic steatohepatitis: a randomized, phase 2 trial. Hepatology. 2018;67:549–559. doi: 10.1002/hep.29514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Friedman SL, Ratziu V, Harrison SA, Abdelmalek MF, Aithal GP, Caballeria J, Francque S, Farrell G, Kowdley KV, Craxi A, et al. A randomized, placebo-controlled trial of cenicriviroc for treatment of nonalcoholic steatohepatitis with fibrosis. Hepatology. 2018;67:1754–1767. doi: 10.1002/hep.29477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tsuchida T, Friedman SL. Mechanisms of hepatic stellate cell activation. Nat Rev Gastroenterol Hepatol. 2017;14:397–411. doi: 10.1038/nrgastro.2017.38. [DOI] [PubMed] [Google Scholar]
- 9.Puche JE, Saiman Y, Friedman SL. Hepatic stellate cells and liver fibrosis. Compr Physiol. 2013;3:1473–1492. doi: 10.1002/cphy.c120035. [DOI] [PubMed] [Google Scholar]
- 10.Schwabe RF, Tabas I, Pajvani UB. Mechanisms of fibrosis development in nonalcoholic steatohepatitis. Gastroenterology. 2020;158:1913–1928. doi: 10.1053/j.gastro.2019.11.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu B, Tian Y, Chen M, Shen H, Xia J, Nan J, Yan T, Wang Y, Shi L, Shen B, et al. CircUBAP2 promotes MMP9-mediated oncogenic effect via sponging miR-194-3p in hepatocellular carcinoma. Front Cell Dev Biol. 2021;9:675043. doi: 10.3389/fcell.2021.675043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shen H, Liu B, Xu J, Zhang B, Wang Y, Shi L, Cai X. Circular RNAs: characteristics, biogenesis, mechanisms and functions in liver cancer. J Hematol Oncol. 2021;14:134. doi: 10.1186/s13045-021-01145-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shi L, Liu B, Shen DD, Yan P, Zhang Y, Tian Y, Hou L, Jiang G, Zhu Y, Liang Y, et al. A tumor-suppressive circular RNA mediates uncanonical integrin degradation by the proteasome in liver cancer. Sci Adv. 2021;7(13):eabe5043. doi: 10.1126/sciadv.abe5043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jin X, Gao J, Zheng R, Yu M, Ren Y, Yan T, Huang Y, Li Y. Antagonizing circRNA_002581-miR-122-CPEB1 axis alleviates NASH through restoring PTEN-AMPK-mTOR pathway regulated autophagy. Cell Death Dis. 2020;11:123. doi: 10.1038/s41419-020-2293-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu X, Wang X, Li J, Hu S, Deng Y, Yin H, Bao X, Zhang QC, Wang G, Wang B, et al. Identification of mecciRNAs and their roles in the mitochondrial entry of proteins. Sci China Life Sci. 2020;63:1429–1449. doi: 10.1007/s11427-020-1631-9. [DOI] [PubMed] [Google Scholar]
- 16.Liu X, Yang Y, Shan G. Identification and detection of mecciRNAs. Methods. 2021;196:147–152. doi: 10.1016/j.ymeth.2021.02.006. [DOI] [PubMed] [Google Scholar]
- 17.Zhao Q, Liu J, Deng H, Ma R, Liao JY, Liang H, Hu J, Li J, Guo Z, Cai J, et al. Targeting mitochondria-located circRNA SCAR alleviates NASH via reducing mROS output. Cell. 2020;183(76–93):e22. doi: 10.1016/j.cell.2020.08.009. [DOI] [PubMed] [Google Scholar]
- 18.Miranda KC, Huynh T, Tay Y, Ang YS, Tam WL, Thomson AM, Lim B, Rigoutsos I. A pattern-based method for the identification of MicroRNA binding sites and their corresponding heteroduplexes. Cell. 2006;126:1203–1217. doi: 10.1016/j.cell.2006.07.031. [DOI] [PubMed] [Google Scholar]
- 19.Kozomara A, Birgaoanu M, Griffiths-Jones S. miRBase: from microRNA sequences to function. Nucleic Acids Res. 2019;47:D155–D162. doi: 10.1093/nar/gky1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Glazar P, Papavasileiou P, Rajewsky N. circBase: a database for circular RNAs. RNA. 2014;20:1666–1670. doi: 10.1261/rna.043687.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hsu SD, Lin FM, Wu WY, Liang C, Huang WC, Chan WL, Tsai WT, Chen GZ, Lee CJ, Chiu CM, et al. miRTarBase: a database curates experimentally validated microRNA-target interactions. Nucleic Acids Res. 2011;39:D163–169. doi: 10.1093/nar/gkq1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vergoulis T, Vlachos IS, Alexiou P, Georgakilas G, Maragkakis M, Reczko M, Gerangelos S, Koziris N, Dalamagas T, Hatzigeorgiou AG. TarBase 60: capturing the exponential growth of miRNA targets with experimental support. Nucleic Acids Res. 2012;40:D222–229. doi: 10.1093/nar/gkr1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xiao F, Zuo Z, Cai G, Kang S, Gao X, Li T. miRecords: an integrated resource for microRNA-target interactions. Nucleic Acids Res. 2009;37:D105–110. doi: 10.1093/nar/gkn851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu X, Wang S, Meng F, Wang J, Zhang Y, Dai E, Yu X, Li X, Jiang W. SM2miR: a database of the experimentally validated small molecules' effects on microRNA expression. Bioinformatics. 2013;29:409–411. doi: 10.1093/bioinformatics/bts698. [DOI] [PubMed] [Google Scholar]
- 25.Rukov JL, Wilentzik R, Jaffe I, Vinther J, Shomron N. Pharmaco-miR: linking microRNAs and drug effects. Brief Bioinform. 2014;15:648–659. doi: 10.1093/bib/bbs082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jiang Q, Wang Y, Hao Y, Juan L, Teng M, Zhang X, Li M, Wang G, Liu Y. miR2Disease: a manually curated database for microRNA deregulation in human disease. Nucleic Acids Res. 2009;37:D98–104. doi: 10.1093/nar/gkn714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ruepp A, Kowarsch A, Theis F. PhenomiR: microRNAs in human diseases and biological processes. Methods Mol Biol. 2012;822:249–260. doi: 10.1007/978-1-61779-427-8_17. [DOI] [PubMed] [Google Scholar]
- 28.Dai E, Yu X, Zhang Y, Meng F, Wang S, Liu X, Liu D, Wang J, Li X, Jiang W. EpimiR: a database of curated mutual regulation between miRNAs and epigenetic modifications. Database. 2014;2014:023. doi: 10.1093/database/bau023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Keerthikumar S, Chisanga D, Ariyaratne D, Al Saffar H, Anand S, Zhao K, Samuel M, Pathan M, Jois M, Chilamkurti N, et al. ExoCarta: a web-based compendium of exosomal cargo. J Mol Biol. 2016;428:688–692. doi: 10.1016/j.jmb.2015.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Szklarczyk D, Gable AL, Nastou KC, Lyon D, Kirsch R, Pyysalo S, Doncheva NT, Legeay M, Fang T, Bork P, et al. The STRING database in 2021: customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021;49:D605–D612. doi: 10.1093/nar/gkaa1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wilkerson MD, Hayes DN. ConsensusClusterPlus: a class discovery tool with confidence assessments and item tracking. Bioinformatics. 2010;26:1572–1573. doi: 10.1093/bioinformatics/btq170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bindea G, Mlecnik B, Tosolini M, Kirilovsky A, Waldner M, Obenauf AC, Angell H, Fredriksen T, Lafontaine L, Berger A, et al. Spatiotemporal dynamics of intratumoral immune cells reveal the immune landscape in human cancer. Immunity. 2013;39:782–795. doi: 10.1016/j.immuni.2013.10.003. [DOI] [PubMed] [Google Scholar]
- 33.Misir S, Wu N, Yang BB. Specific expression and functions of circular RNAs. Cell Death Differ. 2022;29(3):481–491. doi: 10.1038/s41418-022-00948-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Koliaki C, Szendroedi J, Kaul K, Jelenik T, Nowotny P, Jankowiak F, Herder C, Carstensen M, Krausch M, Knoefel WT, et al. Adaptation of hepatic mitochondrial function in humans with non-alcoholic fatty liver is lost in steatohepatitis. Cell Metab. 2015;21:739–746. doi: 10.1016/j.cmet.2015.04.004. [DOI] [PubMed] [Google Scholar]
- 35.Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, Marzluff WF, Sharpless NE. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19:141–157. doi: 10.1261/rna.035667.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen LL. The expanding regulatory mechanisms and cellular functions of circular RNAs. Nat Rev Mol Cell Biol. 2020;21:475–490. doi: 10.1038/s41580-020-0243-y. [DOI] [PubMed] [Google Scholar]
- 37.Bosson AD, Zamudio JR, Sharp PA. Endogenous miRNA and target concentrations determine susceptibility to potential ceRNA competition. Mol Cell. 2014;56:347–359. doi: 10.1016/j.molcel.2014.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Denzler R, Agarwal V, Stefano J, Bartel DP, Stoffel M. Assessing the ceRNA hypothesis with quantitative measurements of miRNA and target abundance. Mol Cell. 2014;54:766–776. doi: 10.1016/j.molcel.2014.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu CX, Chen LL. Circular RNAs: characterization, cellular roles, and applications. Cell. 2022;185(12):2016–2034. doi: 10.1016/j.cell.2022.04.021. [DOI] [PubMed] [Google Scholar]
- 40.Zhang W, Liu Y, Min Z, Liang G, Mo J, Ju Z, Zeng B, Guan W, Zhang Y, Chen J, et al. circMine: a comprehensive database to integrate, analyze and visualize human disease-related circRNA transcriptome. Nucleic Acids Res. 2022;50:D83–D92. doi: 10.1093/nar/gkab809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fan Y, Siklenka K, Arora SK, Ribeiro P, Kimmins S, Xia J. miRNet - dissecting miRNA-target interactions and functional associations through network-based visual analysis. Nucleic Acids Res. 2016;44:W135–141. doi: 10.1093/nar/gkw288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhu C, Tabas I, Schwabe RF, Pajvani UB. Maladaptive regeneration - the reawakening of developmental pathways in NASH and fibrosis. Nat Rev Gastroenterol Hepatol. 2021;18:131–142. doi: 10.1038/s41575-020-00365-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang L, Roh YS, Song J, Zhang B, Liu C, Loomba R, Seki E. Transforming growth factor beta signaling in hepatocytes participates in steatohepatitis through regulation of cell death and lipid metabolism in mice. Hepatology. 2014;59:483–495. doi: 10.1002/hep.26698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huang M, Kim HG, Zhong X, Dong C, Zhang B, Fang Z, Zhang Y, Lu X, Saxena R, Liu Y, et al. Sestrin 3 protects against diet-induced nonalcoholic steatohepatitis in mice through suppression of transforming growth factor beta signal transduction. Hepatology. 2020;71:76–92. doi: 10.1002/hep.30820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Winkler M, Staniczek T, Kurschner SW, Schmid CD, Schonhaber H, Cordero J, Kessler L, Mathes A, Sticht C, Nessling M, et al. Endothelial GATA4 controls liver fibrosis and regeneration by preventing a pathogenic switch in angiocrine signaling. J Hepatol. 2021;74:380–393. doi: 10.1016/j.jhep.2020.08.033. [DOI] [PubMed] [Google Scholar]
- 46.Ramirez T, Li YM, Yin S, Xu MJ, Feng D, Zhou Z, Zang M, Mukhopadhyay P, Varga ZV, Pacher P, et al. Aging aggravates alcoholic liver injury and fibrosis in mice by downregulating sirtuin 1 expression. J Hepatol. 2017;66:601–609. doi: 10.1016/j.jhep.2016.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kisseleva T, Brenner D. Molecular and cellular mechanisms of liver fibrosis and its regression. Nat Rev Gastroenterol Hepatol. 2021;18:151–166. doi: 10.1038/s41575-020-00372-7. [DOI] [PubMed] [Google Scholar]
- 48.Xu Y, Sun X, Zhang R, Cao T, Cai SY, Boyer JL, Zhang X, Li D, Huang Y. A positive feedback loop of TET3 and TGF-beta1 promotes liver fibrosis. Cell Rep. 2020;30(1310–1318):e1315. doi: 10.1016/j.celrep.2019.12.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gao J, Wei B, de Assuncao TM, Liu Z, Hu X, Ibrahim S, Cooper SA, Cao S, Shah VH, Kostallari E. Hepatic stellate cell autophagy inhibits extracellular vesicle release to attenuate liver fibrosis. J Hepatol. 2020;73:1144–1154. doi: 10.1016/j.jhep.2020.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Isaac R, Reis FCG, Ying W, Olefsky JM. Exosomes as mediators of intercellular crosstalk in metabolism. Cell Metab. 2021;33:1744–1762. doi: 10.1016/j.cmet.2021.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wruck W, Kashofer K, Rehman S, Daskalaki A, Berg D, Gralka E, Jozefczuk J, Drews K, Pandey V, Regenbrecht C, et al. Multi-omic profiles of human non-alcoholic fatty liver disease tissue highlight heterogenic phenotypes. Sci Data. 2015;2:150068. doi: 10.1038/sdata.2015.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bai J, Xia M, Xue Y, Ma F, Cui A, Sun Y, Han Y, Xu X, Zhang F, Hu Z, et al. Thrombospondin 1 improves hepatic steatosis in diet-induced insulin-resistant mice and is associated with hepatic fat content in humans. EBioMedicine. 2020;57:102849. doi: 10.1016/j.ebiom.2020.102849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.van Koppen A, Verschuren L, van den Hoek AM, Verheij J, Morrison MC, Li K, Nagabukuro H, Costessi A, Caspers MPM, van den Broek TJ, et al. Uncovering a predictive molecular signature for the onset of NASH-related fibrosis in a translational NASH mouse model. Cell Mol Gastroenterol Hepatol. 2018;5(83–98):e10. doi: 10.1016/j.jcmgh.2017.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jung KH, Yoo W, Stevenson HL, Deshpande D, Shen H, Gagea M, Yoo SY, Wang J, Eckols TK, Bharadwaj U, et al. Multifunctional effects of a small-molecule STAT3 inhibitor on NASH and hepatocellular carcinoma in mice. Clin Cancer Res. 2017;23:5537–5546. doi: 10.1158/1078-0432.CCR-16-2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gomes AL, Teijeiro A, Buren S, Tummala KS, Yilmaz M, Waisman A, Theurillat JP, Perna C, Djouder N. Metabolic inflammation-associated IL-17A causes non-alcoholic steatohepatitis and hepatocellular carcinoma. Cancer Cell. 2016;30:161–175. doi: 10.1016/j.ccell.2016.05.020. [DOI] [PubMed] [Google Scholar]
- 56.Wang Y, Wen H, Fu J, Cai L, Li PL, Zhao CL, Dong ZF, Ma JP, Wang X, Tian H, et al. Hepatocyte TNF receptor-associated factor 6 aggravates hepatic inflammation and fibrosis by promoting lysine 6-linked polyubiquitination of apoptosis signal-regulating kinase 1. Hepatology. 2020;71:93–111. doi: 10.1002/hep.30822. [DOI] [PubMed] [Google Scholar]
- 57.Sutti S, Albano E. Adaptive immunity: an emerging player in the progression of NAFLD. Nat Rev Gastroenterol Hepatol. 2020;17:81–92. doi: 10.1038/s41575-019-0210-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ferreyra Solari NE, Inzaugarat ME, Baz P, De Matteo E, Lezama C, Galoppo M, Galoppo C, Chernavsky AC. The role of innate cells is coupled to a Th1-polarized immune response in pediatric nonalcoholic steatohepatitis. J Clin Immunol. 2012;32:611–621. doi: 10.1007/s10875-011-9635-2. [DOI] [PubMed] [Google Scholar]
- 59.Rolla S, Alchera E, Imarisio C, Bardina V, Valente G, Cappello P, Mombello C, Follenzi A, Novelli F, Carini R. The balance between IL-17 and IL-22 produced by liver-infiltrating T-helper cells critically controls NASH development in mice. Clin Sci. 2016;130:193–203. doi: 10.1042/CS20150405. [DOI] [PubMed] [Google Scholar]
- 60.Rinella ME, Elias MS, Smolak RR, Fu T, Borensztajn J, Green RM. Mechanisms of hepatic steatosis in mice fed a lipogenic methionine choline-deficient diet. J Lipid Res. 2008;49:1068–1076. doi: 10.1194/jlr.M800042-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.McCuskey RS, Ito Y, Robertson GR, McCuskey MK, Perry M, Farrell GC. Hepatic microvascular dysfunction during evolution of dietary steatohepatitis in mice. Hepatology. 2004;40:386–393. doi: 10.1002/hep.20302. [DOI] [PubMed] [Google Scholar]
- 62.Younossi Z, Tacke F, Arrese M, Chander Sharma B, Mostafa I, Bugianesi E, Wai-Sun Wong V, Yilmaz Y, George J, Fan J, Vos MB. Global perspectives on nonalcoholic fatty liver disease and nonalcoholic steatohepatitis. Hepatology. 2019;69:2672–2682. doi: 10.1002/hep.30251. [DOI] [PubMed] [Google Scholar]
- 63.Yuan X, Diao J, Du A, Wen S, Zhou L, Pan Y. Circular RNA expression profiles and features in NAFLD mice: a study using RNA-seq data. J Transl Med. 2020;18:476. doi: 10.1186/s12967-020-02637-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang G, Chen HW, Oktay Y, Zhang J, Allen EL, Smith GM, Fan KC, Hong JS, French SW, McCaffery JM, et al. PNPASE regulates RNA import into mitochondria. Cell. 2010;142:456–467. doi: 10.1016/j.cell.2010.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cheng Y, Liu P, Zheng Q, Gao G, Yuan J, Wang P, Huang J, Xie L, Lu X, Tong T, et al. Mitochondrial trafficking and processing of telomerase RNA TERC. Cell Rep. 2018;24:2589–2595. doi: 10.1016/j.celrep.2018.08.003. [DOI] [PubMed] [Google Scholar]
- 66.Smirnov A, Comte C, Mager-Heckel AM, Addis V, Krasheninnikov IA, Martin RP, Entelis N, Tarassov I. Mitochondrial enzyme rhodanese is essential for 5 S ribosomal RNA import into human mitochondria. J Biol Chem. 2010;285:30792–30803. doi: 10.1074/jbc.M110.151183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ruprecht JJ, Kunji ERS. The SLC25 mitochondrial carrier family: structure and mechanism. Trends Biochem Sci. 2020;45:244–258. doi: 10.1016/j.tibs.2019.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Herai RH, Negraes PD, Muotri AR. Evidence of nuclei-encoded spliceosome mediating splicing of mitochondrial RNA. Hum Mol Genet. 2017;26:2472–2479. doi: 10.1093/hmg/ddx142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chen L, Wang C, Sun H, Wang J, Liang Y, Wang Y, Wong G. The bioinformatics toolbox for circRNA discovery and analysis. Brief Bioinform. 2021;22:1706–1728. doi: 10.1093/bib/bbaa001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mansouri A, Gattolliat CH, Asselah T. Mitochondrial dysfunction and signaling in chronic liver diseases. Gastroenterology. 2018;155:629–647. doi: 10.1053/j.gastro.2018.06.083. [DOI] [PubMed] [Google Scholar]
- 71.Hotamisligil GS. Inflammation, metaflammation and immunometabolic disorders. Nature. 2017;542:177–185. doi: 10.1038/nature21363. [DOI] [PubMed] [Google Scholar]
- 72.Ruan X, Li P, Chen Y, Shi Y, Pirooznia M, Seifuddin F, Suemizu H, Ohnishi Y, Yoneda N, Nishiwaki M, et al. In vivo functional analysis of non-conserved human lncRNAs associated with cardiometabolic traits. Nat Commun. 2020;11:45. doi: 10.1038/s41467-019-13688-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hebbard L, George J. Animal models of nonalcoholic fatty liver disease. Nat Rev Gastroenterol Hepatol. 2011;8:35–44. doi: 10.1038/nrgastro.2010.191. [DOI] [PubMed] [Google Scholar]
- 74.Ma J, Tan X, Kwon Y, Delgado ER, Zarnegar A, DeFrances MC, Duncan AW, Zarnegar R. A novel humanized model of NASH and its treatment with META4, a potent agonist of MET. Cell Mol Gastroenterol Hepatol. 2022;13:565–582. doi: 10.1016/j.jcmgh.2021.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Supplementary Figure. 1 mecciRNAs Regulate Fibrosis-related Signaling Pathways in HSCs
Additional file 2: Supplementary Figure. 2 A Novel Immunotyping of NASH Based on mecciRNA-related Network
Additional file 3: Supplementary Figure Legends and Primer Sequence.
Additional file 4: Supplementary Table. 1 The 2000 mRNAs Targeted by 26 miRNAs in miRNet Database.
Additional file 5: Supplementary Table. 2 The 1343 mRNAs Targeted by 7 exosomal miRNAs in miRNet Database.
Additional file 6: Supplementary Table. 3 . The Evidence Indicates The Presence of These Following 7 miRNAs in Exosomes.
Additional file 7: Supplementary Table. 4 The 419 mRNAs Regulated by mecciRNA-miRNA Network in Hepatocytes.
Additional file 8: Supplementary Table. 5 The 273 Differentially Expressed Genes Between Cluster 1 and Cluster 2.
Data Availability Statement
The data sets analysed during this study are available in public, open access repositories listed in this article.