Abstract
Background:
Campylobacter species are the zoonotic bacteria and the most common cause of foodborne gastroenteritis around the world. The link between human campylobacteriosis and infected poultry consumption has been well established.
Aims:
In this study, we aimed to isolate Campylobacter spp. from chicken and characterize them with molecular methods.
Methods:
Totally, 241 chicken caecal mucosal scrapings were collected from five districts of Tamil Nadu. Bacterial isolation was done by plating on blood-free Campylobacter selective medium with supplements. Campylobacter species were identified by multiplex PCR and Campylobacter coli isolates were tested for 11 virulence genes by PCR. C. coli isolates were typed based on seven housekeeping genes multilocus sequence typing (MLST) scheme. The antimicrobial susceptibility was determined by a microdilution resazurin assay.
Results:
The prevalence of C. coli and C. jejuni were 14.94% (36/241), and 3.32% (8/241), respectively. The virulence genes flaA, flaB, cadF, cdtA, cdtB, cdtC, ciaB, and ceuE were present in all 36 C. coli isolates, pldA and racR genes were present in 58.33% (21/36), and 16.67% (6/36) of the isolates, respectively, and dnaJ was present in only one isolate. Two novel sequence types (ST-10872, ST-11031) were found in this study. Though different STs were identified, all the STs belonged to the same clonal complex of ST-828. All 14 C. coli isolates showed 100% resistance to nalidixic acid, and higher resistance to tetracycline (92.8%), erythromycin (71.4%), clindamycin (71.4%), and azithromycin (64.2%) was noticed. All C. coli isolates were sensitive to chloramphenicol, and higher sensitivity to ciprofloxacin (78.5%), and gentamicin (71.4%) was observed.
Conclusion:
The present study demonstrated that C. coli is more prevalent in broilers than C. jejuni in Tamil Nadu. The presence of C. jejuni and C. coli in chicken caecal samples from the slaughterhouse are indicative of the possibility of public health hazards.
Key Words: Antimicrobial resistance, Campylobacter coli, Multilocus sequence typing, Multiplex PCR, Virulence genes
Introduction
India is one of the world’s countries with the largest poultry population, and in this country, poultry meat consumption is commonly high due to Indian culture (Khan et al., 2018). The poultry population in Tamil Nadu had a tremendous growth rate and reached a total population of 120.8 million in 2019, the highest poultry population in India (Livestock Census of India, 2019). There is a risk of transmission of diseases from poultry in such an intensive state. Campylobacteriosis is a zoonotic disease that is predominantly spread from poultry and poultry products. The prevalence of Campylobacter spp. in chicken and its products were reported to be 4.9% to 100% globally (Kaakoush et al., 2015).
All the domestic poultry species like chickens, turkeys, ducks, and geese harbours Campylobacter spp., but the highest colonisation and prevalence was seen in chickens (Byrd et al., 2001). Despite the highest colonisation rate, there were no clinical symptoms or reduced life span in chickens (Sahin et al., 2001). Consumption of poultry meat was incriminated for 20% to 30% of campylobacteriosis cases in the European Union (Skarp et al., 2016). There are currently 17 species, and six subspecies in the Campylobacter genus. C. jeuni and C. coli are the most common species causing gastrointestinal infections in humans, but C. lari and C. upsaliensis are rarely associated with human infection (Skarp et al., 2016). In recent studies, C. coli was found to be more prevalent than C. jejuni in most countries (Walker et al., 2019; Wieczorek et al., 2020). Because of the phenotypic similarities between Campylobacter spp., identification of the individual species is difficult. Various PCR-based detection methods have been proposed. Multiplex PCR and LAMP assay could help in the fast and effective diagnosis of Campylobacter spp. (Wang et al., 2002; Toplak et al., 2012; Romero et al., 2018).
The Campylobacter genus has complex multifactorial systems that include motility, chemotaxis, adherence, invasion, antioxidant resistance, heat shock, and the ability to enter the viable but not cultivable (VBNC) state (to replicate in chickens), survive during food processing, and cause virulence in humans (Bolton et al., 2015). The virulence capacity of Campylobacter spp. was found to be increased in humans when passed through the poultry (Wysok et al., 2020).
Antimicrobial resistance is a worldwide threat. Antibiotics such as fluoroquinolones, macrolides, and tetracycline are widely used to treat campylobacteriosis. However, resistance to these antibiotics develops quickly (Engberg et al., 2001). Campylobacter’s resistance to fluoroquinolones had been identified even in the absence of their use (Caffrey et al., 2021).
Campylobacteriosis is a sporadic disease, and the detection of the bacterial sources is epidemiologically difficult. Multilocus sequence typing (MLST) of C. jejuni/coli assists in the recognition of its sequence types and clonal complex associated with the source of isolation. MLST could aid in effective public-health measures to control campylobacteriosis (Vasiliki et al., 2013). There is no previous study on the prevalence of C. jejuni/coli sequence types or the most common clonal complexes in India. Hence, this research aimed to detect virulence factors, type the C. coli isolates from chickens, and determine the minimum inhibitory concentration (MIC) of C. coli isolates against various antimicrobials.
Materials and Methods
Sample collection and Campylobacter spp. isolation
A total of 241 chicken intestine samples were collected from five different districts of Tamil Nadu. Among the 241 samples, 97 intestine samples were collected from five various areas of the Chennai district, and 36 samples each from six separate areas of Namakkal, Krishnagiri, Erode, and Coimbatore districts. Chicken caecal contents were cleared, and the mucosal layer was scraped with the help of a glass slide. Scraped contents of samples from the Chennai district were directly streaked on the sterile blood-free Campylobacter broth base (HiMEDIA catalogue #M1318) with 2% Agar agar type 1, Campylobacter growth supplement IV (HiMEDIA catalogue #FD042), and CCDA supplement (HiMEDIA catalogue #FD135), then the plates were incubated at 42°C for 48 h in microaerophilic condition. Scraped caecal contents from other districts were transferred into a 1.5 ml sterile microcentrifuge tube containing 1 ml of blood free Campylobacter broth with CCDA, and Campylobacter growth supplement IV, then transported to the laboratory for Campylobacter isolation as mentioned above.
Colonies on the selective media were checked by staining with dilute carbol fuchsin, oxidase, and catalase tests for presumptive identification as Campylobacter.
Multiplex PCR
To confirm Campylobacter spp. isolates, multiplex PCR was used. Bacterial genomic DNA was extracted by boiling method (Ananda Chitra et al., 2015). Briefly, each colony was suspended in nuclease-free water and boiled at 100°C for 10 min. The supernatant was separated by centrifugation at 11,200 × g for 10 min and later used as a template in the amplification reactions. The details of primers are given in Table 1. Multiplex PCR was performed in 20 μL reaction volume containing 100-150 ng of DNA, 0.2 μM of 23S rRNA primers, 0.5 μM of C. jejuni and C. lari primers, 1 μM of C. coli primers and 2 μM C. upsaliensis primers in 2X master mix (Ampliqon, Denmark) with the following conditions: initial denaturation at 95°C for 5 min, followed by 30 cycles of denaturation at 95°C for 30 s, annealing at 59°C for 30 s and elongation at 72°C for 30 s, and a final elongation of 72°C for 7 min. Amplified PCR products were electrophoresed in 2% agarose gel stained with ethidium bromide, visualized under UV, and documented.
Campylobacter virulence genes and virulence gene profiling
The Campylobacter isolates were tested for the presence of 11 virulence genes using PCR. The virulence genes include those involved in motility (flaA, flaB), cell adhesion (cadF, dnaJ, pldA and racR), invasion (ciaB, ceuE), and cytotoxin production (cdtA, cdtB, cdtC). The details of the PCR primers are shown in Table 2. For virulence gene profile analysis, binary code 1 was used for gene presence and 0 for gene absence. Hierarchical clustering was carried out using Displayr software (https://app.displayr.com/).
Table 1.
Multiplex PCR primers along with amplicon size, and references used for identification of Campylobacter spp.
| S. No. | Gene/species | Primer sequences (5´-3´) | References | Product size bp |
|---|---|---|---|---|
| 1 | 23S rRNA | F-TATACCGGTAAGGAGTGCTGGAG | Wang et al. (2002) | 650 |
| R-ATCAATTAACCTTCGAGCACCG | ||||
| 2 | hipO | F-ACTTCTTTATTGCTTGCTGC | Wang et al. (2002) | 323 |
| C. jejuni | R-GCCACAACAAGTAAAGAAGC | |||
| 3 | glyA | F-GTAAAACCAAAGCTTATCGTG | Wang et al. (2002) | 126 |
| C. coli | R-TCCAGCAATGTGTGCAATG | |||
| 4 | glyA | F-TAGAGAGATAGCAAAAGAGA | Wang et al. (2002) | 251 |
| C. lari | R-TACACATAATAATCCCACCC | |||
| 5 | glyA | F-AATTGAAACTCTTGCTATCC | Wang et al. (2002) | 204 |
| C. upsaliensis | R-TCATACATTTTACCCGAGCT |
Table 2.
Primers along with amplicon size, annealing temperature, and references used for the detection of virulence genes in Campylobacter coli
| S. No. | Gene | Primer sequence (5´-3´) | References | Annealing Temp. °C |
Product size bp |
|---|---|---|---|---|---|
| 1 | flaA | F-AATAAAAATGCTGATAAAACAGGTG | Datta et al. (2003) | 53 | 855 |
| R-TACCGAACCAATGTCTGCTCTGATT | |||||
| 2 | flaB | F-AAGGATTTAAAATGGGTTTTAGAATAAACACC | Goon et al. (2003) | 57 | 260 |
| R-GCTCATCCATAGCTTTATCTGC | |||||
| 3 | cadF | F-TTGAAGGTAATTTAGATATG | Konkel et al. (1999) | 48 | 400 |
| R-CTAATACCTAAAGTTGAAAC | |||||
| 4 | dnaJ | F-AAGGCTTTGGCTCATC | Datta et al. (2003) | 41 | 720 |
| R-CTTTTTGTTCATCGTT | |||||
| 5 | pldA | F-AAGAGTGAGGCGAAATTCCA | Zheng et al. (2006) | 55 | 384 |
| R-GCAAGATGGCAGGATTATCA | |||||
| 6 | racR | F-GATGATCCTGACTTTG | Datta et al. (2003) | 45 | 584 |
| R-TCTCCTATTTTTACCC | |||||
| 7 | cdtA | F-TGTCCCACCTGTAATCACTCC | Denis et al. (2017) | 57 | 245 |
| R-CTCTTGCATCTCCAAAAGGTCT | |||||
| 8 | cdtB | F-GAGTGGATGTAGGAGCAAATCG | Denis et al. (2017) | 57 | 332 |
| R-CGTAGAAGAAGGCGGAACAAC | |||||
| 9 | cdtC | F-AGCTTGGATGAATTAGCAGACT | Denis et al. (2017) | 57 | 403 |
| R-TGGCGATACTAGAGTCAGGAAA | |||||
| 10 | ciaB | F-TGCGAGATTTTTCGAGAATG | Denis et al. (2017) | 57 | 284 |
| R-TGCCCGCCTTAGAACTTACA | |||||
| 11 | CeuE | F-ATGAAAAAATATTTAGTTTTTGCA | Gonzalez et al. (1997) | 49 | 894 |
| R-ATTTTATTATTTGTAGCAGCG |
Multilocus sequence typing (MLST)
C. coli isolates were typed based on seven housekeeping genes including, aspA (aspatase), glnA (glutamine synthetase), gltA (citrate synthase), glyA (serine hydroxy methyl transferase), pgm (phospho glucomutase), tkt (transketolase), and uncA (ATP synthase alpha subunit). Primers details used for MLST are given in Table 3. PCR was performed in 20 µL reaction volume for each gene of each isolate. The reaction mixture containing 100-150 ng of DNA, and 1 μM of each primer in 2X PrimeStar master mix (Takara Bio Inc., Japan) which contained high fidelity hot-start DNA polymerase. PCR was performed under the thermal conditions of initial denaturation at 98°C for 2 min, followed by 34 cycles of denaturation at 98°C for 10 s, annealing at 53°C for 30 s and elongation at 72°C for 30 s, and a final elongation of 72°C for 5 min. Amplified PCR products were loaded in 1.5% agarose gel stained with ethidium bromide, and electrophoresed, then visualized by a transilluminator. The PCR products were purified by a gel purification kit (Favorgen, catalogue #FAGC 001) according to the manufacturer’s instructions. Purified PCR products were sequenced by Sanger’s DNA sequencing method in a commercial firm (Eurofins Genomics India Pvt. Ltd.). Forward and reverse sequences for each gene were aligned using the MEGAX program. Allele numbers and profiles were documented by submitting the aligned sequences on the PubMLST website (https://pubmlst.org/organisms/ campylobacter-jejunicoli/) (Jolley et al., 2018).
Minimum inhibitory concentration (MIC)
The antimicrobial susceptibility was determined by a microdilution resazurin assay. Briefly, the antibiotic powders containing, azithromycin, chloramphenicol, clindamycin, ciprofloxacin, erythromycin, gentamicin, nalidixic acid, and tetracycline were individually diluted in suitable diluents to make 1 mg/ml stock solutions. The colonies were directly transferred from the plate into Brucella broth with 10% horse serum, and the suspension was matched to the 0.5 McFarland standard. The antibiotics starting from 100 µg/ml concentration, were diluted at two folds in Brucella broth with 10% horse serum in flat bottom sterile cell culture plate, and 100 µL of bacterial suspension was added to all wells. After 24 h incubation at 42°C under microaerophilic conditions, 10 µL (0.2%) of resazurin dye was added to each well, and plates were further incubated at 42°C for 3 h, then observed for the colour change. CLSI breakpoints (2020) were used for determining the MIC of erythromycin, ciprofloxacin, and tetracycline, and NARMS breakpoints (2020) were used for the other tested antimicrobials.
Table 3.
Primers along with amplicon size, and references used for MLST amplification and sequencing of C. coli isolates
| S. No. | Gene | Primer sequence (5´-3´) | References | Product size (bp) |
|---|---|---|---|---|
| 1 | aspA | F-CAACTTCAAGATGCAGTACC | Dingle et al. (2001) | 594 |
| R-ATCTGCTAAAGTATGCATTGC | ||||
| 2 | glnA | F-TTCATGGATGGCAACCTATTG | Dingle et al. (2001) | 615 |
| R-GCTTTGGCATAAAAGTTGCAG | ||||
| 3 | gltA | F-GATGTAGTGCATCTTTTACTC | Dingle et al. (2001) | 528 |
| R-AAGCGCTCCAATACCTGCTG | ||||
| 4 | glyA | F-TCAAGGCGTTTATGCTGCAC | Dingle et al. (2001) | 627 |
| R-CCATCACTTACAAGCTTATAC | ||||
| 5 | Pgm | F-TTATAAGGTAGCTCCGACTG | Dingle et al. (2001) | 649 |
| R-GTTCCGAATAGCGAAATAACAC | ||||
| 6 | Tkt | F-AGGCTTGTGTTTTCAGGCGG | Dingle et al. (2001) | 581 |
| R-TGACTTCCTTCAAGCTCTCC | ||||
| 7 | uncA | F-AAGCACAGTGGCTCAAGTTG | Dingle et al. (2001) | 614 |
| R-CTACTTGCCTCATCCAATCAC |
Results
Prevalence of Campylobacter spp. in chicken
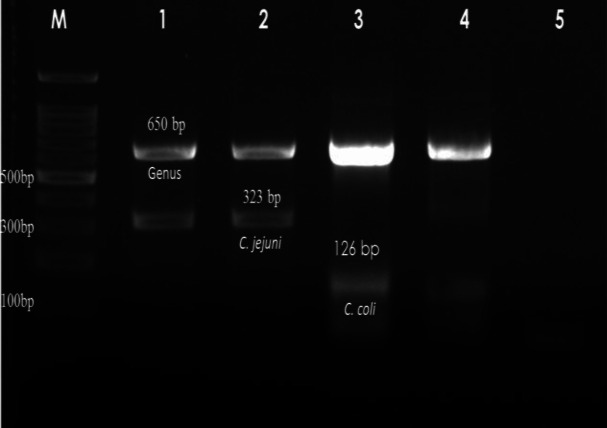
Out of 241 samples, 14.94% (36/241), and 3.32% (8/241) were found positive for C. coli, and C. jejuni, respectively, by multiplex PCR (Fig. 1). None of the samples was positive for C. lari and C. upsaliensis. Among 36 C. coli isolates, 35 were isolated from Chennai district, and one was isolated from Namakkal district of Tamil Nadu.
Fig. 1.
Agarose gel showing the multiplex PCR products of Campylobacter spp., C. jejuni and C. coli. M: 100 bp DNA marker. Lanes 1-2: C. jejuni, Lane 3: C. coli, Lane 4: Campylobacter spp., and Lane 5: Non-template control
Virulence genes profile
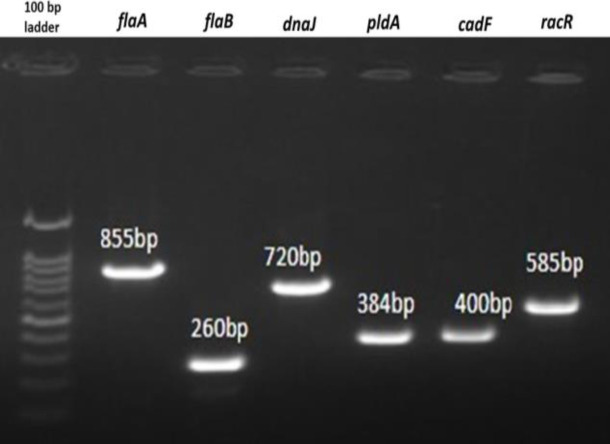
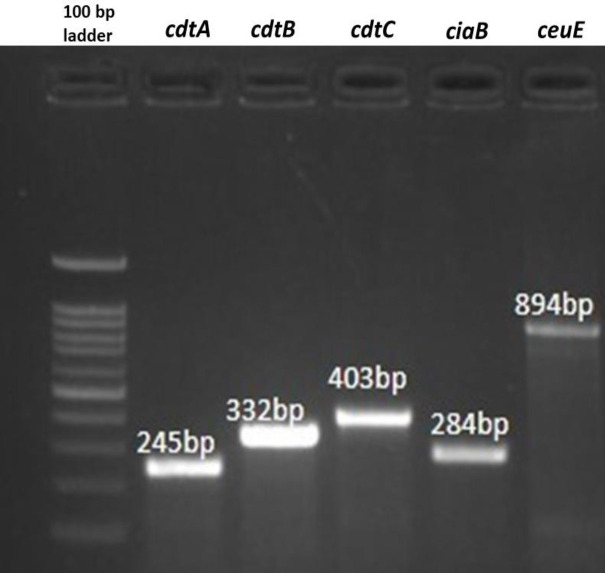
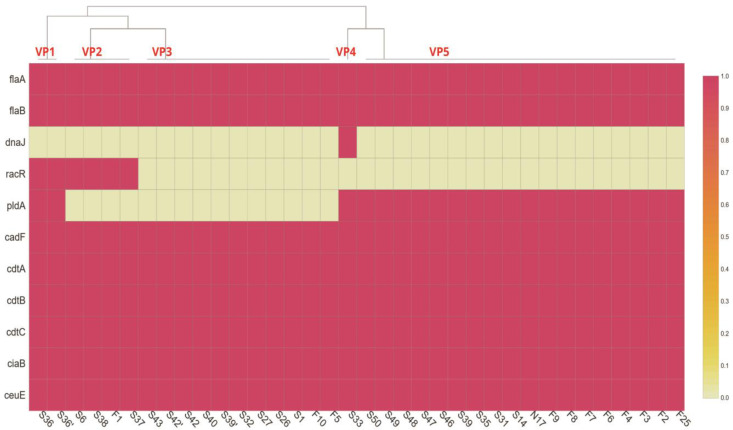
Virulence genes detection (Figs. 2 and 3) in 36 C. coli isolates showed that genes involved in motility (flaA and flab), invasion (ciaB, ceuE), and cytotoxin production (cdtA, cdtB, cdtC) were present in all the isolates, whereas the prevalence of genes involved in cell adhesion varies in C. coli isolates. The cadF, pldA, and racR genes were present in 100% (36/36), 58.33% (21/36), and 16.67% (6/36) of the isolates, respectively. The cell adhesion factor dnaJ was found in only one of the isolates (2.78%). A total of four different clusters and one individual virulence profile (VPs) were observed (Fig. 4). Virulence genes profile flaA-flaB-pldA-cadF-cdtA-cdtB-cdtC-ciaB-ceuE (VP5) had the maximum number of strains (50%), followed by VP3 with 31% (11/36), and VP2 with 11% (4/36) of the isolates.
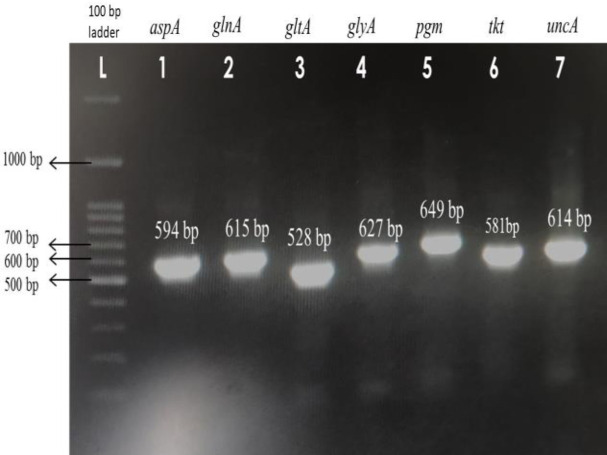
Multilocus sequence typing
Among the 36 C. coli isolates, five randomly selected C. coli isolates were subjected to the MLST sequencing method (Fig. 5). The details of the allele number, sequence type (ST), and clonal complex (CC) are given in Table 4. Allele numbers for glnA, gltA, and uncA were the same for all the five C. coli isolates. In contrast, tkt gene sequences were highly variable, which was evident by displaying four different alleles within five isolates. In this study, two novel STs were found in the Chennai area, and the newly assigned sequence types were ST-10872 and ST-11031. However, all the STs, including two new ones, belonged to the same clonal complex ST-828.
Antimicrobial resistance of C. coli isolates
All the 14 tested C. coli isolates were resistant to nalidixic acid (100%). Higher resistance was shown to tetracycline (92.8%), erythromycin (71.4%), and clindamycin (71.4%). For azithromycin, 64.2% of the isolates were resistant. On the contrary, all the C. coli isolates (100%) were sensitive to chloramphenicol. For ciprofloxacin, and gentamicin, 78.5%, and 71.4% of the isolates were respectively sensitive. In the present study, all the C. coli isolates were resistant to at least one tested antimicrobial. Three isolates exhibited an azithromycin-clindamycin-erythromycin-nalidixic acid-tetracycline antimicrobial resistant pattern. Five isolates were resistant to 5 tested antimicrobials, and three isolates were resistant to six different antimicrobials. One isolate was found to be sensitive to all antimicrobials except for nalidixic acid. Results of antimicrobial sensitive patterns are shown in Table 5.
Discussion
Campylobacter spp. are zoonotic bacteria that cause food poisoning. Conventional identification approaches are troublesome to some extent, since it necessitates fastidious growth criteria. Rapid diagnosis of Campylobacter spp. can be aided by molecular approaches such as multiplex PCR, and LAMP assay. Also, with the MLST system, the prevalent clone of Campylobacter in a region can be identified, and the sources of transmission could be traced. Additionally, the MIC is used to assess the sensitivity of Campylobacter spp. to various antimicrobials which will help the clinician to determine the drug dose for effective treatment of campylobacteriosis.
Fig. 2.
Agarose gel showing the PCR products of virulence genes flaA, flaB, dnaJ, pldA, cadF, and racR of C. coli isolates
Fig. 3.
Agarose gel showing the PCR products of virulence genes cdtA, cdtB, cdtC, ciaB, and ceuE of C. coli isolates
Fig. 4.
Heat map showing the virulence genes profiles of Campylobacter coli isolated from chickens. Virulence profiles are marked in red bold font as VP1, VP2, VP3, VP4, and VP5
Table 4.
Details of MLST of C. coli isolates and clonal complex
| S. No. | Isolate ID | Type of chicken | Genes-Allele number | ST | CC | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| aspA | glnA | gltA | glyA | pgm | tkt | uncA | |||||
| 1 | F1 | Broiler | 33 | 39 | 30 | 82 | 113 | 35 | 17 | 899 | 828 |
| 2 | F25 | Desi | 124 | 39 | 30 | 79 | 113 | 43 | 17 | 10872 | 828 |
| 3 | N17 | Giriraja | 33 | 39 | 30 | 82 | 113 | 44 | 17 | 872 | 828 |
| 4 | S1 | Broiler | 33 | 39 | 30 | 82 | 908 | 47 | 17 | 9108 | 828 |
| 5 | S6 | Broiler | 124 | 39 | 30 | 82 | 113 | 47 | 17 | 11031 | 828 |
ST: Sequence type, and CC: Clonal complex
Fig. 5.
Agarose gel showing PCR products of MLST genes - aspA, glnA, gltA, glyA, pgm, tkt, and uncA of C. coli isolates
Table 5.
Antimicrobial resistance profiles of C. coli isolates
| S. No. | Antimicrobial resistance profile | No. of antimicrobials | No. of isolates |
|---|---|---|---|
| 1 | NAL | 1 | 1 |
| 2 | AZI-NAL-TET | 3 | 1 |
| 3 | AZI-GEN-NAL-TET | 4 | 1 |
| 4 | AZI-CIP-NAL-TET | 4 | 1 |
| 5 | CLI-ERY-NAL-TET | 4 | 2 |
| 6 | CLI-ERY-GEN-NAL-TET | 5 | 2 |
| 7 | AZI-CLI-ERY-NAL-TET | 5 | 3 |
| 8 | AZI-CIP-CLI-ERY-NAL-TET | 6 | 2 |
| 9 | AZI-CLI-ERY-GEN-NAL-TET | 6 | 1 |
NAL: Nalidixic aid, AZI: Azithromycin, GEN: Gentamicin, CLI: Clindamycin, ERY: Erythromycin, TET: Tetracycline, and CIP: Ciprofloxacin
In the present study, the prevalence of C. coli (7%) was found in the districts of Tamil Nadu, India. Wieczorek et al. (2020) assessed the prevalence of Campylobacter spp. in chicken carcasses from 2014 to 2018 in Poland. Out of 2637 swab samples collected, 738 (28%) C. coli and 525 (22.2%) C. jejuni were isolated. Though C. coli isolates were more than C. jejuni, they were not predominant as observed in the present study. Walker et al. (2019) investigated the prevalence of C. jejuni and C. coli in retail chicken, beef, lamb, and pork products in Australia over two years. They found that C. coli were the most commonly isolated species from chicken meat, and offal. Whereas C. jejuni was primarily isolated from beef, lamb, and pork offal and meat. Few other studies also reported the predominance of C. coli over C. jejuni (Torralbo et al., 2015; Vinueza-Burgos et al., 2017; Rossler et al., 2020).
On the contrary, a study on the prevalence of Campylobacter spp. in chicken and chicken by-products in Japan revealed that out of 341 isolates, 278 (81.5%), and 63 (18.5%) were C. jejuni, and C. coli, respectively (Sallam et al., 2007). A very recent study by Wangroongsarb et al. (2021) also reported the predominance of C. jejuni (33.5%) over C. coli (18.2%). Prevalence of 38.6% and 24% of C. jejuni in chicken meat, and chicken intestine, respectively was recorded in Northern India (Khan et al., 2018).
However, the prevalence study of Campylobacter spp. in broiler chickens from rearing to slaughter in China revealed an almost equal prevalence of C. jejuni (66.3%), and C coli (60.4%) from 1534 samples (Tang et al., 2020).
In the present study, direct plating of caecal content yielded more Campylobacter isolates than plating after transportation in enrichment media. A similar experience was reported by Musgrove et al. (2001), who collected 128 caecal samples from broiler processing units, then plated them directly on Campy Cefex plates, and the same samples were also enriched in Bolton broth for 20 h at 42°C then plated on Campy Cefex agar plates. They could isolate Campylobacter spp. from all the directly plated agar plates (100%) but only from 63% of enriched caecal samples. These suggest that transportation in enrichment may lead to loss of Campylobacter, and direct plating is an efficient method for isolation of Campylobacter spp. However, Ugarte-Ruiz et al. (2012) reported that pre-enrichment step increased the isolation rate, and Kuana et al. (2008) reported that there was no statistical difference between the direct plating and enrichment method for isolation of Campylobacter spp.
There are several factors responsible for the survival and virulence of Campylobacter spp. Among the motility factors, Koolman et al. (2015) found that the prevalence of flaA was higher than that of flaB. However, in our study, both of these genes were present in all of the C. coli isolates. Gahamanyi et al. (2021) studied the virulence genes present in C. jejuni, and C. coli isolated from layer chickens in South Korea. They reported that the virulence genes flaA, cdtB, cadF, and dnaJ were found in all the C. coli isolates. In the present study, all the above-mentioned genes were present in all the C. coli isolates except dnaJ which was found in only one C. coli isolate.
Rossler et al. (2020) reported that flaA (100%), and cadF (92%) genes were highly prevalent in their Campylobacter spp. isolates which are similar to this study. Garcia-Sanchez et al. (2020) reported that cdtA and cdtC genes were absent in their C. coli isolates but were present in all C. coli isolates in the present study.
Out of five C. coli isolates MLST profiles, two new STs were found. However, all isolates belonged to ST-828 clonal complex. The exclusive predominance of ST-828 clonal complex was reported from Turkey by Kashoma et al. (2014), and Ecuador by Vinueza-Burgos et al. (2017). The presence of ST-899 in commercial broiler flocks was also reported by Ladely et al. (2017) in Georgia. ST-872 C. coli was reported from human campylobacteriosis cases in Luxembourg (Mossong et al., 2016), but it was isolated from broiler chicken in this study. All these studies proved that the ST-828 clonal complex was found to be predominant among C. coli isolates all over the world, and this clonal complex ST-828 was found in C. coli isolated from different sources like poultry, ruminants, and even humans. Since there is no previous study on the MLST profile of Indian Campylobacter species neither from humans nor from animals, the significance of the ST profile of this study could not be ascertained, and we could not even validate our findings. Hence, to the best of our knowledge, this is the first MLST study of Campylobacter isolates in India.
Most of the C. coli isolates in the present study were highly resistant to nalidixic acid, tetracycline, erythromycin, and clindamycin. Nevertheless, the study of Khan et al. (2018) on the antimicrobial resistance of C. jejuni isolates from North India showed that 59.4% of isolates were resistant to tetracycline, and even lower resistance (6.9% to 8.9%) was observed against nalidixic acid, ciprofloxacin, erythromycin, gentamicin, and azithromycin. In this study, 100% resistance to nalidixic acid was found in all C. coli isolates. A study in Thailand also disclosed that C. coli isolates from retail chickens were 100% resistant to quinolones, and they showed 76.8% resistance to cyclines, 37.7% resistance to macrolides, 36.2% resistance to clindamycin, and 13% resistance to gentamicin (Wangroongsarb et al., 2021). Wieczorek et al. (2020) reported that C. coli isolates from chicken were 93.9% resistant to ciprofloxacin and 93.8% resistant to nalidixic acid. However, in the present study, C. coli isolates were 100% resistant to nalidixic acid and 78.5% sensitive to ciprofloxacin. The possible reason for this kind of resistance could be due to the point mutation in the Quinolone determining region (QRDR) of gyrA gene of C. coli and C. jejuni, which confers resistance only to nalidixic acid without conferring resistance to ciprofloxacin. Thr86Ala mutation was the one that bestowed resistance to only nalidixic acid, but not to ciprofloxacin (Bachoual et al., 2001; Jesse et al., 2006). The higher resistance of Campylobacter isolates to nalidixic acid and tetracycline might be due to their usage in feed.
We conclude that C. coli is more prevalent in broilers than C. jejuni in Tamil Nadu. The present study also demonstrated that C. jejuni and C. coli in chicken caecal samples from the slaughterhouse are indicative of the possibility of public health hazards. Hence, necessary precautions, and hygienic measures should be followed in slaughterhouses and chicken shops to prevent campylobacteriosis outbreaks. Though two new sequence types were identified in this study, all the five C. coli isolates belonged to the same clonal complex ST-828. To our knowledge, this is the first report with the characterization of C. coli isolates by MLST in India.
Conflict of interest
The authors declare that they have no conflict of interest.
Acknowledgment
Authors thank the Vice-Chancellor, TANUVAS for providing the infrastructure to carry out this research.
References
- Ananda Chitra M, Jayanthy C, Nagarajan B. Detection and sequence analysis of accessory gene regulator genes of Staphylococcus pseudintermedius isolates. Vet. World. . 2015;8:902–907. doi: 10.14202/vetworld.2015.902-907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachoual R, Ouabdesselam S, Mory F, Lascols C, Soussy CJ, Tankovic J. Single or double mutational alterations of gyrA associated with fluoroquinolone resistance in Campylobacter jejuni and Campylobacter coli. Microb. Drug Resist. 2001;7:257–261. doi: 10.1089/10766290152652800. [DOI] [PubMed] [Google Scholar]
- Bolton DJ. Campylobacter virulence and survival factors. Food Microbiol. 2015;48:99–108. doi: 10.1016/j.fm.2014.11.017. [DOI] [PubMed] [Google Scholar]
- Byrd JA, Hargis BM, Caldwell DJ, Bailey RH, Herron KL, McReynolds JL, Brewer RL, Anderson RC, Bischoff KM, Callaway TR. Effect of lactic acid administration in the drinking water during preslaughter feed withdrawal on Salmonella and Campylobacter contamination of broilers. Poult. Sci. 2001;80:278–283. doi: 10.1093/ps/80.3.278. [DOI] [PubMed] [Google Scholar]
- Caffrey N, Agunos A, Gow S, Liljebjelke K, Waldner CL, Mainali C, Checkley SL. A cross-sectional study of the prevalence factors associated with fluoroquinolone resistant Campylobacter jejuni in broiler flocks in Canada. Prev. Vet. Med. 2021;186:105–164. doi: 10.1016/j.prevetmed.2020.105164. [DOI] [PubMed] [Google Scholar]
- CLSI. Performance standards for antimicrobial susceptibility testing CLSI supplement M100. 30th Edn. Wayne, PA: Clinical and Laboratory Standards Institute; 2020. [Google Scholar]
- Datta S, Niwa H, Itoh K. Prevalence of 11 pathogenic genes of Campylobacter jejuni by PCR in strains isolated from humans, poultry meat and broiler and bovine faeces. J. Med. Microbiol. 2003;52:345–348. doi: 10.1099/jmm.0.05056-0. [DOI] [PubMed] [Google Scholar]
- Denis M, Nagard B, Rose V, Bourgoin K, Cutimbo M, Kerouanton A. No clear differences between organic or conventional pig farms in the genetic diversity or virulence of Campylobacter coli isolates. Front. Microbiol. 2017;8:1016. doi: 10.3389/fmicb.2017.01016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dingle KE, Colles FM, Wareing DRA, Ure R, Fox AJ, Bolton FE, Bootsma HJ, Willems RJL, Urwin R, Maiden MCJ. Multilocus sequence typing system for Campylobacter jejuni. J. Clin. Microbiol. 2001;39:14–23. doi: 10.1128/JCM.39.1.14-23.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engberg J, Aarestrup FM, Taylor DE, Gerner-Smidt P, Nachamkin I. Quinolone and macrolide resistance in Campylobacterjejuni and C coli resistance mechanisms and trends in human isolates. Emerg. Infect. Dis. 2001;7:24–34. doi: 10.3201/eid0701.010104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gahamanyi N, Song DG, Yoon KY, Mboera LE, Matee MI, Mutangana D, Amachawadi RG, Komba EV, Pan CH. Antimicrobial resistance profiles, virulence genes, and genetic diversity of thermophilic Campylobacter species isolated from a layer poultry farm in Korea. Front. Microbiol. 2021;12:554. doi: 10.3389/fmicb.2021.622275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Sánchez L, Melero B, Diez AM, Jaime I, Canepa A, Rovira J. Genotyping, virulence genes and antimicrobial resistance of Campylobacter spp isolated during two seasonal periods in Spanish poultry farms. Prev. Vet. Med. 2020;176:104935. doi: 10.1016/j.prevetmed.2020.104935. [DOI] [PubMed] [Google Scholar]
- Gonzalez I, Grant KA, Richardson PT, Park SF, Collins MD. Specific identification of the enteropathogens Campylobacter jejuni and Campylobacter coli by using a PCR test based on the ceuE gene encoding a putative virulence determinant. J. Clin. Microbiol. 1997;35:759–763. doi: 10.1128/jcm.35.3.759-763.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goon S, Kelly JF, Logan SM, Ewing CP, Guerry P. Pseudaminic acid, the major modification on Campylobacter flagellin, is synthesized via the Cj1293 gene. Mol. Microbiol. 2003;50:659–671. doi: 10.1046/j.1365-2958.2003.03725.x. [DOI] [PubMed] [Google Scholar]
- Jesse TW, Englen MD, Pittenger-Alley LG, Fedorka-Cray PJ. Two distinct mutations in gyrA lead to ciprofloxacin and nalidixic acid resistance in Campylobacter coli and Campylobacter jejuni isolated from chickens and beef cattle. J. Appl. Microbiol. 2006;100:682–688. doi: 10.1111/j.1365-2672.2005.02796.x. [DOI] [PubMed] [Google Scholar]
- Jolley KA, Bray JE, Maiden MCJ. Open-access bacterial population genomics: BIGSdb software, the PubMLS org website and their applications. Wellcome Open Res. 2018;3:124. doi: 10.12688/wellcomeopenres.14826.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaakoush NO, Mitchell HM, Man SM. Campylobacter. Mol. Med. Microbiol. 2015 [Google Scholar]
- Kashoma IP, Kumar A, Sanad YM, Gebreyes W, Kazwala RR, Garabed R, Rajashekara G. Phenotypic and genotypic diversity of thermophilic Campylobacter spp in commercial turkey flocks: a longitudinal study. Foodborne Pathog. Dis. 2014;11:850–860. doi: 10.1089/fpd.2014.1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan JA, Rathore RS, Abulreesh HH, Qais FA, Ahmad I. Prevalence and antibiotic resistance profiles of Campylobacter jejuni isolated from poultry meat and related samples at retail shops in Northern India. Foodborne Pathog. Dis. 2018;15:218–225. doi: 10.1089/fpd.2017.2344. [DOI] [PubMed] [Google Scholar]
- Konkel ME, Gray SA, Kim BJ, Garvis SG, Yoon J. Identification of the enteropathogens Campylobacter jejuni and Campylobacter coli based on the cadF virulence gene and its product. J. Clin. Microbiol. 1999;37:510–517. doi: 10.1128/jcm.37.3.510-517.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koolman L, Whyte P, Burgess C, Bolton D. Distribution of virulence-associated genes in a selection of Campylobacter isolates. Foodborne Pathog. Dis. 2015;12:424–432. doi: 10.1089/fpd.2014.1883. [DOI] [PubMed] [Google Scholar]
- Kuana SL, Santos LR, Rodrigues LB, Borsoi A, Moraes HLS, Salle CTP, Nascimento VP. Occurrence and characterization of Campylobacter in the Brazilian production and processing of broilers. Avian Dis. 2008;52:680–684. doi: 10.1637/8296-032608-Reg.1. [DOI] [PubMed] [Google Scholar]
- Ladely SR, Berrang ME, Meinersmann RJ, Cox NA. Campylobacter multi-locus sequence types and antimicrobial susceptibility of broiler cecal isolates: A two year study of 143 commercial flocks. J. Food Saf. 2017;37:e12366. [Google Scholar]
- Mossong J, Mughini-Gras L, Penny C, Devaux A, Olinger C, Losch S, Cauchie HM, van Pelt W, Ragimbeau C. Human campylobacteriosis in Luxembourg, 2010-2013: a case-control study combined with multilocus sequence typing for source attribution and risk factor analysis. Sci. Rep. 2016;6:1–12. doi: 10.1038/srep20939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musgrove MT, Berrang ME, Byrd JA, Stern NJ, Cox NA. Detection of Campylobacter spp in ceca and crops with and without enrichment. Poult. Sci. 2001;80:825–828. doi: 10.1093/ps/80.6.825. [DOI] [PubMed] [Google Scholar]
- Romero MR, Cook N. A rapid LAMP-based method for screening poultry samples for Campylobacter without enrichment. Front. Microbiol. 2018;9:2401. doi: 10.3389/fmicb.2018.02401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossler E, Olivero C, Soto LP, Frizzo LS, Zimmermann J, Rosmini MR, Sequeira GJ, Signorini ML, Zbrun MV. Prevalence, genotypic diversity and detection of virulence genes in thermotolerant Campylobacter at different stages of the poultry meat supply chain. Int. J. Food Microbiol. 2020;326:108641. doi: 10.1016/j.ijfoodmicro.2020.108641. [DOI] [PubMed] [Google Scholar]
- Sahin O, Zhang Q, Meitzler JC, Harr BS, Morishita TY, Mohan R. Prevalence, antigenic specificity, and bactericidal activity of poultry anti-Campylobacter maternal antibodies. Appl. Environ. Microbiol. 2001;67:3951–3957. doi: 10.1128/AEM.67.9.3951-3957.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sallam KI. Prevalence of Campylobacter in chicken and chicken by-products retailed in Sapporo area, Hokkaido, Japan. Food Control. . 2007;18:1113–1120. [Google Scholar]
- Skarp CPA, Hänninen ML, Rautelin HIK. Campylobacteriosis: the role of poultry meat. Clin. Microbiol. Infect. 2016;22:103–109. doi: 10.1016/j.cmi.2015.11.019. [DOI] [PubMed] [Google Scholar]
- Tang Y, Jiang Q, Tang H, Wang Z, Yin Y, Ren F, Kong L, Xinan Jiao X, Huang J. Characterization and prevalence of Campylobacter spp from broiler chicken rearing period to the slaughtering process in eastern China. Front. Vet. Sci. 2020;7:227. doi: 10.3389/fvets.2020.00227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toplak N, Kovač M, Piskernik S, Možina SS, Jeršek B. Detection and quantification of Campylobacter jejuni and Campylobacter coli using real-time multiplex PCR. J. Appl. Microbiol. 2012;112:752–764. doi: 10.1111/j.1365-2672.2012.05235.x. [DOI] [PubMed] [Google Scholar]
- Torralbo A, Borge C, García-Bocanegra I, Méric G, Perea A, Carbonero A. Higher resistance of Campylobacter coli compared to Campylobacter jejuni at chicken slaughterhouse. Comp. Immunol. Microbiol. Infect. Dis. 2015;39:47–52. doi: 10.1016/j.cimid.2015.02.003. [DOI] [PubMed] [Google Scholar]
- Ugarte-Ruiz M, Gomez-Barrero S, Porrero MC, lvarez JA, Garci M, Comeron MC, Wassenaar TM, Dominguez L. Evaluation of four protocols for the detection and isolation of thermophilic Campylobacter from different matrices. J. Appl. Microbiol. 2012;113:200–208. doi: 10.1111/j.1365-2672.2012.05323.x. [DOI] [PubMed] [Google Scholar]
- Vasiliki I, Ioannidis A, Magiorkinis E, Bagos P, Nicolaou C, Legakis N, Chatzipanagiotou S. Multilocus sequence typing (and phylogenetic analysis) of Campylobacter jejuni and Campylobacter coli strains isolated from clinical cases in Greece. BMC Res. Notes. 2013;. 6:359. doi: 10.1186/1756-0500-6-359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinueza-Burgos C, Wautier M, Martiny D, Cisneros M, Van Damme I, De Zutter L. Prevalence, antimicrobial resistance and genetic diversity of Campylobacter coli and Campylobacter jejuni in Ecuadorian broilers at slaughter age. Poult. Sci. 2017;96:2366–2374. doi: 10.3382/ps/pew487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker LJ, Wallace RL, Smith JJ, Graham T, Saputra T, Symes S, Stylianopoulos A, Polkinghorne BG, Kirk MD, Glass K. Prevalence of Campylobacter coli and Campylobacter jejuni in retail chicken, beef, lamb, and pork products in three Australian states. J. Food Prot. 2019;82:2126–2134. doi: 10.4315/0362-028X.JFP-19-146. [DOI] [PubMed] [Google Scholar]
- Wang G, Clifford GC, Tracy M, Pucknell C, Barton C, Price L, Woodward DL, Rodgers FG. Colony Multiplex PCR assay for identification and differentiation of Campylobacter jejuni, upsaliensis, and C fetus subsp fetus. J. Clin. Microbiol. 2002;40:4744–4747. doi: 10.1128/JCM.40.12.4744-4747.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wangroongsarb P, Cheunban N, Jittaprasatsin C, Kamthalang T, Saipradit N, Chaichana P, Chaiwat P, Sittiporn P, Orapan S. Prevalence and antimicrobial susceptibility of Campylobacter isolated from retail chickens in Thailand. Int. J. Food Microbiol. 2021;339:109017. doi: 10.1016/j.ijfoodmicro.2020.109017. [DOI] [PubMed] [Google Scholar]
- Wieczorek K, Bocian Ł, Osek J. Prevalence and antimicrobial resistance of Campylobacter isolated from carcasses of chickens slaughtered in Poland–a retrospective study. Food Control. . 2020;11:107159. [Google Scholar]
- Wysok B, Wojtacka J, Kivistö R. Pathogenicity of Campylobacter strains of poultry and human origin from Poland. Int. J. Food Microbiol. 2020;334:108830. doi: 10.1016/j.ijfoodmicro.2020.108830. [DOI] [PubMed] [Google Scholar]
- Zheng J, Meng J, Zhao S, Singh R, Song W. Adherence to and invasion of human intestinal epithelial cells by Campylobacter jejuni and Campylobacter coli isolates from retail meat products. J. Food Prot. 2006;69:768–774. doi: 10.4315/0362-028x-69.4.768. [DOI] [PubMed] [Google Scholar]