Abstract
U/G and T/G mismatches commonly occur due to spontaneous deamination of cytosine and 5-methylcytosine in double-stranded DNA. This mutagenic effect is particularly strong for extreme thermophiles, since the spontaneous deamination reaction is much enhanced at high temperature. Previously, a U/G and T/G mismatch-specific glycosylase (Mth-MIG) was found on a cryptic plasmid of the archaeon Methanobacterium thermoautotrophicum, a thermophile with an optimal growth temperature of 65°C. We report characterization of a putative DNA glycosylase from the hyperthermophilic archaeon Pyrobaculum aerophilum, whose optimal growth temperature is 100°C. The open reading frame was first identified through a genome sequencing project in our laboratory. The predicted product of 230 amino acids shares significant sequence homology to [4Fe-4S]-containing Nth/MutY DNA glycosylases. The histidine-tagged recombinant protein was expressed in Escherichia coli and purified. It is thermostable and displays DNA glycosylase activities specific to U/G and T/G mismatches with an uncoupled AP lyase activity. It also processes U/7,8-dihydro-oxoguanine and T/7,8-dihydro-oxoguanine mismatches. We designate it Pa-MIG. Using sequence comparisons among complete bacterial and archaeal genomes, we have uncovered a putative MIG protein from another hyperthermophilic archaeon, Aeropyrum pernix. The unique conserved amino acid motifs of MIG proteins are proposed to distinguish MIG proteins from the closely related Nth/MutY DNA glycosylases.
Hydrolytic deamination of cytosine and 5-methylcytosine in double-stranded DNA results in U/G and T/G mismatches, which in the next round of replication produce C/G to T/A transition mutations (18). DNA glycosylases that excise uracil or thymine at the N-glycosylic bond can be generally classified into two major types according to their primary amino acid sequences and enzyme functions (3). The first type is uracil-DNA glycosylase (UDG), which excises uracil from both single- and double-stranded DNA (U/G and U/A mispairs). However, it does not excise thymine from T/G mismatches. UDG is widely present in many organisms. There is 56% amino acid sequence identity between Escherichia coli UDG and human UDG, indicating that the primary sequences of UDGs are highly conserved during evolution (27). The second type includes mismatch-specific uracil-DNA glycosylase (MUG), found in E. coli and Serratia marcescens, and thymine-DNA glycosylase (TDG) from humans (8, 24). MUG and TDG recognize the mismatched base pairs in double-stranded DNA and remove both mismatched uracil and thymine. MUG shares 32% amino acid sequence identity with the central part of human TDG (about 165 amino acids in length). While TDG recognizes and repairs U/G and T/G equally, MUG is primarily U/G mismatch specific. However, it does display activity on T/G mismatches at high enzyme concentrations. It is interesting that MUG and UDG show only low sequence identity, but their tertiary structures are remarkably similar (3).
Recently, two novel DNA glycosylases with low sequence identity to the known UDG and MUG/TDG were identified. SMUG1 protein from Xenopus and humans cleaves uracil residues from DNA with preference for single-stranded DNA containing U (11). Human MBD4, a 70-kDa methyl-CpG-binding protein, contains a glycosylase domain at the C terminus (about 126 amino acid residues) which repairs T/G and U/G mismatches (12).
The spontaneous hydrolysis of cytosine and 5-methylcytosine is greatly enhanced at high temperatures, indicating that thermo- and hyperthermophilic organisms are at a high risk of mutagenesis as a consequence of this reaction (18, 19). However, despite the completion of several thermophile genome sequences, no UDG or MUG/TDG homologs have been detected by sequence comparisons in thermo- and hyperthermophiles. Instead, activities of novel glycosylases with unrelated or distantly related sequences are being detected.
UDG-like activities have been detected in the crude cell extracts of hyperthermophilic microorganisms (15), and a recent study reported the characterization of a novel type of UDG (TMUDG) from the thermophilic bacteria Thermotoga maritima (30). Homologs of this apparently new family of UDGs can be detected by sequence comparisons in several genomes of bacteria (mesophiles and thermophiles) as well as in members of the domain Archaea (30).
A functional analog of the MUG/TDG glycosylases has been identified in an archaeal thermophile. It is a mismatch glycosylase (Mth-MIG) encoded on cryptic plasmid pFV1 of the archaeon Methanobacterium thermoautotrophicum, a thermophile with an optimal growth temperature of 65°C (13). Mth-MIG processes U/G and T/G but not U (on a single strand). However, Mth-MIG has very little amino acid sequence similarity to MUG/TDG and UDG. Instead Mth-MIG has significant sequence similarity to the [4Fe-4S]-containing Nth/MutY DNA glycosylase family which catalyzes N-glycosylic reactions on DNA substrates other than U/G and T/G mispairs (5, 13, 23). These DNA glycosylases include DNA endonuclease III (Nth, thymine glycol DNA glycosylase), MutY DNA glycosylase (A/G-specific adenine glycosylase), UV endonuclease (UVendo), and methylpurine DNA glycosylase II (MpgII). These unique structural and functional characteristics of Mth-MIG suggest that it is a new type of U/G and T/G mismatch-specific glycosylase.
Little is known about the repair mechanism for U/G and T/G mismatches in hyperthermophiles, whose optimal growth temperatures are around 100°C. Here, we report the identification and characterization of a DNA glycosylase encoded on the single circular chromosome DNA of the hyperthermophilic archaeon Pyrobaculum aerophilum (35). Through detailed structural and functional analysis, we found that this DNA glycosylase processes U/G and T/G mismatches. Therefore, it was designated Pa-MIG. Further sequence analysis of complete bacterial and archaeal genomes identified one additional putative MIG homolog, APE0875, in another hyperthermophilic archaeon, Aeropyrum pernix. The conserved amino acid motifs in the MIG proteins were analyzed and compared with the [4Fe-4S]-containing Nth/MutY DNA glycosylases, whose sequences were closely related to that of MIG.
MATERIALS AND METHODS
Identification of the candidate protein.
Candidate protein coding region pag5_3199 was identified in the recently completed genome sequence of P. aerophilum (7; S. Fitz-Gibbon et al., unpublished data) by sequence similarity using TFASTA (1).
Expression and purification of Pa-MIG.
The pag5_3199 DNA was cloned between the SphI and SalI sites of the bacterial expression vector pQE30 (Qiagen, Chatsworth, Calif.) after PCR amplification. Transformants of E. coli CC104mutY/pREP4 were grown at 37°C in 1.5 liters of Luria-Bertani medium with ampicillin (200 μg/ml) and kanamycin (25 μg/ml). When the culture grew to an optical density at 600 nm of 2.5, isopropyl-β-d-thiogalactopyranoside (IPTG) was added to the final concentration of 0.1 mM to induce the expression of Pa-MIG protein overnight. Bacterial lysate was prepared by French press in buffer A (50 mM sodium phosphate [pH 8.0], 300 mM NaCl) plus 0.5 mM phenylmethylsulfonyl fluoride. After clarification by centrifugation, the lysate was mixed with 5 ml of Ni-nitrilotriacetic acid (NTA) matrix (Qiagen) for 2 h at 4°C with gentle shaking. Then the mixture was poured into a column, washed with buffer A containing 60 mM imidazole, and eluted with buffer A containing 0.5 M imidazole. A dark olive-colored band eluted; the fractions containing this color were dialyzed overnight in buffer B (50 mM Tris-HCl [pH 7.5], 1 mM EDTA, 1 mM dithiothreitol, 30 mM NaCl, 50% glycerol). A clear protein sample was obtained after centrifugation of the dialyzed sample and was stored at −80°C.
Electrospray mass spectrometry.
A Perkin-Elmer Sciex (Thornhill, Ontario, Canada) API III triple-quadrupole mass spectrometer fitted with an ion spray source was tuned and calibrated as previously described (9), and positive ion protein spectra were analyzed. Molecular weights from the series of multiply charged ions found in the protein spectra, and deconvolution of the ion series into a molecular weight spectrum, were calculated with the program MacSpec, and theoretical protein molecular weights were calculated with the program MacBiospec (17).
Heat treatment of Pa-MIG.
The purified Pa-MIG protein was diluted to 1 mg/ml in buffer B. About 50 μl of the sample was transferred to a microcentrifuge tube and incubated at the indicated temperature for 10 min. A clear supernatant was obtained after centrifugation. For the DNA glycosylase activity assay (see below), a 70°C heat-treated Pa-MIG protein sample was used.
Oligonucleotide substrates.
Oligonucleotides (96-mers, sequence 1 and sequence 2 [modified from reference 37]) were synthesized and purified by urea-polyacrylamide gel electrophoresis (PAGE) (Gibco BRL, Grand Island, N.Y.). The sequence 2 oligonucleotide containing 7,8-dihydro-8-oxoguanine (GO) was synthesized and PAGE purified by DNAgency (Malvern, Pa.). Sequences of the oligonucleotides (nucleotides involved in mispair formation are in boldface) are as follows: sequence 1, 5′ CCG GGG CCG GAT CGG AAC CCT AAA GGG AGC CCC CGA TTT AGA GCT TGA CGG GGA AAG CCX AAT TCG GCG AAC GTG GCG AGA AAG GAA GGG AAG GAC 3′ (X = A, C, G, T, or U); sequence 2, 5′ AAT TGT CCT TCC CTT CCT TTC TCG CCA CGT TCG CCG AAT TYG GCT TTC CCC GTC AAG CTC TAA ATC GGG GGC TCC CTT TAG GGT TCC GAT CCG GCC 5′ (Y = A, C, G, T, or GO).
Sequence 1 was 32P labeled and annealed with an excess of unlabeled sequence 2 to form double-stranded DNA containing the base pair X/Y at position 60, using protocols as previously described (22).
Glycosylase activity assay.
The glycosylase activity assay detects the combined action of the glycosylase and the subsequent cleavage of DNA at apurinic/apyrimidinic (AP) sites (21, 22). The standard reaction mixture contained 20 mM Tris-HCl (pH 7.5), 1 mM dithiothreitol, 1 mM EDTA, 3% glycerol, 80 mM NaCl, and 20 fmol of labeled double-stranded DNA in a total volume of 20 μl. Unless otherwise stated, the 70°C heat-treated Pa-MIG protein, diluted in buffer B, was added to the reaction mixture and incubated at 70°C for 15 min. Unless otherwise stated, the reaction products were treated with 4 μl of 1 M NaOH and were heated at 90°C for 4 min to complete the cleavage of DNA at AP sites before electrophoresis. The reaction products were mixed with 8 μl of loading buffer (95% formamide, 20 mM EDTA, 5% bromophenol blue, xylene cyanol FF) at 94°C for 2 min and then analyzed on 15% polyacrylamide denaturing gels.
AP lyase activity assay.
The double-stranded DNA containing an AP site was prepared essentially by the method of Horst and Fritz (13). Twenty fentomoles of labeled DNA substrate containing a U/G mismatch was incubated with 1 U of thermolabile UDG (Epicenter, Madison, Wis.) in a standard glycosylase assay buffer at 37°C for 30 min to produce AP/G substrate. The AP/G substrate was incubated with 5 or 50 ng of 70°C heat-treated Pa-MIG protein or with buffer B alone at 70°C for 15 min without subsequent alkaline treatment. The reaction products were mixed with 8 μl of loading buffer at 94°C for 2 min and were analyzed on 15% polyacrylamide denaturing gels.
Ugi inhibition assay.
Uracil-DNA glycosylase inhibitor (Ugi) and E. coli UDG were gifts from Dale W. Mosbaugh (Department of Agricultural Chemistry and Biochemistry and Biophysics and Environmental Health Sciences Center, Oregon State University, Corvallis). The Ugi inhibition assay was carried out as previously described (36). Ugi of various amounts (0, 3, and 10 U; 0.1 pmol/U) was incubated with 1 U of E. coli UDG or 25 ng of Pa-MIG at 37°C for 10 min following addition of the double-stranded DNA substrates containing T/G or U/G mismatches or single-stranded DNA containing U. The glycosylase activity assay was carried out at 50°C for 15 min. The reaction products were treated with 4 μl of 1 M NaOH to cleave the AP sites before electrophoresis. The reaction products were mixed with 8 μl of loading buffer at 94°C for 2 min and were analyzed on 15% polyacrylamide denaturing gels.
Phylogenetic analysis.
Putative homologs of Nth, MutY, MIG, MpgII, and UV endo glycosylases in the complete bacterial and archaeal genomes were identified using the program BLAST (28). Characterized and putative homologs were multiply aligned using the program ClustalW (34). Distance analysis was performed using neighbor joining in the PAUP program (32).
RESULTS
Homology between Pa-MIG protein and Nth/MutY DNA glycosylases.
Open reading frame (ORF) pag5_3199 from hyperthermophilic archaeon P. aerophilum was identified as a putative Nth/MutY DNA glycosylase through a genome sequencing project in our laboratory. It encodes a predicted product of 230 amino acid residues that is homologous to several [4Fe-4S]-containing DNA glycosylases. It has 34 and 30% amino identity to M. thermoautotrophicum Mth-MIG and E. coli MutY, respectively. Further down the list were E. coli endonuclease III (28% amino acid identity) and human MBD4 (21% amino acid identity in the glycosylase domain). Subsequent biochemical analysis demonstrated that the protein encoded by this ORF was indeed a U/G and T/G mismatch glycosylase (see below). Therefore, this protein was designated a MIG homolog from P. aerophilum (Pa-MIG). The database search also reveals another putative MIG homolog from A. pernix, for which the predicted 286-amino-acid product APE0875 (accession no. BAA79857) has 57% amino acid identity to Pa-MIG. This ORF was annotated as a putative A/G-specific adenine glycosylase by the original researchers (14).
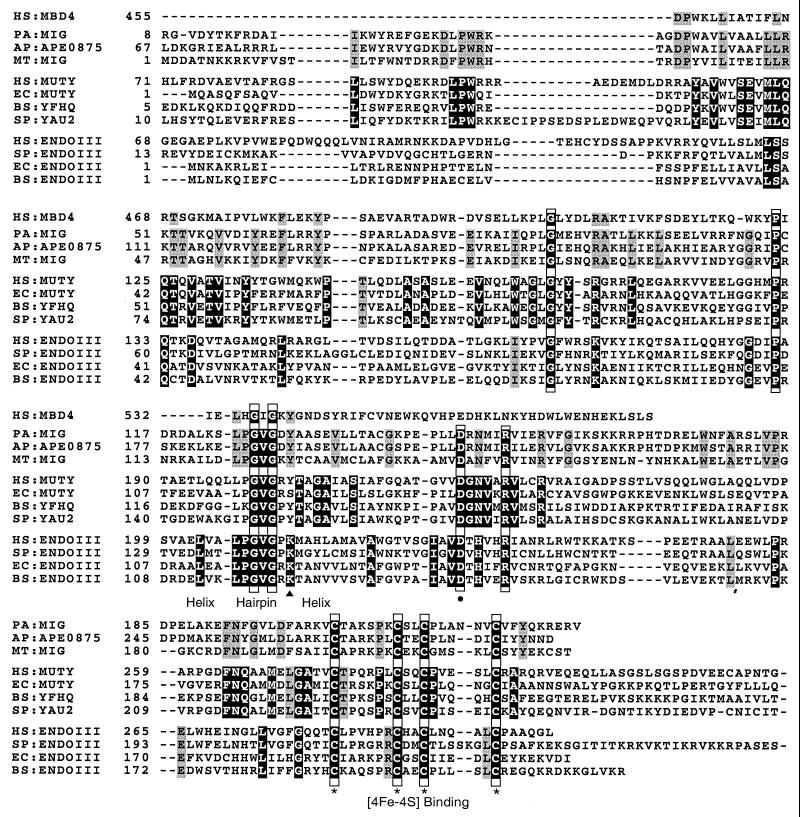
The amino acid sequence alignments are shown in Fig. 1. Pa-MIG protein has the general features which are conserved in [4Fe-4S]-containing Nth/MutY DNA glycosylase family (2, 12, 20, 23, 29, 33). It contains a helix-hairpin-helix motif and a [4Fe-4S] binding motif, which are important for DNA binding. It also contains four amino acid residues that are strictly conserved in the Nth/MutY glycosylase family: Gly 90, Pro 115, Asp 148, and Arg 153. Among them, the aspartic acid residue corresponding to Asp 138 of E. coli endonuclease III is known to be important for the catalytic activity of the Nth/MutY enzymes.
FIG. 1.
Alignment of sequences of the Pa-MIG protein and the following Nth/MutY glycosylase family members (species code and GenBank accession number in parentheses): MBD4 protein of Homo sapiens (HS, AAD22195); MIG-like proteins of A. pernix (AP, BAA79857) and M. thermoautotrophicum (MT, P29588); MutY-like proteins of H. sapiens (HS, U63329), E. coli (EC, P17802), Bacillus subtilis (BS, CAB12691), and Schizosaccharomyces pombe (SP, AF053340); endonuclease III (Nth)-like proteins of H. sapiens (HS, U79718), S. pombe (SP, Q09907), E. coli (EC, P20625), and B. subtilis (BS, P39788). The conserved amino acid residues within each MIG, MutY, and Nth family are shaded. The strictly conserved amino acid residues among all three families are shaded black and boxed. The MutY-specific and Nth-specific ones that are consistent with previous publications are shaded black (20, 29). The conserved amino acid residues within the presented sequences are shaded gray. The highly conserved helix-hairpin-helix motif is indicated. The cysteine residues involved in binding the [4Fe-4S] cluster are marked with asterisks. The strictly conserved aspartic acid residue is marked with a dot. The conserved lysine residue within the Nth family is marked with a triangle.
Pa-MIG protein is similar to E. coli MutY and Mth-MIG but different from Nth proteins in having a tyrosine residue at the corresponding position of Lys 120 in E. coli endonuclease III (Nth). This residue, conserved among all Nth proteins, is critical for their AP lyase activities (33). However, Pa-MIG protein varies from MutY in MutY family-specific motifs (10, 20). Pa-MIG, Mth-MIG, and APE0875 proteins have their own unique conserved amino acid residues. Most noticeable differences are the amino acid residues corresponding to the active-site pocket of MutY, the minor groove reading α2-α3 (10). Similar to Mth-MIG and APE0875, Pa-MIG protein has 48-LLRKTTV-54, corresponding to 39-MLQQTQV-45 of E. coli MutY. Also similar to Mth-MIG and APE0875, Pa-MIG lacks about 130 amino acids corresponding to the C-terminal domain of E. coli MutY, which has been shown to enhance the binding of MutY to A/7,8-dihydro-8-oxoguanine (GO [25rsqb;), a physiological relevant substrate.
Expression of Pa-MIG protein in E. coli and purification of the recombinant protein.
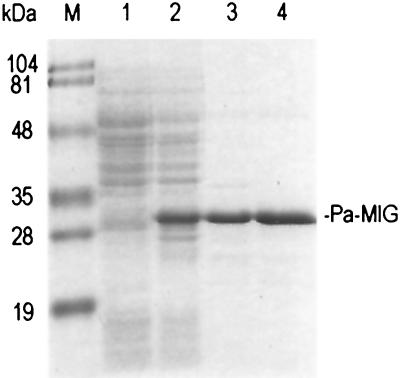
The DNA encoding the Pa-MIG was subcloned into the pQE30 vector behind a six-histidine tag and was expressed in E. coli. The hexahistidine-tagged Pa-MIG protein was largely soluble and was purified to near homogeneity by affinity chromatography on an Ni2+-NTA column (Fig. 2, lane 3). The molecular mass of the purified protein was determined by mass spectrometry to be 27.9 kDa, consistent with the molecular mass of 27.9 kDa predicted from the hexahistidine-tagged Pa-MIG protein sequence. Also consistent with the presence of a binding motif for the [4Fe-4S] cluster (2), the purified protein had a dark olive color at high concentrations (10 mg/ml) and an absorption spectrum with a peak at ≈414 nm (data not shown).
FIG. 2.
Purification of Pa-MIG recombinant protein from E. coli. A Coomassie blue-stained SDS-polyacrylamide gel (10%) contained a lysate of CC104mutY/pREP4 cells containing either pQE30 vector (lane 1) or pQE30/Pa-MIG (lane 2) after induction by IPTG, Ni-NTA column Pa-MIG protein fraction after dialysis (lane 3), and 70°C heat-treated Pa-MIG sample (lane 4). Pa-MIG protein is indicated on the right. Lane M contains molecular mass standards (Bio-Rad) as indicated on the left.
Thermostability of Pa-MIG.
The purified Pa-MIG protein in buffer B was incubated for 10 min at different temperatures, and the amount of remaining soluble protein was determined by the Bradford protein assay. About 87% of the Pa-MIG protein remained soluble after 10 min at 70°C (Fig. 2, lane 4). The protein became unstable at temperatures higher than 70°C. At 90°C, only about 56% of the protein remained soluble (data not shown). Subsequent glycosylase activity assays were carried out with 70°C heat-treated protein samples to reduce the possible contaminated proteins from E. coli.
U/G, U/GO, T/G, and T/GO mismatch glycosylase activities of Pa-MIG protein.
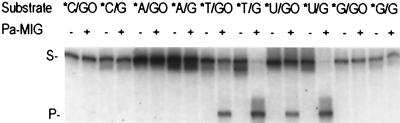
We carried out the DNA glycosylase activity assay using double-stranded DNA substrates containing X/G and X/GO (X = A, T, G, C, and U) (Fig. 3). Pa-MIG protein could efficiently process both T/G and U/G substrates. It could also process T/GO and U/GO. However, Pa-MIG protein could not process mismatches A/G and A/GO, the substrates for MutY. It also could not process G/G or G/GO substrates. These results suggest that Pa-MIG is a MIG homolog in P. aerophilum.
FIG. 3.
T/G and U/G mismatch-specific DNA glycosylase activity of Pa-MIG in the absence (−) or presence (+) of 50 ng of Pa-MIG on 96-bp double-stranded DNA containing X/G or X/GO (X = A, C, G, T, and U). The uncut 96-mer DNA substrate (S) and cleaved 60-mer product (P) are indicated on the left. Asterisks indicate the 5′-end-labeled strand.
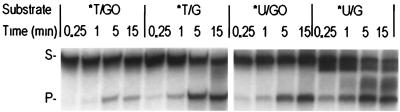
To determine whether a guanine moiety in the complementary strand was crucial in this repair process, we tested Pa-MIG glycosylase activity using double-stranded DNA substrates containing U/Y or T/Y (Y = A, G, C, or T) (Fig. 4). Pa-MIG processes T/G or U/G but not other combinations, indicating that guanine moiety in the complementary strand is necessary for Pa-MIG activity. When the T/G and U/G substrates were labeled at the 5′ end of the DNA strand containing guanine, no product was observed (data not shown). These results indicate that the strand containing guanine remains intact during the cleavage of thymine or uracil in the opposite strand. We also tested the other possible double-stranded DNA mismatch (A/C, A/A, C/C, C/A, C/T, G/A, and G/T) substrates and detected no activity (data not shown).
FIG. 4.
Requirement of guanine in the complementary strand, determined by analysis of the products of the glycosylase activity in the absence (−) or presence (+) of 50 ng of Pa-MIG on 96-bp double-stranded DNA containing U/Y and T/Y (Y = A, C, G, or T). The uncut 96-mer DNA substrate (S) and cleaved 60-mer product (P) are indicated on the left. Asterisks indicate the 5′-end-labeled strand.
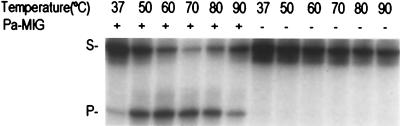
To compare the glycosylase activities of Pa-MIG at different temperatures, the glycosylase assay was carried out at 37, 50, 60, 70, 80, and 90°C. Pa-MIG exhibited greater activity at higher temperatures, as shown in Fig. 5. While small amounts of the product were generated at 37°C, at 70°C most of the substrates were converted to the products. At temperatures higher than 70°C, the amount of product decreased. The reduction was probably due to the heat instability of the protein at temperatures higher than 70°C, although it could also have been due to the heat instability of the double-stranded DNA template at higher temperatures.
FIG. 5.
Temperature dependency of Pa-MIG glycosylase activity in the absence (−) or presence (+) of 25 ng of Pa-MIG incubated with a 96-bp heteroduplex DNA containing a U/G mismatch at different temperatures for 15 min. The strand containing U was 5′ end labeled. The uncut 96-mer DNA substrate (S) and cleaved 60-mer product (P) are indicated on the left.
The time course of the reaction is shown in Fig. 6. In the presence of 5 ng of Pa-MIG protein, it has greater activities on U/G and T/G but weaker activities on U/GO and T/GO.
FIG. 6.
Time course of Pa-MIG glycosylase activity on substrates T/G, T/GO, U/G, and U/GO. Five nanograms of Pa-MIG was incubated with 96-bp heteroduplex DNA containing T/G, T/GO, U/G, or U/GO mismatches. The uncut 96-mer DNA substrate (S) and cleaved 60-mer product (P) are indicated on the left. Asterisks indicate the 5′-end-labeled strand.
AP lyase activity of Pa-MIG.
That Pa-MIG has a tyrosine residue at position 130 instead of lysine suggested that it might have an inefficient AP lyase activity uncoupled with the glycosylase activity (6, 10, 16, 20, 38). To test this idea, the glycosylase activity assay was performed in the absence of alkali cleavage of the AP sites (Fig. 7A). Nicking products in the absence of NaOH treatment were observed, suggesting that the Pa-MIG may have AP lyase activity (Fig. 7A, lanes 3 and 5). The amount of the nicking products was less than that of the NAOH-treated sample, indicating that the weak AP lyase activity was likely to be uncoupled with the glycosylase activity (Fig. 7A, lanes 3 to 6).
FIG. 7.
AP lyase activity of Pa-MIG. Products of the glycosylase activity of 5 or 50 ng of Pa-MIG on a 96-bp heteroduplex DNA containing a U/G mismatch (A) or AP/G (B) were analyzed. After incubation at 70°C for 15 min, samples were either directly used for electrophoresis or treated with 4 μl of 1 M NaOH at 90°C for 4 min to cleave DNA at AP sites before electrophoresis. The uncut 96-mer DNA substrate (S) and cleaved 60-mer product (P) are indicated on the left. Asterisks indicate the 5′-end-labeled strand.
To verify the observed AP lyase activity, we performed a similar experiment using double-stranded DNA containing an AP site in the opposite of G (AP/G substrate). The AP/G substrate was prepared from thermolabile UDG-treated U/G substrate. The complete conversion of U/G to AP/G was visualized by the alkali sensitivity of the formerly U-containing DNA strand (Fig. 7B, lanes 4 and 5). The results showed that significant amounts of the product were produced in the absence of NaOH treatment (Fig. 7B, lanes 2 and 3), suggesting that Pa-MIG has AP lyase activity.
Differences between Pa-MIG and UDG.
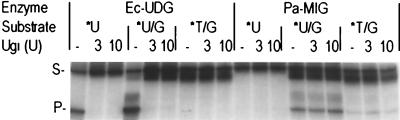
We have demonstrated that Pa-MIG can process both U/G and T/G mismatches. To distinguish the enzymatic activities of Pa-MIG and UDG, we tested the Pa-MIG activity on single-stranded DNA containing uracil and the effect of Ugi. The DNA glycosylase assay was performed with or without Ugi, using E. coli UDG as a control. The assay was done at 50°C due to the heat stability of E. coli UDG. As shown in Fig. 8, no product was detected with Pa-MIG on single-stranded DNA containing U, indicating that Pa-MIG could not process this substrate (a similar result was obtained using 50 ng of Pa-MIG at 70°C for 15 min [data not shown]). Ugi (up to 10 U, or 1 pmol) did not inhibit the activity of 25 ng (∼1 pmol) of Pa-MIG on U/G and T/G, while it totally inhibited activities of E. coli UDG on U or U/G substrates.
FIG. 8.
Ugi does not inhibit Pa-MIG activity. Products of the glycosylase activity of 25 ng of Pa-MIG on a 96-bp heteroduplex DNA containing a U/G or T/G mismatch or a 5′-end-labeled 96-mer single-stranded DNA (20 fmol) containing U in the presence of Ugi were analyzed. One unit of E. coli UDG (Ec-UDG) or 25 ng of 70°C heat-treated Pa-MIG was incubated with Ugi (0, 3, and 10 U) at 37°C for 10 min before addition of the DNA substrates. After incubation at 50°C for 15 min, samples were treated with 4 μl of 1 M NaOH at 90°C for 4 min to cleave DNA at AP sites before electrophoresis. The uncut 96-mer DNA substrate (S) and cleaved 60-mer product (P) are indicated on the left. Asterisks indicate the 5′-end-labeled strand.
Phylogenetic analysis of Nth/MutY/MIG/MpgII/UVendo glycosylase superfamily.
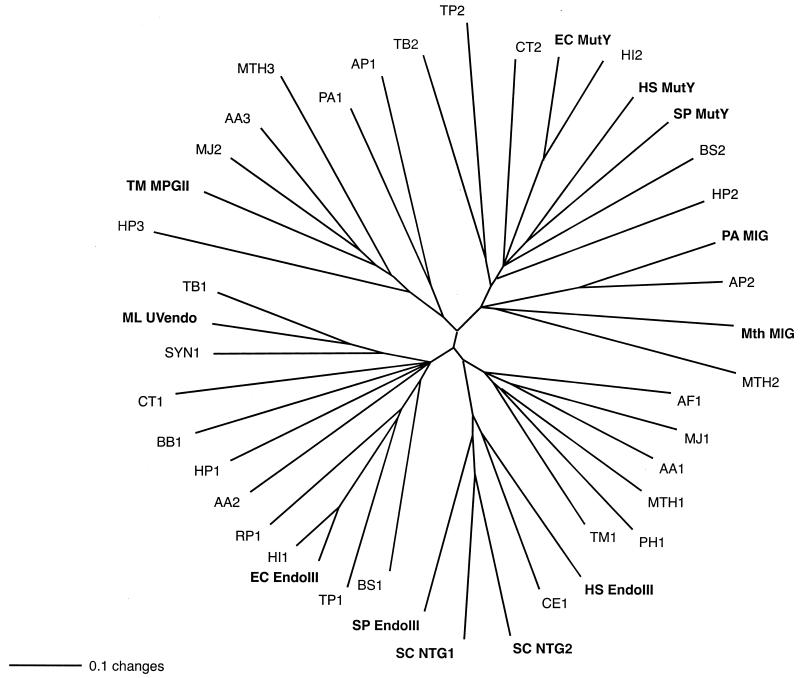
Amino acid sequences of characterized and putative homologs of the Nth/MutY/MIG/MpgII/UVendo glycosylases from complete bacterial and archaeal genomes were compared using the programs ClustalW (34) and PAUP (32). As shown in Fig. 9, there are roughly five major branches which contain one or more characterized DNA glycosylases. The branch containing MIG proteins is closer to MutY proteins than other DNA glycosylases, such as endonuclease III or MpgII. This indicates that MutY and MIG may have had a common ancestor during evolution. A putative DNA glycosylase encoded by the chromosome of M. thermoautotrophicum, MTH496 (or MTH2), is also on the MIG branch, indicating that it could potentially have MIG activity. Notably, although all of the proteins are homologs, glycosylases with related functions cluster near each other in the tree.
FIG. 9.
Phylogenetic analysis of members of the Nth/MutY/MIG/MpgII/UVendo glycosylase superfamily for the following organisms (protein code and GenBank accession number in parentheses). Archaea include A. pernix (AP1, BAA79061; AP2 [APE0875], BAA79857), Archaeoglobus fulgidus (AF1, AAB89556), M. thermoautotrophicum (Mth MIG, P29588; MTH1, AAB85267; MTH2, AAB85002 with extra 42 amino acid residues at the N terminus; MTH3, AAB85250), Methanococcus jannaschii (MJ1, E64376; MJ2, A64479), P. aerophilum (PA1, AF222334; PA MIG, AF222335), and Pyrococcus horikoshii (PH1, BAA30606). Bacteria include Aquifex aeolicus (AA1, AAC06594; AA2, AAC06742; AA3, AAC06526), Bacillus subtilis (BS1, P39788; BS2, CAB12691), Borrelia burgdorferi (BB1, AAC67089), Chlamydia trachomatis (CT1, AAC68292; CT2, AAC67698), E. coli (EC EndoIII, P20625; EC MutY, P17802), Haemophilus influenzae (HI1, P44319; HI2, P44320), Helicobacter pylori (HP1, AAD07651; HP2, AAD07210; HP3, AAD07668), Micrococcus luteus (ML UVendo, P46303), Mycobacterium tuberculosis (TB1, CAA17996; TB2, CAA17858), Rickettsia prowazekii (RP1, O05956), Synechocystis strain PCC6803 (SYN1, P73715), Treponema pallidum (TP1, AAC65744; TP2, AAC65331), and T. maritima (TM1, AAD35453; TM MpgII, AAD35467). Eukaryota include Caenorhabditis elegans (CE1, P54137), Saccharomyces cerevisiae (SC NTG1, AAC04942; SC NTG2, CAA99045), Schizosaccharomyces pombe (SP EndoIII, Q09907; SP MutY, AAC36207), and Homo sapiens (HS EndoIII, U79718; HS MutY, U63329). The bar scale represents number of substitutions per site. Proteins with biochemically determined functions are in boldface.
DISCUSSION
In this study, we analyzed a candidate ORF in the extreme thermophilic archaeon P. aerophilum, identified by the genome sequencing project. We designated it Pa-MIG, for it displays a U/G and T/G mismatch-specific glycosylase activity. The purified recombinant Pa-MIG is stable at 70°C and has a dark olive color due to the presence of [4Fe-4S] cluster. Pa-MIG could process both U/G and T/G mispairs, with better efficiency for U/G mispair. In comparison with reported activities of Mth-MIG (13), Pa-MIG seems to be more specific on its substrates. Among the substrates tested, it cleaves only U/G, U/GO, T/G, and T/GO, with no detectable activities on A/G, T/C, and U/C mispairs, which are the minor substrates for Mth-MIG (13). With the Pa-MIG protein sample, we observed a weak but definite AP lyase activity which is absent in Mth-MIG (4, 13). The AP lyase activity was not inhibited by 10 mM EDTA, indicating that the lyase activity was not from contaminating AP endonuclease IV from E. coli (data not shown). Although the Pa-MIG sample was 70°C heat treated and the assays were done at 70°C, we cannot rule out the possibility that some other heat-resistant contaminating E. coli proteins contributed to the observed AP lyase activity. Pa-MIG shares very little sequence homology with either UDG or MUG/TDG. Functionally Pa-MIG is similar to MUG/TDG and differs from UDG both by lack of activity against single-stranded DNA containing U and by lack of inhibition by Ugi.
MIG proteins and [4Fe-4S]-containing Nth/MutY DNA glycosylases share close sequence homology. Conserved elements include several amino acid residues, the helix-hairpin-helix motif, and the [4Fe-4S] cluster. The biochemical properties of these conserved amino acid residues and motifs in Nth/MutY proteins may be similar in the MIG proteins, which perform their activities at high temperature.
The amino acid motifs conserved specifically among Mth-MIG, Pa-MIG, and APE0875 provide a means to distinguish MIG proteins from the Nth/MutY glycosylases, useful for annotation of uncharacterized DNA glycosylases. These unique conserved amino acid residues could also be candidates for further study of the specificity of substrate binding as well as thermostability of MIG glycosylases. As more MIG homologs are discovered, the consensus sequence determined in this work may change.
The MIG family is remotely related to the recently identified human MBD4 thymine glycosylase, which also repairs T/G and U/G mismatches in double-stranded DNA (12). MIGs have the [4Fe-4S] binding motif, which is missing in MBD4. MBD4, on the other hand, has about 400 amino acid residues at the N terminus, including a methyl-CpG binding domain (12). The sequence identity of the glycosylase domain between MIGs and MBD4 is only approximately 20%; however, the shared conserved residues suggest that these protein domains are homologous and may help to further specify the critical residues for the activity.
The oxidized form of guanine (GO) is the most frequent oxidative lesion on DNA. It is premutagenic by forming A/GO mispair during DNA replication (31). It is worth noting that MIG shares close sequence homology with MutY, the A/G-specific adenine glycosylase with high specificity to A/GO mispairs. Interestingly, Pa-MIG can also process T/GO and U/GO mispairs. The efficiency is lower for GO-containing substrates than for the corresponding G-containing substrates. The significance of this activity remains to be determined. It is unclear whether T/GO or U/GO mispairs could result from the errors of DNA replication of C/GO in P. aerophilum, as in the case of the resulting A/GO mispair in E. coli. On the other hand, the E. coli UDG can also process U/GO mispair (data not shown), suggesting that GO-containing mispairs can be the substrates simply by virtue of the structural similarity of G and GO. In the case of MutY, whose physiologically relevant substrate is A/GO, there are about 130 additional amino acid residues at the C-terminal domain to enhance the binding to GO (25).
The physiological significance of MIG remains to be studied. However, Nölling et al. (26) noted a GGCC-recognizing restriction-modification system (a restriction enzyme and a DNA cytosine C5-methyltransferase) along with Mth-MIG on the pFV1 plasmid. Comparison of the DNA sequences adjacent to the coding regions of Pa-MIG and APE0875 reveal that the two neighboring ORFs upstream of both Pa-MIG and APE0875 are highly conserved. One ORF (APE0872) encodes a putative DNA cytosine C5-methyltransferase; the other (APE0874) encodes a protein of unknown function. The conserved close arrangement between these three ORFs suggests that they may function as part of the same biochemical pathway, perhaps a DNA restriction-modification system. MIG activity may be particularly important in thermophiles and hyperthermophiles to maintain the thermolabile 5-methylcytosine in DNA (13, 26).
Many questions remain. How important is MIG in reducing the mutation rate in DNA? Is MIG Archaea-specific? How did the Nth/MutY/MIG/MpgII/UVendo family evolve? The database search of the existing complete archaeal genomes reveals that some thermophilic archaea do not have MIG sequence homologs, which suggests the existence of other mechanisms for U/G and T/G mismatch repair. The increasing number of the completed bacterial and archaeal genomes certainly provide a powerful tool for studying putative DNA glycosylases and their structural and functional relationship during evolution, as well as for searching for novel DNA glycosylases.
ACKNOWLEDGMENTS
We thank Dale W. Mosbaugh for providing UDG inhibitor and E. coli UDG. We thank Malgorzata M. Slupska, Wendy M. Luther, Claudia Baikalov, and Ju-Huei Chiang for technical assistance.
This work was supported by Tumor Immunology Training Grant 5-T32-CA009120 (to H.Y.) and by National Institutes of Health grant GM57917 (to J.H.M.). Mass spectrometry was done in the UCLA Pasarow Mass Spectrometry laboratory by Kym F. Faull. Financial support from the W. M. Keck Foundation is acknowledged.
REFERENCES
- 1.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aspinwall R, Rothwell D G, Roldán-Arjona T, Anselmino C, Ward C J, Cheadle J P, Sampson J R, Lindahl T, Harris P C, Hickson I D. Cloning and characterization of a functional human homolog of Escherichia coli endonuclease III. Proc Natl Acad Sci USA. 1997;94:109–114. doi: 10.1073/pnas.94.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barrett T E, Savva R, Panayotou G, Barlow T, Brown T, Jiricny J, Pearl L H. Crystal structure of a G:T/U mismatch-specific DNA glycosylase: mismatch recognition by complementary-strand interactions. Cell. 1998;92:117–129. doi: 10.1016/s0092-8674(00)80904-6. [DOI] [PubMed] [Google Scholar]
- 4.Begley T J, Cunningham R P. Methanobacterium thermoautotrophicum thymine DNA mismatch glycosylase: conversion of an N-glycosylase to an AP lyase. Protein Eng. 1999;12:333–340. doi: 10.1093/protein/12.4.333. [DOI] [PubMed] [Google Scholar]
- 5.Begley T J, Haas B J, Noel J, Shekhtman A, Williams W A, Cunningham R P. A new member of the endonuclease III family of DNA repair enzymes that removes methylated purines from DNA. Curr Biol. 1999;9:653–656. doi: 10.1016/s0960-9822(99)80288-7. [DOI] [PubMed] [Google Scholar]
- 6.Dodson M L, Michaels M L, Lloyd R S. Unified catalytic mechanism for DNA glycosylases. J Biol Chem. 1994;269:32709–32712. [PubMed] [Google Scholar]
- 7.Fitz-Gibbon S, Choi A J, Miller J H, Stetter K O, Simon M I, Swanson R, Kim U J. A fosmid-based genomic map and identification of 474 genes of the hyperthermophilic archaeon Pyrobaculum aerophilum. Extremophiles. 1997;1:36–51. doi: 10.1007/s007920050013. [DOI] [PubMed] [Google Scholar]
- 8.Gallinari P, Jiricny J. A new class of uracil-DNA glycosylases related to human thymine-DNA glycosylase. Nature. 1996;383:735–738. doi: 10.1038/383735a0. [DOI] [PubMed] [Google Scholar]
- 9.Glasgow B J, Abduragimov A R, Yusifov T N, Gassymov O K, Horwitz J, Hubbell W L, Faull K F. A conserved disulfide motif in human tear lipocalins influences ligand binding. Biochemistry. 1998;37:2215–2225. doi: 10.1021/bi9720888. [DOI] [PubMed] [Google Scholar]
- 10.Guan Y, Manuel P C, Arvai A S, Parikh S S, Mol C D, Miller J H, Lloyd R S, Tainer J A. MutY catalytic core, mutant and bound adenine structures define specificity for DNA repair enzyme superfamily. Nat Struct Biol. 1998;5:1058–1064. doi: 10.1038/4168. [DOI] [PubMed] [Google Scholar]
- 11.Haushalter K A, Todd Stukenberg M W, Kirschner M W, Verdine G L. Identification of a new uracil-DNA glycosylase family by expression cloning using synthetic inhibitors. Curr Biol. 1999;9:174–185. doi: 10.1016/s0960-9822(99)80087-6. [DOI] [PubMed] [Google Scholar]
- 12.Hendrich B, Hardeland U, Ng H-H, Jiricny J, Bird A. The thymine glycosylase MBD4 can bind to the product of deamination at methylated CpG sites. Nature. 1999;401:301–304. doi: 10.1038/45843. [DOI] [PubMed] [Google Scholar]
- 13.Horst J P, Fritz H J. Counteracting the mutagenic effect of hydrolytic deamination of DNA 5-methylcytosine residues at high temperature: DNA mismatch N-glycosylase Mig.Mth of the thermophilic archaeon Methanobacterium thermoautotrophicum THF. EMBO J. 1996;15:5459–5469. [PMC free article] [PubMed] [Google Scholar]
- 14.Kawarabayasi Y, Hino Y, Horikawa H, Yamazaki S, Haikawa Y, Jin-no K, Takahashi M, Sekine M, Baba S, Ankai A, Kosugi H, Hosoyama A, Fukui S, Nagai Y, Nishijima K, Nakazawa H, Takamiya M, Masuda S, Funahashi T, Tanaka T, Kudoh Y, Yamazaki J, Kushida N, Oguchi A, Kikuchi H, et al. Complete genome sequence of an aerobic hyper-thermophilic crenarchaeon Aeropyrum pernix K1. DNA Res. 1999;6:83–101. doi: 10.1093/dnares/6.2.83. , 145–152. [DOI] [PubMed] [Google Scholar]
- 15.Koulis A, Cowan D A, Pearl L H, Savva R. Uracil-DNA glycosylase activities in hyperthermophilic micro-organisms. FEMS Microbiol Lett. 1996;143:267–271. doi: 10.1111/j.1574-6968.1996.tb08491.x. [DOI] [PubMed] [Google Scholar]
- 16.Labahn J, Schärer O D, Long A, Ezaz-Nikpay K, Verdine G L, Ellenberger T E. Structural basis for the excision repair of alkylation-damaged DNA. Cell. 1996;86:321–329. doi: 10.1016/s0092-8674(00)80103-8. [DOI] [PubMed] [Google Scholar]
- 17.Lee T D, Vemuri S. MacProMass: a computer program to correlate mass spectral data to peptide and protein structures. Biomed Environ Mass Spectrom. 1990;19:639–645. doi: 10.1002/bms.1200191103. [DOI] [PubMed] [Google Scholar]
- 18.Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993;362:709–715. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- 19.Lindahl T, Nyberg B. Heat-induced deamination of cytosine residues in deoxyribonucleic acid. Biochemistry. 1974;13:3405–3410. doi: 10.1021/bi00713a035. [DOI] [PubMed] [Google Scholar]
- 20.Lu A-L, Fawcett W P. Characterization of the recombination MutY homolog, an adenine DNA glycosylase, from yeast Schizosaccharomyces pombe. J Biol Chem. 1998;273:25098–25105. doi: 10.1074/jbc.273.39.25098. [DOI] [PubMed] [Google Scholar]
- 21.Lu A-L, Tsai-Wu J J, Cillo J. DNA determinants and substrate specificities of Escherichia coli MutY. J Biol Chem. 1995;270:23582–23588. doi: 10.1074/jbc.270.40.23582. [DOI] [PubMed] [Google Scholar]
- 22.Michaels M L, Cruz C, Grollman A P, Miller J H. Evidence that MutY and MutM combine to prevent mutations by an oxidation damaged form of guanine in DNA. Proc Natl Acad Sci USA. 1992;89:7022–7025. doi: 10.1073/pnas.89.15.7022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nash H M, Bruner S D, Schärer O D, Kawate T, Addona T A, Spooner E, Lane W S, Verdine G L. Cloning of a yeast 8-oxoguanine DNA glycosylase reveals the existence of a base-excision DNA-repair protein superfamily. Curr Biol. 1996;6:968–980. doi: 10.1016/s0960-9822(02)00641-3. [DOI] [PubMed] [Google Scholar]
- 24.Neddermann P, Gallinari P, Lettieri T, Schmid D, Truong O, Hsuan J J, Wiebauer K, Jiricny J. Cloning and expression of human G/T mismatch-specific thymine-DNA glycosylase. J Biol Chem. 1996;271:12767–12774. doi: 10.1074/jbc.271.22.12767. [DOI] [PubMed] [Google Scholar]
- 25.Noll D M, Gogos A, Granek J A, Clarke N D. The C-terminal domain of the adenine-DNA glycosylase MutY confers specificity for 8-oxoguanine adenine mispairs and may have evolved from MutT, an 8-oxo-dGTPase. Biochemistry. 1999;38:6374–6379. doi: 10.1021/bi990335x. [DOI] [PubMed] [Google Scholar]
- 26.Nölling J, van Eeden F J M, Eggen R I L, de Vos W M. Modular organization of related archaeal plasmids encoding different restriction-modification systems in Methanobacterium thermoformicicum. Nucleic Acids Res. 1992;20:6501–6507. doi: 10.1093/nar/20.24.6501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Olsen L C, Aasland R, Wittwer C U, Krokan H E, Helland D E. Molecular cloning of human uracil-DNA glycosylase, a highly conserved DNA repair enzyme. EMBO J. 1989;8:3121–3125. doi: 10.1002/j.1460-2075.1989.tb08464.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pearson W R, Lipman D J. Improved tools for biological sequence comparison. Proc Natl Acad Sci USA. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roldán-Arjona T, Anselmino C, Lindahl T. Molecular cloning and functional analysis of a Schizosaccharomyces pombe homologue of Escherichia coli endonuclease III. Nucleic Acids Res. 1996;24:3307–3312. doi: 10.1093/nar/24.17.3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sandigursky M, Franklin W A. Thermostable uracil-DNA glycosylase from Thermotoga maritima a member of a novel class of DNA repair enzymes. Curr Biol. 1999;9:531–534. doi: 10.1016/s0960-9822(99)80237-1. [DOI] [PubMed] [Google Scholar]
- 31.Shibutani S, Takeshita M, Grollman A P. Insertion of specific bases during DNA synthesis past the oxidation-damaged base 8-oxodG. Nature. 1991;349:431–434. doi: 10.1038/349431a0. [DOI] [PubMed] [Google Scholar]
- 32.Swofford D L. PAUP*. Phylogenetic analysis using parsimony (*and other methods). Version 4. Sunderland, Mass: Sinauer Associates; 1999. [Google Scholar]
- 33.Thayer M M, Ahern H, Xing D, Cunningham R P, Tainer J A. Novel DNA binding motif in the DNA repair enzyme endonuclease III crystal structure. EMBO J. 1995;14:4108–4120. doi: 10.1002/j.1460-2075.1995.tb00083.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Völkl P, Huber R, Drobner E, Rachel R, Burggraf S, Trincone A, Stetter K O. Pyrobaculum aerophilum sp. nov., a novel nitrate-reducing hyperthermophilic archaeum. Appl Environ Microbiol. 1993;59:2918–2926. doi: 10.1128/aem.59.9.2918-2926.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang Z, Mosbaugh D W. Uracil-DNA glycosylase inhibitor gene of bacteriophage PBS2 encodes a binding protein specific for uracil-DNA glycosylase. J Biol Chem. 1989;264:1163–1171. [PubMed] [Google Scholar]
- 37.Yeh Y C, Chang D Y, Masin J, Lu A-L. Two nicking enzyme systems specific for mismatch-containing DNA in nuclear extracts from human cells. J Biol Chem. 1991;266:6480–6484. [PubMed] [Google Scholar]
- 38.Zharkov D O, Grollman A P. MutY DNA glycosylase: base release and intermediate complex formation. Biochemistry. 1998;37:12384–12394. doi: 10.1021/bi981066y. [DOI] [PubMed] [Google Scholar]