Abstract
Background
In separate phase 2 trials, 120 patients received maribavir for cytomegalovirus (CMV) infection failing conventional therapy (trial 202) and 119 received maribavir for asymptomatic infection (trial 203). Overall, 172 cleared their CMV infection (CMV DNA <200 copies/mL) within 6 weeks.
Methods
Baseline and posttreatment plasma samples were tested for mutations in viral genes UL97, UL54, and/or UL27. Selected viral mutants were phenotyped for drug susceptibility.
Results
Baseline samples revealed UL54 mutations newly phenotyped as conferring resistance to standard DNA polymerase inhibitor(s), including K493N, P497S, K513T, L565V, V823A, A987V, and E989D. Of 29 patients (including 25 from trial 202) who cleared but later experienced recurrent CMV infection while on maribavir, 23 had available UL97 genotyping data; 17 had known resistance mutations (T409M or H411Y) and 5 additional had UL97 C480F alone. The newly phenotyped mutation C480F conferred high-grade maribavir resistance and low-grade ganciclovir resistance. Among 25 who did not respond to >14 days of therapy, 9 showed T409M or H411Y and 4 others showed C480F alone.
Conclusions
After maribavir therapy (400–1200 mg twice daily), UL97 mutations T409M, H411Y, or C480F emerge to confer maribavir resistance in patients with recurrent CMV infection while on therapy or no response to therapy.
Clinical Trials Registration
NCT01611974 and EudraCT 2010-024247-32.
Keywords: cytomegalovirus, maribavir, antiviral therapy, antiviral drug resistance, kinase
In phase 2 trials of maribavir therapy, viral UL97 mutations T409M, H411Y, or C480F emerged to confer maribavir resistance in patients with recurrent CMV infection while on therapy or no response to therapy.
The treatment of systemic cytomegalovirus (CMV) infection and end-organ disease has long relied on the nucleoside analog ganciclovir and other compounds targeting the DNA polymerase encoded by the viral UL54 gene. Although successful in many cases, particularly when used before the onset of symptomatic disease [1], toxicity and the emergence of drug resistance have prompted the development of alternative therapies with different antiviral targets and improved tolerability. An example is the terminase inhibitor letermovir, which was recently approved for the prevention of CMV infection in stem cell recipients [2]. The role of letermovir as treatment of active CMV infection has yet to be defined.
The UL97 kinase inhibitor maribavir is a benzimidazole l-riboside that showed high antiviral potency in vitro [3] and favorable properties during preclinical and early clinical testing [4], including a benign toxicity profile without myelosuppression or nephrotoxicity and notable mainly for dysgeusia (taste disturbance). The UL97 kinase has important roles in the replication of CMV, such that genetic knockout or maribavir suppression of its function severely impairs viral growth but does not prevent it entirely [5]. Phase 3 trials showed that low-dose maribavir (100 mg twice a day) lacked efficacy for prevention of CMV infection in transplant recipients [6, 7]. These were followed by phase 2 trials of higher doses (400, 800, and 1200 mg twice a day) for the treatment of CMV infection resistant or refractory to conventional therapy (trial 202, NCT01611974) [8], or treatment of asymptomatic CMV infection in immunocompromised transplant recipients (trial 203, EudraCT 2010-024247-32) [9]. The recently published reports of these trials indicate that 67%–77% of patients successfully cleared their plasma CMV DNA (<200 copies/mL) within 6 weeks, although some of them later went on to develop recurrent CMV infection after initially clearing.
This analysis reports on the viral mutations and associated drug-resistance phenotypes detected at baseline and after maribavir therapy in patients enrolled in trials 202 and 203.
METHODS
Clinical Trial Samples
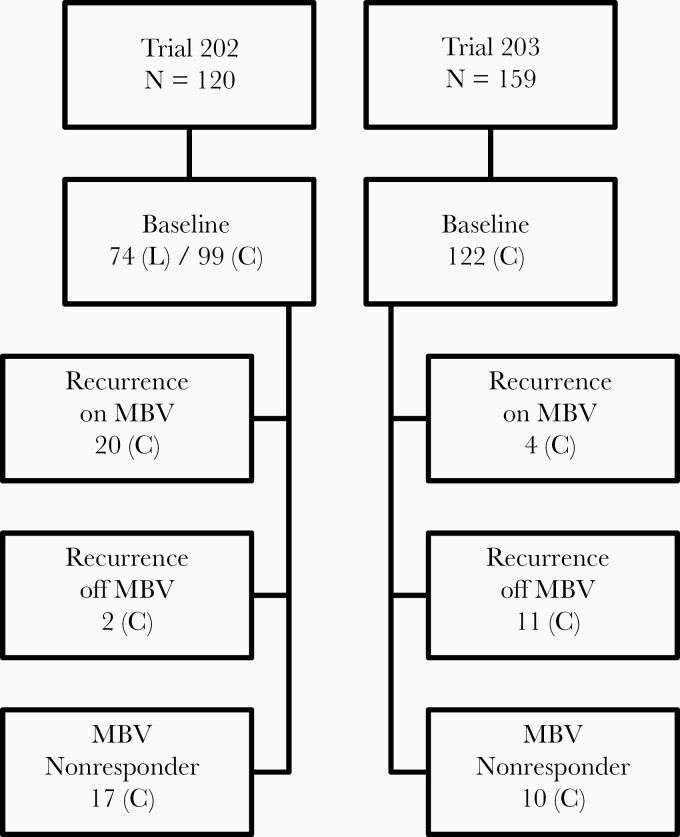
The characteristics and treatment outcomes of 120 patients (trial 202) and 119 patients (trial 203) who received maribavir at doses of 400, 800, or 1200 mg twice daily have been published [8, 9]. Trial 203 included 40 additional patients randomized to valganciclovir. Plasma specimens for genotyping were scheduled at the time of study entry (baseline) in all patients, and at follow-up in those who had a detectable posttreatment plasma CMV load representing scenarios of (1) initial viral clearance after receiving maribavir as judged by 2 consecutive undetectable viral loads (<200 copies/mL) at least 5 days apart, followed by recurrence of CMV infection while remaining on maribavir therapy, as judged by 2 consecutive measurable viral loads (≥200 copies/mL) at least 5 days apart; (2) initial viral clearance after receiving maribavir and recurrence of infection after drug discontinuation; and (3) no documented CMV clearance by the end of the maribavir treatment period (nonresponder). The number of patients in each of these categories with available genotype information is shown in Figure 1. Baseline genotyping was not performed on all patients, not all patients had follow-up genotyping as intended, and genotyping was done on only 1 posttreatment specimen in most cases. In trial 202, the study sites often submitted locally acquired genotyping data to support an entry criterion of CMV infection resistant to conventional therapy [8].
Figure 1.
The genotyping study population. In top level boxes, N is the number of patients who received study drug, including 40 given valganciclovir in trial 203. Numbers in lower level boxes are the count of patients with available genotypic data at baseline (all patients) or with different outcomes after maribavir (MBV) therapy. Nonresponder group received maribavir for >14 days. Viral DNA sequencing was performed at the central laboratory (C), supplemented by local laboratory testing (L) at baseline. Baseline genotypic data included portions of genes UL27, UL54, and UL97, and posttreatment data included UL27 and UL97. Not all patients had coverage of all targeted genes.
Genotyping Protocol
The genotyping protocol and viral gene sequencing targets were developed before study initiation without input from the present authors [8, 9]. Frozen DNA extracts of plasma samples were sent to a designated central laboratory where nested polymerase chain reaction (PCR) was performed to amplify regions of viral genes as follows: (1) UL97 from codon 270 to 482, (2) UL54 from codon 389 to 852 and from 943 to 1058, and (3) UL27 from codon 108 to 424, using primers as listed in Supplementary Table 1. After fluorescent dideoxy DNA sequencing of the PCR products (Supplementary Table 1), results were compiled by the central laboratory and reported as amino acid substitutions relative to CMV strain AD169. Because the same primers were used for sequencing as for template preparation, bidirectional reads were feasible 15–20 codons inside each of the ranges listed above, while codons at the extremes of the amplified range were covered in only 1 direction of sequencing. For example, the UL97 codon range targeted for bidirectional coverage was 288–468, which notably excludes most of the canonical ganciclovir-resistance mutations that are beyond UL97 codon 468. Therefore, meaningful genotypic detection of emerging resistance was impossible for the patients in trial 203 who received valganciclovir as a comparator.
Sequencing Interpretation and Quality Control
Important mutation readouts provided by the central laboratory, including the well-known maribavir-resistance mutations at UL97 codons 409 and 411, as well as uncharacterized mutations, were reassessed by manual review of the original sequencing chromatograms. Known resistance mutations required at least 1 visually distinct readout with a quality value score of 30 or better in the read vicinity and a compatible readout in the opposite direction, while previously undocumented mutations (absent in published literature and genetic databases) required confirmation on both DNA strands with a quality value score of 30 or better. Readouts of unclear authenticity were recommended for independent PCR and resequencing, but this was feasible in only a few cases because of loss of the original samples or their DNA extracts.
Interpretation of amino acid substitutions as sequence polymorphisms was based on their presence among genomic CMV sequences posted in the Genbank database without indication of antiviral drug exposure. UL54 variants between codons 600 and 695 were tentatively classified as polymorphisms because no drug-resistance mutations have mapped to this region that contains many documented polymorphisms [10, 11]. Because of the abundance of polymorphisms in UL27, baseline variants (present before receiving maribavir) were also tentatively classified as polymorphisms. Known resistance mutations are those confirmed by a recombinant phenotype, while suspected resistance mutations and candidates for phenotyping are those that emerged as a change from baseline, especially at codons already implicated in drug resistance or at the same or similar location as known resistance mutations.
Recombinant Phenotyping
This process for assigning a drug susceptibility profile to a specific mutation consists of recombining the desired mutation into a baseline susceptible cloned laboratory CMV strain expressing a secreted alkaline phosphatase (SEAP) reporter gene for viral growth quantitation, confirming the transferred mutation by sequencing, and performing yield reduction susceptibility testing in ARPEp cell cultures with 8 or more replicates over at least 4 setup dates, all as previously described and standardized [12, 13]. Susceptibility is expressed as the drug concentration required to reduce SEAP growth by 50% (half maximum effective concentration, EC50).
RESULTS
The genotyping study population is outlined in Figure 1. At follow-up, more patients in trial 202 than in trial 203 had genotyping done after nonresponse to therapy or recurrence of infection while still receiving maribavir, consistent with a history of treatment-refractory or drug-resistant CMV infection.
Validation of Genotyping Data
Review of 2353 amino acid substitutions reported from the central laboratory revealed 283 that were not verifiable from original sequence chromatograms according to quality control criteria described in the “Methods” section (Supplementary Table 2). These rejected data involved 58 distinct substitutions, 34 in UL27, 21 in UL54, and 3 in UL97. In many cases, the amino acid substitution was unspecified (marked “X”) because no clear readout was possible from the multiple peaks and unstable baselines. There were 200 unexplainable readouts of UL54 I833M across trials 202 and 203. Certain substitutions such as UL27 S352R, A397P, R399G, M418I, and Q424H were reported in multiple samples, where no results from any of the samples supported the readout. A limited set of 7 samples was independently amplified and sequenced in another reference laboratory, with results shown in Supplementary Table 3. Substitutions that appeared in Supplementary Table 2, such as UL27 Y168C and L169W, or UL54 I833M, were not reproducible on resequencing.
Sequence Variants Observed at Baseline
Baseline UL97 sequence findings in trial 202 have been separately published [13]. In addition to the expected abundance of canonical ganciclovir-resistance mutations after prior therapy as reported by the local study sites, a P-loop mutation F342Y conferring ganciclovir-maribavir cross-resistance and atypical ganciclovir-resistance mutations K359E and K359Q were documented. In trial 203, 118 patients had baseline UL97 sequencing done at the central laboratory, and no known or suspected resistance mutations were detected in the designated codon range (288–468).
Baseline UL54 mutations reported by local study sites for trial 202 are shown in Supplementary Table 4. Most of the 14 distinct mutations were confirmed by the central laboratory for the same patients, although in several cases the concordant baseline mutations were detected only on reinspection of the sequence chromatograms. The central laboratory tested for UL54 variants in baseline samples from 86 patients in trial 202 and 93 patients in trial 203. After filtering out known and suspected polymorphisms (Supplementary Table 4), 29 variants of interest are shown in Table 1, of which 15 were known drug-resistance mutations [11], 8 were selected for recombinant phenotyping, and 6 remain uncharacterized. Among the latter, the variant V998I was observed at baseline in a valganciclovir trial without subsequent treatment failure [14], and no mutations at codons beyond 1000 have yet been shown to confer drug resistance.
Table 1.
UL54 DNA Polymerase Variants Detected at Baseline (Central Laboratory)
| Trial | Substitution | n | Polymerase Domain | Mutation Category | Resistance Conferreda |
|---|---|---|---|---|---|
| 202 | N408K | 1 | ExoII | Known | CDV, GCV |
| 202 | F412C | 1 | ExoII | Known | CDV, GCV |
| 202 | F412L | 1 | ExoII | Known | CDV, GCV |
| 202 | K493N | 1 | Exo | Newly phenotyped | CDV, GCV, FOS |
| 202 | N495K | 1 | Exo | Known | FOS |
| 202 | P497S | 1 | Exo | Newly phenotyped | CDV, (GCV) |
| 202 | T503I | 2 | ExoIII | Known | CDV, GCV |
| 202 | K513N | 1 | ExoIII | Known | CDV, GCV |
| 202 | K513T | 1 | ExoIII | Newly phenotyped | CDV, GCV |
| 202 | L545S | 1 | ExoIII | Known | CDV, GCV |
| 202 | L565V | 1 | NH2 Terminal 2 | Newly phenotyped | (CDV), (GCV), FOS |
| 202 | Q578H | 1 | NH2 Terminal 2 | Known | CDV, GCV, FOS |
| 202 | Q578R | 1 | NH2 Terminal 2 | Uncharacterized | … |
| 202 | D594N | 1 | NH2 Terminal 2 | Newly phenotyped | None |
| 202 | T700A | 1 | Palm 1 | Known | FOS |
| 202 | V715M | 2 | Palm 1 | Known | FOS |
| 203 | V715M | 1 | Palm 1 | Known | FOS |
| 202 | E756Q | 1 | Palm 1 | Known | FOS |
| 202 | L773V | 1 | Finger | Known | CDV, GCV, FOS |
| 203 | V781I | 1 | Finger | Known | GCV, FOS |
| 202 | V823A | 1 | Finger | Newly phenotyped | CDV, GCV |
| 202 | A834P | 2 | Palm 2 | Known | CDV, GCV, FOS |
| 203 | A987G | 1 | Thumb | Known | CDV, GCV |
| 202 | A987V | 1 | Thumb | Newly phenotyped | FOS |
| 203 | E989D | 1 | Thumb | Newly phenotyped | (CDV), GCV, FOS |
| 202 | V998I | 2 | Thumb | Uncharacterized | … |
| 203 | R1017H | 1 | Thumb | Uncharacterized | … |
| 202 | R1037C | 1 | Thumb | Uncharacterized | … |
| 203 | V1040M | 1 | Thumb | Uncharacterized | … |
| 203 | H1049Y | 1 | Thumb | Uncharacterized | … |
n = number of patients with the listed substitution detected.
Abbreviations: CDV, cidofovir; EC50, half maximum effective concentration; FOS, foscarnet; GCV, ganciclovir.
aParentheses denote borderline resistance (<2-fold increased EC50).
Baseline UL27 sequence variants were reported by the central laboratory for 89 patients in trial 202 and 100 patients in trial 203. These are listed in Supplementary Table 5. Among over 1000 readouts of UL27 variation reported in these specimens, 98% were known polymorphisms [15, 16] and the remainder were presumptive polymorphisms as they did not reflect any prior exposure to maribavir.
Sequence Variants Observed After Maribavir Therapy
Follow-up UL97 genotyping results are shown in Table 2 for trial patients who successfully cleared their plasma CMV DNA (<200 copies/mL) but later developed a recurrence of CMV DNA while still receiving maribavir. Genotyping was attempted on all 29 patients across both trials and was successful in 23 cases. The previously characterized UL97 mutations [12, 17], T409M that confers 75 to 90-fold increased maribavir EC50, and H411Y that confers 12 to 18-fold increased EC50, were well represented with 14 and 3 patients, respectively, testing positive. A previously uncharacterized substitution, UL97 C480F, was discovered in 6 patients on review of original chromatogram data (Supplementary Figure 1). Both C480F and T409M were detected in 1 patient. Overall, 22 patients in this category tested positive for T409M, H411Y, or C480F, after 42 to 168 days of maribavir therapy, representing 96% of the 23 with UL97 genotyping data and 76% of those with CMV recurrence while on maribavir.
Table 2.
UL97 Genotyping Follow-up on Patients With CMV Clearance and Recurrence While on Maribavir
| Both Trials | Trial 202 | Trial 203 | ||||||
|---|---|---|---|---|---|---|---|---|
| Characteristic | All Doses | All Doses | 400 mg | 800 mg | 1200 mg | All Doses | 400 mg | 800 mg |
| No. of patients with recurrence on maribavir after clearance | 29 | 25 | 6 | 9 | 10 | 4 | 2 | 2 |
| No. of patients with UL97 follow-up genotyping data | 23 | 19 | 5 | 9 | 5 | 4 | 2 | 2 |
| Median treatment duration, d | 95 | 111 | 95 | 160 | 76 | 70 | 74 | 57 |
| Median starting CMV load, copies/mL | 10 000 | 10 000 | 10 000 | 10 000 | 20 000 | 5500 | 5500 | 6500 |
| Median study day at follow-up genotyping | 84 | 86 | 106 | 87 | 83 | 71 | 75 | 62 |
| Study day range at follow-up | 52–169 | 70–169 | 85–122 | 71–169 | 70–161 | 52–80 | 69–80 | 52–72 |
| UL97 mutation detected, No.a | ||||||||
| T409M | 14 (61%) | 10 | 2 | 5 | 3b | 4 | 2 | 2 |
| H411Y | 3 (13%) | 3 | 2 | 1 | 0 | 0 | 0 | 0 |
| C480F | 6 (26%) | 6 | 0 | 3 | 3b | 0 | 0 | 0 |
| T409M, H411Y, or C480F | 22 (96%) | 18 | 4 | 9 | 5 | 4 | 2 | 2 |
Abbreviations: CMV, cytomegalovirus.
aPercentages based on number with available UL97 genotyping data.
bOne patient had UL97 C480F and T409M detected.
Table 3 shows the UL97 genotyping results of nonresponders who did not clear their CMV DNA after receiving more than 14 days of maribavir. Overall, 13 of 25 patients tested positive for UL97 mutation T409M, H411Y, or C480F after 46 to 116 days of maribavir therapy, which is 52% of those with genotype data and 33% of nonresponder patients. One patient tested positive for both T409M and C480F; the sample containing a mixture of C480F as a minority subpopulation was independently sequenced in a second reference laboratory with the same finding (Supplementary Table 3).
Table 3.
UL97 Genotyping in Nonresponders Who Received >14 Days of Maribavir
| Both Trials | Trial 202 | Trial 203 | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Characteristic | All Doses | All Doses | 400 mg | 800 mg | 1200 mg | All Doses | 400 mg | 800 mg | 1200 mg |
| Nonresponders who received maribavir for >14 d | 39 | 26 | 11 | 11 | 4 | 13 | 5 | 5 | 3 |
| Patients with UL97 follow-up genotyping data | 25 | 16 | 7 | 6 | 3 | 9 | 3 | 4 | 2 |
| Median treatment duration, d | 55 | 62 | 61 | 59 | 70 | 46 | 45 | 58 | 49 |
| Median starting CMV load, copies/mL | 40 000 | 50 000 | 50 000 | 30 000 | 200 000 | 14 500 | 14 500 | 155 000 | 11 500 |
| Median study day at follow-up genotyping | 58 | 59 | 59 | 59 | 58 | 54 | 46 | 68 | 50 |
| Study day range at follow-up | 15–140 | 15–140 | 15–140 | 47–116 | 52–91 | 21–83 | 46–53 | 54–83 | 21–78 |
| UL97 mutation detected, Noa | |||||||||
| T409M | 7 (28%) | 5 | 1 | 3b | 1 | 2 | 0 | 2 | 0 |
| H411Y | 2 (8.3%) | 2 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| C480F | 5 (21%) | 3 | 0 | 3b | 0 | 2 | 1 | 0 | 1 |
| T409M, H411Y or C480F | 13 (52%) | 9 | 2 | 6 | 1 | 4 | 1 | 2 | 1 |
| F342Y | 1 | 1c | |||||||
Abbreviation: CMV, cytomegalovirus.
aPercentages based on number with available UL97 genotyping data.
bOne patient had both UL97 C480F and T409M detected.
cThis mutation was present at baseline and later detected in combination with H411YJ.
Patients with CMV recurrence after maribavir discontinuation showed no evidence of UL97 mutation in available follow-up samples from 2 of 5 patients in trial 202 and 11 of 18 patients in trial 203, including reinspection of sequencing chromatograms for C480F. In 1 patient from trial 202 who initially responded to maribavir, a day 57 plasma sample was positive for UL97 mutation H411L that confers approximately 70-fold increased maribavir EC50 [17]. When maribavir was stopped at day 58, the case did not meet protocol criteria for recurrence while on maribavir (2 positive viral loads at least 5 days apart), although this criterion would have been met using locally obtained viral loads.
Follow-up UL27 genotyping was performed on 39 patients in trial 202 and 43 patients in trial 203. The only uncharacterized UL27 variant detected that was not among those detected at baseline (Supplementary Table 5) was G344D, found in a day 15 sample from a patient whose viral load increased from 500 copies/mL at baseline to 10 000 copies/mL. There was no baseline UL27 sequencing in this patient but the G344D readout at day 15 was confirmed in a second laboratory (Supplementary Table 3).
Recombinant Phenotyping
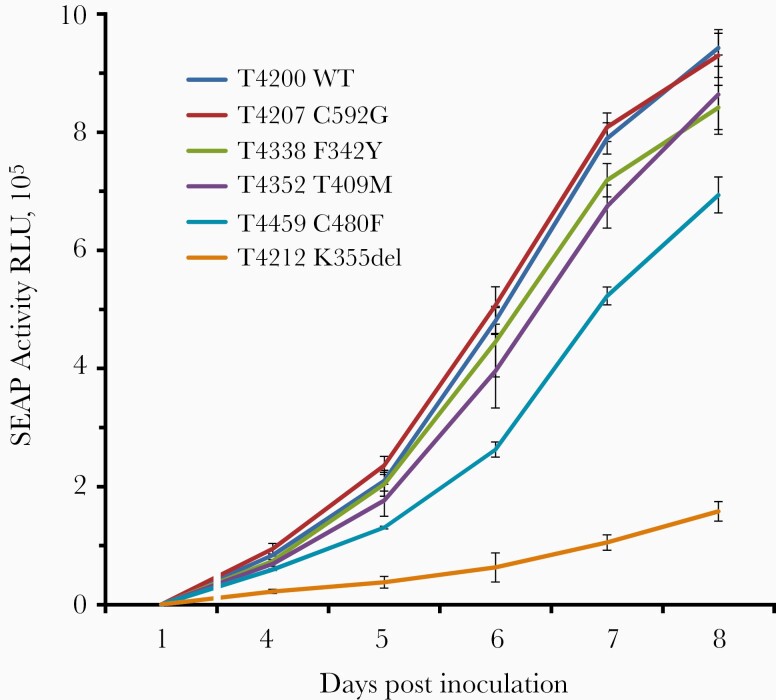
Uncharacterized mutations selected for phenotyping included UL27 G344D and UL97 C480F in Table 4 and 8 UL54 mutations in Table 5. UL27 G344D conferred no maribavir resistance, UL97 C480F conferred high-grade maribavir resistance and low-grade ganciclovir resistance. The C480F mutant was growth retarded by about 1 day at days 6 and 7 (Figure 2), a slower growth than the T409M mutant but better than the kinase knockout mutant K355del.
Table 4.
Genotypes and Phenotypes of UL27 and UL97 Recombinant Viruses
| Maribavir EC50, µM | Ganciclovir EC50, µM | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Straina | Genotypeb | Mean | SD | n | Ratio | Mean | SD | n | Ratio |
| Control strains | |||||||||
| 4200 | UL97 WT | 0.12 | 0.029 | 16 | 1.35 | 0.16 | 22 | ||
| 4344 | UL27 WT | 0.12 | 0.033 | 13 | |||||
| 4207 | UL97 C592G | 4.14 | 0.68 | 21 | 3.1 | ||||
| 4352 | UL97 T409M | 10 | 1.9 | 10 | 78 | ||||
| 4353 | UL97 H411Y | 1.9 | 0.40 | 6 | 15 | ||||
| New recombinants | |||||||||
| 4375 | UL27 G344D | 0.11 | 0.03 | 13 | 0.9 | ||||
| 4459 | UL97 C480F | 28 | 3.6 | 10 | 224 | 3.10 | 0.53 | 19 | 2.3* |
Ratio = EC50 of mutant virus/EC50 of wild type control. Bold indicates drug resistance. WT control including silent Frt motif upstream of indicated gene in all strains.
Abbreviations: EC50, half maximum effective concentration; n, number of replicates; WT, wild type.
aSerial number of recombinant CMV strain.
bGene and amino acid substitution.
*Significantly elevated from WT control (P < 5 × 10−12, Student t test, 2-tailed, unequal variance).
Table 5.
Genotypes and Phenotypes of UL54 Recombinant Viruses
| Cidofovir EC50, µM | Ganciclovir EC50, µM | Foscarnet EC50, µM | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Straina | Genotypeb | Mean | SD | n | Ratio | Mean | SD | n | Ratio | Mean | SD | n | Ratio |
| Control strains | |||||||||||||
| 4198 | UL54 WT | 0.24 | 0.06 | 27 | 1.22 | 0.22 | 44 | 37 | 7 | 43 | |||
| 4376 | D981del2 | 0.90 | 0.18 | 24 | 3.7 | 7.64 | 1.82 | 25 | 6.2 | 115 | 19 | 34 | 3.1 |
| New recombinants (n = 8) | |||||||||||||
| 4460 | K493N | 0.50 | 0.10 | 15 | 2.0 | 5.05 | 1.28 | 15 | 4.1 | 124 | 25 | 10 | 3.3 |
| 4484 | P497S | 0.58 | 0.14 | 12 | 2.4 | 2.30 | 0.40 | 16 | 1.9 | 29 | 4 | 10 | 0.8 |
| 4485 | K513T | 4.71 | 1.16 | 12 | 19 | 5.98 | 0.94 | 16 | 4.9 | 36 | 4 | 8 | 1.0 |
| 4471 | L565V | 0.39 | 0.04 | 12 | 1.6 | 2.22 | 0.30 | 14 | 1.8 | 138 | 31 | 14 | 3.7 |
| 4465 | D594N | 0.24 | 0.07 | 10 | 1.0 | 1.28 | 0.48 | 11 | 1.0 | 36 | 5 | 12 | 1.0 |
| 4404 | V823A | 0.87 | 0.27 | 11 | 3.6 | 3.00 | 0.59 | 15 | 2.4 | 20 | 6 | 11 | 0.5 |
| 4473 | A987V | 0.22 | 0.05 | 10 | 0.9 | 1.06 | 0.24 | 12 | 0.9 | 116 | 17 | 12 | 3.1 |
| 4474 | E989D | 0.46 | 0.08 | 13 | 1.9 | 3.91 | 0.70 | 10 | 3.2 | 213 | 31 | 8 | 5.7 |
Ratio = EC50 of mutant virus/EC50 of wild type control. Bold indicates EC50 >1.5 × WT control (all significant at P < 2 × 10−8, Student t test, 2-tailed, unequal variance).
Abbreviations: EC50, half maximum effective concentration; n, number of replicate assays; SD, standard deviation of the EC50 values; WT, wildtype.
aSerial number of recombinant CMV strain
bUL54 amino acid substitution shown. D981del2 = in frame deletion of codons 981–982.
Figure 2.
Comparative growth curves of viral strains. WT control virus and UL97 mutants were inoculated at equal multiplicity of infection of 0.02 as calibrated by culture supernatant SEAP activity at 24 hours. At each of 4 to 8 days post inoculation, culture supernatants were assayed for SEAP activity (RLU) as a measure of viral growth. Data points are the mean and standard deviation of 4 replicates set up simultaneously. Abbreviations: SEAP, secreted alkaline phosphatase; RLU, relative light unit; WT, wild type.
The UL54 DNA polymerase K513T mutant has the cidofovir-ganciclovir resistant and foscarnet-sensitive phenotype characteristic of exonuclease domain mutations, including K513N [18], K513R [19], and K513E [20]. The K493N and P497S mutations, while located within the broadly defined exonuclease region, are not within specific ExoI to ExoIII domains [11] and have divergent phenotypes. The foscarnet resistance conferred by K493N is similar to that of nearby N495K [21] but is accompanied by a low-grade ganciclovir and cidofovir resistance, while P497S has only the low-grade ganciclovir-cidofovir resistance without foscarnet resistance.
The foscarnet resistance and borderline ganciclovir-cidofovir resistance of mutant L565V is compatible with phenotypes of mutations Q578H and D588N in the same polymerase amino terminal domain [22]. Unlike D588N, D594N did not confer significant resistance. The finger domain mutant V823A is close to many other foscarnet-resistance mutations but was found instead to have a low-grade ganciclovir-cidofovir–resistance phenotype. The foscarnet-resistant thumb domain mutant A987V has the opposite phenotype of the well-known mutant A987G that shows ganciclovir and cidofovir resistance without foscarnet resistance [20, 23]. Finally, the thumb domain mutant E989D is severely growth retarded (by about 3 days) and shows a triple-resistant phenotype.
DISCUSSION
This combined genotypic analysis of samples from clinical trials 202 and 203 confirms the known UL97 mutations at codons 409 and 411 as major causes of maribavir resistance in those who did not clear or had recurrence of CMV while on prolonged therapy. In addition, UL97 C480F is newly characterized as an important resistance mutation in those receiving maribavir as dosed in these trials, despite some technical limitations in the genotypic analysis. Taken together, T409M, H411Y, or C480F were frequently found in those with recurrence of CMV infection while on maribavir therapy (Table 2) and in a lower proportion of those who did not clear their CMV infection (Table 3), perhaps consistent with the shorter median treatment duration at the time of genotyping and varying reasons for drug discontinuation. The shortest maribavir exposure associated with a resistance mutation (T409M, H411Y, or C480F) was 6 weeks.
The status of UL97 T409M and H411Y as principal genotypic markers of maribavir resistance was anticipated by in vitro selection data [17] and initial case reports [24, 25]. These mutations confer moderate (H411Y) to moderately high (T409M) maribavir resistance, with no ganciclovir cross-resistance, and map to a hinge region of the ATP-binding site of the UL97 kinase that is the target of competitive inhibition by maribavir [17]. Statistics for T409M and H411Y among those with CMV recurrence in trial 202 (Table 2) are the same as in the primary publication [8], with additional instances as shown in Table 3 for nonresponders. The primary publication of trial 203 [9] mentioned 2 cases of T409M among CMV recurrences at study week 6 (800 mg dose in Table 2) but additional cases exist, as shown in Table 2 and Table 3. The H411L mutation, observed once in trial 202, has also been selected in vitro to confer moderately high maribavir resistance.
UL97 C480F lies in a conserved kinase catalytic loop outside the UL97 codon range specified for analysis by the central laboratory and was not described in the primary publication for either trial 202 or 203 [8, 9], but was retrospectively identified with a frequency between those of T409M and H411Y. It was fortuitously included in the PCR product amplified for sequencing but not verified with bidirectional sequencing. The available chromatograms (Supplementary Figure 1) and confirmatory sequencing on 1 sample, however, make it very likely that the finding is clinically significant. C480F was previously reported in a clinical specimen from a maribavir-treated patient [26] and C480R was identified in vitro as conferring maribavir and ganciclovir resistance [27]. C480F confers the highest degree of maribavir resistance (224-fold; Table 4) of any single mutation encountered so far in vivo, along with low-grade (2.3-fold) ganciclovir cross-resistance. Emergence of C480F may be favored at higher maribavir doses.
A concern about cross-resistance is that prior ganciclovir therapy may select for mutant subpopulations such as C480F or the recently characterized F342Y [13] to facilitate the development of clinical maribavir resistance. There is currently insufficient evidence to address this possibility. Relevant factors include the growth fitness of the cross-resistant mutants, which is fairly well preserved for F342Y and C480F (Figure 2), and the relative frequency of selection of such mutants compared with the well-established ganciclovir-resistance mutations at UL97 codons 460, 520, and 590–607, which do not confer maribavir cross-resistance. Baseline UL97 sequencing in trial 202 detected only 1 case of F342Y potentially impairing response to maribavir therapy, but the sensitivity of detection of minor mutant subpopulations by Sanger sequencing is inadequate. Further evidence may emerge from deep sequencing analyses of maribavir pretreatment samples.
Limited sequencing data did not suggest a major clinical role for UL27 mutations in maribavir resistance. The variant G344D that was detected only in postbaseline specimens was phenotyped as maribavir susceptible. In theory, any of the multitude of known and suspected baseline polymorphisms could influence maribavir susceptibility, but the low-level effects of UL27 mutation [28] are expected to have less impact than the resistance phenotypes documented for the main UL97 mutations.
Despite the large number of UL54 resistance mutations already mapped, the 8 newly characterized amino acid substitutions listed in Table 5 offer new insights. These include the foscarnet-resistance conferred by K493N, the differential susceptibilities of L565V and D594N that are on either side of known mutations Q578H and D588N, and the opposite phenotypes of A987G and A987V. The findings reinforce the difficulty of estimating the phenotypes of newly observed UL54 mutations from similar or nearby mutations and the continuing need for accurate phenotypes to guide treatment decisions.
There were technical limitations in this genotypic analysis. A single timepoint of follow-up genotyping may be suboptimal. The limited range of UL97 codons (288–468) targeted for sequencing omitted coverage of important resistance mutations and did not allow for comparison with valganciclovir in trial 203. Many central laboratory readouts, including those listed as ambiguous and multiple occurrences of specific ones (Supplementary Table 1), were unsupportable on reinspection of original chromatograms, and some were demonstrably irreproducible (Supplementary Table 2). There were falsely negative readouts of some resistance mutations (Supplementary Table 4 and UL97 C480F). Resequencing to confirm the authenticity of questionable mutations was not possible in most cases without adequate archived specimens. More rigorous genotypic testing planned for the ongoing phase 3 maribavir treatment trials should further elucidate the genetics of maribavir resistance in clinical use, including the relative frequency of C480F and other mutations.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Acknowledgments. We thank Ronald Ercolani and Anh Le-Cook for technical assistance with phenotyping at Portland VA Medical Center, and Anna Wijatyk and Minggeng Gao (former employees of Shire, now a Takeda Company) for earlier analyses of genotypic data from these trials. We thank Marc Uknis MD (Transplant Infectious Disease fellow and former employee of ViroPharma/Shire, a Takeda company) for his contribution to studies 202 and 203.
Financial support. This work was supported by the National Institutes of Health, National Institute of Allergy and Infectious Diseases (grant number R01-AI116635); Shire Human Genetic Therapies, Inc., a Takeda Company (VA-Shire Cooperative Research and Development Agreement); Shire Development LLC, a Takeda Company (clinical trials 202 and 203); and by the Department of Veterans Affairs for use of facilities and resources.
Potential conflicts of interest. S. C. is principal investigator on separate Cooperative Research and Development Agreements between Portland VA Medical Center and (1) Merck, and (2) Shire Human Genetic Therapies, Inc., Lexington, MA, a Takeda Company, providing for costs of recombinant phenotyping without payments to the principal investigator. C. C. received consultant fees from Shire Human Genetic Therapies, Inc., a Takeda Company. T. B., K. S., and J. W. are employees of Shire Human Genetic Therapies, Inc., a Takeda Company.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
Data sharing. The clinical trials datasets, including redacted study protocol, redacted statistical analysis plan, and individual participants’ data behind the results reported in this article, will be available within 3 months from initial request to researchers who provide a methodologically sound proposal. The data will be provided after its deidentification, in compliance with applicable privacy laws, data protection, and requirements for consent and anonymization.
Contributor Information
Sunwen Chou, Oregon Health and Science University and Veterans Affairs Health Care System, Portland, Oregon, USA.
Kening Song, Shire Human Genetic Therapies Inc., a Takeda Company, Lexington, Massachusetts, USA.
Jingyang Wu, Shire Human Genetic Therapies Inc., a Takeda Company, Lexington, Massachusetts, USA.
Tien Bo, Shire Human Genetic Therapies Inc., a Takeda Company, Lexington, Massachusetts, USA.
Clyde Crumpacker, Harvard Beth Israel-Deaconess Medical Center, Boston, Massachusetts, USA.
References
- 1. Kotton CN, Kumar D, Caliendo AM, et al. The third international consensus guidelines on the management of cytomegalovirus in solid-organ transplantation. Transplantation 2018; 102:900–31. [DOI] [PubMed] [Google Scholar]
- 2. Marty FM, Ljungman P, Chemaly RF, et al. Letermovir prophylaxis for cytomegalovirus in hematopoietic-cell transplantation. N Engl J Med 2017; 377:2433–44. [DOI] [PubMed] [Google Scholar]
- 3. Biron KK, Harvey RJ, Chamberlain SC, et al. Potent and selective inhibition of human cytomegalovirus replication by 1263W94, a benzimidazole L-riboside with a unique mode of action. Antimicrob Agents Chemother 2002; 46:2365–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Winston DJ, Young JA, Pullarkat V, et al. Maribavir prophylaxis for prevention of cytomegalovirus infection in allogeneic stem cell transplant recipients: a multicenter, randomized, double-blind, placebo-controlled, dose-ranging study. Blood 2008; 111:5403–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Prichard MN. Function of human cytomegalovirus UL97 kinase in viral infection and its inhibition by maribavir. Rev Med Virol 2009; 19:215–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Marty FM, Ljungman P, Papanicolaou GA, et al. Maribavir prophylaxis for prevention of cytomegalovirus disease in recipients of allogeneic stem-cell transplants: a phase 3, double-blind, placebo-controlled, randomised trial. Lancet Infect Dis 2011; 11:284–92. [DOI] [PubMed] [Google Scholar]
- 7. Winston DJ, Saliba F, Blumberg E, et al. Efficacy and safety of maribavir dosed at 100 mg orally twice daily for the prevention of cytomegalovirus disease in liver transplant recipients: a randomized, double-blind, multicenter controlled trial. Am J Transplant 2012; 12:3021–30. [DOI] [PubMed] [Google Scholar]
- 8. Papanicolaou GA, Silveira FP, Langston AA, et al. Maribavir for refractory or resistant cytomegalovirus infections in hematopoietic-cell or solid-organ transplant recipients: a randomized, dose-ranging, double-blind, phase 2 study. Clin Infect Dis 2019; 68:1255–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Maertens J, Cordonnier C, Jaksch P, et al. Maribavir for preemptive treatment of cytomegalovirus reactivation. N Engl J Med 2019; 381:1136–47. [DOI] [PubMed] [Google Scholar]
- 10. Chou S, Lurain NS, Weinberg A, Cai GY, Sharma PL, Crumpacker CS. Interstrain variation in the human cytomegalovirus DNA polymerase sequence and its effect on genotypic diagnosis of antiviral drug resistance. Adult AIDS Clinical Trials Group CMV Laboratories. Antimicrob Agents Chemother 1999; 43:1500–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chou S. Advances in the genotypic diagnosis of cytomegalovirus antiviral drug resistance. Antiviral Res 2020; 176:104711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chou S. Foscarnet resistance mutations mapping to atypical domains of the cytomegalovirus DNA polymerase gene. Antiviral Res 2017; 138:57–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chou S, Wu J, Song K, Bo T. Novel UL97 drug resistance mutations identified at baseline in a clinical trial of maribavir for resistant or refractory cytomegalovirus infection. Antiviral Res 2019; 172:104616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chou S, Boivin G, Ives J, Elston R. Phenotypic evaluation of previously uncharacterized cytomegalovirus DNA polymerase sequence variants detected in a valganciclovir treatment trial. J Infect Dis 2014; 209:1209–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hantz S, Couvreux A, Champier G, et al. Conserved domains and structure prediction of human cytomegalovirus UL27 protein. Antivir Ther 2009; 14:663–72. [PubMed] [Google Scholar]
- 16. Sijmons S, Thys K, Mbong Ngwese M, et al. High-throughput analysis of human cytomegalovirus genome diversity highlights the widespread occurrence of gene-disrupting mutations and pervasive recombination. J Virol 2015; 89:7673–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chou S, Marousek GI. Accelerated evolution of maribavir resistance in a cytomegalovirus exonuclease domain II mutant. J Virol 2008; 82:246–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cihlar T, Fuller MD, Mulato AS, Cherrington JM. A point mutation in the human cytomegalovirus DNA polymerase gene selected in vitro by cidofovir confers a slow replication phenotype in cell culture. Virology 1998; 248:382–93. [DOI] [PubMed] [Google Scholar]
- 19. Chou S, Ercolani RJ, Sahoo MK, Lefterova MI, Strasfeld LM, Pinsky BA. Improved detection of emerging drug-resistant mutant cytomegalovirus subpopulations by deep sequencing. Antimicrob Agents Chemother 2014; 58:4697–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Cihlar T, Fuller MD, Cherrington JM. Characterization of drug resistance-associated mutations in the human cytomegalovirus DNA polymerase gene by using recombinant mutant viruses generated from overlapping DNA fragments. J Virol 1998; 72:5927–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ducancelle A, Champier G, Alain S, Petit F, Le Pors MJ, Mazeron MC. A novel mutation in the UL54 gene of human cytomegalovirus isolates that confers resistance to foscarnet. Antivir Ther 2006; 11:537–40. [PubMed] [Google Scholar]
- 22. Chou S. Phenotypic diversity of cytomegalovirus DNA polymerase gene variants observed after antiviral therapy. J Clin Virol 2011; 50:287–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sullivan V, Biron KK, Talarico C, et al. A point mutation in the human cytomegalovirus DNA polymerase gene confers resistance to ganciclovir and phosphonylmethoxyalkyl derivatives. Antimicrob Agents Chemother 1993; 37:19–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Schubert A, Ehlert K, Schuler-Luettmann S, Gentner E, Mertens T, Michel D. Fast selection of maribavir resistant cytomegalovirus in a bone marrow transplant recipient. BMC Infect Dis 2013; 13:330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Strasfeld L, Lee I, Villano S, Chou S. Virologic characterization of multidrug-resistant cytomegalovirus infection in 2 transplant recipients treated with maribavir. J Infect Dis 2010; 202:104–8. [DOI] [PubMed] [Google Scholar]
- 26. Houldcroft CJ, Bryant JM, Depledge DP, et al. Detection of low frequency multi-drug resistance and novel putative maribavir resistance in immunocompromised pediatric patients with cytomegalovirus. Front Microbiol 2016; 7:1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Komazin-Meredith G, Chou S, Prichard MN, et al. Human cytomegalovirus UL97 kinase is involved in the mechanism of action of methylenecyclopropane analogs with 6-ether and -thioether substitutions. Antimicrob Agents Chemother 2014; 58:274–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chou S. Diverse cytomegalovirus UL27 mutations adapt to loss of viral UL97 kinase activity under maribavir. Antimicrob Agents Chemother 2009; 53:81–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.