Abstract
Background
Congenital cytomegalovirus (CMV) infection is the most common infectious cause of birth defects and neurological damage in newborns. Despite a well-established role for natural killer (NK) cells in control of CMV infection in older children and adults, it remains unknown whether fetal NK cells can sense and respond to CMV infection acquired in utero.
Methods
Here, we investigate the impact of congenital CMV infection on the neonatal NK-cell repertoire by assessing the frequency, phenotype, and functional profile of NK cells in cord blood samples from newborns with congenital CMV and from uninfected controls enrolled in a birth cohort of Ugandan mothers and infants.
Results
We find that neonatal NK cells from congenitally CMV infected newborns show increased expression of cytotoxic mediators, signs of maturation and activation, and an expansion of mature CD56− NK cells, an NK-cell subset associated with chronic viral infections in adults. Activation was particularly prominent in NK cell subsets expressing the Fcγ receptor CD16, indicating a role for antibody-mediated immunity against CMV in utero.
Conclusions
These findings demonstrate that NK cells can be activated in utero and suggest that NK cells may be an important component of the fetal and infant immune response against CMV.
Clinical Trials Registration
Keywords: NK cells, congenital CMV, cytomegalovirus, CD56-negative NK cells, NKG2C, neonatal immunity, cord blood, flow cytometry
NK cells from cord blood of congenitally CMV-infected newborns show increased expression of cytotoxic mediators, signs of maturation and activation, and an expansion of the CD56-negative subset, suggesting a potential role in the fetal and neonatal defense against CMV.
(See the Editorial Commentary by Permar et al, on pages 563–5.)
Cytomegalovirus (CMV) is the most common congenital infection in humans, impacting 1%–5% of newborns [1]. Infection with CMV in utero can lead to poorly controlled viremia and devastating clinical consequences, including poor intrauterine growth, neurologic impairment, and hearing loss [2]. In contrast, primary infection with CMV in early childhood is common and generally asymptomatic. While the unique vulnerability of the fetus to severe CMV disease is due in part to ongoing organogenesis, immunologic factors including the tolerogenic in utero environment and developmental differences in immune effector mechanisms needed for viral control likely contribute to the poor outcomes [3]. Because the fetus and newborn infant lack memory responses to previously encountered pathogens, they have limited ability to mount defenses against acute viral infections. Innate immune cells, including natural killer (NK) cells, likely comprise an important first line of defense to protect the infant upon pathogen encounter.
NK cells kill virally infected cells via release of lytic granules containing granzyme B and perforin. This cytotoxic function can be triggered through direct recognition via activating NK receptors (NKRs) or indirectly via engagement of the low-affinity immunoglobulin G (IgG) receptor CD16 (FcRγIIIa), enabling antibody-dependent cellular cytotoxicity (ADCC). The crucial role NK cells play in the host defense against CMV is demonstrated by cases of severe and even fatal CMV infection among children with genetic NK-cell deficiency [4, 5]. However, the fetal NK-cell response to congenital CMV (cCMV) infection has not been characterized.
Murine models suggest that NK cells may play a particularly important and nonredundant role in controlling CMV infection during early life. While murine CMV infection is fatal in neonatal mice, infected neonates can be rescued from lethality by adoptive transfer of NK cells from adult mice [6]. Whether NK cells play an equally essential role during CMV infection of human infants is unclear, as neonatal mice are profoundly immunodeficient at birth compared to newborn humans. Furthermore, murine CMV encodes a ligand that can be directly sensed by the activating Ly49h receptor on murine NK cells. While the UL40 protein of human CMV influences interactions between HLA-E and NKG2 receptors, it is unclear whether human NK cells recognize and become activated by CMV through other means [7, 8]. Here, we investigated the ability of fetal NK cells to sense and respond to CMV infection prenatally. We compared the frequency and phenotype of NK-cell subsets, including expression of activation markers and antiviral cytotoxic mediators, in cord blood from Ugandan infants with and without congenital CMV infection. We found that cCMV infection resulted in prenatal expansion, activation, and maturation of NK cells with robust upregulation of cytotoxic mediators. These findings were particularly striking in the CD56dim and CD56neg NK-cell subsets, which express CD16. These findings suggest that NK cells, especially those capable of ADCC, may play an important role in the immune response to CMV in utero.
METHODS
Study Population and Samples
Cord blood mononuclear cells (CBMCs) were obtained from a subset of infants (n = 85) enrolled in a clinical trial of prenatal malaria chemoprevention conducted in the Busia District, a highly malaria endemic area in Eastern Uganda (PROMOTE-BC3: NCT02793622). All mothers in this trial screened negative for human immunodeficiency virus (HIV) at enrollment. Clinical and epidemiologic details of this cohort have been previously published [9]. Approximately two-thirds of infants in our cohort had histologic evidence of placental malaria (68.6% of cCMV-positive infants, 66.6% of cCMV-negative infants; Supplementary Table 3). Umbilical cord blood was collected at delivery using cord blood collection kits (Pall Medical) and an aliquot of whole cord blood preserved in RNAlater (ThermoFisher). CBMCs were promptly isolated from the remaining cord blood using density gradient centrifugation (Ficoll-Histopaque; GE Life Sciences) and cryopreserved in liquid nitrogen.
Ethical Approval
Written informed consent was obtained from study participants upon enrollment. The study protocol was approved by Makerere University School of Biomedical Sciences Ethics Committee, Uganda National Council of Science and Technology, and University of California, San Francisco Research Ethics Committee.
Identification and Evaluation of Infants With Congenital CMV Infection
To identify congenitally CMV-infected newborns, DNA was extracted from whole cord blood preserved in RNAlater using QIAamp DNA Blood Mini Kit (Qiagen) according to manufacturer’s instructions. Presence of CMV was determined by quantitative polymerase chain reaction (qPCR) targeting the viral UL123 and UL55 genes using custom primers and SYBR Green chemistry [10]. Growth parameters were reviewed for the 16 infants who were CMV positive at birth. Infants were considered symptomatic if severely microcephalic at birth (below third percentile for head circumference) or severely small for gestational age (SGA; below third percentile for birth weight for gestational age). Of 16 cCMV-positive newborns in this study, 5 were symptomatic (2 with microcephaly, 2 with SGA, and 1 with both).
Flow Cytometry
CBMCs were thawed, counted, evaluated for viability, and stained for extra- and intracellular targets with antibodies listed in Supplementary Tables 1 and 2 [11, 12]. CBMCs were stained with LIVE/DEAD Fixable Aqua or Near-IR (ThermoFisher) to exclude dead cells. For intracellular staining, CBMCs were fixed using Cytofix/Cytoperm kit (BD) and stained in Perm/Wash buffer (BD) per manufacturer’s instructions. Data was acquired on an LSR II (BD) using FACS DIVA software. Compensation was performed using single-color stained UltraComp beads (Invitrogen) and minimum 500 000 events were recorded from each sample. SPHERO Rainbow Calibration Particles (BD) were used to normalize instrument settings across batches to ensure validity of median fluorescence intensity (MFI) comparisons. Flow cytometry data was analyzed using FlowJo version 10.8 (Tree Star). Fluorescence minus one (FMO) controls were used to set gates for the markers shown in Supplementary Figure 3. Coexpression analysis was calculated in FlowJo using Boolean gating and visualized in SPICE version 6.1 (National Institute of Allergy and Infectious Diseases).
Statistical Analysis
Statistical analyses were performed in R. Two group comparisons were performed using Wilcoxon rank sum test; P values < .05 were considered significant and not adjusted for multiple comparisons. Boxplots display median values with 25th/75th percentiles and show all data points. Analyses stratified by placental malaria exposure (± placental histopathology) were performed using the Kruskal-Wallis test and if significant, Dunn post hoc test.
RESULTS
CMV Infection in Utero Induces Expansion of CD56-Negative NK Cells in Cord Blood
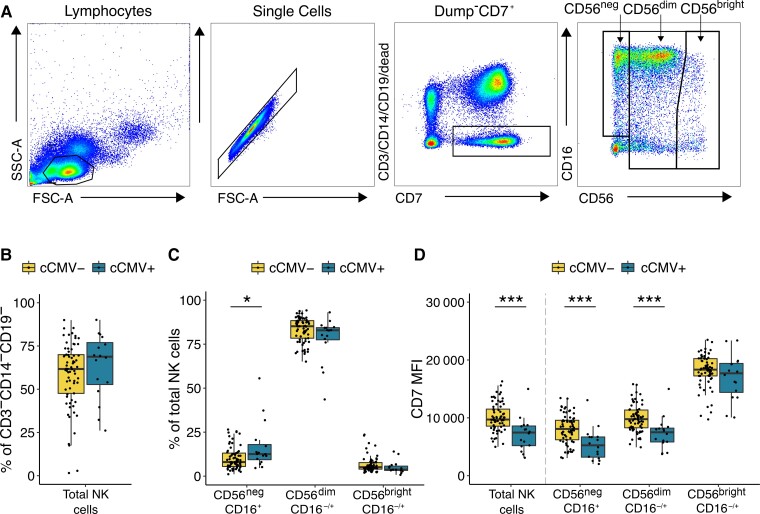
To evaluate the impact of congenital CMV infection on the newborn NK-cell repertoire, we compared the frequency of NK-cell subsets in cord blood samples derived from 16 congenitally CMV infected (cCMV-positive) and 69 uninfected (cCMV-negative) newborns using flow cytometry. NK cells were defined as CD3−/CD14−/CD19−/CD7+ lymphocytes positive for CD56 and/or CD16. CD7 is an early lymphoid marker whose inclusion aids in separating NK cells, particularly those lacking CD56 expression, from nonclassical myeloid cells [13]. We defined 3 major NK subsets based on relative expression of CD56 and CD16 (Figure 1A): CD56dimCD16−/+ cells, which are more mature/cytotoxic and constitute the majority of peripheral NK cells; CD56brightCD16−/+ cells, which are considered developmentally less mature; and finally CD56negCD16+ NK cells which are increasingly recognized as an additional mature NK subset that expands during several chronic viral infections [14]. Overall, we did not observe any difference in the frequency of total NK cells between cCMV-positive and cCMV-negative newborns (P = .31; Figure 1B). However, cCMV-positive newborns displayed a skewed distribution towards more mature/differentiated NK cells, with a significantly higher frequency of CD56neg NK cells (P = .02; Figure 1C). Because infants in this cohort had a high prevalence of placental malaria, we assessed whether malaria might also impact the neonatal NK-cell repertoire. We compared the frequency of NK subsets in cCMV-negative infants with placental malaria by histopathology to those without. Interestingly, we observed that placental malaria, in contrast to cCMV, was associated with a lower proportion of CD56negCD16+ NK cells (P = .03) and higher proportion of CD56dimCD16−/+ cells (P = .04; Supplementary Figure 1A). When stratified by placental malaria exposure, cCMV-positive infants still displayed a significantly higher proportion of CD56neg NK cells (P = .01), suggesting cCMV has an influence distinct from that of placental malaria (Supplementary Figure 1B). In all analyses of NK subsets and markers that follow, no influence of placental malaria was observed.
Figure 1.
CD56neg NK cells expand in newborns with congenital CMV infection. A, Gating strategy for total NK cells and NK-cell subsets. NK cells are defined as lymphocytes > single cells > CD3−/CD14−/CD19−/CD7+ > CD56+ and/or CD16+. NK subsets are defined based on their relative expression of CD56 and CD16. Total NK cells is the sum of the 3 gated subsets. B, Frequency of total NK cells in CMV-infected (cCMV-positive, n = 16) and uninfected (cCMV-negative, n = 69) newborns. C, Frequency of NK-cell subsets out of the total NK cells in cord blood derived from cCMV-positive (n = 16) and cCMV-negative (n = 69) newborns. D, Density of CD7 expression on NK cells measured as normalized MFI (median) between cCMV-positive (n = 16) and cCMV-negative (n = 69) infants. *P < .05, ***P < .001, Wilcoxon rank sum test. Abbreviations: SSC-A, side scatter area; FSC-A, forward scatter area; cCMV, congenital CMV; CMV, cytomegalovirus; MFI, mean fluorescence intensity; NK cell, natural killer cell.
We further observed that NK cells from cCMV-positive infants displayed a striking downregulation of CD7 measured by MFI, although they remained clearly distinguishable from the CD7-negative population (Supplementary Figure 2A). This downregulation was particularly evident among the more mature/differentiated subsets (CD56negP < .001; CD56dimP < .001; Figure 1D). NK cells have been shown to downregulate CD7 upon in vitro stimulation with interleukin 2 (IL-2) or IL-12 plus IL-18 [15, 16]. Thus, the reduced CD7 expression on cord blood NK cells may suggest recent activation-induced downregulation. Together, these data indicate that even during fetal life, human NK cells are activated and differentiate in response to viral infection.
NK Cells in Congenitally CMV-Infected Newborns Show Increased Expression of Cytotoxic Mediators
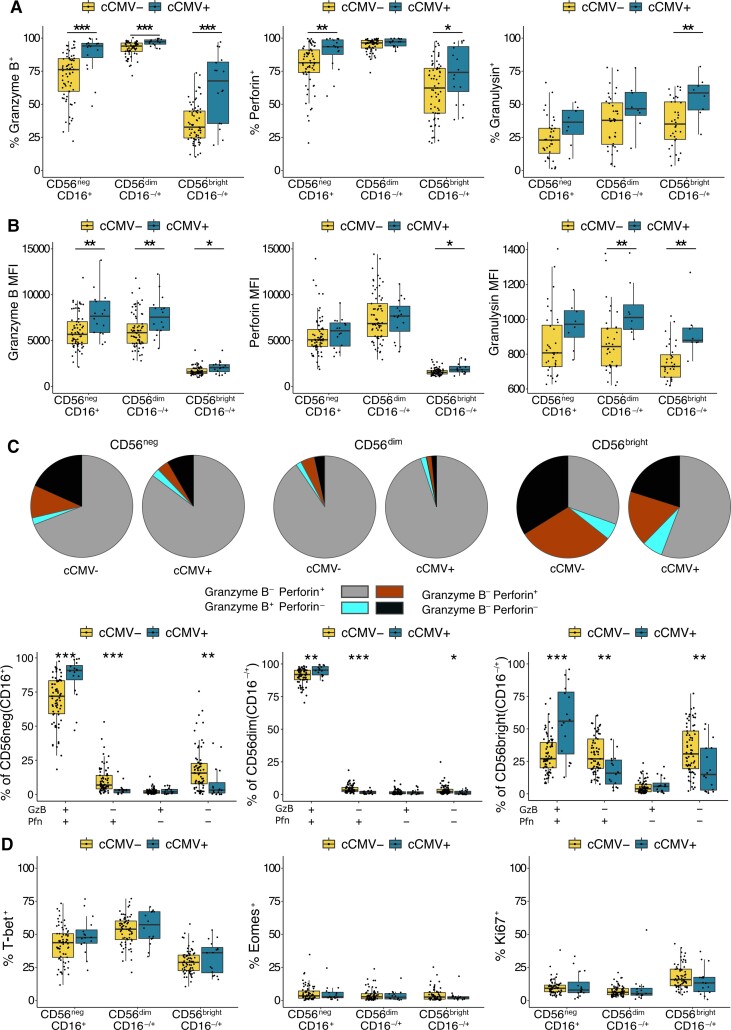
To assess the functional capacity of fetal NK cells, we measured the expression of cytotoxic mediators granzyme B, perforin, and granulysin in cord blood from cCMV-positive and cCMV-negative newborns. Overall, a high percentage of NK cells in all infants expressed granzyme B and perforin, suggesting high functional capacity of newborn NK cells, consistent with previous findings [17]. Among cCMV-positive infants, we observed a significantly higher proportion of granzyme B expression among all NK subsets (CD56negP < .001; CD56dimP = .002; CD56brightP = .008), higher perforin expression on CD56neg (P = .003) and CD56bright (P = .04) NK cells, and higher granulysin expression on CD56bright NK cells (P = .01) with a trend toward higher expression on CD56neg cells (Figure 2A) as compared to cCMV-negative controls. In addition, among cCMV-positive infants the MFI of granzyme B was higher on all NK-cell subsets (CD56negP = .005; CD56dimP = .006; CD56brightP = .007), as was the MFI of perforin on CD56bright NK cells (P = .01) and granulysin on CD56dim (P = .009) and CD56bright (P = .001) NK cells (Figure 2B). Coexpression analysis not only confirmed that the majority of CD56dim and CD56neg NK cells coexpress both granzyme B and perforin, but revealed that NK cells from cCMV-positive individuals more frequently express at least one cytotoxic mediator and are more likely to express more than one (Figure 2C).
Figure 2.
NK cells from cCMV-positive neonates express higher levels of cytotoxic mediators. A, Frequency of cord blood NK cells expressing granzyme B, perforin, and granulysin from cCMV-positive newborns (n = 16) and cCMV-negative controls (n = 69). Granulysin expression was evaluated on a smaller subset of cord blood samples (cCMV-positive, n = 8; cCMV-negative, n = 36). NK cells are defined as lymphocytes > single cells >CD3−/CD14−/CD19− > CD56+ and/or CD16+. Note that CD7 was not included in the flow cytometry panel used to evaluate markers listed in Figure 2. See details in Supplementary Figure 2B. B, Density of granzyme B, perforin, and granulysin expression on NK-cell subsets measured as normalized MFI from cCMV-positive newborns (n = 16) and cCMV-negative controls (n = 69). C, Proportion of NK-cell subsets expressing the listed combination of granzyme B and perforin in cCMV-negative (n = 69) and cCMV-positive infants (n = 16). Coexpression was calculated using Boolean gating in FlowJo and pie graphs, depicting average proportions, were generated in SPICE. D, Frequency of NK cells expressing T-bet, eomesodermin, and Ki67 in cCMV-positive infants (n = 16) and cCMV-negative controls (n = 69). *P < .05, **P < .01, ***P < .001, Wilcoxon rank sum test. Abbreviations: cCMV, congenital cytomegalovirus; GzB, granzyme B; MFI, mean fluorescence intensity; NK cell, natural killer cell; Pfn, perforin.
While CD56neg NK cells overall had lower cytotoxic granule content than CD56dim NK cells, among cCMV-positive infants this difference was diminished, with coexpression of perforin and granzyme B on CD56neg NK cells approaching that of CD56dim cells, implying functional competence of fetal CD56neg NK cells (Figure 2C). Together these data suggest that neonatal NK cells are fully equipped with cytotoxic mediators that enable direct cytolysis and/or ADCC function and contribute to antiviral immunity in utero.
We additionally evaluated expression of T-bet and eomesodermin, transcription factors that govern NK-cell maturation, development, and function, along with the proliferation marker Ki67 [18]. Consistent with previous findings, we found a sizeable proportion of cord blood NK cells expressing T-bet, with higher expression on the more mature NK subsets, and minimal expression of eomesodermin across all NK subsets (Figure 2D) [19]. We found no difference in T-bet or eomesodermin expression between cCMV-positive and cCMV-negative samples, nor did we see any difference in Ki67 aside from a trend towards less expression among CD56bright NK cells (P = .06; Figure 2D).
NK Cells From cCMV-Positive Neonates Show Altered Expression of NKRs
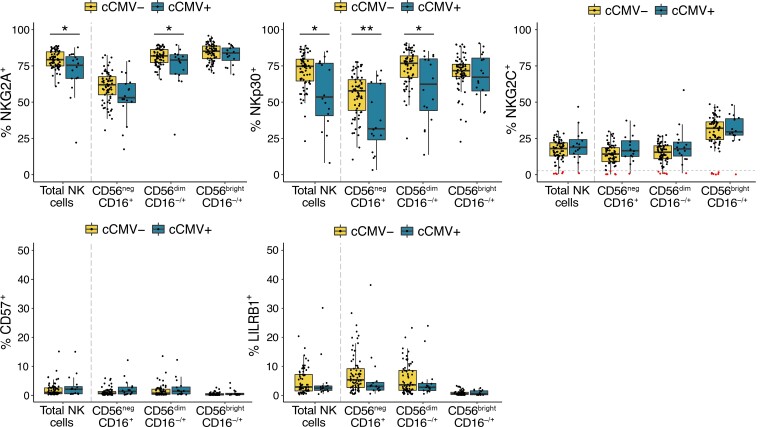
NK-cell activity is regulated through a complex interplay between germline-encoded activating and inhibitory receptors [20]. CMV infection is known to dramatically alter the NKR repertoire in children and adults, leading most notably to an expansion of NK cells expressing the activating receptor NKG2C, often together with the differentiation marker CD57 and the inhibitory receptor LILRB1 [21]. To determine how CMV infection in utero influences the infant NKR repertoire, we compared expression of NKRs on NK cells from cCMV-positive newborns and uninfected controls. Overall, we saw high expression of the inhibitory receptor NKG2A across all subsets, supporting previous findings that NKG2A is more highly expressed on neonatal than adult NK cells and decreases with maturation [22, 23]. Consistent with findings in CMV infected children and adults, cCMV-positive newborns had decreased expression of NKG2A, particularly on more mature subsets (CD56dimP = .04, CD56negP = .07; Figure 3). They also had a lower frequency of CD56neg and CD56dim NK cells expressing the natural cytotoxicity receptor NKp30 (CD56negP = .009; CD56dimP = .03; Figure 3) [21, 24]. Notably, cCMV was not associated with higher expression of NKG2C, LILRB1, or CD57, which were expressed at low levels in nearly all infants (Figure 3). Six infants, including 1 with cCMV infection, completely lacked NKG2C on all NK cells, consistent with the known approximately 10% frequency of NKG2C (KLRC2) gene deletion in African populations [25] (Figure 3, highlighted in red). The cCMV-positive infant lacking NKG2C+ NK cells was asymptomatic and born at full term with normal growth parameters and an unremarkable NK-cell profile. Together, our data indicate that the neonatal NK-cell response to CMV differs somewhat from that described in adults, particularly with respect to expansion of NKG2C+ NK cells, which are a hallmark of the NK response to CMV infection later in life.
Figure 3.
NK cells show altered expression of NKRs in cCMV-positive newborns. Frequency of NK cells expressing NKG2A, NKp30, NKG2C, CD57, and LILRB1 in cord blood derived from cCMV-positive (n = 16) and cCMV-negative (n = 69) newborns. NK cells are defined as lymphocytes > single cells > CD3−/CD14−/CD19−/CD7+ > CD56+ and/or CD16+. For NK-cell gating strategy refer to Figure 1A. Individuals with < 2% of their total NK cells expressing NKG2C+ are highlighted in red/below dotted line in the NKG2C panel. Axes for % CD57+ and % LILRB1+ range from 0% to 50%. *P < .05, **P < .01, Wilcoxon rank sum test. Abbreviations: cCMV, congenital cytomegalovirus; NK cell, natural killer cell; NKR, NK receptor.
NKG2C Expression Is Elevated in Newborns With Symptomatic CMV Infection
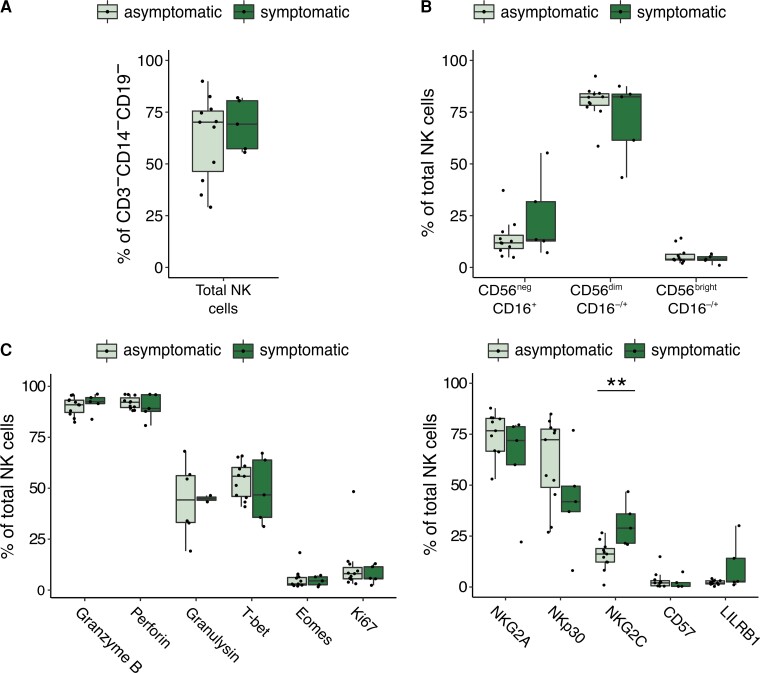
Finally, we examined whether there is a relationship between cord blood NK-cell phenotypes and clinical manifestations of congenital CMV. Among the 16 cCMV-positive infants in our cohort, 5 had severe growth abnormalities at birth that were consistent with symptomatic infection, whereas the other 11 infants were asymptomatic. We compared the frequency of NK-cell subsets and expression of cytotoxic mediators and NKRs between symptomatic and asymptomatic cCMV-positive infants. We found no difference in the frequency of total NK cells (Figure 4A) or NK-cell subsets (Figure 4B), nor in expression of cytotoxic markers, proliferation markers, or transcription factors (Figure 4C). However, infants with symptomatic cCMV had a statistically higher percentage of NK cells expressing NKG2C (P = .005; Figure 4D). This could suggest that the higher in utero inflammation associated with severe CMV disease fosters an expansion of NKG2C+ NK cells.
Figure 4.
NK-cell phenotype in symptomatic and asymptomatic cCMV cases. A, Frequency of total NK cells defined as CD3−/CD14−/CD19−/CD7+ lymphocytes in cord blood from symptomatic (n = 5, dark green) and asymptomatic (n = 11) cCMV-positive newborns. Newborns are defined as symptomatic if they demonstrated severe microcephaly and/or were small for gestational age at birth. B, Frequency of NK-cell subsets out of total NK cells in cord blood derived from symptomatic (n = 5) and asymptomatic (n = 11, light green) cCMV-positive newborns. C, Frequency of NK cells expressing granzyme B, perforin, granulysin, T-bet, eomesodermin, and Ki67 in symptomatic (n = 5) and asymptomatic (n = 11) cCMV-positive newborns. D, Frequency of NK cells expressing NKG2A, NKp30, NKG2C, CD57, and LILRB1 in symptomatic (n = 5) and asymptomatic (n = 11) cCMV-positive newborns. **P < .01. For gating strategy of NK-cell subsets refer to Figure 1A for (A, B, and D), and Supplementary Figure 2B for (C). Abbreviations: cCMV, congenital cytomegalovirus; NK cell, natural killer cell.
DISCUSSION
The fetal immune system is prone towards tolerance to prevent fetal-maternal alloreactivity, which poses a challenge during in utero viral infection. While NK cells play a critical role in host defense against CMV in adults, little is known about how fetal NK cells respond to viral infection [26]. Because NK cells develop by gestational week 6 and are the dominant lymphocyte population in the fetal liver and lung, they are poised to play an important role in fetal immunity [27, 28]. Here, we show that neonatal NK cells mature, differentiate, and upregulate production of cytotoxic mediators in response to CMV infection in utero. This is, to our knowledge, the first study to demonstrate in utero expansion and maturation of NK cells in response to a congenital infection. These findings strongly suggest that NK cells may be an important component of fetal and neonatal host defense against CMV and other viral pathogens.
Fetal and neonatal NK cells were previously thought to be functionally impaired, but it has subsequently been shown that while hyporesponsive towards MHC-devoid cells, they can be readily activated by cytokine and antibody-mediated stimulation [17, 22, 27, 29]. During gestation, maternal antibodies are transferred transplacentally to the fetus via an active transport mechanism mediated by FcRn, the neonatal Fc receptor [30]. We speculate that fetal NK cells are preferentially activated via CD16 engagement by IgG, rather than by cytokines or MHC-devoid cells, enabling them to harness the breadth and specificity of maternal-origin IgG. Indeed, we found NK activation and expansion to be particularly evident in the more mature NK-cell subsets dominated by CD16+ cells: the CD56neg subset (100% CD16+) and the CD56dim subset (approximately 90% CD16+). Notably, it has recently been shown that the placenta selectively transfers maternal antibodies with a glycosylation pattern that enhances binding to both FcRn and CD16 [31]. This suggests that the placenta may preferentially sieve IgG with an Fc-profile skewed towards activating fetal NK cells. The ability of CD16+ NK cells to engage maternally derived anti-CMV IgG may serve as a critical early defense, enabling the infant to, in a sense, “borrow” immune memory from the mother. Enhanced antibody-mediated NK-cell killing could help explain the much lower rate of adverse sequelae seen in congenitally infected infants born to mothers with preexisting anti-CMV antibodies compared to seronegative pregnant women who develop primary CMV infection during pregnancy [32]. This is supported by the finding of Semmes et al that high-avidity maternal CMV-specific nonneutralizing antibodies correlate with protection against congenital CMV transmission, suggesting that Fc-mediated immune functions are important factors in protection against fetal infection [33].
In this study, we demonstrate that CD56neg NK cells, an NK subset associated with chronic viral infections in adults, expand in the cord blood of CMV infected newborns [14]. It is intriguing that this skewing toward CD56neg cells was the opposite effect of that observed in infants with placental malaria (in whom CD56neg NK cells were decreased and CD56dim were increased). CD56neg NK cells are less well studied than their CD56-expressing counterparts, partly due to the absence of a canonical marker to define the NK lineage. Here, we used CD7 to aid in discerning the CD56neg NK subset [13]. CD56neg NK cells have been reported to have decreased cytolytic, replicative, and antiviral potential compared to their CD56-expressing counterparts [34–36]. In contrast to adult peripheral blood, cord blood contains a sizeable population of CD56neg cells, even in the absence of infection. Like adult CD56neg NK cells, cord blood CD56neg NK cells have been described as functionally impaired [29, 37, 38]. Nonetheless, we observed that CD56neg NK cells from cCMV-positive newborns downregulated CD7 and gained cytotoxic mediators at levels approaching that of the mature CD56dim subset. Additionally, more recent studies have shown that CD56neg NK cells in fact display high transcriptional and proteomic resemblance to the more mature and cytotoxic CD56dim NK cells, suggesting their functional competence may be greater than initially thought [39, 40]. Specifically, Forconi et al suggested that CD56neg NK cells are not well adapted for direct natural cytotoxicity because of their downregulation of cytotoxic and activating receptors, but rather rely on antibody-dependent mechanisms to kill target cells, as illustrated by high expression of both CD16 (FcRγIIIA/B) and CD32 (FcRγIIA/B) [40].
The phenotypic maturation of NK cells in cCMV-positive infants resembled reports in CMV-positive adults in some ways but diverged in others. In particular, we did not observe elevated frequencies of NKG2C+ cells in congenitally CMV-infected newborns, nor of CD57 or LILRB1, both of which are frequently coexpressed on NKG2C+ NK cells [21, 41, 42]. We did, however see lower frequencies of NK cells expressing NKG2A, the inhibitory counterpart of NKG2C. NKG2C/A recognize the nonclassical class I MHC molecule HLA-E on target cells [43]. HLA-E is normally stabilized at the cell surface by a conserved leader peptide derived from classical class I HLA molecules. However, in CMV-infected cells, which downregulate class I HLA, a peptide derived from the CMV UL40 protein can stabilize HLA-E, leading to NKG2C-mediated NK-cell activation [16, 44]. This expansion of NKG2C+ NK cells appears unique to CMV infection and is not reported in other herpesvirus infections [24, 41]. However, NKG2C+ NK-cell expansion also requires costimulation in the form of proinflammatory cytokines, particularly IL-12 [16, 45]. IL-12 is under tight epigenetic control in the tolerogenic immune environment maintained throughout gestation [46], and stimulation of cord blood NK cells with IL-12 and IL-15 has been shown to rapidly increase their functional capacity to levels approaching that of adult NK cells [29, 47, 48]. Thus, it is possible that the restricted cytokine environment in utero hinders expansion of fetal NKG2C+ NK cells.
We did observe an increase in NKG2C specifically among symptomatic CMV cases, indicating increased inflammation or viral load might drive fetal expression of NKG2C. Notably, Noyola et al assessed NK cells in congenitally CMV-infected children 1 month to 7 years after birth and found that children with symptomatic cCMV infection had higher NKG2C+ NK cell frequencies than asymptomatic children, further supporting an association of NKG2C upregulation with severe infection [49].
Our study cohort was limited in size and ability to comprehensively evaluate congenital CMV symptomology, particularly hearing loss and neurocognitive disabilities, which can manifest years after birth. It will be important to determine in larger cohorts whether NK-mediated antiviral function correlates with clinical outcomes following cCMV infection. Additionally, we did not formally adjust for multiple comparisons as we anticipated many of the phenotypic features to be highly correlated. While many of the differences noted between CMV-positive and negative infants were statistically robust, other more marginal differences will need to be confirmed in future studies encompassing larger numbers. Future studies should additionally investigate the functional capacity of NK cells to mediate ADCC through maternal IgG engagement and to control viral replication.
CONCLUSION
In summary, we have demonstrated that despite the tolerogenic environment in utero, fetal NK cells expand, differentiate, and functionally mature in response to CMV infection prior to birth. This provides a critical innate first line of defense at a time when the fetus lacks acquired immune memory. Along with prior studies demonstrating potent neonatal NK cell activation by antibody-mediated stimulation [31], these findings strongly suggest that NK cells may be an important component of fetal and neonatal host defense against CMV, and perhaps other viral pathogens. Furthermore, they suggest that vaccination strategies to optimize maternal titers of anti-CMV antibodies that favor transplacental transfer and FcR engagement could be of benefit in protecting the fetus from adverse outcomes following congenital CMV infection.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online (http://jid.oxfordjournals.org/). Supplementary materials consist of data provided by the author that are published to benefit the reader. The posted materials are not copyedited. The contents of all supplementary data are the sole responsibility of the authors. Questions or messages regarding errors should be addressed to the author.
Supplementary Material
Contributor Information
Anna V Vaaben, Department of Medicine, University of California San Francisco, San Francisco, California, USA.
Justine Levan, Department of Medicine, University of California San Francisco, San Francisco, California, USA.
Catherine B T Nguyen, Department of Medicine, University of California San Francisco, San Francisco, California, USA.
Perri C Callaway, Department of Medicine, University of California San Francisco, San Francisco, California, USA; Infectious Diseases and Immunity Graduate Group, University of California Berkeley, California, Berkeley, USA.
Mary Prahl, Department of Pediatrics, University of California San Francisco, San Francisco, California, USA.
Lakshmi Warrier, Department of Medicine, University of California San Francisco, San Francisco, California, USA.
Felistas Nankya, Infectious Disease Research Collaboration, Kampala, Uganda.
Kenneth Musinguzi, Infectious Disease Research Collaboration, Kampala, Uganda.
Abel Kakuru, Infectious Disease Research Collaboration, Kampala, Uganda.
Mary K Muhindo, Infectious Disease Research Collaboration, Kampala, Uganda.
Grant Dorsey, Department of Medicine, University of California San Francisco, San Francisco, California, USA.
Moses R Kamya, Infectious Disease Research Collaboration, Kampala, Uganda; Department of Medicine, Makerere University College of Health Sciences, Kampala, Uganda.
Margaret E Feeney, Department of Medicine, University of California San Francisco, San Francisco, California, USA; Department of Pediatrics, University of California San Francisco, San Francisco, California, USA.
Notes
Acknowledgments. The authors are very grateful to all the study participants and their families, and additionally sincerely thank all the study team members for their efforts and contributions.
Financial support . This work was supported by the National Institutes of Health (grant numbers 5P01HD059454 to G. D., M. R. K., and M. E. F.; and R01AI093615, U01AI168390, and K24AI113002 to M. E. F.); and the American Association of Immunologists (Intersect Fellowship Program for Computational Scientists and Immunologists to J. L.).
Potential conflicts of interest. All authors: No reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed. Funding to pay the Open Access publication charges for this article was provided by the National Institutes of Health.
References
- 1. Lanzieri TM, Dollard SC, Bialek SR, Grosse SD. Systematic review of the birth prevalence of congenital cytomegalovirus infection in developing countries. Int J Infect Dis 2014; 22:44–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Boppana SB, Ross SA, Fowler KB. Congenital cytomegalovirus infection: clinical outcome. Clin Infect Dis 2013; 57:S178–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Gantt S, Orem J, Krantz EM, et al. Prospective characterization of the risk factors for transmission and symptoms of primary human herpesvirus infections among Ugandan infants. J Infect Dis 2016; 214:36–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Orange JS. Natural killer cell deficiency. J Allergy Clin Immunol 2013; 132:515–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Mace EM, Paust S, Conte MI, et al. Human NK cell deficiency as a result of biallelic mutations in MCM10. J Clin Invest 2020; 130:5272–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bukowski JF, Woda BA, Welsh RM. Pathogenesis of murine cytomegalovirus infection in natural killer cell-depleted mice. J Virol 1984; 52:119–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Arase H, Mocarski ES, Campbell AE, Hill AB, Lanier LL. Direct recognition of cytomegalovirus by activating and inhibitory NK cell receptors. Science 2002; 296:1323–6. [DOI] [PubMed] [Google Scholar]
- 8. Smith HRC, Heusel JW, Mehta IK, et al. Recognition of a virus-encoded ligand by a natural killer cell activation receptor. Proc Natl Acad Sci 2002; 99:8826–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kajubi R, Ochieng T, Kakuru A, et al. Monthly sulfadoxine-pyrimethamine versus dihydroartemisinin-piperaquine for intermittent preventive treatment of malaria in pregnancy: a double-blind, randomised, controlled, superiority trial. Lancet 2019; 393:1428–39. [DOI] [PubMed] [Google Scholar]
- 10. Boeckh M, Huang M, Ferrenberg J, et al. Optimization of quantitative detection of cytomegalovirus DNA in plasma by real-time PCR. J Clin Microbiol 2004; 42:1142–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lamoreaux L, Roederer M, Koup R. Intracellular cytokine optimization and standard operating procedure. Nat Protoc 2006; 1:1507–16. [DOI] [PubMed] [Google Scholar]
- 12. Maecker HT, Trotter J. Flow cytometry controls, instrument setup, and the determination of positivity. Cytometry A 2006; 69:1037–42. [DOI] [PubMed] [Google Scholar]
- 13. Milush JM, Long BR, Snyder-Cappione JE, et al. Functionally distinct subsets of human NK cells and monocyte/DC-like cells identified by coexpression of CD56, CD7, and CD4. Blood 2009; 114:4823–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Björkström NK, Ljunggren HG, Sandberg JK. CD56 Negative NK cells: origin, function, and role in chronic viral disease. Trends Immunol 2010; 31:401–6. [DOI] [PubMed] [Google Scholar]
- 15. Rabinowich H, Pricop L, Herberman RB, Whiteside TL. Expression and function of CD7 molecule on human natural killer cells. J Immunol 1994; 152:517–26. [PubMed] [Google Scholar]
- 16. Hammer Q, Rückert T, Borst EM, et al. Peptide-specific recognition of human cytomegalovirus strains controls adaptive natural killer cells. Nat Immunol 2018; 19:453–63. [DOI] [PubMed] [Google Scholar]
- 17. Dalle J-H, Menezes J, Wagner É, et al. Characterization of cord blood natural killer cells: implications for transplantation and neonatal infections. Pediatr Res 2005; 57:649–55. [DOI] [PubMed] [Google Scholar]
- 18. Zhang J, Le Gras S, Pouxvielh K, et al. Sequential actions of EOMES and T-BET promote stepwise maturation of natural killer cells. Nat Commun 2021; 12:5446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Collins A, Rothman N, Liu K, Reiner SL. Eomesodermin and T-bet mark developmentally distinct human natural killer cells. JCI Insight 2017; 2:e90063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lanier LL. Up on the tightrope: natural killer cell activation and inhibition. Nat Immunol 2008; 9:495–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Muntasell A, Vilches C, Angulo A, López-Botet M. Adaptive reconfiguration of the human NK-cell compartment in response to cytomegalovirus: a different perspective of the host-pathogen interaction. Eur J Immunol 2013; 43:1133–41. [DOI] [PubMed] [Google Scholar]
- 22. Strauss-Albee DM, Liang EC, Ranganath T, Aziz N, Blish CA. The newborn human NK cell repertoire is phenotypically formed but functionally reduced. Cytometry B Clin Cytom 2017; 92:33–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Béziat V, Descours B, Parizot C, Debré P, Vieillard V. NK cell terminal differentiation: correlated stepwise decrease of NKG2A and acquisition of KIRs. PLoS One 2010; 5:e11966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gumá M, Angulo A, Vilches C, Gómez-Lozano N, Malats N, López-Botet M. Imprint of human cytomegalovirus infection on the NK cell receptor repertoire. Blood 2004; 104:3664–71. [DOI] [PubMed] [Google Scholar]
- 25. Goodier MR, White MJ, Darboe A, et al. Rapid NK cell differentiation in a population with near-universal human cytomegalovirus infection is attenuated by NKG2C deletions. Blood 2014; 124:2213–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Goodier MR, Riley E. Regulation of the human NK cell compartment by pathogens and vaccines. Clin Transl Immunol 2021; 10:e1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ivarsson MA, Loh L, Marquardt N, et al. Differentiation and functional regulation of human fetal NK cells. J Clin Invest 2013; 123:3889–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Phillips JH, Hori T, Nagler A, Bhat N, Spits H, Lanier LL. Ontogeny of human natural killer (NK) cells: Fetal NK cells mediate cytolytic function and express cytoplasmic CD3 epsilon, delta proteins. J Exp Med 1992; 175:1055–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gaddy J, Risdon G, Broxmeyer HE. Cord blood natural killer cells are functionally and phenotypically immature but readily respond to interleukin-2 and interleukin-12. J Interferon Cytokine Res 1995; 15:527–36. [DOI] [PubMed] [Google Scholar]
- 30. Callaway PC, Farrington LA, Feeney ME. Malaria and early life immunity: competence in context. Front Immunol 2021; 12:634749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Jennewein MF, Goldfarb I, Dolatshahi S, et al. Fc glycan-mediated regulation of placental antibody transfer. Cell 2019; 178:202–15.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Fowler KB, Stagno S, Pass RF, Britt WJ, Boll TJ, Alford CA. The outcome of congenital cytomegalovirus infection in relation to maternal antibody status. N Engl J Med 1992; 326:663–7. [DOI] [PubMed] [Google Scholar]
- 33. Semmes EC, Miller IG, Jenks JA, et al. Maternal Fc-mediated non-neutralizing antibody responses correlate with protection against congenital human cytomegalovirus infection. medRxiv, 7December2021, preprint: not peer reviewed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mavilio D, Lombardo G, Benjamin J, et al. Characterization of CD56−/CD16+ natural killer (NK) cells: a highly dysfunctional NK subset expanded in HIV-infected viremic individuals. Proc Natl Acad Sci U S A 2005; 102:2886–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Milush JM, López-Vergès S, York VA, et al. CD56negCD16+ NK cells are activated mature NK cells with impaired effector function during HIV-1 infection. Retrovirology 2013; 10:158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Forconi CS, Cosgrove CP, Saikumar-Lakshmi P, et al. Poorly cytotoxic terminally differentiated CD56negCD16pos NK cells accumulate in Kenyan children with Burkitt lymphomas. Blood Adv 2018; 2:1101–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Gaddy J, Broxmeyer HE. Cord blood CD16+56− cells with low lytic activity are possible precursors of mature natural killer cells. Cell Immunol 1997; 180:132–42. [DOI] [PubMed] [Google Scholar]
- 38. Jacobson A, Bell F, Lejarcegui N, Mitchell C, Frenkel L, Horton H. Healthy neonates possess a CD56-negative NK cell population with reduced anti-viral activity. PloS One 2013; 8:e67700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Voigt J, Malone DFG, Dias J, et al. Proteome analysis of human CD56neg NK cells reveals a homogeneous phenotype surprisingly similar to CD56dim NK cells. Eur J Immunol 2018; 48:1456–69. [DOI] [PubMed] [Google Scholar]
- 40. Forconi CS, Oduor CI, Oluoch PO, et al. A new hope for CD56negCD16pos NK cells as unconventional cytotoxic mediators: an adaptation to chronic diseases. Front Cell Infect Microbiol 2020; 10:162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hendricks DW, Balfour HH, Dunmire SK, Schmeling DO, Hogquist KA, Lanier LL. Cutting edge: NKG2C(hi)CD57+ NK cells respond specifically to acute infection with cytomegalovirus and not Epstein-Barr virus. J Immunol 2014; 192:4492–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lopez-Vergès S, Milush JM, Schwartz BS, et al. Expansion of a unique CD57+ NKG2Chi natural killer cell subset during acute human cytomegalovirus infection. Proc Natl Acad Sci 2011; 108:14725–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Braud VM, Allan DSJ, O’Callaghan CA, et al. HLA-E binds to natural killer cell receptors CD94/NKG2A, B and C. Nature 1998; 391:795–9. [DOI] [PubMed] [Google Scholar]
- 44. Tomasec P, Braud VM, Rickards C, et al. Surface expression of HLA-E, an inhibitor of natural killer cells, enhanced by human cytomegalovirus gpUL40. Science 2000; 287:1031. [DOI] [PubMed] [Google Scholar]
- 45. Rölle A, Pollmann J, Ewen EM, et al. IL-12-producing monocytes and HLA-E control HCMV-driven NKG2C+ NK cell expansion. J Clin Invest 2014; 124:5305–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Goriely S, Van Lint C, Dadkhah R, et al. A defect in nucleosome remodeling prevents IL-12(p35) gene transcription in neonatal dendritic cells. J Exp Med 2004; 199:1011–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Lau AS, Sigaroudinia M, Yeung MC, Kohl S. Interleukin-12 induces interferon-γ expression and natural killer cytotoxicity in cord blood mononuclear cells. Pediatr Res 1996; 39:150–5. [DOI] [PubMed] [Google Scholar]
- 48. Pollara J, Edwards RW, Jha S, et al. Redirection of cord blood T cells and natural killer cells for elimination of autologous HIV-1-infected target cells using bispecific DART® molecules. Front Immunol 2020; 11:713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Noyola DE, Fortuny C, Muntasell A, et al. Influence of congenital human cytomegalovirus infection and the NKG2C genotype on NK-cell subset distribution in children. Eur J Immunol 2012; 42:3256–66. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.