Abstract
The visual pigments known as opsins are the primary molecular basis for colour vision in animals. Insects are among the most diverse of animal groups and their visual systems reflect a variety of life histories. The study of insect opsins in the fruit fly Drosophila melanogaster has led to major advances in the fields of neuroscience, development and evolution. In the last 25 years, research in D. melanogaster has improved our understanding of opsin genotype–phenotype relationships while comparative work in other insects has expanded our understanding of the evolution of insect eyes via gene duplication, coexpression and homologue switching. Even so, until recently, technology and sampling have limited our understanding of the fundamental mechanisms that evolution uses to shape the diversity of insect eyes. With the advent of genome editing and in vitro expression assays, the study of insect opsins is poised to reveal new frontiers in evolutionary biology, visual neuroscience, and animal behaviour.
This article is part of the theme issue ‘Understanding colour vision: molecular, physiological, neuronal and behavioural studies in arthropods’.
Keywords: opsin, visual pigments, photoreceptor specification, photoreceptor differentiation, colour vision, insects
1. Introduction
Opsins are the most widely used photopigments across animals. Present in basal lineages Ctenophora, Placozoa and Cnidaria, opsins have been widely studied to understand the evolution of eyes and vision [1–5]. In the past 25 years, research into the molecular evolution, expression and function of insect opsins has revealed extraordinary diversity of insect visual systems owing to gene duplication and loss, coexpression and spectral tuning of opsins [6–9]. Since von Frisch established that honeybees could see in colour over 100 years ago, the study of insect colour vision has provided fundamental insights into sensory physiology and animal behaviour [10].
The canonical role of opsins is that of light detection. Animal opsins are a monophyletic clade of seven transmembrane G-protein-coupled receptors [5]. Visual opsins bind a vitamin A-derived molecule called the chromophore (11-cis-retinal in vertebrates and 3-hydroxy-11-cis-retinal in the fruit fly Drosophila melanogaster) [11] (see box 1 for a glossary of terms). The chromophore binds to the opsin protein at a conserved lysine residue, forming a functional rhodopsin visual pigment molecule. Light activates rhodopsin through a chromophore conformational change from 11-cis to all-trans, which initiates the G-protein-mediated phototransduction cascade, leading to ion exchange and amplification of light information into a cellular signal [12]. Opsin protein sequences, opsin coexpression and filtering effects determine the wavelength of light to which photoreceptor cells respond. Opsins are particularly useful in the study of genotype–phenotype relationships because their sequence and expression are directly related to cell physiology and animal behaviour. The degree of overlap in spectral sensitivities allows animals to discriminate between wavelengths of light and underlies their colour vision (see also non-spectral colours [13]).
Box 1. Glossary of terms.
| Bolwig organ | name for the larval eye in D. melanogaster |
| BRh, B opsin | blue-absorbing opsin homologue shared by insects |
| cell differentiation | in development, when a cell phenotypically specializes and expresses the functional proteins of its specific adult cell type, e.g. Rh3-expressing photoreceptor cell |
| cell specification | in development, the point at which a cell is committed to a particular fate, e.g. photoreceptor cell precursor |
| chromophore | vitamin A derived molecule that absorbs photons of light |
| cilliary opsin (c-opsin) | monophyletic clade of opsins historically associated with the outer segment (a modified cilium) of vertebrate photoreceptor cells, though now known to be found also in invertebrates |
| Cnidaria | animal group including jellyfish, anemones, and corals |
| Coleoptera | insect group including beetles |
| colour vision | the ability to discriminate between light stimuli of differing wavelengths |
| compound eye | the main visual organ of insects, made up of many repeating ommatidia |
| Ctenophora | early-branching animal group including comb jellies |
| Diptera | insect group including flies and mosquitoes |
| DRA | the dorsal-most row(s) of ommatidia in the compound eye, typically expressing more ultraviolet or B opsin, associated with polarized light detection and navigation |
| ERG | method of recording a sum of photoreceptor sensitivities from a region of the eye extracellularly |
| eyelet | photosensitive visual structure in the brain of adult insects |
| gene regulatory network | a group of genes that interact with each other, affecting downstream gene expression and phenotype |
| holometabolous insects | insects that undergo complete metamorphosis |
| homologue | gene found in multiple groups with a shared common ancestor, can be an orthologue or paralogue |
| homologue/paralogue switching | process by which one homologue/paralogue of a gene is swapped for another, achieving a similar function with a new gene |
| Hymenoptera | insect group including ants, bees and wasps |
| intracellular recording | a method of recording from individual neurons, the best way to measure individual photoreceptor cell physiology |
| lamina | distal-most layer of the insect optic lobe, associated with motion and contrast processing |
| Lepidoptera | insect group including moths and butterflies |
| LWRh, LW opsin | long-wavelength- or green-absorbing opsin homologue in insects |
| medulla | more proximal layer of the insect optic lobe, associated with colour vision processing |
| microspectrophotometry | a method measuring opsin absorbance spectra in the eye |
| morphogenetic furrow | a physical groove of differentiating cells that sweeps in a wave across the developing D. melanogaster eye |
| non-visual opsins | opsins that are not historically associated with vision, outside of rhabdomeric and ciliary opsin clades |
| non-spectral colour | a colour that is perceived by sampling from non-overlapping photoreceptors |
| ocellus | single chambered eye found on the dorsal head of most insects, with circadian and navigation-related functions |
| Odonata | insect group including dragonflies, mayflies and damselflies |
| ommatidium | a single unit of the compound eye containing photoreceptor cells, pigment cells and a focusing apparatus |
| opsin | visual pigment protein |
| orthologue | gene found in multiple animal groups related by inheritance from a single common ancestor, e.g. UVRh |
| Orthoptera | insect group including crickets and grasshoppers |
| paralogue | one of two or more genes in a species related by duplication within a single lineage, such as LWRh1 and LWRh2 |
| Pax6 | transcription factor expressed early in development that determines eye tissue in many animals |
| photoreceptor cell | light-sensitive cell typically expressing opsin, typically sensory neural cell |
| phototaxis | an animal's movement toward or away from light |
| phototransduction | a G-protein-mediated signalling pathway by which light information is transduced into a biochemical signal in photoreceptor cells |
| Placozoa | early-branching group of small, free living, simple animals |
| polarized light | photons of light that travel in a wave along a single plane |
| precursor cell | cell that undergoes a limited number of divisions to form daughter cells that differentiate |
| retrotransposition | mechanism of gene duplication by which a transposable element inserts intronless mRNA encoding a gene back into the genome as DNA |
| rhabdomere | structure formed by photoreceptor cell microvilli through which light is guided and absorbed by expressed opsins |
| rhabdomeric opsin (r-opsin) | monophyletic clade of opsins historically associated with expression in the microvillar rhabdom of invertebrate photoreceptor cells, though now known to be found also in vertebrates |
| rhodopsin | functional visual pigment complex of opsin protein and vitamin A-derived chromophore |
| sexual dimorphism | distinct phenotype between males and females; can be gene expression, physiology, behaviour or morphology |
| spectral sensitivity | the degree of response of a photoreceptor cell over wavelengths of light |
| spectral tuning | process by which the sensitivity of a photoreceptor cell is shifted to detect new wavelengths of light, either evolutionarily or via physiological plasticity |
| stemmata | simple larval eyes with a limited number of opsins expressed |
| tetrachromacy | having four independent sensory channels for colour vision |
| trichromacy | having three independent sensory channels for colour vision (similar to normal human vision) |
| turbinate eye | specialized dorsal compound eye enlarged on vertical stalks and unique to mayflies, in addition to the usual compound eye |
| UVRh, UV opsin | ultraviolet-absorbing opsin homologue shared by insects |
In addition to opsin biochemical function, understanding the developmental processes leading to when and where opsins are expressed is important to understanding colour vision. Linking Pax6 (Eyeless) to eyes in all animals was first recognized and functionally validated in D. melanogaster, and the genetic tractability of this organism led to many advances in how gene regulation is related to a complex organ like the eye [14]. Studies of photoreceptor cell development uniquely connect early development through terminal differentiation to mature cell and tissue phenotype. New research has revealed the complexity of the regulatory networks that specify photoreceptor cell fate and differentiation in D. melanogaster, and the relationship between these regulatory networks and terminal cell differentiation is perhaps one of the best understood of any complex trait in biology. We first review what we have learned in D. melanogaster and examine how this gene regulation has evolved in other insects. We relate this to how the evolution of colour vision has proceeded via opsin gene duplication and to changes in opsin gene expression. While descriptions of opsin expression patterns have expanded in other insects over the past 25 years, much work remains to characterize the developmental genetics behind opsin expression in other species. To conclude, we suggest future avenues of research in insect opsins as models for neuroscience, evolutionary biology and development.
2. What have we learned about opsin regulation from Drosophila melanogaster?
(a) . Genetic regulation of specification and terminal differentiation of insect photoreceptor cells
Much of what we know about opsin-expressing photoreceptor cell development in insects comes from D. melanogaster. Over the past 25 years, work in D. melanogaster has shown that an intricate process involving both stochastic and deterministic mechanisms has led to the retinal mosaic in the compound eye. Opsin regulation is intertwined with photoreceptor cell differentiation and has taught us about cell differentiation more broadly. While many reviews summarize eye development (such as early determination by the retinal determination network) in D. melanogaster, we focus on photoreceptor cell specification (when the cell becomes committed to a particular fate) and terminal differentiation (when the cell matures and expresses markers of its specific adult function), synthesizing previous information with the most recent work [15–18].
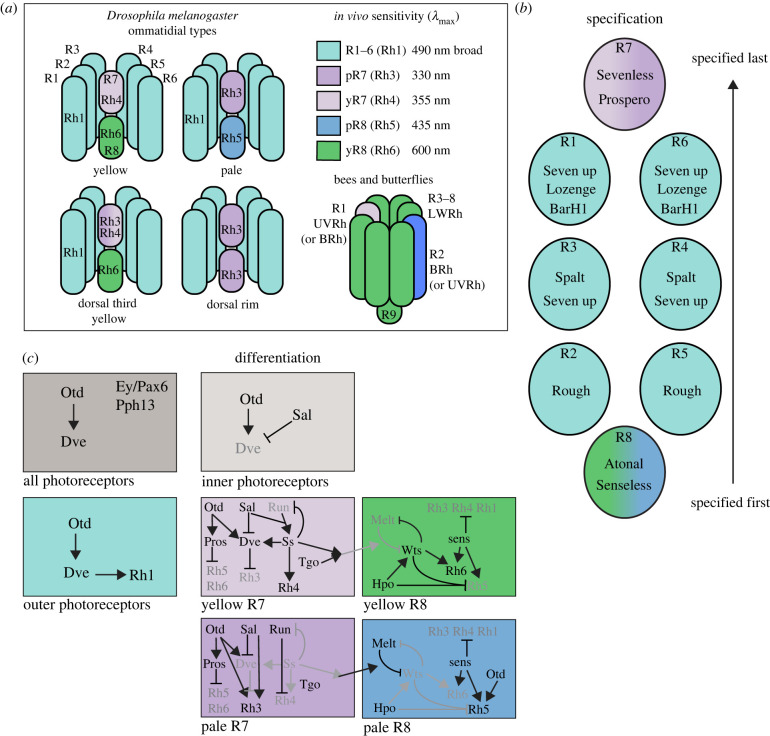
In D. melanogaster, like other insects, the compound eye is made up of many repeating unit-eyes called ommatidia. Each ommatidium is made up of eight photoreceptor cells labelled R1–R8. The outer R1–6 cells express the opsin Rh1 in every ommatidium in the retina and are broadband contrast and motion-sensing cells that project to the lamina, while R7/R8 are the ‘inner’ photoreceptors that project to the medulla, known to be involved in colour processing (figure 1a). The opsin-expressing microvilli of these two cells are stacked along the proximo-distal axis (R7 more distal) in the ommatidium, sampling from the same point in space. The typical R7 cell expresses either Rh3 or Rh4 (ultraviolet (UV)) opsin and induces R8 to express either Rh5 (blue) or Rh6 (long-wavelength or LW) opsin, respectively [20,21]. The ommatidia expressing Rh3 in R7 and Rh5 in R8 are known as pale ommatidia while ommatidia expressing Rh4 in R7 and Rh6 in R8 are known as yellow ommatidia. The pale and yellow types are stochastically found throughout the retina at a ratio of 35 : 65 in typical laboratory fly stocks [17,22]. In D. melanogaster, like in other insects [23], the dorsal-most row of ommatidia is known as the dorsal rim area (DRA) and expresses only Rh3 in both R7 and R8 cells. Insect DRA ommatidia are typically enriched with UV or blue-sensing cells and are involved in polarized light detection for navigation [24,25]. A specialized yellow ommatidial type in D. melanogaster is found only in the dorsal third of the eye and in addition to Rh4 also expresses Rh3 in the R7 cell [17] (figure 1a).
Figure 1.
Insect photoreceptor cell arrangements, specification and regulation of differentiation. (a) Top, schematic representation of D. melanogaster ommatidial types. R1–R6 labels outer photoreceptor cells while R7 and R8 are inner photoreceptor cells involved in colour vision. Distinct ommatidia vary by opsin expression in inner photoreceptor cells. Right, corresponding maximum sensitivities (λmax) of photoreceptor cells measured in vivo incorporate pigment filtering and other photoreceptor cell dynamics (from [19]). Bottom, Hymenoptera and Lepidoptera have an additional photoreceptor cell in their ommatidia. The R1/R2 cells in butterflies and R1/R5 cells in bees are the ‘inner’ photoreceptors and contribute to colour vision, and both are R7-like in the D. melanogaster numbering scheme. The small R9 cell in bees and butterflies is equivalent to the R8 cell in D. melanogaster. The additional photoreceptors have allowed for three main ommatidial types with regard to UVRh opsin expression in the retinas of bees and butterflies as opposed to two in D. melanogaster. (b) The specification of photoreceptor cells in each ommatidium is stereotyped and proceeds via temporal differences in the expression of transcription factors. R8 is specified first, followed by R2/R5, then R3/R4, then R1/R6, with R7 specified last. Transcription factors known to be important for subtype specification are shown within each cell. (c) Once cells are specified an interconnected series of transcription factors are expressed to terminally differentiate each photoreceptor cell. These networks are characterized by a series of feedforward bistable loops of regulation, mutual inhibition and a combination of stochastic and deterministic choices. Arrows are positive regulatory relationships while T's are negative regulatory relationships. Black shows active pathways while grey indicates pathways not activated in each subtype. Otd, Orthodenticle; Dve, Defective proventriculus; Ey, Eyeless; Pph13, PvuII-PstI homology 13; Sal, Spalt complex; Pros, Prospero; Run, Runt; Ss, Spineless; Tgo, Tango; Melt, Melted; Wts, Warts; Sens, Senseless; Hpo, Hippo pathway.
In D. melanogaster, photoreceptor cell specification begins first with the R8 cell. Atonal increases Senseless expression, inhibiting Rough, leading to R8 cell fate (figure 1b). The cell-adhesion protein Hibris is also involved in signalling for proper R8 specification [26]. R2 and R5 are specified next, where Rough is highly expressed and represses Senseless leading to R2 and R5 cell fate. Rough suppresses Seven up in R2/R5, but Rough is not expressed and Seven up is upregulated in R3/R4 and R1/R6 which are next to be specified. R3/R4 cell fate requires the Spalt complex [27] while R1/R6 cell fate requires Lozenge which in turn upregulates BarH1 in R1/R6. Finally R7 is specified last, where the receptor tyrosine kinase Sevenless is expressed and binds to Boss in the R8 cell. Epidermal growth factor receptor (EGFR) signalling from R1/R6 activates the Ras pathway which in turn upregulates Prospero in R7. Notch is also involved in blocking differentiation in R7 until Sevenless is upregulated highly enough to overcome this, in a distinct mechanism from EGFR signalling in R1–R6 [28] (figure 1b).
Next, photoreceptor cell differentiation involves a complex series of overlapping events that work in concert to ensure proper and robust opsin expression in each cell. Rh1 has been shown to be directly activated by Pax6/Eyeless [29]. Defective proventriculus (Dve) is also expressed in R1–R6 and activates Rh1. Differentiating R7 and R8 inner photoreceptors, both of which mediate colour vision (the former of which also contributes to motion vision [30]), involves the precise restriction of specific opsins and activation of others. Following specification, the Spalt complex is a marker for inner photoreceptor cell differentiation (R7/R8) [31] where it blocks Dve and thus Rh1 expression. R7 expresses Prospero, which inhibits Rh5 and Rh6 opsin expression [32]. R8 expresses Senseless which promotes expression of these opsins and blocks Rh3/Rh4 expression [33]. Then, a stochastic choice in each R7 results in Spineless expression in a subset of cells across the retina as differentiation proceeds [34,35]. Spineless leads to Rh4 expression and yellow ommatidial fate. Together with Tango, Spineless induces the R8 cell to express Rh6. In the absence of Spineless, Rh3 is expressed in R7 and Rh5 is expressed in R8 (figure 1c).
A variety of interdependent regulatory loops pattern the retina and dictate opsin expression. A bistable loop of the mutually inhibiting proteins Melted and Warts ensures robust expression of opsins in R8 cells. Whereas Melted promotes Rh5 and blocks Warts, Warts promotes Rh6 and blocks Melted [36]. Another feedforward loop involves Orthodenticle (Otd/Crx). In the absence of Dve, Otd is a permissive factor that leads to pale ommatidial fate by activating Rh3 and Rh5 expression in R7 and R8 cells. Spalt complex blocks Dve in pale ommatidia, but Dve expression in yellow ommatidia represses this permissive Otd, blocking Rh3 and Rh5 expression. In the dorsal third retina, lower Dve expression and Iroquois complex expression activate Rh3 coexpression with Rh4 in dorsal yellow ommatidia [16,37,38]. Recently, another feedback loop was discovered, showing that interaction with stereotyped upstream Runt expression contributes to the stochastic Spineless expression that determines pale or yellow ommatidial types [39] (figure 1c). Feedforward, feedback and bistable loops build redundancy and modularity into this system and have become hallmarks of robust developmental processes in other complex traits. The study of these genetic regulatory programmes has contributed to our modern view of development, and further study will continue to yield new insights that explain how complex traits develop.
The cis regulatory biochemical interactions of some of these genetic relationships have been elucidated. As mentioned above, Pax6/Eyeless binds directly to the Rh1 promoter and activates expression. There is also evidence that Pax6 is a general activator for all D. melanogaster rhodopsins [29,40]. In R8 cells which are competent to express Rh5 throughout their lifetime, expression and functional activity of Rh6 directly inhibits Rh5 expression [41]. A conserved 11 bp palindromic motif is found in the proximal promoter of phototransduction genes expressed in all D. melanogaster photoreceptors but is modified in rhodopsins with restricted photoreceptor cell expression domains, such as Rh1 found only in R1–6 cells or Rh5 found only in R8 cells. Differences in rhodopsin expression are owing to specific base-pair mutations that break the palindrome, resulting in changes to this site's affinity for binding repressors and activators, PvuII-PstI homology 13 (Pph13), Dve and Otd [42]. Recent work swapping promoter domains from multiple rhodopsins has revealed a complex regulatory landscape in addition to this palindromic sequence that delineates photoreceptor subset specific expression. Each rhodopsin-specific promoter landscape evolved by duplication from a simple single pan-photoreceptor rhodopsin followed by subsequent divergence for specificity of expression [43]. This highlights the importance of duplication as a mechanism for generating novel genetic material (reviewed below), and that by linking development and evolutionary biology we can more deeply understand how visual systems work [43].
3. Genome editing
Advances in CRISPR/Cas9 editing technology have allowed researchers to validate gene functions and discover genetic interactions. In terms of insect eye development, in Drosophila, CRISPR deletion of an eyeless enhancer site resulted in a small eye phenotype [44]. This enhancer was not only important for eyeless expression, but also for Decapentaplegic expression and proper formation of the morphogenetic furrow [44]. In terms of photoreceptor cell fate, CRISPR knockouts revealed the genes and interactions necessary for compound eye retinal mosaic choices [45]. While eye development genes are difficult to knock out owing to their deleterious effects, opsin and eye colour gene knockouts have revealed genes important for visually guided behaviour [46], phototaxis [47] and eye pigmentation [48–51].
4. Opsin regulation in other insects
In other insects, relatively little is known about development and opsin regulation, despite Hymenoptera and Lepidoptera being major visual ecology models [52–54]. One key theme to emerge in the last 25 years is the understanding of homology between the inner colour photoreceptor cells between D. melanogaster, Lepidoptera, and Hymenoptera. In butterflies and bees but not beetles, the equivalent of D. melanogaster inner photoreceptor R7 was independently ‘duplicated’ with two R7-like cells spanning the length of the ommatidium, while the R8 cell has been miniaturized and sits at the proximal-most part of the ommatidium. In Lepidoptera and Hymenoptera, this proximal cell is called R9. R1/R2 and R9 project to the medulla similar to R7/R8 in D. melanogaster, all presumably contributing to colour vision [55]. The difference is that R1/R2 (R7-like) in bees and butterflies alter their particular opsin expression with either blue- or UV-absorbing opsins, unlike in D. melanogaster where R7/R8 do this. The R9 cell (R8 homologue) so far has only been shown to express long-wavelength opsin (LWRh) in all ommatidial types although more sampling is required [18,56]. In most other insects, including crickets, locusts, dragonflies and beetles, no such cell type diversification occurred, and the arrangement of photoreceptors in ommatidia is similar to D. melanogaster. This suggests the additional R7-like cell evolved independently in bees and butterflies (figure 1a, bottom).
Until recently, the molecular logic, which led to three ommatidial types in the main retina of bees and butterflies and only two main types in D. melanogaster, was unknown. As mentioned above, in D. melanogaster, Spineless is expressed stochastically in a subset of R7 cells and directs the expression of particular opsins in both R7 and R8 yielding either pale or yellow ommatidia. In the swallowtail butterfly Papilio xuthus, Spineless is also expressed stochastically in a subset of R1 and R2 cells, both of which are homologous to the D. melanogaster R7, with ON expression leading to blue opsin (BRh) expression and OFF yielding ultraviolet opsin (UVRh) expression [45]. As a result, there are two Spineless decisions within each ommatidium yielding three combinations of UV/UV UV/blue or blue/blue ommatidial types [45]. (This pattern is complicated in nymphalid butterflies with the recent discovery of the coexpression of BRh and LWRh opsins in a subset of R1/R2 cells in Heliconius, producing three additional ommatidial classes, and the existence of R1/R2 photoreceptors with both blue- and LW sensitivities across a variety of nymphalid species [57,58].) In Heliconius, which are rare among Lepidoptera and other insects for duplicating a UV opsin [8], yet more combinations of ommatidial types exist where a choice between BRh, UVRh1 or UVRh2 is made [59]. Counts of ommatidial types in multiple species within this genus have shown stereotyped percentages for these novel ommatidial types, suggesting there may be an additional stochastic yet well-controlled step similar to what we see with Spineless [60]. Other than this, there is no experimental evidence showing that genetic regulation in D. melanogaster opsin-expressing cells is conserved in either Hymenoptera or Lepidoptera.
Although little functional data exist, orthologous genes known to be involved in D. melanogaster eye development have been identified in the visual systems of many other insects and are thought to be involved in similar ways to D. melanogaster differentiation. Recent work has suggested at least some transcription factor orthologues (Pax2/5/8, Optix, Ocelliless and Araucan/Caupolican) involved in D. melanogaster photoreceptor differentiation are potentially regulating UV opsin expression in Heliconius butterflies [57]. In Tribolium, Pph13 and Otd are required for proper regulation of both rhabdomeric opsin transcription and rhabdomere morphogenesis [61]. The Glass gene is also required for proper photoreceptor cell differentiation and for Pph13 expression in both D. melanogaster and Tribolium [62]. Unlike D. melanogaster and Tribolium, in scarabaeid beetles, RNAi-mediated knockdown of Otd induces functional ectopic eyes [63]. These ectopic eyes are located in the medial dorsal head, where the ocelli (simple eyes) are found in other insects, but nearly all beetles have lost their ocelli [64], so this may be a reactivation of some vestigial pathway. So far only a handful of insect orders are represented within the opsin regulation literature, and the next years should bring functional studies in more diverse taxa.
5. Larval photoreceptors and ocelli
Holometabolous insects have evolved complete metamorphosis from ancestral insect lineages that do not have such an extreme transformation [65]. The larvae of holometabolous insects typically have simple eyes called stemmata with a limited number of photoreceptors and opsins compared to the adult compound eye. Other insects that do not exhibit complete metamorphosis have compound eyes in larval stages, suggesting stemmata are derived evolutionarily from compound eyes [66]. Although most stemmata are simple, some lineages have elaborated these structures considerably, especially among holometabolous larvae [66]. The molecular differences between larval eyes and adult structures are poorly studied in most insects, obscuring conclusions about the homology of eyes within arthropods and across the animal tree of life.
Nearly all functional molecular data come from D. melanogaster. The larval eye, also known as the Bolwig organ, is made up of 12 photoreceptor cells that express the same opsins as R8 cells in adults (Rh5, four cells or Rh6, eight cells) [67]. Primary precursor cells are specified by Atonal [68], then these cells recruit secondary precursors via EGFR signalling [67]. The primary precursors differentiate into Rh5-expressing cells while the secondary precursors express Rh6. Despite a similarity to the 35 : 65 ratio seen in adult ommatidia, this process is deterministic and not driven by stochastic choices [68]. Spalt and Otd are required for Rh5 expression and repression of Seven up while Seven up blocks Spalt and thus Rh5 in Rh6-expressing cells [67,68].
During metamorphosis, the Rh6-expressing cells of the Bolwig organ die, and the Rh5 cells switch to Rh6 expression forming an adult extraretinal photosensitive structure known as the eyelet, which mediates circadian entrainment [69,70]. Senseless also acts to maintain Rh5 in larval photoreceptor cells, while it blocks apoptosis during metamorphosis and promotes Rh6 expression in the adult eyelet [71]. Furthermore, Pph13 (Hazy) is required for Rh5 and Rh6 expression in the larva but during metamorphosis blocks Senseless expression in Rh6-expressing cells, leading to apoptosis [71]. Not much is known about the stemmata of other insects; however in the adult butterfly, Vanessa cardui, eyelets exist and express both UVRh and LWRh [72], unlike in D. melanogaster where only the UV-absorbing Rh5 is expressed.
The adult forms of winged insects also contain medial ocelli, single chambered, optical structures on the dorsal side of the head [73]. These ocelli and compound eyes are thought to have been split from some ancestral eye prior to the rise of Arthropoda [74]. Subsequent duplication of opsins allowed for differential expression and thus separation of distinct visual tasks [75]. In D. melanogaster, Rh2 is expressed only in the ocelli, resulting in violet-sensitive photoreceptor cells [76]. It has recently been shown that Homothorax (Hth) is expressed in ocelli and represses Rh1 expression while promoting Rh2 in these photoreceptor cells [77]. Hth appears to cause a fate switch between Rh1 and Rh2 because Rh2 expression can only be induced by ectopic Hth in outer photoreceptor cells of the compound eye, which normally express Rh1. This observation further supports that Rh1 and Rh2 are the result of a gene duplication that allowed for neofunctionalization of light-mediated tasks [77]. Ocellus-specific duplicates of opsins are common in most insect orders: specific UV and LW opsins are found only in ocelli in the cricket Gryllus bimaculatus [78], in multiple dragonfly species [7], and in bees [79,80] (figure 2). Together this further supports that opsin gene duplication has coincided with the evolution of novel sensory structures and allowed for neofunctionalization of light-mediated tasks.
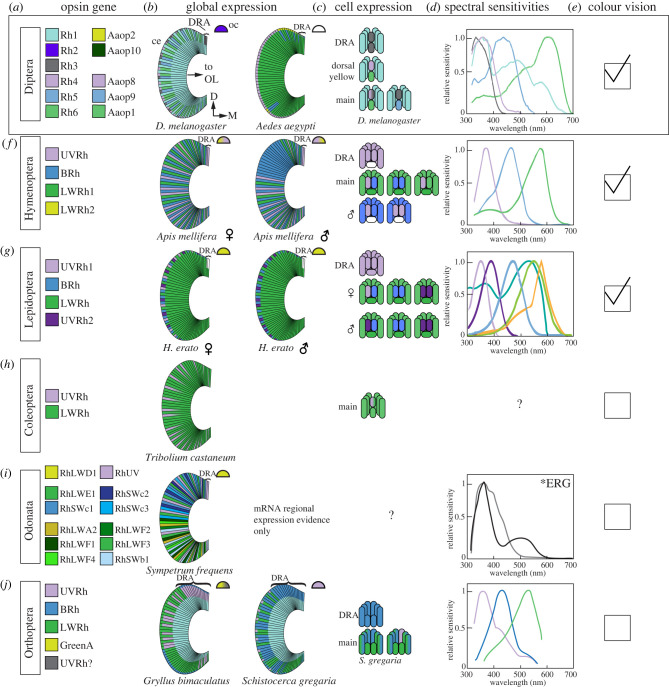
Figure 2.
Holistic opsin sequence, expression pattern and visual system function is only known in a few insects. Cartoons of exemplar systems are shown from major insect orders. (a) Opsin sequences in example dipterans are coloured to match the general wavelength of absorption. Use of the same colour indicates orthologous sequences. Names are specific to literature in each species. (b) Broad opsin expression patterns in the compound eyes (ce) and ocelli (oc) are shown. Dorsal rim area (DRA) expression is distinct in the dorsal-most cell layers, if present. For all cartoons, colours correspond to sequences in the first column, gradient colours indicate co-expression, dorsal is up, medial is to the right. White, expression is unknown. Tiered retinal opsin expression is shown if known. Photoreceptor axons project to the optic lobe (OL) from the proximal side of the eye. (c) Expression in single cells is known for fewer insect examples. Opsin-expressing cells are arranged as ommatidial types present in the compound eye. (d) Single-cell resolution of photoreceptor spectral sensitivities is known in still fewer insect species. Relative sensitivity is shown over the visible light spectrum. (e) The presence of colour-discrimination behavioural data is indicated by a tick. Data in (e) and (d) are unknown in many insects. (f) In Hymenoptera, the honeybee Apis mellifera is best studied for colour vision behaviour, although other well-studied hymenopteran models (such as bumblebees) also exist. The honeybee eye is sexually dimorphic with respect to opsin expression. (g) Lepidoptera is one of the best-studied insect groups for colour vision. Molecular, physiological and behavioural examples exist in moths and butterflies. The Heliconius erato eye is sexually dimorphic and Heliconius is one of the few insect genera where colour vision has been studied from molecular genetics to behaviour. (h) Beetles have lost the BRh opsin, and the retina is dominated by LWRh and UVRh opsins. (i) Dragonflies are known to have greatly expanded their opsin repertoires based on transcriptomics but single-cell level expression data remain to be gathered. (j) Crickets and locusts are classic models for studying the DRA owing to its conspicuousness in these species. Single-cell expression and physiology has been worked out but few colour-specific behavioural studies exist. References: (a–e) [17,19,81,82], (f) [83,84], (g) [45,56,57,59,60,85–92], (h) [93], I) [7] and (j) [78,94].
6. Opsin expression patterns in insect photoreceptor cells
In contrast with the dearth of genetic regulatory studies of opsins across insects, studies of opsin expression have expanded to include multiple insect orders and have contributed to shifts in thinking about receptor patterning and function. The number of opsins and their spatial patterns of expression in the eye and/or their inferred expression based on physiological measurements has been reviewed across insects [6,17,85]. Regions, bands and spots of specialized ommatidia expressing unique combinations of opsins are found throughout insects and can be sex-specific [17] (figure 2). The DRA is found in many insects, and evidence from D. melanogaster (above) suggests that distinct regulatory pathways lead to UV or blue opsins expressed in all or a subset of photoreceptor cells of the ommatidia in the DRA. Dorsal/ventral expression and sex-specific regions of specialized ommatidia in bees and flies are used for locating prey or mates in flight. These patterns of opsin expression remain interesting from both developmental and behavioural perspectives and merit further study [17].
One way our understanding of sensory neurons has changed significantly is the ‘one cell one receptor’ rule, stating that in sensory cells, a single molecular receptor is expressed to the exclusion of all other possible receptors in any individual cell [95]. It was thought until recently that the expression of two opsins in a single photoreceptor cell would diminish colour vision capability by the generation of a broadband receptor. In contrast with this view, multiple insects have independently evolved opsin coexpression in single photoreceptor cells (figure 2) and new, though limited, evidence suggests coexpression contributes to colour vision in some instances [57,58,96]. In D. melanogaster, the dorsal yellow ommatidial type expresses both Rh3 and Rh4 in the R7 cell. The Rh1-expressing outer cells, though not coexpressed with another opsin, are modified broadband receptors which participate in colour vision [97]. In butterflies, a variety of different opsin coexpression patterns is common, with examples of coexpressing photoreceptor cells found in nearly all major families. In the lycaenid Lycaena rubidus, one BRh duplicate is coexpressed with LWRh opsin messenger RNA (mRNA) in the dorsal eye of females only in all R3-8 (outer) photoreceptor cells [98]. In Pieridae, Colias erate expresses three distinct opsin mRNAs in a single cell [99]. Papilio xuthus (Papilionidae) has been shown to coexpress two green-absorbing opsins. Both opsins participate in phototransduction, generating a double-peaked green cell [96], which is used in the butterfly's tetrachromatic colour vision system [86]. Recently, a LW+B opsin cell type has been found in Heliconius butterfly R1/R2 cells, and intracellular recordings have identified a blue-green broadband cell across Nymphalidae that could be this coexpressing cell type [57,58]. In the desert locust Schistocerca gregaria, most photoreceptor cells in the main retina coexpress blue and green opsin mRNAs [100]. While not all photoreceptor cells with opsin coexpression are involved in colour vision, some probably are. Future work including behavioural assays should focus on how photoreceptors with opsin coexpression contribute to colour vision.
(a) . A note of caution: opsin messenger RNA does not always equal opsin protein
Most studies of opsin expression in non-Drosophila insects examine opsin mRNA expression using transcriptomics instead of opsin protein expression using immunohistochemistry. We, along with other researchers, have found that quantifiable opsin mRNA expression levels do not always correspond to opsin protein expression. For instance in the butterfly Heliconius melpomene, UVRh2 is expressed at low levels in the eye and brain as assayed using RNA-Seq but no protein is detected [60,101]. A similar finding of opsin mRNA expression in the absence of opsin protein expression has been found in bats and in the swallowtail butterfly Papilio xuthus [102,103]. These findings suggest more mechanistic studies of post-transcriptional opsin regulation are needed to better understand the relationship between opsin genotype and phenotype.
7. Opsin duplication
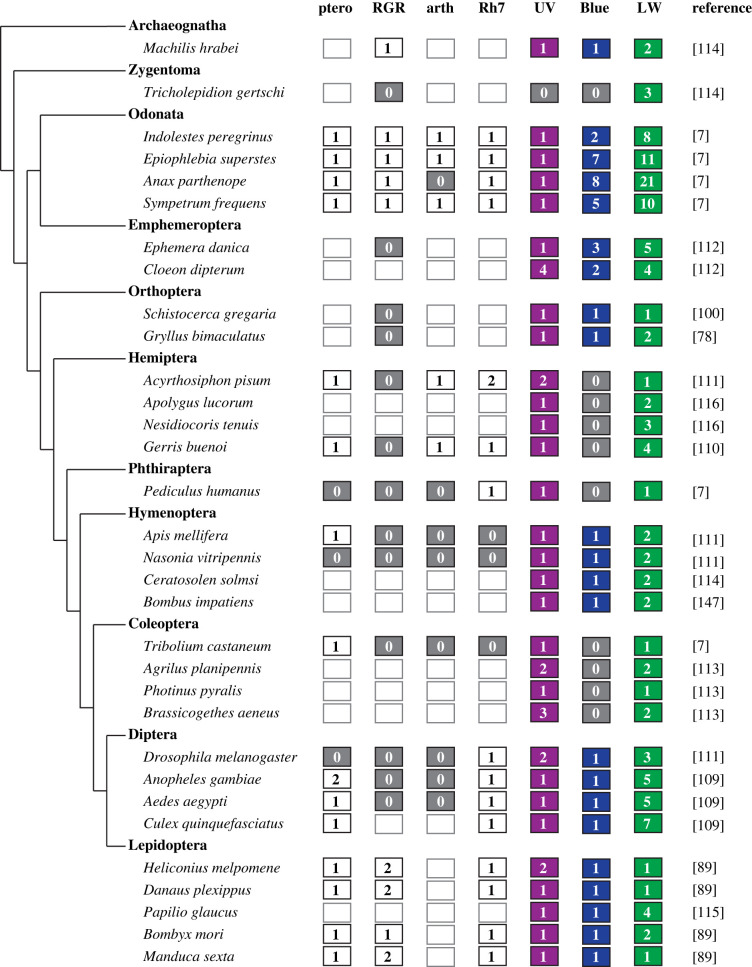
Studies of opsins can tell us about general trends and fates of gene duplicates. Current knowledge indicates that duplications can occur by a variety of mechanisms even in closely related groups [104]. Similarly, selection can act on different sites to achieve almost identical outcomes and selective pressure weakens with additional duplication events [105,106]. The vast majority of recent findings in insect opsins have come from surveys of opsin genes or mRNAs together with phylogenetic studies. Opsin genes evolve by coding sequence variation, gene duplication and gene loss. The insect opsin gene family includes four non-visual opsin and three visual opsin clades, roughly corresponding to mRNAs encoding UV-, blue- and LW-absorbing photopigments (summarized in figure 3 and considered below). General trends are that UVRh is duplicated less often than BRh and LWRh; BRh has been lost in multiple insect orders and LWRh is most variable with multiple duplications in most insect orders. Overall UVRh, BRh and LWRh duplicates have resulted in diversity in insect visual system function including modifications of sensitivity to light via spectral tuning mutations and the evolution of sexual dimorphisms of the eye (figures 2 and 3).
Figure 3.
Opsin diversity in representative insects. The arthropod opsin complement includes canonical c-opsins, canonical r-opsins, retinal pigment epithelium G protein-coupled receptor (RGR) and neuropsin [5]. Within arthropods, it is estimated that there were anywhere between 1 and 3 UV/B/LW rhodopsin, 1 c-opsin, 1 arthropsin and 1–2 peropsin before the Chelicerate split [107,108]. The ancestral insect eye was probably trichromatic through the use of an ultraviolet (UVRh), blue (BRh) and long-wavelength (LWRh) rhodopsin [6]. Empty boxes represent missing data. The numbers in the boxes show number of gene copies present in that species. Figure made using data from [7,78,100,101,109–116].
(a) . Long-wavelength opsins and tandem duplication
In insects, the most duplications and largest diversity in opsins are that of LWRh, a finding foreshadowed by the first publication describing butterfly opsins in 1998 [117]. While there are many hypotheses linking LWRh duplications to ecology, the significance of LWRh duplications and expansions across most insects remains unclear because few studies have examined the behavioural context in which a particular receptor is used (e.g. [88,118]). Most Hymenoptera have two LWRh opsins (one of which is expressed in ocelli) [119–121]. A comprehensive study in Lepidoptera found that opsin duplications are more common in diurnal species and that LWRh was duplicated more often than other opsins; specifically, LWRh has had approximately 10 duplication events, while six have occurred in BRh and three in UVRh [8]. In Diptera, the current LWRh complement in mosquitoes has been produced by an estimated 18 or 19 duplication events [109,122]. At the genetic level, signatures of gene evolution mechanisms include genes located in tandem owing to unequal crossing over or a lack of introns by retrotransposition [123]. In insects, we see LWRh evolution by both of these mechanisms and probably others that are not as easy to identify. In the water strider Gerris buenoi and in Anopheles gambiae, four and five LWRh genes, respectively, are located in tandem [110,122]. Some LWRh paralogues in moths and Anopheles are intronless and are proposed to have evolved by retrotransposition [109,111,124]. In mayflies, four LWRh genes are in genomic clusters that vary in size and have undergone rearrangement between species [112]. A limitation in investigating the molecular mechanisms by which LWRh genes are undergoing expansions is a lack of high-quality contiguous insect genomes. With chromosome-level genome assemblies, searches for other signatures of gene evolution via retrotransposition and other mechanisms become possible.
(b) . Blue duplications, expansions and localized expression in butterflies, bees and Paleoptera
Independent duplications of BRh have been found in butterflies. Pieris rapae and Lycaena rubidus have two BRh that underlie sexual dimorphism in their compound eyes [98,125]. In Pieris rapae, males have a double peak in blue sensitivity; in Lycaena rubidus females, BRh is coexpressed with LWRh in R3-8 cells while in males, BRh has entirely replaced LWRh in the R3-8 cells in the dorsal region of the compound eye [98,125]. In both of these cases, the sexually dimorphic eye is believed to be important for male behaviour and possibly for vision in the blue spectrum. A related pattern of BRh opsin expression is that of the honeybee Apis mellifera. Honeybee drones have different ommatidia and photoreceptor morphology in their dorsal eye region compared to workers [126]; at late pupal stages, drones express a larger proportion of UVRh and BRh compared to LWRh mRNAs while worker bees predominantly express LWRh in their developing compound eye [127].
Paleoptera (damselflies and dragonflies) has multiple independent expansions in the number of BRh copies. Damselfly and dragonfly adults are diurnal, brightly coloured, territorial and possess large eyes. Larvae are aquatic. In 12 species surveyed, all species had 1 to 8 BRh genes [7]. Opsin regional expression in adult eyes, ocelli and larval eyes varies between species [7]. In Odonata, BRh transcripts are divided into three subgroups. Interestingly, BRh from group a are upregulated in larval eyes, BRh from group b are upregulated in the ventral adult eye, and BRh from group c (which has the most duplications) are expressed in the dorsal region of the adult eye [7]. Comparative opsin studies in mayflies have not been as in-depth. Cloeon dipterum has two BRh opsin genes which are located in tandem; one copy is also specific to male turbinate eyes [112]. Variation in Paleoptera visual systems is speculated to be related to differences in behaviour, light ecology and microhabitat.
(c) . Polarized light detection and paralogue switching in Orthoptera
Although BRh, to our knowledge, is not duplicated in Orthoptera, it is probably functioning in polarized light detection. Insects use polarized light for orientation and navigation (see reviews [128,129]). The specialized DRA found in many insects often expresses only UVRh to detect polarized light [23,128,130]. However, in Orthoptera, BRh is expressed in the DRA and hypothesized to be important for polarized light detection [131,132]. In the cricket, Gryllus bimaculatus BRh and UVRh are expressed in the DRA while in the desert locust Schistocerca gregaria, only BRh is expressed in all photoreceptor cells of the DRA [78,100]. This is an example of paralogue switching where Orthoptera have swapped UVRh typically found in most insect DRAs for the BRh paralogue. Studies thus far have shown that both UVRh and BRh can be used for polarized light detection. While LWRh is not typically used in polarized light detection, butterflies furnish an exception [86,89,133–135].
(d) . Ultraviolet opsin duplications
UVRh duplications are rarer among surveyed insects compared to BRh and LWRh duplications. The mayfly Cloeon dipterum has four copies of UVRh, and one of the UVRh genes is only expressed in male-specific turbinate eyes [112]. UVRh duplications in the pea aphid (Acyrthosiphon pisum) and in beetles (order Coleoptera) appear to have evolved to overcome the loss of BRh [113,136]. A survey of 175 moths and butterflies found only two independent UV duplications, in Triodia sylvina (Hepialidae) and Chilo suppressalis (Crambidae), in addition to that of Heliconius, described below [8]. Another lepidopteran with multiple UVRh copies is the armyworm Mythimna separata. Mythimma separata has three UVRh genes but only one copy is expressed at a level similar to other visual opsin genes while the other two copies are very lowly expressed [137]. In Lepidoptera where opsin evolution has been extensively studied, the first discovery of a UVRh duplication was made in the butterfly genus Heliconius, famous for Müllerian mimicry [90]. In one of the duplicates, UVRh2, some of the sites evolved under positive selection within Heliconius compared to UVRh1, and in other systems, these sites are associated with spectral tuning [90]. Physiological recordings indicate UVRh1 encodes a UV receptor, while UVRh2 encodes a violet receptor. Following duplication of UVRh, a remarkable diversity in opsin expression patterns, including several forms of sexually dimorphic UV opsin expression, has evolved within the genus [60]. These opsin expression pattern differences, filtering effects, and spectral tuning correspond to a diversity of photoreceptor cell sensitivities [57,59,60]. Recent behavioural experiments show that the two UV opsins are used in UV colour vision in foraging Heliconius erato females, while H. erato males, which express only one UV opsin, are colour blind to the same UV discrimination task [89].
8. The role of opsins in colour vision: physiology and behaviour
Studies of opsin presence, absence and expression establish an experimental framework connecting molecular evolution to expression level changes, photoreceptor sensitivity, and colour visual behaviour. In Heliconius butterflies, UV opsin duplication and divergence [90] led to sexually dimorphic expression differences and photoreceptor cell sensitivities [59,60], which has resulted in differences in adult colour visual behaviour in the UV [89]. Transcriptomics and genomics have been used to identify UVRh, BRh and LWRh genes while electroretinograms, microspectrophotometry and intracellular recordings have been used to determine eye and photoreceptor cell spectral sensitivities [85]. However, in only a limited number of species and orders have opsins been connected molecularly to sensory neuron phenotype, and in even fewer to organismal behaviour (figure 2).
(a) . In vitro expression systems to study insect visual pigment absorption spectra
A major contribution to the field of visual ecology is the refinement of an in vitro insect opsin-expressing system which permits the measurement of their absorption spectrum [9]. Vertebrate ciliary-type opsins are often expressed and purified in in vitro systems, and this has greatly aided in understanding the specific contribution of opsin absorbance to photoreceptor cell physiology, especially when in vivo electrophysiology is a challenge [138,139]. These systems also allow for targeted mutagenesis experiments to measure the effect on opsin absorbance of spectral tuning site changes [140]. Although insect rhabdomeric-opsins are ancestrally related to vertebrate opsins, in vitro expression systems have proven challenging [81,141] (but see [87,142]). The new expression system established by Liénard et al. [9] should greatly facilitate future research on insect opsin phenotypes.
(b) . A need for behavioural studies
Behavioural studies are crucial for addressing the link between opsin genotype and phenotype yet continue to be relatively rare. Among hymenopterans, ants are believed to have UV, LW and possibly B photoreceptors. Yet, behavioural studies have found them to be dichromatic, where some are capable of discriminating between UV and green wavelengths of light, while others can discriminate UV from blue, and UV from green, but not between blue and green [143–145]. In Lepidoptera, behavioural studies have found that gene duplications and filtering pigments drive shifts in butterfly sensitivities allowing them to discriminate in the UV, blue, green and red [86,88,89,133]. It is clear from available evidence that there is a direct relationship between opsin genes and phenotypes, but it is important to remember that much of what we know about insect opsins is based on limited sequence data, and that much work remains in order to build a more complete picture of opsin and eye evolution in insects.
9. Concluding remarks
Colour visual systems with three–four colour receptor channels should be sufficient to adequately sample over the entire visual spectrum in bright daylight. Nonetheless cycles of opsin duplications occur, leading to increased numbers of colour channels in the visual systems of insects. The number of channels subsequently gets pruned, which explains the high level of turnover we see among opsins throughout insect evolution [146]. Insects that make transitions between light environments, such as fireflies, diving beetles and some moths, have additional selective constraints shifting opsin repertoires in predictable ways [8,147,148]. However we still do not fully understand the selective pressures for particular sensory arrangements and further physiological and behavioural studies are needed to better understand the role of selection and drift in the evolution and diversity of sensory systems. Lepidoptera and Hymenoptera are rich with examples of unique visual ecology and evolutionary transitions such as switches from nocturnal to diurnal flying, and these transitions change opsin repertoires [8,149]. With CRISPR/Cas9 genome editing becoming more accessible in these insect orders, and with refinements of in vitro opsin expression systems, investigating opsin spectral tuning, expression and gene regulation in these classical visual ecology models will be a fruitful avenue for future research.
Data accessibility
This article has no additional data.
Authors' contributions
K.J.M.: conceptualization, data curation, formal analysis, writing—original draft, writing—review and editing; A.D.B.: conceptualization, funding acquisition, writing—review and editing; A.M.-M.: conceptualization, formal analysis, writing—original draft, writing—review and editing.
All authors gave final approval for publication and agreed to be held accountable for the work performed therein.
Conflict of interest declaration
We declare we have no competing interests.
Funding
This study was funded by National Science Foundation (grant no. IOS-1257627).
References
- 1.Suga H, Schmid V, Gehring WJ. 2008. Evolution and functional diversity of jellyfish opsins. Curr. Biol. 18, 51-55. ( 10.1016/j.cub.2007.11.059) [DOI] [PubMed] [Google Scholar]
- 2.Schnitzler CE, et al. 2012. Genomic organization, evolution, and expression of photoprotein and opsin genes in Mnemiopsis leidyi: a new view of ctenophore photocytes. BMC Biol. 10, 107. ( 10.1186/1741-7007-10-107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Feuda R, Hamilton SC, McInerney JO, Pisani D. 2012. Metazoan opsin evolution reveals a simple route to animal vision. Proc. Natl Acad. Sci. USA 109, 18 868-18 872. ( 10.1073/pnas.1204609109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Feuda R, Rota-Stabelli O, Oakley TH, Pisani D. 2014. The comb jelly opsins and the origins of animal phototransduction. Genome Biol. Evol. 6, 1964-1971. ( 10.1093/gbe/evu154) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ramirez MD, Pairett AN, Pankey MS, Serb JM, Speiser DI, Swafford AJ, Oakley TH. 2016. The last common ancestor of most bilaterian animals possessed at least nine opsins. Genome Biol. Evol. 8, 3640-3652. ( 10.1093/gbe/evw248) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Briscoe AD, Chittka L. 2001. The evolution of color vision in insects. Annu. Rev. Entomol. 46, 471-510. ( 10.1146/annurev.ento.46.1.471) [DOI] [PubMed] [Google Scholar]
- 7.Futahashi R, Kawahara-Miki R, Kinoshita M, Yoshitake K, Yajima S. 2015. Extraordinary diversity of visual opsin genes in dragonflies. Proc. Natl Acad. Sci. 112, E1247-E1256. ( 10.1073/pnas.1424670112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sondhi Y, Ellis EA, Bybee SM, Theobald JC, Kawahara AY. 2021. Light environment drives evolution of color vision genes in butterflies and moths. Commun. Biol. 4, 177. ( 10.1038/s42003-021-01688-z) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liénard MA, et al. 2021. The evolution of red color vision is linked to coordinated rhodopsin tuning in lycaenid butterflies. Proc. Natl Acad. Sci. USA 118, 1-12. ( 10.1073/pnas.2008986118) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.von Frisch K. 1914. Der farbensinn und Formensinn der Biene. Jena, Germany: Fischer ( 10.5962/bhl.title.11736) [DOI]
- 11.Vogt K, Kirschfeld K. 1984. Chemical identity of the chromophores of fly visual pigment. Sci. Nat. 71, 211-213. ( 10.1007/bf00490436) [DOI] [Google Scholar]
- 12.Terakita A. 2005. The opsins. Genome Biol. 6, 213. ( 10.1186/gb-2005-6-3-213) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stoddard MC, Eyster HN, Hogan BG, Morris DH, Soucy ER, Inouye DW. 2020. Wild hummingbirds discriminate nonspectral colors. Proc. Natl Acad. Sci. USA 117, 15 112-15 122. ( 10.1073/pnas.1919377117) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Halder G, Callaerts P, Gehring WJ. 1995. Induction of ectopic eyes by targeted expression of the eyeless gene in Drosophila. Science 267, 1788-1792. ( 10.1126/science.7892602) [DOI] [PubMed] [Google Scholar]
- 15.Rister J, Desplan C, Vasiliauskas D. 2013. Establishing and maintaining gene expression patterns: insights from sensory receptor patterning. Development 140, 493-503. ( 10.1242/dev.079095) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tsachaki M, Sprecher SG. 2012. Genetic and developmental mechanisms underlying the formation of the Drosophila compound eye. Dev. Dyn. 241, 40-56. ( 10.1002/dvdy.22738) [DOI] [PubMed] [Google Scholar]
- 17.Wernet MF, Perry MW, Desplan C. 2015. The evolutionary diversity of insect retinal mosaics: common design principles and emerging molecular logic. Trends Genet. 31, 316-328. ( 10.1016/j.tig.2015.04.006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Friedrich M, Wood EJ, Wu M. 2011. Developmental evolution of the insect retina: insights from standardized numbering of homologous photoreceptors. J. Exp. Zool. B Mol. Dev. Evol. 316, 484-499. ( 10.1002/jez.b.21424) [DOI] [PubMed] [Google Scholar]
- 19.Sharkey CR, Blanco J, Leibowitz MM, Pinto-Benito D, Wardill TJ. 2020. The spectral sensitivity of Drosophila photoreceptors. Sci. Rep. 10, 18242. ( 10.1038/s41598-020-74742-1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chou WH, Huber A, Bentrop J, Schulz S, Schwab K, Chadwell LV, Paulsen R, Britt SG. 1999. Patterning of the R7 and R8 photoreceptor cells of Drosophila: evidence for induced and default cell-fate specification. Development 126, 607-616. ( 10.1242/dev.126.4.607) [DOI] [PubMed] [Google Scholar]
- 21.Chou WH, Hall KJ, Wilson DB, Wideman CL, Townson SM, Chadwell LV, Britt SG. 1996. Identification of a novel Drosophila opsin reveals specific patterning of the R7 and R8 photoreceptor cells. Neuron 17, 1101-1115. ( 10.1016/s0896-6273(00)80243-3) [DOI] [PubMed] [Google Scholar]
- 22.Anderson C, et al. 2017. Natural variation in stochastic photoreceptor specification and color preference in Drosophila. Elife 6, e29593. ( 10.7554/eLife.29593) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sauman I, Briscoe AD, Zhu H, Shi D, Froy O, Stalleicken J, Yuan Q, Casselman A, Reppert SM. 2005. Connecting the navigational clock to sun compass input in monarch butterfly brain. Neuron 46, 457-467. ( 10.1016/j.neuron.2005.03.014) [DOI] [PubMed] [Google Scholar]
- 24.Reppert SM, Zhu H, White RH. 2004. Polarized light helps monarch butterflies navigate. Curr. Biol. 14, 155-158. ( 10.1016/j.cub.2003.12.034) [DOI] [PubMed] [Google Scholar]
- 25.Stalleicken J, Labhart T, Mouritsen H. 2006. Physiological characterization of the compound eye in monarch butterflies with focus on the dorsal rim area. J. Comp. Physiol. A Neuroethol. Sens. Neural Behav. Physiol. 192, 321-331. ( 10.1007/s00359-005-0073-6) [DOI] [PubMed] [Google Scholar]
- 26.Tan H, Fulton RE, Chou WH, Birkholz DA, Mannino MP, Yamaguchi DM, Aldrich JC, Jacobsen TL, Britt SG. 2020. Drosophila R8 photoreceptor cell subtype specification requires hibris. PLoS ONE 15, e0240451. ( 10.1371/journal.pone.0240451) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Domingos PM, Mlodzik M, Mendes CS, Brown S, Steller H, Mollereau B. 2004. Spalt transcription factors are required for R3/R4 specification and establishment of planar cell polarity in the Drosophila eye. Development 131, 5695-5702. ( 10.1242/dev.01443) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tomlinson A, Mavromatakis YE, Arias R. 2019. The role of Sevenless in Drosophila R7 photoreceptor specification. Dev. Biol. 454, 181-189. ( 10.1016/j.ydbio.2019.06.007) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sheng G, Thouvenot E, Schmucker D, Wilson DS, Desplan C. 1997. Direct regulation of rhodopsin 1 by Pax-6/eyeless in Drosophila: evidence for a conserved function in photoreceptors. Genes Dev. 11, 1122-1131. ( 10.1101/gad.11.9.1122) [DOI] [PubMed] [Google Scholar]
- 30.Longden KD, Rogers EM, Nern A, Dionne H, Reiser MB. 2021. Synergy of color and motion vision for detecting approaching objects in Drosophila. bioRxiv 2021.11.03.467132. ( 10.1101/2021.11.03.467132) [DOI] [PMC free article] [PubMed]
- 31.Domingos PM, Brown S, Barrio R, Ratnakumar K, Frankfort BJ, Mardon G, Steller H, Mollereau B. 2004. Regulation of R7 and R8 differentiation by the spalt genes. Dev. Biol. 273, 121-133. ( 10.1016/j.ydbio.2004.05.026) [DOI] [PubMed] [Google Scholar]
- 32.Cook T, Pichaud F, Sonneville R, Papatsenko D, Desplan C. 2003. Distinction between color photoreceptor cell fates is controlled by Prospero in Drosophila. Dev. Cell 4, 853-864. ( 10.1016/s1534-5807(03)00156-4) [DOI] [PubMed] [Google Scholar]
- 33.Xie B, Charlton-Perkins M, McDonald E, Gebelein B, Cook T. 2007. Senseless functions as a molecular switch for color photoreceptor differentiation in Drosophila. Development 134, 4243-4253. ( 10.1242/dev.012781) [DOI] [PubMed] [Google Scholar]
- 34.Thanawala SU, et al. 2013. Regional modulation of a stochastically expressed factor determines photoreceptor subtypes in the Drosophila retina. Dev. Cell 25, 93-105. ( 10.1016/j.devcel.2013.02.016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wernet MF, Mazzoni EO, Çelik A, Duncan DM, Duncan I, Desplan C. 2006. Stochastic spineless expression creates the retinal mosaic for colour vision. Nature 440, 174-180. ( 10.1038/nature04615) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mikeladze-Dvali T, Wernet MF, Pistillo D, Mazzoni EO, Teleman AA, Chen YW, Cohen S, Desplan C. 2005. The growth regulators warts/lats and melted interact in a bistable loop to specify opposite fates in Drosophila R8 photoreceptors. Cell 122, 775-787. ( 10.1016/j.cell.2005.07.026) [DOI] [PubMed] [Google Scholar]
- 37.Johnston RJ Jr, et al. 2011. Interlocked feedforward loops control cell-type-specific rhodopsin expression in the Drosophila eye. Cell 145, 956-968. ( 10.1016/j.cell.2011.05.003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mazzoni EO, Celik A, Wernet MF, Vasiliauskas D, Johnston RJ, Cook TA, Pichaud F, Desplan C. 2008. Iroquois complex genes induce co-expression of rhodopsins in Drosophila. PLoS Biol. 6, e97. ( 10.1371/journal.pbio.0060097) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miller AC, Urban EA, Lyons EL, Herman TG, Johnston RJ Jr. 2021. Interdependent regulation of stereotyped and stochastic photoreceptor fates in the fly eye. Dev. Biol. 471, 89-96. ( 10.1016/j.ydbio.2020.12.008) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Papatsenko D, Nazina A, Desplan C. 2001. A conserved regulatory element present in all Drosophila rhodopsin genes mediates Pax6 functions and participates in the fine-tuning of cell-specific expression. Mech. Dev. 101, 143-153. ( 10.1016/s0925-4773(00)00581-5) [DOI] [PubMed] [Google Scholar]
- 41.Vasiliauskas D, Mazzoni EO, Sprecher SG, Brodetskiy K, Johnston RJ Jr, Lidder P, Vogt N, Celik A, Desplan C. 2011. Feedback from rhodopsin controls rhodopsin exclusion in Drosophila photoreceptors. Nature 479, 108-112. ( 10.1038/nature10451) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rister J, Razzaq A, Boodram P, Desai N, Tsanis C, Chen H, Jukam D, Desplan C. 2015. Single-base pair differences in a shared motif determine differential rhodopsin expression. Science 350, 1258-1261. ( 10.1126/science.aab3417) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Poupault C, Choi D, Lam-Kamath K, Dewett D, Razzaq A, Bunker J, Perry A, Cho I, Rister J. 2021. A combinatorial cis-regulatory logic restricts color-sensing rhodopsins to specific photoreceptor subsets in Drosophila. PLoS Genet. 17, e1009613. ( 10.1371/journal.pgen.1009613) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Baker LR, Weasner BM, Nagel A, Neuman SD, Bashirullah A, Kumar JP. 2018. Eyeless/Pax6 initiates eye formation non-autonomously from the peripodial epithelium. Development 145, dev163329. ( 10.1242/dev.163329) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Perry M, Kinoshita M, Saldi G, Huo L, Arikawa K, Desplan C. 2016. Molecular logic behind the three-way stochastic choices that expand butterfly colour vision. Nature 535, 280-284. ( 10.1038/nature18616) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhan Y, Alonso San Alberto D, Rusch C, Riffell JA, Montell C. 2021. Elimination of vision-guided target attraction in Aedes aegypti using CRISPR. Curr. Biol. 31, 4180-4187. ( 10.1016/j.cub.2021.07.003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen SP, et al. 2021. CRISPR/Cas9-mediated knockout of LW-opsin reduces the efficiency of phototaxis in the diamondback moth Plutella xylostella. Pest Manag. Sci. 77, 3519-3528. ( 10.1002/ps.6405) [DOI] [PubMed] [Google Scholar]
- 48.Xu X, Harvey-Samuel T, Yang J, Alphey L, You M. 2020. Ommochrome pathway genes kynurenine 3-hydroxylase and cardinal participate in eye pigmentation in Plutella xylostella. BMC Mol. Cell Biol. 21, 63. ( 10.1186/s12860-020-00308-8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Khan SA, Reichelt M, Heckel DG. 2017. Functional analysis of the ABCs of eye color in Helicoverpa armigera with CRISPR/Cas9-induced mutations. Sci. Rep. 7, 40025. ( 10.1038/srep40025) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Perera OP, Little NS, Pierce CA 3rd. 2018. CRISPR/Cas9 mediated high efficiency knockout of the eye color gene Vermillion in Helicoverpa zea (Boddie). PLoS ONE 13, e0197567. ( 10.1371/journal.pone.0197567) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xue WH, Xu N, Yuan XB, Chen HH, Zhang JL, Fu SJ, Zhang CX, Xu HJ. 2018. CRISPR/Cas9-mediated knockout of two eye pigmentation genes in the brown planthopper, Nilaparvata lugens (Hemiptera: Delphacidae). Insect Biochem. Mol. Biol. 93, 19-26. ( 10.1016/j.ibmb.2017.12.003) [DOI] [PubMed] [Google Scholar]
- 52.Hempel de Ibarra N, Vorobyev M, Menzel R. 2014. Mechanisms, functions and ecology of colour vision in the honeybee. J. Comp. Physiol. A Neuroethol. Sens. Neural Behav. Physiol. 200, 411-433. ( 10.1007/s00359-014-0915-1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Arias M, Elias M, Andraud C, Berthier S, Gomez D. 2020. Transparency improves concealment in cryptically coloured moths. J. Evol. Biol. 33, 247-252. ( 10.1111/jeb.13560) [DOI] [PubMed] [Google Scholar]
- 54.Hausmann AE, Kuo CY, Freire M, Rueda MN, Linares M, Pardo-Diaz C, Salazar C, Merrill RM. 2021. Light environment influences mating behaviours during the early stages of divergence in tropical butterflies. Proc. R. Soc. B 288, 20210157. ( 10.1098/rspb.2021.0157) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kinoshita M, Shimohigasshi M, Tominaga Y, Arikawa K, Homberg U. 2015. Topographically distinct visual and olfactory inputs to the mushroom body in the swallowtail butterfly, Papilio xuthus. J. Comp. Neurol. 523, 162-182. ( 10.1002/cne.23674) [DOI] [PubMed] [Google Scholar]
- 56.Briscoe AD. 2008. Reconstructing the ancestral butterfly eye: focus on the opsins. J. Exp. Biol. 211, 1805-1813. ( 10.1242/jeb.013045) [DOI] [PubMed] [Google Scholar]
- 57.McCulloch KJ, Macias-Muñoz A, Mortazavi A, Briscoe AD. 2022. Multiple mechanisms of photoreceptor spectral tuning in Heliconius butterflies. Mol. Biol. Evol. 39, msac067. ( 10.1093/molbev/msac067) [DOI]
- 58.Belušič G, Ilić M, Meglič A, Pirih P. 2021. Red-green opponency in the long visual fibre photoreceptors of brushfoot butterflies (Nymphalidae). Proc. R. Soc. B 288, 20211560. ( 10.1098/rspb.2021.1560) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.McCulloch KJ, Osorio D, Briscoe AD. 2016. Sexual dimorphism in the compound eye of Heliconius erato: a nymphalid butterfly with at least five spectral classes of photoreceptor. J. Exp. Biol. 219, 2377-2387. ( 10.1242/jeb.136523) [DOI] [PubMed] [Google Scholar]
- 60.McCulloch KJ, Yuan F, Zhen Y, Aardema ML, Smith G, Llorente-Bousquets J, Andolfatto P, Briscoe AD. 2017. Sexual dimorphism and retinal mosaic diversification following the evolution of a violet receptor in butterflies. Mol. Biol. Evol. 34, 2271-2284. ( 10.1093/molbev/msx163) [DOI] [PubMed] [Google Scholar]
- 61.Mahato S, Morita S, Tucker AE, Liang X, Jackowska M, Friedrich M, Shiga Y, Zelhof AC. 2014. Common transcriptional mechanisms for visual photoreceptor cell differentiation among Pancrustaceans. PLoS Genet. 10, e1004484. ( 10.1371/journal.pgen.1004484) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liang X, Mahato S, Hemmerich C, Zelhof AC. 2016. Two temporal functions of glass: ommatidium patterning and photoreceptor differentiation. Dev. Biol. 414, 4-20. ( 10.1016/j.ydbio.2016.04.012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zattara EE, Macagno ALM, Busey HA, Moczek AP. 2017. Development of functional ectopic compound eyes in scarabaeid beetles by knockdown of orthodenticle. Proc. Natl Acad. Sci. USA 114, 12 021-12 026. ( 10.1073/pnas.1714895114) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Leschen RAB, Beutel RG. 2004. Ocellar atavism in Coleoptera: plesiomorphy or apomorphy? J. Zool. Syst. Evol. Res. 42, 63-69. ( 10.1046/j.0947-5745.2003.00241.x) [DOI] [Google Scholar]
- 65.Singh A, Kango-Singh M. 2020. Molecular genetics of axial patterning, growth and disease in the Drosophila eye. Switzerland: Springer Nature. ( 10.1007/978-3-030-42246-2) [DOI] [Google Scholar]
- 66.Buschbeck EK. 2014. Escaping compound eye ancestry: the evolution of single-chamber eyes in holometabolous larvae. J. Exp. Biol. 217, 2818-2824. ( 10.1242/jeb.085365) [DOI] [PubMed] [Google Scholar]
- 67.Sprecher SG, Pichaud F, Desplan C. 2007. Adult and larval photoreceptors use different mechanisms to specify the same rhodopsin fates. Genes Dev. 21, 2182-2195. ( 10.1101/gad.1565407) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Daniel A, Dumstrei K, Lengyel JA, Hartenstein V. 1999. The control of cell fate in the embryonic visual system by atonal, tailless and EGFR signaling. Development 126, 2945-2954. ( 10.1242/dev.126.13.2945) [DOI] [PubMed] [Google Scholar]
- 69.Sprecher SG, Desplan C. 2008. Switch of rhodopsin expression in terminally differentiated Drosophila sensory neurons. Nature 454, 533-537. ( 10.1038/nature07062) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Helfrich-Förster C, Edwards T, Yasuyama K, Wisotzki B, Schneuwly S, Stanewsky R, Meinertzhagen IA, Hofbauer A. 2002. The extraretinal eyelet of Drosophila: development, ultrastructure, and putative circadian function. J. Neurosci. 22, 9255-9266. ( 10.1523/JNEUROSCI.22-21-09255.2002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mishra AK, Tsachaki M, Rister J, Ng J, Celik A, Sprecher SG. 2013. Binary cell fate decisions and fate transformation in the Drosophila larval eye. PLoS Genet. 9, e1004027. ( 10.1371/journal.pgen.1004027) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Briscoe AD, White RH. 2005. Adult stemmata of the butterfly Vanessa cardui express UV and green opsin mRNAs. Cell Tissue Res. 319, 175-179. ( 10.1007/s00441-004-0994-3) [DOI] [PubMed] [Google Scholar]
- 73.Yoon CS, Hirosawa K, Suzuki E. 1996. Studies on the structure of ocellar photoreceptor cells of Drosophila melanogaster with special reference to subrhabdomeric cisternae. Cell Tissue Res. 284, 77-85. ( 10.1007/s004410050568) [DOI] [PubMed] [Google Scholar]
- 74.Paulus HF. 2000. Phylogeny of the Myriapoda – Crustacea – Insecta: a new attempt using photoreceptor structure. J. Zool. Syst. Evol. Res. 38, 189-208. ( 10.1046/j.1439-0469.2000.383152.x) [DOI] [Google Scholar]
- 75.Friedrich M. 2006. Continuity versus split and reconstitution: exploring the molecular developmental corollaries of insect eye primordium evolution. Dev. Biol. 299, 310-329. ( 10.1016/j.ydbio.2006.08.027) [DOI] [PubMed] [Google Scholar]
- 76.Friedrich M. 2008. Opsins and cell fate in the Drosophila Bolwig organ: tricky lessons in homology inference. Bioessays 30, 980-993. ( 10.1002/bies.20803) [DOI] [PubMed] [Google Scholar]
- 77.Mishra AK, Fritsch C, Voutev R, Mann RS, Sprecher SG. 2021. Homothorax controls a binary rhodopsin switch in Drosophila ocelli. PLoS Genet. 17, e1009460. ( 10.1371/journal.pgen.1009460) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Henze MJ, Dannenhauer K, Kohler M, Labhart T, Gesemann M. 2012. Opsin evolution and expression in arthropod compound eyes and ocelli: insights from the cricket Gryllus bimaculatus. BMC Evol. Biol. 12, 163. ( 10.1186/1471-2148-12-163) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Velarde RA, Sauer CD, Walden KKO, Fahrbach SE, Robertson HM. 2005. Pteropsin: a vertebrate-like non-visual opsin expressed in the honey bee brain. Insect Biochem. Mol. Biol. 35, 1367-1377. ( 10.1016/j.ibmb.2005.09.001) [DOI] [PubMed] [Google Scholar]
- 80.Spaethe J, Briscoe AD. 2005. Molecular characterization and expression of the UV opsin in bumblebees: three ommatidial subtypes in the retina and a new photoreceptor organ in the lamina. J. Exp. Biol. 208, 2347-2361. ( 10.1242/jeb.01634) [DOI] [PubMed] [Google Scholar]
- 81.Hu X, Whaley MA, Stein MM, Mitchell BE, O'Tousa JE. 2011. Coexpression of spectrally distinct rhodopsins in Aedes aegypti R7 photoreceptors. PLoS ONE 6, e23121. ( 10.1371/journal.pone.0023121) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hu X, Leming MT, Whaley MA, O'Tousa JE. 2014. Rhodopsin coexpression in UV photoreceptors of Aedes aegypti and Anopheles gambiae mosquitoes. J. Exp. Biol. 217, 1003-1008. ( 10.1242/jeb.096347) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Dyer AG, Neumeyer C. 2005. Simultaneous and successive colour discrimination in the honeybee (Apis mellifera). J. Comp. Physiol. A Neuroethol. Sens. Neural Behav. Physiol. 191, 547-557. ( 10.1007/s00359-005-0622-z) [DOI] [PubMed] [Google Scholar]
- 84.Autrum H, et al. 1979. Comparative physiology and evolution of vision in invertebrates. A: invertebrate photoreceptors. Berlin, Germany: Springer. ( 10.1007/978-3-642-66999-6) [DOI] [Google Scholar]
- 85.van der Kooi CJ, Stavenga DG, Arikawa K, Belušič G, Kelber A. 2021. Evolution of insect color vision: from spectral sensitivity to visual ecology. Annu. Rev. Entomol. 66, 435-461. ( 10.1146/annurev-ento-061720-071644) [DOI] [PubMed] [Google Scholar]
- 86.Koshitaka H, Kinoshita M, Vorobyev M, Arikawa K. 2008. Tetrachromacy in a butterfly that has eight varieties of spectral receptors. Proc. R. Soc. B 275, 947-954. ( 10.1098/rspb.2007.1614) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wakakuwa M, Terakita A, Koyanagi M, Stavenga DG, Shichida Y, Arikawa K. 2010. Evolution and mechanism of spectral tuning of blue-absorbing visual pigments in butterflies. PLoS ONE 5, e15015. ( 10.1371/journal.pone.0015015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zaccardi G, Kelber A, Sison-Mangus MP, Briscoe AD. 2006. Color discrimination in the red range with only one long-wavelength sensitive opsin. J. Exp. Biol. 209, 1944-1955. ( 10.1242/jeb.02207) [DOI] [PubMed] [Google Scholar]
- 89.Finkbeiner SD, Briscoe AD. 2021. True UV color vision in a female butterfly with two UV opsins. J. Exp. Biol. 224, jeb.242802. ( 10.1242/jeb.242802) [DOI] [PubMed] [Google Scholar]
- 90.Briscoe AD, Bybee SM, Bernard GD, Yuan F, Sison-Mangus MP, Reed RD, Warren AD, Llorente-Bousquets J, Chiao CC. 2010. Positive selection of a duplicated UV-sensitive visual pigment coincides with wing pigment evolution in Heliconius butterflies. Proc. Natl Acad. Sci. USA 107, 3628-3633. ( 10.1073/pnas.0910085107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kuenzinger W, Kelber A, Weesner J, Travis J, Raguso RA, Goyret J. 2019. Innate colour preferences of a hawkmoth depend on visual context. Biol. Lett. 15, 20180886. ( 10.1098/rsbl.2018.0886) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Dahake A, Stöckl AL, Foster JJ, Sane SP, Kelber A. 2018. The roles of vision and antennal mechanoreception in hawkmoth flight control. Elife 7, e37606. ( 10.7554/eLife.37606) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Jackowska M, Bao R, Liu Z, McDonald EC, Cook TA, Friedrich M. 2007. Genomic and gene regulatory signatures of cryptozoic adaptation: loss of blue sensitive photoreceptors through expansion of long wavelength-opsin expression in the red flour beetle Tribolium castaneum. Front. Zool. 4, 24. ( 10.1186/1742-9994-4-24) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Vishnevskaia TM, Cherkasov AD, Shura-Bura TM. 1986. Spectral sensitivity of photoreceptors of the compound eye of the locust. Neurophysiology 18, 69-76. ( 10.1007/bf01052492) [DOI] [PubMed] [Google Scholar]
- 95.Mazzoni EO, Desplan C, Celik A. 2004. ‘One receptor’ rules in sensory neurons. Dev. Neurosci. 26, 388-395. ( 10.1159/000082281) [DOI] [PubMed] [Google Scholar]
- 96.Arikawa K, Mizuno S, Kinoshita M, Stavenga DG. 2003. Coexpression of two visual pigments in a photoreceptor causes an abnormally broad spectral sensitivity in the eye of the butterfly Papilio xuthus. J. Neurosci. 23, 4527-4532. ( 10.1523/JNEUROSCI.23-11-04527.2003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Schnaitmann C, Garbers C, Wachtler T, Tanimoto H. 2013. Color discrimination with broadband photoreceptors. Curr. Biol. 23, 2375-2382. ( 10.1016/j.cub.2013.10.037) [DOI] [PubMed] [Google Scholar]
- 98.Sison-Mangus MP, Bernard GD, Lampel J, Briscoe AD. 2006. Beauty in the eye of the beholder: the two blue opsins of lycaenid butterflies and the opsin gene-driven evolution of sexually dimorphic eyes. J. Exp. Biol. 209, 3079-3090. ( 10.1242/jeb.02360) [DOI] [PubMed] [Google Scholar]
- 99.Ogawa Y, Awata H, Wakakuwa M, Kinoshita M, Stavenga DG, Arikawa K. 2012. Coexpression of three middle wavelength-absorbing visual pigments in sexually dimorphic photoreceptors of the butterfly Colias erate. J. Comp. Physiol. A Neuroethol. Sens. Neural Behav. Physiol. 198, 857-867. ( 10.1007/s00359-012-0756-8) [DOI] [PubMed] [Google Scholar]
- 100.Schmeling F, Wakakuwa M, Tegtmeier J, Kinoshita M, Bockhorst T, Arikawa K, Homberg U. 2014. Opsin expression, physiological characterization and identification of photoreceptor cells in the dorsal rim area and main retina of the desert locust, Schistocerca gregaria. J. Exp. Biol. 217, 3557-3568. ( 10.1242/jeb.108514) [DOI] [PubMed] [Google Scholar]
- 101.Macias-Muñoz A, Rangel Olguin AG, Briscoe AD. 2019. Evolution of phototransduction genes in Lepidoptera. Genome Biol. Evol. 11, 2107-2124. ( 10.1093/gbe/evz150) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Sadier A, et al. 2018. Multifactorial processes underlie parallel opsin loss in neotropical bats. Elife 7, e37412. ( 10.7554/eLife.37412) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Arikawa K, Iwanaga T, Wakakuwa M, Kinoshita M. 2017. Unique temporal expression of triplicated long-wavelength opsins in developing butterfly eyes. Front. Neural Circuits 11, 96. ( 10.3389/fncir.2017.00096) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Cortesi F, et al. 2015. Ancestral duplications and highly dynamic opsin gene evolution in percomorph fishes. Proc. Natl Acad. Sci. USA 112, 1493-1498. ( 10.1073/pnas.1417803112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Schott RK, Refvik SP, Hauser FE, López-Fernández H, Chang BSW. 2014. Divergent positive selection in rhodopsin from lake and riverine cichlid fishes. Mol. Biol. Evol. 31, 1149-1165. ( 10.1093/molbev/msu064) [DOI] [PubMed] [Google Scholar]
- 106.Suvorov A, Jensen NO, Sharkey CR, Fujimoto MS, Bodily P, Wightman HMC, Ogden TH, Clement MJ, Bybee SM. 2017. Opsins have evolved under the permanent heterozygote model: insights from phylotranscriptomics of Odonata. Mol. Ecol. 26, 1306-1322. ( 10.1111/mec.13884) [DOI] [PubMed] [Google Scholar]
- 107.Koyanagi M, Nagata T, Katoh K, Yamashita S, Tokunaga F. 2008. Molecular evolution of arthropod color vision deduced from multiple opsin genes of jumping spiders. J. Mol. Evol. 66, 130-137. ( 10.1007/s00239-008-9065-9) [DOI] [PubMed] [Google Scholar]
- 108.Eriksson BJ, Fredman D, Steiner G, Schmid A. 2013. Characterisation and localisation of the opsin protein repertoire in the brain and retinas of a spider and an onychophoran. BMC Evol. Biol. 13, 186. ( 10.1186/1471-2148-13-186) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Feuda R, et al. 2021. Phylogenomics of opsin genes in Diptera reveals lineage-specific events and contrasting evolutionary dynamics in Anopheles and Drosophila. Genome Biol. Evol. 13, evab170. ( 10.1093/gbe/evab170). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Armisén D, et al. 2018. The genome of the water strider Gerris buenoi reveals expansions of gene repertoires associated with adaptations to life on the water. BMC Genomics 19, 832. ( 10.1186/s12864-018-5163-2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Feuda R, Marle F, Bentley MA, Holland PWH. 2016. Conservation, duplication, and divergence of five opsin genes in insect evolution. Genome Biol. Evol. 8, 579-587. ( 10.5287/bod-leian) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Almudi I, et al. 2020. Genomic adaptations to aquatic and aerial life in mayflies and the origin of insect wings. Nat. Commun. 11, 2631. ( 10.1038/s41467-020-16284-8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Sharkey CR, Fujimoto MS, Lord NP, Shin S, McKenna DD, Suvorov A, Martin GJ, Bybee SM. 2017. Overcoming the loss of blue sensitivity through opsin duplication in the largest animal group, beetles. Sci. Rep. 7, 8. ( 10.1038/s41598-017-00061-7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Henze MJ, Oakley TH. 2015. The dynamic evolutionary history of pancrustacean eyes and opsins. Integr. Comp. Biol. 55, 830-842. ( 10.1093/icb/icv100) [DOI] [PubMed] [Google Scholar]
- 115.Briscoe AD. 2000. Six opsins from the butterfly Papilio glaucus: molecular phylogenetic evidence for paralogous origins of red-sensitive visual pigments in insects. J. Mol. Evol. 51, 110-121. ( 10.1007/s002390010071) [DOI] [PubMed] [Google Scholar]
- 116.Xu P, Lu B, Chao J, Holdbrook R, Liang G, Lu Y. 2021. The evolution of opsin genes in five species of mirid bugs: duplication of long-wavelength opsins and loss of blue-sensitive opsins. BMC Ecol. Evol. 21, 66. ( 10.1186/s12862-021-01799-5) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Briscoe AD. 1998. Molecular diversity of visual pigments in the butterfly Papilio glaucus. Naturwissenschaften 85, 33-35. ( 10.1007/s001140050448) [DOI] [PubMed] [Google Scholar]
- 118.Kelber A. 1999. Ovipositing butterflies use a red receptor to see green. J. Exp. Biol. 202, 2619-2630. ( 10.1242/jeb.202.19.2619) [DOI] [PubMed] [Google Scholar]
- 119.Spaethe J, Briscoe AD. 2004. Early duplication and functional diversification of the opsin gene family in insects. Mol. Biol. Evol. 21, 1583-1594. ( 10.1093/molbev/msh162) [DOI] [PubMed] [Google Scholar]
- 120.Wang B, Xiao JH, Bian SN, Niu LM, Murphy RW, Huang DW. 2013. Evolution and expression plasticity of opsin genes in a fig pollinator, Ceratosolen solmsi. PLoS ONE 8, 1-11. ( 10.1371/journal.pone.0053907) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Zhang SF, Kong XB, Wang HB, Zhou G, Yu JX, Liu F, Zhang Z. 2016. Sensory and immune genes identification and analysis in a widely used parasitoid wasp Trichogramma dendrolimi (Hymenoptera: Trichogrammatidae). Insect Sci. 23, 417-429. ( 10.1111/1744-7917.12330) [DOI] [PubMed] [Google Scholar]
- 122.Giraldo-Calderón GI, Zanis MJ, Hill CA. 2017. Retention of duplicated long-wavelength opsins in mosquito lineages by positive selection and differential expression. BMC Evol. Biol. 17, 84. ( 10.1186/s12862-017-0910-6) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Zhang J. 2003. Evolution by gene duplication: an update. Trends Ecol. Evol. 18, 292-298. ( 10.1016/S0169-5347(03)00033-8) [DOI] [Google Scholar]
- 124.Xu P, Feuda R, Lu B, Xiao H, Graham RI, Wu K. 2016. Functional opsin retrogene in nocturnal moth. Mob. DNA 7, 18. ( 10.1186/s13100-016-0074-8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Arikawa K, Wakakuwa M, Qiu X, Kurasawa M, Stavenga DG. 2005. Sexual dimorphism of short-wavelength photoreceptors in the small white butterfly, Pieris rapae crucivora. J. Neurosci. 25, 5935-5942. ( 10.1523/JNEUROSCI.1364-05.2005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Menzel JG, Wunderer H, Stavenga DG. 1991. Functional morphology of the divided compound eye of the honeybee drone (Apis mellifera). Tissue Cell 23, 525-535. ( 10.1016/0040-8166(91)90010-q) [DOI] [PubMed] [Google Scholar]
- 127.Lichtenstein L, Grübel K, Spaethe J. 2018. Opsin expression patterns coincide with photoreceptor development during pupal development in the honey bee, Apis mellifera. BMC Dev. Biol. 18, 1-11. ( 10.1186/s12861-018-0162-8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Labhart T, Meyer EP. 1999. Detectors for polarized skylight in insects: a survey of ommatidial specializations in the dorsal rim area of the compound eye. Microsc. Res. Tech. 47, 368-379. () [DOI] [PubMed] [Google Scholar]
- 129.Roberts NW, Porter ML, Cronin TW. 2011. The molecular basis of mechanisms underlying polarization vision. Phil. Trans. R. Soc. B 366, 627-637. ( 10.1098/rstb.2010.0206) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Wernet MF, Labhart T, Baumann F, Mazzoni EO, Pichaud F, Desplan C. 2003. Homothorax switches function of Drosophila photoreceptors from color to polarized light sensors. Cell 115, 267-279. ( 10.1016/s0092-8674(03)00848-1) [DOI] [PubMed] [Google Scholar]
- 131.Labhart T, Hodel B, Valenzuela I. 1984. The physiology of the cricket's compound eye with particular reference to the anatomically specialized dorsal rim area. J. Comp. Physiol. A 155, 289-296. ( 10.1007/BF00610582) [DOI] [Google Scholar]
- 132.Herzmann D, Labhart T. 1989. Spectral sensitivity and absolute threshold of polarization vision in crickets: a behavioral study. J. Comp. Physiol. A Neuroethol. Sens. Neural Behav. Physiol. 165, 315-319. ( 10.1007/bf00619350) [DOI] [PubMed] [Google Scholar]
- 133.Sison-Mangus MP, Briscoe AD, Zaccardi G, Knuttel H, Kelber A. 2008. The lycaenid butterfly Polyommatus icarus uses a duplicated blue opsin to see green. J. Exp. Biol. 211, 361-369. ( 10.1242/jeb.012617) [DOI] [PubMed] [Google Scholar]
- 134.Kelber A, Thunell C, Arikawa K. 2001. Polarisation-dependent colour vision in Papilio butterflies. J. Exp. Biol. 204, 2469-2480. ( 10.1242/jeb.204.14.2469) [DOI] [PubMed] [Google Scholar]
- 135.Kelber A. 1999. Why ‘false’ colours are seen by butterflies. Nature 402, 251. ( 10.1038/46204) [DOI] [PubMed] [Google Scholar]
- 136.Collantes-Alegre JM, Mattenberger F, Barberà M, Martínez-Torres D. 2018. Characterisation, analysis of expression and localisation of the opsin gene repertoire from the perspective of photoperiodism in the aphid Acyrthosiphon pisum. J. Insect Physiol. 104, 48-59. ( 10.1016/j.jinsphys.2017.11.009) [DOI] [PubMed] [Google Scholar]
- 137.Liu Z, Wang X, Lei C, Zhu F. 2017. Sensory genes identification with head transcriptome of the migratory armyworm, Mythimna separata. Sci. Rep. 7, 46033. ( 10.1038/srep46033) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Dungan SZ, Kosyakov A, Chang BSW. 2016. Spectral tuning of killer whale (Orcinus orca) rhodopsin: evidence for positive selection and functional adaptation in a cetacean visual pigment. Mol. Biol. Evol. 33, 323-336. ( 10.1093/molbev/msv217) [DOI] [PubMed] [Google Scholar]
- 139.Bhattacharyya N, Darren B, Schott RK, Tropepe V, Chang BSW. 2017. Cone-like rhodopsin expressed in the all-cone retina of the colubrid pine snake as a potential adaptation to diurnality. J. Exp. Biol. 220, 2418-2425. ( 10.1242/jeb.156430) [DOI] [PubMed] [Google Scholar]
- 140.van Hazel I, Dungan SZ, Hauser FE, Morrow JM, Endler JA, Chang BSW. 2016. A comparative study of rhodopsin function in the great bowerbird (Ptilonorhynchus nuchalis): spectral tuning and light-activated kinetics. Protein Sci. 25, 1308-1318. ( 10.1002/pro.2902) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Frentiu FD, Yuan F, Savage WK, Bernard GD, Mullen SP, Briscoe AD. 2015. Opsin clines in butterflies suggest novel roles for insect photopigments. Mol. Biol. Evol. 32, 368-379. ( 10.1093/molbev/msu304) [DOI] [PubMed] [Google Scholar]
- 142.Terakita A, Tsukamoto H, Koyanagi M, Sugahara M, Yamashita T, Shichida Y. 2008. Expression and comparative characterization of Gq-coupled invertebrate visual pigments and melanopsin. J. Neurochem. 105, 883-890. ( 10.1111/j.1471-4159.2007.05184.x) [DOI] [PubMed] [Google Scholar]
- 143.Camlitepe Y, Aksoy V. 2010. First evidence of fine colour discrimination ability in ants (Hymenoptera, Formicidae). J. Exp. Biol. 213, 72-77. ( 10.1242/jeb.037853) [DOI] [PubMed] [Google Scholar]
- 144.Aksoy V, Camlitepe Y. 2012. Behavioural analysis of chromatic and achromatic vision in the ant Formica cunicularia (Hymenoptera: Formicidae). Vision Res. 67, 28-36. ( 10.1016/j.visres.2012.06.013) [DOI] [PubMed] [Google Scholar]
- 145.Yilmaz A, Dyer AG, Rössler W, Spaethe J. 2017. Innate colour preference, individual learning and memory retention in the ant Camponotus blandus. J. Exp. Biol. 220, 3315-3326. ( 10.1242/jeb.158501) [DOI] [PubMed] [Google Scholar]
- 146.Kondrashov FA. 2012. Gene duplication as a mechanism of genomic adaptation to a changing environment. Proc. R. Soc. B 279, 5048-5057. ( 10.1098/rspb.2012.1108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Sander SE, Hall DW. 2015. Variation in opsin genes correlates with signalling ecology in North American fireflies. Mol. Ecol. 24, 4679-4696. ( 10.1111/mec.13346) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Tierney SM, Friedrich M, Humphreys WF, Jones TM, Warrant EJ, Wcislo WT. 2017. Consequences of evolutionary transitions in changing photic environments. Austral. Entomol. 56, 23-46. ( 10.1111/aen.12264) [DOI] [Google Scholar]
- 149.Sasagawa H, Narita R, Kitagawa Y, Kadowaki T. 2003. The expression of genes encoding visual components is regulated by a circadian clock, light environment and age in the honeybee (Apis mellifera). Eur. J. Neurosci. 17, 963-970. ( 10.1046/j.1460-9568.2003.02528.x) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This article has no additional data.