Abstract
Phosphoglucosamine mutase (GlmM) catalyzes the formation of glucosamine-1-phosphate from glucosamine-6-phosphate, an essential step in the pathway for UDP-N-acetylglucosamine biosynthesis in bacteria. This enzyme must be phosphorylated to be active and acts according to a ping-pong mechanism involving glucosamine-1,6-diphosphate as an intermediate (L. Jolly, P. Ferrari, D. Blanot, J. van Heijenoort, F. Fassy, and D. Mengin-Lecreulx, Eur. J. Biochem. 262:202–210, 1999). However, the process by which the initial phosphorylation of the enzyme is achieved in vivo remains unknown. Here we show that the phosphoglucosamine mutase from Escherichia coli autophosphorylates in vitro in the presence of [32P]ATP. The same is observed with phosphoglucosamine mutases from other bacterial species, yeast N-acetylglucosamine-phosphate mutase, and rabbit muscle phosphoglucomutase. Labeling of the E. coli GlmM enzyme with [32P]ATP requires the presence of a divalent cation, and the label is subsequently lost when the enzyme is incubated with either of its substrates. Analysis of enzyme phosphorylation by high-pressure liquid chromatography and coupled mass spectrometry confirms that only one phosphate has been covalently linked to the enzyme. Only phosphoserine could be detected after acid hydrolysis of the labeled protein, and site-directed mutagenesis of serine residues located in or near the active site identifies the serine residue at position 102 as the site of autophosphorylation of E. coli GlmM.
In bacteria, UDP-N-acetylglucosamine (UDP-GlcNAc) is the precursor of essential cell envelope components, namely, peptidoglycan, lipopolysaccharides, and teichoic acids (17, 23, 33, 38, 39). Its formation from fructose-6-phosphate requires the successive actions of three enzymes, the glmS, glmM, and glmU gene products, respectively (12, 15, 28–30, 40). As expected, the inhibition of any of these enzymes in vivo results in dramatical morphological changes and subsequent cell lysis (28, 30, 36, 42). As these enzymes represent potential targets in the search for new antibacterial compounds, increased attention has recently been paid to their reaction mechanism and structure (2, 20, 32).
Phosphoglucosamine mutase (GlmM) catalyzes the interconversion of glucosamine-6-phosphate (GlcN-6-P) and GlcN-1-P isomers. It has been characterized for the first time in Escherichia coli, and the corresponding gene glmM has been identified in the 71-min region of the chromosome (9, 30). Phosphoglucosamine mutases from other bacterial species have now been identified (11, 21), and this family is rapidly growing due in particular to the completion of various genome sequencing projects. Most interestingly, the pure E. coli enzyme has been shown to be active only in its phosphorylated form (20, 30). It is thus tempting to speculate that enzyme phosphorylation could be a factor regulating the flow of metabolites in this pathway.
The interconversion of GlcN-6-P and GlcN-1-P isomers catalyzed by GlmM occurs by a two-step ping-pong reaction mechanism (20) in which GlcN-1,6-diP acts as both the first product and the second substrate:GlcN-6-P + phosphorylated enzyme ⇓⇑GlcN-1,6-diP + dephosphorylated enzyme ⇓⇑GlcN-1-P + phosphorylated enzyme
As observed previously with the more extensively studied phosphoglucomutases (PGM) and phosphomannomutases (PMM) (8, 22, 34), the GlmM enzyme requires only the substrate diphosphate (GlcN-1,6-diP) as a cofactor to remain in an active phosphorylated form (20, 30). Furthermore, the amino acid sequences of all members of this class of hexosephosphate mutases contain the particular motif (GA)(LIVM)X(LIVM)(ST)(PGA)S*HXPX4(GN) (S* represents the phosphorylated residue involved in the catalytic process) found in the PROSITE database and considered their specific signature. This sequence appears as GIVISAS*HNPFYDNG in the E. coli GlmM enzyme, with the putative active-site serine being located at the position 102 in the 444-amino-acid polypeptide (30). Using site-directed mutagenesis, we have recently confirmed that the serine S102 is essential for catalysis and is the site of phosphorylation (20).
The phosphoglucosamine mutase is synthesized in an inactive, dephosphorylated form (30). How this enzyme is then activated (phosphorylated) in vivo is not known. In fact, this appears as a general problem which surprisingly has never been investigated in the case of previously characterized hexosephosphate mutases. At least two different routes can be proposed for this initial phosphorylation (6): (i) a kinase-dependent phosphorylation (or an autophosphorylation) of the mutase, with a nucleoside triphosphate as phosphoryl group donor, or (ii) a phosphorylation by GlcN-1,6-diP, which is also the reaction intermediate, a hypothesis implying the presence in E. coli cells of a specific enzyme catalyzing the formation of the diphosphate compound. We here show that purified E. coli phosphoglucosamine mutase is capable of phosphorylating itself in vitro in the presence of ATP and that the site of phosphorylation is active-site serine residue S102.
MATERIALS AND METHODS
Materials.
DNA restriction and modification enzymes and synthetic oligonucleotides were obtained from Eurogentec or New England Biolabs. PCR amplification of DNA was performed in a Thermocycler 60 apparatus (Bio-med) using Taq polymerase from Appligene. DNA fragments were purified with a Wizard purification system from Promega, and DNA sequencing was performed using a T7 sequencing kit from Pharmacia. [α-35S]dATP (37 TBq/mmol), [γ-32P]ATP (110 TBq/mmol), and [14C]ATP (1.9 GBq/mmol) were bought from Amersham. GlcN-6-P, GlcN-1-P, Glc-1,6-diP, phosphoamino acids, and rabbit PGM were bought from Sigma.
Bacterial strains, plasmid vectors, and growth conditions.
E. coli strains JM83 (ara Δ[lac-proAB] rpsL thi φ80 dlacZΔM15) (43) and GPM83 (JM83 glmM::kan [pGMM]) (30) were used as hosts for plasmids and for the preparation of the overproduced wild-type and mutant GlmM enzymes. The mutant strain GPM83 carries an inactivated copy of the glmM gene on the chromosome and a wild-type copy of glmM on a plasmid (pGMM) whose replication is thermosensitive. Strain BMH71-18 mutS, defective in mismatch repair, was used in site-directed mutagenesis experiments (10). The pTrc99A plasmid vector was from Pharmacia; its pTrcHis60 derivative was described recently (32). 2YT (31) was used as a rich medium, and growth was monitored at 600 nm with a Shimadzu UV-1601 spectrophotometer. For strains carrying drug resistance genes, antibiotics were used at concentrations of 100 (ampicillin), 35 (kanamycin), and 25 (chloramphenicol) μg/ml.
General DNA techniques and E. coli cell transformation.
Small- and large-scale plasmid isolations were carried out by the alkaline lysis method, and standard procedures for endonuclease digestions, ligation, and agarose electrophoresis were used (35). E. coli cells were made competent and transformed with plasmid DNA by the method of Dagert and Ehrlich (7) or by electroporation. Competent cells of the thermosensitive mutant GPM83 were used to test the different plasmids for functional complementation as reported previously (30).
Construction of plasmids and site-directed mutagenesis.
A plasmid suitable for high-level overproduction of wild-type GlmM was constructed as follows. PCR primers were designed to incorporate a BspHI site (in boldface) 5′ to the initiation codon (underlined) of glmM (5′-AAACGTCATGAGTAATCGTAAATATTTC-3′) and a PstI site (in boldface) 3′ to the gene after the stop codon (5′-TTATCTGCAGCTTTAAACGGCTTTTACTGC-3′). These primers were used to amplify the glmM gene from E. coli chromosome; the resulting material was treated with BspHI and PstI and was ligated between the compatible NcoI and PstI sites of vector pTrc99A. The ligation mixture was then used to transform by electroporation strain GPM83, and clones were selected for both ampicillin resistance and growth at 42°C. All transformants isolated in this way carried the expected plasmid named pMLJ1, allowing expression of the GlmM enzyme under the control of the strong IPTG (isopropyl-β-d-thiogalactopyranoside)-inducible trc promoter. Plasmid pMLJ4 allowing expression of the enzyme under a C-terminal His6-tagged form was described recently (20). Plasmids pMLJ5 and pMLJ7 for expression of the S100A and S102A mutant GlmM enzymes were constructed recently by site-directed mutagenesis of plasmid pMLJ4 (20), using the method of Deng and Nickoloff (10). Plasmids pMLJ8 and pMLJ9 for expression of the S327A and S413A mutant enzymes (His6 tagged) were constructed by the same procedure, using 5′-ATCGGTGCAGAGAATGCCGGTCATGTGATC-3′ and 5′-GTGTTGCTGCGTAAAGCCGGCAACGAACCG-3′, respectively, as mutagenesis oligonucleotides. For amplification of the yeast AGM1 gene encoding N-acetylglucosamine-phosphate mutase (19), PCR primers were designed to incorporate a BspHI site (in boldface) 5′ to the initiation codon (underlined) of the gene (5′-GAGATCATGAAGGTTGATTACGAG-3′) on the forward primer and a BamHI site (in boldface) 3′ to the end of the gene without its stop codon (5′-TAATGGGATCCAGCAGATGCCTTAACGTGCTCC-3′) on the reverse primer. After amplification from the yeast genome, the DNA was treated with BspHI and BamHI, and the resulting fragment was ligated into the compatible NcoI and BglII sites of the expression vector pTrcHis60 (32). The resulting plasmid, pMLD133, allowed expression of the Agm1p enzyme (C-terminal His6-tagged form) under control of the trc promoter. Plasmids pMLJ2 (21) and pMLD106 (11) for expression of the glmM gene products from Staphylococcus aureus and Helicobacter pylori, respectively, were previously described.
Preparation of crude protein extracts and enzyme purification.
E. coli cells (JM83 or JM83 glmM::kan) carrying plasmids described in this work were grown exponentially at 37°C in 2YT-ampicillin medium (0.5-liter cultures). When the optical density (OD) of the culture reached 0.1, IPTG was added at a final concentration of 1 mM, and growth was continued for 2 to 3 h (final OD = 1). Harvested cells were disrupted by sonication, and crude protein extracts (5 ml, 10 to 12 mg of protein/ml) were prepared as described previously (30). Wild-type GlmM protein was purified as previously described (30). The different His6-tagged proteins (GlmM and Agm1p) were purified on Ni2+-nitrilotriacetate-agarose as reported recently (20). Sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) analysis of proteins was performed with 12% polyacrylamide gels (24). Protein concentration was determined by the method of Bradford (1), using bovine serum albumin as a standard.
Phosphoglucosamine mutase assay.
The specific activities of wild-type and mutant enzymes reported in this work were determined by the coupled assay (6-P to 1-P) in which GlcN-1-P synthesized from GlcN-6-P by the mutase is quantitatively converted into UDP-GlcNAc by the pure bifunctional GlmU enzyme (20).
In vitro protein phosphorylation.
Unless otherwise noted, protein extracts to be assayed were dialyzed against 25 mM HEPES buffer containing 0.1% β-mercaptoethanol and 0.5 mM MgCl2. Crude protein extract (soluble fraction, 100 to 150 μg of protein) or purified protein (2 μg) was incubated for 30 min at 37°C in a reaction mixture (25 μl) containing 25 mM HEPES buffer (pH 7.3), 5 mM MgCl2, 1 mM dithiothreitol, 1 mM EDTA, and 50 μM [γ-32P]ATP (100 kBq) (14); 25 μl of a buffer containing 125 mM Tris-HCl (pH 6.8), 4% SDS, 1% β-mercaptoethanol, and 20% (vol/vol) glycerol was added, and the mixture was heated for 5 min at 100°C. Proteins were separated by one-dimensional gel electrophoresis. After migration, gels were treated for 15 min with 16% trichloroacetic acid (TCA) at 90°C to eliminate phosphate-containing contaminants and then stained with Coomassie blue R250 in 30% methanol–10% acetic acid and dried. Labeled proteins were visualized by overnight autoradiography at −80°C using BioMax MS films and BioMax Transcreen (Kodak). The radioactive bands were excised, and radioactivity was counted in a Betamatic IV liquid scintillation spectrophotometer (Kontron) with a solvent system consisting of 2 ml of water and 13 ml of Aqualyte mixture (J. T. Baker Chemicals, Deventer, The Netherlands).
Analysis of phosphorylated amino acids.
Crude protein extracts or purified proteins were incubated with [γ-32P]ATP as described above. To eliminate the excess of radioactive ATP and by-products such as phosphate, the phosphorylated proteins were precipitated with 5% TCA and the pellet was washed several times with chloroform. Labeled proteins were then subjected to acid hydrolysis in 6 M HCl for 2 h at 110°C (4, 14, 26). The authentic phosphoamino acids O-phospho-l-serine, O-phospho-l-threonine, and O-phospho-l-tyrosine were added to the hydrolysate (4 to 400 nmol of each, depending on the method used for detection). Two different techniques were used for the analysis of labeled phosphoamino acids (3, 4, 13, 26). In most cases, they were separated by high-voltage electrophoresis on Schleicher & Schuell 3469 paper at pH 1.9, for 90 min at 40 V/cm. They were then stained with ninhydrin, and the radioactivity was detected by overnight autoradiography as described above. Alternatively, phosphoamino acids were separated on the column (DC6A Dionex, 0.6 by 30 cm) of an amino acid analyzer (Biotronik model LC-2000). Elution was with a 67 mM sodium citrate buffer, for 25 min at pH 1.2 and then 40 min at pH 2.95, at a flow rate of 0.5 ml/min. Under these conditions, phosphoserine, phosphothreonine, and phosphotyrosine eluted in 16, 18, and 55 min, respectively. They were detected after post-column derivatization, using o-phthaldialdehyde and β-mercaptoethanol as reagents. Radioactivity in fractions corresponding to phosphoamino acids was measured as described above.
Separation of dephosphorylated and phosphorylated forms of GlmM.
The two enzyme forms were separated by high-pressure liquid chromatography (HPLC) using a previously described procedure (20), slightly modified as follows: a Vydac C4 (2.1 by 150 mm) column was used, and elution was performed with a gradient of acetonitrile in 0.1% trifluoroacetic acid (15% acetonitrile at t [time] = 0, 30% at t = 6.5 min, 70% at t = 14 min and up to t = 16 min, 15% at t = 18 min) at a flow rate of 0.3 ml/min. Peaks were detected by absorbance at 214 nm and analyzed by mass spectrometry (MS) on an LCQ mass spectrometer fitted with a Finnigan ESI source. Under these conditions, phosphorylated and dephosphorylated forms of GlmM were eluted in 7.8 and 10.6 min, respectively. For the preparation of phosphorylated enzyme, pure GlmM protein (12 μg) was incubated for 1 h at 30°C in a reaction mixture (60 μl) containing 0.1 M Tris-HCl, (pH 8), 2.5 mM MgCl2, 10 mM KCl, and either 0.32 mM Glc-1,6-diP or 0.25 mM ATP.
RESULTS AND DISCUSSION
Phosphoglucosamine mutase and other members of the hexosephosphate mutase family (PGM and PMM) are known to be active only in their phosphorylated forms, but the way by which these enzymes are initially activated (phosphorylated) in vivo remains unknown. The kinase-dependent phosphorylation or autophosphorylation of various bacterial proteins has been recently demonstrated in vitro, using either [γ-32P]ATP or [32P]pyrophosphate as phosphate donor (4–6, 14, 16). The hypothesis that such a mechanism could be responsible for the phosphorylation of the GlmM protein was thus envisaged.
In vitro phosphorylation of GlmM.
Crude extracts from wild-type JM83 cells were incubated with [γ-32P]ATP, and the phosphorylated proteins were detected by autoradiography after one-dimensional SDS-PAGE. We detected a limited number of labeled proteins, but none of them apparently corresponded to GlmM (Fig. 1, lane A). However, when a crude extract was prepared from cells overproducing to high levels the glmM gene product, either wild type or His6 tagged (JM83[pMLJ1] or JM83[pMLJ4] cells induced with IPTG, respectively), a new major radioactive band comigrating with GlmM was observed (Fig. 1). The slower migration of the His6-tagged enzyme (lane C) than of the wild-type enzyme (lane B) was consistent with its slightly higher molecular mass due to the C-terminal extension Arg-Ser-Arg-His6 (20). The finding that the latter band was absent when cells were grown in the absence of IPTG (data not shown) confirmed its identification as the GlmM protein.
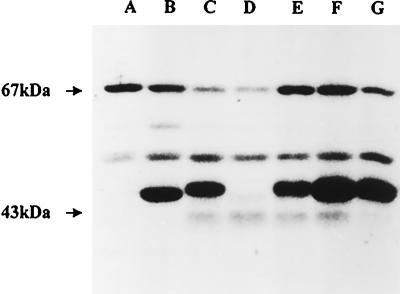
FIG. 1.
In vitro phosphorylation of proteins from crude protein extracts of E. coli cells overexpressing the glmM gene product. Cells of JM83 carrying either of the plasmids described in the text were grown and induced with IPTG. Crude extracts were prepared, and soluble fractions were assayed for protein phosphorylation in the presence of [γ-32P]ATP. Proteins were separated by SDS-PAGE and stained with Coomassie blue, and the labeled proteins were detected by autoradiography. Lane A, extract from wild-type JM83 cells; lane B, extract from induced JM83(pMLJ1) cells overproducing the wild-type GlmM enzyme; lanes C to G, extracts from induced cells overproducing either the wild-type His6-GlmM enzyme (lane C) or one of the mutants His6-GlmM S102A (lane D), His6-GlmM S100A (lane E), His6-GlmM S327A (lane F), and His6-GlmM S413A (lane G). Molecular size standards indicated on the left are bovine serum albumin (67 kDa) and ovalbumin (43 kDa).
To distinguish between a kinase-dependent phosphorylation and an autophosphorylation, the enzyme was purified to near homogeneity. Whatever the source of pure enzyme used (wild type or histidine tagged), GlmM was clearly capable of phosphorylating itself from [γ-32P]ATP (Fig. 2a, lane A). The labeling was similar to that obtained with crude extracts (for the same amount of GlmM), suggesting that no other protein was required for its phosphorylation. However, a possibility exists that the phosphorylation of GlmM was carried out by small amounts of a contaminating kinase tightly bound to the protein, but it seems unlikely.
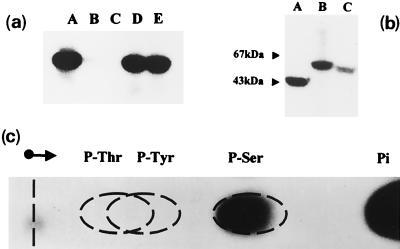
FIG. 2.
In vitro phosphorylation of purified hexosephosphate mutases. The pure enzymes were assayed for in vitro phosphorylation in the presence of [γ-32P]ATP, and reaction mixtures were analyzed by SDS-PAGE as described in the legend to Fig. 1. (a) Autophosphorylation of wild-type and mutant GlmM enzymes. Lane A, wild-type GlmM; lane B, mutant S100A; lane C, mutant S102A; lane D, mutant S327A; lane E, mutant S413A. (b) Autophosphorylation of other hexosephosphate mutases. Lane A, E. coli GlmM; lane B, rabbit muscle PGM; lane C, yeast N-acetylglucosamine-phosphate mutase Agm1p. (c) Identification of the labeled phosphoamino acid. A pure sample of wild-type GlmM enzyme was incubated with [γ-32P]ATP and acid hydrolyzed as described in Materials and Methods. Authentic standards of phosphothreonine (P-Thr), phosphotyrosine (P-Tyr), and phosphoserine (P-Ser) (200 nmol of each) were added to the protein hydrolysate, and the mixture was subjected to high-voltage paper electrophoresis. Phosphoamino acids were detected by staining with ninhydrin, and labeled compounds were detected by autoradiography. Pi, inorganic phosphate.
Autophosphorylation was dependent on the presence of divalent cations, as the addition of 1 mM EDTA to the reaction mixture containing 0.5 mM MgCl2 clearly abolished GlmM phosphorylation (Fig. 3a). The latter inactivation was reversible, as the capability to autophosphorylate was completely restored by the addition of 5 mM MgCl2. Surprisingly, addition of Zn2+ instead of Mg2+ greatly improved the labeling of the protein, but Mn2+ and Ca2+ appeared much less effective than Mg2+ (Fig. 3a). It was noteworthy that the ability of the GlmM enzyme to autophosphorylate paralleled in most cases its enzymatic activity (enzyme activity was 80% lower when Mg2+ was replaced by either Mn2+ or Ca2+ [data not shown]). In the presence of Zn2+, however, we observed efficient autophosphorylation but a complete absence of catalytic activity. Under our in vitro conditions, incorporation of 32P into GlmM increased linearly with time (Fig. 3b) but reached a plateau value after about 1 h. When a 20-fold excess of unlabeled ATP was added at t = 20 min, no further incorporation of 32P into the protein occurred, and the radioactivity stayed at the level of the addition of unlabeled ATP (data not shown). This suggested that the plateau value was not due to an equilibrium between phosphorylation and dephosphorylation reactions. Measurements of the radioactivity present in labeled protein bands after their excision from SDS-polyacrylamide gels showed that at most 2,000 cpm of 32P had been incorporated per μg of GlmM when ATP was used at a concentration of 50 μM. Increasing the concentration of ATP to 0.5 mM resulted in a twofold-increased level of enzyme phosphorylation (a fivefold-reduced amount of incorporated radioactivity), and the Km for ATP was estimated at approximately 60 μM.
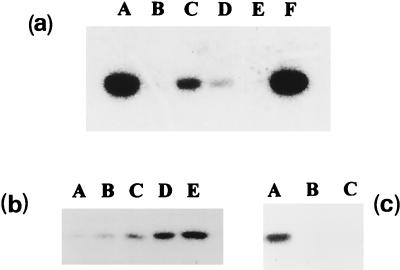
FIG. 3.
(a) Effect of divalent cations on the in vitro phosphorylation of E. coli GlmM. Phosphorylation of the pure wild-type GlmM enzyme by [γ-32P]ATP was tested in the absence (lane A) or presence (lane B) of 1 mM EDTA (the concentration of MgCl2 was 0.5 mM). The EDTA-treated sample was then tested after addition of either MgCl2 (lane C), MnCl2 (lane D), CaCl2 (lane E), or ZnCl2 (lane F) at 5 mM (final concentration). (b) Kinetics of GlmM autophosphorylation. In vitro phosphorylation of the pure wild-type GlmM enzyme was observed after 5, 10, 20, 30, and 60 min of incubation with [γ-32P]ATP (lanes A to E, respectively). (c) Transfer of GlmM covalently linked radioactivity to substrates. After incubation of pure wild-type E. coli GlmM with [γ-32P]ATP for 30 min, substrates (1 mM) were eventually added and incubation was continued for 5 min. Lane A, no addition; lanes B and C, addition of GlcN-6-P and GlcN-1-P, respectively. In all cases, reaction mixtures were analyzed by SDS-PAGE as described in the legend to Fig. 1.
Inorganic phosphate strongly inhibited the phosphorylation. Consequently, previous enzyme preparations made in 20 mM potassium phosphate buffer were first considered inactive for autophosphorylation but clearly regained this capability once dialyzed against other buffers such as morpholinepropanesulfonic acid or HEPES. Addition of unlabeled GTP or CTP did not inhibit the incorporation of 32P from ATP into the protein when used at a high concentration (a 10-fold excess with respect to ATP). UTP, however, inhibited protein labeling greatly at 50 μM and completely at 250 μM (data not shown). Further experiments with radiolabeled UTP are required to determine whether this nucleotide could also phosphorylate the enzyme.
No incorporation of radioactivity into the protein was observed when [γ-32P]ATP was replaced by [14C]ATP or [α-32P]ATP. In addition, treatment of the labeled protein by alkaline phosphatase resulted in the release of inorganic phosphate, indicating that the phosphoryl group in this protein was bound to an amino acid as an O-phosphomonoester and therefore was not due to nucleotidylation or ADP-ribosylation reactions (18).
Identification of the site of phosphorylation.
The resistance of the 32P-labeled residue(s) from GlmM (on electrophoresis gels) to hot TCA treatment already suggested that it was an O-phosphomonoester. Its precise nature was further investigated by using previously described procedures based on the acid hydrolysis of the radioactive protein and analysis of the released phosphoamino acids (as detailed in Materials and Methods). The GlmM protein appeared to be phosphorylated exclusively at serine residue(s), no label being detected in phosphothreonine or phosphotyrosine, even after long exposure of films and regardless of the source of enzyme used, purified wild-type and histidine-tagged enzymes or crude extracts from a GlmM-overproducing strain (Fig. 2c).
The amino acid sequences of all members of the hexosephosphate mutase family contain a highly conserved motif considered their specific signature. It appears as GIVISAS*HNPFYDNG (S* represents the S102 residue that is phosphorylated during the catalytic process [20]) in the E. coli GlmM enzyme. The E. coli GlmM protein contains 20 serine residues, 4 of which (S100, S102, S327, and S413) appeared strictly conserved in alignments with other GlmM sequences. To determine whether S102 or another serine residue was involved in the ATP-dependent autophosphorylation process, each was individually replaced by an alanine residue in the protein sequence by site-directed mutagenesis. From the four mutant GlmM proteins thus generated, only S102A lost all ability to phosphorylate itself (Fig. 1 and 2). The incorporation of 32P from ATP into the mutant S100A was severely diminished, but levels in the S327A and S413A mutants and the wild-type enzyme were quite similar (Fig. 2). Here also, autophosphorylation and phosphoglucosamine mutase activity were clearly correlated features: the S102A and S100A mutant proteins appeared as completely inactive and 50-fold less active (0.06 μmol/min/mg of protein) than the wild-type enzyme, respectively (20). The phosphoglucosamine mutase activity of the S327A and S413A mutant proteins (about 3 μmol/min/mg of protein) was similar to that of the wild-type enzyme, and plasmids pMLJ8 and pMLJ9 expressing these mutated genes fully complemented the glmM defect of the thermosensitive mutant strain GPM83. However, it is noteworthy that the S100A protein was more efficiently phosphorylated in crude protein extracts than when assayed after its purification to homogeneity (compare Fig. 1, lane E, with Fig. 2a, lane B). As the same is not observed with the wild-type and the other mutant proteins, it is likely due to some instability of the purified S100A mutant protein rather than to the loss of some tightly bound contaminating kinase occurring specifically in the case of this mutant protein species.
Autophosphorylation as a general feature in the hexosephosphate mutase family.
Two other recently characterized bacterial phosphoglucosamine mutases, from H. pylori (11) and S. aureus (21, 41), were assayed for in vitro phosphorylation. Incorporation of 32P from ATP into these two proteins was similarly observed, using crude extracts from overproducing strains (JM83 cells carrying plasmids pMLD106 and pMLJ2, respectively) as enzyme source (data not shown). Moreover, this property was not restricted to the sole GlmM enzymes, as labeling of pure yeast N-acetylglucosamine-phosphate mutase Agm1p (prepared in this work) and rabbit muscle PGM (commercial) proteins was also observed in our assay conditions (Fig. 2b). Hydrolysis of these different labeled proteins generated only phosphoserine (data not shown), suggesting that the site of phosphorylation would most likely be in each case the active-site serine residue.
Physiological role of GlmM autophosphorylation.
The data obtained in this study show that phosphoglucosamine mutases and possibly all members of the hexosephosphate mutase family can autophosphorylate in vitro, using ATP or eventually another nucleoside triphosphate as phosphoryl group donor. This is the first time such a property for this class of enzymes has been described. As shown by the complete loss of enzyme label that follows incubation of [32P]phospho-GlmM with either of its substrates, GlcN-6-P or GlcN-1-P (Fig. 3c), and further demonstration that the generated radioactive compound behaves as GlcN-P in thin-layer chromatography on polyethyleneimine-cellulose plates (data not shown), the phosphoserine residue generated by enzyme autophosphorylation is clearly capable of transferring its phosphoryl group to a substrate molecule, as expected for a ping-pong reaction mechanism (20). It is thus tempting to speculate that this process could act as a starter of enzyme phosphorylation that is required to initiate the catalytic activity of the enzyme in vivo. However, as emphasized by Smith et al. (37), it would probably be an error to assume that protein autophosphorylation has relevance in all instances, and the physiological significance of the feature described here remains to be demonstrated.
From measurements of the amount of radioactivity detected in protein bands after their excision from SDS-polyacrylamide gels, we calculated that at most 2 to 3% of the molecules of GlmM had been phosphorylated by ATP under the in vitro conditions used (2,000 cpm of 32P incorporated per μg of enzyme when ATP was used at 50 μM). However, significant losses of radioactivity could have occurred in the steps preceding detection and quantitation of protein bands, in particular during treatment of gels with hot TCA. Perhaps also the in vitro assay conditions are far from optimal and physiological ones. In particular, the pool level of ATP in exponentially growing cells (about 1 mM) is much higher than the concentration used in the in vitro experiments involving radioactivity, and other cellular factors could also influence the extent of phosphorylation. We recently showed that His6-tagged GlmM enzymes could be rapidly extracted and purified in one step from bacterial cell contents and then appear to be 30 to 70% phosphorylated when analyzed by an HPLC procedure allowing separation of the dephosphorylated and phosphorylated forms of the enzyme (20). The same HPLC technique has been now used to test the phosphorylation of GlmM by unlabeled ATP. As shown in Fig. 4, incubation of dephosphorylated wild-type GlmM (12 μg) for 1 h at 30°C with 0.25 mM ATP results in the appearance of a peak of phosphorylated protein which accounts for 15 to 20% of the total protein. Incubation with Glc-1,6-diP has the same effect, but the final yield of phosphorylated protein is higher, about 90%. In both cases, molecular weights of 47,489 and 47,409 have been determined by coupled MS analysis for the material present in peaks corresponding to the phosphorylated and dephosphorylated enzyme forms, respectively. The increment of weight of 80 also indicates that only one phosphate has been incorporated into the protein. ATP-dependent phosphorylation of the enzyme was also correlated with activation of the enzyme, as we measured a low but detectable activity, about fivefold lower than that of the 50% phosphorylated His6-tagged wild-type GlmM enzyme, in the absence of hexose diphosphate. The extent of enzyme phosphorylation revealed by this technique is much higher than that estimated in experiments involving radioactive ATP, but as discussed above, several factors (e.g., assay conditions or recovery of protein label) could explain this difference. The yield of enzyme phosphorylation with ATP is lower than that observed with Glc-1,6-diP (Fig. 4) or GlcN-1,6-diP (20). However, it should be noted that the latter compounds are natural intermediates in the phosphoglucosamine mutase-catalyzed reaction (20) which by evidence should phosphorylate the enzyme much better. In fact, it is not yet clear whether hexose diphosphates exist only as intermediates of PGM- and GlmM-catalyzed reactions or if additional enzymatic activities also catalyze their specific formation in vivo. The existence of such enzymes and of significant pools of these hexose diphosphates in bacteria remains to be demonstrated. It was previously reported that once phosphorylated, PGM did not require Glc-1,6-diP for activity (27, 34). The same is observed with E. coli phosphoglucosamine mutase (20, 30). In both cases, addition of the hexose diphosphate enhances the enzyme activity, due simply to a stimulation of the second step in the reaction mechanism (20, 27). In particular, we have found that the wild-type GlmM enzyme is present in exponentially growing cells under both dephosphorylated and phosphorylated forms, in roughly equivalent amounts, and that its activity could be increased by a factor of about 10 to 20 in the presence of a saturating concentration of GlcN-1,6-diP (20, 30). Taking into account the maximal activity value, it has been concluded that the GlmM enzyme is present in a great (about 50-fold) excess in E. coli cells compared to the specific requirements in UDP-GlcNAc molecules of the pathways for peptidoglycan and lipopolysaccharide biosyntheses (30). In fact, the basal activity of the phosphorylated enzyme (estimated in absence of GlcN-1,6-diP) is quite sufficient to sustain these specific cell requirements, and it can thus be hypothesized that GlcN-1,6-diP acts only as an intermediate in the reaction and is a priori not required for enzyme activity in vivo. The demonstration that Glc-1,6-diP rarely dissociates from the PGM active site during catalysis, once for every 20 or more catalytic cycles (25, 34), is consistent with a model in which the hexose diphosphate is required only to maintain the enzyme in its phosphorylated state. This raises the question of the in vivo mechanism by which the initial activation (phosphorylation) of the enzyme is achieved. The discovery that hexosephosphate mutases could autophosphorylate in the presence of ATP is very important in this respect. Interestingly, we have recently observed that the yeast N-acetylglucosamine-phosphate mutase (the AGM1 gene product [19]) is functional when expressed in E. coli cells (from the pMLD133 plasmid), as it has been shown to efficiently catalyze the formation of GlcNAc-1-P from GlcNAc-6-P in vivo (data not shown). As this enzyme does not normally exist in E. coli, host cells theoretically do not contain GlcNAc-1,6-diP (its reaction intermediate) before expressing the yeast enzyme. Consequently, the in vivo phosphorylation of the yeast mutase could not have been achieved by this compound and probably results from a more general mechanism such as phosphorylation by ATP. The demonstration in the present work that this enzyme from yeast could also autophosphorylate in vitro is consistent with this hypothesis.
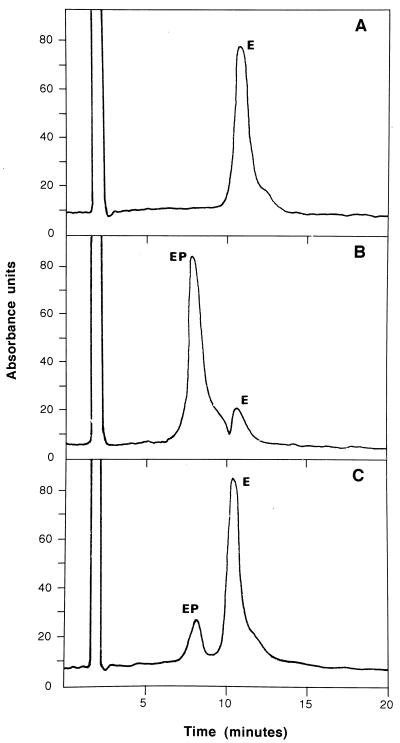
FIG. 4.
Separation of phosphorylated and dephosphorylated forms of GlmM by HPLC. Pure samples of wild-type GlmM enzyme were analyzed by the HPLC procedure described in Materials and Methods, before (A) or after incubation in the presence of either Glc-1,6-diP (B) or ATP (C). The phosphorylated (EP) and dephosphorylated (E) forms of enzyme eluted in 7.8 and 10.6 min, respectively. Molecular weights of 47,489 and 47,409 were determined by coupled MS for the material present in these two peaks.
As indicated by the transfer of its γ-phosphoryl group to the active-site serine residue, the molecule of ATP could fit the active site of phosphoglucosamine mutase. The large size of this nucleotide and the absence of obvious structural homology with the substrates GlcN-P and GlcN-1,6-diP suggest that the binding of ATP to the enzyme active site should be a specific process and that enzyme autophosphorylation should have some physiological significance. No crystallographic structure of a GlmM enzyme is available to date, and it is thus difficult to speculate on how the active site is organized and what the structure and size of compounds that could fit it are. However, the crystal structure of another member of the hexosephosphate mutase family, rabbit muscle PGM, was earlier reported at 2.7-Å resolution (8). In the latter, the active site was identified on the basis of the position of residue S116, within a deep cleft extending from one side of the molecule to the other and lined by 58 residues. The estimated volume of the PGM active-site cleft, 4,000 to 6,000 Å3, was clearly large enough to accommodate a molecule of ATP, as demonstrated in the present study.
ACKNOWLEDGMENTS
We thank Hélène Rey for preparation of the phosphorylated enzyme, Bruno Genet for HPLC-MS analyses, and Patricia Doublet for helpful discussions.
This work was supported by a grant from the Centre National de la Recherche Scientifique (EP1088 CNRS) and a grant “Biotechnologies” from the Ministère de l'Education Nationale, de la Recherche et de la Technologie (97.C.0177). Financial support by Hoechst Marion Roussel AG to L.J. and F.P. is greatly acknowledged.
REFERENCES
- 1.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein using the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 2.Brown K, Pompeo F, Dixon S, Mengin-Lecreulx D, Cambillau C, Bourne Y. Crystal structure of the bifunctional N-acetylglucosamine 1-phosphate uridyltransferase from Escherichia coli: a paradigm for the related pyrophosphorylase superfamily. EMBO J. 1999;18:4096–4107. doi: 10.1093/emboj/18.15.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Capony J P, Demaille J G. A rapid microdetermination of phosphoserine, phosphothreonine and phosphotyrosine in proteins by automatic cation exchange on a conventional amino acid analyzer. Anal Biochem. 1983;128:206–212. doi: 10.1016/0003-2697(83)90365-2. [DOI] [PubMed] [Google Scholar]
- 4.Cortay J-C, Nègre D, Cozzone A J. Analyzing protein phosphorylation in prokaryotes. Methods Enzymol. 1991;200:214–227. doi: 10.1016/0076-6879(91)00141-i. [DOI] [PubMed] [Google Scholar]
- 5.Cortay J-C, Rieul C, Duclos B, Cozzone A J. Characterization of the phosphoproteins of Escherichia coli cells by electrophoretic analysis. Eur J Biochem. 1986;159:227–237. doi: 10.1111/j.1432-1033.1986.tb09858.x. [DOI] [PubMed] [Google Scholar]
- 6.Cozzone A J. Protein phosphorylation in prokaryotes. Annu Rev Microbiol. 1988;42:97–125. doi: 10.1146/annurev.mi.42.100188.000525. [DOI] [PubMed] [Google Scholar]
- 7.Dagert M, Ehrlich S D. Prolonged incubation in calcium chloride improves the competence of Escherichia coli cells. Gene. 1979;6:23–28. doi: 10.1016/0378-1119(79)90082-9. [DOI] [PubMed] [Google Scholar]
- 8.Dai J-B, Liu Y, Ray W J, Jr, Konno M. The crystal structure of muscle phosphoglucomutase refined at 2.7-angstrom resolution. J Biol Chem. 1992;267:6322–6337. [PubMed] [Google Scholar]
- 9.Dallas W S, Dev I K, Ray P H. The dihydropteroate synthase gene, folP, is near the leucine tRNA gene, leuU, on the Escherichia coli chromosome. J Bacteriol. 1993;175:7743–7744. doi: 10.1128/jb.175.23.7743-7744.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deng W P, Nickoloff J A. Site-directed mutagenesis of virtually any plasmids by eliminating a unique site. Anal Biochem. 1992;200:81–88. doi: 10.1016/0003-2697(92)90280-k. [DOI] [PubMed] [Google Scholar]
- 11.De Reuse H, Labigne A, Mengin-Lecreulx D. The Helicobacter pylori ureC gene codes for a phosphoglucosamine mutase. J Bacteriol. 1997;179:3488–3493. doi: 10.1128/jb.179.11.3488-3493.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dobrogosz W J. Effect of amino sugars on catabolite repression in Escherichia coli. J Bacteriol. 1968;95:578–584. doi: 10.1128/jb.95.2.578-584.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Duclos B, Mercandier S, Cozzone A J. Chemical properties and separation of phosphoamino acids by thin-layer chromatography and/or electrophoresis. Methods Enzymol. 1991;201:10–21. doi: 10.1016/0076-6879(91)01004-l. [DOI] [PubMed] [Google Scholar]
- 14.Duclos B, Vaganay E, Dadssi M, Cozzone A J. Pyrophosphate is a source of phosphoryl groups for Escherichia coli protein phosphorylation. FEMS Microbiol Lett. 1996;145:49–54. doi: 10.1111/j.1574-6968.1996.tb08555.x. [DOI] [PubMed] [Google Scholar]
- 15.Dutka-Malen S, Mazodier P, Badet B. Molecular cloning and overexpression of the glucosamine synthetase gene from Escherichia coli. Biochimie. 1988;70:287–290. doi: 10.1016/0300-9084(88)90073-9. [DOI] [PubMed] [Google Scholar]
- 16.Enami M, Ishihama A. Protein phosphorylation in Escherichia coli and purification of a protein kinase. J Biol Chem. 1984;259:526–533. [PubMed] [Google Scholar]
- 17.Fischer W. Bacterial phosphoglycolipids and lipoteichoic acids. In: Kates M, editor. Glycolipids, phosphoglycolipids, and sulfoglycolipids. New York, N.Y: Plenum Press; 1990. pp. 123–234. [Google Scholar]
- 18.Foster R, Thorner J, Martin G S. Nucleotidylation, not phosphorylation, is the major source of the phosphotyrosine detected in enteric bacteria. J Bacteriol. 1989;171:272–279. doi: 10.1128/jb.171.1.272-279.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hofman M, Boles E, Zimmermann F K. Characterization of the essential yeast gene encoding N-acetylglucosamine-phosphate mutase. Eur J Biochem. 1994;221:741–747. doi: 10.1111/j.1432-1033.1994.tb18787.x. [DOI] [PubMed] [Google Scholar]
- 20.Jolly L, Ferrari P, Blanot D, van Heijenoort J, Fassy F, Mengin-Lecreulx D. Reaction mechanism of phosphoglucosamine mutase from Escherichia coli. Eur J Biochem. 1999;262:202–210. doi: 10.1046/j.1432-1327.1999.00373.x. [DOI] [PubMed] [Google Scholar]
- 21.Jolly L, Wu S, van Heijenoort J, de Lencastre H, Mengin-Lecreulx D, Tomasz A. The femR315 gene from Staphylococcus aureus, the interruption of which results in reduced methicillin resistance, encodes a phosphoglucosamine mutase. J Bacteriol. 1997;179:5321–5325. doi: 10.1128/jb.179.17.5321-5325.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Knowles J R. Enzyme-catalyzed phosphoryl transfer reactions. Annu Rev Biochem. 1980;49:877–919. doi: 10.1146/annurev.bi.49.070180.004305. [DOI] [PubMed] [Google Scholar]
- 23.Kuhn H-M, Meier-Dieter U, Mayer H. ECA, the enterobacterial common antigen. FEMS Microbiol Rev. 1988;54:195–222. doi: 10.1111/j.1574-6968.1988.tb02743.x. [DOI] [PubMed] [Google Scholar]
- 24.Laemmli U K, Favre M. Maturation of the head of bacteriophage T4. J Mol Biol. 1973;80:575–599. doi: 10.1016/0022-2836(73)90198-8. [DOI] [PubMed] [Google Scholar]
- 25.Lowry O H, Passonneau J. Phosphoglucomutase kinetics with the phosphates of fructose, glucose, mannose, ribose, and galactose. J Biol Chem. 1969;244:910–916. [PubMed] [Google Scholar]
- 26.Martensen T M. Chemical properties, isolation, and analysis of O-phosphates in proteins. Methods Enzymol. 1984;107:3–23. doi: 10.1016/0076-6879(84)07003-8. [DOI] [PubMed] [Google Scholar]
- 27.McCoy E E, Najjar V A. The purification and mechanism of action of yeast phosphoglucomutase. J Biol Chem. 1959;234:3017–3021. [Google Scholar]
- 28.Mengin-Lecreulx D, van Heijenoort J. Identification of the glmU gene encoding N-acetylglucosamine-1-phosphate uridyltransferase in Escherichia coli. J Bacteriol. 1993;175:6150–6157. doi: 10.1128/jb.175.19.6150-6157.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mengin-Lecreulx D, van Heijenoort J. Copurification of glucosamine-1-phosphate acetyltransferase and N-acetylglucosamine-1-phosphate uridyltransferase activities of Escherichia coli: characterization of the glmU gene product as a bifunctional enzyme catalyzing two subsequent steps in the pathway for UDP-N-acetylglucosamine synthesis. J Bacteriol. 1994;176:5788–5795. doi: 10.1128/jb.176.18.5788-5795.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mengin-Lecreulx D, van Heijenoort J. Characterization of the essential gene glmM encoding phosphoglucosamine mutase in Escherichia coli. J Biol Chem. 1996;271:32–39. doi: 10.1074/jbc.271.1.32. [DOI] [PubMed] [Google Scholar]
- 31.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 32.Pompeo F, van Heijenoort J, Mengin-Lecreulx D. Probing the role of cysteine residues in glucosamine-1-phosphate acetyltransferase activity of the bifunctional GlmU protein from Escherichia coli: site-directed mutagenesis and characterization of the mutant enzymes. J Bacteriol. 1998;180:4799–4803. doi: 10.1128/jb.180.18.4799-4803.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Raetz C R H. Bacterial lipopolysaccharides: a remarkable family of bioactive macroamphiphiles. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 1035–1063. [Google Scholar]
- 34.Ray W J, Jr, Peck E J., Jr . Phosphomutases. In: Boyer P D, editor. The enzymes. 3rd ed. Vol. 6. N.Y: Academic Press; 1972. pp. 407–477. [Google Scholar]
- 35.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 36.Sarvas M. Mutant of Escherichia coli K-12 defective in d-glucosamine biosynthesis. J Bacteriol. 1971;105:467–471. doi: 10.1128/jb.105.2.467-471.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smith J A, Francis S H, Corbin J D. Autophosphorylation: a salient feature of protein kinases. Mol Cell Biochem. 1993;127:51–70. doi: 10.1007/BF01076757. [DOI] [PubMed] [Google Scholar]
- 38.Stevenson G, Neal B, Liu D, Hobbs M, Packer N H, Batley M, Redmond J W, Lindquist L, Reeves P. Structure of the O antigen of Escherichia coli K-12 and the sequence of its rfb gene cluster. J Bacteriol. 1994;176:4144–4156. doi: 10.1128/jb.176.13.4144-4156.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van Heijenoort J. Murein synthesis. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella, cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 1025–1034. [Google Scholar]
- 40.White R J. Control of amino sugar metabolism in Escherichia coli and isolation of mutants unable to degrade amino sugars. Biochem J. 1968;106:847–858. doi: 10.1042/bj1060847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu S, de Lencastre H, Sali A, Tomasz A. A phosphoglucomutase-like gene essential for the optimal expression of methicillin resistance in Staphylococcus aureus: molecular cloning and DNA sequencing. Microb Drug Resist. 1996;2:277–286. doi: 10.1089/mdr.1996.2.277. [DOI] [PubMed] [Google Scholar]
- 42.Wu H C, Wu T C. Isolation and characterization of a glucosamine-requiring mutant of Escherichia coli K-12 defective in glucosamine-6-phosphate synthetase. J Bacteriol. 1971;105:455–466. doi: 10.1128/jb.105.2.455-466.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]