Abstract
Background
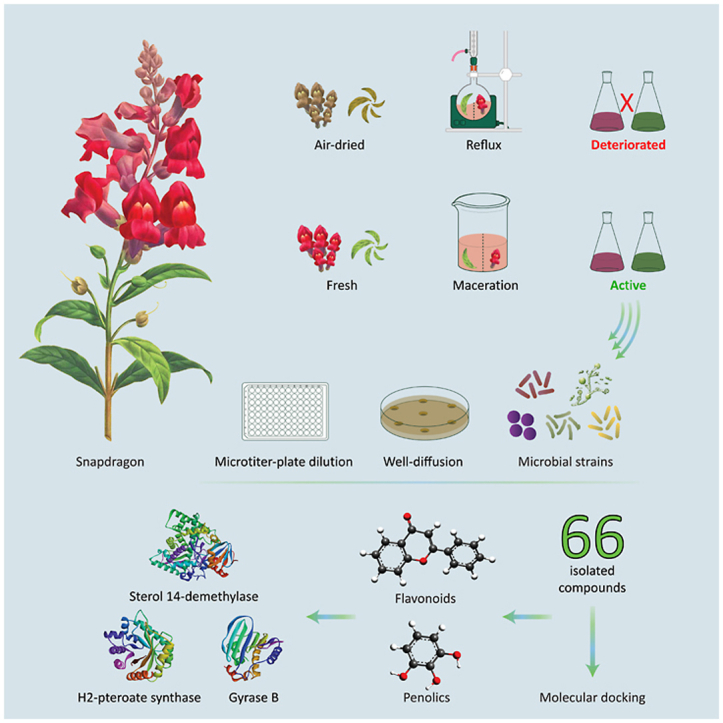
Antirrhinum majus (Snapdragon) is a perennial Mediterranean-native plant that is commonly used for mass display. Few reports acknowledged the traditional use of A. majus for its medicinal and therapeutic effects. Herein, we assess the impact of A. majus’s sample preparation and extraction methods on the plant-aerial parts’ phytochemical contents and antimicrobial activity. Furthermore, the microbial targets of the extracts’ secondary metabolites are inspected using molecular docking simulations.
Methods
The leaves and flowers of A. majus were prepared as fresh and air-dried samples, then extracted using cold maceration and hot reflux, respectively. Extracts with the best phytochemical profiles were selected to test their antimicrobial activities against Bacillus subtilis, Staphylococcus aureus, Enterobacter aerogenes, Escherichia coli and Candida albicans. Besides, molecular docking of 66 reported isolated compounds was conducted against various microbial targets.
Results
The dried-refluxed samples revealed a massive deterioration in their phytochemical profiles, whereas the macerated flowers extract exhibited the highest total phenolic content and antimicrobial activity against all tested bacterial strains. However, both flowers and leaves extracts showed similar minimum inhibitory and lethal concentrations against C. albicans. Molecular docking studies revealed that chlorogenic acid, chalcononaringenin 4’-glucoside, 3,4,2’,4’,6’-pentahydroxy-chalcone 4’-glucoside, apigenin-7-glucuronide, and luteolin-7-glucuronide were the lead compounds in expressing the antimicrobial activity. Yet, A. majus’s compounds could neither inhibit the 30S ribosomal subunit nor muramyl ligase E.
Conclusion
Our results suggest that cold maceration of A. majus fresh aerial parts gave higher flavonoid and phenolic content contributing to its antimicrobial properties. These flavonoids and phenolic compounds are predicted to have a crucial role in inhibiting fungal sterol 14-demethylase, and bacterial dihydropteroate synthase and gyrase B subunit proteins.
Keywords: Antirrhinum majus, Reflux extraction, Maceration extraction, Total phenolic content, Antimicrobial activity, Molecular docking
Graphical abstract
Highlights
-
•
Air-drying of A. majus’s aerial parts deteriorates its phytochemical composition, affecting its antimicrobial activity.
-
•
A. majus’s fresh-flowers macerate exhibited the highest total phenolic content and antibacterial activity.
-
•
The antimycotic activity of A. majus was the same for flowers and leaves macerates.
-
•
In-silico results showed that some phenolics, chalcones, and flavonoids are responsible for the antimicrobial activity.
-
•
A.majus’s components act on fungal sterol 14-demethylase, and bacterial dihydropteroate synthase and gyrase B enzymes.
Antirrhinum majus; Reflux extraction; Maceration extraction; Total phenolic content; Antimicrobial activity; Molecular docking.
1. Introduction
Herbal medicine constructs the base of today’s therapy and medicines, as the plant kingdom provides an infinite source of herbs and plants which can be used as crude drugs or in the form of nutraceuticals (Abdelhalim et al., 2017). The earliest records of employing herbal medicines in managing health issues and microbial infections refer back to the Sumerian, Egyptian, and Chinese civilisations (Süntar, 2020; Luo et al., 2021). Thus, the search for new anti-infectives, especially antimicrobial drugs, is still in demand. Plants’ antimicrobial activity can be related to their essential oils (Varghese et al., 2020) or other isolated compounds such as alkaloids (Casciaro et al., 2020), flavonoids, tannins, phenolic acids (Takó et al., 2020), among other chemical classes including naturally occurring peptides (Datta and Roy, 2021; Li et al., 2021). These compounds possess their antimicrobial activity through various mechanisms such as depressing the nuclear or ribosomal enzyme(s) synthesis, altering the membrane structure and the electron flow, or affecting the metabolic activity of the microbial cell, as well as inhibiting the secretion of their toxins (Redo et al., 1989; Ultee and Smid, 2001; Porras et al., 2021). It was also found that natural products and their derivatives can aid in combating the microbial resistome through different mechanisms which were reviewed thoroughly by Hobson et al. (2021).
Antirrhinum majus, family Plantaginaceae, is a perennial plant native to the Mediterranean region (Tank et al., 2006; Lim, 2014; Ferrer-Gallego and Güemes, 2020). In Jordan, A. majus is used as an ornamental garden plant for mass display, pots and borders (Taifour and El-Oqlah, 2014). In Iraqi culture and traditions, the decoction of the whole plant is used as a detergent, astringent, diuretic, and for treating liver ailments (Al-Douri and Al-Essa, 2010; Al-Snafi, 2015). The leaves and flowers of A. majus were reported to be used as resolvent, stimulant, and anti-inflammatory (Al-Snafi, 2015). The decoction of the whole plant, including the root, is used to resolve epiphora (overflow of tears) (Harkiss, 1971). Yet, A. majus’s seeds are a rich source of fixed oil used as an alternative to olive oil in cooking and diet, with an abundance of neutral lipids, glycolipids, and phospholipids (Ramadan and El-Shamy, 2013).
Few studies have reported this plant to have a high antioxidant capacity as well as potential antimicrobial activity (Riaz et al., 2013; González-Barrio et al., 2018; Saqallah et al., 2018; Shahtalebi et al., 2018; Stefaniak and Grzeszczuk, 2019). However, to the best of our knowledge, no studies have been conducted correlating the effect of extraction methods on the biological activity of A. majus. Thus, our present study aims to analyse the phytochemical composition of A. majus’s leaves and flowers, separately, their antimicrobial activity, and the possible mechanisms by which the plant’s isolated compounds exert their antimicrobial effect.
2. Materials and methods
2.1. Collection and preparation of plant samples
Flowering A. majus shrubs were purchased from a local greenhouse in Amman, Jordan, between February and March 2017. The plant was identified and authenticated by The Royal Society for the Conservation of Nature (RSCN), Jordan. The aerial parts were divided into two parts, flowers and leaves (without stem). Both parts were cut, washed twice with water to remove soil and dust, and divided into two groups; one was air-dried at room temperature (RT) under shade, while the other was kept fresh for immediate extraction.
2.2. Extraction of Antirrhinum majus
Cold maceration and hot reflux extraction methods were used for this study. For the hot reflux, the air-dried leaves (122.6 g) were finely ground using a bar blender (Waring BB1050, USA) and refluxed in 500 mL of 80% methanol (Tedia, USA) for 3 h at 45 °C. After filtration, the extract was concentrated using a rotary evaporator (30 °C, 90 rpm) (Heidolph VV 2000, Germany) and lyophilised (Edwards, UK) to get the crude extract LR3. Meanwhile, the air-dried flowers (39.2 g) were finely ground using a bar blender and refluxed in 750 mL 99% ethyl acetate (S.D. Fine-Chem Ltd., India) for 60 min at 45 °C. This sample was then separated into filtrate and mark. The mark was air-dried and refluxed once again in 500 mL 80% methanol for 30 min. Both filtrates were concentrated separately using a rotary evaporator and lyophilised to get crude extracts FR1 and FR½, respectively. All extracts were kept at -20 °C for later use.
For cold extraction, fresh leaves (1284 g) were blended using a bar blender and macerated in 2.5 L of 80% methanol at RT in a dark place for five consecutive days over a magnetic stirrer (Heidolph, Germany). On the other hand, the fresh flowers (275 g) were blended and macerated in 1.0 L 80% methanol under the same conditions and time. After maceration, both samples were filtered, the residual solvent was removed, and lyophilised to achieve crude extracts LM and FM, respectively. Similarly, the extracts were kept at -20 °C for later use.
2.3. Qualitative phytochemical analysis of Antirrhinum majus
All extracts were prepared at a concentration of 50 mg mL−1 in DMSO:methanol (1:5). Qualitative phytochemical screening of each extract was performed according to our previously reported method (Saqallah et al., 2018).
2.4. Total phenolic content determination
Samples with confirmed presence of flavonoids and phenolics during the qualitative phytochemical analysis were selected for this part of the study. The total phenolic content of LM and FM extracts was determined according to the Folin-Ciocalteu procedure (Saqallah et al., 2018; Savaya et al., 2020). A stock solution of each extract was prepared in distilled water with a concentration of 10 mg mL−1, followed by a series of dilutions (1.0, 0.5, 0.25, 0.125 mg mL−1). Briefly, 12.5 μL aliquots of each dilution were mixed with 250 μL of 2% sodium carbonate solution in 96-well microplates and allowed to stand for 5 min. Then, 12.5 μL of 50% Folin-Ciocalteu reagent was added, and the mixtures were allowed to stand for another 30 min. The absorbance was read using a plate reader (Biotek EL-x800, USA) at 630 nm. The assay was carried out in triplicate. The results were plotted against gallic acid standard curve. Data are expressed as milligram equivalent of gallic acid per one gram extract.
2.5. Antimicrobial activity
2.5.1. Microbial strains
Two Gram-positive (G+ve) (Bacillus subtilis ATCC 6633, Staphylococcus aureus ATCC 25923), two Gram-negative (G-ve) (Enterobacter aerogenes ATCC 13048, Escherichia coli ATCC 15224) bacteria, and a yeast strain (Candida albicans ATCC 10231) were employed in this part of the study. These strains were a generous gift from Hamdi Mango Centre for Scientific Research, The University of Jordan. An inoculum of each strain was transferred using an inoculation loop to 10 mL of sterile growth media in glass tubes. Bacterial strains were cultivated in Mueller-Hinton II Broth (MHB) (BioLab, Hungary), while yeast strain was cultivated in MHB and Sabouraud dextrose broth (SDB) (BioLab, Hungary). The tubes were incubated overnight at 37 °C. Prior to the assay, strains-turbidity was adjusted to 0.5 McFarland standard (1.5×108 CFU mL−1).
2.5.2. Sample preparation
A stock solution (1000 mg mL−1) of each extract (LM and FM) was prepared in MHB. These stock solutions were filtrated using 0.22 μm syringe-filter (EuroClone, Italy) and diluted into a series of dilutions (100, 50, 20, 10, 7.5, 5, and 2.5 mg mL−1) in MHB under sterile conditions. The antimicrobial activity of the extracts was determined using well-diffusion and microtiter-plate dilution methods.
2.5.3. Well-diffusion method
The assay was done based on Magaldi et al. (2004) method with some modifications. Thirty-five mL of sterile Muller Hinton II agar (MHA) (BioLab, Hungary) was filled into 150 mm Petri dishes. After the media had solidified, 100 μL of each microbial strain was transferred and spread using a sterile glass inoculation spreader. Four to six wells (6 mm in diameter) were made into the agar using a sterile metal cork-borer. A volume of 50 μL of extracts’ stocks and their dilutions was filled into the designated wells and incubated overnight at 37 °C. MHB alone was used as a negative control, whereas 2 mg mL−1 gentamicin (Medochemie, Cyprus) and 20 mg mL−1 fluconazole (Pfizer, France) were used as positive controls against bacteria and yeast, respectively. The assay was conducted in duplicate.
2.5.4. Microtiter-plate dilution method
This assay was done to determine the minimum inhibitory concentration (MIC) and the minimum lethal concentration (MLC), based on Talib et al. (2010) method with some modifications. Using 96-well microplates, each well contained 100 μL of sample solution (0.8–100 mg mL−1), where the series of two-fold dilution was made directly from the first row downwards. Adjusted microbial cultures (100 μL; 1.5×108 CFU mL−1) were added to each well. MHB was used as negative control while gentamicin (2 mg mL−1) and fluconazole (20 mg mL−1) were used as positive controls. Plates were covered and incubated overnight at 37 °C. The turbidity was read using a plate reader at 630 nm to determine the MIC. The assay was conducted in duplicate.
Furthermore, aliquots of 10 μL from wells without visible growth were transferred into 2 mL of sterile MHB in Eppendorf microtubes to determine the MLC. The tubes were incubated for 24 h at 37 °C, and the visual turbidity was examined. The lowest concentration that maintained clear (no visible growth) was marked as the MLC (Murray and Hospenthal, 2004).
2.6. Molecular docking
2.6.1. Ligands preparation
Molecular docking simulations were carried out to inspect the possible targets of previously reported isolated compounds from A. majus. A total of 78 compounds have been retrieved successfully from literature (Table S1, Supplementary Information). These compounds included 9 phenols (P01–P09), 2 chalcones (C01 and C02), 22 flavonoids (F01–F22), 10 steroids (S01–S10), 6 alkaloids (A01-A06), 7 iridoids (I01–I07), 10 terpenes and terpenoids (T01-T10), 3 fatty acids (FA01-FA03), and 9 hydrocarbons (H01–H09). Molecular docking was performed for all compounds, excluding fatty acids and the hydrocarbons (a total of 66 compounds were employed). Chemical structures were downloaded from NCBI PubChem (pubchem.ncbi.nlm.nih.gov) and were subjected to MM2 energy minimisation using PerkinElmer® Chem3D® 16.0.
2.6.2. Proteins selection and preparation
In order to inspect the antifungal activity of the isolated compounds, the sterol 14-demethylase crystal structure (PDB: 5TZ1) (Hargrove et al., 2017) was downloaded from RCSB Protein Data Bank (rcsb.org). Sterol 14-demethylase is an enzyme that mediates the synthesis of ergosterol (Zhang et al., 2019), a vital sterol for the fungal cell wall to maintain its permeability and fluidity, and is the target for the azole antifungals (Rodrigues, 2018). The choice of C. albicans’ sterol 14-demethylase was made to validate whether the reported isolated compounds are able to inhibit this enzyme in a similar way to fluconazole, which was used as a control drug in the in-vitro assessment. Additionally, discovering naturally isolated compounds could aid in minimising the highly increasing antifungals resistance in some mycotic strains, such as C. albicans (Krishnasamy et al., 2018; Ganeshkumar et al., 2020).
Antibacterial activity was inspected against five targets in G+ve and G-ve bacteria. S. aureus-targets were chosen to represent the possible mechanisms in G+ve, while E. coli-targets to represent G-ve. Bacterial targets included 30S ribosome (PDB’s: 5TCU and 4V53, respectively) (Borovinskaya et al., 2007; Belousoff et al., 2017), dihydropteroate synthase (PDB’s: 1AD4 and 5V7A, respectively) (Hampele et al., 1997; Dennis et al., 2018), gyrase B (PDB’s: 4URN and 1KZN, respectively) (Lafitte et al., 2002; Lu et al., 2014), muramyl ligase E (MurE) (PDB’s: 4C13 and 1E8C, respectively) (Gordon et al., 2001; Ruane et al., 2013), and transpeptidase (PDB’s: 5TW8 and 6NTW, respectively) (Alexander et al., 2018; Caveney et al., 2019) (Table 1). These targets were chosen to cover the most famous bacterial targets that antibiotics act on or to test for the possible targets that can be used to minimise microbial resistance to certain antimicrobial agents.
Table 1.
Molecular docking targets of yeast and bacterial strains, PDB ID’s, active site coordinates, and control ligand.
| Classifi-cation | Target | PDB ID | Coordinates |
Control Ligand | |||
|---|---|---|---|---|---|---|---|
| x | Y | z | |||||
| Yeast | Sterol 14-demethlase | 5TZ1 | 67.215 | 67.596 | 3.789 | Fluconazole | |
| Bacteria | 30S Ribosome | G+ve | 5TCU | 157.277 | 220.301 | 199.687 | Gentamicin |
| G-ve | 4V53 | 102.490 | 3.090 | -32.973 | Gentamicin | ||
| Dihydropteroate synthase | G+ve | 1AD4 | 33.106 | 8.125 | 41.463 | H4K6WCP5DQ | |
| G-ve | 5V7A | -17.836 | 7.522 | 103.740 | 8Y7 | ||
| Gyrase B | G+ve | 4URN | -31.684 | 8.021 | -4.598 | Novobiocin | |
| G-ve | 1KZN | 19.150 | 30.393 | 34.745 | Clorobiocin | ||
| Muramyl ligase E (MurE) | G+ve | 4C13 | -23.122 | 2.508 | 9.873 | C05892 | |
| G-ve | 1E8C | 46.162 | 37.112 | 75.288 | DB02314 | ||
| Transpeptidase | G+ve | 5TW8 | 21.390 | -62.210 | 36.200 | Ceftaroline | |
| G-ve | 6NTW | 21.480 | -32.370 | 42.150 | Vimirogant | ||
Bacterial 30S ribosomal subunit’s functionality lies in providing a binding site for mRNA and monitoring the base-pairing between the codon of mRNA and the anticodon of tRNA (Arenz and Wilson, 2016). Bacterial 30S ribosome was chosen to predict the ability of A. majus’s isolated compounds to inhibit this target, thus evaluating their potential activity to act as the aminoglycoside antibiotics (such as gentamicin, neomycin, amikacin, etc.) and inhibit protein synthesis in the bacterial cell (Prokhorova et al., 2017).
Besides, dihydropteroate synthase functions as a catalyst in the condensation reaction between dihydropteridine pyrophosphate and p-aminobenzoic acid to generate tetrahydrofolate (THF). THF is an essential co-factor for synthesising amino acids and nitrogen bases, thus proteins (Capasso and Supuran, 2019; Satuluri et al., 2020). Dihydropteroate synthase was selected as its inhibition would suppress the synthesis of nucleic acids through the inhibition of folate synthesis, acting like sulphonamide antibiotics (such as sulfamethoxazole and sulfisoxazole) (Griffith et al., 2018; Capasso and Supuran, 2019).
Gyrase B is a subunit of DNA gyrase which is a topoisomerase type II (Skok et al., 2020). Gyrase B subunits’ hydrolysis generates energy that can be consumed during the ligation process of the cleaved strands by the act of gyrase A subunits (Tiz et al., 2019). Gyrase B is responsible for the ATP-dependent negative supercoiling of DNA. Inhibiting the ATPase of gyrase B would dramatically affect the replication of bacteria in a similar approach to fluoroquinolone antibiotics (such as nadifloxacin, ciprofloxacin, moxifloxacin, etc.) (Henderson et al., 2020).
Furthermore, MurE ligase is an essential enzyme for the biosynthesis of peptidoglycan of the bacterial cell wall. It aids in the aminoacylation process of UDP-N-acetylmuramic acid with L-alanine, D-glutamate, and either L-lysine or meso-diaminopimelic acid to produce UDP-MurNAc-tripeptide, which is a precursor for prostaglandin biosynthesis (Billones and Bangalan, 2019). Its inhibition can suppress peptidoglycan synthesis in the bacterial cell wall. MurE’s high substrate specificity for L-LYS (in G+ve) and meso-A2pm (in G-ve) makes it an attractive target for the discovery of antibacterial agents. To date, several MurE inhibitors have been reported, including phosphinates, peptidosulfonamides, natural compounds, and some quinolone-derivatives, among others. Despite that, none of them demonstrated potent activity in both G+ve and G-ve (Saha and Azam, 2020). Inhibition of MurE ligase through phytochemical isolated compounds could contribute to minimising bacterial resistance to known antibiotics by offering a new target to disturb the bacterial cell wall integrity (Osman et al., 2012; Kouidmi et al., 2014).
Lastly, transpeptidase is an enzyme that forms a domain in class A and B of penicillin-binding proteins. It catalyses the transpeptidation reaction between pentapeptide chains with nearby peptide chains in bacterial cell wall peptidoglycan (Cochrane and Lohans, 2020). The most famous antibiotics of β-lactams, including the penicillins, cephalosporins, and the carbapenems which act as bacterial transpeptidase inhibitors. This inhibition affects the cell wall of bacteria of both Gram-stains (Cochrane and Lohans, 2020; Lima et al., 2020).
It is predicted that inhibiting one of these enzymes can dramatically impacts the microbial life cycle by affecting its nucleic acids cleavage, assembly and replication, proteins, or by disturbing its cell wall components and functions as described earlier.
All protein structures were prepared using BIOVIA® Discovery Studio® 16.1 by removing water molecules and complexed co-structures. Complexed inhibitors were separated from the crystal structures to be used as control ligands, except for the fungal sterol 14-demethylase and the bacterial 30S ribosomal subunit where fluconazole and gentamicin, respectively, were employed as control molecules. Using AutoDockTools 1.5.6, Kollman charges and polar hydrogen atoms were assigned to the proteins.
2.6.3. Simulations parameters and execution
Using AutoDockTools 1.5.6, Gasteiger charges were added for all chemical structures. A cubic grid box (60×60×60, 0.375 Å spacing) was created at the active site of each protein with the coordinates as shown earlier in Table 1. Simulations were carried out using AutoDock 4.2.6 with 250 Lamarckian Genetic Algorithm runs with the default parameters. Conformations with the lowest free energy of binding (LEB) and the most populated cluster were selected for further analysis. Interactions’ analyses were carried out using BIOVIA® Discovery Studio® 16.1.
3. Results
3.1. Extraction yields of Antirrhinum majus
Extracted yields of each sample from different sample types and extraction methods are presented in Table 2. In general, the percentage yields between the freshly extracted and the air-dried samples are incomparable due to the high water content in the earlier which built up their initial bulk weight. However, comparing the air-dried samples, the leaves (LR3) have given a percentage yield of 6.15% compared to its initial weight (122.60 g), which is much lower than the overall yield of air-dried flowers samples (FR1 and FR½, combined) of 16.94% in comparison with their initial weight (39.20 g). In contrast, the fresh leaves (LM) yielded 1.4% extract from their initial weight (1284 g), whereas the fresh flowers (FM) yielded 3.19% from their initial weight (275 g).
Table 2.
Yield (g) and percentage yield (%) of Antirrhinum majus flowers and leaves following different sample preparation and extraction methods.
| Extraction Method | Sample Type, Extract | Sample Weight (g) | Solvent Type | Extract Yield (g) | % Yield |
|---|---|---|---|---|---|
| Reflux | Air-dried Leaves, LR3 | 122.6 | 80% methanol | 7.54 | 6.15 |
| Air-dried Flowers, FR1 | 39.2 | 99% ethyl acetate | 1.8 | 4.59 | |
| Air dried Flowers, FR½ | 33.7 | 80% methanol | 4.84 | 14.36 | |
| Cold Maceration | Fresh Leaves, LM | 1284.0 | 80% methanol | 17.92 | 1.4 |
| Fresh Flowers, FM | 275.0 | 80% methanol | 8.76 | 3.19 |
LR3: Leaves reflux for 3 h. FR1: Flowers reflux for 1 h. FR½: Flowers reflux for half an hour. LR1: Leaves reflux for 1 h. LM: Leaves macerate. FM: Flowers macerate.
3.2. Qualitative phytochemical analysis of Antirrhinum majus
Our results show that the fresh samples (LM and FM) extracted using a cold maceration technique exhibited a wider variation of the secondary metabolites than the air-dried samples (LR3, FR1 and FR½) with the hot reflux method. Flavonoids, phenolics (tannins, phenols, anthraquinones), terpenoids, and steroids were not present in the refluxed extracts, while glycosides and alkaloids were detected at both methods of extraction. No qualitative difference in the phytochemical composition was found between the flowers and the leaves. Table 3 illustrates the phytochemicals analysis of A. majus extracts.
Table 3.
Qualitative phytochemical screening of various extracts of Antirrhinum majus.
| Phytochemical Class | Test | LR3 | FR1 | FR½ | LM | FM |
|---|---|---|---|---|---|---|
| Flavonoids | Sodium hydroxide | – | – | – | + | + |
| Lead acetate | – | – | – | + | + | |
| Glycosides | Sodium hydroxide | + | + | + | + | + |
| Kellar Killani | + | + | + | + | + | |
| Saponin | Froth | – | – | – | – | – |
| Haemolysis | – | – | – | – | – | |
| Alkaloids | Dragendroff’s | + | + | + | + | + |
| Hager’s | + | + | + | + | + | |
| Tannins | Braemer’s | – | – | – | + | + |
| Phenols | Ferric chloride | – | – | – | + | + |
| Anthraquinones | Borntrager’s | + | – | – | + | + |
| Terpenoids | Liebermann–Burchardt | – | – | – | + | + |
| Salkowski | – | – | – | + | + | |
| Steroids | Liebermann–Burchardt | – | – | – | + | + |
LR3: Leaves reflux for 3 h. FR1: Flowers reflux for 1 h. FR½: Flowers reflux for half an hour. LR1: Leaves reflux for 1 h. LM: Leaves macerate. FM: Flowers macerate. +: Present. –: Absent.
3.3. Total phenolic content determination
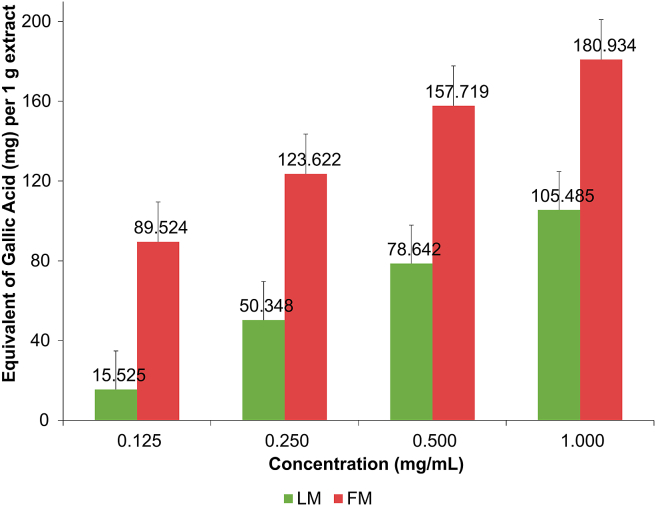
Generally, flavonoids, phenolic acids, and polyphenolics will significantly contribute to the total phenolic content of a plant (Soobrattee et al., 2005). The total phenolic content of A. majus extracts was determined only for extracts that showed positive results for the presence of flavonoids and phenolics. Total phenolic content of LM and FM extracts was examined at a concentration range of 0.125–1.0 mg mL−1. Results were plotted against gallic acid standard calibration curve (0.1–1.0 mg mL−1; y = 0.6892x+0.0668, R2 = 0.9998). Figure 1 shows a linear relationship between extracts’ concentrations and their total phenolic content for leaves (LM) and flowers (FM) of A. majus. The flowers extract expressed a better total phenolic content than the leaves extract, about 1.7 times higher.
Figure 1.
Total phenolic content (mg g−1) of fresh leaves macerate (LM) and fresh flowers macerate (FM) of Antirrhinum majus. The total phenolic content was expressed in mg gallic acid equivalent per 1 g extract (mg GAeq g−1).
3.4. Antimicrobial activity
Well-diffusion and microtiter-plate dilution methods were performed to determine the antimicrobial activity of phytochemicals-rich A. majus extracts (FM and LM). After 24 h incubation, no zones of inhibition appeared around the wells of the tested extracts using the well-diffusion method. However, both positive controls (gentamicin and fluconazole) showed inhibition zones of 26±4 mm. In contrast, the microtiter-plate dilution assay revealed the antimicrobial activity of FM and LM extracts of A. majus against all tested strains (Table 4). FM extract displayed a stronger antibacterial activity than LM extract against both G+ve and G-ve bacterial strains, indicated by the low values of MIC and MLC. Both FM and LM extracts showed moderate to weak antifungal activity against C. albicans.
Table 4.
Minimum inhibitory concentration (MIC, in mg mL−1) and minimum lethal concentration (MLC, in mg mL−1) of fresh flowers macerate (FM) and fresh leaves macerate (LM) of Antirrhinum majus and positive controls against selected microbial strains.
| Microbial Strain | FM |
LM |
Positive Control∗ |
|||
|---|---|---|---|---|---|---|
| MIC | MLC | MIC | MLC | MIC | MLC | |
| Bacillus subtilis | 6.25 | 25.00 | 100.00 | 100.00 | 0.125 | 0.25 |
| Staphylococcus aureus | 25.00 | 50.00 | 50.00 | 100.00 | 0.125 | 0.25 |
| Enterobacter aerogenes | 12.50 | 25.00 | 50.00 | 100.00 | 0.063 | 0.125 |
| Escherichia coli | 25.00 | 50.00 | 50.00 | 100.00 | 0.125 | 0.25 |
| Candida albicans | 50.00 | 100.00 | 50.00 | 100.00 | 2.50 | 2.50 |
Positive controls used were gentamicin (2 mg mL−1) for the bacterial strains and fluconazole (20 mg mL−1) for C. albicans.
3.5. Molecular docking
3.5.1. Overview
A total of 66 previously reported isolated compounds from A. majus have been successfully docked against the crystal structures of C. albicans’ sterol 14-demethylase and five bacterial targets representing both G+ve and G-ve bacteria. These targets included the 30S ribosome, dihydropteroate synthase, gyrase B, muramyl ligase E (MurE), and transpeptidase. Findings are expressed as the lowest energy of binding (LEB) for each compound. The lower the LEB value, the higher the binding affinity. Hydrogen bonds, the most durable interactions, and hydrophobic bonds, i.e., carbon-hydrogen, van der Waals, Pi-sigma, Pi-Pi, etc., were also inspected.
3.5.2. Compounds’ binding affinities towards C. albicans sterol 14-demethylase
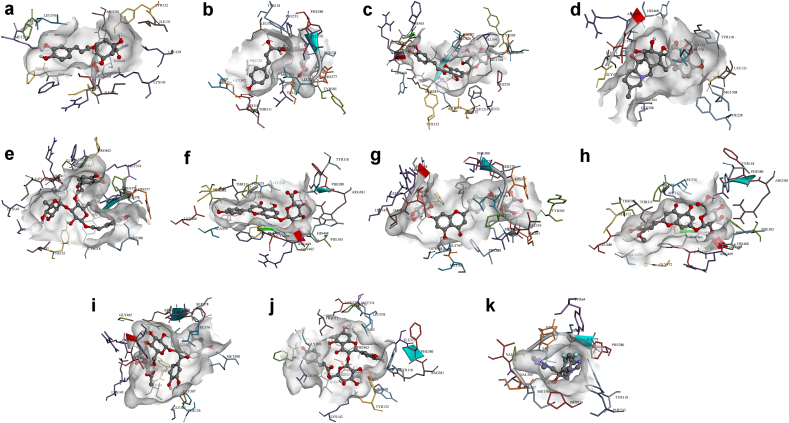
Molecular docking of A. majus’s isolated compounds against the active site of C. albicans’ sterol 14-demethylase showed LEB’s ranging between -3.49 to -12.88 kcal mol−1 (Figure 2 and Table S2, Supplementary Information). Two-dimensional representations of compounds’ binding interactions can be viewed in Figure S1 (Supplementary Information). Among these, sterols (S01–S10) displayed the lowest binding energies (−11.67 kcal mol−1 for sitostanol, S06, to -12.88 kcal mol−1 for 7-avenasterol, S02) compared to other phytochemical classes. Despite sterols’ high binding affinities, they only managed to form mostly one hydrogen interaction with SER378, TYR64, or TYR505 that can sustain and maintain their binding. The low LEB’s may be due to the abundance of hydrophobic interactions with various amino acids at the active site.
Figure 2.
3D representations of compounds conformations at the binding pocket of C. albicans sterol 14-demethylase. (a) Chlorogenic acid (P01), (b) Chalcononaringenin 4’-glucoside (C01), (c) 3,4,2’,4’,6’-pentahydroxy-chalcone 4’-glucoside (C02), (d) Protoverine (A06), (e) Verbascoside (I07). (f) Apigenin-7-glucuronide (F04), (g) Apigenin-7,4’-diglucuronide (F05), (h) Chrysoeriol-7-glucuronide (F08), (i) Cyanidine-3-rutinoside (F09), (j) Quercetin-3-(6’’-coumaroyl)-β-galactoside (F17), and (k) Fluconazole. Compounds chemical structures are coloured per element, whereas the interacting amino acid residues are coloured according to their type. A more detailed analysis of the binding interactions is availbale in Table S2 and Figure S1 (Supplementary Information). Pictures were generated using BIOVIA Discovery Studio 16.1.
Chlorogenic acid (P01) among the phenols showed good activity with a LEB of -7.83 kcal mol−1 and six hydrogen bonds with LYS143, LEU376, PRO462, HIS468, and ILE471. The four isomers of tocopherol; α, β, γ, and δ (P05–P09), showed relatively similar affinity to the target as to that of sterols.
Flavonoids (F01–F22), on the other hand, rank second in terms of the binding affinity with a LEB range between -7.95 kcal mol−1 for p-coumarylglucose (F01) and quercetin-3-arabinofuranoside (F18), and -10.96 kcal mol−1 for apigenin-7,4’-diglucuronide (F05). Flavonoids were found to form up to eight conventional hydrogen interactions (for F05, F17 (quercetin-3-(6’’-coumaroyl)-β-galactoside), and F18) mostly with TYR118, TYR132, HIS377, SER378, and ARG381.
Chalcones were found to bind to sterol 14-demethylase efficiently. Chalcononaringenin 4’-glucoside (C01) can inhibit the enzyme at -9.87 kcal mol−1, forming hydrogen bonds with MET306, THR311, HIS377, SER378, and SER507. Similarly, 3,4,2’,4’,6’-pentahydroxy-chalcone 4’-glucoside (C02) obtained a LEB of -9.84 kcal mol−1 while interacting with HIS377, SER378, PHE463, and SER507.
Among alkaloids, only protoverine (A06) was found to bind and inhibit the receptor at -8.22 kcal mol−1, forming hydrogen interactions with TYR132, HIS468, ARG469, and ILE471. In contrast, verbascoside (I07) iridoid demonstrated binding energy of -9.40 kcal mol−1 and formed two hydrogen interactions with HIS468 and MET508. At the same time, antirrhinoside (I03) exhibited a moderate LEB of -6.85 kcal mol−1 while interacting through eight hydrogen bonds with TYR118, LYS143, ARG381, HIS468, ARG469, CYS470, and ILE471.
Finally, terpenes (T01-T10) did not interact well with the enzyme, judging by their relatively higher LEB values and lack of hydrogen bond interactions. Fluconazole, the positive control, a known inhibitor of sterol 14-demethylase had a LEB of -7.12 kcal mol−1, which agrees with the previous finding (Jović and Šmuc, 2020), and was able to form hydrogen bonds with HIS377, SER378, PHE380, MET508.
Overall, chlorogenic acid (P01), chalcononaringenin 4’-glucoside (C01), 3,4,2’,4’,6’-pentahydroxy-chalcone 4’-glucoside (C02), protoverine (A06), verbascoside (I07), apigenin-7-glucuronide (F04), apigenin-7,4’-diglucuronide (F05), chrysoeriol-7-glucuronide (F08), cyanidine-3-rutinoside (F09), and quercetin-3-(6’’-coumaroyl)-β-galactoside (F17) can be concluded to be responsible for the antimycotic activity by inhibiting sterol 14-demethylase. Figure 2 shows the aforementioned compounds’ binding conformations and interacting residues at the binding pocket.
3.5.3. Compounds’ binding affinities towards bacterial targets
3.5.3.1. Compounds’ binding affinities towards bacterial 30S ribosome
As for the bacterial targets, all compounds have failed to demonstrate good binding affinity compared to the control, gentamicin, in G+ve and G-ve bacteria. Gentamicin binds to the active site of the ribosomal subunit with a LEB of -36.55 and -32.78 kcal mol−1 in G+ve and G-ve, respectively. A. majus’s isolated compounds demonstrated weak LEB’s with the ranges of +1.79 to -11.65 kcal mol−1 against G+ve ribosome, and +0.08 to -10.39 kcal mol−1 against G-ve ribosome when compared to gentamicin’s results (Table S3, Supplementary Information). As such, interactions of A. majus compounds with the ribosomes were excluded from further analysis.
3.5.3.2. Compounds’ binding affinities towards bacterial dihydropteroate synthase
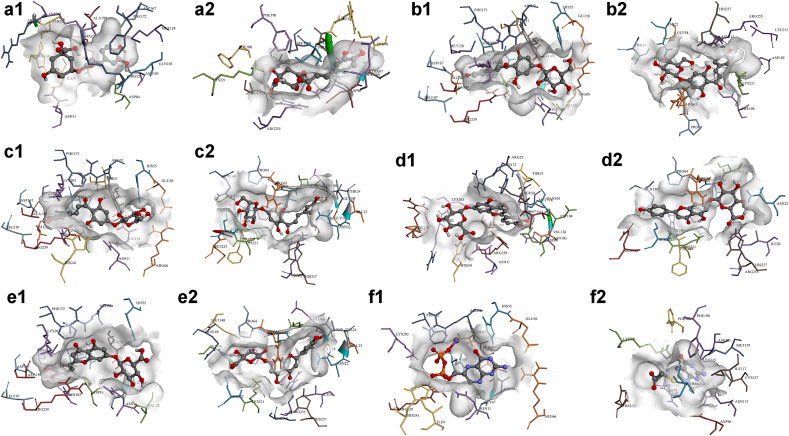
A. majus’s isolated compounds were found to have binding energies ranging between -3.07 to -9.14 kcal mol−1 against S. aureus’s dihydropteroate synthase, and between -3.65 to -9.92 kcal mol−1 against that of E. coli (Table S4, Supplementary Information). Two-dimensional representations of compounds’ binding interactions can be viewed in Figure S2 (Supplementary Information), while the three-dimensional conformations of the best compounds can be viewed in Figure 3.
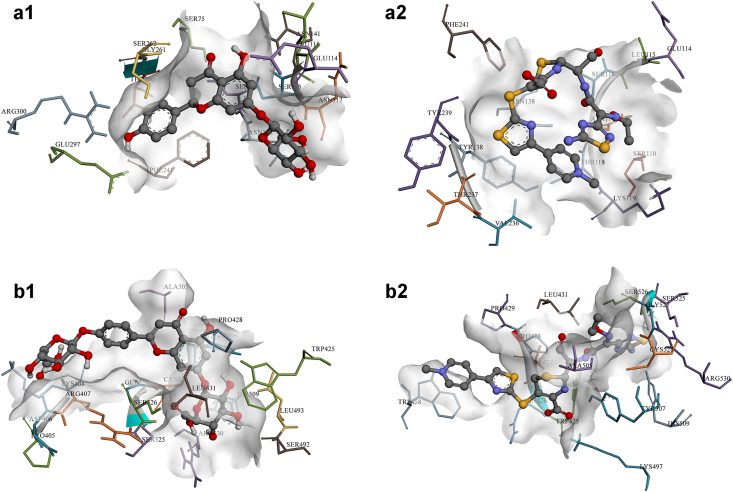
Figure 3.
3D representations of compounds conformations at the binding pocket of S. aureus (letter-1) and E. coli (letter-2) dihydropteroate synthases. (a1 and a2) Chlorogenic acid (P01), (b1 and b2) Chalcononaringenin 4’-glucoside (C01), (c1 and c2) 3,4,2’,4’,6’-pentahydroxy-chalcone 4’-glucoside (C02), (d1 and d2) Apigenin-7-glucuronide (F04), (e1 and e2) Luteolin-7-glucuronide (F12), and (f1 and f2) control molecules. Compounds chemical structures are coloured per element, whereas the interacting amino acid residues are coloured according to their type. A more detailed analysis of the binding interactions is availbale in Table S4 and Figure S2 (Supplementary Information). Pictures were generated using BIOVIA Discovery Studio 16.1.
As in the case of C. albicans, sterols (S01–S10) displayed high binding energies in the range of -7.19 (sitostanol, S06) to -9.17 kcal mol−1 (brassicasterol, S03) for G+ve, and -9.07 (lanosterol, S05) to -9.92 kcal mol−1 (stigmasterol, S10) for G-ve. Similar findings can be seen for the four isomers of tocopherol (P05–P09).
Chlorogenic acid (P01) was found to bind with a relatively moderate LEB of -7.35 kcal mol−1 forming five hydrogen interactions (with ASN11, ARG52, ASP84, ASN103, and SER201) in G+ve’s dihydropteroate synthase, and -7.68 kcal mol−1 with six hydrogen bonds (with SER98, ASN115, ASP116, ILE117, GLY217, and ARG255) against G-ve’s enzyme. Other chalcones, chalcononaringenin 4’-glucoside (C01) and 3,4,2’,4’,6’-pentahydroxy-chalcone 4’-glucoside (C02), displayed favourable binding energies of -8.22 and -7.76 kcal mol−1, respectively, against G+ve, forming hydrogen bonds with several amino acids (SER50, ARG52, ASP167, ARG239, among others), and of -7.56 and -7.78 kcal mol−1, respectively, against G-ve with hydrogen interactions with ASN22, THR62, ASN115 for C01, and with ASN22, GLY189, LYS221, SER222, ARG255 for C02.
Of the flavonoids, aureusidin-6-glucoside (F06), naringenin-7-glucoside (F13), bracteatin-6-glucoside (F07), luteolin-7-glucuronide (F12), and apigenin-7-glucuronide (F04) demonstrated favourable LEB with no significant differences in terms of the interactions between G+ve and G-ve enzymes. In contrast, alkaloids (A01-A06), iridoids (I01–I07), and terpenes and terpenoids (T01-T10), all showed relatively weak binding affinities towards dihydropteroate synthase of both G+ve and G-ve bacteria compared with the control molecules 6-hydroxymethylpterin-diphosphate (H4K6WCP5DQ; -7.59 kcal mol−1) and [(2-amino-9-methyl-6-oxo-6,9-dihydro-1H-purin-8-yl)sulfanyl]acetic acid (8Y7; -7.00 kcal mol−1).
Overall, chlorogenic acid (P01), chalcononaringenin 4’-glucoside (C01), 3,4,2’,4’,6’-pentahydroxy-chalcone 4’-glucoside (C02), apigenin-7-glucuronide (F04), and luteolin-7-glucuronide (F12) can be concluded to be able to express their antibacterial activity by inhibiting dihydropteroate synthase enzyme in both S. aureus and E. coli bacterial strains. Figure 3 shows the aforementioned compounds’ binding conformations and interacting residues at the binding pocket.
3.5.3.3. Compounds’ binding affinities towards bacterial DNA gyrase B subunit
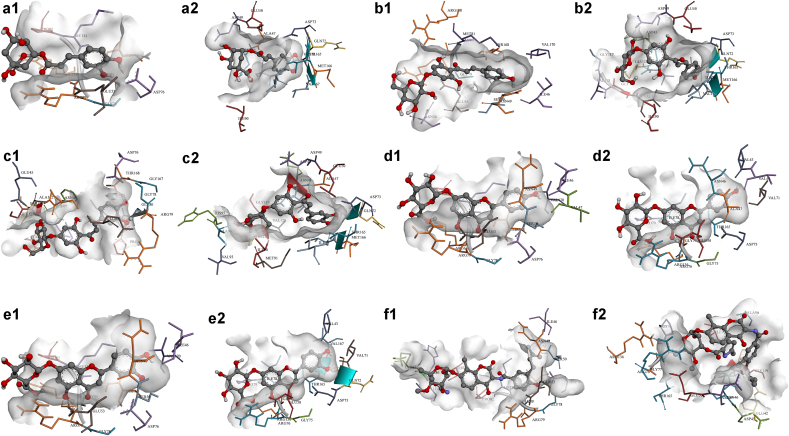
Compounds’ binding energies against gyrase B subunit range between -3.32 and -10.05 kcal mol−1 when docked to that of S. aureus, and between -3.72 and -10.97 kcal mol−1 against that of E. coli (Figure 4, Figure S3 and Table S5-Supplementary Information). Sterols (S01–S10) were found to achieve the lowest binding energies compared to other phytochemical classes ranging between -8.94 kcal mol−1 (lanosterol, S05) and -10.05 kcal mol−1 (5-avenasterol, S01, and 5,24-stigmastadinol, S08) for G+ve and -8.94 kcal mol−1 (lanosterol, S05) and -10.97 kcal mol−1 (campesterol, S04) for G-ve.
Figure 4.
3D representations of compounds conformations at the binding pocket of S. aureus (letter-1) and E. coli (letter-2) gyrase B subunit. (a1 and a2) Chlorogenic acid (P01), (b1 and b2) Chalcononaringenin 4’-glucoside (C01), (c1 and c2) 3,4,2’,4’,6’-pentahydroxy-chalcone 4’-glucoside (C02), (d1 and d2) Apigenin-7-glucuronide (F04), (e1 and e2) Luteolin-7-glucuronide (F12), and (f1 and f2) control molecules. Compounds chemical structures are coloured per element, whereas the interacting amino acid residues are coloured according to their type. A more detailed analysis of the binding interactions is availbale in Table S5 and Figure S3 (Supplementary Information). Pictures were generated using BIOVIA Discovery Studio 16.1.
Chlorogenic acid (P01) and both chalcones (chalcononaringenin 4’-glucoside, C01 and 3,4,2’,4’,6’-pentahydroxy-chalcone 4’-glucoside, C02) showed LEB’s which are lower than the control molecules (−8.34 and -8.74 kcalmol−1 for G+ve and G-ve, respectively). However, they formed more hydrogen bond interactions with the binding pockets-lining amino acids such as ASP76, GLY80 and ARG138 for G+ve, and ASN46, ASP49 and VAL71 for G-ve.
Nine flavonoids (apigenin-7-glucuronide, F04, chrysoeriol-7-glucuronide, F08, cyanidine-3-rutinoside, F09, luteolin-7-glucuronide, F12, quercetin-3-(6’’-benzoyl)-β-galactoside, F16, quercetin-3-(6’’-coumaroyl)-β-galactoside, F17, quercetin-3-glucoside, F20, quercetin-3-rhamnoside, F21 and quercetin 3-rutinoside, F22) were found to have comparable LEB’s to that of the control ligand in S. aureus’s gyrase B subunit. They also were found to form more interactions at the active sites of both enzymes. F08 and F09 formed eight hydrogen bonds with ILE46, ASN49, GLU53, ASN56, ARG79, GLY80, ARG138 and THR168 (for F08 with S. aureus’s gyrase B subunit), and with ASN49, SER50, ASP52, ASP76, GLY80, ILE96, VAL99 and ALA122 (for F09 with S. aureus’s gyrase B subunit). Whereas, for E. coli’s gyrase B subunit, only five compounds showed LEB’s which are lower than -8.00 kcal mol−1 (kaempferol-3-glucoside, F10, quercetin-3-(6’’-benzoyl)-β-galactoside, F16, quercetin-3-(6’’-coumaroyl)-β-galactoside, F17, quercetin-3-glucoside, F20 and quercetin 3-rutinoside, F22). These five flavonoids were able to bind through an average of four hydrogen bonds. F22 had a superior binding energy of -10.67 kcal mol−1, interacting with ASN46, GLU50, ILE90, and SER121.
In contrast, alkaloids (A01-A06), iridoids (I01–I07), and terpenes and terpenoids (T01-T10), showed relatively weak binding affinities towards the gyrase B subunit of both G+ve and G-ve bacteria. This indicates that they probably do not contribute to the antibacterial activity on this specific enzyme. LEB’s of both control molecules agree to previous findings (Pingaew et al., 2017; RK et al., 2018).
In a similar case to dihydropteroate synthase, chlorogenic acid (P01), chalcononaringenin 4’-glucoside (C01), 3,4,2’,4’,6’-pentahydroxy-chalcone 4’-glucoside (C02), apigenin-7-glucuronide (F04), and luteolin-7-glucuronide (F12) can be concluded to be able to express their antibacterial activity by inhibiting DNA gyrase B subunit in both S. aureus and E. coli bacterial strains. Figure 4 shows the aforementioned compounds’ binding conformations and interacting residues at the binding pocket.
3.5.3.4. Compounds’ binding affinities towards bacterial muramyl ligase E (MurE)
All A. majus’s isolated compounds displayed weaker binding affinities towards the active site of both the G+ve and G-ve MurE. The LEB values were found within the range of -3.41 to -9.09 kcal mol−1 for S. aureus’s enzyme, and between -3.83 and -9.71 kcal mol−1 for that of E. coli (Table S6, Supplementary Information). In contrast, both control molecules binding energies were -12.89 and -11.32 kcal mol−1, respectively, forming hydrogen interactions with THR28, TYR45, THR46, VAL47, ASN151, THR152, SER179, TYR351, ARG383, and GLU460 in G+ve, and with LEU26, SER28, GLN44, ALA45, ASN156, THR157, THR158, SER184, and GLN190 in G-ve.
Although some flavonoids like caffeylglucose (F02), ferulylglucose, (F03), apigenin-7,4’-diglucuronide (F05), bracteatin-6-glucoside (F07), and chrysoeriol-7-glucuronide (F08) were able to form multiple hydrogen bonds, their binding energies are much higher than the control. This indicates lower binding affinities of these flavonoids towards the MurE ligase. The two-dimensional representations of compounds’ binding interactions can be viewed in Figure S4 (Supplementary Information).
3.5.4. Compounds’ binding affinities towards bacterial transpeptidase
Compounds’ binding energies against the transpeptidases were found to range between -3.05 and -9.90 kcal mol−1 against S. aureus’s transpeptidase, and between -2.82 and -10.17 kcal mol−1 against that of E. coli (Figure 5, Figure S5, Table S7-Supplementary Information). Sterols (S01–S10) displayed the lowest binding energies among other phytochemical classes, where their ranges were between -8.86 kcal mol−1 (7-stigmastenol, S09) and -9.90 kcal mol−1 (lanosterol, S05) for G+ve, and -8.51 kcal mol−1 (Lanosterol, S05) and -9.43 kcal mol−1 (5-avenasterol, S01) for G-ve. These high affinities towards the transpeptidases might be predominantly due to hydrophobic interactions as on an average only one hydrogen bond can be observed for each of these compounds.
Figure 5.
3D representations of (a1) Naringenin-7-glucoside (F13) and (a2) Ceftaroline (control molecule) binding conformations at the binding pocket of S. aureus transpeptidase, and (b1) Apigenin-7,4’-diglucuronide (F05) and (b2) Vimirogant (control molecule) binding conformations at the binding pocket of E. coli transpeptidase. Compounds chemical structures are coloured per element, whereas the interacting amino acid residues are coloured according to their type. A more detailed analysis of the binding interactions is availbale in Table S7 and Figure S5 (Supplementary Information). Pictures were generated using BIOVIA Discovery Studio 16.1.
On the other hand, p-coumaric acid (P02), ferulic acid (P03), and syringic acid (P04) were able to bind through five to six hydrogen interactions with G+ve transpeptidase. Still, their binding affinities (−6.50, -6.81 and -6.14 kcal mol−1, respectively) appear to be modest compared to that of the control molecule (ceftaroline, -8.80 kcal mol−1). Likewise, their binding affinities towards G-ve’s transpeptidase (−7.15, -7.21 and -7.35 kcal mol−1, respectively) were much less than that of the control molecule (vimirogant, -10.97 kcal mol−1).
However, chlorogenic acid (P01) appeared to inhibit G+ve’s transpeptidase more favourably compared to that of G-ve, while forming a higher number of hydrogen bonds (with GLU114, SER262, TYR291, and GLU297) and achieving a lower binding energy (−8.94 kcal mol−1) compared to the control molecules.
Both chalcones, C01 and C02, appeared to show similar binding affinities to those of P02 and P03, through multiple hydrogen bonds. Chalcononaringenin 4’-glucoside, C01, was found to interact with ASN72, SER75, GLU114, GLU183, and TYR291 of G+ve’s transpeptidase (−7.71 kcal mol−1), and with TRP425, THR430, SER492, TYR507, and SER526 of G-ve’s (−8.00 kcal mol−1). On the other hand, 3,4,2’,4’,6’-pentahydroxy-chalcone 4’-glucoside, C02, interacted with SER75, LEU115, ASN117, LYS119, ASN138, and SER262 in G+ve’s transpeptidase (−7.60 kcal mol−1), and with ASP406, ALA505, SER526, and CYS528 of G-ve’s (−7.16 kcal mol−1).
Of the flavonoids, naringenin-7-glucoside (F13) had the lowest binding energy of -9.79 kcal mol−1 towards G+ve’s transpeptidase, forming five hydrogen bonds with SER75, GLU114, ASN117, GLU297, and ARG300 (Figure 5, a1). In contrast, apigenin-7,4’-diglucuronide (F05) had the lowest binding energy of -10.17 kcal mol−1 towards G-ve’s transpeptidase, with five hydrogen bonds with ASP406, SER492, HIS509, SER526, and CYS528 (Figure 5, b1).
Alkaloids (A01-A06), iridoids (I01–I07), and terpenes and terpenoids (T01-T10), showed relatively weak binding affinities towards the transpeptidase of both G+ve and G-ve bacteria; indicating that their contributions towards the antibacterial might not specifically be through this enzyme.
Overall, naringenin-7-glucoside (F13) can be assumed as the compound which expresses the antibacterial activity by inhibiting transpeptidase enzyme in S. aureus. Whereas apigenin-7,4’-diglucuronide (F05) is the compound that inhibits E. coli’s transpeptidase. Figure 5 shows the aforementioned compounds’ binding conformations and interacting residues at the binding pocket.
4. Discussion
In this study, methanol (80%) and ethyl acetate (99%) were used to extract different parts of A. majus. Methanol is known to extract almost all of the sample contents, polar and the less polar (Tshilanda et al., 2015). For that, 80:20 methanol:water would cover the extraction of the polar to the semi-polar components. On the other hand, ethyl acetate would be responsible for extracting the semi-polar to the non-polar compounds (Soni et al., 2013; Sembiring et al., 2015). Grinding and blending of the dried and fresh samples were conducted to permit maximum contact of the samples to the extractants. Extraction from fresh samples appeared to give a lower percentage yield than the dried samples. This lower yield of the fresh samples can be attributed to the high content of water in the starting material. Although cold maceration gave lesser yields of extractable materials, more phytochemical variation can be observed in the macerated samples compared to the hot-refluxed extracts. Our results also showed no qualitative difference in the phytochemical composition between extraction from fresh flowers (FM) and the fresh leaves (LM).
The drying process is generally intended to preserve the quality of the sample from degradation due to microbial contamination and/or enzymatic hydrolysis (Mediani et al., 2014), but our findings showed fewer phytochemical variations in the dried sample. In the case of A. majus, air-drying of the plant parts seems to affect the qualitative and quantitative results due to the moisture content, which affected the stability of the phytochemical components (Bernard et al., 2014). Furthermore, high temperature during the hot-reflux process most likely deteriorated or altered some phytochemicals (San Chang et al., 2013; Alabri et al., 2014), which eventually affected the phenolic content of the extracts. The higher total phenolic content of the flowers is probably due to the presence of anthocyanins, carotenoids, and aurones that do not commonly exist in the leaves (Kaufmann and El Baya, 1969; González-Barrio et al., 2018). Although there is no difference in the qualitative phytochemical variation between the flowers and the leaves, there are apparent quantitative differences that contributed to the total phenolic content.
The absence of the antimicrobial activity shown with the well-diffusion method was possibly due to solubility and diffusion issues. Both FM and LM extracts were not easily soluble in the MHB, thus, it would be difficult to diffuse or penetrate through the agar medium (MHA) and express an antimicrobial effect (Valgas et al., 2007). Moreover, some of the extracts’ compounds were suspended in the microtiter-plate assay. This would allow the extracts and their compounds to diffuse into the microbes through the media, thus producing the desired antimicrobial effect. The antimicrobial activity of A. majus has already been reported previously from its ethanolic extract towards Bacillus cereus, Bacillus brevis, Staphylococcus epidemitus, Micrococcus luteus, Mycobacterium phlei, and Bortidelia bronchoseptica (Shabana et al., 1987). In addition, Riaz et al. (2013) reported the presence of carvomenthone as one of the active antimicrobial compounds in A. majus that has the ability to disturb the lipid fraction of the bacterium plasma membrane, resulting in rupture and leakage of the contained organelles (Trombetta et al., 2005; Riaz et al., 2013).
In our attempt to scrutinise the possible mechanisms behind the antimicrobial activity of A. majus’s phytochemicals, molecular docking was carried out. Chlorogenic acid (P01), chalcones (C01–C02), flavonoids (F01–F22), protoverine (A06), and verbascoside (I07) were predicted to exert strong inhibition against sterol 14-demethylase. This can be proposed as a probable mechanism where the activity of this enzyme can be suppressed to disturb the fungal cell wall functions and permeability. A recent study has demonstrated the effect of adding phenolic compounds to azole antifungals against azole-resistant C. albicans strains. Their findings indicate that phenolic compounds, including chlorogenic acid (P01), were able to express fungicidal effects alone and in combination with the azoles. This can be an efficient approach to oppose and prevent azole resistance in some fungi strains (Rhimi et al., 2020). Likewise, chalcones and flavonoids activity against C. albicans has been studied extensively and confirmed (Seleem et al., 2017; de Andrade Monteiro & dos Santos, 2019). Protoverine (A01) was previously reported to have hypotensive and cytotoxic activities (Kukula-Koch and Widelski, 2017). However, and according to our knowledge, its antimicrobial effect has not been studied adequately. Oyourou et al. (2013) has confirmed the in-vivo antifungal activity of verbascoside (I07) from Lantana camara leaves, where they suggested its potential use as an alternative fungicide in agriculture against Penicillium digitatum infections. The synergistic antifungal activity of some verbascoside (I07) acyl esters derivatives and amphotericin B against C. albicans has been studied, and up to a four-fold reduction in the MIC was demonstrated (Khazir et al., 2015).
None of A. majus’s isolated compounds were predicted to inhibit bacterial 30S ribosomal subunit and the MurE. Despite that, all sterols (S01–S10) showed a strong binding affinity towards dihydropteroate synthase, gyrase B, and transpeptidase. Sterols are predicted not to have a strong influence on the antibacterial activity as no hydrogen interactions at the active sites were observed. Yet, sterols are believed to slow down bacterial growth, but not to express an actual bactericidal activity. This mechanism might allow a synergistic effect between them and other active phytochemicals (Burčová et al., 2018).
Most likely, the antibacterial activity of A. majus might be due to the inhibition of dihydropteroate synthase and gyrase B enzymes. P01, C01, C02, F04, and F12 were able to inhibit both enzymes mutually with varying binding affinities. In addition, F06, F07, F12, and F13 were also found to bind well at the active site of dihydropteroate synthase, inhibiting further protein synthesis in the bacterium. While F08–F09, F12, F16–F17, and F20–F22 are predicted to inhibit gyrase B. This, in turn, may affect the replication of the bacterial genetic matter. Docking studies also revealed that only F13 was able to competitively inhibit G+ve transpeptidase, while F05 towards G-ve transpeptidase. Terpenes’ antimicrobial activity has been studied and confirmed (Perveen, 2018; Guimarães et al., 2019). Yet, terpenes and terpenoids of A. majus remain to be investigated on other mycotic and bacterial targets. On the other hand, alkaloids and iridoids did not appear to exert any favourable activities on the studied enzymes. For this, the biological importance of A. majus’s alkaloids and iridoids can be furtherly investigated for possible cytotoxic activities.
5. Conclusions
A. majus’s aerial parts are very delicate as they cannot withstand harsh extraction conditions as observed in the preliminary phytochemical screening where hot-reflux extraction has deteriorated most of the plant’s components. Similarly, air-drying of the aerial parts of this plant seems to affect the qualitative and quantitative results. Quantitatively, FM exhibited the highest total phenolic content, possibly due to the presence of flavonoids and phenols. The antimicrobial evaluation demonstrated that FM exhibited potent activity with the lowest MIC’s and MLC’s against G+ve and G-ve bacterial strains, while the antimycotic activity was the same for FM and LM. Based on the molecular docking results, chlorogenic acid (P01), chalcononaringenin 4’-glucoside (C01), 3,4,2’,4’,6’-pentahydroxy-chalcone 4’-glucoside (C02), apigenin-7-glucuronide (F04), and luteolin-7-glucuronide (F12) were found to be responsible for the antimicrobial activity affecting fungal sterol 14-demethylase, and bacterial dihydropteroate synthase and the gyrase B subunit.
Declarations
Author contribution statement
Fadi G. Saqallah: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Wrote the paper.
Wafaa M. Hamed; Wamidh H. Talib: Conceived and designed the experiments.
Roza Dianita: Contributed reagents, materials, analysis tools or data.
Habibah A. Wahab: Conceived and designed the experiments; Contributed reagents, materials, analysis tools or data.
Funding statement
Habibah A. Wahab was supported by Universiti Sains Malaysia [1001/PFARMASI/870031].
Data availability statement
Data included in article/supplementary material/referenced in article.
Declaration of interests statement
The authors declare no conflict of interest.
Additional information
Electronic Supplementary Information (ESI) available. See https://doi.org/10.1016/j.heliyon.2022.e10391.
Acknowledgements
Authors would like to acknowledge Hamdi Mango Centre for Scientific Research, Amman, Jordan. This work was supported by Universiti Sains Malaysia’s Research University Grant (1001/PFARMASI/870031).
Footnotes
This article is a part of the "Ethnopharmacology – challenges for the future.
Contributor Information
Wafaa M. Hamed, Email: hamedwafaa@yahoo.fr.
Habibah A. Wahab, Email: habibahw@usm.my.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
References
- Abdelhalim A., Aburjai T., Hanrahan J., Abdel-Halim H. Medicinal plants used by traditional healers in Jordan, the tafila region. Phcog. Mag. 2017;13(49):95. doi: 10.4103/0973-1296.203975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Douri N., Al-Essa L. A survey of plants used in Iraqi traditional medicine. Jordan J. Pharma. Sci. 2010;3(2):100–108. [Google Scholar]
- Al-Snafi A. The pharmacological importance of Antirrhinum majus - a review. Asian J. Pharmaceut. Sci. Technol. 2015;5(4):313–320. [Google Scholar]
- Alabri T.H.A., Al Musalami A.H.S., Hossain M.A., Weli A.M., Al-Riyami Q. Comparative study of phytochemical screening, antioxidant and antimicrobial capacities of fresh and dry leaves crude plant extracts of Datura metel L. J. King Saud Univ. Sci. 2014;26(3):237–243. [Google Scholar]
- Alexander J.A.N., Chatterjee S.S., Hamilton S.M., Eltis L.D., Chambers H.F., Strynadka N.C.J. Structural and kinetic analyses of penicillin-binding protein 4 (PBP4)-mediated antibiotic resistance in Staphylococcus aureus. J. Biol. Chem. 2018;293(51):19854–19865. doi: 10.1074/jbc.RA118.004952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arenz S., Wilson D.N. Bacterial protein synthesis as a target for antibiotic inhibition. Cold Spring Harbor Perspectives Med. 2016;6(9):a025361. doi: 10.1101/cshperspect.a025361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belousoff M.J., Eyal Z., Radjainia M., Ahmed T., Bamert R.S., Matzov D., Bashan A., Zimmerman E., Mishra S., Cameron D. Structural basis for linezolid binding site rearrangement in the Staphylococcus aureus ribosome. mBio. 2017;8(3) doi: 10.1128/mBio.00395-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard D., Asare I.K., Ofosu D.O., Daniel G.A., Elom S.A., Sandra A. The effect of different drying methods on the phytochemicals and radical scavenging activity of ceylon cinnamon (Cinnamomum zeylanicum) plant parts. Eur. J. Med. Plants. 2014;4(11):1324. [Google Scholar]
- Billones J.B., Bangalan M.A.T. Structure-based discovery of inhibitors against MurE in methicillin-resistant Staphylococcus aureus. Orient. J. Chem. 2019;35(2):618–625. [Google Scholar]
- Borovinskaya M.A., Pai R.D., Zhang W., Schuwirth B.S., Holton J.M., Hirokawa G., Kaji H., Kaji A., Cate J.H.D. Structural basis for aminoglycoside inhibition of bacterial ribosome recycling. Nat. Struct. Mol. Biol. 2007;14(8):727–732. doi: 10.1038/nsmb1271. [DOI] [PubMed] [Google Scholar]
- Burčová Z., Kreps F., Greifová M., Jablonský M., Ház A., Schmidt Š., Šurina I. Antibacterial and antifungal activity of phytosterols and methyl dehydroabietate of Norway spruce bark extracts. J. Biotechnol. 2018;282:18–24. doi: 10.1016/j.jbiotec.2018.06.340. [DOI] [PubMed] [Google Scholar]
- Capasso C., Supuran C.T. Dihydropteroate synthase (sulfonamides) and dihydrofolate reductase inhibitors. Bacterial Resist. Antibiotics–From Mole. Man. 2019:163–172. [Google Scholar]
- Casciaro B., Mangiardi L., Cappiello F., Romeo I., Loffredo M.R., Iazzetti A., Calcaterra A., Goggiamani A., Ghirga F., Mangoni M.L., et al. Naturally-occurring alkaloids of plant origin as potential antimicrobials against antibiotic-resistant infections. Molecules. 2020;25(16):3619. doi: 10.3390/molecules25163619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caveney N.A., Caballero G., Voedts H., Niciforovic A., Worrall L.J., Vuckovic M., Fonvielle M., Hugonnet J.-E., Arthur M., Strynadka N.C.J. Structural insight into YcbB-mediated beta-lactam resistance in Escherichia coli. Nat. Commun. 2019;10(1):1–11. doi: 10.1038/s41467-019-09507-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cochrane S.A., Lohans C.T. Breaking down the cell wall: strategies for antibiotic discovery targeting bacterial transpeptidases. Eur. J. Med. Chem. 2020;194 doi: 10.1016/j.ejmech.2020.112262. [DOI] [PubMed] [Google Scholar]
- Datta S., Roy A. Antimicrobial peptides as potential therapeutic agents: a review. Int. J. Pept. Res. Therapeut. 2021;27(1):555–577. [Google Scholar]
- de Andrade Monteiro C., dos Santos J.R.A. 2019. Phytochemicals and Their Antifungal Potential against Pathogenic Yeasts Phytochemicals In Human Health: IntechOpen; pp. 1–31. [Google Scholar]
- Dennis M.L., Lee M.D., Harjani J.R., Ahmed M., DeBono A.J., Pitcher N.P., Wang Z.C., Chhabra S., Barlow N., Rahmani R. 8-Mercaptoguanine derivatives as inhibitors of dihydropteroate synthase. Chem.--Eur. J. 2018;24(8):1922–1930. doi: 10.1002/chem.201704730. [DOI] [PubMed] [Google Scholar]
- Ferrer-Gallego P.P., Güemes J. Typification of three names in Antirrhinum (Plantaginaceae: antirrhineae) Nord. J. Bot. 2020;38(6) [Google Scholar]
- Ganeshkumar A., Suvaithenamudhan S., Rajaram R. In vitro and in silico analysis of ascorbic acid towards lanosterol 14-α-Demethylase enzyme of fluconazole-resistant Candida albicans. Curr. Microbiol. 2020:1–11. doi: 10.1007/s00284-020-02269-9. [DOI] [PubMed] [Google Scholar]
- González-Barrio R., Periago M.J., Luna-Recio C., Garcia-Alonso F.J., Navarro-González I. Chemical composition of the edible flowers, pansy (Viola wittrockiana) and snapdragon (Antirrhinum majus) as new sources of bioactive compounds. Food Chem. 2018;252:373–380. doi: 10.1016/j.foodchem.2018.01.102. [DOI] [PubMed] [Google Scholar]
- Gordon E., Flouret B., Chantalat L., van Heijenoort J., Mengin-Lecreulx D., Dideberg O. Crystal structure of UDP-N-Acetylmuramoyl-L-Alanyl-D-Glutamate: meso-diaminopimelate ligase from Escherichia coli. J. Biol. Chem. 2001;276(14):10999–11006. doi: 10.1074/jbc.M009835200. [DOI] [PubMed] [Google Scholar]
- Griffith E.C., Wallace M.J., Wu Y., Kumar G., Gajewski S., Jackson P., Phelps G.A., Zheng Z., Rock C.O., Lee R.E. The structural and functional basis for recurring sulfa drug resistance mutations in Staphylococcus aureus dihydropteroate synthase. Front. Microbiol. 2018;9:1369. doi: 10.3389/fmicb.2018.01369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guimarães A.C., Meireles L.M., Lemos M.F., Guimarães M.C.C., Endringer D.C., Fronza M., Scherer R. Antibacterial activity of terpenes and terpenoids present in essential oils. Molecules. 2019;24(13):2471. doi: 10.3390/molecules24132471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampele I.C., D’Arcy A., Dale G.E., Kostrewa D., Nielsen J., Oefner C., Page M.G.P., Schönfeld H.-J., Stüber D., Then R.L. Structure and function of the dihydropteroate synthase from Staphylococcus aureus. J. Mol. Biol. 1997;268(1):21–30. doi: 10.1006/jmbi.1997.0944. [DOI] [PubMed] [Google Scholar]
- Hargrove T.Y., Friggeri L., Wawrzak Z., Qi A., Hoekstra W.J., Schotzinger R.J., York J.D., Guengerich F.P., Lepesheva G.I. Structural analyses of Candida albicans sterol 14α-demethylase complexed with azole drugs address the molecular basis of azole-mediated inhibition of fungal sterol biosynthesis. J. Biol. Chem. 2017;292(16):6728–6743. doi: 10.1074/jbc.M117.778308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harkiss K.J. Studies in the scrophulariaceae - Part III investigation of the occurrence of alkaloids in Antirrhinum majus L. Planta Med. 1971;20(4):108–113. doi: 10.1055/s-0028-1099673. [DOI] [PubMed] [Google Scholar]
- Henderson S.R., Stevenson C.E.M., Malone B., Zholnerovych Y., Mitchenall L.A., Pichowicz M., McGarry D.H., Cooper I.R., Charrier C., Salisbury A.-M. Structural and mechanistic analysis of ATPase inhibitors targeting mycobacterial DNA gyrase. J. Antimicrob. Chemother. 2020;75(10):2835–2842. doi: 10.1093/jac/dkaa286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobson C., Chan A.N., Wright G.D. The antibiotic resistome: a guide for the discovery of natural products as antimicrobial agents. Chem. Rev. 2021;121(6):3464–3494. doi: 10.1021/acs.chemrev.0c01214. [DOI] [PubMed] [Google Scholar]
- Jović O., Šmuc T. Combined machine learning and molecular modelling workflow for the recognition of potentially novel fungicides. Molecules. 2020;25(9):2198. doi: 10.3390/molecules25092198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann E., El Baya A. Pro-und Antioxydantien auf dem Fettgebiet XXIV: ihr Einfluß auf die Auron-Biosynthese in der Blüte. Fette Seifen Anstrichm. 1969;71(1):25–28. [Google Scholar]
- Khazir J., Ali I., Khan I.A., Sampath Kumar H.M. Enzyme mediated-transesterification of verbascoside and evaluation of antifungal activity of synthesised compounds. Nat. Prod. Res. 2015;29(8):727–734. doi: 10.1080/14786419.2014.984181. [DOI] [PubMed] [Google Scholar]
- Kouidmi I., Levesque R.C., Paradis-Bleau C. The biology of mur ligases as an antibacterial target. Mol. Microbiol. 2014;94(2):242–253. doi: 10.1111/mmi.12758. [DOI] [PubMed] [Google Scholar]
- Krishnasamy L., Krishnakumar S., Kumaramanickavel G., Saikumar C. Molecular mechanisms of antifungal drug resistance in Candida species. J. Clin. Diagn. Res. 2018;12(9) [Google Scholar]
- Kukula-Koch W.A., Widelski J. In: Pharmacognosy. Badal S., Delgoda R., editors. Academic Press; Boston: 2017. Chapter 9 - alkaloids; pp. 194–195. [Google Scholar]
- Lafitte D., Lamour V., Tsvetkov P.O., Makarov A.A., Klich M., Deprez P., Moras D., Briand C., Gilli R. DNA gyrase interaction with coumarin-based inhibitors: the role of the hydroxybenzoate isopentenyl moiety and the 5 ‘-Methyl group of the noviose. Biochemistry. 2002;41(23):7217–7223. doi: 10.1021/bi0159837. [DOI] [PubMed] [Google Scholar]
- Li J., Hu S., Jian W., Xie C., Yang X. Plant antimicrobial peptides: structures, functions, and applications. Botanical Studies. 2021;62(1):5. doi: 10.1186/s40529-021-00312-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim T. Vol. 8. Springer Netherlands; Dordrecht: 2014. pp. 633–639. (Antirrhinum majus Edible Medicinal and Non Medicinal Plants). [Google Scholar]
- Lima L.M., da Silva B.N.M., Barbosa G., Barreiro E.J. β-Lactam antibiotics: an overview arom a medicinal chemistry perspective. Eur. J. Med. Chem. 2020 doi: 10.1016/j.ejmech.2020.112829. [DOI] [PubMed] [Google Scholar]
- Lu J., Patel S., Sharma N., Soisson S.M., Kishii R., Takei M., Fukuda Y., Lumb K.J., Singh S.B. Structures of kibdelomycin bound to Staphylococcus aureus GyrB and ParE showed a novel U-shaped binding mode. ACS Chem. Biol. 2014;9(9):2023–2031. doi: 10.1021/cb5001197. [DOI] [PubMed] [Google Scholar]
- Luo L., Yang J., Wang C., Wu J., Li Y., Zhang X., Li H., Zhang H., Zhou Y., Lu A., et al. Science China Life Sciences; 2021. Natural Products for Infectious Microbes and Diseases: an Overview of Sources, Compounds, and Chemical Diversities. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magaldi S., Mata-Essayag S., De Capriles C., Perez C., Colella M., Olaizola C., Ontiveros Y. Well diffusion for antifungal susceptibility testing. Int. J. Infect. Dis. 2004;8(1):39–45. doi: 10.1016/j.ijid.2003.03.002. [DOI] [PubMed] [Google Scholar]
- Mediani A., Abas F., Tan C., Khatib A. Effects of different drying methods and storage time on free radical scavenging activity and total phenolic content of Cosmos caudatus. Antioxidants. 2014;3(2):358–370. doi: 10.3390/antiox3020358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray C., Hospenthal D. Broth microdilution susceptibility testing for. Leptospira spp. Antimicrobial Agents and Chemotherapy. 2004;48(5):1548–1552. doi: 10.1128/AAC.48.5.1548-1552.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osman K., Evangelopoulos D., Basavannacharya C., Gupta A., McHugh T.D., Bhakta S., Gibbons S. An antibacterial from Hypericum acmosepalum inhibits ATP-dependent MurE ligase from Mycobacterium tuberculosis. Int. J. Antimicrob. Agents. 2012;39(2):124–129. doi: 10.1016/j.ijantimicag.2011.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oyourou J.N., Combrinck S., Regnier T., Marston A. Purification, stability and antifungal activity of verbascoside from Lippia javanica and Lantana camara leaf extracts. Ind. Crop. Prod. 2013;43:820–826. [Google Scholar]
- Perveen S. Introductory chapter: terpenes and terpenoids. Perveen S., Al-Taweel A., editors. Terpenes and Terpenoids (Vol. 1): IntechOpen. 2018:1–12. [Google Scholar]
- Pingaew R., Sinthupoom N., Mandi P., Prachayasittikul V., Cherdtrakulkiat R., Prachayasittikul S., Ruchirawat S., Prachayasittikul V. Synthesis, biological evaluation and in silico study of bis-thiourea derivatives as anticancer, antimalarial and antimicrobial agents. Med. Chem. Res. 2017;26(12):3136–3148. [Google Scholar]
- Porras G., Chassagne F., Lyles J.T., Marquez L., Dettweiler M., Salam A.M., Samarakoon T., Shabih S., Farrokhi D.R., Quave C.L. Ethnobotany and the role of plant natural products in antibiotic drug discovery. Chem. Rev. 2021;121(6):3495–3560. doi: 10.1021/acs.chemrev.0c00922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prokhorova I., Altman R.B., Djumagulov M., Shrestha J.P., Urzhumtsev A., Ferguson A., Chang C.-W.T., Yusupov M., Blanchard S.C., Yusupova G. Aminoglycoside interactions and impacts on the eukaryotic ribosome. Proc. Natl. Acad. Sci. USA. 2017;114(51):E10899–E10908. doi: 10.1073/pnas.1715501114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramadan M.F., El-Shamy H. Snapdragon (Antirrhinum majus) seed oil: characterization of fatty acids, bioactive lipids and radical scavenging potential. Ind. Crop. Prod. 2013;42:373–379. [Google Scholar]
- Redo M., Rios J., Villar A. A review of some antimicrobial compounds isolated from medicinal plants reported in the literature 1978–1988. Phytother Res. 1989;3(4):117–125. [Google Scholar]
- Rhimi W., Aneke C.I., Annoscia G., Otranto D., Boekhout T., Cafarchia C. Effect of chlorogenic and gallic acids combined with azoles on antifungal susceptibility and virulence of multidrug-resistant Candida spp. and Malassezia furfur isolates. Med. Mycol. 2020;2020:1–11. doi: 10.1093/mmy/myaa010. [DOI] [PubMed] [Google Scholar]
- Riaz M., Rasool N., Rasool S., Rashid U., Bukhari I.H., Zubair M., Noreen M., Abbas M. Chemical analysis, cytotoxicity and antimicrobial studies by snapdragon: a medicinal plant. Asian J. Chem. 2013;25(10):5479. [Google Scholar]
- Rk M., Arifa Begum S.B., Begum S., Bharathi K., Sujatha D. Molecular docking studies of 4-benzylidene-2-(4-hydroxy-3-methoxystyryl) oxazol-5(4H)-ones; interactions with ARG-136 and ASP-73 of DNA gyrase. J. Global Trends Pharmaceut. Sci. 2018;9(2):5172–5180. [Google Scholar]
- Rodrigues M.L. The multifunctional fungal ergosterol. mBio. 2018;9(5) doi: 10.1128/mBio.01755-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruane K.M., Lloyd A.J., Fülöp V., Dowson C.G., Barreteau H., Boniface A., Dementin S., Blanot D., Mengin-Lecreulx D., Gobec S. Specificity determinants for lysine incorporation in Staphylococcus aureus peptidoglycan as revealed by the structure of a MurE enzyme ternary complex. J. Biol. Chem. 2013;288(46):33439–33448. doi: 10.1074/jbc.M113.508135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha N., Azam M.A. MurE inhibitors as antibacterial agents: a review. J. Inclusion Phenom. Macrocycl. Chem. 2020:1–10. [Google Scholar]
- San Chang J., Wang K.C., Yeh C.F., Shieh D.E., Chiang L.C. Fresh ginger (Zingiber officinale) has anti-viral activity against human respiratory syncytial virus in human respiratory tract cell lines. J. Ethnopharmacol. 2013;145(1):146–151. doi: 10.1016/j.jep.2012.10.043. [DOI] [PubMed] [Google Scholar]
- Saqallah F.G., Hamed W., Talib W. In vivo evaluation of Antirrhinum majus’ wound-healing activity. Sci. Pharm. 2018;86(4):45. doi: 10.3390/scipharm86040045. [DOI] [PubMed] [Google Scholar]
- Satuluri S.H., Katari S.K., Pasala C., Amineni U. Novel and potent inhibitors for dihydropteroate synthase of Helicobacter pylori. J. Recept. Signal Transduction. 2020;40(3):246–256. doi: 10.1080/10799893.2020.1731533. [DOI] [PubMed] [Google Scholar]
- Savaya N.S.A., Issa R.A., Talib W.H. In vitro evaluation of the antioxidant, anti-Propioni bacterium acne and antityrosinase effects of Equisetum ramosissimum (Jordanian horsetail) Trop. J. Pharmaceut. Res. 2020;19(10):2147–2152. [Google Scholar]
- Seleem D., Pardi V., Murata R.M. Review of flavonoids: a diverse group of natural compounds with anti-Candida albicans activity in vitro. Arch. Oral Biol. 2017;76:76–83. doi: 10.1016/j.archoralbio.2016.08.030. [DOI] [PubMed] [Google Scholar]
- Sembiring H., Barus T., Marpaung L., Simanjuntak P. Antioxidant and antibacterial activity of some leaves extracts (methanol, ethyl acetate and n-hexane) of Scurrula fusca G. Don. Int. J. Pharm. Tech. Res. 2015;8(9):24–30. [Google Scholar]
- Shabana M.M., Doss S.L., Mousa O. A phytochemical study of Antirrhinum majus L. Cultivated in Egypt. Bulletin Faculty Pharm. (Cairo University) 1987;25(1):23–32. [Google Scholar]
- Shahtalebi M.A., Asghari G.R., Rahmani F., Shafiee F., Jahanian-Najafabadi A. Formulation of herbal gel of Antirrhinum majus extract and evaluation of its anti-Propionibacterium acne effects. Adv. Biomed. Res. 2018;7:53. doi: 10.4103/abr.abr_99_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skok Z.i., Barančoková M., Benek O.e., Cruz C.D., Tammela P.i., Tomašič T., Zidar N., Mašič L.P., Zega A., Stevenson C.E.M. Exploring the chemical space of benzothiazole-based DNA gyrase B inhibitors. ACS Med. Chem. Lett. 2020 doi: 10.1021/acsmedchemlett.0c00416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soni R., Mehta N., Srivastava D. Wound repair and regenerating effect of ethyl acetate soluble fraction of ethanolic extract of Cinnamomum tamala leaves in diabetic rats. Eur. J. Exp. Biol. 2013;3(3):43–47. [Google Scholar]
- Soobrattee M.A., Neergheen V.S., Luximon-Ramma A., Aruoma O.I., Bahorun T. Phenolics as potential antioxidant therapeutic agents: mechanism and actions. Mutation Res./Fundamen. Mole. Mech. Mutagenesis. 2005;579(1-2):200–213. doi: 10.1016/j.mrfmmm.2005.03.023. [DOI] [PubMed] [Google Scholar]
- Stefaniak A., Grzeszczuk M.E. Nutritional and biological value of five edible flower species. Not. Bot. Horti Agrobot. Cluj-Napoca. 2019;47(1):128–134. [Google Scholar]
- Süntar I. Importance of ethnopharmacological studies in drug discovery: role of medicinal plants. Phytochemistry Rev. 2020;19(5):1199–1209. [Google Scholar]
- Taifour H., El-Oqlah A. 1 ed. Royal Botanic Garden; 2014. Jordan Plant Red List; p. 95. [Google Scholar]
- Takó M., Kerekes E.B., Zambrano C., Kotogán A., Papp T., Krisch J., Vágvölgyi C. Plant phenolics and phenolic-enriched extracts as antimicrobial agents against food-contaminating microorganisms. Antioxidants. 2020;9(2) doi: 10.3390/antiox9020165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talib W., Mahasneh A. Antimicrobial, cytotoxicity and phytochemical screening of Jordanian plants used in traditional medicine. Molecules. 2010;15(3):1811–1824. doi: 10.3390/molecules15031811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tank D., Beardsley P., Kelchner S., Olmstead R. Review of the cystematics of scrophulariaceae s.l. and their current disposition. Aust. Syst. Bot. 2006;19(4):289–307. [Google Scholar]
- Tiz D.B., Skok Ž., Durcik M., Tomašič T., Mašič L.P., Ilaš J., Zega A., Draskovits G., Révész T., Nyerges Á. An optimised series of substituted N-phenylpyrrolamides as DNA gyrase B inhibitors. Eur. J. Med. Chem. 2019;167:269–290. doi: 10.1016/j.ejmech.2019.02.004. [DOI] [PubMed] [Google Scholar]
- Trombetta D., Castelli F., Sarpietro M., Venuti V., Cristani M., Daniele C., Saija A., Mazzanti G., Bisignano G. Mechanisms of antibacterial action of three monoterpenes. Antimicrob. Agents Chemother. 2005;49(6):2474–2478. doi: 10.1128/AAC.49.6.2474-2478.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tshilanda D., Onyamboko D., Tshibangu D., Ngbolua K., Tsalu P., Mpiana P. In vitro antioxidant activity of essential oil and polar and non-polar extracts of Ocimum canum from mbuji-mayi (DR Congo) J. Advancement Med. Life Sci. 2015;3(3):1–5. [Google Scholar]
- Ultee A., Smid E. Influence of carvacrol on growth and toxin production by Bacillus cereus. Int. J. Food Microbiol. 2001;64(3):373–378. doi: 10.1016/s0168-1605(00)00480-3. [DOI] [PubMed] [Google Scholar]
- Valgas C., Souza S.M.d., Smânia E.F.A., Smânia A., Jr. Screening methods to determine antibacterial activity of natural products. Braz. J. Microbiol. 2007;38(2):369–380. [Google Scholar]
- Varghese S.A., Siengchin S., Parameswaranpillai J. Essential oils as antimicrobial agents in biopolymer-based food packaging - a comprehensive review. Food Biosci. 2020;38 [Google Scholar]
- Zhang J., Li L., Lv Q., Yan L., Wang Y., Jiang Y. The fungal CYP51s: their functions, structures, related drug resistance, and inhibitors. Front. Microbiol. 2019;10:691. doi: 10.3389/fmicb.2019.00691. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data included in article/supplementary material/referenced in article.