Abstract
The whiD locus, which is required for the differentiation of Streptomyces coelicolor aerial hyphae into mature spore chains, was localized by map-based cloning to the overlap between cosmids 6G4 and D63 of the minimal ordered library of Redenbach et al. (M. Redenbach et al., Mol. Microbiol. 21:77–96, 1996). Subcloning and sequencing showed that whiD encodes a homologue of WhiB, a protein required for the initiation of sporulation septation in S. coelicolor. WhiD and WhiB belong to a growing family of small (76- to 112-residue) proteins of unknown biochemical function in which four cysteines are absolutely conserved; all known members of this family are found in the actinomycetes. A constructed whiD null mutant showed reduced levels of sporulation, and those spores that did form were heat sensitive, lysed extensively, and were highly irregular in size, arising at least in part from irregularity in septum placement. The whiD null mutant showed extreme variation in spore cell wall deposition; most spores had uniformly thin (20- to 30-nm) walls, but spore chains were frequently observed in which there was irregular but very pronounced (up to 170 nm) cell wall thickening at the junctions between spores. whiD null mutant spores were frequently partitioned into irregular smaller units through the deposition of additional septa, which were often laid down in several different planes, very close to the spore poles. These “minicompartments” appeared to be devoid of chromosomal DNA. Two whiD promoters, whiDp1 and whiDp2, were identified, and their activities were analyzed during development of wild-type S. coelicolor on solid medium. Both promoters were developmentally regulated; whiDp1 and whiDp2 transcripts were detected transiently, approximately at the time when sporulation septa were observed in the aerial hyphae.
Streptomycetes are gram-positive soil bacteria with a mycelial growth habit (7). Germination of spores gives rise to a vegetative mycelium consisting of a branching network of multigenomic hyphae. In order to disperse themselves, streptomycetes develop specialized aerial hyphae that grow out of the aqueous environment of the vegetative mycelium into the air, giving the colonies a characteristic fuzzy appearance (23). These aerial hyphae differentiate into chains of exospores, a process that begins with the synchronous deposition of 50 or more sporulation septa at ∼1 μm intervals at the tips of the hyphae (7). The formation of sporulation septa creates, for the first time in the developing colony, unigenomic cells, referred to as prespores. These cylindrical prespore compartments subsequently mature to form chains of thick-walled, ovoid spores, during which time the colonies develop a characteristic color, gray in the case of the model species Streptomyces coelicolor A3(2), due to the synthesis of a polyketide spore pigment (11).
Hopwood et al. (19) identified sporulation-deficient mutants of S. coelicolor by virtue of their inability to synthesize the gray spore pigment, thereby remaining white, even on prolonged incubation on plates. Fifty of these white (whi) mutants have been mapped genetically, and current information suggests that they represent eight separate loci: whiA, whiB, whiD, whiE, whiG, whiH, whiI, and whiJ (6, 8, 38). Mutations in six of these loci, referred to as the early whi genes (whiA, whiB, whiG, whiH, whiI, and whiJ), essentially abolish the formation of sporulation septa (1, 6, 8, 9, 12, 13, 38, 39). whiE specifies the spore pigment itself, and whiE mutants do not appear to be morphologically defective (6, 11, 26, 32). Although other loci required for sporulation have subsequently been described in S. coelicolor (31, 35, 40), some of which affect the late stages of sporulation, of the originally described whi strains only the whiD mutant formed sporulation septa but failed to go on to produce mature spores. The whiD mutant formed spores at wild-type abundance but these were unpigmented, were thin-walled, and showed frequent lysis (6, 32). Here we describe the map-based cloning of whiD, show that WhiD is a member of a growing family of actinomycete proteins of unknown biochemical function that includes WhiB (in which four cysteine residues are absolutely conserved), describe the phenotype of a constructed whiD null mutant, and show that whiD is developmentally regulated at the level of transcription.
MATERIALS AND METHODS
Bacterial strains, plasmids, growth conditions, and protoplast transformation.
S. coelicolor strains used are summarized in Table 1 and were cultured on R5 (20), MM (20) containing 0.5% (wt/vol) mannitol as the carbon source, or MS agar (mannitol plus soya flour) (18), supplemented with uracil and histidine where necessary. For transformation of S. coelicolor, strains were cultured in YEME liquid medium (20) and protoplasts were prepared and transformed as described previously (20). To bypass the methyl-group specific restriction system of S. coelicolor during protoplast transformation, unmethylated plasmid and cosmid DNA was isolated from the dam dcm hsdS E. coli strain ET12567 (30). To stimulate integration by homologous recombination when using cosmid or plasmid replicons that do not replicate in Streptomyces, double-stranded DNA was alkaline denatured before protoplast transformation (33). The vectors used were pDH5 (17), pKC1132 (4), and pSET152 (4).
TABLE 1.
Derivatives of S. coelicolor A3(2) used in this study
| Strain | Genotype | Source or reference |
|---|---|---|
| Wild type | Pgl+ SCP1+ SCP2+ | 20 |
| M145 | Pgl+ SCP1− SCP2− | 20 |
| J1915 | ΔglkA119 Pgl+ SCP1− SCP2− | 24 |
| J2152 | ΔglkA119 whiD::hyg Pgl+ SCP1− SCP2− | This work |
| J1508a | hisA1 strA1 uraA1 Pgl− SCP1NF SCP2− | 22 |
| J1979a | hisA1 strA1 uraA1 sigF::tsr Pgl− SCP1NF SCP2− | 35 |
| J1942 | strA1 uraA1 Pgl+ SCP1+ SCP2+ | This work |
| J243 | strA1 uraA1 whiD16 Pgl+ SCP1+ SCP2+ | K. F. Chater |
NF, normal fertility. The SCP1 plasmid is integrated into the 9-o'clock region of the chromosome.
Conjugation from E. coli into Streptomyces.
Because whiD mutant spores were found to be temperature sensitive (see Results), the method of Flett et al. (14) was adapted to avoid heat shocking; high frequencies of exconjugants were obtained without difficulty. pSET152, pKC1132, or their derivatives were introduced by transformation into E. coli ET12567 containing the RK2 derivative pUZ8002 (34). pUZ8002 supplies transfer functions to oriT-carrying plasmids, such as pSET152 and pKC1132, but is not efficiently transferred itself because of a mutation in its own oriT. E. coli containing pSET152, pKC1132, or their derivatives was grown in L broth to A600 of 0.4 to 0.6, washed twice with an equal volume of fresh medium, and resuspended in 1/10th the volume of L broth. S. coelicolor J243 was grown on one plate of MS agar and spores were harvested after 5 days and resuspended in 2 ml of 20% (vol/vol) glycerol. Then, 0.5 ml of fresh spores and 0.5 ml of fresh E. coli suspension were mixed and spread on one plate of MS agar containing 10 mM MgCl2. After incubation for 16 to 20 h at 30°C, exconjugants were selected by overlaying the plate with 1 ml of water containing 0.5 mg of nalidixic acid (to kill E. coli) and 1 mg of apramycin (to select Streptomyces exconjugants).
Construction of a whiD null mutant.
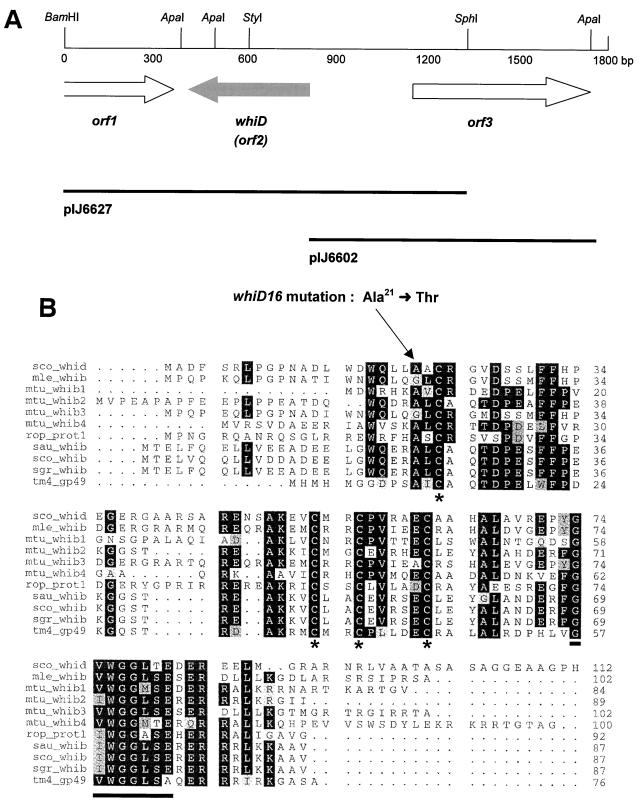
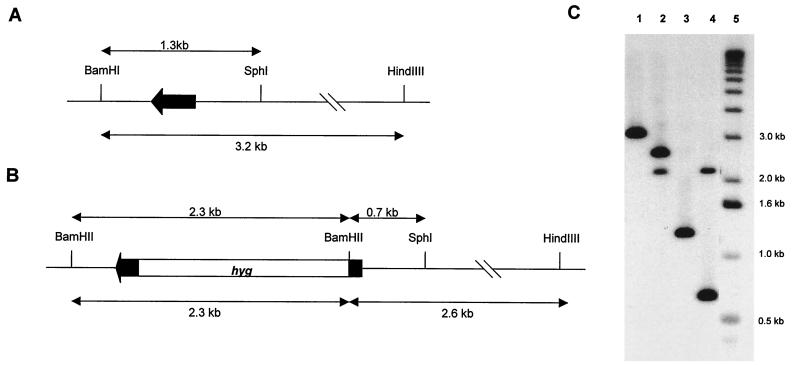
A 1.3-kb BamHI-SphI fragment carrying whiD was cloned into pUC19 digested with BamHI and SphI, and a whiD null mutant allele was created by blunt-end cloning a 1.8-kb hyg cassette (46) into the StyI site internal to whiD (see Fig. 3A and 4). Additional flanking sequences were added to this plasmid by inserting a 2-kb SphI-HindIII fragment upstream of whiD and a 3-kb BamHI-KpnI fragment downstream to create pIJ6628. This reconstructed a contiguous 6.3-kb segment of the chromosome but carrying the 1.8-kb hyg insertion in whiD. The entire 8.1-kb insert was removed from pIJ6628 as a PvuII fragment and cloned into the EcoRV site of pKC1132. Finally, a 1.3-kb BglII fragment carrying the counterselectable glkA gene was ligated into the unique BglII site in the vector to create pIJ6629.
FIG. 3.
(A) Genetic organization of the 1.8-kb segment of DNA containing whiD. The positions of the three protein-coding regions are indicated by arrows, and restriction sites referred to in the text are marked. The extent of the subclones used in complementation tests is shown below. (B) Alignment of the predicted amino acid sequence of WhiD with related proteins. The four absolutely conserved cysteines are marked by asterisks, and the highly conserved C-terminal motif G(V/I)WGGLSE is underlined. The alanine-to-threonine substitution arising from the whiD16 mutation is indicated. The proteins and their corresponding gene accession numbers are as follows: sco_whid, S. coelicolor WhiD (AJ010601); mle_whib, M. leprae WhiB (U00015); mtu_whib1, M. tuberculosis WhiB1 (Z95120); mtu_whib2, M. tuberculosis WhiB2 (AL021840); mtu_whib3, M. tuberculosis WhiB3 (Z77165); mtu_whib4, M. tuberculosis WhiB4 (AL022121); rop_whib, Rhodococcus opacus WhiB (AF030176); sau_whib, Streptomyces aureofaciens WhiB (L22864); sco_whib, S. coelicolor WhiB (X62287); sgr_whib, Streptoverticillium griseocarneum WhiB (X68708); tm4_gp49, mycobacterial phage TM4 GP49 (AF068845).
FIG. 4.
Construction of a whiD null mutant. (A and B) Restriction maps of the wild-type and whiD::hyg null mutant alleles, respectively; the black arrow represents the whiD gene. (C) Southern blot analysis of chromosomal DNA from J1915 (whiD+; lanes 1 and 3) and J2152 (whiD::hyg; lanes 2 and 4) digested with BamHI and HindIII (lanes 1 and 2) or BamHI and SphI (lanes 3 and 4). The size markers (lane 5) are the 1-kb ladder (Bethesda Research Laboratories). The probe used was the 1.3-kb BamHI-SphI whiD fragment (A).
pIJ6629 was introduced into S. coelicolor J1915 (ΔglkA119) by mating from E. coli, and exconjugants in which the plasmid had presumptively integrated at the whiD locus by single-crossover homologous recombination were selected with apramycin. After one round of nonselective growth, putative whiD::hyg null mutants in which the delivery plasmid had been lost were selected on MM containing 100 mM 2-deoxyglucose and 50 μg of hygromycin per ml.
RNA isolation and S1 nuclease protection analysis.
The developmental time course of RNA samples described by Kelemen et al. (25) was used to analyze expression of whiD. The whiD promoter region was mapped with a 0.6-kb probe generated by PCR from pIJ6626, using a 5′-end-labeled oligonucleotide primer internal to whiD (5′-CCAGGAGCTGCCAGTCCCAC-3′) and the universal sequencing primer. Oligonucleotide primers were labelled and S1 nuclease protection assays were set up as described earlier (25).
Microscopy.
For light microscopy, coverslips were placed gently on the surface of S. coelicolor colonies grown on solid medium for 4 days and then lifted and analyzed by phase-contrast light microscopy with oil immersion (Carl Zeiss; 100/2.0 objective).
For transmission electron microscopy, colonies were fixed in 2% (vol/vol) glutaraldehyde in 0.05 M sodium cacodylate (16), stained with osmic acid, and embedded in LR White resin according to the manufacturer's instructions (The London Resin Co.). Sections were cut on an Ultracut Microtome (Reichart) and examined in a JEOL 1200 EX electron microscope.
For scanning electron microscopy, colonies were mounted on the surface of an aluminum stub with O.C.T. compound (BDH Laboratory Supplies, Poole, United Kingdom), plunged into liquid nitrogen slush at approximately −210°C to cryopreserve the material, and transferred to the cryostage of a CT1500HF cryo-transfer system (Oxford Instruments, Oxford, United Kingdom), attached to a Phillips XL30 FEG scanning electron microscope (Phillips Electron Optics, FEI UK Ltd, Cambridge, United Kingdom). Surface frost was sublimated at −95°C for 3 min before sputter coating the sample with platinum for 2 min at 10 mA at colder than −110°C. Finally, the sample was moved onto the cryostage in the main chamber of the microscope, held at approximately −140°C, and viewed at 3 kV. Photographs were taken using Ilford FP4 120 roll film in a Linhof camera.
RESULTS
Localization of the whiD gene on the physical map of the S. coelicolor chromosome.
Redenbach et al. (36) constructed a minimal, ordered cosmid library covering the S. coelicolor chromosome by using the E. coli vector Supercos-1. Although these cosmids cannot replicate autonomously in S. coelicolor, selection for kanamycin resistance after protoplast transformation results in the recovery of isolates in which the cosmid has integrated into the chromosome via insert-directed homologous recombination. The cosmids can therefore be used to clone genes by complementation (36), and we used this approach to isolate whiD.
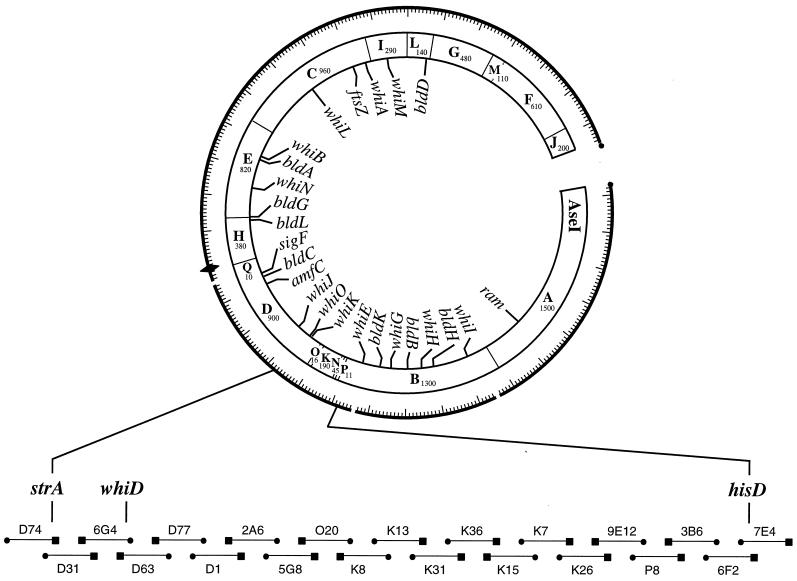
Chater (6) mapped whiD genetically to a position between strA and uvsB, in the “7-o'clock” region of the chromosome, and while strA has been mapped physically (to the overlap between cosmids D31 and D74 [36]), uvsB has not. However, hisD, the next locus counterclockwise of uvsB, has also been mapped physically (to the overlap between cosmids 6F2 and 7E4 [36]). Therefore, we reasoned that whiD must reside on one or two of the twenty overlapping cosmids covering the strA-hisD interval (Fig. 1). Each of these cosmids was passaged through the methylation-deficient E. coli strain ET12567 and introduced individually into the whiD strain J243 by protoplast transformation. Transformants were patched onto MS agar, and potentially complementing cosmids were identified initially by restoration of spore pigment production and confirmed by phase-contrast examination of spore morphology. Only the overlapping cosmids 6G4 and D63 complemented the whiD phenotype of J243; a small percentage of transformants carrying D63 remained white, presumably due to cosmid instability or gene conversion.
FIG. 1.
A simplified version of the combined physical and genetic map of the S. coelicolor chromosome in the 7-o'clock region, showing the locations of developmental genes and the 21 ordered cosmids that span the strA to hisD interval (adapted from references 36 and 40). The location of whiD is shown on the overlap between cosmids 6G4 and D63, as determined by complementation. The locations of other cloned developmental genes on the cosmid contig are shown on the inside of the circle. The cosmids and their overlaps are arbitrarily shown to be of equal length (the average insert size of the cosmids is 37.5 kb and of the overlaps 12.5 kb [36]). The sizes of the AseI fragments are given in kilobases (27). Symbols: ⧫, oriC; ●, T3 end; ■, T7 end.
Construction of a whiD-whiD+ congenic pair.
Because the original whiD mutant, C16, has been lost, and J243 arose from a cross used in the original genetic mapping of whiD, there existed no whiD-whiD+ congenic pair to allow valid phenotypic comparison. To construct such a pair, we took advantage of the slight instability of J243 carrying cosmid 6G4 or D63 in the absence of selection. Such strains carry tandem duplications of approximately 40 kb of DNA, and we found that they gave rise to kanamycin-sensitive (Kans) colonies at readily detectable frequencies, presumptively due to the excision of the cosmid by homologous recombination. A gray 6G4 transformant of J243 was cultured nonselectively through two rounds of growth and sporulation, plated for single colonies on MS agar, and replica plated to identify Kans derivatives. Both gray and white Kans derivatives were identified, presumably reflecting the nature of the remaining whiD allele; gray colonies retained the wild-type allele originally present on the cosmid, while white colonies retained the mutant allele from J243. The structure of the whiD region of the chromosome of a representative gray colony was confirmed by Southern blot analysis, and this strain was designated J1942. It subsequently turned out that, although the insert in cosmid 6G4 is approximately 41 kb in size, whiD lies only 1 kb from the end of the insert (see below), making it relatively unlikely that homologous recombination would occur at a significant frequency in the short interval. It therefore seems likely that gene conversion, in which the wild-type allele acted as a template for repair synthesis of the whiD16 allele, might have occurred in the generation of J1942 and the other gray Kans derivatives of J243/6G4.
whiD spores are heat sensitive.
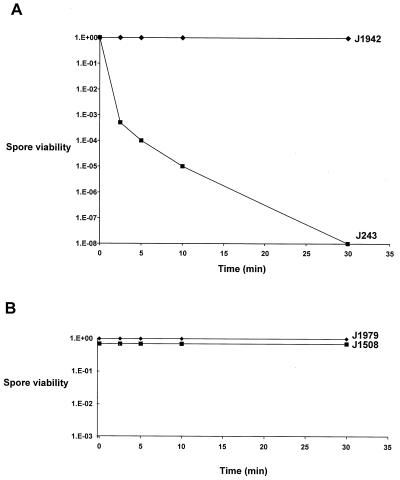
Our initial attempts to introduce plasmids into J243 from E. coli by mating yielded extremely low frequencies of exconjugants. Preliminary control experiments designed to determine the reason for these low frequencies suggested that whiD spores might be less able to survive the 10-min 50°C heat shock used to induce germination in our standard mating protocol (14). To investigate this possibility, spores of J243 and the congenic whiD+ strain J1942 were incubated in 2xYT or 20% (vol/vol) glycerol at 50°C for various times, and the spore survival was assessed by plating serial dilutions on MS agar containing uracil. Whereas spore viability in J1942 (whiD+) was virtually unaffected by a 30-min incubation at 50°C, J243 (whiD) spore viability dropped by a factor of 108 in the same period (Fig. 2A). This loss of viability was not simply an osmotic effect, since spores of J243 (whiD) showed no loss of viability when incubated in 2xYT or 20% (vol/vol) glycerol at 25°C (data not shown).
FIG. 2.
Heat inactivation curves for spores of J243 (whiD) and J1942 (whiD+) (A) and spores of J1979 (sigF) and J1508 (sigF+) (B). Spores were incubated in 20% (vol/vol) glycerol at 50°C.
One possible explanation for the heat sensitivity of whiD spores is that they fail to develop the thick spore wall characteristic of the wild type (32). To investigate this possibility, we examined the heat sensitivity of another strain that has thin-walled spores. sigF encodes a spore-specific RNA polymerase sigma factor, ςF, that is required for spore maturation and, like whiD mutants, sigF mutants are defective in spore wall thickening (25, 35, 44). However, when spores of the sigF null mutant J1979 were subjected to the same heat treatment as those of whiD, we found that they were as heat resistant as those of the congenic sigF+ parent strain, J1508 (Fig. 2B).
Subcloning of whiD from cosmid 6G4.
Cosmid 6G4 was digested with BamHI, generating seven fragments ranging in size from 2 to 16 kb. Each fragment was subcloned into the E. coli vector pDH5, introduced into J243, and thiostrepton-resistant transformants were selected in which the plasmid had presumptively integrated into the chromosome via insert-directed homologous recombination. Only the largest BamHI fragment from cosmid 6G4, fragment I, complemented J243. Further analysis showed that fragment I comprised the entire Supercos-1 vector with 1.8 kb of Streptomyces DNA from the “T3 end” of the 6G4 insert and 7 kb of DNA from the “T7 end.” Self-ligation of fragment I (in the absence of pDH5) gave rise to pIJ5900, which also complemented J243. Because the T3 end and not the T7 end of cosmid 6G4 overlaps D63 (36), the other cosmid that complemented the whiD mutant, we reasoned that the 1.8-kb fragment of DNA must contain whiD, and this was confirmed by deleting the 7-kb fragment from the T7 end of pIJ5900 to create pIJ5908, which also complemented J243.
Because the whiD mutant formed spore chains, albeit of a somewhat irregular nature, it was not trivial to judge complementation of J243 in the phase-contrast microscope. The discovery that whiD spores were heat sensitive gave us a useful additional phenotype by which to assess complementation of the mutant. Exploiting this discovery, we went back to J243 carrying cosmid D63 or cosmid 6G4 and through all the successive rounds of subcloning of whiD to assess complementation by the heat sensitivity of the spores. In every case, the fragments that complemented the morphological defects of J243 also restored wild-type levels of heat resistance to the spores. Apart from its utility in scoring complementation, these results provided strong evidence that both phenotypes arise from the same mutation.
whiD is a member of a family of genes that includes whiB.
The nucleotide sequence of the 1.8-kb fragment containing whiD was determined, and protein-coding sequences were predicted with the aid of the FRAME program (3). One incomplete (orf1) and two complete potential protein coding sequences (orf2 and orf3) were identified (Fig. 3A). orf2 and orf3 were each subcloned into the vector pSET152, which integrates site specifically into the S. coelicolor chromosome at the phage ΦC31 attB site (4), to create pIJ6627 and pIJ6602, respectively (Fig. 3A), and these two plasmids were introduced into J243 by mating from E. coli. orf3 had no effect on the whiD phenotype of J243, whereas orf2 fully complemented both the morphological defects and the spore temperature sensitivity of the strain. As a consequence, orf2 was designated whiD. To confirm that a significant base change was indeed present in whiD in J243, the entire 1.8-kb region was amplified from J243 by PCR and sequenced; a single nucleotide difference from the wild type was identified. whiD16 has a CG to TA transition (at position 700 in database submission AJ010601), giving rise to an alanine to threonine substitution at position 21 in the primary amino acid sequence of WhiD (Fig. 3B). In addition, the whiD allele of J1942, the morphologically wild-type strain derived from J243/6G4 by homogenotisation, was amplified by PCR and shown by sequencing to have lost the whiD16 mutation.
Global similarity searches of the NCBI databases showed that the incomplete open reading frame, orf1, encodes the C-terminal 118 residues of a member of the LysR family of transcriptional regulators, with highest similarity to GltC, involved in the regulation of glutamate biosynthesis in Bacillus subtilis. The predicted product of orf3 is 203 amino acids long and is a typical member of the two-component response regulator family of proteins, showing, for example, 32% identity with DegU, which is involved in the control of competence and degradative enzyme biosynthesis in B. subtilis (10).
WhiD is 112 amino acids long and is a member of a family of proteins that includes WhiB, required for sporulation septum formation in S. coelicolor and Streptomyces aureofaciens (12, 28, 29), six proteins revealed by genome sequencing in the related actinomycete pathogen Mycobacterium tuberculosis, proteins from two other actinomycetes, Rhodococcus opacus (41) and Streptoverticillium griseocarneum (42), and one protein encoded by the mycobacterial phage TM4 (15). Several genes encoding WhiD-related proteins are also present in the unfinished genome sequence of Mycobacterium leprae. An alignment of some of these proteins, shown in Fig. 3B, reveals several striking features. First, all of the proteins are small, varying from 76 to 112 residues in length. Second, four cysteine residues are completely conserved in all the members of the family and, third, the motif G(V/I)WGGLSE is highly conserved close to the C terminus.
Construction and phenotypic characterization of a whiD null mutant, J2152.
A whiD null mutant allele was constructed in vitro by inserting a hygromycin resistance gene (hyg) at a unique StyI restriction site internal to whiD (Fig. 3A and 4B). This mutant allele was used to replace the wild-type allele in J1915, a plasmid-free, glkA derivative of the wild-type strain, using the method of Buttner et al. (5). This method makes use of the counterselectable glucose kinase gene (glkA) which allows a positive selection to be made for gene replacement, provided that the mutations are constructed in a strain carrying a deletion of glkA. The genomic structures of five independently isolated whiD mutants were confirmed by Southern blot analysis (e.g., Fig. 4C), and one was designated J2152.
On plates, colonies of J2152 (whiD::hyg) produced wild-type levels of aerial mycelium but remained completely white, even on prolonged incubation. In the scanning and transmission electron microscopes, it was immediately apparent that the phenotype of J2152 (whiD::hyg) was more severe than that of J243 (whiD16), implying that whiD16 is not a null allele (consistent with the relatively conservative substitution of threonine for alanine in WhiD16). However, it should be noted that the ΔglkA119 allele, present in the genetic background of J2152 (whiD::hyg) but absent from that of J243 (whiD16), causes ectopic sporulation in the substrate hyphae (the Esp phenotype) on certain media (24). Spore chains in Esp mutants are morphologically normal, and the available evidence suggests that the ΔglkA119 deletion simply relieves suppression of sporulation in substrate hyphae (24). However, the formal possibility that the ΔglkA119 genetic background could potentiate the whiD null mutant phenotype has not been excluded.
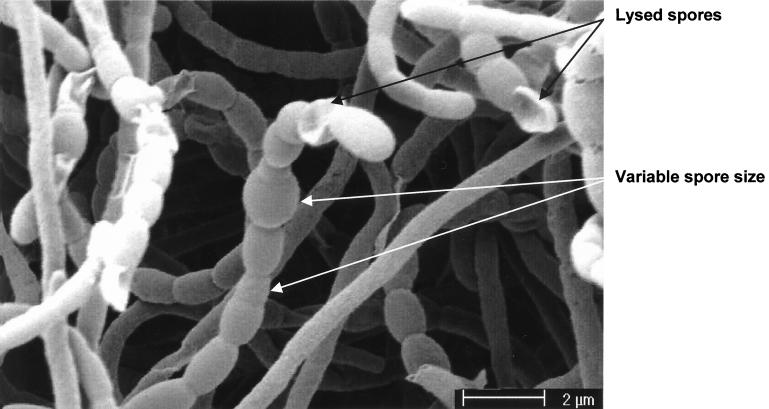
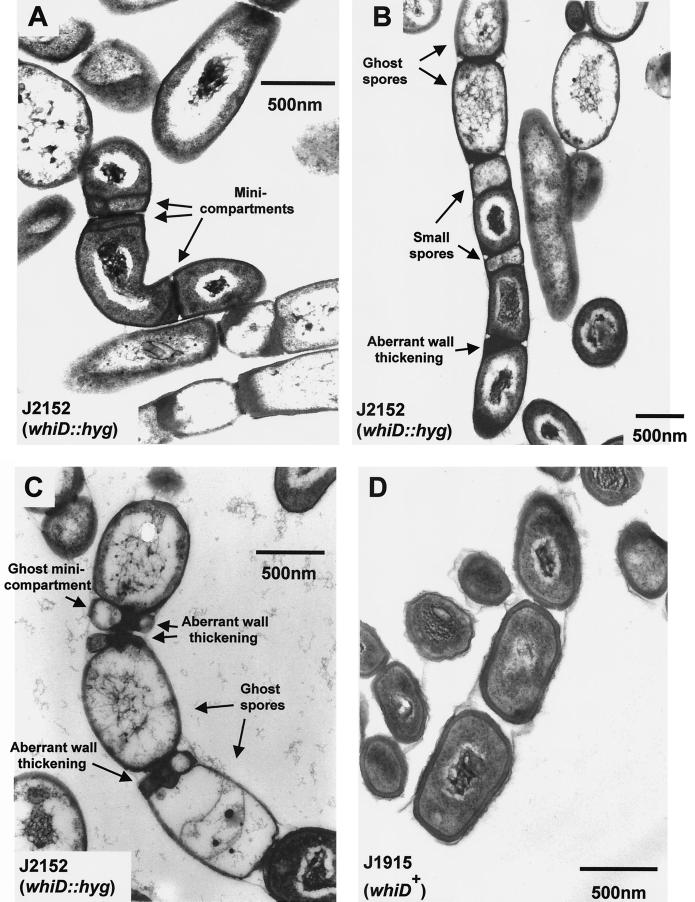
Although J243 (whiD16) forms defective spores, the level of sporulation in this strain is the same as in its congenic whiD+ parent, J1942. In contrast, based on visual inspection in the scanning electron microscope, the level of sporulation in J2152 (whiD::hyg) was approximately only 25% of that found in its congenic whiD+ parent, J1915. In addition, there was considerable irregularity in spore size (Fig. 5), arising at least in part from irregularity in sporulation septum placement (Fig. 6B) but perhaps also from spore swelling and lysis (Fig. 6C). Strikingly, we frequently observed additional, aberrant sporulation septa within spores, deposited close to the poles and often laid down in several planes (Fig. 6A). These additional septa further divided the spores into irregular “minicompartments” apparently devoid of chromosomal DNA (Fig. 6A). In young spore chains these minicompartments were usually box-like in appearance (Fig. 6A), but in “mature” spore chains they rounded and lysed to form ghost minicompartments analogous to the larger ghost spores seen in the same chains (Fig. 6C). Another striking feature was the extreme variation in cell wall deposition between and within spores. In most mature J2152 spores the walls were uniformly thin (20 to 30 nm, compared to 60 to 80 nm in J1915; Fig. 6D), but spore chains were frequently observed in which there was pronounced but irregular cell wall thickening at the junctions between spores (Fig. 6B and C). In certain cases these deposits were 170-nm thick (Fig. 6B). After 5 days of growth, the vast majority of spores that had formed were lysed and often swollen (Fig. 6C). Thermosensitivity tests were carried out on J2152 spores and yielded results very similar to those obtained with spores from the whiD point mutant J243 (data not shown). J2152 was fully complemented for all aspects of its phenotype by pIJ6627, the pSET152 derivative carrying whiD alone.
FIG. 5.
Scanning electron micrograph showing the variability in spore size and spore lysis associated with the constructed whiD null mutant J2152. Colonies were grown on MS agar.
FIG. 6.
Transmission electron micrographs showing young (A), intermediate (B), and mature (C) spore chains of the constructed whiD null mutant J2152 and a mature spore chain of the congenic whiD+ strain J1915 (D). Colonies were grown on MS agar.
Transcription of whiD is developmentally regulated.
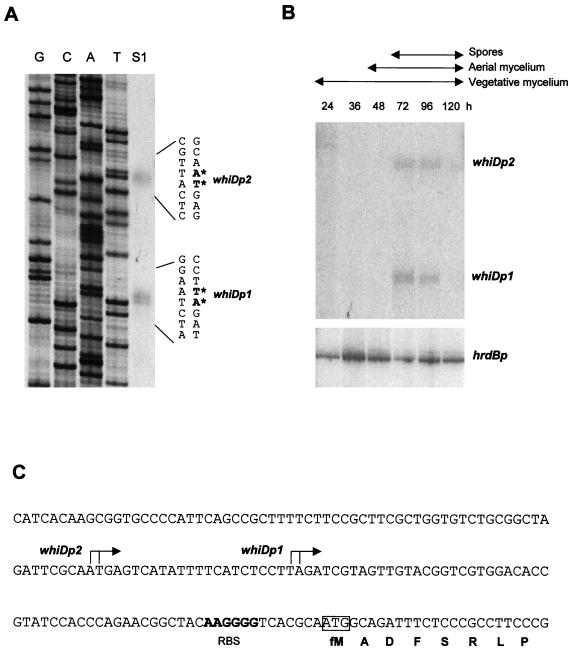
High resolution S1 nuclease mapping of the whiD promoter region was performed by using a PCR-generated probe and RNA isolated from wild-type S. coelicolor grown for 72 h on solid medium. Two promoters were identified, initiating transcription 62 to 63 bp (whiDp1) and 84 to 85 bp (whiDp2) upstream of the whiD ATG start codon (Fig. 7A and C).
FIG. 7.
Transcriptional analysis of whiD. (A) High-resolution S1 nuclease mapping of the 5′ ends of the whiDp1 and whiDp2 transcripts. The lane labeled “S1” represents the DNA fragments protected by RNA initiating from whiDp1 and whiDp2. The most likely transcription start points are indicated by the asterisks. Lanes labeled G, C, A, and T represent a dideoxy sequencing ladder generated by using the same oligonucleotide that was used to generate the S1 mapping probe. The RNA used was from the 72-h time point from panel B. (B) S1 nuclease protection analysis of transcription from whiDp1, whiDp2, and hrdBp during development. RNA was isolated from wild-type S. coelicolor grown on cellophane discs on MM containing mannitol as carbon source. The time points (in hours) at which mycelium was harvested for RNA isolation, and the presence of vegetative mycelium, aerial mycelium, and spores as judged by microscopic examination, are shown. The hrdB panel from this figure was published previously (25, 26, 39) and is shown here for comparison with the whiD data. (C) Nucleotide sequence of the whiD promoter region indicating the whiDp1 and whiDp2 transcription start points, the putative ribosome binding site (RBS), and the start of the whiD coding sequence.
The pattern of transcription from whiDp1 and whiDp2 during development of wild-type S. coelicolor on solid medium was monitored by S1 nuclease protection. Following previous work (25, 26, 39), we used a time course of RNA samples that had already been used to assess the developmental pattern of transcription of a number of sporulation genes, including the early sporulation gene whiH (39), the early sporulation-specific sigma factor gene whiG (25), the late sporulation-specific sigma factor gene sigF (25), and whiE (26), the locus specifying the polyketide spore pigment. As a positive internal control for these experiments, transcription of hrdB, encoding the principal (essential) sigma factor of S. coelicolor, was also analyzed. The data for hrdB have been published previously (25, 26, 39) and are reproduced here for comparison. No attempt was made to fractionate the harvested cell material used for RNA preparation; thus, for example, the late samples contained vegetative and aerial mycelium as well as spores (25, 26).
The whiDp1 and whiDp2 promoters were found to be developmentally regulated: both transcripts first appeared at 72 h, the time at which sporulation was first detected in the culture, and were present at similar levels 24 h later (Fig. 7B). The whiD transcripts were not seen during vegetative growth or during aerial mycelium formation and were almost undetectable at 120 h in mature colonies. At the low level of resolution of these experiments, the developmental profiles of the two whiD transcripts were very similar to those of the sigF (25) and whiE transcripts (26), but the whiD transcripts clearly appeared later than the whiH transcript (39). whiD transcription was undetectable in RNA isolated from submerged culture, conditions that do not support sporulation of S. coelicolor (data not shown).
DISCUSSION
Taking advantage of the minimal, ordered cosmid library of Redenbach et al. (36), we have used a map-based cloning strategy to isolate the whiD gene. This strategy (36) relies on the fact that, although these cosmids cannot replicate autonomously in S. coelicolor, they integrate reasonably efficiently into the chromosome via insert-directed homologous recombination. As an alternative to the construction of shotgun libraries, this is an attractive approach for isolating any S. coelicolor locus for which an approximate genetic map position has been determined, and it has also been used to clone two other developmental genes, whiI (2) and bldC (G. H. Kelemen and A. C. Hunt, personal communication).
Sequencing of whiD showed that it encodes a homologue of WhiB, another protein required for sporulation in S. coelicolor and also in S. aureofaciens. S. coelicolor whiB mutants produce abnormally long, tightly coiled aerial hyphae and are completely unable to form sporulation septa (6, 12, 13). Genome sequencing and directed approaches have identified a growing number of whiD-related genes in the actinomycete genera Streptomyces, Mycobacterium, Streptoverticillium, and Rhodococcus (Fig. 3B [43]). This includes, in addition to whiB and whiD, at least four further genes in S. coelicolor itself (43). To date, no members of the WhiB-WhiD family of proteins have been identified outside the actinomycetes. It is interesting to note that, although it does not sporulate, Mycobacterium tuberculosis has likely orthologues of both WhiD and WhiB. This conclusion is based on the level of amino acid sequence similarity between the S. coelicolor and M. tuberculosis proteins and the conservation of gene organization around the corresponding genes. The M. tuberculosis WhiD orthologue (mtu_whib3, Fig. 3B) shows 60% amino acid sequence identity to WhiD, and the M. tuberculosis WhiB orthologue (mtu_whib2, Fig. 3B) shows 71% amino acid sequence identity to WhiB. As in S. coelicolor, genes encoding an extracytoplasmic function (ECF) sigma factor and inosine-5′-monophosphate dehydrogenase are found upstream of the M. tuberculosis whiD orthologue and groELS is found downstream, although other genes intervene in one species or the other such that the absolute gene organization is not identical. Similarly, the two genes upstream of whiB in S. coelicolor, encoding proteins of unknown function, are also present upstream of the M. tuberculosis whiB orthologue. Apart from whiB and whiD in S. coelicolor, there is only one other report of the disruption of a whiD-related gene in any organism; inactivation of whiB3, the whiD orthologue of Mycobacterium smegmatis (64% identity), did not affect growth or the dormancy response (21).
The biochemical function of the WhiB-WhiD family of proteins remains unknown, although it has been suggested that they may function as transcription factors (12, 43). The most striking aspect of their primary amino acid sequence is the perfect conservation of four cysteines. This conservation suggests that these residues may act as ligands for a metal cofactor such as zinc, copper or an iron-sulfur cluster or, alternatively, that they may be involved in intramolecular disulfide bond formation. Some of these speculative possibilities would be consistent with redox regulation of the activity of WhiD and other members of the family.
The constructed whiD null mutant, J2152, had a more severe phenotype than J243, the strain carrying the whiD16 point mutation that originally defined this locus. In contrast to the situation in J243, the level of sporulation in J2152 was considerably lower than in the congenic whiD+ parent, suggesting that whiD not only influences prespore maturation but also affects the initiation of sporulation septation. Consistent with this, there was considerable variation in spore size in the whiD null mutant (Fig. 5). This variation arose in part from the swelling that seemed to accompany the lysis of “mature” spores (Fig. 6C), but it also clearly reflected irregularity in septum placement (Fig. 6B). In addition to variation in spore size, one of the most striking aspects of the whiD null mutant phenotype was that spore-sized compartments were frequently partitioned into irregular, smaller units through the deposition of additional septa, often laid down in several different planes. The additional septa that defined these minicompartments were usually found very close to the poles of the spores and the minicompartments appeared to be devoid of chromosomal DNA (Fig. 6A). These observations are somewhat reminiscent of minicell formation in unicellular bacteria. In E. coli (1) and B. subtilis (37) a system for preventing division at old cell poles was discovered through the characterization of min mutants. These mutants often divide correctly at midcell to produce equal-sized daughter cells, but with approximately equal frequency they divide close to an existing cell pole to produce a small, usually anucleoidal minicell (45).
ACKNOWLEDGMENTS
We thank Maureen Bibb, Mervyn Bibb, Keith Chater, and David Hopwood for their helpful comments on the manuscript, Gabriella Kelemen for the gift of the developmental time course of RNA samples, Sue Bunnewell for hand-printing the transmission electron micrographs, Tobias Kieser for help in preparing Fig. 1, and Julian Parkhill for helpful discussion.
This work was supported by a John Innes Foundation studentship (to V.M.), by a BBSRC studentship (to W.J.P.), by a Lister Institute Research Fellowship (to M.J.B.), and by grants-in-aid to the John Innes Centre from the BBSRC and the John Innes Foundation.
REFERENCES
- 1.Adler H I, Fisher W D, Cohen A, Hardigree A A. Miniature Escherichia coli cells deficient in DNA. Proc Natl Acad Sci USA. 1967;57:321–326. doi: 10.1073/pnas.57.2.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aínsa J A, Parry H D, Chater K F. A response regulator-like protein that functions at an intermediate stage of sporulation in Streptomyces coelicolor A3(2) Mol Microbiol. 1999;34:607–619. doi: 10.1046/j.1365-2958.1999.01630.x. [DOI] [PubMed] [Google Scholar]
- 3.Bibb M J, Findlay P R, Johnson M W. The relationship between base composition and codon usage in bacterial genes and its use for the simple and reliable identification of protein-coding sequences. Gene. 1984;30:157–166. doi: 10.1016/0378-1119(84)90116-1. [DOI] [PubMed] [Google Scholar]
- 4.Bierman M, Logan R, O'Brien K, Seno E T, Rao R N, Schoner B E. Plasmid cloning vectors for the conjugal transfer of DNA from Escherichia coli to Streptomyces spp. Gene. 1992;116:43–49. doi: 10.1016/0378-1119(92)90627-2. [DOI] [PubMed] [Google Scholar]
- 5.Buttner M J, Chater K F, Bibb M J. Cloning, disruption, and transcriptional analysis of three RNA polymerase sigma factor genes of Streptomyces coelicolor A3(2) J Bacteriol. 1990;172:3367–3378. doi: 10.1128/jb.172.6.3367-3378.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chater K F. A morphological and genetic mapping study of white colony mutants of Streptomyces coelicolor. J Gen Microbiol. 1972;72:9–28. doi: 10.1099/00221287-72-1-9. [DOI] [PubMed] [Google Scholar]
- 7.Chater K F. Taking a genetic scalpel to the Streptomyces colony. Microbiology. 1998;144:1465–1478. doi: 10.1099/00221287-144-6-1465. [DOI] [PubMed] [Google Scholar]
- 8.Chater K F, Merrick M J. Approaches to the study of differentiation in Streptomyces coelicolor A3(2) In: MacDonald K D, editor. Second international symposium on the genetics of industrial microorganisms. London, United Kingdom: Academic Press; 1976. pp. 583–593. [Google Scholar]
- 9.Chater K F, Bruton C J, Plaskitt K A, Buttner M J, Méndez C, Helmann J D. The developmental fate of Streptomyces coelicolor hyphae depends on a gene product homologous with the motility sigma factor of Bacillus subtilis. Cell. 1989;59:133–143. doi: 10.1016/0092-8674(89)90876-3. [DOI] [PubMed] [Google Scholar]
- 10.Dahl M K, Msadek T, Kunst F, Rapoport G. The phosphorylation state of the DegU response regulator acts as a molecular switch allowing either degradative enzyme synthesis or expression of genetic competence in Bacillus subtilis. J Biol Chem. 1992;267:14509–14514. [PubMed] [Google Scholar]
- 11.Davis N K, Chater K F. Spore colour in Streptomyces coelicolor A3(2) involves the developmentally regulated synthesis of a compound biosynthetically related to polyketide antibiotics. Mol Microbiol. 1990;4:1679–1691. doi: 10.1111/j.1365-2958.1990.tb00545.x. [DOI] [PubMed] [Google Scholar]
- 12.Davis N K, Chater K F. The Streptomyces coelicolor whiB gene encodes a small transcription factor-like protein dispensable for growth but essential for sporulation. Mol Gen Genet. 1992;232:351–358. doi: 10.1007/BF00266237. [DOI] [PubMed] [Google Scholar]
- 13.Flärdh K, Findlay K C, Chater K F. Association of early sporulation genes with suggested developmental decision points in Streptomyces coelicolor A3(2) Microbiology. 1999;145:2229–2243. doi: 10.1099/00221287-145-9-2229. [DOI] [PubMed] [Google Scholar]
- 14.Flett F, Mersinias V, Smith C P. High efficiency intergeneric conjugal transfer of plasmid DNA from Escherichia coli to methyl DNA-restricting streptomycetes. FEMS Microbiol Lett. 1997;155:223–229. doi: 10.1111/j.1574-6968.1997.tb13882.x. [DOI] [PubMed] [Google Scholar]
- 15.Ford M E, Stenstrom C, Hendrix R W, Hatfull G F. Mycobacteriophage TM4: genome structure and gene expression. Tubercle Lung Dis. 1998;79:63–73. doi: 10.1054/tuld.1998.0007. [DOI] [PubMed] [Google Scholar]
- 16.Gordon G B, Miller L R, Bensch K G. Fixation of tissue culture cells for ultrastructural cytochemistry. Exp Cell Res. 1963;31:440–443. doi: 10.1016/0014-4827(63)90024-7. [DOI] [PubMed] [Google Scholar]
- 17.Hillemann D, Pühler A, Wohlleben W. Gene disruption and gene replacement in Streptomyces via single-stranded DNA transformation of integration vectors. Nucleic Acids Res. 1991;19:727–731. doi: 10.1093/nar/19.4.727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hobbs G, Frazer C, Gardner D C J, Cullum J, Oliver S G. Dispersed growth of Streptomyces in liquid culture. Appl Microbiol Biotechnol. 1989;31:272–277. [Google Scholar]
- 19.Hopwood D A, Wildermuth H, Palmer H M. Mutants of Streptomyces coelicolor defective in sporulation. J Gen Microbiol. 1970;61:397–408. doi: 10.1099/00221287-61-3-397. [DOI] [PubMed] [Google Scholar]
- 20.Hopwood D A, Bibb M J, Chater K F, Kieser T, Bruton C J, Kieser H M, Lydiate D J, Smith C P, Ward J M, Schrempf H. Genetic manipulation of Streptomyces: a laboratory manual. Norwich, United Kingdom: The John Innes Foundation; 1985. [Google Scholar]
- 21.Hutter B, Dick T. Molecular genetic characterisation of whiB3, a mycobacterial homologue of a Streptomyces sporulation factor. Res Microbiol. 1999;150:295–301. doi: 10.1016/s0923-2508(99)80055-2. [DOI] [PubMed] [Google Scholar]
- 22.Ikeda H, Seno E T, Bruton C J, Chater K F. Genetic mapping, cloning and physiological aspects of the glucose kinase gene of Streptomyces coelicolor. Mol Gen Genet. 1984;196:501–507. doi: 10.1007/BF00436199. [DOI] [PubMed] [Google Scholar]
- 23.Kelemen G H, Buttner M J. Initiation of aerial mycelium formation in Streptomyces. Curr Opin Microbiol. 1998;1:656–662. doi: 10.1016/s1369-5274(98)80111-2. [DOI] [PubMed] [Google Scholar]
- 24.Kelemen G H, Plaskitt K A, Lewis C G, Findlay K, Buttner M J. Deletion of DNA lying close to the glkA locus induces ectopic sporulation in Streptomyces coelicolor A3(2) Mol Microbiol. 1995;17:221–230. doi: 10.1111/j.1365-2958.1995.mmi_17020221.x. [DOI] [PubMed] [Google Scholar]
- 25.Kelemen G H, Brown G L, Kormanec J, Potúčková L, Chater K F, Buttner M J. The positions of the sigma factor genes, whiG and sigF, in the hierarchy controlling the development of spore chains in the aerial hyphae of Streptomyces coelicolor A3(2) Mol Microbiol. 1996;21:593–603. doi: 10.1111/j.1365-2958.1996.tb02567.x. [DOI] [PubMed] [Google Scholar]
- 26.Kelemen G H, Brian P, Flärdh K, Chamberlin L, Chater K F, Buttner M J. Developmental regulation of transcription of whiE, a locus specifying the polyketide spore pigment in Streptomyces coelicolor A3(2) J Bacteriol. 1998;180:2515–2521. doi: 10.1128/jb.180.9.2515-2521.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kieser H M, Kieser T, Hopwood D A. A combined genetic and physical map of the chromosome of Streptomyces coelicolor A3(2) J Bacteriol. 1992;174:5496–5507. doi: 10.1128/jb.174.17.5496-5507.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kormanec J, Homerova D. Streptomyces aureofaciens whiB gene encoding putative transcription factor essential for differentiation. Nucleic Acids Res. 1993;21:2512. doi: 10.1093/nar/21.10.2512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kormanec J, Ševčikova B, Sprušanský O, Benada O, Kofroňová O, Nováková R, Řežuchová B, Potúčková L, Homérová D. The Streptomyces aureofaciens homologue of the whiB gene is essential for sporulation; its expression correlates with the developmental stage. Folia Microbiol. 1998;43:605–612. doi: 10.1007/BF02816376. [DOI] [PubMed] [Google Scholar]
- 30.MacNeil D J, Occi J L, Gewain K M, MacNeil T, Gibbons P H, Ruby C L, Danis S L. Complex organization of the Streptomyces avermitilis genes encoding the avermectin polyketide synthase. Gene. 1992;115:119–125. doi: 10.1016/0378-1119(92)90549-5. [DOI] [PubMed] [Google Scholar]
- 31.McCormick J R, Su E P, Driks A, Losick R. Growth and viability of Streptomyces coelicolor mutant for the cell division gene ftsZ. Mol Microbiol. 1994;14:243–254. doi: 10.1111/j.1365-2958.1994.tb01285.x. [DOI] [PubMed] [Google Scholar]
- 32.McVittie A. Ultrastructural studies on sporulation in wild-type and white colony mutants of Streptomyces coelicolor. J Gen Microbiol. 1974;81:291–302. doi: 10.1099/00221287-81-2-291. [DOI] [PubMed] [Google Scholar]
- 33.Oh S-H, Chater K F. Denaturation of circular or linear DNA facilitates targeted integrative transformation of Streptomyces coelicolor A3(2): possible relevance to other organisms. J Bacteriol. 1997;179:122–127. doi: 10.1128/jb.179.1.122-127.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Paget M S B, Chamberlin L, Atrih A, Foster S J, Buttner M J. Evidence that the extracytoplasmic function sigma factor, ςE, is required for normal cell wall structure in Streptomyces coelicolor A3(2) J Bacteriol. 1999;181:204–211. doi: 10.1128/jb.181.1.204-211.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Potúčková L, Kelemen G H, Findlay K C, Lonetto M A, Buttner M J, Kormanec J. A new RNA polymerase sigma factor, ςF, is required for the late stages of morphological differentiation in Streptomyces sp. Mol Microbiol. 1995;17:37–48. doi: 10.1111/j.1365-2958.1995.mmi_17010037.x. [DOI] [PubMed] [Google Scholar]
- 36.Redenbach M, Kieser H M, Denapaite D, Eichner A, Cullum J, Kinashi H, Hopwood D A. A set of ordered cosmids and a detailed genetic and physical map for the 8Mb Streptomyces coelicolor A3(2) chromosome. Mol Microbiol. 1996;21:77–96. doi: 10.1046/j.1365-2958.1996.6191336.x. [DOI] [PubMed] [Google Scholar]
- 37.Reeve J N, Mendelson N H, Coyne S I, Hallock L L, Cole R M. Minicells of Bacillus subtilis. J Bacteriol. 1973;114:860–873. doi: 10.1128/jb.114.2.860-873.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ryding N J. Analysis of sporulation genes in Streptomyces coelicolor A3(2). Ph.D. thesis. Norwich, United Kingdom: University of East Anglia; 1995. [Google Scholar]
- 39.Ryding N J, Kelemen G H, Whatling C A, Flärdh K, Buttner M J, Chater K F. A developmentally regulated gene encoding a repressor-like protein is essential for sporulation in Streptomyces coelicolor A3(2) Mol Microbiol. 1998;29:343–357. doi: 10.1046/j.1365-2958.1998.00939.x. [DOI] [PubMed] [Google Scholar]
- 40.Ryding N J, Bibb M J, Molle V, Findlay K C, Chater K F, Buttner M J. New sporulation loci in Streptomyces coelicolor A3(2) J Bacteriol. 1999;181:5419–5425. doi: 10.1128/jb.181.17.5419-5425.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Seibert V, Kourbatova E M, Golovleva L A, Schlömann M. Characterization of the maleylacetate reductase MacA of Rhodococcus opacus 1CP and evidence for the presence of an isofunctional enzyme. J Bacteriol. 1998;180:3503–3508. doi: 10.1128/jb.180.14.3503-3508.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Soliveri J, Vijgenboom E, Granozzi C, Plaskitt K A, Chater K F. Functional and evolutionary implications of a survey of various actinomycetes for homologues of two Streptomyces coelicolor sporulation genes. J Gen Microbiol. 1993;139:2569–2578. doi: 10.1099/00221287-139-11-2569. [DOI] [PubMed] [Google Scholar]
- 43.Soliveri, J. A., J. Gomez, W. R. Bishai, and K. F. Chater. Multiple paralogous genes related to the Streptomyces coelicolor developmental regulatory gene whiB are present in Streptomyces and other actinomycetes. Microbiology, in press. [DOI] [PubMed]
- 44.Sun J, Kelemen G H, Fernández-Abalos J M, Bibb M J. Green fluorescent protein as a reporter for spatial and temporal gene expression in Streptomyces coelicolor A3(2) Microbiology. 1999;145:2221–2227. doi: 10.1099/00221287-145-9-2221. [DOI] [PubMed] [Google Scholar]
- 45.Teather R M, Collins J F, Donachie W D. Quantal behaviour of a diffusible factor which initiates septum formation at potential division sites in Escherichia coli. J Bacteriol. 1974;118:407–413. doi: 10.1128/jb.118.2.407-413.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zalacaín M, González A, Guerrero M C, Mattaliano R J, Malpartida F, Jiménez A. Nucleotide sequence of the hygromycin B phosphotransferase gene from Streptomyces hygroscopicus. Nucleic Acids Res. 1986;14:1565–1581. doi: 10.1093/nar/14.4.1565. [DOI] [PMC free article] [PubMed] [Google Scholar]