Abstract
Background
An epidemic of the Omicron variant of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) began in March 2022, and over 600,000 cases were confirmed until early May 2022 in Shanghai, China. Data on Omicron infections are available in other countries, but the clinical features of patients in the Chinese population, especially in Shanghai, are still lacking. We collected data from a subset of asymptomatic and mildly ill patients to learn about the age and sex disparity of Omicron infection based on changes in cycle threshold values.
Methods
The basic information of 325 patients who were consecutively admitted to the Shanghai Geriatrics Center was collected through medical records, and patients were tested for viral nucleic acid carriage using nasal swab samples during hospitalization. SAS 9.4 was used for data analysis, and a p value < 0.05% was considered statistically significant.
Results
Among the 325 included patients, 58.8% were males, with a mean age of 47.2 years and 13.6 days of hospitalization on average. The average number of nucleic acid tests among female patients was 4.7, which was higher than that among male patients (4.1). The median value of the slope for cycle threshold (Ct) changes in the nucleic acid detection (NAD) test was 1.4. Logistic regression indicated that the proportion of slope for Ct changes >1.5 was slightly higher among male patients than among female patients (odds ratio (OR) = 1.06, 95% confidence interval (CI): 0.68–1.66), and patients aged <45 years and 45–59 years had a higher proportion of slope for Ct changes >1.5 than patients aged ≥60 years. Ct values were more variable in the early stages of infection and stabilized in the later stages of infection.
Conclusion
Among patients with mild illness or asymptomatic infection, the Ct value is a good, timely, and cost-effective method to reflect the recovery progress of patients. The slope of Ct changes was steeper among younger patients and male patients, which indicates faster disease recovery.
Keywords: COVID-19, SARS-CoV-2, cycle threshold, Omicron, RT‒PCR, nucleic acid tests
Introduction
By May 2022, more than 500 million people were infected with coronavirus disease 2019 (COVID-19), with approximately 6 million deaths worldwide. In China, since the successful control of a localized epidemic in Wuhan in 2020, sporadic cases were reported in the following two years, and an epidemic of COVID-19 began in Shanghai in March 2022, with more than 600,000 infections in total. During this epidemic, the strains of the virus that was prevalent in Shanghai belonged to the Omicron BA.2 and BA.2.2 variants, which was the most serious challenge in combating COVID-19 since 2020. Since Omicron was identified in South Africa on November 9, 2021,1 it has induced a pandemic, including the current epidemic in Shanghai. Strict public health measures were taken in Shanghai, such as large-scale viral nucleic acid and antigen screening, quarantine of infected cases and reduced close contact in shelter hospitals and hotels, lockdowns of districts with severe outbreaks. The strict control strategies adopted in Shanghai have prevented the continuous spread of the virus and saved many lives. The number of new confirmed cases decreased from a peak of more than 20,000 per day in April to approximately 1000 per day in May.
Data on Omicron infections are available in other countries around the world. In general, patients infected Omicron also presented most probably with, dry cough, fever, and fatigue. But data on the clinical features of patients in the Chinese population, especially in Shanghai, are still lacking. When the COVID-19 epidemic began in 2019, the reverse transcription-polymerase chain reaction (RT‒PCR) assay was developed and was highly sensitive for virus detection; therefore, RT‒PCR has been treated as a mainstay of laboratory diagnosis for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection. Due to the weak pathogenicity of Omicron variants and patients who are mainly asymptomatic or have mild illness, RT‒PCR is more suitable for Omicron infection detection than chest X-ray and chest computed tomography (CT). There is a consensus that cycle threshold (Ct) values can, to some extent, reflect viral load and subsequently indicate the infection condition. Pujadas found that viral load can predict the mortality of patients with COVID-19.2 Magleby indicated that a lower Ct value was associated with a higher risk of intubation among COVID-19 patients.3 Wang found that nasopharyngeal swabs were more sensitive than oropharyngeal swabs for COVID-19 diagnosis and monitoring the SARS-CoV-2 load by using Ct values.4 Heald-Sargent investigated the relationship between viral load and age range and found that young children have equivalent or more viral nucleic acid in their upper respiratory tract than older children and adults.5 The findings of the abovementioned investigation indicated some controversy regarding whether Ct values can represent the real viral load.6 Based on a previous meta-analysis,7 cell culture methods can provide more reliable results to distinguish infectious samples from noninfectious samples. However, considering the widespread epidemic caused by the Omicron variant, rapid and accurate RT‒PCR is still the first-line choice for virus infection confirmation.
Shanghai continues adopting the “zero-COVID-19 policy”, which is an effective measure to reduce health damage and curb the transmission of the virus in the long term in addition to vaccination.8,9 In this study, we analyzed the features of patients with mild and asymptomatic infections admitted to the isolation ward in the Shanghai Geriatrics Center and aimed to explore how Ct values obtained through RT‒PCR assays during the recovery process reflect the recovery from Omicron variants among patients in different age and sex groups.
Methods
Study Population
In this observational study, we included 325 mild and asymptomatic patients admitted sequentially to wards G8 and G9 of the Shanghai Geriatrics Center from March 15 to April 14, 2022. A patient with mild illness was defined as an RT‒PCR-confirmed case without pulmonary involvement, and an asymptomatic patient was defined as a confirmed case without any discomfort that was detected through nucleic acid testing. We collected information on patients’ age, sex, time of hospital admission and discharge, time of the first nucleic acid test, and the results of each nucleic acid test of infectious respiratory specimens (nasal swab) during hospitalization (Ct values). All collected medical records were reviewed independently and cross-checked by a second reviewer.
Nucleic Acid Testing
SARS-CoV-2 was amplified and detected by the reverse transcription-polymerase chain reaction (RT‒PCR) method. The assay equipment was a Life Technologies 7500 (Life Technologies Corporation, USA). Nasal swab specimens were collected by nurses from Yueyang Hospital of Integrated Traditional Chinese and Western Medicine. Specimens were tested in the laboratory of the Geriatrics Center. To ensure the comparability of Ct values, we used coronavirus nucleic acid detection kits (fluorescent PCR method, Shanghai Fosun Long March Medical Science Co.) on patient nasal swab samples for ORF1ab (ORF1/a, a conserved region in the nonstructural region), the E gene (used for specific detection of SARS-CoV-2, for pan-sarbecovirus detection), and the nucleocapsid (N). The Ct value for subsequent analysis was recorded as the mean value of the abovementioned three values. Most patients underwent nucleic acid testing on the 8th day after hospital admission and had subsequent tests every 2 days. The hospital discharge criterion was a Ct value >35 two consecutive times or two consecutive negative results on the nucleic acid test.
Definition and Index Calculation
In this study, days of hospitalization were calculated as the differences between the time of hospital discharge and the time of hospital admission and then were classified into ≤15 days and >15 days. Censors after hospital discharge were defined as patients who were lost to follow-up and could not be contacted after hospital discharge. In this study, the patients’ age was stratified into <45, 45–59, and ≥60 years age groups, and the nucleic acid detection (NAD) test times were classified into ≤5 times and >5 times. The slope for Ct value changes for each patient was calculated based on the linear regression model (y=a+bx, where b is defined as the slope for Ct change) and was established to show the linear correlation between each Ct value and its test time, with a higher slope value representing a quicker change in Ct value during the recovery process among patients. The slope value for Ct changes was then divided into ≤1.5 and >1.5 groups.
Statistical Analysis
In this study, data analysis was performed by using SAS 9.4 (Cary, NC, USA). Quantitative data with a normal distribution are expressed as the means and standard deviations (SDs), and Student’s t test was used to test the significance of differences between groups. Quantitative data with skewed distribution are expressed as medians and interquartile ranges (IQRs), and a nonparametric rank-sum test was applied to examine the differences between groups. Qualitative data are expressed as frequency counts and percentages (%), and a chi-square test was used to determine the significant differences between groups. Univariate and multivariate logistic regression were used to calculate the odds ratios (ORs) and 95% confidence intervals (CIs) and to explore the relationship between the slope of Ct values and demographic features of patients, the total number of virus NAD tests, and days of hospitalization. In this study, a p value of less than 0.05 (two-tailed) was considered statistically significant.
Results
Demographic Features of Patients
In this study, a total of 325 patients were enrolled, with a proportion of 58.8% male patients (n=191). The mean age was 47.2 years (SD: 16.8), and the mean number of days of hospitalization was 13.6 (SD: 4.5). The majority of patients were enrolled in March (80.3%), and 28.6% of patients were censored after hospital discharge. In this study, male patients were younger than female patients, and the proportion of male patients over 60 years of age (22.0%) was lower than that of female patients (38.1%); the difference was statistically significant (P<0.05). There were no significant differences in the month of admission, days of hospitalization, or censor proportion among patients of different sexes (Table 1).
Table 1.
The Demographic Features of Patients with COVID-19 in a Hospital in Shanghai, China
| Variables | Total Patients (n=325) | Patients by Sex | t/χ2 | P | |
|---|---|---|---|---|---|
| Male (n=191) | Female (n=134) | ||||
| Age (years)†, mean (SD) | 47.2 (16.8) | 45.2 (16.9) | 50.1 (16.4) | −2.61 | 0.001 |
| Age (years)†, median (IQR) | 48.0 (32.5–61.0) | 45.0 (31.0–57.0) | 52.0 (38.0–64.0) | −2.84 | 0.004 |
| Age group (years)†, n (%) | 10.07 | 0.006 | |||
| <45 | 146 (44.9) | 95 (49.7) | 51 (38.1) | ||
| 45–59 | 86 (26.5) | 54 (28.3) | 32 (23.9) | ||
| ≥60 | 93 (28.6) | 42 (22.0) | 51 (38.1) | ||
| Month of hospital admission, n (%) | 1.71 | 0.191 | |||
| March | 261 (80.3) | 158 (82.7) | 103 (76.9) | ||
| April | 64 (19.7) | 33 (17.3) | 31 (23.1) | ||
| Days of hospitalization, mean (SD) | 13.6 (4.5) | 13.4 (4.3) | 13.9 (4.8) | −0.99 | 0.325 |
| Days of hospitalization, n (%) | 2.86 | 0.091 | |||
| ≤15 days | 230 (70.8) | 142 (74.3) | 88 (65.7) | ||
| >15 days | 95 (29.2) | 49 (25.7) | 46 (34.3) | ||
| Censored after hospital discharge, n (%) | 0.01 | 0.93 | |||
| Yes | 93 (28.6) | 55 (28.8) | 38 (28.4) | ||
| No | 232 (71.4) | 136 (71.2) | 96 (71.6) | ||
Note: †The differences between males and females were statistically significant (P<0.05).
Abbreviations: SD, standard deviation; IQR, interquartile range.
Nucleic Acid Testing Among Patients of Different Sexes
During hospitalization, the average number of nucleic acid tests among female patients was 4.7, which was higher than that among male patients (4.1), and female patients had a higher proportion of having NAD tests >5 times; the difference was statistically significant. The median duration for the 1st NAD test and hospitalization was 5 days among males and 3 days among females, but this difference was not statistically significant. There was a significant difference between male and female patients in the first and second Ct test values after hospital admission (mean value was 30.0 for males and 27.3 for females for the first test and 33.1 for males and 29.9 for females for the second test). The duration between the first and last NAD test was significantly different between male and female patients (4 days for males and 6.5 days for females). The median value of slope for Ct changes in the NAD test was 1.4 for both male and female patients, and the proportion of slope for Ct changes >1.5 was also nearly the same between male and female patients (Table 2).
Table 2.
The Virus Nucleic Acid Detection (NAD) Tests for COVID-19 Patients in a Hospital in Shanghai, China
| Variables | Total Patients (n=325) | Patients by Sex | t/χ2 | P | |
|---|---|---|---|---|---|
| Male (n=191) | Female (n=134) | ||||
| Times of NAD test†, mean (SD) | 4.3 (2.2) | 4.1 (2.1) | 4.7 (2.4) | −2.44 | 0.015 |
| NAD test times†, n (%) | 5.39 | 0.021 | |||
| ≤5 times | 238 (73.2) | 149 (78.0) | 89 (66.4) | ||
| >5 times | 87 (26.8) | 42 (22.0) | 45 (33.6) | ||
| Duration for 1st NAD test and hospitalization, median (IQR) | 3.0 (2.0–6.0) | 5.0 (2.0–7.0) | 3.0 (2.0–6.0) | 3.79 | 0.052 |
| Ct value of virus NAD test each time, median (IQR) | |||||
| Time 1 (n=325)† | 28.4 (23.2–35.8) | 30.0 (24.3–36.6) | 27.3 (21.5–34.3) | 7.01 | 0.008 |
| Time 2 (n=325)† | 31.9 (23.8–37.2) | 33.1 (26.2–37.9) | 29.9 (21.8–35.7) | 9.70 | 0.002 |
| Time 3 (n=245) | 33.3 (26.9–37.7) | 33.7 (28.7–37.9) | 31.3 (25.6–37.2) | 2.69 | 0.101 |
| Time 4 (n=170) | 36.1 (31.0–38.7) | 36.4 (32.9–39.3) | 35.7 (30.0–38.1) | 1.96 | 0.162 |
| Time 5 (n=133) | 37.5 (34.6–40.0) | 37.7 (34.5–40.0) | 37.2 (34.9–39.4) | 0.61 | 0.435 |
| Time 6 (n=87) | 37.2 (35.6–40.0) | 37.2 (36.1–39.2) | 37.5 (35.3–40.0) | 0.02 | 0.898 |
| Time 7 (n=52) | 37.3 (36.2–39.4) | 38.1 (36.2–38.9) | 37.1 (36.1–38.7) | 1.19 | 0.275 |
| Time 8 (n=26) | 36.6 (35.1–38.9) | 36.1 (34.8–38.9) | 36.8 (35.5–38.4) | 0.29 | 0.584 |
| Time 9 (n=17) | 37.3 (36.1–38.8) | 38.3 (34.7–39.6) | 36.9 (36.1–38.3) | 0.73 | 0.393 |
| Time 10 (n=14) | 37.9 (36.4–40.0) | 37.3 (33.8–39.9) | 39.5 (37.2–40.0) | 2.25 | 0.133 |
| Time 11 (n=8) | 37.2 (36.1–39.4) | 37.1 (35.4–40.0) | 37.4 (36.7–38.8) | 0.03 | 0.881 |
| Duration between first and last NAD test†, median (IQR) | 5.0 (3.0–10.0) | 4.0 (3.0–9.0) | 6.5 (3.0–11.0) | 6.29 | 0.012 |
| Slope value for Ct changes in NAD tests, median (IQR) | 1.4 (0.5–2.2) | 1.4 (0.3–2.2) | 1.4 (0.8–2.1) | 0.17 | 0.677 |
| Proportion of slope value for Ct changes, n (%) | 0.07 | 0.796 | |||
| ≤1.5 | 184 (56.6) | 107 (56.1) | 77 (57.5) | ||
| >1.5 | 141 (43.4) | 84 (43.9) | 57 (42.5) | ||
Note: †The differences between males and females were statistically significant (P<0.05).
Abbreviations: SD, standard deviation; IQR, interquartile range; NAD, nucleic acid detection; Ct, cycle threshold.
Factors Associated with the Rate of Ct Value Changes Among COVID-19 Patients
Univariate logistic regression and multivariate logistic regression were applied to explore factors associated with the slope of Ct value changes among patients with COVID-19. The proportion of slope for Ct changes >1.5 was slightly higher among male patients than among female patients, but without statistical significance (OR=1.06, 95% CI: 0.68–1.66), even with the adjustment for potential confounders (OR=1.16). Compared with patients ≥60 years of age, patients aged <45 years and 45–59 years had a higher proportion of slope for Ct changes >1.5, and the ORs were 2.49 (95% CI: 1.41–4.41) and 2.58 (95% CI: 1.34–4.95), respectively. With the adjustment for confounders, the proportion of slope for Ct changes >1.5 was higher among patients who were hospitalized ≤15 days than among those who were hospitalized >15 days, and the OR was 3.81 (95% CI: 1.67–8.70) (Table 3).
Table 3.
The Influencing Factors Associated with the Slope Value for Ct Changes Based on Linear Regression for Nucleic Acid Detection (NAD) Tests Among COVID-19 Patients in Shanghai, China
| Variables | Percentage of Slope Value for Ct Changes>1.5 | Model 1 | Model 2 | ||
|---|---|---|---|---|---|
| OR | 95% CI | OR | 95% CI | ||
| Sex, n (%) | |||||
| Male | 84 (44.0) | 1.06 | 0.68–1.66 | 1.16 | 0.72–1.87 |
| Female | 57 (42.5) | Ref | Ref | ||
| Age (years), n (%) | |||||
| <45 | 73 (50.0) | 2.32 | 1.34–4.02 | 2.49 | 1.41–4.41 |
| 45–59 | 40 (46.5) | 2.02 | 1.09–3.73 | 2.58 | 1.34–4.95 |
| ≥60 | 28 (30.1) | Ref | Ref | ||
| Total virus NAD test times, n (%) | |||||
| ≤5 times | 108 (45.4) | 1.35 | 0.82–2.25 | 1.78 | 0.78–4.08 |
| >5 times | 33 (37.9) | Ref | Ref | ||
| Days of hospitalization, n (%) | |||||
| ≤15 days | 112 (48.7) | 2.16 | 1.30–3.59 | 3.81 | 1.67–8.70 |
| >15 days | 29 (30.5) | Ref | Ref | ||
Notes: Model 1: univariate logistic regression, Model 2: multivariate logistic regression. Bold & italicized values indicate that there was a significant difference between groups.
Abbreviations: OR, odds ratio; CI, confidence interval; NAD, nucleic acid detection; Ct, cycle threshold.
Time Intervals of the Nucleic Acid Test for Patients of Different Sexes and Ages
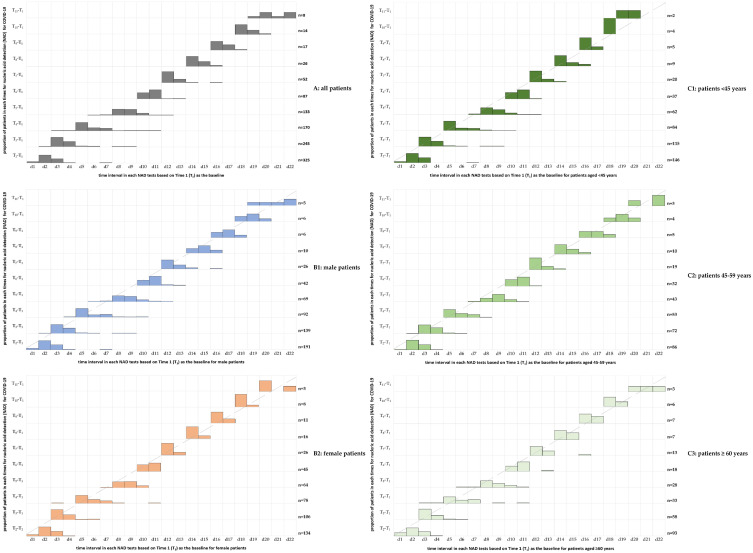
In this study, the NAD test time was scheduled as a consecutive test at a frequency of 2 days. Figure 1 indicates that the majority of patients who underwent the NAD test followed the scheduled time, especially for the last 4 times of the NAD test. In comparison with male patients, female patients were more likely to receive NAD tests strictly by the scheduled time. Moreover, patients aged 45–59 years old were also more likely to receive NAD tests strictly following the scheduled time than patients aged <45 years and ≥60 years. Overall, the number of patients participating in NAD testing decreased with the attainment of Ct values for nucleic acid testing (two consecutive negative nucleic acid test results 24 hours apart or two consecutive CT values >35 for nucleic acid testing), indicating that more patients recovered and reached the discharge criteria during the NAD tests (Figure 1).
Figure 1.
The distribution of the time interval for each nucleic acid detection (NAD) test was based on setting Time 1 (T1) as the baseline among patients of different sexes and ages with COVID-19 infection.
Ct Values in the NAD Tests for Patients with COVID-19
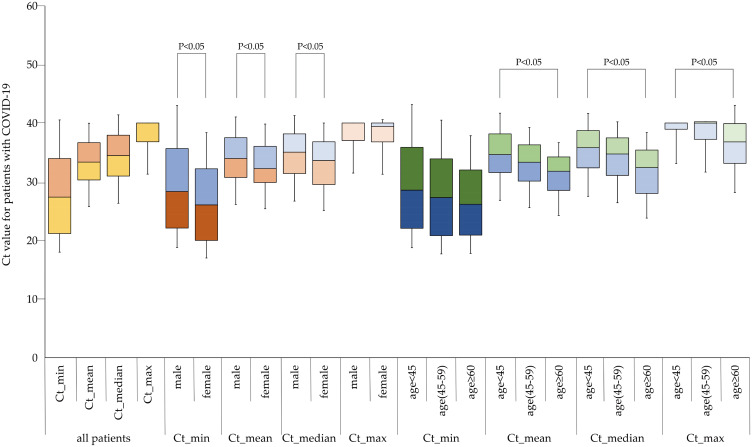
For the Ct value in the overall NAD tests during hospitalization for each patient, we calculated the minimum, mean, median, and maximum value for each patient based on their individual Ct values of each NAD test, and then we used a box plot to describe the distribution features among patients of different ages and sexes. Figure 2 shows that female patients had lower Ct values than male patients in comparisons of the minimum, mean and median values. Likewise, in comparison with patients aged <45 years, patients aged 45–59 years or ≥ 60 years had lower Ct values in comparisons of the maximum, mean, and median values (Figure 2).
Figure 2.
Comparison of minimum, maximum, median, and mean individual cycle threshold (Ct) values among COVID-19 patients of different sexes and ages.
The Trend for Ct Value Changes with Test Times Among Patients
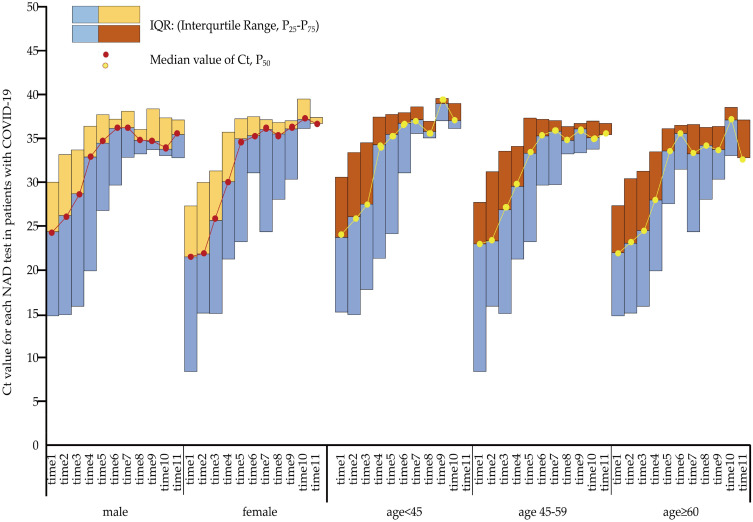
Figure 3 indicates the trend of Ct value changes with the time of the NAD tests. The Ct value of all patient groups showed a relatively rapid increase at the beginning of the test but fluctuated once the Ct value reached 35. In comparison with female patients, male patients tended to have a steeper trend, which demonstrated a larger slope value. Moreover, compared with patients aged ≥ 60 years, patients aged <45 years and 45–59 years also tended to have a steeper trend.
Figure 3.
The cycle threshold (Ct) value for each nucleic acid detection (NAD) test among patients of different sexes and ages with COVID-19 infection.
Discussion
In this descriptive study, we preliminarily analyzed the features of asymptomatic and mildly ill patients with Omicron variant infection in Shanghai and the disease evolution among patients of different ages, sexes, and with different clinical features. We confirmed that the Ct value could be a good indicator to reflect the SARS-CoV-2 infection status among asymptomatic and mildly ill patients. In the study of SARS-CoV-2, the cell culture approach enables the identification of infectious viruses and noninfectious viruses in a highly accurate manner. However, virus cultivation in cell cultures requires biosafety level 3 laboratories and is relatively time-consuming and complex. Compared to previous variants of SARS-CoV-2,10,11 Omicron has a shorter incubation period and similar or milder clinical symptoms12 but an approximately 10 times higher infectivity13 that limits the protection imparted by conventional surgical masks.14 To curb the quick spread of the virus in the population, RT‒PCR has now become a widely used gold standard method for SARS-CoV-2 infection detection in various countries.15 A smaller number of detectable cycles of the fluorescent signal (Ct threshold) represents a high viral load. Although some studies suggest that the interpretation of viral load by Ct values is inaccurate,6 the Ct value has been employed in most studies for viral load assessment. A great deal of meaning has been attributed to the Ct value, including a negative correlation between the Ct level and the risk of deterioration into severe illness and mortality16,17 and a negative correlation with the degree of infectivity.18 Subsequent tests can be performed to monitor the dynamics of rising Ct values as a sign of a decrease in viral load over time to determine the possibility of return to work of health care workers who are infected and in isolation,19 to determine a response to antiviral therapy, and to associate symptoms with viral load.20
The data in this study, although collected from only one hospital with a relatively small number of patients, still allow for a more consistent conclusion due to the high quality of the data collection during the same period with the same instrument test and same data sample collection staff (the same nurses performed the nasal swabs). China’s zero-COVID-19 policy has resulted in fewer cases overall, allowing for the research of mild and asymptomatic patients. The RT‒PCR test was based on nasal swabs in this study, which can more accurately reflect the amount of SARS-CoV-2 than oropharyngeal swabs.21
In terms of the overall findings, this study corroborates that the Ct value changes by days and possesses the ability to reflect the viral clearance rate. In the early stage of the COVID-19 epidemic in 2020, Elisabet et al found that SARS-CoV-2 viral load predicted COVID-19 mortality after controlling for the influence of age, sex, coexisting disease, and ethnicity.2 Soa et al proposed that Ct values can reflect the current epidemiological trends and dynamics of a certain region. When the sample size is large enough, the overall mean Ct value can assist in the identification of the epidemic stage, including the beginning, the peak, and the end of the epidemic.22 Therefore, the findings in this study can provide basic disease duration patterns of patients with different features, which can provide a reference for decision-making under the dynamic epidemic strategy in China.
Few studies have focused on the difference in Ct values among SARS-CoV-2-infected patients with different features.23 Gupta et al found considerable differences in oral microbiology among infected individuals by age and sex. Rehman et al found that there was no significant difference in viral loads between age groups. Males tended to have a higher viral load than females.24 In our study, patients aged 45–59 years or over 60 years had lower Ct values than patients aged <45 years in terms of the minimum, mean, and median total Ct values. Male patients tended to have a steeper trend in Ct value changes with the time of the NAD test than females, which demonstrated a larger slope value. Moreover, compared with patients aged over 60 years, patients aged <45 years and 45–59 years also had a steeper trend. The proportion of slope for Ct changes >1.5 was slightly higher among male patients than among female patients. Compared with patients over 60 years, patients aged <45 years and 45–59 years had a higher proportion of slope for Ct changes >1.5, and the ORs were 2.49 and 2.58, respectively, which was in line with previous studies. Taylor compared children of different ages5 and found that although symptoms were usually milder after COVID-19 infection, viral replication and the number of viral counts detected were similar to those of adults; Ct values were lower among patients aged <5 years than among those aged 5–17 and ≥18 years. Damien found a relatively high viral load in older adults.12 In an earlier study with a smaller number of cases in 2020 (30), Kelvin roughly plotted a positive correlation curve between age and viral load.25 Mohammad found that a younger patient group had a shorter hospital stay.26 Overall, for SARS-CoV-2, similar to SARS-CoV, older age was found to be an independent factor associated with higher viral load, which may be related to decreased immune function with ageing.27 Moreover, all patients in this study in Shanghai were SARS-CoV-2 BA.2.2 patients; thus, the findings in this study added to the evidence that the severe form of COVID-19 was most prevalent among older patients. For differences in Ct values by sex and age, we demonstrated a faster recovery among male patients and those aged <45 years (faster to reach a fluctuating interval around the mean value of 35), which might be due to different disease conditions among patients with different ages and sexes, so a large controlled study with a proper follow-up time could be used to validate this hypothesis in the future.
COVID-19 is an emerging infection, especially the highly infectious Omicron variant, which poses a challenge to population health and economic development worldwide.28 In this study, some patients had an extremely long recovery time, which reached 22 days for some patients (n=8, 2.5%), and the prolonged viral load poses a challenge to the facilities of isolation sites. In China, due to the imbalance in health care resources and a rapidly ageing society, strict adherence to the zero-COVID-19 policy will do more good than harm to the overall health of the population.29 The progress of this study reveals viral kinetic data from asymptomatic and mildly ill patients in Shanghai, which has the largest number of current Omicron infections in China. Considering that data on mildly ill and asymptomatic patients with Omicron infection are limited in other countries due to different policies, this study provides valuable information and complements the clinical features of patients with nonsevere infections.
Limitations
This study has some limitations. First, the clinical information was incomplete. Body mass index, body temperature, heart rate, blood pressure, and oxygen saturation were not collected in our study. None of the patients were tested for blood samples, so routine blood test, liver, and kidney function values were not obtained, and there were no complete records of patients’ vaccination and times, so we missed the chance to explore the association between COVID-19 vaccination and the disease course, disease severity, and viral load. Second, due to the different instruments and reagents, the findings in this study may not be comparable to those of other studies regarding specific parameters of Ct values, but the trend among patients can be used as a reference for subsequent researchers. Third, due to limited medical resources, we could not sample each patient every day. However, the retrospective data in this study provide first-hand evidence for reference to adjust the implementation of strategies and measures for epidemic control.
Conclusion
Among patients with mild illness or asymptomatic infection, the Ct value based on RT‒PCR is a good, timely, and cost-effective method to reflect the recovery progress of patients with COVID-19. The slope of Ct changes was steeper among younger patients and male patients, indicating a faster recovery among them. More research is needed in the future to confirm these conclusions.
Acknowledgments
We thank the nurses from the First Anti-COVID-19 Medical Team in Yueyang Hospital of Integrated Traditional Chinese and Western Medicine for their hard work in nasal swab collection in this study. We thank the medical and technical staff of the Department of Laboratory Medicine, Shanghai Geriatrics Center, for providing accurate nucleic acid results for this study.
Funding Statement
This work was supported by a National Natural Science Foundation of China (NSFC) grant (82074266, 81904007), the Science and Technology Commission of Shanghai Municipality (STCSM) Research Fund (21JC1405300, 202040306), Yueyang Hospital Research Projects (2018YJ05), Shanghai Talent Development Fund (2021073), and China Fund for Medical Equipment (IIT2022-01).
Data Sharing Statement
The original contributions from the current study are presented in the article. Further inquiries can be directed to the corresponding authors.
Ethics Approval and Consent to Participate
This study was reviewed and approved by the Review Board of Yueyang Hospital of Integrated Traditional Chinese and Western Medicine affiliated with Shanghai University of Traditional Chinese Medicine (2022-004). In light of the data collection in this study was conducted as a retrospective quality assessment, the collection of patients’ information was thereafter anonymized and maintained with confidentiality, so the informed consent to review their medical records among patients was waived by the Review Board of Yueyang Hospital of Integrated Traditional Chinese and Western Medicine affiliated with Shanghai University of Traditional Chinese Medicine. All procedures were performed in accordance with the ethical standards of the national research committee and the 1964 Helsinki Declaration and its later amendments.
Author Contributions
All authors contributed substantially to the work reported, whether in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Disclosure
The authors declare no conflicts of interest in this article.
References
- 1.Team CC-R. SARS-CoV-2 B.1.1.529 (Omicron) Variant - United States, December 1–8, 2021. MMWR Morb Mortal Wkly Rep. 2021;70(50):1731–1734. doi: 10.15585/mmwr.mm7050e1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pujadas E, Chaudhry F, McBride R, et al. SARS-CoV-2 viral load predicts COVID-19 mortality. Lancet Respir Med. 2020;8(9):e70. doi: 10.1016/S2213-2600(20)30354-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Magleby R, Westblade LF, Trzebucki A, et al. Impact of severe acute respiratory syndrome coronavirus 2 viral load on risk of intubation and mortality among hospitalized patients with coronavirus disease 2019. Clin Infect Dis. 2021;73(11):e4197–e4205. doi: 10.1093/cid/ciaa851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang H, Liu Q, Hu J, et al. Nasopharyngeal swabs are more sensitive than oropharyngeal swabs for COVID-19 diagnosis and monitoring the SARS-CoV-2 load. Front Med. 2020;7:334. doi: 10.3389/fmed.2020.00334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Heald-Sargent T, Muller WJ, Zheng X, Rippe J, Patel AB, Kociolek LK. Age-related differences in nasopharyngeal severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) levels in patients with mild to moderate coronavirus disease 2019 (COVID-19). JAMA Pediatr. 2020;174(9):902–903. doi: 10.1001/jamapediatrics.2020.3651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miranda RL, Guterres A, De azeredo lima CH, Filho PN, Gadelha MR. Misinterpretation of viral load in COVID-19 clinical outcomes. Virus Res. 2021;296:198340. doi: 10.1016/j.virusres.2021.198340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fall A, Eldesouki RE, Sachithanandham J, et al. The displacement of the SARS-CoV-2 variant Delta with Omicron: an investigation of hospital admissions and upper respiratory viral loads. EBioMedicine. 2022;79:104008. doi: 10.1016/j.ebiom.2022.104008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Malik JA, Mulla AH, Farooqi T, Pottoo FH, Anwar S, Rengasamy KRR. Targets and strategies for vaccine development against SARS-CoV-2. Biomed Pharmacother. 2021;137:111254. doi: 10.1016/j.biopha.2021.111254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wan Z, Lu R, Zhao Y, Zhang C. Diagnostic strategy of SARS-CoV-2 for containment under China’s zero-COVID-19 policy. J Infect. 2022;85:e7–e9. doi: 10.1016/j.jinf.2022.04.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Farooqi T, Malik JA, Mulla AH, et al. An overview of SARS-COV-2 epidemiology, mutant variants, vaccines, and management strategies. J Infect Public Health. 2021;14(10):1299–1312. doi: 10.1016/j.jiph.2021.08.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Malik JA, Ahmed S, Mir A, et al. The SARS-CoV-2 mutations versus vaccine effectiveness: new opportunities to new challenges. J Infect Public Health. 2022;15(2):228–240. doi: 10.1016/j.jiph.2021.12.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jacot D, Greub G, Jaton K, Opota O. Viral load of SARS-CoV-2 across patients and compared to other respiratory viruses. Microbes Infect. 2020;22(10):617–621. doi: 10.1016/j.micinf.2020.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen J, Wang R, Gilby NB, Wei GW. Omicron variant (B.1.1.529): infectivity, vaccine breakthrough, and antibody resistance. J Chem Inf Model. 2022;62(2):412–422. doi: 10.1021/acs.jcim.1c01451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Khandia R, Singhal S, Alqahtani T, et al. Emergence of SARS-CoV-2 Omicron (B.1.1.529) variant, salient features, high global health concerns and strategies to counter it amid ongoing COVID-19 pandemic. Environ Res. 2022;209:112816. doi: 10.1016/j.envres.2022.112816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Malik JA, Aroosa M, Ahmed S, et al. SARS-CoV-2 vaccines: clinical endpoints and psychological perspectives: a literature review. J Infect Public Health. 2022;15(5):515–525. doi: 10.1016/j.jiph.2022.03.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rao SN, Manissero D, Steele VR, Pareja J. A systematic review of the clinical utility of cycle threshold values in the context of COVID-19. Infect Dis Ther. 2020;9(3):573–586. doi: 10.1007/s40121-020-00324-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Asai N, Sakanashi D, Ohashi W, et al. Could threshold cycle value correctly reflect the severity of novel coronavirus disease 2019 (COVID-19)? J Infect Chemother. 2021;27(1):117–119. doi: 10.1016/j.jiac.2020.09.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.La Scola B, Le Bideau M, Andreani J, et al. Viral RNA load as determined by cell culture as a management tool for discharge of SARS-CoV-2 patients from infectious disease wards. Eur J Clin Microbiol Infect Dis. 2020;39(6):1059–1061. doi: 10.1007/s10096-020-03913-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Domeracki S, Clapp RN, Taylor K, Lu CM, Lampiris H, Blanc PD. Cycle threshold to test positivity in COVID-19 for return to work clearance in health care workers. J Occup Environ Med. 2020;62(11):889–891. doi: 10.1097/JOM.0000000000001996 [DOI] [PubMed] [Google Scholar]
- 20.Dergaa I, Abubaker M, Souissi A, et al. Age and clinical signs as predictors of COVID-19 symptoms and cycle threshold value. Libyan J Med. 2022;17(1):2010337. doi: 10.1080/19932820.2021.2010337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Patel MR, Carroll D, Ussery E, et al. Performance of oropharyngeal swab testing compared with nasopharyngeal swab testing for diagnosis of coronavirus disease 2019-United States, January 2020-February 2020. Clin Infect Dis. 2021;72(3):403–410. doi: 10.1093/cid/ciaa759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Andriamandimby SF, Brook CE, Razanajatovo N, et al. Cross-sectional cycle threshold values reflect epidemic dynamics of COVID-19 in Madagascar. Epidemics. 2022;38:100533. doi: 10.1016/j.epidem.2021.100533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gupta A, Bhanushali S, Sanap A, et al. Oral dysbiosis and its linkage with SARS-CoV-2 infection. Microbiol Res. 2022;261:127055. doi: 10.1016/j.micres.2022.127055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ur Rehman M, Sajjad Naqvi S, Ullah R, et al. Elucidation of correlation between SARS-CoV-2 RdRp and N gene cycle threshold (Ct) by RT-PCR with age and gender. Clin Chim Acta. 2022;533:42–47. doi: 10.1016/j.cca.2022.06.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.To KK, Tsang OT, Leung WS, et al. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: an observational cohort study. Lancet Infect Dis. 2020;20(5):565–574. doi: 10.1016/S1473-3099(20)30196-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Albanghali M, Alghamdi S, Alzahrani M, et al. Clinical characteristics and treatment outcomes of mild to moderate COVID-19 patients at tertiary care hospital, al baha, Saudi Arabia: a single centre study. J Infect Public Health. 2022;15(3):331–337. doi: 10.1016/j.jiph.2022.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen WJ, Yang JY, Lin JH, et al. Nasopharyngeal shedding of severe acute respiratory syndrome-associated coronavirus is associated with genetic polymorphisms. Clin Infect Dis. 2006;42(11):1561–1569. doi: 10.1086/503843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ahmad Malik J, Ahmed S, Shinde M, et al. The impact of COVID-19 on comorbidities: a review of recent updates for combating it. Saudi J Biol Sci. 2022;29(5):3586–3599. doi: 10.1016/j.sjbs.2022.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang X, Zhang W, Chen S. Shanghai’s life-saving efforts against the current omicron wave of the COVID-19 pandemic. Lancet. 2022;399:2011–2012. doi: 10.1016/S0140-6736(22)00838-8 [DOI] [PMC free article] [PubMed] [Google Scholar]