Abstract
This study assessed the prevalence of plague bacterium (Yersinia pestis) among rodents captured in Umzingwane and Nkayi districts, south-western Zimbabwe. A total of 44 rodents were captured on three consecutive days per trapping session in the study sites using a removal trapping method in April 2018. Captured rodents were euthanized, and blood samples were collected. The Giemsa stain method was used to detect plague bacteria. The trapping success was not significantly different (χ² = 1.50, df = 1, P = 0.221), 8.5% for the Nkayi district, while in the Umzingwane district, it was 8%. Overall, only one rodent species, i.e., Mastomys natalensis, tested positive for Y. pestis in the Umzingwane district, thus yielding a prevalence rate of 2.3% for the entire study area. This was the most important finding of a Y. pestis-positive rodent in a non-endemic wild area in the Umzingwane district. These results point to a low prevalence of Y. pestis in the study area and the importance of an active plague disease surveillance and monitoring system.
Keywords: plague prevalence, rodents, species diversity, trapping success
1. Introduction
Plague, a disease caused by the bacilli bacterium, Yersinia pestis, primarily affects rodents, mainly transmitted from one host to another through bites of infective fleas [1]. However, there are alternative transmission routes, such as inhalation and direct contact, which do not normally play a role in the maintenance of plague bacterium in the animal reservoirs [2]. Fleas are generalists in their feeding behavior; thus, in the absence of rodents, they seek alternative hosts, such as wild and domestic animals, as well as people [3]. Thus, some flea species, for instance, Xenopsylla brasiliensis, are implicated as effective plague transmitters [4]. Alternatively, humans can acquire plague by handling infected rodents or by droplet infection, although humans do not play a role in the long-term survival of the plague bacterium [2]. In most cases, a plague outbreak causes a great population decrease in rodents of some species within their local and established ranges (Barnes, 1993, cited in Thiagarajan et al. [5]). However, some rodent hosts act as reservoirs of the disease in the enzootic environment allowing low levels of the pathogen circulation [3].
The first cases of human plague in Zimbabwe were recorded in 1974. The cases showed a complicated combination of plagues [6]. From then up to 1985, there were 89 cases noted, of which 23 deaths occurred [7]. Subsequent plague human cases were recorded in 1994 and 1997, where the highest cases of plague were noted, with about 400 cases and 30 deaths confirmed in Nkayi and Lupane districts, Matabeleland North [7,8]. In 2012 (latest information), plague was detected in Zimbabwe, but there was no full report of the disease [6]. Unavailability of active plague surveillance may have adverse repercussions, as an epidemic can occur undetected [9]. Plague foci are dynamic and keep on emerging and re-emerging [10], hence the importance of research to establish any potential new plague foci.
Plague bacterium can be hosted by a number of rodents. It was detected in multimammate mouse (Mastomys natalensis) and Gerbilliscus species in Tanzania [11]. Furthermore, small mammals, such as rabbits (Sylvilagus floridanus), marmots (Marmota), and chipmunks (Eutamias spp.), were shown to maintain plague in the wilderness [3]. Surveying diseases among free ranging wild animal populations may provide an early warning system for the presence of a disease. There is generally less active plague surveillance in Zimbabwe [4], except the few publications on rodents and fleas which largely focus on rodent and flea species diversity [12,13]. Therefore, this study aims to fill the gap in knowledge on plague prevalence in Zimbabwe by assessing the prevalence of plague bacteria among rodent species in Nkayi and Umzingwane districts. These two districts are in the same agro-ecological zone but differ in their plague occurrence: one is plague endemic (Nkayi district) while the other is plague non-endemic (Umzingwane district) [6,7].
2. Materials and methods
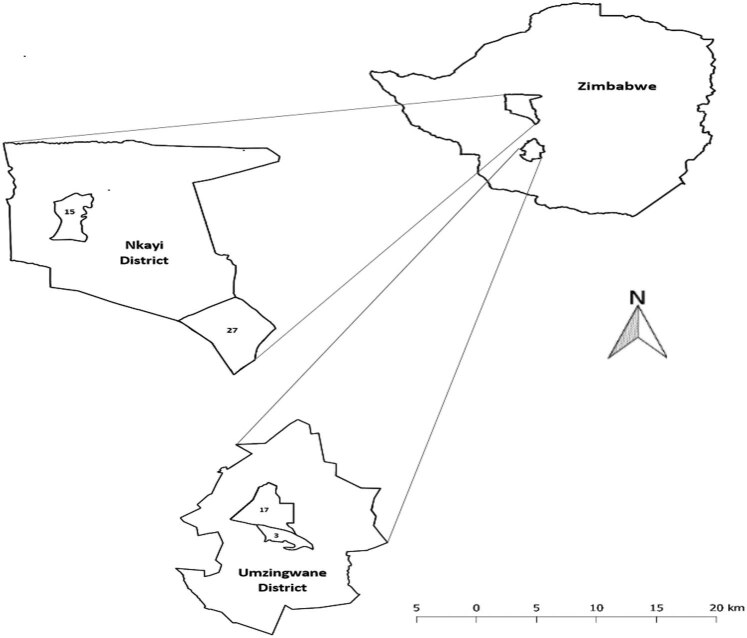
The study was conducted in south-western Zimbabwe in Nkayi and Umzingwane districts’ located in natural region IV (Figure 1). Nkayi district has deep Kalahari sands occupying 60% of the area whereas Umzingwane district’s soils are derived from granite rocks being coarse, sandy, and low in fertility [14]. The most common type of vegetation in Nkayi district is broad leafed woodlands, teak (Baikiaea plurijuga), and Brachystegia spp. [15]. Umzingwane district is characterized by three types of vegetation, which are bushveld, mainly covered with Acacia ranging between 1 and 5 m high, wooded grassland, and woodland covered by Terminalia and Combretum genus trees. The grasslands are the main source of grazing land [14].
Figure 1.
Location of Nkayi and Umzingwane Districts in south-western Zimbabwe. Notes: 15, 27, 3, and 17 represent Monki, Mathoba, Nhlekiyane, and Crocodile wards, respectively.
In the Nkayi district, most of the local rural community members are engaged in extensive livestock production and cultivation of some drought-tolerant crops, such as sorghum (Sorghum vulgare), finger millet (Eleusine coracana) and pearl millet (Pennisetum glaucum). However, farmers do sometimes grow some short-season maize (Zea mays) varieties. Nkayi district’s population was reported to be 109,135 people, while the Umzingwane district had 62,990 people in 2012 [16].
This study followed a quasi-experimental design comprising two strata, i.e., districts and then two villages in each district. In the Nkayi district, the two villages studied were Mathoba and Monki villages while in the Umzingwane district were Crocodile and Nhlekiyane villages. All the trapping procedures and further processing of rodents were carried out following Dennis et al. [7], except that traps were placed 10 m apart.
Rodent trapping was conducted in April 2018 by first observing rodent activity, such as maize cob consumption and/or clearly constructed tracks and warrens. Rodents were captured in villages, i.e., bushes habitats using 15 Sherman live traps placed about 10 m apart in transects [17]. Three transects were placed in uncultivated places near fields in each study village. On the first day of sampling, traps were set late afternoon and inspected the following morning between 6 am and 7 am before it was too hot to reduce stress in captured rodents. Traps were left open for three consecutive days per village, i.e., 45 traps.
Productive traps were replaced in the morning following Kimaro et al. [17]. Traps with captures were taken to a central processing point in each study village, in an open area, for euthanization of rodents using chloroform, species identification and blood sample collection. Rodents were identified according to the illustrations and descriptions by du Plessis et al. [18]. Rodents identified were confirmed with the Natural History Museum, Bulawayo, Zimbabwe. Rodents’ cadavers were buried in a pit of about 50 cm deep.
After about 2–3 min after introducing the rodent to chloroform, blood was collected from the euthanized rodent using a hypodermic needle with a connected syringe. Blood was collected from the heart blood on which blood smears were made. Slides with Giemsa stain were examined at ×1,000 for 5–10 min, searching for bipolar coccobacilli [19] in the Wildlife Laboratory at Chinhoyi University of Technology, Chinhoyi, Zimbabwe.
2.1. Data analysis
Trapping success was calculated as the number of animals caught divided by the trapping period divided by the number of traps set per period multiplied by 100 (7). The Chi-square (χ²) test of independence or cross-tabulation was used to determine if there were differences among the trapping success in STATISTICA version 10 for Windows [20]. Diversity of rodents in each district was calculated following the Shannon–Wiener (H′) index measure of diversity [21]. The prevalence of Y. pestis in rodents’ blood samples was calculated as the number of Y. pestis-positive rodent individuals divided by the total number of rodents examined multiplied by 100.
3. Results
A total of 44 rodents were captured in the two study districts, i.e., Nkayi (H′ = 0.89) and Umzingwane (H′ = 0.62). There was no significant difference in the trapping success between the study districts, i.e., Nkayi (8.5%) and Umzingwane (8%) (Table 1) (χ² = 1.50, df = 1, P = 0.221). Twenty-three (23) rodents were caught in the Nkayi district and 21 in the Umzingwane district, i.e., 17 M. natalensis (Smith, 1834), 6 Gerbilliscus brantsi (Smith, 1836), 4 Gerbilliscus leucogaster (Peters, 1852), and 17 Saccostomys campestris (Peters, 1846). Only one rodent, i.e., M. natalensis, caught in the Umzingwane district tested positive for the plague bacterial, indicating an overall prevalence of plague of 2.4% in the study area (Table 2).
Table 1.
Rodents trapping success in the study area
| Variable | Nkayi district | Umzingwane district | ||
|---|---|---|---|---|
| Mathoba | Monki | Crocodile | Nhlekiyane | |
| No. rodents per site | 10 | 13 | 13 | 8 |
| Trapping success (%) | 7 | 10 | 10 | 6 |
| Mean trapping success (%) | 8.50 | 8.00 | ||
Table 2.
Rodents captured species diversity and prevalence of bacterial plague in the study area
| Rodent species | Nkayi district | Umzingwane district | Overall prevalence (%) | ||||
|---|---|---|---|---|---|---|---|
| # of rodents captured | # of rodents that are positive for Y. pestis | Prevalence (%) | # of rodents captured | # of rodents that positive for Y. pestis | Prevalence (%) | ||
| M. natalensis | 0 | 0 | 17 | 1 | 5.9 | 5.9 | |
| S. campestris | 4 | 0 | 0 | 2 | 0 | 0 | 0 |
| G. brantsi | 4 | 0 | 0 | 2 | 0 | 0 | 0 |
| G. leucogaster | 15 | 0 | 0 | 0 | 0 | 0 | 0 |
| Species diversity (H′) | 0.89 | — | — | 0.62 | — | — | — |
| Total/average (%) | 23 | 0 | 0 | 21 | 1 | 4.8 | 2.4 |
Note: – denotes not applicable; # – denotes number.
4. Discussion
This study is the first to report the presence of plague bacteria among rodents in Umzingwane district, even though a low prevalence of plague disease exists in the study area. Elsewhere, plague bacteria were reported as being difficult to detect in rodents and fleas associated with prairie dog colonies (Cynomys) at Thunder Basin National Grassland in Wyoming, USA [5]. Thus, it was suggested that where possible plague determination investigations could be conducted in places where there are noticeable rodent die-offs [5]. Since time immemorial M. natalensis was observed not to easily succumb to plague bacteria, thus termed an enzootic host [11,22]. Thus, some of the caught species may not be ideal host for Y. pestis, for instance T. leucogaster was observed to easily succumb to Y. pestis closely related bacilli Y. pestis tuberculosis subspecies pestis [23].
Since no die-offs were observed during the sampling time, it may imply that an inter-epizootic period could have existed during which Y. pestis cannot be recovered from fleas, rodents, or any other host [1]. Instead, it was proposed in Iran and Madagascar that the bacteria could remain in existence in the soil. However, one study recommended that the transmission route by exposure of susceptible mice to Y. pestis-contaminated soil seems doubtful under natural conditions because the infectious period was short-lived and the transmission efficiency was low [24]. Thus, during plague quiescent times, it is likely that detecting Y. pestis is by mere chance.
In northern Tanzania, 517 wild, peri-domestic, and small commensal mammals, including rodents and wild carnivores, were captured, but only three tested positive for Y. pestis. This is an indication for the need for large samples to fully get a good picture of the infection status of a rodent population [11]. There can, however, be possibilities that Y. pestis is concentrated on certain rodent body organs as was reported in a study in Mongolia, where Y. pestis was detected in spleen samples, while liver samples from rodents tested negative using Polymerase Chain Reaction [25]. This therefore points to the importance of a large sample associated with diversified samples from the rodent body parts to enhance the chances of detection of the representative infection status of populations.
5. Conclusion
The study provides the first evidence of plague bacteria infection in a rodent species in Umzingwane district, a non-endemic region for the plague disease. It is recommended that future studies on plague bacteria assessment should involve a larger sample both for rodent populations and areas covering endemic and non-endemic regions, cover a longer sampling timeframe, and take samples from other body parts of rodents (liver and spleen) for testing beyond the blood.
Acknowledgments
Our sincere gratitude goes to the following village heads, Mr. R. Ncube and Mr. R. Tshuma (Umzingwane district) and Mr. A Mlotshwa and Mr. J. M. Gwayi (Nkayi district) for permission to conduct the study in their areas of jurisdiction. We are also indebted to Nkayi Principal Environmental Health Technician, Mr. J. Sibanda, and the Health Technicians, Mr. G. Mabhena and Mr. N. Ncube, for providing us with plague historical information.
Footnotes
Funding information: Authors state no funding involved.
Author contributions: Annabel Banda conceived and designed the experiments, performed the experiments, analyzed the data, contributed reagents/materials/analysis tools, prepared figures and/or tables, authored or reviewed drafts of the manuscript, and approved the final draft. Edson Gandiwa conceived and designed the experiments, analyzed the data, contributed analysis tools, authored or reviewed drafts of the manuscript, and approved the final draft. Never Muboko and Victor K. Muposhi authored or reviewed drafts of the manuscript, and approved the final draft.
Conflict of interest: Authors state no conflict of interest.
Data availability statement: The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
- [1].Rivière-Cinnamond A, Santandreu A, Luján A, Mertens F, Espinoza JO, Carpio Y, et al. Identifying the social and environmental determinants of plague endemicity in Peru: Insights from a case study in Ascope, la Libertad. BMC Public Health. 2018;18(1):1–11. [DOI] [PMC free article] [PubMed]
- [2].Perry RD, Fetherston JD. Yersinia pestis – Etiologic agent of plague. Clin Microbiol Rev. 1997;10(1):35–66. [DOI] [PMC free article] [PubMed]
- [3].Gage KL, Kosoy MY. Natural history of plague: Perspectives from more than a century of research. Annu Rev Entomol. 2005;50:505–28. [DOI] [PubMed]
- [4].Chikwenhere G. Rodents and human diseases in Zimbabwe. In: Rats and human health in Africa: Proceedings of an international workshop on rodent-borne diseases and the RatZooMan research project. Malelane, Republic of South Africa: Natural Resources Institute of the University of Greenwich; 2006. p. 1–46.
- [5].Thiagarajan B, Bal Y, Gage KL, Cully JF. Prevalence of Yersinia pestis in rodents and fleas associated with black-tailed prairie dogs (Cynomys ludovicianus) at Thunder Basin National Grassland, Wyoming. J Wildl Dis. 2008;44(3):731–6. [DOI] [PubMed]
- [6].Munyenyiwa A, Zimba M, Nhiwatiwa T, Barson M. Plague in Zimbabwe from 1974 to 2018: A review article. PLoS Negl Trop Dis. 2019;13(11):1–17. [DOI] [PMC free article] [PubMed]
- [7].Dennis DT, Gage KL, Gratz N, Poland JD, Tikhomirov E. Plague manual-epidemiology, distribution, surveillance and control. In: Plague manual: Epidemiology, distribution, surveillance and control; Geneva, Switzerland: WHO; 1999. p. 447.
- [8].Manungo P, Peterson D, Todd C, Mthamo N, Pazvakavambwa B. Risk factors for contracting plague in Nkayi district, Zimbabwe. Cent Afr J Med. 1998;44:173–6. [PubMed]
- [9].WHO. Plague around the world, 2010–2015. Wkly Epidemiol Rec/Heal Sect Secr Leag Nations. 2016;91(8):89–93. https://www.who.int/wer/2016/wer9108.pdf?ua=1.
- [10].Andrianaivoarimanana V, Kreppel K, Elissa N, Duplantier JM, Carniel E, Rajerison M, et al. Understanding the persistence of plague foci in madagascar. PLoS Negl Trop Dis. 2013;7(11):1–8. [DOI] [PMC free article] [PubMed]
- [11].Ziwa MH, Matee MI, Hang’Ombe BM, Lyamuya EF, Kilonzo BS. Plague in Tanzania: An overview. Tanzan J Health Res. 2013;15(4):252–8. [PubMed]
- [12].Zimba M, Pfukenyi D, Mukaratirwa S. Seasonal abundance of plague vector xenopsylla brasiliensis from rodents captured in three habitat types of periurban suburbs of harare, Zimbabwe. J Vec-Bor and Zoon Dis. 2010;11(8):1187–92. [DOI] [PubMed]
- [13].Zimba M, Loveridge J, Pfukenyi DM, Mukaratirwa S. Seasonal abundance and epidemiological indices of potential plague vectors dinopsyllus lypusus (Siphonaptera: Hystrichopsyllidae) and ctenophthalmus calceatus (Siphonaptera: Ctenophthalmidae) on rodents captured from three habitat types of Hatcliffe and Dzivarasekwa suburbs of Harare, Zimbabwe. J Med Entomol. 2012;49(6):1453–9. [DOI] [PubMed]
- [14].Renaudin B, Patinet J. Environmental impact assessment (EIA)-ACF Zimbabwe. La Fontaine des Marins; 2010.
- [15].Dube T, Homann-Kee Tui S, Van Rooyen AF, Rodriguez D. Baseline and situation analysis report: Integrating crop and livestock production for improved food security and livelihoods in rural Zimbabwe. ICRISAT – Socioecon Discuss Pap Ser. 2014;29:8–9.
- [16].Zimstat C. Census 2012: Preliminary report. 2013. p. 123.
- [17].Kimaro DN, Msanya BM, Meliyo J, Hieronimo P, Mwango S, Kihupi NI, et al. Anthropogenic soils and land use patterns in relation to small mammal and flea abundance in plague endemic area of Western Usambara Mountains, Tanzania. Tanzan J Health Res. 2014;16(3):1–12. [DOI] [PubMed]
- [18].du Plessis J, Swanepoel L, McDonough M, Schoeman C. A conservation assessment of Gerbilliscus leucogaster. South Africa: The Red list of Mammals of South Africa, Swaziland and Lesotho; 2016.
- [19].WHO. Interregional meeting on prevention and control of plague. In: Antananarivo, Madagascar 1–11 April 2006. Antananarivo: World Health Organisation; 2008. p. 1–65. http://www.who.int/csr/resources/publications/WHO_HSE_EPR_2008_3/en/.
- [20].StatSoft. STATISTICA for Windows. 2300 Tulsa: StaSoft Inc; 2010.
- [21].Krebs CJ. Ecology: The experimental analysis of distribution. 4th edn. New York, USA: HarperCollins College; 1994. p. 801.
- [22].Neerinckx SB, Peterson AT, Gulinck H, Deckers J, Leirs H. Geographic distribution and ecological niche of plague in sub-Saharan Africa. Int J Health Geogr. 2008;7:54. [DOI] [PMC free article] [PubMed]
- [23].Isaacson M, Taylor P, Arntzen L. Ecology of plague in Africa: Response of indigenous wild rodents to experimental plague infection. Bull World Health Organ. 1983;61(2):339–44. [PMC free article] [PubMed]
- [24].Boegler KA, Graham CB, Montenieri JA, McMillan K, Holmes JL, Petersen JM, et al. Evaluation of the infectiousness to mice of soil contaminated with yersinia pestis-infected blood. Vector Borne Zoonotic Dis. 2012;12(11):948–52. [DOI] [PMC free article] [PubMed]
- [25].Riehm JM, Tserennorov D, Kiefer D, Stuermer IW, Tomaso H, Zöller L, et al. Yersinia pestis in small rodents, Mongolia. Emerg Infect Dis. 2011;17(7):1320–2. [DOI] [PMC free article] [PubMed]