Abstract
Candidate partitioning genes (parA and parB) for the linear chromosome of Streptomyces coelicolor were identified by DNA sequencing in a series of seven genes located between rnpA and trxA near the chromosomal replication origin. The most likely translation start point of parB overlapped the parA stop codon, suggestive of coregulation, and transcription analysis suggested that the two genes formed an operon. Deletion of part of parB had no effect on the growth or appearance of colonies but caused a deficiency in DNA partitioning during the multiple septation events involved in converting aerial hyphae into long chains of spores. At least 13% of spore compartments failed to inherit the normal DNA allocation. The same phenotype was obtained with a deletion removing a segment of DNA from both parA and parB. Reinforcing the idea of a special role for the par locus during sporulation, the stronger of two parAB promoters was greatly upregulated at about the time when sporulation septation was maximal in colonies. Three copies of a 14-bp inverted repeat (GTTTCACGTGAAAC) were found in or near the parAB genes, and at least 12 more identical copies were identified within 100 kb of oriC from the growing genome sequence database. Only one perfect copy of the 14-bp sequence was present in approximately 5 Mb of sequence available from the rest of the genome. The 14-bp sequence was similar to sequences identified as binding sites for Spo0J, a ParB homologue from Bacillus subtilis believed to be important for DNA partitioning (D. C.-H. Lin and A. D. Grossman, Cell 92:675–685, 1998). One of these sites encompassed the transcription start point of the stronger parA promoter.
Streptomyces coelicolor is the best-characterized member of an actinomycete genus renowned for its ability to produce secondary metabolites. Streptomycetes are also of great interest because of their complex morphological differentiation and their unusually large linear chromosome (ca. 8 Mb [40]). Spores germinate to form a vegetative mycelium of branching multigenomic hyphae. This coherent structure achieves dispersal by developing aerial branches that metamorphose into chains of unigenomic spores (6). Replication of the DNA appears to involve a conventional origin (oriC) located in the center of the genome, from which bidirectional replication proceeds to the telomeres (34). It is postulated that a protein attached to the ends of the DNA is involved in the maintenance and replication of chromosome ends, possibly priming patch replication of the recessed 5′ ends of the lagging strands (8, 39). We are interested to understand how correct chromosomal partitioning is achieved in a mycelial organism with a linear chromosome, since the limited existing knowledge of bacterial chromosome partitioning has been obtained exclusively from more or less rod-shaped organisms that grow by binary fission and possess circular chromosomes (e.g., see references 16, 27, 31, and 43). Moreover, within the life cycle of Streptomyces, different partitioning requirements are likely to be associated with normal tip extension (in which most replication events are not associated with septation), branching, occasional vegetative septation, and the organized multiple septation that eventually gives rise to spore chains.
In bacteria as diverse as Caulobacter crescentus and Bacillus subtilis (though not in Escherichia coli), components of the partitioning apparatus are encoded by a pair of genes, parA and parB, located close to oriC (in B. subtilis these genes are called soj and spo0J, respectively) (44). Similar genes in plasmids such as R1, P1 prophage, and F have been extensively studied (see reference 25 for references). In brief, it is thought that ParB forms aggregates on the oriC-proximal region of the chromosome following its binding to a moderate number of dispersed repeats of a particular sequence (29, 31). Studies of the location and movement of these aggregates in cells of various bacteria suggest that they could be involved in determining chromosome position (27, 28, 31). ParA-like proteins are also DNA-binding proteins and have ATPase activity that, at least in plasmid R1, is essential for partitioning (24). The significance of parAB varies among bacteria—in C. crescentus, mutations in either gene are lethal (31), whereas in B. subtilis vegetative growth is little affected by soj or spo0J mutations, though the frequency of anucleate cells is increased from about 0.01 to about 1 to 2% in spo0J mutants (22); however, spo0J mutants are blocked at an early stage of sporulation, because Soj, in the absence of Spo0J, can bind to and repress certain early sporulation-specific promoters (3).
The origin of replication of the Streptomyces chromosome is located downstream of dnaA (an arrangement that is frequently conserved in bacterial chromosomes) (2, 47). This region has been shown to replicate autonomously (47). In other bacteria, the partitioning genes are often located upstream of dnaA. Accordingly, a 7-kb region of DNA upstream of the S. coelicolor dnaA gene was cloned and sequenced, revealing homologues of parA and parB. (During the preparation of the manuscript, a similar analysis of this DNA region was described [15].) Here we outline features of this region, with some differences of interpretation or emphasis from reference 15, describe the construction and phenotypes of mutants affected in parB and parAB, and provide evidence of a possible sporulation-associated increase in expression from one of two parAB promoters.
(A preliminary report of the isolation of the par genes and their linkage within the trx-dnaA region was presented at the Keystone Symposium on Bacterial Chromosomes in 1995 [J. Cell. Biochem. Suppl. 19:123].)
MATERIALS AND METHODS
Bacterial strains and culture conditions.
E. coli DH5α (17) was the host strain for standard plasmid manipulation, and the nonmethylating E. coli strain ET12567 (30) was used to propagate DNA when it was to be introduced into S. coelicolor A3(2) derivatives, which restrict methylated DNA (26). The S. coelicolor A3(2) derivatives used are listed in Table 1. Conditions and general techniques for the culture and transformation of bacterial strains and for DNA manipulation were as described elsewhere for E. coli (41) and S. coelicolor (21). For the preparation of protoplasts and chromosomal DNA, mycelium of S. coelicolor was grown in YEME liquid medium supplemented with proline, arginine, cysteine, and histidine each at 0.375 g/liter and uracil at 7.5 mg/liter (SuperYEME). Protoplast regeneration was on R5 medium. Minimal medium (MM) containing 1% mannitol was used for RNA isolation and microscopic examination.
TABLE 1.
S. coelicolor A3(2) derivatives used in this work
| Strain no. | Genotype | Reference |
|---|---|---|
| Morphologically wild-type strains | ||
| M145 | Prototrophic, SCP1− SCP2− Pgl+ | 21 |
| J1501 | hisA1 uraA1 strA1 SCP1− SCP2− Pgl− | 7 |
| M145 derivatives | ||
| J2537 | parB::aac(3)IV | This paper |
| J2538 | parAB::aac(3)IV | This paper |
| J2540 | parAB::aac(3)IV pIJ6539 | This paper |
| J2539 | parB::aac(3)IV pIJ6540 | This paper |
| J1501 derivatives | ||
| J2535 | parB::aac(3)IV | This paper |
| J2536 | parAB::aac(3)IV | This paper |
DNA sequence analysis.
Overlapping BamHI and HindIII fragments containing the par region of the S. coelicolor chromosome were subcloned from a previously described cosmid clone (2) for sequence determination. Sequencing was performed on one strand from unidirectional deletions created by the exonuclease III method using a Sequenase 2.0 kit (U.S. Biochemicals, Cleveland, Ohio), and the second strand was determined by primer walking using an ABI 377 sequencer. Analysis used programs provided in the GCG package (Genetics Computer Group, Madison, Wis.).
Disruption of the parAB locus.
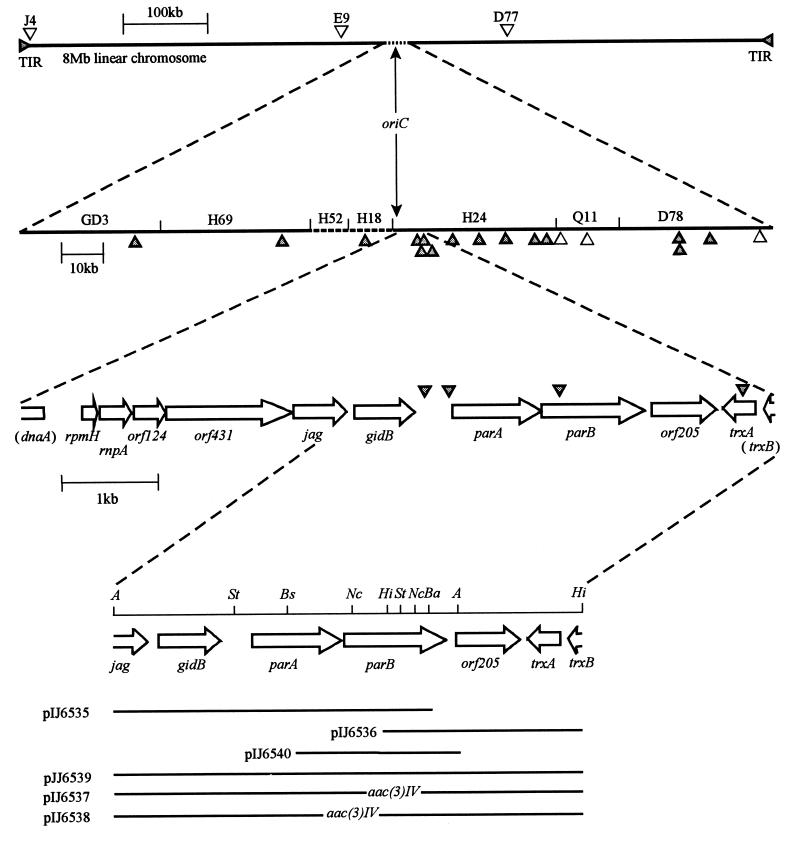
A 3.4-kb Asp718I/BamHI fragment containing parA and the 5′ part of parB and a 2-kb HindIII fragment containing the 3′ part of parB, orf205, trxA, and the 3′ part of trxB were cloned into pTZ18R (Pharmacia) and pBluescript SK(+) (Stratagene) to create pIJ6535 and pIJ6536, respectively (Fig. 1). In order to disrupt parB, a 1.8-kb NcoI/HindIII fragment containing the 3′ end of parB, orf205, trxA, and the 3′ part of trxB was isolated as an NcoI/XbaI fragment from pIJ6536 and cloned into similarly digested pIJ6535, resulting in deletion of a 0.8-kb NcoI fragment internal to parB. A 1.7-kb SmaI fragment containing aac(3)IV was inserted into the NcoI site which had been blunted with Klenow enzyme, creating pTZ45a. To construct a plasmid with a parAB disruption, a 2.3-kb StuI/HindIII fragment containing the 3′ part of parB, orf205, trxA, and the 3′ part of trxB was isolated as a StuI/XbaI fragment from pIJ6536 and cloned into similarly digested pIJ6535, resulting in deletion of a 1.7-kb StuI fragment containing parA and the 5′ part of parB. A 1.7-kb SmaI fragment containing aac(3)IV was cloned into the StuI site, yielding pTZ53a. The whole inserts from pTZ45a and pTZ53a were isolated as 6.1- and 5.2-kb EcoRI fragments, respectively, and cloned into similarly digested pIJ566 (36), an integrational vector containing tsr for selection in Streptomyces, yielding pIJ6537 and pIJ6538, respectively (Fig. 1).
FIG. 1.
Features of DNA referred to in this work. Top line, linear chromosome of S. coelicolor (TIR, terminal inverted repeats); second line, cosmids around the oriC region (solid lines, sequences deposited in EMBL database; dashed lines, sequencing not completed); third line, arrangement of genes in the region sequenced in this work; fourth line, restriction map of parAB region (A, Asp718I; St, StuI; Bs, BsiWI; Nc, NcoI; Hi, HindIII; Ba, BamHI); lower bars, subclones and disruption cassettes described in this work. The positions of 14-mer inverted repeats (GTTTCACGTGAAAC) are marked by filled triangles, and open triangles mark the positions of sequences diverging from the 14-mer sequence by only 1 bp.
pIJ6537 and pIJ6538 isolated from ET12567 were denatured with alkali (37) and used to transform J1501. Transformants were selected on R5 medium containing apramycin (100 μg/ml), and then putative parB or parAB null mutants that had lost the vector were identified by their sensitivity to thiostrepton (50 μg/ml). Total DNA (38) from each of two independent recombinants of each putative mutant was digested with Asp718 and BamHI and subjected to Southern analysis using pIJ6535 as a probe to verify the deletion. To generate the same mutations in the prototrophic S. coelicolor strain M145, alkaline-denatured total DNA from the parB::aac(3)IV and parAB::aac(3)IV strains J2535 and J2536 (J1501 derivatives) was used to transform M145 protoplasts (37). The parB and parAB disruptions in J2537 and J2538 (M145 derivatives) were verified by Southern blotting.
Complementation of par disruption mutants.
To reconstitute the wild-type allele of parAB, a 1.9-kb StuI/HindIII fragment containing the 3′ end of parB, orf205, trxA, and the 3′ part of trxB was isolated as a StuI/XbaI fragment from pIJ6536 and cloned into similarly digested pIJ6535, resulting in pTZ5300: the left one of two StuI sites in the insert of pIJ6535 was protected from digestion by Dam methylation. The whole insert, as a 5.3-kb EcoRI fragment, and a 1.8-kb BsiWI/Asp718I fragment from pTZ5300 containing the intact parB gene were cloned into similarly digested pDH5, an integration vector containing tsr for selection in Streptomyces (19), yielding pIJ6539 and pIJ6540, respectively. pIJ6539 and pIJ6540 were introduced into J2537 [parB::aac(3)IV] and J2538 [parAB::aac(3)IV] at the par locus by single-crossover recombination, and this was verified by Southern blot analysis.
Microscopy.
Mycelium was grown against a sterile coverslip by inoculating the surface of the agar medium adjacent to the half-submerged coverslip (4). Coverslips were lifted from the medium after 2 to 4 days of incubation at 30°C and cleaned with paper tissue without disturbing the mycelium. Further processing and examination were done using staining with DAPI (4′,6-diamino-2-phenylindole) to stain DNA, as described in reference 11. Images presented in figures were obtained by scanning negative film and processing with Adobe Photoshop.
RNA analysis.
Mycelium grown on cellophane disks placed on MM containing mannitol was collected with a spatula into a sterile grinder containing liquid N2 and ground vigorously. The samples were then kept frozen until use for RNA extraction. RNA was extracted using the acid phenol procedure (42). S1 nuclease protection experiments were based on the procedure in the work of Bibb et al. (1). Fifty micrograms of RNA was used in each experiment, and hybridization with a probe was at 45°C in NaTCA buffer (33) after denaturation (75°C, 10 min). Probes were prepared by PCR amplification using cold forward primers and reverse primers labeled with [γ-32P]ATP (3,000 Ci/mmol) using T4 polynucleotide kinase. For low- and high-resolution S1 nuclease mapping of the parA transcription start point, combinations of primers, A1 and A2, and A1 and A3, were used to amplify probes of 655 and 360 bp, respectively (see Fig. 4). To try to find a parB transcription start site, a 570-bp probe was used, extending from a position 241 bp inside the parA coding region to a position 333 bp inside the parB coding region (not shown).
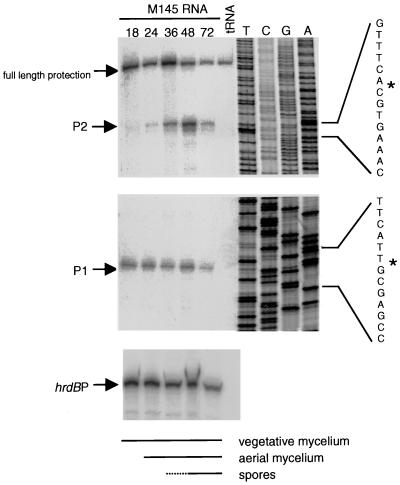
FIG. 4.
Mapping of the parAB transcription start points and detection of the parAB transcript level during development in surface-grown culture of the wild-type S. coelicolor strain M145. S1 mapping of parAB mRNA was carried out with RNA isolated from M145 grown on MM containing mannitol for different periods (18, 24, 36, 48, and 72 h). The presence of vegetative mycelium, aerial mycelium, and spores at each time of harvest was judged by microscopic examination. The first and second panels were from the same gel, and thus the proportion of two transcripts is valid for comparison. The sequence of the template strand is shown on the right, and the two transcription start points are indicated by asterisks. The levels of hrdB mRNA in the same RNA samples were measured by S1 mapping as an internal control.
Nucleotide sequence accession number.
The nucleotide sequence has been deposited in GenBank under accession no. AF 187159.
RESULTS
Analysis of DNA proximal to dnaA on the S. coelicolor chromosome.
Sequencing between the previously sequenced rpmH and trxAB regions revealed seven complete open reading frames (ORFs) oriented in the same direction as rnpA and rpmH, followed by the trxAB region (GenBank accession no. X92105) (Fig. 1). Each of the seven ORFs resembled genes found in the equivalent regions of other bacterial chromosomes. Here we briefly summarize the features of the first four of these genes before focusing on parA and parB, which are the main subject of this paper. Our interpretation of the sequence generally, but not entirely, coincides with that reported independently (15).
Apparently translationally coupled to rnpA is orf124, homologues of which are found in the equivalent location in most bacterial chromosomes, though not in that of Mycobacterium leprae (14, 15). The predicted S. coelicolor protein is larger (13.6 kDa) than the homologues in other bacteria (9 kDa) and contains no predicted transmembrane domains and is therefore probably cytoplasmic. The next gene, orf431, is 3 nucleotides (nt) downstream of orf124. Similarly located related genes, including spoIIIJ of B. subtilis, which is required for sporulation (10), are found in most bacteria (15). All of these genes encode proteins predicted to bind nucleoside triphosphate (14) and to contain several transmembrane domains, consistent with a membrane location. Downstream (by 15 nt) of orf431 is orf170, whose product of 170 amino acids has extensive homology to the jag gene product of B. subtilis and, particularly, its orthologues in mycobacteria. The function of Jag proteins is not known, although Jag is associated with spoIIIJ in B. subtilis and plays a role in sporulation (10). The absence of detectable transmembrane domains in Jag proteins suggests a cytoplasmic location. In our interpretation, the next gene, gidB, is located 71 nt downstream of the jag homologue, whereas in reference 15 an intergenic region of 124 residues was suggested. GidB was originally described as the product of the glucose-inhibited division gene in E. coli, but its precise function is unknown (35). A gidB-like gene is present in all eubacterial genomes sequenced to date.
The parA homologue of S. coelicolor is separated from gidB by several hundred nucleotides, depending on the allocation of the parA translation initiation codon. parA is the first of a pair of ORFs, parA and parB, which are candidates for chromosome partitioning genes of S. coelicolor. The designations soj and spo0J (used in reference 15) are avoided here in view of the mutational analysis detailed below. The four candidate parA translation initiation codons indicated in Fig. 2 would result in ParA proteins of 319, 307, 294, and 274 amino acid residues, and two other candidate ATG start codons are present a little further upstream. Sequence comparisons suggest that the 319-residue protein is probably made (but do not rule out the possibility of a nested set of translation start points). The ParA protein from S. coelicolor (ParASc) is most similar to that from mycobacteria (e.g., 61% identity over 290 residues to the M. leprae protein) (14), although the homologues from B. subtilis (22) and C. crescentus (31) are also very similar (55 and 49% identity, respectively). The S. coelicolor and mycobacterial ParA proteins are likely to have N-terminal extensions compared to the B. subtilis and C. crescentus proteins. ParASc also has evident homology to ParA-like proteins encoded by plasmid or prophage replicons, including ParA of prophage P1 and SopA of the F plasmid. All of these proteins contain putative ATP-binding sites (32), which are well conserved in ParASc, and may provide energy for replicon segregation (for a review, see reference 20).
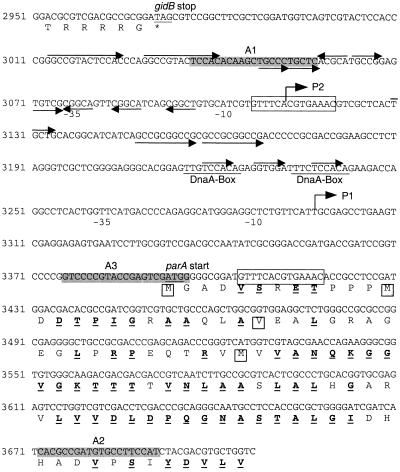
FIG. 2.
Features of the gidB-parA untranslated region from S. coelicolor. The gidB stop codon and the favored possible start codon for parA are underlined. The four possible translation starts discussed in the text are indicated by a boxed amino acid residue, and the amino acids that are identical between the S. coelicolor and M. leprae reading frames are in boldface and underlined. Two copies of the 14-nt palindrome that may function as the DNA-binding site for ParB are boxed. Two putative DnaA boxes are shown (horizontal line). Two transcription start points (P1 and P2) with −35 and −10 positions are shown. The sequences of primers (A1, A2, and A3) used to amplify probes for S1 mapping are shaded. Other local repeat features are indicated by arrows.
Downstream of parA is parB. Based on sequence comparison to other ParB homologues, the length of ParBSc would be 368 amino acid residues, and the parB start codon and the parA stop codon would overlap, indicating a possible translational coupling mechanism to permit the equal expression of each subunit. (In reference 15, 131 nt intervene between parA and parB.) The parA-parB intergenic region is only 3 nt in the M. leprae chromosome. ParBSc protein has extensive identity with chromosomally encoded orthologues from several bacteria (e.g., 57% identity to ParB of Mycobacterium tuberculosis and 41% identity to Spo0J of B. subtilis), in addition to more distant resemblance to paralogues encoded by other replicons, for example, ParB of prophage P1 and SopB of the F plasmid. Both of the latter, and the chromosomally encoded ParB from C. crescentus and Spo0J from B. subtilis, bind centromere-like DNA sequences in the cognate replicon (references 20, 22, and 29 and references therein).
A further gene, orf205 (of unknown function), is separated from parB by 337 nt. Similar sequences have been detected only in the related actinomycetes M. leprae and M. tuberculosis. orf205 converges on the genes encoding thioredoxin (trxA) and thioredoxin reductase (trxB) (9).
As pointed out by others (15, 29), a 14-bp palindromic sequence (GTTTCACGTGAAAC) is present four times within the sequenced region (Fig. 1 and 2). There is one copy in the 5′ regions of both parA and parB. Another copy is found upstream of parA within an untranslated region, plus a copy immediately downstream of trxA. A database search with this palindrome led to the identification of the sequence in a similar trxA distal position in the Streptomyces clavuligerus chromosome. A further 13 perfect copies of the palindrome were found in the S. coelicolor genome sequence database, which at the time of the analysis (July 1999) comprised about 60% of the 8-Mb genome (http://www.sanger.ac.uk/Project /S_coelicolor), and all but one of them were located within 100 kb of the origin region (Fig. 1). The 14-mer sequence is closely similar to the consensus Spo0J-binding site (29), and so this repeated motif close to putative partitioning genes may have a role as a chromosome partitioning site (29).
In addition to the 14-bp repeated sequences, the comparatively long untranslated region between gidB and parA contains two copies of the Streptomyces DnaA box (23): one “optimal” copy and one with a single permitted deviation from optimal consensus (Fig. 2).
Disruption of the parAB locus does not visibly affect growth, viability, or spore development.
In the gram-negative C. crescentus, the parAB genes are essential for colony formation (31), whereas in the low-G+C gram-positive B. subtilis they can be disrupted without detectably affecting growth or colony formation (22). However, B. subtilis spo0J (= parB) mutants cannot sporulate, an effect suppressible by a mutation in soj (= parA) (22). In order to evaluate the importance of parA and parB for growth and differentiation of S. coelicolor, which is phylogenetically distant and developmentally different from C. crescentus and B. subtilis, the cloned parAB cluster was first disrupted in two ways, using the aac(3)IV cassette to replace (a) most of parB (in pIJ6537) and (b) the whole of parA and part of parB (in pIJ6538) (Fig. 1) (the latter construct was made principally to find out whether any aspects of the parB mutant phenotype might be suppressed by a parA deficiency, as is the case for the sporulation defect of spo0J [= parB] mutants of B. subtilis [22]). When these plasmids were used to transform S. coelicolor J1501 to apramycin resistance to construct J2535 [parB::aac(3)IV] and J2536 [parAB::aac(3)IV], it proved straightforward to find colonies that were sensitive to thiostrepton and were therefore candidates for double-crossover replacement of the chromosomal parAB locus with the disrupted versions. Southern blotting verified this. Alkali-denatured chromosomal DNA from J2535 and J2536 was used to transform the prototrophic strain M145 to generate J2537 [parB::aac(3)IV] and J2538 [parAB::aac(3)IV] (the auxotrophic strain J1501 is usually the first choice as a recipient due to its better transformation efficiency, but the morphological phenotypes of developmental mutants are preferably examined in the M145 derivatives). The disruption mutants showed no obvious macroscopic change in colony size, blue pigmentation (actinorhodin production), aerial mycelium formation, or the development of grey spore pigment, and they produced abundant spores as visualized by phase-contrast microscopy. Therefore, ParASc and ParBSc do not play important roles in growth (in contrast to the situation in C. crescentus), and their interface with sporulation, if any, is different from that of Soj-Spo0J in B. subtilis.
An important role for parB in accurate chromosome partitioning during sporulation.
In view of evidence that the parB (spo0J) product is physically associated with the origin region of the chromosomes of C. crescentus (31) and B. subtilis (29) and that its absence from B. subtilis causes an increase in the number of anucleate cells (22), it seemed possible that the ParB protein of S. coelicolor might play a contributory role in chromosome partitioning. Accordingly, the two par locus-disrupted strains were examined by epifluorescence microscopy after staining with the DNA-specific agent DAPI. Partitioning was difficult to analyze in vegetative hyphae, in which chromosomes are not readily visualized as discrete bodies, but the production of long chains of unigenomic spores from multigenomic tip compartments of aerial hyphae provided more favorable material for the detection of possible partitioning abnormalities. The results are shown in Fig. 3.
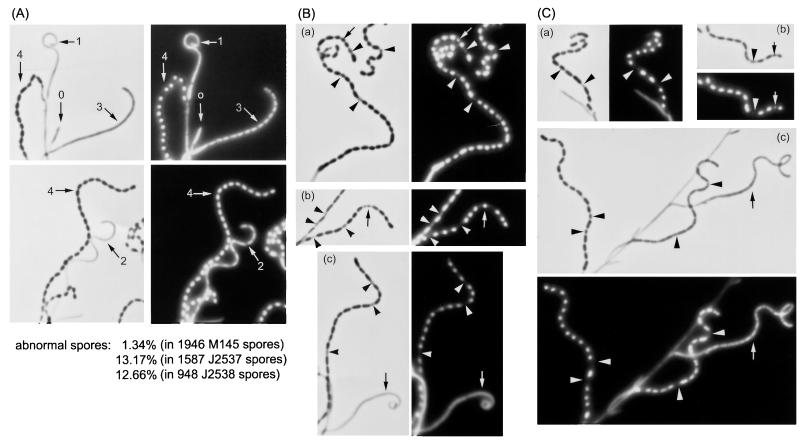
FIG. 3.
Phenotypes of the parB and parAB null mutants. Four-day-old cultures on MM containing mannitol were sampled on coverslips and stained with DAPI, and phase-contrast and fluorescent images of the same fields were taken using a Zeiss Axiophot microscope. (A) M145. Each arrow indicates a hypha at a different developmental stage. 0, aerial hypha just emerging from substrate mycelium; 1, aerial hypha showing typical coiled shape and full of uncondensed chromosomal DNA; 2, aerial hypha before sporulation and showing an early stage of chromosome segregation; 3, young spore chain showing regular septation and segregation of chromosomes; 4, mature spore chain having rounded spores and condensed chromosomes. (B) J2537 [parB::aac(3)IV]. (a) Arrowheads indicate anucleate spores, and arrow indicates a normal-sized spore with an obviously reduced amount of chromosomal DNA; (b) spore chains with several anucleate spores (arrowheads; the arrow indicates a minispore containing DNA, flanked by one which is anucleate and a full-sized but anucleate spore compartment); (c) the spore chain has one anucleate compartment and two minispores (arrowheads), but the arrowed aerial mycelium is full of dispersed chromosomal DNA, which makes it technically difficult to detect any defect in chromosome positioning. (C) J2538 [parAB::aac(3)IV]. (a) Arrowheads indicate a normal-sized anucleate spore and an anucleate part with lysis; (b) arrowhead and arrow indicate a normal-sized anucleate spore and a spore with reduced DNA content; (c) arrowheads indicate anucleate spores in a young and a mature spore chain, and the arrow indicates an aerial hypha in an early stage of sporulation and with no obvious defect in chromosome positioning.
In both mutants, there were abundant spore chains of apparently normal length. However, most chains contained abnormalities. Some “spores” were apparently DNA free (these were also very pale in phase-contrast microscopy). Although many spores were normally sized, some were tiny, implying abnormally short spacing between adjacent sporulation septa. Examples were often seen of spores with small amounts of DNA, presumably because of trapping of an incompletely partitioned chromosome by the ingrowing septum (note that the strains used, J1501 and M145, are both plasmid free, and so plasmid DNA did not contribute to DAPI staining). To verify that the mutant phenotypes were due to the changes in the parAB locus, 1.8- and 5.3-kb fragments in pIJ6540 and pIJ6539 (Fig. 1), incorporating the parB and parAB regions, respectively, were subcloned into pDH5 and introduced into the two disruption mutants by single-crossover integration at the par locus, with selection for the vector's thiostrepton resistance marker. In each case, the wild-type phenotype was restored: most spore chains were completely regular and stained uniformly with DAPI, though—as with the wild type—occasional spore chains contained an example of imperfect DNA partitioning.
In order to provide semiquantitative estimates of the frequency of partitioning defects caused by the parAB disruptions, photomicrographs of DAPI-stained spore chains (each chain containing at least 10 spores) were scored. In all, about 13% of spores of both mutants clearly showed abnormalities of the kinds indicated in Fig. 3. For comparison, only 1.3% of spores in spore chains of the wild type showed such defects, and the complemented mutants exhibited similar low frequencies of obvious partitioning abnormalities (1.0% for the complemented parAB mutant and 0.3% for the complemented parA mutant).
One of two promoters upstream of parA is upregulated at the time of spore development.
In order to find out whether the expression of the par locus showed any changes during colony development, we extracted RNA from surface cultures of S. coelicolor (strain M145) grown for different lengths of time and used S1 nuclease protection analysis to determine par mRNA levels and transcription start points.
Since the transcription start point(s) of the mRNA had not been determined, two fairly long probes were used, one (655 bp) extending from a position immediately downstream of the preceding gene (gidB) to a position 299 bp inside the parA coding sequence, and the other (570 bp) crossing from parB into parA. With the latter probe, only full-length protection was obtained, implying that parB does not have an independent promoter. With the more upstream probe, two mRNA 5′ ends were identified (Fig. 4), both within the gidB-parA intergenic region. These are likely to correspond to transcripts from two promoters, since their ratio changed very markedly during the time course (though changes in processing of a single mRNA during development could be a possible alternative explanation). The more downstream putative promoter (P1) showed little variation in strength during the time course, but the more upstream (P2) was very weak at the earlier time points, becoming stronger than P1 only around the time when the bulk of the aerial hyphae were beginning to form spore chains. This pattern of transcription was consistent with the cytological evidence that the parAB locus fulfills a particular role during sporulation.
DISCUSSION
The parAB genes of S. coelicolor are located about 6 kb from the origin of chromosomal replication, downstream of a series of five genes whose shared orientation and close spacing suggest that they may be cotranscribed. However, the parAB gene pair are at least partially transcribed as a bicistronic operon from two promoters located in the large gidB-parA intergenic interval (we cannot yet rule out some readthrough from further upstream). When this work was initiated, no partitioning genes had been identified for a linear chromosome. However, during the course of this work, the complete sequence of the Borrelia burgdorferi chromosome, which is linear but has quite different ends from those of S. coelicolor, was determined, revealing that homologues of ParA and ParB together with an analogous partitioning site are present in this organism also (13). Coupling this with our evidence that in S. coelicolor the par genes do have a detectable role in DNA partitioning, we conclude that the ParA-ParB-type partitioning system is widespread in organisms with linear chromosomes as well as in those with circular chromosomes. Not all bacteria use ParAB homologues to bring about chromosome partitioning. For example, E. coli and Haemophilus influenzae do not contain such homologues. In E. coli, mukB, mukE, and mukF have been identified as genes involved in chromosome partitioning (20, 46). Homologues of each of these genes are present in H. influenzae. Interestingly, the two sequenced mycoplasma genomes contain parA but not parB homologues (12, 18). The small linear plasmid genome of pSCL1 from S. clavuligerus is similar in this respect (45). In addition to their role in partitioning, the B. subtilis par homologues appear to serve as a developmental checkpoint during endospore formation (3). This does not seem to be the case for S. coelicolor sporulation.
Not only are ParASc and ParBSc proteins highly conserved in relation to functionally identifiable protein sequence motifs of other bacterial homologues, but at least 16 identical copies of an inverted repeat DNA sequence related to the Spo0J (= ParB)-binding site of B. subtilis are present in a ca.-200-kb segment centered on oriC, three being in the parAB locus itself. Only one other copy of this sequence was found in more than 5 Mb of sequence from various regions of the genome, indicating that the sequences are probably at least 100 times more abundant near the replication origin. In B. subtilis, eight Spo0J-binding sites have been recognized in a broad region (about 20%) of the chromosome that encompasses oriC, and they are believed to play centromere-like roles in a ParB-mediated aspect of chromosome partitioning (29). It seems likely that ParB interfaces between the chromosome and cellular architecture in essentially the same way at the molecular level in S. coelicolor as in B. subtilis and C. crescentus (possibly via interaction with ParA, by analogy with the ParM protein of plasmid R1 [25]). However, whereas in the unicellular organisms the chromosomally attached ParB moves toward cell poles during cell division, in Streptomyces hyphae most replicating chromosomes are not located near a cell pole, raising an interesting question about the possible nature of the cellular architectural feature involved in (ParA and ParB-mediated?) anchoring of the oriC region.
When the search for putative ParB-binding sites was extended to permit a single-base mismatch, only five more sites were found, of which four were also in the origin region. The other was in cosmid J4, which is separated from the left chromosome end by only one cosmid (1). Sequence from the right end is not yet available. The presence of this telomere-proximal site raises the possibility that the telomeres and the replication origin could be spatially associated via ParB.
The observed pattern of partitioning aberrations in spore chains of the parAB mutants suggests that the Par proteins have a role in partitioning during sporulation septation, which appears to take place only in hyphae that have essentially stopped extension growth (11). The increase in levels of the mRNA from promoter P2 at the time of sporulation is consistent with the idea that there is a need for a greater amount of Par proteins at this stage and implies that P2 is subject to developmental regulation. Further analysis will be needed to establish that the increase in P2 expression is spatially, as well as temporally, associated with sporulation and to find out whether it is affected by mutations in the (at least) six regulatory genes known to be needed for normal sporulation septation (5). Since the two par genes are expressed at a somewhat lower level throughout growth, they may also contribute to other chromosome partitioning events. Perhaps these are associated with vegetative septation. The absence of chromosome condensation and the low frequency of vegetative septa during normal growth will make it necessary to use devices such as ParB-green fluorescent protein fusions or multiple lac operator technology, if vegetative partitioning is to be studied.
Remarkably, one of four likely ParB-binding sites in the parAB region is centered very close to the P2 transcription start point. One would expect that binding of ParB to this site would provide an autorepressing regulatory loop. The upregulation of P2 might be entirely accounted for by release from such a feedback loop, in a model that would dispense with any direct role of early sporulation regulatory gene products. In this model, a basal low level of P2 expression would be set by autorepression. However, at the end of aerial growth the multiple incipient sporulation septa would—directly or indirectly—bind ParB and therefore reduce the amount available for autorepression. After a period of derepression, the cell division sites and the ParB-binding sites in the chromosome would become saturated with ParB, restoring repression. The subcellularly localized ParB would then bind to the DNA-bound ParB, attaching the chromosomes to the developing sporulation septa. The signal for ParB sequestration by incipient sporulation septa might originate from the blockade of further rounds of DNA replication: for example, the presence of DnaA box sequences between P1 and P2 raises the possibility that the DNA in this region might form alternative DNA-protein complexes with DnaA or ParB. This region of the chromosome also contains other repeat features, and it may well be a rich playground for DNA-protein interaction.
ACKNOWLEDGMENTS
We thank Klas Flärdh for help with fluorescence microscopy and Abraham Eisenstark for his support during this project. We thank David Hopwood, Gabriella Kelemen, and Tobias Kieser for useful comments on the manuscript; Celia Bruton for drawing Fig. 1; and Tobias Kieser for helping with computer analysis.
H.-J.K. was funded by an ORS award from the Committee of Vice-Chancellors of the UK. Work in the laboratory of K.F.C. is supported by the Biotechnology and Biological Research Council via a grant to the John Innes Centre and by the John Innes Foundation.
REFERENCES
- 1.Bibb M J, Janssen G R, Ward J M. Cloning and analysis of the promoter region of the erythromycin resistance gene (ermE) of Streptomyces erythraeus. Gene. 1986;41:E357–E368. doi: 10.1016/0378-1119(85)90220-3. [DOI] [PubMed] [Google Scholar]
- 2.Calcutt M J, Schmidt F J. Conserved gene arrangement in the origin region of the Streptomyces coelicolor chromosome. J Bacteriol. 1992;174:3220–3226. doi: 10.1128/jb.174.10.3220-3226.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cervin M A, Spiegelman G B, Raether B, Ohlsen K, Perego M, Hoch J A. A negative regulator linking chromosome segregation to developmental transcription in Bacillus subtilis. Mol Microbiol. 1998;29:85–95. doi: 10.1046/j.1365-2958.1998.00905.x. [DOI] [PubMed] [Google Scholar]
- 4.Chater K F. A morphological and genetic mapping study of white colony mutants of Streptomyces coelicolor. J Gen Microbiol. 1972;72:9–28. doi: 10.1099/00221287-72-1-9. [DOI] [PubMed] [Google Scholar]
- 5.Chater K F. Taking a genetic scalpel to the Streptomyces colony. Microbiology. 1998;144:1465–1478. doi: 10.1099/00221287-144-6-1465. [DOI] [PubMed] [Google Scholar]
- 6.Chater K F, Losick R. The mycelial life-style of Streptomyces coelicolor A3(2) and its relatives. In: Shapiro J A, Dworkin M, editors. Bacteria as multicellular organisms. New York, N.Y: Oxford University Press; 1997. pp. 149–182. [Google Scholar]
- 7.Chater K F, Bruton C J, King A A, Suárez J E. The expression of Streptomyces and Escherichia coli drug resistance determinants cloned into the Streptomyces phage φC31. Gene. 1982;19:21–32. doi: 10.1016/0378-1119(82)90185-8. [DOI] [PubMed] [Google Scholar]
- 8.Chen C W. Complications and implications of linear bacterial chromosomes. Trends Genet. 1996;12:192–196. doi: 10.1016/0168-9525(96)30014-0. [DOI] [PubMed] [Google Scholar]
- 9.Cohen G, Yanko M, Mislovati M, Argaman A, Schreiber R, Av-Gay Y, Aharonowitz Y. Thioredoxin-thioredoxin reductase system of Streptomyces clavuligerus; sequences, expression, and organization of the genes. J Bacteriol. 1993;175:5159–5167. doi: 10.1128/jb.175.16.5159-5167.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Errington J, Abbleby L, Daniel R A, Goodfellow H, Partridge S R, Yudkin M D. Structure and function of the spoIIIJ gene of Bacillus subtilis: a vegetatively expressed gene that is essential for ςG activity at an intermediate stage of sporulation. J Gen Microbiol. 1992;138:2609–2618. doi: 10.1099/00221287-138-12-2609. [DOI] [PubMed] [Google Scholar]
- 11.Flärdh K, Findlay K C, Chater K F. Association of early sporulation genes with suggested developmental decision points in Streptomyces coelicolor A3(2) Microbiology. 1999;145:2229–2243. doi: 10.1099/00221287-145-9-2229. [DOI] [PubMed] [Google Scholar]
- 12.Fraser C M, Gocayne J D, White O, Adams M D, Clayton R A, et al. The minimal gene complement of Mycoplasma genitalium. Science. 1995;270:397–403. doi: 10.1126/science.270.5235.397. [DOI] [PubMed] [Google Scholar]
- 13.Fraser C M, Casjens S, Huang W M, Sutton G G, Clayton R, et al. Genomic sequence of a Lyme disease spirochaete, Borrelia burgdorferi. Nature. 1997;390:580–586. doi: 10.1038/37551. [DOI] [PubMed] [Google Scholar]
- 14.Fsihi H, De Rossi E, Salazar L, Cantoni R, Labo M, Riccardi G, Takiff H E, Eiglmeier K, Bergh S, Cole S T. Gene arrangement and organization in a ∼76 kb fragment encompassing the oriC region of the chromosome of Mycobacterium leprae. Microbiology. 1996;142:3147–3161. doi: 10.1099/13500872-142-11-3147. [DOI] [PubMed] [Google Scholar]
- 15.Gal-Mor O, Brorvok I, Av-Gay Y, Cohen G, Aharonowitz Y. Gene organization in the trxA/B-oriC region of the Streptomyces coelicolor chromosome and comparison with other eubacteria. Gene. 1998;217:83–90. doi: 10.1016/s0378-1119(98)00357-6. [DOI] [PubMed] [Google Scholar]
- 16.Gordon G G, Sitnikov D, Webb C D, Teleman A, Straight A, Losick R, Murray A W, Wright A. Chromosome and low copy plasmid segregation in E. coli: visual evidence for distinct mechanisms. Cell. 1997;90:1113–1121. doi: 10.1016/s0092-8674(00)80377-3. [DOI] [PubMed] [Google Scholar]
- 17.Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 18.Hilbert H, Himmelreich R, Plagens H, Herrmann R. Sequence analysis of 56 kb from the genome of the bacterium Mycoplasma pneumoniae comprising the dnaA region, the atp operon and a cluster of ribosomal protein genes. Nucleic Acids Res. 1996;15:628–639. doi: 10.1093/nar/24.4.628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hillemann D, Puhler A, Wohlleben W. Gene disruption and gene replacement in Streptomyces via single stranded DNA transformation of integration vectors. Nucleic Acids Res. 1991;19:727–731. doi: 10.1093/nar/19.4.727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hiraga S. Chromosome and plasmid partition in Escherichia coli. Annu Rev Biochem. 1992;61:283–306. doi: 10.1146/annurev.bi.61.070192.001435. [DOI] [PubMed] [Google Scholar]
- 21.Hopwood D A, Bibb M J, Chater K F, Kieser T, Bruton C J, Kieser H M, Lydiate D J, Smith C P, Ward J M, Schrempf H. Genetic manipulation of Streptomyces. A laboratory manual. Norwich, United Kingdom: John Innes Foundation; 1985. [Google Scholar]
- 22.Ireton K, Gunther N W, Grossman A D. spo0J is required for normal chromosome segregation as well as the initiation of sporulation in Bacillus subtilis. J Bacteriol. 1994;176:5320–5329. doi: 10.1128/jb.176.17.5320-5329.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jakimowicz D, Majka J, Messer W, Speck C, Fernandez M, Martin M C, Sanchez J, Schauwecker F, Keller U, Schrempf H, Zakrzewska-Czerwinska J. Structural elements of the Streptomyces oriC region and their interactions with the DnaA protein. Microbiology. 1998;144:1281–1290. doi: 10.1099/00221287-144-5-1281. [DOI] [PubMed] [Google Scholar]
- 24.Jensen R B, Gerdes K. Partitioning of plasmid R1. The ParM protein exhibits ATPase activity and interacts with the centromere-like ParR-parC complex. J Mol Biol. 1997;269:505–513. doi: 10.1006/jmbi.1997.1061. [DOI] [PubMed] [Google Scholar]
- 25.Jensen R B, Gerdes K. Mechanism of DNA segregation in prokaryotes: ParM partitioning protein of plasmid R1 co-localizes with its replicon during the cell cycle. EMBO J. 1999;18:4076–4084. doi: 10.1093/emboj/18.14.4076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kieser T, Hopwood D A. Genetic manipulation of Streptomyces: new integrating vectors and methods for gene replacement. Methods Enzymol. 1991;204:430–458. doi: 10.1016/0076-6879(91)04023-h. [DOI] [PubMed] [Google Scholar]
- 27.Lewis P J, Errington J. Direct evidence for active segregation of oriC regions of the Bacillus subtilis chromosome and co-localization with the Spo0J partitioning protein. Mol Microbiol. 1997;25:945–954. doi: 10.1111/j.1365-2958.1997.mmi530.x. [DOI] [PubMed] [Google Scholar]
- 28.Lin D C-H, Levin P A, Grossman A D. Bipolar localization of a chromosome partition protein in Bacillus subtilis. Proc Natl Acad Sci USA. 1997;94:4721–4726. doi: 10.1073/pnas.94.9.4721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lin D C-H, Grossman A D. Identification and characterization of a bacterial chromosome partitioning site. Cell. 1998;92:675–685. doi: 10.1016/s0092-8674(00)81135-6. [DOI] [PubMed] [Google Scholar]
- 30.MacNeil D J, Gewain K M, Ruby C L, Dezeny G, Gibbons P H, MacNeil T. Analysis of Streptomyces avermitilis genes required for avermectin biosynthesis utilising a novel integrating vector. Gene. 1992;111:61–68. doi: 10.1016/0378-1119(92)90603-m. [DOI] [PubMed] [Google Scholar]
- 31.Mohl D A, Gober J W. Cell cycle-dependent polar localization of chromosome partitioning proteins in Caulobacter crescentus. Cell. 1997;88:675–684. doi: 10.1016/s0092-8674(00)81910-8. [DOI] [PubMed] [Google Scholar]
- 32.Motallebi-Veshareh M, Rouch D A, Thomas C M. A family of ATPases involved in active partitioning of diverse bacterial plasmids. Mol Microbiol. 1990;4:1455–1463. doi: 10.1111/j.1365-2958.1990.tb02056.x. [DOI] [PubMed] [Google Scholar]
- 33.Murray M G. Use of sodium trichloroacetate and mung bean nuclease to increase sensitivity and precision during transcript mapping. Anal Biochem. 1986;158:165–170. doi: 10.1016/0003-2697(86)90605-6. [DOI] [PubMed] [Google Scholar]
- 34.Musialowski M S, Flett F, Scott G B, Hobbs G, Smith C P, Oliver S G. Functional evidence that the principal DNA replication origin of the Streptomyces coelicolor chromosome is close to the dnaA-gyrB region. J Bacteriol. 1994;176:5123–5125. doi: 10.1128/jb.176.16.5123-5125.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ogasawara N, Yoshokawa H. Genes and their organization in the replication origin of the bacterial chromosome. Mol Microbiol. 1992;6:629–634. doi: 10.1111/j.1365-2958.1992.tb01510.x. [DOI] [PubMed] [Google Scholar]
- 36.Oh S H. Development of efficient systems for integrative transformation of Streptomyces coelicolor A3(2) and their use in molecular analysis of whiB. Ph.D. thesis. Norwich, United Kingdom: University of East Anglia; 1998. [Google Scholar]
- 37.Oh S H, Chater K F. Denaturation of circular or linear DNA facilitates targeted integrative transformation of Streptomyces coelicolor A3(2): possible relevance to other organisms. J Bacteriol. 1997;179:122–127. doi: 10.1128/jb.179.1.122-127.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pospiech A, Neumann B. A versatile quick-prep of genomic DNA from Gram-positive bacteria. Trends Genet. 1995;11:217–218. doi: 10.1016/s0168-9525(00)89052-6. [DOI] [PubMed] [Google Scholar]
- 39.Qin Z, Cohen S N. Replication at the telomeres of the Streptomyces linear plasmid pSLA2. Mol Microbiol. 1998;28:893–903. doi: 10.1046/j.1365-2958.1998.00838.x. [DOI] [PubMed] [Google Scholar]
- 40.Redenbach M, Kieser H M, Denapaite D, Eichner A, Cullum J, Kinashi H, Hopwood D A. A set of ordered cosmids and a detailed genetic and physical map for the 8 Mb Streptomyces coelicolor A3(2) chromosome. Mol Microbiol. 1996;21:77–96. doi: 10.1046/j.1365-2958.1996.6191336.x. [DOI] [PubMed] [Google Scholar]
- 41.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 42.Tsui H C T, Pease A J, Koehler T M, Winkler M E. Detection and quantitation of RNA transcribed from bacterial chromosomes and plasmids. Methods Mol Genet. 1994;3:179–204. [Google Scholar]
- 43.Webb C D, Teleman A, Gordon S, Straight A, Belmont A, Lin D C-H, Grossman A D, Wright A, Losick R. Bipolar localization of the replication origin regions of chromosomes in vegetative and sporulating cells of B. subtilis. Cell. 1997;88:667–674. doi: 10.1016/s0092-8674(00)81909-1. [DOI] [PubMed] [Google Scholar]
- 44.Wheeler R T, Shapiro L. Bacterial chromosome segregation: is there a mitotic apparatus? Cell. 1997;88:577–579. doi: 10.1016/s0092-8674(00)81898-x. [DOI] [PubMed] [Google Scholar]
- 45.Wu X, Roy K L. Complete nucleotide sequence of a linear plasmid from Streptomyces clavuligerus and characterization of its RNA transcripts. J Bacteriol. 1993;175:37–52. doi: 10.1128/jb.175.1.37-52.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yamanaka K, Ogura T, Niki H, Higara S. Identification of two new genes. mukE and mukF, involved in chromosome partitioning in Escherichia coli. Mol Gen Genet. 1996;250:241–251. doi: 10.1007/BF02174381. [DOI] [PubMed] [Google Scholar]
- 47.Zakrzewska-Czerwinska J, Schrempf H. Characterization of an autonomously replicating region from the Streptomyces lividans chromosome. J Bacteriol. 1992;174:2688–2693. doi: 10.1128/jb.174.8.2688-2693.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]