The chronic activation of pDCs is a key feature in multiple autoimmune diseases. This article reports that pDCs from autoimmune patients have defects in the regulation of metabolic pathways and that blocking the TCA cycle abrogates chronic IFN-I responses.
Abstract
Plasmacytoid dendritic cells (pDCs) chronically produce type I interferon (IFN-I) in autoimmune diseases, including systemic sclerosis (SSc) and systemic lupus erythematosus (SLE). We report that the IRE1α-XBP1 branch of the unfolded protein response (UPR) inhibits IFN-α production by TLR7- or TLR9-activated pDCs. In SSc patients, UPR gene expression was reduced in pDCs, which inversely correlated with IFN-I–stimulated gene expression. CXCL4, a chemokine highly secreted in SSc patients, downregulated IRE1α-XBP1–controlled genes and promoted IFN-α production by pDCs. Mechanistically, IRE1α-XBP1 activation rewired glycolysis to serine biosynthesis by inducing phosphoglycerate dehydrogenase (PHGDH) expression. This process reduced pyruvate access to the tricarboxylic acid (TCA) cycle and blunted mitochondrial ATP generation, which are essential for pDC IFN-I responses. Notably, PHGDH expression was reduced in pDCs from patients with SSc and SLE, and pharmacological blockade of TCA cycle reactions inhibited IFN-I responses in pDCs from these patients. Hence, modulating the IRE1α-XBP1–PHGDH axis may represent a hitherto unexplored strategy for alleviating chronic pDC activation in autoimmune disorders.
Introduction
The sensing of nucleic acids, from pathogens or self, by TLR7 or TLR9 triggers secretion of extraordinary levels of type I IFN (IFN-I) by plasmacytoid dendritic cells (pDCs; Barrat and Su, 2019; Liu, 2005; Reizis, 2019; Swiecki and Colonna, 2015). IFN-α binds to its receptor IFNAR (IFN-I receptor), which is broadly expressed on cells, and induces the expression of hundreds of IFN-stimulated genes (ISGs; Barrat and Su, 2019; Liu, 2005; Reizis, 2019; Swiecki and Colonna, 2015). This response is central to the ability of pDCs to contribute to the control of viral infections (Cervantes-Barragan et al., 2012; Swiecki et al., 2010); however, it can also promote autoimmune diseases (Barrat and Su, 2019; Reizis, 2019). Elevation of IFN-I responses has been associated with autoimmunity (Crow et al., 2019), and the role of pDCs in multiple autoimmune diseases associated with the chronic presence of an IFN signature, particularly in the skin, has been well documented (Ah Kioon et al., 2018; Conrad et al., 2018; Guiducci et al., 2010; Nestle et al., 2005; Rowland et al., 2014; Sisirak et al., 2014; Wenzel and Tuting, 2008). Recently, we and others have reported that levels of CXCL4 are elevated in the blood of systemic sclerosis (SSc) patients, and that this chemokine contributes to the induction of skin fibrosis (van Bon et al., 2014; Volkmann et al., 2016; Ah Kioon et al., 2018; Lande et al., 2019; Affandi et al., 2022). Strikingly, pDCs isolated from patients with SSc exhibit an activated phenotype, as they chronically secrete IFN-α and CXCL4 and have aberrant expression of TLR8 (Ah Kioon et al., 2018; Lande et al., 2019; van Bon et al., 2014). Targeting these cells using two different approaches has shown promise in recent clinical trials for systemic lupus erythematosus (SLE; Furie et al., 2019; Karnell et al., 2021). However, the mechanism underlying the chronic activation of pDCs in autoimmune diseases is unclear.
Responding and adapting to the activation of innate immune sensors, such as TLRs, demands high levels of protein folding, modification, and secretion, which are events coordinated by the ER. During this process, accumulation of misfolded proteins in this organelle can cause “ER stress” and subsequent activation of the unfolded protein response (UPR). Three distinct sensors of protein folding stress located at the ER membrane orchestrate the UPR: IRE1α (inositol-requiring enzyme 1α), ATF6 (Activating Transcription Factor 6), and PERK (protein kinase R-like endoplasmic reticulum kinase ; Bettigole and Glimcher, 2015; Hetz et al., 2020). IRE1α regulates the most evolutionarily conserved arm of the UPR. Upon autophosphorylation and activation under ER stress, IRE1α uses its endoribonuclease domain to excise a 26-nucleotide fragment from XBP1 (X-box binding protein 1) mRNA, enabling expression of the functionally active transcription factor XBP1 that operates to induce multiple factors that restore ER proteostasis (Bettigole and Glimcher, 2015). Additionally, we and others have identified that activation of IRE1α-XBP1 can govern diverse metabolic processes that dictate immune cell function in health and disease. IRE1α-XBP1 signaling controls DC and T cell activity in the tumor microenvironment by altering lipid metabolism and mitochondrial function, respectively (Cubillos-Ruiz et al., 2015; Song et al., 2018). XBP1s has also been implicated in the survival of some DC subsets (Iwakoshi et al., 2007; Tavernier et al., 2017) and in the regulation of multiple inflammatory mediators (Chopra et al., 2019; Reverendo et al., 2019). In mouse pDCs, IFN-α promotes fatty acid oxidation and regulates ATP levels, which is required for optimal IFN-I signaling (Wu et al., 2016). A recent study reported metabolic dysregulation in skin biopsies obtained from patients with lupus (Kingsmore et al., 2021). Yet, the underlying defect or alteration in metabolic reprogramming of specific subsets of immune cells and whether IRE1α-XBP1 activation shapes the metabolism and function of pDCs in patients with autoimmunity has not been examined.
Here, we report that ER stress regulates IFN-I responses in TLR-activated pDCs. We uncover that IRE1α-XBP1 signaling governs pDC metabolic programming, and we highlight the relevance of this pathway in chronically activated pDCs from patients with autoimmune diseases.
Results
Activation of the UPR inhibits IFN-α production by human pDCs
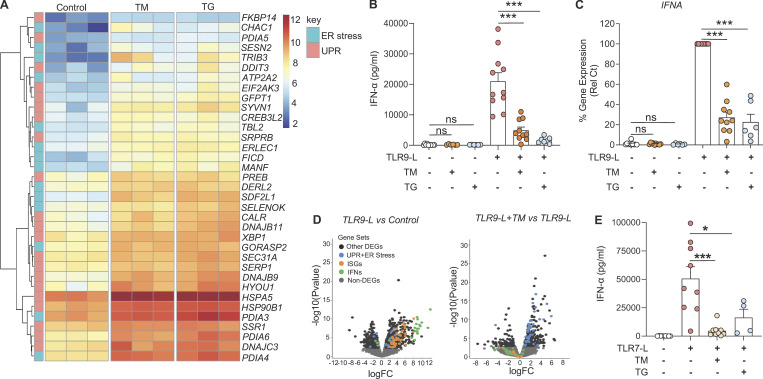
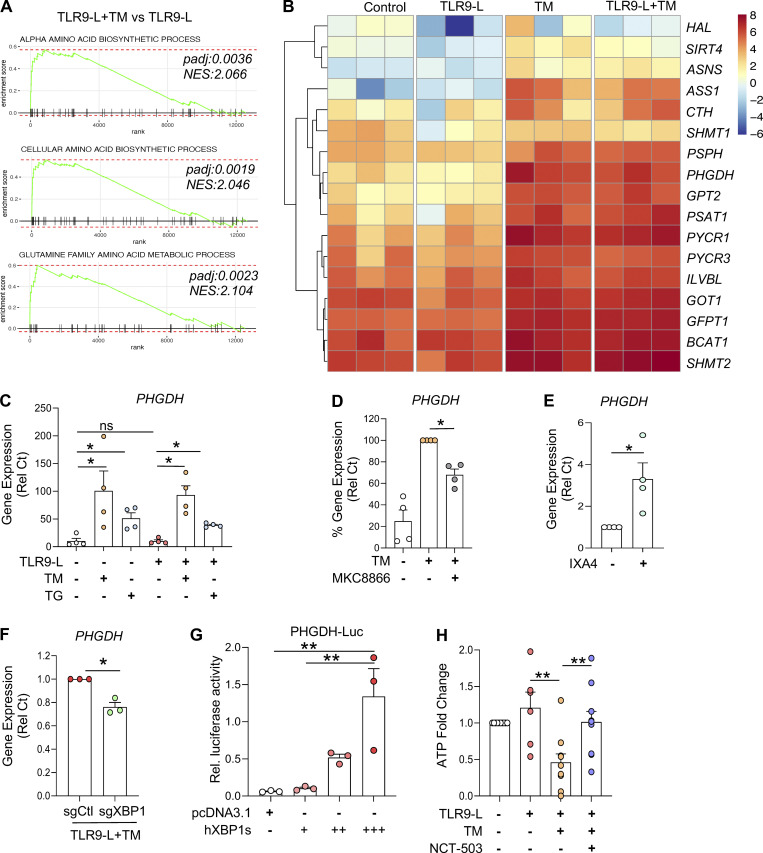
The contribution of IFN-I produced by pDCs to a series of autoimmune indications has been well established (Barrat and Su, 2019; Reizis, 2019), and the UPR has been shown to modulate TLR signaling in multiple cell types. However, little is known about whether the UPR may modulate pDC responses to nucleic acid sensing by TLR7 or TLR9. To define the effect of the UPR on pDC activity, we used two well-characterized pharmacological inducers of ER stress: tunicamycin (TM) and thapsigargin (TG; Bettigole and Glimcher, 2015; Chopra et al., 2019; Song et al., 2018). As expected, RNA sequencing (RNA-seq) analyses revealed potent induction of key genes associated with the ER stress response and the UPR in pDCs treated with these compounds (Fig. 1 A and Fig. S1, A–C). Surprisingly, these ER stressors significantly dampened the expression of IFN-α in TLR9-activated pDCs, at both the protein and mRNA levels, whereas the expression of IFN-α was not impacted when these inducers were used alone (Fig. 1, B and C). This result is in contrast with previous reports using mouse macrophages where the UPR induces STING-dependent secretion of IFN-α (Moretti et al., 2017). Furthermore, both TM and TG had negligible effects on viability or IL-6 expression in pDCs (Fig. S1, A and B). Consistently, our transcriptomic analyses revealed that administration of TM in addition to the TLR9 agonist CpG inhibited IFNA and ISGs while inducing a strong UPR (Fig. 1 D). Similarly, ER stress also inhibited IFN-α secretion from pDCs exposed to a TLR7 agonist (Fig. 1 E). Hence, ER stress inhibits IFN-I responses in pDCs sensing nucleic acids via TLR7 or TLR9.
Figure 1.
ER stress response inhibits IFN-α in pDCs. (A) Purified pDCs from HDs (n = 3) were cultured for 8 h in medium alone or medium with TM (at 3 µg/ml) or TG (at 0.5 µM), and RNA was analyzed by RNA-seq. Heatmaps of genes induced by ER stress as part of the UPR, identified by RNA-seq analysis of pDCs cultured as indicated. (B and C) Purified pDCs from HDs (n = 10–11) were first cultured with medium only or medium with TM at 3 µg/ml or TG at 0.5 µM for 3 h, before addition of the TLR9 agonist (CpG-C274 at 0.075 µM). Secreted IFN-α was quantified in conditioned medium after 13 h of culture (B) and gene expression level of IFNA was quantified at 5 h and normalized to TLR9 agonist treatment (C). (D) Volcano plot comparing gene expression analyzed by RNA-seq from pDCs of HDs (n = 3) cultured for 8 h with the TLR9 agonist CpG-C274 versus medium (left) or with CpG-C274 and TM versus CpG-C274 alone (right). Colors on all graphs indicate differentially expressed genes (DEGs) and ER stress genes (in blue); IFN genes (in green) and ISGs (in orange) are indicated. (E) pDCs (n = 9) were cultured in medium alone or with TM or TG for 3 h, before addition of a TLR7 agonist (influenza virus FLU at 0.5 pfu/cell). Secreted IFN-α was quantified in conditioned medium by ELISA after 13 h of culture. Individual donors are indicated; all results are represented as mean ± SEM; and statistical significance was evaluated using Mann–Whitney U test. ns, P > 0.05; *, P < 0.05; ***, P < 0.001.
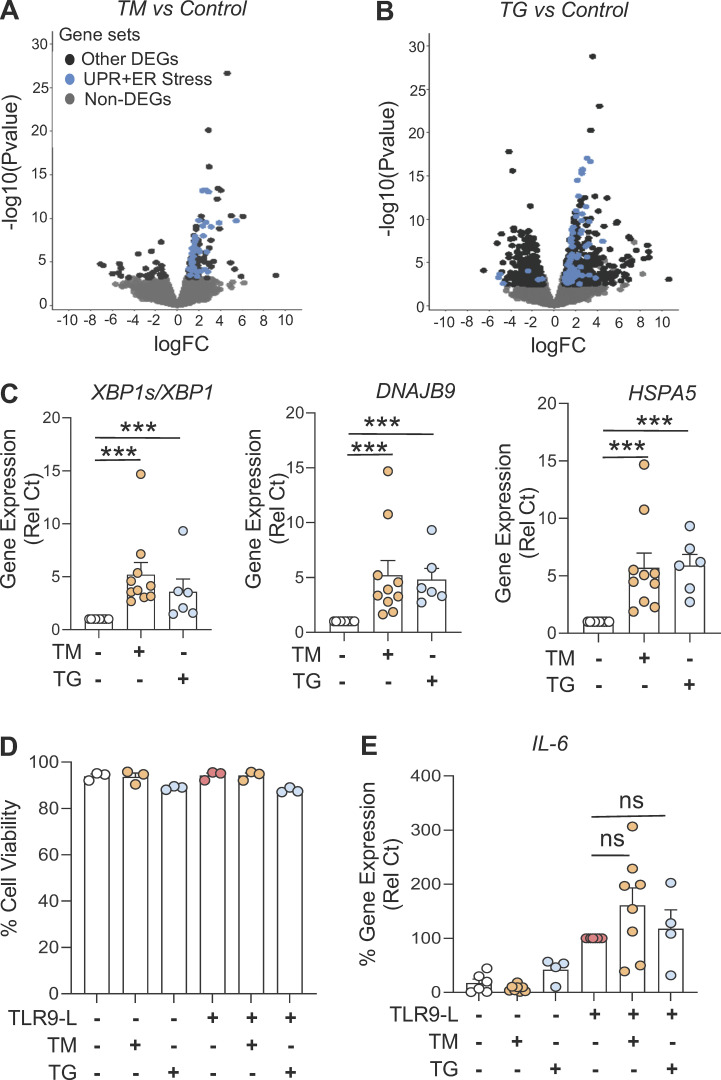
Figure S1.
Activated UPR impacts pDC response by reducing IFN-regulated genes. (A and B) Volcano plot comparing gene expression analyzed by RNA-seq of human pDCs (n = 3) from HDs cultured for 8 h in medium alone or with either TM (at 3 µg/ml; A) or TG (at 0.5 µM; B). Colors on all graphs indicate differentially expressed genes (DEGs), and UPR+ER stress genes (in blue) are indicated. logFC, log(fold-change). (C) pDCs (n = 10) were cultured in medium alone or with either TM or TG. Gene expression levels of XBP1s/XBP1, DNAJB9, and HSPA5 were quantified by qPCR after 8 h of culture and normalized to medium. (D and E) pDCs (n = 3–8) were cultured in medium alone or with either TM or TG for 3 h, before addition of TLR9 agonist. (D) Cell viability was quantified via flow cytometer. (E) Gene expression level of IL-6 was quantified at 5 h and normalized to TLR9 agonist treatment. Individual donors are indicated, all results are represented as mean ± SEM, and statistical significance was evaluated using Mann–Whitney U test. ns, P > 0.05; ***, P < 0.001.
IRE1α-XBP1 activation inhibits IFN-α production by pDCs by controlling ATP levels
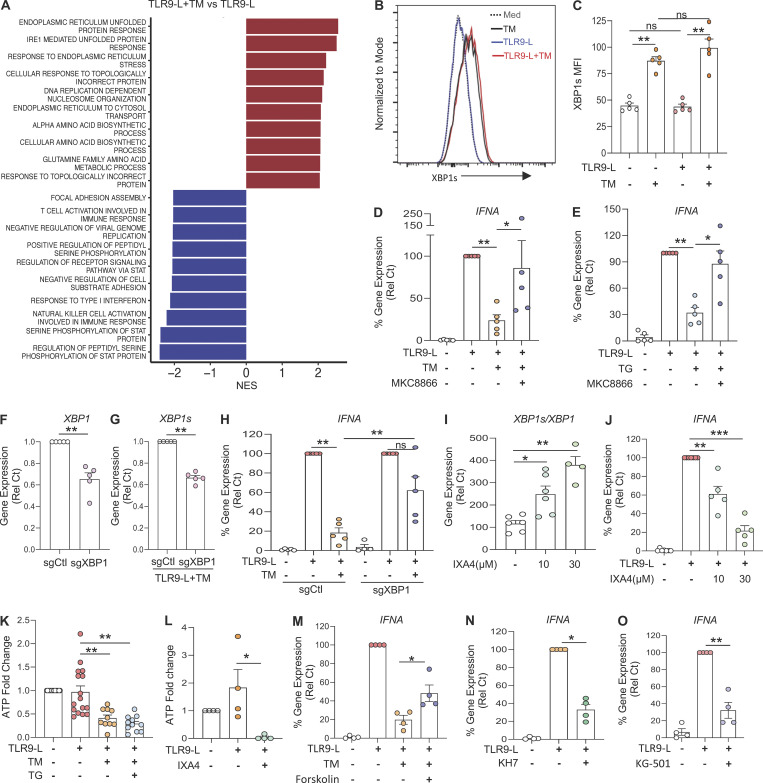
Our RNA-seq analysis revealed engagement of the IRE1α arm of the UPR in TLR9-activated pDCs under ER stress (Fig. 2 A). Hence, we assessed IRE1α-mediated XBP1s protein expression and confirmed that, in contrast to TM, TLR9 stimulation alone was not sufficient to induce robust XBP1s expression, and TLR9 signaling did not augment XBP1s levels in pDCs already facing ER stress (Fig. 2, B and C; and Fig. S2 E). Furthermore, we used two well-characterized independent inhibitors of the RNase domain of IRE1α, 4μ8c and MKC8866 (Chopra et al., 2019; Song et al., 2018), and showed that the blockade of IRE1α dampened IRE1α-dependent XBP1 splicing in TM- or TG-treated pDCs (Fig. S2, A and B). Strikingly, both inhibitors of IRE1α alleviated the ER stress–driven inhibition of IFN-α in TLR9-activated pDCs (Fig. 2, D and E; and Fig. S2, C and D). Confirming these results, reduced expression of XBP1 in pDCs via CRISPR/Cas9-mediated editing (Fig. 2, F and G) also rescued IFNA expression in TLR9-stimulated pDCs facing ER stress (Fig. 2 H). In contrast, pharmacologic inhibition of the other UPR branches, namely PERK or ATF6, had no effect on TM-induced XBP1 splicing (Fig. S2 H) or on the inhibition of TLR9-induced IFN-α (Fig. S2, F and H). To further confirm the effect of IRE1α-XBP1 on pDC responses, we used a gain-of-function approach and incubated pDCs with IXA4, a small molecule that we have recently demonstrated to selectively activate IRE1α-XBP1 signaling without inducing RIDD (regulated IRE1-dependent decay) or affecting other arms of the UPR (Grandjean et al., 2020; Madhavan et al., 2022). We observed that IXA4 enhanced XBP1 splicing (Fig. 2 I) while dose-dependently abrogating IFNA expression in TLR9-activated pDCs (Fig. 2 J), establishing that canonical IRE1α-XBP1 signaling regulates IFN-I responses in TLR-activated pDCs. It is well documented that ER stress is associated with the induction of ROS (Cao and Kaufman, 2014; Cui et al., 2022; Maamoun et al., 2019). To investigate the role of oxidative stress, we used N-acetyl-L-cysteine (NAC), an antioxidant known to inhibit ROS production (Halasi et al., 2013), and observed that the addition of NAC did rescue IFNA expression in TLR9-activated pDCs facing ER stress (Fig. S2 L), suggesting a role for ROS in the regulation of IFNA response by ER stress in pDCs.
Figure 2.
ER stress response inhibits ATP production to reduce IFN-α in pDCs via the IRE1⍺-XBP pathway. (A) All differentially expressed genes identified by RNA-seq of pDCs from HDs (n = 3) cultured for 8 h with the TLR9 agonist CpG-C274 (0.075 µM) and TM versus CpG-C274 alone, were analyzed for pathway analysis using gene set enrichment analysis. NES, normalized enrichment score. (B and C) pDCs from HDs (n = 5) were cultured in medium only or medium with TM (at 3 µg/ml) for 3 h, before addition of the TLR9 agonist. XBP1s protein levels were assessed after 3 h of culture via flow cytometry. MFI, mean fluorescence intensity. (D and E) pDCs from HDs (n = 5) were cultured in medium only, medium with TM (at 3 µg/ml), or medium with TG (at 0.5 µM) alone or in combination with IRE1α inhibitors (MKC8866 at 1 µM) for 3 h, before addition of the TLR9 agonist. Gene expression levels of IFNA were quantified at 5 h and normalized to TLR9 agonist treatment. (F and G) pDCs (n = 5) were electroporated with Cas9–sgRNA complex targeting XBP1 and cultured with IL-3 (20 ng/ml) for 72 h. TM was added to culture for 3 h, before addition of the TLR9 agonist (CpG-C274 at 0.3 µM) for 5 h. Gene expression levels of XBP1 and XBP1s were quantified. (H) pDCs (n = 5) were electroporated with Cas9–sgRNA complex targeting XBP1 and cultured with IL-3 (20 ng/ml) for 72 h. TM was added to culture for 3 h, before addition of the TLR9 agonist (CpG-C274 at 0.3 µM) for 5 h. Gene expression levels of IFNA were quantified and normalized to TLR9 agonist treatment. (I and J) pDCs (n = 6–7) were cultured in medium only or medium with the IRE1α-XBP1 agonist (IXA4 at 10 and 30 μM) for 6 h (I) or 1 h (J), before addition of the TLR9 agonist for 5 h. In I, XBP1 splicing was quantified by qPCR and shown as XBP1s/XBP1. In J, IFNA gene expression was quantified at 5 h and normalized to TLR9 agonist treatment. (K) pDCs (n = 17) were cultured with TM or TG alone for 3 h, before addition of TLR9 agonist. Intracellular ATP was quantified after 2.5 h of TLR9 activation and normalized to medium. (L) pDCs (n = 4) were cultured in medium only or medium with IRE1α-XBP1 agonist (IXA4 at 30 μM) for 1 h, before addition of TLR9 agonist for 4 h. Intracellular ATP was then quantified and normalized to medium. (M) pDCs (n = 4) were cultured in medium only or medium with TM in combination with adenylyl cyclase activator (Forskolin at 5 µM) for 3 h before addition of TLR9 agonist. Gene expression levels of IFNA were quantified at 5 h and normalized to TLR9 agonist treatment. (N and O) pDCs (n = 4) were cultured in medium only or medium with the adenylyl cyclase inhibitor (KH7 at 40 µM; N) or CREB inhibitor (KG-501 at 20 µM; O) for 1 h, before addition of TLR9 agonist. IFNA gene expression was quantified at 5 h and normalized to TLR9 agonist treatment. Individual donors are indicated, all results are represented as mean ± SEM, and statistical significance was evaluated using Mann–Whitney U test. ns, P > 0.05; *, P < 0.05; **, P < 0.01; ***, P < 0.001.
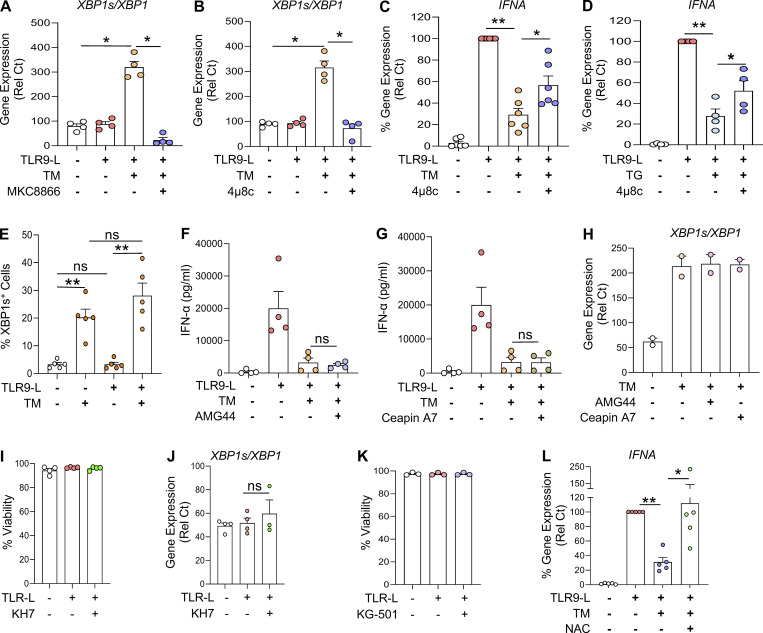
Figure S2.
ER stress inhibits TLR9-induced IFN-α in pDCs via IRE1⍺-XBP1–cAMP axis. (A–D) pDCs from HDs (n = 4–6) were cultured in medium alone or with either TM or with TG in combination with IRE1α inhibitors (MKC8866 at 1 µM or 4µ8c at 10 µM) for 3 h, before addition of TLR9 agonist. Gene expression levels of XBP1s/XBP1 (A and B) and IFNA (C and D) were quantified at 5 h by qPCR. IFNA was normalized to TLR9 agonist treatment. (E) pDCs from HDs (n = 5) were cultured in medium alone or with TM for 3 h, before addition of TLR9 agonist. XBP1 splicing positive pDCs were quantified after 3 h of culture via flow cytometer. (F–H) pDCs (n = 2–4) were cultured in medium alone or with TM, either alone or with a PERK inhibitor (AMG44 at 1 μM; F) or ATF6 inhibitor (Ceapin A7 at 5 μM; G) for 3 h, then the TLR9 agonist was added to culture. Secreted IFN-α was quantified by ELISA after 13 h of culture. (H) After 8 h, expression of the XBP1 isoforms quantified by qPCR. (I–K) pDCs (n = 3–4) were cultured in medium alone or with either adenylyl cyclase inhibitor (KH7 at 40 µM; I and J) or CREB inhibitor (KG-501 at 20 µM; K) for 1 h, before addition of TLR9 agonist. (I and K) Cell viability was quantified at 5 h. (J) Gene expression for XBP1s/XBP1 was quantified. (L) pDCs (n = 5) were cultured in medium only or with TM in combination with NAC (10 mM) for 3 h when a TLR9 agonist was added to the culture. Gene expression levels of IFNA were quantified at 5 h and normalized to TLR9 agonist treatment. Individual donors are indicated, all results are represented as mean ± SEM, and statistical significance was evaluated using Mann–Whitney U test. ns, P > 0.05; *, P < 0.05; **, P < 0.01.
It was previously reported that an increase in ATP levels induced by IFN-α can further enhance IFN-α production in mouse pDCs (Wu et al., 2016). Therefore, we evaluated whether IRE1α-XBP1 signaling controls ATP levels in TLR9-activated pDCs. Strikingly, we observed that both TM and TG significantly reduced the levels of intracellular ATP in TLR9-activated pDCs (Fig. 2 K). Corresponding with IFNA suppression (Fig. 2 J), we observed that the IRE1α-XBP1 activator IXA4 also dampened intracellular ATP levels in TLR9-activated pDCs (Fig. 2 L). It has been shown that cAMP can be synthesized from ATP by adenylyl cyclase to act as a second messenger and activate the transcription factor CREB (Acin-Perez et al., 2009; Delghandi et al., 2005; Dessauer and Gilman, 1997; Steegborn, 2014; Zhang et al., 2020). Hence, we activated the adenylyl cyclase using Forskolin (Robbins et al., 1996; Rodriguez et al., 2013; Steegborn, 2014) and observed that IFNA expression was rescued in TLR9-activated pDCs facing ER stress (Fig. 2 M). In contrast, inhibiting this adenylyl cyclase with KH7 (Hu et al., 2021; Wu et al., 2020) led to the reduced expression of IFNA in TLR9-activated pDCs (Fig. 2 N) without affecting cell viability (Fig. S2 I) or XBP1 splicing (Fig. S2 J). Furthermore, and consistent with these data, the inhibition of CREB activation with KG-501 (Best et al., 2004) led to reduced expression of IFNA in similar culture conditions (Fig. 2 O) without affecting cell viability (Fig. S2 K). These results suggest that IRE1α-XBP1 signaling inhibits IFN-I response via an ATP–cAMP–CREB axis.
ER stress–driven IRE1α-XBP1 signaling controls ATP levels in pDCs by inducing phosphoglycerate dehydrogenase (PHGDH)
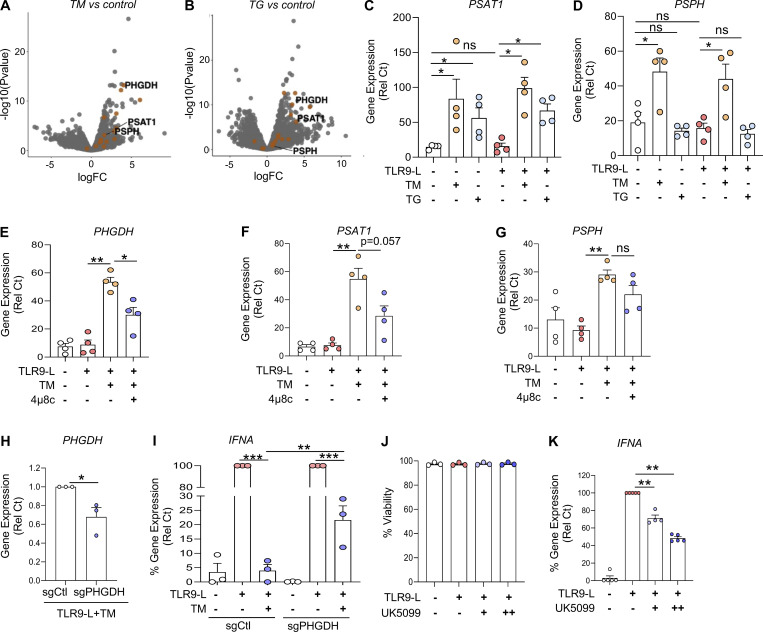
The spliced XBP1 isoform generated by IRE1α encodes the functional transcription factor XBP1, which restores ER proteostasis while also controlling diverse metabolic programs (Chen and Cubillos-Ruiz, 2021). Using an unbiased gene set enrichment analysis, we observed that the transcriptional networks implicated in amino acid biosynthesis were markedly induced in pDCs experiencing ER stress, with or without TLR9 agonist (Fig. 2 A and Fig. 3, A and B). An in-depth analysis revealed that gene programs related to serine amino acid biosynthesis were highly enriched among the amino acid biosynthetic pathways (Fig. S3, A and B). Hence, TM or TG significantly induced the expression of genes associated with serine amino acid biosynthesis pathway in pDCs, irrespective of TLR9 signaling (Fig. S3, C and D). Both TM and TG markedly induced the gene encoding PHGDH in pDCs (Fig. 3 C). This enzyme transforms 3-phosphoglycerate into phosphohydroxypyruvate, which is subsequently converted to serine via transamination and phosphate ester hydrolysis reactions driven by PSAT1 and phosphoserine phosphatase (PSPH), respectively (Locasale et al., 2011; Possemato et al., 2011; Spillier and Frédérick, 2021). Of note, ER stress response–driven induction of PHGDH, PSAT1, and PSPH was markedly inhibited upon abrogation of IRE1α-XBP1 signaling (Fig. 3 D and Fig. S3, E–G). The IRE1α-XBP1 activator IXA4 also led to the induction of PHGDH expression in pDCs (Fig. 3 E), whereas targeting XBP1 reduced PHGDH expression in TLR9-activated pDCs facing ER stress (Fig. 3 F). Importantly, we found the canonical XBP1s binding site CCACGT (Chopra et al., 2019; Glimcher, 2010; Liou et al., 1990) in the promoter sequence of PHGDH, and luciferase reporter assays in HEK293T cells demonstrated activation of the PHGDH promoter by XBP1s (Fig. 3 G). Because the ER stress response affected ATP levels in pDCs (Fig. 3 K), we assessed whether PHGDH could control ATP production using the specific inhibitor NCT-503 (Pacold et al., 2016). Blocking PHGDH activity restored ATP levels in pDCs facing ER stress (Fig. 3 H), demonstrating that ER stress reduces ATP generation in pDCs via transcriptional regulation of PHGDH.
Figure 3.
ER stress response regulates ATP production via induction of PHGDH. (A and B) Purified pDCs from HDs (n = 3) were cultured in medium alone or with TM for 3 h, before addition of TLR9 agonist for 5 h. (A) Gene enrichment score of amino acid biosynthesis. (B) Heatmaps of genes involved in amino acid biosynthesis. NES, normalized enrichment score. (C) pDCs (n = 4) were cultured in medium alone or with TM or TG for 3 h, before addition of TLR9 agonist for 5 h. Gene expression of PHGDH was quantified. (D) pDCs (n = 4) were cultured with TM alone or in combination with an IRE1α inhibitor (MKC8866 at 1 μM) for 8 h. Gene expression level of PHGDH was quantified and normalized to TM treatment. (E) pDCs (n = 4) were cultured in medium alone or with IRE1α-XBP1 agonist (IXA4 at 30 µM) for 6 h. Gene expression level of PHGDH was quantified and normalized to medium. (F) pDCs (n = 3) were electroporated with Cas9–sgRNA complex targeting XBP1 and cultured with IL-3 (20 ng/ml) for 72 h. TM was added to culture for 3 h, before addition of the TLR9 agonist (CpG-C274 at 0.3 µM) for 5 h. Gene expression levels of PHGDH were quantified. Statistical significance was evaluated using two-tailed paired t test. *, P < 0.05. (G) HEK293T cells (n = 3) were cultured for 24 h in a 96-well plate and cotransfected with an PHGDH-Luc reporter plasmid and Renilla reporter plasmid with either expression vectors pcDNA3.1 or expression vectors with spliced XBP1 proteins (18 ng [+], 54 ng [++], or 90 ng [+++]). After 48 h of transfection, dual-luciferase reporter assay was performed, and PHGDH-Luc activity was normalized to Renilla Luc activity. (H) pDCs (n = 10) were cultured with TM in combination with PHGDH inhibitor (NCT-503 at 2 μM) for 3 h, before addition of the TLR9 agonist for 2.5 h. Intracellular ATP was quantified and normalized to medium. Individual donors are indicated, all results are represented as mean ± SEM, and statistical significance was evaluated using Mann–Whitney U test. ns, P > 0.05; *, P < 0.05; **, P < 0.01.
Figure S3.
ER stress induces serine biosynthesis in pDCs via PHGDH. (A and B) Volcano plots comparing gene expression analyzed by RNA-seq of pDCs (n = 3) cultured for 8 h with either TM or TG vs. medium. (C and D) pDCs (n = 4) were cultured in medium alone or with either TM or TG for 3 h, before addition of TLR9 agonist for 5 h. Gene expression of PHGDH, PSAT1, and PSPH was quantified by qPCR. (E–G) pDCs (n = 4) were cultured in medium alone or with TM, either alone or in combination with an IRE1⍺ inhibitor (4μ8c at 10 µM) for 3 h, before addition of TLR9 agonist for 5 h. Gene expression of PHGDH, PSAT1, and PSPH was quantified by qPCR. (H and I) pDCs (n = 3) were electroporated with Cas9–sgRNA complex targeting PHGDH and cultured with IL-3 (20 ng/ml) for 72 h. TM was added to culture for 3 h, before addition of the TLR9 agonist (CpG-C274 at 0.3 µM) for 5 h. (H) Gene expression levels of PHGDH were quantified. (I) Gene expression levels of IFNA were quantified and normalized to TLR9 agonist treatment. (J and K) pDCs (n = 3–5) were cultured in medium alone or with inhibitor for pyruvate transporter (UK-5099 at 20 µM [+] or 40 µM [++]) for 1 h, before addition of the TLR9 agonist. (J) Cell viability was quantified at 5 h via flow cytometer. (K) Gene expression level IFNA was quantified and normalized to TLR9 agonist. Individual donors are indicated, all results are represented as mean ± SEM, and statistical significance was evaluated using Mann–Whitney U test, two-tailed paired t test, or one-way ANOVA with Turkey’s correction. ns, P > 0.05; *, P < 0.05; **, P < 0.01; ***, P < 0.001.
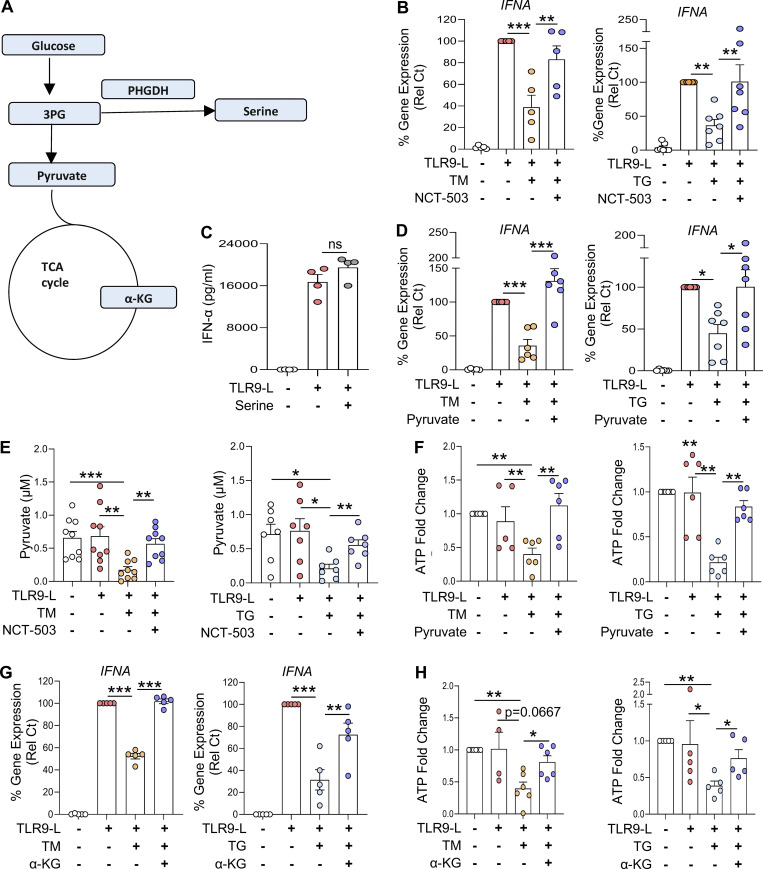
Induction of PHGDH by ER stress redirects pyruvate away from the tricarboxylic acid (TCA) cycle
A key enzyme of serine biosynthesis, PHGDH integrates serine biosynthesis pathway with glycolysis (Fig. 4 A; Locasale, 2013; Locasale et al., 2011). Because PHGDH regulated ATP levels (Fig. 3 H), we next evaluated the impact of PHGDH on pDC activation. Similar to IRE1α inhibition, abrogating PHGDH activity restored IFNA expression by TLR9-activated pDCs experiencing ER stress (Fig. 4 B). Consistently, we also observed that reduced expression of PHGDH in pDCs via CRISPR/Cas9-mediated editing (Fig. S3 H) rescued IFNA expression in TLR9-stimulated pDCs facing ER stress (Fig. S3 I). In addition to the role of PHGDH in serine biosynthesis, upregulation of PHGDH may also shunt glycolysis away from pyruvate synthesis, causing a deficiency in pyruvate levels in the cells. Therefore, we evaluated the impact of serine and pyruvate metabolism on the activity of pDCs. First, we observed that IFN-α secretion by TLR9-activated pDCs was unaltered upon exogenous supplementation with L-serine (Fig. 4 C), suggesting that elevated L-serine is not involved in ER stress–mediated IFN-α inhibition. However, exogeneous pyruvate supplementation was sufficient to restore IFNA expression by TLR9-activated pDCs under ER stress (Fig. 4 D).
Figure 4.
ER stress response rewires pyruvate to control IFN-I response by pDCs. (A) Graphical representation of the role of PHGDH in glucose metabolism. (B) pDCs (n = 5–7) were cultured in medium alone or with TM or TG in combination with PHGDH inhibitor (NCT-503 at 2 µM) for 3 h, before addition of TLR9 agonist. Gene expression level of IFNA was quantified at 5 h and normalized to TLR9 agonist treatment. (C) pDCs (n = 4) were cultured in medium alone or with L-serine (1 mg/ml) for 1 h, before addition of TLR9 agonist. Secreted IFN-α was quantified by ELISA after 13 h of culture. (D) pDCs (n = 6–7) were cultured with TM (left) or TG (right) in combination with PHGDH inhibitor (NCT-503 at 2 μM) for 3 h, when TLR9 agonist was added to the culture for 2.5 h. Intracellular pyruvate was quantified. (E and F) pDCs (n = 6–9) were cultured with TM or TG alone or with sodium pyruvate (pyruvate at 10 mM) for 3 h, before addition of TLR9 agonist. (E) Intracellular ATP was quantified using ATP assay kit after 2.5 h of culture and normalized to medium. (F) Gene expression level of IFNA was quantified at 5 h and normalized to TLR9 agonist treatment. (G and H) pDCs (n = 5–6) were cultured with TM or TG alone or with α-KG at 10 mM for 3 h, before addition of TLR9 agonist. (G) Intracellular ATP was quantified after 2.5 h of culture and normalized to medium. (H) Gene expression level of IFNA was quantified at 5 h and normalized to TLR9 agonist treatment. Individual donors are indicated, all results are represented as mean ± SEM, and statistical significance was evaluated using Mann–Whitney U test. ns, P > 0.05; *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Supporting these observations, intracellular pyruvate levels were significantly reduced in ER-stressed pDCs, which could be restored by blocking PHGDH activity (Fig. 4 E). Of note, pyruvate is generated by pyruvate kinase, which can also be activated by serine (Chaneton et al., 2012); however, our data suggest a disconnect of such phenomena in pDCs, as the increased expression of PHGDH in pDCs by ER stress (Fig. 4 E) did not result in an increase but rather a decrease in pyruvate levels. These data directly link the increased expression of PHGDH to reduced levels of pyruvate observed in ER-stressed pDCs. Notably, reduced intracellular ATP levels by ER stress were also restored in pDCs upon pyruvate supplementation (Fig. 4 F). It is well known that pyruvate enters the mitochondria to fuel the TCA cycle, where it is converted to acetyl-CoA and other TCA cycle substrates such as α-ketoglutarate (α-KG), to ultimately produce ATP by the electron transport chain (Martinez-Reyes and Chandel, 2020; Pearce and Everts, 2015). This process has been demonstrated to mediate optimal immune cell activation (Everts et al., 2014). α-KG in the TCA cycle can also be synthesized from glutaminolysis (Yang et al., 2017), but we did not observe any change in the expression of the genes involved in glutaminolysis in our RNA-seq analysis. Blocking the mobilization of pyruvate into the mitochondria inhibited the expression of IFNA (Fig. S3 K) without impacting cell viability (Fig. S3 J). However, we observed that treatment with a cell-permeable analog of α-KG (Kee et al., 2013) rescued IFNA expression by TLR9-activated pDCs undergoing ER stress (Fig. 4 G). Consistent with these findings, exogenous supplementation with this analog normalized intracellular ATP levels in pDCs upon ER stress (Fig. 4 H). These data uncover that pyruvate and α-KG are key intermediate metabolites in the TCA cycle required for optimal IFN-α responses in TLR9-activated pDCs, and that this process is curtailed by ER stress–driven activation of the IRE1α-XBP1–PHGDH axis.
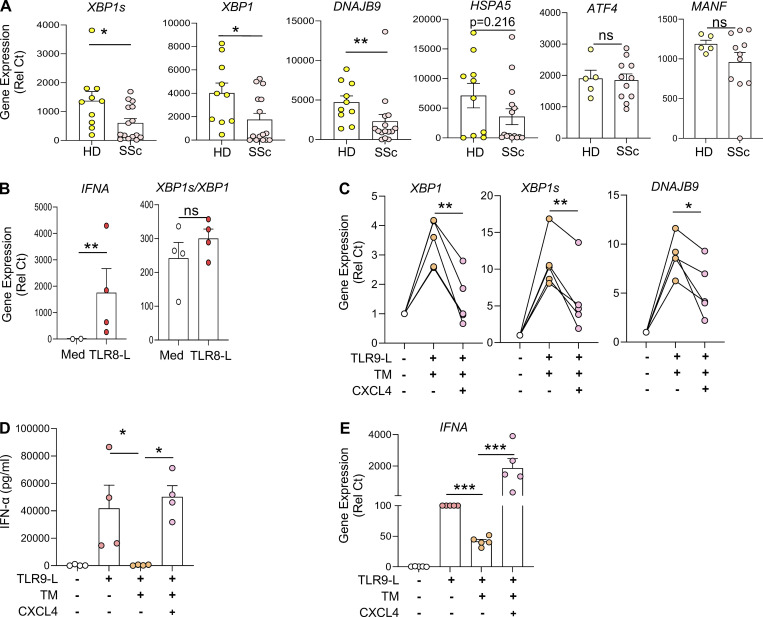
Expression of UPR-related genes is downregulated in pDCs of patients with SSc and is impacted by CXCL4
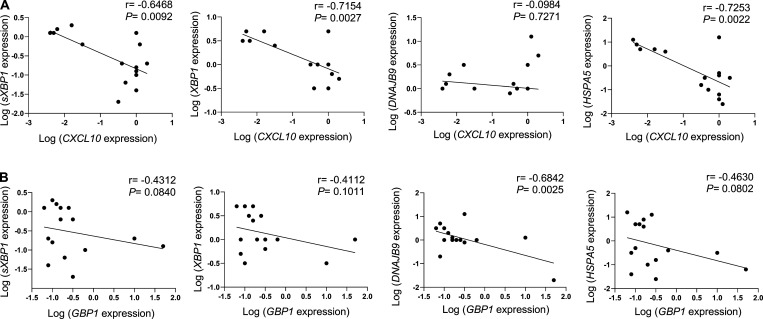
pDCs are key players in the induction of skin fibrosis, since their depletion prevents disease in a mouse model (Ah Kioon et al., 2018). Yet the mechanisms underlying the chronic activation of pDCs in autoimmune disorders remain elusive. We found that pDCs from SSc patients had decreased expression of several UPR marker genes, including XBP1, its spliced isoform XBP1s, and DNAJB9, the latter two being regulated by the IRE1α pathway. In contrast, the PERK-eIF2α and ATF6 branches of the UPR appear unaltered, as evidenced by the expression status of their canonical genes ATF4 and MANF, respectively (Adamson et al., 2016), which remained unaffected (Fig. 5 A). Notably, we observed an inverse correlation between the expression of these UPR genes with ISGs such as CXCL10 (Fig. S4 A) and GBP1 (Fig. S4 B) in pDCs from patients with SSc. We previously showed that pDCs from patients with SSc exhibit an aberrant expression of TLR8, and activation of this ligand resulted in secretion of CXCL4 and IFN-α (Ah Kioon et al., 2018). We examined the effect of TLR8 activation and observed an induction of IFNA expression in pDCs from SSc patients, but we did not detect differences in XBP1s expression (Fig. 5 B), suggesting that the downregulation of the UPR in cells of these patients is not related to the aberrant TLR8 expression in their pDCs. However, CXCL4, which has been shown to be elevated in patients with SSc (van Bon et al., 2014; Volkmann et al., 2016), had a significant impact on the UPR, suppressing expression of UPR genes such as XBP1, XBP1s, and DNAJB9 in activated pDCs facing ER stress (Fig. 5 C). Notably, we recently showed that CXCL4, as well as other chemokines, can superinduce IFN-I from TLR9-activated pDCs (Du et al., 2022), and we observed that CXCL4 rescued the expression and production of IFN-α that was inhibited upon ER stress induction (Fig. 5, D and E). These data indicate that the UPR, particularly the IRE1α arm of the UPR, is downregulated in pDCs of patients with SSc and identify CXCL4 as a potential mediator of the reduced IRE1α-XBP1 activation observed in chronically activated pDCs from patients with SSc.
Figure 5.
CXCL4-mediated dysregulation of ER stress response in pDCs isolated from SSc patients promotes IFN-I production. (A) pDCs were isolated from the blood of HDs (n = 10) and patients with SSc (n = 15). pDCs were lysed, and RNA was collected for gene expression of XBP1s, XBP1, DNAJB9, HSPA5, ATF4, and MANF. (B) pDCs (n = 4) were isolated from the blood of patients with SSc and cultured in medium alone or with the TLR8 agonist (ORN8L at 130 μg/ml). After 6 h of culture, RNA was collected and quantified for gene expression of IFNA and XBP1 splicing (measuring XBP1s/XBP1). (C–E) pDCs from HDs (n = 4–5) were cultured in medium alone or with TM for 3 h, before addition of TLR9 agonist alone or with CXCL4 (3 μg/ml). (C) Gene expression level of XBP1, XBP1s, and DNAJB9 quantified after 5 h of culture. (D) Secreted IFN-α was quantified in conditioned medium by ELISA after 13 h of culture. (E) Gene expression level of IFNA after 5 h of culture and normalized to TLR9 treatment. Individual donors are indicated, and all results are represented as mean ± SEM. Statistical significance was evaluated using two-tailed paired t test or Mann–Whitney U test. ns, P > 0.05; *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Figure S4.
ER stress gene levels negatively correlate with ISG expression in pDCs from SSc patients. (A and B) pDCs were isolated from freshly isolated blood of patients with SSc (n = 15). pDCs were lysed, and RNA was collected for gene expression of CXCL10, GBP1, XBP1, XBP1s, DNAJB9, and HSPA5. Expression of ER stress genes such as XBP1, XBP1s, DNAJB9, and HSPA5 linked with the expression of CXCL10 (A) and GBP1 (B). Pairwise correlation analysis was calculated using Spearman’s correlation coefficient (r) with P values (two-tailed), 95% confidence intervals for all correlation analysis.
Reduced PHGDH expression in pDCs of SSc and SLE patients promotes their chronic IFN-I response through TCA cycle activation
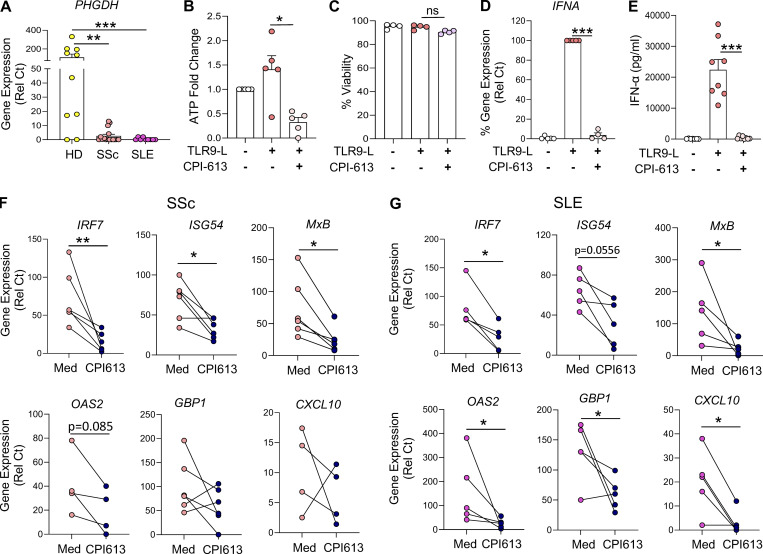
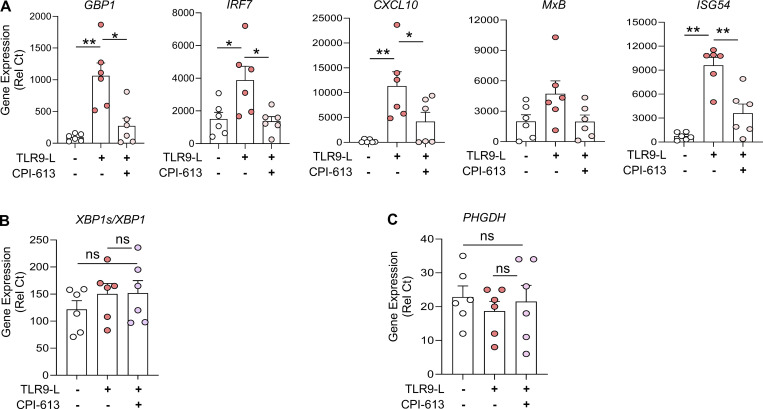
Several studies have reported that pDCs from patients with autoimmune diseases such as SSc or SLE are chronically activated, hence constantly secreting IFN-α and expressing ISGs (Barrat and Su, 2019; Reizis, 2019). In addition, increased levels of acetate in the serum of SLE patients and succinic acid and fumaric acid in the serum of SSc patients, all of which are end products of TCA cycles, have been reported (Bengtsson et al., 2016; Guleria et al., 2016). Intriguingly, we observed a drastic reduction in the basal expression of PHGDH in pDCs from both SSc and SLE patients (Fig. 6 A). These data suggest a dysregulation of the metabolic landscape in pDCs of patients with SSc or SLE. To determine whether it is possible to attenuate the chronic activation of pDCs in autoimmune patients, and as our data revealed, the significance of pyruvate and α-KG for optimal IFN-α responses in TLR9-activated pDCs (Fig. 4), we used a small-molecule inhibitor of both pyruvate dehydrogenase (PDH) and α-KG dehydrogenase (KGDH), named CPI-613 (Stuart et al., 2014; Yang et al., 2018; Zachar et al., 2011), to test whether disrupting the TCA cycle could impact the chronic IFN-I responses present in patients’ pDCs. The specificity of this drug has been well characterized, and it is currently being tested in clinical trials for pancreatic cancer (Philip et al., 2019; Stuart et al., 2014). We observed that CPI-613 also reduced intracellular ATP levels in TLR9-activated pDCs (Fig. 6 B). Further, CPI-613 inhibited the secretion and expression of IFN-α (Fig. 6, D and E) and the expression of ISGs such as GBP1, IRF7, CXCL10, MxB, and ISG54 (Fig. S5 A) in pDCs from healthy donors (HDs), while the drug had no detrimental effect on cell viability (Fig. 5 C), XBP1 splicing (Fig. S5 B), or the expression of PHGDH (Fig. S5 C). These findings demonstrate that the activated TCA cycle in pDCs is integral for IFN-I responses induced by TLR9.
Figure 6.
PHGDH reduction in pDCs of patients with SSc and SLE is associated with chronic IFN-I signatures. (A) pDCs were isolated from the blood of either HDs (n = 10) or patients with SSc (n = 15) or SLE (n = 9). pDCs were lysed, and RNA was collected for gene expression of PHGDH. (B–E) pDCs (n = 4–8) were cultured in medium alone or with the inhibitor for both PDH and α-KGDH (CPI-613 at 200 µM) for 1 h, before the addition of TLR9 agonist. (B) Intracellular ATP was quantified after 4 h of culture and normalized to medium. (C) Cell viability was quantified at 5 h via flow cytometer. (D) Gene expression level of IFNA was quantified at 5 h and normalized to TLR9 agonist treatment. (E) Secreted IFN-α was quantified by ELISA after 12 h of culture. (F and G) pDCs (n = 5–6) were isolated from patients with SSc (F) or SLE (G) and cultured in medium with CPI-613 at 200 μM for 12 h. Gene expression levels of ISGs such as GBP1, IRF7, ISG54, MxB, OAS2, and CXCL10 were quantified. Individual donors are indicated, all results are represented as mean ± SEM, and statistical significance was evaluated using Mann–Whitney U test. ns, P > 0.05; *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Figure S5.
The TCA inhibitor CPI-613 has little to no effect on viability or ER stress gene levels in human pDCs. (A–C) pDCs (n = 6) were cultured in medium alone or with an inhibitor for both PDH and α-KGDH (CPI-613 at 200 µM for 1 h), before addition of the TLR9 agonist for 5 h. (A) Gene expression level of ISGs such as GBP1, IRF7, ISG54, MxB, OAS2, and CXCL10 were quantified at 5 h. (B and C) Gene expression levels of XBP1s/XBP1 and PHGDH were quantified by qPCR. Individual donors are indicated, all results are represented as mean ± SEM, and statistical significance was evaluated using Mann–Whitney U test. ns, P > 0.05; *, P < 0.05; **, P < 0.01.
There are no good in vivo models to study the chronic activation of pDCs in the mouse. However, we successfully purified pDCs from patients with SSc or SLE and tested the effect of blocking TCA with CPI-613. As previously reported by us and others, pDCs from these patients had high basal expression of a set of IFN-inducible genes (Fig. 6, F and G). Remarkably, we observed that culturing the pDCs with the PDH and KGDH dual inhibitor CPI-613 significantly reduced the expression of ISGs in the pDCs from patients with SSc (Fig. 6 F) and SLE (Fig. 6 G). These data suggest that the chronic activation status of pDCs is associated with a dysregulation in both the ER stress and metabolic responses and that drugs, such as CPI-613 or others, by disrupting the TCA cycle in these cells, can thwart IFN-I responses that are chronically present in pDCs of patients with SSc and SLE.
Discussion
The contribution by pDCs to the chronic presence of the IFN-I signature observed in patients with multiple autoimmune diseases has been well documented, in particular in conditions associated with interface dermatitis, where targeting pDCs has shown clinical benefit (Barrat and Su, 2019; Crow et al., 2019; Furie et al., 2019; Karnell et al., 2021; Reizis, 2019; Wenzel and Tuting, 2008). However, the mechanism underlying the chronic activation of pDCs in these diseases is not well defined. Here, we present experimental evidence indicating that the rewiring of pyruvate mobilization mediated by the IRE1α-XBP1–PHGDH axis regulates IFN-I responses by TLR-activated pDCs. We show that IRE1α-XBP1 signaling controls pDC activation by impacting ATP production and cAMP signaling via induction of PHGDH. We identified that CXCL4 dampens the expression of genes regulated by IRE1α-XBP1 signaling and promotes IFN-α production in pDCs. Finally, we determined that PHGDH expression is dramatically reduced in pDCs isolated from patients with SLE or SSc, suggesting that a dysregulation of metabolic pathways is responsible for the chronic activation of pDCs in these patients. Hence, the inhibition of the TCA cycle using the drug CPI-613 can revert the chronic activation of pDCs in both patient populations, which suggests that the disruption of the TCA cycle may be a new method to reduce the hyperactivation of pDCs in autoimmune patients.
ER stress can alter a variety of cellular functions, and multiple drugs that modulate the UPR are currently in clinical development (Hetz et al., 2019). Previous studies have shown that ER stress can modulate the activation of immune cells including T cells, macrophages, DCs, and natural killer cells (Chopra et al., 2019; Cubillos-Ruiz et al., 2015; Dong et al., 2019). The link between IFN-I regulation and ER stress has been previously shown in murine macrophages; however, the ER stress response led to a STING-dependent increased IFN-I response mediated by the PERK arm of the UPR (Moretti et al., 2017). Using murine microglia with impaired proteasome activity, it was also shown that ER stress can mediate the production of IFN-β (Studencka-Turski et al., 2019). In sharp contrast, we demonstrate herein that IRE1α-XBP1 activation inhibits IFN-I production in TLR-stimulated pDCs facing ER stress. The difference in response by pDCs to ER stress response, compared with murine myeloid cell populations, may be due to the inherent properties of pDCs, which are professional IFN-I producers. Another potential factor is the dependence on TLR7 and TLR9 signaling for the induction of IFN-I by pDCs compared with the cGAS/STING pathway (Barrat et al., 2016), which involves different adaptors and transcription factors (Barrat and Su, 2019; Reizis, 2019).
Although ER stress has been studied in the context of autoimmune diseases (Burman et al., 2018; Kropski and Blackwell, 2018; Tanjore et al., 2013), the effect of this cellular state on specific immune cells that contribute to the pathogenesis of these indications remains poorly understood. Here, we report that IRE1α-XBP1–regulated genes are downregulated in pDCs isolated from SSc patients, which leads to chronic production of IFN-I and activation of ISGs, such as CXCL10 and GBP1, all of which are part of the so-called IFN signature present in patients with SLE, SSc, and other autoimmune diseases (Barrat and Su, 2019; Reizis, 2019). Our analyses also revealed that CXCL10 and GBP1 are inversely correlated with the expression status of UPR genes in pDCs from SSc patients. In addition, we identified that CXCL4, which is produced at high levels in the blood of SSc patients, dampens IRE1α-XBP1 signaling and enhances IFN-α production in pDCs from these patients. This uncovers a novel immunopathogenic mechanism in SSc whereby reduced IRE1α-XBP1 signaling in pDCs promotes chronic production of IFN-I and subsequent ISG activation.
Recent studies have shown that reduction in mitochondrial ATP induces ER stress via Ca2+ accumulation in the cells (Costa et al., 2019; Tanimura et al., 2018), whereas we observed that activation of the UPR reduces intracellular ATP in pDCs. Previous studies have demonstrated that IFN-α induces fatty acid oxidation in murine pDCs, which increases intracellular ATP and further IFN-I signaling (Wu et al., 2016). This directly fits with our hypothesis that pDCs facing ER stress have reduced levels of intercellular ATP and IFN-α. Intracellular ATP can be used by cells for enzymatic reaction or to synthesize intracellular cAMP by adenylyl cyclase that acts as second messenger for cell signaling and activates the transcriptional factor CREB (Acin-Perez et al., 2009; Delghandi et al., 2005; Steegborn, 2014; Zhang et al., 2020). We did not observe changes in the expression of adenylyl cyclase in the pDCs facing ER stress but found that cAMP is required for IFN-I expression during TLR9 activation in the presence or absence of ER stress. We also identified that CREB activation is required for TLR9 signaling to induce IFN-I production. Thus, our study reveals a key mechanism by which ATP regulates IFN-I in pDCs. We have previously reported that persistent IRE1α-XBP1 activation induces lipid accumulation in intratumoral DCs, a process that ultimately blunts adaptive anticancer immunity (Cubillos-Ruiz et al., 2015). Interestingly, this arm of the UPR did not control transcriptional profiles involved in lipid metabolism in human pDCs. Instead, we observed that IRE1α-XBP1 signaling in pDCs led to marked upregulation of genes predominantly involved in serine biosynthesis. Consistently, activation of this UPR branch in pDCs transcriptionally induced key enzymes in the serine biosynthetic pathway, such as PHGDH, PSAT1, and PSPH. Although previous studies have shown TLR4-mediated attenuation of PHGDH expression in macrophages (Wilson et al., 2020), TLR9 signaling did not significantly impact the expression of PHGDH. This is suggestive of immune cell–specific effects in response to varying TLR signaling pathways. Interestingly, we also noted decreased cellular pyruvate levels in pDCs upon activation of the IRE1α-XBP1 pathway, which could be restored upon blockade of PHGDH activity. Serine is the product of serine biosynthesis and is known to be synthesized into glycine (Locasale, 2013; Locasale et al., 2011). In addition, serine was also shown using a cell-free system to activate pyruvate kinase (Chaneton et al., 2012); however, this pathway does not seem to be significantly at play in pDCs, as pyruvate levels are decreased in pDCs facing ER stress. Pyruvate produced from glycolysis enters the mitochondria to fuel the TCA cycle, where it is converted to acetyl-CoA and other intermediates, such as α-KG (Martinez-Reyes and Chandel, 2020; Pearce and Everts, 2015). We observed that the exogenous supplementation of pyruvate and α-KG restored IFN-I production in pDCs under ER stress conditions by reestablishing cellular ATP levels. Of note, we previously showed that α-KG supplementation could restore mitochondrial respiration and the effector function of T cells under ER stress (Song et al., 2018). Thus, our study uncovers the novel significance of the TCA cycle and its substrates in the functioning of pDCs. This presents new avenues of intervention in the TCA cycle to regulate pDC functioning in autoimmune disorders.
The expression of PHGDH was drastically reduced in pDCs purified from both SSc and SLE patients, which may explain the high levels of products of the TCA cycle, such as acetate, succinic acid, or fumaric acid, observed in the sera of autoimmune patients (Bengtsson et al., 2016; Guleria et al., 2016). However, in contrast to the situation in SSc, we did not observe any significant changes in the expression of ER stress–regulated genes in pDCs of patients with SLE, suggesting that additional signals received by the SSc pDCs, possibly such as CXCL4, may differentially impact these cells in patients with SSc. However, regardless of the mechanism at play, these data directly link the dysregulation of the immune metabolism of pDCs with the exacerbated activation of pDCs in autoimmune patients, and further studies will be needed to comprehensively explore the regulation of PHGDH in pDCs in these and other autoimmune diseases. Our findings that both pyruvate and α-KG are important for optimal induction of IFN-I in pDCs suggest that modulating the IRE1α-XBP1–PHGDH axis or impacting the TCA cycle to prevent the chronic activation of pDCs in patients may provide a new translational approach to intervene for the benefit of these patient populations.
Materials and methods
Patients
Participants were recruited, and blood samples were obtained after informed consent of donors under protocols approved by the Institutional Review Board of the Hospital for Special Surgery and the Institutional Biosafety Committee of Weill Cornell Medicine (protocol #2014-221 and #2014-276). All participants provided written informed consent before enrollment. All patients fulfilled the 2013 American College of Rheumatology/European League Against Rheumatism Classification Criteria for SSc (van den Hoogen et al., 2014). Patients were categorized as having limited SSc or diffuse subtype (early diffuse or late diffuse) SSc according to LeRoy and Medsger (2001). Disease duration was defined as the time from the first SSc-related symptom apart from Raynaud phenomenon and was classified as early if disease duration was ≤2 yr. All SLE patients fulfilled the 1997 American College of Rheumatology criteria (Hochberg, 1997) and their complementary criteria, the 2012 Systemic Lupus International Collaborating Clinics criteria (Petri et al., 2012) for SLE patients. The clinical and demographic characteristics of the patients with SSc and SLE are described in Table S1 and Table S2.
Purification and culture of pDCs from HDs and patients
Enriched leukocytes were obtained from the New York Blood Center (Long Island City, NY) after informed consent of donors who were deemed healthy by the New York Blood Center’s criteria and used under a protocol approved by the Institutional Review Board of the Hospital for Special Surgery and the Institutional Biosafety Committee of Weill Cornell Medicine (protocol #2014-221). Peripheral blood mononuclear cells were prepared using Ficoll-Paque density gradient, and pDCs were isolated using BDCA4+ positive selection (130-097-415; Miltenyi Biotech) as previously described (Guiducci et al., 2006). pDCs were cultured at 40,000 cells (for HDs) or 10,000–20,000 cells (for patients with SSc and SLE) per well in a 96-well round-bottom plate and incubated at 37°C, 5% CO2, and 95% humidity. For TLR7 and TLR9 activation assays, pDCs were stimulated with 2 multiplicity of infection heat-inactivated H1N1 VR-95 influenza A virus (ATCC) and 0.075 μM of CpG C274 (Guiducci et al., 2006), respectively.
In some culture conditions, cells were cultured with the TM (654380; Thermo Fisher Scientific), TG (T9033; Sigma-Aldrich), 4μ8c (412512; EMD Millipore), MKC8866 (HY-104040; Medchem Express), IXA4 (131171.1; Chembridge), AMG PERK44 (5517; R&D), Ceapin-A7 (SML2330; Sigma-Aldrich), NCT-503(2623; Axon Medchem), L-serine (S4500; EMD Millipore), sodium pyruvate (8636; Sigma-Aldrich), α-ketoglutaric acid disodium salt hydrate (K3752; Sigma-Aldrich), CPI-613 (S2776; Selleckchem), UK5099 (HY-15475; Medchemexpress), NAC (A7250; Sigma-Aldrich),KH7 (3834; Torcis), Forskolin (HY-15371; Medchemexpress), NAC (A7250-5G; Sigma-Aldrich), or KG-501 (HY-103299; Medchemexpress).
Flow cytometry
For XBP1s staining, pDCs were cultured for 6 h, washed in PBS, and fixed with fixation and permeabilization kit (00-5123-43; eBioscience) for 30 min at room temperature. Fixed pDCs were permeabilized with 0.05% digitonin in FACS buffer for 20 min at room temperature and stained with sXBP1 antibody (562821; 1:70; BD Pharmingen) for 1.5 h at room temperature. Cells were washed with FACS buffer and acquired by FACS. For cell viability, pDCs were cultured for 6–8 h, washed in PBS, resuspended in FACS buffer, and stained with DAPI or propidium iodide (V13243B; Invitrogen). Cells were acquired by FACS, and analysis was performed using FlowJo analysis software. The gating strategy for viable cells involved progressively measuring total cells without uptake of DAPI.
RNA extraction and RT-PCR
After 6–13 h of cell culture, pDCs were lysed for total RNA extraction using the Qiagen RNeasy Plus Mini Kit. Quantity of RNA was measured by Nanodrop, and high-capacity cDNA Reverse Transcription kit (Thermo Fisher Scientific) was used to generate cDNA. Quantitative PCR (qPCR) reactions were performed. Gene expression levels were calculated based on relative threshold cycle (Rel Ct) values as described (Barrat et al., 2005). This was done using the formula relative Ct = 100 or 1,000 × 1.8 (HSK−GENE), where HSK is the mean Ct of duplicate housekeeping gene runs (Ubiquitin), GENE is the mean Ct of duplicate runs of the gene of interest, and 100/1,000 is arbitrarily chosen as a factor to bring all values >0. Primers are given in Table S3.
RNA-seq analysis
Total RNA was extracted from cells using the Qiagen RNeasy Plus Mini Kit. All samples were examined for RNA quality by Agilent Bioanalyzer 2100. Illumina libraries were constructed using NEB low input library preparation kit. Multiplexed libraries were generated and pooled at equimolar concentration, and pair-end reads were sequenced on an Illumina HiSeq 4000 in the Weill Cornell Epigenomics Core Facility at a depth of 21 million to 37 million fragments per sample. Sequencing quality was measured with fastp (Chen et al., 2018). Reads mapped in genes were counted against the human genome (hg38) with STAR aligner and Gencode v21. Differential gene expression analysis was performed in R (R Core Team, 2020) using the edgeR package (Robinson et al., 2010; Gu et al., 2016). Genes with low expression levels (<3 cpm) were filtered from all downstream analyses. The Benjamini–Hochberg false discovery rate (FDR) procedure was used to calculate FDR. Genes with FDR <0.05 and log2(fold-change) >1 were considered significant. Volcano plots and heatmaps were generated by complex heatmap packages. Pathways analysis for differential regulated genes were performed in R using the fgsea package, and normalized gene enrichment scores were used for plotting.
Chemokine and cytokine measurement
Secreted cytokines such as IFN-α (3425-1H-20; Mabtech) and IL-6 (3460-1H-20; Mabtech) were quantified in the supernatant of pDC cultures using ELISA according to the manufacturer’s protocol.
Metabolism assay
Intracellular pyruvate levels were determined in pDCs by pyruvate detection kit (700470; Cayman Chemicals) per the manufacturer’s protocol. pDCs cultured in RPMI were washed in 1 ml of PBS and centrifuged at 10,000 g for 5 min at 4οC. Supernatants were removed, and cells were deproteinated in 0.5 ml of 0.5 M metaphosphoric acid on ice for 5 min, followed by centrifugation at 10,000 g for 5 min at 4οC. The deproteinated samples were neutralized with 25 μl of potassium carbonate and then centrifuged at 10,000 g for 5 min at 4οC. The supernatant was removed, and deproteinated samples were used for the pyruvate assay. For ATP determination, an ATP determination kit (A22066; Sigma-Aldrich) was used in pDC extracts as per the manufacturer’s protocol.
Gene editing in human pDCs
Human pDCs isolated from peripheral blood mononuclear cells were electroporated by adding 150 nM sgRNA (single guide RNA)-CAS9 ribonucleoprotein complexes to 2 × 105 cells in suspension using the Neon transfection system (MPK5000; Thermo Fisher Scientific). All materials for sgRNA-Cas9 complex generation were purchased from Integrated DNA Technologies and prepared as instructed (https://sfvideo.blob.core.windows.net/sitefinity/docs/default-source/user-guide-manual/alt-r-crispr-cas9-user-guide-ribonucleoprotein-transfections-recommended.pdf?promo_name=Reference%20Protocol&promo_id=1d&promo_creative=CRISPR-Cas9_System—RNP-transfections&promo_position=Get%20Started%20Box). 80 h after transfection, genetic ablation of target genes was assessed via qRT-PCR. The 20-nucleotide CRISPR-RNA (crRNA) targeting human XBP1 (Homo sapiens chromosome 22, GRCh38.p12, GenBank accession no. NC_000022.11) is directed at the genomic sequence 5′-CGGTGCGTAGTCTGGAGCTACGG-3′, and human PHGDH (Homo sapiens chromosome 22, GRCh38.p12, GenBank accession no. NC_000022.11) is directed at the genomic sequence 5′-TGGATCTGGAGGCCGCAACAAGG-3′ (the three additional nucleotides highlighted in bold represent the protospacer adjacent motif). This target sequence corresponds to exon 1 of the human XBP1 transcript, and exon 2 of the human PHGDH transcript and was manually chosen by identifying a 20-bp fragment immediately upstream of the highlighted protospacer adjacent motif (Ran et al., 2013). The most likely on- and off-target effects of the manually selected CRISPR sequence were then analyzed using the Broad Institute’s Genetic Perturbation Platform (https://portals.broadinstitute.org/gpp/public/analysis-tools/%20sgrna-design). To validate the genomic editing capacity of the crRNA, qRT-PCR was performed on total RNA isolated from cells transfected with sgRNA–Cas9 complexes containing the XBP1 crRNA described above. The primers for evaluating deletion efficacy are listed in Table S3.
Luciferase reporter assays and plasmid constructs
The expression construct used for luciferase-based reporter assays is pcDNA3.1 XBP1s (RefSeq accession no. NM_001079539.1), while the reporter construct used is pGL3-PHGDH promoter region with XBP1 binding site CCACGT. For dual luciferase reporter assays, 2 × 104 HEK-293T cells were plated overnight in a 96-well plate and transfected with the indicated plasmids using jetPRIME (Polyplus-transfection) according to the manufacturer’s protocol. Briefly, 18 ng of reporter and 2 ng of Renilla plasmid were cotransfected with various ratios (wt:wt) of expression plasmids (reporter:expression plasmid = 1:1, 1:3, or 1:5) and pcDNA3.1, which was added to reach a total of 200 ng of plasmid/well. After 48 h, cells were washed with PBS and lysed in Passive Lysis Buffer according to the manufacturer's protocol (dual luciferase reporter assay system, catalog #E1960; Promega). Firefly and Renilla luciferase activities were measured in white-bottom 96-well plates using an automated luminometer (SpectraMax iD3 Multi-Mode Microplate Reader; Molecular Devices). The reporter activity (Firefly) was normalized to its own Renilla luciferase activity.
Statistical analysis
All statistical analyses were performed using GraphPad Prism 9 software. Significance for pairwise correlation analysis was calculated using Spearman’s correlation coefficient (r) with P values (two-tailed). Comparisons between two groups were assessed using unpaired or paired (for matched comparisons) two-tailed Student’s t test, non-parametric Mann–Whitney U test, or one-way ANOVA with Tukey’s correction, wherever applicable. Each dot indicates an individual donor. Data are presented as mean ± SEM. P values of <0.05 were considered to be statistically significant.
Online supplemental material
Fig. S1 shows that activated UPR impacts pDC response by reducing IFN-regulated genes. Fig. S2 shows that ER stress inhibits TLR9-induced IFN-α in pDCs via IRE1⍺-XBP1–cAMP axis. Fig. S3 shows that ER stress induces serine biosynthesis in pDCs via PHGDH. Fig. S4 shows that ER stress gene levels negatively corelate with ISG expression in pDCs from SSc patients. Fig. S5 shows that TCA inhibitor CPI-613 has little to no effect on viability or on ER stress gene levels in human pDCs. Table S1 shows clinical and demographic characteristics of the patients with SSc. Table S2 shows clinical and demographic characteristics of patients with SLE. Table S3 lists primers.
Data availability
The bulk RNA-seq data assessing the transcriptional effect of TM, TG, TLR9-L (C274), and TLR9-L + TM are available in the GEO database (accession no. GSE211230).
Supplementary Material
shows clinical and demographic characteristics of the patients with SSc.
shows clinical and demographic characteristics of patients with SLE.
lists primers.
Acknowledgments
We thank the Weill Cornell Epigenomics Core Facility as well as David Oliver for help with the genomic analysis.
This work was supported by the National Institutes of Health grant 1R01AI132447 (F.J. Barrat), the Scleroderma Research Foundation (F.J. Barrat), and the Scleroderma Foundation (F.J. Barrat). J.R. Cubillos-Ruiz was supported by National Institutes of Health grants R01 NS114653 and R21 CA248106, U.S. Department of Defense grants OC200224 and OC200166, and the Mark Foundation for Cancer Research ASPIRE Award. R.L. Wiseman was supported by National Institutes of Health grant AG046495 for the development of IXA4.
Author contributions: V. Chaudhary and F.J. Barrat designed the study; V. Chaudhary, S-M. Hwang, and M.D. Ah Kioon performed experiments; K.S., K.A. Kirou, J. Zhang-Sun, M.K. Crow, R.F. Spiera, J.K. Gordon, and R.L. Wiseman followed the cohort of patients and provided clinical samples or essential reagents; J.R. Cubillos-Ruiz shared reagents and provided conceptual advice; V. Chaudhary, J.R. Cubillos-Ruiz, and F.J. Barrat wrote the manuscript.; F.J. Barrat supervised the project.
References
- Acin-Perez, R., Salazar E., Kamenetsky M., Buck J., Levin L.R., and Manfredi G.. 2009. Cyclic AMP produced inside mitochondria regulates oxidative phosphorylation. Cell Metabol. 9:265–276. 10.1016/j.cmet.2009.01.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adamson, B., Norman T.M., Jost M., Cho M.Y., Nunez J.K., Chen Y., Villalta J.E., Gilbert L.A., Horlbeck M.A., Hein M.Y., et al. 2016. A multiplexed single-cell CRISPR screening Platform enables systematic dissection of the unfolded protein response. Cell. 167:1867–1882.e21. 10.1016/j.cell.2016.11.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Affandi, A.J., Carvalheiro T., Ottria A., de Haan J.J., Brans M.A.D., Brandt M.M., Tieland R.G., Lopes A.P., Fernandez B.M., Bekker C.P.J., et al. 2022. CXCL4 drives fibrosis by promoting several key cellular and molecular processes. Cell Rep. 38:110189. 10.1016/j.celrep.2021.110189 [DOI] [PubMed] [Google Scholar]
- Ah Kioon, M.D., Tripodo C., Fernandez D., Kirou K.A., Spiera R.F., Crow M.K., Gordon J.K., and Barrat F.J.. 2018. Plasmacytoid dendritic cells promote systemic sclerosis with a key role for TLR8. Sci. Transl. Med. 10:eaam8458. 10.1126/scitranslmed.aam8458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrat, F.J., Elkon K.B., and Fitzgerald K.A.. 2016. Importance of nucleic acid recognition in inflammation and autoimmunity. Annu. Rev. Med. 67:323–336. 10.1146/annurev-med-052814-023338 [DOI] [PubMed] [Google Scholar]
- Barrat, F.J., and Su L.. 2019. A pathogenic role of plasmacytoid dendritic cells in autoimmunity and chronic viral infection. J. Exp. Med. 216:1974–1985. 10.1084/jem.20181359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrat, F.J., Meeker T., Gregorio J., Chan J.H., Uematsu S., Akira S., Chang B., Duramad O., and Coffman R.L.. 2005. Nucleic acids of mammalian origin can act as endogenous ligands for Toll-like receptors and may promote systemic lupus erythematosus. J. Exp. Med. 202:1131–1139. 10.1084/jem.20050914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bengtsson, A.A., Trygg J., Wuttge D.M., Sturfelt G., Theander E., Donten M., Moritz T., Sennbro C.J., Torell F., Lood C., et al. 2016. Metabolic profiling of systemic lupus erythematosus and comparison with primary sjogren's syndrome and systemic sclerosis. PloS One. 11:e0159384. 10.1371/journal.pone.0159384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Best, J.L., Amezcua C.A., Mayr B., Flechner L., Murawsky C.M., Emerson B., Zor T., Gardner K.H., and Montminy M.. 2004. Identification of small-molecule antagonists that inhibit an activator: Coactivator interaction. Proc. Natl. Acad. Sci. USA. 101:17622–17627. 10.1073/pnas.0406374101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bettigole, S.E., and Glimcher L.H.. 2015. Endoplasmic reticulum stress in immunity. Annu. Rev. Immunol. 33:107–138. 10.1146/annurev-immunol-032414-112116 [DOI] [PubMed] [Google Scholar]
- Burman, A., Tanjore H., and Blackwell T.S.. 2018. Endoplasmic reticulum stress in pulmonary fibrosis. Matrix Biol. 68-69:355–365. 10.1016/j.matbio.2018.03.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao, S.S., and Kaufman R.J.. 2014. Endoplasmic reticulum stress and oxidative stress in cell fate decision and human disease. Antioxid. Redox Signal. 21:396–413. 10.1089/ars.2014.5851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cervantes-Barragan, L., Lewis K.L., Firner S., Thiel V., Hugues S., Reith W., Ludewig B., and Reizis B.. 2012. Plasmacytoid dendritic cells control T-cell response to chronic viral infection. Proc. Natl. Acad. Sci. USA. 109:3012–3017. 10.1073/pnas.1117359109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaneton, B., Hillmann P., Zheng L., Martin A.C.L., Maddocks O.D.K., Chokkathukalam A., Coyle J.E., Jankevics A., Holding F.P., Vousden K.H., et al. 2012. Serine is a natural ligand and allosteric activator of pyruvate kinase M2. Nature. 491:458–462. 10.1038/nature11540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, X., and Cubillos-Ruiz J.R.. 2021. Endoplasmic reticulum stress signals in the tumour and its microenvironment. Nat. Rev. Cancer. 21:71–88. 10.1038/s41568-020-00312-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, S., Zhou Y., Chen Y., and Gu J.. 2018. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 34:i884–i890. 10.1093/bioinformatics/bty560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chopra, S., Giovanelli P., Alvarado-Vazquez P.A., Alonso S., Song M., Sandoval T.A., Chae C.S., Tan C., Fonseca M.M., Gutierrez S., et al. 2019. IRE1alpha-XBP1 signaling in leukocytes controls prostaglandin biosynthesis and pain. Science. 365:eaau6499. 10.1126/science.aau6499 [DOI] [PubMed] [Google Scholar]
- Conrad, C., Di Domizio J., Mylonas A., Belkhodja C., Demaria O., Navarini A.A., Lapointe A.K., French L.E., Vernez M., and Gilliet M.. 2018. TNF blockade induces a dysregulated type I interferon response without autoimmunity in paradoxical psoriasis. Nat. Commun. 9:25. 10.1038/s41467-017-02466-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa, R., Peruzzo R., Bachmann M., Monta G.D., Vicario M., Santinon G., Mattarei A., Moro E., Quintana-Cabrera R., Scorrano L., et al. 2019. Impaired mitochondrial ATP production downregulates wnt signaling via ER stress induction. Cell Rep. 28:1949–1960.e6. 10.1016/j.celrep.2019.07.050 [DOI] [PubMed] [Google Scholar]
- Crow, M.K., Olferiev M., and Kirou K.A.. 2019. Type I interferons in autoimmune disease. Annu. Rev. Pathol. 14:369–393. 10.1146/annurev-pathol-020117-043952 [DOI] [PubMed] [Google Scholar]
- Cubillos-Ruiz, J.R., Silberman P.C., Rutkowski M.R., Chopra S., Perales-Puchalt A., Song M., Zhang S., Bettigole S.E., Gupta D., Holcomb K., et al. 2015. ER stress sensor XBP1 controls anti-tumor immunity by disrupting dendritic cell homeostasis. Cell. 161:1527–1538. 10.1016/j.cell.2015.05.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui, X., Zhang Y., Lu Y., and Xiang M.. 2022. ROS and endoplasmic reticulum stress in pulmonary disease. Front. Pharmacol. 13:879204. 10.3389/fphar.2022.879204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delghandi, M.P., Johannessen M., and Moens U.. 2005. The cAMP signalling pathway activates CREB through PKA, p38 and MSK1 in NIH 3T3 cells. Cell Signal. 17:1343–1351. 10.1016/j.cellsig.2005.02.003 [DOI] [PubMed] [Google Scholar]
- Dessauer, C.W., and Gilman A.G.. 1997. The catalytic mechanism of mammalian adenylyl cyclase. Equilibrium binding and kinetic analysis of P-site inhibition. J. Biol. Chem. 272:27787–27795. 10.1074/jbc.272.44.27787 [DOI] [PubMed] [Google Scholar]
- Dong, H., Adams N.M., Xu Y., Cao J., Allan D.S.J., Carlyle J.R., Chen X., Sun J.C., and Glimcher L.H.. 2019. The IRE1 endoplasmic reticulum stress sensor activates natural killer cell immunity in part by regulating c-Myc. Nat. Immunol. 20:865–878. 10.1038/s41590-019-0388-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du, Y., Ah Kioon M.D., Laurent P., Chaudhary V., Pierides M., Yang C., Oliver D., Ivashkiv L.B., and Barrat F.J.. 2022. Chemokines form nanoparticles with DNA and can superinduce TLR-driven immune inflammation. J. Exp. Med. 219:e20212142. 10.1084/jem.20212142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everts, B., Amiel E., Huang S.C., Smith A.M., Chang C.H., Lam W.Y., Redmann V., Freitas T.C., Blagih J., van der Windt G.J., et al. 2014. TLR-driven early glycolytic reprogramming via the kinases TBK1-IKKɛ supports the anabolic demands of dendritic cell activation. Nat. Immunol. 15:323–332. 10.1038/ni.2833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furie, R., Werth V.P., Merola J.F., Stevenson L., Reynolds T.L., Naik H., Wang W., Christmann R., Gardet A., Pellerin A., et al. 2019. Monoclonal antibody targeting BDCA2 ameliorates skin lesions in systemic lupus erythematosus. J. Clin. Invest. 129:1359–1371. 10.1172/JCI124466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glimcher, L.H. 2010. XBP1: The last two decades. Ann. Rheum. Dis. 69 Suppl 1:i67–i71. 10.1136/ard.2009.119388 [DOI] [PubMed] [Google Scholar]
- Grandjean, J.M.D., Madhavan A., Cech L., Seguinot B.O., Paxman R.J., Smith E., Scampavia L., Powers E.T., Cooley C.B., Plate L., et al. 2020. Pharmacologic IRE1/XBP1s activation confers targeted ER proteostasis reprogramming. Nat. Chem. Biol. 16:1052–1061. 10.1038/s41589-020-0584-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu, Z., Eils R., and Schlesner M.. 2016. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics. 32:2847–2849. 10.1093/bioinformatics/btw313 [DOI] [PubMed] [Google Scholar]
- Guiducci, C., Ott G., Chan J.H., Damon E., Calacsan C., Matray T., Lee K.-D., Coffman R.L., and Barrat F.J.. 2006. Properties regulating the nature of the plasmacytoid dendritic cell response to Toll-like receptor 9 activation. J. Exp. Med. 203:1999–2008. 10.1084/jem.20060401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guiducci, C., Gong M., Xu Z., Gill M., Chaussabel D., Meeker T., Chan J.H., Wright T., Punaro M., Bolland S., et al. 2010. TLR recognition of self nucleic acids hampers glucocorticoid activity in lupus. Nature. 465:937–941. 10.1038/nature09102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guleria, A., Pratap A., Dubey D., Rawat A., Chaurasia S., Sukesh E., Phatak S., Ajmani S., Kumar U., Khetrapal C.L., et al. 2016. NMR based serum metabolomics reveals a distinctive signature in patients with Lupus Nephritis. Sci. Rep. 6:35309. 10.1038/srep35309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halasi, M., Wang M., Chavan T.S., Gaponenko V., Hay N., and Gartel A.L.. 2013. ROS inhibitor N-acetyl-L-cysteine antagonizes the activity of proteasome inhibitors. Biochem. J. 454:201–208. 10.1042/BJ20130282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetz, C., Axten J.M., and Patterson J.B.. 2019. Pharmacological targeting of the unfolded protein response for disease intervention. Nat. Chem. Biol. 15:764–775. 10.1038/s41589-019-0326-2 [DOI] [PubMed] [Google Scholar]
- Hetz, C., Zhang K., and Kaufman R.J.. 2020. Mechanisms, regulation and functions of the unfolded protein response. Nat. Rev. Mol. Cell Biol. 21:421–438. 10.1038/s41580-020-0250-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochberg, M.C. 1997. Updating the American College of Rheumatology revised criteria for the classification of systemic lupus erythematosus. Arthritis Rheum. 40:1725. 10.1002/art.1780400928 [DOI] [PubMed] [Google Scholar]
- Hu, J., Wang C., Huang X., Yi S., Pan S., Zhang Y., Yuan G., Cao Q., Ye X., and Li H.. 2021. Gut microbiota-mediated secondary bile acids regulate dendritic cells to attenuate autoimmune uveitis through TGR5 signaling. Cell Rep. 36:109726. 10.1016/j.celrep.2021.109726 [DOI] [PubMed] [Google Scholar]
- Iwakoshi, N.N., Pypaert M., and Glimcher L.H.. 2007. The transcription factor XBP-1 is essential for the development and survival of dendritic cells. J. Exp. Med. 204:2267–2275. 10.1084/jem.20070525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karnell, J.L., Wu Y., Mittereder N., Smith M.A., Gunsior M., Yan L., Casey K.A., Henault J., Riggs J.M., Nicholson S.M., et al. 2021. Depleting plasmacytoid dendritic cells reduces local type I interferon responses and disease activity in patients with cutaneous lupus. Sci. Transl. Med. 13:eabf8442. 10.1126/scitranslmed.abf8442 [DOI] [PubMed] [Google Scholar]
- Kee, J.M., Oslund R.C., Perlman D.H., and Muir T.W.. 2013. A pan-specific antibody for direct detection of protein histidine phosphorylation. Nat. Chem. Biol. 9:416–421. 10.1038/nchembio.1259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingsmore, K.M., Bachali P., Catalina M.D., Daamen A.R., Heuer S.E., Robl R.D., Grammer A.C., and Lipsky P.E.. 2021. Altered expression of genes controlling metabolism characterizes the tissue response to immune injury in lupus. Sci. Rep. 11:14789. 10.1038/s41598-021-93034-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kropski, J.A., and Blackwell T.S.. 2018. Endoplasmic reticulum stress in the pathogenesis of fibrotic disease. J. Clin. Invest. 128:64–73. 10.1172/JCI93560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lande, R., Lee E.Y., Palazzo R., Marinari B., Pietraforte I., Santos G.S., Mattenberger Y., Spadaro F., Stefanantoni K., Iannace N., et al. 2019. CXCL4 assembles DNA into liquid crystalline complexes to amplify TLR9-mediated interferon-alpha production in systemic sclerosis. Nat. Commun. 10:1731. 10.1038/s41467-019-09683-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeRoy, E.C., and T.A. Medsger, Jr.. 2001. Criteria for the classification of early systemic sclerosis. J. Rheumatol. 28:1573–1576. [PubMed] [Google Scholar]
- Liou, H.C., Boothby M.R., Finn P.W., Davidon R., Nabavi N., Zeleznik-Le N.J., Ting J.P., and Glimcher L.H.. 1990. A new member of the leucine zipper class of proteins that binds to the HLA DR alpha promoter. Science. 247:1581–1584. 10.1126/science.2321018 [DOI] [PubMed] [Google Scholar]
- Liu, Y.J. 2005. IPC: Professional type 1 interferon-producing cells and plasmacytoid dendritic cell precursors. Annu. Rev Immunol. 23:275–306. 10.1146/annurev.immunol.23.021704.115633 [DOI] [PubMed] [Google Scholar]
- Locasale, J.W. 2013. Serine, glycine and one-carbon units: Cancer metabolism in full circle. Nat. Rev. Cancer. 13:572–583. 10.1038/nrc3557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locasale, J.W., Grassian A.R., Melman T., Lyssiotis C.A., Mattaini K.R., Bass A.J., Heffron G., Metallo C.M., Muranen T., Sharfi H., et al. 2011. Phosphoglycerate dehydrogenase diverts glycolytic flux and contributes to oncogenesis. Nat. Genet. 43:869–874. 10.1038/ng.890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maamoun, H., Benameur T., Pintus G., Munusamy S., and Agouni A.. 2019. Crosstalk between oxidative stress and endoplasmic reticulum (ER) stress in endothelial dysfunction and aberrant angiogenesis associated with diabetes: A focus on the protective roles of heme oxygenase (HO)-1. Front. Physiol. 10:70. 10.3389/fphys.2019.00070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madhavan, A., Kok B.P., Rius B., Grandjean J.M.D., Alabi A., Albert V., Sukiasyan A., Powers E.T., Galmozzi A., Saez E., and Wiseman R.L.. 2022. Pharmacologic IRE1/XBP1s activation promotes systemic adaptive remodeling in obesity. Nat. Commun. 13:608. 10.1038/s41467-022-28271-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Reyes, I., and Chandel N.S.. 2020. Mitochondrial TCA cycle metabolites control physiology and disease. Nat. Commun. 11:102. 10.1038/s41467-019-13668-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moretti, J., Roy S., Bozec D., Martinez J., Chapman J.R., Ueberheide B., Lamming D.W., Chen Z.J., Horng T., Yeretssian G., et al. 2017. STING senses microbial viability to orchestrate stress-mediated autophagy of the endoplasmic reticulum. Cell. 171:809–823.e13. 10.1016/j.cell.2017.09.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nestle, F.O., Conrad C., Tun-Kyi A., Homey B., Gombert M., Boyman O., Burg G., Liu Y.J., and Gilliet M.. 2005. Plasmacytoid predendritic cells initiate psoriasis through interferon-alpha production. J. Exp. Med. 202:135–143. 10.1084/jem.20050500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacold, M.E., Brimacombe K.R., Chan S.H., Rohde J.M., Lewis C.A., Swier L.J., Possemato R., Chen W.W., Sullivan L.B., Fiske B.P., et al. 2016. A PHGDH inhibitor reveals coordination of serine synthesis and one-carbon unit fate. Nat. Chem. Biol. 12:452–458. 10.1038/nchembio.2070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce, E.J., and Everts B.. 2015. Dendritic cell metabolism. Nat. Rev. Immunol. 15:18–29. 10.1038/nri3771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petri, M., Orbai A.-M., Alarcón G.S., Gordon C., Merrill J.T., Fortin P.R., Bruce I.N., Isenberg D., Wallace D.J., Nived O., et al. 2012. Derivation and validation of the Systemic Lupus International Collaborating Clinics classification criteria for systemic lupus erythematosus. Arthritis Rheum. 64:2677–2686. 10.1002/art.34473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philip, P.A., Buyse M.E., Alistar A.T., Rocha Lima C.M., Luther S., Pardee T.S., and Van Cutsem E.. 2019. A Phase III open-label trial to evaluate efficacy and safety of CPI-613 plus modified FOLFIRINOX (mFFX) versus FOLFIRINOX (FFX) in patients with metastatic adenocarcinoma of the pancreas. Future Oncol. 15:3189–3196. 10.2217/fon-2019-0209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Possemato, R., Marks K.M., Shaul Y.D., Pacold M.E., Kim D., Birsoy K., Sethumadhavan S., Woo H.K., Jang H.G., Jha A.K., et al. 2011. Functional genomics reveal that the serine synthesis pathway is essential in breast cancer. Nature. 476:346–350. 10.1038/nature10350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team . 2020. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. http://www.r-project.org/
- Ran, F.A., Hsu P.D., Wright J., Agarwala V., Scott D.A., and Zhang F.. 2013. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 8(11):2281–2308. 10.1038/nprot.2013.143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reizis, B. 2019. Plasmacytoid dendritic cells: Development, regulation, and function. Immunity. 50:37–50. 10.1016/j.immuni.2018.12.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reverendo, M., Mendes A., Arguello R.J., Gatti E., and Pierre P.. 2019. At the crossway of ER-stress and proinflammatory responses. FEBS J. 286:297–310. 10.1111/febs.14391 [DOI] [PubMed] [Google Scholar]
- Robbins, J.D., Boring D.L., Tang W.J., Shank R., and Seamon K.B.. 1996. Forskolin carbamates: Binding and activation studies with type I adenylyl cyclase. J. Med. Chem. 39:2745–2752. 10.1021/jm960191+ [DOI] [PubMed] [Google Scholar]
- Robinson, M.D., McCarthy D.J., and Smyth G.K.. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 26:139–140. 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez, G., Ross J.A., Nagy Z.S., and Kirken R.A.. 2013. Forskolin-inducible cAMP pathway negatively regulates T-cell proliferation by uncoupling the interleukin-2 receptor complex. J. Biol. Chem. 288:7137–7146. 10.1074/jbc.M112.408765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowland, S.L., Riggs J.M., Gilfillan S., Bugatti M., Vermi W., Kolbeck R., Unanue E.R., Sanjuan M.A., and Colonna M.. 2014. Early, transient depletion of plasmacytoid dendritic cells ameliorates autoimmunity in a lupus model. J. Exp. Med. 211:1977–1991. 10.1084/jem.20132620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sisirak, V., Ganguly D., Lewis K.L., Couillault C., Tanaka L., Bolland S., D'Agati V., Elkon K.B., and Reizis B.. 2014. Genetic evidence for the role of plasmacytoid dendritic cells in systemic lupus erythematosus. J. Exp. Med. 211:1969–1976. 10.1084/jem.20132522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, M., Sandoval T.A., Chae C.S., Chopra S., Tan C., Rutkowski M.R., Raundhal M., Chaurio R.A., Payne K.K., Konrad C., et al. 2018. IRE1α-XBP1 controls T cell function in ovarian cancer by regulating mitochondrial activity. Nature. 562:423–428. 10.1038/s41586-018-0597-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spillier, Q., and Frédérick R.. 2021. Phosphoglycerate dehydrogenase (PHGDH) inhibitors: A comprehensive review 2015-2020. Expert Opin. Ther. Pat. 31:597–608. 10.1080/13543776.2021.1890028 [DOI] [PubMed] [Google Scholar]
- Steegborn, C. 2014. Structure, mechanism, and regulation of soluble adenylyl cyclases - similarities and differences to transmembrane adenylyl cyclases. Biochim. Biophys. Acta. 1842:2535–2547. 10.1016/j.bbadis.2014.08.012 [DOI] [PubMed] [Google Scholar]
- Stuart, S.D., Schauble A., Gupta S., Kennedy A.D., Keppler B.R., Bingham P.M., and Zachar Z.. 2014. A strategically designed small molecule attacks alpha-ketoglutarate dehydrogenase in tumor cells through a redox process. Cancer Metabol. 2:4. 10.1186/2049-3002-2-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studencka-Turski, M., Cetin G., Junker H., Ebstein F., and Kruger E.. 2019. Molecular insight into the IRE1α-mediated type I interferon response induced by proteasome impairment in myeloid cells of the brain. Front. Immunol. 10:2900. 10.3389/fimmu.2019.02900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swiecki, M., and Colonna M.. 2015. The multifaceted biology of plasmacytoid dendritic cells. Nat. Rev. Immunol. 15:471–485. 10.1038/nri3865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swiecki, M., Gilfillan S., Vermi W., Wang Y., and Colonna M.. 2010. Plasmacytoid dendritic cell ablation impacts early interferon responses and antiviral NK and CD8(+) T cell accrual. Immunity. 33:955–966. 10.1016/j.immuni.2010.11.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanimura, A., Miyoshi K., Horiguchi T., Hagita H., Fujisawa K., and Noma T.. 2018. Mitochondrial activity and unfolded protein response are required for neutrophil differentiation. Cell Physiol. Biochem. 47:1936–1950. 10.1159/000491464 [DOI] [PubMed] [Google Scholar]
- Tanjore, H., Lawson W.E., and Blackwell T.S.. 2013. Endoplasmic reticulum stress as a pro-fibrotic stimulus. Biochim. Biophys. Acta. 1832:940–947. 10.1016/j.bbadis.2012.11.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavernier, S.J., Osorio F., Vandersarren L., Vetters J., Vanlangenakker N., Van Isterdael G., Vergote K., De Rycke R., Parthoens E., van de Laar L., et al. 2017. Regulated IRE1-dependent mRNA decay sets the threshold for dendritic cell survival. Nat. Cell Biol. 19:698–710. 10.1038/ncb3518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Bon, L., Affandi A.J., Broen J., Christmann R.B., Marijnissen R.J., Stawski L., Farina G.A., Stifano G., Mathes A.L., Cossu M., et al. 2014. Proteome-wide analysis and CXCL4 as a biomarker in systemic sclerosis. N. Engl. J. Med. 370:433–443. 10.1056/NEJMoa1114576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkmann, E.R., Tashkin D.P., Roth M.D., Clements P.J., Khanna D., Furst D.E., Mayes M., Charles J., Tseng C.H., Elashoff R.M., and Assassi S.. 2016. Changes in plasma CXCL4 levels are associated with improvements in lung function in patients receiving immunosuppressive therapy for systemic sclerosis-related interstitial lung disease. Arthritis Res. Ther. 18:305. 10.1186/s13075-016-1203-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenzel, J., and Tuting T.. 2008. An IFN-associated cytotoxic cellular immune response against viral, self-or tumor antigens is a common pathogenetic feature in “interface dermatitis”. J. Invest. Dermatol. 128:2392–2402. 10.1038/jid.2008.96 [DOI] [PubMed] [Google Scholar]
- Wilson, J.L., Nagele T., Linke M., Demel F., Fritsch S.D., Mayr H.K., Cai Z., Katholnig K., Sun X., Fragner L., et al. 2020. Inverse data-driven modeling and multiomics analysis reveals phgdh as a metabolic checkpoint of macrophage polarization and proliferation. Cell Rep. 30:1542–1552.e7. 10.1016/j.celrep.2020.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, D., Sanin D.E., Everts B., Chen Q., Qiu J., Buck M.D., Patterson A., Smith A.M., Chang C.H., Liu Z., et al. 2016. Type 1 interferons induce changes in Core metabolism that are critical for immune function. Immunity. 44:1325–1336. 10.1016/j.immuni.2016.06.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, Y., Hu Y., Wang B., Li S., Ma C., Liu X., Moynagh P.N., Zhou J., and Yang S.. 2020. Dopamine uses the DRD5-ARRB2-PP2A signaling Axis to block the TRAF6-mediated NF-κB pathway and suppress systemic inflammation. Mol. Cell. 78:42–56.e6. 10.1016/j.molcel.2020.01.022 [DOI] [PubMed] [Google Scholar]
- Yang, L., Venneti S., and Nagrath D.. 2017. Glutaminolysis: A hallmark of cancer metabolism. Annu. Rev. Biomed. Eng. 19:163–194. 10.1146/annurev-bioeng-071516-044546 [DOI] [PubMed] [Google Scholar]
- Yang, Z., Wang Y., Zhang Y., He X., Zhong C.Q., Ni H., Chen X., Liang Y., Wu J., Zhao S., et al. 2018. RIP3 targets pyruvate dehydrogenase complex to increase aerobic respiration in TNF-induced necroptosis. Nat. Cell Biol. 20:186–197. 10.1038/s41556-017-0022-y [DOI] [PubMed] [Google Scholar]
- Zachar, Z., Marecek J., Maturo C., Gupta S., Stuart S.D., Howell K., Schauble A., Lem J., Piramzadian A., Karnik S., et al. 2011. Non-redox-active lipoate derivates disrupt cancer cell mitochondrial metabolism and are potent anticancer agents in vivo. J. Mol. Med. 89:1137–1148. 10.1007/s00109-011-0785-8 [DOI] [PubMed] [Google Scholar]
- Zhang, H., Kong Q., Wang J., Jiang Y., and Hua H.. 2020. Complex roles of cAMP-PKA-CREB signaling in cancer. Exp. Hematol. Oncol. 9:32. 10.1186/s40164-020-00191-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
shows clinical and demographic characteristics of the patients with SSc.
shows clinical and demographic characteristics of patients with SLE.
lists primers.
Data Availability Statement
The bulk RNA-seq data assessing the transcriptional effect of TM, TG, TLR9-L (C274), and TLR9-L + TM are available in the GEO database (accession no. GSE211230).