Abstract
Fragile X syndrome (FXS) is the most common inherited cause of intellectual disability. Fragile X mental retardation protein, a putative translation suppressor, is significantly reduced in FXS. The prevailing hypothesis is that rates of cerebral protein synthesis (rCPS) are increased by the absence of this regulatory protein. We have previously reported increased rCPS in the Fmr1 knockout mouse model of FXS. To address the hypothesis in human subjects, we measured rCPS in young men with FXS with L-[1-11C]leucine PET. In previous studies we had used sedation during imaging, and we did not find increases in rCPS as had been seen in the mouse model. Since mouse measurements were conducted in awake animals, we considered the possibility that sedation may have confounded our results. In the present study we used a modified and validated PET protocol that made it easier for participants with FXS to undergo the study awake. We compared rCPS in 10 fragile X participants and 16 healthy controls all studied while awake. Contrary to the prevailing hypothesis and findings in Fmr1 knockout mice, results indicate that rCPS in awake participants with FXS are decreased in whole brain and most brain regions by 13–21% compared to healthy controls.
Keywords: Fragile X syndrome, FMRP, protein synthesis, PET, translation
Introduction
It is thought that in fragile X syndrome (FXS) in the absence of the translation suppressor fragile X mental retardation protein (FMRP), rates of protein synthesis are increased. In previous studies in awake and functioning animals (Fmr1 knockout (KO) mice), regional rates of cerebral protein synthesis (rCPS) were significantly elevated over those measured in wildtype (WT) mice, most remarkably in hippocampus, thalamus, hypothalamus, and some parts of the cortex.1–4 Based on findings in the Fmr1 KO mouse model, we hypothesized that dysregulation of protein synthesis is a core phenotype of this disease and that in participants with FXS we should also be able to detect increases in rCPS.
In our first study of FXS in humans, we applied the L-[1-11C]leucine PET method for measurement of rCPS5–7 to the study of young adult men with FXS and age-matched healthy controls. It was necessary to sedate participants with FXS for these studies because they could not tolerate placement of an arterial catheter and were unable to remain motionless for a 90-minute scan. We used the hypnotic, propofol, for sedation, and we compared rCPS in FXS participants with healthy controls all of whom were sedated with propofol. We found that rCPS in the FXS participants were not increased, but rather rates were reduced in the whole brain, cerebellum, and cortex 4 despite our previous finding that propofol did not affect rCPS in healthy control subjects. 8 In a follow-up study of the effects of propofol sedation in mice, rCPS were decreased in sedated Fmr1 KO mice compared with awake but were only slightly affected in WT. 4 Propofol acts primarily through effects on GABAA receptors, and it is known that the GABAA system is affected in Fmr1 KO mice,9–12 in dfmr1 flies 12 and in humans with FXS. 13 Next, we measured rCPS in another group of participants with FXS and healthy controls sedated with dexmedetomidine which, unlike propofol, is an α2-adrenergic agonist. Again, we did not find increased rates of protein synthesis in the participants with FXS. 14
Since the elevated rCPS had been found in awake and functioning Fmr1 KO mice, we considered the possibility that the sedating agents may have confounded our results in human subjects. We subsequently developed a modified PET procedure to make it easier for participants with FXS to tolerate it while awake. In the modified method, PET data are acquired over a shorter (60-min) scan interval 15 and venous is substituted for arterial blood sampling. 16 We validated this modified procedure in healthy controls studied both awake and under sedation and in participants with FXS studied under sedation. 16 In the present study the modified procedure was applied to healthy controls and participants with FXS all in the awake state in order to evaluate the hypothesis that the rate of protein synthesis is increased in fragile X compared to healthy controls.
Materials and methods
All procedures on human subjects were carried out as described in a protocol (06-M-0214, NCT00362843) approved by the National Institutes of Health Combined Neurosciences Institutional Review Board, the National Institutes of Health Radioactive Drug Research Committee, and the National Institutes of Health Radiation Safety Committee. All participants or legal guardians gave written informed consent prior to study enrollment. Participants were monetarily compensated.
Participants
Male full mutation FXS participants 18–24 years of age who had taken no psychotropic medication during the previous year and who had no recent history of seizures were recruited to participate. We used the number of CGG repeats in the 5′-untranslated region of FMR1 as a criterion for diagnosis. If the number of CGG repeats exceeds 200 the gene is silenced. The number of CGG repeats in FMR1 was confirmed by DNA analysis (Mayo Clinic, Rochester, MN). Control participants were healthy males 18 to 24 years old recruited from local universities and post-baccalaureate training positions at the NIH. Pre-enrollment screening for controls was clinical history, physical examination and an abbreviated version of the Structured Clinical Interview for DSM-IV-TR. 17 Inclusion criteria were: [1] no current or past diagnoses of psychiatric, neurologic or chronic medical condition, [2] no history of neurologic disorders, [3] no family history of genetically transmissible neurologic syndrome, [4] no history of harmful/paradoxical reactions to general and/or local anesthetics, [5] no allergy to egg and soy proteins, and [6] HIV negative. Participants also all negatively tested for drugs of abuse at screening and again on the day of PET scanning. Although 16 healthy controls and 12 FXS participants met the inclusion criteria and enrolled, one participant with fragile X was unable to complete the awake study and data from another FXS participant was excluded because of uncorrectable motion during the scan, so study groups presented here include 16 in the control group and 10 in the FXS group. In the healthy control group 12 identified as white, two as African American, one as white/native American, and one as Asian. In the FXS group eight identified as white, one as Hispanic, and one did not identify with any racial group.
Psychological testing
Participants with FXS were administered the Wechsler Adult Intelligence Scale 4th edition (WAIS-IV) 18 for Intelligence Quotients (IQs) to assess intellectual disability. All participants tested had full scale IQ scores in the disability range (Perceptual Reasoning Standard Scores 50–69, Full Scale IQ 44–63). Psychologists assessed for the presence of autism symptoms by means of the Autism Diagnostic Interview, Revised 19 as well as the Autism Diagnostic Observation Schedule, 2nd Edition (ADOS-2), 20 and parent questionnaires, including the Social Responsiveness Scale. None of the subjects tested met diagnostic criteria for autism spectrum disorder.
Brain magnetic resonance imaging (MRI)
All subjects underwent a noncontrast T1-weighted MRI of the brain for region of interest (ROI) placement. MRI examinations were performed on a 3.0 Tesla scanner (Phillips Healthcare, Cleveland, OH, USA). Images were reconstructed with voxel dimensions of 0.94 × 0.94 × 1 mm and interpolated to isotropic voxel dimensions of (0.94 mm) 3 .
Brain regions of interest
Whereas our PET studies could not be blind to the diagnosis and they could not be randomized, placement of regions of interest (ROIs) on MRIs followed assignment of a participant to a study number and ROI placement was according to anatomical landmarks; in this sense analysis was blinded to diagnosis.
ROIs were manually drawn with custom software written in Matlab (Mathworks, Natick, MA, USA) on each MRI by visually identifying anatomic landmarks as follows:
The parietal cortex was drawn in the coronal plane. The entire parietal cortex was not delineated; an inferior parietal lobule, which mainly consisted of the supra marginal and angular gyri, was chosen. It was separated from the temporal lobe by the lateral fissure, from the frontal lobe by the central sulcus, and from the superior lobule of the parietal lobe by the intraparietal sulcus.
The hippocampus and amygdala were drawn in the sagittal plane. From lateral to medial, the hippocampus began at the appearance of the tail of the caudate, under the inferior horn of the lateral ventricle, and continued medially about 28 mm. Similarly, the amygdala began at about 2 mm medial to the first appearance of the hippocampus. It is rostral to the hippocampus and sandwiched between the claustrum and the inferior horn of the lateral ventricle. Further medial, the amygdala lies between the lateral sulcus and the hippocampus and below the globus pallidus. The amygdala extends about 24 mm in the sagittal plane.
All other regions were drawn in the transverse plane. Approximately 80 slices were chosen as the frontal cortex ROI. It was caudally limited at the disappearance of the medial orbital cortex, coincident laterally with the lateral curvature of the frontal lobe and separated from parietal lobe by the central sulcus and from the temporal lobe by the lateral fissure. The cingulate cortex ROI was about 35–45 slices. Proceeding from ventral to dorsal, the first slice was at the level of the first appearance of the ungulate sulcus and the last slice was at the level of the intersection of coronal sulcus and medial plane. We sampled the corona radiata as an oval placed in the center in ten slices bilaterally. The first slice was located by the disappearance of the putamen. The ROI was limited medially by the caudate and thalamus and laterally by the insula. The caudate was defined as the distinctly dark, oval-shaped structure above medial orbital cortex. It was limited laterally by the walls of the first and second ventricles and medially by lumen of the ventricles. The putamen was marked at the level where it first separated from the head of caudate and limited by the insula laterally and internal capsule medially. Thalami were limited medially by the lumen of third ventricle and by the globus pallidus laterally. In addition to the above regions, the whole brain was also outlined on the transverse slices.
PET studies
The acquisition protocol was identical to that previously described 21 except for blood sampling procedures. In all participants in this study an intravenous catheter was placed in the antecubital fossa of the dominant arm for injection of tracer and an intravenous catheter was placed in the contralateral antecubital fossa for venous blood sampling. In ten healthy control subjects, an arterial catheter was also placed in the radial artery of the non-dominant arm for sampling arterial blood for use in separate studies.14,15 In this group, PET data were acquired for 90 min but analyzed over a 60-min scan interval, and in this study, only venous blood samples were used for input function and unlabeled leucine determinations. In the remaining six healthy control subjects and in all FXS subjects, only venous blood samples were collected, and PET data were acquired over a 60-min scan interval.
PET scans were performed on the ECAT High Resolution Research Tomograph (CPS Innovations, Knoxville, TN). After optimal positioning of the subject within the field of view, a six-minute transmission scan was obtained for attenuation correction. The emission scan was initiated coincident with the intravenous infusion of 7.3–11.8 MBq/kg (mean, 11.1 MBq/kg) of L-[1-11C]leucine administered by a computer-controlled infusion pump at a constant rate over 2 minutes. Data were acquired in list mode and reconstructed by means of the motion-compensated 3D ordinary Poisson ordered subset expectation maximization (OSEM) algorithm (30 subsets, 2 iterations). 22 Spatial resolution after reconstruction was approximately 2.6 mm full width at half maximum in radial and transverse directions. 22 Three-dimensional data were reconstructed to 207 slices 1.23 mm thick with a pixel size of 1.21 × 1.21 mm. Images were reconstructed as 36 frames of data for the 60 min interval of analysis (16 × 15 s, 4 × 30 s, 4 × 60 s, 4 × 150 s, 8 × 300 s). With this reconstruction, motion-correction was based on position data collected throughout the scan with the Polaris system, 23 and the data were corrected for attenuation based on a single attenuation-correction map (MuMap) applied to all frames of data. To correct for any residual motion, we proceeded as follows: [1] Summed Frames 1–17 (0–4.5 min) and aligned the summed image to the MuMap; [2] Determined 3D rigid body spatial transformation matrices for Frames 18–36 (4.5–60 min post injection) by aligning each frame to the summed image; [3] Applied the inverse of each transformation from the previous step to the original MuMap to obtain individual MuMaps for Frames 18–36; [4] Reconstructed the data again utilizing the original motion-correction together with individual MuMaps for each frame; [5] Aligned Frames 18–36 with the sum of the first 17 frames using the transformations defined in Step 2. These steps assured not only that the data from each frame were aligned to the initial frames, but also that an accurate attenuation correction was applied.
Analysis of blood samples
Venous samples were collected a minimum of 4 times for all participants in both diagnostic groups. In the first 10 healthy control and 6 FXS participants, 4 venous samples were collected (15, 30, 45 and 60 min after tracer injection). Later we modified the blood collection protocol slightly, and in the remaining 6 healthy controls and 5 participants with FXS, venous samples were collected in duplicate at 10, 20, 30, 40, 50 and 60 min after tracer injection. The collection of these few additional blood samples would have no effect on computed rCPS, they served as replicates for the blood curve. Concentrations of unlabeled and labeled leucine in plasma and total 11C activity in blood were measured according to the methods detailed previously. 5 Control venous samples were also taken prior to tracer injection to measure the initial concentration of plasma leucine. Input functions for each subject were determined by use of the venous data collected between 30 and 60 min to calibrate arterial input functions measured in a separate population of 25 healthy control subjects, as previously validated. 16
PET data analysis
For each scan, a 3D volume was constructed from the average of the emission data acquired between 30 and 60 minutes. This volume was isotropically smoothed with a Gaussian filter (full width at half maximum 3 mm) and aligned to the MRI volume by use of the Flexible Image Registration Toolbox22 with a 3D rigid body transformation. The resliced average 30- to 60-min PET image was visually reviewed for correct alignment with the MRI by use of Vinci (Volume Imaging in Neurological Research, CoRegistration and ROIs Included; the Max Planck Institute for Neurological Research, Cologne, Germany). The transformation parameters were then applied to each frame of the PET study (without prior smoothing) to effect its alignment with the MRI volume.
The kinetic model for the behavior of leucine in brain has been described previously. 24 The parameters of the model were estimated for each voxel in the whole brain volume by means of the Basis Function Method 24 with a slightly modified algorithm to avoid negative parameter estimates (See Electronic Supplementary Information 25 ) PET data beginning at the time of tracer injection and continuing for 60 min were used in all analyses. Use of voxelwise estimation helps to reduce errors in model parameter estimates because of kinetic heterogeneity in the ROIs. Images of each parameter were constructed, and ROIs drawn on MRIs were transferred to parametric images to compute average values of all parameters of the model.
Statistical analyses
A power analysis preceded the study. It was based on variance in rCPS measured in large cortical regions (6–11%) and computed by the same analysis method used in the present study. 24 We hypothesized that rCPS would be 10–15% higher in the FXS subjects compared to healthy controls; this was based on our results in the Fmr1 KO mouse model. 3 For α = 0.05, with 90% power we estimated that we would need to scan six subjects per group if the effect were 15% (Cohen’s d = 2.3). If the effect size were smaller, i.e., 10% (Cohen’s d = 1.6) we would need to scan 10 per group.
Before conducting the principal statistical analyses, we asked if results of studies completed between 10/3/2014 and 12/22/2015 (Cohort I, 10 healthy controls and 5 participants with FXS) and results of studies completed between 6/6/2018 and 12/11/2019 (Cohort II, 6 healthy controls and 5 participants with FXS) were different. Was there an effect of the 2.5 year hiatus between cohorts? We first analyzed the rCPS results of the two cohorts by means of a mixed model ANOVA with both diagnosis and cohort as between subject variables. For rCPS measurements in brain regions we also included brain region as a within subject variable. Results, presented in the Supplement (Supplemental Table 1) indicate no differences between the two cohorts. The region × diagnosis × cohort (F8,176 = 1.463, p = 0.217) interaction was not statistically significant. Neither the region × cohort (F8,176 = 0.875, p = 0.494) nor the diagnosis × cohort (F1,22 = 1.152, p = 0.338) interactions were statistically significant. Assured that there was no evidence of an unknown drift in our procedures, we combined the two cohorts and analyzed the study for effects of diagnosis. Regional volumes and rCPS measurements were analyzed by means of a mixed model ANOVA with region as a within subject variable and diagnosis as a between subject variable. Results (volume and rCPS) in whole brain and venous plasma leucine concentrations were analyzed separately by means of unpaired t-tests.
Results
Study groups characteristics
The group of 10 participants with FXS were well-matched for age (18–24 y) and size with the group of 16 healthy controls (Table 1). Whereas mean values for weight were similar for the two groups, the variance was somewhat greater in the participants with FXS in keeping with the tendency for young men with this disease to be overweight. CGG repeat numbers were evaluated in all FXS participants and in 13 of the healthy controls. In healthy controls, values ranged from 20–31. Of the subjects with FXS, two were mosaic with one allele in the premutation range and the other in the >200 range. In one mosaic participant, alleles in the premutation range were methylated. 26 All other FXS participants had only alleles with CGG repeat lengths exceeding 200. Full scale IQs of participants with FXS ranged from 44–63, indicating mild to moderate cognitive impairment. Mean venous plasma leucine concentrations were 16% lower in subjects with FXS (Table 1).
Table 1.
Participants.
| Healthy controls (16) | Fragile Xsubjects (10) | |
|---|---|---|
| Age (yr) | 22.2 ± 1.5 | 21.7 ± 2.4 |
| Height (m)a | 1.78 ± 0.05 | 1.77 ± 0.11 |
| Weight (kg) | 77.6 ± 9.5 | 79.8 ± 11.1 |
| FMR1 CGG repeat numberb | 28 ± 4 | >200c |
| Full Scale IQ | ND | 53 ± 6 |
| Venous plasma leucine (nmol/mL)d | 114 ± 18 | 95 ± 16 |
| Injected L-[1-11C]leucine dose (MBq/kg)e | 11.3 ± 0.3 | 10.7 ± 1.3 |
Values are means ± SD for the number of subjects indicated in parentheses.
aHeight was not measured in two controls, so n = 14.
bFMR1 CGG repeat number was analyzed in all FXS participants and 13 controls.
cThree of the participants with FXS were mosaic with one allele in the > 200 CGG repeats and the other in the premutation range. All other subjects were full mutation FXS with >200 CGG repeats.
dPlasma leucine concentration in each subject is the average over the 60 min interval of the sample measurements weighted by the interval of time between samples. Mean venous plasma leucine concentrations were 16% lower in participants with FXS compared with healthy controls (unpaired t-test; t = 2.668, df = 24, p = 0.013).
eSpecific activity of injectate was >100 MBq/nmol.
Volumes
Except for the corona radiata, all regions encompassed the whole of the area, so each measurement of rCPS is a weighted average for that region. The corona radiata was sampled with an oval placed at the center of the region. Volumes of the nine brain regions were analyzed by means of a mixed model ANOVA with region as a within subject variable and diagnosis as a between subject variable (Table 2). The region × diagnosis (F8,192 = 4.238, p = 0.042) interaction was statistically significant. Post-hoc multiple comparisons indicate that volume in frontal cortex was statistically significantly lower (−9%) in participants with FXS. Whole brain volumes were analyzed separately by means of an unpaired t-test. Whereas the mean whole brain volume in the FXS group was 7% lower than the healthy controls, this difference was not statistically significant (t = 1.676, df = 24, p = 0.107).
Table 2.
Whole brain and regional volumes (mL).
| Healthy controls (16) | Fragile Xsubjects (10) | |
|---|---|---|
| Whole brain | 1433.9 ± 170.4 | 1337.4 ± 77.2 |
| Frontal cortex | 181.4 ± 19.0 | 165.1 ± 18.2* |
| Parietal cortex(Inferior parietal lobule) | 29.5 ± 3.3 | 29.4 ± 4.0 |
| Cingulate cortex | 19.7 ± 2.8 | 18.2 ± 3.2 |
| Hippocampus | 8.2 ± 1.0 | 7.8 ± 1.4 |
| Amygdala | 1.3 ± 0.3 | 1.2 ± 0.2 |
| Thalamus | 14.5 ± 2.1 | 13.9 ± 2.1 |
| Caudate | 8.5 ± 1.5 | 9.3 ± 1.5 |
| Putamen | 8.8 ± 1.2 | 8.5 ± 0.8 |
| Corona radiata (sampled volume) | 5.5 ± 0.7 | 5.4 ± 0.7 |
Values are the means ± SD for the number of subjects indicated in parentheses.
Data from the regional analysis were subjected to a mixed model ANOVA with region as a within subject variable and diagnosis as a between subject variable. The region x diagnosis (F8,192 = 4.238, p = 0.042) interaction was statistically significant. *, Post-hoc Bonferroni-corrected multiple comparison results indicate that volume in frontal cortex was statistically significantly lower (−9%) in subjects with FXS. Whole brain volumes were analyzed separately by means of an unpaired t-test. Difference was not statistically significant (t = 1.676, df = 24, p = 0.107).
rCPS
Mean protein synthesis rates in the brain as a whole were 17% lower in participants with FXS than healthy controls (Table 3, Supplemental Figure S1) (t = 3.872, df = 24, p = 0.0007). We measured rCPS in nine brain regions: frontal cortex, parietal cortex, cingulate cortex, thalamus, caudate, putamen, hippocampus, amygdala, and corona radiata (Table 3). The regional data were subjected to a mixed model ANOVA with diagnosis as a between subject variable and region as a within subject variable. The region × diagnosis interaction (F8,192 = 12.395, p < 0.001) was statistically significant. Bonferroni-corrected post-hoc t-tests indicate that differences between healthy controls and participants with FXS were statistically significant in frontal cortex (−20%, p < 0.001), parietal cortex (−21%, p < 0.001), cingulate cortex (−21%, p < 0.001), thalamus (−18%, p = 0.001), caudate (−17%, p < 0.001), putamen (−15%, p = 0.001) and hippocampus (−13%, p = 0.003) (Table 3; Supplementary Fig. S2). Effects in frontal cortex and in hippocampus are illustrated in parametric images of typical subjects (Figures 1 and 2, respectively). Our results indicate that in most brain regions, rCPS were lower in young adults with FXS compared to age-matched healthy controls.
Table 3.
rCPS (nmol/g/min).a
| Healthy controls (16) | Fragile Xsubjects (10) | |
|---|---|---|
| Whole brain | 1.44 ± 0.16 | 1.20 ± 0.15*** |
| Frontal cortex | 1.68 ± 0.19 | 1.34 ± 0.15*** |
| Parietal cortex | 1.77 ± 0.21 | 1.39 ± 0.17*** |
| Cingulate cortex | 1.54 ± 0.17 | 1.21 ± 0.13*** |
| Hippocampus | 1.37 ± 0.15 | 1.19 ± 0.11** |
| Amygdala | 1.21 ± 0.16 | 1.15 ± 0.21 |
| Thalamus | 1.36 ± 0.20 | 1.07 ± 0.13** |
| Caudate | 0.93 ± 0.07 | 0.77 ± 0.11*** |
| Putamen | 1.15 ± 0.11 | 0.97 ± 0.12** |
| Corona radiata | 0.61 ± 0.05 | 0.56 ± 0.13 |
Values are the means ± SD for the number of subjects indicated in parentheses.
Data from the regional analysis were subjected to a mixed model ANOVA with region as a within subject variable and diagnosis as a between subject variable. The region x diagnosis (F8,192 = 12.395, p < 0.001) interaction was statistically significant. Bonferroni-corrected post-hoc t-tests for region x diagnosis indicate that rCPS were statistically significantly lower in subjects with FXS in most regions as indicated in the table: *, 0.05 ≥ p ≥ 0.01; **, 0.01 ≥ p ≥ 0.001; ***, p < 0.001. Whole brain rCPS were analyzed by means of an unpaired t-test. Whole brain rCPS was 17% lower in subjects with FXS compared with healthy controls (t = 3.872, df = 24, p = 0.0070.
aWe assumed a tissue density of 1.0 g/mL.
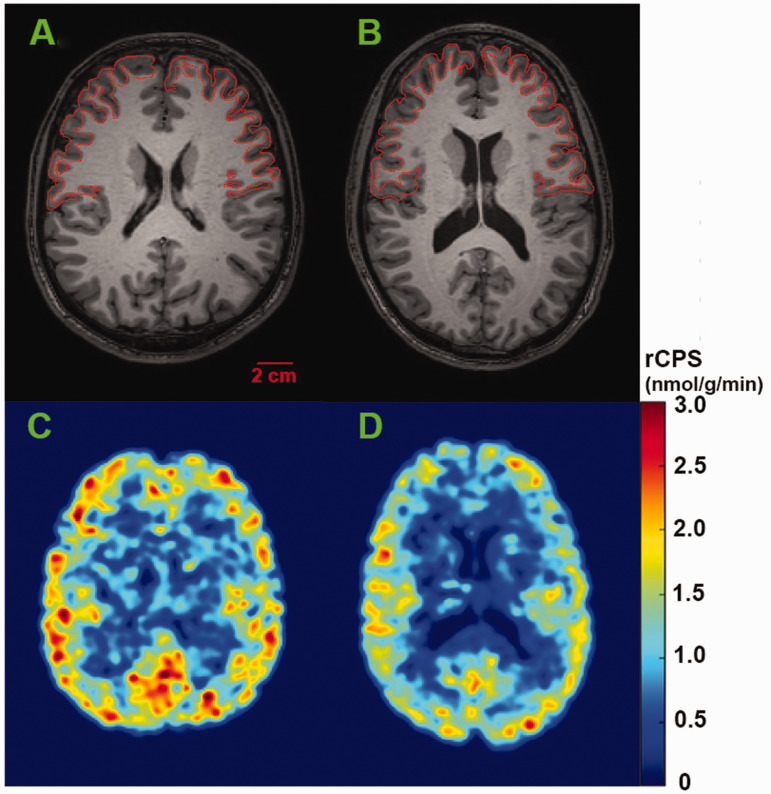
Figure 1.
Representative parametric images of rCPS at the level of the frontal cortex in awake subjects. Magnetic resonance images (upper row) from a healthy control (A) and a subject with FXS (B) correspond to rCPS images shown below (C, D, respectively). Parametric rCPS images were smoothed with a 3D Gaussian filter (kernel 4 mm full-width-at half-maximum) and color-coded for rCPS (colorbar on the right). Images are in the transverse plane. Scalebar on lower right in A applies to all four images. We assumed a tissue density of 1.0 g/mL. Frontal cortex outlines are shown on the MR images.
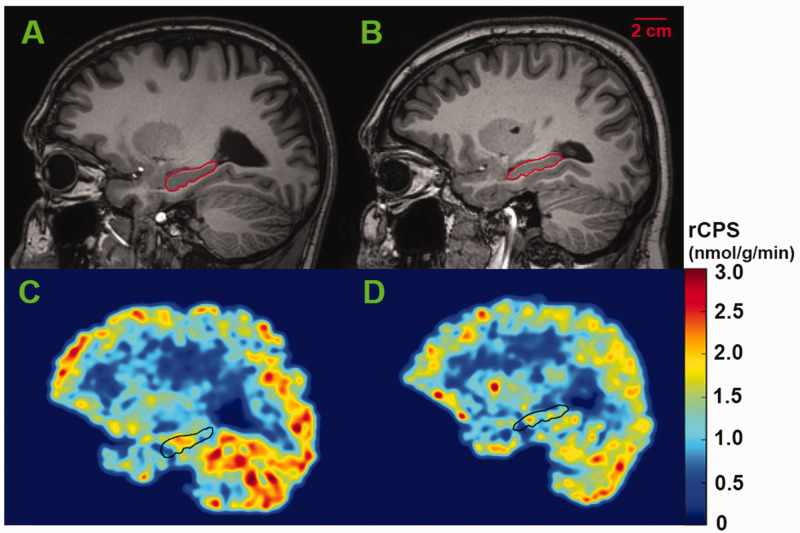
Figure 2.
Representative parametric images of rCPS at the level of the hippocampus in awake subjects. Magnetic resonance images (upper row) from a healthy control (A) and a subject with FXS (B) correspond to rCPS images shown below (C, D, respectively). Parametric rCPS images were smoothed with a 3D Gaussian filter (kernel 4 mm full-width-at half-maximum) and color-coded for rCPS (colorbar on the right). Images are in the sagittal plane. Scalebar on upper right in B applies to all four images. We assumed a tissue density of 1.0 g/mL. Outlines of the hippocampus are shown on all images.
To address any concern that differences in mean plasma leucine concentrations in the two diagnostic groups may have affected the group differences in rCPS, we compared subsets of healthy controls and participants with FXS with matched plasma leucine concentrations (Supplement, Fig. S3). We analyzed for differences in rCPS between healthy controls and participants with FXS with plasma leucine concentrations between 90 and 130 nmol/ml. With these limits on plasma leucine concentration, we excluded three healthy controls (plasma leucine concentrations of 84, 135, 150 nmol/ml) and four participants with FXS (plasma leucine concentrations of 75, 78, 79 and 83 nmol/ml). The groups of included subjects were well matched with respect to plasma leucine concentrations (Supplement Fig. S3A) (healthy controls (n = 13), 111.3 ± 13.1 nmol/ml; FXS (n = 6), 105.3 ± 12.0 nmol/ml. Even in this smaller number of subjects with reduced statistical power, rCPS was statistically significantly lower (13%, p = 0.028) in participants with FXS in whole brain (Supplement Fig. S3B): healthy controls, 1.44 ± 0.16 nmol/g/min; FXS, 1.26 ± 0.13 nmol/g/min (a tissue density of 1.0 g/mL was assumed). Data from the regional analysis (Supplementary Fig. S4) were subjected to a mixed model ANOVA with region as a within subject variable and diagnosis as a between subject variable. The region x diagnosis (F8,136 = 7.59, p < 0.001) interaction was statistically significant. Bonferroni-corrected post-hoc t-tests indicate that rCPS were statistically significantly lower in participants with FXS in most regions. Statistically significant percent differences were: frontal cortex, 16% (p = 0.006); parietal cortex, 16% (p = 0.006); cingulate cortex, 20% (p = 0.001); thalamus, 14% (p = 0.017); caudate, 12% (p = 0.014); putamen, 11% (p = 0.032). Mean value in hippocampus was 9% lower in subjects with FXS, but this effect did not reach statistical significance (p = 0.096). In amygdala and corona radiata, mean values were similar in both groups.
Discussion
PET measurements of rCPS reported in this study indicate that, in many brain regions and in the brain as a whole, rates are significantly lower in young adult men with FXS. To our knowledge, ours is the only study to report rates of mRNA translation in brain in awake human subjects with FXS. Whereas results in animal models appear to be consistent with the theories about the functions of FMRP as a translation suppressor, studies, both in vivo and in vitro, may be limited by certain experimental conditions and assumptions. Moreover, findings in animal models do not always translate to human subjects.
We used a fully validated modification of the original L-[1-11C]leucine PET method15,16 to make it more feasible for participants to undergo the procedure in the awake state. The method requires participants to lie motionless with the head positioned in the scanner for about 70 min while a few timed venous blood samples are collected. Healthy controls were able to complete this procedure with no difficulty as were most of the participants with FXS. A few of the FXS subjects became anxious in the scanner but were able to continue the procedure with coaching from family. Likely the FXS participants who enrolled in our study were high functioning, based on their participation in vocational and educational activities, although genetic and IQ testing confirmed the full mutation diagnosis and below average IQ scores.
In vivo studies in a mouse model of FXS, Fmr1 KO, have reported that rCPS are higher in model mice compared with WT.1–4 Mice in these studies were awake and freely moving at the time of the study, but they had previously undergone surgical implantation of vascular catheters under isoflurane anesthesia 24 h prior to the study. Differences from WT ranged from 10–20% in cortical areas in these mouse studies. Higher protein synthesis rates were also reported in hippocampal slices from Fmr1 KO mice compared to WT,27–28 but these studies were performed under amino acid starvation conditions, known to have profound effects on translation possibly through the integrated stress response (ISR). 29 Elevated puromycin binding (an index of translation) in fibroblasts harvested and cultured from subjects with FXS has also been reported, 30 although variability was very large in the FXS cells. In another study of neurons derived from isogenic human stem cells, puromycin binding was reported to be elevated in cells deficient in FMRP compared to cells with FMRP. In accord with these results, ERK1/2 and Akt signaling were increased in FMRP-deficient cells. 31 It is possible that mRNA translation in isolated neurons is regulated quite differently than in neurons integrated into CNS networks. In the intact nervous system, it is expected that synaptic activity may have a significant regulatory effect on rates of translation. Moreover, in FXS it is the effects of synaptic activity that may be dysregulated.
One of the interesting differences between healthy controls and participants with FXS is the difference between the response of rCPS to sedation. In healthy controls, we found no difference in rCPS between awake and dexmedetomidine-sedated states. 14 This was also true for propofol 8 anesthesia. Comparison of rCPS in healthy controls and participants with FXS all under propofol anesthesia showed that rCPS was decreased by about 10% in whole brain and cortex of FXS subjects. 4 In studies of subjects anesthetized with dexmedetomidine, we found no statistically significant differences in rCPS between healthy controls and FXS subjects. 14 In the awake state, rCPS in subjects with FXS was decreased by 13–21% compared with healthy controls. Taken together these results suggest that sedation diminishes the effect of the absence of FMRP on rCPS (Supplement, Supplement Fig. S5). Our data suggest that when synapses are active (as in the awake state), mRNA translation is suppressed in the absence of FMRP, and such suppression does not occur in healthy controls. Under sedation when synaptic activity is reduced, effects of the absence on FMRP on translation are less apparent.
One possible explanation is an activation of the ISR in the absence of FMRP. General cap-dependent translation is suppressed in response to diverse forms of cellular stress through the phosphorylation of eIF2α. 32 eIF2α is part of the regulatory subcomplex of eIF2B, a guanine nucleotide exchange factor, that regulates the formation of the ternary complex (eIF2 · GTP · Met-tRNA) and therefore initiation of cap-dependent translation. Phosphorylation of eIF2α by one of several kinases generates the ISR which in addition to decreasing general translation also increases translation of a subset of transcripts containing upstream open reading frames (uORF). In a study of hippocampal slices incubated in amino acid replete conditions, synaptic activation by dihydroxyphenylglycine (DHPG) in the medium did not affect eIF2α in WT, but in slices from Fmr1 KO, DHPG treatment resulted in phosphorylation of eIF2α. 29 Consistent with these findings in the hippocampal slice is the observation in L-[1-11C]leucine PET studies that effects on rCPS in anesthetized subjects with FXS are smaller than effects in awake subjects.
FMRP is thought to act as a translation suppressor with its action primarily at the synapse. We posit that this could lead to an accumulation of synaptic proteins and an induction of the ISR with its consequent inhibition of formation of the ternary complex and suppression of general mRNA translation. In conditions of reduced synaptic activity as in sedation this cascade would be reduced and mRNA translation less affected.
Results of the present measurements of rates of mRNA translation in awake subjects with FXS challenge previously held ideas about the consequences of a lack of FMRP. Whereas there is ample evidence that FMRP can stall translation of specific messages, how this affects the regulation of overall translation rates in the intact nervous system was not previously appreciated. That the regulation of general translation is complex is inarguable. In nervous tissue in which translation is intimately involved in synaptic function and plasticity, it is imperative that its regulation be temporally and spatially precise. The absence of FMRP in human subjects results in decreases in overall rates of translation and in changes in sensitivity to activity state.
Supplemental Material
Supplemental material, sj-pdf-1-jcb-10.1177_0271678X221090997 for Decreased rates of cerebral protein synthesis in conscious young adults with fragile X syndrome demonstrated by L-[1-11C]leucine PET by Kathleen C Schmidt, Inna Loutaev, Thomas V Burlin, Audrey Thurm, Carrie Sheeler and Carolyn Beebe Smith in Journal of Cerebral Blood Flow & Metabolism
Acknowledgements
We gratefully acknowledge the participation of the NIH PET Department, in particular, M. Channing, K. Chung, W. Kong, S. Conant, and S. Thada. We also acknowledge the help of the NIH Department of Perioperative Medicine, in particular, A. Mannes and J. Labovsky. We acknowledge the help of R. Hommer, D. Picchioni, A. Morrow, K. Turetsky, B. Evans, D. Vesselinovitch, Z. Xia, and T. Huang during the studies and R. Saré for helpful advice on the manuscript.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This research was supported by the Intramural Research Program, National Institute of Mental Health.
Declaration of conflicting interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Authors’ contributions: KCS, AT, and CBS and made substantial contribution to the study design and data interpretation, and critically revised the article.
KCS, IL, TB, AT, CS, and CBS made substantial contributions to the data analysis.
KCS and CBS drafted the manuscript.
All authors approved the final draft submitted for publication.
Supplemental material: Supplemental material for this article is available online.
References
- 1.Liu ZH, Huang T, Smith CB. Lithium reverses increased rates of cerebral protein synthesis in a mouse model of fragile X syndrome. Neurobiol Dis 2012; 45: 1145–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Qin M, Huang T, Kader M, et al. R-Baclofen reverses a social behavior deficit and elevated protein synthesis in a mouse model of fragile X syndrome. IJNPPY 2015; 18: pyv034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Qin M, Kang J, Burlin TV, et al. Postadolescent changes in regional cerebral protein synthesis: an in vivo study in the FMR1 null mouse. J Neurosci 2005; 25: 5087–5095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Qin M, Schmidt KC, Zametkin Aj, et al. Altered cerebral protein synthesis in fragile X syndrome: studies in human subjects and knockout mice. J Cerebral Blood Flow Metab 2013; 33: 499–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bishu S, Schmidt KC, Burlin T, et al. Regional rates of cerebral protein synthesis measured with L-[1-11C]leucine and PET in conscious, young adult men: normal values, variability, and reproducibility. J Cereb Blood Flow Metab 2008; 28: 1502–1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schmidt KC, Cook MP, Qin M, et al. Measurement of regional rates of cerebral protein synthesis with L-[1-11C]leucine and PET with correction for recycling of tissue amino acids: I. Kinetic modeling approach. J Cereb Blood Flow Metab 2005; 25: 617–628. [DOI] [PubMed] [Google Scholar]
- 7.Smith CB, Schmidt KC, Qin M, et al. Measurement of regional rates of cerebral protein synthesis with L-[1-11C]leucine and PET with correction for recycling of tissue amino acids: II. Validation in rhesus monkeys. J Cereb Blood Flow Metab 2005; 25: 629–640. [DOI] [PubMed] [Google Scholar]
- 8.Bishu S, Schmidt KC, Burlin TV, et al. Propofol anesthesia does not alter regional rates of cerebral protein synthesis measured with L-[1-(11)C]leucine and PET in healthy male subjects. J Cereb Blood Flow Metab 2009; 29: 1035–1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Olmos-Serrano JL, Corbin JG, Burns MP. The GABA(A) receptor agonist THIP ameliorates specific behavioral deficits in the mouse model of fragile X syndrome. Dev Neurosci 2011; 33: 395–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Olmos-Serrano JL, Paluszkiewicz SM, Martin BS, et al. Defective GABAergic neurotransmission and pharmacological rescue of neuronal hyperexcitability in the amygdala in a mouse model of fragile X syndrome. J Neurosci 2010; 30: 9929–9938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Curia G, Papouin T, Seguela P, et al. Downregulation of tonic GABAergic inhibition in a mouse model of fragile X syndrome. Cereb Cortex 2009; 19: 1515–1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.D'Hulst C, De Geest N, Reeve SP, et al. Decreased expression of the GABAA receptor in fragile X syndrome. Brain Res 2006; 1121: 238–245. [DOI] [PubMed] [Google Scholar]
- 13.D'Hulst C, Heulens I, Van der Aa N, et al. Positron Emission Tomography (PET) quantification of GABAA receptors in the brain of fragile X patients. PLoS One 2015; 10: e0131486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schmidt KC, Loutaev I, Quezado Z, et al. Regional rates of brain protein synthesis are unaltered in dexmedetomidine sedated young men with fragile X syndrome: a L-[1-C-11]leucine PET study. Neurobiol Dis 2020; 143: 104978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tomasi G, Veronese M, Bertoldo A, et al. Effects of shortened scanning intervals on calculated regional rates of cerebral protein synthesis determined with the L-[1-11C]leucine PET method. PLoS One 2018; 13: e0195580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tomasi G, Veronese M, Bertoldo A, et al. Substitution of venous for arterial blood sampling in the determination of regional rates of cerebral protein synthesis with L-[1-(11)C]leucine PET: a validation study. J Cereb Blood Flow Metab 2019; 39: 1849–1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gibbon M, Spitzer RL, Williams JB, et al. Structured clinical interview for DSM-IV axis II personality disorders (SCID-II). 1997.
- 18.Wechsler D. Wechsler adult intelligence scale. Fourth edition. San Antonio, TX: Psychological Corporation, 2008. [Google Scholar]
- 19.Rutter M, LeCouteur AL, Lore C. Autism diagnostic Interview-Revised (ADI-R). LosAngeles, CA: Western Psychological Services, 2003. [Google Scholar]
- 20.Lord C, Rutter M, Dilavore PC, et al. Autism diagnostic observation schedule. Second edition (ADOS-2) manual (part 1): modules 1–4. Torrance, CA: Western Psychological Services, 2012.
- 21.Veronese M, Bertoldo A, Bishu S, et al. A spectral analysis approach for determination of regional rates of cerebral protein synthesis with the L-[1-(11)C]leucine PET method. J Cereb Blood Flow Metab 2010; 30: 1460–1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carson RE, Wu Y, Lang L, et al. Brain uptake of the acid metabolites of F-18-labeled WAY 100635 analogs. J Cereb Blood Flow Metab 2003; 23: 249–260. [DOI] [PubMed] [Google Scholar]
- 23.Bloomfield PM, Spinks TJ, Reed J, et al. The design and implementation of a motion correction scheme for neurological PET. Phys Med Biol 2003; 48: 959–978. [DOI] [PubMed] [Google Scholar]
- 24.Tomasi G, Bertoldo A, Bishu S, et al. Voxel-based estimation of kinetic model parameters of the L-[1-(11)C]leucine PET method for determination of regional rates of cerebral protein synthesis: validation and comparison with region-of-interest-based methods. J Cereb Blood Flow Metab 2009; 29: 1317–1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Veronese M, Bertoldo A, Tomasi G, et al. Impact of tissue kinetic heterogeneity on PET quantification: case study with the L-[1-(11)C]leucine PET method for cerebral protein synthesis rates. Sci Rep 2018; 8: 931–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hayward B, Loutaev I, Ding X, et al. Fragile X syndrome in a male with methylated premutation alleles and no detectable methylated full mutation alleles. Am J Med Genet A 2019; 179: 2132–2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dolen G, Osterweil E, Rao BS, et al. Correction of fragile X syndrome in mice. Neuron 2007; 56: 955–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Michalon A, Sidorov M, Ballard TM, et al. Chronic pharmacological mGlu5 inhibition corrects fragile X in adult mice. Neuron 2012; 74: 49–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cooke SK, Russin J, Moulton K, et al. Effects of the presence and absence of amino acids on translation, signaling, and long-term depression in hippocampal slices from Fmr1 knockout mice. J Neurochem 2019; 151: 764–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jacquemont S, Pacini L, Jonch AE, et al. Protein synthesis levels are increased in a subset of individuals with fragile X syndrome. Hum Mol Genet 2018; 27: 2039–2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Utami KH, Yusof N, Kwa JE, et al. Elevated de novo protein synthesis in FMRP-deficient human neurons and its correction by metformin treatment. Mol Autism 2020; 11: 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pain VM. Initiation of protein synthesis in eukaryotic cells. Eur J Biochem 1996; 236: 747–771. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-pdf-1-jcb-10.1177_0271678X221090997 for Decreased rates of cerebral protein synthesis in conscious young adults with fragile X syndrome demonstrated by L-[1-11C]leucine PET by Kathleen C Schmidt, Inna Loutaev, Thomas V Burlin, Audrey Thurm, Carrie Sheeler and Carolyn Beebe Smith in Journal of Cerebral Blood Flow & Metabolism