Abstract
The biosynthesis of unsaturated fatty acids is involved in the initiation and progression of colon adenocarcinoma (COAD). In this study, we aimed to investigate the multi-omics characteristics of unsaturated fatty acid biosynthesis-related genes and explore their prognostic value in colon cancer by analyzing the data from The Cancer Genome Atlas (TCGA) and Gene Expression Omnibus (GEO) databases. An unsaturated fatty acid biosynthesis pathway related-genes enrichment score (BUFAS) was constructed utilizing the single sample gene set enrichment analysis (ssGSEA). We discovered that a high BUFAS was associated with longer overall survival (OS) in both the training and the validation sets. Multivariable analysis including the clinical characteristics further verified the independent prognostic value of the BUFAS in both the TCGA-COAD and the GSE39582 datasets. In addition, GSEA analysis revealed that BUFAS was positively associated with several signaling pathways, including MTORC1, peroxisome, and pathways related to fatty acid metabolism, while was negatively associated with other signaling pathways, such as hedgehog, NOTCH, and Wnt/beta-catenin pathway. Furthermore, in the COAD cell lines of the Genomics of Drug Sensitivity in Cancer (GDSC) database, we found that BUFAS was positively correlated with the drug sensitivities of cisplatin, gemcitabine, camptothecin, lapatinib, and afatinib, while was negatively correlated with that of ponatinib. Moreover, in the COAD single-cell transcriptomic dataset (GSE146771), the BUFAS varied among different cell types and was enriched in mast cells and fibroblasts. Taken together, the BUFAS we constructed could be used as an independent prognostic signature in predicting the OS and drug resistance of colon cancer. Unsaturated fatty acid biosynthesis pathway might serve as potential therapeutic targets for cancer treatment.
Keywords: Unsaturated fatty acids biosynthesis, colon cancer, prognostic signature, overall survival, multi-omics characterization
Introduction
Colon cancer is one of the leading causes of cancer-related deaths worldwide, accounting for almost 10% of cancer mortality in 2020 [1]. In the newly diagnosed colon cancer patients, one in five has already developed metastasis, and more than a quarter of them will experience regional or distant metastases later even with the initial diagnosis of localized disease [2], attributing to the dismal clinical outcomes [3]. Thus, stratifying patients at the time of diagnosis is critical in selecting the most appropriate treatment options and cancer management. However, except the generally accepted clinical features such as age and tumor stage, biomarkers for predicting the prognosis of colon cancer are yet limited. Therefore, identifying novel and reliable signatures for improving the overall outcome of colon cancer are urgently needed.
Lipid metabolism is known to be involved in tumorigenesis. As the product of lipid metabolism, triacylglycerol is served as a substrate in energy metabolism to ensure adequate energy supply for tumor cell growth [4]. The biosynthesis of other lipids, such as cholesterol, phospholipids, and sphingolipids, provides indispensable materials for cytomembrane generation during cancer cell proliferation [5]. Hence, the abnormal lipid metabolism is a distinct feature of malignant cells [6], which is supported by the findings of altered lipid component in tumors, including breast [7], lung [8], renal [9], and liver cancers [10]. Consistently, the activated expression of genes encoding lipid synthesis-related enzymes has been found in colon cancer cells [11]. Furthermore, in vitro studies have demonstrated the potential of using lipid synthesis inhibitors in cancer therapy [12], suggesting the pivotal role of lipid metabolism in colon cancer.
Consistent with the importance of lipid metabolism in tumor initiation and progression, fatty acids, as one of the most important productions of lipid synthesis, have been suggested to associate with the cellular survival, malignancy, metastasis, and immune phenotype of different tumors, including glioma [13], cervical cancer [14], and myeloid leukemia [15]. In addition, extra intake of unsaturated fatty acids has been demonstrated to have significant benefits in cardiovascular diseases [16], inflammatory diseases [17], and diet-related tumors including colon cancer [18], whereas an excessive dietary intake of saturated fatty acids is an important risk factor for colon cancer [19]. Furthermore, unsaturated fatty acid supplementation helps sensitize the multi-drug resistant colon cancer cells to chemo treatment [20], although the functional mechanism of unsaturated fatty acids in colon cancer development remains elusive. Collectively, unsaturated fatty acids may have potential protective functions in colon cancer development, which provides the rationale for further investigations on the role of unsaturated fatty acid biosynthesis-related genes in tumorigenesis, which may provide new insights into colon cancer management.
Using public databases, including The Cancer Genome Atlas (TCGA) project and the Gene Expression Omnibus (GEO) database, we depicted the multi-omics characteristics of unsaturated fatty acid biosynthesis pathways in colon cancer, and further constructed a prognostic signature, which might reveal the potential molecular mechanisms underlying the function of unsaturated fatty acids in the development of colon cancer.
Methods and materials
Data collection
Three cohorts were included in this study. HTSeq-FPKM gene expression matrix of The Cancer Genome Atlas Colon Adenocarcinoma (TCGA-COAD) was obtained via the R package “TCGAbiolinks”, together with the survival and clinical information [21]. Mutation annotation format, copy number variation (CNV) information, and methylation sequencing results by Illumina HumanMethylation450 BeadChip (450K) were retrieved from UCSC XENA database (https://xenabrowser.net/). Survival and clinical of COAD samples in the GSE39582 cohort were acquired via the R package “GEOquery” [22]. Raw expression microarray “CEL” data files of the GSE39582 cohort were directly downloaded from the Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo), which was further normalized by the R package “affy” [23]. The transcriptomic data of single-cell sequencing (Smart-seq2) in colon cancer patients (GSE146771) were downloaded from the GEO database [24]. The unsaturated fatty acid biosynthesis-related gene set (hsa01040) was downloaded from the Kyoto Encyclopedia of Genes and Genomes (KEGG) database via the R package “KEGGREST”, and 27 genes involved in the biosynthesis of unsaturated fatty acid were obtained from the gene set. The 50 gene sets of cancer hallmark pathways were obtained from the Molecular Signature Database v.7.4 (MSigDB). In the TCGA-COAD and GSE39582 cohorts, patients were excluded if their survival status was unknown, or survival time was 0. Patients with both clinical information and expression profiling data were included for further analysis.
Multi-omics characterization of unsaturated fatty acid biosynthesis in colon cancer
The expression and methylation levels of the 27 genes related to the biosynthesis of unsaturated fatty acids were extracted from the expression matrix and methylation matrix, respectively. Mann-Whitney test was applied to evaluate the difference in expression and methylation levels of these 27 genes between normal and tumor tissues. The CNV amplification and deletion frequency of unsaturated fatty acid biosynthesis-related genes were extracted and calculated from the sequencing results, while the somatic single-nucleotide variation of these genes was calculated via the R package “maftools” based on sequencing results [25].
Development of a prognostic score via single sample gene set enrichment analysis (ssGSEA)
The unsaturated fatty acid biosynthesis pathway gene set enrichment score (BUFAS) of every sample was calculated via the R package “GSVA” through “ssGSEA” method in the TCGA-COAD cohort [26,27]. Samples were further divided into high- and low-BUFAS groups according to the median of BUFAS. Kaplan-Meier analysis was utilized to calculate the overall survival (OS), and the log-rank test was applied to compare the difference in survival curves between the high- and low-BUFAS groups through the R package “survival”. BUFAS as an independent prognostic factor was verified by univariable and multivariable Cox regression analyses, which was applied by the coxph function in the R package “survival”. Furthermore, model validation was conducted by applying the same methods to the GSE39582 cohort. The consensus molecular subtypes (CMSs) of colon cancer patients in both the TCGA-COAD and GSE39582 cohorts were assessed through the R package “CMScaller” [28]. The BUFAS of different clinical characteristics was compared to evaluate the association between the BUFAS and the clinical information of the TCGA-COAD cohort, as well as the estimated CMS subtypes.
Pathway correlation analysis
The enrichment scores for many hallmarks of cancer were calculated by the ssGSEA analysis based on the gene expression matrix in the TCGA-COAD cohort. The correlation between the BUFAS and 50 hallmarks was quantified by the Spearman’s correlation analysis according to their ssGSEA scores. The corAndPvalue function in the R package “WGCNA” was exploited to evaluate the correlation between each gene in the unsaturated fatty acid biosynthesis pathway gene set and 50 hallmarks of cancer based on single gene expression and the ssGSEA scores of these hallmarks [29]. GSEA analysis was conducted utilizing the java GSEA Desktop Application (http://software.broadinstitute.org/gsea/index.jsp) to associate the signaling pathway enrichment in c2.cp.kegg.v7.5.symbols.gmt in the BUFAS-high and BUFAS-low groups. The normalized enrichment score (NES) was examined to identify significant pathways with a P-value <0.05, which was adjusted by the false discovery rate (FDR) method.
Drug sensitivity analysis
The pRRophetic algorithm was originally exploited to predict drug response based on gene expression microarray data and the half maximal inhibitory concentration (IC50) information from the Genomics of Drug Sensitivity in Cancer (GDSC) database [30,31]. The R package “pRRophetic” was applied to estimate the drug sensitivity of 138 drugs based on the mRNA expression data of those samples in the TCGA-COAD cohort, which was shown as IC50 prediction. The correlation between the estimated drug sensitivity and the BUFAS was estimated for every drug through the Spearman’s correlation analysis, and a P<0.05 was considered statistically significant. Furthermore, the corAndPvalue function in the R package “WGCNA” was utilized for conducting the Pearson’s correlation analysis between the drug susceptibility and gene expression [29]. Genes that were potentially related to the drug response were selected by the Pearson’s correlation coefficient (r) with a P-value <0.05.
Single-cell transcriptomic analysis
Each single cell in the GSE146771 dataset was assigned with a BUFAS score by the ssGSEA method via the R package “GSVA”, together with the annotation by the Tumor Immune Single-cell Hub (TISCH) database (http://tisch.comp-genomics.org). Uniform manifold approximation and projection (UMAP) plots of the BUFAS for single cell in the GSE146771 dataset were displayed under two resolutions: “Cell type malignancy” and “Cell type major lineage”. The differences in the BUFAS among cell types were calculated by the Kruskal-Wallis test, and P-value <0.05 was considered statistically significant. Finally, the BUFAS in each cell type was compared with the others for calculation of the log2 Fold Change (log2FC), which was used to predict the cell subpopulations on which unsaturated fatty acid biosynthesis pathway mainly targeted.
Statistical analysis
Statistical analyses, as well as analyses that used various R packages, were all conducted in R (version 4.1.2). Visualization of the results was achieved by R and GraphPad Prism (version 9). The median value (range) and the exact number (percentage) were presented for continuous variables and categorical variables, respectively. The distribution of continuous variables was compared among different groups by the Wilcoxon test. The proportions of categorical variables were examined by the Chi-Square test or the Fisher’s exact test. Statistical significance was considered as P-value <0.05.
Results
Multi-omics characterization of unsaturated fatty acid biosynthesis-related genes in colon cancer
A total of 27 gene candidates involved in the unsaturated fatty acid biosynthesis were derived from the KEGG hsa01040 gene set (Supplementary Table 1). The expression and methylation levels of the unsaturated fatty acid biosynthesis-related genes were analyzed in 453 cancer and 41 normal samples from the TCGA-COAD dataset (Figure 1A and 1B). Since the methylation information was only available in 23 genes, the correlation between the gene expression and the methylation levels was only analyzed in these genes. Six genes showed negative regulation of the methylated modification in gene expression, including ACOT4 and ELOVL4 (hypermethylated and down-regulated in tumor tissues), and ACOT7, SCD, ELOVL3, and HSD17B12 (hypomethylated and up-regulated in tumor tissues), while an inverted correlation was found in another six genes, including FADS1, FADS2, and BAAT (hypermethylated and up-regulated in tumors), and ACOT1, ACOX1, and ACOX3 (hypomethylated and down-regulated in tumors), suggesting additional gene expression regulatory mechanisms other than methylation were involved. Furthermore, the CNV and mutational characteristics of these genes were also included in the multi-omics analyses. Copy number amplification was observed in ELOVL1 and ACOX1, while ACAA1, ACOT7, HSP17B14, ELOVL2, FADS2, SCD, and ELOVL3 exhibited significant copy number deletion (Figure 1C). Various gene mutations were detected in 77 (17.0%) tumor samples, and three genes, ELOVL2 (14%), ACOX1 (12%), and FADS2 (12%) exhibited the high mutational frequency (Figure 1D).
Figure 1.
Characterization of unsaturated fatty acid biosynthesis-related genes in the TCGA-COAD cohort. (A, B) Boxplots depicting the expression (A) and methylation status (B) of the genes involved in unsaturated fatty acid biosynthesis. (C, D) Histogram of the copy number variation (CNV) features (C) and oncoprint showing the mutation characteristics (D) of the unsaturated fatty acid biosynthesis-related genes in tumor tissues. *P<0.05; **P<0.01; ***P<0.001; and ****P<0.0001 by Mann-Whitney test.
Prognostic value of unsaturated fatty acid biosynthesis score in colon cancer
Based on the ssGSEA algorithm, an unsaturated fatty acid biosynthesis-related gene enrichment score (BUFAS) was calculated and assigned to each tumor sample in the TCGA-COAD dataset. To investigate the potential prognostic value of BUFAS, samples were divided into the high- and low-BUFAS groups by the median value for Kaplan-Meier analysis. In the TCGA-COAD cohort, a high BUFAS was associated with longer overall survival (OS, log-rank P=0.003; HR, 0.54; 95% CI, 0.35-0.81, Figure 2A). To verify the independent prognostic value of the BUFAS, univariable and multivariable analyses were performed on the clinical features, including age, sex, TNM stage, location of the primary tumor, microsatellite status, and consensus molecular subtype (CMS). Patients’ age and clinical stage exhibited statistical significance (P<0.05, Figure 2B) in the univariable models and were integrated into the following multivariable model, in which BUFAS remained to be significantly associated with OS (HR, 0.59, 95% CI, 0.38-0.90; P=0.01, Figure 2C), suggesting BUFAS as an independent prognostic marker in colon cancer.
Figure 2.
Prognostic value of BUFAS in COAD. (A) The overall survival (OS) of colon cancer patients compared between the high- and low-BUFAS groups, with the Kaplan-Meier survival curves showing the longer OS in patients with high-BUFAS (3042 vs. 1881 days). (B, C) The univariable (B) and multivariable (C) analyses of the BUFAS and the clinical characteristics in the TCGA-COAD dataset. (D) The OS of colon cancer patients compared between the high- and low-BUFAS groups, with the Kaplan-Meier survival curves showing the longer OS in patients with high-BUFAS (4350 vs. 3180 days). (E, F) The univariable (B) and multivariable (C) analyses of the BUFAS and clinical characteristics in the GSE39582 dataset. HR, hazard ratio; 95% CI, 95% confidence interval.
Importantly, similar results were obtained in the validation set (GSE39582). A high BUFAS was associated with longer OS (HR, 0.72, 95% CI, 0.54-0.96; P=0.03, Figure 2D), and, moreover, this prognostic effect remained significant in the multivariable model (HR, 0.73, 95% CI, 0.55-0.98; P=0.03, Figure 2E, 2F), confirming BUFAS as an independent prognostic factor for predicting the OS of colon cancer.
Association between BUFAS and clinical features in COAD
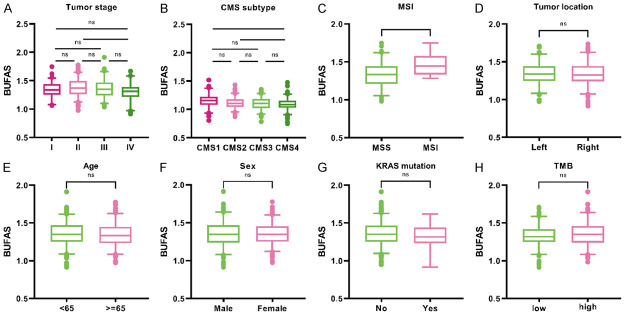
The BUFAS was compared with different clinical features, including age, sex, tumor site, TNM stage, CMS, microsatellite status, KRAS mutation status, and tumor mutational burden among patients (TMB, Figure 3). A relatively lower BUFAS was observed in the patients with stage IV (Figure 3A), CMS4 subtype (Figure 3B), and microsatellite stability (Figure 3C), while the BUFAS was not associated with tumor sites (Figure 3D), age (Figure 3E), sex (Figure 3F), KRAS mutations (Figure 3G), and TMB (Figure 3H).
Figure 3.
The association between BUFAS and clinical characteristics. (A-H) Boxplot showing the distribution of BUFAS in in different groups of TNM stages (A), consensus molecular subtypes (CMS, B), microsatellite status (C), tumor sites (D), age (E), sex (F), K-Ras mutations (G), and tumor mutational burden (TMB, H). ns (non-significant) P>0.05, *P<0.05; **P<0.01; ***P<0.001; and ****P<0.0001 by Wilcoxon test.
Functional mechanisms of unsaturated fatty acid biosynthesis
Considering that other cancer-related pathways may be involved in the unsaturated fatty acid biosynthesis, the correlation between the BUFAS and other pathways was determined on the samples in the TCGA-COAD cohort by assigning a group of cancer hallmark ssGSEA scores. We found that the BUFAS was positively associated with MTORC1 signaling, fatty acid metabolism, and peroxisome, while BUFAS was negatively correlated with such signaling pathways as hedgehog, NOTCH, and Wnt/beta-catentin (Figure 4A and Supplementary Table 2). Besides, all the genes constructing the BUFAS were analyzed. The samples with upregulated expression of HACD4 and ELOVL4 showed higher enrichment scores in the pathways of KRAS signaling upregulation, angiogenesis, and TGF-beta signaling, but showed lower enrichment scores in the pathways of DNA repair and MYC targets (Figure 4B). These results suggested that HACD4 and ELOVL4 might be the core genes in unsaturated fatty acid biosynthesis. In addition, GSEA analysis further revealed the prominent enrichment of signatures in the high-BUFAS group, which were related to oxidative phosphorylation, steroid biosynthesis, fatty acid metabolism, nucleotide excision repair, DNA replication, and mismatch repair (Figure 4C). Together, these data suggested the relationship between unsaturated fatty acid synthesis and other metabolism pathways, as well as the pathways related to authentic DNA replication.
Figure 4.
Potential cancer hallmarks associated with unsaturated fatty acids biosynthesis. (A) Volcano plot showing the cancer hallmark pathways that were most correlated with BUFAS score. (B) GSEA enrichment of KEGG pathways in the BUFAS-high group. (C) The association of cancer hallmark pathways with the genes involved in unsaturated fatty acid biosynthesis. *P<0.05; **P<0.01; ***P<0.001; and ****P<0.0001 by spearman correlation.
To explore the potential association between immune response and the BUFAS, the faction of 22 types of tumor-infiltrating immune cells, which reflected the immune microenvironment of tumor samples, was estimated by utilizing the R package “CIBERSORT”. A lower proportion of Treg cells and higher infiltration levels of activated CD4+ memory T cells and macrophage M1 were observed in the high-BUFAS groups. The proportions of the other 19 immune cell types were not significantly differed between the high- and the low-BUFAS groups (Supplementary Figure 1). Furthermore, among the 14 immune checkpoints, only the expression of TNFRSF9 was upregulated in the high-BUFAS group (Supplementary Figure 2). These findings suggested a weak association between unsaturated fatty acid biosynthesis and immunoregulation.
Drug sensitivity
To help in drug selection for cancer treatment, the association between the BUFAS and the drug sensitivity was evaluated using GDSC database. A total of 69 drugs were identified to have significantly different IC50 levels between the low- and high-BUFAS groups. We further analyzed the potential association between the BUFAS score and the IC50 of agents with analogues that are currently used for colon cancer treatment. Significant results were obtained as we found that the BUFAS was negatively associated with the IC50 of several chemotherapeutics, including cisplatin (analogues of oxaliplatin) (Figure 5A), gemcitabine (analogue of capecitabine) (Figure 5B), and camptothecin (analogue of irinotecan) (Figure 5C). In addition, increased sensitivity to Epidermal growth factor receptor (EGFR) tyrosine kinase inhibitors (TKIs) such as lapatinib (Figure 5D) and afatinib (BIBW2992) (Figure 5E) was associated with increased BUFAS, suggesting the beneficial effect of these drugs in patients with higher BUFAS. In contrast, the IC50 of ponatinib (AP.24534) (Figure 5F), a tumor angiogenesis inhibitor, exhibited a positive correlation with BUFAS, suggesting a better response to ponatinib in patients with lower BUFAS who had poor overall survival.
Figure 5.
Association between BUFAS and drug sensitivity. (A-F) The correlation between BUFAS of cell samples and the estimated IC50 value of medications, including cisplatin (A), gemcitabine (B), camptothecin (C), lapatinib (D), BIBW2992 (E), and AP.24534 (F). (G) The association of IC50 value estimated in 69 medications with the genes involved in unsaturated fatty acid production. *P<0.05; **P<0.01; ***P<0.001; and ****P<0.0001 by spearman correlation.
Moreover, we investigated the association between IC50 values and the expression of all the unsaturated fatty acid biosynthesis-related genes and identified a significant association between drug sensitivity and the expression of 8 genes: ACOT7, HACD2, ELOVL4, HACD4, ACOX1, SCP2, HSD17B12, and HACD3 (P<0.05 and |correlation| >0.5) (Figure 5G). These genes might be the potential core genes of unsaturated fatty acid biosynthesis involved in drug response.
Dysregulated unsaturated fatty acid biosynthesis
Since the dysregulation of unsaturated fatty acid biosynthesis plays important roles during oncogenesis, we explored the possible origin of abnormal unsaturated fatty acid biosynthesis by utilizing the single-cell sequencing dataset (GSE146771). Classification of the cell types was conducted based on the annotation information from the TISCH database, and the cells were classified as malignant cells, immune cells, and stromal cells (Figure 6A), or presented as 13 detailed subsets (Figure 6B). The expression of the unsaturated fatty acid biosynthesis-related genes was analyzed in these cells, and the BUFAS was assigned to each cell type by ssGSEA (Figure 6C). The BUFAS was different among different cell subsets (P<0.001), as the B cells and plasma cells exhibited lower BUFAS distribution, while the BUFAS score was higher in mast cells and fibroblasts (Figure 6D). A log2FC value of BUFAS distribution was calculated in each cell subset for enrichment analysis of the score, and a significant enrichment of BUFAS was observed in mast cells and fibroblasts, suggesting that the unsaturated fatty acid biosynthesis alteration might originate from or accumulate on specific cell types (Figure 6E).
Figure 6.
Biosynthesis of unsaturated fatty acids in single cell cohorts. (A, B) The tumor microenvironment (TMB) in the GSE146771 datasets identified by the single cell sequencing markers and the calculation of uniform manifold approximation and projection (UMAP). The cells are displayed in the malignant-immune-stromal sequencing (A) or more detailed subsets of TMB (B). (C) The BUFAS distribution of each cell sample in the GSE146771 dataset. (D) The Kuskal-Wallis test revealed a varied distribution of BUFAS among 13 cell subgroups. (E) Log2 fold change (log2FC) of BUFAS in cell subsets.
Discussion
As early as in the 1920s, the German physiologist Otto Warburg and his colleagues first reported metabolic alterations in tumors [32]. Over the past decade, the idea of cancer cell metabolic reprogramming has become one of the focuses in cancer research [33] and has been considered as one of the hallmarks of cancer by facilitating tumorigenesis [34-36]. In recent years, growing evidence has revealed that the cancer metabolism is complex with heterogeneity and plasticity to facilitate the rapid proliferation of cancer cells [37-39]. In support this, fatty acid metabolism is involved in many aspects of cancer cell biology. Fatty acid metabolism influences the synthesis of lipid building blocks of membrane and the signaling molecules. Fatty acids are also the substrate for ATP and NADH synthesis. Additionally, the acceleration of polysaturated fatty acid (PUFA) can induce ferroptosis in different cancer types [40-42]. All studies suggest that fatty acid synthesis and metabolism have multifaceted roles in cancer [43,44].
Cancer cells can acquire fatty acids through intracellular and extracellular sources under different circumstances. For example, fatty acid de novo synthesis uses non-lipid substrates to produce palmitate, which can be further converted into unsaturated fatty acids through desaturases and elongases, such as fatty acid desaturase 1 (FADS1) and very long-chain fatty acid protein 5 (ELOVL5). Based on the annotation information in KEGG database [45], a total of 27 genes were identified to be involved in the biosynthesis of unsaturated fatty acids, such as in the production of long-chain omega-3 fatty acid, eicosapentaenoic acid (EPA), docosahexaenoic acid (DHA). These genes, including acyl-CoA thioesterase (ACOT) [46], acyl-CoA oxidase (ACOX) [47], and elongation of very-long-chain-fatty acids (ELOVL) [48] gene families, regulate both lipid metabolism and unsaturated fatty acid biosynthesis; however, little is known about the integrated roles of these unsaturated fatty acid biosynthesis-related genes in the initiation and progression of colon cancer. To tackle these questions and further understand the multifaceted roles of unsaturated fatty acids in colon cancer, in this study, we developed a prognostic model, BUFAS, based on the 27 unsaturated fatty acid biosynthesis-related genes retrieved from the KEGG hsa01040 gene set by ssGSEA algorithm.
To date, this is the first study on developing a prognostic factor based on unsaturated fatty acid biosynthesis-related genes enrichment score in colon cancer. Although there were existing prognostic signatures based on different signaling pathways or biological molecules in colon cancer [49], most studies focused on immune-related biological functions. For example, a tumor microenvironment risk score (TMRS) was established by LASSO Cox regression in the GEO cohort, which was a robust tool not only for prognostic prediction but also for the tailored therapy in colon cancer [50]. Xu et al. identified 11 key immune-related genes and constructed a prognostic model by random forest algorithm. They further validated their model in an independent validation set and investigated the molecular mechanism of their model in a single-cell sequencing dataset, which proved that their model was accurate and reliable [51].
However, so far, very few prognostic models in colon cancer are based on the fatty acid synthesis or metabolism-related genes. Ding et al. extracted the differentially expressed fatty acid metabolism-related genes between colon cancer and normal tissues. They further developed a prognostic model based on these fatty acid-related genes using LASSO Cox regression analysis [52]. Nevertheless, given the multifaced roles and different categories of fatty acids, our study particularly focused on the function and prognostic value of unsaturated fatty acid biosynthesis-related genes in colon cancer. Previous studies have revealed the important roles of PUFA in different cancer types. For example, it has been reported that the biosynthesis of arachidonic acid (AA) and adrenic acid (AdA) by ELOVL5 and FADS1 may be a critical checkpoint in the ferroptosis pathway in gastric cancer [40]. This study supports the notion that the PUFA biosynthesis pathway plays an essential role in ferroptosis which may, in turn, predict the efficacy and prognosis after ferroptosis-mediated cancer therapy. In colon cancer, the intake of long-chain omega-3 fatty acid, a type of PUFA, could improve the DFS in patients with high COX2 expression [53]. Therefore, in our study, we constructed our prognostic factor based on the unsaturated fatty acid biosynthesis pathway-related gene enrichment score in the TCGA cohort and validated it in the GSE39582 cohort, which was proved to be an independent prognostic signature for the OS of colon cancer.
Pathway correlation analysis revealed that the unsaturated fatty acid biosynthesiswas strongly correlated with some important hallmarks of cancer, such as the peroxisome pathway and the mTOR complex 1 pathway. Specifically, in peroxisome pathway, Peroxisome Proliferator-Activated Receptor γ (PPARγ) is a nuclear receptor and is involved in regulating lipid metabolism-related gene expression. Several PUFAs are the natural ligands of this receptor [54,55]. Importantly, colon cancer patients with PPARγ expressing in the tumor tissues have better overall survival, which is consistent with our prognostic factor and correlation analysis results [56]. Furthermore, PUFA, such as those in fish oil, can activate PPARγ, and their consumption is associated with the prevention of colon cancer [57]. It has been reported that Eicosapentaenoic acid (EPA), one type of PUFA, can inhibit the growth of human colon cancer HT-29 cells, and this process may be associated with the downregulation of PI3K/Akt/mTOR signaling pathway [58]. Several genes in our gene set, such as ELVOL5 and FADS1, were also reported to be associated with ferroptosis in cancer, and PUFA is was reported to induce ferroptosis in various cancer cells [59-61]. According to these studies, we further investigated whether our BUFAS was associated with the ssGSEA score of the ferroptosis signaling pathway by using Spearman correlation analysis. We found that our BUFAS was positively associated with the ferroptosis pathway in colon cancer (Rho=0.35, P<0.001, Supplementary Figure 3).
The immune microenvironment is critically involved in the tumor formation, development, and therapeutic efficacy in various cancer types including colon cancer [62,63]. Therefore, the association of immune cell infiltration in tumor microenvironment with the BUFAS was evaluated in this study (Supplementary Figure 1). There was a significantly higher infiltration proportion of activated CD4+ memory T cells and macrophage M1 in the high-BUFAS group, while the fraction of Treg cells was lower in the high-BUFAS group. Interestingly, a low intraepithelial CD3+/FoxP3+ cell ratio is associated with poor survival in patients with colon cancer, which is consistent with our results [64]. Finally, our analyses into the previous single-cell sequencing dataset indicated that the biosynthesis of the unsaturated fatty acid pathway might intervene in the behaviors and activities of mast cells and macrophages in colon cancer, which is in consistent with the findings of increased M1 macrophages infiltration in the high-BUFAS group.
There are several limitations in our study. First, we developed our BUFAS prognostic factor via ssGSEA to estimate the overall enrichment score of a certain gene set, which made it difficult to identify key genes of prognostic value. Second, the analyses were mainly conducted under in silico circumstances, which needs further experimental validation and clinical validation. Finally, since the signature was constructed in retrospective cohorts, further validation on its performance in prospective studies should be conducted.
Conclusions
In this study, a novel unsaturated fatty acid biosynthesis-related signature was constructed as an independent prognostic signature in COAD. This signature was correlated with the sensitivity to chemotherapeutic drugs and was identified to originate from mast cells and macrophages. These results may provide rationale for the stratification and therapeutic treatment selection in the management of colon cancer.
Acknowledgements
This work was supported by the Youth Scientific Research Project of Fujian Provincial Health, Family Planning Commission (grant number 2018-2-5) and the Sail Fund of Fujian Medical University (grant number 2017XQ1151), and Foundation of 2020 Fujian Provincial Department of Finance Health and Health Provincial Special Subsidy.
Disclosure of conflict of interest
None.
Abbreviations
- TCGA
The Cancer Genome Atlas
- GEO
Gene Expression 21 Omnibus database
- BUFAS
unsaturated fatty acid biosynthesis pathway gene set enrichment score (BUFAS)
- ssGSEA
single sample gene set enrichment analysis
- OS
overall survival
- IC50
half maximal inhibitory concentration
- CMS
consensus molecular subtypes
- NES
normalized enrichment score
- FDR
false discovery rate
- GDSC
Genomics of Drug Sensitivity in Cancer database
- TISCH
Tumor Immune Single-cell Hub database
- UMAP
Uniform manifold approximation and projection
- CNV
copy number variations
- TMB
tumor mutational burden
- EGFR
Epidermal growth factor receptor
- TKIs
tyrosine kinase inhibitors
- VEGFR
vascular endothelial growth factor receptor
- PUFA
polysaturated fatty acids
- PPARγ
Peroxisome Proliferator Activated Receptor γ
Supporting Information
References
- 1.Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Biller LH, Schrag D. Diagnosis and treatment of metastatic colorectal cancer: a review. JAMA. 2021;325:669–685. doi: 10.1001/jama.2021.0106. [DOI] [PubMed] [Google Scholar]
- 3.Sagaert X, Vanstapel A, Verbeek S. Tumor heterogeneity in colorectal cancer: what do we know so far? Pathobiology. 2018;85:72–84. doi: 10.1159/000486721. [DOI] [PubMed] [Google Scholar]
- 4.Santos CR, Schulze A. Lipid metabolism in cancer. FEBS J. 2012;279:2610–2623. doi: 10.1111/j.1742-4658.2012.08644.x. [DOI] [PubMed] [Google Scholar]
- 5.Matsushita Y, Nakagawa H, Koike K. Lipid metabolism in oncology: why it matters, how to research, and how to treat. Cancers (Basel) 2021;13:474. doi: 10.3390/cancers13030474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vriens K, Christen S, Parik S, Broekaert D, Yoshinaga K, Talebi A, Dehairs J, Escalona-Noguero C, Schmieder R, Cornfield T, Charlton C, Romero-Pérez L, Rossi M, Rinaldi G, Orth MF, Boon R, Kerstens A, Kwan SY, Faubert B, Méndez-Lucas A, Kopitz CC, Chen T, Fernandez-Garcia J, Duarte JAG, Schmitz AA, Steigemann P, Najimi M, Hägebarth A, Van Ginderachter JA, Sokal E, Gotoh N, Wong KK, Verfaillie C, Derua R, Munck S, Yuneva M, Beretta L, DeBerardinis RJ, Swinnen JV, Hodson L, Cassiman D, Verslype C, Christian S, Grünewald S, Grünewald TGP, Fendt SM. Evidence for an alternative fatty acid desaturation pathway increasing cancer plasticity. Nature. 2019;566:403–406. doi: 10.1038/s41586-019-0904-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cífková E, Holčapek M, Lísa M, Vrána D, Gatěk J, Melichar B. Determination of lipidomic differences between human breast cancer and surrounding normal tissues using HILIC-HPLC/ESI-MS and multivariate data analysis. Anal Bioanal Chem. 2015;407:991–1002. doi: 10.1007/s00216-014-8272-z. [DOI] [PubMed] [Google Scholar]
- 8.Eggers LF, Müller J, Marella C, Scholz V, Watz H, Kugler C, Rabe KF, Goldmann T, Schwudke D. Lipidomes of lung cancer and tumour-free lung tissues reveal distinct molecular signatures for cancer differentiation, age, inflammation, and pulmonary emphysema. Sci Rep. 2017;7:11087. doi: 10.1038/s41598-017-11339-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Melana JP, Mignolli F, Stoyanoff T, Aguirre MV, Balboa MA, Balsinde J, Rodriguez JP. The hypoxic microenvironment induces stearoyl-CoA desaturase-1 overexpression and lipidomic profile changes in clear cell renal cell carcinoma. Cancers (Basel) 2021;13:2962. doi: 10.3390/cancers13122962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nakagawa H, Hayata Y, Kawamura S, Yamada T, Fujiwara N, Koike K. Lipid metabolic reprogramming in hepatocellular carcinoma. Cancers (Basel) 2018;10:447. doi: 10.3390/cancers10110447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zaytseva YY, Harris JW, Mitov MI, Kim JT, Butterfield DA, Lee EY, Weiss HL, Gao T, Evers BM. Increased expression of fatty acid synthase provides a survival advantage to colorectal cancer cells via upregulation of cellular respiration. Oncotarget. 2015;6:18891–18904. doi: 10.18632/oncotarget.3783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Peck B, Schug ZT, Zhang Q, Dankworth B, Jones DT, Smethurst E, Patel R, Mason S, Jiang M, Saunders R, Howell M, Mitter R, Spencer-Dene B, Stamp G, McGarry L, James D, Shanks E, Aboagye EO, Critchlow SE, Leung HY, Harris AL, Wakelam MJO, Gottlieb E, Schulze A. Inhibition of fatty acid desaturation is detrimental to cancer cell survival in metabolically compromised environments. Cancer Metab. 2016;4:6. doi: 10.1186/s40170-016-0146-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Qi Y, Chen D, Lu Q, Yao Y, Ji C. Bioinformatic profiling identifies a fatty acid metabolism-related gene risk signature for malignancy, prognosis, and immune phenotype of glioma. Dis Markers. 2019;2019:3917040. doi: 10.1155/2019/3917040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang C, Liao Y, Liu P, Du Q, Liang Y, Ooi S, Qin S, He S, Yao S, Wang W. FABP5 promotes lymph node metastasis in cervical cancer by reprogramming fatty acid metabolism. Theranostics. 2020;10:6561–6580. doi: 10.7150/thno.44868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tabe Y, Konopleva M, Andreeff M. Fatty acid metabolism, bone marrow adipocytes, and AML. Front Oncol. 2020;10:155. doi: 10.3389/fonc.2020.00155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chang CL, Deckelbaum RJ. Omega-3 fatty acids: mechanisms underlying ‘protective effects’ in atherosclerosis. Curr Opin Lipidol. 2013;24:345–350. doi: 10.1097/MOL.0b013e3283616364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Simopoulos AP. Omega-3 fatty acids in inflammation and autoimmune diseases. J Am Coll Nutr. 2002;21:495–505. doi: 10.1080/07315724.2002.10719248. [DOI] [PubMed] [Google Scholar]
- 18.Maskrey BH, Megson IL, Rossi AG, Whitfield PD. Emerging importance of omega-3 fatty acids in the innate immune response: molecular mechanisms and lipidomic strategies for their analysis. Mol Nutr Food Res. 2013;57:1390–1400. doi: 10.1002/mnfr.201200723. [DOI] [PubMed] [Google Scholar]
- 19.Bairey O, Shaklai M, Inbal A. Haemarthrosis in patients with mild coagulation factor deficiency. Blood Coagul Fibrinolysis. 1991;2:669–671. doi: 10.1097/00001721-199110000-00014. [DOI] [PubMed] [Google Scholar]
- 20.Gelsomino G, Corsetto PA, Campia I, Montorfano G, Kopecka J, Castella B, Gazzano E, Ghigo D, Rizzo AM, Riganti C. Omega 3 fatty acids chemosensitize multidrug resistant colon cancer cells by down-regulating cholesterol synthesis and altering detergent resistant membranes composition. Mol Cancer. 2013;12:137. doi: 10.1186/1476-4598-12-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Colaprico A, Silva TC, Olsen C, Garofano L, Cava C, Garolini D, Sabedot TS, Malta TM, Pagnotta SM, Castiglioni I, Ceccarelli M, Bontempi G, Noushmehr H. TCGAbiolinks: an R/Bioconductor package for integrative analysis of TCGA data. Nucleic Acids Res. 2016;44:e71. doi: 10.1093/nar/gkv1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Davis S, Meltzer PS. GEOquery: a bridge between the Gene Expression Omnibus (GEO) and BioConductor. Bioinformatics. 2007;23:1846–1847. doi: 10.1093/bioinformatics/btm254. [DOI] [PubMed] [Google Scholar]
- 23.Gautier L, Cope L, Bolstad BM, Irizarry RA. Affy--analysis of Affymetrix GeneChip data at the probe level. Bioinformatics. 2004;20:307–315. doi: 10.1093/bioinformatics/btg405. [DOI] [PubMed] [Google Scholar]
- 24.Zhang L, Li Z, Skrzypczynska KM, Fang Q, Zhang W, O’Brien SA, He Y, Wang L, Zhang Q, Kim A, Gao R, Orf J, Wang T, Sawant D, Kang J, Bhatt D, Lu D, Li CM, Rapaport AS, Perez K, Ye Y, Wang S, Hu X, Ren X, Ouyang W, Shen Z, Egen JG, Zhang Z, Yu X. Single-cell analyses inform mechanisms of myeloid-targeted therapies in colon cancer. Cell. 2020;181:442–459. e429. doi: 10.1016/j.cell.2020.03.048. [DOI] [PubMed] [Google Scholar]
- 25.Mayakonda A, Lin DC, Assenov Y, Plass C, Koeffler HP. Maftools: efficient and comprehensive analysis of somatic variants in cancer. Genome Res. 2018;28:1747–1756. doi: 10.1101/gr.239244.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hänzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-Seq data. BMC Bioinformatics. 2013;14:7. doi: 10.1186/1471-2105-14-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Barbie DA, Tamayo P, Boehm JS, Kim SY, Moody SE, Dunn IF, Schinzel AC, Sandy P, Meylan E, Scholl C, Fröhling S, Chan EM, Sos ML, Michel K, Mermel C, Silver SJ, Weir BA, Reiling JH, Sheng Q, Gupta PB, Wadlow RC, Le H, Hoersch S, Wittner BS, Ramaswamy S, Livingston DM, Sabatini DM, Meyerson M, Thomas RK, Lander ES, Mesirov JP, Root DE, Gilliland DG, Jacks T, Hahn WC. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature. 2009;462:108–112. doi: 10.1038/nature08460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Eide PW, Bruun J, Lothe RA, Sveen A. CMScaller: an R package for consensus molecular subtyping of colorectal cancer pre-clinical models. Sci Rep. 2017;7:16618. doi: 10.1038/s41598-017-16747-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Geeleher P, Cox NJ, Huang RS. Clinical drug response can be predicted using baseline gene expression levels and in vitrodrug sensitivity in cell lines. Genome Biol. 2014;15:R47. doi: 10.1186/gb-2014-15-3-r47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Geeleher P, Cox N, Huang RS. pRRophetic: an R package for prediction of clinical chemotherapeutic response from tumor gene expression levels. PLoS One. 2014;9:e107468. doi: 10.1371/journal.pone.0107468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Warburg O, Wind F, Negelein E. The metabolism of tumors in the body. J Gen Physiol. 1927;8:519–530. doi: 10.1085/jgp.8.6.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Faubert B, Solmonson A, DeBerardinis RJ. Metabolic reprogramming and cancer progression. Science. 2020;368:eaaw5473. doi: 10.1126/science.aaw5473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pavlova Natalya N, Thompson Craig B. The emerging hallmarks of cancer metabolism. Cell Metab. 2016;23:27–47. doi: 10.1016/j.cmet.2015.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhu J, Thompson CB. Metabolic regulation of cell growth and proliferation. Nat Rev Mol Cell Biol. 2019;20:436–450. doi: 10.1038/s41580-019-0123-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 37.Fendt SM, Frezza C, Erez A. Targeting metabolic plasticity and flexibility dynamics for cancer therapy. Cancer Discov. 2020;10:1797–1807. doi: 10.1158/2159-8290.CD-20-0844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Martinez-Reyes I, Chandel NS. Cancer metabolism: looking forward. Nat Rev Cancer. 2021;21:669–680. doi: 10.1038/s41568-021-00378-6. [DOI] [PubMed] [Google Scholar]
- 39.Wu H, Han Y, Rodriguez Sillke Y, Deng H, Siddiqui S, Treese C, Schmidt F, Friedrich M, Keye J, Wan J, Qin Y, Kuhl AA, Qin Z, Siegmund B, Glauben R. Lipid droplet-dependent fatty acid metabolism controls the immune suppressive phenotype of tumor-associated macrophages. EMBO Mol Med. 2019;11:e10698. doi: 10.15252/emmm.201910698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee JY, Nam M, Son HY, Hyun K, Jang SY, Kim JW, Kim MW, Jung Y, Jang E, Yoon SJ, Kim J, Kim J, Seo J, Min JK, Oh KJ, Han BS, Kim WK, Bae KH, Song J, Kim J, Huh YM, Hwang GS, Lee EW, Lee SC. Polyunsaturated fatty acid biosynthesis pathway determines ferroptosis sensitivity in gastric cancer. Proc Natl Acad Sci U S A. 2020;117:32433–32442. doi: 10.1073/pnas.2006828117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Blomme A, Ford CA, Mui E, Patel R, Ntala C, Jamieson LE, Planque M, McGregor GH, Peixoto P, Hervouet E, Nixon C, Salji M, Gaughan L, Markert E, Repiscak P, Sumpton D, Blanco GR, Lilla S, Kamphorst JJ, Graham D, Faulds K, MacKay GM, Fendt SM, Zanivan S, Leung HY. 2,4-dienoyl-CoA reductase regulates lipid homeostasis in treatment-resistant prostate cancer. Nat Commun. 2020;11:2508. doi: 10.1038/s41467-020-16126-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Perez MA, Magtanong L, Dixon SJ, Watts JL. Dietary lipids induce ferroptosis in caenorhabditiselegans and human cancer cells. Dev Cell. 2020;54:447–454. e444. doi: 10.1016/j.devcel.2020.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hoy AJ, Nagarajan SR, Butler LM. Tumour fatty acid metabolism in the context of therapy resistance and obesity. Nat Rev Cancer. 2021;21:753–766. doi: 10.1038/s41568-021-00388-4. [DOI] [PubMed] [Google Scholar]
- 44.Rohrig F, Schulze A. The multifaceted roles of fatty acid synthesis in cancer. Nat Rev Cancer. 2016;16:732–749. doi: 10.1038/nrc.2016.89. [DOI] [PubMed] [Google Scholar]
- 45.Kanehisa M, Sato Y, Kawashima M, Furumichi M, Tanabe M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 2016;44:D457–462. doi: 10.1093/nar/gkv1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xie X, Chen C, Feng S, Zuo S, Zhao X, Li H. Acyl-CoA thioesterase 7 is transcriptionally activated by kruppel-like factor 13 and promotes the progression of hepatocellular carcinoma. J Hepatocell Carcinoma. 2021;8:1623–1641. doi: 10.2147/JHC.S338353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Perfilyev A, Dahlman I, Gillberg L, Rosqvist F, Iggman D, Volkov P, Nilsson E, Riserus U, Ling C. Impact of polyunsaturated and saturated fat overfeeding on the DNA-methylation pattern in human adipose tissue: a randomized controlled trial. Am J Clin Nutr. 2017;105:991–1000. doi: 10.3945/ajcn.116.143164. [DOI] [PubMed] [Google Scholar]
- 48.Ofman R, Dijkstra IM, van Roermund CW, Burger N, Turkenburg M, van Cruchten A, van Engen CE, Wanders RJ, Kemp S. The role of ELOVL1 in very long-chain fatty acid homeostasis and X-linked adrenoleukodystrophy. EMBO Mol Med. 2010;2:90–97. doi: 10.1002/emmm.201000061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ahluwalia P, Kolhe R, Gahlay GK. The clinical relevance of gene expression based prognostic signatures in colorectal cancer. Biochim Biophys Acta Rev Cancer. 2021;1875:188513. doi: 10.1016/j.bbcan.2021.188513. [DOI] [PubMed] [Google Scholar]
- 50.Zhou R, Zeng D, Zhang J, Sun H, Wu J, Li N, Liang L, Shi M, Bin J, Liao Y, Huang N, Liao W. A robust panel based on tumour microenvironment genes for prognostic prediction and tailoring therapies in stage I-III colon cancer. EBioMedicine. 2019;42:420–430. doi: 10.1016/j.ebiom.2019.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xu J, Dai S, Jiang K, Xiao Q, Yuan Y, Ding K. Combining single-cell sequencing to identify key immune genes and construct the prognostic evaluation model for colon cancer patients. Clin Transl Med. 2021;11:e465. doi: 10.1002/ctm2.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ding C, Shan Z, Li M, Chen H, Li X, Jin Z. Characterization of the fatty acid metabolism in colorectal cancer to guide clinical therapy. Mol Ther Oncolytics. 2021;20:532–544. doi: 10.1016/j.omto.2021.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Blarigan EV, Fuchs CS, Niedzwiecki D, Ye X, Zhang S, Song M, Saltz L, Mayer RJ, Mowat RB, Whittom R, Hantel A, Benson AB, Atienza DM, Messino MJ, Kindler HL, Venook AP, Ogino S, Willett WC, Giovannucci EL, Meyerhardt JA. Long-chain omega-3 fatty acid and fish intake after colon cancer diagnosis and disease-free, recurrence-free, and overall survival in CALGB 89803 (Alliance) 2017;35:585. [Google Scholar]
- 54.Koeffler HP. Peroxisome proliferator-activated receptor γ and cancers. Clin Cancer Res. 2003;9:1. [PubMed] [Google Scholar]
- 55.Forman BM, Chen J, Evans RM. Hypolipidemic drugs, polyunsaturated fatty acids, and eicosanoids are ligands for peroxisome proliferator-activated receptors alpha and delta. Proc Natl Acad Sci U S A. 1997;94:4312–4317. doi: 10.1073/pnas.94.9.4312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ogino S, Shima K, Baba Y, Nosho K, Irahara N, Kure S, Chen L, Toyoda S, Kirkner GJ, Wang YL, Giovannucci EL, Fuchs CS. Colorectal cancer expression of peroxisome proliferator-activated receptor gamma (PPARG, PPARgamma) is associated with good prognosis. Gastroenterology. 2009;136:1242–1250. doi: 10.1053/j.gastro.2008.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kliewer SA, Sundseth SS, Jones SA, Brown PJ, Wisely GB, Koble CS, Devchand P, Wahli W, Willson TM, Lenhard JM, Lehmann JM. Fatty acids and eicosanoids regulate gene expression through direct interactions with peroxisome proliferator-activated receptors alpha and gamma. Proc Natl Acad Sci U S A. 1997;94:4318–4323. doi: 10.1073/pnas.94.9.4318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tang FY, Cho HJ, Pai MH, Chen YH. Concomitant supplementation of lycopene and eicosapentaenoic acid inhibits the proliferation of human colon cancer cells. J Nutr Biochem. 2009;20:426–434. doi: 10.1016/j.jnutbio.2008.05.001. [DOI] [PubMed] [Google Scholar]
- 59.Dierge E, Debock E, Guilbaud C, Corbet C, Mignolet E, Mignard L, Bastien E, Dessy C, Larondelle Y, Feron O. Peroxidation of n-3 and n-6 polyunsaturated fatty acids in the acidic tumor environment leads to ferroptosis-mediated anticancer effects. Cell Metab. 2021;33:1701–1715. e1705. doi: 10.1016/j.cmet.2021.05.016. [DOI] [PubMed] [Google Scholar]
- 60.Stockwell BR, Friedmann Angeli JP, Bayir H, Bush AI, Conrad M, Dixon SJ, Fulda S, Gascón S, Hatzios SK, Kagan VE, Noel K, Jiang X, Linkermann A, Murphy ME, Overholtzer M, Oyagi A, Pagnussat GC, Park J, Ran Q, Rosenfeld CS, Salnikow K, Tang D, Torti FM, Torti SV, Toyokuni S, Woerpel KA, Zhang DD. Ferroptosis: a regulated cell death nexus linking metabolism, redox biology, and disease. Cell. 2017;171:273–285. doi: 10.1016/j.cell.2017.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zou Y, Henry WS, Ricq EL, Graham ET, Phadnis VV, Maretich P, Paradkar S, Boehnke N, Deik AA, Reinhardt F, Eaton JK, Ferguson B, Wang W, Fairman J, Keys HR, Dančík V, Clish CB, Clemons PA, Hammond PT, Boyer LA, Weinberg RA, Schreiber SL. Plasticity of ether lipids promotes ferroptosis susceptibility and evasion. Nature. 2020;585:603–608. doi: 10.1038/s41586-020-2732-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schmitt M, Greten FR. The inflammatory pathogenesis of colorectal cancer. Nat Rev Immunol. 2021;21:653–667. doi: 10.1038/s41577-021-00534-x. [DOI] [PubMed] [Google Scholar]
- 63.Ganesh K, Stadler ZK, Cercek A, Mendelsohn RB, Shia J, Segal NH, Diaz LA Jr. Immunotherapy in colorectal cancer: rationale, challenges and potential. Nat Rev Gastroenterol Hepatol. 2019;16:361–375. doi: 10.1038/s41575-019-0126-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sinicrope FA, Rego RL, Ansell SM, Knutson KL, Foster NR, Sargent DJ. Intraepithelial effector (CD3+)/regulatory (FoxP3+) T-cell ratio predicts a clinical outcome of human colon carcinoma. Gastroenterology. 2009;137:1270–1279. doi: 10.1053/j.gastro.2009.06.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.