Abstract
Coagulation regulates angiogenesis in cancer, and is associated with tumor development and metastasis. To date, there have been no studies quantifying the state of intra-tumoral coagulation. We measured intra-tumoral coagulation gene expression using the “Hallmark-COAGULATION” gene set in the MSigDB, performing gene set variation analysis and then assigning a “coagulation score” to quantify gene expression. Clinical, histologic, and genetic data were analyzed in 807 gastric cancer patients from the TCGA_STAD and GSE84437 databases. Tumors with increased expression of pro-coagulation genes were consistently associated with higher AJCC T-categories (p = 0.018), lymph node metastasis (p = 0.036), and stage (p = 0.006) in both cohorts. Patients with high coagulation scores were found to have worse disease-specific survival and overall survival (OS) (p = 0.019 and 0.011, respectively) in TCGA, and worse OS in GSE84437 cohort (p = 0.012). Higher expression of pro-coagulation genes correlated with increased intra-tumoral angiogenesis, as well as increased proportions of lymphatic and microvascular endothelial cells, endothelial cells, and pericytes, calculated by xCell algorithm. High coagulation scores were significantly associated with low tumor mutation burden, but not with intratumor heterogeneity and homologous recombination deficiency. Gastric cancers with high coagulation scores contained higher amounts of M1 macrophages and dendritic cells, and low numbers of Th1 cells (all P<0.001). Genes for epithelial mesenchymal transition (EMT), myogenesis, apical junction, transforming growth factor (TGF)-β signaling, and angiogenesis were enriched in high coagulation score-gastric cancers (all false discovery rate <0.25). In conclusion, gastric cancers expressing higher levels of pro-coagulation genes demonstrate increased angiogenesis, EMT, TGF-β signaling and worse patient prognosis.
Keywords: Coagulation, gastric cancer, prognosis, signaling, angiogenesis, EMT
Introduction
Hypercoagulable state is the second leading cause of death among patients with advanced cancer, only after organ failure by metastatic disease [1]. It is also known as cancer-associated thrombosis and may manifest as Trousseau’s syndrome (thrombophlebitis migrans) or thromboembolic events such as venous thromboembolism (VTE) or stroke. The mechanism of cancer-associated thrombosis is not fully understood, but there are several known interactions between tumor cells and host cells that influence the host coagulation cascade [2].
Interestingly, several recent studies have demonstrated that the reverse of this relationship is also true, and that coagulation factors also promote tumor metastasis and cancer progression via various proposed pathways [3,4]. For example, platelets contribute to metastasis by forming a physical shield around tumor cells which protects them from host natural killer (NK) cells. Platelets have also been shown to promote an epithelial to mesenchymal transition (EMT) in tumor cells, which is associated with an invasive phenotype [5,6]. Further, thrombin has been shown to increase cancer proliferation, migration and angiogenesis in preclinical models [7,8]. Cancer cells promote the secretion of von Willebrand Factor from blood endothelial cells [9,10], which also has been reported to contribute to the process of EMT [11]. With that said, these findings were made in in vitro and in vivo systems, and their clinical relevance remains unclear.
Gene set variation analysis (GSVA) is a well-validated method of characterizing gene expression by organizing genes into sets according to functional pathways, rather than measuring activity of individual genes [12]. This method provides a relevant clinical context for interpreting gene expression data, and our group has extensive experience using the GSVA algorithm to predict clinical outcomes using large transcriptome databases. For example, we found that increased expression of the G2M checkpoint pathway genes in pancreatic cancer is predictive of treatment response and overall survival [13-15]. In patients with hepatocellular carcinoma we found that tumor growth and patient survival correlated with unfolded protein response signaling, which has been implicated in tumorigenesis [16]. Currently, GSVA scoring is widely used to estimate the activity of several pathways in cancer research [17-20].
Here, we hypothesize that higher intra-tumoral expression of pro-coagulation genes predicts increased angiogenesis, cancer cell proliferation, EMT, and poorer survival in gastric cancer patients.
Materials and methods
Patients
Transcriptomic as well as clinical demographic data of The Cancer Genome Atlas Stomach Adenocarcinoma project (TCGA-STAD, n = 375) was obtained through The Genomic Data Commons portal [21,22]. The Pan-Cancer Clinical Data Resource, which is a standardized dataset of clinical outcomes of patients included in TCGA, was used to obtain information on survival endpoints [23]. A second cohort, GSE84437 (n = 432, Yoon et al.) [24] was obtained from the Gene Expression Omnibus (GEO) repository and used to validate our findings. All analyses were performed using log2-transformed gene expression.
Coagulation score
We utilized the gene set “HALLMARK_COAGULATION” from the Molecular Signatures Database (MSigDB) hallmark collection [25], which is comprised of genes encoding components of blood coagulation, and calculated a “coagulation score” using GSVA as published previously [15,19,20,26,27]. A high score represents increased expression of pro-coagulation genes. The median of the score was used as a cut-off to split high vs. low score groups.
Hallmark pathway gene set enrichment analysis
The Gene Set Enrichment Analysis (GSEA) Java application (vers. 4.0) was used to analyze Hallmark collection gene sets in the Molecular Signatures Database (MSigDB). GSEA [28] was used to identify differences in gene expression between the high- and low-score groups as we previously described [29-34]. A false discovery rate (FDR) <0.25 was chosen as the cutoff indicative of statistical significance, as recommended for the GSEA method.
Tumor immune microenvironment analysis
xCell software was used to quantify the relative abundance of stromal and immune cells in each sample from their transcriptome profile [35], as we previously reported [36-41].
Other statistical analyses
R software (version 4.0.2) was used for all analyses and figures generated in this study. Each boxplot illustrates the median and interquartile range (IQR). Kruskal-Wallis and Mann Whitney U tests were used for group comparisons, and Spearman’s correlation coefficient was used for two factor comparisons. Groups were separated into “high” vs. “low” scores relative to the median score. Survival curves were plotted using the Kaplan-Meier method with log-rank test. P-values <0.05 were regarded as statistically significant.
Results
Increased expression of pro-coagulation genes predicts advanced gastric cancer stage and poorer prognosis
Transcriptomic data from two large gastric cancer cohorts, TCGA and GSE84437, were analyzed using GSVA, as we have previously described [22]. First, we studied the relationship between the intra-tumoral expression of pro-coagulation genes and clinical aggressiveness, using American Joint Committee on Cancer (AJCC) T-category/N-category/stage, pathological grade, and patient survival in two independent cohorts, TCGA and GSE84437. In TCGA, tumors with high coagulation scores had advanced AJCC T-category and increased likelihood of lymph node metastasis (Figure 1A, P = 0.018 and 0.036, respectively). This association was validated by GSE84437 (P = 0.006 and P<0.001, respectively). High scores consistently predicted advanced stage in TCGA (P = 0.006) as well, but did not correlate with pathological grade (P = 0.753).
Figure 1.

Relationship between the coagulation score and clinical cancer aggressiveness. A. Boxplots of the coagulation score by AJCC T- and N-category, in the TCGA and GSE84437, and AJCC stage and Nottingham histological grade in the TCGA cohort. Kruskal-Wallis and Mann Whitney U tests were used to determine p values. B. Kaplan-Meier curves of disease specific (DSS) and overall survival (OS) in the TCGA (n = 368) and OS in the GSE84437 (n = 432) cohorts of Low (blue line) and High (red line) in coagulation score. Log-rank test was used to determine p values.
Consequently, TCGA patients with high coagulation scores also had poorer disease specific survival (DSS) and overall survival (OS) (Figure 1B, P = 0.019 and 0.011, respectively). This association between coagulation score and OS was validated by GSE84437 (P = 0.012). These results indicate that intra-tumoral coagulation was significantly associated with larger, more aggressive gastric cancers and with poor patient prognosis. Next, we further investigated whether previously proposed mechanisms have clinical relevance in large gastric cancer patient cohorts.
A high coagulation score correlated with angiogenesis in gastric tumors
Multiple previous studies have demonstrated an association between pro-coagulation factors and angiogenesis in various tumors [2,42]. To this end, we investigated the relationship between the coagulation and angiogenesis scores, which were analyzed by the same methodology, GSVA. We found that coagulation strongly correlated with angiogenesis, but not with tissue factor (TF) expression (Figure 2A; r = 0.810 and 0.838, respectively, both P<0.01 for angiogenesis). Gastric cancers with high coagulation scores demonstrated significant enrichment of angiogenesis gene sets in both the TCGA and GSE84437 cohorts (Figure 2B; normalized enrichment score (NES) = 2.24 and 1.71, respectively). Congruently, high-coagulation gastric tumors contained significantly higher proportions of angiogenesis-related vascular cells, including endothelial cells, microvascular endothelial cells, pericytes, and lymphatic endothelial cells across both cohorts (Figure 2C; all P<0.001). Tumors with higher coagulation scores also demonstrated increased expression of genes related to angiogenesis (von Willebrand Factor (vWF) and CD31), endothelial cells, and vascular stability (VE-cadherin, ANGPT1, TIE2, TIE1, JAM2 and Claudin 5) (Figure 2D and 2E; all P<0.001). In summary, the expression of coagulation and angiogenesis gene sets were closely linked in malignant gastric tumors.
Figure 2.
Relationship between the coagulation score and angiogenesis-related genes in the GSE84437 and TCGA cohorts. (A) Scatter plots of the Spearman’s rank correlation test (r) between coagulation score and angiogenesis score or TF (tissue factor) gene expression. (B) Angiogenesis gene set enrichment plots with false discovery rate (FDR) and normalized enrichment score (NES). Classical gene set enrichment analysis (GSEA) method was used to determine FDR and NES. Boxplots of the comparison between low and high coagulation score in (C) angiogenesis-related cells; endothelial cells, microvascular endothelial cells (mvE), lymphatic endothelial cells (lyE), and pericytes. (D) Endothelial cell markers; CD31, von-Willebrand factor (VWF), (E) vascular stability-related genes; TIE1, TIE2, ANGPT1, E-cadherin, Claudin5 and JAM2. P value was analyzed with Mann Whitney U test.
High coagulation score did not correlate with immune cell infiltration or host immune response
Given that coagulopathy often occurs with vascular injury in clinical practice [43], we next investigated the relationship of coagulation and immune response in the tumor microenvironment. We found that increased expression of pro-coagulation genes was not predictive of either tumor infiltrating lymphocyte (TIL) regional fraction score nor interferon (IFN)-γ activity in the TCGA cohort (Figure 3A). Gastric cancers with high coagulation scores did not enrich IFN-α nor IFN-γ signaling by GSEA in the GSE84437 cohort (Figure 3B). Next, we investigated the relationship between coagulation and immune cell infiltration using xCell algorithm. Although high coagulation gastric cancers contained higher proportions of dendritic cells (DC) and M1 macrophages (all p<0.001) and relatively lower proportions of T helper type 1 (Th1) cells, no other immune cells consistently correlated with coagulation score across both TCGA and GSE96058 cohorts (Figure 3C). Therefore, we conclude that expression of pro-coagulation genes does not correlate with the host immune response nor immune cell infiltration in the gastric cancer tumor microenvironment.
Figure 3.
Association between the coagulation score and immune function and cell fractions in gastric cancer. A. Box plots of tumor infiltrating lymphocytes (TIL) regional fraction score and interferon (IFN)-γ response in the TCGA cohort. B. GSEA plots with FDR and NES for the IFN-α and IFN-γ signaling gene sets in the GSE84437 cohort. The classical GSEA method was used to determine FDR and NES. C. Boxplots of infiltrating immune cells including CD8+ T cells, CD4+ memory T cells, T helper type 1 (Th1), dendritic cells (DCs), M1 macrophages, regulatory T cells (Tregs), and Th2 cells, and M2 macrophages, by low and high coagulation score in the TCGA and GSE84437 cohorts. P value was analyzed with Mann Whitney U test.
Higher expression of pro-coagulation genes correlated with low mutation load in gastric tumors
Since mutation load is known to correlate with aggressiveness of cancer [41,44-47], we examined the association between the coagulation score and tumor mutation load. Although low- and high-coagulation score groups did not show significant differences with regard to intratumor heterogeneity nor homologous recombination deficiency (Figure 4A; P = 0.134 and 0.243, respectively), gastric cancers with high coagulation scores demonstrated high levels of SNV neoantigens, fraction of genome altered, and both silent and non-silent mutation rate in TCGA (Figure 4B; P<0.001, P<0.001, P = 0.022, and P = 0.011, respectively).
Figure 4.
Association of the coagulation score with intratumor heterogeneity, homologous recombinant deficient (HRD), and mutation-related score in the TCGA cohort. (A) Box plots of intratumor heterogeneity, HRD, and (B) mutation-related score, including silent and non-silent mutation rate, fraction altered, and single nucleotide variant (SNV) neoantigens by low and high coagulation score groups. P value was analyzed with Mann Whitney U test.
Coagulation score did not correlate with cell proliferation signaling in gastric cancer
Given that highly proliferative cancer results in worse survival [13,48,49], we next queried whether a high coagulation score predicts increased expression of cell proliferation pathways. The score did not correlate with MKI67 expression, a cell proliferation marker, in two cohorts. None of the cell proliferation-related gene sets in Hallmark collection, including those for mitotic spindle, MYC Targets v1 and v2, G2M Checkpoint, and E2F Targets, were highly expressed in high-coagulation gastric cancers (Figure 5A and 5B).
Figure 5.
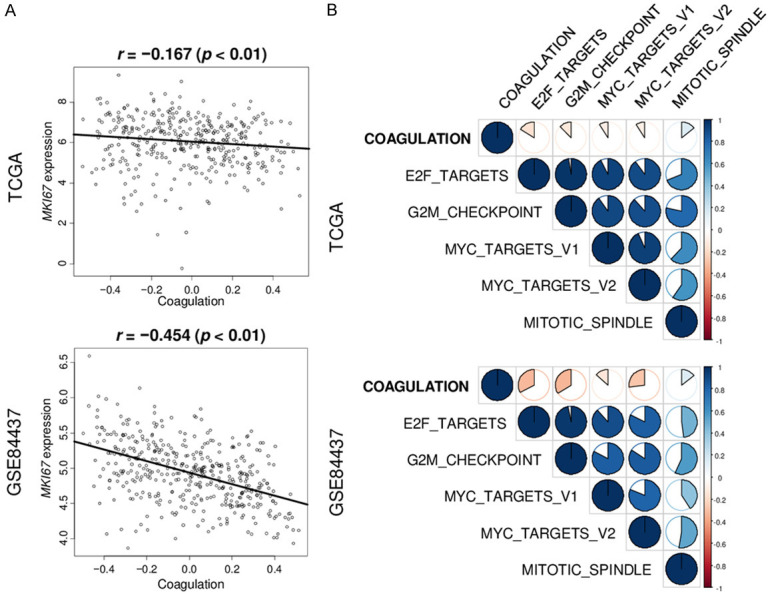
Correlation of the coagulation score with MKi67 expression and cell proliferation-related gene sets score in the TCGA and GSE84437 cohorts. A. Scatter plots between the coagulation score and MKi67 gene expression. B. Correlation plots between coagulation score and cell proliferation-related gene sets score, including MITOTIC spindle E2F targets, G2M checkpoint, MYC targets v1, and MYC targets v2. A Spearman’s rank correlation test (r) was used where blue indicates positive and red indicates negative correlation.
Genes for transforming growth factor (TGF)-β signaling epithelial mesenchymal transition (EMT), and myogenesis, apical junction, were highly expressed in high-coagulation gastric cancers
Finally, we examined the correlation between coagulation and pathways related to transition and transformation of malignant cells, an important factor in tumor progression. The high-coagulation score group significantly enriched EMT, as evidenced by the highest NES across both cohorts (Figure 5; NES = 2.25 and 1.75, respectively). Furthermore, they also demonstrated higher expression of genes encoding myogenesis, apical junction, and TGF-β signaling across both cohorts (Figure 6).
Figure 6.
Gene Set Enrichment Analysis (GSEA) demonstrating enrichment gene sets in high coagulation score gastric cancer in the GSE84437 and TCGA cohorts. Epithelial mesenchymal transition (EMT), myogenesis, apical junction, and TGF-β signaling gene sets demonstrated significant enrichment consistently in both cohorts. NES and FDR were determined with the classical gene set enrichment analysis (GSEA) method.
Discussion
Bleeding tendency and thrombotic events are very common in patients with advanced cancer [43,50]. Most cancer cells constitutively express tissue factor (TF) on the cell surface that generates thrombin by combining with factor VIIa to activate factor IX to IXa and X to Xa on the activated platelet surface, resulting in the conversion of prothrombin to thrombin. Thus, malignancy initiates a vicious cycle in which greater tumor burden supplies greater thrombin and platelet-tumor interaction. However, it has not been fully elucidated whether tumor expression of these coagulation cascade genes augments the malignant biology of cancer.
In recent years, the use of large transcriptome databases and gene set analysis has facilitated massive growth in the field of translational research [51]. Additionally, the ability to digitally dissect tumor samples and determine their cellular composition allows researchers to gain a broader picture of the tumor microenvironment [35,52,53]. Here, we demonstrated the clinical relevance of intra-tumoral pro-coagulation gene expression in gastric cancer. Using multiple large cohorts of patients with gastric cancer, we found that intratumoral expression of pro-coagulation genes was associated with angiogenesis and EMT, but not consistently with cell proliferation.
Moreover, we discovered that gastric cancers with higher expression of coagulation pathways did not demonstrate enrichment of cell proliferation- nor immune-related gene sets, which have previously been reported to be important for tumor aggressiveness. However, our results indicate that gastric cancers with high coagulation scores did demonstrate EMT, angiogenesis, apical junction, and TGF-β signaling, which play key roles in the transformation and migration of malignant cells [54]. The requirement of angiogenesis for tumor growth and metastasis is well recognized [55]. Mary et al. reported that thrombin, one of the major players in coagulation signaling, activates tumor cell adhesion to platelets, endothelial cells, and subendothelial matrix proteins, which enhances tumor cell growth, and stimulates tumor cell angiogenesis [56]. Our results show that although TF gene expression did not correlate with angiogenesis, coagulation score strongly correlated with angiogenesis score.
Interpreting the clinical relevance of any single gene out of its biological context is challenging and prone to error [57]. Assays which measure gene expression within sets organized according to their function provide a more comprehensive and relevant analysis [58-61]. Thus, the GSVA method is utilized in this study to provide a global picture of the coagulation pathway within gastric cancers. It is difficult to elucidate these pathways in in vitro or in vivo studies because the complex interactions within the tumor microenvironment are not easily replicated in in vitro experiments, and difficult to isolate in in vivo studies. Our results indicate that previously reported relationships between coagulation signaling and tumor growth also occur within tumors. The nature of this type of study does not lend itself to determining causality, but supports previously made observations that factors within the coagulation cascade subsequently promote angiogenesis, EMT and TGF-beta signaling.
By using transcriptomic expression across multiple cohorts, we showed that intratumoral coagulation is linked to outcomes in gastric cancer. Although prognostic biomarkers have been developed in gastric cancer, none have yet been clinically applied. Although further investigation is required, the coagulation score may prove to be a useful biomarker for gastric cancer patients. Additional studies are needed to determine the clinical utility of the intratumoral coagulation score.
There are several limitations to interpreting this study. This is a retrospective study that does not allow us to account for confounding factors that may influence clinical outcomes. Patient outcome is naturally affected by multiple clinical factors, such as comorbidities, functional status, surgical and systemic treatment, for which limited information is available in a transcriptomic database. Additionally, this study does not provide original data on the mechanisms that underlie the associations of tumor coagulation with outcomes and various tumor features of gastric cancer.
In summary, the coagulation score provides a method to quantify the intra-tumoral expression of coagulative factors, which is associated with angiogenesis, EMT and TNF-beta signaling and worse outcomes of gastric cancer patients.
Acknowledgements
This work was supported by the National Institutes of Health (NIH) USA grant number R01CA160688, R37CA248018, R01CA250412, R01CA251545, as well as US Department of Defense BCRP grant number W81XWH-19-1-0674, and W81XWH-19-1-0111 to K.T. Roswell Park Comprehensive Cancer Center is supported by NCI/NIH grant P30-CA016056 grant.
Disclosure of conflict of interest
None.
Abbreviations
- AJCC
American Joint Committee on Cancer
- FDR
false discovery rate
- GSEA
gene set enrichment analysis
- GSVA
gene set variation analysis
- NES
normalized enrichment score
- TCGA
The Cancer Genome Atlas
References
- 1.Elyamany G, Alzahrani AM, Bukhary E. Cancer-associated thrombosis: an overview. Clin Med Insights Oncol. 2014;8:129–137. doi: 10.4137/CMO.S18991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rubio-Jurado B, Sosa-Quintero LS, Guzmán-Silahua S, García-Luna E, Riebeling-Navarro C, Nava-Zavala AH. The prothrombotic state in cancer. Adv Clin Chem. 2021;105:213–242. doi: 10.1016/bs.acc.2021.03.001. [DOI] [PubMed] [Google Scholar]
- 3.Hill CN, Hernández-Cáceres MP, Asencio C, Torres B, Solis B, Owen GI. Deciphering the role of the coagulation cascade and autophagy in cancer-related thrombosis and metastasis. Front Oncol. 2020;10:605314. doi: 10.3389/fonc.2020.605314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Falanga A, Russo L, Milesi V. The coagulopathy of cancer. Curr Opin Hematol. 2014;21:423–429. doi: 10.1097/MOH.0000000000000072. [DOI] [PubMed] [Google Scholar]
- 5.Orellana R, Kato S, Erices R, Bravo ML, Gonzalez P, Oliva B, Cubillos S, Valdivia A, Ibañez C, Brañes J, Barriga MI, Bravo E, Alonso C, Bustamente E, Castellon E, Hidalgo P, Trigo C, Panes O, Pereira J, Mezzano D, Cuello MA, Owen GI. Platelets enhance tissue factor protein and metastasis initiating cell markers, and act as chemoattractants increasing the migration of ovarian cancer cells. BMC Cancer. 2015;15:290. doi: 10.1186/s12885-015-1304-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Labelle M, Begum S, Hynes RO. Direct signaling between platelets and cancer cells induces an epithelial-mesenchymal-like transition and promotes metastasis. Cancer Cell. 2011;20:576–590. doi: 10.1016/j.ccr.2011.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Henrikson KP, Salazar SL, Fenton JW 2nd, Pentecost BT. Role of thrombin receptor in breast cancer invasiveness. Br J Cancer. 1999;79:401–406. doi: 10.1038/sj.bjc.6690063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Remiker AS, Palumbo JS. Mechanisms coupling thrombin to metastasis and tumorigenesis. Thromb Res. 2018;164(Suppl 1):S29–S33. doi: 10.1016/j.thromres.2017.12.020. [DOI] [PubMed] [Google Scholar]
- 9.Xu Y, Pan S, Liu J, Dong F, Cheng Z, Zhang J, Qi R, Zang Q, Zhang C, Wang X, Zhang J, Wang F, Allen TD, Liu J. GATA3-induced vWF upregulation in the lung adenocarcinoma vasculature. Oncotarget. 2017;8:110517–110529. doi: 10.18632/oncotarget.22806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kerk N, Strozyk EA, Pöppelmann B, Schneider SW. The mechanism of melanoma-associated thrombin activity and von Willebrand factor release from endothelial cells. J Invest Dermatol. 2010;130:2259–2268. doi: 10.1038/jid.2010.136. [DOI] [PubMed] [Google Scholar]
- 11.Ling J, Sun Y, Pan J, Wang H, Ma Z, Yin J, Bao Z, Yang H, Liu L. Feedback modulation of endothelial cells promotes epithelial-mesenchymal transition and metastasis of osteosarcoma cells by Von Willebrand Factor release. J Cell Biochem. 2019;120:15971–15979. doi: 10.1002/jcb.28875. [DOI] [PubMed] [Google Scholar]
- 12.Hänzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics. 2013;14:7. doi: 10.1186/1471-2105-14-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oshi M, Patel A, Le L, Tokumaru Y, Yan L, Matsuyama R, Endo I, Takabe K. G2M checkpoint pathway alone is associated with drug response and survival among cell proliferation-related pathways in pancreatic cancer. Am J Cancer Res. 2021;11:3070–3084. [PMC free article] [PubMed] [Google Scholar]
- 14.De Almeida MC, Sanchez-Quintana D, Anderson RH. The membranous septum revisited: a glimpse of our anatomical past. Clin Anat. 2021;34:178–186. doi: 10.1002/ca.23599. [DOI] [PubMed] [Google Scholar]
- 15.Oshi M, Newman S, Tokumaru Y, Yan L, Matsuyama R, Endo I, Katz MHG, Takabe K. High G2M pathway score pancreatic cancer is associated with worse survival, particularly after margin-positive (R1 or R2) resection. Cancers (Basel) 2020;12:2871. doi: 10.3390/cancers12102871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Patel A, Oshi M, Yan L, Matsuyama R, Endo I, Takabe K. The unfolded protein response is associated with cancer proliferation and worse survival in hepatocellular carcinoma. Cancers (Basel) 2021;13:4443. doi: 10.3390/cancers13174443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Murthy V, Oshi M, Tokumaru Y, Endo I, Takabe K. Increased apoptosis is associated with robust immune cell infiltration and cytolytic activity in breast cancer. Am J Cancer Res. 2021;11:3674–3687. [PMC free article] [PubMed] [Google Scholar]
- 18.Gandhi S, Oshi M, Murthy V, Repasky EA, Takabe K. Enhanced thermogenesis in triple-negative breast cancer is associated with pro-tumor immune microenvironment. Cancers (Basel) 2021;13:2559. doi: 10.3390/cancers13112559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Oshi M, Tokumaru Y, Angarita FA, Lee L, Yan L, Matsuyama R, Endo I, Takabe K. Adipogenesis in triple-negative breast cancer is associated with unfavorable tumor immune microenvironment and with worse survival. Sci Rep. 2021;11:12541. doi: 10.1038/s41598-021-91897-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oshi M, Newman S, Tokumaru Y, Yan L, Matsuyama R, Endo I, Takabe K. Inflammation is associated with worse outcome in the whole cohort but with better outcome in triple-negative subtype of breast cancer patients. J Immunol Res. 2020;2020:5618786. doi: 10.1155/2020/5618786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tokumaru Y, Oshi M, Huyser MR, Yan L, Fukada M, Matsuhashi N, Futamura M, Akao Y, Yoshida K, Takabe K. Low expression of miR-29a is associated with aggressive biology and worse survival in gastric cancer. Sci Rep. 2021;11:14134. doi: 10.1038/s41598-021-93681-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oshi M, Satyananda V, Angarita FA, Kim TH, Tokumaru Y, Yan L, Matsuyama R, Endo I, Nagahashi M, Takabe K. Angiogenesis is associated with an attenuated tumor microenvironment, aggressive biology, and worse survival in gastric cancer patients. Am J Cancer Res. 2021;11:1659–1671. [PMC free article] [PubMed] [Google Scholar]
- 23.Liu J, Lichtenberg T, Hoadley KA, Poisson LM, Lazar AJ, Cherniack AD, Kovatich AJ, Benz CC, Levine DA, Lee AV, Omberg L, Wolf DM, Shriver CD, Thorsson V, Hu H. An integrated TCGA pan-cancer clinical data resource to drive high-quality survival outcome analytics. Cell. 2018;173:400–416. e411. doi: 10.1016/j.cell.2018.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yoon SJ, Park J, Shin Y, Choi Y, Park SW, Kang SG, Son HY, Huh YM. Deconvolution of diffuse gastric cancer and the suppression of CD34 on the BALB/c nude mice model. BMC Cancer. 2020;20:314. doi: 10.1186/s12885-020-06814-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liberzon A, Birger C, Thorvaldsdóttir H, Ghandi M, Mesirov JP, Tamayo P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015;1:417–425. doi: 10.1016/j.cels.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oshi M, Kim TH, Tokumaru Y, Yan L, Matsuyama R, Endo I, Cherkassky L, Takabe K. Enhanced DNA repair pathway is associated with cell proliferation and worse survival in Hepatocellular Carcinoma (HCC) Cancers (Basel) 2021;13:323. doi: 10.3390/cancers13020323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oshi M, Tokumaru Y, Angarita FA, Yan L, Matsuyama R, Endo I, Takabe K. Degree of early estrogen response predict survival after endocrine therapy in primary and metastatic ER-positive breast cancer. Cancers (Basel) 2020;12:3557. doi: 10.3390/cancers12123557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Okano M, Oshi M, Butash AL, Asaoka M, Katsuta E, Peng X, Qi Q, Yan L, Takabe K. Estrogen receptor positive breast cancer with high expression of androgen receptor has less cytolytic activity and worse response to neoadjuvant chemotherapy but better survival. Int J Mol Sci. 2019;20:2655. doi: 10.3390/ijms20112655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Okano M, Oshi M, Butash AL, Katsuta E, Tachibana K, Saito K, Okayama H, Peng X, Yan L, Kono K, Ohtake T, Takabe K. Triple-negative breast cancer with high levels of annexin A1 expression is associated with mast cell infiltration, inflammation, and angiogenesis. Int J Mol Sci. 2019;20:4197. doi: 10.3390/ijms20174197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tokumaru Y, Asaoka M, Oshi M, Katsuta E, Yan L, Narayanan S, Sugito N, Matsuhashi N, Futamura M, Akao Y, Yoshida K, Takabe K. High expression of microRNA-143 is associated with favorable tumor immune microenvironment and better survival in estrogen receptor positive breast cancer. Int J Mol Sci. 2020;21:3213. doi: 10.3390/ijms21093213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Oshi M, Katsuta E, Yan L, Ebos JML, Rashid OM, Matsuyama R, Endo I, Takabe K. A novel 4-gene score to predict survival, distant metastasis and response to neoadjuvant therapy in breast cancer. Cancers (Basel) 2020;12:1148. doi: 10.3390/cancers12051148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Takahashi H, Oshi M, Asaoka M, Yan L, Endo I, Takabe K. Molecular biological features of Nottingham histological grade 3 breast cancers. Ann Surg Oncol. 2020;27:4475–4485. doi: 10.1245/s10434-020-08608-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chouliaras K, Tokumaru Y, Asaoka M, Oshi M, Attwood KM, Yoshida K, Ishikawa T, Takabe K. Prevalence and clinical relevance of tumor-associated tissue eosinophilia (TATE) in breast cancer. Surgery. 2021;169:1234–1239. doi: 10.1016/j.surg.2020.07.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Aran D, Hu Z, Butte AJ. xCell: digitally portraying the tissue cellular heterogeneity landscape. Genome Biol. 2017;18:220. doi: 10.1186/s13059-017-1349-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Oshi M, Tokumaru Y, Asaoka M, Yan L, Satyananda V, Matsuyama R, Matsuhashi N, Futamura M, Ishikawa T, Yoshida K, Endo I, Takabe K. M1 macrophage and M1/M2 ratio defined by transcriptomic signatures resemble only part of their conventional clinical characteristics in breast cancer. Sci Rep. 2020;10:16554. doi: 10.1038/s41598-020-73624-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Oshi M, Asaoka M, Tokumaru Y, Angarita FA, Yan L, Matsuyama R, Zsiros E, Ishikawa T, Endo I, Takabe K. Abundance of regulatory T cell (Treg) as a predictive biomarker for neoadjuvant chemotherapy in triple-negative breast cancer. Cancers (Basel) 2020;12:3038. doi: 10.3390/cancers12103038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Oshi M, Newman S, Tokumaru Y, Yan L, Matsuyama R, Kalinski P, Endo I, Takabe K. Plasmacytoid Dendritic Cell (pDC) infiltration correlate with tumor infiltrating lymphocytes, cancer immunity, and better survival in Triple Negative Breast Cancer (TNBC) more strongly than Conventional Dendritic Cell (cDC) Cancers (Basel) 2020;12:3342. doi: 10.3390/cancers12113342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tokumaru Y, Oshi M, Patel A, Tian W, Yan L, Matsuhashi N, Futamura M, Yoshida K, Takabe K. Organoids are limited in modeling the colon adenoma-carcinoma sequence. Cells. 2021;10:488. doi: 10.3390/cells10030488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Oshi M, Huyser MR, Le L, Tokumaru Y, Yan L, Matsuyama R, Endo I, Takabe K. Abundance of microvascular endothelial cells is associated with response to chemotherapy and prognosis in colorectal cancer. Cancers (Basel) 2021;13:1477. doi: 10.3390/cancers13061477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chouliaras K, Oshi M, Asaoka M, Tokumaru Y, Khoury T, Endo I, Ishikawa T, Takabe K. Increased intratumor heterogeneity, angiogenesis and epithelial to mesenchymal transition pathways in metaplastic breast cancer. Am J Cancer Res. 2021;11:4408–4420. [PMC free article] [PubMed] [Google Scholar]
- 42.Zarychta E, Ruszkowska-Ciastek B. Cooperation between angiogenesis, vasculogenesis, chemotaxis, and coagulation in breast cancer metastases development: pathophysiological point of view. Biomedicines. 2022;10:300. doi: 10.3390/biomedicines10020300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Harrington DJ, Western H, Seton-Jones C, Rangarajan S, Beynon T, Shearer MJ. A study of the prevalence of vitamin K deficiency in patients with cancer referred to a hospital palliative care team and its association with abnormal haemostasis. J Clin Pathol. 2008;61:537–540. doi: 10.1136/jcp.2007.052498. [DOI] [PubMed] [Google Scholar]
- 44.Takahashi H, Asaoka M, Yan L, Rashid OM, Oshi M, Ishikawa T, Nagahashi M, Takabe K. Biologically aggressive phenotype and anti-cancer immunity counterbalance in breast cancer with high mutation rate. Sci Rep. 2020;10:1852. doi: 10.1038/s41598-020-58995-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Oshi M, Kawaguchi T, Yan L, Peng X, Qi Q, Tian W, Schulze A, McDonald KA, Narayanan S, Young J, Liu S, Morris LG, Chan TA, Kalinski P, Matsuyama R, Otsuji E, Endo I, Takabe K. Immune cytolytic activity is associated with reduced intra-tumoral genetic heterogeneity and with better clinical outcomes in triple negative breast cancer. Am J Cancer Res. 2021;11:3628–3644. [PMC free article] [PubMed] [Google Scholar]
- 46.Oshi M, Gandhi S, Huyser MR, Tokumaru Y, Yan L, Yamada A, Matsuyama R, Endo I, Takabe K. MELK expression in breast cancer is associated with infiltration of immune cell and pathological compete response (pCR) after neoadjuvant chemotherapy. Am J Cancer Res. 2021;11:4421–4437. [PMC free article] [PubMed] [Google Scholar]
- 47.Satyananda V, Oshi M, Endo I, Takabe K. High BRCA2 gene expression is associated with aggressive and highly proliferative breast cancer. Ann Surg Oncol. 2021;28:7356–7365. doi: 10.1245/s10434-021-10063-5. [DOI] [PubMed] [Google Scholar]
- 48.Oshi M, Takahashi H, Tokumaru Y, Yan L, Rashid OM, Nagahashi M, Matsuyama R, Endo I, Takabe K. The E2F pathway score as a predictive biomarker of response to neoadjuvant therapy in ER+/HER2- breast cancer. Cells. 2020;9:1643. doi: 10.3390/cells9071643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schulze A, Oshi M, Endo I, Takabe K. MYC targets scores are associated with cancer aggressiveness and poor survival in ER-positive primary and metastatic breast cancer. Int J Mol Sci. 2020;21:8127. doi: 10.3390/ijms21218127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sanz MA, Montesinos P. Advances in the management of coagulopathy in acute promyelocytic leukemia. Thromb Res. 2020;191(Suppl 1):S63–S67. doi: 10.1016/S0049-3848(20)30399-6. [DOI] [PubMed] [Google Scholar]
- 51.Roychowdhury S, Chinnaiyan AM. Translating cancer genomes and transcriptomes for precision oncology. CA Cancer J Clin. 2016;66:75–88. doi: 10.3322/caac.21329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Miao YR, Zhang Q, Lei Q, Luo M, Xie GY, Wang H, Guo AY. ImmuCellAI: a unique method for comprehensive T-cell subsets abundance prediction and its application in cancer immunotherapy. Adv Sci (Weinh) 2020;7:1902880. doi: 10.1002/advs.201902880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Newman AM, Liu CL, Green MR, Gentles AJ, Feng W, Xu Y, Hoang CD, Diehn M, Alizadeh AA. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 2015;12:453–457. doi: 10.1038/nmeth.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hapke RY, Haake SM. Hypoxia-induced epithelial to mesenchymal transition in cancer. Cancer Lett. 2020;487:10–20. doi: 10.1016/j.canlet.2020.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Katsuta E, Rashid OM, Takabe K. Clinical relevance of tumor microenvironment: immune cells, vessels, and mouse models. Hum Cell. 2020;33:930–937. doi: 10.1007/s13577-020-00380-4. [DOI] [PubMed] [Google Scholar]
- 56.Nierodzik ML, Karpatkin S. Thrombin induces tumor growth, metastasis, and angiogenesis: evidence for a thrombin-regulated dormant tumor phenotype. Cancer Cell. 2006;10:355–362. doi: 10.1016/j.ccr.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 57.Ein-Dor L, Zuk O, Domany E. Thousands of samples are needed to generate a robust gene list for predicting outcome in cancer. Proc Natl Acad Sci U S A. 2006;103:5923–5928. doi: 10.1073/pnas.0601231103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang X, Sun Z, Zimmermann MT, Bugrim A, Kocher JP. Predict drug sensitivity of cancer cells with pathway activity inference. BMC Med Genomics. 2019;12:15. doi: 10.1186/s12920-018-0449-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lee E, Chuang HY, Kim JW, Ideker T, Lee D. Inferring pathway activity toward precise disease classification. PLoS Comput Biol. 2008;4:e1000217. doi: 10.1371/journal.pcbi.1000217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Khatri P, Sirota M, Butte AJ. Ten years of pathway analysis: current approaches and outstanding challenges. PLoS Comput Biol. 2012;8:e1002375. doi: 10.1371/journal.pcbi.1002375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Su J, Yoon BJ, Dougherty ER. Accurate and reliable cancer classification based on probabilistic inference of pathway activity. PLoS One. 2009;4:e8161. doi: 10.1371/journal.pone.0008161. [DOI] [PMC free article] [PubMed] [Google Scholar]






