Abstract
Lysine-specific demethylase 1 (LSD1) is widely involved in the proliferation, invasion, and metastasis of cancers. However, it is uncertain whether LSD1 plays a role in facilitating colon cancer progression. Here, we have clarified the molecular mechanism by which LSD1 interacts with X-ray repair cross complementing protein 5 (Ku80) to promote colon cancer progression by directly targeting forehead protein transcription factor 2 (FOXF2). First, the interacting proteins of LSD1 were identified by immunoprecipitation and mass spectrometry. The expression of Ku80 and FOXF2 in colon cancer was detected using immunohistochemistry, real-time quantitative transcription polymerase chain reaction, and western blot. Next, the proliferation, invasion, and metastasis of colon cancer in vitro and in vivo were detected by methyl thiazolyl tetrazolium, 5-ethynyl-20-deoxyuridine, colony formation, wound healing, and nude mice xenograft model assays, respectively. Chromatin immunoprecipitation (ChIP) and ChIP-PCR were performed to investigate the molecular mechanism of LSD1 and Ku80 in colon cancer. Our results indicated that Ku80 expression was positively correlated with the invasion and migration of colon cancer cells, and negatively correlated with FOXF2 expression. More importantly, the high expression of Ku80 and the low expression of FOXF2 were particularly associated with driving the progression of colon cancer. Ku80 knockdown and LSD1 silencing inhibited the proliferation, migration, and invasion of colon cancer in vitro and in vivo. Mechanically, LSD1 interacts with Ku80 and also binds directly to the 687-887-bp portion of the FOXF2 promoter region. The upregulated methylation level of H3K4me2 in the FOXF2 promoter region facilitated the transcriptional activation of FOXF2, and downregulated protein expression associated with the Wnt/β-catenin signaling pathway. In conclusion, our study suggests that LSD1 regulates the FOXF2-mediated Wnt/β-catenin signaling pathway by interacting with Ku80, promoting the malignant biological properties of colon cancer, highlighting the binding of LSD1 and Ku80 as a useful anti-cancer target for colon cancer.
Keywords: Lysine-specific demethylase, X-ray repair cross complementing protein 5, forehead protein transcription factor 2, histone modification, tumorigenesis, colon cancer
Introduction
Colorectal cancer is the third most common malignancy and the third leading cause of cancer-associated death worldwide [1]. Metastasis and recurrence of colon cancer are related to the excessive proliferation of cancer cells and an imbalanced regulation mechanism. Cell growth and proliferation are regulated by various post-transcriptional modifications, including the demethylation process regulated by demethylase. Dynamic regulation based on histone covalent modification is critical for maintaining genome integrity and cell function. Recent evidence [2] suggests that histone demethylase is a novel therapeutic target for different types of tumors, including colon cancer.
Lysine-specific demethylase 1 (LSD1) was discovered for the first time in 2004, and emerging evidence suggests that LSD1 also may be a potential target for cancer treatment [3-5]. Studies have found that LSD1 is highly expressed in colon cancer with advanced lymph node metastasis and distant metastasis, whereas E-cadherin expression is significantly decreased. The positive expression of LSD1 and the negative expression of E-cadherin may indicate poor prognosis in patients with colon cancer. Conclusive evidence has shown that LSD1 knockdown inhibits the proliferation and invasion of colon cancer cells, inducing apoptosis in vitro [6]. LSD1 was first isolated as an interaction partner of CoREST [7] transcription repressor complex with histone deacetylase HDAC1/2 [8]. LSD1 can initiate or inhibit the transcription of target genes selectively by demethylating H3K4 or H3K9 in the complex through interactions with various molecular chaperones [8,9]. In addition, demethylation contributes to the metastasis of colon cancer. Compared with histone substrates, LSD1 can demethylate lysine residues on several non-histone substrates, such as p53, Dnmt1, E2F1, and MYPT1 [10], which means that LSD1 plays a complex role in vivo.
Upregulated expression of the forehead box (FOX) transcription factor can lead to the development of malformations, especially malignant tumors [11-13]. As a member of the FOX gene subfamily, forehead protein transcription factor 2 (FOXF2) plays an important role in the synthesis of extracellular matrix and epithelial-mesenchymal transformation in histological development, and its expression disorder is related to cancers of multiple tissue types [14]. Chen et al. [15] found that downstream targets CABYR and CDH1 are regulated directly by LSD1 in colon cancer, whereas downstream targets FOXF2 and TLE4 are regulated indirectly by the same. The underlying mechanism by which LSD1 regulates FOXF2 expression and affects colon cancer metastasis remain unclear.
As a highly conserved DNA binding protein, Ku is found widely in prokaryotes and eukaryotes and plays a vital role in maintaining genome integrity [16]. In addition, Ku is involved in many nuclear processes, including telomere maintenance [18], V(D)J reorganization [19], cell cycle regulation [20], and transcriptional regulation [21,22]. Ku80, a subunit of Ku, repairs DNA double-strand breaks through the non-homologous end joining pathway and plays a key role in maintaining chromosomal stability [17]. Ku80 is usually expressed abnormally in tumor patients and is associated with poor prognosis [23,24].
Given the importance of LSD1-focused protein complexes in tumors, we recognize the importance of investigating the dual-target inhibition of these complexes. Here, we perform a series of experiments to analyze the relationship between the expression of Ku80 and FOXF2 and the malignant biological progression of colon cancer, and to explore the potential role of Ku80, LSD1, and FOXF2 in regulating the development of colon cancer and its underlying molecular mechanisms.
Material and methods
Cell lines and cell culture
Human colon cancer cell lines (HT-29, LOVO, SW480, HCT116) used in this study were purchased from Zhong Qin Xin Zhou Biotechnology Co., Ltd. (Shanghai, China). HT-29 cells were cultured with RPMI-1640 medium, LOVO cells were cultured with F-12K medium, SW480 cells were cultured with L-15 medium, and HCT-116 cells were cultured with McCoy’s5a medium. The four prior media were purchased from Sigma-Aldrich Corp. (St. Louis, MO, USA). The HEK293T cell line was obtained from Kunming Cell Bank (Kunming, China). HEK293T cells were cultured with DMEM medium (HyClone, South Logan, Utah, USA). All media were supplemented with 10% fatal bovine serum (04-001-1ACS; BI, Beit HaEmek, Israel) and 1% antibiotic/antimycotic solution (Biowest, Nuaillé, France). All cell lines were cultured in an incubator with 5% CO2 at 37°C.
Immunoprecipitation (IP) and mass spectrometry (MS)
HCT116 cell lysates were immunoprecipitated using LSD1 monoclonal antibody and following the instructions of BeaverBeads™ Protein A/G Immunoprecipitation Kit [25]. After the IP experiment and electrophoresis, protein gels were placed in fixative solution containing 50% ethanol, 40% water, and 10% acetic acid, and then shaken overnight at room temperature. The protein bands were observed with Coomassie brilliant blue staining and analyzed by MS with EASY-nLC1000 (Shanghai, China).
Real-time quantitative transcription polymerase chain reaction (RT-qPCR) and western blot
Total RNA was obtained by TRIzol reagent, RNA concentration and purity were measured by spectrophotometer (Thermo Fisher), and cDNA synthesis was performed using RevertAid™ First Strand cDNA Synthesis Kit (Takara, Japan). The LSD1 expression was standardized with GAPDH. Primers for amplification of LSD1, FOXF2, and Ku80 cDNA are presented in Table 3. As described in the literature [6], protein lysates were extracted from cells and analyzed by western blot. The antibody dilution ratio was as follows: LSD1, 1:1,000; Ku80, 1:2,000; FOXF2, 1:2,000.
Table 3.
Ku80, LSD1, FOXF2 primer list
| gene | Forward (5’-3’) | Reverse (5’-3’) | size (bp) |
|---|---|---|---|
| Ku80 | GCACTGACAATCCCCTTTCTG | TGTTGAGCTTCAGCTTTAACCTG | 97 |
| FOXF2 | ACTCAGGTGGGAAGATGTGC | TTCAGATTGGGGAACGCTAC | 203 |
| LSD1 | CCTGAAGAACCATCGGGTGT | CCTTCTGGGTCTGTTGTGGT | 124 |
| GAPDH | TGACTTCAACAGCGACACCCA | CACCCTGTTGCTGTAGCCAAA | 121 |
Immunohistochemical staining
Paraffin-embedded tissue microarray (BC051110c Biomax, lnc., USA) used in this study contained a total of 120 samples, including 108 colon cancer tissues and 12 normal colon mucosal tissues. Primary antibodies targeting Ku80 (rabbit monoclonal, 1:100) and FOXF2 (rabbit polyclonal, 1:30), were purchased from Abcam (Cambridge, USA). Staining was then performed using the EnVision + anti-rabbit system (Dako Corporation, Carpinteria, CA, USA). Non-immune rabbit antibody and phosphate-buffered saline were used to replace the primary antibody for negative control. This study was conducted with the approval of the Ethics Committee of Guizhou Provincial People’s Hospital, China.
Lentiviral vector construction and transfection
According to the sequence of Ku80 (NM-021141) retrieved from the GenBank database, three sequences targeting Ku80 were constructed from GeneChem (Shanghai, China). The sequence information was as follows: shRNA#1: TCATATAAGCATAACTAT; shRNA#2: CTTTAACAACTTCCTGAAA; shRNA#3: TGCAATTCTTCTTGCCTTT. After 24 hours of culture, the number of cells increased to about 1×104. Next, the corresponding lentivirus was transfected with lentivirus or a control lentivirus in 5 μg/mL polybrene (MOI: 20:1). After transfection, the cells were further cultured for 72 h. Then, when transfecting plasmids containing green fluorescent marker sequences, we calculated the transfection efficiency by observing the counts of fluorescent cells.
Wound healing assay and transwell assay
We used four untransfected colorectal cancer cell lines (purchased from the ATCC cell bank) to identify the cell lines with the most extraordinary ability to invade and migrate. Untransfected colon cancer cells were plated into 6-well plates with 3×105/well. The cell monolayer was scratched using a 200-ul pipette tip. Scratch assay was observed microscopically and photographed at 0 h, 24 h and 48 h of scribing, respectively, and Image-Pro plus 6.0 software was used to analyze the scratch healing rate. Migration rate was normalized using the 0-h scratch area. The migration and invasion ability of colon cancer cells was estimated by transwell assays. Matrigel (BD Biosciences, NY, USA) was diluted 1:8 with serum-free medium. To the lower chamber was added 400 μl medium containing 10% fetal bovine serum. Colon cancer cells resuspended in 100 ul serum-free medium were added to the upper chamber, which was pre-treated with or without 60 μl Matrigel. Then, four strains of cells were spread on the TRANSWELL chambers of metal matrix gel, cultured for 24 hours, fixed by formaldehyde, and stained with crystal violet. The number of cells crossing the TRANSWELL chamber membrane was observed under the microscope (three fields of view were randomly selected for counting) to observe further which of the four colon cancer cells had the most substantial migration ability. All images were taken with an inverted microscope (Olympus, Tokyo, Japan). All experiments were performed at least three times.
Methyl thiazolyl tetrazolium (MTT) assay and 5-ethynyl-20-deoxyuridine (EdU) assay
MTT assay was performed with MTT Assay kit (Abcam, ab211091) according to the instructions. HCT116 cells in logarithmic growth phase were inoculated into 96-well plates at 5×104 cells per well, and 4 replicate wells were set up for each group, and a blank control was set up at the same time. After 24 h of treatment with RN-1 (10, 30, 50, 70, 70, 90 nM), 5 mg/mL of MTT solution was added and the culture was continued for 4 h. The supernatant was discarded and 150 μL of dimethyl sulfoxide (DMSO) was added to each well and shaken for 10 min to dissolve the crystals completely and mix well. Using the blank wells as control, the absorbance value of each well was read by enzyme marker at A490 nm, and the cell survival rate of each group was calculated at 0, 24, 48 and 72 h, respectively. An EdU assay was performed to estimate the proliferation of colon cancer cells according to the instructions of EdU Assay Kit (Thermo Fisher). Colon cancer cells were seeded in 96-well plates at 4×103 cells/well. DMEM medium was used to make a 1× EdU solution with a concentration of 25 μM. After cell fixation and permeation enhancement, 0.1 mL Click-iT reaction mixture was added to each well, and the culture plate was shaken and incubated at room temperature in darkness for 30 min. After removing the reaction mixture, 0.1 mL 1× Hoechst 33342 solution was added to each well and plates were incubated at room temperature in darkness for 30 min. Next, DNA counterstaining was performed. The nuclei were stained and placed under the microscope to observe three randomly selected fields of view under the fluorescence microscope, and the total number of cells and the number of Edu staining positive cells were counted, where the blue fluorescence was Hoechst 33342 color development, and the red fluorescence was EdU-labeled proliferating cells. Three replicate wells were set up for each group, and the experiment was repeated three times.
Apoptosis assay
After the adherent cells were digested with trypsin, 1× Annexin V binding buffer was added to resuspend the cells into 1×106 cells/mL. Next, 5 uL Annexin V-FITC and 5 uL PI staining solution were added to 100 uL of the above cell re-suspension and it was maintained in the dark at room temperature for 15 min. After the reaction was complete, 400 uL binding buffer was added and mixed gently. The solution was tested using the flow cytometer within 1 h.
Chromatin immunoprecipitation-qPCR
ChIP Kit (Abcam, ab500) was used for chromatin immunoprecipitation. 3×106 HCT116 cells (above 80% viability by MTT) were selected from each intervention group, fixed with formaldehyde, and terminated with glycine. After shaking for 5 min at room temperature, the cells were centrifuged at 4°C and washed with pre-chilled PBS, followed by lysis with 1 mL of buffer containing inhibitor and incubation on ice for 10 min. DNA fragments were interrupted to 200~1000 bp size by sonication on ice (sonication settings: 3 s on, 3 s off, 50 W, treatment time 2, 4, 6, 8, 10, 12 min). One tube was divided into 2 tubes, and 2 μg of IgG antibody was added as a Chip sample. The other tube was added with 2 μg of anti-Ku80 antibody (Abcam, ab236277) and incubated overnight at 4°C. The supernatant was incubated with 5 μg of antibody overnight, and IgG of the corresponding species was used as the negative control. The FOXF2 promoter region of the 1-2,000-bp fragment was amplified by ten pairs of primers (each fragment was divided into 200 bp). The FOXF2 promoter region of the 1-2,000-bp fragment was amplified by ten pairs of primers (each fragment was divided into 200 bp). Primers are shown in Table 4.
Table 4.
Gene FOXF2 promoter sequence segmented primer sequence list
| Gene name | 5’-3’ Sequence | size | interval | |
|---|---|---|---|---|
| FOXF2-1 | Forward | GACGCCTGTTCAGCTAATTT | 243 bp | 102-344 |
| Reverse | GCTGGTGGGGCTGTAGAT | |||
| FOXF2-2 | Forward | CATCTACAGCCCCACCAGC | 120 bp | 326-445 |
| Reverse | GGCAAAGAGCCTTCACAGC | |||
| FOXF2-3 | Forward | ACACTCAGCCAGGAGCAGTC | 245 bp | 345-589 |
| Reverse | GGACCAAACAGGAGGTAGAGC | |||
| FOXF2-4 | Forward | ACCTCGGTCCTTTCAGCC | 240 bp | 531-770 |
| Reverse | CCTCGGGAGCAATCACTTC | |||
| FOXF2-5 | Forward | CTATCCCTGGTCGGACTACATT | 201 bp | 687-887 |
| Reverse | TTAACATTGCCCACCCAAA | |||
| FOXF2-6 | Forward | TTTGGGTGGGCAATGTTAA | 243 bp | 869-1111 |
| Reverse | ACCTGGATCTCATGGGACTTA | |||
| FOXF2-7 | Forward | GGGAAGAAGTGGAAGCAAAT | 236 bp | 1064-1299 |
| Reverse | CCAGACGCCCAAAGGTAA | |||
| FOXF2-8 | Forward | AGGATTGGCACGTTACCTTT | 274 bp | 1270-1543 |
| Reverse | CTCGACCACCTCTGACTTCAT | |||
| FOXF2-9 | Forward | AGTCAGAGGTGGTCGAGTTT | 174 bp | 1467-1640 |
| Reverse | CCTCCCTCTCTTATCTCGCTC | |||
| FOXF2-10 | Forward | AGAGGAATGAAGAGCGAGG | 240 bp | 1644-1833 |
| Reverse | GGACGAGCCCGACGTCTC | |||
Xenograft tumor model
This experiment was approved by the Animal Ethics Committee of Guizhou Provincial People’s Hospital. Four-week-old SPF-grade nude mice (weighing 18-20 g) were purchased from the Department of Laboratory Animal Science, School of Medicine, Shanghai Jiaotong University. These nude mice were housed in SPF standard cages for one week before the experiment. The mice were divided into two experimental groups (HCT116 cells injected with shKu80#2 and shKu80#2 + RN-1 subcutaneously) and a control group (HCT116 cells injected subcutaneously), with five mice in each group. Then, the cells were diluted into 5×107 cells/mL cell suspension with sterile PBS, and 150 ul of cell suspension was injected subcutaneously into the mid-posterior axilla of each nude mouse. The tumor volume was measured once every four days. The formula V=½×a×b2 calculated the tumour volume (a is the long axis, b is the short axis). After feeding for 25 days, mice were sacrificed by cervical dislocation, and each tumor was removed for imaging and immunohistochemical detection.
Statistical analysis
GraphPad Prism 7.0 (San Diego, CA, USA) was used for statistical analysis. Data were presented as mean ± standard deviation (SD). Independent sample Student’s t-tests were used for comparisons between two groups, whereas one-way ANOVA was employed for comparisons among multiple groups. P-values of <0.05 were considered to be statistically significant.
Results
LSD1 interacts with Ku80 in colon cancer
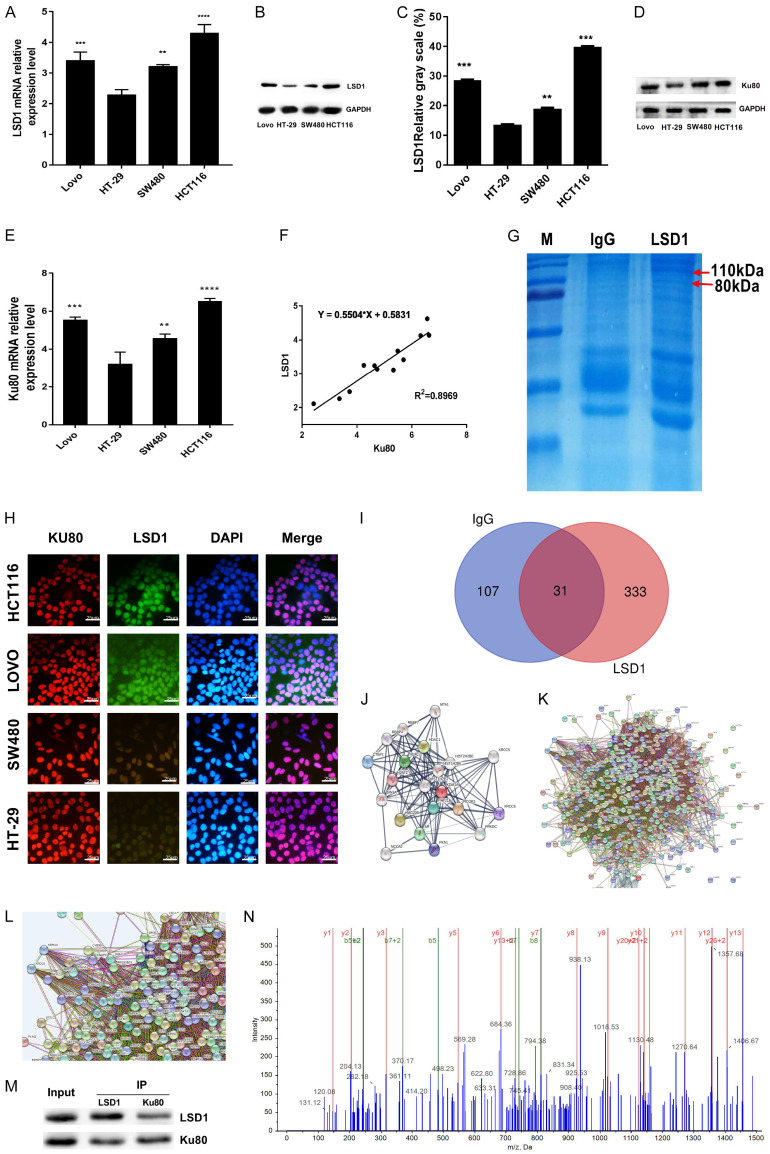
LSD1 acts as a histone demethylase in cells and must bind to other interacting proteins to form a complex and thus perform its biological function. Therefore, we first explored the interacting protein of LSD1 in colon cancer cells. Then, we detected the expression of LSD1 and Ku80 in HT-29, LOVO, SW480, and HCT116 colon cancer cell lines by RT-qPCR and western blot, so as to facilitate the selection of colon cancer cell lines with the highest expression of LSD1 for immunoprecipitation. Protein and mRNA expression of LSD1 and Ku80 were found to be the highest in HCT116 cells (Figure 1A-E). Correlation analysis indicated that the expression of LSD1 and Ku80 was positively correlated (Figure 1F). Immunoprecipitation with specific antibody LSD1 and Coomassie brilliant blue staining after gel electrophoresis showed obvious blue staining at 80 KDa and 110 KDa in the LSD1 group compared with the IgG control group (Figure 1G). Immunofluorescence cell localization revealed that both red fluorescence-labeled Ku80 and green fluorescence-labeled LSD1 overlapped with blue fluorescence-labeled DAPI, suggesting that Ku80 and LSD1 were located mainly in the nucleus (Figure 1H). To identify LSD binding proteins, 364 proteins interacting with LSD1 were identified by MS using gel electrophoresis bands of immunoprecipitation of LSD1 monoclonal antibody. A Venn diagram analysis revealed that 31 proteins were captured by both LSD1 and IgG (Figure 1I; Table S1). Further, string database (https://string-db.org/) was used to visualize the signaling pathways and molecular functions of the protein network interacting with LSD1 (Figure 1J-L). The interaction between Ku80 and LSD1 was confirmed by IP and western blot (Figure 1M). Similarly, Ku80 protein was identified successfully by MS (Figure 1N).
Figure 1.
Ku80 and LSD1 interact in colon cancer cells. LSD1 mRNA levels in the indicated cell lines were measured by RT-qPCR (A). The data were normalized to the expression of the housekeeping gene glyceraldehyde 3-phosphate dehydrogenase (GAPDH). (B) The protein expression of LSD1 in the indicated cells was detected by immunoblot. (C) LSD1 protein gray value statistics. The data were normalized to the expression of the housekeeping gene glyceraldehyde 3-phosphate dehydrogenase (GAPDH). (D) The protein expression of Ku80 in the indicated cells was detected by immunoblot. (E) RT-qPCR measured ku80 mRNA levels in the indicated cell lines. (F) Linear correlation analysis of the expression relationship between Ku80 mRNA and LSD1 mRNA in indicated cells. (G) LSD1 monoclonal antibody was used for immunoprecipitation, and Coomassie brilliant blue staining was performed after gel electrophoresis; Compared with the IgG group, there are obvious bands at 80 kda and 110 kda. (H) LSD1 and Ku80 immunofluorescence subcellular localization in 4 colon cancer cells. (I) Venn diagram of different protein sets between experimental samples, experimental group (exclusive to LSD1) 333. (J-L) Visualize the interaction network between proteins; it can be seen that Ku80 interacts with LSD1. (M) Immunoprecipitation to detect the interaction of Ku80 and LSD1. (N) Mass-to-charge ratio diagram of Ku80 protein identified by mass spectrometry. Three independent assays were performed in triplicate. The data were expressed as mean ± SD. **P<0.01, ***P<0.001.
Migration and invasion ability of colon cancer cells is positively correlated with Ku80 and negatively correlated with FOXF2
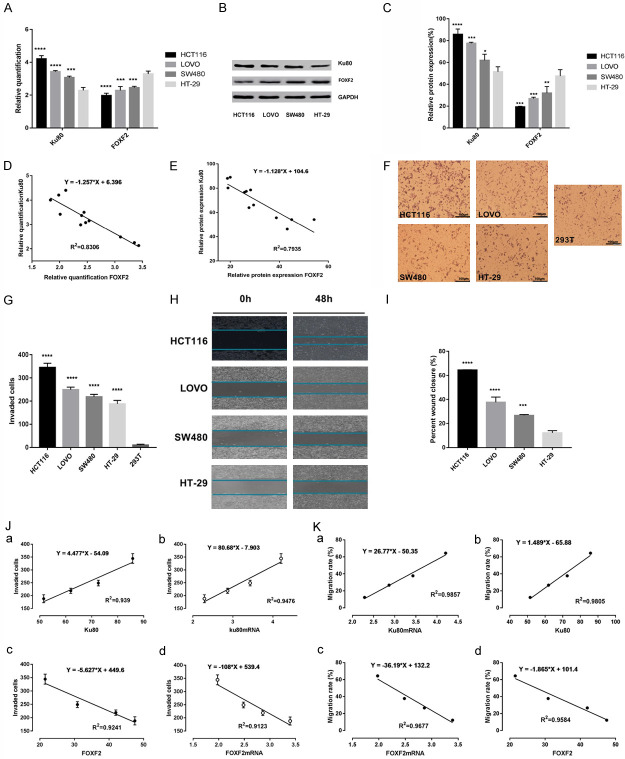
Four colon cancer cell lines were used to detect mRNA and protein expression of Ku80 and FOXF2, as well as to investigate the effect of Ku80 and FOXF2 on migration and invasion ability of colon cancer cells. The results indicated that mRNA and protein expression of Ku80 was highest in HCT116 cells, whereas mRNA and protein expression of FOXF2 was lowest in HCT116 cells (Figure 2A-C). Pearson linear correlation analysis showed that the mRNA and protein expression of Ku80 and FOXF2 were negatively correlated (Figure 2D, 2E). Transwell invasion and wound healing assays were performed to evaluate the invasion and migration ability of colon cancer cells. The results suggested that HCT116 cells displayed the strongest invasion ability in the transwell invasion assay (Figure 2F, 2G) and the fastest migration speed in the wound healing assay (Figure 2H, 2I). Pearson linear correlation analysis revealed that the invasion and migration ability of colon cancer cells was positively correlated with mRNA and protein expression of Ku80, and negatively correlated with mRNA and protein expression of FOXF2 (Figure 2J, 2K).
Figure 2.
Correlation of LSD1, FOXF2 and Ku80 with the invasiveness of colon cancer cell lines. A. RT-qPCR measured ku80, FOXF2 mRNA levels in the indicated cell lines. The data were normalized to the expression of the housekeeping gene glyceraldehyde 3-phosphate dehydrogenase (GAPDH). B. The protein expression of Ku80 and FOXF2 in the indicated cells was detected by immunoblot. C. Ku80, FOXF2 protein gray value statistics. The data were normalized to the expression of the housekeeping gene glyceraldehyde 3-phosphate dehydrogenase (GAPDH). D. Analyzing the correlation between Ku80 and FOXF2 mRNA in the indicated cells was detected by Pearson linear correlation statistics. E. Analyzing the correlation between Ku80 and FOXF2 protein in the indicated cells was detected by Pearson linear correlation statistics. F. Matrigel assay in colon cancer cell lines (HCT116, SW480, LOVO, HT-29) and 293T cell line (magnification, ×100). G. Statistics of the number of cells crossing matrigel in indicated cells. H. Scratch assay to detect the migration distance of 4 colon cancer cell lines at 0 h and 48 h. I. Statistics of the percentage of wound healing in the indicated four cell lines. J. The correlation between the expression levels of Ku80 and FOXF2 with the invasiveness of 4 indicated cells was analyzed by Pearson. K. The correlation between the expression levels of Ku80 and FOXF2 with the migrations of 4 indicated cells was analyzed by Pearson. Three independent assays were performed in triplicate. The data were expressed as mean ± SD. **P<0.01, ***P<0.001.
Abnormal expression of Ku80 and FOXF2 is associated with prognosis in patients with colon cancer
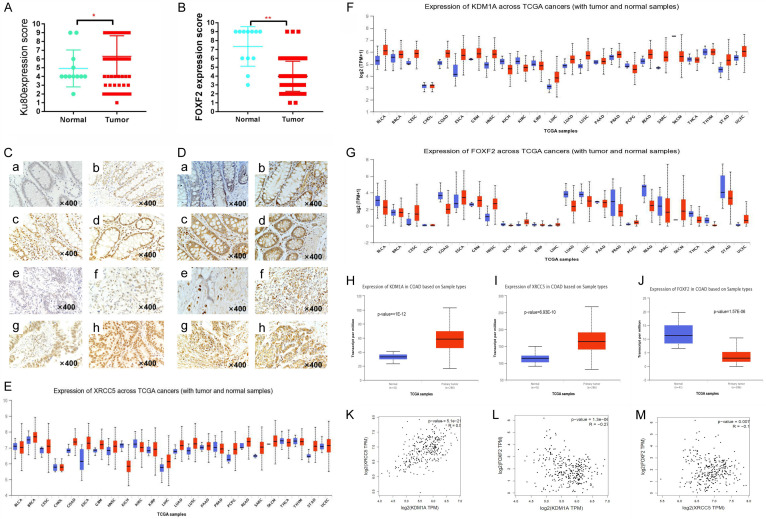
To explore whether abnormal expression of Ku80 and FOXF2 is associated with prognosis in patients with colon cancer, immunohistochemistry was used to detect their expression in 108 colon cancer tissues and 12 para-cancer tissues. Ku80 expression was increased significantly in colon cancer tissues compared with para-cancer tissues (Figure 3A), whereas FOXF2 expression was decreased significantly in colon cancer tissues (Figure 3B). At the same time, we found that Ku80 was stained noticeably in the nucleus, whereas FOXF2 was stained more noticeably in the cytoplasm of colon cancer tissues (Figure 3C, 3D). Among 108 colon cancer samples, 76.93% (82/108) of the cases showed high expression of Ku80, and Ku80 was overexpressed in 58.33% (7/12) of normal colon tissues. No statistically significant differences were found in the expression of Ku80 in colon cancer tissues sorted by gender, age, and Broders’ classification (P>0.05), whereas statistically significant differences were observed when TNM staging (III/IV compared to I/II) and lymph node metastasis (N0 compared to N1) were considered (Table 1). The low expression rate of FOXF2 in colon cancer tissues was 72.22% (78/108) and the high expression rate in normal tissues was 66.67% (8/12). Further analysis found no statistically significant differences in the expression of FOXF2 in colon cancer tissues sorted by gender, age, and Broders’ classification (P>0.05); however, statistically significant expression was observed when TNM staging (comparison between stage III/IV and stage I/II) and lymph node metastasis were considered (Table 2). Furthermore, bioinformatics analysis was used to explore the expression of LSD1, Ku80, and FOXF2 in pan-carcinoma of TCGA by using the UALCAN (http://ualcan.path.uab.edu/) online database. LSD1 (also known as KDM1A) and Ku80 (also known as XRCC5) were found to be increased in colon cancer, whereas FOXF2 was decreased in colon cancer (Figure 3E-G). The expression of LSD1 and Ku80 in the colon adenocarcinoma dataset of TCGA was also higher in primary tumors (n=286) than in the control group (n=41) (Figure 3H, 3I), whereas the expression of FOXF2 was the reverse (Figure 3J). Pearson linear correlation analysis revealed that the expression of LSD1 and Ku80 was positively correlated (r=0.5, P=5.1E-21) (Figure 3K), whereas the expression of LSD1 and Ku80 was negatively correlated with FOXF2 in terms of transcription (Figure 3L, 3M).
Figure 3.
Abnormal expression of Ku80, FOXF2 correlates with clinical characteristics of colon cancer patients. (A) Immunohistochemical scores of Ku80 and (B) FOXF2 expression in colon cancer and normal control group; (C) Ku80 expression in colon tissues: (a) Negative; (b) Weakly positive; (c) Positive; (d) Strongly positive. Expression of Ku80 in colon cancer tissues: (e) Negative; (f) Weakly positive; (g) Positive; (h) Strong positive. (D) FOXF2 expression in colon tissues: (a) Negative; (b) Weakly positive; (c) Positive; (d) Strongly positive. Expression of FOXF2 in colon cancer tissues: (e) Negative; (f) Weakly positive; (g) Positive; (h) Strong positive. SP ×400; (E) The transcriptional expression of Ku80 (XRCC5) in pan-cancer; (F) The transcriptional expression of KDM1A (LSD1) in pan-cancer; (G) The transcriptional expression of FOXF2 in pan-cancer. (H) XRCC5 is expressed at the transcription level in the colon cancer dataset; (I) KDM1A is expressed at the transcription level in the colon cancer dataset; (J) FOXF2 is expressed at the transcription level in the colon cancer dataset; (K) XRCC5 is positively correlated with the expression of KDM1A (LSD1); (L) KDM1A (LSD1) is negatively correlated with FOXF2 expression; (M) XRCC5 is negatively correlated with FOXF2 expression.
Table 1.
Relationship between Ku80 expression in colon cancer and clinicopathological characteristics
| Item | Case (n) | Ku80 expression | χ2 | P-value | |
|---|---|---|---|---|---|
|
| |||||
| Low | High | ||||
| Origin of specimens | |||||
| Colon cancer | 108 | 26 (24.07%) | 82 (76.93%) | ||
| Normal control tissues | 12 | 5 (41.66%) | 7 (58.33%) | 2.521 | 0.0117 |
| Gender | |||||
| Male | 75 | 17 | 58 | ||
| Female | 33 | 9 | 24 | 0.5157 | 0.6060 |
| Age | |||||
| <50 years | 31 | 9 | 22 | ||
| ≥50 years | 77 | 17 | 60 | 0.7647 | 0.4444 |
| TNM stage | |||||
| Stage I/II | 74 | 22 | 52 | ||
| Stage III/IV | 34 | 4 | 30 | 2.028 | 0.0425 |
| Broders’ classification | |||||
| Grade I/II | 86 | 15 | 71 | ||
| Grade III/IV | 22 | 11 | 11 | 3.187 | 0.0513 |
| Distant metastasis | |||||
| Yes | 4 | 0 | 4 | ||
| No | 104 | 26 | 78 | 1.148 | 0.2511 |
| Lymph node metastasis | |||||
| N0 | 73 | 23 | 50 | ||
| N1/N2 | 35 | 3 | 32 | 2.609 | 0.0091 |
Abbreviations: TNM = tumour-node-metastasis; SOFA = Sequential Organ Failure Assessment.
Table 2.
Relationship between FOXF2 expression in colon cancer and clinicopathological characteristics
| Item | Case (n) | FOXF2 expression | χ2 | P-value | |
|---|---|---|---|---|---|
|
| |||||
| Low | High | ||||
| Origin of specimens | |||||
| Colon cancer | 108 | 78 (72.22%) | 30 (27.78%) | ||
| Normal control tissues | 12 | 4 (33.33%) | 8 (66.67%) | 2.747 | 0.0060 |
| Gender | |||||
| Male | 75 | 50 | 25 | ||
| Female | 33 | 28 | 5 | 1.477 | 0.1397 |
| Age | |||||
| <50 years | 31 | 21 | 10 | ||
| ≥50 years | 77 | 57 | 20 | 0.6596 | 0.5095 |
| TNM stage | |||||
| Stage I/II | 74 | 48 | 26 | ||
| Stage III/IV | 34 | 30 | 4 | 2.518 | 0.0118 |
| Broders’ classification | |||||
| Grade I/II | 86 | 66 | 20 | ||
| Grade III/IV | 22 | 12 | 10 | 2.074 | 0.3137 |
| Distant metastasis | |||||
| Yes | 4 | 3 | 1 | ||
| No | 104 | 75 | 29 | 0.1264 | 0.8994 |
| Lymph node metastasis | |||||
| N0 | 73 | 47 | 26 | ||
| N1/N2 | 35 | 31 | 4 | 2.627 | 0.0086 |
Abbreviations: TNM = tumour-node-metastasis.
Effect of interference with Ku80 combined with inhibition of LSD1 on proliferation, invasion, and apoptosis of HCT116 cells
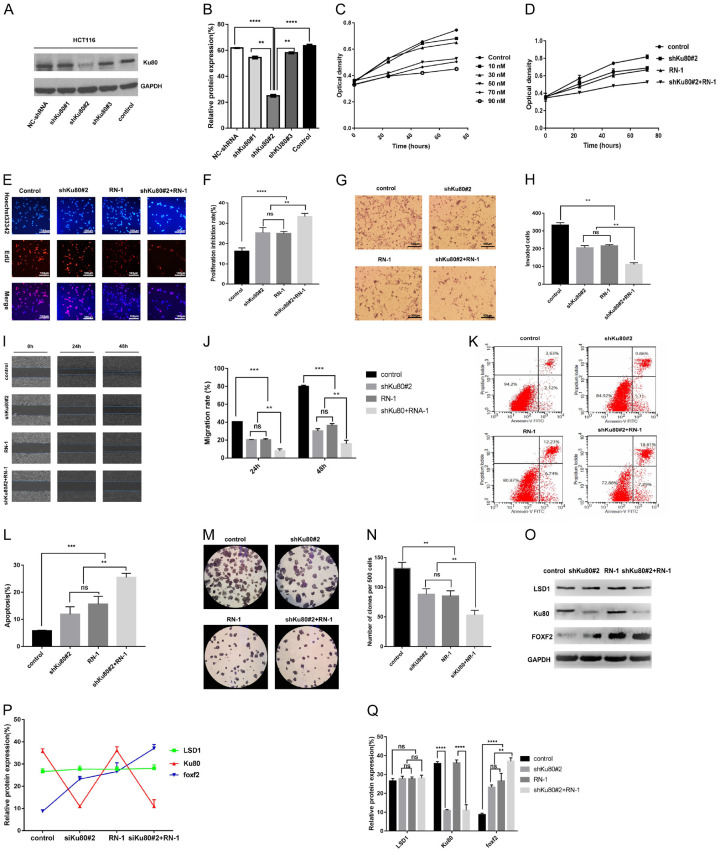
RN-1 (hydrochloride), an LSD1 inhibitor, can effectively and irreversibly inhibit the activity of LSD1 [26,27]. RN-1 was used for experiments to determine the biological function of Ku80 and LSD1 in colon cancer cells. We designed short hairpin RNAs (shRNAs) targeting Ku80 and performed western blot to confirm that transfection of these shNRAs decreased the expression of Ku80 (Figure 4A). Among these, shRNA#2 elicited the best interference efficiency (Figure 4B). Subsequently, the MTT assay was used to detect the effect of different concentrations of RN-1 on the growth of HCT116 cells. We found that a 50-nM RN-1 concentration has the same inhibitory effect on the growth of HCT116 cells as do higher concentrations of RN-1 (Figure 4C). Therefore, we chose 50 nM for follow-up experiments. The results showed that shKu80#2, RN-1, and shKu80#2 + RN-1 inhibited the proliferation of HCT116, and the inhibitory effect on the proliferation of HCT116 cells was time-dependent (Figure 4D). Compared with the control group, the proliferative activity of HCT116 cells in the treatment group (shKu80#2, RN-1, shKu80#2 + RN-1) was inhibited significantly at 24 h, 48 h, and 72 h. In addition, similar results were observed in the EdU assay (Figure 4E, 4F). We next investigated the invasion, apoptosis, and colony formation of HCT116 cells under the above conditions via transwell invasion assay, wound healing assay, and flow cytometry. The results showed that Ku80 knockdown and LSD1 silencing reduced the invasion, migration, proliferation, and colony formation of HCT116 cells, and induced the apoptosis of colon cancer cells (Figure 4G-N). The combined group (shRNA#2 + RN-1) increased the inhibition of the above phenotypes. Furthermore, both Western blot assay and fold plot revealed that knockdown of Ku80 expression and inhibition of LSD1 activity could increase FOXF2 expression (Figure 4O-Q).
Figure 4.
Ku80 knockdown and inhibition of LSD1 repress invasiveness and proliferation and induces apoptosis of colon cancer cells in vitro. A. Western blot detected the shRNA transfection efficiency. B. Statistically significant differences among Different interference sequences. C. Effect of different concentrations of RN-1 on colon cancer HCT116 cell growth. D. At 24 h, 48 h, and 72 h, the proliferation activity of HCT116 cells in the experimental group (shKu80#2, RN-1 group, shKu80#2 + RN-1 group) was significantly inhibited compared with the control group. At 24 h, 48 h, and 72 h, there was no significant difference in the proliferation activity of HCT116 cells in the shKu80#2 and RN-1 groups, but the shKu80#2 + RN-1 group had significant differences compared with the shKu80#2 and RN-1 groups, respectively. E, F. Edu assay also obtained similar results. G-N. Effect of HCT116 cells that transfected shRNA#2 or/and RN-1-treated was observed on the invasion, migration, apoptosis, and clone formation. O-Q. The expression of LSD1, Ku80, and FOXF2 in HCT116 cells that transfected shRNA#2 or/and RN-1-treated were detected by Western blot. Values represent the mean ± standard deviation of 3 independent experiments. **P<0.01, ***P<0.001.
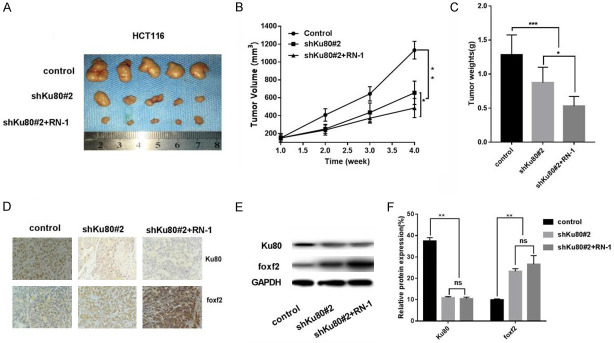
Effect of shKu80#2 and RN-1 on the tumorigenic ability of HCT116 cells in tumorigenic assays
HCT116 cells were injected subcutaneously into nude mice to establish a xenograft tumor model. Using this model, we further evaluated the effect of shKu80#2 or/and RN-1 on the tumorigenesis ability of HCT116 cells. The results revealed that the volume and weight of xenograft tumors in the shKu80#2 group and shKu80#2 + RN-1 group were significantly lower than those in the control group, and the difference in the shKu80#2 + RN-1 group was extremely significant (Figure 5A-C). Western blot and immunohistochemistry of xenograft tumors showed that the expression of FOXF2 was increased and significantly higher in the shKu80#2 + RN-1 group than in the control group. Expression of Ku80 was reduced in xenograft tumors. The protein expression of FOXF2 was restored in colon cancer cells with Ku80 knocked down and LSD1 silenced (Figure 5D-F).
Figure 5.
Effect of shKu80#2 and RN-1 on the tumorigenic ability of HCT116 cells in tumorigenic assays. A. The general picture of xenograft tumor from executed nude mice; B. The volume of xenograft tumor in shKu80#2 group and shKu80#2 + RN-1 group was significantly small than that in the control group, and the volume of xenograft tumor in shKu80#2 + RN-1 group was higher than that in shKu80#. There were also statistical differences between the two groups; C. The statistically significant difference in weight. D. Immunohistochemical detection of Ku80 and foxf2 protein expressions in nude mice xenografts in the control group, shKu80#2 group, shKu80#2 + RN-1 group revealed that FOXF2 protein increased. E. The Ku80 and foxf2 protein expression in transplanted tumors were was detected by Western blot; the shKu80#2 + RN-1 group found that FOXF2 protein was significantly restored. F. FOXF2 gene promoter region 1-2000 bp base sequence.
Ku80 binds to the promoter region of FOXF2 and regulates its expression through histone demethylation modification
Previous studies have found that FOXF2 is a downstream target related to colon cancer metastasis regulated by LSD1 [15]. Based on the above experimental and previous findings, we speculated that LSD1 regulates the expression of FOXF2 through mediating Ku80. Majority of identified FOXF2 binding sites were located in the promoters, of which most localized in proximal (defined as 0-2000 bp upstream of transcription start site (TSS)). To identify binding sites on FOXF2 that are regulated by LSD1, we searched the NCBI website (https://www.ncbi.nlm.nih.gov/gene/?term=FOXF2) (Supplementary Text 1) for the 1-2000 bp base sequences of FOXF2 and designed ten pairs of primers for these sequence segments. Finally, we verified our speculation by chromatin immunoprecipitation combined with PCR technique. At the same time, agarose gel electrophoresis revealed the abundance of the area bound by Ku80. We found that FOXF2 primer pairs 2, 4, 5, 7, 8, and 9 had corresponding amplified fragment bands in agarose gel electrophoresis detection. Finally, primers 2, 4, 5, 7, 8, and 9 were selected for qPCR detection of the amplified fragments, and the fifth pair of primers was found (the amplified region is 687-887 bp in the base sequence of the promoter region of the FOXF2 gene) (Figure 6A). The amplified binding region was significantly higher than the fragment amplified by other primers (Figure 6B). Further, Ku80 was detected by western blot after CHIP (Figure 6C). For these reasons, we concluded that Ku80 can bind to the 687-887-bp part of the FOXF2 promoter region. Subsequently, the eluate after CHIP was used to detect the level of H3K4me2 in the FOXF2 promoter region by western blot. The expression of H3K4me2 in the shKu80#2 + RN-1 group was significantly higher than in the other two groups (Figure 6D, 6E). Thus, Ku80 shRNA effectively reduced Ku80 expression, which significantly reduced the binding of Ku80 to the FOXF2 promoter region and, combined with active inhibitors of LSD1, increased the level of H3K4me2 in the FOXF2 promoter region and inhibited HCT116 cell metastasis.
Figure 6.
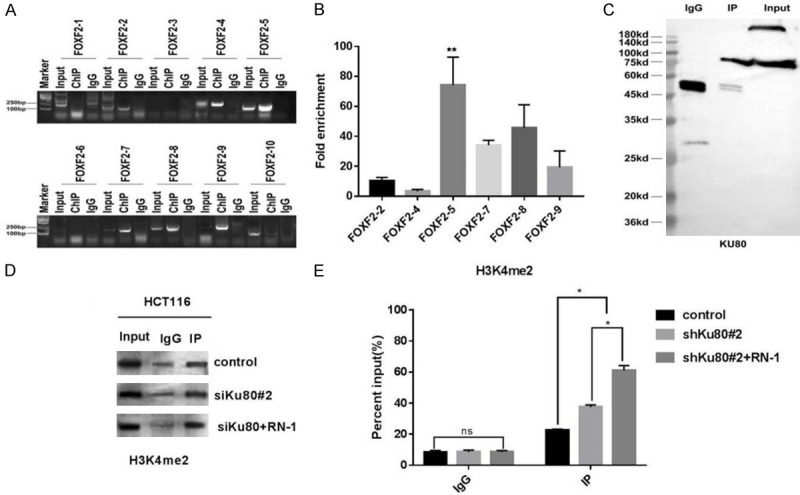
Ku80 knockdown or/and inhibition of LSD1 repress the growth of xenograft tumor in vivo, and CHIP-PCR detection of the abundance of ku80 bound to the FOXF2 promoter region. A. The fifth pair of primers (the amplified region is FOXF2 The 687-887 bp in the base sequence of the promoter region of the gene). B. The amplified binding region was significantly higher than the fragment amplified by other primers. C. After CHIP and detecting the protein in the eluate, Ku80 protein can be detected. D. Western bolt detects the expression level of H3K4me2 in the FOXF2 gene promoter region in Ku80#2 + RN-1 group; E. Comparison of Ku80#2 + RN-1 group with control and shKu80#2 group. **P<0.01, ***P<0.001.
LSD1 regulates the Wnt/β-catenin signaling pathway via demethylation of FOXF2
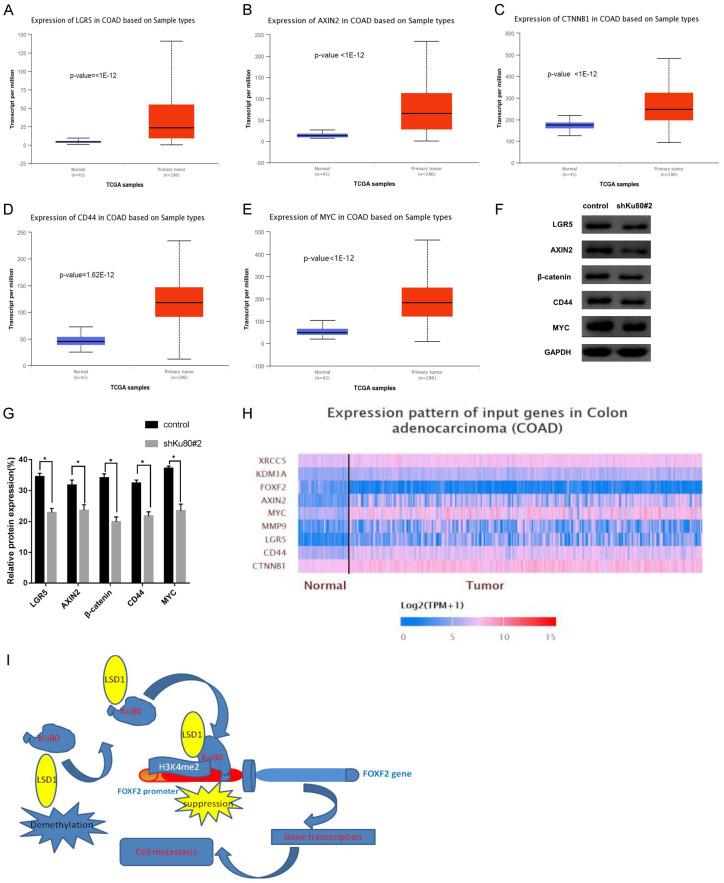
The CHIP-qPCR assay indicated that the FOXF2 promoter region where Ku80 binds to the gene is 687-887 bp, and LSD1 upregulates the methylation level of H3K4me2 in the promoter region to inhibit the transcriptional activation of FOXF2. Previous studies have found that FOXF2 and LSD1 can influence tumor progression through the Wnt/β-catenin signaling pathway [28,29]. Therefore, downstream target genes LGR5, AXIN2, CD44, MYC, and CTNNB1 of the Wnt/β-catenin signaling pathway in the TCGA colon cancer dataset (http://ualcan.path.uab.edu/analysis.) were analyzed using bioinformatics analysis. The transcription level of these target genes was significantly upregulated in primary colon cancer tissues (n=286) compared with normal colon tissues (n=41) (Figure 7A-E). Subsequently, we detected the expression of some of these genes in HCT116 cells transfected with shKu80#2 by western blot. Compared with the control, the expression of these genes was significantly downregulated (Figure 7F, 7G). Heat maps showed that the primary tumor was significantly darker than the normal control, indicating that Ku80, LSD1, and downstream genes of Wnt/β-catenin were overexpressed in the tumor, whereas FOXF2 was underexpressed (Figure 7H).
Figure 7.
LSD1 Regulates Wnt/β-catenin via demethylation of the FOXF2 gene. A. In the TCGA colon cancer dataset, LGR5 is significantly higher in colon cancer tissues than in normal colon tissues; B. AXIN2 is significantly higher in colon cancer tissues than in normal colon tissues; C. β-catenin encoding gene CTNNB1 is significantly higher in colon cancer tissues than in normal colon tissues. D. CD44 is significantly higher in colon cancer tissues than in normal colon tissues; E. MYC is significantly higher in colon cancer tissues than normal colon tissues. F. The expression of LGR5, AXIN2, CD44, MYC, and CTNNB1 genes was detected by western blot in HCT116 cells that interfered with Ku80; G. Compared with control cells, the expression of the above genes was significantly down-regulated. H. The heat map shows the expression of each gene Happening. I. The mechanism diagram of the full text. **P<0.01, ***P<0.001.
The above results indicate that silencing Ku80 or inhibiting the activity of LSD1 can upregulate the expression of FOXF2 and inhibit the activity of the Wnt/β-catenin pathway both in vivo and in vitro, which is consistent with previous results [30-32].
Discussion
It is widely accepted that tumorigenesis is caused by a series of genetic aberrations, including oncogenes and tumor suppressor genes, which have been identified as the cause of tumors [33]. However, in many diseases, including tumors, epigenetic modifications that cooperate with genetic mechanisms regulating transcriptional activity are dysregulated [34,35]. Over time, researchers have gradually realized that histone lysine demethylation is reversible. In particular, the discovery of H3K4 demethylation by LSD1, a member of the amine oxidase family, changed the concept of this field [36]. Abnormal DNA methylation, histone modification, and long non-coding RNA expression are closely related to tumor initiation, progression, and metastasis. In the past decade, cancer epigenetics has allowed us to unearth novel biomarkers and provide therapeutic targets for multiple types of cancer [2]. In order to implement these functions, such as differentiation of normal and cancer cells, cell activity, epithelial-mesenchymal transformation, autophagy, senescence, neurodegenerative diseases, metabolism, and a wide range of biological functions, LSD1 is required as a component of different multi-protein complexes and has been proven to interact with more than 60 regulatory proteins [37]. Using immunoprecipitation, western blot, mass spectrometry and other methods, we found that LSD1 interacts with Ku80. The expression of Ku80 in colon cancer cells and colon tissue is positively correlated with the expression of LSD1. Previous studies found that high expression of LSD1 often is associated with poor prognosis of patients with colon cancer [6,38], indicating that Ku80 not only interacts with LSD1, but may also be related to the progression of colon cancer.
Ku80 is known for its key function in repairing DNA double-strand breaks [39]. Recent studies [17,40] have found that Ku is downregulated in malignant melanoma, lung cancer, and cervical cancer. In contrast, the expression of Ku is elevated in ovarian cancer, bladder cancer, malignant lymphoma, esophageal cancer, breast cancer, non-melanoma skin cancer, and oral cancer tissues, which indicates that the abnormal expression of Ku is closely related to the occurrence and development of these tumors. Thus, overexpression of Ku promotes hyperproliferation and anti-apoptosis, whereas insufficient or low expression of Ku leads to genome instability and tumorigenesis, suggesting that Ku can act as a tumor suppressor or an oncogene [41]. Here, we found that the expression of Ku80 was significantly upregulated in colon cancer cells and tissues, and the expression was positively correlated with the invasion and migration ability of colon cancer cells. The expression of Ku80 is associated with the clinical characteristics, which means that high expression of Ku80 is associated with poor prognosis in patients with colon cancer. Recently, the protein expression of Ku80 has been reported to be significantly higher in thyroid cancer tissues than that of non-neoplastic adjacent tissue samples. Knockdown of Ku80 reduces invasion and colony formation, increases cell apoptosis, and reduces involvement in MAPK signal transduction. The high expression of Ku80 in thyroid cancer is related to the expression of RET/TC and the activation of NF-dB, whereas the knockdown of Ku80 reduces the malignancy of thyroid cancer cells [42]. In the present study, we knocked down Ku80 in colon cancer cells, which can promote apoptosis and inhibit colon cancer cell invasion, migration, and proliferation in vitro and in vivo. At the same time, the expression of downstream targets LGR5, AXIN2, CD44, and MYC in the Wnt/β-catenin pathway was significantly downregulated. These findings indicate that Ku80 is expected to become an important candidate target for the development of anti-cancer drugs.
Our previous study found that FOXF2 is one of the downstream targets of LSD1 regulating the metastasis of colon cancer cells. FOXF2 expression is downregulated in many types of cancer [43-45]. FOXF2 plays an important role in the synthesis of extracellular matrix (ECM). In foxf2+/-mice, due to defects in synthesis, ECM was reduced severely, with cleft lip and abnormal tongue development [46]. Another study demonstrated that FOXF2 reduces and regulates cellular ECM signaling in prostate cancer [47]. Kong et al. [48] found that the decline of FOXF2 mRNA expression in primary breast cancer was negatively correlated with tumor progression, including tumor size, number of metastatic lymph nodes, and clinical stage. In addition, patients with low FOXF2 mRNA expression in their tumors had poorer prognosis. Similarly, we found reduced expression of FOXF2 in colon cancer tissues compared with normal colon tissues. We confirmed these findings by subsequent analysis of the TCGA colon cancer dataset using the online UALCAN tool. Pearson linear correlation analysis revealed that the expression of LSD1 and Ku80 was significantly negatively correlated with FOXF2. FOXF2 expression level is also related to the clinicopathological characteristics of patients with colon cancer.
As a core regulator of transcription, LSD1 is involved in the activation or repression of gene transcription. LSD1 inhibits the transcription process and interacts with members of the SNAG domain of transcription factors, such as SNAIL1/2 and GFI1/B. By interacting with the SNAG domain of SNAIL, the LSD1-CoREST complex is recruited to the E-cadherin promoter to remove the methyl group on histone H3 lysine 4, thereby inhibiting its expression [49]. In addition, LSD1 is also a co-activator of androgen and estrogen receptor-dependent transcription [50,51]. Other research has proposed that LSD1 recruits estrogen or c-Myc’s upper target gene, and that DNA triggers oxidation and base excision repair enzymes that recruit its beneficial chromatin transcriptional activation cycle [50,52]. To further clarify the relationship between Ku80, LSD1, and FOXF2, we speculate that Ku80 interacts with LSD1 and binds to the promoter region of FOXF2 to regulate the expression of FOXF2. This inference was subsequently verified by CHIP-PCR assay. LSD1 demethylates and interacts with Ku80, and binds to the 687-887-bp site of the FOXF2 promoter region. In addition, LSD1-silencing and Ku80 knockdown both lead to the upregulation of FOXF2, and both inhibited the malignant phenotype of colon cancer cells.
Due to the importance of LSD1, some LSD1 inhibitors are already in clinical trials. However, many issues still need to be overcome before LSD1 inhibitors can be applied [4]. Although many LSD1 inhibitors can eliminate LSD1-mediated demethylation, they do not necessarily have effective anti-cancer activity, because many oncogenes or tumor-suppressors are usually regulated by multiple enzymes or proteins. Our results suggest that Ku80 combined with LSD1 selective inhibitor (RN-1) interferes with invasion, migration, and proliferation of colon cancer cells more effectively than RN-1 or Ku80 knockdown in vitro. Ku80 promotes apoptosis, while inhibiting the tumorigenic ability of colon cancer cells in vivo and in vitro. These results suggest that LSD1 inhibitor combined with Ku80 knockdown has a better inhibition on the malignant phenotype of colon cancer cells.
The Wnt/β-catenin signaling pathway is involved in all aspects of embryonic development and controls homeostasis and self-renewal in many adult tissues [53]. The Wnt/β-catenin signaling pathway also mediates a variety of cellular processes, such as proliferation, differentiation, survival, apoptosis, and cell movement [53]. Dysregulation of this pathway can lead to the occurrence of a variety of tumors, including colorectal cancer [29,53-55]. According to these reports, LSD1 downregulates DKK1, which allows free β-catenin to avoid phosphorylation and degradation. Then, free β-catenin accumulates and is transported to the nucleus, where it upregulates the transcription of the downstream target c-Myc. Next, the overexpression of c-Myc can cause the cell to transform into a malignant phenotype. This discovery connects LSD1 and DKK1, and clarifies the role of LSD1 on the activation of Wnt/β-catenin signaling pathway and its target genes [29]. Previous studies have shown that high expression of c-Myc, CyclinD1, MMP9, and LGR5 plays a significant role in promoting the development of tumors [56,57]. With regard to cervical cancer, FOXF2 was found to inhibit the expression of β-catenin in the nucleus, and inhibit the expression of target c-Myc, CyclinD1, MMP9, and Lgr5 in the Wnt/β-catenin signaling pathway, so that FOXF2 regulates the Wnt/β-catenin signaling pathway and inhibits the proliferation, migration, and invasion of Hela cells. Previous studies have found that the fork-shaped transcription factor FoxF2 in intestinal fibroblasts can inhibit paracrine Wnt/β-catenin signaling, and limit the niche of mouse intestinal crypt stem cells. This affects the number of stem cells in mice and the formation and growth of adenomas. Loss of FoXF2 promotes the formation and growth of adenomas [30]. The above reports indicate that LSD1 and FOXF2 are involved in the regulation of tumor cells by the Wnt/β-catenin signaling pathway. Our results illustrate the regulatory relationship between LSD1 and FOXF2. Similarly, we analyzed the TCGA colon cancer dataset through bioinformatics and found that the downstream targets of the Wnt/β-catenin signaling pathway were significantly higher in colon cancer compared with that in normal colon tissues. After knocking down the protein expression of Ku80 in colon cancer cells, the expression of the downstream targets LGR5, AXIN2, CD44, and MYC of the Wnt/β-catenin signaling pathway was significantly downregulated. Therefore, we believe that Ku80, LSD1, and FOXF2 are involved in the malignant phenotype of colon cancer by influencing the Wnt/β-catenin signaling pathway.
In summary, Ku80 interacts with LSD1 and binds to the 687-887-bp site of the promoter region of FOXF2 to demethylate and inhibit FOXF2 expression. The inhibition of Ku80 and LSD1 activity can better inhibit the invasion, migration, proliferation, and allogeneic tumor formation of colon cancer cells, and induce their apoptosis. The mechanism is to significantly upregulate H3K4me2 methylation level in the FOXF2 promoter region, facilitate transcriptional activation of FOXF2, and inhibit the expression of downstream targets through the Wnt/β-catenin signaling pathway, thereby inhibiting the malignant characteristics of colon cancer cells (Figure 7I). The joint development of LSD1 and Ku80 inhibitors may be one of the promising therapeutic targets for colon cancer in the future.
Acknowledgements
This study was supported by the National Natural Science Foundation of China (81360366, 81302169); Guizhou High-level Innovative Talents Foundation (GZSYQCC [2014] 001); Guizhou Outstanding Young Scientific and Technological Talents Foundation ([2017] 5602); Guizhou Innovation and Entrepreneurship Foundation for High-level Overseas Talent ([2018] 04); Guizhou Science and Technology Foundation ([2019] 1198, [2020] 1Z064); Youth Fund of Guizhou Provincial People’s Hospital (GZSYQN [2018] 01).
Disclosure of conflict of interest
None.
Supplementary Text 1
Table S1
References
- 1.Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2021. CA Cancer J Clin. 2021;71:7–33. doi: 10.3322/caac.21654. [DOI] [PubMed] [Google Scholar]
- 2.Park JW, Han JW. Targeting epigenetics for cancer therapy. Arch Pharm Res. 2019;42:159–170. doi: 10.1007/s12272-019-01126-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shi YJ, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, Casero RA, Shi Y. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119:941–953. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 4.Fang Y, Liao GC, Yu B. LSD1/KDM1A inhibitors in clinical trials: advances and prospects. J Hematol Oncol. 2019;12:129. doi: 10.1186/s13045-019-0811-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Karakaidos P, Verigos J, Magklara A. LSD1/KDM1A, a gate-keeper of cancer stemness and a promising therapeutic target. Cancers (Basel) 2019;11:1821. doi: 10.3390/cancers11121821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ding J, Zhang ZM, Xia Y, Liao GQ, Pan Y, Liu S, Zhang Y, Yan ZS. LSD1-mediated epigenetic modification contributes to proliferation and metastasis of colon cancer. Br J Cancer. 2013;109:994–1003. doi: 10.1038/bjc.2013.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Saleque S, Kim J, Rooke HM, Orkin SH. Epigenetic regulation of hematopoietic differentiation by Gfi-1 and Gfi-1b is mediated by the cofactors CoREST and LSD1. Mol Cell. 2007;27:562–572. doi: 10.1016/j.molcel.2007.06.039. [DOI] [PubMed] [Google Scholar]
- 8.Shi YJ, Matson C, Lan F, Iwase S, Baba T, Shi Y. Regulation of LSD1 histone demethylase activity by its associated factors. Mol Cell. 2005;19:857–864. doi: 10.1016/j.molcel.2005.08.027. [DOI] [PubMed] [Google Scholar]
- 9.Lee MG, Wynder C, Cooch N, Shiekhattar R. An essential role for CoREST in nucleosomal histone 3 lysine 4 demethylation. Nature. 2005;437:432–435. doi: 10.1038/nature04021. [DOI] [PubMed] [Google Scholar]
- 10.Gu FY, Lin YX, Wang Z, Wu XX, Ye ZY, Wang YZ, Lan HY. Biological roles of LSD1 beyond its demethylase activity. Cell Mol Life Sci. 2020;77:3341–3350. doi: 10.1007/s00018-020-03489-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coffer PJ, Burgering BM. Forkhead-box transcription factors and their role in the immune system. Nat Rev Immunol. 2004;4:889–899. doi: 10.1038/nri1488. [DOI] [PubMed] [Google Scholar]
- 12.Myatt SS, Lam EW. The emerging roles of forkhead box (Fox) proteins in cancer. Nat Rev Cancer. 2007;7:847–859. doi: 10.1038/nrc2223. [DOI] [PubMed] [Google Scholar]
- 13.Zhu H. Forkhead box transcription factors in embryonic heart development and congenital heart disease. Life Sci. 2016;144:194–201. doi: 10.1016/j.lfs.2015.12.001. [DOI] [PubMed] [Google Scholar]
- 14.Jackson BC, Carpenter C, Nebert DW, Vasiliou V. Update of human and mouse forkhead box (FOX) gene families. Hum Genomics. 2010;4:345–352. doi: 10.1186/1479-7364-4-5-345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen J, Ding J, Wang ZW, Zhu J, Wang XJ, Du JY. Identification of downstream metastasis-associated target genes regulated by LSD1 in colon cancer cells. Oncotarget. 2017;8:19609–19630. doi: 10.18632/oncotarget.14778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pazhanisamy SK. Stem cells, DNA damage, ageing and cancer. Hematol Oncol Stem Cell Ther. 2009;2:375–384. doi: 10.1016/s1658-3876(09)50005-2. [DOI] [PubMed] [Google Scholar]
- 17.Fell VL, Schild-Poulter C. The Ku heterodimer: function in DNA repair and beyond. Mutat Res Rev Mutat Res. 2015;763:15–29. doi: 10.1016/j.mrrev.2014.06.002. [DOI] [PubMed] [Google Scholar]
- 18.Boulton SJ, Jackson SP. Identification of a Saccharomyces cerevisiae Ku80 homologue: roles in DNA double strand break rejoining and in telomeric maintenance. Nucleic Acids Res. 1996;24:4639–4648. doi: 10.1093/nar/24.23.4639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nussenzweig A, Chen C, da Costa Soares V, Sanchez M, Sokol K, Nussenzweig MC, Li GC. Requirement for Ku80 in growth and immunoglobulin V(D)J recombination. Nature. 1996;382:551–555. doi: 10.1038/382551a0. [DOI] [PubMed] [Google Scholar]
- 20.Munoz P, Zdzienicka MZ, Blanchard JM, Piette J. Hypersensitivity of Ku-deficient cells toward the DNA topoisomerase II inhibitor ICRF-193 suggests a novel role for Ku antigen during the G2 and M phases of the cell cycle. Mol Cell Biol. 1998;18:5797–5808. doi: 10.1128/mcb.18.10.5797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Giffin W, Torrance H, Rodda DJ, Prefontaine GG, Pope L, Hache RJ. Sequence-specific DNA binding by Ku autoantigen and its effects on transcription. Nature. 1996;380:265–268. doi: 10.1038/380265a0. [DOI] [PubMed] [Google Scholar]
- 22.Ono M, Tucker PW, Capra JD. Ku is a general inhibitor of DNA-protein complex formation and transcription. Mol Immunol. 1996;33:787–796. doi: 10.1016/0161-5890(96)00030-2. [DOI] [PubMed] [Google Scholar]
- 23.Wang S, Xi JJ, Lin ZW, Hao JT, Yao C, Zhan C, Jiang W, Shi Y, Wang Q. Clinical values of Ku80 upregulation in superficial esophageal squamous cell carcinoma. Cancer Med. 2018;7:1006–1018. doi: 10.1002/cam4.1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang S, Wang Z, Yang YU, Shi MO, Sun ZG. Overexpression of Ku80 correlates with aggressive clinicopathological features and adverse prognosis in esophageal squamous cell carcinoma. Oncol Lett. 2015;10:2705–2712. doi: 10.3892/ol.2015.3635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ye J, Wei X, Shang Y, Pan Q, Yang M, Tian Y, He Y, Peng Z, Chen L, Chen W, Wang R. Core 3 mucin-type O-glycan restoration in colorectal cancer cells promotes MUC1/p53/miR-200c-dependent epithelial identity. Oncogene. 2017;36:6391–6407. doi: 10.1038/onc.2017.241. [DOI] [PubMed] [Google Scholar]
- 26.Jambhekar A, Anastas JN, Shi Y. Histone lysine demethylase inhibitors. Cold Spring Harb Perspect Med. 2017;7:a026484. doi: 10.1101/cshperspect.a026484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cui SY, Lim KC, Shi LH, Lee M, Jearawiriyapaisarn N, Myers G, Campbell A, Harro D, Iwase S, Trievel RC, Rivers A, DeSimone J, Lavelle D, Saunthararajah Y, Engel JD. The LSD1 inhibitor RN-1 induces fetal hemoglobin synthesis and reduces disease pathology in sickle cell mice. Blood. 2015;126:386–396. doi: 10.1182/blood-2015-02-626259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ormestad M, Astorga J, Landgren H, Wang T, Johansson BR, Miura N, Carlsson P. Foxf1 and Foxf2 control murine gut development by limiting mesenchymal Wnt signaling and promoting extracellular matrix production. Development. 2006;133:833–843. doi: 10.1242/dev.02252. [DOI] [PubMed] [Google Scholar]
- 29.Huang ZB, Li SZ, Song W, Li X, Li QS, Zhang ZY, Han YQ, Zhang XD, Miao SY, Du RL, Wang LF. Lysine-specific demethylase 1 (LSD1/KDM1A) contributes to colorectal tumorigenesis via activation of the Wnt/beta-catenin pathway by down-regulating Dickkopf-1 (DKK1) [corrected] PLoS One. 2013;8:e70077. doi: 10.1371/journal.pone.0070077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nik AM, Reyahi A, Ponten F, Carlsson P. Foxf2 in intestinal fibroblasts reduces numbers of Lgr5(+) stem cells and adenoma formation by inhibiting Wnt signaling. Gastroenterology. 2013;144:1001–1011. doi: 10.1053/j.gastro.2013.01.045. [DOI] [PubMed] [Google Scholar]
- 31.Zhang J, Zhang CX, Sang L, Huang L, Du J, Zhao XB. FOXF2 inhibits proliferation, migration, and invasion of Hela cells by regulating Wnt signaling pathway. Biosci Rep. 2018;38:BSR20180747. doi: 10.1042/BSR20180747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Higashimori A, Dong YJ, Zhang YQ, Kang W, Nakatsu G, Ng SSM, Arakawa T, Sung JJY, Chan FKL, Yu J. Forkhead box F2 suppresses gastric cancer through a novel FOXF2-IRF2BPL-beta-catenin signaling axis. Cancer Res. 2018;78:1643–1656. doi: 10.1158/0008-5472.CAN-17-2403. [DOI] [PubMed] [Google Scholar]
- 33.Hoeijmakers JH. Genome maintenance mechanisms for preventing cancer. Nature. 2001;411:366–374. doi: 10.1038/35077232. [DOI] [PubMed] [Google Scholar]
- 34.You JS, Jones PA. Cancer genetics and epigenetics: two sides of the same coin? Cancer Cell. 2012;22:9–20. doi: 10.1016/j.ccr.2012.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Baylin SB, Jones PA. Epigenetic determinants of cancer. Cold Spring Harb Perspect Biol. 2016;8:a019505. doi: 10.1101/cshperspect.a019505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Trojer P, Reinberg D. Histone lysine demethylases and their impact on epigenetics. Cell. 2006;125:213–217. doi: 10.1016/j.cell.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 37.Majello B, Gorini F, Sacca CD, Amente S. Expanding the role of the histone lysine-specific demethylase LSD1 in cancer. Cancers (Basel) 2019;11:324. doi: 10.3390/cancers11030324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jie D, Zhongmin Z, Guoqing L, Sheng L, Yi Z, Jing W, Liang Z. Positive expression of LSD1 and negative expression of E-cadherin correlate with metastasis and poor prognosis of colon cancer. Dig Dis Sci. 2013;58:1581–1589. doi: 10.1007/s10620-012-2552-2. [DOI] [PubMed] [Google Scholar]
- 39.Ross GM, Eady JJ, Mithal NP, Bush C, Steel GG, Jeggo PA, McMillan TJ. DNA strand break rejoining defect in xrs-6 is complemented by transfection with the human Ku80 gene. Cancer Res. 1995;55:1235–1238. [PubMed] [Google Scholar]
- 40.Tuteja R, Tuteja N. Ku autoantigen: a multifunctional DNA-binding protein. Crit Rev Biochem Mol Biol. 2000;35:1–33. doi: 10.1080/10409230091169177. [DOI] [PubMed] [Google Scholar]
- 41.Gullo C, Au M, Feng G, Teoh G. The biology of Ku and its potential oncogenic role in cancer. Biochim Biophys Acta. 2006;1765:223–234. doi: 10.1016/j.bbcan.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 42.Fan YL, Li JY, Wei W, Fang HR, Duan Y, Li NM, Zhang YY, Yu J, Wang JH. Ku80 gene knockdown by the CRISPR/Cas9 technique affects the biological functions of human thyroid carcinoma cells. Oncol Rep. 2019;42:2486–2498. doi: 10.3892/or.2019.7348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kong PZ, Yang F, Li L, Li XQ, Feng YM. Decreased FOXF2 mRNA expression indicates early-onset metastasis and poor prognosis for breast cancer patients with histological grade II tumor. PLoS One. 2013;8:e61591. doi: 10.1371/journal.pone.0061591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.van der Heul-Nieuwenhuijsen L, Dits N, Van Ijcken W, de Lange D, Jenster G. The FOXF2 pathway in the human prostate stroma. Prostate. 2009;69:1538–1547. doi: 10.1002/pros.20996. [DOI] [PubMed] [Google Scholar]
- 45.Zheng YZ, Wen J, Cao X, Yang H, Luo KJ, Liu QW, Huang QY, Chen JY, Fu JH. Decreased mRNA expression of transcription factor forkhead box F2 is an indicator of poor prognosis in patients with resected esophageal squamous cell carcinoma. Mol Clin Oncol. 2015;3:713–719. doi: 10.3892/mco.2015.511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xu JY, Liu H, Lan Y, Aronow BJ, Kalinichenko VV, Jiang RL. A Shh-Foxf-Fgf18-Shh molecular circuit regulating palate development. PLoS Genet. 2016;12:e1005769. doi: 10.1371/journal.pgen.1005769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cai J, Tian AX, Wang QS, Kong PZ, Du X, Li XQ, Feng YM. FOXF2 suppresses the FOXC2-mediated epithelial-mesenchymal transition and multidrug resistance of basal-like breast cancer. Cancer Lett. 2015;367:129–137. doi: 10.1016/j.canlet.2015.07.001. [DOI] [PubMed] [Google Scholar]
- 48.Kong PZ, Yang F, Li L, Li XQ, Feng YM. Decreased FOXF2 mRNA expression indicates early-onset metastasis and poor prognosis for breast cancer patients with histological grade II tumor. PLoS One. 2013;8:e61591. doi: 10.1371/journal.pone.0061591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lin YW, Wu YD, Li JL, Dong CF, Ye XF, Chi YI, Evers BM, Zhou BP. The SNAG domain of Snail1 functions as a molecular hook for recruiting lysine-specific demethylase 1. EMBO J. 2010;29:1803–1816. doi: 10.1038/emboj.2010.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Perillo B, Ombra MN, Bertoni A, Cuozzo C, Sacchetti S, Sasso A, Chiariotti L, Malorni A, Abbondanza C, Avvedimento EV. DNA oxidation as triggered by H3K9me2 demethylation drives estrogen-induced gene expression. Science. 2008;319:202–206. doi: 10.1126/science.1147674. [DOI] [PubMed] [Google Scholar]
- 51.Metzger E, Wissmann M, Yin N, Muller JM, Schneider R, Peters AH, Gunther T, Buettner R, Schule R. LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription. Nature. 2005;437:436–439. doi: 10.1038/nature04020. [DOI] [PubMed] [Google Scholar]
- 52.Amente S, Bertoni A, Morano A, Lania L, Avvedimento EV, Majello B. LSD1-mediated demethylation of histone H3 lysine 4 triggers Myc-induced transcription. Oncogene. 2010;29:3691–3702. doi: 10.1038/onc.2010.120. [DOI] [PubMed] [Google Scholar]
- 53.Clevers H. Wnt/beta-catenin signaling in development and disease. Cell. 2006;127:469–480. doi: 10.1016/j.cell.2006.10.018. [DOI] [PubMed] [Google Scholar]
- 54.Turashvili G, Bouchal J, Burkadze G, Kolar Z. Wnt signaling pathway in mammary gland development and carcinogenesis. Pathobiology. 2006;73:213–223. doi: 10.1159/000098207. [DOI] [PubMed] [Google Scholar]
- 55.Ilyas M. Wnt signalling and the mechanistic basis of tumour development. J Pathol. 2005;205:130–144. doi: 10.1002/path.1692. [DOI] [PubMed] [Google Scholar]
- 56.Cho HS, Suzuki T, Dohmae N, Hayami S, Unoki M, Yoshimatsu M, Toyokawa G, Takawa M, Chen T, Kurash JK, Field HI, Ponder BA, Nakamura Y, Hamamoto R. Demethylation of RB regulator MYPT1 by histone demethylase LSD1 promotes cell cycle progression in cancer cells. Cancer Res. 2011;71:655–660. doi: 10.1158/0008-5472.CAN-10-2446. [DOI] [PubMed] [Google Scholar]
- 57.Lv TF, Yuan DM, Miao XH, Lv YL, Zhan P, Shen XK, Song Y. Over-expression of LSD1 promotes proliferation, migration and invasion in non-small cell lung cancer. PLoS One. 2012;7:e35065. doi: 10.1371/journal.pone.0035065. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.