Abstract
The development of personalized neoantigen-based vaccines in cancer immunotherapy has shown promise. In this study, a large-scale bioinformatics analysis was performed to identify potential GBM-associated neoantigens based on abnormal alternative splicing, and then screen suitable patients for vaccination. Gene expression profiles and clinical information were collected from TCGA. We filtered the percent-spliced-in (PSI) spectrum of alternative splicing events in the dataset to identify abnormal alternative splicing events. MAF package was used to identify and analyse tumour mutation burden (TMB) in cancer samples. Tumour Immune Estimation Resource (TIMER) was used to calculate and visualize the infiltration of antigen presenting cells (APCs). In addition, consistent clustering algorithm utilized to identify immune subtypes of GBM. Five potential tumour neoantigens (LRP1, TCF12, DERL3, WIPI2, and TSHZ3) were identified in GBM by selecting genes both with abnormal alternative splicing (upregulated) and gene frameshift mutations, in which LRP1 was significantly associated with APCs. According to the expressions of five potential tumour neoantigens, 160 patients with GBM were divided into three immune subtypes. Patients in cluster3 exhibited good prognoses. Furthermore, the characteristics, including TMB, abnormal alternative splicing events, immune activity, immune cells proportion, and association with tumour biomarkers, were unique in each immune subtypes. The characteristics of cluster3 illustrated that cluster3 participants were more suitable candidates for vaccination. LRP1 was identified as a potential neoantigen for immunotherapy against GBM, and patients in cluster3 were more suitable for vaccination. Our findings provide important guidance for the development of novel neoantigens and therapeutic targets in patients with GBM.
Keywords: Personalized neoantigen-based vaccines, glioblastoma, abnormal alternative splicing
Introduction
Glioma is one of the most malignant solid tumours, accounting for majority primary malignant brain tumours [1]. Compared with other tumour types, glioblastoma (GBM) has a 5-year survival rate of only 6.8%, and the median overall survival is often < 1 year [2]. Standard treatment approaches for GBM mainly include surgical resection, chemotherapy and radiotherapy [3]. In recent years, with the development of science and technology, the gene therapy and immunotherapy have gradually improved clinical outcomes in patients [4], however, GBM might cause immune dysfunction and immunosuppression, leading to a highly immunosuppressive tumour microenvironment and treatment resistance.
The development and application of individual neoantigen cancer vaccine is a new approach of cancer immunotherapy. Neoantigens are fragments of proteins found only on cancer cells. Because of the unique properties of neoantigens, targeting them allows a patient’s immune system to find and attack cancer cells rather than attacking healthy cells. Neoantigens provide a novel, precise approach to track cancer cells, highly specific to tumours [5]. For example, NeoVax, a tumour neoantigen vaccine, achieved efficacy in eight high-risk melanoma patients. After a mean follow-up of 4 years, all patients were alive and 6 of them remained free of disease activity [6]. Therefore, it is of great significance to find novel and efficient GBM-associated neoantigens to improve the treatment efficiency in patients.
Alternative splicing events (ASEs) play an important role in the post-transcriptional regulation of genes and are one of the biological mechanisms that maintain biodiversity and tissue specificity. Alternative splicing regulates basic biological processes such as cell development, cell differentiation, and response to environmental factors [7]. Abnormal alternative splicing may alter mRNA stability and affect protein interactions, especially in nervous system and immune system [8]. This is closely related to the functional diversity and response sensitivity of nervous and immune systems. Many studies have demonstrated that the abnormal alternative splicing events take part in tumorigenesis and development. Changes in migration, growth regulation, hormone responsiveness, and response to chemotherapy of cancer cell may be all associated with abnormal alternative splicing [9]. In cancer cells, the abnormal transcripts might generate, escape degradation, and accumulate in large quantities when the expression and function of alternative splicing regulatory factors are often destroyed [10-12], directly participating in the occurrence and development of malignant tumours [13]. Interestingly, transcripts containing abnormal alternative splicing patterns can produce antigenic peptides. Furthermore, abnormal RNA splicing represents another potential source of novel neoepitopes in the tumor transcriptome as well, thus, exploring novel neoepitopes of tumors with abnormal alternative splicing will contribute to a more complete understanding of tumor immunity [14].
In this study, we explored for potential GBM-associated neoantigens based on alternative splicing. It was found that LRP1 was significantly associated with poor prognosis and antigen-presenting cells (APCs) infiltration of GBM, suggesting that LRP1 might be an effective GBM-associated neoantigen. Subsequently, in order to identify patients suited for vaccination, 160 patients with GBM were divide into three distinct immune subtypes based on the expression profiles of candidate antigen genes. The GBM patients in cluster3 subtype might be more efficient for vaccination. The findings of this study are expected to provide valuable information and a reliable theoretical basis for the development of potential GBM-associated neoantigens, and selection suitable patients for vaccination.
Methods
Acquisition and processing of TCGA-GBM data
Normalized gene expression and clinical follow-up data from 160 GBM samples were downloaded from The Cancer Genome Atlas (TCGA); detailed information are shown in Tables 1 and 2. The UCSC Xena database (https://gdc-hub.s3.us-east-1.amazonaws.com/download/TCGA-GBM.htseq_fpkm.tsv.gz) was used to obtain RNA-seq data. Ensembl Symbol annotation data was downloaded from the UCSC genome browser database. (https://gdc-hub.s3.us-east-1.amazonaws.com/download/gencode.v22.annotation.gene.probeMap; Full metadata). Survival data and phenotypic data were also obtained from the UCSC database: (https://gdc-hub.s3.us-east-1.amazonaws.com/download/TCGA-GBM.GDC_phenotype.tsv.gz) (https://gdc-hub.s3.us-east-1.amazonaWs.com/download/TCGA-GBM.survival.tsv).
Table 1.
The sample types of 165 samples in TCGA-GBM
| type | number | |
|---|---|---|
| Type | Normal | 5 |
| Cancer | 160 |
Table 2.
The clinical information of 160 samples in TCGA-GBM
| type | number | |
|---|---|---|
| OS | Alive | 29 |
| Dead | 131 | |
| Gender | Female | 57 |
| Male | 103 | |
| Radiation therapy | Asian | 5 |
| Black or African American | 9 | |
| White | 144 | |
| Not reported | 2 |
The Ensembl ID of RNA-seq data was converted to gene Symbols based on the information in the annotation file. First, the new expression value of a gene was assigned by calculating the mean value of the genes associated to the same Symbol. Next, genes with 0 expression in at least 30% of the samples were deleted. Then, samples with a survival time of more than 30 days were extracted, and the intersection of RNA-Seq data, phenotype data and survival data was recorded.
We screened GBM samples with genetic mutations from that National Cancer Institute’s Genomic Data Commons (GDC) and cBioPortal databases: https://gdc.cancer.gov/about-data/publications/mc3-2017/mc3.v0.2.8.PUBLIC.maf.gz and https://www.cbioportal.org/study/clinicalData?id=gbm_tcga. GBM-tumour mutation burden (GBM-TMB) data were also obtained from the GDC database: https://gdc.cancer.gov/about-data/publications/PanCan-CellOfOrigin/mutation-load-updated.txt. GBM-copy number variation (GBM-CNV) data came from the UCSC database: https://gdc-hub.s3.us-east-1.amazonaws.com/download/TCGA-GBM.cnv.tsv.gz. Alternative splicing was recorded from the TCGASpliceSeq database: https://bioinformatics.mdanderson.org/TCGASpliceSeq/PSIdownload.jsp.
Identification of abnormal alternative splicing events
Firstly, PSI spectra of alternative splicing events in TCGA-GBM were screened. We calculated the mean values of Percent-Spliced-In (PSI) of the alternative splicing events in the cancer and normal samples, respectively, and filtered out the alternative splicing events with mean values of 0 or 1 in the cancer samples or normal samples. Because PSI values of these alternative splicing events were discontinuous values in the samples, the T-test of PSI values in the cancer samples and normal samples could not be performed. After filtering, there were 38052 remaining alternative splicing events. Next, we calculated the T-test and logFC of the alternative splicing events individually. In addition, BH correction is performed on the P value of the T-test. We screened FDR < 0.05 and |logFC| > 1 alternative splicing as abnormal alternative splicing events.
Correlation analysis of potential antigen candidate genes and immune cell infiltration
Tumour immune estimate resources (TIMER, https://cistrome.shinyapps.io/timer/) is used for analysis and visualization tumour immune infiltrating cells (TIIC) and GBM intersection potential neoantigen of the correlation between candidate genes. Spearman analysis was used, and a P < 0.05 was considered statistically significant.
Acquisition of immune gene sets and identification of immune subtypes
Immunotyping can be used to identify immune states in tumours and their microenvironments, thus helping to identify patients suitable for therapy. From Immport database (https://s3.immport.org/release/genelists/GeneListGOAnnotation.txt?download=true) (1255) and Charoentong’s study [15] (782), a total of 1894 immune genes were obtained to form a GBM immune gene spectrum. 1451 prognostic genes were screened from GBM immunogene profiles. The download link is as follows: https://s3.immport.org/release/genelists/GeneListGOAnnotation.txt?download=true. http://icbi.at/TCIA/SupplementaryTables.pdf.
We then used the ConsensusClusterPlus package to conduct consistent clustering of immune gene expression profiles. The same analysis was performed for the validation set.
Analysis of immune cells among immune subtypes
Tracking tumour immunophenotype (TIP) has the capability to rapidly analyse and intuitively visualize the activity of anticancer immunity and the extent of tumour-infiltrating immune cells across the seven-step cancer-immunity cycle [16]. To observe immune activity differences among subtypes, we downloaded the immune active points of TCGA-GBM samples from the TIP database (http://biocc.hrbmu.edu.cn/TIP/PancancerSearchAction?cancerType=GBM). The distribution of immune activity of samples in different subtypes was characterized by ggpubr package (ANOVA, P < 0.05).
Analysis of immunosubtypes and identified tumour biomarkers
The existing cancer tumour biomarker and cancer gene were taken from The Cancer Genome Interpreter (CGI; https://www.cancergenomeinterpreter.org/home) database. We analyzed the differential distribution of existing cancer biomarkers and cancer genes in different subtypes in the TCGA data set. An ANOVA test was used for inter-group difference test, and a T-test was used for between-groups comparison, with threshold set at P < 0.05.
Result
Study design
The framework and workflow are described as following. First, the characteristic information and gene expression profiles of GBM cases and healthy controls in were obtained from TCGA database (TCGA-GBM). The alternative splicing and GBM-tumour mutation burden (GBM-TMB) data were downloaded from the TCGASpliceSeq database and GDC database, respectively.
Next, we conducted the conjoint analysis of abnormal alternative splicing events and distribution of mutated genes to explore the potential GBM-associated neoantigens. A total of 34 potential neoantigen candidate genes were identified, among which, LRP1 might be a more suitable neoantigen candidate gene.
Third, in order to screen the appropriate patients that suitable for vaccination, we utilized consensus clustering to classify 160 GBM patients into three immune subtypes according to the expression profiles of candidate neoantigen genes. Based on the conjoint analysis of TMB, abnormal alternative splicing events, immune activity, and immune cells proportion, we found that patients in cluster3 might be more suitable for the personalized neoantigen-based vaccines.
Identification potential neoantigen candidate genes
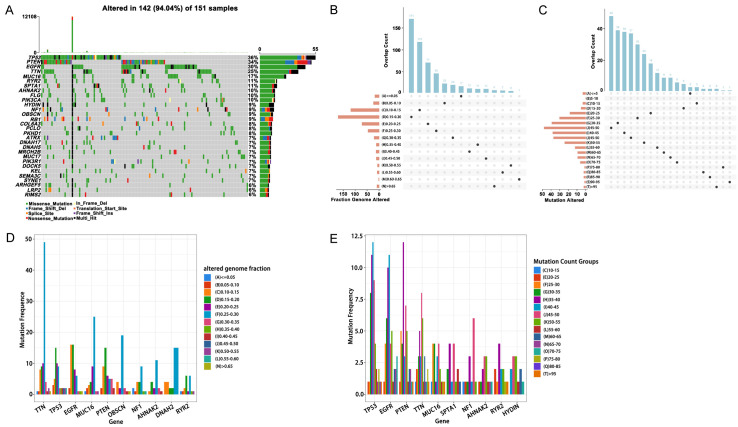
Acquired tumour mutation burden (TMB) may generate and increase the chance of immunogenic neoantigen production. Therefore, we first attempted to explore neoantigens based on mutated genes within GBM samples. Figure 1A revealed the distribution of mutated genes in GBM samples; the gene with the highest mutation rate was TP53. Figure 1B, 1C showed the distribution of frequently mutated genes in each sample in the sections of altered genome fraction and mutation counts. These results indicated that TP53 was frequently mutated gene in the sections of altered genome fraction and mutation counts. Figure 1D, 1E exhibited that most patients have low proportion of genomic changes and mutation counts. According to the above analyses, we speculated that if tumour-associated antigens were explored only based on the mutated genes in GBM, the immunogenicity might be low.
Figure 1.
Analysis of mutant genes. A. Waterfall diagram of mutant genes. B. Distribution of samples in altered genome fraction. C. Distribution of samples in mutation count groups. D. Top 10 high-frequency mutation genes in altered genome fraction. E. High frequency mutation genes in mutation count groups.
Next, we analyzed the abnormal alternative splicing events in GBM samples. In a general survey of the 38052 alternative splicing events from TCGA-GBM cohort, it was found that Exon skip (ES) was the most frequent, totaling 18360 pieces. The least-frequently occurring alternative splicing event was mutually exclusive exons (ME), totaling 184 pieces (Figure 2A, 2B). ME occurred mostly with other alternative splicing events. In addition, 7 kinds of alternative splicing events occurred in the PTK2 gene (Figure 2C). 3789 abnormal alternative splicing events were screened (FDR < 0.05 and |logFC| > 1); up-regulated alternative splicing events total 1699 (including 1320 genes), and down-regulated alternative splicing events total 2090 (including 1706 genes) (Figure 3A, 3B). Figure 3C, 3D showed the distribution of abnormal alternative splicing events in genes.
Figure 2.
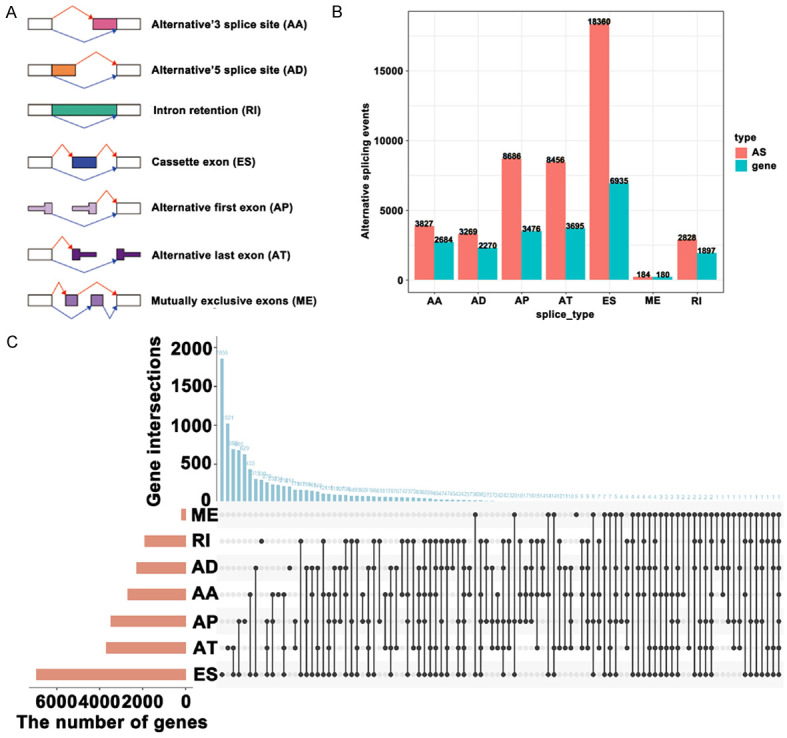
Abnormal alternative splicing events identification and overview. A. The types of abnormal alternative splicing events. B, C. The overview of abnormal alternative splicing events.
Figure 3.
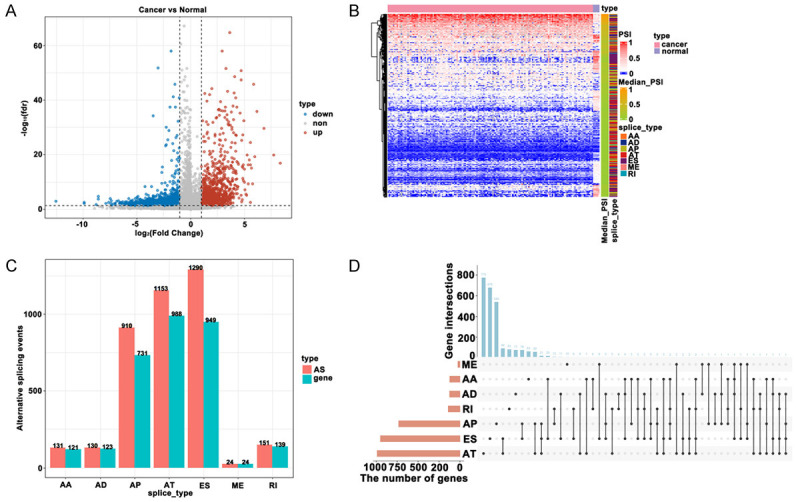
Abnormal alternative splicing events identification (A, B) and overview (C, D).
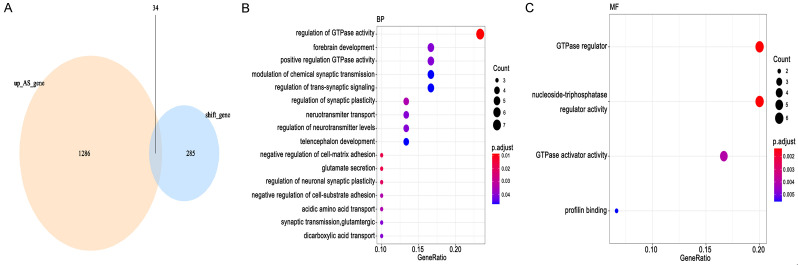
Third, genes with both abnormal alternative splicing events (up-regulation) and frameshift mutations were identified as potential neoantigen candidate genes for GBM. Our results discovered a total of 1320 up-regulated alternative splicing abnormalities and 319 frameshift mutated genes. Genes classified both as up-regulated alternative splicing abnormalities and frameshift mutations were considered for potential neoantigen candidate genes; a total of 34 potential neoantigen candidate genes were identified (Figure 4A). Clusterprofiler package was used for functional enrichment analysis of identified potential neoantigen candidate genes (p.adj < 0.05). We found that potential neoantigen candidate genes were enriched to 16 biological processes and 4 molecular functions (Figure 4B, 4C). These genes were enriched in regulation of GTPase activity, regulation of synaptic plasticity, and negative regulation of cell-matrix adhesion (Figure 4B). The results of molecular function manifested that potential neoantigen candidate genes were mainly targeted at GTPase regulator activity and nucleoside-Triphosphatase regulator activity (Figure 4C).
Figure 4.
Screening of potential antigen candidate genes. A. Identification of potential antigen candidate genes. B, C. Enrichment analysis of potential antigen candidate genes.
Association between potential neoantigen candidate genes and nonsense-mediated mRNA decay (NMD) factors
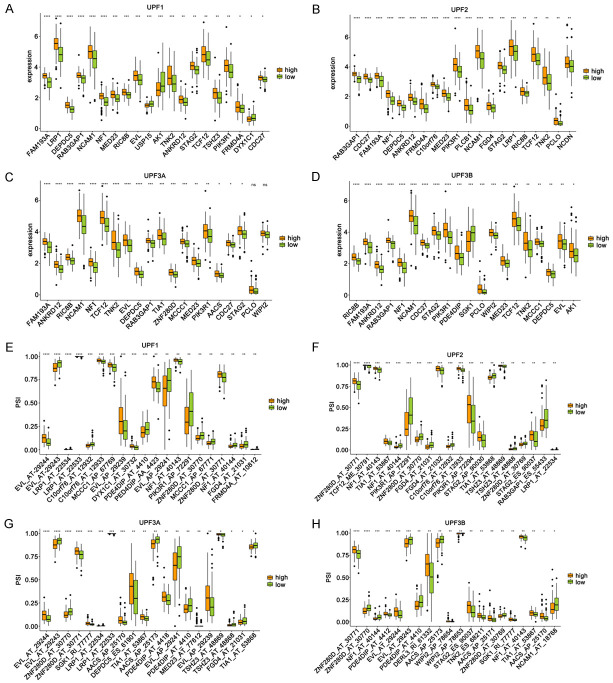
Nonsense-mediated mRNA decay (NMD), a transcriptional monitoring pathway that exists in all eukaryotes, can reduce abnormal transcripts by eliminating mRNA transcripts containing premature termination codons [10]. It is reported that NMD is closely related to tumour immunity [17] and also a core target for tumour therapy [18]. In order to explore the association between NMD regulatory factors and neoantigen candidate genes, we first obtained four confirmed NMD regulatory factors from published studies, namely UPF1, UPF2, UPF3A, and UPF3B [19]. According to the median expression level of these four NMD factors in GBM samples, all GBM samples were divided into two groups - high expression group and low expression group. Ggpubr package was used to analyze the expression level of potential antigen candidate genes and the difference of alternative splicing events between the two groups (T test, P < 0.05).
The expression differences of 34 potential neoantigen candidate genes in the high expression and low expression groups were discovered in Figure 5A-D. We found that the top 20 potential neoantigen candidate genes were consistent with the expression trend of NMD factors. Percent spliced in (PSI) is a quantitative index of variable splicing. PSI enables the comparison of a single sample or multiple samples between groups, calculated as PSI = splice_in/(splice_in + splice_out). We obtained 238 alternative splicing events associated with 34 potential neoantigen candidate genes. The differential distribution of PSI values of 238 alternative splicing events in the high and low expression groups was analyzed. The results discovered that the top 20 alternative splicing events had higher PSI values in the high expression group (Figure 5E-H).
Figure 5.
Relationship between NMD factors and potential antigen candidate genes. A-D. The top 20 most significantly differentially expressed genes in the high-expression and low-expression groups. E-H. The top 20 most significant differences in the distribution of splicing events in the high-expression and low-expression groups. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001.
Association between infiltration of potential neoantigen candidate genes and APCs
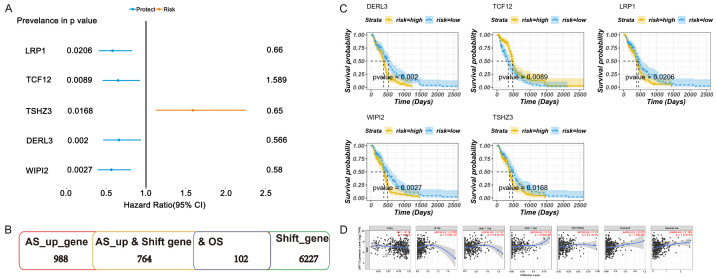
The survival correlation of the above potential candidate neoantigen genes facilitates further screening of prognostic antigens that may have immunostimulative or inhibitory effects as candidate antigens. We used a univariate Cox proportional risk regression model to analyse 34 potential neoantigen candidate genes and screened out five genes related to survival (LRP1, TCF12, DERL3, WIPI2, TSHZ3). LRP1, TCF12, DERL3, and WIPI2 were protective genes (HR < 1). However, TSHZ3 was identified as a risk gene (HR > 1). Figure 6A indicated the prognostic efficacy of these five genes. Figure 6B manifested the number of alternative splicing abnormal upregulation genes, frameshift mutated genes, and prognostic potential antigen candidate genes. Then, multivariate Cox regression analysis was performed on five potential neoantigen candidate genes associated with prognosis. Patients were divided into low-risk and high-risk groups according to the median gene expression value (P < 0.05), and the survival period was analysed. Figure 6C exhibited the longer survival period of the low-risk group. Therefore, the five aforementioned prognostic potential antigen candidate genes possessed potential immune stimulation effects.
Figure 6.
Overall prognostic efficacy of potential antigen candidate genes. A. Overall prognostic efficacy of potential antigen candidate genes. B. An overview abnormal alternative splicing, frameshift, and potential antigen candidate genes. C. Survival curves of five prognostic potential antigen candidate genes. D. Correlation between potential antigen candidate genes and infiltration of APC in antigen presenting cells.
TIMER is a comprehensive resource for systematic analysis of immune infiltration in different cancer types [20]. TIMER was used to observe the correlation between APCs infiltration and expression of identified potential antigen candidate genes (P < 0.05). We found that the expression level of LRP1 was significantly positively correlated with the levels of immune-infiltrating cells, especially Macrophages, B cells, and Dendritic cells (Figure 6D). These results suggested that LRP1 could be processed and presented by APCs to trigger a more sensitive immune response. Therefore, LRP1 might have potential immune stimulation and might be an effective candidate gene as a GBM-associated neoantigen for immunotherapy.
Identification of immune subtypes
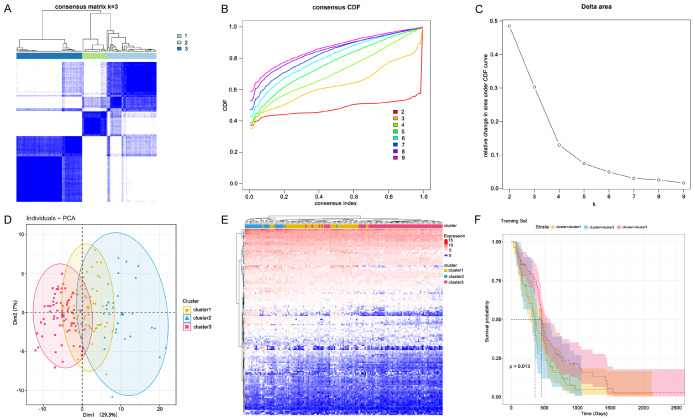
Immune subtypes can be used to reflect the immune status of malignant tumor, thus helping to screen patients suitable for vaccination. Based on the expression profiles of five prognostic candidate neoantigen genes (LRP1, TCF12, DERL3, WIPI2, TSHZ3), we performed consensus clustering on 160 GBM samples from the TCGA-GBM database and identified immune subtypes.
According to the cumulative distribution function and function delta region, we selected k=3 of the stable cluster for immune-related genes (Figure 7A-C), and obtained three immune subtypes named cluster1, cluster2, and cluster3, respectively. Next, principal component analysis (PCA) was used to verify the three subtypes, and the results discovered that three immune subtypes could be well separated (Figure 7D). Figure 7E illustrated the expression profiles of immune genes in cluster1, cluster2, and cluster3. The results of survival analysis suggested that the overall prognosis of these three subtypes was significantly different. Patients in cluster3 had a better prognosis, while cluster2 patients had a lower survival rate (Figure 7F). In summary, we defined three immune subtypes that were associated with clinical outcomes and were able to predict prognosis in GBM patients.
Figure 7.
Identification of immune subtypes. A. Heat map of sample clustering. B. Cumulative distribution function curve. C. Delta area of immune-related genes. D. Principal component analysis (PCA) distribution of GBM sample. E. Heat map of immune-related gene expressions in cluster1, cluster2, and cluster3. F. Survival curves of cluster1, cluster2, and cluster3 in test cohort.
Analysis of TMB and abnormal alternative splicing events among immune subtypes
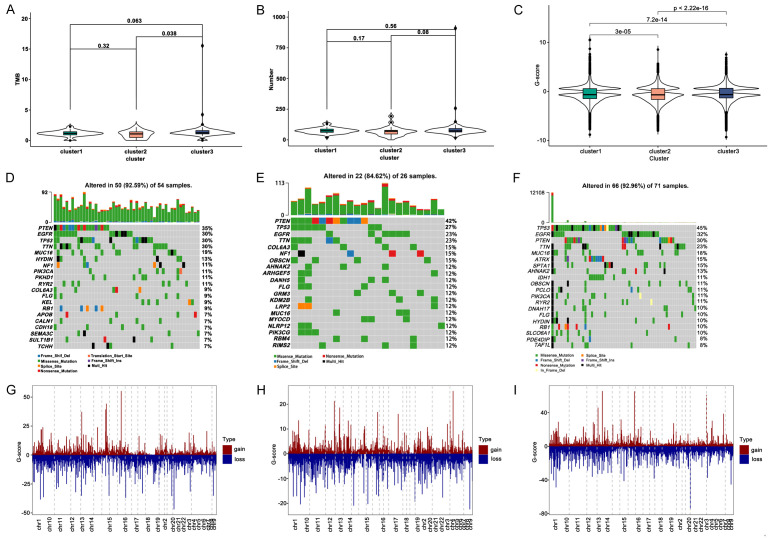
Several studies have reported that the combination of TMB and abnormal alternative splicing events can significantly predict accuracy of immunotherapy responders [13]. Higher TMB and somatic mutation rate are associated with stronger anti-cancer immunity [21]. Therefore, first, we used the mutation dataset to calculate the TMB for each sample and analysed it in three immune subtypes. The number of TMB and mutant genes among the different subtypes was discovered in Figure 8A, 8B.
Figure 8.
Mutation and CNV analysis between immune subtypes. A. Distribution of TMB score in immune subtypes. B. Number of mutation genes in immune subtypes. C. G-score of three immune subtype. D-F. Waterfall diagram of mutation genes in cluster1, cluster2, and cluster3. G-I. Distribution of copy number on chromosomes in cluster1, cluster2, and cluster3.
Figure 8C-E revealed the frequency of mutated genes in different subtypes. We found that TP53, EGFR, PTEN, and TTN were the most frequently mutated genes in each subtype, among which cluster3 was the subtype with the most frequently mutated genes. Then, we selected samples with a copy number with an absolute G-score greater than 0.4, and visualized the G-score of CNV in each subtype. The chromosome distribution of CNV in cluster1, cluster2, and cluster3 was manifested in Figure 8G-I. Totally, we found that significant differences among each subtype and cluster3 possessed higher TMB.
Subsequently, we observed the differences in the abundance of abnormal alternative splicing events in different subtypes and displayed the top 20 abnormal alternative splicing events (up-regulated; P < 0.05). A total of 51 abnormal alternative splicing events were identified among the three immune subtypes, and 9, 20, and 22 abnormal up-regulated alternative splicing events were found in cluster1, cluster2, and cluster3, respectively (Figure 9A-C). We speculated that cluster3 appeared higher TMB and more abnormal alternative splicing events, thus, patients in cluster3 might be more suitable for vaccination.
Figure 9.
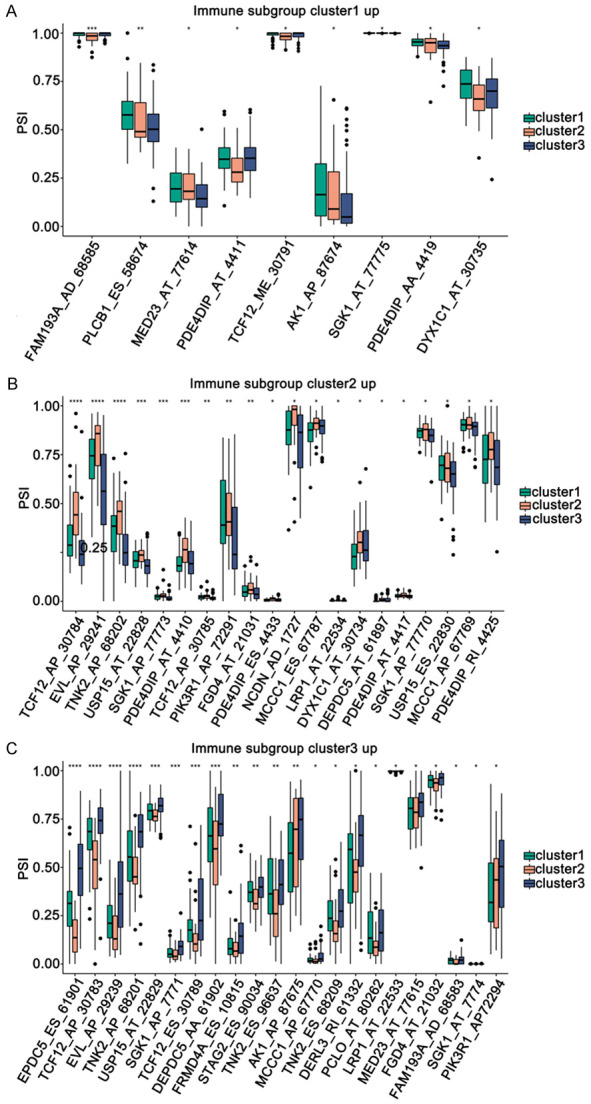
Alternative splicing analysis of potential antigen candidate genes among immune subtypes. A-C. The genes significantly up-regulated in different subtypes. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001.
Analysis of immune activity, immune cells proportion among immune subtypes
It is reported that the immune activity and proportion of immune cells are major components closely associated with prognosis of immunotherapy [22], therefore, we explored the characteristic of immune activity and proportion of immune cells in each immune subtype.
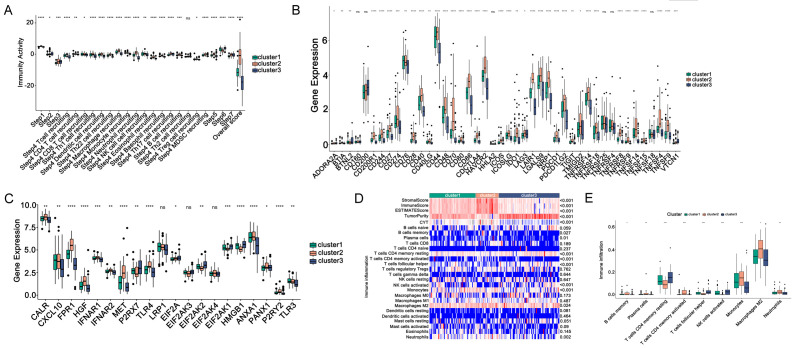
First, to analyse the differences in immune activity among three immune subtypes, we downloaded the immune activity scores of TCGA-GBM samples from TIP database, and observed the immune cell activity of each samples in different subtypes (P < 0.05). Figure 10A revealed the obvious differences in immune activity scores among the three immune subtypes. In particular, the scores of natural killer (NK) cell, T cell, macrophage, T helper type 1 (Th1) cell, and dendritic cell recruiting in cluster3 were significantly lower than those of the other two subtypes, indicating that after receiving vaccination, patients in cluster3 might enhance the greatest immune response.
Figure 10.
Distribution of immune activity scores among immune subtypes. (A) Immune activity, (B) ICPs, (C) ICDs in three immune subtypes, (D) Heat map and (E) box plot of immune cell infiltration in three subtypes. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001.
Second, it is considered that immune checkpoints (ICPs) and immunogenic cell death regulators (ICDs) play important roles in regulating host anti-tumour immunity, and may influence the efficacy of immunotherapy. Therefore, we obtained ICPs and ICDs from previous studies [23], and calculated the differences of their expression levels in three subtypes. We detected the 47 ICPs were differentially expressed among three immune subtypes (Figure 10B). In the TCGA-GBM cohort, CTLA4, PDCD1 (PD-1), and CD274 (PD-L1), the major immune checkpoints in tumours, had the lowest expression levels in cluster3. Similarly, Figure 10C showed that totally 17 ICDs were differentially expressed among three immune subtypes. CALR, FPR1, HGF, IFNAR1, IFNAR2, MET, TLR4, EIF2AK4, ANXA1, PANX1, and P2RY2 were significantly downregulated in cluster3, suggesting GBM-associated neoantigens might be more effective for patients in cluster3.
Third, the proportion of immune cells in each immune subtype was evaluated as well. We utilized the ssGSEA to examine the abundance differences of 28 immune infiltrating cells among three immune subtypes (Figure 10D). Figure 10E discovered the abundance of nine immune infiltrating cells, such as memory B cells, CD4 memory-activated T cells, Monocytes, M2 macrophages, and neutrophils, decreased significantly in cluster3, illustrating patients in cluster3 might be more suitable for vaccination, and possess the potential to produce immune-infiltrating cells.
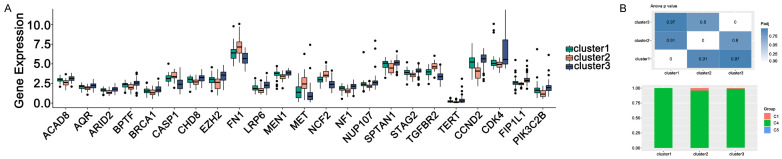
The immune subtypes were compared with the tumour biomarkers and pan-cancer subtypes
Tumour markers can predict cancer progression, prognosis, and recurrence. We identified multiple tumour markers from the TCGA-GBM dataset, and 23 cancer biomarkers showed significant differences among three immune subtypes. As shown in Figure 11A, the expression abundance of 23 cancer biomarkers was significantly different among the three subtypes. Multiple oncogenic genes were up-regulated in cluster2 GBM samples, while more tumour suppressor was present in cluster3. Previous study had identified six pan-cancer subtypes, including C1 (Wound Healing), C2 (IFN-γ Dominant), C3, C4 (Lymphocyte), C5 (Immunologically Quiet) and C6 (TGF-β Dominant) [24]. We then assessed the distribution of these six types among the three immune subtypes. As shown in Figure 11B, C4 occupies the largest proportion in cluster1, cluster2 and cluster3. This result further illustrated patients in cluster3 might be more suitable for vaccination, and administration of the neoantigens might stimulate a stronger immune response in cluster3 patients.
Figure 11.
Analysis of tumor biomarkers (A) and pan-cancer (B) between immune subtypes. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001.
Discussion
Vaccines have traditionally been used to prevent infectious diseases, interestingly, in recent years, personalized neoantigen-based vaccine is a promising new immunotherapy among currently available cancer vaccine strategies. In this study, we constructed a gene profile with abnormal alternative splicing events (up-regulated) and frameshift mutations, from which 34 targetable antigens were identified. Especially, LRP1 is not only closely related to the poor prognosis of GBM, but also related to the high level of APCs and B cell infiltration. Therefore, LRP1 may have strong immune stimulation potential and is expected to be an effective neoantigen candidate gene. Then, by constructing three robust immune subtypes, we determined that the patients in cluster3 were the most suitable to receive personalized neoantigen-based vaccines.
Early therapeutic vaccination strategies focused on abnormally expressed or overexpressed autoantigens called tumor-associated antigens (TAA) in tumors, but such strategies were largely unsuccessful in the clinic due to TAA-specific T cells being influenced by central and/or peripheral tolerance. In addition, this TAAs are also expressed to some extent in non-malignant tissues, which increases the risk of vaccine-induced autoimmune toxicity. Furthermore, vaccines based on neoantigens rather than traditional TAAs have several advantages. First, neoantigens are expressed only by tumor cells and can trigger a true tumor-specific T-cell response, thereby preventing “off-target” damage to non-tumor tissues. Second, the persistence of neoantigen-specific T cell responses offers the potential for post-treatment immune memory, which offers the possibility of long-term prevention of cancer recurrence [5].
At present, many clinical trials of personalized neoantigen-based vaccines are underway. These preliminary studies provide important clues for the immunogenicity and therapeutic potential of personalized neoantigen-based vaccines, including GEN-009 [25], RO7198457 [26], and mRNA-4157 [27]. Therefore, screening novel and effective neoantigens with higher immunogenicity, and overcoming inhibitory tumour microenvironment are the most important future directions for the study of personalized neoantigen-based vaccines. Alternative splicing is involved in the regulation of many cancer genes, which is involved in various aspects of the biological behaviour of cancer cells, especially cell proliferation [28], apoptosis [29], angiogenesis [30], and immune escape [31]. Increasing evidence shows that unbalanced expression of splicing variants or misexpressed isomers are important features of cancer [32]. Specific and inaccurate splicing variants have been studied in many cancers as diagnostic, prognostic and therapeutic targets [33]. Intrigued by this evidence, we screened for genes with aberrant alternative splicing events (upregulation) and frameshift mutations in the hope that they would become strong neoantigens and lead to the development of personalized vaccines. LRP1, TCF12, DERL3, WIPI2, and TSHZ3 were were selected as candidate factors in this study.
Transcription factor 12 (TCF12) is significantly related to invasion and metastasis of colorectal cancer, bladder cancer and breast cancer [34-36]. DERL3 belongs to the Derlin family (DERL1, DERL2, and DERL3). It has been reported that DERL1 is overexpressed in non-small cell lung cancer [37], breast cancer [38], and colon cancer [39]. WD Repeat Domain Phosphoinositide-Interacting Protein 2 (WIPI2) expression was significantly higher in HCC tissues and was associated with low survival [40]. Teashirt zinc finger family member 3 (TSHZ3) was significantly down-regulated in glioma tissues and cell lines. Overexpression of TSHZ3 inhibits the invasiveness of U87 and U251 GBM cells [41].
Low density lipoprotein receptor (LDLR) associated protein-1 (LRP1) is a large multifunctional endocytic cell surface receptor [42]. Low expression of LRP1 is closely related to the low survival rate of liver cancer [43]. However, high expression of LRP1 is associated with advanced tumour staging of endometrial cancer [44], breast cancer [45], and prostate cancer [46]. Therefore, the exact effect of LRP1 on tumour cells may depend on tumour cell type. Whether tumor neoantigens have strong immunogenicity and the ability to target and kill tumor cells depends on whether they can induce T cell response, especially the ability to maximize the activation and amplification of CD8+ T cells. One way to achieve this is to use autologous antigen-presenting cells (APCs), particularly DCs, to stimulate an immune response to tumor neoantigens. In addition, the vaccine can also use DCs as a vaccine delivery system, so that the vaccine can quickly and effectively work [47]. Therefore, we evaluated the relationships between five neoantigen candidate genes and APCs. More interestingly, we found that LRP1 is closely related to antigen presenting cells, especially CD8+ T cells, CD4+ T cells, and DCs, indicating that LRP1 can be directly processed by APCs, then presented to T cells and recognized by B cells to trigger an immune response. Therefore, we speculate that LRP1 may have relatively strong immunogenicity, and can use DCs as a delivery system to kill tumor cells quickly and in a targeted manner. Therefore, LRP1 may be a promising candidate for the development of neoantigen-based vaccine against GBM.
Previous studies have identified multiple potential neoantigen candidate genes for neoantigen-based vaccines based on gene profiles of somatic mutations [48,49]. However, studies have shown that for many types of cancer, the predictive accuracy of tumour mutation burden is inadequate. Abnormal alternative splicing and frameshift mutations have great potential to generate large numbers of neoantigens [50-52]. Therefore, we suggest that these biomarkers with abnormal alternative splicing events (upregulation) and frameshift mutations could more effectively serve as potential tumour-associated antigens for GBM personalized neoantigen-based vaccines. Although the potential of these genes in vaccines for GBM has been predicted, further clinical evaluation and validation are needed.
Due to the different therapeutic effects of vaccines on patients, we identified three immune subtypes based on candidate neoantigen genes profiles to select the appropriate patients for vaccination. All three immune-related subtypes exhibited different molecular, cellular, and clinical characteristics. Patients in cluster3 showed high levels of TMB, and more abnormal alternative splicing events. The combination of nonsense-mediated mRNA decay and tumour mutation burden was reported to significantly improve the predictive accuracy of immunotherapy responders [53], suggesting that cluster3 patients may be the most suitable candidates for vaccination. In addition, because the memory B cells, T.cells. CD4 memory activated T cells, monocytes, M2 macrophages, and neutrophils were lowest in the cluster3, indicating that vaccination of patients in cluster3 might produce immune-infiltrating cells, which make immunotherapy more effective. These results suggest that cluster3 patients may be beneficiaries of vaccination, making personalized neoantigen-based vaccines therapy more responsive and effective.
Overall, LRP1 may be a potential antigen for GBM personalized neoantigen-based vaccines development and may benefit for patients in cluster3. Therefore, our findings provide a valuable analysis of personalized neoantigen-based vaccines against GBM and identify suitable patients for vaccination.
Acknowledgements
We wish to express our strong appreciation to TCGA and GEO for their public data. This study was supported by Wuhan Yellow Crane Talent-Outstanding Young Talent Project (Wenqi Gao and Zhifang Deng).
Disclosure of conflict of interest
None.
Abbreviations
- GBM
Glioblastoma
- TIMER
Tumour Immune Estimation Resource
- APCs
Antigen presenting cells
- TMB
Tumour mutation burden
- PSI
Percent-spliced-in
- TIP
Tracking tumour immunophenotype
- ICPs
Immune checkpoints
- ICD
Immunogenic cell death
References
- 1.Huang S, Song Z, Zhang T, He X, Huang K, Zhang Q, Shen J, Pan J. Identification of immune cell infiltration and immune-related genes in the tumor microenvironment of glioblastomas. Front Immunol. 2020;11:585034. doi: 10.3389/fimmu.2020.585034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ostrom QT, Cioffi G, Gittleman H, Patil N, Waite K, Kruchko C, Barnholtz-Sloan JS. CBTRUS statistical report: primary brain and other central nervous system tumors diagnosed in the United States in 2012-2016. Neuro Oncol. 2019;21:v1–v100. doi: 10.1093/neuonc/noz150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jiang T, Nam DH, Ram Z, Poon WS, Wang J, Boldbaatar D, Mao Y, Ma W, Mao Q, You Y, Jiang C, Yang X, Kang C, Qiu X, Li W, Li S, Chen L, Li X, Liu Z, Wang W, Bai H, Yao Y, Li S, Wu A, Sai K, Li G, Yao K, Wei X, Liu X, Zhang Z, Dai Y, Lv S, Wang L, Lin Z, Dong J, Xu G, Ma X, Zhang W, Zhang C, Chen B, You G, Wang Y, Wang Y, Bao Z, Yang P, Fan X, Liu X, Zhao Z, Wang Z, Li Y, Wang Z, Li G, Fang S, Li L, Liu Y, Liu S, Shan X, Liu Y, Chai R, Hu H, Chen J, Yan W, Cai J, Wang H, Chen L, Yang Y, Wang Y, Han L, Wang Q. Clinical practice guidelines for the management of adult diffuse gliomas. Cancer Lett. 2021;499:60–72. doi: 10.1016/j.canlet.2020.10.050. [DOI] [PubMed] [Google Scholar]
- 4.Foks AC, Engelbertsen D, Kuperwaser F, Alberts-Grill N, Gonen A, Witztum JL, Lederer J, Jarolim P, DeKruyff RH, Freeman GJ, Lichtman AH. Blockade of Tim-1 and Tim-4 enhances atherosclerosis in low-density lipoprotein receptor-deficient mice. Arterioscler Thromb Vasc Biol. 2016;36:456–465. doi: 10.1161/ATVBAHA.115.306860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blass E, Ott PA. Advances in the development of personalized neoantigen-based therapeutic cancer vaccines. Nat Rev Clin Oncol. 2021;18:215–229. doi: 10.1038/s41571-020-00460-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hu Z, Leet DE, Allesoe RL, Oliveira G, Li S, Luoma AM, Liu J, Forman J, Huang T, Iorgulescu JB, Holden R, Sarkizova S, Gohil SH, Redd RA, Sun J, Elagina L, Giobbie-Hurder A, Zhang W, Peter L, Ciantra Z, Rodig S, Olive O, Shetty K, Pyrdol J, Uduman M, Lee PC, Bachireddy P, Buchbinder EI, Yoon CH, Neuberg D, Pentelute BL, Hacohen N, Livak KJ, Shukla SA, Olsen LR, Barouch DH, Wucherpfennig KW, Fritsch EF, Keskin DB, Wu CJ, Ott PA. Personal neoantigen vaccines induce persistent memory T cell responses and epitope spreading in patients with melanoma. Nat Med. 2021;27:515–525. doi: 10.1038/s41591-020-01206-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Le KQ, Prabhakar BS, Hong WJ, Li LC. Alternative splicing as a biomarker and potential target for drug discovery. Acta Pharmacol Sin. 2015;36:1212–1218. doi: 10.1038/aps.2015.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Matera AG, Wang Z. A day in the life of the spliceosome. Nat Rev Mol Cell Biol. 2014;15:108–121. doi: 10.1038/nrm3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lei Y, Shi Y, Duan J, Liu Y, Lv G, Shi R, Zhang F, Yang Q, Zhao W. Identification of alternative splicing and lncRNA genes in pathogenesis of small cell lung cancer based on their RNA sequencing. Adv Clin Exp Med. 2019;28:1043–1050. doi: 10.17219/acem/94392. [DOI] [PubMed] [Google Scholar]
- 10.Wang Y, Zhang L, Xu Z, Miao L, Huang L. mRNA vaccine with antigen-specific checkpoint blockade induces an enhanced immune response against established melanoma. Mol Ther. 2018;26:420–434. doi: 10.1016/j.ymthe.2017.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu L, Wang Y, Miao L, Liu Q, Musetti S, Li J, Huang L. Combination immunotherapy of MUC1 mRNA nano-vaccine and CTLA-4 blockade effectively inhibits growth of triple negative breast cancer. Mol Ther. 2018;26:45–55. doi: 10.1016/j.ymthe.2017.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang Y, Zhang Z, Luo J, Han X, Wei Y, Wei X. mRNA vaccine: a potential therapeutic strategy. Mol Cancer. 2021;20:33. doi: 10.1186/s12943-021-01311-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rausch S, Schwentner C, Stenzl A, Bedke J. mRNA vaccine CV9103 and CV9104 for the treatment of prostate cancer. Hum Vaccin Immunother. 2014;10:3146–3152. doi: 10.4161/hv.29553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smart AC, Margolis CA, Pimentel H, He MX, Miao D, Adeegbe D, Fugmann T, Wong KK, Van Allen EM. Intron retention is a source of neoepitopes in cancer. Nat Biotechnol. 2018;36:1056–1058. doi: 10.1038/nbt.4239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.VanBlargan LA, Himansu S, Foreman BM, Ebel GD, Pierson TC, Diamond MS. An mRNA vaccine protects mice against multiple tick-transmitted flavivirus infections. Cell Rep. 2018;25:3382–3392. doi: 10.1016/j.celrep.2018.11.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hobo W, Novobrantseva TI, Fredrix H, Wong J, Milstein S, Epstein-Barash H, Liu J, Schaap N, van der Voort R, Dolstra H. Improving dendritic cell vaccine immunogenicity by silencing PD-1 ligands using siRNA-lipid nanoparticles combined with antigen mRNA electroporation. Cancer Immunol Immunother. 2013;62:285–97. doi: 10.1007/s00262-012-1334-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaczmarek JC, Patel AK, Kauffman KJ, Fenton OS, Webber MJ, Heartlein MW, DeRosa F, Anderson DG. Polymer-lipid nanoparticles for systemic delivery of mRNA to the lungs. Angew Chem Int Ed Engl. 2016;55:13808–13812. doi: 10.1002/anie.201608450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Beg S, Almalki WH, Khatoon F, Alharbi KS, Alghamdi S, Akhter MH, Khalilullah H, Baothman AA, Hafeez A, Rahman M, Akhter S, Choudhry H. Lipid/polymer-based nanocomplexes in nucleic acid delivery as cancer vaccines. Drug Discov Today. 2021;26:1891–1903. doi: 10.1016/j.drudis.2021.02.013. [DOI] [PubMed] [Google Scholar]
- 19.Oka M, Xu L, Suzuki T, Yoshikawa T, Sakamoto H, Uemura H, Yoshizawa AC, Suzuki Y, Nakatsura T, Ishihama Y, Suzuki A, Seki M. Aberrant splicing isoforms detected by full-length transcriptome sequencing as transcripts of potential neoantigens in non-small cell lung cancer. Genome Biol. 2021;22:9. doi: 10.1186/s13059-020-02240-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pardi N, Hogan MJ, Weissman D. Recent advances in mRNA vaccine technology. Curr Opin Immunol. 2020;65:14–20. doi: 10.1016/j.coi.2020.01.008. [DOI] [PubMed] [Google Scholar]
- 21.Rooney MS, Shukla SA, Wu CJ, Getz G, Hacohen N. Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell. 2015;160:48–61. doi: 10.1016/j.cell.2014.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang X, Zhang G, Tang T, Liang T. Identification of tumor antigens and immune subtypes of pancreatic adenocarcinoma for mRNA vaccine development. Mol Cancer. 2021;20:44. doi: 10.1186/s12943-021-01310-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qi X, Qi C, Qin B, Kang X, Hu Y, Han W. Immune-stromal score signature: novel prognostic tool of the tumor microenvironment in lung adenocarcinoma. Front Oncol. 2020;10:541330. doi: 10.3389/fonc.2020.541330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thorsson V, Gibbs DL, Brown SD, Wolf D, Bortone DS, Ou Yang TH, Porta-Pardo E, Gao GF, Plaisier CL, Eddy JA, Ziv E, Culhane AC, Paull EO, Sivakumar I, Gentles AJ, Malhotra R, Farshidfar F, Colaprico A, Parker JS, Mose LE, Vo NS, Liu J, Liu Y, Rader J, Dhankani V, Reynolds SM, Bowlby R, Califano A, Cherniack AD, Anastassiou D, Bedognetti D, Mokrab Y, Newman AM, Rao A, Chen K, Krasnitz A, Hu H, Malta TM, Noushmehr H, Pedamallu CS, Bullman S, Ojesina AI, Lamb A, Zhou W, Shen H, Choueiri TK, Weinstein JN, Guinney J, Saltz J, Holt RA, Rabkin CS Cancer Genome Atlas Research Network. Lazar AJ, Serody JS, Demicco EG, Disis ML, Vincent BG, Shmulevich I. The immune landscape of cancer. Immunity. 2018;48:812–830. doi: 10.1016/j.immuni.2018.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lam H, McNeil LK, Starobinets H, DeVault VL, Cohen RB, Twardowski P, Johnson ML, Gillison ML, Stein MN, Vaishampayan UN, DeCillis AP, Foti JJ, Vemulapalli V, Tjon E, Ferber K, DeOliveira DB, Broom W, Agnihotri P, Jaffee EM, Wong KK, Drake CG, Carroll PM, Davis TA, Flechtner JB. An empirical antigen selection method identifies neoantigens that either elicit broad antitumor T-cell responses or drive tumor growth. Cancer Discov. 2021;11:696–713. doi: 10.1158/2159-8290.CD-20-0377. [DOI] [PubMed] [Google Scholar]
- 26.Iravani A, Osman MM, Weppler AM, Wallace R, Galligan A, Lasocki A, Hunter MO, Akhurst T, Hofman MS, Lau P, Kee D, Au-Yeung G, Sandhu S, Hicks RJ. FDG PET/CT for tumoral and systemic immune response monitoring of advanced melanoma during first-line combination ipilimumab and nivolumab treatment. Eur J Nucl Med Mol Imaging. 2020;47:2776–2786. doi: 10.1007/s00259-020-04815-w. [DOI] [PubMed] [Google Scholar]
- 27.Burris HA, Patel MR, Cho DC, Clarke JM, Gutierrez M, Zaks TZ, Frederick J, Hopson K, Mody K, Binanti-Berube A, Robert-Tissot C, Goldstein B, Breton B, Sun J, Zhong S, Pruitt SK, Keating K, Meehan RS, Gainor JF. A phase I multicenter study to assess the safety, tolerability, and immunogenicity of mRNA-4157 alone in patients with resected solid tumors and in combination with pembrolizumab in patients with unresectable solid tumors. J. Clin. Oncol. 2019;37(Suppl):2523–2523. [Google Scholar]
- 28.Triulzi T, Bianchi GV, Tagliabue E. Predictive biomarkers in the treatment of HER2-positive breast cancer: an ongoing challenge. Future Oncol. 2016;12:1413–1428. doi: 10.2217/fon-2015-0025. [DOI] [PubMed] [Google Scholar]
- 29.Sveen A, Kilpinen S, Ruusulehto A, Lothe RA, Skotheim RI. Aberrant RNA splicing in cancer; expression changes and driver mutations of splicing factor genes. Oncogene. 2016;35:2413–2427. doi: 10.1038/onc.2015.318. [DOI] [PubMed] [Google Scholar]
- 30.Doherty JK, Bond C, Jardim A, Adelman JP, Clinton GM. The HER-2/neu receptor tyrosine kinase gene encodes a secreted autoinhibitor. Proc Natl Acad Sci U S A. 1999;96:10869–10874. doi: 10.1073/pnas.96.19.10869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Scott GK, Robles R, Park JW, Montgomery PA, Daniel J, Holmes WE, Lee J, Keller GA, Li WL, Fendly BM, et al. A truncated intracellular HER2/neu receptor produced by alternative RNA processing affects growth of human carcinoma cells. Mol Cell Biol. 1993;13:2247–2257. doi: 10.1128/mcb.13.4.2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jackson C, Browell D, Gautrey H, Tyson-Capper A. Clinical significance of HER-2 splice variants in breast cancer progression and drug resistance. Int J Cell Biol. 2013;2013:973584. doi: 10.1155/2013/973584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lindeboom R, Vermeulen M, Lehner B, Supek F. The impact of nonsense-mediated mRNA decay on genetic disease, gene editing and cancer immunotherapy. Nat Genet. 2019;51:1645–1651. doi: 10.1038/s41588-019-0517-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen WS, Chen CC, Chen LL, Lee CC, Huang TS. Secreted heat shock protein 90alpha (HSP90alpha) induces nuclear factor-kappaB-mediated TCF12 protein expression to down-regulate E-cadherin and to enhance colorectal cancer cell migration and invasion. J Biol Chem. 2013;288:9001–9010. doi: 10.1074/jbc.M112.437897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee CC, Chen WS, Chen CC, Chen LL, Lin YS, Fan CS, Huang TS. TCF12 protein functions as transcriptional repressor of E-cadherin, and its overexpression is correlated with metastasis of colorectal cancer. J Biol Chem. 2012;287:2798–2809. doi: 10.1074/jbc.M111.258947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.He J, Shen S, Lu W, Zhou Y, Hou Y, Zhang Y, Jiang Y, Liu H, Shao Y. HDAC1 promoted migration and invasion binding with TCF12 by promoting EMT progress in gallbladder cancer. Oncotarget. 2016;7:32754–32764. doi: 10.18632/oncotarget.8740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dong QZ, Wang Y, Tang ZP, Fu L, Li QC, Wang ED, Wang EH. Derlin-1 is overexpressed in non-small cell lung cancer and promotes cancer cell invasion via EGFR-ERK-mediated up-regulation of MMP-2 and MMP-9. Am J Pathol. 2013;182:954–964. doi: 10.1016/j.ajpath.2012.11.019. [DOI] [PubMed] [Google Scholar]
- 38.Wang J, Hua H, Ran Y, Zhang H, Liu W, Yang Z, Jiang Y. Derlin-1 is overexpressed in human breast carcinoma and protects cancer cells from endoplasmic reticulum stress-induced apoptosis. Breast Cancer Res. 2008;10:R7. doi: 10.1186/bcr1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tan X, He X, Jiang Z, Wang X, Ma L, Liu L, Wang X, Fan Z, Su D. Derlin-1 is overexpressed in human colon cancer and promotes cancer cell proliferation. Mol Cell Biochem. 2015;408:205–213. doi: 10.1007/s11010-015-2496-x. [DOI] [PubMed] [Google Scholar]
- 40.Liu C, Li F, Li X, Cao M, Feng G, Yuan X, Shi X. WIPI2 depletion inhibits the growth of hepatocellular carcinoma cells through the AMPK signaling pathway. Oncol Rep. 2020;43:1467–1478. doi: 10.3892/or.2020.7531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li Y, Huang Y, Qi Z, Sun T, Zhou Y. MiR-338-5p promotes glioma cell invasion by regulating TSHZ3 and MMP2. Cell Mol Neurobiol. 2018;38:669–677. doi: 10.1007/s10571-017-0525-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lillis AP, Van Duyn LB, Murphy-Ullrich JE, Strickland DK. LDL receptor-related protein 1: unique tissue-specific functions revealed by selective gene knockout studies. Physiol Rev. 2008;88:887–918. doi: 10.1152/physrev.00033.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Meng H, Chen G, Zhang X, Wang Z, Thomas DG, Giordano TJ, Beer DG, Wang MM. Stromal LRP1 in lung adenocarcinoma predicts clinical outcome. Clin Cancer Res. 2011;17:2426–2433. doi: 10.1158/1078-0432.CCR-10-2385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Catasus L, Llorente-Cortes V, Cuatrecasas M, Pons C, Espinosa I, Prat J. Low-density lipoprotein receptor-related protein 1 (LRP-1) is associated with highgrade, advanced stage and p53 and p16 alterations in endometrial carcinomas. Histopathology. 2011;59:567–571. doi: 10.1111/j.1365-2559.2011.03942.x. [DOI] [PubMed] [Google Scholar]
- 45.Catasus L, Gallardo A, Llorente-Cortes V, Escuin D, Munoz J, Tibau A, Peiro G, Barnadas A, Lerma E. Low-density lipoprotein receptor-related protein 1 is associated with proliferation and invasiveness in Her-2/neu and triple-negative breast carcinomas. Hum Pathol. 2011;42:1581–1588. doi: 10.1016/j.humpath.2011.01.011. [DOI] [PubMed] [Google Scholar]
- 46.McGarvey T, Hussain MM, Stearns ME. In situ hybridization studies of alpha 2-macroglobulin receptor and receptor-associated protein in human prostate carcinoma. Prostate. 1996;28:311–317. doi: 10.1002/(SICI)1097-0045(199605)28:5<311::AID-PROS7>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 47.Saxena M, van der Burg SH, Melief C, Bhardwaj N. Therapeutic cancer vaccines. Nat Rev Cancer. 2021;21:360–378. doi: 10.1038/s41568-021-00346-0. [DOI] [PubMed] [Google Scholar]
- 48.Ye L, Wang L, Yang J, Hu P, Zhang C, Tong S, Liu Z, Tian D. Identification of tumor antigens and immune subtypes in lower grade gliomas for mRNA vaccine development. J Transl Med. 2021;19:352. doi: 10.1186/s12967-021-03014-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhong H, Liu S, Cao F, Zhao Y, Zhou J, Tang F, Peng Z, Li Y, Xu S, Wang C, Yang G, Li ZQ. Dissecting tumor antigens and immune subtypes of glioma to develop mRNA vaccine. Front Immunol. 2021;12:709986. doi: 10.3389/fimmu.2021.709986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Turajlic S, Litchfield K, Xu H, Rosenthal R, McGranahan N, Reading JL, Wong Y, Rowan A, Kanu N, Al BM, Chambers T, Salgado R, Savas P, Loi S, Birkbak NJ, Sansregret L, Gore M, Larkin J, Quezada SA, Swanton C. Insertion-and-deletion-derived tumour-specific neoantigens and the immunogenic phenotype: a pan-cancer analysis. Lancet Oncol. 2017;18:1009–1021. doi: 10.1016/S1470-2045(17)30516-8. [DOI] [PubMed] [Google Scholar]
- 51.Smart AC, Margolis CA, Pimentel H, He MX, Miao D, Adeegbe D, Fugmann T, Wong KK, Van Allen EM. Intron retention is a source of neoepitopes in cancer. Nat Biotechnol. 2018;36:1056–1058. doi: 10.1038/nbt.4239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kahles A, Lehmann KV, Toussaint NC, Huser M, Stark SG, Sachsenberg T, Stegle O, Kohlbacher O, Sander C, Ratsch G. Comprehensive analysis of alternative splicing across tumors from 8,705 patients. Cancer Cell. 2018;34:211–224. doi: 10.1016/j.ccell.2018.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lindeboom R, Vermeulen M, Lehner B, Supek F. The impact of nonsense-mediated mRNA decay on genetic disease, gene editing and cancer immunotherapy. Nat Genet. 2019;51:1645–1651. doi: 10.1038/s41588-019-0517-5. [DOI] [PMC free article] [PubMed] [Google Scholar]