Abstract
Metabolic reprogramming is critical for the setup of the tumor microenvironment (TME). Glutamine has slipped into the focus of research of cancer metabolism, but its role in clear cell renal cell carcinoma (ccRCC) remains vague. Our study aimed to investigate the regulatory mechanism of glutamine in ccRCC and its prognostic value. Gene expression profiles and clinical data of ccRCC patients were obtained from The Cancer Genome Atlas database (TCGA) and Gene Expression Omnibus (GEO) database. Kaplan-Meier survival analysis was used for survival analysis. Consensus clustering was used to extract differentially expressed genes (DEGs) related to glutamine metabolism. Functional analyses, including gene set variation analysis (GSVA) and gene set enrichment analysis (GSEA), were conducted to elucidate the functions and pathways involved in these DEGs. The single-sample GSEA and Estimation of Stromal and Immune cells in Malignant Tumor tissues using Expression data (ESTIMATE) methods were applied to estimate the immune infiltration in the TMEs of two clusters. The univariate regression and the least absolute shrinkage and selection operator (LASSO) Cox regression were used to construct a prognostic signature. R software was utilized to analyze the expression levels and prognostic values of genes in ccRCC. A total of 19 glutamine metabolic genes (GMGs) were screened out for differential expression analysis of normal and ccRCC tissues. Based on survival-related GMGs, two glutamine metabolic clusters with different clinical and transcriptomic characteristics were identified. Patients in cluster B exhibited worse survivals, higher immune infiltration scores, more significant immunosuppressive cell infiltration, higher expression levels of immune checkpoints, and more enriched oncogenic pathways. Glutamine metabolic index (GMI) was constructed according to the GMGs and survival data. In addition, the expression levels of GMGs were associated with immune cell infiltration and immune checkpoints in the TME of ccRCC. Among the GMGs, PYCR1 was the most powerful regulator of immune TME. Our analysis revealed higher-level glutamine metabolism in ccRCC patients with a worse prognosis. The GMI could predict the prognosis of ccRCC patients with a high accuracy. GMGs, such as PYCR1, may be exploited to design novel immunotherapies for ccRCC.
Keywords: Cancer metabolism, glutamine, tumor microenvironment, PYCR1, immunotherapy
Introduction
Metabolic reprogramming is a hallmark in tumor development. In cancer cells, metabolism is boosted through various pathways to provide bioenergy and reduce oxidative stress for cancer cell proliferation and survival. These pathways are associated with mutations of cancer-driving genes. The abnormal accumulation of metabolites can also facilitate the occurrence of tumors. In the tumor microenvironment (TME), some nutrients can be depleted to maintain cancer cell proliferation.
Growing evidence proves that the progression of some cancers depends on the abnormal metabolism of amino acids [1,2]. Glutamine belongs to a group of conditionally essential amino acids, especially under catabolic stress [3,4]. Previous observations suggest that glutamine is indispensable for the proliferation of cancer cells, and that tumor cells die immediately after glutamine deletion [5-7]. Glutamine can be used for biosynthesis during cell growth and division [8,9]. The high serum glutamine provides rich carbon and nitrogen to support biosynthesis, thus providing energy to maintain the intracellular homeostasis and promote the growth of cancer cells [10]. The expression of enzymes involved in glutamine metabolism varies greatly in across cancer types [11,12]. At present, the role of glutamine metabolism in renal cell carcinoma (RCC) remains unclear.
In the current study, we systematically evaluated the glutamine metabolic profile in clear cell renal cell carcinoma (ccRCC) and screened the glutamine metabolites most related to the immune checkpoints in the TME of ccRCC. The mechanism of PYCR1 in regulating glutamine metabolism in the TME may provide a clue for designing new immunotherapies for ccRCC.
Methods
Data acquisition and study design
The transcriptomic and clinical data of ccRCC patients were obtained from The Cancer Genome Atlas (TCGA-KIRC, https://www.cancer.gov/tcga). A list of 21 genes encoding proteins participating in glutamine metabolic pathways were summarized from previous studies [8]. We defined these genes as glutamine metabolic genes (GMGs). The expression data of GSE73731 [13], GSE66272 [14], and GSE67501 [15] were obtained from the Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) database.
Differential gene expression (DGE) analysis
Differentially expressed GMGs between tumor and normal tissue samples were screened out with the Wilcoxon test and Limma in the R package. The significance was set as a false discovery rate (FDR) <0.05.
Survival analysis
For each differentially expressed gene (DEG), the best cutoff value of the relative mRNA expression level was used to classify patients into the high expression group and low expression group, using the “surv_cutpoint” command. The differences in clinical outcomes were calculated with the Log-rank test through the Kaplan-Meier method. R packages, including KMsurv, survival and survminer, were utilized to perform the prognostic analysis. P-value <0.05 was considered as statistically significant.
Cluster analysis
ccRCC samples were subtyped by the consistent clustering using the R package ConsensusClusterPlus [16]. The datasets were clustered by the Euclidean squared distance metric and the K-means algorithm with k from 2 to 9. The results were presented in heatmaps of the consistency matrix generated by R package pheatmap. The optimal number of clusters should meet the following criteria: high consistency of clustering, moderate number of samples in each cluster, significant difference in survival between clusters, and no significant increase in the area under the cumulative distribution function (CDF) curve.
Biological pathway and functional enrichment analysis
Gene Set Variation Analysis (GSVA) was performed to evaluate the pathways enriched in each cluster with the R package GSVA [17] and “c2.cp.kegg.v7.4.symbols” from the Molecular Signatures Database (MSigDB). Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses were performed based on the GO database (http://geneontology.org) and KEGG database (http://www.genome.jp/kegg/) in R package clusterProfiler. Significance was defined according to a nominal P-value <0.05 and a FDR <0.05.
Immune microenvironment assessment
Estimation of Stromal and Immune cells in Malignant Tumor tissues using Expression data (ESTIMATE) analysis was conducted to quantify the immune scenario in the TME of each sample with R package estimate, including the ESTIMATE score, immune score and stromal score. Single-sample Gene Set Enrichment Analysis (ssGSEA) was conducted to quantify the relative abundance of each immune cell in the TME with the R package GSVA [17] and the immune-related gene set [18]. The Student’s t test was used to evaluate differences in the above parameters between clusters. P-value <0.05 was defined as statistically significant.
Establishment of glutamine metabolic index (GMI)
The differentially expressed GMGs between different clusters were screened by the above method. The univariate Cox regression analysis was performed to identify the prognostic value of these genes for OS, with P-value <0.05 considered statistically significant. Subsequently, the least absolute shrinkage and selection operator (LASSO) Cox regression analysis was conducted to shrink the scope of gene screening, identify highly correlated GMGs, and construct a prognostic gene signature with R package glmnet [17]. Using the linear combination of gene expression weighted regression coefficients, we got the glutamine metabolic index (GMI) formula. The ccRCC patients with survival data were divided into low- and high-GMI groups, according to the median of GMI. The Kaplan-Meier survival analysis was used to evaluate the predictive ability of the prognostic model with R package survminer. The time-dependent receiver operating characteristic (ROC) curve was plotted and the area under the ROC curve (AUC) was also calculated with R package survivalROC. Univariate and multivariate Cox regression analyses were used to evaluate the independence of prognostic gene signature and other clinical parameters (age, gender, grade, clinical stage, T stage, N stage, and M stage).
Correlation analysis
Based on the median expression level of PYCR1, we divided all ccRCC samples into PYCR1 high and low-expression groups. The correlation analysis of key gene expression with clinicopathological characteristics was performed in the TCGA-KIRC samples, and validated in the GSE 73731 and GSE 66272 cohorts. We also explored the correlation of PYCR1 level with immune cell infiltration and common immune checkpoints. The Pearson coefficient (r) between PYCR1 mRNA level and drug response (IC50) was evaluated in ccRCC lines. The data about drug response in cell lines were downloaded from the Genomics of Drug Sensitivity in Cancer (GDSC) [19]. Pearson correlation coefficient and FDR were calculated in the GDSC, in which |r| >0.3 and FDR <0.05 were considered statistically significant.
Statistical analysis
All analyses were performed using R 4.1.0. All statistical tests were two-sided, and P-value <0.05 was considered statistically significant unless otherwise noted. Continuous variables in normal distribution were between-group compared through the independent t test, while continuous variables in skewed distribution through the Mann-Whitney U test. The relationship between hub DEGs and overall survival was analyzed through the Kaplan-Meier curve by log-rank test. The univariate regression model was constructed to analyze the effect of each variable on the survival. The LASSO regression model was constructed to determine the factors independently associated with survival.
Results
Glutamine metabolism with distinct prognostic and biological features in ccRCC
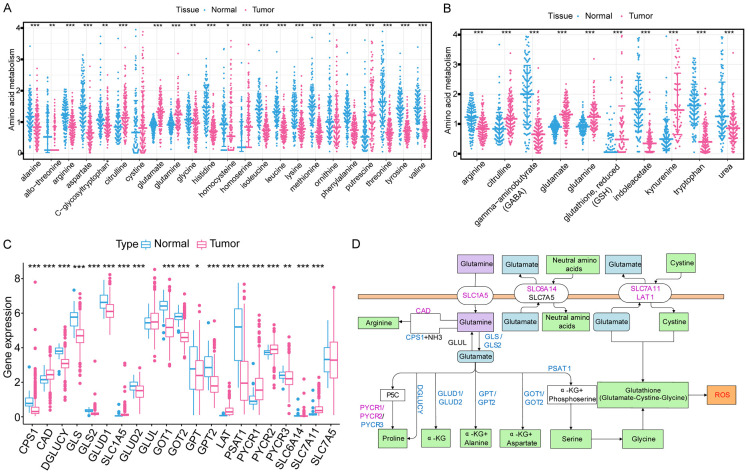
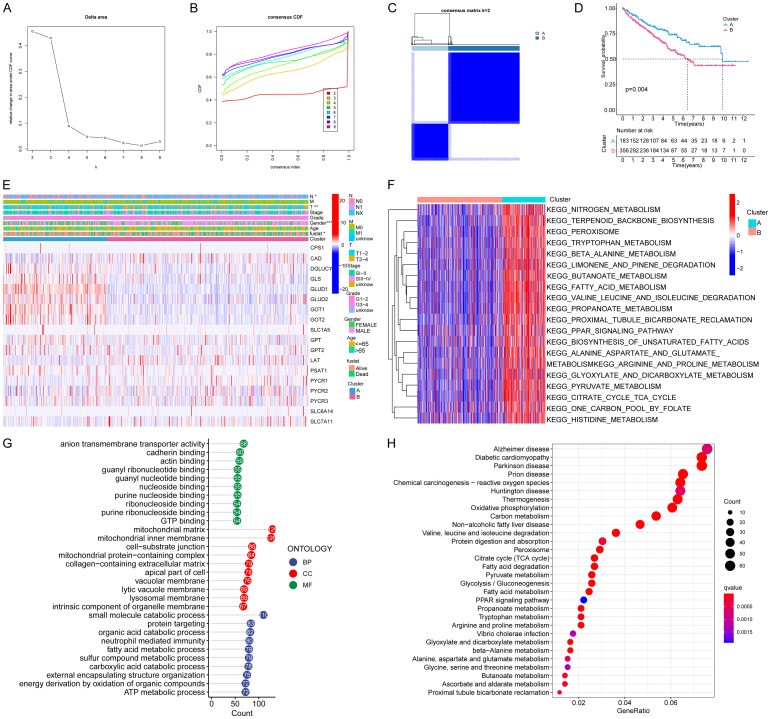
We explored the differentially expressed amino acids and related metabolites between normal kidney and ccRCC tissues (Figure 1A, 1B). The levels of glutamine and its metabolite (glutamate) in ccRCC tissue were significantly higher than those of other amino acids. Thus, we developed a passion for the metabolic pathway of glutamine in ccRCC. Totally, 21 GMGs were included in this study, including 19 differentially expressed (Figure 1C). The expression levels of CAD, SLC1A5, LAT, PYCR1, PYCR2, SLC7A11 were significantly higher, while those of CPS1, DGLUCY, GLS, GLS2, GLUD1, GLUD2, GOT1, GOT2, GPT, GPT2, PSAT1, PYCR3 and SLC6A14 were significantly decreased in tumor tissues. The roles and expressions of these genes varied with the progression of glutamine metabolism in ccRCC cells (Figure 1D). Then we studied the relationships between these GMGs and the prognosis of ccRCC patients. Our analysis revealed that the expression of 19 GMGs was significantly related to the prognosis of ccRCC patients. Patients with high expression levels of CAD, GPT2, LAT, PSAT1, SLC1A5, SLC6A14, PYCR1, PYCR2, PYCR3, SLC7A5 and SLC7A11 had a shorter OS (Figure S1A-K), while patients with higher expression levels of GLUD1, GLUD2, GOT1, CPS1, GLS, GOT2, GPT and DGLUCY expression showed a longer OS (Figure S1L-S). These 18 differentially expressed and survival-related GMGs might exert a synergistic effect on ccRCC, we performed the cluster analysis. As k increased from 2 to 9, the cluster stability kept rising (Figure 2A). However, the CDF plot and delta area plot revealed that k = 4 was also preferable (Figure 2B, 2C). Therefore, we performed survival analysis to investigate the prognostic performance of each cluster. When the 539 ccRCC patients were divided into two clusters, the Kaplan-Meier curves exhibited an evident difference (Figure 2D). When the patients were divided into 3, 4, or 5 clusters, some of them showed no between-cluster differences in the consistency matrix and survival curve (Figure S2A-F). Therefore, we divided the ccRCC patients from TCGA database into two clusters with different metabolic patterns (n = 183 in cluster A; n = 356 in cluster B). The clinicopathological features of two clusters are shown in Figure 2E. Cluster A witnessed a longer survival.
Figure 1.
Differentially expressed amino acids, related metabolites, and glutamine metabolic genes (GMGs) between normal kidney and ccRCC tissues. A. Expression profiles of amino acids in ccRCC and normal tissues. B. Expression profiles of amino acid metabolites in ccRCC and normal tissues. C. Expression profiles of GMGs in ccRCC and normal tissues. Red, tumor; Blue, normal. The upper and lower ends of the boxes represent the interquartile range of values. The lines in the boxes represent the median values. Adjusted P<0.05 was used as the criteria for screening differentially expressed GMGs. The asterisks represent the statistical P value (*P<0.05, **P<0.01, ***P<0.001). D. The flow chart of glutamine metabolism. Red, up-regulated genes; blue, down-regulated genes.
Figure 2.
Identification and comparison of various glutamine metabolic patterns of ccRCC. A. Consensus clustering cumulative distribution function (CDF) for K = 2-9. B. Delta region graph showed the relative change in the area under the CDF curve for K = 2-9. C. Consensus clustering matrix for K = 2. D. Kaplan-Meier overall survival (OS) curves for 539 ccRCC patients in clusters A and B. Patients in cluster A had a significantly longer survival (P = 0.004). E. Based on the results of the cluster analysis, heatmap showed the correlation with clinicopathological characteristics. F. GSVA enrichment analysis showing the biological pathways associated with distinct glutamine metabolic modification patterns. The heatmap was used to visualize these biological processes; red represents activated pathways and blue represents inhibited pathways. G. GO functional annotation analysis of DEGs between clusters A and B, and enriched biological processes (BPs), cellular components (CCs), and molecular functions (MFs). H. KEGG pathway enrichment of DEGs between clusters A and B. The enriched items were analyzed by using gene counts, gene ratio, and adjusted P values.
To screen out the mechanism responsible for the worse prognosis in cluster B, the biological behaviors of two clusters were compared through DGE analysis. Consequently, 1654 DEGs with an adjusted P-value <0.05 and an absolute value of Log2 fold-change (L2FC) >2 were identified. GSVA analysis showed that, cluster A was markedly enriched in pathways associated with amino acid metabolic activation, including tryptophan metabolism, valine leucine and isoleucine degradation, alanine aspartate and glutamate metabolism, arginine and proline metabolism, and histidine metabolism (Figure 2F). In terms of BPs of GO analysis, the DEGs were principally involved in small molecule catabolic process and protein targeting. In terms of CCs, the DEGs were mainly enriched in mitochondrial matrix and mitochondrial inner membrane. In terms of MFs, the DEGs were significantly enriched in anion transmembrane transporter activity and GTP binding (Figure 2G). At the same time, the KEGG pathway analysis showed the DEGs were enriched in valine, leucine and isoleucine degradation and protein digestion and absorption.
TME characteristics in two clusters for ccRCC
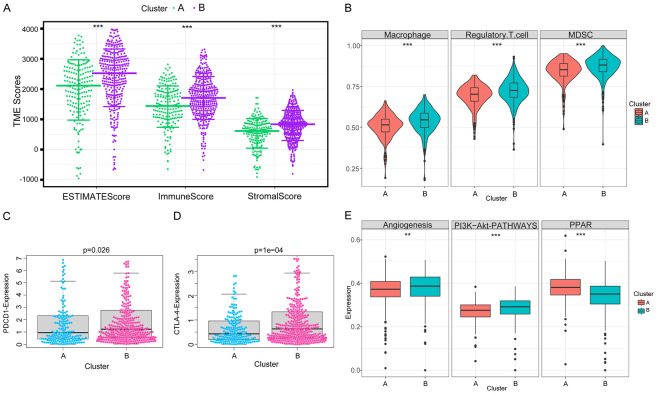
Furthermore, we evaluated the correlation between the glutamine metabolic clusters and the immune infiltration in the TME. Our data showed significant differences in the levels of infiltrating cells (MDSCs, Tregs, macrophages) and the expression of immune checkpoints (PDCD1, CTLA4 and CD274) between the two clusters. Cluster B showed higher immune score, stromal score and tumor purity than cluster A (Figure 3A). Moreover, the levels of MDSCs, Tregs, macrophages and the expression of PDCD1, CTLA4 were higher in cluster B than in cluster A (Figure 3B-D). These results indicated that the immunity was suppressed in the TME of cluster B, which could explain the worse prognosis in this cluster. Enrichment analysis showed that angiogenesis and PI3K-AKT signaling pathways were abnormally involved in cluster B, indicating that antiangiogenic therapy and PI3K inhibitor may improve the prognosis of patients in cluster B.
Figure 3.
Immune-related TME characteristics in two clusters of distinct glutamine metabolic patterns. A. Samples in cluster B exhibited higher ESTIMATE, stromal, and immune scores than cluster A. B. Abundances of main immunosuppressive infiltrating cells in TME in two clusters. Cluster B was classified as the immunosuppressive phenotype, characterized by the suppression of immunity. C, D. Difference in the expression of PDCD1 and CTLA-4, two common immune checkpoint molecules, between clusters A and B (PDCD1, P = 0.026; CTLA-4, P = 0.0001). E. Differences in tumor-related pathways, including angiogenesis, PI3K-Akt, and PPAR signaling pathways between two clusters. Wilcox test was used, and the asterisks represent the statistical P value (*P<0.05, **P<0.01, ***P<0.001).
Construction and validation of the glutamine metabolic signature
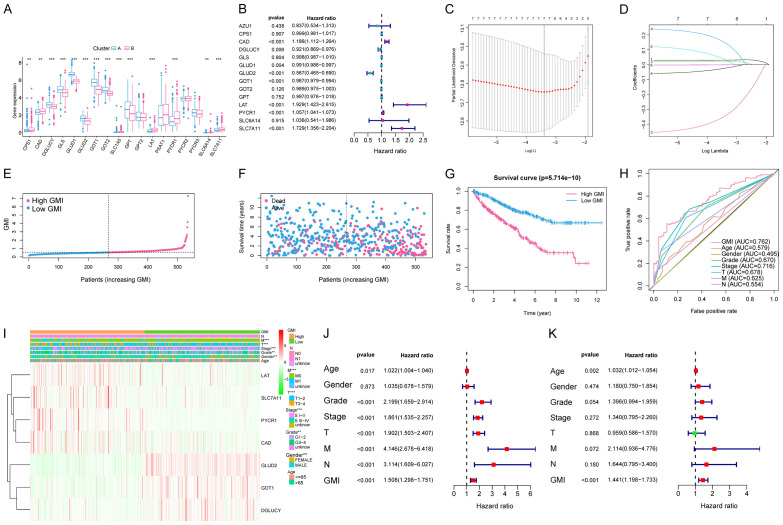
We screened out a total of 14 DEGs regulating glutamine metabolism between two clusters. Among them, 8 prognostic genes (CAD, DGLUCY, GLUD1, GLUD2, GOT1, LAT, PYCR1, and SLC7A11) were selected through the univariate Cox regression analysis (P<0.05, Figure 4A, 4B). The LASSO regression analysis was then applied to obtain 7 robust glutamine metabolic regulators for the construction of glutamine metabolic index (GMI) (Figure 4C, 4D). The GMI was developed on the algorithm below:
Figure 4.
Construction of the GMI model and relationships between the GMI, clinicopathological features and glutamine metabolic patterns. A. 14 GMGs were identified between two clusters. Adjusted P<0.05 was used as the criteria for screening differentially expressed GMGs. The asterisks represent the statistical p value (*P<0.05, **P<0.01, ***P<0.001). B. Eight prognostic genes were selected by the univariate Cox regression analysis. C, D. A 7-mRNA signature was constructed by the LASSO Cox regression. E, F. Prognostic analysis of the 7-gene signature in the TCGA cohort. The dotted line represents the median GMI, and divides the patients into low and high-GMI groups. A higher mortality was correlated to a higher GMI according to the curve of GMI and survival status of the patients. G. Kaplan-Meier survival analysis of the whole ccRCC set from the TCGA. The survival rate of the patients in the high-GMI group was significantly lower than that in the low-GMI group (P = 5.714e-10). H. Time-dependent ROC analysis of the whole ccRCC set from the TCGA. The area under the 5-year ROC curve was 0.762, which was higher than those of current clinical predictive factors. I. Differences in representative gene expression profiles and clinicopathological characteristics between the low and high-GMI groups. J, K. Univariate and multivariate Cox regression analyses of clinicopathological features for predicting the survival in ccRCC patients.
GMI = 0.0302 × exp PYCR1 + (-0.3437) × exp GLUD2 + 0.0227 × exp CAD + 0.1464 × exp SLC7A11 + 0.0328 × exp LAT + (-0.0017) × exp GOT1 + (-0.0242) × exp DGLUCY
According to the median value of the GMI, ccRCC patients with a higher GMI were assigned to the high-risk group, and those with a lower GMI to the low-risk group. The GMI distribution, OS and prognosis in the TCGA-KIRC cohort are shown in Figure 4E-G. The OS was lower and the prognosis was worse in the high-risk group. The ROCs showed that our GMI had good sensitivity and specificity in predicting the 5-year survival in ccRCC patients from the TCGA cohort (AUC = 76.2%). The predictive power of GMI was stronger than other clinicopathological features (Figure 4H). The heatmap visualizes the differences in gene expression profile and clinical characteristics between the two groups (Figure 4F). To explore whether GMI was independent from age, gender, stage and other clinical parameters, we performed univariate and multivariate Cox regression analyses for age, gender, stage, grade, TNM and GMI. In the univariate Cox regression analysis, age, grade, pathological T, M stage, pathological stage and high GMI were associated with poor survival (Figure 4J). In the multivariate Cox analysis, GMI remained an independent prognostic factor (hazard ratio, 1.441; CI, 1.198 to 1.733; P-value <0.001) (Figure 4K).
Clinical significance of PYCR1
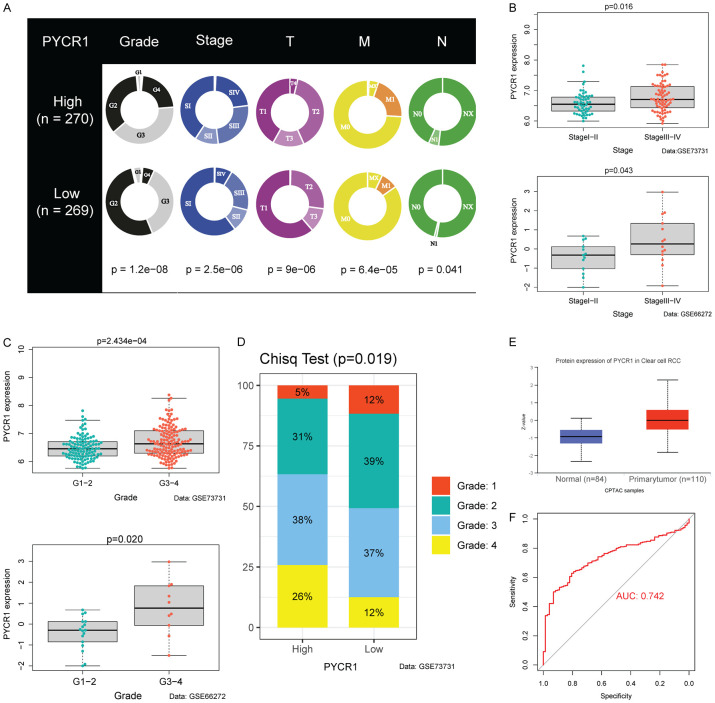
We stratified the patients into high and low-expression groups, based on the median expression value of genes in the signature. Only PYCR1 was closely related to clinical parameters (Figure S3). In the TCGA cohort, higher PYCR1 expression was significantly associated with worse clinical predictors, including histological grade (P<0.001), pathological stage (P<0.001), T stage (P<0.001), M stage (P<0.001), and N stage (P = 0.041) (Figure 5A). We validated this in the GSE66272 and GSE73731 datasets, finding that higher PYCR1 expression predicted more advanced and lower differentiation of tumor (Figure 5B, 5C). In the GSE73731 cohort, we explored the rates of different histological grades in two groups. The patients with high expression of PYCR1 exhibited more advanced histological grades (P = 0.019). The data from the CPTAC showed significant differences in PYCR1 expression between normal and ccRCC tissues, which verified the reliability of the signature (Figure 5E). We also plotted the 1-year ROC curve with PYCR1 as a prognostic predictor (AUC = 0.742), which also verified the potential of PYCR1 as an independent prognostic factor (Figure 5F).
Figure 5.
PYCR1 expression was upregulated in advanced ccRCC. A. Higher PYCR1 expression was associated with worse clinical parameters in ccRCC patients, such as histological grade, stage, T stage, N stage, and M stage according to data from the TCGA. B. PYCR1 expression was significantly higher in high-stage ccRCC in GSE73731 (P = 0.016) and GSE66272 (P = 0.043). C. PYCR1 expression was significantly higher in high-grade ccRCC in GSE73731 (P = 2.434e-04) and GSE66272 (P = 0.020). D. Difference in the proportion of cases with different grades (Chisq Test, P = 0.019) from GSE73731 between the high and low-PYCR1 expression subgroups. E. Differences in the protein expression of PYCR1 between ccRCC and normal tissues (P = 7.69e-19). F. Diagnostic value of PYCR1. An AUC value of 0.742 suggested that PYCR1 had a significant predictive value.
Immunomodulatory effect of PYCR1
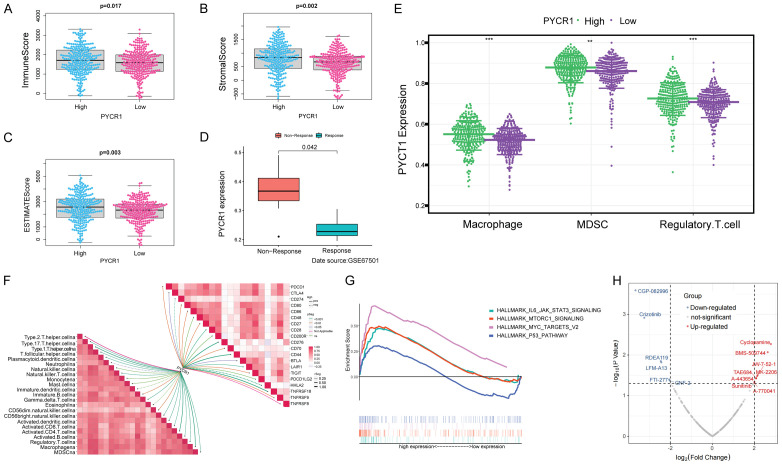
To investigate the relationships between PYCR1 and the immune TME, we scored the immune cells and stromal cells in the TME of 539 ccRCC patients, and found that the patients with higher expression of PYCR1 had greater immune, stromal, and ESTIMATE scores (P = 0.017; P = 0.002; P = 0.003; Figure 6A-C). We then explored the relationship between PYCR1 expression and immunotherapy response using the samples in the GSE67501. As shown in Figure 6D, ccRCC patients with high expression of PYCR1 exhibited a greater probability of non-response to immunotherapy. To investigate the effect of PYCR1 on the immune infiltration in the TME, the abundances of infiltrated immune cells were estimated via ssGSEA in each sample. Besides, the correlations of PYCR1 expression with immune cell infiltration and immune checkpoints were also calculated. The results indicated that high PYCR1 expression was associated with higher-degree infiltration of MDSCs, Tregs and macrophages, as well as higher expression of immune checkpoints, such as PDCD1 and CTLA4 (Figure 6E, 6F). GSEA analysis revealed pathways related to high PYCR1 expression, including IL-6-JAK-STAT3 signaling pathway, mTORC1 signaling pathway, myc-targets-v2 pathway, and p53 pathway (Figure 6G). These were primarily correlated with carcinogenesis, invasion, and the immune microenvironment of tumor cells. We finally explored the correlation between PYCR1 and drugs. The results of the volcano map indicated that PYCR1 was positively correlated with IC50 in 8 drugs (e.g., cyclopamine, BMS-509744) and negatively correlated with IC50 in 6 drugs (e.g., CGP-082996, Crizotinib).
Figure 6.
PYCR1 was associated with the immunity in the TME of ccRCC. A-C. Difference analysis of ImmuneScore, StromalScore, and ESTIMATEScore between the high and low-PYCR1-expression subgroups. High PYCR1 expression corresponded to high ImmuneScore (P = 0.017), StromalScore (P = 0.002), and ESTIMATEScore (P = 0.003). D. Correlation of PYCR1 expression with the response to immunotherapy (data source: GSE67501). PYCR1 expression level was significantly higher in ccRCC patients showing no response to immunotherapy (P = 0.042). E. Higher PYCR1 expression was accompanied by higher-degree infiltration of immunosuppressive cells. Wilcox test was used, and the asterisks represent the statistical P value (*P<0.05, **P<0.01, ***P<0.001). F. Correlation analysis of PYCR1 expression with TME cell infiltration and the levels of immune checkpoint molecules. PYCR1 expression was positively correlated with the levels of many immunosuppressive factors, such as MDSC, macrophage, Tregs, PDCD1, and CTLA-4. G. GSEA analysis of hallmarks in the high and low-PYCR1-expression subgroups. The MYC-TARGRTS, MTORC1, and IL6-JAK-STAT3 signaling pathways were up-regulated in high-PYCR1-expression subgroups, all known as oncogenic pathways. H. Correlation analysis of PYCR1 expression and drug sensitivity. PYCR1 was positively correlated with IC50 in 8 drugs and negatively in 6 drugs.
Discussion
Various pathways maneuver the anabolism and metabolism of cancer cells to supply energy for growth and survival [20]. During the cancer progression from preneoplastic lesions to localized, the metabolic phenotype and metabolic dependence of neighboring mesenchymal and immune cells in the TME is reprogrammed [21]. According to Otto Warburg [22,23], the glucose metabolism in tumor cells shifts from aerobic oxidation to glycolysis, thereby providing sufficient energy and essential macromolecular precursors for the proliferation of tumor cells. During metabolic reprogramming, a hypoxic, acidic, nutrient-deficient TME forms, in which the antitumor immune response is inhibited [24,25]. In other words, the tumor-specific metabolic reprogramming promotes tumor growth and prevents anti-tumor immune response.
As a substrate for protein synthesis, amino acids were second only to glucose as an important source of energy and nutrition within the cell. Apart from a substrate for protein synthesis, amino acids act as metabolites and metabolic regulators in supporting cancer cell growth [26,27]. An abundant supply of amino acids allowed tumor cells to maintain their proliferative drive. Thus, investigation on these amino acids would clarify the underlying molecular events of malignancy and help to open new prospects for cancer diagnosis and treatment.
Glutamine, a non-essential amino acid, serves nitrogen and carbon for the synthesis of amino acids, lipids and nucleic acids. In tumor cells, glutamine addiction contributes carbon and ammonia to supplement the donor of a-ketoglutarate, an intermediate of the tricarboxylic acid cycle (TCA) and enhances nucleotide biosynthesis [28,29]. Here, we found that glutamine metabolism was an indispensable process of ccRCC tumorigenesis, because 19 of 21 genes regulating glutamine metabolism were differentially expressed between normal and ccRCC samples. According to the glutamine metabolic profile, we defined two clusters of ccRCC patients with distinct clinical and biological features, as well as TME immune characteristics. In addition, we generated a GMI based on 7 genes related to the prognosis, showing a favorable prognostic value in different datasets.
Recent studies have found a specific nutrient partitioning mechanism in the TME: glucose is preferentially consumed by immune cells and glutamine by tumor cells [30]. As a result, the uptake and catabolism of glutamine are significantly enhanced in many tumor cells. Cell experiments have shown that external glutamine supply promotes the proliferation, invasion and metastasis of tumor cells [31]. Leone et al. have found that glutamine blockade suppresses cancer cell metabolism and enhances anti-tumor immune response [32]. Glutamine blockade in tumor-bearing mice deactivates Warburg physiology by inhibiting glycolysis, and drives tumor-infiltrating lymphocytes (TILs) towards a long-lived, memory-like phenotype capable of enhancing effector function. Consequently, the malignant phenotype is maintained in ccRCC cells.
Our bioinformatic analysis revealed a significant increase of PYCR1 expression in human ccRCC samples, indicating PYCR1 as the most potent regulator of glutamine metabolism. In the glutamine metabolism, PYCR1, as a mitochondrial inner membrane protein, can catalyze the NAD(P)H-dependent synthesis of pyrroline-5-carboxylates (P5C) and ornithine into proline [33]. Proline is a unique non-essential amino acid with a secondary amine in humans, and its biosynthesis is essential for normal cellular metabolism and redox regulation [34,35]. In normal cells, proline is employed as an osmotic agent or chemical chaperone to directly scavenge intracellular reactive oxygen species; meanwhile, the proline metabolic flux is activated to maintain cellular energy and trigger signaling pathways associated with cell survival [36]. Interestingly, proline starvation impairs the tumorigenicity of tumor cells, because proline can alleviate endoplasmic reticulum stress and strengthen cellular homeostasis and clonogenicity [37]. Moreover, in the formation of collagen, proline activates the microenvironmental reservoir in the extracellular matrix [38], thus protecting tumor cells from immunotherapy [39] and increasing their invasive and migrative capabilities [40].
Given that proline metabolism is essential to the metabolic reprogramming in tumor cells [41], previous studies have evaluated the feasibility of certain enzymes of proline metabolism in antitumor therapy [38,42-45]. Molecular mechanisms of PYCR1 in diverse tumors have been investigated. Nilsson et al. have analyzed the mRNA profiles from 1,981 tumor samples across 19 cancer types, finally identifying PYCR1 as the most consistently over-expressed metabolic gene [46]. Additional studies have highlighted the significant correlation between breast cancer aggressiveness and high PYCR1 expression [47,48]. Similarly, PYCR1 suppression counters the growth of different cancers, such as non-small-cell lung cancer [49,50], prostate cancer [51], hepatocellular carcinoma [52,53], and melanoma [54,55]. In general, PYCR1 leads to metabolic adaptation in ccRCC through glutamine/proline-dependent signaling pathways. In the present study, we found that PYCR1 regulated tumor progression through remodeling the immune TME.
The TME is a complex environment consisting of tumor and native cells. Some native cells, such as CD8+ cytotoxic T cells (CTLs), are capable of killing tumor cells. To facilitate tumor progression, the tumor cells interact with host components and generate a highly immunosuppressive environment to escape from the attack of CTLs for survival, which thwarted CTL cytotoxicity and promoted tumor progression. The immunosuppressive factors include immune checkpoints and immunosuppressive cells [56]. CTLA-4 and PD-1 are two major immune checkpoints that can inhibit T-cell activation and proliferation and induce T-cell anergy [57-59]. MDSCs, Tregs and tumor-associated macrophages (TAMs) are major immunosuppressive cells in the TME. They suppress anti-tumor immunity by releasing immunosuppressive cytokines, seizing raw materials for T cell proliferation, expressing immune checkpoint molecules, releasing toxic metabolites, etc. [60-63]. Notably, the present study was the first to depict the immunosuppressive TME characteristics in ccRCC patients with cluster B glutamine metabolism and high PYCR1 expression. However, how this PYCR1-mediated glutamine- and proline-dependent metabolic reprogramming contributes to the buildup of and immunosuppressive TME?
Glutamine is available in the TME and preferentially partitioned into cancer cells, rather than infiltrating immune cells [30]. However, activation of naïve CD8+ T cells also requires high glutamine uptake and metabolism to satisfy its high demand for energetic and biosynthetic precursors [64]. CD8+ T cell proliferation and cytokine production are significantly hindered upon glutamine starvation [65]. Thus, under this circumstance in which tumor cells compete with activated CD8+ T cells for glutamine, nutrient consumption may promote the proliferation and survival of tumor cells, meanwhile repressing T cell-mediated antitumor immunity. On the contrary, naïve CD4+ T cells, once activated, differentiate into Treg cells under the condition of glutamine deprivation [66]. This brings with a tumor-suppressive activity in vivo. Meanwhile, the oncogenic transcription factor MYC proto-oncogene, whose known function was to induce the expression of mitochondrial glutaminase (GLS) to stimulate the conversion of glutamine to glutamate [67,68], was recently documented to enhance subsequent conversion of glutamate to proline via up-regulating the expression of PYCR1 [69]. PYCR1, as a client of mitochondrial Lon, is up-regulated to increase the production of mitochondrial reactive oxygen species (ROS) and subsequent ROS-dependent production of inflammatory cytokines, such as IL-13 and VEGF-A [70]. These cytokines from tumor cells polarize macrophages towards an M2-like phenotype, which comprises the predominant TAMs having infiltrated into the TME [71]. Afterward, the TME manifests an immunosuppressive state that benefits the progression of tumors [72]. All these mechanisms may explain our findings. However, the mechanism of PYCR1 in immune TME remodeling remains enigmatic. Since the activation of immune cells depends on the distribution of nutrients, including amino acids [64-66,73-75], we suspect that PYCR1 may educate immune cells to create an immunosuppressive microenvironment by producing proline.
Our research also has certain limitations. Our findings need to be validated by in vitro experiments, such as quantitative real-time polymerase chain reaction and Western blotting. Further studies on human tissue samples are required to validate these results. In addition, the biological mechanisms of PYCR1 in remodeling the immune TME still need to be explored in vivo and in vitro.
Conclusions
Glutamine metabolism plays a critical role in shaping the immune TME of ccRCC. The GMI is efficient in discriminating the risk and predicting the survival of ccRCC patients. PYCR1 poses an immunosuppressive effect in the TME, and may serve as a potential target in treating ccRCC.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (grant number 82071638).
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Hosios AM, Hecht VC, Danai LV, Johnson MO, Rathmell JC, Steinhauser ML, Manalis SR, Vander Heiden MG. Amino acids rather than glucose account for the majority of cell mass in proliferating mammalian cells. Dev Cell. 2016;36:540–549. doi: 10.1016/j.devcel.2016.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mullard A. Cancer metabolism pipeline breaks new ground. Nat Rev Drug Discov. 2016;15:735–737. doi: 10.1038/nrd.2016.223. [DOI] [PubMed] [Google Scholar]
- 3.Hensley CT, Wasti AT, DeBerardinis RJ. Glutamine and cancer: cell biology, physiology, and clinical opportunities. J Clin Invest. 2013;123:3678–3684. doi: 10.1172/JCI69600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.DeBerardinis RJ, Cheng T. Q’s next: the diverse functions of glutamine in metabolism, cell biology and cancer. Oncogene. 2010;29:313–324. doi: 10.1038/onc.2009.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Craze ML, Cheung H, Jewa N, Coimbra NDM, Soria D, El-Ansari R, Aleskandarany MA, Wai Cheng K, Diez-Rodriguez M, Nolan CC, Ellis IO, Rakha EA, Green AR. MYC regulation of glutamine-proline regulatory axis is key in luminal B breast cancer. Br J Cancer. 2018;118:258–265. doi: 10.1038/bjc.2017.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Son J, Lyssiotis CA, Ying H, Wang X, Hua S, Ligorio M, Perera RM, Ferrone CR, Mullarky E, Shyh-Chang N, Kang Y, Fleming JB, Bardeesy N, Asara JM, Haigis MC, DePinho RA, Cantley LC, Kimmelman AC. Glutamine supports pancreatic cancer growth through a KRAS-regulated metabolic pathway. Nature. 2013;496:101–105. doi: 10.1038/nature12040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Strohecker AM, Guo JY, Karsli-Uzunbas G, Price SM, Chen GJ, Mathew R, McMahon M, White E. Autophagy sustains mitochondrial glutamine metabolism and growth of BrafV600E-driven lung tumors. Cancer Discov. 2013;3:1272–1285. doi: 10.1158/2159-8290.CD-13-0397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Altman BJ, Stine ZE, Dang CV. From Krebs to clinic: glutamine metabolism to cancer therapy. Nat Rev Cancer. 2016;16:619–634. doi: 10.1038/nrc.2016.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang W, Choi W, Chen Y, Zhang Q, Deng H, He W, Shi Y. A proposed role for glutamine in cancer cell growth through acid resistance. Cell Res. 2013;23:724–727. doi: 10.1038/cr.2013.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bhutia YD, Babu E, Ramachandran S, Ganapathy V. Amino acid transporters in cancer and their relevance to “glutamine addiction”: novel targets for the design of a new class of anticancer drugs. Cancer Res. 2015;75:1782–1788. doi: 10.1158/0008-5472.CAN-14-3745. [DOI] [PubMed] [Google Scholar]
- 11.Curthoys NP, Watford M. Regulation of glutaminase activity and glutamine metabolism. Annu Rev Nutr. 1995;15:133–159. doi: 10.1146/annurev.nu.15.070195.001025. [DOI] [PubMed] [Google Scholar]
- 12.Human genomics. The genotype-tissue expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015;348:648–660. doi: 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wei X, Choudhury Y, Lim WK, Anema J, Kahnoski RJ, Lane B, Ludlow J, Takahashi M, Kanayama HO, Belldegrun A, Kim HL, Rogers C, Nicol D, Teh BT, Tan MH. Recognizing the continuous nature of expression heterogeneity and clinical outcomes in clear cell renal cell carcinoma. Sci Rep. 2017;7:7342. doi: 10.1038/s41598-017-07191-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liep J, Kilic E, Meyer HA, Busch J, Jung K, Rabien A. Cooperative effect of miR-141-3p and miR-145-5p in the regulation of targets in clear cell renal cell carcinoma. PLoS One. 2016;11:e0157801. doi: 10.1371/journal.pone.0157801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ascierto ML, McMiller TL, Berger AE, Danilova L, Anders RA, Netto GJ, Xu H, Pritchard TS, Fan J, Cheadle C, Cope L, Drake CG, Pardoll DM, Taube JM, Topalian SL. The intratumoral balance between metabolic and immunologic gene expression is associated with anti-pd-1 response in patients with renal cell carcinoma. Cancer Immunol Res. 2016;4:726–733. doi: 10.1158/2326-6066.CIR-16-0072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wilkerson MD, Hayes DN. ConsensusClusterPlus: a class discovery tool with confidence assessments and item tracking. Bioinformatics. 2010;26:1572–1573. doi: 10.1093/bioinformatics/btq170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hänzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics. 2013;14:7. doi: 10.1186/1471-2105-14-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Charoentong P, Finotello F, Angelova M, Mayer C, Efremova M, Rieder D, Hackl H, Trajanoski Z. Pan-cancer immunogenomic analyses reveal genotype-immunophenotype relationships and predictors of response to checkpoint blockade. Cell Rep. 2017;18:248–262. doi: 10.1016/j.celrep.2016.12.019. [DOI] [PubMed] [Google Scholar]
- 19.Iorio F, Knijnenburg TA, Vis DJ, Bignell GR, Menden MP, Schubert M, Aben N, Gonçalves E, Barthorpe S, Lightfoot H, Cokelaer T, Greninger P, van Dyk E, Chang H, de Silva H, Heyn H, Deng X, Egan RK, Liu Q, Mironenko T, Mitropoulos X, Richardson L, Wang J, Zhang T, Moran S, Sayols S, Soleimani M, Tamborero D, Lopez-Bigas N, Ross-Macdonald P, Esteller M, Gray NS, Haber DA, Stratton MR, Benes CH, Wessels LFA, Saez-Rodriguez J, McDermott U, Garnett MJ. A landscape of pharmacogenomic interactions in cancer. Cell. 2016;166:740–754. doi: 10.1016/j.cell.2016.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pavlova NN, Thompson CB. The emerging hallmarks of cancer metabolism. Cell Metab. 2016;23:27–47. doi: 10.1016/j.cmet.2015.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Faubert B, Solmonson A, DeBerardinis RJ. Metabolic reprogramming and cancer progression. Science. 2020;368:eaaw5473. doi: 10.1126/science.aaw5473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Warburg O. On the origin of cancer cells. Science. 1956;123:309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- 23.Warburg O. On respiratory impairment in cancer cells. Science. 1956;124:269–270. [PubMed] [Google Scholar]
- 24.Kouidhi S, Ben Ayed F, Benammar Elgaaied A. Targeting tumor metabolism: a new challenge to improve immunotherapy. Front Immunol. 2018;9:353. doi: 10.3389/fimmu.2018.00353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhao E, Maj T, Kryczek I, Li W, Wu K, Zhao L, Wei S, Crespo J, Wan S, Vatan L, Szeliga W, Shao I, Wang Y, Liu Y, Varambally S, Chinnaiyan AM, Welling TH, Marquez V, Kotarski J, Wang H, Wang Z, Zhang Y, Liu R, Wang G, Zou W. Cancer mediates effector T cell dysfunction by targeting microRNAs and EZH2 via glycolysis restriction. Nat Immunol. 2016;17:95–103. doi: 10.1038/ni.3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tsun ZY, Possemato R. Amino acid management in cancer. Semin Cell Dev Biol. 2015;43:22–32. doi: 10.1016/j.semcdb.2015.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sivanand S, Vander Heiden MG. Emerging roles for branched-chain amino acid metabolism in cancer. Cancer Cell. 2020;37:147–156. doi: 10.1016/j.ccell.2019.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krall AS, Christofk HR. Rethinking glutamine addiction. Nat Cell Biol. 2015;17:1515–1517. doi: 10.1038/ncb3278. [DOI] [PubMed] [Google Scholar]
- 29.Fernandez-de-Cossio-Diaz J, Vazquez A. Limits of aerobic metabolism in cancer cells. Sci Rep. 2017;7:13488. doi: 10.1038/s41598-017-14071-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reinfeld BI, Madden MZ, Wolf MM, Chytil A, Bader JE, Patterson AR, Sugiura A, Cohen AS, Ali A, Do BT, Muir A, Lewis CA, Hongo RA, Young KL, Brown RE, Todd VM, Huffstater T, Abraham A, O’Neil RT, Wilson MH, Xin F, Tantawy MN, Merryman WD, Johnson RW, Williams CS, Mason EF, Mason FM, Beckermann KE, Vander Heiden MG, Manning HC, Rathmell JC, Rathmell WK. Cell-programmed nutrient partitioning in the tumour microenvironment. Nature. 2021;593:282–288. doi: 10.1038/s41586-021-03442-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang J, Pavlova NN, Thompson CB. Cancer cell metabolism: the essential role of the nonessential amino acid, glutamine. EMBO J. 2017;36:1302–1315. doi: 10.15252/embj.201696151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Leone RD, Zhao L, Englert JM, Sun IM, Oh MH, Sun IH, Arwood ML, Bettencourt IA, Patel CH, Wen J, Tam A, Blosser RL, Prchalova E, Alt J, Rais R, Slusher BS, Powell JD. Glutamine blockade induces divergent metabolic programs to overcome tumor immune evasion. Science. 2019;366:1013–1021. doi: 10.1126/science.aav2588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bogner AN, Stiers KM, Tanner JJ. Structure, biochemistry, and gene expression patterns of the proline biosynthetic enzyme pyrroline-5-carboxylate reductase (PYCR), an emerging cancer therapy target. Amino Acids. 2021;53:1817–1834. doi: 10.1007/s00726-021-02999-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pandhare J, Donald SP, Cooper SK, Phang JM. Regulation and function of proline oxidase under nutrient stress. J Cell Biochem. 2009;107:759–768. doi: 10.1002/jcb.22174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Phang JM, Liu W, Hancock C, Christian KJ. The proline regulatory axis and cancer. Front Oncol. 2012;2:60. doi: 10.3389/fonc.2012.00060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liang X, Zhang L, Natarajan SK, Becker DF. Proline mechanisms of stress survival. Antioxid Redox Signal. 2013;19:998–1011. doi: 10.1089/ars.2012.5074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sahu N, Dela Cruz D, Gao M, Sandoval W, Haverty PM, Liu J, Stephan JP, Haley B, Classon M, Hatzivassiliou G, Settleman J. Proline starvation induces unresolved er stress and hinders mtorc1-dependent tumorigenesis. Cell Metab. 2016;24:753–761. doi: 10.1016/j.cmet.2016.08.008. [DOI] [PubMed] [Google Scholar]
- 38.D’Aniello C, Patriarca EJ, Phang JM, Minchiotti G. Proline metabolism in tumor growth and metastatic progression. Front Oncol. 2020;10:776. doi: 10.3389/fonc.2020.00776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Choi J, Credit K, Henderson K, Deverkadra R, He Z, Wiig H, Vanpelt H, Flessner MF. Intraperitoneal immunotherapy for metastatic ovarian carcinoma: resistance of intratumoral collagen to antibody penetration. Clin Cancer Res. 2006;12:1906–1912. doi: 10.1158/1078-0432.CCR-05-2141. [DOI] [PubMed] [Google Scholar]
- 40.van Kempen LC, Rijntjes J, Mamor-Cornelissen I, Vincent-Naulleau S, Gerritsen MJ, Ruiter DJ, van Dijk MC, Geffrotin C, van Muijen GN. Type I collagen expression contributes to angiogenesis and the development of deeply invasive cutaneous melanoma. Int J Cancer. 2008;122:1019–1029. doi: 10.1002/ijc.23147. [DOI] [PubMed] [Google Scholar]
- 41.Elia I, Broekaert D, Christen S, Boon R, Radaelli E, Orth MF, Verfaillie C, Grünewald TGP, Fendt SM. Proline metabolism supports metastasis formation and could be inhibited to selectively target metastasizing cancer cells. Nat Commun. 2017;8:15267. doi: 10.1038/ncomms15267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bergers G, Fendt SM. The metabolism of cancer cells during metastasis. Nat Rev Cancer. 2021;21:162–180. doi: 10.1038/s41568-020-00320-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Burke L, Guterman I, Palacios Gallego R, Britton RG, Burschowsky D, Tufarelli C, Rufini A. The Janus-like role of proline metabolism in cancer. Cell Death Discov. 2020;6:104. doi: 10.1038/s41420-020-00341-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Phang JM. Proline metabolism in cell regulation and cancer biology: recent advances and hypotheses. Antioxid Redox Signal. 2019;30:635–649. doi: 10.1089/ars.2017.7350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tanner JJ, Fendt SM, Becker DF. The proline cycle as a potential cancer therapy target. Biochemistry. 2018;57:3433–3444. doi: 10.1021/acs.biochem.8b00215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nilsson R, Jain M, Madhusudhan N, Sheppard NG, Strittmatter L, Kampf C, Huang J, Asplund A, Mootha VK. Metabolic enzyme expression highlights a key role for MTHFD2 and the mitochondrial folate pathway in cancer. Nat Commun. 2014;5:3128. doi: 10.1038/ncomms4128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Possemato R, Marks KM, Shaul YD, Pacold ME, Kim D, Birsoy K, Sethumadhavan S, Woo HK, Jang HG, Jha AK, Chen WW, Barrett FG, Stransky N, Tsun ZY, Cowley GS, Barretina J, Kalaany NY, Hsu PP, Ottina K, Chan AM, Yuan B, Garraway LA, Root DE, Mino-Kenudson M, Brachtel EF, Driggers EM, Sabatini DM. Functional genomics reveal that the serine synthesis pathway is essential in breast cancer. Nature. 2011;476:346–350. doi: 10.1038/nature10350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ding J, Kuo ML, Su L, Xue L, Luh F, Zhang H, Wang J, Lin TG, Zhang K, Chu P, Zheng S, Liu X, Yen Y. Human mitochondrial pyrroline-5-carboxylate reductase 1 promotes invasiveness and impacts survival in breast cancers. Carcinogenesis. 2017;38:519–531. doi: 10.1093/carcin/bgx022. [DOI] [PubMed] [Google Scholar]
- 49.Wang D, Wang L, Zhang Y, Yan Z, Liu L, Chen G. PYCR1 promotes the progression of non-small-cell lung cancer under the negative regulation of miR-488. Biomed Pharmacother. 2019;111:588–595. doi: 10.1016/j.biopha.2018.12.089. [DOI] [PubMed] [Google Scholar]
- 50.Cai F, Miao Y, Liu C, Wu T, Shen S, Su X, Shi Y. Pyrroline-5-carboxylate reductase 1 promotes proliferation and inhibits apoptosis in non-small cell lung cancer. Oncol Lett. 2018;15:731–740. doi: 10.3892/ol.2017.7400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zeng T, Zhu L, Liao M, Zhuo W, Yang S, Wu W, Wang D. Knockdown of PYCR1 inhibits cell proliferation and colony formation via cell cycle arrest and apoptosis in prostate cancer. Med Oncol. 2017;34:27. doi: 10.1007/s12032-016-0870-5. [DOI] [PubMed] [Google Scholar]
- 52.Zhuang J, Song Y, Ye Y, He S, Ma X, Zhang M, Ni J, Wang J, Xia W. PYCR1 interference inhibits cell growth and survival via c-Jun N-terminal kinase/insulin receptor substrate 1 (JNK/IRS1) pathway in hepatocellular cancer. J Transl Med. 2019;17:343. doi: 10.1186/s12967-019-2091-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ding Z, Ericksen RE, Escande-Beillard N, Lee QY, Loh A, Denil S, Steckel M, Haegebarth A, Wai Ho TS, Chow P, Toh HC, Reversade B, Gruenewald S, Han W. Metabolic pathway analyses identify proline biosynthesis pathway as a promoter of liver tumorigenesis. J Hepatol. 2020;72:725–735. doi: 10.1016/j.jhep.2019.10.026. [DOI] [PubMed] [Google Scholar]
- 54.De Ingeniis J, Ratnikov B, Richardson AD, Scott DA, Aza-Blanc P, De SK, Kazanov M, Pellecchia M, Ronai Z, Osterman AL, Smith JW. Functional specialization in proline biosynthesis of melanoma. PLoS One. 2012;7:e45190. doi: 10.1371/journal.pone.0045190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ye Y, Wu Y, Wang J. Pyrroline-5-carboxylate reductase 1 promotes cell proliferation via inhibiting apoptosis in human malignant melanoma. Cancer Manag Res. 2018;10:6399–6407. doi: 10.2147/CMAR.S166711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Binnewies M, Roberts EW, Kersten K, Chan V, Fearon DF, Merad M, Coussens LM, Gabrilovich DI, Ostrand-Rosenberg S, Hedrick CC, Vonderheide RH, Pittet MJ, Jain RK, Zou W, Howcroft TK, Woodhouse EC, Weinberg RA, Krummel MF. Understanding the tumor immune microenvironment (TIME) for effective therapy. Nat Med. 2018;24:541–550. doi: 10.1038/s41591-018-0014-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Waldman AD, Fritz JM, Lenardo MJ. A guide to cancer immunotherapy: from T cell basic science to clinical practice. Nat Rev Immunol. 2020;20:651–668. doi: 10.1038/s41577-020-0306-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lewis AL, Chaft J, Girotra M, Fischer GW. Immune checkpoint inhibitors: a narrative review of considerations for the anaesthesiologist. Br J Anaesth. 2020;124:251–260. doi: 10.1016/j.bja.2019.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pardoll DM. The blockade of immune checkpoints in cancer immunotherapy. Nat Rev Cancer. 2012;12:252–264. doi: 10.1038/nrc3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Condeelis J, Pollard JW. Macrophages: obligate partners for tumor cell migration, invasion, and metastasis. Cell. 2006;124:263–266. doi: 10.1016/j.cell.2006.01.007. [DOI] [PubMed] [Google Scholar]
- 61.Wing JB, Tanaka A, Sakaguchi S. Human FOXP3(+) regulatory T cell heterogeneity and function in autoimmunity and cancer. Immunity. 2019;50:302–316. doi: 10.1016/j.immuni.2019.01.020. [DOI] [PubMed] [Google Scholar]
- 62.Ostrand-Rosenberg S, Sinha P, Beury DW, Clements VK. Cross-talk between myeloid-derived suppressor cells (MDSC), macrophages, and dendritic cells enhances tumor-induced immune suppression. Semin Cancer Biol. 2012;22:275–281. doi: 10.1016/j.semcancer.2012.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lin Y, Xu J, Lan H. Tumor-associated macrophages in tumor metastasis: biological roles and clinical therapeutic applications. J Hematol Oncol. 2019;12:76. doi: 10.1186/s13045-019-0760-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Buck MD, O’Sullivan D, Pearce EL. T cell metabolism drives immunity. J Exp Med. 2015;212:1345–1360. doi: 10.1084/jem.20151159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Carr EL, Kelman A, Wu GS, Gopaul R, Senkevitch E, Aghvanyan A, Turay AM, Frauwirth KA. Glutamine uptake and metabolism are coordinately regulated by ERK/MAPK during T lymphocyte activation. J Immunol. 2010;185:1037–1044. doi: 10.4049/jimmunol.0903586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Klysz D, Tai X, Robert PA, Craveiro M, Cretenet G, Oburoglu L, Mongellaz C, Floess S, Fritz V, Matias MI, Yong C, Surh N, Marie JC, Huehn J, Zimmermann V, Kinet S, Dardalhon V, Taylor N. Glutamine-dependent α-ketoglutarate production regulates the balance between T helper 1 cell and regulatory T cell generation. Sci Signal. 2015;8:ra97. doi: 10.1126/scisignal.aab2610. [DOI] [PubMed] [Google Scholar]
- 67.Wise DR, DeBerardinis RJ, Mancuso A, Sayed N, Zhang XY, Pfeiffer HK, Nissim I, Daikhin E, Yudkoff M, McMahon SB, Thompson CB. Myc regulates a transcriptional program that stimulates mitochondrial glutaminolysis and leads to glutamine addiction. Proc Natl Acad Sci U S A. 2008;105:18782–18787. doi: 10.1073/pnas.0810199105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gao P, Tchernyshyov I, Chang TC, Lee YS, Kita K, Ochi T, Zeller KI, De Marzo AM, Van Eyk JE, Mendell JT, Dang CV. c-Myc suppression of miR-23a/b enhances mitochondrial glutaminase expression and glutamine metabolism. Nature. 2009;458:762–765. doi: 10.1038/nature07823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Liu W, Le A, Hancock C, Lane AN, Dang CV, Fan TW, Phang JM. Reprogramming of proline and glutamine metabolism contributes to the proliferative and metabolic responses regulated by oncogenic transcription factor c-MYC. Proc Natl Acad Sci U S A. 2012;109:8983–8988. doi: 10.1073/pnas.1203244109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kuo CL, Chou HY, Chiu YC, Cheng AN, Fan CC, Chang YN, Chen CH, Jiang SS, Chen NJ, Lee AY. Mitochondrial oxidative stress by Lon-PYCR1 maintains an immunosuppressive tumor microenvironment that promotes cancer progression and metastasis. Cancer Lett. 2020;474:138–150. doi: 10.1016/j.canlet.2020.01.019. [DOI] [PubMed] [Google Scholar]
- 71.Chen D, Zhang X, Li Z, Zhu B. Metabolic regulatory crosstalk between tumor microenvironment and tumor-associated macrophages. Theranostics. 2021;11:1016–1030. doi: 10.7150/thno.51777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mantovani A, Sozzani S, Locati M, Allavena P, Sica A. Macrophage polarization: tumor-associated macrophages as a paradigm for polarized M2 mononuclear phagocytes. Trends Immunol. 2002;23:549–555. doi: 10.1016/s1471-4906(02)02302-5. [DOI] [PubMed] [Google Scholar]
- 73.Geiger R, Rieckmann JC, Wolf T, Basso C, Feng Y, Fuhrer T, Kogadeeva M, Picotti P, Meissner F, Mann M, Zamboni N, Sallusto F, Lanzavecchia A. L-arginine modulates T cell metabolism and enhances survival and anti-tumor activity. Cell. 2016;167:829–842. e13. doi: 10.1016/j.cell.2016.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ron-Harel N, Ghergurovich JM, Notarangelo G, LaFleur MW, Tsubosaka Y, Sharpe AH, Rabinowitz JD, Haigis MC. T cell activation depends on extracellular alanine. Cell Rep. 2019;28:3011–3021. e4. doi: 10.1016/j.celrep.2019.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sinclair LV, Howden AJ, Brenes A, Spinelli L, Hukelmann JL, Macintyre AN, Liu X, Thomson S, Taylor PM, Rathmell JC, Locasale JW, Lamond AI, Cantrell DA. Antigen receptor control of methionine metabolism in T cells. Elife. 2019;8:e44210. doi: 10.7554/eLife.44210. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.